Graphical abstract
Abbreviations: COS, chitooligosaccharides; paCOS, partially acetylated COS; GlcN, D-glucosamine; GlcNAc, N-Acetyl-d-glucosamine; GH, Glycoside Hydrolase; FRAP, Ferric Reducing Antioxidant Power; RSA, Radical Scavenging Activity; DP, degree of polymerization; DD, degree of deacetylation
Keywords: Chitin, Chitosan, Pichia pastoris, Heterologous expression, Bioactive chito-oligomers
Highlights
-
•
Overproduction of endo-chitinase Chit33 to 0.6 g/L in yeast medium.
-
•
Chitinolytic polymers transformed in total/partial acetylated COS by Chit33.
-
•
Production of GlcN-(GlcNAc)1-6 and (GlcN)2-(GlcNAc)1-5 from chitosan by Chit33.
-
•
Production of chitinolytic derivatives with antioxidant activity using Chit33.
Abstract
The biological activity of chitooligosaccharides (COS) has made them targets for industrial and medical sectors. In this work, endo-chitinase Chit33 from Trichoderma harzianum CECT 2413 was expressed in Pichia pastoris GS115 to levels never achieved before (630 mg/L; 3.3 U/mL), without its biochemical characteristics being substantially affected. Chit33 produced a mixture of fully and partially acetylated COS from different chitin derivatives. HPAEC-PAD Chromatography and mass spectrometry analyses showed that (GlcNAc)4 and GlcN-(GlcNAc)2 were mainly produced from colloidal chitin and chitosan, respectively. COS in reaction mixtures were fragmented according to their size and their antioxidant activity analyzed by reducing power and free radical scavenging activity essays. The highest antioxidant activity was achieved with COS in the range of 0.5−2 and 2−10 kDa produced from colloidal chitin and chitosan, respectively, which gives biotechnological potential to both the chitin derivatives of 0.5−10 kDa and the biocatalyst producing them.
1. Introduction
Chitin is the most widespread amino renewable carbohydrate polymer in nature, and the second most abundant polysaccharide after cellulose, constitutes a fundamental component of fungi cell walls, exoskeletons of invertebrates and endoskeletons of mollusks. More than 1010 tons of chitin are produced annually in nature, of which 10 % comes from small marine crustaceans. Indeed, chitin constitutes up to 20–58 % of the wastes dry-weight coming from the shellfish processing industry [1]. This large linear homopolymer is mainly composed of β-(1–4)-linked N-acetyl-d-glucosamine (GlcNAc) units. Deacetylation of chitin produces chitosan, the only known natural polycationic polysaccharide containing GlcNAc and d-glucosamine (GlcN) units with the later usually exceeding 80 % of the residues [2]. Enzymatic or chemical hydrolysis of chitin and chitosan yields chitin oligosaccharides (also chitooligosaccharides; COS). Due to its biocompatibility, non-toxicity and wide availability in nature, chitinolytic materials have uses in food, health, cosmetic and agriculture fields [3]. However, the poor aqueous solubility at neutral pH values of chitin and chitosan limits their use, which makes COS derived from their hydrolysis gain biotechnological interest. Indeed, COS biological activity has been studied for some years, and among others, they show antioxidant, anti-inflammatory and/or anti-tumor properties [4,5]. Although there is no broad consensus on the results obtained, the size (degree of polymerization, DP), degree of deacetylation (DD) and pattern of acetylation (PA) seems to exert a notable influence on COS properties [[6], [7], [8], [9]].
Chitinases are essential glycosyl hydrolases (GH) for the biotransformation of chitin into COS. They cleave at terminal or internal β-(1-4)-glycosidic linkages of the biopolymer generating basically acetylated COS, di-acetyl chitobiose ((GlcNAc)2) and/or GlcNAc [10]. Thus, exo-chitinases (β-N-acetyl hexosaminidases; EC 3.2.1.51) attack the polysaccharide from its reducing or non- reducing end, whereas endo-enzymes (endo-β-N-acetyl glucosaminidase; EC 3.2.1.14) act on random points along the polysaccharide skeleton. According to the Carbohydrate-Active enzyme database (www.cazy.org), the GH family 18 (GH18) contains an ancient type of chitinases found in all kingdoms of life, including yeast and fungi, where are involved in the microbial cell wall degradation and morphogenesis but also in the exogenous chitin decomposition [11]. Thereby, many species of the soil fungi Trichoderma are commonly used in agriculture against plant pathogens because they produce different types of lytic enzymes including chitinases. Among them, T. harzianum and T. atroviride are widely used as biocontrol agents [[12], [13], [14], [15]], both producing different endo- and exo-chitinases since both types of enzymes need to act in a combined and processive way to efficiently degrade the polysaccharide, the so-called “multiple attack”.
Genomic DNA of numerous species of Trichoderma has been sequenced, revealing the presence of a wide chitinolytic machinery. One of the best analyzed genomes was that of T. reesei, which contained at least 18 potential exo- and endo-chitinases, all in the GH18 family [16]. The difference in the mechanism of the polymer attack of these two enzymes a priori making the latter more effective as was previously demonstrated when different endo- and exo-chitinase genes from Trichoderma spp were expressed in apple and maize plants to defend them from pathogenic fungi [17,18]. Recently, exo-chitinases Chit46 (46 kDa) and Chit42 (42 kDa) from T. harzianum were expressed in Pichia pastoris [1,19]. Both proteins produced mainly (GlcNAc)2 from colloidal chitin, but in mixtures containing traces of small fully acetylated (Chit46) or full and partial acetylated COS (Chit42). The effect of DP on the biological activity of COS has been previously discussed. Thus, COS with DP ≥ 4 showed much stronger antimicrobial and immune enhancing activity than COS with DP ≤ 3 [20,21]. This fact together with the mentioned apparent better effectiveness of endo-chitinases to degrade chitin, made us to focus on them. An endo-chitinase from T. harzianum, Chit33 (33 kDa), widely used as biocontrol agent against fungi such as Fusarium spp or Aspergillus flavus was previously reported [13,16,22]. This protein was molecularly characterized [23] and poorly produced in E. coli [24]. Besides, their corresponding orthologous from T. atroviride and T. virens were also expressed in E. coli and P. pastoris, respectively, but no data concerning heterologous protein production levels or any characterization of products obtained from colloidal chitin were indicated [25,26]. In this study, chitinase Chit33 from T. harzianum was expressed in P. pastoris, properties of the heterologous protein were analyzed and its utility for the production of COS evaluated. In addition, the antioxidant activity of COS with different sizes obtained from chitosan and colloidal chitin biopolymers was also studied.
