Significance
Food and drug products are supplemented with small molecules called excipients that are assumed to be inert. In this study, we screened a collection of common oral excipients and identified 24 that inhibit intestinal drug transport, including the common excipient FD&C Red No. 40, which decreased drug absorption in mice. Excipient inhibitors were enriched for azo dyes, which human gut bacteria could metabolize, producing metabolites that no longer inhibit intestinal drug transporter activity. This work demonstrates the unintended consequences of oral excipients and a beneficial role for the gut microbiome in limiting these unfavorable effects.
Keywords: excipients, drug absorption, human gut microbiome, azoreductases, food additives
Abstract
Food and drug products contain diverse and abundant small-molecule additives (excipients) with unclear impacts on human physiology, drug safety, and response. Here, we evaluate their potential impact on intestinal drug absorption. By screening 136 unique compounds for inhibition of the key intestinal transporter OATP2B1 we identified and validated 24 potent OATP2B1 inhibitors, characterized by higher molecular weight and hydrophobicity compared to poor or noninhibitors. OATP2B1 inhibitors were also enriched for dyes, including 8 azo (R−N=N−R′) dyes. Pharmacokinetic studies in mice confirmed that FD&C Red No. 40, a common azo dye excipient and a potent inhibitor of OATP2B1, decreased the plasma level of the OATP2B1 substrate fexofenadine, suggesting that FD&C Red No. 40 has the potential to block drug absorption through OATP2B1 inhibition in vivo. However, the gut microbiomes of multiple unrelated healthy individuals as well as diverse human gut bacterial isolates were capable of inactivating the identified azo dye excipients, producing metabolites that no longer inhibit OATP2B1 transport. These results support a beneficial role for the microbiome in limiting the unintended effects of food and drug additives in the intestine and provide a framework for the data-driven selection of excipients. Furthermore, the ubiquity and genetic diversity of gut bacterial azoreductases coupled to experiments in conventionally raised and gnotobiotic mice suggest that variations in gut microbial community structure may be less important to consider relative to the high concentrations of azo dyes in food products, which have the potential to saturate gut bacterial enzymatic activity.
One of the most notable aspects of life in the developed world is the routine exposure to chemicals through processed foods, pharmaceuticals, cosmetics, and the environment. Recent studies suggest that many of these small molecules have deleterious effects on human health (1), although the mechanisms through which they impact human pathophysiology remain poorly described. Even less is known about the reciprocal interactions between these compounds and the trillions of microorganisms that reside within the gastrointestinal tract (the gut microbiome) (2–4), despite emerging evidence that food and cosmetic additives can influence host–microbiome interactions (1, 5).
In the current study, we focus on the bioactivity of pharmaceutical excipients—defined as substances other than the active pharmaceutical ingredients that are intentionally included in an approved drug delivery system or a finished drug product. On average, excipients make up 90% of a drug formulation and play crucial roles including assisting in stability, bioavailability, manufacturing, and patient acceptability (6). Excipients in oral drug products are present in intestinal fluid together with active ingredients; however, their impact on drug disposition is poorly understood. More broadly, many of the excipients used in pharmaceuticals are also widespread and abundant in processed foods and cosmetics and therefore provide a routine source of chemical exposures for most individuals in the developed world.
Results
We aimed to systematically screen the interactions between common pharmaceutical excipients and the intestinal transporter Organic Anion Transporting Polypeptide 2B1 (OATP2B1) by developing an in vitro assay to identify OATP2B1-inhibiting excipients. OATP2B1 (SLCO2B1) is localized to the apical membrane of the intestine and mediates the absorption of many oral prescription drugs, including the statins fluvastatin and rosuvastatin (7), the antihistamine fexofenadine (8, 9), and the β-adrenoreceptor blocker atenolol (10). Compounds that inhibit OATP2B1 transport may potentially reduce the absorption of many drugs. To identify excipients that interact with OATP2B1, we analyzed a comprehensive collection of molecular excipients identified using the Centers of Excellence in Regulatory Science and Innovation (CERSI) Excipients Browser (11) and assembled a library of unique molecular excipients to screen for OATP2B1 transport inhibition. We excluded excipients that are no longer used, poorly soluble (solubility of <1 mM in H2O, dimethyl sulfoxide [DMSO], or ethanol), commercially unavailable, formulated for delivery by inhalation, or alternative salt forms of other excipients (e.g., sodium acetate was tested while calcium/magnesium acetate were not). This collection totaled 136 oral excipients spanning multiple functional classes including dyes, buffering agents, antimicrobial agents, and flavoring agents (Fig. 1A, SI Appendix, Fig. S1, and Dataset S1). This excipient collection was not exclusive to drugs, as 65% of our collection are also found in food products (Dataset S1).
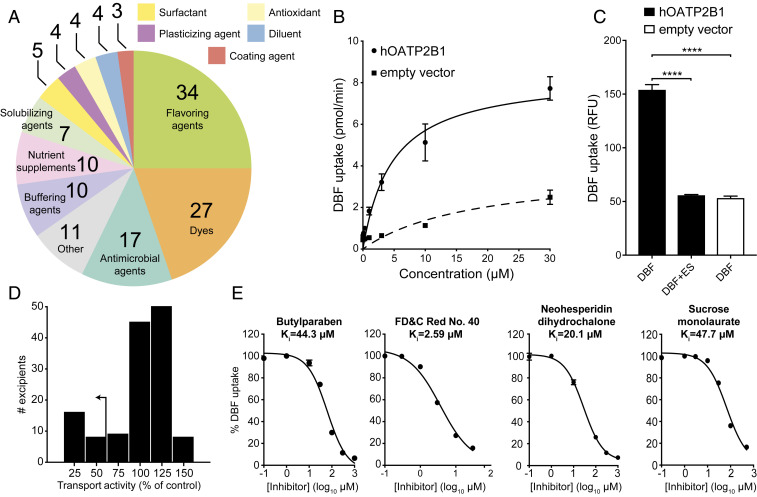
Fig. 1.
Multiple oral excipients inhibit OATP2B1-mediated transport. (A) Functional categories of the 136 oral molecular excipients included in our screen. (B) Kinetics of human OATP2B1-mediated DBF uptake. Human OATP2B1-overexpressing (circles) and empty vector-transfected HEK cells (squares) were incubated with DBF from 0.01 μM to 30 μM for 2 min. Data points represent the mean ± SD of DBF uptake from three or more replicate determinations in three experiments. (C) Human OATP2B1-overexpressing HEK cells (black bars) were incubated in HBSS uptake buffer containing 2 μM DBF for 3 min with or without 200 μM estrone sulfate (ES). Empty vector-transfected HEK cells (white bar) were assayed as above for background DBF uptake determination. Each column represents the mean ± SD of DBF uptake from eight replicate determinations. ****P < 0.0001, ANOVA with Dunnett’s correction. (D) Histogram showing the inhibition results of screening 136 excipients against OATP2B1 uptake. Our <50% transport activity cutoff is indicated by the arrow. (E) Dose–response curves of excipients against OATP2B1 transport. A representative excipient from each functional category is shown. Values represent the mean ± SD of DBF uptake from three replicate determinations in a single experiment.
We developed an in vitro assay to identify potential inhibitors of OATP2B1-mediated uptake. The human OATP2B1 complementary DNA (cDNA) was cloned into the mammalian expression vector pcDNA 5/FRT and transfected into human embryonic kidney (HEK) Flp-In cells to generate a stable OATP2B1-overexpressing cell line. We selected HEK cells for our screen because of their routine and robust use for functional transporter expression assays (12) as well as possessing a low innate rate of drug uptake. The fluorescent molecule 4′,5′-dibromofluorescein (DBF) was used as a substrate of OATP2B1 uptake for screening. We determined a Michaelis constant (Km) = 4.7 μM for OATP2B1-mediated uptake of DBF (Fig. 1B), which is in agreement with the literature (13). The role of OATP2B1 in DBF uptake was confirmed by comparing our overexpressing cell line to a control cell line (HEK Flp-In cells transfected with empty vector pcDNA), revealing a significant (threefold) increase in uptake under OATP2B1-overexpressing conditions (Fig. 1C). DBF uptake was significantly inhibited by the endogenous OATP2B1 substrate estrone sulfate (Fig. 1C). The statistical effect size (Z′-factor) was 0.79, indicating excellent assay quality (14). Together, these results demonstrate that our assay reliably and reproducibly measures the rate of OATP2B1-dependent DBF uptake.
