Significance
The notion of genetic dominance was first introduced by Gregor Mendel and is defined as the relationship between alleles of the same gene in which one allele (dominant) masks the phenotypic contribution of a second allele (recessive). The effect of genetic dominance on bacterial evolution has generally been neglected because most bacteria of human interest carry a single copy of their chromosome. However, bacteria also carry mobile genetic elements (MGE), which produce local islands of polyploidy. This polyploidy arises both from the multicopy nature of MGE and from the allelic redundancy between MGE and chromosomal genes. Here we show that genetic dominance dictates both the evolution and successful spread of genes encoded on MGE, shaping horizontal gene transfer in bacteria.
Keywords: mobile genetic elements, antibiotic resistance, evolution, genetic dominance
Abstract
Mobile genetic elements (MGEs), such as plasmids, promote bacterial evolution through horizontal gene transfer (HGT). However, the rules governing the repertoire of traits encoded on MGEs remain unclear. In this study, we uncovered the central role of genetic dominance shaping genetic cargo in MGEs, using antibiotic resistance as a model system. MGEs are typically present in more than one copy per host bacterium, and as a consequence, genetic dominance favors the fixation of dominant mutations over recessive ones. In addition, genetic dominance also determines the phenotypic effects of horizontally acquired MGE-encoded genes, silencing recessive alleles if the recipient bacterium already carries a wild-type copy of the gene. The combination of these two effects governs the catalog of genes encoded on MGEs. Our results help to understand how MGEs evolve and spread, uncovering the neglected influence of genetic dominance on bacterial evolution. Moreover, our findings offer a framework to forecast the spread and evolvability of MGE-encoded genes, which encode traits of key human interest, such as virulence or antibiotic resistance.
Horizontal gene transfer (HGT) between bacteria is largely mediated by specialized mobile genetic elements (MGEs), such as plasmids and bacteriophages, which provide an important source of genetic diversity and play a fundamental role in bacterial ecology and evolution (1). The repertoire of accessory genes encoded on MGEs and their ability to be phenotypically expressed in different genetic backgrounds are key aspects of MGE-mediated evolution (2–5). There are several factors known to impact the fate of horizontally transferred genes in bacteria, such as the level of gene expression, the degree of protein connectivity, or the biochemical properties of proteins, but the specific parameters that shape the repertoire of genes encoded on MGEs remain largely unknown (6–9).
Genetic dominance is the relationship between alleles of the same gene in which one allele (dominant) masks the phenotypic contribution of a second allele (recessive). In diploid or polyploid organisms, the evolution of new traits encoded by recessive mutations is therefore constricted because the presence of a dominant allele will always block the phenotypic contribution of the recessive allele [an effect known as Haldane’s sieve (10, 11)]. Most bacteria of human interest carry a single copy of their chromosome. In haploid organisms like these, new alleles are able to produce a phenotype regardless of the degree of genetic dominance of the underlying mutations. Therefore, the role of genetic dominance in bacterial evolution has generally been overlooked. However, the bacterial genome consists of more than the single chromosome; a myriad of MGEs populate bacterial cells. Many MGEs, including plasmids and filamentous phages, replicate independently of the bacterial chromosome and are generally present at more than one copy per cell, with copy number ranging from a handful to several hundred (12, 13). Extrachromosomal MGEs thus produce an island of local polyploidy in the bacterial genome (14, 15). Moreover, HGT in bacteria mostly occurs between close relatives (16, 17), and genes encoded on mobile elements can therefore create allelic redundancy with chromosomal genes. In light of these evidences, genetic dominance should strongly affect both the emergence of new mutations in MGE-encoded genes and the phenotypic effects of horizontally transferred alleles.
Results and Discussion
Genetic Dominance Shapes the Emergence of Mutations in MGE-Encoded Genes.
To test whether genetic dominance determines the emergence of mutations in MGE-encoded genes, we used a two-gene synthetic system conceptually similar to the one that F. Jacob and J. Monod used to investigate the regulation of the Lac operon (18). This construct consists of a cI gene, encoding the bacteriophage λ CI repressor, in control of the expression of a contiguous tetA gene, which encodes a tetracycline efflux pump (ref. 19 and Fig. 1A). This system provides tetracycline resistance when tetA transcription is derepressed. Derepression can be achieved through mutations that either inactivate the cI gene or disrupt the CI binding site upstream of tetA (Fig. 1A). The repressor gene provides a large target (714 bp), but mutations inactivating cI should be recessive since in trans copies of wild-type CI will still be able to repress tetA. In contrast, the CI binding site is a short target (166 bp), but mutations in this region are likely to be dominant because they will lead to nonrepressible, constitutive TetA production.
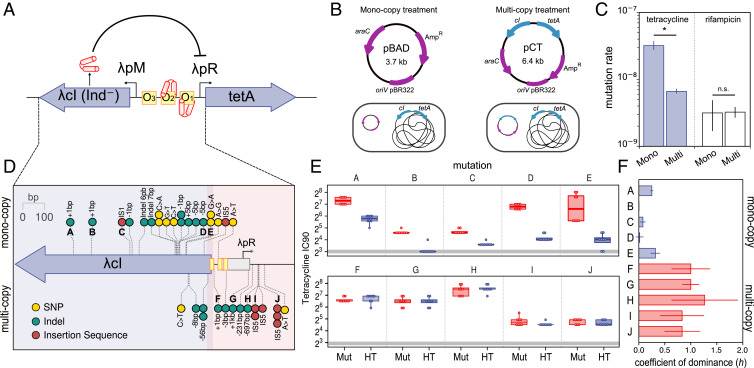
Fig. 1.
Genetic dominance and gene copy number modulate phenotypic mutation rates. (A) The cI-tetA system. Under normal circumstances, transcription from the lambda promoter pM drives the production of the phage repressor CI (red protein) which binds the operator boxes (O1 and O2) located upstream the tetA gene, thus blocking its expression. (B) Schematic representation of plasmids pBAD, pCT, and the experimental model (note the isogenic nature of the clones apart from the dosage of cI-tetA). (C) Tetracycline and rifampicin resistance phenotypic mutation rates in the different clones. Error bars represent 84% confidence intervals. The asterisk denotes statistical significance; n.s., nonsignificant. (D) Location and type of tetracycline resistance mutations in the monocopy (upper part) and multicopy (lower part) treatments. Blue shading denotes the cI coding region, and red shading denotes the CI binding site plus the cI-tetA intergenic region. (E) Tetracycline resistance level of the different clones constructed to measure the coefficient of dominance of tetracycline resistance mutations detailed in D. The tetracycline inhibitory concentration 90 (IC90, in mg/L) of the homozygous mutant clones (Mut) and heterozygous mutant clones (HT) are represented by boxes. The line inside the box marks the median. The upper and lower hinges correspond to the 25th and 75th percentiles, and whiskers extend to 1.5 times the interquartile range. Resistance level of the homozygous wild-type clone is indicated by a horizontal gray line for reference. The letters in the panels correspond to the mutations indicated in D. (F) Coefficient of dominance (h) of 10 mutations described in C and D. Bars represent the median of eight biological replicates; error bars represent the interquartile range.