2. Materials and methods
2.1. Materials
Chitin (from shrimp shells, coarse flakes), glycol chitosan, GlcNAc and biotin were from Sigma Aldrich (St. Louis, MO, USA). Chitosan CHIT100 and CHIT600 (both from shrimp shells) were from Acros Organics (Thermo Fischer Scientific Inc., Waltham, MA). Chitosan QS2 (from Pandalus borealis) was from InFiQuS (Madrid, Spain). Colloidal chitin, glycol chitin and chitosan solutions were obtained as explained previously [20]. ABTS (2,2′-azino-bis (3-ethylbenzothiazoline-6-sulfonate), DPPH (2,2-diphenyl-1-picrylhydrazyl), N,N′, -di-acetyl-glucosamine ((GlcNAc)2), N,N′,Nʺ -tri-acetyl-glucosamine ((GlcNAc)3) and N,N′,Nʺ,N′ʺ -tetra-acetyl-glucosamine ((GlcNAc)4) were from Carbosynth Ltd. (Berkshire, UK). Yeast Nitrogen Base w/o amino acids (YNB) was from Difco (BD, Sparks, MD, USA). All reagents were of the highest purity grade.
2.2. Strains, growth and expression media
Pichia pastoris GS115 (his4-; Invitrogen, Carlsbad, CA, USA) was used as expression host. Yeast transformants were selected on MD (13.4 mg/mL YNB, 4 mg/mL biotin, 20 mg/mL glucose) and Chit33 expression analyzed on BMM after growing in BMG (both MD including 100 mM potassium phosphate pH 6.0 and 0.5 % methanol or 1 % glycerol, respectively) as referred [20]. BMG-F (as BMG but pH 5.0 and 4 % glycerol) was used when high cell density was required. Culture growths were monitored at 600 nm (OD600). Escherichia coli DH5α was used as host for DNA manipulations as previously referred [24].
2.3. DNA amplification and cloning
The gene chit33 from T. harzianum CECT2413 (X80006.1) comprised of 909 bp including the TAA stop triplet and coding for a protein of 321 amino acids with a signal peptide of 19 residues (Q12713), was included in pCHIT33, a pBluescript SK (+) derivative plasmid [23]. In this work, plasmid CHIT33-pIB4, a derivative of pIB4 (His4) including the methanol-regulated alcohol oxidase promoter (AOX1p) was obtained, which makes expression of Chit33 fused to the Saccharomyces cerevisiae MFα1 secretion signal. The plasmid CHIT42-pIB4 previously obtained to express chitinase Chit42 from T. harzianum fused to MFα1 [20] and a restriction-free cloning strategy based in two PCR reactions was used. First, chit33 was amplified from plasmid pCHIT33 using primers: CHIT33F: 5′- tctcgagaaaagagaggctgaagctGGCTGGAATGTGAACTCGA -3′ (MFα signal peptide sequence in lower case) and CHIT33R: 5′- actgaggaacagtcatgtctaagaagcttTTACCTCAAAGCATTGACAACCT -3′ (pIB4 sequence in lower case) and Phusion High-fidelity DNA polymerase (NEB, Ipswich, UK) with the following conditions: (i) 98 °C for 30 s; (ii) 25 cycles of 98 °C for 10 s, 55 °C for 30 s and 72 °C for 30 s; (iii) final extension at 72 °C for 600 s. The PCR product (963 bp) was purified using Wizard SV Gel kit (Promega, Madison, USA), and used as mega-primer in a second PCR reaction with plasmid CHIT42-pIB4 as template. Amplification conditions were as before but in (ii) 35 cycles of 98 °C for 10 s, 55 °C for 30 s and 72 °C for 210 s. PCR reaction was treated with DpnI and transformed into E. coli. Colonies including CHIT33-pIB4 were detected by PCR using primers AOX1 and AOX2 from Sigma Aldrich (St. Louis, MO) that generate a 1216 bp amplification. In plasmid CHIT33-pIB4, last 1167 bp of gene chit42 was cleanly replaced by last 909 bp of chit33 and its integrity was verified by DNA sequencing.
2.4. Pichia pastoris transformation and protein expression
Plasmid CHIT33-pIB4 was linearized with Stu1 (into His4) and transformed into P. pastoris according to manual for protein expression in Pichia (Invitrogen, Carlsbad, CA, USA). Integration of chit33 in transformants was confirmed by PCR using primers CHIT33 F and CHIT33R. Yeasts including empty vector pIB4 were used as controls. Expression of Chit33 was analyzed using BMM medium and heterologous activity evaluated in culture filtrates. Transformants carrying CHIT33-pIB4 were cultivated in BMG during 24 h and then in BMM (1 L flasks) during 6 days, with addition of 1 m L-methanol/day. Yeast growth and pH of cultures were evaluated. Cells were removed (4000×g for 10 min) and extracellular fractions concentrated (if required) using 10000 MWCO PES membranes and a Vivaflow 50 system (Sartorius, Gottingen, Germany). Protein concentration was determined by NanoDrop at 280 nm.
Transformants of P. pastoris were grown in a 5-L bioreactor (Biostart BPluss Sartorius Ltd., Gottingen, Germany) as referred [20]. Yeasts expressing Chit33 were cultivated in 500 mL-BMG-F during 24 h and then used to inoculate the bioreactor containing 3.5 L of a batch medium to initial OD600 of ∼0.3 units. Fermentation parameters: 30 °C, 600 rpm agitation, 20 % dissolved O2 and pH controlled at 5.0 units with NH4OH during 24 h (∼44 UOD600). Then methanol was added during 4 days at 20 μL/min/L (final ∼220 UOD600), cells were removed by centrifugation and protein concentrated as above.
2.5. Enzyme and kinetic analysis
Unless otherwise indicated, chitinase activity was determined by detection of reducing sugars obtained from chitinolytic materials. Reactions were performed by addition of 100 μL of the enzymatic solution (in 70 mM potassium phosphate pH 5.5) to 400 μL of 1 % (w/v) substrates and were incubated at 900 rpm in Thermo Shaker TS-100 (Boeco, Hamburg, Germany) during 30 min. Colloidal chitin at pH 6.0 was used in the temperature dependence assay. Reactions were boiled 10 min and one volume of 0.2 M NaOH was added to precipitate remaining polysaccharides by centrifugation (12000×g; 5 min). To quantify reducing sugars in supernatants the 3,5-dinitrosalicylic acid (DNS) method adapted to 96-well microplate was used as described [20], with GlcN 0−3 mg/mL as calibration curve. One unit of chitinase activity (U) was defined as that corresponding to the release of 1 μmol of reducing sugar per minute.
