Summary
Multiple protocols have been published for generation of iMGLs from hESCs/iPSCs. To date, there are no guides to assist researchers to determine the most appropriate methodology for microglial studies. To establish a framework to facilitate future microglial studies, we first performed a comparative transcriptional analysis between iMGLs derived using three published datasets, which allowed us to establish the baseline protocol that is most representative of bona fide human microglia. Secondly, using CRISPR to tag the classic microglial marker CX3CR1 with nanoluciferase and tdTomato, we generated and functionally validated a reporter ESC line. Finally, using this cell line, we demonstrated that co-culture of iMGL precursors with human glia and neurons enhanced transcriptional resemblance of iMGLs to ex vivo microglia. Together, our comprehensive molecular analysis and reporter cell line are a useful resource for neurobiologists seeking to use iMGLs for disease modeling and drug screening studies.
Keywords: microglia, in-vitro-derived microglia, microglia genetic reporter, PSC differentiation, synapse internalization, RNA-seq, integrative molecular analysis
Highlights
-
•
Integrated molecular characterization reveals differences in four iMGL protocols
-
•
A dual tdTomato and nanoluciferase reporter tool was generated to track iMGLs via CX3CR1
-
•
Kinetics of surface marker expression during iMGL differentiation
-
•
Co-culture with glia/neurons restores ex vivo microglial transcription factors
In this article, Jose Polo and colleagues molecularly compare existing microglia differentiation methods from stem cells, generate a new dual fluorescent and enzymatic reporter tool to study microglia, and show that human glia/neuron co-culture pushes the regulatory landscape of in vitro microglia-like cells to more closely resemble bona fide microglia.
Introduction
Microglia, the resident macrophages of the central nervous system, are essential for brain development and function (Paolicelli et al., 2011, Thion et al., 2018), and have been genetically, epigenetically (Gjoneska et al., 2015), and transcriptionally (Zhang et al., 2013) shown to be directly involved in neurodegenerative diseases, including Alzheimer's disease and multiple sclerosis (Skene and Grant, 2016).
iPSC/hESC-derived cells are a useful research platform, potentially more representative of human development and systems compared with cell lines or animal models. Recently, multiple protocols described differentiation of human embryonic or human induced pluripotent stem cells (hESCs or iPSCs, respectively) toward microglia-like cells (iMGLs) (Abud et al., 2017, Brownjohn et al., 2018, Douvaras et al., 2017, Garcia-Reitboeck et al., 2018, Haenseler et al., 2017, Konttinen et al., 2019, Muffat et al., 2016, Ormel et al., 2018, Pandya et al., 2017, Takata et al., 2017). The differences between these protocols inherently result in transcriptomic and functional variation between the iMGLs generated, thus it is critical to understand which most closely resemble in vivo microglia.
Primary microglia rapidly downregulate key signature genes upon in vitro culture (Gosselin et al., 2017), indicating that growth factors currently utilized for in vitro culture are insufficient for establishment or maintenance of microglial identity. To study microglia and examine their interactions with other cells, it is useful to track permanent reporter expression targeted onto a key microglial gene.
In this brief report, we performed a molecular comparison of three existing iMGL differentiation strategies to identify the baseline protocol most similar to ex vivo microglia. Next, we used a dual CRISPR/Cas9-nickase system to selectively target one allele of the microglial marker CX3CR1 in the H9 hESC line, tagging the gene with a dual fluorescent/enzymatic construct, while ensuring physiological expression of CX3CR1 protein. We functionally validated iMGLs derived from this reporter cell line, demonstrating expression of key microglial markers, functional cytokine responses, and internalization of synaptosome fragments. Finally, we demonstrated that co-culture of iMGLs with human glia and neurons improves the transcriptional identity of iMGLs. Our reporter line and integrative transcriptional analysis can be utilized by researchers worldwide to further improve iMGL molecular signatures, with the ultimate aim of accurately recapitulating in vivo microglia for disease modeling and drug screening applications.