Next, we used our validated assay to screen the full 136-member excipient library for inhibition of DBF uptake. Considering that the amount of excipients in an oral drug product can outweigh the active ingredient by 10- to 100-fold, we selected an excipient screening concentration of 200 µM [10-fold above the standard 20 µM used in drug screens (15); Dataset S1]. We classified hits as excipients that inhibited OATP2B1 uptake by >50% in order to focus on a manageable number of excipients for follow-up dose–response studies. Using this criterion, we identified 24 inhibitors in our primary screen (17.6% of the library; Fig. 1D). Next, we validated each of the 24 excipients by performing dose–response inhibition assays. This analysis validated 100% (24/24) of the inhibitors identified in our primary screen (Fig. 1E and SI Appendix, Table S1), providing additional support for our >50% inhibition threshold and suggesting that additional excipient inhibitors remain to be discovered. Six excipients were potent inhibitors of OATP2B1-mediated uptake with an inhibition constant (Ki) ≤1.0 µM (SI Appendix, Table S1). We further validated two excipient uptake inhibitors, FD&C Red No. 40 and FD&C Yellow No. 6, using the endogenous OATP2B1 substrate estrone sulfate (SI Appendix, Fig. S2). An aggregation test on the 24 excipient inhibitors demonstrated that the Ki values of nine excipients were >10 times their aggregation concentration, suggesting specific inhibition of OATP2B1 (SI Appendix, Table S2). These results indicate that the ability of oral food and drug excipients to inhibit drug uptake is far broader than previously appreciated (16).
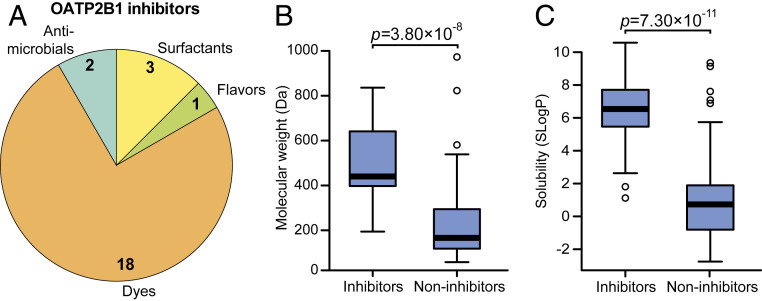
The identified OATP2B1 transport inhibitors were chemically and functionally diverse, which included dyes, surfactants, antimicrobials, and flavoring agents (Fig. 2A). Despite this diversity, we were able to identify characteristic signatures of OATP2B1 inhibitors relative to the overall library. Multiple physicochemical properties of excipient inhibitors were distinct relative to noninhibitors, including increased molecular weight (P = 3.80 10−8, Student’s t test; Fig. 2B) and lipophilicity (P = 7.30 10−11, Student’s t test; Fig. 2C and SI Appendix, Fig. S3 and Table S3). We also found that OATP2B1 inhibitors were significantly enriched for dyes relative to the overall panel, representing 18/24 (75%) of the inhibitors (P < 0.0001, Fisher’s exact test; Fig. 2A and SI Appendix, Table S1).
Fig. 2.
Excipient inhibitors of OATP2B1 uptake have distinct physicochemical and functional properties compared to noninhibitors. (A) Composition of the OATP2B1-inhibiting excipients identified from our screen. Excipient inhibitors were significantly enriched for dyes (P < 0.0001, Fisher’s exact test). Excipient inhibitors display higher molecular weight (B) and solubility (SLogP; Crippen’s log of the octanol/water partition coefficient) (C) compared to noninhibitors. Box plots are shown for data in B and C (n = 24 inhibitors versus 112 noninhibitors) with the bold line representing the median. Points outside the bars are shown as open circles. P values represent Student’s t tests.
OATP2B1 plays an important role in fexofenadine absorption (8, 17) and is a target of food–drug interactions (9, 18). We selected the azo dye excipient FD&C Red No. 40 for in vivo studies because it is the dye with the highest approved amount in the United States by the Food and Drug Administration (FDA) and widely used in both food and drug products (19). The inhibitory effect of FD&C Red No. 40 on fexofenadine bioavailability was examined in P-glycoprotein (Pgp)-deficient (mdr1a/b−/−) mice since fexofenadine is also a substrate of Pgp, which reduces absorption of fexofenadine (20). Concomitant administration of 25 mg/kg FD&C Red No. 40 (10 mM estimated intestinal concentration) significantly reduced fexofenadine area under the plasma concentration–time curve (AUC0–360) (n = 9) by 48% compared to administration of the vehicle (n = 8 mice per group, P = 0.0026, ANOVA with Tukey’s correction; Fig. 3 A and C and SI Appendix, Table S4). We also observed a trend toward decreased peak plasma concentration (Fig. 3D). In contrast, administration of 2.5 mg/kg FD&C Red No. 40 (1 mM estimated intestinal concentration) (n = 4) resulted in fexofenadine levels comparable to vehicle controls (Fig. 3 A–D).
Fig. 3.
Oral administration of the OATP2B1 transport excipient inhibitor FD&C Red No. 40 reduces fexofenadine bioavailability in mice. (A) Plasma concentration–time profiles of fexofenadine after oral administration of 15 mg/kg fexofenadine with saline (blue circles, n = 8), 2.5 mg/kg FD&C Red No. 40 (green squares, n = 4), or 25 mg/kg FD&C Red No. 40 (red triangles, n = 9) to Pgp-deficient (mdr1a/b−/−) mice. Each point represents the mean ± SEM. (B–D) Pharmacokinetic parameters calculated using Phoenix WinNonlin v8.1: (B) plasma fexofenadine AUC0–120min, (C) plasma fexofenadine AUC0–360min, and (D) maximum plasma concentration (Cmax). Each point represents the data from an individual mouse administered with 15 mg/kg fexofenadine plus saline (blue; n = 8), fexofenadine plus 2.5 mg/kg FD&C Red No. 40 (green; n = 4), and fexofenadine plus 25 mg/kg FD&C Red No. 40 (red; n = 9). *P < 0.05, **P < 0.01; ANOVA with Tukey’s correction.
Of the 18 excipient dyes identified as inhibitors of OATP2B1 transport, 8 belong to the azo dye family: synthetic dyes with one or more azo bonds (the functional group R−N=N−R′). The reductive cleavage of the azo bond is facilitated by azoreductases, which are encoded by phylogenetically diverse bacteria, including multiple bacterial taxa prevalent in the human gastrointestinal tract (21). Because azo dye excipients are orally administered, they have the opportunity of encountering and being cleaved by bacterial azoreductases, thus altering their chemical structure and potentially also their bioactivity.
Despite sharing an azo bond, azo dyes are structurally diverse and display variable susceptibility to reduction by bacteria (22, 23). Furthermore, the ability of gut bacteria to metabolize the specific azo dyes identified in our screen was poorly understood (22, 24, 25). To test the ability of complex human gut microbiotas to metabolize the eight identified azo dye OATP2B1 inhibitors, we performed an ex vivo screen wherein each of the azo dye excipients was anaerobically incubated with human fecal samples from three unrelated healthy individuals. One dye was removed from this analysis for technical reasons. Following azo bond reduction, these dyes lose their chromogenic properties and therefore become colorless. All of the tested dyes were cleared by human gut bacteria (Fig. 4A). The extent of dye elimination varied between dyes, ranging from 90.5 ± 2.9 to 48.2 ± 7.3%, consistent with prior data suggesting that these enzymes display some degree of substrate specificity (22, 23).
