Significance
The syndrome of cytotrophoblast invasion beyond the normal boundary (in the superficial myometrium) is collectively termed placenta accreta spectrum. The incidence of this condition is rising. However, little is known about the underlying molecular changes. Global transcriptomic profiling of cytotrophoblasts isolated from these cases, as compared to gestational age-matched controls, revealed numerous changes in gene expression involving diverse pathways, including cell signaling, migration, and immune functions. DOCK4 was the most highly up-regulated mRNA in the cases. Mutations in this gene are mechanistically linked to cancer progression. Overexpression of DOCK4 in primary cytotrophoblasts increased their invasiveness. This study provides molecular insights into the pathways driving placenta accreta spectrum and suggests numerous future directions.
Keywords: placenta accreta spectrum, cytotrophoblast, transcriptomics, DOCK4
Abstract
In humans, a subset of placental cytotrophoblasts (CTBs) invades the uterus and its vasculature, anchoring the pregnancy and ensuring adequate blood flow to the fetus. Appropriate depth is critical. Shallow invasion increases the risk of pregnancy complications, e.g., severe preeclampsia. Overly deep invasion, the hallmark of placenta accreta spectrum (PAS), increases the risk of preterm delivery, hemorrhage, and death. Previously a rare condition, the incidence of PAS has increased to 1:731 pregnancies, likely due to the rise in uterine surgeries (e.g., Cesarean sections). CTBs track along scars deep into the myometrium and beyond. Here we compared the global gene expression patterns of CTBs from PAS cases to gestational age-matched control cells that invaded to the normal depth from preterm birth (PTB) deliveries. The messenger RNA (mRNA) encoding the guanine nucleotide exchange factor, DOCK4, mutations of which promote cancer cell invasion and angiogenesis, was the most highly up-regulated molecule in PAS samples. Overexpression of DOCK4 increased CTB invasiveness, consistent with the PAS phenotype. Also, this analysis identified other genes with significantly altered expression in this disorder, potential biomarkers. These data suggest that CTBs from PAS cases up-regulate a cancer-like proinvasion mechanism, suggesting molecular as well as phenotypic similarities in the two pathologies.
During pregnancy, a subset of placental cells—the villous cytotrophoblasts (CTBs)—differentiate, acquiring the ability to invade the uterus and its vasculature (1–3). Normally, this CTB subpopulation migrates through the uterine lining (decidua), stopping forward progress at the inner third of the organ’s muscular layer (myometrium). Along the way, they invade and remodel the spiral arteries. Completion of this process produces vessels with a unique architecture; placental CTBs of embryonic/fetal origin replace the maternal endothelium and intercalate within their muscular walls. Consequently, these arteries undergo the very significant expansion that is required to carry the ever-increasing amounts of maternal blood that the fetus requires to sustain growth (4). In contrast, CTBs open the termini of uterine veins, which passively expand to accommodate larger and larger amounts of blood flowing back to the maternal circulation. Thus, CTB invasion achieves the physical and physiologic integration of the maternal-fetal unit that is required for normal pregnancy.
More is known about the mechanisms that CTBs use to invade the uterus than why they stop at a circumscribed location. In the majority of early gestation chorionic villi, CTBs form a polarized monolayer with basal attachments to the trophoblast basement membrane and apical contacts with the fused syncytiotrophoblasts (STBs) that are in direct contact with maternal blood. However, at numerous sites, often near the tips of chorionic villi, repulsive mechanisms—including tenascin-integrin and Eph-ephrin interactions (3)—propel CTBs away from the placenta into columns of cells that attach to the inner surface of the uterine cavity. The columns conduct the passage of the CTB subpopulation that is destined to invade the decidua, the myometrium, and the vasculature that traverses these regions.
Consistent with this dramatic change in fate, invasive CTBs undergo a unique molecular switch, involving mechanisms that have been implicated in the evolution of tumor cells from benign to malignant (5, 6). They execute an epithelial to vascular transformation, involving numerous adhesion, angiogenic/vasculogenic, and signaling molecules, which has also been described in aggressive forms of melanoma and breast cancer (7–9). Simultaneously they up-regulate matrix-degrading proteinases and many immune regulators, including human leukocyte antigen G (HLA-G), which together affect maternal tolerance of the semiallogeneic placental cells (7).
Unlike cancer cells, CTB penetration of the uterus is tightly regulated to prevent over-invasion. Placenta accreta spectrum (PAS) involves an apparent breakdown in the mechanisms that normally restrict advancing CTBs to the inner third of the myometrium (10, 11). As a result, they invade more deeply into the muscle layer of the uterus (placenta accreta), traverse it entirely (placenta increta), or reach the uterine serosa and beyond (placenta percreta) (12). PAS occurs when the decidua is thinner than normal or largely absent. Consequently, CTB invasion to the normal depth produces a deeply adherent placenta. Along the way, CTBs can remodel the spiral arteries to the level of the serosa. As a result, the normal process of placental separation from the uterine lining after delivery of the infant is not possible and attempts to do so can result in significant maternal hemorrhage and the risk of maternal mortality. These cases are often referred to tertiary care centers where surgical removal of the uterus is frequently necessary (13–15).
Placenta accreta was first described in 1937, when Cesarean sections (C-sections) were becoming more common (16). Epidemiological studies have identified prior uterine surgery, with C-sections being the most common type, as the greatest risk factor for the development of PAS (17, 18). In this case, invasive CTBs track along the scar, gaining access to the deeper portions of the uterus from which they are normally excluded. With C-section rates rising, the number of PAS cases is increasing in parallel, from an estimated incidence of 1:30,000 pregnancies in 1950 to more recent estimates of 1:731 (10).
Despite the rapidly increasing incidence, the disease process driving PAS remains poorly understood. For example, it is not known whether CTBs use their normal invasion mechanisms to penetrate the uterus to a greater depth or up-regulate other cancer-associated molecules in the setting of an abnormal uterine environment. To begin addressing this question, we isolated CTBs from PAS cases and performed global transcriptional profiling. The results identified genes and pathways that were dysregulated in samples from the cases versus gestational age-matched controls. They included the guanidine nuclear exchange factor, DOCK4, mutations of which promote cancer cell migration and invasion (19, 20). When overexpressed in CTBs, DOCK4 increased invasion, evidence that PAS may involve a gain in the function of a molecule that promotes cancer progression as well as a loss of the normal architecture that limits placental penetration of the uterus.