For estimation of chitinase activity at different pH values colloidal chitin in 70 mM potassium phosphate at the pH range of 3.5–8.0 was used. Unless otherwise indicated, activity was tested at 45 °C. To compare the chitinase activity using colloidal chitin and chitosan, sodium acetate 100 mM pH 5.0 was used. The thermostability refers to the temperature required for 50 % activity inactivation after maintaining the enzyme at 40−90 °C during 10−90 min, removing samples at regular intervals and estimating the residual activity. All reactions were performed in triplicate. Kinetic constants were determined using 0.1−15 mg/mL of substrates. The plotting and analysis of the curves was carried out using GraphPad Prism software (version 6.0), and the kinetic parameters were calculated fitting the initial rate values to the Michaelis-Menten equation. Standard errors were obtained by fitting the normalized equation as v = (kcat/Km)[S]/(1 + [S]/Km).
2.6. SDS-PAGE and zymogram analyses
InstantBlue protein stain (Expedeon, Cambridge, UK)-sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE 12 %) were used to analyze proteins with the Precision Plus Protein Standards 10−250 kDa (Bio-Rad, CA, USA) markers. Chitinolytic activity was detected by zymogram analysis using PAGE including glycol chitin as referred [20]. Chitinase activity produced a clear halo in a dark purple background.
2.7. COS production, characterization and quantification by HPAEC-PAD and mass spectrometry
Reactions were performed as described above using 100 μL of enzymatic solution (∼1.2 U/mL), 400 μL of 1% substrates, 45 °C and 30 min. Aliquots of 0.2 mL were mixed with 0.2 M NaOH and centrifuged as referred. Supernatants were diluted with 2.5 mM NaOH (final concentration) and analyzed by HPAEC-PAD as described [20]. A chromatograph ICS3000 (Dionex, Thermo Fischer Scientific Inc., Waltham, MA, USA) with anion-exchange Carbo-Pack PA-200 column (4 × 250 mm) connected to CarboPac PA-200 (4 × 50 mm) was used. Standards of fully acetylated COS with DP 1–4 were used for the calibration curves that were adjusted to cubic or quadratic regressions using Chromeleon Software.
The molecular weight of COS was assessed by MALDI-TOF-MS using a mass spectrometer with Ultraflex III TOF/TOF (Bruker, Billerica, MA, USA) and NdYAG laser. Registers were taken in positive reflector mode within the mass interval 40−5000 Da, external calibration and 20 mg/mL 2,5-dihydroxybenzoic in acetonitrile (3:7) as matrix. Samples were mixed with the matrix in a 4:1 ratio and 0.5 μL were analyzed.
2.8. Fragmentation of COS produced according to size
Reactions including 20 mL of Chit33 (0.74 U/mL) and (i) 130 mL of colloidal chitin or chitosan QS2 in 500 mL flasks or (ii) 500 mL chitosan CHIT100 or CHIT600 in 1 L flasks (all 1% (w/v)) were incubated at 45 °C and 250 rpm during 4 days. Reactions were stopped at 80 °C during 10 min and centrifuged at 4000×g for 10 min. Non-precipitated material of a size ≤10 kDa was obtained using 10000 MWCO PES membranes as referred above. COS of 2−10 kDa were obtained by passing the ≤10 kDa fraction through 2000 MWCO PES membranes using a VivaSpin 15R system (Sartorius, Gottingen, Germany), and COS of 0.5−2 kDa when the excluded from the last fractionation passed through dialysis membranes of 500 Da MWCO cellulose acetate (Sigma Aldrich). All fractions were frozen at -70 °C, lyophilized and weighed. Degree of deacetylation (DD) of COS was estimated using UV spectroscopic methods, 0.3 M HCl solutions and a calibration curve of GlcNAc (16−120 μg/mL) as previously reported [27].
2.9. Antioxidant activity assays
The antioxidant capacity was evaluated using Ferric Reducing Antioxidant Power (FRAP) and free-Radical Scavenging Activity (RSA) using ABTS and DPPH methods as previously referred [[28], [29], [30]]. Briefly, in FRAP analysis, 0.25 mL of potassium phosphate 0.2 M pH 6.6, 0.25 mL of 1 % K3Fe(CN)6 and 7.5 mg/mL of chitinolytic materials (final concentration) were incubated at 50 °C for 30 min. Then 0.5 mL of 10 % trichloroacetic acid was added, mixture was centrifuged (12000xg for 10 min); 0.25 mL of supernatant was mixed with 0.25 mL of water and 0.05 mL 0.1 % FeCl3 and absorbance measured at 595 nm. Ascorbic acid (0−30 μg/mL) was used for calibration curve. The antioxidant activity was expressed as μmol of Ascorbic Equivalents per gram of Analyzed Compound (AEAC). The greater the absorbance, the greater the reducing power and antioxidant capacity.
The RSA was evaluated using 7 mM ABTS in 2.45 mM potassium persulfate maintained in dark for 16 h and then diluted in water to 0.7 units at 734 nm (OD734). Reactions containing 400 μL of the ABTS solution and 100 μL of each chitinolytic material (final concentration 1.0 mg/mL) were incubated at room temperature for 10 min and then OD734 measured. Ascorbic acid (0−0.6 μg/mL) was used as calibration curve. The antioxidant activity was expressed as % of RSA using the equation: RSA (%) = [1-(Absorbance of the sample including chitinolytic material/Absorbance of the control including water)] × 100. The less absorbance of samples, the greater RSA % and antioxidant capacity. The RSA was tested using DPPH as described before [30] with few modifications. Briefly, 0.2 mL of 0.1 mM DPPH (in ethanol and homogenized in ultrasonic bath for 30 s) were mixed vigorously with 0.2 mL of the COS solution (10 mg/mL, final concentration) and then incubated in dark for 30 min and 24 °C. Samples were centrifuged briefly (1 min) to precipitate non-soluble chitinolytic materials and the absorbance measured at 515 nm. The DPPH radical scavenging activity (%) was calculated as [(A0 – A1)/A0] × 100, where A0 is the absorbance of the DPPH solution (in which the sample including the chitinolitic material was replaced with ethanol) and A1 the absorbance of the sample to be evaluated.
2.10. Statistical analysis
All samples were prepared and measured in triplicates and standard errors were indicated.