Results and Discussion
Molecular Comparison of Existing Microglia Differentiation Protocols
Since the first description of a directed differentiation protocol yielding IBA1+CD11b+CD45+ cells from a hiPSC or hESC lineage in 2016 (Muffat et al., 2016), to date at least ten differentiation protocols have been described to generate iPS-derived microglia-like cells (iMGLs, Table 1) (Abud et al., 2017, Brownjohn et al., 2018, Douvaras et al., 2017, Garcia-Reitboeck et al., 2018, Haenseler et al., 2017, Muffat et al., 2016, Ormel et al., 2018, Pandya et al., 2017, Takata et al., 2017, Konttinen et al., 2019). However, the transcriptomes generated by these protocols have only been compared with primary microglia cultured in vitro, and bona fide ex vivo microglia rapidly change identity upon in vitro culture resulting in ∼6,000 genes deregulated over 2-fold (Gosselin et al., 2017). Thus, there is a need for microglia researchers to determine which of these protocols to adopt or adapt for their own studies. The protocols differ primarily by the method used to generate microglial progenitors, with some methods relying on embryoid body formation to generate mesoderm (Brownjohn et al., 2018, Garcia-Reitboeck et al., 2018, Haenseler et al., 2017, Muffat et al., 2016, Takata et al., 2017), whereas others follow a 2D induction of mesoderm myeloid differentiation (Abud et al., 2017, Douvaras et al., 2017, Pandya et al., 2017, Konttinen et al., 2019), and some protocols purify intermediates by fluorescence-activated cell sorting (FACS) (Abud et al., 2017, Douvaras et al., 2017) or magnetic-activated cell sorting (Pandya et al., 2017). A recent study also detected native iMGL development within cerebral organoids (Ormel et al., 2018), previously found to be devoid of myeloid cells. The difficulty of comparing protocols is further confounded by the different, although partially overlapping, functional validation experiments used. We, therefore, utilized two recent landmark publications that for the first time transcriptionally profiled ex vivo FACS-isolated microglia from fresh postmortem or surgery-resected human brain (Galatro et al., 2017, Gosselin et al., 2017), to compare with the bona fide microglial transcriptional signature. In our analysis, we included all studies containing iMGLs that were profiled by RNA sequencing (RNA-seq), and that contained at least one common group with any other dataset, for the purpose of cross-study normalization (Abud et al., 2017, Douvaras et al., 2017, Muffat et al., 2016) (Table 1). Thus, we excluded datasets with only microarray data (Haenseler et al., 2017, Pandya et al., 2017), no RNA-seq for hiMGLs (Garcia-Reitboeck et al., 2018, Takata et al., 2017), and datasets containing no additional common sequencing group other than the iMGLs generated in that study (Brownjohn et al., 2018, Konttinen et al., 2019). Our results revealed that ex vivo microglia clustered close together irrespective of the study or fresh postmortem compared with surgery-resected origin of the cells, providing confidence in the method used for normalization (Figure 1A). Similarly, the brain lysate groups sequenced in both studies clustered together. Our results suggest that the first MDS dimension was dominated by the transition from non-myeloid to myeloid cells, and that the second dimension represented the differences in environment ex vivo to in vivo. The third dimension separated cells present in the brain from peripheral cells, as ex vivo monocytes and dendritic cells separated from ex vivo microglia primarily in this dimension (Figure 1B). These results show that there is a component of environment, and particularly of brain environment, in addition to the myeloid lineage that needs to be faithfully recapitulated for a molecularly representative model of microglia. Of the iMGL protocols compared in this study, the protocol of Abud et al. (2017) most closely resembled ex vivo microglia transcriptionally, and clustered with bona fide microglia after at least 24 h in vitro culture (Figures 1A and 1B). The additional iMGL protocols examined here clustered more closely with in-vitro-cultured fetal microglia (pFMGLs), and thus may require further maturation. Furthermore, the higher internal variability of differentiations in (Muffat et al., 2016) compared with other protocols may suggest that either embryoid body formation or multiple sequential collections of progenitors over several weeks may inherently generate more variability than multi-step synchronized directed differentiation or FACS isolation of pure target populations.
Table 1.