Results
Global Alterations in Decidualization Were Observed in the PAS Cases.
The photomicrographs in Fig. 1 illustrate the relative lack of decidua in biopsies of the maternal-fetal interface from PAS cases (n = 7). As comparators, we analyzed the distribution of decidual cells in biopsies of the maternal-fetal interface from pregnancies complicated by preterm birth with no signs of infection (nPTB, n = 5). Previously we showed that the distribution of CTBs and decidual cells is normal in this subset (21). Tissue sections from each case were: 1) stained with hematoxylin and eosin (H&E) (Fig. 1 A, D, G, and J); 2) reacted with anti-cytokeratin (CK) to identify CTBs (Fig. 1 B, E, H, and K); or reacted with anti-vimentin to identify decidual cells (Fig. 1 C, F, I, and L).
Fig. 1.
CTB over-invasion and faulty decidualization at the maternal-fetal interface in PAS cases. Tissue sections from each case were: 1) stained with H&E (A, D, G, and J); 2) reacted with anti-CK to identify CTBs (B, E, H, and K); or reacted with anti-vimentin to identify decidual cells (C, F, I, and L). Gestational age-matched samples from nPTB cases served as controls. In nPTB cases, CK-positive CTBs were intercalated among vimentin-expressing cells of the decidua basalis (A–C). In equivalent PAS samples, CK-positive CTBs were evident in the near absence of vimentin-positive cells (D–F). The results from analyses of the basal plate region of additional case and control samples is shown in SI Appendix, Fig. S1. In nPTB, CTBs of the smooth chorion (scCTB) were juxtaposed with vimentin-positive cells of the decidua parietalis (G–I). In comparable specimens from PAS cases, the CK-positive CTB layer had a normal appearance with no signs of invasion (J–L). The results from analyses of the fetal membranes and adjacent decidua of additional case and control samples is shown in SI Appendix, Fig. S2. However, the adjacent maternal cells had low-to-no vimentin signals. PV, placental villi; BP, basal plate; FV, floating villi; FM, fetal membranes; SC, smooth chorion; MYO, myometrium. (Scale bar, 100 μm.)
First, we analyzed the basal plate, which underlies the site of placental attachment. Consistent with our published analyses of this region in nPTB cases, CK-positive CTBs were intercalated among vimentin-expressing cells of the decidua basalis (Fig. 1 A–C). In equivalent PAS samples, CK-positive CTBs were evident in the near absence of vimentin-positive cells (Fig. 1 D–F). Comparable data from additional cases are included in SI Appendix, Fig. S1.
Second, we examined the maternal-fetal interface at the junction of the fetal membranes and the decidua parietalis. In nPTB, CTBs of the smooth chorion were juxtaposed with vimentin-positive cells of the decidua parietalis (Fig. 1 G–I). In comparable specimens from PAS cases, the CK-positive CTB layer had a normal appearance with no signs of invasion (Fig. 1 J–L). However, the adjacent maternal cells had low-to-no vimentin signals. Thus, the decidual defect associated with PAS may be more widespread than previously thought, occurring well beyond the site of placental attachment. Comparable data from additional cases are included in SI Appendix, Fig. S2.
The deeply invasive CTBs of PAS still expressed typical markers. To begin molecular analyses of CTB invasion in PAS cases, we examined the cells’ expression of HLA-G, a stage-specific antigen that is up-regulated in the basal plate and the adjacent myometrium (n = 7). As was evident from H&E staining (Fig. 2A), many of the samples contained the entire uterine wall as they were from hysterectomies done at the time of delivery. Adjacent sections were double immunostained with anti-CK and anti-alpha smooth muscle actin (α-SMA), which enabled visualization of CTBs and uterine muscle cells, respectively (Fig. 2B). As illustrated by this case, placental cells were found throughout the muscle layer of the uterus. Despite the abnormal depth to which they invaded, the cells appropriately expressed HLA-G, which is up-regulated by CTBs within the uterine wall (Fig. 2C). Comparable data from additional cases are included in SI Appendix, Fig. S3.
Fig. 2.
In PAS cases, invasive CTBs up-regulated HLA-G expression in the basal plate and the adjacent myometrium (n = 7). The montages in A–C were created from photomicrographs of tissue sections that were combined into a single image. (A) H&E staining showed that many of the samples contained the entire uterine wall as they were from hysterectomies done at the time of delivery. (B) Adjacent sections were double immunostained with anti-CK and anti-α-SMA), which enabled visualization of CTBs and uterine muscle cells, respectively. As illustrated by this case, placental cells were found throughout the muscle layer of the uterus. (C) Despite abnormally deep invasion, CTBs throughout the uterine wall up-regulated the expression of HLA-G. MYO, myometrium; BV, blood vessel; FV, floating villi. (Scale bar, 100 µm.)
CTBs Isolated from PAS Cases Had a Unique Transcriptomic Signature.
Next, we asked whether PAS impacted CTB gene expression. The maternal characteristics of PAS cases used in these experiments are summarized in SI Appendix, Table S1. Consistent with epidemiologic data, all patients had had at least one prior Cesarean section (17). In these experiments, we isolated CTBs from the placentas of gestational age-matched PAS and nPTB cases, and compared their transcriptomes at a global level. In total, 118 genes were differentially expressed (DE) between the sample types; red dots denote up-regulated and blue dots denote down-regulated in PAS (Fig. 3A). Hierarchical clustering of the DE genes separated the two groups (Fig. 3B). The expression of 69 genes was higher in CTBs from PAS vs. nPTB cases; the expression of 49 genes was lower. The 25 most highly up-regulated known genes included a protocadherin cell adhesion molecule, PCDHGA10, a cell cycle regulator, CACUL1 (22), and cancer-associated molecules: DOCK4 (19), PTEN (23, 24), LTB (25), FTX (26), ATP5F1A (27), and ABl1 (28) (Fig. 3 A and C). The 25 most highly down-regulated genes included PRL, an endogenous retroviral element (ERVV2), a transcriptional activator that mediates fibroblast growth factor (FGF) signaling, CHURC1 (29), an insulin-like growth factor 1 (IGF) binding protein that inhibits growth, IGFBP6 (30), and cancer-associated molecules, PICK1 and LRRC15 (31) (Fig. 3 A and C). Thus, we identified differences in gene expression that correlated with overly aggressive CTB invasion, including several cancer-associated molecules.