3. Results and discussion
3.1. Heterologous production of the protein Chit33
Chitinase Chit33 from T. harzianum has been expressed in P. pastoris using the MFα1 secretion sequence and the AOX1p, a strategy that was previously used to successfully express an exo-chitinase of the same fungus [20]. The highest chitinase activity, 216 mU/mL, was measured in the extracellular medium of yeasts cultivated during 144 h in a methanol-based medium (Fig. 1A). As expected, only a clear protein band of ∼30 kDa was detected in SDS-PAGE, with more intensity at expression times increased (Fig. 1B) and that showed chitinolytic activity (Fig. 1C). At the maximum protein expression point, an extracellular protein concentration of 43 μg/mL was quantified, thus representing a specific chitinase activity of ∼5.0 U/mg. Production of Chit33 was increased by almost 15-times, to 0.63 mg/mL (3.3 U/mL; 5 U/mg), by growing yeasts and inducing the protein expression in a 5 L-bioreactor. This is the largest production of the endo-chitinase Chit33 reported so far. Thus, only 10 mg/L of Chit33 was previously recovered after refolding of the overexpressed protein from the E. coli cytoplasm [24] and the total uncharacterized extracellular protein concentration increased from 4.5 mg/L to 26 mg/L in T. harzianum transformants overexpressing this protein [31]. Moreover, only 3 mg/L of the also endo-chitinase ENCI (44 kDa) from T. harzianum T25-1 was produced in S. cerevisiae [32] and 186 mg/L of Ech42 from T. atroviride in P. pastoris [33]. Generally, much better results have been obtained by expressing fungi exo-chitinases in P. pastoris, 1.6 g/L of Chit46 from T. harzianum GIM3.442 [1] and 3 g/L of Chit42 (orthologous to Ech42) also from T. harzianum [20] are clear proof of that.
Fig. 1.
Activity profile and PAGE analyses of P. pastoris culture expressing chitinase Chit33. (A) The yeast transformant including plasmid CHIT33-pIB4 was cultivated in BMM using 1 L flask. The extracellular chitinase activity (red line; big circles), cell growth (OD600; blue line; squares) and pH (black line; small circles) were measured at the indicated times. Each point of activity represents the average of three independent measurements and standard errors are indicated. (B) Culture filtrates (11 μL) were evaluated after 0, 24, 48, 72, 96, 120 h and 144 h of methanol induction (lane 1, 2, 3, 4, 5, 6, 7, respectively) using SDS-PAGE. (C) Culture filtrate-144 h induction (500 μL) was 25-times concentrated and revealed in situ (lane 1). (D) Culture filtrate (1 μL) from yeast grown in fed-batch and induced with methanol during 4 days. Numbers on the left of panels (B, C, D) indicate positions of molecular mass standards (lane M) in kDa.
In addition, with the strategy used in this work, the Chit33 purification process was simplified to a single fractional concentration step of a yeast extracellular medium (Fig. 1D).
3.2. Biochemical and kinetic characteristics of Chit33 expressed in Pichia pastoris
The protein Chit33 purified from P. pastoris exhibited a maximum activity (≥80 %) on colloidal chitin at ∼40−50 °C and pH ∼5−6 (Fig. 2A), very similar values to that previously obtained when it was purified from T. harzianum [13]. However, when it was incubated without substrate at 60 °C and then chitinase activity was assayed, that formed by the natural producer retained 50 % of their activity after 30 min while the heterologous protein reached this value after almost 90 min. Thus, pointing towards the greater thermostability of the protein expressed in P. pastoris, which retained 50 % of activity at 60−66 °C in the range of 10−90 min (Fig. 2B).
Fig. 2.
Temperature, pH and thermostability dependence profiles of the heterologous Chit33 activity. (A) The effect of temperature (black line) and pH (light blue line) on the heterologous Chit33 activity was evaluated using colloidal chitin as substrate at pH 6.0 and 45 °C, respectively. (B) Chitinase was incubated for the indicated temperatures and times prior to the addition of substrate. Remaining activity was determined at 45 °C. Lines from top to botton corresponding progressively to the intervals ranging from 10 to 90 min.
The heterologous enzyme released reducing sugars from colloidal chitin and commercial chitosan of different DP and DD (Table 1). Although the viscosity and heterogeneity of the chitinolytic solutions used as substrate only allows obtaining apparent catalytic constants, clearly the enzyme expressed in P. pastoris showed maximum activity, and apparent catalytic efficiency (ratio kcat/Km), on colloidal chitin. In addition, it was about 10-times and 3-times less efficient on substrates showing molecular weights lower than 300 and 800 kDa, respectively (Table 1).
Table 1.
Chit33 hydrolytic activity and apparent catalytic constants on referred substrates.
| Substrate | MW (kDa) | DD (%) | Activity (%) | Km (mg/mL) | kcat (s−1) | kcat/Km(mg−1 s−1 mL) |
|---|---|---|---|---|---|---|
| Colloidal Chitin | n.d. | 8a | 100 ± 6 | 3.4 ± 0.6 | 10.0 ± 0.1 | 2.9 ± 0.4 |
| QS2 | 31 | 77 | 18 ± 4 | 0.9 ± 0.1 | 0.3 ± 0.004 | 0.3 ± 0.06 |
| CHIT100 | 100−300 | >90 | 15 ± 3 | 2.8 ± 0.8 | 0.8 ± 0.01 | 0.3 ± 0.08 |
| CHIT600 | 600−800 | >90 | 29 ± 3 | 0.2 ± 0.02 | 0.2 ± 0.002 | 1.0 ± 0.1 |
100 % activity: 0.74 U/mL; activity data are average of 3 independent experiments and standard errors were indicated; n.d. not determined; a DD of initial chitin flakes; apparent Km and kcat values were obtained; kcat were calculated from Vmax considering a Chit33 protein molecular mass of 33 kDa.