Comparison of the Protocols and Validation Strategies of Currently Available Protocols for the Generation of Human iMGLs
| Study | Protocol |
Validation |
Co-culture? | GEO | |||||
|---|---|---|---|---|---|---|---|---|---|
| Mesoderm | FACS/MACS | Maturation | Yield, ×106 iMGLs per 106 iPSCs | Molecular | Molecular Comparisona | Functional | |||
| Muffat et al., 2016, Nature Medicine | EB | No | M-CSF, IL-34 | 0.5–4 | RNA-seq, FACS, IF | pFMGL | phagocytosis: beads; cytokine secretion; motility: scratch test, migration to injury | hiPS-derived neurons + glia: 2D, 3D | GEO: GSE85839 |
| Pandya et al., 2017, Nature Neuroscience | 2D | MACS d15 CD34+/CD43+ | human astrocyte co-culture + GM-CSF, M-CSF, IL-3 | 0.8–3 | microarray, FACS, IF | pFMGL | phagocytosis: pHrodo E. coli; ROS production | human astrocytesb | GEO: GSE78116 |
| Abud et al., 2017, Neuron | 2D | FACS d10 CD43+ | M-CSF, IL-34, TGF-β1, insulin; D35-38 + CD200, CX3CL1 | 30–40 | RNA-seq, FACS, IF | pFMGL, pAMGL | phagocytosis: synaptosomes, Aβ tau oligomers; cytokine secretion; migration; Ca response to ADP | rat hippocampal neurons, hiPS-derived organoids, in vivo transplant | GEO: GSE89189 |
| Douvaras et al., 2017, Stem Cell Reports | 2D | FACS d25 CD14+ CX3CR1+ | IL-34, GM-CSF | 2.24 ± 0.42 | RNA-seq, FACS, IF | pAMGL, pFMGL, pAMGL + Douvaras media | phagocytosis: microspheres; Ca response to ADP | no | GEO: GSE97744 |
| Takata et al., 2017, Immunity | 2D | No | co-culture with iPSC-derived neurons | 10–20 | FACS, CyTOF | iMacs | phagocytosis: Aβ, latex beads | hiPS-derived neurons | |
| Haenseler et al, 2017, Stem Cell Reports | EB | No | IL-34 | 10–43 | microarray, FACS, IF | exMGL (fetal); exMGL (qPCR only, n = 1) | motility; response to LPS: clustering; morphology; cytokine secretion; neurons + iMGLs: electrophysiology, neuron morphology, synapse IF | hiPS-derived cortical neurons | GEO: GSE89795 |
| Brownjohn et al., 2018, Stem Cell Reports | EB | No | GM-CSF, IL-34 | 23–52 | RNA-seq, IF | none | phagocytosis: pHrodo E. coli; cytokine secretion | hiPS-derived organoid | GEO: GSE110952 |
| Garcia-Reitboeck et al., 2018, Cell Reports | EB | No | M-CSF | NA | qPCR, FACS, IF | PBMC, pMac | phagocytosis: pHrodo E. coli, pHrodo zymosan, apoptotic neurons; motility: scratch test, transwell assay; cytokine secretion | no | |
| Ormel et al., 2018, Nature Communications | organoid | No | within organoid | N/A | RNA-seq, FACS, IF | ex vivo CD45+, CD11b+ | phagocytosis: iC3b; cytokine secretion; PSD95 internalization (confocal) | organoid | GEO: GSE102335 |
| Konttinen et al., 2019, Stem Cell Reports | 2D | No | M-CSF, IL-34 | 20 | RNA-seq, FACS, IF | none | Ca response to ADP/ATP; scratch test; phagocytosis: pHrodo, FITC, Aβ; cytokine secretion; mitochondrial metabolism | iPS-derived neurons in 3D, organoids | GEO: GSE135707 |
EB, embryoid body; CyTOF, cytometry by time of flight; FITC, fluorescein isothiocyanate; GM-CSF, granulocyte-macrophage colony-stimulating factor; IF, immunofluorescence; M-CSF, macrophage colony-stimulating factor; MACS, magnetic-activated cell sorting; PBMC, peripheral blood mononuclear cell; ROS, reactive oxygen species; TGF, transforming growth factor.
Sequenced in that study.
Source of astrocytes is not specified.
Figure 1.
Comparison of Existing Differentiation Protocols and Differentiation of iPSCs to iMGLs
(A and B) Multidimensional scaling (MDS) analysis of the integrated datasets, presented as MDS dimension 1 versus 2 (A) and 2 versus 3 (B) showing separation of ex vivo microglia (exMGL), fetal or adult primary microglia—pFMGL or pAMGL cultured without, or in the presence of serum; +serum), iMGLs generated using various published protocols (_abud, _douvaras, _muffat), and ex vivo brain lysate (exBrain). For comparison and normalization, we also included various in-vitro-generated stem cell transcriptomes (iPSCs, iHPCs, and iNPCs) and primary peripheral blood mononuclear cells (pPBMCs), or other ex vivo myeloid cells (ex vivo monocytes, exM, and ex vivo dendritic cells, exDC).
(C) Bright-field micrographs showing colony and cell morphology at various points throughout the differentiation process, representative of >30 independent differentiations using eight independent iPSC lines. Scale bars, 100 μm.
(D) iMGLs phagocytose pHrodo E. coli particles basally and after LPS stimulation, representative of n = 3 independent experiments. Scale bars, 50 μm.
