Significance
Ribosome biogenesis is a complicated but efficient process requiring numerous steps of protein binding and RNA conformational rearranging. The methylation of specific nucleotides in functional regions by methyltransferases is crucial for both assembly and function of ribosomes. We characterized the structures and function of immature 50S particles from an rrmJ gene (methyltransferase for U2552 of the 23S rRNA) deletion Escherichia coli strain. With the absence of the 2′-O-methylation of U2552, the assembly of the 50S subunit is delayed at multiple late stages. Loss of this methylation also results in compromised translation activities, particularly in the initiation and the elongation steps. These results illustrate an example of chemical modification of rRNA playing a role in both ribosome biogenesis and protein translation.
Keywords: ribosome assembly, rRNA methylation, RrmJ, cryoelectron microscopy, fast kinetics
Abstract
Ribosome biogenesis is a complex process, and dozens of factors are required to facilitate and regulate the subunit assembly in bacteria. The 2′-O-methylation of U2552 in 23S rRNA by methyltransferase RrmJ is a crucial step in late-stage assembly of the 50S subunit. Its absence results in severe growth defect and marked accumulation of pre50S assembly intermediates. In the present work, we employed cryoelectron microscopy to characterize a set of late-stage pre50S particles isolated from an Escherichia coli ΔrrmJ strain. These assembly intermediates (solved at 3.2 to 3.8 Å resolution) define a collection of late-stage particles on a progressive assembly pathway. Apart from the absence of L16, L35, and L36, major structural differences between these intermediates and the mature 50S subunit are clustered near the peptidyl transferase center, such as H38, H68-71, and H89-93. In addition, the ribosomal A-loop of the mature 50S subunit from ΔrrmJ strain displays large local flexibility on nucleotides next to unmethylated U2552. Fast kinetics-based biochemical assays demonstrate that the ΔrrmJ 50S subunit is only 50% active and two times slower than the WT 50S subunit in rapid subunit association. While the ΔrrmJ 70S ribosomes show no defect in peptide bond formation, peptide release, and ribosome recycling, they translocate with 20% slower rate than the WT ribosomes in each round of elongation. These defects amplify during synthesis of the full-length proteins and cause overall defect in protein synthesis. In conclusion, our data reveal the molecular roles of U2552 methylation in both ribosome biogenesis and protein translation.
The ribosome is a molecular machine responsible for protein biosynthesis in all organisms. In prokaryotes, the ribosome is made up of two unequal subunits. The large subunit (50S) in Escherichia coli consists of the 23S rRNA (2904 nt), the 5S rRNA (120 nt), and 33 different ribosomal proteins, while the small subunit (30S) consists of the 16S rRNA (1,542 nt) and 21 proteins (1). Previous work has shown that active ribosomal subunits could be reconstituted in vitro using individual ribosomal proteins and rRNAs. But the in vitro assembly requires nonphysiological conditions, such as high temperature and high Mg2+ concentration, and the efficiency is very low (1–4).
In contrast, ribosome biogenesis in vivo is an efficient, tightly regulated, and highly ordered process, with the help of a collection of assembly factors with diverse molecular functions (1, 4–6). Among these factors, rRNA modification enzymes, which usually modify specific nucleotides in the conserved functional regions of the ribosome (1, 7), have been shown to directly participate in the subunit assembly. There are 25 modifications in the 23S rRNA, including 13 methylations, 1 methylated pseudouridylation, 1 dihydro-uridylation, 1 hydroxy-cytidylation, and 9 pseudouridylation (7–9). It is generally believed that rRNA modification may facilitate the conformational rearrangement of rRNA. However, the functional details of these modifications in ribosome assembly and their potentially more profound roles in translation are still unclear.
The methyltransferase RrmJ (FtsJ, RlmE), which is conserved from bacteria to humans, plays a vital role in the late-stage assembly of large ribosomal subunit in bacteria (10–13). With S-adenosylmethionine (AdoMet) as the methyl group donor, RrmJ, which has three catalytic active residues—Lys-38, Lys-164, and Asp-124 (11)—catalyzes the 2′-O-methylation on U2552 of the 23S rRNA in E. coli (14). This methylation occurs at the ribosomal A-loop (helix 92), which is a critical region related to the A-site tRNA binding and transpeptidation (15, 16). The rrmJ knockout strains exhibit severe growth defect and cellular accumulation of immature pre50S particles, together with a reduction of 70S ribosomes (10, 13, 17). Pre50S particles from rrmJ-deletion cells migrate at ∼45S under low Mg2+ condition, and are deficient in ribosomal proteins L5, L6, L16, L18, L19, L25, L27, L30, L33, L35, and L36 (11, 13, 18). Upon the supply of RrmJ into the crude ribosomal fractions, the 45S particles could further mature into 50S subunits that are capable of binding to 30S subunits (13).
Earlier genetic data showed that the slow growth phenotype of the ΔrrmJ strain could be partially rescued by overexpressing two assembly factors EngA and ObgE (19), but not by enzymatically compromised mutants of RrmJ (11, 12). In addition, a mutant E. coli strain of U2552C mutation shows similar defects as the ΔrrmJ strain, including the cold-sensitive and slow-growth phenotypes and the cellular accumulation of 45S precursors (13). This U2552C mutation resulted in sharply deceased 2′-O-methylation at the position of 2552, with no apparent effects on the binding of RrmJ to the pre50S particles. Therefore, the methylation of U2552 has a direct role in the assembly of the 50S subunits.
To dissect the molecular role of U2552 methylation in ribosome assembly, we determined the structures of a collection of late-stage pre50S and functional mature 50S particles from a ΔrrmJ E. coli strain. We further demonstrated using an in vitro reconstituted translation system that U2552 methylation also affects various major aspects of translation.
Results
The Immature Large Subunits from the ΔrrmJ Strain Are Highly Sensitive to Mg2+ Depletion.
Previous reports showed that immature pre50S particles from the ΔrrmJ strain migrate at 45S fraction at 0.5 mM Mg2+ concentration (13), and such 45S particles were isolated and subjected to further assembly and methylation experiments (13). In E. coli cells, the concentration of intracellular free magnesium ions is between 1 mM and 5 mM (20, 21). A certain level of Mg2+ is essential for the structural integrity of large ribonucleoprotein complexes, such as the ribosome. The extremely low Mg2+ was able to cause irreversible structural distortions and activity reduction of peptidyl transferase (22, 23). We suspect that these 45S particles might have been disrupted by low Mg2+ exposure.
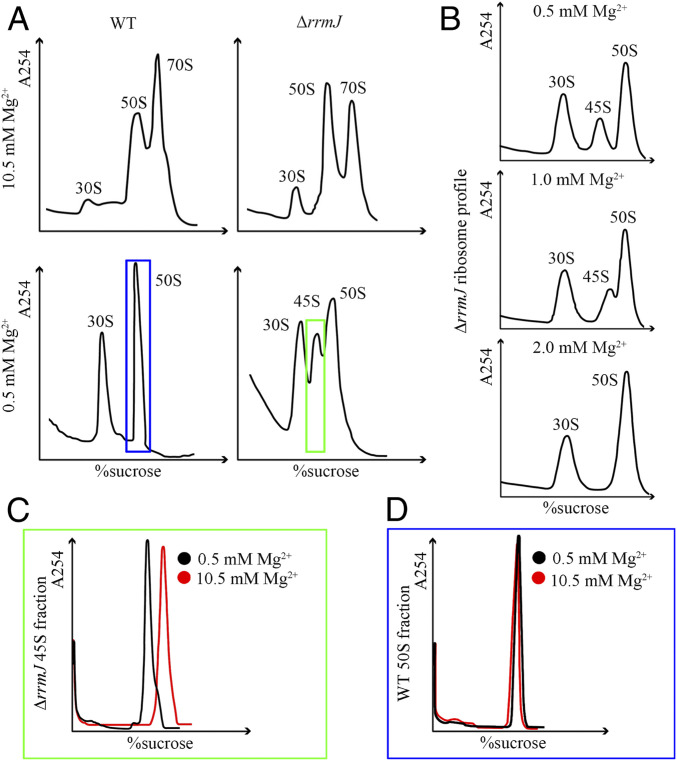
Therefore, we constructed an rrmJ-deletion strain of E. coli and repeated the sedimentation experiment using cellular extracts from ΔrrmJ cells. Consistent with previous reports (10, 12, 13), ΔrrmJ cells exhibited a marked accumulation of 50S fractions (named pre50SH hereafter) compared with the WT cells when the experiment was carried out at 10.5 mM Mg2+ (Fig. 1A). With Mg2+ concentration lowered to 0.5 mM, which is sufficient to convert all 70S ribosomes into subunits, a 45S peak (named pre50SL hereafter) appeared in the sedimentation profile of ΔrrmJ cells (Fig. 1 A and B). Also similar to the previous report (13), the 45S peak was highly sensitive to Mg2+ concentration: At 2 mM Mg2+, no 45S could be detected in cellular extracts of ΔrrmJ cells (Fig. 1B). A natural question is whether these 45S particles have been disturbed by low Mg2+ treatment. Subsequently, we collected ΔrrmJ pre50SL and WT 50SL (see Materials and Methods for details) fractions at 0.5 mM Mg2+, and examined the changes of their sedimentation coefficients with the Mg2+ concentration in the buffer changed to 10.5 mM. Our data show that while WT 50SL had no change in the sedimentation profile (Fig. 1D), the pre50SL from ΔrrmJ cells migrated at the 50S position upon increasing Mg2+ concentration (Fig. 1C). This clearly indicates that the pre50SL particles are sensitive to Mg2+ concentration, and low Mg2+ exposure might have changed their structures.
Fig. 1.
The sedimentation coefficients of in vivo 50S assembly intermediates from WT and ΔrrmJ cells under different Mg2+ concentrations. (A) Ribosome profile of WT and rrmJ-deletion cells at 0.5 mM or 10.5 mM Mg2+. (B) Ribosome profile of ΔrrmJ cells at varying concentration of Mg2+. (C and D) Sedimentation analysis of purified ΔrrmJ 45S fractions (green box in A) and WT 50S fractions (blue box in A) with a buffer containing low (0.5 mM, black curves) or high level of Mg2+ (10.5 mM, red curves). The sedimentation coefficient of purified ΔrrmJ 45S particles increases to 50S under high Mg2+ concentration.