We produced two otherwise isogenic Escherichia coli MG1655 clones carrying the cI-tetA system either as a single chromosomal copy (monocopy treatment) or also present on a pBAD plasmid with ∼20 copies per cell (pCT, multicopy treatment). To maintain an identical genetic background between the clones, we included an empty pBAD plasmid in the monocopy treatment (Fig. 1B). We next calculated the tetracycline resistance phenotypic mutation rate, defined as the rate at which mutations able to produce a tetracycline resistant phenotype emerge. Results revealed a 4.84-fold lower phenotypic mutation rate in the multicopy treatment, despite the higher cI-tetA copy number (likelihood ratio test statistic 55.0, P < 10−12; Fig. 1C). As the underlying mutation rate is equal for both clones (i.e., both clones produce mutations at the same rate; likelihood ratio test statistic 0.02, P ∼ 1; Fig. 1C), we reasoned that the observed difference in the emergence of tetracycline-resistant mutants should be due to the different phenotypic effects of dominant and recessive mutations in each treatment.
To test this possibility, we first analyzed the mutations in the cI-tetA system and confirmed that they were located in different regions in each treatment (Wilcoxon signed-rank test, W = 313, P = 10−06; Fig. 1D and Dataset S1). Most mutants isolated from the monocopy treatment carried mutations in the cI gene, whereas multicopy treatment mutations commonly targeted the intergenic region between cI and tetA genes (Fig. 1D and Dataset S1). Next, we measured the level of dominance of a random subset of these mutations. To this end, we determined tetracycline resistance level (measured as the antibiotic concentration that inhibits 90% of growth [IC90]) for clones carrying only the mutant allele (homozygous mutant), equal number of mutant and wild-type alleles (heterozygous), and only wild-type alleles (homozygous wild type; see a scheme of the experimental design in SI Appendix, Fig. S1). We reasoned that in heterozygous clones, wild-type copies of the cI-tetA system would mask the resistance level conferred by recessive mutations but not by dominant mutations. Results showed that the resistance level of heterozygous clones is lower than that of the homozygous mutant clones for mutations isolated in the monocopy treatment but not in the multicopy treatment (paired Student’s t test t = 6.96, df = 39, P = 2.5 × 10−8 and t = −0.85, df = 39, P = 0.398 for monocopy and multicopy treatments, respectively; Fig. 1E and Dataset S2). Finally, we used tetracycline resistance data to calculate the coefficient of dominance (h) of the mutations under study. h represents the relative resistance level of heterozygous clones compared to homozygous wild-type and mutant clones and ranges from 0 (completely recessive mutation) to 1 (completely dominant) (11). As predicted, mutations recovered from the monocopy treatment were mostly recessive, showing low or intermediate h values, while mutations from the multicopy treatment showed high levels of dominance (ANOVA effect of treatment; F = 70.02, df = 1, P∼ 3 × 10−5; Fig. 1F).
The Interplay between Genetic Dominance and Gene Copy Number Determines the Emergence of Antibiotic Resistance Mutations.
Our results indicate that genetic dominance determines the rate at which MGE-encoded mutations emerge in bacteria. The increased gene dosage provided by MGEs improves the chances of a beneficial mutation being acquired but simultaneously masks the phenotypic contribution of the newly acquired allele if it is recessive. To study the general effect of this interplay on the evolution of MGE-encoded genes, we developed a computational model based on the classic fluctuation assay (ref. 20 and Fig. 2A and SI Appendix, Figs. S2 and S3). This model allows us to simulate the acquisition and segregation of mutations located in an extrachromosomal MGE, in this case a plasmid, with a given copy number. With this information, we can explore the frequency of antibiotic resistant mutants in the bacterial population at any time point; this frequency will depend on the distribution of mutated and wild-type alleles in each individual cell and on the coefficient of dominance of those mutations (SI Appendix, Fig. S3). The simulations showed that the frequency of phenotypic mutants increases with plasmid copy number for mutations of high dominance but decreases for mutations of low dominance (Fig. 2A).
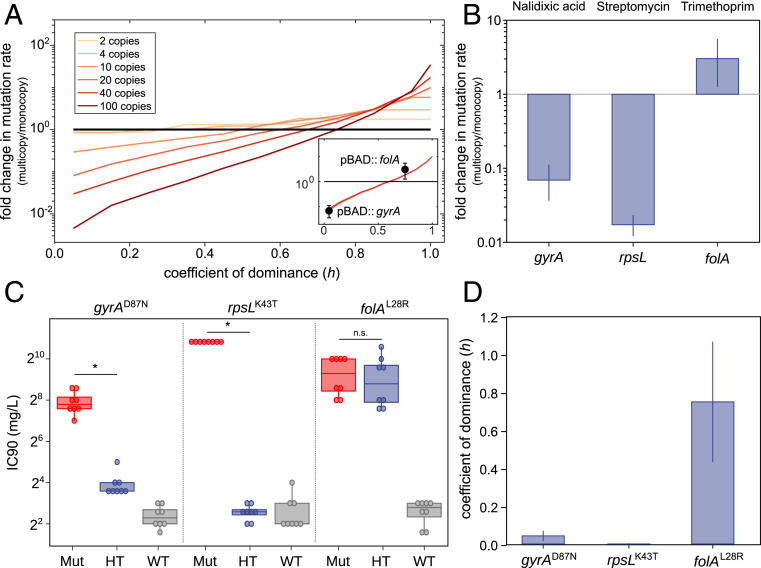
Fig. 2.
Interplay between genetic dominance and gene copy number. (A) Results of simulations analyzing the effect of plasmid copy number and genetic dominance of a mutation on the emergence of phenotypic mutants. The chart shows fold changes in phenotypic mutation rate for a plasmid-carried gene at different copy numbers compared with a chromosomal copy of the same gene (black line). (Inset) The comparison of experimental results obtained for gyrA and folA (presented in B and C) with the prediction for a plasmid of 20 copies. (B) Fold change of antibiotic resistance phenotypic mutation rates in E. coli, comparing multicopy and monocopy treatments for gyrA, rpsL, and folA. Error bars indicate 84% confidence intervals. Note that result for rpsL is an upper bound due to the absence of phenotypic mutants in the multicopy treatment (Materials and Methods). (C) Antibiotic resistance phenotypes of the clones constructed to measure the coefficient of dominance of gyrAD87N (nalidixic acid), rpsLK43T (streptomycin), and folAL28R (trimethoprim) resistance mutations. The inhibitory concentration 90 (IC90, in mg/L) of the homozygous mutant clones (Mut), heterozygous mutant clones (HT), and homozygous wild-type clones (WT) are represented by boxes. The line inside the box marks the median. The upper and lower hinges correspond to the 25th and 75th percentiles, and whiskers extend to 1.5 times the interquartile range. Asterisks denote statistically significant differences (Student’s t test P < 0.0002 in all cases); n.s., nonsignificant. (D) Coefficient of dominance of gyrAD87N, rpsLK43T, and folAL28R. Bars represent the median of eight biological replicates; error bars represent the interquartile range.