Fig. 3.
Transcriptomic profiling revealed PAS-associated CTB gene signatures. We compared the gene expression profiles of CTBs from PAS (over-invasion; n = 6) and gestation-matched nPTB cases (normal invasion; n = 3). There was no statistically significant difference in the gestational ages between the two sample sets (P = 0.11). (A) The statistical significance versus gene expression fold change is displayed as a volcano plot. In total, 118 genes were DE between the sample types (blue dots down-regulated and red dots up-regulated; P < 0.05). (B) Hierarchical clustering of the DE genes separated the two groups. (C) The 25 most highly up-regulated (Top cluster) and down-regulated genes (Bottom cluster) are shown. OE, overexpressed.
Numerous Biologic Processes Were Dysregulated in PAS.
Gene ontology (GO) analysis of DE genes identified 23 enriched biological processes (P < 0.05; ≥ 3 DE genes/term; Dataset S1. Enriched terms included viral defense response, female pregnancy, negative regulation of cell proliferation, cell surface receptor signaling, neuron development, negative regulation of cell migration as well as metabolic processes (e.g., organophosphate and carbohydrate derivative biosynthetic processes; Fig. 4A). The results suggested PAS was associated with alterations in a diverse array of disease-relevant biological processes.
Fig. 4.
Mapping of the PAS-associated DE genes into GO Biological Processes. (A) DAVID was used to identify enriched GO Biological Processes for the DE genes (criteria: P ≤ 0.05; number of DE genes associated with enriched term ≥ 3). GO enrichment scores (−log10 P value) are displayed for selected terms that were associated with the up-regulated or down-regulated DE gene subsets. Clustering of DE genes within terms is shown for: (B) neuron development, (C) response to virus, and (D) cell surface receptor signaling pathway. The Z-scores represent the relative deviation in expression as compared to the average expression levels of all samples.
In keeping with our previous work that showed molecules with important roles in neurogenesis pattern CTB invasion (3), the process of neuron development was populated by DE genes, including PTEN (↑), ABl1(↑), WNK1 (↑), CNR1 (↓), OMD (↓) and OGN (↓) (Fig. 4B). The category defense response to virus contained several genes with immune functions—B2M (↑), RNF216 (↑), MICA(↑), IFIT1 (↓), and BST2 (↓) (Fig. 4C). Cell surface receptor signaling was also well represented among the DE genes: IGFBP5 (↓) and IGFBP6 (↓), ANGPT2 (↓), CHRDL1 (↓), DKK1 (↓), and PRL (↓) (Fig. 4D).
qRT-PCR Confirmed a Subset of the Microarray Results.
We used qRT-PCR to validate dysregulated expression of specific genes identified in the microarray analysis (Fig. 5A). To examine the generalizability of our initial findings, we assayed two of the original PAS samples and five that were subsequently collected (n = 7). An equal number of CTBs isolated from nPTB cases served as controls. The results confirmed higher expression (P < 0.05) of DOCK4, B2M, PAPD4, and ANKRD44 in PAS vs. nPTB; PRL was down-regulated. In general, the microarray and the qRT-PCR data were highly correlated (R2 = 0.78; SI Appendix, Fig. S4.
Fig. 5.
qRT-PCR validation of a subset of PAS-associated DE genes and extending analysis of B2M overexpression in PAS to the protein level. (A) We used qRT-PCR to validate expression, at the mRNA level, of specific genes identified as DE in the microarray analysis. For these experiments, we assayed two of the original PAS samples and five that were subsequently collected (n = 7). An equal number of CTB samples isolated from nPTB cases served as controls. The results confirmed higher expression of DOCK4, B2M, PAPD4, and ANKRD44 in accreta vs. nPTB; PRL was down-regulated. Circles represent nPTB samples and triangles represent PAS samples. Asterisks indicate significant differences between nPTB and PAS samples within each experiment (P < 0.05). (B) To confirm that differences in mRNA expression were also observed at the protein level, we immunolocalized B2M in biopsies from nPTB (n = 5) and PAS cases (n = 7). Lower immunoreactivity with anti-B2M was observed in association with CK-positive CTBs from nPTB (A and B) as compared to a PAS (increta; ICT) case (C and D). (C and D) Immunoblot analyses of CTB lysates corroborated these observations (n = 3 technical replicates). On average, B2M was expressed at fivefold higher levels by CTBs isolated from PAS vs. nPTB cases (P < 0.01). dd, ∆∆; FV, floating villi; STBs, syncytiotrophoblasts; ACT accreta; PCT, percreta. (Scale bars, 25 μm.)
B2M Overexpression in PAS Was Confirmed at the Protein Level.
In two cases, we took the extra step of confirming that differences in mRNA expression were also observed at the protein level, which would increase the probability of the changes having functional relevance. Thus, we immunolocalized B2M in biopsies from PAS (n = 7) and nPTB (n = 5) cases. Representative results are shown in Fig. 5B with data from additional cases included in SI Appendix, Fig. S5. Higher immunoreactivity with anti-B2M was observed in association with CK-positive CTBs from PAS cases as compared to pregnancies that resulted in nPTB. Immunoblot analyses of CTB lysates corroborated these observations (n = 3 technical replicates; Fig. 5C; uncropped gels are shown in SI Appendix, Fig. S6A. On average, B2M was expressed at fivefold higher levels by CTBs isolated from PAS vs. nPTB cases (P < 0.01; Fig. 5D).
DOCK4 Overexpression in PAS Was Also Confirmed at the Protein Level.
