Significance
We here describe the essential roles of RB and MRPS18-2 (S18-2) in homeostasis of cell stemness. Mouse primary cells expressing both S18-2 and RB (designated as RH18RB) exhibited a stem cell phenotype. As a proof of principle, we demonstrated decreased expression of stem-cell–related genes in human mesenchymal stem cells upon down-regulation of S18-2 and RB. Notably, loss of the S18-2 protein resulted in embryonic lethality in zebrafish. To reveal putative molecular mechanisms, we demonstrated that S18-2, in a multimeric complex with RB, enhanced the E3-ubiquitin ligase activity of RNF2 toward histone H2A. Finally, based on our observation that RH18RB cells generated tumors in severe combined immunodeficiency mice, S18-2 could be a promising target in the development of cancer therapies.
Keywords: stemness and differentiation, cell immortalization, embryogenesis, tumorigenesis
Abstract
Stemness encompasses the capability of a cell for self-renewal and differentiation. The stem cell maintains a balance between proliferation, quiescence, and regeneration via interactions with the microenvironment. Previously, we showed that ectopic expression of the mitochondrial ribosomal protein S18-2 (MRPS18-2) led to immortalization of primary fibroblasts, accompanied by induction of an embryonic stem cell (ESC) phenotype. Moreover, we demonstrated interaction between S18-2 and the retinoblastoma-associated protein (RB) and hypothesized that the simultaneous expression of RB and S18-2 is essential for maintaining cell stemness. Here, we experimentally investigated the role of S18-2 in cell stemness and differentiation. Concurrent expression of RB and S18-2 resulted in immortalization of Rb1−/− primary mouse embryonic fibroblasts and in aggressive tumor growth in severe combined immunodeficiency mice. These cells, which express both RB and S18-2 at high levels, exhibited the potential to differentiate into various lineages in vitro, including osteogenic, chondrogenic, and adipogenic lineages. Mechanistically, S18-2 formed a multimeric protein complex with prohibitin and the ring finger protein 2 (RNF2). This molecular complex increased the monoubiquitination of histone H2ALys119, a characteristic trait of ESCs, by enhanced E3-ligase activity of RNF2. Furthermore, we found enrichment of KLF4 at the S18-2 promoter region and that the S18-2 expression is positively correlated with KLF4 levels. Importantly, knockdown of S18-2 in zebrafish larvae led to embryonic lethality. Collectively, our findings suggest an important role for S18-2 in cell stemness and differentiation and potentially also in cancerogenesis.
Stemness refers to the molecular processes underlying the fundamental stem cell (SC) properties of self-renewal and generation of differentiated daughter cells. Highly organized and strictly controlled signaling systems regulate these two characteristics of SCs. The self-renewal of these cells is controlled through the activation of a defined set of genes that sustain a balance between self-renewal and differentiation (1). Under normal physiological conditions, controlled shifts in the balance of such signaling pathways induce differentiation. Abnormalities in signaling cascades can initiate and promote cellular transformation (2).
We showed previously that the ectopic expression of the mitochondrial ribosomal protein S18-2 (MRPS18-2, herein referred to as S18-2) (see SI Appendix, Table S1, for gene names) led to the immortalization of primary rat fibroblasts. Surprisingly, and importantly, these cells exhibited an embryonic stem cell (ESC) phenotype (3, 4). Furthermore, terminally differentiated skin fibroblasts were transformed into malignant cells upon S18-2 overexpression (5). The common features of these S18-2–overexpressing cells included high telomerase activity, anchorage-independent growth, and the ability to form aggressive tumors in severe combined immunodeficiency mice (SCID) mice. However, the mechanism via which S18 immortalizes and transforms cells remains unexplored. Recently, other researchers have used S18-2 as a tool to immortalize chicken embryonic liver cells (6).
The use of a model system of cell immortalization, i.e., infection of primary B cells by the Epstein–Barr virus, led to the targeting of the normally cytoplasmic S18-2 to the nucleus by interacting with the transforming viral protein EBNA-6 in the immortalized cells. Furthermore, we showed that S18-2 forms a bridge between the EBNA-6 and RB proteins (7).
The retinoblastoma-associated protein (RB) exerts its function primarily by inhibiting the E2F family of transcription factors (TFs) through direct binding, thus regulating cell-cycle progression (8). The RB-E2F1 interaction relies on the phosphorylation status of RB, which is mediated by cyclin-dependent kinases (CDKs) (9). Importantly, S18-2 can bind RB directly, regardless of the phosphorylation status of this protein (10). Consequently, E2F1 is released from a complex with RB, thereby promoting cell-cycle progression (7).
Because of the constitutive activity of CDKs in ESCs, RB is hyperphosphorylated, and only a very small fraction of RB binds to E2F1, suggesting a different mode of action of RB in SCs and their differentiated counterparts (11). Conversely, deletion of RB1 led to the loss of SC self-renewal characteristics (11). Generally, ESCs proliferate rapidly and have a distinct cell cycle with truncated gap phases (12). They may remain in a quiescent state but reenter the cell cycle upon induction of proliferation via extrinsic signals (13). The quiescent state must be finely regulated; otherwise, ESCs can be directed toward differentiation or senescence (14). However, the molecular mechanisms underlying the function of RB in SCs are largely unknown (15). To study the role of RB in cell stemness, we developed a model of mouse embryonic fibroblasts (MEFs) derived from homozygous knockout Rb1 embryos. The Rb1−/− MEFs exhibited rapid proliferation with an anchorage-dependent growth pattern. After passage 11, the proliferative rate of the cells diminished, and they became senescent (16). The rationale of the present work was to use the Rb1−/− MEF model to analyze the manner in which high expression levels of RB and S18-2 cooperate to control cell fate. We hypothesized that the simultaneous expression of these two proteins at a high level supports stemness (17).
Results
Overexpression of S18-2 Leads to Immortalization of Rb1−/− MEFs.
To analyze whether expression of RB is needed for S18-2-induced cell immortalization, we transfected Rb1 knockout MEFs (designated as RH1301) with plasmids encoding S18-2 and RB, both individually (RH18, RHRB) and sequentially (RH18RB), as well as with an empty control vector (RH) (Materials and Methods). The RH18 and RH18RB cells rapidly lost contact inhibition and showed anchorage-independent growth in contrast to control RH and RHRB cells which lacked these abilities (Fig. 1 and SI Appendix, Fig. S1 A–D). Moreover, the morphology of RH18RB cells resembled ESCs, as they formed well-defined colonies in ordinary cell culture flasks (Fig. 1 A, Upper) as well as in bacterial petri dishes (Fig. 1 A, Middle). RH18 cells did not show this phenotype (Fig. 1 A, Lower).
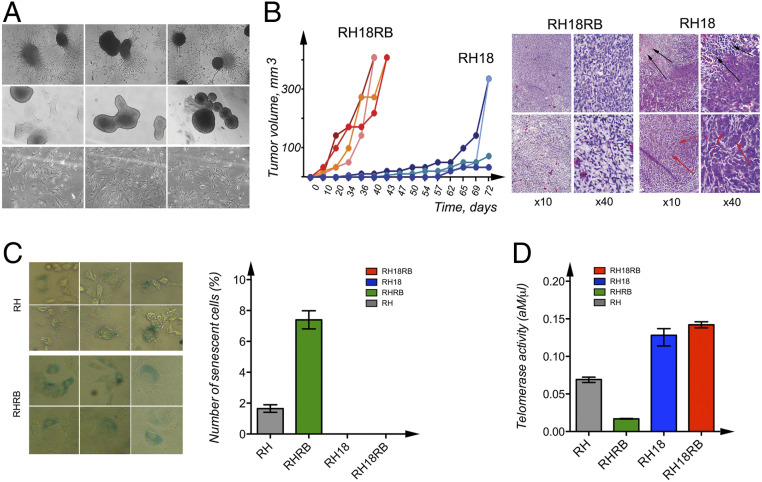
Fig. 1.
Characterization of cell lines generated from Rb1−/− MEFs. (A) Phase-contrast images of RH18RB cells in cell culture dishes (Upper) and in bacterial petri dishes (Middle). Phase-contrast images of RH18 cells in cell culture dishes (Lower). (B) The kinetics of tumor growth of RH18 and RH18RB cells as indicated in SCID mice (Left). Sections of the tumors produced by RH18 and RH18RB cells were stained with hematoxylin and eosin. The RH18RB cells generated aggressive fibrosarcomas. The RH18 tumors contained large necrotic areas (Right, black arrows). A proportion of the cells in these tumors resembled epithelium-like cells (Right, red arrows). (C) Staining with β-galactosidase for the identification of senescent RHRB (Top Left) and RH (Bottom Left) cultures. Arrows indicate cells producing β-galactosidase. The number of senescent cells was analyzed using column analysis and an unpaired t test (Right). (D) The telomerase activity in RH, RHRB, RH18, and RH18RB cells was determined via the QTD method using qPCR. Each bar represents the mean telomerase activity of each cell culture. Reactions were performed twice, with each cell line assessed in triplicate. Statistical analysis was performed as in C.