(E) Schematic of microglia differentiation protocol (adapted from Abud et al., 2017) and microglia reporter validation strategy used in this study.
See also Figure S1.
We first examined whether we could generate iMGLs that molecularly and functionally resembled iMGLs in our hands, and thus whether the protocol of (Abud et al., 2017) was robust to be adopted in multiple different labs. As the astrocyte-derived factors necessary for microglial survival and ramification in vitro were interleukin-34 (IL-34), colony-stimulating factor 1 (CSF1), transforming growth factor β1 (TGF-β1), and cholesterol (Bohlen et al., 2017), and the iMGL maturation media described in Abud et al. (2017) contained all but cholesterol, we also added cholesterol to the differentiation protocol. We showed that, in our hands in two independent labs, the differentiation stages morphologically followed those initially described (Figure 1C) and was consistent for iMGLs generated from eight iPSC lines tested comprising three control lines, three AD patient lines, and two CRISPR-corrected lines (Oksanen et al., 2017). We showed that D12 HPCs lost TRA-1-60 and gained CD43 expression and that, at D38, iMGLs uniformly expressed CD45 and CD11b (Figure S1A). iMGLs also expressed IBA1 as shown by immunofluorescence (IF) (Figure S1B). Moreover, iMGLs phagocytosed pHrodo Red-labeled E. coli, with enhanced phagocytosis (Figure 1D) and increased cytokine responses (Figure S1C) in response to lipopolysaccharide (LPS) stimulation. These data suggest that the differentiation protocol described by Abud et al. (2017) was readily adaptable in multiple labs. Thus, using this protocol as a baseline, we designed an approach to generate a tool for the microglial community that would facilitate established and emerging microglial researchers alike to examine microglial identity and functions in vitro and in vivo, in the context of physiology and disease (Figure 1E).
CRISPR Generation and Validation of CX3CR1-tdTomato H9 ESCs for Tracking Microglial Differentiation
We reasoned that a microglial reporter line would be a useful tool for the derivation of microglia and to allow efficient identification and live cell tracking of microglial cells in vitro and in vivo allowing rapid cell sorting without the need for additional labeling. Thus, we used a CRISPR/Cas9-derived method to facilitate insertion of a dual fluorescent (tdTomato) and enzymatic (nanoLuc) reporter into H9 cells. We chose to use a CRISPR system described previously (Mali et al., 2013, Ran et al., 2013), which utilizes dual Cas9-nickase constructs, to reduce the incidence of off-target double-strand breaks. Our donor vector contained two selection cassettes, a diphtheria toxin A (DTA) cassette, a neomycin/kanamycin resistance cassette, as well as two homology arms for the CX3CR1 gene (Figure 2A; Experimental Procedures). The donor vector enabled replacement of the stop codon of the CX3CR1 open reading frame with an IRES-tdTomato-T2A-Nanoluc-polyA-FRT-Neo-FRT construct. The DTA cassette was designed so that correct insertion of the donor vector into the host CX3CR1 locus resulted in excision of the DTA cassette. Clones were screened by PCR and Southern blot to confirm targeting (Figures S1D and S1E), confirmed to be free of chromosomal aberrations via karyotyping (Figure S1F), and confirmed to be pluripotent by teratoma assay (Figure S1G).
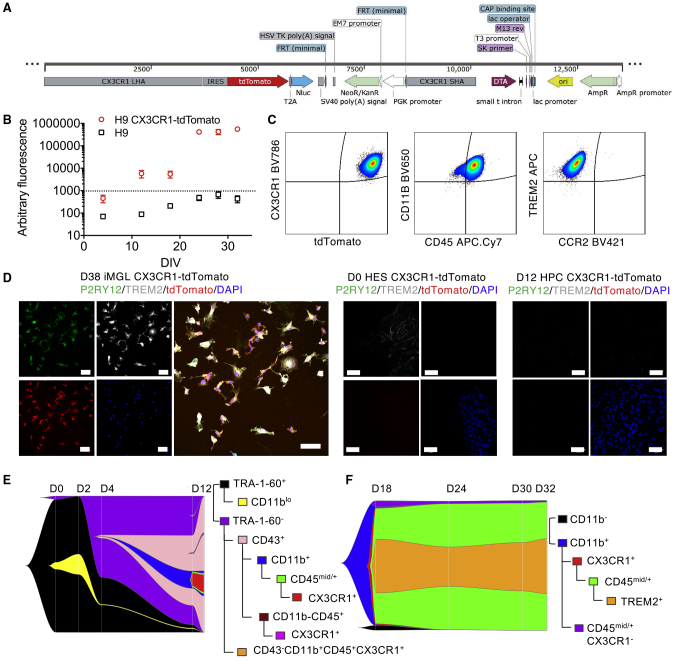
Figure 2.