Since the microarray analysis identified DOCK4 as the most highly PAS-associated DE gene in CTBs, we also confirmed up-regulated expression of this molecule at the protein level. Immunolocalization failed to detect binding of anti-DOCK4 to STBs or CTBs in floating villi from either cases or controls (Fig. 6A). Analysis of basal plate biopsies from pregnancies complicated by nPTB (n = 5) revealed a relatively faint but consistent signal that was associated with invasive CTBs, which was significantly stronger in equivalent PAS samples (n = 7). Data from additional cases are included in SI Appendix, Figs. S7 and S8. Immunoblotting confirmed and enabled quantification of this result (Fig. 6 B and C; uncropped gels are shown in SI Appendix, Fig. S6B. CTB lysates from the increta (ICT) and percreta (PCT) cases had an immunoreactive band that corresponded to the molecular weight of DOCK4 (225 kDa); the additional bands are likely splice variants (32). CTB lysates from the accreta (ACT) and nPTB cases were immunonegative. Overall, DOCK4 expression was ∼threefold higher in the setting of PAS as compared to nPTB (P < 0.05; Fig. 6C).
Fig. 6.
Validation of CTB overexpression of DOCK4 in PAS versus nPTB cases. (A) We confirmed up-regulation of DOCK4 at the protein level. Immunolocalization failed to detect binding of anti-DOCK4 to STBs or CTBs in floating villi (FV) from either controls or cases (A and C). Analysis of basal plate biopsies from pregnancies complicated by nPTB (n = 5) revealed a relatively faint but consistent signal that was associated with invasive CTBs (B), which was significantly stronger in equivalent PAS samples (D, n = 7). (B and C) Immunoblotting confirmed and enabled quantification of this result. Four of the six CTB lysates isolated from PAS cases had an immunoreactive band that corresponded to the molecular weight of DOCK4 (225 kDa) and lower molecular weight splice variants. Notably, these bands were not observed in the two accreta samples. DOCK4 expression was ∼threefold higher in the setting of PAS as compared to PTB. ICT, increta; ACT accreta; PCT, percreta; IB, immunoblot. (Scale bars, 25 μm.)
Overexpression of DOCK4 Increased CTB Invasion.
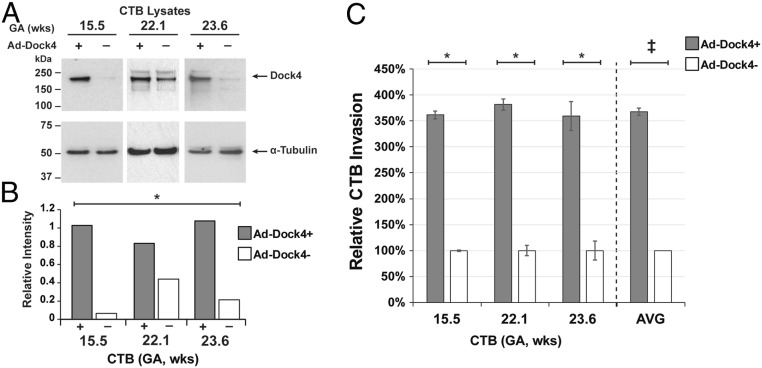
Given the positive correlation between DOCK4 expression/activity and the progression of some cancers (33, 34), we overexpressed this molecule (∼7.5-fold, P < 0.01) in primary CTBs and analyzed its function in terms of CTB invasion (Fig. 7A, quantified in Fig. 7B; uncropped gels are shown in SI Appendix, Fig. S6C). Up-regulation of DOCK4 increased CTB invasion in all three independent experiments (P < 0.01) and on average by ∼3.7-fold as compared to the level observed in the control cultures (P < 0.001; Fig. 7C).
Fig. 7.
Overexpression of DOCK4-increased CTB invasion. Cells for immunoblotting or quantification of invasion were processed at the conclusion of the experiment (36 h). (A and B) Immunoblotting and scanning densitometry showed significantly higher DOCK4 expression (P < 0.01) associated with the CTBs that were transduced with the DOCK4-containing vector as compared to control cells that received the empty vector (n = 3 biological replicates). (C) Up-regulation of DOCK4 increased CTB invasion, on average, by ∼3.7-fold (double cross; ANOVA; P < 0.001) as compared to the level observed in the control cultures (n = 3 biological replicates). Asterisks and double dagger symbols indicate significant differences between DOCK4+ and control within each experiment (P < 0.01).
Discussion
Here we report the results of using a global transcriptional profiling approach to identify the potential mechanisms driving overly aggressive CTB invasion in PAS. In total, we identified 118 DE genes as compared to gestational age-matched control nPTB cases in which CTB invasion and uterine decidualization were ostensibly normal. GO analyses suggested that they participated in numerous biological processes that could contribute to the cancer-like properties of the cells, which traverse the myometrium and sometimes encroach on neighboring organs such as the bladder. In this context, it was logical that the enriched terms were related to processes that are integral to cell migration/invasion, cell signaling, and maternal-fetal immune tolerance.
This analysis identified DOCK4 as the most highly DE/up-regulated gene in CTBs isolated from PAS placentas. This membrane-associated guanine nucleotide exchange factor enhances the formation of adherens junctions between cells (19). Mutations that interfere with the molecule’s ability to activate RhoGTPases, required for the assembly of these specialized attachment sites, are associated with the progression of many tumor types (19, 20). DOCK4 also augments tumor angiogenesis by enabling the complex process by which the lumen of a blood vessel forms (35). The primary mechanism involves a pathway in which RhoG acting through DOCK4 triggers a signaling cascade involving Rac1, DOCK9, and Cdc42. Here we discovered CTB overexpression of DOCK4 in the cancer-like setting of PAS. Whether this is in response to mutations such as those described in certain cancer types remains to be determined. If this is not the case, it appears that DOCK4 must have unique functions in CTBs as compared to other cells as its overexpression significantly increased invasion rather than cell-cell adhesion, which should limit this process. In this regard, our findings are more in line with recent reports that up-regulation of DOCK4 expression is associated with breast and lung cancer metastases (33, 34).