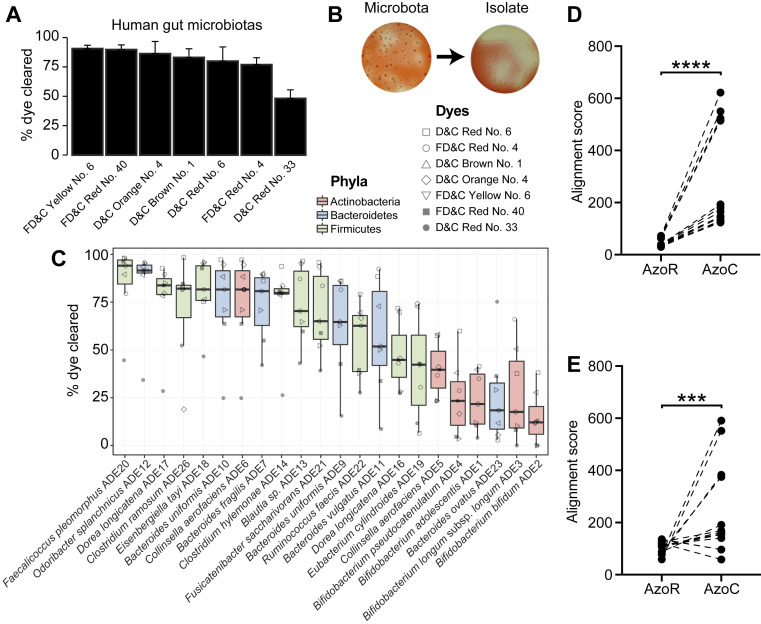
Fig. 4.
Human gut bacteria metabolize azo dye excipients. (A) Excipient azo dyes are cleared following the ex vivo incubation with human fecal samples. Dilutions of human fecal suspensions from three healthy unrelated individuals were incubated anaerobically for 24 h in BHI+ media supplemented with various excipient azo dyes and conditioned media were sampled and analyzed for residual dye concentration spectrophotometrically. Data are normalized to uninoculated controls. Values represent mean ± SEM. (B) Isolation of human gut bacteria capable of clearing FD&C Red No. 40. Dilutions of human fecal suspensions were used to inoculate BHI+ agar media supplemented with various excipient azo dyes and incubated anaerobically, with FD&C Red No. 40 shown here. Azo dye-depleting colonies were identified by a zone of decolorization. Isolates capable of azo dye clearance were picked and restreaked on BHI+ agar media supplemented with the same azo dye to confirm the phenotype. (C) Human gut bacterial isolates show variable abilities to deplete azo dye excipients. Human fecal bacterial isolates previously identified as azo dye metabolizers were incubated in triplicate in BHI+ media supplemented with various excipient azo dyes and incubated anaerobically for 24 h. Residual dye was quantified spectrophotometrically from conditioned media samples and normalized to uninoculated controls. Each point represents the mean dye clearance and the shape corresponds to the excipient azo dye. Colored boxes correspond to bacterial phyla. (D–E) Needleman–Wunsch alignment scores of the azoreductase homologs from the nine newly sequenced genomes (D) and previously sequenced reference genomes (E) when aligned to AzoR from E. coli or AzoC from C. perfringens. Each point represents one gene. Paired Student’s t test, ***P < 0.001, ****P < 0.0001.
Next, we developed an agar plate-based assay to identify human gut bacterial isolates capable of excipient azo dye metabolism. Dilutions of human fecal suspensions were applied on agar plates supplemented with azo dyes and incubated anaerobically. We identified metabolizers by inspecting agar plates for colonies that produced a zone of dye clearance, indicative of azo bond cleavage (Fig. 4B and SI Appendix, Fig. S4A). Representative positive isolates were selected and restreaked on azo dye-containing media to confirm their phenotype (Fig. 4B and SI Appendix, Fig. S4 B and C). A PCR-based fingerprinting method (26) was used to dereplicate strains from the same subject; the same bacterial fingerprint from a single human subject was observed across multiple dyes (SI Appendix, Table S5).
To more definitively test for the ability of each isolate to metabolize multiple dyes, we selected 26 unique azo dye excipient-metabolizing bacterial isolates for 16S ribosomal RNA (rRNA) gene sequencing. We identified bacteria from the three major phyla found in the gut: Bacteroidetes (n = 9), Firmicutes (n = 11), and Actinobacteria (n = 6) (SI Appendix, Table S5). All of the bacterial strains tested (n = 22) were capable of clearing multiple azo dyes (Fig. 4C). The efficiency of dye elimination was primarily determined by strain, not by dye (55% versus 19% of total variation; both factors P < 0.0001, two-way ANOVA). This analysis also revealed a phylogenetic signature of azo dye excipient clearance, with bacteria belonging to the Firmicutes and Bacteroidetes phyla significantly more active than Actinobacteria (both comparisons P < 0.0001, ANOVA with Tukey’s correction; Fig. 4C and SI Appendix, Fig. S5). Together, these results indicate that human gut bacteria can metabolize multiple excipient dyes and the extent of metabolism is influenced by bacterial taxonomy.
Next, we sought to gain insight into the genes and enzymes responsible for gut bacterial azo dye excipient metabolism. We selected nine human gut bacterial isolates from the two most active phyla (SI Appendix, Fig. S5) for sequencing: Firmicutes (one Clostridium ramosum and one Ruminococcus lactaris) and Bacteroidetes (two Bacteroides ovatus, two Bacteroides uniformis, two Bacteroides vulgatus, and one Odoribacter splanchnicus). Illumina sequencing and assembly resulted in high-quality draft genomes (>99% completion; <5% contamination) for all nine isolates (SI Appendix, Table S6). We then searched for predicted proteins with significant sequence homology to a set of five previously characterized gut bacterial azoreductases originating from the Firmicutes and Proteobacteria phyla (27–31). As expected, all nine of the isolate genomes encoded at least one azoreductase homolog (Dataset S2). Surprisingly, all of the top matches were to AzoC, a noncanonical azoreductase from the strict anaerobe Clostridium perfringens that uses flavin adenine dinucleotide (FAD) as a cofactor as opposed to flavin mononucleotide (27) (Fig. 4D).
This observation was generalizable across a broader panel of gut bacterial reference genomes. We retrieved 22 additional reference genomes from the Integrated Microbial Genome Database (32) that belonged to six of the bacterial species identified in our screen (Dataset S2), including Bifidobacterium bifidum (seven genomes), Collinsella aerofaciens (one genome), Bacteroides fragilis (eight genomes), B. ovatus (two genomes), B. vulgatus (two genomes), and O. splanchnicus (two genomes). Consistent with our newly sequenced genomes, the top matches for all but one of the species were to AzoC from C. perfringens (Dataset S2). The one exception, B. bifidum, was the least active isolate of the 22 we characterized (Fig. 4C) and was more closely related to AzoR from Escherichia coli (Fig. 4E). Taken together, these results suggest that the well-characterized AzoR from E. coli may not be representative of the dominant class of azoreductases found within the human gut, even for other Gram-negative phyla like the Bacteroidetes.