The results of the simulation prompted us to test our hypothesis in a more realistic and meaningful experimental system. To study the effect of genetic dominance and gene copy number on the emergence of phenotypic mutants, we used bacterial housekeeping essential genes known to confer antibiotic resistance through mutations. The genes studied were gyrA (DNA gyrase subunit A), rpsL (30S ribosomal protein S12), and folA (dihydrofolate reductase). Mutations in these genes, which are present as single copies in the chromosome, confer resistance to quinolone, aminoglycoside, and trimethoprim antibiotics (21, 22).
We expected resistance mutations in gyrA and rpsL to be recessive and resistance mutations in folA to be dominant. The recessive nature of mutations in gyrA and rpsL can be explained by the toxic effect produced by the combination of antibiotic and wild-type alleles. In the case of gyrA, when the quinolone antibiotic binds to the gyrase, double-strand DNA breaks occur, leading to cell death (23). In the case of streptomycin and rpsL, binding of the antibiotic to RpsL in the ribosome leads to mistranslation events producing a toxic effect in the cell (24). Therefore, even if resistance mutations reduce or avoid the binding of the antibiotic to the mutated target, the presence of wild-type enzymes in the cell will still produce a toxic effect, explaining the predicted recessive nature of mutations in these two genes (25, 26).
FolA is a key enzyme in folate metabolism. Specifically, it catalyzes the reduction of dihydrofolate to tetrahydrofolate. Trimethoprim binds to wild-type FolA and inhibits its activity, blocking folate metabolism and producing a bacteriostatic effect. Mutations in FolA confer trimethoprim resistance through reduction of FolA binding affinity to the drug and/or by increasing FolA activity [both by increasing expression or by specific mutations that increase enzymatic activity (21, 27)]. In this case, the binding of trimethoprim to FolA does not produce a toxic effect per se. Therefore, in the presence of trimethoprim a cell carrying an antibiotic resistant version of the enzyme will be able to reduce dihydrofolate and grow even if there are wild-type copies of FolA, explaining the predicted dominance of these mutations.
For each gene under study, we produced two otherwise isogenic E. coli MG1655 clones with either one copy of the gene (chromosomal) or multiple copies (chromosomal + plasmid = 20 copies; SI Appendix, Fig. S4). We calculated phenotypic mutation rates for each clone using fluctuation assays with the appropriate antibiotics and sequenced the target genes in the resistant clones to confirm the presence of mutations (Fig. 2B and Dataset S1). As expected, the frequency of gyrA and rpsL mutants in the multicopy treatment was lower than in the monocopy treatment (likelihood ratio test statistic 49.48, P < 10−12). Conversely, the mutation rate for folA in the multicopy treatment increased threefold (likelihood ratio test statistic 5.38, P = 0.02).
To confirm that the genetic dominance is responsible for these results, we experimentally determined the level of resistance of homozygous mutant, homozygous wild-type and heterozygous clones of a common mutation for each gene. We found that for folAL28R mutation the resistance level of heterozygous clones is similar to that of the homozygous mutant clones, while for gyrAD87N and rpsLK43T mutations the resistance level of heterozygous clones is similar to that of the homozygous wild-type clones (Fig. 2C). We calculated the coefficient of dominance and confirmed that mutations in gyrAD87N (h = 0.043) and rpsLK43T (h = 0) were recessive, whereas mutation in folAL28R (h = 0.748) was dominant (Fig. 2D).
Genetic Dominance Limits the Spread of Antibiotic Resistance.
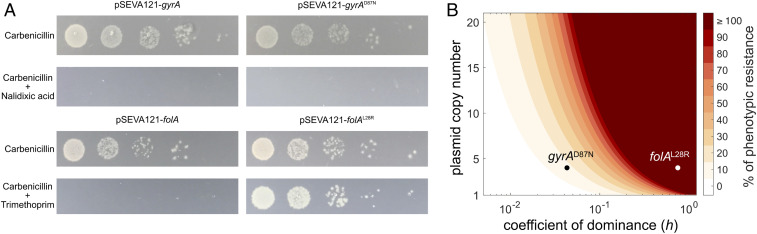
While our results showed that genetic dominance shapes the emergence of mutations in MGE-encoded genes, they leave open the question of how genetic dominance affects the phenotypic effects of horizontally transferred genes in recipient bacteria. HGT in bacteria preferentially occurs between close relatives, which leads to the transfer of redundant genes between cells (16, 17). We hypothesized that phenotypic expression of recessive alleles encoded on MGEs will be masked if the recipient bacterium possesses a chromosomal copy of the dominant allele, effectively hindering transfer of the recessive allele. We tested this hypothesis in an experimental assay of bacterial conjugation, using the low-copy number mobilizable plasmid pSEVA121 (28). The plasmid was modified by independent insertion of mutant, resistance-conferring gyrAD87N (recessive) and folAL28R (dominant) alleles and their wild-type counterparts (SI Appendix, Fig. S5). We transferred these plasmids from E. coli β3914 to E. coli MG1655 (Fig. 3A). After conjugation, resistance conferred by folAL28R was readily expressed in the recipient cells, whereas resistance conferred by gyrAD87N was masked by the resident wild-type allele, preventing phenotypic expression of resistance in transconjugants (Fig. 3A). A simple model of the general effect of genetic dominance on the phenotypic outcome of transferred housekeeping genes predicted that the presence of a dominant allele in the recipient cell would reduce phenotypic expression of recessive alleles over a wide range of plasmid copy numbers (Fig. 3B). Crucially, this effect will be particularly marked for conjugative plasmids, due to their low copy number in the host cell (typically ranging from 1 to 5).
Fig. 3.
Genetic dominance limits the phenotypic contribution of horizontally transferred recessive alleles. (A) Pictures of a representative replicate of the conjugation assays. Overnight cultures of spots inoculated from 10-fold dilutions of conjugation mixes (100 to 10−4, from left to right) on plates selecting for transconjugants. Selection on carbenicillin reveals the actual number of transconjugants; selection on carbenicillin plus nalidixic acid or trimethoprim reveals the number of transconjugants carrying gyrA and folA alleles and expressing the resistant phenotype. (B) Antibiotic resistance level conferred by a plasmid-encoded resistance allele in the recipient bacterium (when a wild-type copy of the gene is present in the chromosome), assuming phenotypic resistance as the product of plasmid copy number and the coefficient of dominance of the allele. Experimental data are presented for gyrAD87N and folAL28R in plasmid pSEVA121.
Genetic Dominance Shapes the Repertoire of Antibiotic Resistance Genes on MGE.