Beyond DOCK 4, our analyses suggested PAS-associated dysregulation of other genes with suspected roles in cell signaling and migration/invasion in the context of cancer, including BST2 (36, 37), ANGPT2 (38, 39), and IGFBP5 (40). While the specific roles of these genes in CTBs are not yet understood, these molecules may drive the hyperinvasive state of CTBs in PAS placentas and will be an interesting area of investigation for future studies. If a less restrictive model is used for the transcriptomic analysis, additional pathways of interest are identified (Dataset S2). For example, a subset of the genes involved in microtubule movement are highlighted. These proteins have diverse roles in the primary cilium, including ARL3 [signal transduction and function (41)]; DYNC1LI1 and DYNC1I2 [protein transport (42)]; VAMP7 [cilia length (43)]; and spastin [microtubule cleavage (44)]. Although the specifics are not yet understood, the primary cilium has a role in regulating the cell cycle and dysfunction has been observed in multiple cancers (45). These cell surface specializations are reported to facilitate invasion of a transformed trophoblast cell line (46). Given our findings, the functions of primary cilia in CTBs from PAS placentas is another potential area of investigation.
One explanation for the increased CTB invasiveness in PAS is lack of decidualization at the placental attachment site. We confirmed this longstanding observation in the cases that were collected for this study. Thus, the decidual signals that may normally constrain invasion are missing in PAS. Here we provide preliminary evidence that this defect may be more widespread than previously reported as there was also a lack of vimentin-positive decidual cells adjacent to the CTBs that formed the smooth chorion layer of the fetal membranes. We speculate that one reason could be an abnormal wound healing response to uterine surgery—e.g., C-section or myomectomy—a risk factor for PAS. This could explain why only some women who undergo these procedures subsequently have overly invasive placentas. The fact that the smooth chorion CTB subpopulation, unlike those of the placenta proper, fails to invade highlights the cells’ fundamental lack of cancer-like potential despite their extravillous location.
Our data suggested that in addition to a PAS-associated decidual defect, there were also CTB aberrations that manifested as changes in gene expression. Whether this is evidence of autocrine dysfunction or a paracrine response to absent, deficient, or deranged decidual signals remains to be determined. Of note, the CTBs we analyzed were primarily (but not exclusively) from the placenta; therefore, largely a villous rather than extravillous/invasive population. The fact that differences in gene expression as compared to the control cells were readily apparent prior to invasion suggested that “precancerous” changes may be taking place before the CTBs leave the placenta and enter the uterine wall. We interpret these data as favoring the theory that CTBs from PAS placentas have cancer-like features that include overexpression of genes that play a role in tumor progression.
In this analysis, we considered CTBs from PAS cases to be one experimental group as we had too few of each subtype to draw additional conclusions. Relative quantitation of DOCK4 immunoblot data showed lower CTB expression in the less invasive subtype (accreta) as compared to the more aggressive forms of PAS (increta and percreta). These preliminary data suggest the value of comparing the different forms of this syndrome in terms of both CTB and decidual effects.
The molecular differences we identified between the cases and controls are possible evidence of biomarkers that could be used to more reliably screen for and identify pregnancies complicated by PAS. Overall, this diagnosis has significant implications for the mother and fetus. Most often, this includes a preterm delivery to decrease the risk of spontaneous labor and hemorrhage as well as a hysterectomy at the time of C-section. Prenatal diagnosis is crucial so that affected women can deliver at a tertiary care center and with proper planning (14). This decision is commonly made based on prenatal imaging, such as ultrasound or magnetic resonance imaging (MRI). A recent meta-analysis showed the sensitivity of ultrasound to be 81 to 93% (47). Thus, there remains room for improvement, a critical issue given the repercussions of a missed diagnosis. As the incidence of PAS rises, the importance of developing a diagnostic biomarker panel that could be assayed using a maternal blood or urine sample increases. This seems feasible given our discovery of numerous CTB genes that are dysregulated in this condition and the fact that excessive invasion of the uterus and its arteries should result in higher circulating levels of placental proteins. Such a test would improve outcomes by allowing appropriate planning in women with PAS and avoiding unnecessary anxiety and procedures in women who do not.
Materials and Methods
Collection of Placentas from Cases and Controls.
The University of California San Francisco Institutional Review Board (Committee on Human Research) and the Stanford Institutional Review Board approved this study. Written informed consent was obtained from all donors. Women with preexisting medical conditions such as (but not limited to) thyroid insufficiency, chronic hypertension, or diabetes mellitus were excluded from the case and the control groups. Other exclusion criteria included premature rupture of membranes and/or fetal anomaly. Cases were identified based on prenatal imaging—ultrasound or MRI—consistent with an invasive placenta (47). Intraoperatively, findings were consistent with PAS as the placenta did not separate after delivery. Final confirmation of this diagnosis was made by a gynecologic pathologist. Gestational age-matched control samples were obtained from women who had a preterm birth (PTB), diagnosed according to the criteria recommended by Herron et al., including regular uterine contractions at >20 or <37 wk of gestation, which were 5 to 8 min apart and accompanied by one of the following events (1): progressive changes in the cervix (2), cervical dilation ≥ 2 cm, and/or (3) cervical effacement ≥ 80% (48). Patients with evidence of inflammation were excluded on the basis of the following criteria: maternal fever >100.4F, uterine tenderness, fetal tachycardia (fetal heart rate >160 beats per min), and (placental) histologic criteria compatible with inflammation.
CTB Isolation.
CTBs were isolated as previously described (1). The basic steps included extensive washing of the chorionic villi, sequential digestion with collagenase/trypsin, and isolation via Percoll density gradient centrifugation. For the first three isolations, CTB purity was confirmed by immunostaining for the CTB marker, cytokeratin 7, which showed ≥ 90% purity. The morphology of the cells in subsequent preparations was consistent with this result. A portion of each CTB preparation was transferred to TRIzol for RNA extraction or to medium salt lysis buffer plus 1:100 protease inhibitor (Roche) and 200 mM diothiothreitol (DTT) for preparation of protein lysates.
Histology.
Placental and placental bed biopsies from cases and controls were fixed in paraformaldehyde, embedded in optimal cutting temperature compound (Fisher Scientific), and stored at −80 °C. Sections (6 μm) were stained with H&E. Images were obtained using a Leica inverted microscope and processed in Photoshop (Adobe).
Global RNA Expression Profiling.
