Significance
The first proteins emerged some 4 billion y ago, and understanding how they came about is a daunting challenge. Further complicating matters, the rules of protein structure and function derived from modern proteins may be irrelevant to their earliest ancestors. We report an integrated approach in which protein sequence, structure, and function are considered. We show that a simple function (phase separation) may have served as the basis for a complex function (specific double-stranded DNA binding), and that disordered polypeptides can give rise to structured, well-packed domains. Finally, we demonstrate that functional proteins may arise from short and simple sequences that include ornithine, an amino acid likely present in early proteins yet absent in modern proteins.
Keywords: protein evolution, abiotic amino acids, prebiotic chemistry, protein synthesis, helix-hairpin-helix
Abstract
De novo emergence demands a transition from disordered polypeptides into structured proteins with well-defined functions. However, can polypeptides confer functions of evolutionary relevance, and how might such polypeptides evolve into modern proteins? The earliest proteins present an even greater challenge, as they were likely based on abiotic, spontaneously synthesized amino acids. Here we asked whether a primordial function, such as nucleic acid binding, could emerge with ornithine, a basic amino acid that forms abiotically yet is absent in modern-day proteins. We combined ancestral sequence reconstruction and empiric deconstruction to unravel a gradual evolutionary trajectory leading from a polypeptide to a ubiquitous nucleic acid-binding protein. Intermediates along this trajectory comprise sequence-duplicated functional proteins built from 10 amino acid types, with ornithine as the only basic amino acid. Ornithine side chains were further modified into arginine by an abiotic chemical reaction, improving both structure and function. Along this trajectory, function evolved from phase separation with RNA (coacervates) to avid and specific double-stranded DNA binding. Our results suggest that phase-separating polypeptides may have been an evolutionary resource for the emergence of early proteins, and that ornithine, together with its postsynthesis modification to arginine, could have been the earliest basic amino acids.
The very first, founding members of today’s protein families emerged de novo. Such de novo emergence is thought to begin with coincidental expression of polypeptides that had no prior physiological role (1). If such polypeptides happen to provide some benefit, they may ultimately, through a series of duplications and fusions and mergings with other peptide fragments, yield a folded and functional protein (2–5). While also an ongoing process (6), this mechanism of emergence applies foremost to the earliest proteins, regardless of whether the precursor polypeptides were formed by a primitive translation machinery (7), by other template-driven processes (8), or by spontaneous assembly (9). Crossing the peptide–protein divide, however, remains a poorly understood evolutionary process. Specifically, an evolutionary trajectory leading from a primordial polypeptide to a modern protein involves three dimensions—sequence, structure, and function—that coevolve and need to be satisfactorily accounted for.
Changes in sequence involve an increase in length as well as expansion of the amino acid alphabet. It is widely accepted that early proteins were comprised predominately of primordial amino acids, amino acids formed by spontaneous abiotic synthesis (10–13). With time, this abiotic alphabet converged to the canonical 20-amino acid set known today. It has also been postulated that the early proteins were statistical—that is, being the products of a primordial synthetic machinery, they comprised a mixture of related sequences (7, 14). Structure is presumed to have evolved from polypeptides with partial or complete disorder into ordered, tightly-packed globular domains (4, 15). Finally, the refinement of sequence and structure allowed function to develop, for example, from ligand binding with low affinity and specificity to highly avid and selective binding.
Insights into how a folded, biochemically active protein can emerge from a simpler polypeptide are rare (16–18), certainly when it comes to the natural protein world, for two reasons. First, biochemical functions, such as binding of small ligands, demand a globular protein with a preorganized active site; thus, we lack knowledge of the meaningful functions that simple precursor polypeptides that lack a defined 3D structure could fulfill. Polypeptides with various biochemical functions have been identified (16, 19, 20), but the functions described so far have limited evolutionary relevance. Second, given a simple polypeptide with a rudimentary yet evolutionarily relevant function, how can a contemporary protein with a proficient and specific function arise?
Given the foregoing questions, we attempted to construct a trajectory that starts with a simple polypeptide made of abiotic amino acids and eventually leads to a contemporary folded domain. Specifically, we explored the emergence of a nucleic acid-binding element. Early polypeptides, those emerging well before the last universal common ancestor (LUCA), likely interacted with polynucleic acids such as RNA (21–23) and possibly also with DNA (24). However, the early emergence of nucleic acid binding presents a conundrum. Basic amino acids are critical for nucleic acid binding and must have been part of primordial proteins (21), but the presence of the current proteogenic basic amino acids (Lys, Arg, and His) is questionable. Arginine is generally considered the earliest canonical basic amino acid (11, 21); however, it has not been found in simple abiotic synthesis experiments or in meteorites, although its precursor has been synthesized under abiotic conditions (25). Here we propose that arginine was preceded by ornithine, an abiotic amino acid that exists in today’s cells but not in today’s proteins. We further show that statistical chemical conversion of ornithine side chains to arginine promotes both function and folding. Finally, we show that avid and selective double-stranded (ds) DNA binding can emerge from a polypeptide that exerts a function presumed to be one of the earliest peptide-protein functions: formation of coacervates with RNA (26).
Methods
Reconstruction of Ancestor-(HhH)2.
Sequences for each of the helix-hairpin-helix (HhH)2 protein families were collected from Pfam (27) and filtered to 50% identity using CD-HIT (28). Initial alignments were generated with MAFFT L-INS-i (29) and then curated manually (sequence alignment in Dataset S1). An unrooted phylogenetic tree for the (HhH)2 protein family was built using IQ-TREE (30) with automatic model selection. Ancestral state inference was performed with CODE-ML from the PAML Suite package (31). Plots of posterior probabilities were generated using the R package GGLogo (32).
Surface Plasmon Resonance.
Binding to 29-bp or 101-bp dsDNA (SI Appendix, Table S1) was monitored by surface plasmon resonance (SPR) on a Biacore T200 system. Since the (HhH)2 variants are positively charged at neutral pH, a C1 chip (GE Life Sciences), which carries less negative charge than the standard CM5 chip, was used. Streptavidin was conjugated to the chip surface using EDC/NHS chemistry, as outlined in the C1 sensor chip manual. Approximately 2,000 RU of streptavidin was stably conjugated to the chip surface, which was then blocked by ethanolamine. Subsequently, 300 RU of biotinylated 29-bp or 101-bp dsDNA was stably associated to the surface. Before data collection, a normalization cycle followed by three priming cycles were run to stabilize the instrument. Binding assays were performed in SPR binding buffer (50 mM Tris, 150 mM NaCl, and 0.05% Tween-20, pH 7.5) with a flow rate of 20 µL/min at 25 °C. In most cases, a 1,000-s contact time was required to achieve steady-state binding, the notable exception being Primordial-(HhH)2-Orn, which required only 250 s of contact time. Regeneration of the chip surface was achieved by a 60-s injection of 2 M NaCl in SPR binding buffer.
Circular Dichroism.
