Significance
Single nucleus RNA sequencing revealed gene expression changes during repair after acute kidney injury. We describe a small population of proximal tubule cells that fail to repair (FR-PTCs). Since this subpopulation expresses abundant proinflammatory and profibrotic genes, it may represent a new therapeutic target to improve repair and reduce fibrosis after AKI.
Keywords: AKI, injury, transcriptomics, epithelia
Abstract
After acute kidney injury (AKI), patients either recover or alternatively develop fibrosis and chronic kidney disease. Interactions between injured epithelia, stroma, and inflammatory cells determine whether kidneys repair or undergo fibrosis, but the molecular events that drive these processes are poorly understood. Here, we use single nucleus RNA sequencing of a mouse model of AKI to characterize cell states during repair from acute injury. We identify a distinct proinflammatory and profibrotic proximal tubule cell state that fails to repair. Deconvolution of bulk RNA-seq datasets indicates that this failed-repair proximal tubule cell (FR-PTC) state can be detected in other models of kidney injury, increasing during aging in rat kidney and over time in human kidney allografts. We also describe dynamic intercellular communication networks and discern transcriptional pathways driving successful vs. failed repair. Our study provides a detailed description of cellular responses after injury and suggests that the FR-PTC state may represent a therapeutic target to improve repair.
Kidneys maintain fluid and electrolyte balance, excrete waste products, and regulate blood pressure. Composed of approximately one million functional units called nephrons, the kidneys receive ∼20% of cardiac output. Nephrons have high metabolic activity rendering them susceptible to injury from toxins or reduced blood flow. These insults cause acute kidney injury (AKI) characterized by decreased kidney function. In early stages of AKI, epithelial cells die, and surviving epithelia dedifferentiate, accompanied by inflammation. Dedifferentiated epithelial cells then proliferate and redifferentiate to repair the damaged nephron (1, 2). There are no specific treatments for AKI, but, with supportive care, the kidney’s intrinsic repair capacity usually allows functional recovery over a period of days to weeks.
After repair, kidney function may not return back to baseline due to residual subclinical inflammation and fibrosis. Survivors of AKI are at high risk of developing future chronic kidney disease (CKD) and even kidney failure (3). The mechanisms for failed repair are not well understood, but a subpopulation of injured proximal tubule (PT) epithelia (the segment most susceptible to injury) are proposed to become arrested at the G2/M cell cycle phase and adopt a senescence-associated secretory phenotype (4). This may prevent complete repair, driving inflammation and fibrosis, and mouse ischemia–reperfusion injury (IRI) models this process well (5). The aim of our study was to understand the cellular events underlying both recovery from AKI as well as the transition to CKD. Bulk transcriptional profiling has successfully characterized kidney injury and recovery (5, 6), but these approaches describe a transcriptional average across cell populations, which may hide or skew signals of interest. We hypothesized that understanding transcriptional changes in single cell types over the course of AKI, repair, and fibrosis would provide unique insights into disease pathogenesis and potentially identify new therapeutic strategies.
Results
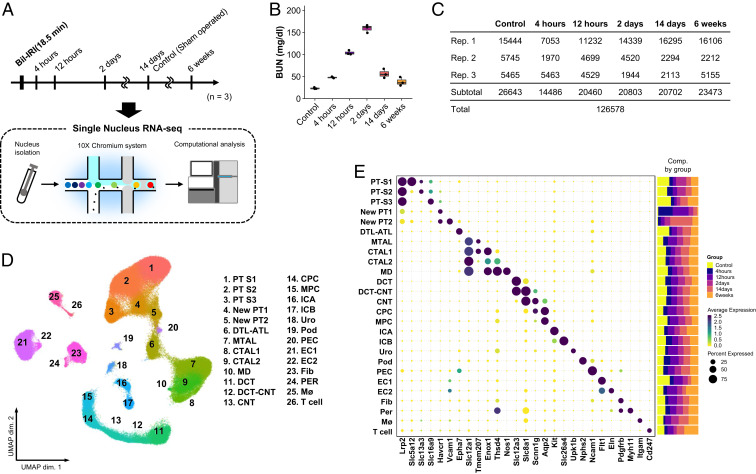
We performed single nucleus RNA-sequencing (snRNA-seq) on cryopreserved mouse kidney (7). Mice were euthanized at 4 and 12 h and 2, 14, and 42 d after bilateral ischemia–reperfusion injury (IRI) (Fig. 1A). Both histologic changes (SI Appendix, Fig. S1) and blood urea nitrogen (BUN) (Fig. 1B) levels confirmed acute injury and its resolution in mouse.
Fig. 1.
Single nucleus RNA-seq atlas of mouse IRI kidney. (A) Summary of experimental strategy. n = 3 mice per group. (B) BUN (mg/dL) after sham and IRI. Data are shown as the mean ± SEM. (C) Table with details of group, replicates, and cell numbers of mouse IRI datasets present in this figure. (D) UMAP plots of all mouse IRI kidney datasets integrated with Harmony. ATL, thin ascending limb of loop of Henle; Bil, bilateral; CNT, connecting tubule; CPC, principle cells of collecting duct in cortex; CTAL, thick ascending limb of loop of Henle in cortex; DCT, distal convoluted tubule; DTL, descending limb of loop of Henle; EC, endothelial cells; Fib, fibroblasts; ICA, type A intercalated cells of collecting duct; ICB, type B intercalated cells of collecting duct; MD, macula densa; Mø, macrophages; MPC, principle cells of collecting duct in medulla; MTAL, thick ascending limb of loop of Henle in medulla; PEC, parietal epithelial cells; Per, pericytes; Pod, podocytes; PT-S1, S1 segment of proximal tubule; PT-S2, S2 segment of proximal tubule; PT-S3, S3 segment of proximal tubule; Uro, urothelium. (E) Dot plot displaying gene expression patterns of cluster-enriched markers, and bar plot displaying composition of clusters by groups.