Structural Characterization of pre50SL (45S) Particles from ΔrrmJ Cells.
To examine the impact of extremely low Mg2+ exposure on these assembly intermediates, we used cryoelectron microscopy (cryo-EM) to explore the structures of pre50SL particles. After multiple rounds of 2D classification, several class averages did display 50S-like structures with the L1 stalk, L7/L12 stalk, and central protuberance (CP), but no high-resolution features could be resolved (SI Appendix, Fig. S1A). Following 3D classification of these particles only revealed a number of low-resolution maps with comparable size to the 50S subunit, and only two of them (14% particles) have discernable 50S features (SI Appendix, Fig. S1B). These data suggest that the 0.5 mM Mg2+ treatment has turned pre50SL particles into loose, nonnative structures that probably do not have a stable conformation.
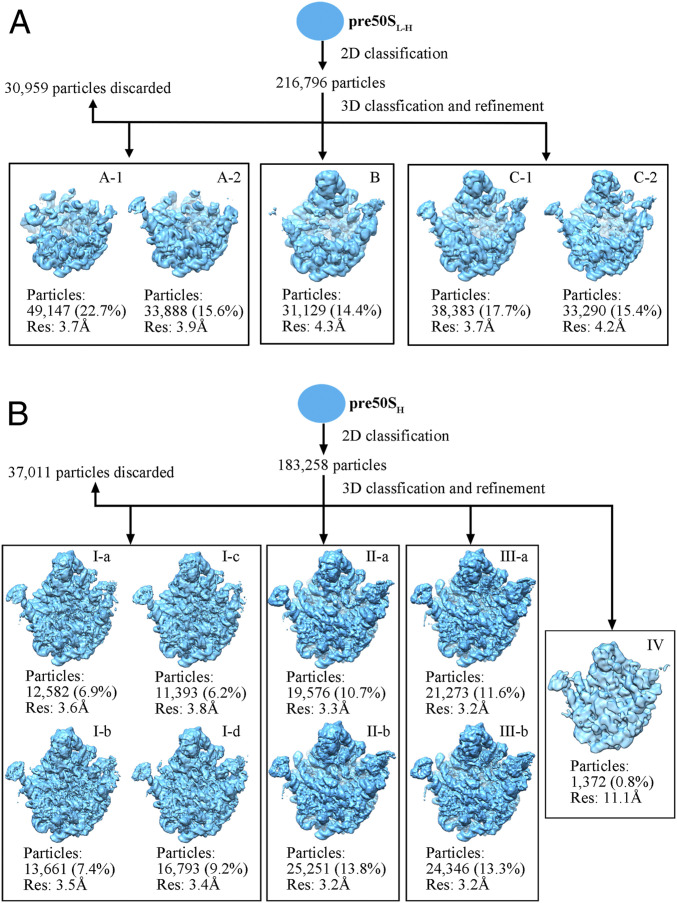
Next, we set to examine whether these structural collapses are reversible. The pre50SL particles from ΔrrmJ cells were replenished with Mg2+ to 5 mM (named pre50SL-H hereafter), and subjected to structural analysis. In sharp contrast to the pre50SL particles, 2D classification of pre50SL-H particles resulted in well-defined class averages with secondary structural features resolved (SI Appendix, Fig. S1C). Further 3D classification separated them into five groups (Fig. 2A). According to their structural features, they were grouped into three states (A, B, C). State A (A-1, A-2), which contains 38.3% particles, has highly flexible L1 and L7/L12 stalks, and lacks densities for the entire region of the CP, such as L5, L16, L18, L25, L27, L33, L35, and the 5S rRNA (L36 was also absent). This observation is consistent with previous compositional data of low Mg2+ exposed ΔrrmJ 45S particles (11, 13, 18). This state A is structurally similar to early assembly intermediates formed during in vitro reconstitution (24). State C (C-1, C-2) accounts for 33.1% particles, and its general structure is close to the mature 50S subunit. State B (14.4%) exhibits flexibility in the L1 stalk and H38, appearing to be a transition state between states A and C. Notably, in the maps of states B and C, rRNA helices H68–71 and H89–93 are completely unresolved, and densities of L16, L35, and L36 are also missing or highly underrepresented.
Fig. 2.
Cryo-EM 3D classification of pre50SL-H and pre50SH particles from ΔrrmJ cells. (A) Structures of ΔrrmJ pre50SL-H particles (purified at 0.5 mM Mg2+, and transferred into a buffer with 5 mM Mg2+). (B) Structures of ΔrrmJ pre50SH particles (purified at 10.5 mM Mg2+, sedimentation coefficient is 50S). Contour level at 3σ to 5σ. Res, resolution of the final refinement.
The contrasting structures of the pre50SL and pre50SL-H particles indicate that low Mg2+ treatment has impaired the native folding of the 23S rRNA. The structural disruption by Mg2+ depletion, at least for the 23S rRNA, is reversible to some extent. However, as shown in a previous study that low Mg2+ exposure affects the protein composition of pre50S particles and causes protein dissociation (25), both pre50SL and pre50SL-H do not reflect the actual composition and structure of the genuine in vivo intermediates. Therefore, it suggests that the interpretation of compositional and structural data based on these 45S particles (low Mg2+) must be cautious, and questions their functional relevance to RrmJ.
Cryo-EM Structures of the pre50SH Particles from ΔrrmJ Cells.
To obtain structural information relevant to RrmJ function, we characterized the structures of pre50S particles (50S fractions) isolated at 10.5 mM Mg2+ from ΔrrmJ cells (named pre50SH hereafter). After image processing, pre50SH particles were sorted into nine well-defined groups. According to their structural features, they were further grouped into four major conformational states (I, II, III, and IV) (Fig. 2B and SI Appendix, Table S1). In contrast to the pre50SL-H particles, all of the structures of the pre50SH particles are apparently at late assembly stages, with the CP region well resolved. Moreover, only three proteins—L16, L35, and L36—are dramatically reduced in structures of pre50SH particles. This composition of pre50SH particles is also in sharp contrast to the previous data on the 45S particles isolated under the low Mg2+ condition from ΔrrmJ cells (11, 13, 18), which showed a much larger number of proteins, including all CP proteins, is underrepresented in 45S particles. These differences between pre50SH and pre50SL-H particles indicate that Mg2+ depletion has caused the dissociation of ribosomal proteins, especially CP-binding proteins, from the ΔrrmJ pre50S particles. In support of this conclusion, a previous work (13) demonstrated that a combination of 0.5 mM Mg2+ and 500 mM NH4Cl was able to further dissociate L5, L6, L9, L14, L16, L25, L27, and L28 from purified 45S particles. Altogether, our data clarify that low Mg2+ alone is sufficient to strip ribosomal proteins from the pre50S particles during purification, and prove that 45S particles obtained with 0.5 mM (pre50SL) are not genuine in vivo assembly intermediates.
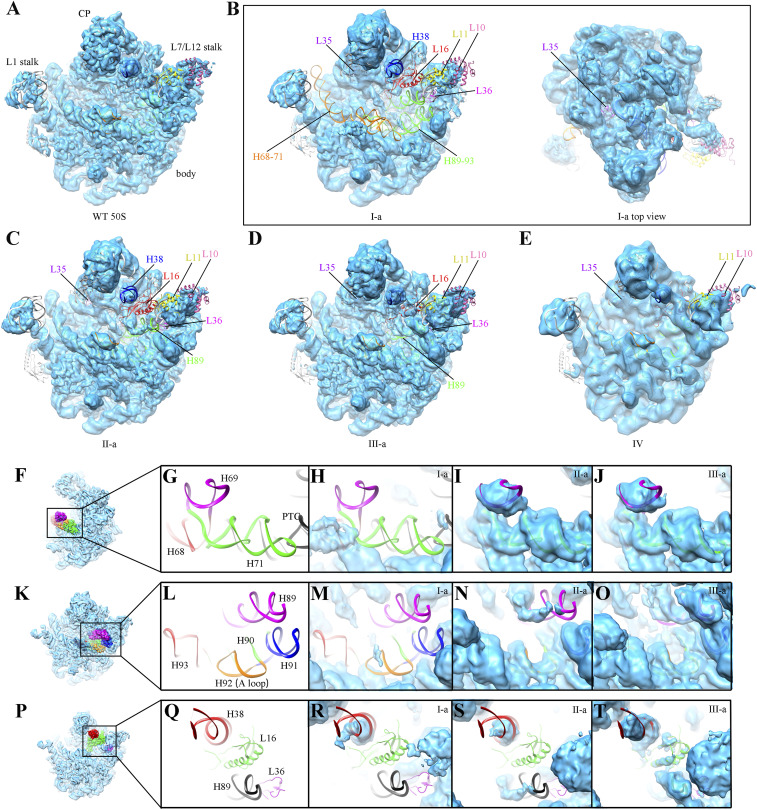
Importantly, these pre50SH maps display typical features of late-assembly intermediates (24, 26–29), including highly flexible H68–71, H89–93, and H38 (Fig. 3 A–E). Thus, our structural data indicate that RrmJ functions very late during the large subunit assembly.
Fig. 3.
Structural analysis of the ΔrrmJ pre50SH assembly intermediates. (A) Density map of WT mature 50S subunit (EMD-20353) (61). (B–E) Four representative density maps of pre50SH intermediates from rrmJ-deletion cells, including state I-a (B), II-a (C), III-a (D), and IV (E). Flexible rRNA helices and underrepresented ribosomal proteins in these intermediates are color-coded and labeled, including H68–71, H89–93, H38, L10, L11, L16, L35, and L36. For clarification, the unsharpened maps at contour level from 3σ to 5σ are shown. (F) Thumbnail of mature WT 50S subunit, viewed from L7/L12 stalk side. (G) Close-up view of the boxed region in F, helices H68–71. The PTC, H68, H69, and H71 are colored black, red, magenta, and green, respectively. All density maps of ΔrrmJ pre50SH particles in H–J are fitted with an atomic model of WT 50S subunit (PDB ID code 5H5U) for comparison. (K) Thumbnail of WT 50S subunit, viewed from the intersubunit side. (L) Close-up view of the boxed region in K to highlight the PTC helices. H89, H90, H91, H92, and H93 are colored magenta, green, blue, orange, and red, respectively. All density maps of ΔrrmJ pre50SH particles in M–O are fitted with an atomic model of WT 50S subunit for comparison. (P) Thumbnail of WT 50S subunit, viewed from the intersubunit side. (Q) Close-up view of the boxed region in P. H38, L16, L36, and H89 are colored red, green, magenta, and black, respectively. All density maps of ΔrrmJ pre50SH particles in R–T are fitted with an atomic model of WT 50S subunit for comparison.