Circular dichroism (CD) spectra were collected on a Chirascan CD spectrometer (Applied Photophysics). Samples containing 10 μM protein in 5 mM Tris⋅HCl and 25 mM NaCl, pH 7.5, were loaded into a 1-mm pathlength quartz cuvette. Spectra were collected from 190 to 260 nm with a data pitch of either 0.5 or 1 nm at 25 °C and a slit width of 1 nm. The photomultiplier tube voltage during measurement was kept below 700 V, and data points exceeding this value were discarded. All reported spectra have been buffer-subtracted. In the case of dsDNA titration experiments (29-bp dsDNA; SI Appendix, Table S1), the spectrum of the dsDNA alone was subtracted from that of the sample containing both dsDNA and protein. Because dsDNA absorbs in the far UV, concentrations of both the dsDNA and the protein were kept relatively low (2 μM and 10 μM, respectively) and samples with a large excess of dsDNA could not be studied. Reliable data could be collected only down to ∼195 nm, a region in which both the protein and the dsDNA exhibit significant CD signal, although of opposite signs. At these low wavelengths, the CD signal of the protein was approximately twofold greater than that of the dsDNA. In the case of trifluoroethanol (TFE) titration experiments, CD spectra spanning 190 to 260 nm of 10 µM polypeptide in the presence of various TFE concentrations (up to 20%) were collected. The spectrum of the TFE alone was subtracted from the spectrum of both TFE and protein and corrected for dilution. As before, measurements that exceeded a photomultiplier tube voltage of 700 V were discarded.
13C, 15N Protein Preparation.
An insert encoding Primordial-(HhH)2-Arg with a TEV-cleavable GB1 solubility tag (33) at the C terminus was cloned into pET-21a, an Escherichia coli expression vector (Merck Millipore) with a C-terminal 6xHis tag. Plasmids were then transformed into chemically competent E. coli Rosetta cells (New England BioLabs) and grown in M9 medium supplemented with 1 g/L 15NH4Cl and 2 g/L 13C-glucose. Protein expression was then induced with 1 mM isopropyl β-d-1-thiogalactopyranoside. After overnight expression at 20 °C, cells were collected by centrifugation and lysed by sonication. The lysate was cleared by high-speed centrifugation, and the protein was purified by nickel affinity chromatography. To ensure complete removal of nonspecifically bound DNA, a 4 M guanidium hydrochloride wash was performed before elution with imidazole. The GB1 tag was removed by TEV protease cleavage (New England BioLabs) and separated by an additional nickel affinity purification step. Sample aggregation was assessed by size-exclusion chromatography (Superdex 75 10/300 GL; GE Healthcare). Exchange into Tris-D11 buffer was achieved by successive rounds of concentration and dilution using 10-kDa/3-kDa molecular weight cutoff Amicon Ultra-15 centrifugal filter units (Millipore Sigma).
NMR Experiments.
All NMR experiments were performed at 25 °C on a Bruker Avance 800-MHz spectrometer equipped with TXI triple-resonance cryogenic probes. All NMR data were processed using Topspin (www.bruker.com) and analyzed using Sparky (34). For NMR analysis, samples of 200 μM protein in 95% H2O/5% D2O, 20 mM Tris-D11 (Cambridge Isotope Laboratories) at pH 7.5, and 250 mM NaCl were prepared. A high salt concentration and a relatively low protein concentration were required to minimize sample precipitation. For backbone resonance assignments of unbound Primordial-(HhH)2-Arg, 2D 15N/13C HSQC and 3D HNCO, HNCA, HNCACB, CBCA(CO)NH, HN(CO)CA, and HN(CO)CACB were collected. Despite duplication of the HhH motif, nearly all HN-NH correlations were obtained, except for flexible terminal residues and the linker regions where residues were broadened by exchange. Secondary structure was predicted by uploading N, HN, C, CA, CB, and HA chemical shifts in TALOS+ (35).
1H-15N HSQC titrations were collected in tandem by first optimizing conditions with unbound protein, followed by molar ratio titrations of a 12-bp dsDNA fragment (SI Appendix, Table S1). DNA base-pairing was confirmed by detection of imino resonance in H2O NOESY spectra.
Fluorescein Labeling of Peptides.
Equimolar amounts of peptide and NHS-fluorescein (0.3 to 3 mM; Thermo Fisher Scientific) were resuspended in 100 µL of acetonitrile (Bio-Lab Ltd.). The pH was adjusted to ∼9 by the addition of triethanolamine, and the reactions were incubated for 1 h. Since the peptides were not soluble in acetonitrile, the unreacted NHS-fluorescein was removed by washing the pellet several times in fresh acetonitrile until the solution was clear. After washing, the residual acetonitrile was removed by vacuum evaporation for 30 min (Concentrator plus; Eppendorf). Dried peptides were dissolved in water, and the concentration of the fluorescein-labeled peptide was estimated by measuring A493nm by spectrophotometry (NanoDrop 2000; Thermo Fisher Scientific).
Phase Separation.
Peptides and polyuridylic acid (polyU; Sigma-Aldrich, P9528) were dissolved in Milli-Q water (Millipore Sigma). Peptide concentrations were measured using the Pierce BCA Protein Assay Kit (Thermo Fisher Scientific). Stock solutions of 10 mg/mL polyU and 500 mM MES pH 5.6 were prepared. Phase separation was induced by mixing the peptide and polyU solutions. The final composition of the phase separation reaction mixture was 50 mM MES pH 5.6, 1.0 to 1.4 mg/mL polyU, and 190 to 240 µM peptide. For imaging by fluorescent microscopy, fluorescein-labeled peptide was added (final concentration 1 to 20 µM, based on the degree of labeling). Microscopy glass slides were PEG-silanized as described previously (36). Capillary channel slides for microscopy were custom-made using PEGylated slides (24 × 40 mm, 0.13 to 0.16 mm thick), microscope slides (24 mm × 60 mm, 0.15 to 0.19 thick) and coverslips (24 × 24 mm, 0.13 to 0.16 mm thick). Typically, 20 to 30 µL of the phase separation reaction mixture was loaded into chambers and observed using an inverted microscope (Nikon Eclipse Ti-S) with a 20× objective (Plan Fluor, 20×/0.45) or an oil-immersion 100× objective (Plan Apo, 100×/1.40 oil). Fluorescent images were obtained with a GFP filter (λexc = 470 ± 20 nm; λem = 525 ± 25 nm) and analyzed using the Fiji platform (37).
Data Availability.
All relevant data are included in the main text and SI Appendix.
Results
Our Model Case: The (HhH)2 Fold.