After quality control filtering, we obtained 26,643 cells from healthy mouse kidneys. Visualization of single nucleus transcriptomes in Uniform Manifold Approximation and Projection (UMAP) space resolved 26 separate clusters (SI Appendix, Fig. S2A and Dataset S1). Subclustering of both epithelial (descending loop of Henle, thin ascending limb) and nonepithelial cells (immune, endothelial, stromal) revealed additional cell clusters (SI Appendix, Fig. S2B). For example, five separate endothelial clusters were identified, including arterial, lymphatic, descending vasa recta and cortical vs. medullary endothelium. Eight stromal clusters were detected, including mesangium and Ren1-positive juxtaglomerular apparatus cells (SI Appendix, Fig. S2B). Major cell types and subclusters were identified based on cell type-specific markers (SI Appendix, Fig. S2 C and D and Dataset S1) (8–10). Each nephron segment performs unique reabsorptive and secretory functions to transform filtrate into urine, and this is reflected by segment-specific expression of all detected solute-linked carriers, ATPases, and channels (SI Appendix, Fig. S3).
Proximal Tubule Responses to Acute Injury.
We generated 99,935 mouse AKI single cell transcriptomes (Fig. 1C) and integrated these with the healthy datasets using the Harmony algorithm to reduce batch effects (Fig. 1D) (11). We could define unique anchor genes for all clusters in the integrated datasets and defined the relative abundance of each cluster in healthy vs. injured kidney. For example, the AKI kidneys contributed a much larger fraction of leukocytes than healthy kidneys (Fig. 1 D and E). All original clusters were retained, but two new clusters in mouse (Fig. 1D) (cluster 4: New PT1; and cluster 5: New PT2) appeared in injury, and these were located adjacent to healthy proximal tubule in UMAP space. Analysis of marker gene expression showed that these new clusters expressed the proximal tubule marker Lrp2 encoding Megalin, but also the injury marker Havcr1 encoding Kim1, indicating that these clusters represented an injured proximal tubule state in mouse.
We focused our analysis on proximal tubule, since this segment suffers the most injury due to high metabolic activity. Unsupervised subclustering of all mouse proximal tubule cells across time points yielded three healthy subclusters (the S1, S2, and S3 segments of the proximal tubule), one repairing subcluster, and three injured subclusters (Fig. 2A). Differential gene expression and gene set enrichment analysis (GSEA) were performed to characterize the subclusters (Dataset S2). Assigning time points to these clusters in UMAP space helps visualize temporal changes in proximal tubule gene expression (SI Appendix, Fig. S4A).
Fig. 2.
Time course analysis of proximal tubular cells revealed new cell state, failed repair proximal tubular cells. (A) UMAP displaying the clustering of proximal tubular cells without Harmony integration and dot plot displaying gene expression patterns of cluster-enriched markers. (B) Bar plot displaying composition of groups by clusters. (C) Representative images of immunofluorescence staining for VCAM1 (red), Kim1 (green), and LTL (white). (Scale bars: 50 μm.) (D) Deconvolution analysis of bulk RNA-seq mouse kidney IRI dataset using gene sets specific for healthy PT, injured PT, and failed repair PT. *P < 0.05; **P < 0.01; ***P < 0.001, one-way ANOVA with post hoc Dunnett’s multiple comparisons test. (E) Monocle2 pseudotime trajectory of proximal tubular cells colored by cluster identity. (F) Gene expression dynamics on the trajectories. The expression dynamics of DEGs were cataloged into three clusters across pseudotime shown as red lines (successful repair) and blue lines (failed repair). Thick lines indicate the average gene expression patterns in each cluster. The top six enriched GO terms for each cluster are shown on the right.
There were three categories of injured proximal tubule cells—we annotated these as “injured S1/2,” “injured S3,” and “severe injured PT,” respectively. “Injured S1/2” and “injured S3” were primarily composed of cells from the 4- and 12-h time points and expressed Myc, which encodes c-Myc playing a role in cell cycle progression, and Havcr1, and also shared a part of differentially expressed genes (DEGs) of “healthy S1” and “healthy S2” as its DEGs (Fig. 2A and Dataset S2), respectively. “Severe injured PT” shared expression of many injured PT genes but additionally expressed the tubule injury markers Krt20 (5, 6), as well as genes encoding heat shock protein, suggesting a more severe injury to these cells. GSEA showed that these proximal tubule injury states had enrichment of response to stress and damage, and “severe injured PT” additionally had “cell cycle arrest” (SI Appendix, Fig. S4B).