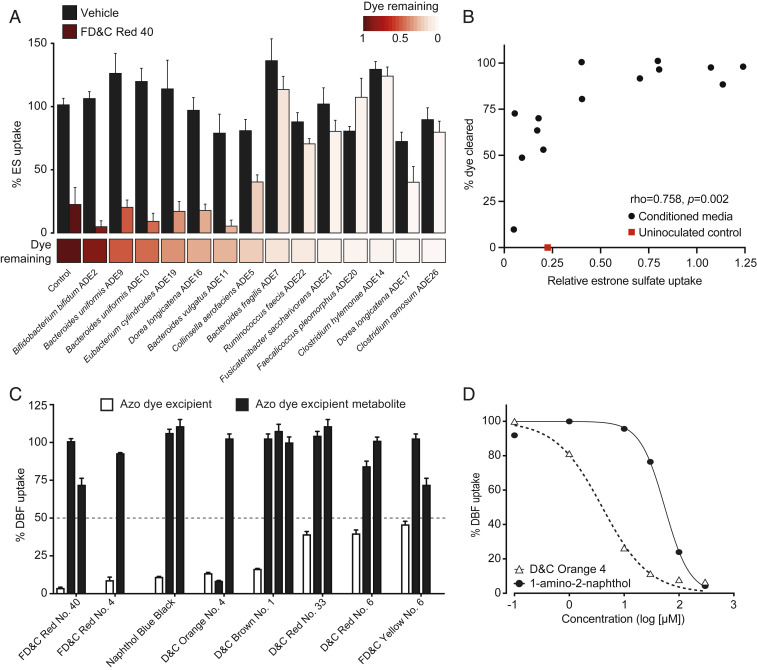
We hypothesized that bacterial metabolism of azo dye excipients would decrease their ability to inhibit OATP2B1 transport based on our previous observation that lower molecular weight is a characteristic feature distinguishing OATP2B1 inhibitors versus noninhibitors (Fig. 2B). To test this hypothesis, we assayed conditioned media of human bacterial isolates grown in the presence of the azo dye FD&C Red No. 40 for OATP2B1 uptake inhibition. This representative azo dye excipient was selected due to our previous in vivo results (Fig. 3) and because it is the dye with the highest amount certified in the United States by the FDA and widely used in both food and drug products (19). Samples of conditioned media that showed complete clearance of FD&C Red No. 40 failed to inhibit OATP2B1-mediated estrone sulfate uptake, whereas samples with high levels of FD&C Red No. 40 remaining inhibited uptake (Fig. 5A). Liquid chromatography–high-resolution mass spectrometry (LC-HRMS) analysis confirmed the depletion of FD&C Red No. 40 from conditioned media samples (SI Appendix, Fig. S6). As expected, the level of clearance of FD&C Red No. 40 was significantly correlated with OATP2B1-mediated estrone sulfate uptake (Fig. 5B).
Fig. 5.
Microbial metabolism of azo dye excipients rescues OATP2B1 uptake inhibition. (A) Conditioned media samples of gut bacteria grown in the presence of 200 μM FD&C Red No. 40 or vehicle (DMSO) were assayed for the ability to inhibit OATP2B1 uptake. Human OATP2B1-overexpressing cells were incubated with HBSS uptake buffer containing 10 nM [3H]estrone sulfate and 10% (vol/vol) bacterial conditioned media supplemented with FD&C Red No. 40 or vehicle (DMSO). Values represent the normalized mean ± SD of [3H]estrone sulfate uptake from three replicate determinations. The heat map below the bar graph shows relative FD&C Red No. 40 dye remaining in conditioned media samples, which was determined spectrophotometrically. (B) FD&C Red No. 40 levels are associated with OATP2B1-mediated [3H]estrone sulfate uptake (Spearman’s correlation). (C) Inhibition of OATP2B1-mediated DBF uptake by azo dyes and their corresponding reduced metabolites. Human OATP2B1-overexpressing HEK cells were incubated in HBSS uptake buffer containing 2 μM DBF for 3 min with the designated concentrations of each azo dye and their metabolites. Concentrations assayed are specified in Dataset S1 and SI Appendix, Table S7. Values represent the mean ± SD of normalized DBF uptake from n ≥ 3 replicate determinations. (D) Dose–response curves of D&C Orange No. 4 and its metabolite, 1-amino-2-naphthol, against OATP2B1-mediated DBF uptake. Values represent the mean ± SD of DBF uptake from three replicate determinations.
To more definitively test if this decrease in OATP2B1 inhibition was due to the biotransformation of azo dye excipients to their downstream microbial metabolites, we tested 12 unique metabolites from each of the eight azo dye inhibitors. In contrast to the azo dye substrates, all but one of the corresponding reduced metabolites resulted in levels of DBF uptake above our 50% cutoff, demonstrating their decreased ability to inhibit OATP2B1 transport (Fig. 5C and SI Appendix, Table S7). The one exception (a metabolite of D&C Orange No. 4) had a much higher inhibition constant: the Ki value for D&C Orange No. 4 was 2.11 μM and the Ki values were 62.5 μM and >200 μM for 1-amino-2-naphthol and sulfanilic acid, respectively (Fig. 5D). These in vitro data suggest that gut bacterial azoreductase activity could rescue the inhibition of OATP2B1 uptake by azo dye excipients.
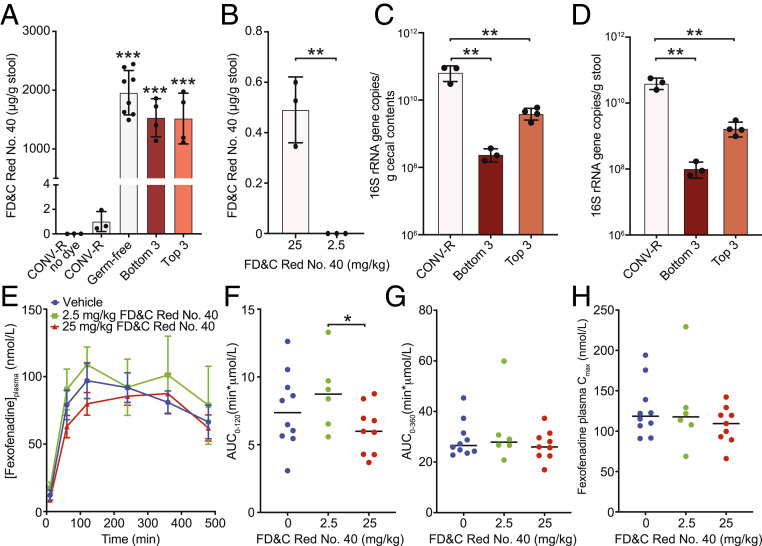
Experiments in mouse models provided additional support for the physiological relevance of azo dye excipient FD&C Red No. 40 and the gut microbiome for the absorption of the OATP2B1 substrate fexofenadine. As expected, conventionally raised (CONV-R) wild-type BALB/c mice excreted significantly less FD&C Red No. 40 (oral dose of 5 mg per mouse or ∼250 mg/kg) relative to germ-free controls (Fig. 6A; n = 3 to 8 mice per group). No dye was detected in untreated CONV-R mice. FD&C Red No. 40 was also detectable in stool samples at the more modest dose (25 mg/kg) that led to reduced fexofenadine bioavailability but not at the lowest dose tested (2.5 mg/kg) that had plasma fexofenadine levels indistinguishable from vehicle controls (Figs. 3 and 6B). These observations led us to hypothesize that gut bacterial azoreductase activity could be saturated at high azo dye levels, which would lead to OATP2B1 inhibition even in the presence of azo dye excipient-metabolizing strains. Consistent with this hypothesis, colonization of germ-free mice with two distinct three-member synthetic communities composed of the top three and bottom three FD&C Red No. 40 metabolizers (SI Appendix, Fig. S4C) led to levels of dye excretion that were indistinguishable from germ-free controls (Fig. 6A; n = 3 to 8 mice per group). The redundancy of azoreductases across diverse bacterial taxa together with the higher levels of colonization (Fig. 6 C and D) may explain why CONV-R mice clear azo dyes better than the gnotobiotic mice tested here (Fig. 6A). Alternatively, the broader differences in intestinal permeability (33) and gene expression (34, 35) in germ-free and gnotobiotic mice may disrupt the complex interactions between excipients, the gut microbiome, and drug transporters. We rederived the Pgp-deficient mouse model under germ-free conditions (Materials and Methods) and repeated our fexofenadine pharmacokinetics experiment; however, unlike in CONV-R mice (Fig. 3), the high dose of FD&C Red No. 40 had a more modest impact on fexofenadine bioavailability in germ-free Pgp-deficient animals (Fig. 6 E–H and SI Appendix, Table S4). More work is needed to determine the mechanisms responsible, including differences in both drug and excipient disposition in the absence of a gut microbiome.