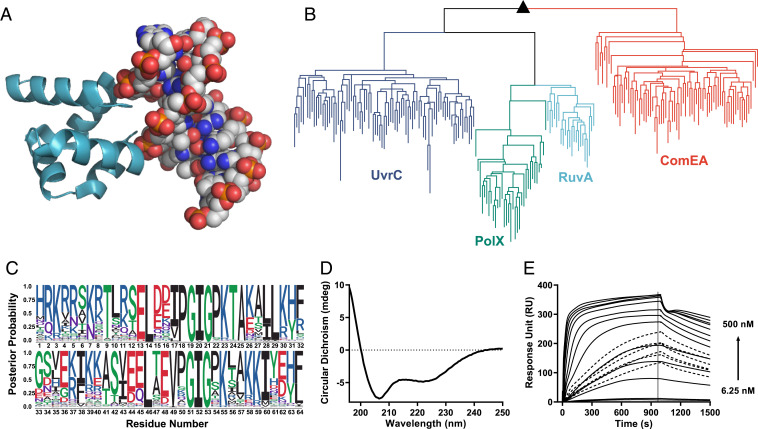
The HhH motif is a pre-LUCA nucleic acid-binding element found in at least eight protein superfamilies (38), including ribosomal protein S13 (39), with broad distribution across the tree of life. Indeed, ribosomal proteins are likely relics of the RNA-peptide world and of the earliest stages of protein evolution (4, 22). The HhH binding loop is simple and glycine-rich (consensus sequence: PGIGP) and can interact with both single- and double-stranded RNA and DNA (38). Furthermore, the HhH-binding loop forms a direct interaction with the phosphate backbone via the N terminus of an α-helix — an ancient mode of phosphate binding that can be realized with short, abiotic sequences (40). HhH motifs can function as a single element within a larger domain or as a standalone four-helix bundle, the (HhH)2 fold, in which two HhH motifs are symmetrically juxtaposed (Fig. 1A). The pre-LUCA history, along with the ability to form an independently folded and functional protein, motivated us to select the (HhH)2 fold as the starting point for our study. We performed a systematic simplification of the (HhH)2 fold, a deconstruction akin to a retrosynthetic analysis in organic chemistry (41), by generating experimental models of ancient evolutionary states.
Fig. 1.
Reconstruction of the (HhH)2 ancestor. (A) The characteristic (HhH)2 fold and its binding to the minor groove of dsDNA (shown is RuvA crystal structure; Protein Data Bank ID code 1C7Y). (B) A midpoint-rooted tree of the four contemporary (HhH)2 protein families used for ancestor state inference. Crucially, sequences associated with the different (HhH)2 protein families segregated to monophyletic clades (dark blue, UvrC; green, PolX; light blue, RuvA; orange, ComEA) are shown; sequences are listed in Dataset S1). The ancestor of all known (HhH)2 protein families is denoted with a black triangle. (C) A sequence logo representing the inferred ancestral amino acids for the common ancestor of the (HhH)2 lineage. Character height scales with posterior probability. The most probable sequence, dubbed Ancestor-(HhH)2, is listed in Table 1. The logo was modified to follow the topology of UvrC with respect to the linker connecting the two HhH motifs and other gaps. (D) CD spectrum of Ancestor-(HhH)2 showing the expected α-helical character. (E) Binding of synthesized Ancestor-(HhH)2 to a 101-bp dsDNA fragment measured by SPR; see Methods for experimental details and SI Appendix, Table S1 for the sequence of the 101-bp dsDNA fragment. The association kinetics are biphasic, with fast initial binding followed by a second, slower step (likely related to a structural rearrangement). At moderate concentrations, there appears to be a change in the rate-limiting step, perhaps from initial binding being rate-limiting at low concentrations to structural isomerization becoming rate-limiting at higher concentrations. Fig. 3C shows a plot of steady-state binding. At these transition concentrations, even a contact time of 1,000 s was insufficient to reach steady state (dotted lines).
Reconstruction of the Last (HhH)2 Common Ancestor.
Four contemporary (HhH)2 protein families are known—UvrC, PolX, RuvA, and ComEA—that comprise two fused HhH motifs and bind nucleic acids, usually dsDNA. A phylogenetic tree was constructed indicating, as expected, four monophyletic clades corresponding to these four known families (Fig. 1B). The last (HhH)2 common ancestor was then inferred using maximum likelihood methods (Fig. 1C). For the core domain, the ancestor sequence was derived, as is customary, from the most probable amino acid at each position (42). However, the linker that connects the two HhH subdomains differs between (HhH)2 families in both sequence and length, and thus its ancestral state could not be reliably inferred. Although ComEA seems like the closest clade to the common (HhH)2 ancestor, its linker represents an outlier with respect to the other three (HhH)2 families and is also more complex, requiring additional N- and C- terminal segments to cap the hydrophobic core (SI Appendix, Fig. S1A). Thus, we opted for the UvrC linker, which is eight residues long and resembles the PolX and RuvA linkers. This linker was chosen due to its relative simplicity and also because the last four residues of the linker (positions 37 to 40 in Fig. 1C) happen to align well with the sequence of the first α-helix (positions 5 to 8). This reduced the effective linker length to just four residues (positions 33 to 36) and further increased the symmetry of the ancestral sequence. The resulting protein represents the progenitor of the modern (HhH)2 families and is referred to as Ancestor-(HhH)2 (Table 1).
Table 1.
Sequences of (HhH)2 proteins and precursor polypeptides
| Construct name | Sequence* | Length, no. of residues† | Internal symmetry, % | Amino acid types |
| Contemporary (HhH)2 proteins‡ | Representative sequences from the UvrC, PolX, RuvA, and ComEA families§ | 63 ± 3 | 21 ± 8 | 16 ± 1 |
| Ancestor-(HhH)2 | HRKRRSKRTLRSELDDIPGIGPKTAKALLKHFGSVE | 64 | 46 | 15 |
| KIKKASVEELTEVPGIGPKLAKKIYEHF | ||||
| Symmetric-(HhH)2 | KIKKASVEELTEVPGIGPKLAKKIYEHFGSVE | 60 | 100 | 13 |
| KIKKASVEELTEVPGIGPKLAKKIYEHF | ||||
| Primordial-(HhH)2-Arg | RIRRASVEELTEVPGIGPRLARRILERLASIE | 60 | 100 | 10 |
| RIRRASVEELTEVPGIGPRLARRILERL | ||||
| Primordial-(HhH)2-Orn | OIOOASVEELTEVPGIGPOLAOOILEOLASIE | 60 | 100 | 10 |
| OIOOASVEELTEVPGIGPOLAOOILEOL | ||||
| Precursor-Arg | RIRRASVEELTEVPGIGPRLARRILERLA | 29 | — | 10 |
| Precursor-Orn | OIOOASVEELTEVPGIGPOLAOOILEOLA | 29 | — | 10 |
The conserved PGIGP motif is shown in bold and the four-residue linker that joins two HhH subdomains is shown in italics. The single-letter amino acid code for ornithine is taken to be O.
The specified length relates to chemically synthesized variants. Protein variants expressed in E. coli include C-terminal linker (LE) and 6xHis tag (SI Appendix, Table S2).
Parameters describing these proteins are reported as mean ± standard deviation.
Ancestor-(HhH)2, along with other relevant constructs, was chemically synthesized to allow a direct comparison of its properties with constructs that are incompatible with heterologous expression due to the presence of noncanonical amino acids (SI Appendix, Fig. S1 B–F). This simple core domain was soluble, adopted the expected helical structure (Fig. 1D), and avidly bound dsDNA (Fig. 1E).
Ancestor-(HhH)2 turned out to be significantly more symmetric than contemporary sequences, with 46% identity between the two HhH subdomains compared with 21 ± 8% identity in extant (HhH)2 proteins (Table 1). Ancestral sequence reconstruction has no inherent bias for sequences with internal symmetry. Although the linker was also chosen to promote symmetry, this increased the internal sequence identity by only a single residue, or by 3.6%. Therefore, the increased symmetry supports the proposed scenario of emergence of the (HhH)2 domain via duplication and fusion of a polypeptide precursor (2). To further support this mechanism of emergence, we attempted to construct an (HhH)2 intermediate with 100% sequence identity between the two HhH subdomains—that is, a functional protein in which both subdomains have an identical sequence.