A “repairing PT” cluster arose 2 d after injury and had enrichment of “mitotic cell cycle” and “meiotic cell cycle” terms, including up-regulation of Top2a, which is essential for proliferation. Cell cycle status analysis revealed that “repairing PT” had the highest proportion of cycling cells (SI Appendix, Fig.S4 C and D). In contrast, the proximal tubule injury clusters had almost disappeared by 2 d (Fig. 2B), and a new distinct cell cluster arose, growing and reaching nearly 30% of all proximal tubule states at 14 d after injury, and remaining ∼8% of total proximal tubule by 6 wk after injury (Fig. 2B). This cluster expressed a distinct set of genes not observed in either healthy or acutely injured mouse proximal tubule. These included Vcam1, Dcdc2a, and Sema5a (Fig. 2A and Dataset S2). Because this cluster additionally down-regulated expression of terminal differentiation markers such as Slc5a12, Slc22a30, and Slc7a13 even at late time points, we annotated this cluster as “failed repair proximal tubule cells,” or FR-PTCs. We recently reported that ∼20% of injured proximal tubule cells fail to repair at 2 wk after AKI, and the presence of the FR-PTC cluster in the current analysis supports and extends those results (12). GSEA of FR-PTC revealed terms such as “positive regulation of lymphocyte activation,” “NIK NFκB signaling,” and “cell–cell signaling by Wnt,” suggesting FR-PTCs are proinflammatory (SI Appendix, Fig. S4B). We localized FR-PTCs after IRI by immunofluorescence. Vcam1-positive FR-PTCs emerged within Kim1-positive injured tubule in a scattered manner at 2 d after IRI and then expanded and remained within atrophic tubules with or without Kim1 expression at late time points (Fig. 2C).
Mapping the PT subclusters back onto the entire dataset revealed that the “New PT1” cluster was primarily composed of the three acute injury states and the “New PT2” cluster was primarily composed of FR-PTCs (SI Appendix, Fig. S4 E and F). We therefore annotated these clusters as “Injured PT” and “FR-PTC,” respectively, in further analyses. We asked whether these distinct PT states could be detected in other bulk RNA-seq datasets by applying the BSEQ-sc deconvolution algorithm (13). We assessed the fraction of healthy PT, injured PT, and FR-PTC in an independent mouse IRI 1-y time course (5). This showed that healthy PT decreased after injury but gradually recovered with time, injured PT increased after injury but resolved by 7 d, and FR-PTCs appeared beginning 1 wk after injury and persisted (Fig. 2D). Similar trends were observed in a mouse folic acid injury model (SI Appendix, Fig. S5A) (14). In human protocol biopsies from kidney transplants, FR-PTCs increased at 1 y after transplant compared to pretransplant (SI Appendix, Fig. S5B) (15). The proportion of FR-PTCs also increased with age in rat kidneys, increasing from ∼5% at 6 mo to ∼12% at 27 mo (SI Appendix, Fig. S5C) (16).
We reconstructed proximal tubule lineage relationships during repair by pseudotemporal ordering. The mouse trajectory began with injury, and most cells progressed to healthy S1/S2 or S2/S3 proximal tubule segments, but FR-PTCs formed an alternate branch point off the successful repair trajectory, indicating that FR-PTCs represent a distinct cell state (Fig. 2E). Gene ontology (GO) analysis across the pseudotime trajectory showed that the successful repair trajectory included terms that would be expected in cells that are redifferentiating, such as “organic acid metabolic process” and “carboxylic acid metabolic process” (Fig. 2F). The FR-PTC arm included terms like “cell motility” and “cell migration.” These results define FR-PTC as a distinct state after injury characterized by a unique set of markers and that persists after resolution of injury.
We next used single cell regulatory network inference and clustering (SCENIC) to map the gene regulatory networks governing these proximal tubule cell states (17). We discovered marked differences in regulon activity between FR-PTC, and either healthy or acutely injured states, providing further evidence that these are distinct proximal tubule cell states (Fig. 3A and Dataset S3). The FR-PTC cluster had regulon activity for both Relb and NFkB, suggesting a proinflammatory status for these cells (Fig. 3B). Also specific to the FR-PTC cluster was the Tcf7l1 regulon, which mediates Wnt signaling, consistent with the strong Wnt GSEA terms in this cluster. Proximal tubule canonical Wnt signaling is important both in specification and development, but also in disease (18, 19).
Fig. 3.
Gene regulatory network analysis of proximal tubular cells predicts transcription factors for successful repair and failing repair. (A) Heat map depicting the average regulon activity in each cluster of proximal tubular cells. Representative transcription factors are highlighted along with corresponding DNA-binding motifs. Cluster identities are according to Fig. 2. (B) Regulon activity dynamics on the pseudotime trajectory. The regulon activity dynamics were cataloged into three clusters across pseudotime shown as blue lines (successful repair) and red lines (delayed repair). Thick lines indicate the average gene expression patterns in each cluster. Pseudotime trajectories are according to Fig. 2. Transcription factors that have downstream CKD GWAS hit genes are displayed with their target genes expression patterns by dot plots. TF, transcription factor.
We mapped the relationship between transcription factors identified by this analysis and their regulation of genome-wide association study (GWAS) genes associated with CKD. In the successful repair cluster, Hnf4a, Hnf1b, and Pbx1 drive expression of multiple differentiation-associated genes that are also GWAS hits for CKD, including a variety of solute-linked carriers plus Plxdc2, Gas2, and Dab2 (Fig. 3B) (20). By contrast, both injury and the FR-PTC clusters had strong gene regulatory network signals for transcription factors regulating the expression of GWAS genes that were not expressed in healthy proximal tubule but rather in the injured state. Examples include the nonmuscle myosin gene Myh9, present in both injured and delayed repair clusters, and Nrg1 encoding the epidermal growth factor ligand neuregulin, present primarily in the injured clusters. In particular, NFkB and Relb regulons were specific to FR-PTC, and we could map downstream CKD GWAS genes to specific clusters, both healthy and injured. These results provide functional annotations of cell state-specific transcription factor-mediated regulatory networks, helping to elucidate the cellular context for susceptibility loci identified in CKD GWAS studies.