Generation of a Dual Microglia Reporter ESC Line and Kinetics of Differentiation to iMGLs
(A) Schematic illustrating the CRISPR vector used for insertion of the tdTomato and nanoluciferase gene into the genome. Long homology arm (LHA) and short homology arm (SHA) for CX3CR1 were designed, using an IRES linker for tdTomato expression. cDNAs encoding tdTomato and nanoluciferase were linked with a T2A fragment, allowing translation of both proteins. A neomycin/kanamycin resistance cassette under the control of a PGK promoter was included for positive selection of correctly targeted clones. A DTA coding cassette was also included for negative selection of cells that do not correctly integrate the donor vector.
(B) Detection of luciferase secretion at various differentiation stages in H9 CX3CR1-tdTomato or H9 iMGLs. Error bars are SEM.
(C) Expression of CX3CR1 on iMGLs corresponds to tdTomato expression, as demonstrated by FACS. H9 CX3CR1-tdTomato iMGLs express CD11b, CD45, and TREM2, but not CCR2 as determined by FACS.
(D) iMGLs, but not H9 CX3CR1-tdTomato ESCs or H9 CX3CR1-tdTomato HPCs, express P2RY12, TREM2, as well as tdTomato. Scale bars, 50 μm.
(E and F) River plot showing the kinetics of cell identity transitions during differentiation of iPSCs to iHPCs (E) and iHPCs to iMGLs (F), as measured by expression of the markers TRA-1-60, CD43, CD11b, CD45, CX3CR1-tdTomato, and TREM2. Population proportions are presented as gated on the markers shown. Ungated FACS plots of the live cell populations are in Figures S2A and S2B.
See also Figures S1 and S2.
To validate that the nanoluciferase reporter was functional and not silenced in iMGLs, we measured nanoluciferase enzymatic activity in media harvested from H9 and H9-CX3CR1-tdTomato iMGLs at different time points throughout differentiation and showed increased luminescence in H9-CX3CR1-tdTomato but not H9 cells from D12 (Figure 2B). To verify that the fluorescent reporter construct was also functional, we generated iMGLs from the targeted H9 reporter cell line and examined the expression of the fluorescent reporter compared with endogenous CX3CR1 by FACS. tdTomato expression correlated with expression of CX3CR1 by FACS, and insertion of the reporter did not interfere with expression of CD11b, CD45, or TREM2, and iMGLs remained negative for the macrophage-specific marker CCR2 (Figure 2C). We confirmed that H9-CX3CR1-tdTomato iMGLs expressed P2RY12 and TREM2, as well as the tdTomato reporter and CX3CR1 (Figures 2D, S1H, and S1I). Together, our data suggest that both the fluorescent and enzymatic reporters are functional in this cell line and can be used to track iMGL differentiation via CX3CR1 expression in cells and media using FACS, IF, or luminescence approaches.
We next used this line to track kinetics of surface marker expression changes during differentiation and to determine the order of changes as iPSCs differentiate to iMGL. Thus, we followed the loss of the pluripotency marker TRA-1-60, which was lost in over 50% of cells from D4, and was entirely absent from non-adherent iHPCs by D12 of differentiation (Figures 2E and S2A). Loss of TRA-1-60 preceded acquisition of primitive lymphoid/myeloid marker CD43, which was present on 98% of non-adherent cells (iHPCs) by D12. Approximately 15% of iHPCs also co-expressed CD11b and CD45, and most of these cells (84.6%) were also positive for the CX3CR1 tdTomato reporter (Figures 2E and S2A) by FACS. A proportion of CD43+ cells at D12 (30.9%) also upregulated CD235a independently of CX3CR1 expression (Figure S2A). Of the microglial markers CD11b, CD45, CX3CR1, and TREM2, 73%–99% of cells expressed CD11b, CD45, and CX3CR1 by D18, and over 90% of cells expressed these markers by D24 (Figures 2F, S2B–D). Of the surface markers we examined, TREM2 was the last to be upregulated and most variably expressed between individual clones and individual differentiations using the same clone (Figure S2D). The pattern of TREM2 expression during iMGL maturation tended to either show a progressive increase in TREM2 staining from D18 to D32, or show highest TREM2 expression at D18, before a transient decrease in TREM2 expression, followed by increase at D32 (Figure S2D). At most, ∼90% of iMGLs were TREM2+ by D32, although for some differentiations this number was as small as 12%. Together, our data show the kinetics of surface marker expression during iMGL differentiation and demonstrate how the reporter cell line generated in this study can be used to track differentiation kinetics in response to different stimuli or for drug screening.