The sublines generated were injected subcutaneously (s.c.) into SCID mice in order to analyze their in vivo growth ability. As expected, only the RH18 and RH18RB cells gave rise to tumors. The RH18RB cells generated tumors that were growing faster and that produced a larger tumor mass compared to the RH18 cells (Fig. 1 B, Left). Staining of tissue sections showed that the tumors produced by RH18RB cells could be described as aggressive fibrosarcomas (Fig. 1 B, Right). In contrast, RH18 cells formed tumors with large necrotic areas (indicated with black arrows, Fig. 1 B, Right). These tumor cells showed epithelial cell-like features (indicated with red arrows, Fig. 1 B, Right).
We next picked four clones each from the RH18 and RH18RB cultures for further studies. To confirm cell immortalization, one of the clones of both RH18 and RH18RB cells were passaged for more than 350 population doublings; thus, these two clones were cultured in vitro for more than 2 y. The remaining three clones of each cell type were kept in culture for 2 mo (SI Appendix, Fig. S1 E and F). In contrast, control RH and RHRB cells became senescent quite rapidly and died after 4 to 6 wk, and no cultures could be passaged beyond this time point. The RHRB cells showed contact inhibition and a significantly higher percentage of senescent cells (up to 12% β-galactosidase–positive cells) compared with RH cultures (with 2% β-galactosidase–positive cells). In contrast, very few senescent cells were detected in the RH18 and RH18RB cultures (Fig. 1C and SI Appendix, Fig. S1 G and H and Table S2).
To explain the unlimited growth of RH18 and RH18RB cells, telomerase activity was quantified based on the number of added telomere repeats, as assessed by qPCR. The RB18RB and RH18 cells showed high telomerase activity (up to 20 amole/µL), which differed significantly (P = 0.0001) from the telomerase activity of RH or RHRB cells (<2 amole/µL). The RHRB cells exhibited the lowest telomerase activity (Fig. 1D and SI Appendix, Fig. S1 I and J), which was in accordance with the largest number of senescent cells observed in these cultures.
Together, these data indicate that simultaneous expression of S18-2 and RB proteins resulted in immortalization of −/−Rb1 MEFs. Moreover, these R18RB cells showed an ESC phenotype.
A Stem-Cell–Related Gene Expression Program Follows the Expression of S18-2 and RB.
To confirm our observations, the levels of S18-2 were analyzed in SCs and differentiated cells using StemMapper, a manually curated database (18). We compared the expression of S18-2 between undifferentiated and differentiated mouse ESCs as well as between induced pluripotent stem cells (iPSCs) and differentiated iPSCs. The genes encoding three of the Yamanaka factors (Oct4, Sox2, and Klf4) were used as controls. The data showed that the overall expression of S18-2 was higher in mouse ESCs (Fig. 2A, red) than in their differentiated counterparts (Fig. 2B, green). Sox2, Oct4 (Pou5f1), and Klf4 showed a similar expression pattern. As expected, changes in the levels of Sox2, Oct4, and Klf4 were more pronounced in iPSCs (Fig. 2B), as most iPSCs are generated by the overexpression of these very genes. Importantly, S18-2 messenger RNA (mRNA) levels also exhibited similar expression trends; i.e., higher levels were detected in undifferentiated iPSCs versus their differentiated counterparts (Fig. 2).
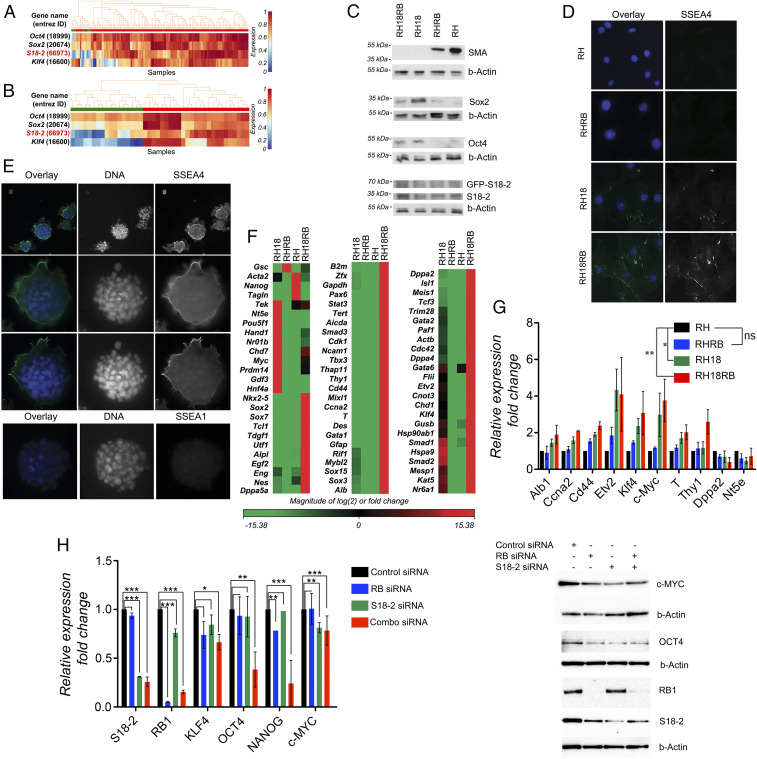
Fig. 2.
Induction of stem cell markers in Rb1−/− MEF sublines expressing RB and S18-2. (A) Analysis of S18-2 mRNA expression in mouse ESCs and in differentiated cells using the StemMapper database. Red: mouse ESCs; green: differentiated mouse cells. (B) Analysis of S18-2 mRNA expression in iPSCs and differentiated iPSCs using the StemMapper database. Red: iPSCs; green: differentiated iPSCs. (C) Western blotting of cell extracts using the indicated antibodies. Beta-Actin (b-Actin) was used as a loading control. (D) Immunostaining with anti-SSEA4 antibody. DNA is stained blue. (E) RH18RB cells were grown in bacterial petri dishes and cultured overnight on glass slides, followed by staining with antibodies against SSEA1 and SSEA4. DNA is stained in blue. (F) Heat map of data obtained from an expression array of 84 genes associated with mouse ESCs: red indicates genes that were expressed at higher levels and green indicates genes that were expressed at lower levels in RH18, RHRB, and RH18RB cells compared with RH cells. (G) The expression of the indicated set of genes was assessed by qPCR using TBP, beta-actin, or gapdh as endogenous controls and is presented as fold change compared to the internal controls. (H) Expression pattern of stemness-related genes or proteins in human mesenchymal stem cells after siRNA treatment. (Left) qPCR of the indicated genes presented as fold change compared to GAPDH which served as the internal control. *0.03 < P < 0.05; **0.01 < P < 0.03; ***P < 0.01. (Right) Western blot analysis using the antibodies for proteins encoded by the genes analyzed in the qPCR as indicated. b-Actin was used as a loading control.
The RH, RHRB, RH18, and RH18RB cells were analyzed for the expression of the fibroblast-specific markers smooth muscle actin (SMA) and vimentin, as well as for ESC markers, including Oct4, Sox2, and mouse-specific stage-specific embryonic antigen-4 (SSEA4). As shown by Western blotting and immunostaining, RH18 and RH18RB cells exhibited loss of SMA and partially also vimentin expression, while Sox2, Oct4, and SSEA4 levels were present (Fig. 2 C and D). Notably, cells that expressed S18-2 at high levels showed loss of SMA expression, regardless of the presence or absence of RB. We further observed that the RH18RB cells that formed foci in a petri dish expressed SSEA4 at a very high level (Fig. 2E).
Next, we analyzed the mRNA expression levels of 84 genes specific to mouse ESCs using the RT2 profiler assay (Fig. 2F). The comparison between RHRB cells (with endogenous levels of S18-2) and RH control cells showed that the overall gene expression pattern was unaltered, with the exception of the down-regulation of Acta2, Nanog, and Tagln and the up-regulation of Gsc.
Simultaneous expression of RB and S18-2 in RH18RB cells robustly altered the gene expression pattern; 58 of the 84 analyzed genes were expressed at high levels, whereas only 4 genes were down-regulated (Gsc, Acta2, Nanog, and Tagin) in RH18RB cells; in contrast, the expression of 22 genes remained unchanged compared to RH cells. Notably, the following genes were expressed at significantly higher levels in RH18 cells compared with the remaining three cell lines: Tek, Nt5e, Pou5f1 (Oct4), Hand1, Nr01b, Chd7, Myc, Prdm14, Gdf3, and Hnf4a. A comparison of the expression profile between RH18 and RH18RB cells revealed that 59 genes were up-regulated in the latter cells, while 15 genes were unchanged. To confirm this expression pattern, 10 genes that were highly expressed in RH18 and RH18RB cells compared to RH and RHRB cells were selected for further analysis. To assess early changes in the expression of the selected genes, transient transfections of RH cells with S18-2 and RB-encoding plasmids were performed. The gene expression pattern was evaluated 48 h posttransfection in triplicates of each of the three types of transfectants, i.e., RHRB, RH18, and RH18RB (Fig. 2G). Of note, Myc expression was higher in RH18 and RH18RB cells than in RH and RHRB cells. A similar trend was observed for Thy, Etv2, and Klf4.