Fig. 6.
Interactions between the gut microbiota, azo dye excipients, and drug absorption in CONV-R and gnotobiotic mice. (A and B) Recovery of residual FD&C Red No. 40 dye in fecal collections over 24 h. (A) CONV-R, germ-free (GF), and gnotobiotic mice were orally dosed with 5 mg of FD&C Red No. 40 (∼250 mg/kg; n = 3 to 8 mice per group). CONV-R mice without dye are also included as a control (n = 3). Bottom 3 indicates mice colonized with the three worst FD&C Red No. 40 metabolizers: B. bifidum ADE2, B. pseudocatenulatum ADE4, and B. adolescentis ADE1. Top 3 indicates mice colonized with the best 3 FD&C Red No. 40 metabolizers: F. pleomorphus ADE20, C. hylemonae ADE14, and C. ramosum ADE26. (B) Fecal FD&C Red No. 40 dye levels in CONV-R mice depend upon oral dose (n = 3 mice per group). (C and D) Total colonization is significantly higher in the (C) cecum and (D) stool of CONV-R mice relative to both gnotobiotic groups. (E) Plasma concentration–time profiles of fexofenadine after oral administration of 15 mg/kg fexofenadine plus saline (blue circles, n = 10), 2.5 mg/kg FD&C Red No. 40 (green squares, n = 6), or 25 mg/kg FD&C Red No. 40 (red triangles, n = 9) to Pgp-deficient (mdr1a/b−/−) germ-free mice. Each point represents the mean ± SEM. (F–H) Pharmacokinetic parameters calculated using Phoenix WinNonlin v8.1: (F) plasma fexofenadine AUC0–120min, (G) plasma fexofenadine AUC0–360min, and (H) maximum plasma concentration (Cmax). Each point represents the data from an individual mouse administered with 15 mg/kg fexofenadine plus saline (blue; n = 10), 2.5 mg/kg FD&C Red No. 40 (green; n = 6), or 25 mg/kg FD&C Red No. 40 (red; n = 9). Statistics: (A, C, and D) ANOVA with Tukey’s correction, **P < 0.01, ***P < 0.001; (B and F) unpaired Student’s t test, *P < 0.05, **P < 0.01. The reported contrasts in A are relative to CONV-R mice either with or without dye administration.
Discussion
Pharmaceutical excipients constitute, on average, 90% of a drug formulation and yet they are often assumed to be inactive despite not being explicitly tested. Nevertheless, the similarity ensemble approach (36) reveals that some excipients are structurally similar to bioactive molecules, including drugs, and may share their on-target activities (11). Our results confirm and extend several recent in vitro studies suggesting that excipients can inhibit intestinal drug absorption (37–39). By comprehensively screening a collection of 136 oral molecular excipients, we report the discovery of 24 inhibitors of the intestinal drug-absorptive transporter OATP2B1. These excipients belong to several functional classes including dyes, surfactants, antimicrobial preservatives, and flavoring agents, six of which are predicted to achieve clinically relevant intestinal concentrations (SI Appendix, Table S8). Our study, along with others, suggests that pharmaceutical excipients are more bioactive than assumed, potentially affecting drug therapy, human biology, and pathophysiology, which are all important areas for future study. Although clinical trials are needed to determine the in vivo effects of these excipients, these findings have potential clinical implications due to the key role of OATP2B1 in drug absorption (8, 9, 17, 18). On the other hand, our data provide multiple strategies for mitigating the bioactivity of food and drug additives. These include identifying prescription drugs that are substrates of OATP2B1 and additives that are inhibitors of OATP2B1. Current formulation strategies can be modified accordingly to mechanistic-understanding-based formulation, which is of particular importance for the generic drug industry to develop bioequivalent drug product formulations (40).
To further assess the potential clinical relevance of the OATP2B1-inhibiting excipients, we estimated the maximal intestinal concentrations of excipients based on the maximum allowable amount from the World Health Organization and the Code of Federal Regulations Title 21. According to the FDA perspective about the role of transporters in drug–drug interactions (41), it is possible that excipients with [I]/Ki ≥10 ([I], estimated intestinal concentration) could inhibit OATP2B1-mediated drug absorption at clinically relevant concentrations. The [I]/Ki values for six excipients exceed 10 if the maximum allowable amount is administered as a single bolus (SI Appendix, Table S8). In addition, a survey reported that the amounts of artificial food colors certified for use in the United States by the FDA was 62 mg per capita per d in 2010, of which FD&C Red No. 40 accounts for 40%, FD&C Yellow No. 6 for 24%, FD&C Blue No.1 for 4%, and FD&C Red No. 3 for 4% (19). Based on 62 mg/d, the [I]/Ki for FD&C Red No. 40 is 84.9 (SI Appendix, Table S8) and thus could have clinically meaningful effects on drug absorption.
Most oral drug products do not contain sufficient concentrations of the identified excipients, especially dyes. Larger amounts are present in food; however, the dyes we consume as part of our diet are not usually delivered as a single bolus together with drugs, lowering the risk of food–drug interactions. High levels of single excipients (e.g., FD&C Red No. 40) can be found in baked food products, up to 1,240 mg/kg (42). Considering a 100-g cupcake, a single serving of baked food products may contain as much as 124 mg of FD&C Red No. 40. For an adult, the fasted small bowel contains a total volume of 43 ± 14 mL of water (43), suggesting that the potential intestinal concentration of FD&C Red No. 40 could reach as high as 10 mM following a single serving of a baked food product. Taken together, these calculations suggest that while drug–dye interactions within a drug formulation may be below physiologically relevant concentrations, dyes consumed through select food products may reach high intestinal concentrations, resulting in the inhibition of OATP2B1-mediated absorption of prescription drugs.
Despite these projections, we found that mice administered with 2.5 mg/kg (1 mM estimated intestinal concentration) of FD&C Red No. 40 did not show a reduction of fexofenadine plasma levels. A likely explanation for this lack of inhibition in vivo is that 2.5 mg/kg FD&C Red No. 40 falls within the metabolic capacity of the gut microbiome’s azoreductase activity, leaving no intact dye and therefore no OATP2B1 interaction. This is supported by a lack of dye detected in the stool (Fig. 6B). On the other hand, 25 mg/kg (10 mM estimated intestinal concentration) of FD&C Red No. 40 appears to saturate the gut microbiome’s azoreductase capacity with detectable levels of FD&C Red No. 40 in the stool (Fig. 6B), resulting in an inhibition of OATP2B1 transport and thus decreased fexofenadine absorption (Fig. 3).
Our data show that excipient inhibitors of OATP2B1 transport are enriched for azo dyes. These dyes are susceptible to metabolism by human gut bacterial azoreductases, producing metabolites that no longer inhibit OATP2B1 transport (SI Appendix, Fig. S7). While our bacterial isolate data provide some support for the possibility that differences in the gut microbiome could alter azo dye metabolism, the ubiquity and functional redundancy of this enzymatic activity across microbial communities and diverse bacterial taxonomic groups (44) suggest that microbiome shifts may not contribute in a meaningful way to interindividual differences in excipient–transporter interactions.
Interestingly, our pharmacokinetic data demonstrate that plasma fexofenadine levels are largely unaffected by FD&C Red No. 40 administration in germ-free mice, which lack bacterial azoreductase activity. We did observe a significant decrease in the fexofenadine AUC0–120 at 25 mg/kg versus 2.5 mg/kg FD&C Red No. 40 or vehicle. On the contrary, despite possessing bacterial azoreductases, CONV-R mice are sensitive to inhibition of fexofenadine absorption at higher FD&C Red No. 40 (25 mg/kg) doses. Despite serving as a powerful tool for investigating the microbial contributions toward the host, germ-free animals display gross developmental and physiological differences compared to their CONV-R counterparts, including broad differences in the expression of genes involved in drug metabolism and transport (34, 35). Although OATP2B1 gene expression does not appear to be differentially expressed in germ-free versus CONV-R mice (35), other factors such as OATP2B1 protein levels and microbial metabolite substrates or inhibitors of OATP2B1 could also contribute to the observed fexofenadine transport inhibition at a high dose of FD&C Red No. 40. Further research is needed to address the mechanisms responsible for the observed differences in fexofenadine absorption during FD&C Red No. 40 administration in CONV-R versus germ-free mice.