Pre50SH Particles Are Delayed in Successive Late-Assembly Stages.
Next, we performed detailed structural comparison of four states (I to IV) of pre50SH particles. State I (I-a, I-b, I-c, I-d, ∼30% particles) has more unstable regions in the density maps, presumably representing the earliest state among the four. In state I, the entire peptidyl transferase center (PTC; H89–93) and H68–71 are extremely flexible (Fig. 3B), resulting in a total loss of corresponding densities in the maps. Furthermore, the tip of H38 is highly mobile. As to the proteins, L16, L35, and L36 are completely missing. In addition, the L7/L12 stalk base is in an immature state, with L10 and L11 unresolved. Both state II (II-a, II-b, 24.5% particles) and state III (III-a, III-b, 24.9% particles) are progressed more toward the mature structure, as they display an already accommodated H68–71 and H90–93 (Fig. 3 C and D). But the protein content is different in these two states: L35, L16, and L36 are completely absent in state II but only mildly underrepresented in state III (Fig. 3 C and D and SI Appendix, Fig. S3 D and E). State IV (0.8% particles) seemed to be in a mature-like state, with almost all of the components ready and in place (Fig. 3E and SI Appendix, Fig. S3F). The CP was resolved in all of the four states, but the CP is in slightly rotated positions in states I and II compared to its native position (SI Appendix, Fig. S4). These structural observations allowed us to analyze the temporal order for these assembly events.
The folding of H68–71 appears to be coupled with the maturation of H90–93 during the late-stage assembly. The densities corresponding to these two regions are hardly observed in state I (Fig. 3H), whereas both of them are relatively ordered in states II and III (Fig. 3 I and J). H69 is essential for the formation of intersubunit bridge B2a through the interaction with h44 of 16S rRNA in the 30S subunit (30). H68 and H71 also contribute to intersubunit bridges B2b and B7a (30). The large flexibility of these helices in state I of pre50SH particles readily explains the inability of these pre50S particles in the 30S subunit association, even with a high Mg2+ concentration.
As mentioned above, while the folding of H90–93 is synchronized with that of H68–71, H89 follows these helices during the rRNA maturation. H89–93 is the main component of the PTC, which is responsible for catalyzing transpeptidation reaction and other translation processes by binding to the CCA-ends of A-site and P-site tRNA. For these PTC helices (H89–93), all of them are nearly invisible in the map of state I, H90–93 become ordered in states II to IV, and H89 is generally stable in state III and eventually fully ordered in state IV (Fig. 3 E and M–O). This indicates an order for the conformational maturation of these helices, with H89 being the latest in the PTC region.
H38 is the last in terms of rRNA maturation and its positioning is coupled with L16 binding. H38, known as the A-site finger, contributes to the formation of intersubunit bridge B1a. The terminal half of H38 is highly unstable in states I and II, and turns into a relatively mature conformation in state III (Fig. 3 R–T), coincident with the incorporation of L16 in state III. It has to be noted that the tip of H38 remains unstable even in the mature 50S subunit, and only becomes fully ordered after the formation of the 70S ribosome. L16 binds into a cleft created by H38 and H89, and this is likely one of the reasons that these two helices are among the latest to mature (Fig. 3 P–T) (24, 26, 28).
Overall, these structural comparisons show that pre50SH particles are delayed in successive late-assembly stages, likely reflecting a set of kinetic barriers for the folding of these functional rRNA helices due to the deficiency in U2552 methylation.
Local Conformational Differences of the PTC in the Mature 50S Subunit from ΔrrmJ Cells.
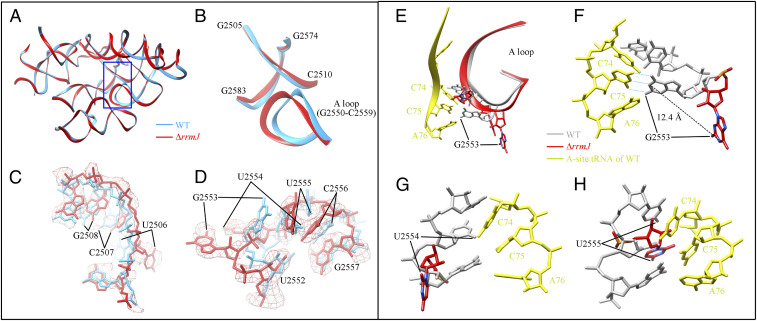
ΔrrmJ strains are viable, indicating that the defects in ribosome assembly from the absence of U2552 methylation could be tolerated. However, it is unknown to what extent the lack of U2552 methylation could affect the conformation of the PTC in a fully matured 50S subunit. We therefore purified the ΔrrmJ mature 50S subunits (isolated from 70S ribosomes) for cryo-EM analysis. The density map of the mature 50S subunits was solved at 3.14 Å (SI Appendix, Fig. S5). Compared to the WT 70S ribosome (PDB ID code 5H5U) (31), the mutant 50S subunit displays local structural variations in several regions of the PTC (Fig. 4A), including G2550-G2559 (A-loop), G2505-C2510, C2579-U2585, and A2572-G2574. Since C2579-U2585 and A2572-G2574 also show considerable differences between the cryo-EM (PDB ID code 5H5U) and crystal structures (PDB ID code 4YBB) of the WT 50S subunit (31, 32), we focused our analysis only on the A-loop and G2505-C2510 regions (Fig. 4B).
Fig. 4.
Structural differences of the mature 50S subunits from WT and ΔrrmJ cells. The differences are clustered in the blue boxed region in A and shown in detail in B–D. (A) The atomic model of the PTC from the ΔrrmJ mature 50S subunit (red) in comparison to that (light blue) of the WT 70S (PDB ID code 5H5U). (B–D) Zoom-in views of the atomic models or density maps of selected regions. (E) The structural alignment of selected nucleotides in the A-loop and A-site tRNA. The A-loop of ΔrrmJ mature 50S, WT mature 50S (PDB ID code 5JTE) and A-site tRNA (PDB ID code 5JTE) are colored red, gray, and yellow, respectively. (F–H) Detailed comparison of selected nucleotides in the A-loop. Distances between selected residue pairs are represented in dashed lines. H-bond between C75 of the A-site tRNA and G2553 of the A-loop is displayed in light blue dashed lines.
For nucleotides G2505-C2510, the largest change is on the base of U2506, which is seen to have a rotation by ∼70° in the mutant 50S subunit (Fig. 4C). Next to this region is the A-loop, which has an apparent distortion at its terminal loop (Fig. 4B). Although the absence of methylation is on U2552, there is no dramatic structural change on this nucleotide. Instead, drastic changes are on adjacent residues G2553, U2554, and U2555 (Fig. 4D). Alignment of the A-site tRNA from the cryo-EM structure of WT 70S ribosome (PDB ID code 5JTE) (33) into the mutant 50S subunit led to an interesting observation. The large displacement of G2553 and U2554 bases, ∼90° and ∼120° rotation, respectively, would dramatically increase their distance with the CCA-end of A-site tRNA (Fig. 4 E–H). Importantly, G2553 is directly involved in hydrogen-bonding with the C75 of A-site tRNA during translation (34, 35).
It was previously reported that the translation efficiency of S30 extract from the ΔrrmJ strain was significantly lower than that of the WT strain (17). Because the A-loop is involved in tRNA binding and recognition of translation factors in various steps of translation, these large changes on the A-loop residues (Fig. 4 E–H) might affect the translation activity of the mutant 70S ribosome.
∆rrmJ 70S Ribosomes Are Defective in Initiation and Elongation of Protein Synthesis.
The structural defects in the PTC region due to the lack of U2552 methylation could have profound effects in various steps of the translation cycle. To explore in which steps of translation the ∆rrmJ ribosomes are defective, we performed a set of fast-kinetics–based translational assays covering all steps of translation, using an in vitro reconstituted translation system with purified components from E. coli. These precision assays included steps of, with in vivo-like time resolution, ribosomal subunit association, initiator fMet-tRNAfMet binding, di- and tripeptide formation, mRNA translocation, peptide release, and ribosome recycling.