3.3. Production and characterization of products formed by Chit33 using chitinolytic biopolymers
To analyze the potential application of Chit33 in the COS production from colloidal chitin, HPAEC-PAD chromatography and mass spectrometry analyses were performed. Masses corresponding to all members of the fully acetylated series of COS between 1–7 units of GlcNAc were detected in the hydrolytic reactions (Fig. 3A). However, only molecules (GlcNAc) 1-4 were identify and quantified by HPAEC-PAD due to the corresponding standard availability (Fig. 3B) and just small traces of (GlcNAc)6-7 were detected (Supplementary Fig. 1 and Supplementary Table 1). As referred before [20], the order of elution in the chromatographic system/conditions used did not correlate with the increased DP of the chitin oligosaccharides. The enzyme produced about 0.43 g/L of (GlcNAc) 1-4 after 24 h reaction, which implies that only 5.4 % of the initial biopolymer suspension was apparently transformed into these small size acetylated oligosaccharides. The tetrasaccharide was the main product, with ∼0.15 g/L (Fig. 3B). In fact, this was the main COS produced at any time of the 24 h-reaction evaluated, pointing to Chit33 hydrolyzed chitinolytic substrates of at least 5 units of GlcNAc. In addition, masses of two different series of partially acetylated COS (paCOS) containing GlcN-(GlcNAc)1-6 and (GlcN)2-(GlcNAc) 1-5 units were also detected, with GlcN-(GlcNAc)3 apparently being the main paCOS (Supplementary Fig. 1 and Supplementary Table 1), and suggesting that initial chitin was not fully acetylated and/or was partially deacetylated during the colloidal suspension preparation. Production of paCOS was previously referred by using colloidal chitin and exo-chitinase Chit42, which mainly produced (GlcNAc)2 in a mixture including low amounts of GlcNAc, (GlcNAc)3 and only traces of GlcN-(GlcNAc) 2-3 and GlcN2-(GlcNAc)3 [20].
Fig. 3.
Analyses of reaction based on Chit33 and colloidal chitin. (A) HPAEC-PAD-chromatogram of the 24 h reaction. Peaks: (1) GlcNAc; (2) (GlcNAc)2; (3) (GlcNAc)3; (4) (GlcNAc)4; (5) Possible GlcN-(GlcNAc)2; (*) Unknown due to lack of the corresponding commercial standard. A schematic representation of DP and composition of reaction products predicted from mass spectrometry data is presented. Blue hexagons; dark symbols): GlcN. Green hexagons (clear symbols): GlcNAc. Peaks correspondence in brackets. (B) Evolution of the referred COS in the reaction mixture. Only the identified products (fully acetylated COS with DP 1-4) were quantified and their evolution represented. Each point represents the average of two measurements and standard errors are indicated. Lines from top to bottom corresponding to (GlcNAc)4, (GlcNAc)2, (GlcNAc)3, GlcNAc.
Similar HPAEC-PAD profiles, including a large number of peaks, were obtained using different types of chitosan (Fig. 4), but only three products, (GlcNAc)1-3, could be identified. Because the high DD of the used chitosan, signals corresponding to non-identified COS (Fig. 4A) must be due to paCOS, as was suggested by mass spectrometry assays, in which masses corresponding to (GlcN)1-2-GlcNAc, (GlcN)8-GlcNAc, (GlcN)4-7-(GlcNAc)2 and GlcN-(GlcNAc)2 were detected. Based on mass spectra analyses (Supplementary Fig. 2 and Table 2), it is feasible to think that highest peak in the HPAEC-PAD chromatogram ((5) in Fig. 4) might correspond to the trisaccharide GlcN-(GlcNAc)2, which would make Chit33 a potential biocatalyst for the production of this paCOS. The mass corresponding to this trisaccharide and a chromatographic signal with the same retention time that peak 5 were also detected when used colloidal chitin as substrate, and consequently this peak was also numbered as (5) in Fig. 3.
Fig. 4.
Analyses of reactions based on Chit33 and different chitosan types. (A) HPAEC-PAD-chromatograms of 24 h reactions including (Lines from bottom to top) chitosan CHIT600 (black), QS2 (red) and CHIT100 (blue), all 0.8 % (w/v) final concentration. Peaks: (1) GlcNAc; (2) (GlcNAc)2; (3) (GlcNAc)3; (4) (GlcNAc)4; (5) Possible GlcN-(GlcNAc)2; (*) Unknown. A schematic representation of DP and composition of reaction products predicted from mass spectrometry data is presented. Symbols and colours as in Fig. 3. (B) Evolution of the referred COS produced from chitosan QS2. Values are means of two measurements and standard errors are indicated. Lines from top to bottom: GlcN-(GlcNAc)2 and (GlcNAc) in clear and dark lines, respectively; (GlcNAc)3; (GlcNAc)2.
The substrate-binding site of fungal chitinases usually accommodates at least five sugars units, being the sugar binding subsites denominated as -3, -2, -1, +1, +2 and cleave occurring between sugars -1 and +1. In addition, chitinases from family GH18 show a substrate-assisted catalytic mechanism where a conserved glutamate residue (in their catalytic site) protonates the glycosidic bond to be hydrolyzed, and the oxygen of the N-acetyl group (of GlcNAc) in the subsite -1 sugar acts as nucleophile [12,20]. Because all products generated by chitinase Chit33 from colloidal chitin and chitosan (excluding GlcN) contained at least one unit of GlcNAc and the catalytic mechanism of chitinases-GH18 require a mandatory GlcNAc in the substrate sugar -1, it is plausible that Chit33 cut all chitinolytic sequences after a GlcNAc residue. Concerning the reactions based on chitosan and because all used substrates were mostly deacetylated, low yields in COS production was expected. In addition, none of the possible paCOS produced, including the main product (GlcN-(GlcNAc)2), could be quantified in terms of concentration due to the lack of the corresponding standard, so only peak areas could be estimated using the Chromeleon software (Fig. 4B).
3.4. Characterization of fragmented COS and analysis of their antioxidant activity
Fractions containing different sizes of COS produced from chitinolytic materials were obtained after filtering the 96-h hydrolytic reactions through varied cut-off membranes. Thus, theoretically, fully acetylated COS mixtures in the range of 3–9 and 1045 GlcNAc units would be detected in the fractions of 0.5−2 and 2−10 kDa, respectively, whereas paCOS that basically contained 1–10 and 6–54 GlcN units would be in these two fractions, respectively. These data were confirmed in mass spectrometry analyses within the limits of the methodology used (Supplementary Fig. 3).
Antioxidant activity of biological materials has important applications in medical and food industries due to free radicals attack macromolecules such as lipids, proteins or DNA, which are related with the age-associated diseases development and food quality deterioration or self-life shortening [34]. Chitinolytic polymer chains and their derivatives can be used as antioxidant and reducing agents to control free-radical oxidation because their amino/acetamido and hydroxyl functional groups can react with these radicals [35,36]. However, in this work neither chitosan nor colloidal chitin polymers showed significant reducing power (FRAP analyses), whereas molecules derived from hydrolysis of these polymers displayed this activity (Table 2), being that of 2−10 kDa derived from chitosan QS2 (31 kDa, 77 % DD) and CHIT100 (100−300 kDa, >90 % DD) the best iron (III) reducers and therefore, a priori the best antioxidants. Curiously, all COS mixtures obtained from chitosan in the range of 0.5−2 kDa sowed and average DD of about 50 % and those in the range 2−10 kDa of about 81–90 % (81, 88 and 90 % DD from CHI600, CHIT100 and QS2, respectively), whereas the two mixtures of chitooligosaccharides obtained from colloidal chitin showed DD of about 25 %.