In conclusion, these findings emphasize the importance of considering the chemical milieu in which drugs are taken and provide a mechanism through which the human gut microbiome ameliorates the impact of chemical exposures. This study also has clear translational implications, providing a step toward the rational selection of excipients to minimize their off-target impacts on human and microbial cells.
Materials and Methods
Oral Molecular Excipient Library and Reagents.
As of March 2017, 292 oral molecular excipients listed on the University of California San Francisco (UCSF)–Stanford CERSI Excipients Browser (http://excipients.ucsf.bkslab.org/excipients/molecular/?route=oral) were considered for screening (11). This collection was curated by excluding excipients that were duplicates, no longer used, experienced solubility issues, commercially unavailable, or formulated for delivery by inhalation. In total, 136 excipients were screened for OATP2B1-mediated transport inhibition (Dataset S1). DBF was purchased from Sigma-Aldrich. Tritium-labeled estrone sulfate was purchased from PerkinElmer with a specific activity of 54 Ci/mmol.
Establishment of Stable OATP2B1-Overexpressing Cell Line and Cell Culture.
The human OATP2B1 cDNA was cloned into Thermo Fisher Scientific pcDNA 5/FRT Mammalian Expression Vector and transfected into Life Technologies Flp-In 293 cells using Lipofectamine LTX according to the manufacturer’s protocol. Stably transfected cells were maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum, penicillin (100 U/mL), streptomycin (100 μg/mL), sodium pyruvate (110 μg/mL), and hygromycin B (100 μg/mL) at 37 °C in a humidified incubator with 5% CO2.
DBF Uptake Kinetics Assay.
The assay was based on a previously reported protocol (45) with minor modifications. Cells (8 × 104 per well) were seeded in black wall poly-d-lysine–coated 96-well plates 24 h prior to experiments. The DBF uptake kinetics study was conducted by incubating cells in Hank’s balanced salt solution (HBSS) containing 0.001 μM to 30 μM DBF at 37 °C for 2 min. Fluorescence in cells was measured using a fluorescence microplate reader with excitation and emission wavelengths at 485 nm and 560 nm, respectively. OATP2B1-mediated uptake of DBF was determined by subtracting background uptake from the empty vector-transfected control cells at each substrate concentration. The Z′ assay factor was calculated according to the equation: Z′ = 1 to 3(δcontrol+ + δcontrol−)/(μcontrol+ − μcontrol−), where δ and μ are the SD and the mean, respectively (14).
Screening Excipient Library for OATP2B1 Uptake Inhibition and IC50 Determinations.
A total of 136 excipients were screened at 200 μM with 1% (vol/vol) of vehicle (DMSO), except for several surfactants and those with limited solubility, which were tested at lower concentrations (Dataset S1). Downstream reduced metabolites of selected azo dye excipients were synthesized by Advinus Therapeutics and tested at 200 μM, except for those with limited solubility, which were tested at lower concentrations (SI Appendix, Table S7). The screen was performed in triplicate with 2 μM DBF. The estrone sulfate uptake assay was conducted by incubating cells in HBSS containing 0.02 μM [3H]estrone sulfate for 3 min at 37 °C. The cells were then washed twice with ice-cold HBSS followed by lysis using lysis buffer (0.1 N NaOH and 0.1% sodium dodecyl sulfate). [3H]estrone sulfate uptake was determined by radioactivity in the lysate using a liquid scintillation counter. IC50 values (concentration that inhibits transporter activity by 50%) for compounds were determined by fitting the uptake results to the Hill equation by nonlinear regression using GraphPad Prism. The Ki was estimated using the following equation (12):
| [1] |
The Km value of DBF for OATP2B1 is 4.7 μM and S is 2 μM.
Analysis of Physicochemical Properties of OATP2B1 Excipient Inhibitors.
Molecular structure files in structure-data format (SDF) were obtained from the CERSI Excipients Browser at http://excipients.ucsf.bkslab.org/ and the PubChem database (46). MayaChemTools software (47) was used to compute eight one-dimensional and two-dimensional descriptors, such as molecular weight (MW), number of heavy atoms (HA), number of rotatable bonds (RB), molecular volume (MV), number of hydrogen bond donors (HBD), number of hydrogen bond acceptors (HBA), Crippen’s log of the octanol/water partition coefficient (including implicit hydrogens) (SLogP), and total polar surface area (TPSA). Boxplots were prepared using R software. Differences between distributions of molecular descriptors were assessed by the Student’s t test using R software. The R package FSelector (48) was used to select two nonredundant and informative physicochemical properties from the eight descriptors (e.g., MW and SLogP). A description of the aggregation test can be found in SI Appendix.
Fexofenadine Pharmacokinetic Studies.
All animal experiments were approved by the UCSF Institutional Animal Care and Use Committee (IACUC). All pharmacokinetic experiments were performed using 8- to 10-wk-old male Pgp-deficient (mdr1a/b−/−) mice (1487-M; Taconic Biosciences). Germ-free Pgp-deficient mice were derived by cesarean delivery into the UCSF Gnotobiotic Core Facility and maintained in germ-free isolators. Germ-free status of mice were confirmed by monthly screening of fecal pellets by PCR for amplification of the 16S rRNA (for detection of bacteria) and internal transcribed spacer (for detection of fungi) gene sequences. Mice were fasted overnight prior to drug and excipient oral administration. For the vehicle control group, saline was administered by oral gavage immediately followed by 15 mg/kg fexofenadine by oral administration. For test groups, FD&C Red No. 40 was administered at a concentration of 2.5 mg/kg or 25 mg/kg by oral gavage immediately followed by 15 mg/kg fexofenadine by oral administration. Blood samples (∼25 μL per time point) were obtained from the mouse tail vein at 10, 30, 60, 120, 240, 360, and 480 min after drug administration. Pharmacokinetic experiments with germ-free mice were conducted in a class II biosafety cabinet to ensure sterility and blood samples were obtained at 10, 60, 120, 240, 360, and 480 min after drug administration. Blood samples were collected in Fisherbrand heparinized glass microhematocrit capillary tubes and centrifuged at 3,500 g for 15 min to recover blood plasma, which was stored at −80 °C until analysis. Methods describing the analytical quantification of fexofenadine from plasma can be found in SI Appendix. Pharmacokinetic parameters were computed via noncompartmental analysis using Phoenix WinNonlin v8.1 (Certara). Area under the curve (AUC) was calculated using the linear-up, log-down method. Exact sample collection times were used for pharmacokinetic analysis.
Isolation and Screening Human Gut Bacteria for Azo Dye Excipient Metabolism.
Our full study protocol for the human fecal samples used in this work received approval from the UCSF Institutional Review Board with informed consent from volunteers. A detailed description of methods can be found in SI Appendix.
Assaying Bacterial Conditioned Azo Dye-Containing Media for Inhibition of OATP2B1 Uptake.
Human gut bacterial isolates were grown as described in SI Appendix, Screening Human Fecal Samples and Bacterial Isolates for Azo Dye Metabolism in Liquid Cultures. Following 48 h of incubation, 96-well plates were centrifuged to pellet bacterial cells and the resulting conditioned media were recovered. Inhibition of OATP2B1-mediated [3H]estrone sulfate uptake in OATP2B1-transfected HEK cells was performed as described in Screening Excipient Library for OATP2B1 Uptake Inhibition and IC50 Determinations with the addition of conditioned media samples at 10% (vol/vol). A detailed description of the analytical analysis of conditioned media samples by LC-HRMS can be found in SI Appendix.