The microarray analysis used methods that we published (49). Total RNA was isolated using the TRIzol reagent according to the manufacturer’s directions. The RNA concentration was determined via a Nanodrop spectrophotometer (Thermo Scientific), and quality was assessed using an Agilent 2100 Bioanalyzer. Only samples with a RNA integrity number (RIN) greater than 7 were used in subsequent analyses. We used Affymetrix Human Gene 2.0 ST arrays to profile the global gene expression patterns of CTBs isolated from gestational age-matched PAS cases (n = 6) vs. placentas from pregnancies that ended in nPTB (n = 3). Sample processing and hybridization was performed by the University of California San Francisco (UCSF) Gladstone Institute as previously described (49). Affymetrix CEL files were processed and annotated using the Oligo software package (50). Raw values were normalized via the Robust Multi-Array Average (RMA) algorithm. Raw and normalized data were deposited in the Gene Expression Omnibus (accession no. GSE136048). We applied linear models for microarray data (LIMMA) to identify DE genes. If there were multiple probes per gene, we used the probe with the lowest P value (differences between cases and controls) for downstream analyses. We selected probes with average log2 intensities > 3 to remove nonspecific probes/low abundant transcripts (lower ∼15th percentile), and to limit the detection of false positives in our downstream analyses. In addition, only probes linked to an Official Gene Symbol [HUGO Gene Nomenclature Committee (HGNC)] were included. In total, we evaluated the expression of 20,289 unique genes. We calculated the ratio of average log2 intensities between PAS and nPTB to determine the average fold change (FC) difference between groups. DE genes were defined by applying a cutoff (uncorrected P < 0.05) and an absolute fold change > 0.5 (log2 scale), which distinguished the two groups. Clustering of relative expression values was performed by Pearson correlation. We utilized the Database for Annotation, Visualization and Integrated Discovery (DAVID) to perform GO enrichment analysis of DE genes. Functional enrichment analysis of Gene Ontology (GO) Biological Processes was performed for all DE genes via DAVID (uncorrected P < 0.05; number of DE genes associated with enriched term ≥ 3) (51). DAVID enables evaluation of enrichment of GO terms at five levels (1 to 5) of ontology based on size and specificity (51). Enrichment analysis was performed at all levels, although we only reported Level 4 Biological Processes—a category of moderate to high specificity within the GO organizational structure—to reduce redundancy of categories with similar functions.
Validation of DE Genes Using TaqMan qRT-PCR.
cDNA libraries were prepared with 500 ng RNA using the iScript Kit (Bio-Rad) and diluted 20-fold in water. For qRT-PCR, we employed TaqMan Universal Master Mix II, no AmpErase Uracil N-Glycosylase (UNG) (Life Technologies), and TaqMan primers for DOCK4, PRL, ANKRD44, B2M, PAPD4, GAPDH, and ACTB (SI Appendix, Table S2). Reactions were carried out for 40 cycles. Seven biological and three technical replicates were compared for all targets. Differences among target expression levels were estimated by the ΔΔCT method with normalization to the geometric mean of two housekeeping genes, GAPDH and ACTB. Differences between means were assessed using a 2-tailed Student’s t test (P = 0.05), assuming unequal variance.
Immunolocalization and Immunoblotting.
The experiments were performed as previously described (52, 53). Sections of seven case and five control samples were examined for the expression of each antigen. Antibody reactivity enabled identification of the following cell types using our published methods: 1) anti-cytokeratin (7D3), CTBs (43); 2) anti-HLA-G, invasive CTBs (43); and 3) anti-vimentin, decidual cells (54). Anti-B2-microglobulin (sc-13565) was purchased from Santa Cruz and used at a dilution of 1:100. Anti-Dock4 (21861-1-AP) was purchased from Proteintech and used at a dilution of 1:500.
Briefly, the primary antibodies were diluted in blocking buffer containing 3% bovine serum albumin (BSA), 0.05% Tween in phosphate-buffered saline (PBS). Tissue sections were incubated with the primary antibody for 1 h at 37 °C. Immunoreactivity was detected with species-specific secondary antibodies diluted in blocking buffer. After washing three times with PBS, sections were stained with 4′,6-diamidino-2-phenylindole (DAPI) (Vector Bio-Labs) to detect nuclei. Representative images were acquired and processed as described above for the morphological analyses.
For immunoblotting, 5 μg samples of CTB lysates were electrophoretically separated on 4 to 12% Bis-Tris protein gels (1.5 mm, Novex by Invitrogen) and transferred to nitrocellulose membranes, followed by blocking for 1 hour with 5% nonfat powdered milk in PBS-Tween-20 before incubating overnight at 4 °C with the primary antibodies: anti-B2M (sc-13565, Santa Cruz, 1:500), anti-DOCK4 (21861-1-1AP, Proteintech, 1:500), anti-α-tubulin (T6199, Sigma, 1:500), or anti-histone H3 (ab1781m Abcam, 1:1,000). Then the membranes were washed three times in PBS-Tween-20 and incubated with a secondary antibody (peroxidase AffiPure donkey anti-rabbit immunoglobulin G (IgG) or peroxidase AffiPure donkey anti-mouse IgG from Jackson ImmunoResearch) at a 1:5,000 dilution for 1 h. Immunoreactive bands were detected with enhanced chemiluminescence 2 (ECL 2) Western Blotting Substrate (Thermo Scientific, Pierce) and ECL high-performance chemiluminescence film (Amersham, GE Healthcare). Image J was used for quantification based on relative intensity of the control samples or loading controls.
DOCK4 Overexpression and CTB Invasion.
Invasion assays were performed as previously described (53). Briefly, 250,000 CTBs isolated from second trimester placentas as described above were plated in Transwell inserts (6.5 mm; Costar Corp.) on Matrigel-coated polycarbonate filters (8-μm pores). To up-regulate DOCK4 expression, the open-reading frame of DOCK4 (construct from ORIGENE) was inserted into an adenovirus vector (Vector Biolabs). At the time of plating, the CTBs were transduced with the DOCK4 adenovirus vector or an empty adenovirus vector, which served as a negative control. An aliquot (2.5 μL) of Ad-hDOCK4 or the empty vector (1010 plaque forming units/mL) was added in medium (final volume = 0.25 mL) to each well (multiplicity of infection = 100). After 36 h, the Transwell filters were stained with an anti-cytokeratin antibody (7D3), which enabled the counting of CTBs that reached the filter’s underside.