Table 2.
FRAP of referred chitinolytic materials.
| Substrate | AEAC (μmol/g) |
||
|---|---|---|---|
| 0.5−2 kDa | 2−10 kDa | Polymers | |
| Colloidal chitin | 0.66±0.01 | 0.17±0.004 | 0 |
| QS2 | 0 | 1.16±0.03 | 0 |
| CHIT100 | 0.07±0.002 | 0.92±0.02 | 0 |
| CHIT600 | 0.23 ± 0.008 | 0.02±.0.001 | 0 |
AEAC: μmol of ascorbic acid equivalents per gram of the analyzed substrate. Values are mean of three determinations ± standard errors.
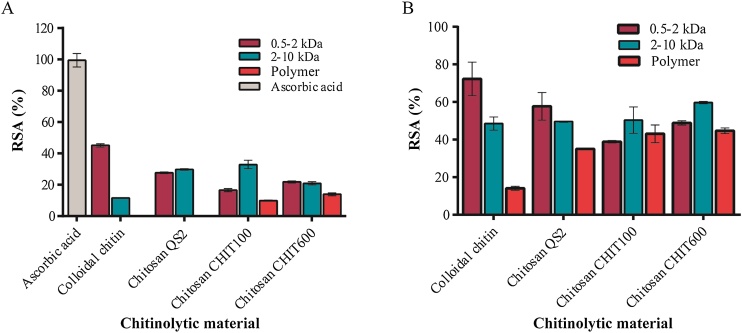
The free radical scavenging activity (RSA) of the different chitinolytic materials was also compared with that of ascorbic acid using ABTS and DPPH (Fig. 5). It has been previously reported that chitosan of high molecular weights showed high intramolecular electrostatic repulsive forces, which increased their hydrodynamic volume resulting in a reduction of their antioxidant activity. The antioxidant scavenging activities of chitosan increasing with chitosan decreasing MW [[37], [38], [39]]. However, in this work it was only clearly maintained for the colloidal chitin derivatives since best scavenging activity, both using ABTS and DPPH, as well as reducing power (Table 2) was obtained with the smallest COS (0.5−2 kDa). This may be very likely because the different COS mixtures obtained from chitin show a very similar pattern of DD. In addition, this effect was also detected with the QS2 derivatives but only when using DPPH and more slightly with CHIT600 and ABTS (Fig. 5, Table 2). In contrast, biological macromolecules of 2−10 kDa derived from chitosan CHIT100 (when using FRAP and RSA with ABTS or DPPH) showed better antioxidant activity than their corresponding smaller derivatives (Table 2 and Fig.5). It seems plausible that COS mixtures derived from chitosan are more complex, including molecules with more heterogeneous patters of acetylation, than those derived from colloidal chitin. Therefore, relating its antioxidant activity with aspects other than size.
Fig. 5.
Free Radical Scavenging Activity of the referred chitinolytic materials. Ascorbic acid was used as positive control. (A) Percentage of radical scavenging activity (RSA) using ABTS, relative to ascorbic acid 8 μg/mL (100 % RSA) used as control is showed. RSA of ascorbic acid at 4.2 μg/mL (50 % RSA) is indicated. (B) Percentage of RSA %) using DPPH. All data are means of three independent assays and standard errors are indicated. For each substrate 3 bars representing values obtaines with 0.5–2 kDa, 2–10 kDa and polymeric samples, from left to right, respectively.
The used methods in this work FRAP and RSA (with ABTS and DPPH) have different reaction mechanisms, and whereas FRAP assay is based on electron transfer reaction, RSA assay is on electron and H atom transfer [39,40]. These assays differ from each other in the reaction conditions and the total antioxidant capacity is dependent of a multitude of factors, including steric accessibility, solubility or distribution of reactive groups [40]. All factors that could have contributed to the differences obtained by using different protocols to analyse the antioxidant activity of the heterogeneous mixtures of COS derived from chitosan produced in this work. However, these methods clearly indicated that the studied chitinolytic materials possess considerable antioxidant activities and that full acetylated COS and paCOS of 0.5−10 kDa could be promising candidates as antioxidant agents, which gives biotechnological potential to the biocatalyst that produces them. Further research into Chit33 structure-specificity relationship will help to redesign it using molecular bioengineering techniques to try to improve the use of chitin wastes and to bias the enzyme activity towards the production of a narrow group of products.
4. Conclusion
Chitooligosaccharides (COS) have a large number of applications in food and medical areas, which increases the industrial interest to convert chitinolytic waste into these higher value derivatives. Production and analyses of biocatalysts that can be used to obtain COS from chitin biopolymers have enormous biotechnological interest. In this work, the fungal endo-chitinase Chit33 has been successful overproduced in Pichia pastoris, the protein purification process being reduced to a simple concentration of the yeast extracellular medium. The enzyme transformed colloidal chitin and different types of chitosan to fully and partial acetylated COS, which were later fragmented according to their size. Analysis of the reducing power and free radical scavenging activity of these biological products suggest that full and partial acetylated COS in the range of 0.5−10 kDa could be considered as antioxidant agents. For the possible industrial application of Chit33, its properties should be improved to transform it into a more efficient biocatalyst.
Funding
This work was supported by Spanish Ministry of Economy and Competitiveness [BIO2016-76601-C3-1/-2], Fundación Ramón Areces [XIX Call of Research Grants in Life and Material Sciences], EU EMFF-Blue Economy-2018 [Fish4Fish-863697] and by an institutional grant from Fundación Ramón Areces to the Centro de Biología Molecular Severo Ochoa.
CRediT authorship contribution statement
Peter Elias Kidibule: Formal analysis, Investigation, Writing - original draft. Paloma Santos-Moriano: Formal analysis, Investigation. Francisco Jose Plou: Supervision, Writing - review & editing. María Fernández-Lobato: Supervision, Funding acquisition, Project administration, Writing - review & editing.
Declaration of Competing Interest
The authors do not have any conflict of interest.
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.btre.2020.e00500.
Contributor Information
Peter Elias Kidibule, Email: pkidibule@cbm.csic.es.
Paloma Santos-Moriano, Email: palomacarmen.santos@universidadeuropea.es.
Francisco Jose Plou, Email: fplou@icp.csic.es.