Quantification of FD&C Red No. 40 from Murine Fecal Samples.
All animal experiments were approved by the UCSF IACUC and conducted with support from the UCSF Gnotobiotic Core Facility (https://gnotobiotics.ucsf.edu/). CONV-R mice were housed in a specific-pathogen-free facility. Female BALB/c mice (BALB-F; Taconic Biosciences) were used between 8 and 10 wk of age. Germ-free female BALB/c mice between 8 and 10 wk of age were bred in-house in sterile breeding isolators (Class Biologically Clean) and transferred to experimental isolators prior to FD&C Red No. 40 dosing. Mice were fed a standard autoclaved chow diet for ad libitum consumption (Lab Diet 5021) with free access to water at room temperature (24 ± 1 °C) and a 12/12-h light/dark cycle (7:00 AM to 7:00 PM). Gnotobiotic mice were colonized by gavage of a pool of overnight cultures of the top three (Faecalicoccus pleomorphus ADE20, Clostridium hylemonae ADE14, and Clostridium ramosum ADE26) or bottom three (Bifidobacterium bifidum ADE2, Bifidobacterium pseudocatenulatum ADE4, and Bifidobacterium adolescentis ADE1) FD&C Red No. 40 metabolizers. Following colonization, gnotobiotic mice acclimated for a week before FD&C Red No. 40 dosing. Metabolic chambers were housed in experimental isolators for the germ-free and gnotobiotic animals to ensure sterility. Mice were orally dosed (0.2 mL) with 5 mg of FD&C Red No. 40 dissolved in saline and filter (0.2 μm)-sterilized and immediately individually housed in a metabolic chamber for mice (72-7060; Harvard Apparatus) for 24 h. A control group of CONV-R mice were dosed with saline instead of FD&C Red No. 40. In addition to the 5 mg of FD&C Red No. 40 dose, CONV-R mice were also orally dosed (0.2 mL) with 25 mg/kg or 2.5 mg/kg of FD&C Red No. 40. Fecal pellets were collected and stored at −80 °C. Fecal pellets were lyophilized overnight to dryness and accurately weighed, then 300 μL of methanol was added and FD&C Red No. 40 was extracted for 40 min by sonication at room temperature. The supernatant was analyzed by LC-HRMS as described in SI Appendix.
Bacterial colonization levels from gnotobiotic and CONV-R mice were determined using qPCR. Total DNA was extracted from cecal and fecal samples using the QIAamp DNA Fast Stool Mini Kit according to the manufacturer’s protocol. qPCR of total 16S rRNA gene copies was carried out in triplicate in 10-µL reactions with 200 nM of 340F (5′-ACTCCTACGGGAGGCAGCAGT-3′) and 514R (5′-ATTACCGCGGCTGCTGGC-3′) primers. Reactions were performed using a BioRad CFX384 thermocycler with Life Technologies SYBRSelect for CFX Master Mix following the manufacturer’s protocol and using a 60 °C annealing temperature. Absolute quantification was determined against a standard curve of E. coli MG1655 genomic DNA. Mean Ct values from each sample were used for analysis. Absolute 16S rRNA gene copies were calculated by adjusting for dilutions and dividing by sample weight used for DNA extraction.
Estimation of Maximum Intestinal Concentrations of Excipients.
The accepted daily intake, maximum allowable amount, or estimated daily intake of each excipient added for oral consumption was used to estimate the maximum intestinal concentration [I] as shown in Eq. 2. The FDA perspective about the role of transporters in drug–drug interactions indicates that drugs exhibiting [I]/IC50 (or Ki) ≥10 should be evaluated for potential clinical drug–drug interactions (41). [I] is the theoretical maximal intestinal concentration of a drug after oral administration calculated as the highest clinical dose (milligrams) in a volume of 250 mL:
| [2] |
Data and Materials Availability.
Data are shown in the main text and figures and SI Appendix. Genome sequencing data has been deposited at the NCBI SRA (BioProject accession no. PRJNA613498) and 16S rRNA gene sequences at GenBank (accession nos. MT268979–MT269002, MT579590, and MT579591).
Supplementary Material
Acknowledgments
We thank Dr. Jordan Bisanz for comments on the manuscript and help with bacterial genome sequencing and assembly, Dr. Jessie Turnbaugh in the UCSF Gnotobiotic Core for technical assistance, and Hayarpi Torosyan and Parnian Lak for their help and expertise in the aggregation assays. This work was supported by the FDA (U01FD004979/U01FD005978) Office of Generic Drugs, which supports the UCSF-Stanford CERSI, the National Institutes of Health (R01HL122593 and R21CA227232), and the Searle Scholars Program (SSP-2016-1352). P.J.T. holds an Investigators in the Pathogenesis of Infectious Disease Award from the Burroughs Wellcome Fund, is a Chan Zuckerberg Biohub investigator, and is a Nadia’s Gift Foundation Innovator supported, in part, by the Damon Runyon Cancer Research Foundation (DRR-42-16). Fellowship support was provided by the Canadian Institutes of Health Research (P.S.) and the National Science Foundation (L.M.P.). The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official views of the Department of Health and Human Services or the US FDA.
Footnotes
Competing interest statement: P.J.T. is on the scientific advisory boards for Kaleido, Pendulum, Seres, and SNIPRbiome; there is no direct overlap between the current study and these consulting duties.
This article is a PNAS Direct Submission.
Data deposition: Genome sequencing data has been deposited at the NCBI SRA, https://www.ncbi.nlm.nih.gov/sra (BioProject accession no. PRJNA613498) and 16S ribosomal RNA gene sequences have been deposited at GenBank, https://www.ncbi.nlm.nih.gov/genbank/ (accession nos. MT268979–MT269002, MT579590, and MT579591).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1920483117/-/DCSupplemental.