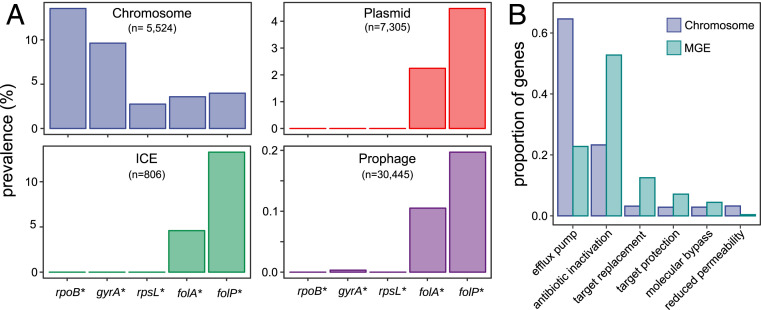
Our results strongly suggest that genetic dominance may be an important factor determining the frequency of antibiotic resistance alleles of housekeeping genes in MGE. This is of critical relevance as many antibiotic resistance genes of clinical concern were originally resistant variants of chromosomal genes that were mobilized by MGE (29, 30). To investigate this further, we analyzed the Comprehensive Antibiotic Resistance Database (CARD), which includes detailed information about antibiotic resistance genes from thousands of bacterial chromosomes and plasmids (31). To extend our analysis to other MGEs, we also examined databases for information on integrative and conjugative elements (ICEs) and prophages. We predicted that housekeeping alleles conferring antibiotic resistance and contained in MGEs would be more frequently dominant than recessive. We investigated the genes in our experimental system plus two additional housekeeping genes that, according to results from previous works, should confer antibiotic resistance through dominant (folP, dihydropteroate synthase, sulfonamide resistance) and recessive (rpoB, RNA polymerase subunit beta, rifampicin resistance) mutations (refs. 32–34 and SI Appendix, SI Text). Results confirmed that mobile resistance-conferring alleles of folA and folP (dominant) were ubiquitous in naturally occurring MGEs, whereas resistance alleles of rpoB, gyrA, and rpsL (recessive) were almost never present in MGEs (Fig. 4A). We note, however, that alternative explanations to genetic dominance, such as protein connectivity or the fitness effects produced by these genes in recipient bacteria (SI Appendix, Figs. S6 and S7), could also contribute to explain the observed bias in gene distribution. As a matter of fact, RpoB and RpsL show a high degree of protein connectivity, whereas GyrA is involved in a similar number of protein–protein interactions to FolA and FolP (SI Appendix, Fig. S7). Additionally, effects typically associated with increased ploidy levels such as changes in gene expression and pleiotropic protein–protein interactions between chromosomal and mobile genes can also play a significant role in the observed results (35).
Fig. 4.
Genetic dominance shapes the bacterial mobilome. (A) Prevalence of antibiotic resistance-conferring alleles of rpoB, rpsL, gyrA, folA, and folP (indicated with an asterisk) in chromosomes and MGEs across bacteria. (B) Proportion of antibiotic resistance genes belonging to each resistance category on MGE (n = 6,053) and chromosomes (n = 26,147; see also SI Appendix, Fig. S8).
To obtain a more general view of the effect of genetic dominance on the distribution of resistance genes on MGE/chromosomes, we further extended our analysis to all possible antibiotic resistance genes according to their Antibiotic Resistance Ontology (ARO, grouped by “determinant of antibiotic resistance,” data obtained from CARD). We calculated the proportion of resistance genes belonging to each of the categories in MGE and chromosomes. Notably, efflux pumps represent more than 60% of all resistance determinants located in chromosomes, probably as a consequence of their role in bacterial physiology beyond antimicrobial resistance (36). As the extreme overrepresentation of pumps in chromosomes might bias our results, we performed the analyses including and excluding efflux pumps and obtained qualitatively similar results (see below and SI Appendix, Fig. S8).
Although the evolutionary forces driving this divergence are likely to be complex (4–7), we predicted that categories enriched in the few dominant resistance alleles of chromosomal genes or in those genes with no resident counterpart in the host bacterium (xenogeneic genes) should be overrepresented in MGE compared to chromosomes, whereas the opposite should be true for categories enriched in recessive alleles. We compared the proportion of resistance determinants in each category for MGE and chromosomes under the null hypothesis of equal proportions. In agreement with our predictions, we found that genes belonging to the antibiotic inactivation, target protection, and target replacement categories were consistently overrepresented in MGE (Bonferroni corrected Fisher’s exact test P < 0.003 in all cases; Fig. 4B and SI Appendix, Fig. S8). These categories include mostly dominant alleles, because they are composed of genes encoding enzymes of xenogeneic origin that are known to confer resistance in virtually all gram-positive and gram-negative bacteria (e.g., antibiotic inactivating β-lactamases or target protection Qnr proteins) (37, 38) or of dominant alleles of housekeeping genes that replace the sensitive antibiotic target such as folP (sul) and folA (dfr). The molecular by-pass category was also significantly overrepresented in MGE when accounting for efflux pumps but underrepresented when efflux pumps were removed from the analysis (Bonferroni corrected Fisher’s exact test P < 10-7 in both cases). Despite this discrepancy, most MGE-encoded genes from this category are dominant vancomycin-resistant van alleles, which are able to confer resistance in the presence of the susceptible chromosomal cell wall synthesis pathway (39).
On the other hand, genes belonging to the reduced permeability to antibiotic category are underrepresented in MGEs (Fisher's exact test; Bonferroni corrected P < 10−15; Fig. 4B and SI Appendix, Fig. S8). This category includes mainly housekeeping porin genes carrying loss of function mutations, which are recessive alleles, as the presence of wild-type alleles will render the cell sensitive. A prediction that stems from this result is that loss of function mutations should be scarce in MGE compared to chromosomes. This might include mutations in efflux pumps regulators and is certainly the case for mutations in AmpC β-lactamases regulators (ampR and ampD), which are common in chromosomal ampC systems but extremely infrequent on plasmid-encoded ones (40, 41).
In light of these results, we propose that genetic dominance contributes to explain the commonly observed divergence between chromosomal and MGE-mediated antibiotic resistance genes (42, 43). Our results suggest that chromosomal singletons are free to explore their mutational landscape in a host bacterium, but only dominant alleles are able to provide a selectable phenotype, reach fixation, and spread successfully on MGEs.
Conclusion
In this work we demonstrate that genetic dominance strongly influences evolution through MGEs in bacteria, revealing an extra layer of complexity in the forces governing HGT. The impact of genetic dominance on MGEs is twofold, affecting both 1) the emergence of new mutations in MGE-encoded genes and 2) the phenotypic effects of horizontally transferred genes. The first effect is driven by the polyploid nature of extrachromosomal MGEs, which induces the filtering of recessive mutations through Haldane’s sieve. However, elevated gene copy number is not an exclusive property of MGEs and can arise physiologically in haploid bacteria in multiple circumstances. For example, gene copy number is increased by gene duplication events and by transient polyploidy during fast bacterial growth (44, 45), suggesting a potential broader influence of genetic dominance on bacterial evolution. The second effect is determined by the dominance relationships that emerge when a new allele arrives in a bacterium that already carries a copy of that gene. Given that HGT in bacteria is strongly favored between close relatives (16, 17), genetic redundancy of this type must be extremely common, underlining the importance of genetic dominance in determining if a given allele is likely to spread horizontally. Together, our findings shed light on the forces that shape MGE-mediated evolution and offer a framework to forecast which genes will spread on MGE.