Deconstructing a Symmetric (HhH)2 Ancestor.
Traditional phylogenetic approaches do not enable reconstruction beyond the last common (HhH)2 ancestor or pre-LUCA sequences in general. Thus, we applied an experimental deconstruction approach by performing successive rounds of simplification to generate plausible models, and proofs of principle, of early evolutionary states. In the first instance, three alternative approaches were applied to achieve complete symmetrization (SI Appendix, Table S2). First, we inferred the ancestral HhH motif from a tree comprising the first and second HhH subdomains of natural (HhH)2 proteins. However, the resulting sequence, when duplicated and joined by the UvrC linker, did not bind dsDNA.
Next, the first and second subdomains of Ancestor-(HhH)2 were duplicated while retaining the UvrC linker (and thereby symmetrizing positions 5 to 8 and 37 to 40; see above). Of these two variants, duplication of the second subdomain of Ancestor-(HhH)2 yielded a functional protein, dubbed Symmetric-(HhH)2 (Table 1 and SI Appendix, Fig. S2). Finally, a third variant was constructed in which the most probable amino acid across the two symmetric halves was chosen. This third approach also yielded a functional protein (SI Appendix, Table S2), indicating that multiple sequences can satisfy the symmetry constraint.
Symmetric-(HhH)2 has 100% sequence identity between subdomains and thus is the conceptual product of duplication and fusion of a precursor polypeptide of 28 amino acids. It also uses only 13 amino acid types, compared with the 16 ± 1 types used by natural (HhH)2 proteins (Table 1). Indeed, it is widely accepted that early proteins were composed predominately of “primordial amino acids,” that is, amino acids formed by spontaneous abiotic synthesis. Foremost, of the current 20 canonical amino acids, Gly, Ala, Ser, Thr, Asp, Glu, Pro, Val, Ile, and Leu are considered abiotic (10–12). However, Symmetric-(HhH)2, includes several amino acids that likely emerged with enzyme-based biosynthesis, specifically His, Phe, Tyr, and Lys. Can a functional variant of the (HhH)2 fold be constructed that is both fully symmetric and composed of an abiotic amino acid alphabet?
A Primordial-(HhH)2 Ancestor.
Removal of His, Phe, and Tyr from Symmetric-(HhH)2 was guided by three considerations: 1) the second most probable amino acid from ancestor inference (Fig. 1C), 2) the consensus amino acid of the UvrC protein family, and 3) abiotic amino acids with similar properties (e.g., leucine or isoleucine instead of aromatic amino acids). In practice, these three approaches often yielded overlapping sequences (SI Appendix, Table S3). Alternative core mutations based on these three considerations were also tested to ensure stable core packing. In all cases, 100% sequence identity between subdomains was preserved.
While phenylalanine and tyrosine were readily eliminated, the only viable substitution of histidine that we could identify was to lysine, which is not considered abiotic. Furthermore, Symmetric-(HhH)2 already contains 12 lysine residues. We thus obtained a protein with 14 lysine residues, composing >20% of its sequence (14 of 60 residues), underscoring the crucial need for basic amino acids in nucleic acid binding (21). Can these lysine residues be substituted to another, more ancient basic amino acid?
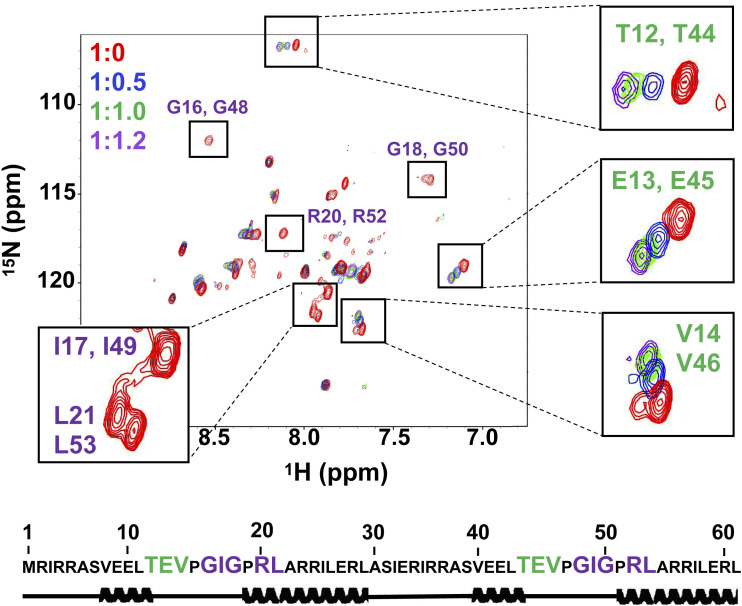
As arginine is thought to predate lysine (11, 21), we attempted to replace the lysine residues with arginine. We constructed a series of variants with increasing degrees of lysine-to-arginine exchange, ultimately replacing all 14 lysines with arginine. These exchanges not only were tolerated, but also improved the affinity for dsDNA (SI Appendix, Table S3). The product of complete exchange, Primordial-(HhH)2-Arg (Table 1) adopted a helical structure and bound specifically to dsDNA (Fig. 2 and SI Appendix, Fig. S3). Therefore, structure and function were achieved despite being constructed from only 10 different amino acid types (nine abiotic amino acid types plus arginine). As dsDNA was titrated in, NMR cross-peaks associated with the glycine residues in the canonical PGIGP binding motif broadened, while nearby residues experienced progressive chemical shift changes. This result suggests that the canonical PGIGP loops and neighboring residues mediate dsDNA binding (Fig. 2), as further confirmed by deactivation mutations in the PGIGP loops of Symmetric-(HhH)2 (SI Appendix, Table S4).
Fig. 2.
NMR characterization of Primordial-(HhH)2-Arg. 1H-15N HSQC titration of Primordial-(HhH)2-Arg with 12-bp dsDNA. Protein:dsDNA ratios tested were 1:0 (red spectrum), 1:0.5 (blue spectrum), 1:1.0 (green spectrum), and 1:1.2 (magenta spectrum). On dsDNA binding, several peaks shifted due to a fast exchange regime (green lettering) or broadened due to an intermediate exchange regime (purple lettering). Notably, glycine peaks associated with the canonical PGIGP binding motif exhibited intermediate exchange binding behavior, suggesting that the interaction with dsDNA is centered around the PGIGP motif. Secondary structure prediction was performed by TALOS+ using chemical shift alignments and confirmed the helical structure of Primordial-(HhH)2-Arg (see also SI Appendix, Fig. S3C). For NMR studies, E. coli-expressed tag-free Primordial-(HhH)2-Arg was used. Numbering is based on the E. coli-expressed protein, in which residue 1 is methionine. A 12-bp dsDNA oligonucleotide was used for NMR experiments to minimize avidity effects in binding and to retain fast molecular tumbling during dsDNA titration. The DNA sequences for all experiments are provided in SI Appendix, Table S1.
Overall, Primordial-(HhH)2-Arg differs from the last common (HhH)2 ancestor at 32 out of 64 positions, including 12 lysine residues and 2 histidine residues replaced by arginine. Compared with modern (HhH)2 domains, and considering only positions that align to the last common (HhH)2 ancestor, Primordial-(HhH)2-Arg differs at 45 ± 4 out of 64 positions. Nonetheless, its folding and binding mode are consistent with those of contemporary (HhH)2 domains.