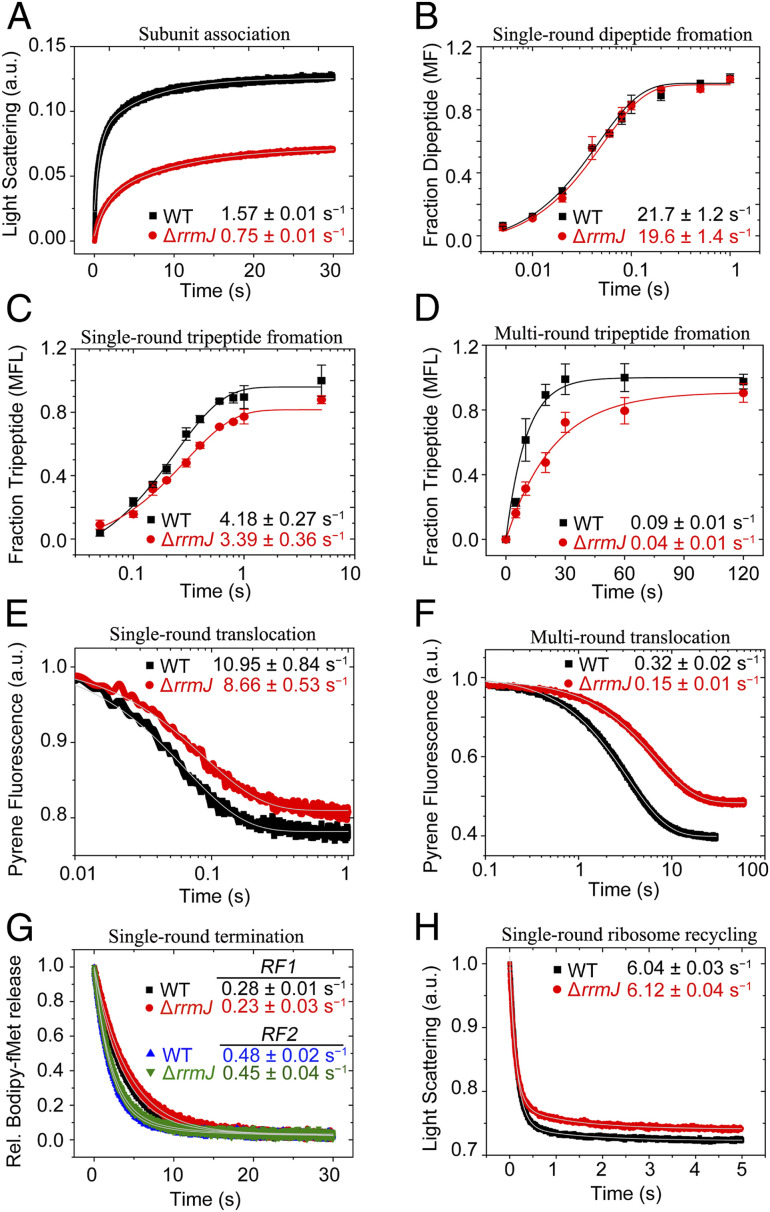
In the subunit association assay, the formation of 70S initiation complex (70S IC) is monitored by a time-dependent increase in Rayleigh light scattering (365 nm) by rapid mixing of the 50S and 30S preinitiation complex (30S pre-IC) in a stopped-flow instrument (36). Our results demonstrate that the ∆rrmJ 50S subunit (obtained by dissociating ∆rrmJ 70S ribosomes) forms 70S IC with a rate of 0.75 ± 0.01 s−1, which is two times slower than the WT 50S subunit (1.57 ± 0.01 s−1) (Fig. 5A). Moreover, compared to the WT, only 50% 70S IC is formed with the same concentration of the ∆rrmJ 50S subunit under the single turnover condition. It suggests that only half of the ∆rrmJ 50S subunits are able to execute fast subunit association. Similar defects in both rate and yield are also observed when only naked 50S and 30S subunits were subjected to subunit association (SI Appendix, Fig. S6A). These results together indicate that the defects in association with 30S are inherent to the ∆rrmJ 50S subunit and not imposed by interaction with any initiation factors (IFs).
Fig. 5.
Characterization of the ∆rrmJ ribosomes lacking U2552 methylation in fast kinetics-based translation assays. All data are fitted using single or double exponential functions (as indicated) and the rates are estimated using Origin 8.0. All experiments were done at least three times. The rates are presented with SDs. (A) Kinetics of association of the 30S preIC with the 50S subunits from the WT (black square) and the ΔrrmJ (red circle) cells. The formation of the 70S IC is followed by increase in Rayleigh light scattering at 365 nm in a stopped-flow instrument. The curves are fitted with double exponential function and the rates are estimated from the fast phase. (B) Kinetics of fMet-Phe dipeptide formation in quench flow from 70S IC from WT (black square) and ΔrrmJ (red circle) cells under the single-turnover condition. The curves are fitted with a single exponential function to obtain the reaction rates. (C and D) Kinetics of fMet-Phe-Leu tripeptide formation from 70S IC (WT,black square and ΔrrmJ, red circle) under single-turnover (C) and multiple-turnover (D) conditions, done in quench flow (C) and manual mixing (D), respectively. The curves are fitted with single exponential function and the rates are estimated. (E and F) Kinetics of EF-G mediated translocation with the ribosomes from WT (black square) and ΔrrmJ (red circle) cells under single-turnover (E) and multiple-turnover (F) conditions. The 3′ pyrene labeled MFL mRNA was used in this assay, where decrease in the pyrene fluorescence representing mRNA movement upon translocation is monitored in stopped-flow (excitaton 343 nm and emission 360 nm). The curves are fitted with single exponential function and the rates are estimated. (G) Kinetics of BOP-Met release from the RC programmed with Met-Stop (UAA) mRNA by RF1 (WT, black square and ΔrrmJ, red circle) and RF2 (WT, blue triangle and ΔrrmJ, green triangle). The rates are estimated from the fast phase by fitting the curves with double exponential function. (H) Kinetics of RRF and EF-G mediated splitting of the posttermination ribosomal complex from the WT (black square) and the ΔrrmJ (red circle) cells, followed by monitoring decrease in Rayleigh light scattering at 365 nm in a stopped-flow instrument. The curves are fitted with single exponential function and the rates are estimated.
The ∆rrmJ 70S ribosomes are, however, not defective in binding the initiator fMet-tRNAfMet. Using fluorescent BODIPY 576/589 (BOP) labeled-tRNAfMet, we obtained identical rates of initiator tRNA binding to the 70S IC containing either ∆rrmJ or WT 70S ribosomes (SI Appendix, Fig. S6B). Thus, we conclude that the ∆rrmJ 50S subunits are primarily defective in subunit association during initiation. Only 50% of the ∆rrmJ 50S subunits are capable of rapid subunit association, yet at a rate 50% slower than the WT 50S.
To estimate the active fraction of the ribosomes capable of peptide bond formation, we have titrated the ∆rrmJ 70S ribosomes, using OD260-based concentration, in a single-round fMet-Phe dipeptide formation assay using a preformed 70S IC. Our results of dipeptide formation at a single time point (10 s), demonstrate that the fraction of the ribosomes active in peptide bond formation is similar for both ∆rrmJ and the WT 70S (SI Appendix, Fig. S6C). Furthermore, in our quench-flow–based single-turnover kinetics assay the rate of dipeptide formation with the ∆rrmJ 70S ribosomes (19.6 ± 1.4 s−1) matches closely with the rate obtained with WT 70S (21.7 ± 1.2 s−1) (Fig. 5B). Thus, we conclude that once assembled into 70S IC, the ∆rrmJ ribosomes are fully capable of decoding and peptide bond formation as the WT 70S. Interestingly, the ∆rrmJ ribosomes are ∼20% slower than the WT ribosomes in fMet-Phe-Leu tripeptide formation under single-turnover condition (Fig. 5C). The rate of tripeptide formation reduces from 4.18 ± 0.27 s−1 with the WT 70S to 3.39 ± 0.36 s−1 with the ∆rrmJ 70S. The defect amplifies under the multiple EF-G turnover condition, when the ∆rrmJ ribosomes form tripeptides at a rate (0.04 ± 0.01 s−1), ∼55% lesser than the rate obtained with the WT ribosomes (0.09 ± 0.01 s−1) (Fig. 5D). Since the ∆rrmJ 70S ribosomes are not defective in peptide bond formation (Fig. 5B), we suspect that the defect in tripeptide formation likely originates from defective tRNA translocation or EF-G release from the ∆rrmJ ribosomes. To check that, ∆rrmJ 70S ribosomes were subjected to a real-time mRNA translocation assay in stopped-flow, using 3′ pyrene-labeled MFL mRNA (37), under single- and multiple-turnover conditions. Coherent with the tripeptide formation assay, under single-turnover condition, the ∆rrmJ 70S ribosomes translocate with a rate 8.66 ± 0.53 s−1, which is about 20% lesser than the rate of translocation of the WT ribosomes (10.95 ± 0.84 s−1) (Fig. 5E). Under a multiple-turnover condition, similar to multiple-turnover tripeptide formation, the ∆rrmJ ribosomes translocate at a rate of 0.15 ± 0.01s− 1, about 55% lesser than the rate obtained with the WT ribosome (0.32 ± 0.02 s−1) (Fig. 5F). Our results thus demonstrate that the ∆rrmJ ribosomes are defective in ribosomal translocation and EF-G turnover. Since translocation is one of the key steps of the repetitive elongation cycle of mRNA translation, larger delay with ∆rrmJ ribosomes can be expected in synthesis of the full-length proteins, which involves multiple rounds of elongation.
The ∆rrmJ 70S ribosomes are further tested in peptide release assay using a ribosomal release complex (RC) carrying BOP-labeled Met-tRNAfMet on a Met-stop (UAA) coding mRNA. Single-round peptide release is monitored by following the decrease in BOP fluorescence with time, due to release of the BOP-Met from the RC with release factors RF1 and RF2. Our result shows that the rate of the peptide release is similar between ∆rrmJ and WT 70S ribosome, both for RF1 (0.28 ± 0.01 s−1 for WT and 0.23 ± 0.03 s−1 for ∆rrmJ) and RF2 (0.48 ± 0.02 s−1 for WT and 0.45 ± 0.04 s−1 for ∆rrmJ) (Fig. 5G).
We have also tested the ∆rrmJ 70S ribosomes in the ribosome recycling assay, for which a posttermination complex containing a deacylated tRNA in the P site is mixed with RRF, EF-G, and IF3 in a stopped-flow instrument. The time course of subunit splitting is followed by monitoring the decrease in Rayleigh light scattering (at 365 nm) with time. Our results demonstrate that the ∆rrmJ 70S ribosomes dissociate into subunits with the rates essentially same as the WT 70S. The rate of 70S dissociation for ∆rrmJ 70S ribosomes is 6.12 ± 0.04 s−1, while the rate for the WT 70S ribosomes is 6.04 ± 0.03 s−1 (Fig. 5H). Thus, we conclude that the ∆rrmJ ribosomes are not defective in peptide release or ribosome recycling.
Finally, we tested the ΔrrmJ 70S ribosomes in a S100 extract-based translation assay. E. coli BL21 cells were used to prepare the S100 extract (the factor mixture), and purified WT and ΔrrmJ 70S ribosomes were assayed using an pET21b plasmid harboring an EGFP gene. Our data show that the overall translation activity of ΔrrmJ 70S is indeed much lower than that of the WT 70S (SI Appendix, Fig. S7). On the basis of multiple rounds of translation over 2 h, WT 70S ribosomes produced significantly more EGFP proteins, about two times of the mutant 70S at a given time.
In summary, these data clearly show that the ΔrrmJ 70S ribosomes are compromised in translation, with defects in initiation and elongation of protein synthesis.
Discussion
The Role of U2552 Methylation in Late-Stage Assembly of the 50S Subunit.