Functional Testing of H9 CX3CR1-tdTomato-Derived iMGLs
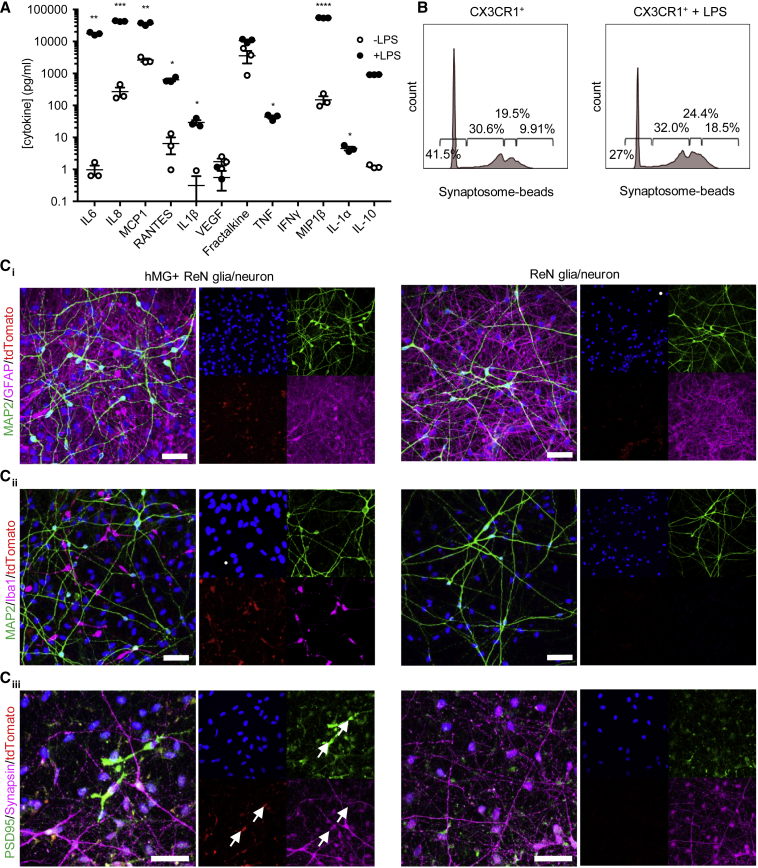
For our reporter line to be a useful tool for microglia, neuroscience, and drug discovery researchers, it must perform robustly in a variety of experimental setups, including imaging, co-culture and functional assays, as well as for a variety of readouts, including those that require either cells or media for analysis. Thus, we sought to validate our line in these settings. Cytokine responses and phagocytosis of cells expressing damage or danger signals, as well as interactions with synapses, are critical in vivo microglial functions. We confirmed that H9 CX3CR1-tdTomato secrete the appropriate range of cytokines and chemokines in response to LPS (Figure 3A) and phagocytose fluorescently labeled synaptosomes isolated from human brain (Figures 3B and S3A). We also showed that H9 CX3CR1-tdTomato iMGLs can internalize native synaptic material immunostained with PSD95 when co-cultured with human ReN cell cultures containing neurons and glia (Figure 3C). Together these data show that CX3CR1-tdTomato-derived iMGLs possess the functional properties of microglia in vitro.
Figure 3.
H9.CX3CR1-tdTomato iMGLs Secrete Cytokines, Internalize Native Synaptic Material, and Can Be Readily Tracked in Co-cultures
(A) Cytometric bead array for the cytokines and chemokines shown in H9.CX3CR1-tdTomato iMGLs basally (open circles), or stimulated for 24 h with LPS (closed circles, 100 ng/mL). Points represent the average of three individual wells from independent differentiations and are expressed as mean ± SEM. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001 by student's t test.
(B) Histograms showing the percentage of live tdTomato+ iMGLs phagocytosing one, two, or more synaptosome-conjugated fluorescent blue carboxylate 2.0-μm microspheres after 1.5 h co-incubation, as determined by FACS. Cells were pre-treated for 22.5 h with LPS (100 ng/mL). Histograms are representative of three independent experiments performed in triplicate wells.