A Functional Ornithine-Based (HhH)2 Ancestor.
Arginine likely emerged before lysine and histidine, yet it is often not considered among the earliest abiotic amino acids. This led us to examine whether the (HhH)2 fold could possibly function with a basic amino acid that might have predated arginine. Ornithine has been identified in a volcanic spark discharge experiment (12) and in meteorites (43) and is a metabolic precursor of arginine (44). Could the first basic amino acid be ornithine?
Since ornithine cannot be incorporated by standard protein expression approaches, we resorted to chemical protein synthesis. Two half-peptides were synthesized and then joined using a native chemical ligation and a deselenization approach (45, 46). To permit deselenization chemical approaches, the glycine in the linker connecting the two HhH motifs of Primordial-(HhH)2 was changed to alanine, with no effect on folding or binding, as tested on the arginine version (SI Appendix, Table S3). For each construct, the N-terminal half-peptide was synthesized with a C-terminal thioester, which was reacted with the C-terminal peptide bearing an N-terminal selenocysteine. Following ligation of the two peptides, the selenocysteine residue was deselenized to yield alanine (SI Appendix, Methods).
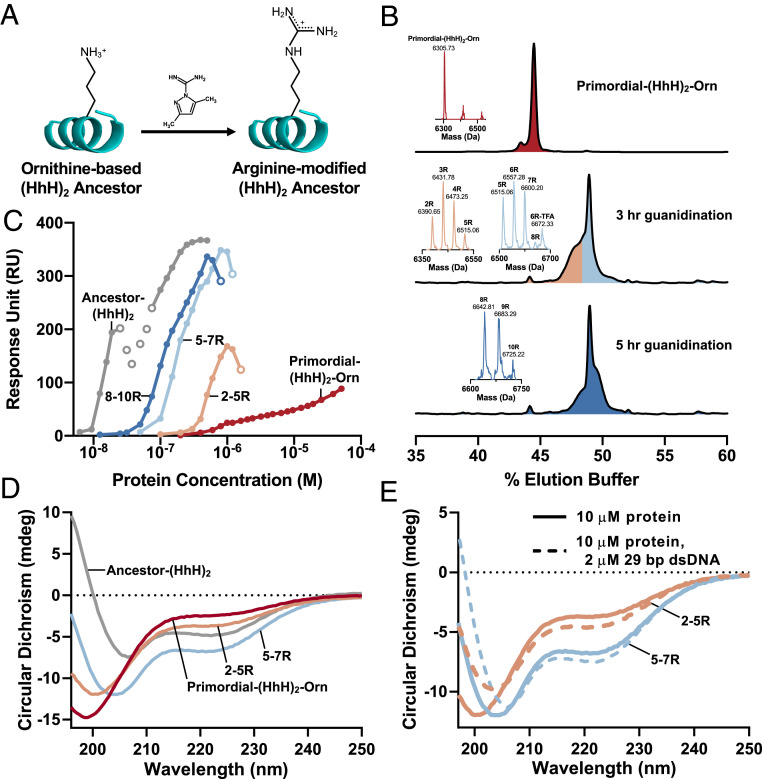
Remarkably, Primordial-(HhH)2-Orn (Table 1 and SI Appendix, Fig. S4), in which all 14 arginine residues have been replaced by ornithine, weakly bound dsDNA (SI Appendix, Fig. S4F and Fig. 3C), demonstrating that in principle, functional dsDNA binding proteins could be produced in the absence of either arginine or lysine. However, CD spectroscopy indicated that Primordial-(HhH)2-Orn is largely unfolded, although the addition of dsDNA seems to induce some helical structure (SI Appendix, Fig. S4G). Exchanging lysine for ornithine in the context of Symmetric-(HhH)2 yielded a functional protein with a higher affinity for dsDNA compared with Primordial-(HhH)2-Orn (SI Appendix, Fig. S5). Indeed, the poor foldability of Primordial-(HhH)2, which is devoid of aromatic residues in the hydrophobic core, is also responsible for its comparatively low affinity. An inevitable result of folding upon binding is lower binding affinity.
Fig. 3.
Statistical conversion of ornithine side chains to arginine promotes structure and dsDNA binding. (A) A simple chemical reaction, guanidination, performed in water can convert the ornithine side chains of a chemically synthesized protein into arginine side chains (see also SI Appendix, Figs. S6 and S7). (B) Synthesized Primordial-(HhH)2-Orn (Top) and three mixtures of Primordial-(HhH)2-Orn with different degrees of guanidination of its 14 ornithine residues (Middle and Bottom) analyzed by reverse-phase chromatography and mass spectrometry (observed masses shown in the figure and calculated and observed are reported below). The three mixtures of Primordial-(HhH)2-Orn with different degrees of guanidination were prepared by varying the reaction time and fractionation by reverse-phase chromatography. These mixtures contained 2 to 5 guanidinations per polypeptide (orange), 5 to 7 guanidinations per polypeptide (light blue), and 8 to 10 guanidinations per polypeptide (dark blue). The compositions of these mixtures were determined by mass spectrometry analysis. Note that a single mass represents a mixture of proteins guanidinylated to the same degree but at different positions. (C) The binding affinity for dsDNA increases monotonically with the extent of guanidination, as indicated by SPR binding experiments with an immobilized 101-bp dsDNA fragment. (SI Appendix, Fig. S8D provides an equivalent plot of binding to 29-bp dsDNA.) Open circles indicate concentrations at which a change in the rate-limiting step occurred (additional details in Fig. 1E). (D) The α-helical character of the protein increases with increasing guanidination, as demonstrated by CD spectroscopy; the 195-nm signal becomes increasingly positive, the signal at 222 nm becomes increasingly negative, and the global minimum shifts toward 208 nm. (E) An increase in α-helicity is observed on mixing of protein with 29-bp dsDNA, suggesting folding on binding. Plotted spectra are background-subtracted to remove the signal associated with dsDNA. Mass spectrometry confirmation: Primordial-(HhH)2-Orn, mcalc = 6,307.57 Da, mobs = 6,305.73 ± 0.56 Da; Primordial-(HhH)2-2R, mcalc = 6,391.65 Da, mobs = 6,390.65 ± 2.02 Da; Primordial-(HhH)2-3R, mcalc = 6,433.69 Da, mobs = 6,431.78 ± 0.30 Da; Primordial-(HhH)2-4R, mcalc = 6,475.73 Da, mobs = 6,473.25 ± 0.99 Da; Primordial-(HhH)2-5R, mcalc = 6,517.77 Da, mobs = 6,515.06 ± 0.81 Da; Primordial-(HhH)2-6R, mcalc = 6,559.81 Da, mobs = 6,557.28 ± 1.80 Da; Primordial-(HhH)2-6R-TFA, mcalc = 6,673.83 Da, mobs = 6,672.33 ± 3.66 Da; Primordial-(HhH)2-7R, mcalc = 6,601.85 Da, mobs = 6,600.20 ± 1.43 Da; Primordial-(HhH)2-8R, mcalc = 6,643.89 Da, mobs = 6,642.81 ± 1.00 Da; Primordial-(HhH)2-9R, mcalc = 6,685.93 Da, mobs = 6,683.29 ± 1.32 Da; Primordial-(HhH)2-10R, mcalc = 6,727.97 Da, mobs = 6,725.22 ± 2.12 Da. Masses were determined with an ion trap mass spectrometer, and the deviations from the theoretical masses are within the measurement error.