Materials and Methods
Strains, Plasmids, and Media.
All of the strains and plasmids used in this study are detailed in Dataset S3. Experiments were performed using Muller Hinton II agar or cation adjusted broth (Becton Dickinson) unless specified. Antibiotics were supplied by Sigma-Aldrich and were used at the following concentrations: carbenicillin 100 µg/mL, chloramphenicol 30 µg/mL, nalidixic acid 30 µg/mL, streptomycin 100 µg/mL, rifampicin 100 µg/mL, trimethoprim 2 µg/mL and 16 µg/mL, and tetracycline 15 µg/mL. Cultures were routinely grown at 37 °C with shaking (225 rpm). Plasmids were extracted using a commercial miniprep kit (Macherey-Nagel) and were transformed into TSS competent cells (46).
Cloning and Site-Directed Mutagenesis.
The gyrA, rpsL, and folA wild-type alleles were PCR amplified using HS Taq Mix (PCR Biosystems) polymerase, using the primers listed in Dataset S4 and MG1655 chromosomal DNA as template. The cI-tetA system was amplified similarly but using IBDS1 as a template (19). Purified PCRs were subsequently cloned into the pBAD TOPO TA Expression kit (Thermo-Fisher) following manufacturer instructions. We selected clones in which the insert was cloned in the opposite direction from that of the PBAD promoter to ensure that expression is driven only from the cognate promoter. Correct clones were identified by PCR and Sanger sequenced to give rise to pBAD-gene and pCT plasmids (Dataset S3).
Site-directed mutagenesis was performed on the pBAD plasmids carrying wild-type alleles to construct the gyrAD87N, rpsLK43T, and folAL28R mutated alleles by using the Q5 site-directed mutagenesis kit (New England Biolabs) and the primers listed in Dataset S4. Correct cloning was assessed by PCR and Sanger sequencing. Next, we generated pBDSM plasmid variants by Gibson assembling PCR amplified (Phusion Hot Start II DNA Polymerase, Thermo-Fisher; see Dataset S4 for primers) mutated and wild-type alleles into the pBGC backbone (SI Appendix, Fig. S4 and Dataset S3)
Tetracycline resistance mutations in the pCT plasmid isolated in the fluctuation assays were purified by plasmid extraction and retransformed into MG1655, selecting in carbenicillin and tetracycline plates to isolate pCT mutated plasmids from the wild-type plasmids that could be coexisting under heteroplasmy (14). Isolated pCT plasmids and chromosomal mutants carrying cI-tetA mutated cassette were PCR amplified (Phusion Hot Start II DNA Polymerase; Thermo-Fisher) and simultaneously Gibson assembled (NEBuilder HiFi DNA Assembly kit; New England Biolabs) into the pBAD and pBGC backbones using the primers listed in Dataset S4 to give rise to pBAD and pBDSM plasmids carrying mutated alleles of the cI-tetA system (SI Appendix, Fig. S1 and Dataset S3).
Fluctuation Assays for Mutation Rate Determination.
Briefly, independent cultures containing carbenicillin were inoculated with ∼103 cells and allowed to grow during 18 h at 37 °C with shaking (225 rpm). The following day, appropriate aliquots were plated in antibiotic-containing agar plates to select for spontaneous mutants and in nonselective plates to determine the number of viable bacteria. After incubation at 37 °C for 24 h (48 h for rifampicin and streptomycin plates), colonies were enumerated. The genes suspected of carrying mutations were PCR amplified and Sanger sequenced using mutant colonies coming from independent cultures as template. In some cases, Sanger reads from the multicopy treatment showed mixed reads and/or unclear results. In those cases, plasmids from the resistant colonies were extracted and retransformed into MG1655 competent cells. Transformants were selected on plates containing carbenicillin (selecting for plasmids) and carbenicillin plus tetracycline (selecting for plasmids carrying tetracycline resistance mutations). The following day, transformant colonies from plates containing carbenicillin plus tetracycline were PCR amplified and Sanger sequenced. This simple procedure allows us to separate plasmids carrying dominant resistance mutations from wild-type plasmids coexisting within the same cell under heteroplasmy (14). Phenotypic mutation rates, defined as the rate at which mutations producing a resistance phenotype emerge, 84% confidence intervals, and likelihood ratio tests to assess statistical significance were then calculated using the newton.LD.plating, confint.LD.plating and LRT.LD.plating functions of the Rsalvador package for R (47). We used 84% confidence intervals because it has been demonstrated that they convey statistical significance better than 95% confidence intervals (48).
Monocopy and multicopy strains of gyrA and rpsL presented the same antibiotic resistance levels, indicating that target overexpression did not increase the level of resistance for these genes (Dataset S2). However, and as previously reported (49), folA overexpression led to an eightfold increase in the trimethoprim resistance level in the multicopy treatment. To perform the mutation rate assay, we used the lowest antibiotic concentration possible that inhibits the growth of nonmutant colonies and maximizes the recovery of mutant cells. These concentrations were 2 and 16 µg/mL for the monocopy and multicopy treatments, respectively, which correspond to 4 times the IC90 of trimethoprim for each clone. Sequencing of folA PCR products revealed mutations in all of the colonies tested from the monocopy treatment but only for ∼10% of the ones from the multicopy treatment. This result could be due to the lack of fixation of folA mutations, which can be maintained at a low frequency in heteroplasmy and not be evident in the chromatogram, or due to the access to different resistance mutations in this treatment. To solve this problem and recover only folA mutants, we introduced an extra step in the fluctuation assays for the multicopy treatment, where we restreaked the colonies from 16 to 50 µg/mL trimethoprim plates. This step allowed us to recover colonies carrying folA mutations only (avoiding false positives) while at the same time allowing the recovery of colonies carrying folA mutations even if the frequency of folA mutated alleles in heteroplasmy would have been too low to initially grow on this concentration during a fluctuation assay (avoiding false negatives) (Dataset S1).
For rpsL, we were not able to recover a single resistant colony in the fluctuation assays for the multicopy treatment (Dataset S1). Subsequent analyses revealed that the coefficient of dominance of the most common rpsL mutation isolated in the chromosome is 0. The complete recessiveness of rpsL streptomycin resistance mutations explains the absence of resistant mutants in the multicopy treatment because even if the plasmid-mediated copy of rpsL mutates and reaches fixation prior to plating, the wild-type chromosomal copy of the gene masks the resistance phenotype. Therefore, for phenotypic resistant mutants to appear, both chromosomal and plasmid copies of the gene should mutate (and reach fixation in the cell), which is extremely unlikely. In Fig. 2B we present the fold change of streptomycin resistance phenotypic mutation rate using the mutation rate calculated for the monocopy treatment and the limit of detection of the mutation rate for the multicopy treatment. Rifampicin mutation rates were performed in all cases to confirm that the underlying mutation rate is equal for all clones used in this study (Fig. 1 and SI Appendix, Fig. S9).
Coefficient of Dominance.