María Fernández-Lobato, Email: mfernandez@cbm.csic.es.
Appendix A. Supplementary data
The following is supplementary data to this article:
References
- 1.Deng J.J., Shi D., hua Mao H., wei Li Z., Liang S., Ke Y., chun Luo X. Heterologous expression and characterization of an antifungal chitinase (Chit46) from Trichoderma harzianum GIM 3.442 and its application in colloidal chitin conversion. Int. J. Biol. Macromol. 2019;134:113–121. doi: 10.1016/j.ijbiomac.2019.04.177. [DOI] [PubMed] [Google Scholar]
- 2.Thadathil N., Velappan S.P. Recent developments in chitosanase research and its biotechnological applications: a review. Food Chem. 2014;150:392–399. doi: 10.1016/j.foodchem.2013.10.083. [DOI] [PubMed] [Google Scholar]
- 3.Khalil A.M., Abdel-Monem R.A., Darwesh O.M., Hashim A.I., Nada A.A., Rabie S.T. Synthesis, characterization, and evaluation of antimicrobial activities of chitosan and carboxymethyl chitosan Schiff-base/silver nanoparticles. J. Chem. 2017:1–12. doi: 10.1155/2017/1434320. [DOI] [Google Scholar]
- 4.Rathore A.S., Gupta R.D. Chitinases from bacteria to human: properties, applications, and future perspectives. Enzyme Res. 2015:1–9. doi: 10.1155/2015/791907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hamed I., Özogul F., Regenstein J.M. Industrial applications of crustacean by-products (chitin, chitosan, and chitooligosaccharides): a review. Trends Food Sci. Technol. 2016;48:40–50. doi: 10.1016/j.tifs.2015.11.007. [DOI] [Google Scholar]
- 6.Hamer S.N., Cord-Landwehr S., Biarnés X., Planas A., Waegeman H., Moerschbacher B.M., Kolkenbrock S. Enzymatic production of defined chitosan oligomers with a specific pattern of acetylation using a combination of chitin oligosaccharide deacetylases. Sci. Rep. 2015;5:1–9. doi: 10.1038/srep08716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Santos-Moriano P., Fernandez-Arrojo L., Mengibar M., Belmonte-Reche E., Peñalver P., Acosta F.N., Ballesteros A.O. Enzymatic production of fully deacetylated chitooligosaccharides and their neuroprotective and anti-inflammatory properties. Biocatal. Biotransf. 2018;36:57–67. doi: 10.1080/10242422.2017.1295231. [DOI] [Google Scholar]
- 8.Lodhi G., Kim Y.S., Hwang J.W., Kim S.K., Jeon Y.J., Je J.Y. Chitooligosaccharide and its derivatives: preparation and biological applications. Biomed Res. Int. 2014:1–12. doi: 10.1155/2014/654913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Santos-Moriano P., Kidibule P., Míguez N., Fernández-Arrojo L., Ballesteros A.O., Fernández-Lobato M., Plou F.J. Tailored enzymatic synthesis of chitooligosaccharides with different deacetylation degrees and their anti-inflammatory activity. Catalysts. 2019;9:405–417. doi: 10.3390/catal9050405. [DOI] [Google Scholar]
- 10.Santos-Moriano P., Kidibule P.E., Alleyne E., Ballesteros A.O., Heras A., Fernandez-Lobato M., Plou F.J. Efficient conversion of chitosan into chitooligosaccharides by a chitosanolytic activity from Bacillus thuringiensis. Process Biochem. 2018;73:102–108. doi: 10.1016/j.procbio.2018.07.017. [DOI] [Google Scholar]
- 11.Arnold N.D., Brück W.M., Garbe D., Brück T.B. Enzymatic modification of native chitin and conversion to specialty chemical products. Mar. Drugs. 2020;18:93–120. doi: 10.3390/md18020093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hartl L., Zach S., Seidl-Seiboth V. Fungal chitinases: diversity, mechanistic properties and biotechnological potential. Appl. Microbiol. Biotechnol. 2012;93:533–543. doi: 10.1007/s00253-011-3723-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De J., La Cruz, Hidalgo‐Gallego A., Lora J.M., Benitez T., Pintor‐Toro J.A., Llobell A. Isolation and characterization of three chitinases from Trichoderma harzianum. Eur. J. Biochem. 1992;206:859–867. doi: 10.1111/j.1432-1033.1992.tb16994.x. [DOI] [PubMed] [Google Scholar]
- 14.Prasetyawan S., Sulistyowati L., Aulanni’Am Glucanase and chitinase from some isolates of endophytic fungus Trichoderma spp. IOP Conf. Ser. Mater. Sci. Eng. 2018;299:1–7. doi: 10.1088/1757-899X/299/1/012026. [DOI] [Google Scholar]
- 15.Harman G.E., Howell C.R., Viterbo A., Chet I., Lorito M. Trichoderma species - opportunistic, avirulent plant symbionts. Nat. Rev. Microbiol. 2004;2:43–56. doi: 10.1038/nrmicro797. [DOI] [PubMed] [Google Scholar]
- 16.Wu Q., Bai L.Q., Liu W.C., Li Y.Y., Lu C.G., Li Y.Q. Construction of Streptomyces lydicus A01 transformant with the chit33 gene from Trichoderma harzianum CECT2413 and its biocontrol effect on Fusaria. Chin. Sci. Bull. 2013;58:3266–3273. doi: 10.1007/s11434-013-5860-9. [DOI] [Google Scholar]
- 17.Martinez D., Berka R.M., Henrissat B., Saloheimo M., Arvas M., Baker S.E. Genome sequencing and analysis of the biomass-degrading fungus Trichoderma reesei (syn. Hypocrea jecorina) Nat. Biotechnol. 2008;26:553–560. doi: 10.1038/nbt1403. [DOI] [PubMed] [Google Scholar]
- 18.Bolar J.P., Norelli J.L., Harman G.E., Brown S.K., Aldwinckle H.S. Synergistic activity of endo-chitinase and exo-chitinase from Trichoderma atroviride (T. harzianum) against the pathogenic fungus (Venturia inaequalis) in transgenic apple plants. Transgenic Res. 2001;10:533–543. doi: 10.1023/A:1013036732691. [DOI] [PubMed] [Google Scholar]