Statistical Conversion of Ornithine to Arginine Promotes Folding and dsDNA Binding.
Ornithine has another distinctly attractive feature. In contemporary metabolism, free ornithine is guanidinated by a series of enzymatic reactions to yield arginine, which is incorporated into proteins (44). Furthermore, guanidination of ornithine to give arginine can also be performed chemically (47), even by simple reagents such as cyanamide (H2N-CN) (48), which were likely present in primordial environments. Cyanamide is of particular prebiotic importance as it can mediate other abiotic reactions, such as phosphorylation and peptide bond formation (49, 50). Foremost, chemical guanidination can be readily performed on polypeptides that contain ornithine as a postsynthesis modification that leads to arginine (Fig. 3A). This reaction has been reported previously (47), but its products were not fully characterized. We tested this protocol on model polypeptides and observed complete conversion of ornithine into arginine in aqueous solution. Conversion was also selective, with slow modification of the peptide’s N-terminal amine group, and no modification of the histidine side chain, observed (SI Appendix, Figs. S6 and S7).
The guanidination reaction was then applied to Primordial-(HhH)2-Orn to achieve partial conversion (Fig. 3B), and guanidinated mixtures were isolated. Although these mixtures were random with respect to which ornithine positions were converted to arginine, they had discrete compositions with respect to the total number of modified ornithine residues (Fig. 3B). We found that greater conversion of ornithine to arginine resulted in progressive improvement in both the affinity for dsDNA and folding toward the expected (HhH)2 helical structure, as judged by SPR (Fig. 3C and SI Appendix, Fig. S8) and CD (Fig. 3 D and E), respectively.
A Single, Primordial HhH Motif Forms Coacervates with RNA.
The results described above demonstrate that the (HhH)2 fold likely emerged from duplication of a short, simple polypeptide composed of just a single HhH motif. In principle, the precursor polypeptide could emerge by chance and, once duplicated, become evolutionarily advantageous. However, emergence from an already functional single HhH motif polypeptide is far more likely, as nascent activity is a viable starting point for evolutionary optimization (3, 18). Thus, we tested preduplicated 29-residue polypeptides derived from Primordial-(HhH)2 containing either ornithine (“Precursor-Orn”) or arginine (“Precursor-Arg”) (Table 1). Neither of these polypeptides showed reproducible dsDNA binding.
The functional and structural properties of ancient polypeptides have been subjects of intense interest. In principle, small-molecule ligand binding and catalysis demand a degree of structural volume, complexity, and preorganization that polypeptides rarely provide. It has been postulated that self-assembly could endow primordial polypeptides with such properties. Specifically, both amyloid formation (51) and peptide-RNA condensates (26) have been proposed as ancient forms of self-assembly. Could the formation of liquid condensates with RNA be an early function predating nucleic acid binding in modern proteins?
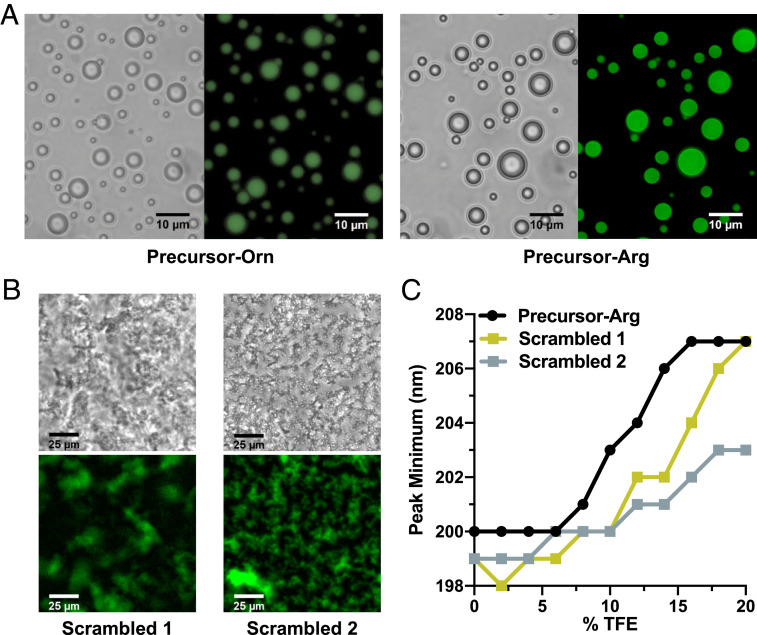
On mixing with polyU, both Precursor-Orn and -Arg formed coacervates (Fig. 4A and SI Appendix, Fig. S9), as did Ancestor-(HhH)2 (SI Appendix, Fig. S10). Neither polypeptide formed droplets in the absence of polyU. Further, precursor-Arg consistently made larger droplets than Precursor-Orn, and at lower concentrations (SI Appendix, Fig. S11). Thus, by increasing the coacervate-forming potential, statistical modification of ornithine to arginine could also provide an advantage at this early stage.
Fig. 4.
Coacervate formation by precursor polypeptides and RNA. (A) Coacervates form on the addition of polyU to both Precursor-Orn (240 μM peptide, 1.2 μM fluorescein-labeled peptide, 1.2 mg/mL polyU) and Precursor-Arg (190 μM peptide, 20 μM fluorescein-labeled peptide, 1.4 mg/mL polyU). (B) Complete scrambling of the sequence of Precursor-Arg (“Scrambled 1”), as well as scrambling of all residues except arginine (“Scrambled 2”), abolishes droplet formation (200 μM peptide, 8 μM fluorescein-labeled peptide, 1 mg/mL polyU); polypeptide sequences are provided in SI Appendix, Table S5. (C) TFE titrations of 10 μM polypeptide in buffer reveal that Precursor-Arg has a higher intrinsic α-helical propensity than the scrambled versions. This difference is manifest by a shift in the CD peak minimum from ∼199 nm (random coil-like) to 207 nm (α-helix-like) at lower TFE concentrations. Raw CD spectra are provided in SI Appendix, Fig. S12.
Phase separation of these polypeptides is perhaps unsurprising, given that arginine-rich peptides have been shown to phase-separate on the addition of RNA or crowding agents (52). However, scrambling the sequence of the Precursor-Arg—either completely or while preserving only the positions of the arginine residues (scrambled 1 and 2, respectively)—resulted in amorphous aggregates on polyU addition (Fig. 4B). Thus, basic amino acids are likely necessary but not sufficient to encode phase separation.
Finally, TFE titration experiments with the precursor polypeptides in solution demonstrated that Precursor-Arg has a higher propensity to form α-helices compared with the scrambled variants (Fig. 4C and SI Appendix, Fig. S12). This result suggests that sequences that evolved to phase-separate may have a greater propensity to fold into a helical structure on duplication.