To confirm the role of S18-2 and RB in the maintenance of stemness, we next used human mesenchymal stem cells (MSCs) derived from bone marrow and down-regulated the S18-2 and RB gene expression using a mixture of small interfering RNAs (siRNAs). Notably, c-MYC levels decreased significantly upon introduction of siRNA against S18-2 while treatment of cells with siRNA against RB1 resulted in significant down-regulation of NANOG expression. Application of a mixture of siRNA against both RB1 and S18-2 resulted in down-regulation to different extents of all stemness-related genes analyzed, with a strong synergistic effect on OCT4 and NANOG (Fig. 2H). These data are in accordance with our results in RH18RB cells where the combined expression of S18-2 and RB induced expression of stem-cell–related genes (Fig. 2 F and G).
Based on these data, we concluded that simultaneous expression of RB and overexpression of S18-2 are necessary and required for a stem-cell–specific gene expression pattern.
Multilineage Differentiation of RH18RB Cells Is Induced In Vitro.
To demonstrate the stemness of RH18RB cells, we induced osteogenic, chondrogenic, and adipogenic differentiation using specific chemical mixtures. For the induction of osteogenic differentiation, cells were treated with dexamethasone, ascorbic acid, and glycerol-2-phosphate. As a measure of osteogenic differentiation, Ca2+-ion deposits were observed in culture with the help of Alizarin Red S staining. Only the RH18 and RH18RB cells showed red signals with different intensities. The expression of the osteogenic differentiation markers Runx2 and Spp1, which encode osteopontin, was markedly elevated in RH18 and RH18RB cells compared with untreated RH18RB or RH and RHRB cells (Fig. 3B). Of note, RH18 cells exhibited a red signal with a lower intensity than observed in differentiated RH18RB cells. Moreover, the up-regulation of Runx2 and Spp1 was not as prominent in RH18 as it was in RH18RB cells. This demonstrated that RH18RB cells differentiated into osteoblast-like cells (Fig. 3 A, Bottom Right, and SI Appendix, Fig. S2, first and second rows), while neither RH nor RHRB cells responded to this treatment.
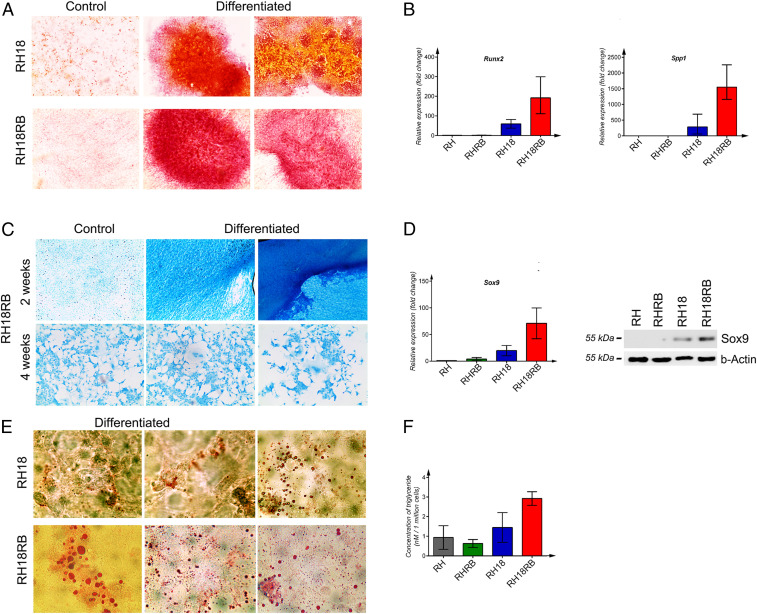
Fig. 3.
Multilineage differentiation of RH18 and RH18RB cells induced in vitro. (A) Alizarin Red S was used to qualitatively assess the presence of cells producing Ca2+ ions to monitor osteogenic lineage differentiation in the indicated cell lines. (B) mRNA expression of Runx2 (Left) and Spp1 (Right) was assessed by qPCR. The qPCRs were performed in triplicate, and data with SE mean values are shown. (C) Representation of control cells and cells differentiated into the chondrogenic lineage in RH18RB cells at 2 and 4 wk, respectively. (D) Expression of the Sox9 gene was assessed at the mRNA and protein levels by qPCR (Left) and Western blotting (Right), respectively. The qPCR conditions were as indicated in B. b-Actin was used as a loading control for Western blot. (E) Qualitative reaction with Oil Red O stain to monitor lipid droplets in cells differentiated into the adipogenic lineage in the indicated cell lines. (F) Lipid production was quantified using a colorimetric analysis in the indicated cell lines. The Kruskal–Wallis test was performed for four groups with three values for each group.
To induce chondrogenic differentiation, cells were treated with sodium pyruvate, β-mercapto-ethanol, and H89 agent. Staining negatively charged molecules by Alcian Blue has been used to monitor chondrogenic differentiation qualitatively. The RH18 cells died within 2 wk of the treatment while the RH and RHRB cells did not respond to the induction medium. In contrast, RH18RB cells showed extensive staining with Alcian Blue (Fig. 3C and SI Appendix, Fig. S2, third row), indicating chondrogenic lineage. All four cell types were treated for only 1 wk before RH18 cells started to die and the expression of Sox9, a specific marker of chondrogenic differentiation, was assessed. Sox9 was expressed at high levels in RH18RB cells, both at the mRNA (Fig. 3 D, Left) and protein (Fig. 3 D, Right) levels. In contrast, Sox9 was barely detectable in RHRB and RH cells and exhibited only low expression levels in RH18 cells (Fig. 3D).
Cells were also cultured in medium supplemented with 3-isobutyl-1-methylxanthine, indomethacin, dexamethasone, and insulin for 1 or 2 wk for the induction of adipogenic differentiation. Lipid droplets were observed mainly in the cytoplasm of RH18 and RH18RB cells, as qualitatively shown by Oil Red O staining (Fig. 3E). In contrast, no lipid droplets were observed in RH or RHRB cells. RH18RB cells produced high amounts of lipids and released triglycerides (Fig. 3 E, Lower, and SI Appendix, Fig. S2, fourth row). To quantify the production of lipids, the concentration of triglycerides was measured in cultures after the induction of adipogenic differentiation. High quantities of triglycerides were recorded in RH18RB cells (mean value, 8.6 nM/1⋅106 cells), which was significantly higher (P = 0.008) than the moderate levels observed in RH18 cells (3.38 nM/1⋅106 cells) and the low concentrations detected in RHRB (1.66 nM/1⋅106 cells) and RH (2.00 nM/1⋅106 cells) cells (Fig. 3F).
Collectively, these data show that the RH18RB cells exhibit the ability to differentiate into various lineages, a characteristic of ESCs.
S18-2 Interacts with Prohibitin and RNF2.
Next, we performed immunoprecipitation followed by a mass spectrometry analysis to identify the binding partners of S18-2. Only a few proteins were detected and included prohibitin 2 (PHB2). The binding between S18-2 and PHB2 was confirmed by a GST pull-down assay. Whole-cell lysates of MCF7 breast carcinoma cells were used for the source of PHB2. The PHB2 signal was exclusively detected on the surface of GST-S18-2 beads and not on control GST beads (Fig. 4A). Notably, PHB2 was expressed at its highest levels in RHRB cells (Fig. 4B). It is known that PHB2 can bind to RB and RNF2, which functions as an E3-ubiquitin ligase that ubiquitinates histone 2A (H2A) (19). Therefore, we investigated whether RNF2 could be detected in the S18-2-RB protein complex.
Fig. 4.
Interaction of S18-2 with prohibitin and RNF2, which shows high activity in RH18RB cells. (A) GST pull-down assay of PHB2 with GST and the GST–S18-2 fusion protein using extracts from MCF7 cells (5% of the whole-cell lysate [WCL] input is shown). (B) Western blotting for Phb2 in extracts from the cells indicated. b-Actin was used as a loading control. (C) RNF2 expression as detected by Western blotting using extracts from the indicated cells. b-Actin was used as a loading control. (D) GST pull-down assay of whole-cell lysates of RH18 and RH18RB cells followed by Western blot analysis for RNF2 (5% of the WCL input is shown). (E) The cellular distribution of RB and S18-2 was analyzed by Western blotting in nuclear (N) and cytoplasmic (C) fractions from RHRB cells. (F) The cellular distribution of RB and S18-2 was analyzed by Western blotting in nuclear (N) and cytoplasmic (C) fractions of RH18RB cells. An antibody against Lamin A/C was used as a nuclear marker. (G) The E3-ligase activity of Rnf2 toward the Lys119 residue of histone H2A was analyzed by Western blotting using an antibody specific for monoubiquitinated histone H2A at Lys119 (Ub-HALys119). (H) Western blot analysis of Ub-H2ALys119 levels in cytoplasmic (C) and nuclear (N) fractions of RH18RB (Left) and RHRB (Right) cells.