- 19.Shoresh M., Harman G.E. Differential expression of maize chitinases in the presence or absence of Trichoderma harzianum strain T22 and indications of a novel exo- endo-heterodimeric chitinase activity. BMC Plant Biol. 2010;10:136–146. doi: 10.1186/1471-2229-10-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kidibule P.E., Santos-Moriano P., Jiménez-Ortega E., Ramírez-Escudero M., Limón M.C., Remacha M. Use of chitin and chitosan to produce new chitooligosaccharides by chitinase Chit42: enzymatic activity and structural basis of protein specificity. Microb. Cell Fact. 2018;17:895–907. doi: 10.1186/s12934-018-0895-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tsai G.J., Wu Z.Y., Su W.H. Antibacterial activity of a chitooligosaccharide mixture prepared by cellulase digestion of shrimp chitosan and its application to milk preservation. J. Food Prot. 2000;63:747–752. doi: 10.4315/0362-028X-63.6.747. [DOI] [PubMed] [Google Scholar]
- 22.Liaqat F., Eltem R. Chitooligosaccharides and their biological activities: a comprehensive review. Carbohydr. Polym. 2018;184:243–259. doi: 10.1016/j.carbpol.2017.12.067. [DOI] [PubMed] [Google Scholar]
- 23.Limón M.C., Lora J.M., García I., de la Cruz J., Llobell A., Benítez T., Pintor-Toro J.A. Primary structure and expression pattern of the 33-kDa chitinase gene from the mycoparasitic fungus Trichoderma harzianum. Curr. Genet. 1995;28:478–483. doi: 10.1007/BF00310819. [DOI] [PubMed] [Google Scholar]
- 24.Boer H., Simolin H., Cottaz S., Söderlund H., Koivula A. Heterologous expression and site-directed mutagenesis studies of two Trichoderma harzianum chitinases, Chit33 and Chit42, in Escherichia coli. Protein Expr. Purif. 2007;51:216–226. doi: 10.1016/j.pep.2006.07.020. [DOI] [PubMed] [Google Scholar]
- 25.Matroudi S., Zamani M.R., Motallebi M. Molecular cloning of chitinase 33 (Chit33) gene from Trichoderma atroviride. Braz. J. Microbiol. 2008;39:433–437. doi: 10.1590/S1517-83822008000300005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Al-Rashed S., Abu Bakar F., Said M., Hassan O., Rabu A., Illias R.Md., Abdul Murad A. Expression and characterization of the recombinant Trichoderma virens endo-chitinase Cht2. Afr. J. Microbiol. Res. 2010;4:1758–1767. [Google Scholar]
- 27.Jiang Y., Fu C., Wu S., Liu G., Guo J., Su Z. Determination of the deacetylation degree of chitooligosaccharides. Mar. Drugs. 2017;15:332–345. doi: 10.3390/md15110332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arabshahi-Delouee S., Urooj A. Antioxidant properties of various solvent extracts of mulberry (Morus indica L.) leaves. Food Chem. 2007;102:1233–1240. doi: 10.1016/j.foodchem.2006.07.013. [DOI] [Google Scholar]
- 29.Ye C.L., Liu X.G., Huang Q. Antioxidant activity and protection of human umbilical vein endothelial cells from hydrogen peroxide-induced injury by DMC, a chalcone from buds of Cleistocalyx operculatus. S. Afr. J. Bot. 2013;86:36–40. doi: 10.1016/j.sajb.2013.01.010. [DOI] [Google Scholar]
- 30.Barros L., Baptista P., Ferreira I.C.F.R. Effect of Lactarius piperatus fruiting body maturity stage on antioxidant activity measured by several biochemical assays. Food Chem. Toxicol. 2007;45(9):1731–1737. doi: 10.1016/j.fct.2007.03.006. [DOI] [PubMed] [Google Scholar]
- 31.Limón M.C., Pintor-Toro J.A., Benítez T. Increased antifungal activity of Trichoderma harzianum transformants that overexpress a 33-kDa chitinase. Phytopathology. 1999;89:254–261. doi: 10.1094/PHYTO.1999.89.3.254. [DOI] [PubMed] [Google Scholar]
- 32.Draborg H., Kauppinen S., Dalbøge H., Christgau S. Molecular cloning and expression in S. cerevisiae of two exochitinases from Trichoderma harzianum. Biochem. Mol. Biol. Int. 1995;36:781–791. http://europepmc.org/abstract/MED/8528140 [PubMed] [Google Scholar]
- 33.Pérez-Martínez A.S., De León-Rodríguez A., Harris L.J., Herrera-Estrella A., Barba de la Rosa A.P. Overexpression, purification and characterization of the Trichoderma atroviride endochitinase, Ech42, in Pichia pastoris. Protein Expr. Purif. 2007;55:183–188. doi: 10.1016/j.pep.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 34.Kerch G. The potential of chitosan and its derivatives in prevention and treatment of Age-related diseases. Mar. Drugs. 2015;13:2158–2182. doi: 10.3390/md13042158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ngo D.H., Kim S.K. 1st ed. Elsevier Inc.; 2014. Antioxidant Effects of Chitin, Chitosan, and Their Derivatives; pp. 15–31. [DOI] [PubMed] [Google Scholar]
- 36.Avelelas F., Horta A., Pinto L.F.V., Marques S.C., Nunes P.M., Pedrosa R., Leandro S.M. Antifungal and antioxidant properties of chitosan polymers obtained from nontraditional Polybius henslowii sources. Mar. Drugs. 2019;17:1–15. doi: 10.3390/md17040239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chang S., Wu C., Tsai G. Effects of chitosan molecular weight on its antioxidant and antimutagenic properties. Carbohydr. Polym. 2018;181:1026–1032. doi: 10.1016/j.carbpol.2017.11.047. [DOI] [PubMed] [Google Scholar]
- 38.Sun X., Zhang J., Mi Y., Chen Y., Tan W., Li Q. Synthesis, characterization, and the antioxidant activity of the acetylated chitosan derivatives containing sulfonium salts. Int. J. Biol. Macromol. 2020;152:349–358. doi: 10.1016/j.ijbiomac.2020.02.177. [DOI] [PubMed] [Google Scholar]
- 39.El Jemli M., Kamal R., Marmouzi I., Zerrouki A., Cherrah Y., Alaoui K. Radical-Scavenging activity and ferric reducing ability of Juniperus thurifera (L.), J. oxycedrus (L.), J. phoenicea (L.) and Tetraclinis articulata (L.) Adv. Pharmacol. Sci. 2016:6392656–6392662. doi: 10.1155/2016/6392656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Magalhães L.M., Segundo M.A., Reis S., Lima J.L. Methodological aspects about in vitro evaluation of antioxidant properties. Anal. Chim. Acta. 2008;613:1–19. doi: 10.1016/j.aca.2008.02.047. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.