We could detect FR-PTC marker expression in apparently healthy human kidneys. Consistent with a prior report, these cells are located in a “scattered” fashion, adjacent to normal proximal tubule cells, throughout the proximal tubule (21). Examination of images from the Human Protein Atlas (22) shows scattered cells in healthy human kidney that express VCAM1 and DCDC2 (SI Appendix, Fig. S6A). We could also detect evidence for down-regulation of differentiation markers in isolated cells scattered throughout the nephron as well (SI Appendix, Fig. S6B). These results suggest that a conserved injury response occurs in individual, isolated cells even during homeostasis.
Stromal Cell Responses to Injury.
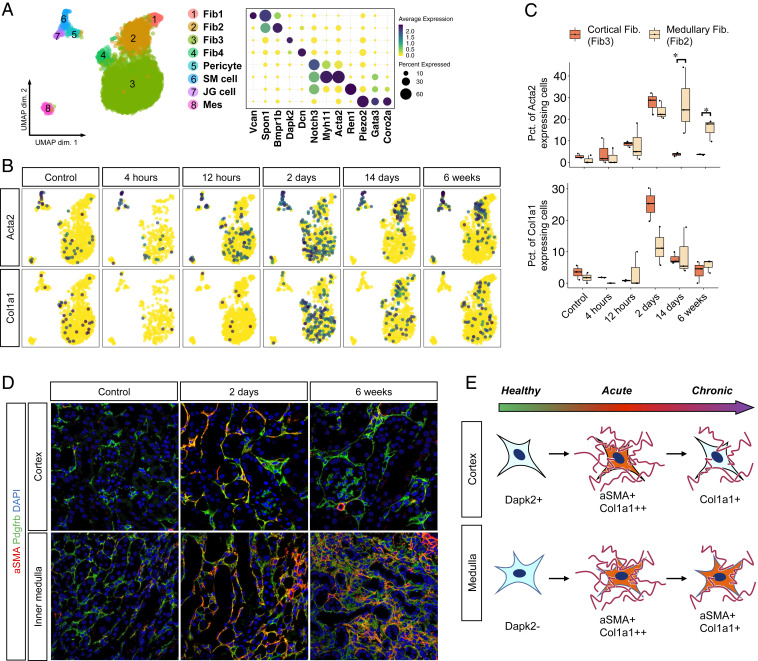
Recent single cell RNA-seq (scRNA-seq) analyses have revealed unexpected stromal heterogeneity in both developing and adult kidney (8, 9, 23). With Harmony integration, we combined the stromal clusters from all time points to identify eight stromal cell subclusters (Fig. 4A). These included four fibroblast populations that differed according to their cortical or papillary site of origin. We could also detect a pericyte and vascular smooth muscle cell population that were both characterized by strong expression of Notch pathway constituents such as Notch3 and Jag1, consistent with important roles for this pathway in pericyte development and angiogenesis (24, 25). We identified renin-secreting juxtaglomerular cells as well as mesangial cells. Several stromal clusters differed according to kidney region. For example, the cortical fibroblast marker Dapk2 was expressed in cluster 3 and 4 but not in cluster 1 and 2 (Fig. 4A) (26), suggesting that cluster 3 and 4 are cortical fibroblasts and cluster 1 and 2 are medullary fibroblasts.
Fig. 4.
Stromal subtypes are identified including a population with reversible expression of αSMA. (A) UMAP displaying the clustering of all stromal cells with Harmony integration and dot plot displaying gene expression patterns of cluster-enriched markers. SM, smooth muscle; JG, juxtaglomerular; Mes, mesangial. (B) UMAP displaying expression levels of typical myofibroblast markers in each group of the datasets. (C) Box plot displaying percentages (Pct.) of myofibroblast markers expressing cells in mouse fibroblast3 (cortical fibroblast) and fibroblast2 (medullary fibroblast) at each time point. (D) Representative images of immunofluorescence staining for αSMA (red) and Pdgfrb (green). (Scale bars: 50 μm.) (E) Diagram of the fate of fibroblasts after IRI.
Myofibroblasts secrete matrix proteins and are critical for fibrogenesis (27). Two marker genes for myofibroblasts are Acta2 and Col1a1. In healthy kidney, Acta2 expression was largely restricted to smooth muscle cells. After IRI, there was strong up-regulation of Acta2 across all stromal clusters, with the exception of mesangial cells, and Col1a1 was also strongly induced in fibroblasts (Fig. 4B). We could observe that cortical fibroblasts only transiently up-regulated Acta2 and Col1a1, with a peak at day 2 after IRI, whereas medullary fibroblasts showed sustained expression of Acta2 but not Col1a1 at 6 wk (Fig. 4 B and C). Medullary fibroblasts also increased as a fraction of the total stromal cells over time (SI Appendix, Fig. S7). These results suggest an unappreciated plasticity of kidney stroma. We could verify the injury-induced transient up-regulation of α-smooth muscle actin (αSMA), the protein encoded by Acta2, in cortical fibroblasts but not medullary fibroblasts, by immunofluorescence analysis (Fig. 4D). These results suggest regional differences in the response of fibroblasts to injury, with medullary fibroblasts progressing to a myofibroblast cell state and cortical fibroblasts reverting to their prior quiescent state (Fig. 4E).