To address this question, a GST pull-down assay using RH18 and RH18RB cell lysates was performed. The expression levels of Rnf2 were similar in all four cell lines studied (Fig. 4C). Importantly, S18-2 interacted with Rnf2 regardless of the presence of RB (Fig. 4D). The Rnf2 protein was detected exclusively in the nucleus of RH and RHRB cells (SI Appendix, Fig. S4 A, Top). A cytoplasmic Rnf2 signal was observed in the presence of overexpression of S18-2 (SI Appendix, Fig. S4 A, Bottom Left) and the signal was stronger when both S18-2 and RB were expressed at high levels (SI Appendix, Fig. S4 A, Bottom Right). Surprisingly, overexpression of S18-2 retained RB in the cytoplasm of RH18RB cells (SI Appendix, Fig. S3 B, Bottom row, Right). To confirm this finding, we performed Western blotting on nuclear and cytoplasmic fractions. As shown in Fig. 4E, a nuclear RB signal was detected in RHRB cells, which expressed endogenous S18-2 at low levels. Overexpression of S18-2 as a GFP-fusion protein led to the presence of both nuclear and cytoplasmic RB signals (Fig. 4F). Moreover, in addition to the cytoplasmic signal, S18-2 also showed a weak nuclear signal (Fig. 4F). Hence, the RB protein was retained in the cytoplasm upon S18-2 overexpression. Next, we evaluated whether the formation of a complex containing RB, S18-2, and prohibitin affected the E3-ligase activity of RNF2. Importantly, Rnf2 showed the highest E3-ligase activity in RH18RB cells compared to the other cell lines (Fig. 4G). This was associated with a monoubiquitinated histone H2A signal in the cytoplasm (Fig. 4H).
We conclude that RB in complex with S18-2 is essential for RNF2 function. Importantly, we observed enhanced monoubiquitination of histone H2A, a characteristic of SCs.
The Transcription Factor KLF4 Binds to the S18-2 Promoter and Activates Its Expression.
Based on our findings, the expression of S18-2 should be finely controlled in SCs and upon differentiation. Thus, we aimed to identify the cellular factors that regulate S18-2 expression at the mRNA level. To this end, we performed bioinformatic analyses to map the S18-2 promoter and the putative transcription factors that bind to this region. Three binding sites for KLF4 and one for KLF6 were identified in the promoter region of human S18-2, while only one Klf4-binding site was identified in the mouse S18-2 promoter (SI Appendix, Fig. S4B). To analyze the functionality of these sites, we generated biotin-labeled DNA fragments by PCR amplification of the human and mouse S18-2 promoter regions. After incubation of these DNA fragments with whole-cell lysates, pull-down assays were performed using iron-streptavidin beads. Lysates of prostate adenocarcinoma PC3 and RH18RB cells were used to precipitate human and mouse transcription factors, respectively. To determine the feasibility of the protocol, E2F1 binding to the adenoviral E1A promoter region was assessed in parallel (SI Appendix, Fig. S4C). As expected, we detected a robust signal using a specific antibody against E2F1 (Fig. 5A). We next precipitated human KLF4 from PC3 and PC3-S18-2 cells, a PC3 subline that constitutively expresses high levels of S18-2 (Fig. 5B) (20). Mouse Klf4 was also precipitated to the S18–2 promoter region from RH18RB cell lysates (Fig. 5C).
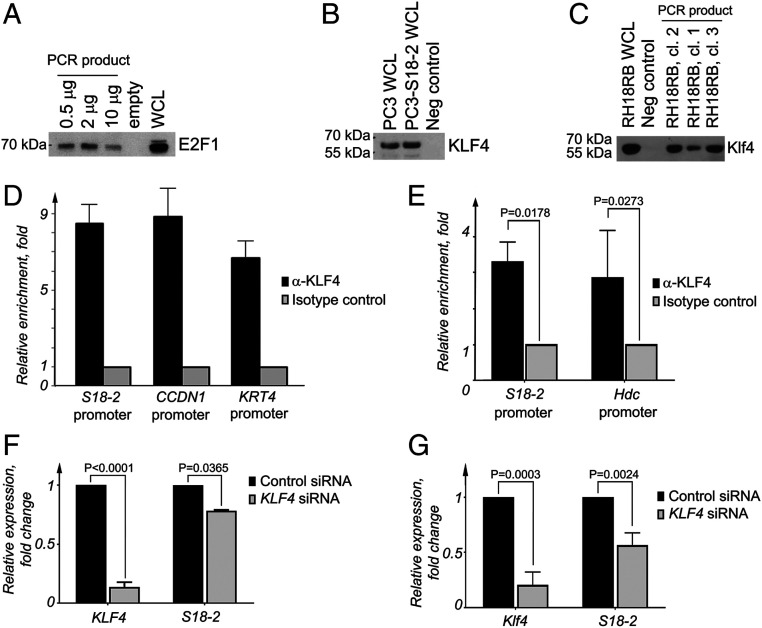
Fig. 5.
KLF4 binds to the S18-2 promoter and regulates its expression. (A) Precipitation of E2F1 from WCLs of PC3 cells using a biotinylated PCR fragment amplified from the adenoviral E1A promoter region. (B) Promoter pull-down assay to show the interaction between KLF4 and a biotin-labeled PCR fragment amplified from the human S18-2 promoter region. WCLs from PC3 cells and a PC3 subline that express constitutive high levels of S18-2 (PC3-S18-2) were used as source of KLF4. (C) Interaction between KLF4 and a biotin-labeled PCR fragment amplified from the mouse S18-2 promoter. WCLs from freshly established RH18RB clones were used as a source of Klf4. (D) ChIP assay using KLF4 antibodies and isotype-specific control antibodies followed by PCR analysis of the human S18-2 promoter region from PC3 cell extracts. The promoter regions of CCND1 and KRT4 were amplified from the same eluted DNA as positive controls. The median values of the relative enrichment in anti-KL4 immunoprecipitations versus the isotype control (which was performed in triplicate) are shown. (E) ChIP assay using anti-KLF4 antibodies and isotype-specific control antibodies for PCR analysis of the mouse S18-2 promoter region from RH18RB cell extracts. As a positive control, the promoter regions of hdc were amplified from the same eluted DNA. The median value of three experiments performed using three clones as the relative median enrichment values of the relative enrichment in anti-KL4 immunoprecipitations vs. isotype control is shown. (F) qPCR analysis of KLF4 and S18-2 mRNA levels after KLF4-specific siRNA treatment in PC3 cells. The mRNA expression in these cells was compared with that of cells transfected with the control siRNA. The relative expression was calculated using the expression of TBP as an endogenous control. (G) Mouse Klf4 and S18-2 were analyzed by qPCR in extracts from RH cells after Klf4-specific siRNA treatment. The mRNA expression in these cells was compared with that of cells transfected with the control siRNA. The relative mRNA expression was calculated using the expression of TBP as an endogenous control.
To confirm these observations, we performed a chromatin immunoprecipitation (ChIP) assay. DNA fragments were precipitated from PC3 cells and three RH18RB clones using an anti-KLF4 antibody coupled to magnetic beads. An isotype-specific antibody was used as a negative control, and enrichment of KLF4 binding to CCND1 and KRT4, which are known targets for KLF4 (21), were used as positive controls. After the elution of protein–DNA complexes, the relative promoter enrichment was assessed in comparison to the levels detected in the negative and positive controls. Our data demonstrated a ninefold enrichment of the S18-2 promoter fragments in the KLF4 ChIP from PC3 cells (Fig. 5D). We also observed binding of KLF4 to the S18-2 promoter in RH18RB cells (Fig. 5E), although at a lower level compared with human PC3 cells. Of note, only one binding site for Klf4 was predicted in the promoter region of mouse S18-2, in contrast to the three predicted binding sites in the promoter of the human gene. The Hdc promoter was used as a positive control, as Klf4 binding to this element was reported previously (21). We found similar binding values of Klf4 at the S18-2 and Hdc promoters (Fig. 5E).
To further study the manner in which KLF4 affects the regulation of S18-2 expression, we knocked down KLF4 using a pool of four specific siRNAs. Cells were also transfected with a nonspecific, control siRNA. We demonstrated that both human KLF4 (in PC3 cells) and mouse Klf4 (in RH cells) were down-regulated after transfection with a mixture of siRNAs specific to KLF4 (Fig. 5 F and G). Notably, S18-2 expression was significantly decreased in RH (P = 0.0024) and PC3 cells (P = 0.036) treated with a KLF4-specific siRNA (Fig. 5G) compared to the controls. Taken together, our data suggest that KLF4 is involved in the transactivation of both mouse and human S18-2 expression.
A Vital Role of S18-2 during Zebrafish Development.
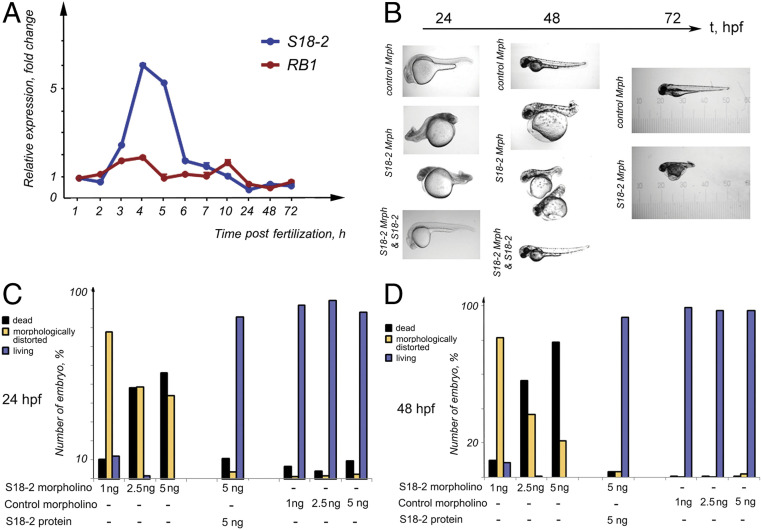
Next, we addressed the role of S18-2 during embryogenesis using the well-characterized zebrafish model system. As a first step, we assessed the mRNA expression of S18-2 and Rb1 during 1 to 72 h postfertilization (hpf). The expression pattern of Rb1 did not change markedly during this time. Maximal Rb1 expression (a twofold increase) was observed at 3 to 4 hpf, i.e., at the blastula stage (Fig. 6A and SI Appendix, Fig. S5A). In contrast, S18-2 exhibited a well-defined expression peak at 4 to 6 hpf. The S18-2 mRNA increased at 3 hpf and remained high during the entire blastula stage (at 3 to 5 hpf), then decreased dramatically during the gastrula stage (at 6 hpf), and remained at a constant low level throughout the experiment (Fig. 6A and SI Appendix, Fig. S5A).