By comparing the structures of pre50SL, pre50SL-H, and pre50SH particles, we showed that Mg2+ depletion leads to ribosomal protein dissociation and structural distortion of native assembly intermediates. Therefore, biochemical results conducted with low Mg2+-exposed assembly intermediates would require reexamination. In the present study, we characterized the structures of immature ΔrrmJ pre50S particles (pre50SH) that are functionally more relevant to the role of RrmJ in ribosome assembly. Our data show that these pre50SH intermediates are trapped in very late stages, with most of the 50S regions fully assembled except the PTC region and its vicinity. This pinpoints a direct role of U2552 methylation in the folding of rRNA helices in the PTC region. This functional relevance of RrmJ is supported by early genetic data that overexpression of EngA or ObgE could partially rescued the slow growth of the ΔrrmJ strain. In fact, both ObgE and EngA directly interact with the PTC helices (38, 39). Therefore, as assembly factors, they might be able to chaperone the local folding of the PTC to compensate the defects due to the lack of U2552 methylation.
These affected rRNA helices, including H89–93 in the PTC and H68–71 and H38 from its surrounding regions. The pre50S assembly intermediates from the ΔrrmJ strain differ from each other in the stability of one or two of these rRNA helices. According to our data, H68–71/H90–93, H89, and H38 take turn to reach its mature or near mature conformation (SI Appendix, Fig. S8). We also observed a coordination between the folding of H68–71 and H90–93 in the pre50SH particles, which could be explained by a three-way hydrogen bond network between nucleotides from H71 (C1955) and H92 (U2552 and C2556) (13, 24). Our data show that similar interactions among these three nucleotides could still form in pre50SH (states II and III) and in mature 50S (SI Appendix, Fig. S9) from ΔrrmJ cells. This indicates that U2552 methylation is not necessary for the formation of this three-way interaction. Instead, this modification may facilitate the establishment of this interface by changing its folding kinetics.
Our structural finding that assembly defects of late-stage particles are clustered in the PTC is consistent with the structural studies of the in vitro-reconstituted pre50S particles (24) and various in vivo pre50S intermediates obtained by genetic perturbations of bacterial strains (such as assembly factor or ribosomal protein deletion/depletion) (26–29, 40, 41). Collectively, these studies reveal that the maturation of the PTC region is a very late event and the most critical step in the 50S assembly. Nevertheless, the assembly of the 50S subunit is inherently a parallel process, and rRNA-folding and protein-binding events are organized into folding blocks and display positive cooperativity within the individual blocks (27). This explains why many assembly factors are not essential and indicates that disruption of a certain factor could direct the assembly into other branches. Indeed, in terms of the order of rRNA folding, our data show subtle differences with the previous in vitro 50S reconstitution, where helices H71 and H89 were shown to be established earlier than H90–93 (24).
For ribosomal protein assembly, only L16, L35, and L36 are highly underrepresented in our ΔrrmJ 50S assembly intermediates. All of them are in the latest binding group, according to the 50S assembly map revealed by the time-resolved pulse-labeling coupled quantitative mass spectometry technique (42). The three proteins are missing from structures of states I and II. It was previously shown that the interaction between L35 and L33 stabilizes the CP in its mature position during assembly (24), which agrees with our observation that the CP of states I and II (both are L35-deficient) is in slightly rotated positions (SI Appendix, Fig. S4). Our data show that L35 and L16 start to appear in state III, suggesting that L36 might be the last to assemble (Fig. 3D and SI Appendix, Table S1). This was supported by the in vitro 50S reconstitution data that a persistent absence of L36 was found in the final product of reconstitution (24). Nevertheless, Nikolay et al. (24) also found that the binding of L35 was prior to L16 in the in vitro reconstitution experiment. The seeming discrepancy is not surprising in the view of parallel assembly pathway (27), as these three proteins could bind alternative orders under different conditions.
Taken together, our and previous structural data reveal general features of the 50S assembly. The most important one is that functional areas—such as H38, H68–71, and H89–93 (PTC center)—are formed at very late stages. This could be conveniently used as an internal quality control for 50S production, because these rRNA elements directly participate in subunit association, tRNA and translation factor binding.
The Impact of U2552 Methylation on Protein Synthesis and Bacterial Growth.
Although several rRNA modifications on the ribosome are known, how they influence different steps of protein synthesis is largely unknown. Our results from the fast-kinetics–based precise translation assays elucidate the exact role of U2552 methylation in 23S rRNA in protein synthesis. The main finding is that the U2552 methylation-lacking ribosomes are defective in two vital steps of protein synthesis, initiation and elongation. Only 50% of the ∆rrmJ 50S subunits are capable in fast subunit association, and also with a compromised rate about half of the WT 50S subunit. While the WT 50S subunit associates with the 30S pre-IC to form 70S IC in 159 ms, the ∆rrmJ 50S subunit takes 333 ms (at 1-µM concentration). The ∆rrmJ 70S ribosomes, however, are not defective in peptide bond formation; in this vital step they show comparable rate and activity with the WT 70S. These two results together indicate that given enough time all ∆rrmJ 50S subunits can associate to form elongation-competent 70S IC.
The other defect in the ∆rrmJ ribosomes occurs in the crucial translocation step of elongation. The ∆rrmJ ribosomes translocate at a rate 20% slower than the WT 70S under a single turn-over condition. As indicated by the structure of the ∆rrmJ 50S subunit, this defect might arise from the lack of proper interaction between the A-site bound tRNA and the displaced A-loop of the 23S rRNA in the ∆rrmJ 70S ribosome. Because of this defect, the ∆rrmJ ribosomes take ∼60 ms longer than the WT ribosomes (mean time 240 ms) in every round of elongation. The turnover of EF-G is also defective as the ∆rrmJ ribosomes take two times longer than the WT ribosomes for making tripeptide under multiple-turnover condition. Due to the repetitive occurrence of the elongation step, this apparent small delay in single round of elongation amplifies and slows down the overall rate of synthesis of the full-length proteins as shown in the S100-based assay.
What causes the growth defect in the ∆rrmJ cells in vivo? Although RrmJ is a well-documented heat-shock protein (10, 14, 43), previous data also showed that the methylation of U2552 in WT E. coli cells under normal growth condition is from 75 to 90% (13, 44). This suggests that U2552 methylation is likely a housekeeping function of RrmJ. Our data show that the growth defects in the ∆rrmJ cells have originated from two major sources, the delayed 50S subunit assembly and compromised 70S translation activity. Our kinetic analysis indicates that translation initiation is affected the most in the in vitro condition. Since only 50% of the ∆rrmJ 50S subunits actively participate in 70S IC formation, a large number of free 50S subunits probably exist in the ∆rrmJ cells, which do not participate in every round of translation. This is exactly consistent with the observations that the ∆rrmJ cells are associated with sharply elevated 50S and reduced 70S fractions (Fig. 1) (10, 13, 17). More importantly, initiation is the primary regulatory step in translation, and often a bottleneck for the translation of many mRNAs. This pronounced delay the ∆rrmJ 50S subunits in initiation alone is capable of causing significant slow-growth phenotype. In addition, the translationally active ∆rrmJ 70S ribosomes are also defective in another crucial step of translation (i.e., elongation, in both translocation and EF-G turnover steps). Although the kinetic defect is not large in a single round of elongation, due to repetitive occurrence of the elongation step the apparent small delay in the single-elongation step amplifies, thereby further slowing down the overall rate of protein synthesis in the cell.
In summary, the cellular defects of ∆rrmJ cells in both ribosome assembly and translation would have a profound effect in production of all housekeeping proteins, including the translation factors and the DNA/RNA polymerases. Even ribosome production would be affected as it is a constellation of more than 50 proteins together with rRNA. Since bacterial growth is largely dependent on the number of active ribosomes and the rate of protein synthesis, a lesser number of translationally active ribosomes and slower rate of protein synthesis in the absence of U2552 methylation are likely the main reasons of the slower growth phenotype of the ∆rrmJ cells. Other defects involving accuracy of protein synthesis (45) and proper protein folding may also incur, which remains to be checked in future. Our results thus demonstrate at the molecular level the importance of the U2552 methylation for bacterial growth.
Materials and Methods
E. coli Strains, Plasmid Construction.
E. coli BW25113 was used as the source of the WT 50S subunit and 70S ribosomes; BW25113 ΔrrmJ is a derivative in which the rrmJ gene was substituted by the kanamycin resistance gene between the FRT sequence through the two-step homologous recombination (46), and confirmed with PCR.
The EGFP gene was cloned by PCR and inserted into the pET21b plasmid. The plasmid was then transformed into DH5α E. coli strain for amplification. Cells were cultured at 37 °C in 200 mL LB media to OD600 ∼1.0 and collected for plasmids extraction. The recombinant plasmids were purified by ethanol precipitation for further in vitro translation assay.
Ribosome Profile Analysis.
Ribosome profile analysis was used to detect the sedimentation coefficients of ribosomal particles from WT BW25113 and ΔrrmJ strains under different Mg2+ concentrations. Cells were cultivated to OD600 ∼0.8 at 37 °C in LB medium, collected by centrifugation, resuspended with buffer I (20 mM Tris⋅HCl [pH 7.5], 100 mM NH4Cl, 10.5 mM MgCl2, 1 mM DTT) or buffer II (20 mM Tris⋅HCl [pH 7.5], 100 mM NH4Cl, 0.5 mM MgCl2, 1 mM DTT) containing 1 mM PMSF, and disrupted by ultrasonication (4 °C). The lysate was centrifuged at 15,000 rpm (Avanti J-26 XP, Beckman Coulter) for 1 h and the supernatants were collected at 4 °C.
For the high Mg2+ condition (10.5 mM Mg2+ group in Fig. 1A), supernatants prepared with buffer I were loaded onto a 10 to 40% sucrose gradient (prepared in buffer I) and centrifuged at 39,000 rpm in an SW41 rotor (Beckman Coulter) for 3.5 h at 4 °C. The fractions were analyzed by a Teledyne ISCO fractionation system with A254 absorbance. For the low Mg2+ condition (0.5 mM Mg2+ group in Fig. 1A), supernatants (cells were disrupted in buffer II) were similarly analyzed (gradient prepared with buffer II).
For low to high Mg2+ exchange experiment, fractions corresponding to the blue (WT 50SL) or green (ΔrrmJ pre50SL) boxes in Fig. 1A were collected, centrifuged in an SW41 rotor, and analyzed similarly (Fig. 1 D or C).
For Mg2+-sensitivity analysis in ΔrrmJ cells (Fig. 1B), supernatants (prepared with buffer II) were subjected to the same sucrose gradient prepared with buffer III (20 mM Tris⋅HCl [pH 7.5], 100 mM NH4Cl, 1 mM DTT, and 0.5 mM Mg2+, 1.0 mM Mg2+ or 2.0 mM Mg2+), centrifuged at 30,000 rpm for 8 h in an SW32 rotor (Beckman Coulter) at 4 °C, and analyzed for the A254 absorbance.