Ligand–Receptor Interactions during Injury and Repair.
Finally, we leveraged our datasets to explore how injury affects intercellular communication within the kidney. We performed ligand–receptor analysis across all time points with simplified global clustering (see Fig. 6A). We highlight the tubulointerstitial compartment, comprising proximal tubule, endothelium, stroma, and leukocytes, because interstitial fibrosis in the kidney cortex (where proximal tubule is located) best predicts future kidney failure (28). Although we could identify six distinct leukocyte clusters (three macrophage subtypes, dendritic cells, and T and B cells) (SI Appendix, Fig. S8), we performed the ligand–receptor analysis on the combined leukocyte cluster to increase robustness of the analysis. Ccl2 and its receptor Ccr2 play important roles in AKI by recruiting monocytes and T cells (29). We used a standardized ligand–receptor score to quantitate signaling from Ccl2 in tubulointerstitium to Ccr2 in leukocytes across time (30). This revealed a temporal progression whereby fibroblasts and endothelial cells were the first cell type to signal to leukocytes, followed by leukocyte–leukocyte signaling at day 2, and finally increasing Ccl2–Ccr2 signaling from FR-PTC (Fig. 5 B and C). Compared to FR-PTC, proximal tubule destined for successful repair minimally up-regulated Ccl2 even in acute injury, emphasizing the proinflammatory nature of FR-PTC.
Fig. 6.
Signaling from proximal tubule to interstitial cell types in health and injury. Heat maps displaying scaled ligand–receptor interaction scores between proximal tubular cells and interstitial cells. (A) PT to endothelial cells. (B) PT to fibroblasts. (C) PT to leukocytes.
Fig. 5.
Ligand–receptor (L-R) analysis reveals dynamics of leukocyte (Leu) stimulating signaling networks during AKI-to-CKD transition. TAL, thick ascending limp. (A) UMAP of the integrated datasets with recategorized cell type names for ligand–receptor analysis. (B) Changes in the standardized interaction scores for Ccl2-Ccr2 ligand–receptor pair between injured proximal tubular cells and interstitial cells. (C) UMAP displaying expression levels of Ccl2 and Ccr2 in each group. (D) Heat map displaying sum of the leukocyte chemotaxis relating L-R interaction scores from each cell type to leukocytes.
To examine leukocyte chemotactic signaling across cell types more globally, we extracted genes from the “leukocyte chemotaxis” GO term and summed the ligand-interaction score for all cell types across time (Fig. 5D). The strongest scores were seen in endothelium and fibroblasts, with increasing values over time—suggesting ongoing leukocyte signaling even after repair was apparently nearly complete, at 6 wk. Consistent with our analysis of Ccl2 signaling, we only observed significant leukocyte chemotactic signaling from epithelia at late time points in the injured or FR-PTC clusters (Fig. 5D). These results highlight striking differences in cell types that are promoting inflammation in kidney after injury. In the acute phase, proinflammatory fibroblasts and endothelium predominate, but, in the chronic phase, FR-PTCs drive ongoing inflammation.
We then compared proinflammatory and profibrotic signaling from successful repair proximal tubule vs. FR-PTC across time. The pattern was similar whether proximal tubule was signaling to endothelial cells, fibroblasts, or leukocytes. For proximal tubule destined to successfully repair, there was very early up-regulation of growth factors or cytokines followed by down-regulation beginning 2 d after injury (Fig. 6 A–C). By contrast, the FR-PTC cluster arising at 2 d after injury up-regulated a distinct set of secreted proteins whose expression rose and continued to increase 6 wk after injury. Ligands from FR-PTC targeting endothelium include Edn1, encoding the potent vasoconstrictor endothelin-1, as well as Tgfb2 encoding transforming growth factor beta-2, which promotes fibrosis and Ltb, encoding lymphotoxin-β, which drives inflammatory lymphangiogenesis (31). FR-PTC signaling to fibroblasts included the profibrotic genes Pdgfrb and Pdgfrd (32). FR-PTC signaling to leukocytes included a variety of proinflammatory and profibrotic cytokines, including Csf1, Il34, Ccl5, Tnf, Ccl2, Ccl7, Ccl8, and Cxcl10. Consistent with a proinflammatory role for FR-PTC, Vcam1+ tubules were surrounded by F4/80+ macrophages at late time points after IRI (SI Appendix, Fig. S9).
Discussion
This single nucleus atlas of mouse AKI will serve as a resource for future studies aimed at understanding cellular responses to kidney injury. Our ability to differentiate between proximal tubule cells that are undergoing successful vs. failed repair allowed the molecular dissection of ligand–receptor interactions, signaling pathways, and gene regulons that determine whether an injured epithelial cell repairs successfully or not. Whether similar failed repair cell states are shared across organs will be an important question for future studies. Deconvolution of bulk RNA-seq datasets suggests that FR-PTCs also exist in human kidney and increase with age. Whether these proinflammatory cells contribute to the well-described age-associated decline in kidney function is another open question. Our results suggest that targeting these proinflammatory FR-PTCs may reduce chronic inflammation and fibrosis after injury, improving repair.
Materials and Methods
Animals.
All mouse experiments were performed according to the animal experimental guidelines issued by the Animal Care and Use Committee at Washington University in St. Louis. C57BL/6J (JAX Stock no. 000664) were purchased from The Jackson Laboratory (Bar Harbor, ME).
Surgery.