Fig. 6.
A vital role for S18-2 during zebrafish embryogenesis. (A) The mRNA expression levels of RB1 (blue line) and S18-2 (red line) were assessed at different stages of zebrafish development using qPCR. The mean values of three qPCR experiments are shown. The SD was not more than 20% of the mean. β-Actin was used as a housekeeping gene, and the expression levels detected in eggs at 0 hpf were taken as the reference expression level. (B) A specific morpholino that inhibited S18-2 translation, a control morpholino, or a S18-2–specific morpholino, together with in vitro-translated S18-2 protein, were introduced into fish eggs within 1 hpf. The morphology of the larvae was observed over 1 to 72 hpf using bright-field microscopy. (C) Quantification of embryos showing morphological changes at 24 h (see details in SI Appendix, Table S3) after the indicated treatments. (D) Quantification of embryos showing morphological changes at 48 h after the indicated treatments. Each bar in C and D represents the mean of four to five measurements; the deviation was less than 30%.
Based on these observations, we evaluated whether S18-2 is essential for normal embryogenesis. For this purpose, specific morpholino oligomers that inhibit S18-2 protein synthesis were injected into zebrafish eggs prior to the cleavage period, when the fertilized egg was at the two-to-four-cell stage. Nonspecific morpholino oligomers were used as controls. Western blot analysis confirmed the specificity of the morpholino (SI Appendix, Fig. S5B). Treatment with morpholinos designed to knock down S18-2 prevented the normal development of fish, as swimming larvae were not observed at 72 hpf (Fig. 6B). Moreover, embryos with S18-2 knockdown did not exhibit proper segmentation. We titrated the morpholinos and found that introduction of 1 ng of the S18-2 morpholino resulted in embryonic death and that up to 20% of embryos had died by day 2, with 92% of living larvae morphologically distorted (Fig. 6 C and D and SI Appendix, Table S3). A similar effect was observed when 2.5 ng of the S18-2 morpholino was used. Fish development was delayed for 2 to 2.5 hpf compared with that observed in normal embryos, and this delay could be observed as early as 12 hpf. Five nanograms of the S18-2-morpholino led to the death of >90% of the injected embryos on day 2 after fertilization compared with the death of ∼10% of embryos injected with control oligomers. Importantly, the control morpholino did not affect the morphology of zebrafish at any of the developmental stages analyzed (Fig. 6B). Importantly, the injection of in vitro-translated human S18-2 together with the S18-2–specific morpholino rescued embryos, and fish developed normally (Fig. 6 B–D and SI Appendix, Table S3). Based on our findings, we concluded that S18-2 is necessary for normal zebrafish development.
Discussion
Effect of S18-2 Overexpression in the Presence of RB on ESC-Associated Characteristics.
Active telomerase can prevent cellular senescence, and replication-dependent loss of telomeres does not occur in rapidly proliferating cells such as SCs derived from germ-line or cancer cells (22). The telomerase is a complex between the reverse transcriptase encoded by the telomerase reverse transcriptase (TERT) gene and an RNA template (telomerase RNA component). By adding telomeric repeats onto the ends of chromosomes, the telomerase ensures the maintenance of telomere length. ESCs express high levels of hTERT and exhibit telomerase activity, both of which decline rapidly during differentiation (23). We observed the induction of telomerase activity upon overexpression of S18-2 in RH18 and RH18RB cells. Moreover, no senescent cells were observed in these cultures, in contrast with the control RH cells. Concomitant expression of RB with endogenous S18-2 in RHRB cells resulted in a marked decrease in telomerase activity and an increase in the number of senescent cells in agreement with a previous report (16). Replicative senescence is linked to RB and its interacting partners (24). Importantly, in the present study, we demonstrate that overexpression of S18-2 results in enhanced telomerase activity, regardless of the presence of RB. The high telomerase activity and low number of senescent cells in RH18RB culture might explain their aggressive characteristics during in vivo growth in SCID mice. The RH18RB cells resembled mouse ESCs both phenotypically and genetically. Specifically, we detected up-regulation of Sox2, Oct4, and SSEA4 in RH18RB cells. Notably, when both RB1 and S18-2 were down-regulated in human MSCs, expression of the stemness-related genes, OCT-4, KLF-4, NANOG, and c-MYC, was reduced as well (Fig. 2H). Data extracted from the StemMapper database (Fig. 2B) also suggest the involvement of S18-2 in the induction of the SC phenotype. RH18RB cells exhibited a gene expression pattern that was similar to that of mouse ESCs, and these cells could differentiate into specific lineages including osteogeneic, chondrogenic, and adipogenic lineages in vitro. RB is involved in the differentiation of multiple lineages by regulating the activity of master transcription factors, such as MyoD in muscles (25), Runx2 in bones (26), and PGC-1 in adipocytes (27). Our data are in accordance with these previous reports.
S18-2 and RB Maintain Cell Stemness by Enhancing RNF2 Activity.
In addition to the well-known function of RB in cell-cycle regulation, several studies have reported the role of the RB protein in chromatin remodeling via an interaction with chromatin-modifying enzymes. For example, RB binds to the HDAC1 (28) and SIRT1 (29) histone deacetylases, the methyltransferase DNMT1 (30), and the lysine-specific demethylase KMD5A (31). Moreover, RB can control the transcription of a set of genes that are involved in cell differentiation by interacting with the ring-finger protein RING1 (32, 33). RING1 is a member of the polycomb repressive complex 1 (PRC1) and interacts with RNF2 and prohibitin (PHB) (34, 35). Of note, PHB can bind RB (36) and E2F1 (37).
Both PHB and its homolog PHB2 play important roles in mitochondrial biogenesis by controlling the morphogenesis of cristae and the fusion of mitochondria (38). The two prohibitins were found only in multimeric complexes, even in the inner mitochondrial membrane. We found that the S18-2 protein, which is usually localized in the mitochondrial matrix, interacted with PHB2. However, prohibitins are also involved in the regulation of gene expression through their interactions with the PRC1 subunits. Importantly, RING1 acts as an adapter for RNF2 by enhancing the E3-ubiquitin ligase activity (39) for histone H2A (34). This histone modification results in a stem-cell–specific gene expression pattern through transcriptional silencing (34, 40). Rnf2 in complex with Oct4 and Nanog bound to the promoter region of 212 common genes in mouse ESCs and the down-regulation of the Rnf2 protein led to an altered expression of 25 genes, 18 of which were de-repressed (41–43). These data suggest that Rnf2 is required for the maintenance of undifferentiated and pluripotent SCs by repressing specific subsets of genes that are transactivated by Oct4 and Nanog. The Gadd45g, Fgf15, Bmp7, Col4a2, Podxl, Gata3, Bmi1, Msx2, Gja1, and Eif4g3 genes were among the 18 genes that were co-occupied by Nanog and Oct4 (43). Interestingly, as shown here, S18-2 bound to RNF2. Furthermore, we demonstrated that Rnf2 exhibited E3-ubiquitin ligase activity only in the presence of RB. Ectopic expression of both RB and S18-2 led to the enrichment of monoubiquitinated histone H2ALys119. We suggest the formation of a multimeric protein complex consisting of RB, S18-2, PHB2, and RNF2 that enables RNF2 for efficient histone H2A ubiquitination, thereby controlling SC-specific gene expression.
Unexpectedly, we found that RB was retained in the cytoplasm of cells that expressed high levels of ectopic S18-2, while RNF2 was detected not only in the nucleus, but also in the cytoplasm of these cells. The cytoplasmic localization of RNF2 has been reported (44). A small proportion of monoubiquitinated histone H2A was detected in the cytoplasm of RH18RB cells, which is in agreement with previous observations (45). We suggest that hyperactivation of RNF2 by interaction within a protein complex containing S18-2 and RB leads to the ubiquitination of histone H2ALys119 in the cytoplasm prior to translocation of histones to the nucleus.
Functional Consequences of the RB-S18-2 Interaction in the Regulation of Cell Stemness.
Rnf2 is required for the normal development of mouse embryos, as deficiency of Rnf2 resulted in embryonic lethality, caused gastrulation arrest, and inhibited cell-cycle progression (46). Furthermore, Rnf2 is required for chondrocyte and osteocyte differentiation in zebrafish by stabilizing the expression of RUNX2 and SOX9, which are both needed for the differentiation of these lineages (47). Here, we showed that, upon differentiation, the expression of Sox9 and Runx2 was markedly induced in RH18RB cells in concordance with previous reports (47, 48). We further demonstrated that KLF4 is involved in the induction of pluripotency and regulates the expression of S18-2 via direct binding to the S18-2 promoter. This finding might explain the similar expression pattern of S18-2 and KLF4 obtained in the StemMapper analysis (Fig. 2 A and B).