Purification of ΔrrmJ pre50S and Mature 50S Particles.
The ΔrrmJ cells were grown to OD600 ∼0.8 at 37 °C in LB medium, collected by centrifugation, resuspended with buffer II or buffer I containing 1 mM PMSF, and disrupted by ultrasonication at 4 °C. The lysate was centrifuged at 15,000 rpm for 1 h (4 °C) and the supernatants were collected.
For ΔrrmJ pre50SL and pre50SL-H particles, supernatants (in buffer II) were placed onto a 10 to 40% sucrose gradient prepared with buffer II, and centrifuged at 4 °C with an SW32 rotor for 8 h at 30,000 rpm. The peak fractions corresponding to pre50SL were collected. Half of them were concentrated and stored in buffer II (named ΔrrmJ pre50SL); the other half were concentrated with buffer II and then changed to buffer III with 5 mM MgCl2 (named ΔrrmJ pre50SL-H particles).
For ΔrrmJ pre50SH and mature 50S particles, cells were disrupted and centrifuged in buffer I. Supernatants were applied onto a 5-mL sucrose cushion (20 mM Tris⋅HCl [pH 7.5], 500 mM NH4Cl, 10.5 mM MgCl2, 0.5 mM EDTA, 1.1 M sucrose, 1 mM DTT) and centrifuged at 28,000 rpm for 19 h with a 70Ti rotor (Beckman Coulter) at 4 °C. The pellets were washed by 2-mL washing buffer (20 mM Tris⋅HCl [pH 7.5], 500 mM NH4Cl, 10.5 mM MgCl2, 0.5 mM EDTA, 1 mM DTT), resuspended in buffer I, and applied onto a 10 to 40% sucrose gradient (prepared with buffer I). After centrifugation at 30,000 rpm for 8 h with an SW32 rotor (4 °C), the fractions of pre50SH or 70S particles were collected. To get the mature 50S particles, 70S fractions were concentrated with buffer IV (20 mM Tris⋅HCl [pH 7.5], 100 mM NH4Cl, 2.5 mM MgCl2, 1 mM DTT), and subjected to another round of centrifugation in an SW32 rotor (with sucrose gradient prepared with buffer IV). The peak fractions corresponding to ΔrrmJ mature 50S particles were collected and stored in buffer IV.
Chemicals and Buffers for Kinetics Experiments.
The analytical grade chemicals were purchased from Sigma and Merck. Fluorescent reagents BODIPY 576/589 were purchased from ThermoFisher. Nucleoside triphosphates were purchased from GE Healthcare. All biochemical experiments were conducted in Hepes-polymix buffer (pH 7.5) containing: 100 mM KCl, 5 mM NH4Cl, 5 mM Mg(OAc)2, 5 mM CaCl2, 5 mM Hepes-KOH pH 7.5, 8 mM putrescine, 1 mM spermidine, and 1 mM dithioerythritol.
Translation Components for the Kinetic Assays.
E. coli translation factors (IF1, IF2, IF3, EF-Tu, EF-Ts, EF-G, and RRF) cloned in pET vectors were overexpressed in the E. coli BL21(DE3) strain. The class I release factors RF1 and RF2 (T246A) were coexpressed with the methylase hemK in E. coli BL21(DE3) GOLD strain (the constructs were kind gifts from Valérie Heurgué-Hamard from Institut de Biologie Physico-Chimique, Paris, France.) to obtain uniform GGQ methylation of RF1 and RF2 (47). The overexpression of the factors was induced by addition of 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) at OD600 = 0.4. All translation factors contain a (His)6-tag at the C terminus. The purification of the factors was carried out using the HisTrap column (GE Healthcare) on an AKTA-prime Plus chromatography system. The concentration of the factors was determined accurately with the Pierce 660-nm protein assay kit.
f[3H]Met-tRNAfMet and tRNAPhe were purified using standard laboratory protocol (48). Labeling of the tRNAfMet with BOP ester was carried out in NaHCO3 (100 mM) solution following the manufacturer’s protocol. After incubation for 16 h at 4 °C, the free dye was removed by phenol and chloroform extraction of the tRNAfMet. BOP-Met-tRNAfMet was further precipitated with 70% ethanol and purified using Lichospher WP 300 RP-18 column (Merck) connected to RP-HPLC system (Waters) with in line UV and fluorescence detectors.
The 3′-Pyrene–labeled mRNA, coding for Met-Phe-Leu (MFL-pyrene) with sequence UAACAAUAAGGGAGUAUUAAAUGUUCCUGC-pyrene, was purchased from IBA. All XR7 mRNAs were transcribed in vitro from the corresponding DNA with T7 RNA polymerase. These are, Met-Phe-Leu (MFL, UAAGGAGGUAUUAAAUGUUCCUGC), Met-Leu-Leu (MLL, UAAGGAGGUAUUAAAUGCUGCUGC), and Met-stop (UAAGGAGGUAUUAAAUGUAA) mRNAs. For all mRNA sequences, the ribosome binding site is underlined and the coding sequences are written in bold. The mRNAs were purified on a Sephadex G-75 column (GE Healthcare).
Ribosome and RC.
The 70S ribosomes were purified from E. coli BW25113 and ∆rrmJ strain using a standard protocol (36). BOP-labeled RCs were prepared by incubating 2 μM E. coli 70S ribosome, 2 μM IF1, 4 μM IF2, 2 μM IF3, 20 μM XR7 mRNA M-stop, and 2 μM BOP-Met-tRNAfMet at 37 °C for 20 min. The RCs were purified by centrifugation at 258,000 × g on a 1.1 M sucrose cushion for 3.5 h at 4 °C. The pelleted RCs are resuspended in Hepes-polymix buffer (pH 7.5) and stored at −80 °C.
Subunit Association Assay.
The 50S and 30S ribosomal subunits were purified from intact 70S according to the published method (36). The 30S pre-IC and 50S mixture were prepared and incubated separately at 37 °C for 10 min. The 30S pre-IC mixtire contains 0.5 μΜ 30S subunits, 5 μΜ XR7 MLL mRNA, 2 μΜ IF1, 2 μΜ IF2, 2 μΜ IF3, 1 μΜ f[3H]Met-tRNAfMet, and 2 μΜ GTP (1 mM). The 50S ribosomes mixture contains 0.5 μΜ 50S subunits. ATP (1 mM) was added to both mixes to reduce free concentration of Mg2+. The reaction mixes were rapidly added in a stopped-flow instrument (BioLogic uSF4000) at 37 °C. The extent of 70S IC formation was monitored by following the Rayleigh light scattering at 365 nm using 320-nm cut-off filter (36). The time course was obtained by averaging three to five traces from each subunit association experiment. The experiments are done at least in triplicates. The rate of subunit association was estimated from the fast phase, by fitting the data with double exponential function using Origin 8.0.
Di- and Tripeptide Formation Assay.
For dipeptide formation experiments, a 70S IC mixture and elongation mixture were formed by incubating them at 37 °C for 15 min separately. IC contains 2 μM 70S ribosomes, 5 μM XR7 MFL mRNA, 2 μM f[3H]Met-tRNAfMet, and all three IFs (2 μM). The elongation mixture was prepared with 5 μM EF-Tu, 5 μM EF-Ts, 5 μM tRNAPhe, 0,2 mM Phe amino acid, and 1 unit Phe tRNA synthetase. For single-turnover tripeptide formation experiments, 40 μM EF-Tu, 5 μM EF-G, 5 μM tRNALeu, Leu amino acid (0.2 mM), and 1 unit Leu tRNA synthetase were added to the elongation mixture. Both mixtures contained energy pump components 1 mM GTP, 1 mM ATP, 10 mM phosphoenolpyruvate, 0.05 mg/mL pyruvate kinase, and 0.002 mg/mL myokinase. The reaction was started by rapidly mixing equal volumes of the IC and elongation complex (EC) at 37 °C in quench-flow instrument (BioLogic QFM-400). For multiround tripeptide formation assay, 100 times less EF-G than 70S ribosomes (2 μM) was used and the reactions were conducted by manual mixing. The reactions were quenched by the addition of 17% formic acid at definite time points and the peptides were released by KOH treatment. The peptides were separated on C18 column connected to a RP-HPLC (Waters) and in line β-RAM radioactive detector. The relative amount of di- and tripeptides were plotted against time. The data points are fitted with single exponential function using Origin 8.0. The experiments are done at least in triplicates.
mRNA Translocation Assay.
IC and EC were prepared in the same way as the single- and multiple-turnover tripeptide formation assays, except that the mRNA was substituted by MFL-pyrene mRNA. Equal volumes of IC and EC were rapidly mixed in the stopped-flow (BioLogic uSF4000) at 37 °C and the decrease of pyrene fluorescence was monitored by using 360-nm long-pass filter after exciting at 343 nm as shown earlier (37). The fluorescence traces were fitted to single exponential function using Origin 8.0 and the rates of mRNA movement were estimated. All experiments were done minimum in triplicates.
Peptide Release Assay.
The RC was prepared as described above. RC (0.2 μM) and release factor RF1 or RF2 (2 μM) were rapidly mixed in a stopped-flow apparatus (BioLogic uSF4000) at 37 °C. The release of BOP-Met was followed by the decrease in fluorescence of BOP by using a 590-nm cut-off filter. The excitation was done at 575 nm. Each time course was obtained by averaging three to five individual traces. The fluorescence traces were fitted with double exponential function in Origin 8.0. The rates were estimated from the fast phase (amplitude ∼80%).
Ribosome Recycling Assay.
A posttermination complex and factor mixture (FM) was formed by incubating separately at 37 °C for 15 min. The posttermination complex contains 1 μM 70S ribosomes, 5 μM MLL mRNA, 1 μM tRNALeu. The FM contains 2 μM EF-G, 20 μM RRF, and 2 μM IF3. All reactions contained energy pump components as mentioned in the di- and tripeptide assay. An equal volume of posttermination complex and FM were rapidly mixed in the stopped-flow instrument (BioLogic uSF4000) at 37 °C. The kinetics of the 70S splitting into subunits was followed by the decrease in Rayleigh light scattering at 365 nm, using a 320-nm cut-off filter (49). The rate of 70S dissociation was estimated by fitting the data with single exponential function in Origin 8.0. The assays were repeated at least three times.