For bilateral IRI, 8- to 10-wk-old male mice were anesthetized with isoflurane, and buprenorphine SR was administered for pain control. Body temperature was monitored and maintained at 36.5 to 37.5 °C throughout the procedure. Bilateral flank incisions were made, and the kidneys were exposed. Ischemia was induced by clamping the renal pedicle with a nontraumatic microaneurysm clamp (Roboz, Rockville, MD) for 18 min. The clamps were subsequently removed, and kidneys were returned to the peritoneal cavity. The peritoneal layer was closed with absorbable suture, and the flank incisions were closed with wound clips. Control mice underwent sham surgery.
Mouse Kidney Samples.
Mice were euthanized with isoflurane, blood was collected, and the left ventricle was perfused with phosphate-buffered saline (PBS). For snRNA-seq, kidneys were snap-frozen with liquid nitrogen. For frozen sections, kidneys were fixed with 4% paraformaldehyde for 2 h on ice, incubated in 30% (vol/vol) sucrose at 4 °C overnight, and embedded in optimum cutting temperature compound (Sakura FineTek) to cut 7-μm sections. For paraffin sections, kidneys were fixed with 10% (vol/vol) formalin and paraffin-embedded to cut 4-μm sections. Immunofluorescence protocols and antibodies are detailed below.
BUN Measurement.
BUN measurement was done using the QuantiChrom Urea Assay kit as per the manufacturer’s protocol.
snRNA-seq.
Single nuclei isolation from tissue was performed as previously described (33). Briefly, nuclei were isolated with Nuclei EZ Lysis buffer (NUC-101; Sigma) supplemented with protease inhibitor (5892791001; Roche) and RNase inhibitor (N2615, Promega; AM2696, Life Technologies). Samples were cut into <2-mm pieces and homogenized using a Dounce homogenizer (885302-0002; Kimble Chase) in 2 mL of ice-cold Nuclei EZ Lysis buffer and incubated on ice for 5 min with an additional 2 mL of lysis buffer. The homogenate was filtered through a 40-µm cell strainer (43-50040-51; pluriSelect) and then centrifuged at 500 × g for 5 min at 4 °C. The pellet was resuspended and washed with 4 mL of the buffer and incubated on ice for 5 min. After another centrifugation, the pellet was resuspended in Nuclei Suspension Buffer (1x PBS, 1% bovine serum albumin, 0.1% RNase inhibitor), filtered through a 5-µm cell strainer (43-50005; pluriSelect). Nuclei were counted on hemocytometers (InCYTO C-chip) and partitioned into each droplet with a barcoded gel bead using the 10× Chromium instrument (10× Genomics, Pleasanton, CA). Single nuclei were lysed, and RNAs were reverse-transcribed into complementary DNA (cDNA) within each droplet. After breaking the emulsion, cDNAs were amplified and fragmented, followed by the addition of Illumina adapters using Single Cell 3′ Library & Gel Bead Kit (v2). Samples were indexed and sequenced on the S4 flow cell of NovaSeq 6000 (Illumina).
Data Processing of snRNA-seq Libraries.
snRNA-seq data were processed with zUMIs as previously described (34). Briefly, low-quality barcodes and unique molecular identifiers (UMIs) were filtered out using the internal read-filtering algorithm and then mapped to the mouse reference genome (mm10) using STAR 2.5.3a. Next, zUMIs quantified the reads that were uniquely mapped to the exonic, intronic, or intergenic region of the genome and inferred the true barcodes that mark nuclei by fitting a k-dimensional multivariate normal distribution with mclust package. Finally, a UMI count table utilizing both exonic and intronic reads was generated for downstream analysis. The whole data processing was executed by running the script on the facilities of the Washington University Center for High Performance Computing.
General Strategy of snRNA-seq Data Analysis.
Seurat v3 was used for downstream analyses, including normalization, scaling, and clustering of nuclei. We analyzed each batch of mouse sample separately and excluded nuclei with less than 150 or more than 8,000 genes detected. We also excluded nuclei with a relatively high percentage of UMIs mapped to mitochondrial genes (≥0.01) and ribosomal genes (≥0.06). Subsequently, we applied SoupX (35) to remove ambient RNA because different batches can be affected by different levels of ambient RNA. Briefly, ambient RNA expression is estimated from the empty droplet pool (10 UMIs or less) with setting “nonExpressedGeneList” to hemoglobin genes, followed by removing ambient RNA counts using the “adjustCounts” function with default parameters, according to the SoupX package (https://github.com/constantAmateur/SoupX). In parallel, we used Scrublet (36) to remove doublets. After merging all mouse data, we log-normalized and scaled the data to remove unwanted sources of variation driven by the number of detected UMIs and performed dimension reduction, clustering, and subclustering. In addition, after clustering or subclustering, we performed curated doublet removal based on known lineage-specific markers.
Clustering and Dimension Reduction.
We first combined all mouse datasets. The highly variable genes for principal component analysis were obtained by identifying the top 500 variable genes from each dataset with FindVariableFeatures and merging the list; we then performed principal component analysis (“RunPCA” function). Combined mouse datasets were integrated using the “RunHarmony” function in the Harmony package. Clustering and UMAP were performed in Seurat using the “harmony” data type as the dimensional reduction type (i.e., reduction.type=“harmony”). Marker genes were identified from each aligned cell type using the FindAllMarkers function in Seurat. Cluster reassignment was performed based on manual review of lineage-specific marker expression.
Time Course Analysis of Proximal Tubular Cells.