The importance of a precise regulation of S18-2 expression was demonstrated using the zebrafish model, in which knockdown of the S18-2 protein resulted in embryonic lethality. Moreover, the short-time survived larvae exhibited a severely abnormal phenotype, as they were significantly smaller and exhibited an underdeveloped endoderm, suggesting the presence of dysregulated segmentation. It was previously shown that disruption of the S18-2 gene via transposon mutagenesis led to the death of zebrafish at 10 d postfertilization because of impaired cardiac contractility (49).
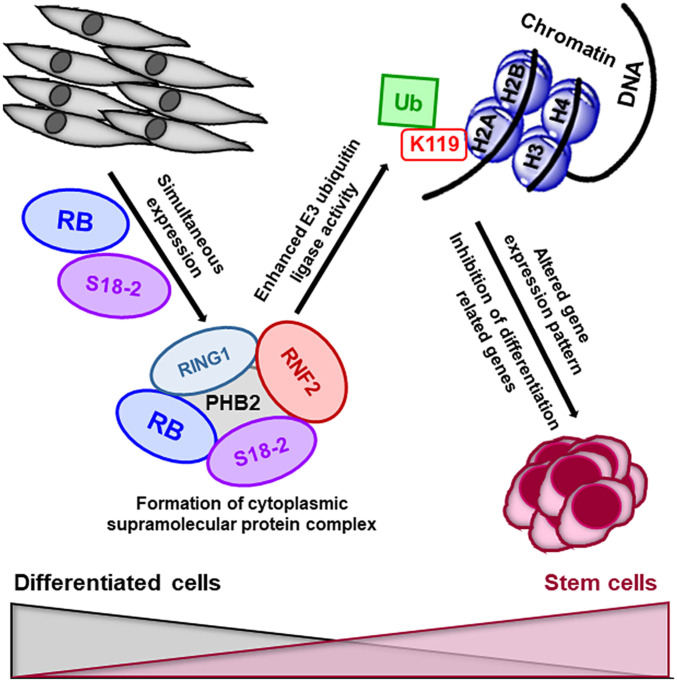
Taken together, our results suggest that Rb1-knockout MEFs exhibit stemness in the presence of RB and simultaneous overexpression of S18-2. In such cells, RB functions beyond the control of the G1-S transition. Importantly, this is in agreement with earlier findings regarding the limited control of RB over S-phase entry during cell-cycle progression in SCs (11). KLF4, which can induce stemness, binds to the S18-2 promoter and can regulate S18-2 expression. Moreover, S18-2 is involved in the formation of supramolecular complexes with subunits of PRC1. This interaction in turn enhances the enzymatic activity of RNF2, the E3-ubiquitin ligase of histone H2A, which is a characteristic of ESCs (Fig. 7).
Fig. 7.
Schematic representation of the molecular mechanisms underlying cellular stemness. The expression of high levels of the RB and S18-2 proteins in Rb1-knockout MEFs leads to the formation of a multiprotein complex containing S18-2, RB, RING1, PHB2, and RNF2. In turn, RNF2 monoubiquitinates histone H2ALys119, leading to the inhibition of differentiation-related genes. Thus, S18-2 together with RB induces or maintains cellular stemness.
The present study reveals functional consequences of the RB-S18-2 interaction in the control of cell stemness and differentiation. Our data show that aberrant levels of S18-2 play an important role in the maintenance of stemness. Importantly, we demonstrate that S18-2 is a potent oncoprotein. Hence, we suggest that decreasing S18-2 levels could be a potential strategy in the development of cancer treatments.
Materials and Methods
Cell Lines and Transfections.
RB1-knockout MEFs, named RH1301, were a kind gift from Hein Te Riele, Netherlands Cancer Institute, Amsterdam, and have been described in detail previously (16). Cells were maintained in a humidified chamber at 37 °C containing 5% CO2 in Iscove’s modified Dulbecco’s medium supplemented with 10% fetal bovine serum and appropriate antibiotics (Sigma-Aldrich, Merck). To perform transfections, RH1301 cells were plated at a density of 2 to 4 × 105 cells per 7.5-cm petri dish. Plasmids expressing a GFP-S18-2 fusion protein or a GFP-vector control were transfected with Lipofectamine 2000 (Life Technologies). Transfected cells were selected with 0.5 mg/mL of G418 (Sigma-Aldrich) for 2 wk; the resulting cell lines were named RH (transfected with the GFP control vector) and RH18 (transfected with GFP-S18-2). Next, the RH and RH18 cells were further transfected with an EX-B0065-M68 plasmid from GeneCopoeia (Source Bioscience) that encoded full-length RB. Individual clones produced by the transfected cells were selected and analyzed for RB expression using Western blotting. Further work was performed on RH cells and three sublines: RH18, expressing S18-2 at high levels; RH18RB, expressing both S18-2 and RB at high levels; and RHRB, overexpressing RB. Initially, a pool of cells was used for each cell type and was maintained for 2 y. Three individual clones from each cell type were later selected and established by performing new transfections. These were grown for no more than 2 mo. Human breast carcinoma cells (MCF7), human prostate adenocarcinoma cells (PC3), and a subclone of PC3 that expressed S18-2 at high levels (PC3-S18-2) were cultured in conditions similar to those described above for the RH subclones.
Human MSCs derived from bone marrow (ATCC PCS-500–012) were acquired from the American Type Culture Collection. These cells were cultured in Basal Medium for adipose, umbilical, and bone-marrow–derived MSCs (ATCC PCS-500–030) supplemented with a Mesenchymal Stem Cell Growth Kit for Bone Marrow-derived MSCs (ATCC PCS-500–041) to maintain their undifferentiated state.
Quantification of Senescent Cells.
To quantify the number of senescent cells, RH cells of each type were cultured in triplicate. A senescence cell histochemical staining kit (Sigma-Aldrich) was used (SI Appendix, Materials and Methods). Multiple comparisons of nonparametric criteria for all experimental data were performed using the GraphPad Prism software (version 6, GraphPad Software). A column analysis was performed using an unpaired t test. Two-tailed P values < 0.05 were considered statistically significant.
Telomerase Activity Assay.
To assess telomerase activity, a quantitative telomerase detection (QTD) kit (Allied Biotech Europe) was used. Cell lysates were prepared from all generated clones (12 cell lysates per cell type) (SI Appendix, Materials and Methods). Reactions were performed in triplicate for each cell line. A column analysis was performed using an unpaired t test. Two-tailed P values < 0.05 were considered statistically significant. Western blotting and cell fractionation are described in SI Appendix, Materials and Methods.
Immunostaining.
Before immunostaining, cells were grown on coverslips and then fixed in a mixture of cold methanol and acetone (1:1) at −20 °C. After rehydration in phosphate-buffered saline (PBS), cells were stained with antibodies. Hoechst 33258 (Sigma-Aldrich) at a concentration of 0.1 mg/mL was added during the secondary antibody incubation to stain DNA. Images were captured on a DAS microscope Leitz DM RB coupled with a C4880 dual-mode cooled charge-coupled device camera (Hamamatsu). The antibodies used are described in SI Appendix, Materials and Methods.
RT2 Profiler Assay.
Cell pellets were resuspended in TRIzol reagent and stored at −80 °C until further use. Total RNA was isolated from cells using a RNeasy Mini kit (Qiagen) according to the manufacturer’s instructions. Two micrograms of total RNA were used to prepare complementary DNA (cDNA) using an RT2 First Strand Kit (Qiagen). The evaluation of the expression profiles of genes associated with the ESC phenotype was carried out using the Mouse Embryonic Stem Cell RT2 Profiler PCR Array (SABiosciences, Qiagen). The cycle threshold values obtained were uploaded onto the manufacturer’s website for online analysis of gene expression.
Directed In Vitro Differentiation.
The experimental setup of each differentiation experiment was the same as that described for the senescence assay. The chemicals that were used to induce osteogenic, chondrogenic, and adipogenic differentiation, in vitro and qualitative reactions, and quantitation of triglycerides are described in SI Appendix, Materials and Methods.
Quantitative PCR.
All four cell types were treated with media to induce osteogenic or chondrogenic differentiation, as described in Directed In Vitro Differentiation. Total RNA was isolated from cells using a RNeasy Mini kit (Qiagen). Approximately 1 μg of total RNA was used for cDNA synthesis using a First Strand Synthesis Kit (Fermentas, Life Technologies) according to the manufacturer’s protocol. The primer concentration was adjusted to a final concentration of 3 μM, and the total reaction volume in all qPCR experiments was 20 μL. qPCR was performed using a SYBR Green master mix on a 7500 PCR system (Applied Biosystems). Relative gene expression was normalized to that of the transcript from the gene encoding the TATA-binding protein (TBP), which served as an endogenous control.
The MSCs were transfected with siRNA specific for S18-2 or RB, both individually as well as in a mixture, using DharmaFECT 1 reagent, according to the manufacturer’s protocol. Forty-eight hours post transfection the RNA was extracted, cDNA was synthesized, and qPCR was carried out as described above. Data were presented as relative gene expression normalized to GAPDH as the internal control.
Cell Tumorigenicity in SCID Mice.