S100/Factor Extract-Based In Vitro Translation Assay.
WT BW25113 and ΔrrmJ strains were grown to OD600 ∼0.8 at 37 °C in LB medium, collected by centrifugation, resuspended with buffer I containing 1 mM PMSF, and disrupted by ultrasonication at 4 °C. The lysates were centrifuged at 15,000 rpm for 1 h and the supernatants were collected. Supernatants were placed onto 10 to 40% sucrose gradient prepared with buffer I, and centrifuged with an SW32 rotor for 8 h at 30,000 rpm (4 °C). The peak fractions corresponding to WT 70S and ΔrrmJ 70S particles were collected and stored in buffer V (40 mM Hepes-KOH [pH 7.5], 100 mM KCl, 10.5 mM MgCl2, 1 mM DTT) for in vitro translation assay. The WT 70S and ΔrrmJ 70S particles were analyzed by SDS/PAGE (SI Appendix, Fig. S7A).
The standard BL21(DE3) E. coli strain was used as the source for the S100/Factor Extract (50). Cells were cultured at 37 °C in 2 L LB media to midlog phase and induced by 1 mM IPTG (Sigma-Aldrich) for 2 h to express the T7 RNA polymerase. Cells were collected and washed with Extract buffer (10 mM Tris·OAc [pH = 7.5], 1 mM DTT, 14 mM Mg(OAc)2, 60 mM KOAc). The pellets were resuspended in 30 mL Extract buffer containing one tablet Protease Inhibitor Mix (Roche) and disrupted using a French press. After centrifugation at 30,000 × g at 4 °C for 30 min twice, the supernatants were separated and centrifuged at 150,000 × g for 4 h using a 70Ti rotor (Beckman Coulter). The upper 80% of the supernatants were recentrifuged at 150,000 × g overnight to purify the S100 Extract, and the top 80% of the mixture was dialyzed with 100 volumes of Extract buffer in a dialysis bag (Mr cut-off = 3,500) for 3 h with four times of buffer change in a cold room (4 °C). The dialyzed S100 Extract was concentrated to A280 ∼ 3 and A260 ∼ 4. The lower 20% of supernatants and pellets resulting from centrifugations in 70Ti rotor were collected to purify the factor extract. The pellets were slowly resuspended overnight by adding NH4Cl to 1 M and then centrifuged at 150,000 × g for 4 h (4 °C). The upper 80% of the supernatant was separated and dialyzed with 100 volumes of Extract buffer in a dialysis bag (Mr cut-off = 3,500) for 3 h with four times of buffer change in a cold room. The dialyzed factor extract was concentrated to A280 ∼ 5 and A260 ∼ 8. Both the S100 and factor extracts were divided into reasonably-sized aliquots and stored at −80 °C.
A standard 60-μL in vitro translation assay contained: 60 mM Hepes-KOH (pH 7.5); 27.5 mM ammonium acetate; 10.7 mM magnesium acetate; 200 mM potassium acetate; 4% PEG8000; 1.7 mM DTT; 1.2 mM ATP, GTP; 1 mM UTP, CTP; 80 mM creatine phosphate dibasic tetrahydrate; 0.64 mM cAMP; 250 μg/mL creatine kinase (Roche); 175 μg/mL total E. coli tRNA (Roche); 0.8 mM each amino acid; some pET21b-egfp plasmids; 2 A260 units ribosomes (WT 70S or ΔrrmJ 70S ribosome); 42 μg S100 extracts; 20 μg S100 factors; 1 mM IPTG; 0.5 U/μL RNase Inhibitor (TaKaRa); 34 μg/mL folinic acid calcium salt hydrate (50, 51). The mixture was then subjected into a black opaque 384-well microplate (Corning) at 30 °C to measure the signal of EGFP every minute using a Multiscan Spectrum (PerkinElmer EnVision) (52). The control group contained all components except the ribosome (Buffer V was added instead) was similarly measured. For WT and ΔrrmJ groups, three different concentrations of pET21b-egfp plasmids (1,500 ng, 2,500 ng, and 3,500 ng, respectively) were added in 60-μL reaction systems, while the control group was done in the presence of 1,500 ng plasmid DNA. Each reaction was repeated three times.
Cryo-EM Sample Preparation, Data Collection.
For cryo-EM 4-μL aliquots of ΔrrmJ pre50SL, pre50SL-H, pre50SH, and mature 50S samples at a concentration of ∼100 nM were applied to glow-discharged 300-mesh Quantifoil R2/2 grids (Quantifoil, Micro Tools) coated with a homemade continuous thin carbon film. After 15 s of waiting, grids were blotted for 2 s and plunged into liquid ethane using an FEI Mark IV Vitrobot operated at 4 °C and 100% humidity.
For pre50SL sample, micrographs were collected on a Tecnai Arctica microscope, operated at 200 kV, with a Falcon II camera. Data acquisition was performed using semiautomatic software AutoEMation II (53), with a nominal magnification of 78,000×, which yields a final pixel size of 1.27 Å at object scale (defocus ranging from –1.0 μm to –2.0 μm).
For other samples, micrographs were captured on a Titan Krios microscope, operated at 300 kV, with a Falcon II camera. Data acquisition was performed using AutoEMation II, with a nominal magnification of 75,000×, which yields a final pixel size of 1.08 Å at object scale (defocus ranging from –1.0 μm to –2.0 μm).
For each micrograph stack, 28 frames were collected, with a total dose of 45 electrons per pixel.
Image Processing.
Motion correction at the micrograph level was done with MotionCorr (29) and MotionCorr2 (54). The program CTFFIND3 was used to estimate the contrast transfer function parameters. Image processing, including micrographs screening, particle picking, 2D and 3D classification, and refinement, was done with RELION 1.4 (55).
For pre50SL particles, a total of 50,226 particles were subject to a cascade of 2D and 3D classification. After 2D classification, 20,516 particles were subjected to 3D classification. Eight conformational states of the pre50SL particles were acquired. For pre50SL-H, a total of 268,000 particles were subject to a cascade of 2D and 3D classification. After 2D classification, 216,796 particles were subjected to 3D classification. Five conformational states of the pre50SL-H intermediates were acquired. For pre50SH particles, a total of 235,600 particles were subject to a cascade of 2D and 3D classification. After two rounds of 2D classification, 183,258 particles were subjected to three rounds of 3D classification. According to the features of class maps, these pre50SH structures were grouped into four major states (I, II, III, and IV) (SI Appendix, Table S1). The details of image processing and map features are summarized in SI Appendix, Table S1.
For ΔrrmJ mature 50S particles, after two rounds of 3D classification, a total number of 98,194 particles were subjected to final refinement. During the final refinement, a soft-edged mask was applied, yielding a 3.14 Å density map (SI Appendix, Table S2). ResMap was used to estimate the local resolution of these maps (56).
Model Building.
A high-resolution crystal structure of the 50S subunit from E. coli (PDB ID code 4YBB) (32) and the cryo-EM structure of E.coli ribosome (PDB ID code 5H5U) (31) were used as the initial models and fitted into the ΔrrmJ mature 50S density map using Chimera (each ribosomal protein, 5S rRNA and 23S rRNA fitted individually as rigid bodies). COOT (57) was used to examine and manually adjust the best fit between the map and the model. Model refinement was first carried out using PHENIX (phenix.real_space_refine) in real space (58) with secondary structure and geometry restraints to prevent overfitting. The final atomic model was refined using REFMAC in reciprocal space (59) with secondary structure restraints, base pair, and planarity restraints applied. To avoid overfitting, different weights of the density map for refinement were tested. The final models were evaluated with MolProbity (60).
Accession Numbers.
Cryo-EM maps of ΔrrmJ mature 50S subunit and pre50SH particles (states I, II, III, and IV) have been deposited in the Electron Microscopy Data Bank (EMDB) with accession codes EMD-30215, EMD-30214, EMD-30213, EMD-30212, and EMD-30211, respectively. Atomic coordinates of ΔrrmJ mature 50S ribosomal subunit have been deposited in the Protein Data Bank with PDB ID code 7BV8. All of the materials generated in the study are available from corresponding authors.
Supplementary Material
Acknowledgments
We thank Dr. Chen Guo from the School of Medicine at Tsinghua University for providing the plasmid containing the egfp sequence, and the Tsinghua University Branch of the China National Center for Protein Sciences (Beijing) for providing resources for cryoelectron microscopy data collection and image processing. This work was supported by National Natural Science Foundation of China Grants 31725007 and 31630087 (to N.G.); Ministry of Science and Technology of China Grant 2019YFA0508900 (to N.G.); China Postdoctoral Science Foundation Grant 1131000065 (to W.L.); Shenzhen Science and Technology Research Funding JCYJ20180302174213122 (to W.L.); Swedish Research Council Grants 2016-06264 and 2018-05498 (to S.S.); Knut and Alice Wallenberg Foundation KAW 2017.0055 (to S.S.); Wenner-Gren Foundation UPD2017-0238 and UPD2018-0306 (to S.S.); and Carl-Tryggers Foundation CTS 18:338 (to S.S.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: Cryoelectron microscopy maps of the ΔrrmJ mature 50S subunit and pre50SH particles (State I, State II, State III and State IV) have been deposited in the Electron Microscopy Data Bank, https://www.ebi.ac.uk/pdbe/emdb (accession nos. EMD-30215, EMD-30214, EMD-30213, EMD-30212 and EMD-30211, respectively). Atomic coordinates of the ΔrrmJ mature 50S ribosomal subunit have been deposited in the Protein Data Bank, http://www.pdb.org (PBD ID code 7BV8).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1914323117/-/DCSupplemental.