References
- 1.Yang H.et al., A common antimicrobial additive increases colonic inflammation and colitis-associated colon tumorigenesis in mice. Sci. Transl. Med. 10, eaan4116 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Spanogiannopoulos P., Bess E. N., Carmody R. N., Turnbaugh P. J., The microbial pharmacists within us: A metagenomic view of xenobiotic metabolism. Nat. Rev. Microbiol. 14, 273–287 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Koppel N., Maini Rekdal V., Balskus E. P., Chemical transformation of xenobiotics by the human gut microbiota. Science 356, eaag2770 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bisanz J. E., Spanogiannopoulos P., Pieper L. M., Bustion A. E., Turnbaugh P. J., How to determine the role of the microbiome in drug disposition. Drug Metab. Dispos. 46, 1588–1595 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chassaing B.et al., Dietary emulsifiers impact the mouse gut microbiota promoting colitis and metabolic syndrome. Nature 519, 92–96 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haywood A., Glass B. D., Pharmaceutical excipients–Where do we begin? Aust. Prescr. 34, 112–114 (2011). [Google Scholar]
- 7.Varma M. V.et al., pH-sensitive interaction of HMG-CoA reductase inhibitors (statins) with organic anion transporting polypeptide 2B1. Mol. Pharm. 8, 1303–1313 (2011). [DOI] [PubMed] [Google Scholar]
- 8.Ming X., Knight B. M., Thakker D. R., Vectorial transport of fexofenadine across Caco-2 cells: Involvement of apical uptake and basolateral efflux transporters. Mol. Pharm. 8, 1677–1686 (2011). [DOI] [PubMed] [Google Scholar]
- 9.Akamine Y.et al., Effects of one-time apple juice ingestion on the pharmacokinetics of fexofenadine enantiomers. Eur. J. Clin. Pharmacol. 70, 1087–1095 (2014). [DOI] [PubMed] [Google Scholar]
- 10.Jeon H.et al., Apple juice greatly reduces systemic exposure to atenolol. Br. J. Clin. Pharmacol. 75, 172–179 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Irwin J. J.et al., A molecular basis for innovation in drug excipients. Clin. Pharmacol. Ther. 101, 320–323 (2017). [DOI] [PubMed] [Google Scholar]
- 12.Giacomini K. M.et al.; International Transporter Consortium , Membrane transporters in drug development. Nat. Rev. Drug Discov. 9, 215–236 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Izumi S.et al., Investigation of fluorescein derivatives as substrates of organic anion transporting polypeptide (OATP) 1B1 to develop sensitive fluorescence-based OATP1B1 inhibition assays. Mol. Pharm. 13, 438–448 (2016). [DOI] [PubMed] [Google Scholar]
- 14.Zhang J. H., Chung T. D., Oldenburg K. R., A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J. Biomol. Screen. 4, 67–73 (1999). [DOI] [PubMed] [Google Scholar]
- 15.Khuri N.et al., Computational discovery and experimental validation of inhibitors of the human intestinal transporter OATP2B1. J. Chem. Inf. Model. 57, 1402–1413 (2017). [DOI] [PubMed] [Google Scholar]
- 16.Panakanti R., Narang A. S., Impact of excipient interactions on drug bioavailability from solid dosage forms. Pharm. Res. 29, 2639–2659 (2012). [DOI] [PubMed] [Google Scholar]
- 17.Medwid S.et al., Fexofenadine and rosuvastatin pharmacokinetics in mice with targeted disruption of organic anion transporting polypeptide 2B1. Drug Metab. Dispos. 47, 832–842 (2019). [DOI] [PubMed] [Google Scholar]
- 18.Dresser G. K.et al., Fruit juices inhibit organic anion transporting polypeptide-mediated drug uptake to decrease the oral availability of fexofenadine. Clin. Pharmacol. Ther. 71, 11–20 (2002). [DOI] [PubMed] [Google Scholar]
- 19.Stevens L. J.et al., Mechanisms of behavioral, atopic, and other reactions to artificial food colors in children. Nutr. Rev. 71, 268–281 (2013). [DOI] [PubMed] [Google Scholar]
- 20.Tahara H., Kusuhara H., Fuse E., Sugiyama Y., P-glycoprotein plays a major role in the efflux of fexofenadine in the small intestine and blood-brain barrier, but only a limited role in its biliary excretion. Drug Metab. Dispos. 33, 963–968 (2005). [DOI] [PubMed] [Google Scholar]
- 21.Haiser H. J., Turnbaugh P. J., Developing a metagenomic view of xenobiotic metabolism. Pharmacol. Res. 69, 21–31 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chung K. T., Fulk G. E., Egan M., Reduction of azo dyes by intestinal anaerobes. Appl. Environ. Microbiol. 35, 558–562 (1978). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chung K. T., Stevens S. E. Jr., Cerniglia C. E., The reduction of azo dyes by the intestinal microflora. Crit. Rev. Microbiol. 18, 175–190 (1992). [DOI] [PubMed] [Google Scholar]
- 24.Wang R.-F., Chen H., Paine D. D., Cerniglia C. E., Microarray method to monitor 40 intestinal bacterial species in the study of azo dye reduction. Biosens. Bioelectron. 20, 699–705 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rafii F., Hall J. D., Cerniglia C. E., Mutagenicity of azo dyes used in foods, drugs and cosmetics before and after reduction by Clostridium species from the human intestinal tract. Food Chem. Toxicol. 35, 897–901 (1997). [DOI] [PubMed] [Google Scholar]
- 26.Versalovic J., Koeuth T., Lupski J. R., Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res. 19, 6823–6831 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morrison J.et al., Structure and stability of an azoreductase with an FAD cofactor from the strict anaerobe Clostridium perfringens. Protein Pept. Lett. 21, 523–534 (2014). [DOI] [PubMed] [Google Scholar]
- 28.Chen H., Xu H., Kweon O., Chen S., Cerniglia C. E., Functional role of Trp-105 of Enterococcus faecalis azoreductase (AzoA) as resolved by structural and mutational analysis. Microbiology 154, 2659–2667 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Macwana S. R., Punj S., Cooper J., Schwenk E., John G. H., Identification and isolation of an azoreductase from Enterococcus faecium. Curr. Issues Mol. Biol. 12, 43–48 (2010). [PubMed] [Google Scholar]
- 30.Nakanishi M., Yatome C., Ishida N., Kitade Y., Putative ACP phosphodiesterase gene (acpD) encodes an azoreductase. J. Biol. Chem. 276, 46394–46399 (2001). [DOI] [PubMed] [Google Scholar]
- 31.Wang C.-J.et al., Molecular cloning, characterisation and ligand-bound structure of an azoreductase from Pseudomonas aeruginosa. J. Mol. Biol. 373, 1213–1228 (2007). [DOI] [PubMed] [Google Scholar]
- 32.Chen I. A.et al., IMG/M v.5.0: An integrated data management and comparative analysis system for microbial genomes and microbiomes. Nucleic Acids Res. 47, D666–D677 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kennedy E. A., King K. Y., Baldridge M. T., Mouse microbiota models: Comparing germ-free mice and antibiotics treatment as tools for modifying gut bacteria. Front. Physiol. 9, 1534 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Björkholm B.et al., Intestinal microbiota regulate xenobiotic metabolism in the liver. PLoS One 4, e6958 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Larsson E.et al., Analysis of gut microbial regulation of host gene expression along the length of the gut and regulation of gut microbial ecology through MyD88. Gut 61, 1124–1131 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Keiser M. J.et al., Predicting new molecular targets for known drugs. Nature 462, 175–181 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sjöstedt N., Deng F., Rauvala O., Tepponen T., Kidron H., Interaction of food additives with intestinal efflux transporters. Mol. Pharm. 14, 3824–3833 (2017). [DOI] [PubMed] [Google Scholar]
- 38.Gurjar R.et al., Inhibitory effects of commonly used excipients on P-glycoprotein in vitro. Mol. Pharm. 15, 4835–4842 (2018). [DOI] [PubMed] [Google Scholar]
- 39.Engel A., Oswald S., Siegmund W., Keiser M., Pharmaceutical excipients influence the function of human uptake transporting proteins. Mol. Pharm. 9, 2577–2581 (2012). [DOI] [PubMed] [Google Scholar]
- 40.Lionberger R. A., FDA critical path initiatives: Opportunities for generic drug development. AAPS J. 10, 103–109 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang L., Zhang Y. D., Strong J. M., Reynolds K. S., Huang S.-M., A regulatory viewpoint on transporter-based drug interactions. Xenobiotica 38, 709–724 (2008). [DOI] [PubMed] [Google Scholar]
- 42.Bastaki M., Farrell T., Bhusari S., Bi X., Scrafford C., Estimated daily intake and safety of FD&C food-colour additives in the US population. Food Addit. Contam. Part A Chem. Anal. Control Expo. Risk Assess. 34, 891–904 (2017). [DOI] [PubMed] [Google Scholar]
- 43.Mudie D. M.et al., Quantification of gastrointestinal liquid volumes and distribution following a 240 mL dose of water in the fasted state. Mol. Pharm. 11, 3039–3047 (2014). [DOI] [PubMed] [Google Scholar]
- 44.Clarke G.et al., Gut reactions: Breaking down xenobiotic-microbiome interactions. Pharmacol. Rev. 71, 198–224 (2019). [DOI] [PubMed] [Google Scholar]
- 45.Erdman A. R.et al., The human organic anion transporter 3 (OAT3; SLC22A8): Genetic variation and functional genomics. Am. J. Physiol. Renal Physiol. 290, F905–F912 (2006). [DOI] [PubMed] [Google Scholar]
- 46.Kim S.et al., PubChem substance and compound databases. Nucleic Acids Res. 44, D1202–D1213 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sud M., MayaChemTools: An open source package for computational drug discovery. J. Chem. Inf. Model. 56, 2292–2297 (2016). [DOI] [PubMed] [Google Scholar]
- 48.Kotthoff L., fselector. https://github.com/larskotthoff/fselector. Accessed 27 August 2019.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.