(C) iMGLs at D21 were co-cultured for 21 days with ReN cell-derived mixture of human glia and neurons, stained with MAP2 and GFAP (i), MAP2 and IBA1 (ii), and synapsin and PSD95 (iii), and visualized using confocal microscopy. Individual channel images are shown as well as the merged images. Arrows represent co-localization between tdTomato, synapsin and PSD95. Scale bars, 50 μm.
See also Figure S3.
Co-culture of iMGLs with Human Neurons and Glia Shifts Transcriptional State toward Ex Vivo Microglial Identity
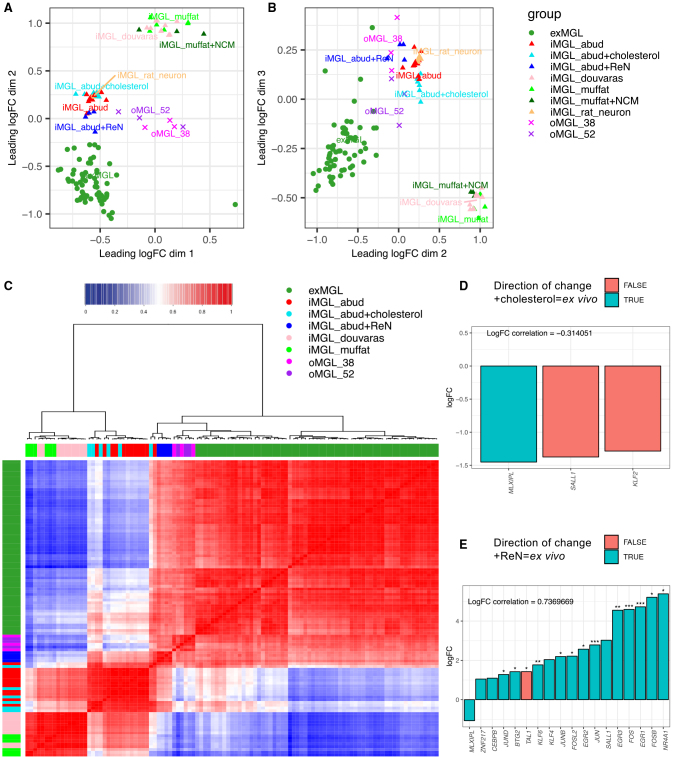
It is becoming clear that the brain niche specifies an independent component of microglial cell fate (Gosselin et al., 2014). To attempt to mimic context-specific functions and cell identity of microglia, to date iMGLs have been co-cultured with rat hippocampal neurons (Abud et al., 2017), hiPSC-derived neurons (Takata et al., 2017), NPC-conditioned medium (Muffat et al., 2016), cerebral organoids (Abud et al., 2017, Brownjohn et al., 2018, Ormel et al., 2018), or astrocytes (Pandya et al., 2017), each reported to partially improve functions and morphology of iMGLs. To further characterize this, we included transcriptional data from iMGLs co-cultured with rat hippocampal neurons (iMGL_rat_neuron; Abud et al., 2017), or NPC-conditioned medium (iMGL_muffat + NCM; Muffat et al., 2016). Curiously, this did not significantly enhance the similarity of iMGLs to ex vivo microglia (Figures 4A and 4B), suggesting that signals provided by direct contact with rat hippocampal neurons or NPC-conditioned medium are not sufficient to significantly shift the transcriptional signature of iMGLs toward ex vivo microglia (Figures 4A and 4B). This may be attributed either to the species differences between rodent and human microglia (Galatro et al., 2017, Gosselin et al., 2017, Smith and Dragunow, 2014), or to the absence of astrocyte-derived signals requisite for microglial maturation (Bohlen et al., 2017). Similarly, our data show that an additional astrocyte-derived metabolite required by microglia (cholesterol) was unable to push the iMGL molecular identity toward ex vivo microglia (Figures 4A and 4B). We thus hypothesized that co-culture of iMGLs or their precursors with human glia and neurons may recapitulate the components of the microglial transcriptional network that are controlled through cell-cell contacts. For this, we utilized ReN VM cells, which are immortalized human NPCs, shown to be electrophysiologically active upon differentiation to a neuron-glia culture (Choi et al., 2014). Indeed, the resultant transcriptomes of FACS-sorted iMGLs co-cultured with ReN-derived neurons and glia (Figures S3B and S3C) were shifted in both the first and second MDS components toward ex vivo microglia, suggesting acquisition of aspects of the ex vivo transcriptional signature (Figures 4A, 4B, and S4A). Similarly, when we integrated transcriptional data from a recent publication showing innate development of microglia in cerebral organoids (Ormel et al., 2018), the iMGL signatures were also shifted in the second MDS component toward an ex-vivo-like state, further highlighting the capacity of direct contact with niche cells to influence microglial molecular identity. Our data thus show two independent components required by microglia to establish an ex vivo identity—the first MDS component defines the developmental trajectory of microglia from iPSCs through iHPCs to iMGLs and thus likely represents maturity of microglia. The second component represents signals provided by the niche, as it separates in vitro and ex vivo microglia. Interestingly, organoid derived microglia (oMGL), both at 38 and 52 days of culture, were not as far shifted along the first MDS component trajectory between iHPCs and iMGL/microglia, suggestive of incomplete maturation toward the microglial lineage. These results are consistent with the role of the developmental trajectory with initial epigenetic priming outside the brain (or brain-like environment) in establishing microglial lineage identity, followed by niche signals for tissue imprinting (Amit et al., 2016).