References
- 1.Shajani Z., Sykes M. T., Williamson J. R., Assembly of bacterial ribosomes. Annu. Rev. Biochem. 80, 501–526 (2011). [DOI] [PubMed] [Google Scholar]
- 2.Traub P., Nomura M., Structure and function of E. coli ribosomes. V. Reconstitution of functionally active 30S ribosomal particles from RNA and proteins. Proc. Natl. Acad. Sci. U.S.A. 59, 777–784 (1968). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nierhaus K. H., Dohme F., Total reconstitution of functionally active 50S ribosomal subunits from Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 71, 4713–4717 (1974). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Davis J. H., Williamson J. R., Structure and dynamics of bacterial ribosome biogenesis. Philos. Trans. R Soc. Lond. B Biol. Sci. 372, 20160181 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Britton R. A., Role of GTPases in bacterial ribosome assembly. Annu. Rev. Microbiol. 63, 155–176 (2009). [DOI] [PubMed] [Google Scholar]
- 6.Kaczanowska M., Rydén-Aulin M., Ribosome biogenesis and the translation process in Escherichia coli. Microbiol. Mol. Biol. Rev. 71, 477–494 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Decatur W. A., Fournier M. J., rRNA modifications and ribosome function. Trends Biochem. Sci. 27, 344–351 (2002). [DOI] [PubMed] [Google Scholar]
- 8.Ofengand J., Del Campo M., Modified nucleosides of Escherichia coli Ribosomal RNA. EcoSal Plus 1 (2004). [DOI] [PubMed] [Google Scholar]
- 9.Sergeeva O. V., Bogdanov A. A., Sergiev P. V., What do we know about ribosomal RNA methylation in Escherichia coli? Biochimie 117, 110–118 (2015). [DOI] [PubMed] [Google Scholar]
- 10.Bügl H.et al., RNA methylation under heat shock control. Mol. Cell 6, 349–360 (2000). [DOI] [PubMed] [Google Scholar]
- 11.Hager J., Staker B. L., Bugl H., Jakob U., Active site in RrmJ, a heat shock-induced methyltransferase. J. Biol. Chem. 277, 41978–41986 (2002). [DOI] [PubMed] [Google Scholar]
- 12.Hager J., Staker B. L., Jakob U., Substrate binding analysis of the 23S rRNA methyltransferase RrmJ. J. Bacteriol. 186, 6634–6642 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arai T.et al., Single methylation of 23S rRNA triggers late steps of 50S ribosomal subunit assembly. Proc. Natl. Acad. Sci. U.S.A. 112, E4707–E4716 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Caldas T.et al., The FtsJ/RrmJ heat shock protein of Escherichia coli is a 23 S ribosomal RNA methyltransferase. J. Biol. Chem. 275, 16414–16419 (2000). [DOI] [PubMed] [Google Scholar]
- 15.Blanchard S. C., Puglisi J. D., Solution structure of the A loop of 23S ribosomal RNA. Proc. Natl. Acad. Sci. U.S.A. 98, 3720–3725 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brunelle J. L., Youngman E. M., Sharma D., Green R., The interaction between C75 of tRNA and the A loop of the ribosome stimulates peptidyl transferase activity. RNA 12, 33–39 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Caldas T., Binet E., Bouloc P., Richarme G., Translational defects of Escherichia coli mutants deficient in the Um(2552) 23S ribosomal RNA methyltransferase RrmJ/FTSJ. Biochem. Biophys. Res. Commun. 271, 714–718 (2000). [DOI] [PubMed] [Google Scholar]
- 18.Sharpe Elles L. M., Sykes M. T., Williamson J. R., Uhlenbeck O. C., A dominant negative mutant of the E. coli RNA helicase DbpA blocks assembly of the 50S ribosomal subunit. Nucleic Acids Res. 37, 6503–6514 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tan J., Jakob U., Bardwell J. C., Overexpression of two different GTPases rescues a null mutation in a heat-induced rRNA methyltransferase. J. Bacteriol. 184, 2692–2698 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alatossava T., Jütte H., Kuhn A., Kellenberger E., Manipulation of intracellular magnesium content in polymyxin B nonapeptide-sensitized Escherichia coli by ionophore A23187. J. Bacteriol. 162, 413–419 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nierhaus K. H., Mg2+, K+, and the ribosome. J. Bacteriol. 196, 3817–3819 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kimes B. W., Morris D. R., Cations and ribosome structure. II. Effects on the 50S subunit of substituting polyamines for magnesium ion. Biochemistry 12, 442–449 (1973). [DOI] [PubMed] [Google Scholar]
- 23.Klein D. J., Moore P. B., Steitz T. A., The contribution of metal ions to the structural stability of the large ribosomal subunit. RNA 10, 1366–1379 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nikolay R.et al., Structural visualization of the formation and activation of the 50S Ribosomal subunit during in vitro reconstitution. Mol. Cell 70, 881–893.e3 (2018). [DOI] [PubMed] [Google Scholar]
- 25.Jiang M.et al., The Escherichia coli GTPase CgtAE is involved in late steps of large ribosome assembly. J. Bacteriol. 188, 6757–6770 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ni X.et al., YphC and YsxC GTPases assist the maturation of the central protuberance, GTPase associated region and functional core of the 50S ribosomal subunit. Nucleic Acids Res. 44, 8442–8455 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Davis J. H.et al., Modular assembly of the bacterial large ribosomal subunit. Cell 167, 1610–1622.e5 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jomaa A.et al., Functional domains of the 50S subunit mature late in the assembly process. Nucleic Acids Res. 42, 3419–3435 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li N.et al., Cryo-EM structures of the late-stage assembly intermediates of the bacterial 50S ribosomal subunit. Nucleic Acids Res. 41, 7073–7083 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu Q., Fredrick K., Intersubunit bridges of the bacterial ribosome. J. Mol. Biol. 428, 2146–2164 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ma C.et al., Mechanistic insights into the alternative translation termination by ArfA and RF2. Nature 541, 550–553 (2017). [DOI] [PubMed] [Google Scholar]
- 32.Noeske J.et al., High-resolution structure of the Escherichia coli ribosome. Nat. Struct. Mol. Biol. 22, 336–341 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Arenz S.et al., A combined cryo-EM and molecular dynamics approach reveals the mechanism of ErmBL-mediated translation arrest. Nat. Commun. 7, 12026 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim D. F., Green R., Base-pairing between 23S rRNA and tRNA in the ribosomal A site. Mol. Cell 4, 859–864 (1999). [DOI] [PubMed] [Google Scholar]
- 35.Zhou J., Lancaster L., Donohue J. P., Noller H. F., How the ribosome hands the A-site tRNA to the P site during EF-G-catalyzed translocation. Science 345, 1188–1191 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ge X., Mandava C. S., Lind C., Åqvist J., Sanyal S., Complementary charge-based interaction between the ribosomal-stalk protein L7/12 and IF2 is the key to rapid subunit association. Proc. Natl. Acad. Sci. U.S.A. 115, 4649–4654 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Studer S. M., Feinberg J. S., Joseph S., Rapid kinetic analysis of EF-G-dependent mRNA translocation in the ribosome. J. Mol. Biol. 327, 369–381 (2003). [DOI] [PubMed] [Google Scholar]
- 38.Feng B.et al., Structural and functional insights into the mode of action of a universally conserved Obg GTPase. PLoS Biol. 12, e1001866 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang X.et al., Structural insights into the function of a unique tandem GTPase EngA in bacterial ribosome assembly. Nucleic Acids Res. 42, 13430–13439 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Seffouh A.et al., Structural consequences of the interaction of RbgA with a 50S ribosomal subunit assembly intermediate. Nucleic Acids Res. 47, 10414–10425 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rabuck-Gibbons J. N.et al., SrmB rescues trapped ribosome assembly intermediates. J. Mol. Biol. 432, 978–990 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen S. S., Williamson J. R., Characterization of the ribosome biogenesis landscape in E. coli using quantitative mass spectrometry. J. Mol. Biol. 425, 767–779 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Richmond C. S., Glasner J. D., Mau R., Jin H., Blattner F. R., Genome-wide expression profiling in Escherichia coli K-12. Nucleic Acids Res. 27, 3821–3835 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ishiguro K., Arai T., Suzuki T., Depletion of S-adenosylmethionine impacts on ribosome biogenesis through hypomodification of a single rRNA methylation. Nucleic Acids Res. 47, 4226–4239 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Widerak M., Kern R., Malki A., Richarme G., U2552 methylation at the ribosomal A-site is a negative modulator of translational accuracy. Gene 347, 109–114 (2005). [DOI] [PubMed] [Google Scholar]
- 46.Baba T.et al., Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: The Keio collection. Mol. Syst. Biol. 2, 2006 0008 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Heurgué-Hamard V., Champ S., Engström A., Ehrenberg M., Buckingham R. H., The hemK gene in Escherichia coli encodes the N(5)-glutamine methyltransferase that modifies peptide release factors. EMBO J. 21, 769–778 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Holm M., Borg A., Ehrenberg M., Sanyal S., Molecular mechanism of viomycin inhibition of peptide elongation in bacteria. Proc. Natl. Acad. Sci. U.S.A. 113, 978–983 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang Y.et al., HflX is a ribosome-splitting factor rescuing stalled ribosomes under stress conditions. Nat. Struct. Mol. Biol. 22, 906–913 (2015). [DOI] [PubMed] [Google Scholar]
- 50.Rasouly A., Davidovich C., Ron E. Z., The heat shock protein YbeY is required for optimal activity of the 30S ribosomal subunit. J. Bacteriol. 192, 4592–4596 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Moll I., Hirokawa G., Kiel M. C., Kaji A., Bläsi U., Translation initiation with 70S ribosomes: An alternative pathway for leaderless mRNAs. Nucleic Acids Res. 32, 3354–3363 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lee K. Y., Lee K. H., Park J. W., Kim D. M., Flexible programming of cell-free protein synthesis using magnetic bead-immobilized plasmids. PLoS One 7, e34429 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lei J., Frank J., Automated acquisition of cryo-electron micrographs for single particle reconstructon on an FEI Tecnai electron microscope. J. Struct. Biol. 150, 60–80 (2005). [DOI] [PubMed] [Google Scholar]
- 54.Zheng S. Q.et al., MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Scheres S. H., A Bayesian view on cryo-EM structure determination. J. Mol. Biol. 415, 406–418 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kucukelbir A., Sigworth F. J., Tagare H. D., Quantifying the local resolution of cryo-EM density maps. Nat. Methods 11, 63–65 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Adams P. D.et al., PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Murshudov G. N., Vagin A. A., Dodson E. J., Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 (1997). [DOI] [PubMed] [Google Scholar]
- 60.Chen V. B.et al., MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stojković V.et al., Assessment of the nucleotide modifications in the high-resolution cryo-electron microscopy structure of the Escherichia coli 50S subunit. Nucleic Acids Res. 48, 2723–2732 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.