To evaluate the data, we applied Student t tests to examine pairwise differences in cell counts between DOCK4+ and control cultures within each experiment. We also assessed differences across experiments utilizing a generalized linear regression model to control for differences in baseline activity (n = 3). Relative % invasion was expressed as the ratio of the average number of cell projections per filter in control vs. DOCK4-transduced wells. All experiments included at least three technical replicates. DOCK4 overexpression was confirmed by immunoblotting as described above.
Data Availability.
Raw and normalized data were deposited in the Gene Expression Omnibus (accession no. GSE136048) as described in the Methods section Global RNA Expression Profiling.
Supplementary Material
Acknowledgments
We are very grateful to the study participants. This work was supported by HD076253 and the UCSF Division of Gynecologic Oncology. Additionally, we acknowledge the contribution of Dr. Deidre Lyell and the Stanford Division of Maternal Fetal Medicine, without whose assistance this work would not have been possible. We thank Dr. Michael McMaster for help editing the manuscript.
Footnotes
Competing interest statement: S.J.F. is a consultant for Novo Nordisk.
This article is a PNAS Direct Submission.
Data deposition: Raw and normalized data were deposited in the Gene Expression Omnibus (accession no. GSE136048).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1920776117/-/DCSupplemental.
References
- 1.Hunkapiller N. M., Fisher S. J., Chapter 12. Placental remodeling of the uterine vasculature. Methods Enzymol. 445, 281–302 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lacroix M. C.et al., Stimulation of human trophoblast invasion by placental growth hormone. Endocrinology 146, 2434–2444 (2005). [DOI] [PubMed] [Google Scholar]
- 3.Red-Horse K.et al., EPHB4 regulates chemokine-evoked trophoblast responses: A mechanism for incorporating the human placenta into the maternal circulation. Development 132, 4097–4106 (2005). [DOI] [PubMed] [Google Scholar]
- 4.Pijnenborg R., Vercruysse L., Hanssens M., The uterine spiral arteries in human pregnancy: Facts and controversies. Placenta 27, 939–958 (2006). [DOI] [PubMed] [Google Scholar]
- 5.Ferretti C., Bruni L., Dangles-Marie V., Pecking A. P., Bellet D., Molecular circuits shared by placental and cancer cells, and their implications in the proliferative, invasive and migratory capacities of trophoblasts. Hum. Reprod. Update 13, 121–141 (2007). [DOI] [PubMed] [Google Scholar]
- 6.Novakovic B., Saffery R., Placental pseudo-malignancy from a DNA methylation perspective: Unanswered questions and future directions. Front. Genet. 4, 285 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maltepe E., Bakardjiev A. I., Fisher S. J., The placenta: Transcriptional, epigenetic, and physiological integration during development. J. Clin. Invest. 120, 1016–1025 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hendrix M. J., Seftor E. A., Hess A. R., Seftor R. E., Vasculogenic mimicry and tumour-cell plasticity: Lessons from melanoma. Nat. Rev. Cancer 3, 411–421 (2003). [DOI] [PubMed] [Google Scholar]
- 9.Wagenblast E.et al., A model of breast cancer heterogeneity reveals vascular mimicry as a driver of metastasis. Nature 520, 358–362 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goh W. A., Zalud I., Placenta accreta: Diagnosis, management and the molecular biology of the morbidly adherent placenta. J. Matern. Fetal Neonatal Med. 29, 1795–1800 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jauniaux E., Collins S., Burton G. J., Placenta accreta spectrum: Pathophysiology and evidence-based anatomy for prenatal ultrasound imaging. Am. J. Obstet. Gynecol. 218, 75–87 (2018). [DOI] [PubMed] [Google Scholar]
- 12.Anonymous , Obstetric care consensus No. 7: Placenta accreta spectrum. Obstet. Gynecol. 132, e259–e275 (2018). [DOI] [PubMed] [Google Scholar]
- 13.Bartels H. C.et al., Association of implementing a multidisciplinary team approach in the management of morbidly adherent placenta with maternal morbidity and mortality. Obstet. Gynecol. 132, 1167–1176 (2018). [DOI] [PubMed] [Google Scholar]
- 14.Eller A. G.et al., Maternal morbidity in cases of placenta accreta managed by a multidisciplinary care team compared with standard obstetric care. Obstet. Gynecol. 117, 331–337 (2011). [DOI] [PubMed] [Google Scholar]
- 15.Shamshirsaz A. A.et al., Outcomes of planned compared with urgent deliveries using a multidisciplinary team approach for morbidly adherent placenta. Obstet. Gynecol. 131, 234–241 (2018). [DOI] [PubMed] [Google Scholar]
- 16.Lurie S., Glezerman M., The history of cesarean technique. Am. J. Obstet. Gynecol. 189, 1803–1806 (2003). [DOI] [PubMed] [Google Scholar]
- 17.Eshkoli T., Weintraub A. Y., Sergienko R., Sheiner E., Placenta accreta: Risk factors, perinatal outcomes, and consequences for subsequent births. Am. J. Obstet. Gynecol. 208, 219.e1-219.e7 (2013). [DOI] [PubMed] [Google Scholar]
- 18.Zeevi G.et al., The risk of placenta accreta following primary cesarean delivery. Arch. Gynecol. Obstet. 297, 1151–1156 (2018). [DOI] [PubMed] [Google Scholar]
- 19.Yajnik V.et al., DOCK4, a GTPase activator, is disrupted during tumorigenesis. Cell 112, 673–684 (2003). [DOI] [PubMed] [Google Scholar]
- 20.Gadea G., Blangy A., Dock-family exchange factors in cell migration and disease. Eur. J. Cell Biol. 93, 466–477 (2014). [DOI] [PubMed] [Google Scholar]
- 21.Zhou Y.et al., Comparative analysis of maternal-fetal interface in preeclampsia and preterm labor. Cell Tissue Res. 329, 559–569 (2007). [DOI] [PubMed] [Google Scholar]
- 22.Chen T. J.et al., CDK-associated Cullin 1 promotes cell proliferation with activation of ERK1/2 in human lung cancer A549 cells. Biochem. Biophys. Res. Commun. 437, 108–113 (2013). [DOI] [PubMed] [Google Scholar]