Discussion
Proteins with defined biochemical function are thought to be exceedingly rare in sequence space, with some probability estimates as low as 10−11 for nucleotide binding (53). However, the trajectory described here suggests a possible solution to this evolutionary conundrum whereby evolution begins with a rudimentary function, such as phase separation, that can be mediated by polypeptides with minimal sequence constraints. Duplication events and expansion of the amino acid alphabet would then enable a smooth transition to more complex protein forms, with both structure and function becoming increasingly defined over time. During simplification, ∼70% of the positions in Ancestor-(HhH)2 were changed to generate Primordial-(HhH)2-Arg. These changes included 14 simultaneous exchanges of the basic amino acids to the currently nonproteogenic ornithine, while maintaining dsDNA binding. Therefore, there is a large cloud of traversable sequences from which the actual, historical ancestor of the HhH element could have emerged. This unusual robustness to sequence variation and emergence by duplication indicate a probability of emergence much higher than 10−11.
Our results also show that polypeptides and proteins can bind polynucleic acids with ornithine as the sole basic amino acid. Nucleic acid binding is a primordial function that likely emerged in a peptide-RNA world (9, 21, 22). Thus, we offer a hypothesis in which ornithine was the main basic amino acid at the earliest stages of protein evolution. The evolutionary change in amino acid composition and the genetic code is often viewed as going from a subset of the current 20 proteogenic amino acids to the complete set; however, many other amino acids are seen in abiotic synthesis experiments and/or meteorites, including amino acids that are ubiquitous in modern biology but are not incorporated into proteins (nonproteogenic). Some of these amino acids are thought to have been included in early proteins, such as norvaline and α-aminobutyric acid (54). Indeed, the earliest hypotheses regarding the emergence of translation and the genetic code assume a large primordial set of amino acids from which the current set of 20 canonical amino acids was selected (7, 14). Thus, the amino acid composition of proteins has likely changed across evolutionary time. As translation fidelity improved, some abiotic amino acids became biologically irrelevant or nonproteogenic (e.g., ornithine), and, conversely, new amino acids were biosynthesized and incorporated into the genetic code.
Ornithine is present in living cells but is not encoded by ribosomal protein synthesis, primarily because of the presumed instability of its tRNA ester due to intramolecular lactam formation (55). However, the fact that ornithine is present in nonribosomal peptides (56) indicates that its esters (and even more reactive thioesters) are sufficiently long-lived to allow its condensation into polypeptides. Foremost, ornithine is the biosynthetic precursor of arginine. There are multiple indications that intermediates of contemporary metabolic pathways served as end-point metabolites at earlier evolutionary stages (44); thus, arginine may have first emerged as a spontaneous, chemical modification of ornithine side chains in already synthesized polypeptides. The superior properties of arginine in promoting coacervate formation, binding to dsDNA, and protein folding on the one hand, and the instability of ornithine esters on the other hand, likely led to the takeover of arginine as a protein building block (57).
Finally, this experimentally validated trajectory, which starts from a polypeptide that forms coacervates with RNA and leads to a contemporary dsDNA-binding protein, lends support to a hypothesis that stems from Oparin’s protocells hypothesis—namely, that peptide-RNA condensates had a role in the earliest life forms (26).
Supplementary Material
Acknowledgments
We thank Brian Ross and Vladimir Kiss for assistance with fluorescence microscopy, Dora Tang for assistance with droplet imaging, Einav Aharon for assistance with molecular cloning, Jelena Cvetičanin for assistance with mass spectrometry, Reem Mousa for assistance with polypeptide guanidination, and Ita Gruić-Sovulj for suggesting the role of ornithine as an alternative basic amino acid. We also thank Andrei Lupas, Donald Hilvert, and James Bardwell for insightful comments on this manuscript. This work was funded by the Israel Science Foundation (Grants 980/14, to D.S.T. and 783/18, to N.M.), an Israel Cancer Research Fund Acceleration Grant (to N.M.), and the NIH (Grant R35 GM126942, to G.V.). D.S.T. is the Nella and Leon Benoziyo Professor of Biochemistry. O.W.-K. acknowledges the Kaete Klausner Fellowship for financial support. L.M.L., D.D., and J.J. acknowledge support from a Systems Biology Innovative Award from the Weizmann Institute of Science.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2001989117/-/DCSupplemental.
References
- 1.Ruiz-Orera J., Verdaguer-Grau P., Villanueva-Cañas J. L., Messeguer X., Albà M. M., Translation of neutrally evolving peptides provides a basis for de novo gene evolution. Nat. Ecol. Evol. 2, 890–896 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Eck R. V., Dayhoff M. O., Evolution of the structure of ferredoxin based on living relics of primitive amino acid sequences. Science 152, 363–366 (1966). [DOI] [PubMed] [Google Scholar]
- 3.Romero Romero M. L., Rabin A., Tawfik D. S., Functional proteins from short peptides: Dayhoff’s hypothesis turns 50. Angew. Chem. Int. Ed. Engl. 55, 15966–15971 (2016). [DOI] [PubMed] [Google Scholar]
- 4.Kovacs N. A., Petrov A. S., Lanier K. A., Williams L. D., Frozen in time: The history of proteins. Mol. Biol. Evol. 34, 1252–1260 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alva V., Lupas A. N., From ancestral peptides to designed proteins. Curr. Opin. Struct. Biol. 48, 103–109 (2018). [DOI] [PubMed] [Google Scholar]
- 6.Baalsrud H. T.et al., De novo gene evolution of antifreeze glycoproteins in codfishes revealed by whole genome sequence data. Mol. Biol. Evol. 35, 593–606 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Woese C. R., On the evolution of the genetic code. Proc. Natl. Acad. Sci. U.S.A. 54, 1546–1552 (1965). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guseva E., Zuckermann R. N., Dill K. A., Foldamer hypothesis for the growth and sequence differentiation of prebiotic polymers. Proc. Natl. Acad. Sci. U.S.A. 114, E7460–E7468 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van der Gulik P. T., Speijer D., How amino acids and peptides shaped the RNA world. Life 5, 230–246 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miller S. L., A production of amino acids under possible primitive earth conditions. Science 117, 528–529 (1953). [DOI] [PubMed] [Google Scholar]
- 11.Trifonov E. N., Consensus temporal order of amino acids and evolution of the triplet code. Gene 261, 139–151 (2000). [DOI] [PubMed] [Google Scholar]
- 12.Johnson A. P.et al., The Miller volcanic spark discharge experiment. Science 322, 404 (2008). [DOI] [PubMed] [Google Scholar]
- 13.Longo L. M., Lee J., Blaber M., Simplified protein design biased for prebiotic amino acids yields a foldable, halophilic protein. Proc. Natl. Acad. Sci. U.S.A. 110, 2135–2139 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yčas M., On earlier states of the biochemical system. J. Theor. Biol. 44, 145–160 (1974). [DOI] [PubMed] [Google Scholar]
- 15.Tokuriki N., Tawfik D. S., Protein dynamism and evolvability. Science 324, 203–207 (2009) [DOI] [PubMed] [Google Scholar]
- 16.Studer S.et al., Evolution of a highly active and enantiospecific metalloenzyme from short peptides. Science 362, 1285–1288 (2018). [DOI] [PubMed] [Google Scholar]