The coefficient of dominance (h) of mutations ranges from 0, completely recessive, to 1, completely dominant. To calculate h, we developed an experimental system based on two compatible plasmids of similar copy number (n ∼ 15 to 20) (50, 51), where we cloned the genes of interest carrying the wild-type or mutant alleles under investigation (SI Appendix, Fig. S1). We then transformed E. coli MG1655 with the single plasmids and the combinations of plasmids to produce the two possible heterozygous clones. To construct the homozygous mutant clones for rpsLK43T and gyrAD87N, we used the MG1655 background carrying the same mutation in the chromosomal copy of the gene under study. We then performed antibiotic susceptibility assays to determine the resistance phenotypes of the different clones (measured as inhibitory concentration 90, IC90). Using the IC90 values we calculated h with the following formula:
where IC90AA, IC90aA, and IC90aa are the IC90 of the homozygous wild-type, heterozygous, and homozygous mutant clones, respectively. Since there are two different heterozygous clones (SI Appendix, Fig. S1), we calculated h as the median value of four independent replicates of each heterozygous clone (n = 8; Dataset S2).
IC90 values were obtained following ref. 52, with some alterations. In short, strains were streaked from freeze stock onto Mueller Hinton II agar plates and incubated overnight at 37 °C. Single colonies were picked and suspended in liquid Mueller Hinton II broth and incubated at 37 °C and 225 rpm. Overnight cultures were diluted 1:10,000 into Mueller Hinton II broth to a final volume of 200 µL per well in microtiter plates. The wells contained increasing concentrations of the appropriate antibiotic in 1.5-fold increments. Plates were incubated at 37 °C for 22 h and 225 rpm. After incubation we ensured homogenous mixture of bacteria by orbital shaking for 1 min (548 rpm with 2-mm diameter) before reading of OD600 in a Synergy HTX (BioTek) plate reader. IC90 is defined as ≥90% inhibition of growth which was calculated using the formula 1 – [ODdrug/ODcontrol], where the control is the no-antibiotic treatment. Wells only containing media were used for background subtraction. Appropriate antibiotic for plasmid maintenance was included in the media throughout the assay.
Conjugation Assays.
We developed an experimental model system to determine the frequencies of conjugation of plasmids pSEVA121-gyrA, pSEVA121-gyrAD87N, pSEVA121-folA, and pSEVA121-folAL28R between E. coli strains (SI Appendix, Fig. S5). pSEVA121 is a vector carrying the IncP-type RK2 (also called RP4) replicon, with a trfA replication initiator protein gene plus the oriV origin of replication and the oriT origin of transfer. pSEVA121 displays, therefore, low copy number in the host cell (circa four copies per cell) (53), representing a great model system to reproduce the features of a natural conjugative plasmid. To mobilize pSEVA121 we used strain β3914 as donor, which is auxothropic for diaminopimelic acid and carries RP4 conjugation machinery inserted in the chromosome. As a recipient we used E. coli MG1655 (Dataset S3).
Precultures of donor and recipient strains were incubated overnight in 2 mL of Mueller Hinton broth with the appropriate antibiotics at 37 °C and 225 rpm. Next day, 1:100 dilution of the overnight cultures in 5 mL of Mueller Hinton broth were incubated until the culture reached midexponential phase at 37 °C and 225 rpm (2.5 h approximately). Cultures were then centrifuged for 15 min at 1,500 × g, and the pellets were resuspended in 200 µL of fresh Mueller Hinton broth; 50 µL of donor and 10 µL of recipient suspensions were mixed and spotted on Mueller Hinton agar plates.
The conjugation mix was incubated for 18 h at 37 °C and then suspended in 2 mL of 0.9% NaCl sterile solution. Dilutions of this suspension were spotted as 5-µL drops on Mueller Hinton agar plates containing antibiotics selecting for donor, recipient, and transconjugant cells. The total number of transconjugants was determined by selecting on carbenicillin, while the number of transconjugants expressing the resistance phenotype conferred by gyrA and folA alleles were selected on carbenicillin plus nalidixic acid or trimethoprim, respectively. We performed four independent biological replicates of each conjugation assay, and we show one representative replicate in Fig. 3A.
Growth Curves.
Growth curves were performed in a Synergy HTX (BioTek) plate reader. Briefly, overnight cultures were diluted 1:1,000 in Mueller Hinton broth supplemented with carbenicillin, and 200 µL of the dilutions were transferred to a 96-well multiwell plate (Thermo Scientific). Plates were incubated at 37 °C with strong orbital shaking before reading absorbance at 600 nm every 15 min. Six biological replicates were included per strain.
Bioinformatic Analysis.
CARD prevalence data containing an analysis of 82 pathogens with more than 89,000 resistomes and more than 175,000 antimicrobial resistance allele sequences were obtained from the CARD website (31) (downloaded on 18 September 2019).
We downloaded all available bacterial ICEs from ICEberg 2.0 database (54), amounting to a total of 806 ICEs from which 662 were conjugative type IV secretion system (T4SS)-type ICEs, 111 were chromosome-borne integrative and mobilizable elements, and 33 were cis-mobilizable elements. A total of 741 perfect and strict hits from 267 ICEs were identified using CARD RGI (version 4.2.2, parameters used: DNA sequence, perfect and strict hits only, exclude nudge, high quality/coverage, and we used DIAMOND to align the CDS against the CARD database).
We downloaded all complete bacterial genomes from NCBI, a total of 13,169 genomes (assembly level: complete, downloaded on 15 February 2019). We used the algorithm phiSpy to identify prophages in the bacterial chromosomes, and we predicted a total of 30,445 prophages in 7,376 genomes (55). To study the presence of genes conferring antibiotic resistance in prophages, we extracted the CDS overlapping with prophages, and we predicted their resistome using the command line tool CARD RGI as above.
We then used the antibiotic resistance data from prophages, ICEs, plasmids, and chromosomes to calculate the prevalence of the following Antibiotic Resistance Ontology (ARO) categories: “antibiotic resistant DNA topoisomerase subunit gyrA” (ARO: 3000273), “antibiotic resistant rpsL” (ARO: 3003419), “sulfonamide resistant dihydropteroate synthase” (ARO: 3000558), “antibiotic resistant rpoB” (ARO:3003276), and “antibiotic resistant dihydrofolate reductase” (ARO:3003425). To test the overrepresentation or underrepresentation of resistance mechanisms in MGEs and chromosomes, we used the following ARO categories to query our database: “antibiotic target replacement protein” (ARO:3000381), “antibiotic inactivation enzyme” (ARO:3000557), “antibiotic target protection protein” (ARO:3000185), “protein modulating permeability to antibiotic” (ARO:3000270), “protein(s) conferring antibiotic resistance via molecular bypass” (ARO:3000012), and “efflux pump complex or subunit conferring antibiotic resistance” (ARO:3000159). We used the number of genes encoded in MGE and chromosomes to build a 2 × 2 contingency table for each category, in which the first column contains the number of genes in a given category in MGE and chromosomes and the second column contains the number of resistance genes that do not belong to that category. We used two-tailed Fisher’s exact test to test whether there are different proportions of chromosomal versus MGE-encoded antibiotic resistance determinants in each category and adjusted the resulting P values for multiple comparisons using Bonferroni’s method.