To analyze the tumorigenicity of RH, RHRB, RH18, and RH18RB cells, 4 to 5 × 106 cells of each subline were injected s.c. into mice with SCID. Each mouse was injected with the same cell line at two sites, in the left and right flanks, in order to reduce the number of animals. Two female mice and thus four injections were used for each cell line.
All animal experiments were performed under ethical permission no.192/14, granted by the Solna court in Stockholm and according to the guidelines and regulations of the Karolinska Institutet and Swedish law.
Immunoprecipitation.
Five micrograms of a mouse anti-GFP antibody was coupled to CN-Br Sepharose 4 Fast Flow (GE Healthcare Bio-Sciences AB), according to the manufacturer’s protocol. On this antibody-coupled CN-Br sepharose support, S18-2–binding proteins were captured from Nonidet P-40 cell lysates from MCF7 breast carcinoma cells transfected with either the GFP-fusion construct or the GFP-vector alone (as a negative control). The initial number of cells was ∼5 × 106. After extensive washes with Nonidet P-40 lysis buffer and PBS, protein complexes were eluted from the Sepharose beads by heating the samples to 94 °C. The obtained protein complexes were separated by sodium dodecyl sulfate/polyacrylamide gel electrophoresis on 8 and 12% gels, which were then fixed in buffer containing methanol and acetic acid and stained with colloidal Coomassie blue (0.2% Coomassie blue, 7.5% acetic acid, and 50% ethanol) overnight. Relevant bands were excised and further analyzed by mass spectrometry.
Mass Spectrometry Analysis.
Samples were prepared as described previously in detail (50). Briefly, after the removal of Coomassie blue from gel bands by washing in buffer containing acetonitrile and ammonium bicarbonate, bands were dried and treated with modified porcine trypsin (Promega). The peptides generated after overnight incubation were analyzed by peptide mass fingerprinting on a matrix-assisted laser desorption/ionization/time-of-flight mass spectrometer (Ultraflex TOF/TOF, Bruker Daltonics). Scanning sequence databases (National Center for Biotechnology Information) and a ProFound (http://prowl.rockefeller.edu/prowl-cgi/profound.exe) search were used to identify peptides. Selected peptides were sequenced using postsource decay after the sulfonation of N termini (51).
Further information about the GST pull-down assay can be found in SI Appendix, Materials and Methods.
Promoter Pull-Down Assay.
A bioinformatics analysis of the S18-2 promoter region was performed. We used the Gene2Promoter prediction software (Genomatix AG 2018) (52) to identify an S18-2 promoter region located −600 bp upstream of the ATG start codon. The obtained sequence was analyzed with the help of the BLASTn software (53). In the promoter sequence, 145 bp of the 5′-untranslated region were not included because of the presence of another ATG codon (SI Appendix, Fig. S4A).
To analyze the putative binding sites of various TFs to the S18-2 promoter region, a manually curated database, TRANSFAC (TRANScription FACtor) (54), was used. Based on the results of the TRANSFAC analysis, the promoter regions of both human and mouse S18-2 were amplified by PCR using specific primers. The 5′ ends of forward primers were labeled with biotin.
PC3 cell lysates were used to precipitate TFs on the human S18-2 promoter, while RH18RB cell lysates were used for the mouse promoter. A mixture of biotin-labeled PCR fragments amplified from the S18-2 promoters and the respective cell lysates were incubated overnight at 4 °C. Iron-conjugated streptavidin beads were then added, and DNA-protein complexes were washed five times with ice-cold PBS. After washing, 20 µL of 2× Laemmli buffer was added to beads and boiled at 98 °C for 10 min. Samples were loaded on a 10% polyacrylamide gel for Western blot analysis. The binding of E2F1 to the E1A promoter was analyzed using same procedure, as a positive control. The synthesized DNA fragment used for E2F1 is described in SI Appendix, Materials and Methods.
ChIP Assay.
To assess Klf4 binding at the mouse S18-2 promoter, three subclones from RH18RB cells were used. Briefly, cells were fixed at room temperature for 10 min in a medium that contained 1% formaldehyde. After fixation, an EZ-Magna ChIP G-Chromatin Immunoprecipitation Kit (Merck Millipore, Darmstadt, Germany) was used. Extracted nuclei were sheared for six to eight cycles with a sonicator set to 30 s “on” and 50 s “off” per cycle. After sonication, each sample was loaded on a 1.5% agarose gel. Samples with a smear size in the range of 250 to 1,000 bp were used for the ChIP assays (SI Appendix, Fig. S4D, related to Fig. 5). After the elution of chromatin-DNA complexes, specific primers for the mouse S18-2 promoter were used for qPCR, and the fold enrichment was determined. To assess the human S18-2 promoter, the same procedure was applied using human prostate cancer PC3 cells.
KLF4 Knockdown.
Control and human- and mouse-specific KLF4 siRNAs were ordered from Dharmacon. Human PC3 and mouse RH cells were transfected in triplicate using DharmaFECT, according to the manufacturer’s protocol. After 48 h of cell culture, cells were harvested, total RNA was isolated, and qPCR was performed as described above.
Expression Patterns in the Zebrafish Model.
All experiments in zebrafish were performed before embryos were 5 d old; therefore, no ethical permission was needed. Work with zebrafish was carried out at the Karolinska Institutet zebrafish facility under the supervision of experienced technicians. For the analysis of mRNA expression, ∼50 embryos were collected at 1, 2, 3, 4, 5, 6, 7, 10, 24, 48, and 72 hpf. Total RNA was isolated, cDNA was synthesized, and qPCR was performed as described above.
Inhibition of S18-2 Protein Synthesis in the Zebrafish Model.
Briefly, eggs were dechorionated and ∼1 to 5 ng of S18-2–specific morpholinos was injected before the cleavage stage of embryos (within 1 hpf). The S18-2–specific morpholino (5′-ATGCGTTGTAAGGAGGCTGCCATTT-3′) and standard control oligomers were purchased from Gene Tool. Embryos were maintained at 28 °C in E3 water. A cDNA encoding the human S18-2 protein was cloned into a pGEM vector possessing a sp6 promoter. To transcribe and translate the S18-2 protein in vitro, the TNT Coupled Wheat Germ Extract System (Promega) was used according to the manufacturer’s protocol.
PCR Primers and Antibodies.
The details and sequences of all primers and all antibodies used in the different analyses are described in SI Appendix.
Data Availability.
All of the data obtained in the present study are provided in the main text and SI Appendix. The described original cell lines, constructs, images, and reagents related to this study are available upon request.
Supplementary Material
Acknowledgments
We thank H. Te Riele (Netherlands Cancer Institute, Amsterdam) for providing Rb1 knockout MEFs, U. Hellman (Ludwig Institute for Cancer Research, Uppsala, Sweden) for mass-spectrometry analysis, and the Karolinska Institutet zebrafish core facility for expertise. This work was supported by the Swedish Cancer Society, a matching grant from the Concern Foundation (Los Angeles), the Cancer Research Institute (New York), the Swedish Institute Visby framework program, the Lillian Sagen and Curt Ericsson Research Foundation, the Emil and Wera Cornell Foundation, and by funds from the Karolinska Institutet.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1922535117/-/DCSupplemental.