- 23.Kuchay S.et al., PTEN counteracts FBXL2 to promote IP3R3- and Ca2+-mediated apoptosis limiting tumour growth. Nature 546, 554–558 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang L.et al., Microenvironment-induced PTEN loss by exosomal microRNA primes brain metastasis outgrowth. Nature 527, 100–104 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haybaeck J.et al., A lymphotoxin-driven pathway to hepatocellular carcinoma. Cancer Cell 16, 295–308 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu F.et al., Long noncoding RNA FTX inhibits hepatocellular carcinoma proliferation and metastasis by binding MCM2 and miR-374a. Oncogene 35, 5422–5434 (2016). [DOI] [PubMed] [Google Scholar]
- 27.Yuan L.et al., A novel correlation between ATP5A1 gene expression and progression of human clear cell renal cell carcinoma identified by co-expression analysis. Oncol. Rep. 39, 525–536 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Greuber E. K., Smith-Pearson P., Wang J., Pendergast A. M., Role of ABL family kinases in cancer: From leukaemia to solid tumours. Nat. Rev. Cancer 13, 559–571 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kok F. O., Shepherd I. T., Sirotkin H. I., Churchill and Sip1a repress fibroblast growth factor signaling during zebrafish somitogenesis. Dev. Dyn. 239, 548–558 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bach L. A., IGFBP-6 five years on; not so “forgotten”? Growth Horm. IGF Res. 15, 185–192 (2005). [DOI] [PubMed] [Google Scholar]
- 31.Purcell J. W.et al., LRRC15 is a novel mesenchymal protein and stromal target for antibody-drug conjugates. Cancer Res. 78, 4059–4072 (2018). [DOI] [PubMed] [Google Scholar]
- 32.Kanai A.et al., Identification of DOCK4 and its splicing variant as PIP3 binding proteins. IUBMB Life 60, 467–472 (2008). [DOI] [PubMed] [Google Scholar]
- 33.Westbrook J. A.et al., Identification and validation of DOCK4 as a potential biomarker for risk of bone metastasis development in patients with early breast cancer. J. Pathol. 247, 381–391 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yu J. R.et al., TGF-β/Smad signaling through DOCK4 facilitates lung adenocarcinoma metastasis. Genes Dev. 29, 250–261 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Abraham S.et al., A Rac/Cdc42 exchange factor complex promotes formation of lateral filopodia and blood vessel lumen morphogenesis. Nat. Commun. 6, 7286 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mahauad-Fernandez W. D., DeMali K. A., Olivier A. K., Okeoma C. M., Bone marrow stromal antigen 2 expressed in cancer cells promotes mammary tumor growth and metastasis. Breast Cancer Res. 16, 493 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu W., Cao Y., Guan Y., Zheng C., BST2 promotes cell proliferation, migration and induces NF-κB activation in gastric cancer. Biotechnol. Lett. 40, 1015–1027 (2018). [DOI] [PubMed] [Google Scholar]
- 38.Qiu L. W.et al., Abnormal expression of angiopoietin-2 associated with invasion, metastasis and prognosis of lung cancer [in Chinese]. Zhonghua Yi Xue Za Zhi 98, 1261–1266 (2018). [DOI] [PubMed] [Google Scholar]
- 39.Kawashima T.et al., Oligodendrocytes up-regulate the invasive activity of glioblastoma cells via the angiopoietin-2 signaling pathway. Anticancer Res. 39, 577–584 (2019). [DOI] [PubMed] [Google Scholar]
- 40.Sureshbabu A.et al., IGFBP5 induces cell adhesion, increases cell survival and inhibits cell migration in MCF-7 human breast cancer cells. J. Cell Sci. 125, 1693–1705 (2012). [DOI] [PubMed] [Google Scholar]
- 41.Wright K. J.et al., An ARL3-UNC119-RP2 GTPase cycle targets myristoylated NPHP3 to the primary cilium. Genes Dev. 25, 2347–2360 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hua K., Ferland R. J., Primary cilia proteins: Ciliary and extraciliary sites and functions. Cell. Mol. Life Sci. 75, 1521–1540 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Szalinski C. M., Labilloy A., Bruns J. R., Weisz O. A., VAMP7 modulates ciliary biogenesis in kidney cells. PLoS One 9, e86425 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sharp D. J., Ross J. L., Microtubule-severing enzymes at the cutting edge. J. Cell Sci. 125, 2561–2569 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sánchez I., Dynlacht B. D., Cilium assembly and disassembly. Nat. Cell Biol. 18, 711–717 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang C. Y., Tsai H. L., Syu J. S., Chen T. Y., Su M. T., Primary cilium-regulated EG-VEGF signaling facilitates trophoblast invasion. J. Cell. Physiol. 232, 1467–1477 (2017). [DOI] [PubMed] [Google Scholar]
- 47.Pagani G.et al., Diagnostic accuracy of ultrasound in detecting the severity of abnormally invasive placentation: A systematic review and meta-analysis. Acta Obstet. Gynecol. Scand. 97, 25–37 (2018). [DOI] [PubMed] [Google Scholar]
- 48.Herron M. A., Katz M., Creasy R. K., Evaluation of a preterm birth prevention program: Preliminary report. Obstet. Gynecol. 59, 452–456 (1982). [PubMed] [Google Scholar]
- 49.Zhou Y.et al., Reversal of gene dysregulation in cultured cytotrophoblasts reveals possible causes of preeclampsia. J. Clin. Invest. 123, 2862–2872 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Team R. C., R: A Language and Environment for Statistical Computing, (R Foundation for Statistical Computing, 2018). [Google Scholar]
- 51.Huang W., Sherman B. T., Lempicki R. A., Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37, 1–13 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhou Y.et al., Vascular endothelial growth factor ligands and receptors that regulate human cytotrophoblast survival are dysregulated in severe preeclampsia and hemolysis, elevated liver enzymes, and low platelets syndrome. Am. J. Pathol. 160, 1405–1423 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Genbacev O.et al., Integrin α4-positive human trophoblast progenitors: Functional characterization and transcriptional regulation. Hum. Reprod. 31, 1300–1314 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Garrido-Gomez T.et al., Severe pre-eclampsia is associated with alterations in cytotrophoblasts of the smooth chorion. Development 144, 767–777 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw and normalized data were deposited in the Gene Expression Omnibus (accession no. GSE136048) as described in the Methods section Global RNA Expression Profiling.