Connectivity data for all protein families were downloaded from the STRING (Search Tool for Retrieval of Interacting Genes/Proteins) database (56), and the number of high-confidence interactions (>700 STRING confidence) was determined for each protein family.
Stochastic Model.
We assume that plasmids replicate randomly throughout the cell cycle until reaching an upper limit determined by the plasmid copy number control mechanism. Therefore, the probability of plasmid replication can be modeled as , where (t) represents the number of copies of a plasmid of type at time and is the maximum number of plasmids. We explicitly consider random mutations occurring during replication events, so if denotes the probability of a mutation occurring in a plasmid, the per-cell mutation rate is . Stochastic simulations of mutation and replication dynamics were performed using a Gillespie algorithm implemented in MATLAB with propensities determined from the distribution of plasmid copies of each allele carried by the cell (code can be downloaded from GitHub; https://github.com/ccg-esb/dominatrix) (SI Appendix, Fig. S2).
We also consider that plasmids segregate randomly during cell division. The probability that each plasmid is inherited to one of the daughter cells is a random process that follows a binomial distribution. By implementing an agent-based extension of the replication–mutation model coupled with the segregation dynamics, we simulated the intracellular plasmid dynamics of individual cells in an exponentially growing population (SI Appendix, Fig. S3). The frequency of phenotypic mutants observed in the population at the end of the numerical experiment was estimated from the fraction of mutated/wild-type alleles in each individual cell and the coefficient of dominance of those mutations (). The results presented in this study were obtained after simulation runs in a range of plasmid copy numbers (Fig. 2A). Finally, we used Rsalvador package (47) to estimate from this synthetic data the fold change in phenotypic mutation rate with respect to chromosomally encoded genes.
Statistical Analyses.
All statistical tests, analysis, and plots were performed using R (v. 3.4.2).
Data Availability.
All data generated during this study are included in this published article (and SI Appendix).
Supplementary Material
Acknowledgments
We thank R. Craig MacLean, José R. Penadés, Patrice Courvalin, and Hildegard Uecker for helpful discussion. We are grateful to Ivan Matic, Jesús Blázquez, Jose A. Escudero, Didier Mazel, Fernando de la Cruz, Raúl Fernández López, and the Standard European Vector Architecture (SEVA) collection for generous gifts of strains and plasmids. This work was supported by the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme (ERC grant agreement 757440-PLASREVOLUTION) and by the Instituto de Salud Carlos III (grant PI16-00860) cofunded by European Development Regional Fund “a way to achieve Europe.” Á.S.M. is supported by a Miguel Servet Fellowship (MS15-00012). J.R.-B. is a recipient of a Juan de la Cierva-Incorporación Fellowship (IJC2018-035146-I) cofunded by Agencia Estatal de Investigación del Ministerio de Ciencia e Innovación. M.T.-R. acknowledges support from the Swiss National Science Foundation (Ambizione grant PZ00P3_161545). R.P.-M. was funded by Consejo Nacional de Ciencia y Tecnología (CONACYT) Ciencia Básica (grant A1-S-32164). V.S. is supported by Northern Norway Regional Health Authority (HNF1494-19) and The National Graduate School in Infection Biology and Antimicrobials (Norwegian Research Council grant 249062).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: The data and code for this paper have been deposited in GitHub, https://github.com/ccg-esb/dominatrix.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2001240117/-/DCSupplemental.
References
- 1.Wiedenbeck J., Cohan F. M., Origins of bacterial diversity through horizontal genetic transfer and adaptation to new ecological niches. FEMS Microbiol. Rev. 35, 957–976 (2011). [DOI] [PubMed] [Google Scholar]
- 2.Sorek R.et al., Genome-wide experimental determination of barriers to horizontal gene transfer. Science 318, 1449–1452 (2007). [DOI] [PubMed] [Google Scholar]
- 3.Jain R., Rivera M. C., Lake J. A., Horizontal gene transfer among genomes: The complexity hypothesis. Proc. Natl. Acad. Sci. U.S.A. 96, 3801–3806 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cohen O., Gophna U., Pupko T., The complexity hypothesis revisited: Connectivity rather than function constitutes a barrier to horizontal gene transfer. Mol. Biol. Evol. 28, 1481–1489 (2011). [DOI] [PubMed] [Google Scholar]
- 5.Porse A., Schou T. S., Munck C., Ellabaan M. M. H., Sommer M. O. A., Biochemical mechanisms determine the functional compatibility of heterologous genes. Nat. Commun. 9, 522 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rankin D. J., Rocha E. P. C., Brown S. P., What traits are carried on mobile genetic elements, and why? Heredity 106, 1–10 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nogueira T.et al., Horizontal gene transfer of the secretome drives the evolution of bacterial cooperation and virulence. Curr. Biol. 19, 1683–1691 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eberhard W. G., Evolution in bacterial plasmids and levels of selection. Q. Rev. Biol. 65, 3–22 (1990). [DOI] [PubMed] [Google Scholar]
- 9.Eberhard W. G., Why do bacterial plasmids carry some genes and not others? Plasmid 21, 167–174 (1989). [DOI] [PubMed] [Google Scholar]
- 10.Haldane J. B. S., A mathematical theory of natural and artificial selection, Part V: Selection and mutation. Math. Proc. Camb. Philos. Soc. 23, 838–844 (1927). [Google Scholar]
- 11.Marad D. A., Buskirk S. W., Lang G. I., Altered access to beneficial mutations slows adaptation and biases fixed mutations in diploids. Nat. Ecol. Evol. 2, 882–889 (2018). [DOI] [PubMed] [Google Scholar]
- 12.del Solar G., Espinosa M., Plasmid copy number control: An ever-growing story. Mol. Microbiol. 37, 492–500 (2000). [DOI] [PubMed] [Google Scholar]
- 13.Rakonjac J., Bennett N. J., Spagnuolo J., Gagic D., Russel M., Filamentous bacteriophage: Biology, phage display and nanotechnology applications. Curr. Issues Mol. Biol. 13, 51–76 (2011). [PubMed] [Google Scholar]
- 14.Rodriguez-Beltran J.et al., Multicopy plasmids allow bacteria to escape from fitness trade-offs during evolutionary innovation. Nat. Ecol. Evol. 2, 873–881 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.San Millan A., Escudero J. A., Gifford D. R., Mazel D., MacLean R. C., Multicopy plasmids potentiate the evolution of antibiotic resistance in bacteria. Nat. Ecol. Evol. 1, 10 (2016). [DOI] [PubMed] [Google Scholar]
- 16.Popa O., Dagan T., Trends and barriers to lateral gene transfer in prokaryotes. Curr. Opin. Microbiol. 14, 615–623 (2011). [DOI] [PubMed] [Google Scholar]
- 17.Gogarten J. P., Townsend J. P., Horizontal gene transfer, genome innovation and evolution. Nat. Rev. Microbiol. 3, 679–687 (2005). [DOI] [PubMed] [Google Scholar]