We extracted mouse proximal tubular cell clusters and then performed clustering without Harmony integration. The highly variable genes for principal component analysis were obtained by identifying the top 300 variable genes from each dataset with FindVariableFeatures and merging the list. We then performed principal component analysis (“RunPCA” function), clustering, and UMAP.
Pseudotemporal Analysis.
Pseudotemporal analysis was performed using Monocle2. We ordered the cells onto a pseudotime trajectory based on the union of highly variable genes in a set of principal components (PCs) that were previously used for time course analysis of proximal tubular cells. Next, we defined the branch-dependent genes by branched expression analysis modeling (BEAM) function in Monocle2 and then cataloged them into two clusters in a pseudotime manner. Finally, we performed GO analysis of each gene cluster.
Gene Regulatory Network Analysis on Proximal Tubular Cells.
We used SCENIC for gene regulatory network analysis. In brief, we generated coexpression networks of mouse proximal tubular nuclei data via GRNBoost2. We then utilized the SCENIC package to generate cell regulatory networks from mouse proximal tubular nuclei data, with the mouse mm10 genome for cis-regulatory analysis. We used two gene-motif rankings: 10 kilobases around the transcription start site (TSS) or 500 base pairs (bp) upstream and 100 bp downstream of the TSS, which were obtained from https://resources.aertslab.org/cistarget/.
Single Cell Deconvolution.
We used BSeq-sc to estimate the proportion of each PT subtype identified from snRNA-seq in the previously reported bulk RNA-seq data as previously described (37). Briefly, the marker genes for each PT subtype and the reads per kilobase million (RPKM) normalized gene expression matrix from bulk RNA-seq were used as input according to the tutorial from BSeq-sc package (https://shenorrlab.github.io/bseqsc/vignettes/bseq-sc.html).
Ligand–Receptor Interaction Analysis.
To study ligand–receptor interactions across cell types, we used a draft network (38) and defined an interaction score as previously described with a slight modification (30). In brief, first we eliminated ligand–receptor pairs that included laminin and collagen genes as ligands and then considered only ligands and receptors expressed in more than 0.5% of the nuclei in the specific cell type. Next, we defined the interaction score as the product of the average expression of a ligand in a cell type at a time point and the average expression of its cognate receptor of another cell type at the same time point. We then standardized each ligand–receptor interaction score by taking the distance between the interaction score and the mean interaction score in units of SDs.
Immunofluorescence.
Kidneys were fixed in 4% paraformaldehyde (Electron Microscopy Services), cryoprotected in 30% sucrose solution overnight, and embedded in optimum cutting temperature (OCT) compound (Tissue Tek). Kidneys were cryosectioned at 7 μm thickness and mounted on Superfrost slides (Thermo Fisher Scientific). Sections were washed with PBS (three times, 5 min each) and then blocked with 10% normal goat serum (Vector Labs), permeabilized with 0.2% Triton X-100 in PBS, and then stained with primary antibody specific for Cy3-conjugated anti-αSMA (C6198, 1:400; Sigma), rat anti-PDGFRβ (16-1402, 1:200; eBioscience), rabbit anti-CD31 (ab28364, 1:200; Abcam), and rat anti-F4/80 (ab6640, 1:200; Abcam). Secondary antibodies included AF488-, Cy3-, or Cy5-conjugated (Jackson ImmunoResearch). Then, sections were stained with DAPI (4′,6-diamidino-2-phenylindole) and mounted in Prolong Gold (Life Technologies). Images were obtained by confocal microscopy (Nikon C2+ Eclipse; Nikon, Melville, NY).
Data Availability.
All relevant data have been deposited in the Gene Expression Omnibus under accession number GSE139107.
Supplementary Material
Acknowledgments
These experiments were funded by NIH/National Institute of Diabetes and Digestive and Kidney Diseases Grants DK103740 and DK103225 and a Chan Zuckerberg Initiative Seed Network Grant.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: All relevant data have been deposited in the Gene Expression Omnibus under accession number GSE139107.
See online for related content such as Commentaries.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2005477117/-/DCSupplemental.