References
- 1.Dalton S., Signaling networks in human pluripotent stem cells. Curr. Opin. Cell Biol. 25, 241–246 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blank U., Karlsson G., Karlsson S., Signaling pathways governing stem-cell fate. Blood 111, 492–503 (2008). [DOI] [PubMed] [Google Scholar]
- 3.Kashuba E.et al., MRPS18-2 protein immortalizes primary rat embryonic fibroblasts and endows them with stem cell-like properties. Proc. Natl. Acad. Sci. U.S.A. 106, 19866–19871 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yenamandra S. P.et al., Stem cell gene expression in MRPS18-2-immortalized rat embryonic fibroblasts. Cell Death Dis. 3, e357 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Darekar S. D.et al., Mitochondrial ribosomal protein S18-2 evokes chromosomal instability and transforms primary rat skin fibroblasts. Oncotarget 6, 21016–21028 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Feng L.et al., Immortalization of chicken embryonic liver-derived cell line by stable expression of hMRP18S-2 for serotype 4 fowl adenovirus propagation. Biologicals 54, 50–57 (2018). [DOI] [PubMed] [Google Scholar]
- 7.Kashuba E.et al., EBV-encoded EBNA-6 binds and targets MRS18-2 to the nucleus, resulting in the disruption of pRb-E2F1 complexes. Proc. Natl. Acad. Sci. U.S.A. 105, 5489–5494 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weinberg R. A., The retinoblastoma protein and cell cycle control. Cell 81, 323–330 (1995). [DOI] [PubMed] [Google Scholar]
- 9.Sherr C. J., Mammalian G1 cyclins. Cell 73, 1059–1065 (1993). [DOI] [PubMed] [Google Scholar]
- 10.Snopok B., Yurchenko M., Szekely L., Klein G., Kashuba E., SPR-based immunocapture approach to creating an interfacial sensing architecture: Mapping of the MRS18-2 binding site on retinoblastoma protein. Anal. Bioanal. Chem. 386, 2063–2073 (2006). [DOI] [PubMed] [Google Scholar]
- 11.Conklin J. F., Baker J., Sage J., The RB family is required for the self-renewal and survival of human embryonic stem cells. Nat. Commun. 3, 1244 (2012). [DOI] [PubMed] [Google Scholar]
- 12.Savatier P., Huang S., Szekely L., Wiman K. G., Samarut J., Contrasting patterns of retinoblastoma protein expression in mouse embryonic stem cells and embryonic fibroblasts. Oncogene 9, 809–818 (1994). [PubMed] [Google Scholar]
- 13.Edlund T., Jessell T. M., Progression from extrinsic to intrinsic signaling in cell fate specification: A view from the nervous system. Cell 96, 211–224 (1999). [DOI] [PubMed] [Google Scholar]
- 14.Galderisi U., Cipollaro M., Giordano A., The retinoblastoma gene is involved in multiple aspects of stem cell biology. Oncogene 25, 5250–5256 (2006). [DOI] [PubMed] [Google Scholar]
- 15.Mushtaq M., Gaza H. V., Kashuba E. V., Role of the RB-interacting proteins in stem cell biology. Adv. Cancer Res. 131, 133–157 (2016). [DOI] [PubMed] [Google Scholar]
- 16.Dannenberg J. H., van Rossum A., Schuijff L., te Riele H., Ablation of the retinoblastoma gene family deregulates G(1) control causing immortalization and increased cell turnover under growth-restricting conditions. Genes Dev. 14, 3051–3064 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kashuba E., Mushtaq M., Do MRPS18-2 and RB proteins cooperate to control cell stemness and differentiation, preventing cancer development? Exp. Oncol. 39, 12–16 (2017). [PubMed] [Google Scholar]
- 18.Pinto J. P.et al., StemMapper: A curated gene expression database for stem cell lineage analysis. Nucleic Acids Res. 46, D788–D793 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee S. J.et al., PHB2 interacts with RNF2 and represses CP2c-stimulated transcription. Mol. Cell. Biochem. 319, 69–77 (2008). [DOI] [PubMed] [Google Scholar]
- 20.Mushtaq M.et al., The MRPS18-2 protein levels correlate with prostate tumor progression and it induces CXCR4-dependent migration of cancer cells. Sci. Rep. 8, 2268 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu G., Zheng H., Ai W., C-terminal binding proteins (CtBPs) attenuate KLF4-mediated transcriptional activation. FEBS Lett. 583, 3127–3132 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blasco M. A., Telomeres and human disease: Ageing, cancer and beyond. Nat. Rev. Genet. 6, 611–622 (2005). [DOI] [PubMed] [Google Scholar]
- 23.Hiyama E., Hiyama K., Telomere and telomerase in stem cells. Br. J. Cancer 96, 1020–1024 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ben-Porath I., Weinberg R. A., The signals and pathways activating cellular senescence. Int. J. Biochem. Cell Biol. 37, 961–976 (2005). [DOI] [PubMed] [Google Scholar]
- 25.Novitch B. G., Spicer D. B., Kim P. S., Cheung W. L., Lassar A. B., pRb is required for MEF2-dependent gene expression as well as cell-cycle arrest during skeletal muscle differentiation. Curr. Biol. 9, 449–459 (1999). [DOI] [PubMed] [Google Scholar]
- 26.Thomas D. M.et al., The retinoblastoma protein acts as a transcriptional coactivator required for osteogenic differentiation. Mol. Cell 8, 303–316 (2001). [DOI] [PubMed] [Google Scholar]
- 27.Scimè A.et al., Rb and p107 regulate preadipocyte differentiation into white versus brown fat through repression of PGC-1alpha. Cell Metab. 2, 283–295 (2005). [DOI] [PubMed] [Google Scholar]
- 28.Brehm A.et al., Retinoblastoma protein recruits histone deacetylase to repress transcription. Nature 391, 597–601 (1998). [DOI] [PubMed] [Google Scholar]
- 29.Wang C.et al., Interactions between E2F1 and SirT1 regulate apoptotic response to DNA damage. Nat. Cell Biol. 8, 1025–1031 (2006). [DOI] [PubMed] [Google Scholar]
- 30.Pradhan S., Kim G. D., The retinoblastoma gene product interacts with maintenance human DNA (cytosine-5) methyltransferase and modulates its activity. EMBO J. 21, 779–788 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim Y. W., Otterson G. A., Kratzke R. A., Coxon A. B., Kaye F. J., Differential specificity for binding of retinoblastoma binding protein 2 to RB, p107, and TATA-binding protein. Mol. Cell. Biol. 14, 7256–7264 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dahiya A., Wong S., Gonzalo S., Gavin M., Dean D. C., Linking the Rb and polycomb pathways. Mol. Cell 8, 557–569 (2001). [DOI] [PubMed] [Google Scholar]
- 33.Endoh M.et al., Polycomb group proteins Ring1A/B are functionally linked to the core transcriptional regulatory circuitry to maintain ES cell identity. Development 135, 1513–1524 (2008). [DOI] [PubMed] [Google Scholar]
- 34.Wang H.et al., Role of histone H2A ubiquitination in Polycomb silencing. Nature 431, 873–878 (2004). [DOI] [PubMed] [Google Scholar]
- 35.Choi D., Lee S. J., Hong S., Kim I. H., Kang S., Prohibitin interacts with RNF2 and regulates E2F1 function via dual pathways. Oncogene 27, 1716–1725 (2008). [DOI] [PubMed] [Google Scholar]
- 36.Wang S., Nath N., Adlam M., Chellappan S., Prohibitin, a potential tumor suppressor, interacts with RB and regulates E2F function. Oncogene 18, 3501–3510 (1999). [DOI] [PubMed] [Google Scholar]
- 37.Wang S., Nath N., Fusaro G., Chellappan S., Rb and prohibitin target distinct regions of E2F1 for repression and respond to different upstream signals. Mol. Cell. Biol. 19, 7447–7460 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Osman C., Merkwirth C., Langer T., Prohibitins and the functional compartmentalization of mitochondrial membranes. J. Cell Sci. 122, 3823–3830 (2009). [DOI] [PubMed] [Google Scholar]
- 39.Lee S. J.et al., E3 ligase activity of RING finger proteins that interact with Hip-2, a human ubiquitin-conjugating enzyme. FEBS Lett. 503, 61–64 (2001). [DOI] [PubMed] [Google Scholar]
- 40.Hammond-Martel I., Yu H., Affar B., Roles of ubiquitin signaling in transcription regulation. Cell. Signal. 24, 410–421 (2012). [DOI] [PubMed] [Google Scholar]
- 41.Boyer L. A.et al., Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature 441, 349–353 (2006). [DOI] [PubMed] [Google Scholar]
- 42.Loh Y. H.et al., The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat. Genet. 38, 431–440 (2006). [DOI] [PubMed] [Google Scholar]
- 43.van der Stoop P.et al., Ubiquitin E3 ligase Ring1b/Rnf2 of polycomb repressive complex 1 contributes to stable maintenance of mouse embryonic stem cells. PLoS One 3, e2235 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rao P. S., Mallya K. B., Srivenugopal K. S., Balaji K. C., Rao U. S., RNF2 interacts with the linker region of the human P-glycoprotein. Int. J. Oncol. 29, 1413–1419 (2006). [PubMed] [Google Scholar]
- 45.Vassilev A. P., Rasmussen H. H., Christensen E. I., Nielsen S., Celis J. E., The levels of ubiquitinated histone H2A are highly upregulated in transformed human cells: Partial colocalization of uH2A clusters and PCNA/cyclin foci in a fraction of cells in S-phase. J. Cell Sci. 108, 1205–1215 (1995). [DOI] [PubMed] [Google Scholar]
- 46.Voncken J. W.et al., Rnf2 (Ring1b) deficiency causes gastrulation arrest and cell cycle inhibition. Proc. Natl. Acad. Sci. U.S.A. 100, 2468–2473 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van der Velden Y. U., Wang L., Querol Cano L., Haramis A. P., The polycomb group protein ring1b/rnf2 is specifically required for craniofacial development. PLoS One 8, e73997 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Takahashi K.et al., Msx2 is a repressor of chondrogenic differentiation in migratory cranial neural crest cells. Dev. Dyn. 222, 252–262 (2001). [DOI] [PubMed] [Google Scholar]
- 49.Ding Y.et al., Trapping cardiac recessive mutants via expression-based insertional mutagenesis screening. Circ. Res. 112, 606–617 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yenamandra S. P.et al., Epstein-Barr virus encoded EBNA-3 binds to vitamin D receptor and blocks activation of its target genes. Cell. Mol. Life Sci. 67, 4249–4256 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hellman U., Bhikhabhai R., Easy amino acid sequencing of sulfonated peptides using post-source decay on a matrix-assisted laser desorption/ionization time-of-flight mass spectrometer equipped with a variable voltage reflector. Rapid Commun. Mass Spectrom. 16, 1851–1859 (2002). [DOI] [PubMed] [Google Scholar]
- 52.Genomatix, Gene2Promoter: Retrieval and analysis of promoters. https://www.genomatix.de/online_help/help_eldorado/Gene2Promoter_Intro.html. Accessed 200.
- 53.Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J., Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990). [DOI] [PubMed] [Google Scholar]
- 54.Matys V.et al., TRANSFAC and its module TRANSCompel: Transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 34, D108–D110 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All of the data obtained in the present study are provided in the main text and SI Appendix. The described original cell lines, constructs, images, and reagents related to this study are available upon request.