- 18.Willson C., Perrin D., Cohn M., Jacob F., Monod J., Non-inducible mutants of the regulator gene in the “lactose” system of Escherichia coli. J. Mol. Biol. 8, 582–592 (1964). [DOI] [PubMed] [Google Scholar]
- 19.Bjedov I.et al., Involvement of Escherichia coli DNA polymerase IV in tolerance of cytotoxic alkylating DNA lesions in vivo. Genetics 176, 1431–1440 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Luria S. E., Delbrück M., Mutations of bacteria from virus sensitivity to virus resistance. Genetics 28, 491–511 (1943). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Toprak E.et al., Evolutionary paths to antibiotic resistance under dynamically sustained drug selection. Nat. Genet. 44, 101–105 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Woodford N., Ellington M. J., The emergence of antibiotic resistance by mutation. Clin. Microbiol. Infect. 13, 5–18 (2007). [DOI] [PubMed] [Google Scholar]
- 23.Cirz R. T.et al., Inhibition of mutation and combating the evolution of antibiotic resistance. PLoS Biol. 3, e176 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nagel R., Chan A., Mistranslation and genetic variability: The effect of streptomycin. Mutat. Res. 601, 162–170 (2006). [DOI] [PubMed] [Google Scholar]
- 25.Hane M. W., Wood T. H., Escherichia coli K-12 mutants resistant to nalidixic acid: Genetic mapping and dominance studies. J. Bacteriol. 99, 238–241 (1969). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lederberg J., Streptomycin resistance; a genetically recessive mutation. J. Bacteriol. 61, 549–550 (1951). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Watson M., Liu J. W., Ollis D., Directed evolution of trimethoprim resistance in Escherichia coli. FEBS J. 274, 2661–2671 (2007). [DOI] [PubMed] [Google Scholar]
- 28.Martínez-García E., Aparicio T., Goñi-Moreno A., Fraile S., de Lorenzo V., SEVA 2.0: An update of the standard European vector architecture for de-/re-construction of bacterial functionalities. Nucleic Acids Res. 43, D1183–D1189 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Davies J., Origins and evolution of antibiotic resistance. Microbiologia 12, 9–16 (1996). [PubMed] [Google Scholar]
- 30.Stokes H. W., Gillings M. R., Gene flow, mobile genetic elements and the recruitment of antibiotic resistance genes into Gram-negative pathogens. FEMS Microbiol. Rev. 35, 790–819 (2011). [DOI] [PubMed] [Google Scholar]
- 31.Jia B.et al., CARD 2017: Expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 45, D566–D573 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hayward, DNA blockade by rifampicin-inactivated Escherichia coli RNA polymerase, and its amelioration by a specific mutation. Eur. J. Biochem. 71, 19–24 (1976). [DOI] [PubMed] [Google Scholar]
- 33.Austin S., Scaife J., A new method for selecting RNA polymerase mutants. J. Mol. Biol. 49, 263–267 (1970). [DOI] [PubMed] [Google Scholar]
- 34.Vedantam G., Guay G. G., Austria N. E., Doktor S. Z., Nichols B. P., Characterization of mutations contributing to sulfathiazole resistance in Escherichia coli. Antimicrob. Agents Chemother. 42, 88–93 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Otto S. P., The evolutionary consequences of polyploidy. Cell 131, 452–462 (2007). [DOI] [PubMed] [Google Scholar]
- 36.Blanco P.et al., Bacterial multidrug efflux pumps: Much more than antibiotic resistance determinants. Microorganisms 4, 14 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rodríguez-Martínez J. M.et al., Plasmid-mediated quinolone resistance: Two decades on. Drug Resist. Updat. 29, 13–29 (2016). [DOI] [PubMed] [Google Scholar]
- 38.Poole K., Resistance to β-lactam antibiotics. Cell. Mol. Life Sci. 61, 2200–2223 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Courvalin P., Vancomycin resistance in gram-positive cocci. Clin. Infect. Dis. 42 (suppl. 1), S25–S34 (2006). [DOI] [PubMed] [Google Scholar]
- 40.Jacoby G. A., AmpC beta-lactamases. Clin. Microbiol. Rev. 22, 161–182 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Philippon A., Arlet G., Jacoby G. A., Plasmid-determined AmpC-type beta-lactamases. Antimicrob. Agents Chemother. 46, 1–11 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Redgrave L. S., Sutton S. B., Webber M. A., Piddock L. J. V., Fluoroquinolone resistance: Mechanisms, impact on bacteria, and role in evolutionary success. Trends Microbiol. 22, 438–445 (2014). [DOI] [PubMed] [Google Scholar]
- 43.Liu Y. Y.et al., Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: A microbiological and molecular biological study. Lancet Infect. Dis. 16, 161–168 (2016). [DOI] [PubMed] [Google Scholar]
- 44.Näsvall J., Sun L., Roth J. R., Andersson D. I., Real-time evolution of new genes by innovation, amplification, and divergence. Science 338, 384–387 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sun L.et al., Effective polyploidy causes phenotypic delay and influences bacterial evolvability. PLoS Biol. 16, e2004644 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chung C. T., Niemela S. L., Miller R. H., One-step preparation of competent Escherichia coli: Transformation and storage of bacterial cells in the same solution. Proc. Natl. Acad. Sci. U.S.A. 86, 2172–2175 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zheng Q., rSalvador: An R package for the fluctuation experiment. G3 (Bethesda) 7, 3849–3856 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zheng Q., Methods for comparing mutation rates using fluctuation assay data. Mutat. Res. 777, 20–22 (2015). [DOI] [PubMed] [Google Scholar]
- 49.Palmer A. C., Kishony R., Opposing effects of target overexpression reveal drug mechanisms. Nat. Commun. 5, 4296 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee C., Kim J., Shin S. G., Hwang S., Absolute and relative QPCR quantification of plasmid copy number in Escherichia coli. J. Biotechnol. 123, 273–280 (2006). [DOI] [PubMed] [Google Scholar]
- 51.Hiszczyńska-Sawicka E., Kur J., Effect of Escherichia coli IHF mutations on plasmid p15A copy number. Plasmid 38, 174–179 (1997). [DOI] [PubMed] [Google Scholar]
- 52.Imamovic L., Sommer M. O. A., Use of collateral sensitivity networks to design drug cycling protocols that avoid resistance development. Sci. Transl. Med. 5, 204ra132 (2013). [DOI] [PubMed] [Google Scholar]
- 53.Figurski D. H., Helinski D. R., Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. U.S.A. 76, 1648–1652 (1979). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu M.et al., ICEberg 2.0: An updated database of bacterial integrative and conjugative elements. Nucleic Acids Res. 47, D660–D665 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Akhter S., Aziz R. K., Edwards R. A., PhiSpy: A novel algorithm for finding prophages in bacterial genomes that combines similarity- and composition-based strategies. Nucleic Acids Res. 40, e126 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Szklarczyk D.et al., STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 47, D607–D613 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated during this study are included in this published article (and SI Appendix).