References
- 1.Chang-Panesso M., Humphreys B. D., Cellular plasticity in kidney injury and repair. Nat. Rev. Nephrol. 13, 39–46 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Kusaba T., Lalli M., Kramann R., Kobayashi A., Humphreys B. D., Differentiated kidney epithelial cells repair injured proximal tubule. Proc. Natl. Acad. Sci. U.S.A. 111, 1527–1532 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Coca S. G., Singanamala S., Parikh C. R., Chronic kidney disease after acute kidney injury: A systematic review and meta-analysis. Kidney Int. 81, 442–448 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang L., Besschetnova T. Y., Brooks C. R., Shah J. V., Bonventre J. V., Epithelial cell cycle arrest in G2/M mediates kidney fibrosis after injury. Nat. Med. 16, 535–543 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu J.et al., Molecular characterization of the transition from acute to chronic kidney injury following ischemia/reperfusion. JCI Insight 2, e94716 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xu K.et al., Unique transcriptional programs identify subtypes of AKI. J. Am. Soc. Nephrol. 28, 1729–1740 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu H., Kirita Y., Donnelly E. L., Humphreys B. D., Advantages of single-nucleus over single-cell RNA sequencing of adult kidney: Rare cell types and novel cell states revealed in fibrosis. J. Am. Soc. Nephrol. 30, 23–32 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Park J.et al., Single-cell transcriptomics of the mouse kidney reveals potential cellular targets of kidney disease. Science 360, 758–763 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stewart B. J.et al., Spatiotemporal immune zonation of the human kidney. Science 365, 1461–1466 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Young M. D.et al., Single-cell transcriptomes from human kidneys reveal the cellular identity of renal tumors. Science 361, 594–599 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Korsunsky I.et al., Fast, sensitive, and accurate integration of single-cell data with Harmony. Nat. Methods 16, 1289–1296 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chang-Panesso M.et al., FOXM1 drives proximal tubule proliferation during repair from acute ischemic kidney injury. J. Clin. Invest. 129, 5501–5517 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baron M.et al., A single-cell transcriptomic map of the human and mouse pancreas reveals inter- and intra-cell population structure. Cell Syst. 3, 346–360.e4 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Craciun F. L.et al., RNA sequencing identifies novel translational biomarkers of kidney fibrosis. J. Am. Soc. Nephrol. 27, 1702–1713 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cippà P. E.et al., Transcriptional trajectories of human kidney injury progression. JCI Insight 3, e123151 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shavlakadze T.et al., Age-related gene expression signature in rats demonstrate early, late, and linear transcriptional changes from multiple tissues. Cell Rep. 28, 3263–3273.e3 (2019). [DOI] [PubMed] [Google Scholar]
- 17.Aibar S.et al., SCENIC: Single-cell regulatory network inference and clustering. Nat. Methods 14, 1083–1086 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marose T. D., Merkel C. E., McMahon A. P., Carroll T. J., Beta-catenin is necessary to keep cells of ureteric bud/Wolffian duct epithelium in a precursor state. Dev. Biol. 314, 112–126 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kawakami T., Ren S., Duffield J. S., Wnt signalling in kidney diseases: Dual roles in renal injury and repair. J. Pathol. 229, 221–231 (2013). [DOI] [PubMed] [Google Scholar]
- 20.Qiu C.et al., Renal compartment-specific genetic variation analyses identify new pathways in chronic kidney disease. Nat. Med. 24, 1721–1731 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smeets B.et al., Proximal tubular cells contain a phenotypically distinct, scattered cell population involved in tubular regeneration. J. Pathol. 229, 645–659 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Uhlén M.et al., Proteomics. Tissue-based map of the human proteome. Science 347, 1260419 (2015). [DOI] [PubMed] [Google Scholar]
- 23.Lindström N. O.et al., Conserved and divergent features of mesenchymal progenitor cell types within the cortical nephrogenic niche of the human and mouse kidney. J. Am. Soc. Nephrol. 29, 806–824 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Y., Pan L., Moens C. B., Appel B., Notch3 establishes brain vascular integrity by regulating pericyte number. Development 141, 307–317 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kofler N. M., Cuervo H., Uh M. K., Murtomäki A., Kitajewski J., Combined deficiency of Notch1 and Notch3 causes pericyte dysfunction, models CADASIL, and results in arteriovenous malformations. Sci. Rep. 5, 16449 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guay J. A., Wojchowski D. M., Fang J., Oxburgh L., Death associated protein kinase 2 is expressed in cortical interstitial cells of the mouse kidney. BMC Res. Notes 7, 345 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kramann R., DiRocco D. P., Humphreys B. D., Understanding the origin, activation and regulation of matrix-producing myofibroblasts for treatment of fibrotic disease. J. Pathol. 231, 273–289 (2013). [DOI] [PubMed] [Google Scholar]
- 28.Nath K. A., Tubulointerstitial changes as a major determinant in the progression of renal damage. Am. J. Kidney Dis. 20, 1–17 (1992). [DOI] [PubMed] [Google Scholar]
- 29.Xu L., Sharkey D., Cantley L. G., Tubular GM-CSF promotes late MCP-1/CCR2-mediated fibrosis and inflammation after ischemia/reperfusion injury. J. Am. Soc. Nephrol. 30, 1825–1840 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schiebinger G.et al., Optimal-transport analysis of single-cell gene expression identifies developmental trajectories in reprogramming. Cell 176, 928–943.e22 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Furtado G. C.et al., Lymphotoxin beta receptor signaling is required for inflammatory lymphangiogenesis in the thyroid. Proc. Natl. Acad. Sci. U.S.A. 104, 5026–5031 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Floege J., Eitner F., Alpers C. E., A new look at platelet-derived growth factor in renal disease. J. Am. Soc. Nephrol. 19, 12–23 (2008). [DOI] [PubMed] [Google Scholar]
- 33.Wilson P. C.et al., The single-cell transcriptomic landscape of early human diabetic nephropathy. Proc. Natl. Acad. Sci. U.S.A. 116, 19619–19625 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parekh S., Ziegenhain C., Vieth B., Enard W., Hellmann I., zUMIs–A fast and flexible pipeline to process RNA sequencing data with UMIs. Gigascience 7, giy059 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Young M. D., Behjati S., SoupX removes ambient RNA contamination from droplet based single cell RNA sequencing data. bioRxiv: 10.1101/303727 (3 February 2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wolock S. L., Lopez R., Klein A. M., Scrublet: Computational identification of cell doublets in single-cell transcriptomic data. Cell Syst. 8, 281–291.e9 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu H.et al., Comparative analysis and refinement of human PSC-derived kidney organoid differentiation with single-cell transcriptomics. Cell Stem Cell 23, 869–881.e8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ramilowski J. A.et al., A draft network of ligand-receptor-mediated multicellular signalling in human. Nat. Commun. 6, 7866 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All relevant data have been deposited in the Gene Expression Omnibus under accession number GSE139107.