- 17.Smock R. G., Yadid I., Dym O., Clarke J., Tawfik D. S., De novo evolutionary emergence of a symmetrical protein is shaped by folding constraints. Cell 164, 476–486 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mutter A. C.et al., De novo design of symmetric ferredoxins that shuttle electrons in vivo. Proc. Natl. Acad. Sci. U.S.A. 116, 14557–14562 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnsson K., Allemann R. K., Widmer H., Benner S. A., Synthesis, structure and activity of artificial, rationally designed catalytic polypeptides. Nature 365, 530–532 (1993). [DOI] [PubMed] [Google Scholar]
- 20.Zozulia O., Dolan M. A., Korendovych I. V., Catalytic peptide assemblies. Chem. Soc. Rev. 47, 3621–3639 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Blanco C., Bayas M., Yan F., Chen I. A., Analysis of evolutionarily independent protein-RNA complexes yields a criterion to evaluate the relevance of prebiotic scenarios. Curr. Biol. 28, 526–537.e5 (2018). [DOI] [PubMed] [Google Scholar]
- 22.Lupas A. N., Alva V., Ribosomal proteins as documents of the transition from unstructured (poly)peptides to folded proteins. J. Struct. Biol. 198, 74–81 (2017). [DOI] [PubMed] [Google Scholar]
- 23.Tagami S., Attwater J., Holliger P., Simple peptides derived from the ribosomal core potentiate RNA polymerase ribozyme function. Nat. Chem. 9, 325–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Poole A. M.et al., The case for an early biological origin of DNA. J. Mol. Evol. 79, 204–212 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Patel B. H., Percivalle C., Ritson D. J., Duffy C. D., Sutherland J. D., Common origins of RNA, protein and lipid precursors in a cyanosulfidic protometabolism. Nat. Chem. 7, 301–307 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hyman T., Brangwynne C., In retrospect: The origin of life. Nature 491, 524–525 (2012). [Google Scholar]
- 27.El-Gebali S.et al., The Pfam protein families database in 2019. Nucleic Acids Res. 47, D427–D432 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fu L., Niu B., Zhu Z., Wu S., Li W., CD-HIT: Accelerated for clustering the next-generation sequencing data. Bioinformatics 28, 3150–3152 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Katoh K., Toh H., Recent developments in the MAFFT multiple sequence alignment program. Brief. Bioinform. 9, 286–298 (2008). [DOI] [PubMed] [Google Scholar]
- 30.Trifinopoulos J., Nguyen L. T., von Haeseler A., Minh B. Q., W-IQ-TREE: A fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 44, W232–W235 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang Z., PAML 4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24, 1586–1591 (2007). [DOI] [PubMed] [Google Scholar]
- 32.Hare E., Heike H., Data from "R package for creating sequence logo plots." Github. https://github.com/heike/gglogo. Accessed 4 June 2020.
- 33.Huth J. R.et al., Design of an expression system for detecting folded protein domains and mapping macromolecular interactions by NMR. Protein Sci. 6, 2359–2364 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee W., Tonelli M., Markley J. L., NMRFAM-SPARKY: Enhanced software for biomolecular NMR spectroscopy. Bioinformatics 31, 1325–1327 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shen Y., Delaglio F., Cornilescu G., Bax A., TALOS+: A hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J. Biomol. NMR 44, 213–223 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alberti S.et al., “A user’s guide for phase separation assays with purified proteins” in J. Mol. Biol., (2018), Vol. 430, pp. 4806–4820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schindelin J.et al., Fiji: An open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alva V., Söding J., Lupas A. N., A vocabulary of ancient peptides at the origin of folded proteins. eLife 4, e09410 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Doherty A. J., Serpell L. C., Ponting C. P., The helix-hairpin-helix DNA-binding motif: A structural basis for non-sequence-specific recognition of DNA. Nucleic Acids Res. 24, 2488–2497 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Longo L. M., Petrović D., Kamerlin S. C. L., Tawfik D. S., Short and simple sequences favored the emergence of N-helix phospho-ligand binding sites in the first enzymes. Proc. Natl. Acad. Sci. U.S.A. 117, 5310–5318 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Corey E. J., The logic of chemical synthesis: Multistep synthesis of complex carbogenic molecules (Nobel Lecture). Angew. Chem. Int. Ed. Engl. 30, 455–465 (1991). [Google Scholar]
- 42.Eick G. N., Bridgham J. T., Anderson D. P., Harms M. J., Thornton J. W., Robustness of reconstructed ancestral protein functions to statistical uncertainty. Mol. Biol. Evol. 34, 247–261 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Meringer M., Cleaves H. J. 2nd, Freeland S. J., Beyond terrestrial biology: Charting the chemical universe of α-amino acid structures. J. Chem. Inf. Model. 53, 2851–2862 (2013). [DOI] [PubMed] [Google Scholar]
- 44.Noda-Garcia L., Liebermeister W., Tawfik D. S., Metabolite–enzyme coevolution: From single enzymes to metabolic pathways and networks. Annu. Rev. Biochem. 87, 187–216 (2018). [DOI] [PubMed] [Google Scholar]
- 45.Reddy P. S., Dery S., Metanis N., Chemical synthesis of proteins with non-strategically placed cysteines using selenazolidine and selective deselenization. Angew. Chem. Int. Ed. Engl. 55, 992–995 (2016). [DOI] [PubMed] [Google Scholar]
- 46.Metanis N., Keinan E., Dawson P. E., Traceless ligation of cysteine peptides using selective deselenization. Angew. Chem. Int. Ed. Engl. 49, 7049–7053 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ariely S., Wilchek M., Patchornik A., Synthesis of poly-l-arginine. Biopolymers 4, 91–96 (1966). [Google Scholar]
- 48.Alonso-Moreno C., Antiñolo A., Carrillo-Hermosilla F., Otero A., Guanidines: From classical approaches to efficient catalytic syntheses. Chem. Soc. Rev. 43, 3406–3425 (2014). [DOI] [PubMed] [Google Scholar]
- 49.Steinman G., Lemmon R. M., Calvin M., Cyanamide: A possible key compound in chemical evolution. Proc. Natl. Acad. Sci. U.S.A. 52, 27–30 (1964). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pasek M. A., Kee T. P., “On the origin of phosphorylated biomolecules” in Origins of Life: The Primal Self-Organization, (2011), pp. 57–84. [Google Scholar]
- 51.Greenwald J., Riek R., On the possible amyloid origin of protein folds. J. Mol. Biol. 421, 417–426 (2012). [DOI] [PubMed] [Google Scholar]
- 52.Boeynaems S., De Decker M., Tompa P., Van Den Bosch L., Arginine-rich peptides can actively mediate liquid-liquid phase separation. Bio Protoc. 7, e2525 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Keefe A. D., Szostak J. W., Functional proteins from a random-sequence library. Nature 410, 715–718 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bilus M.et al., On the mechanism and origin of isoleucyl-tRNA synthetase editing against norvaline. J. Mol. Biol. 431, 1284–1297 (2019). [DOI] [PubMed] [Google Scholar]
- 55.Weber A. L., Miller S. L., Reasons for the occurrence of the twenty coded protein amino acids. J. Mol. Evol. 17, 273–284 (1981). [DOI] [PubMed] [Google Scholar]
- 56.Felnagle E. A.et al., Nonribosomal peptide synthetases involved in the production of medically relevant natural products. Mol. Pharm. 5, 191–211 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Frenkel-Pinter M.et al., Selective incorporation of proteinaceous over nonproteinaceous cationic amino acids in model prebiotic oligomerization reactions. Proc. Natl. Acad. Sci. U.S.A. 116, 16338–16346 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All relevant data are included in the main text and SI Appendix.