Figure 4.
Co-culture with ReN Human Glia/Neurons, but Not Cholesterol alone, Shifts iMGL Transcriptional Profile toward an Ex Vivo Cell State
(A and B) Multidimensional scaling analysis of the integrated datasets as in Figures 1A and 1B presented as MDS dimension 1 versus 2 (A) and 2 versus 3 (B), including the RNA-seq datasets generated in this study. iMGL_abud group includes the original data from Abud et al. (2017) and data from this study, generated using the same protocol. As in Figures 1A and 1B, ex vivo microglia (exMGL) and iMGL_abud are included for comparison. Datasets include iMGLs co-cultured with rat hippocampal neurons, cholesterol, or ReN cells (iMGL_rat_neuron, Abud et al., 2017; iMGL_abud + cholesterol, this study; iMGL_abud + ReN, this study), or NPC conditioned medium (iMGL_muffat + NCM; Muffat et al., 2016), organoid microglia at D38 or 52 (oMGL38_ormel and oMGL52_ormel; Ormel et al., 2018).
(C) Cluster dendrogram (ward.D2) and correlation analysis (Pearson) of TF expression in ex vivo microglia and various in vitro iMGL protocols.
(D) Bar plot showing gene expression changes in ex vivo super enhancer or motif TFs with abs(logFC)>1 between iMGL_abud + cholesterol and iMGL_abud protocols (logFC>0 represents upregulation in iMGL_abud + cholesterol). The color of the bars depicts whether the direction of change for gene expression in iMG_abud + cholesterol is the same as that for ex vivo microglia (teal) or opposite to ex vivo microglia (salmon).
(E) Bar plot as for (D), depicting TF gene expression changes between iMGL_abud + ReN and iMGL_abud protocols. ∗adj. p < 0.05, ∗∗adj. p < 0.001, ∗∗∗adj. p < 0.0001.
To investigate the nature of the molecular transition induced in iMGLs by contact with human glia and neurons further, we focused on changes to transcription factors (TFs) shown to be deregulated in microglia in vitro. TF landscapes govern cell identity transitions and are master regulators of signals transduced from the environment; this is particularly evident in the adaptations of macrophage subpopulations to their environment (Bennett et al., 2018, Gosselin et al., 2014). We thus reasoned that early or partial cell fate changes could be captured by examining whether TF networks deregulated upon in vitro culture may be restored by co-culture with human neurons and glia. We examined whether expression levels of the 63 TFs associated with ex-vivo-specific super enhancers (ATAC-seq open chromatin regions also carrying an H3K27ac mark) or ex-vivo-enriched motifs (Gosselin et al., 2017) were restored to ex vivo levels. We first performed a clustering and correlation analysis of all iMGL transcriptomes based on expression levels of these 63 critical microglial TFs. This analysis showed that iMGLs cultured using the Abud protocol that were co-cultured with neurons/glia clustered with bona fide microglia, and that the expression of microglial TFs was highly correlated between these samples (Figures 4C and S4B). Similarly, the transcriptomes from oMGL that innately develop within organoids (Ormel et al., 2018) also clustered together with ex vivo microglia in this analysis. Importantly, although microglial TFs were unchanged in iMGLs cultured in cholesterol (Figure 4D), 12 microglial TFs were significantly upregulated in iMGL_abud + ReN, including multiple TFs from the JUN, FOS, EGR, and KLF families (Figure 4E). Together, these data show that direct interactions of iMGLs with human neurons and glia leads to a shift in the transcriptional program of iMGLs toward a more ex-vivo-like state.
Nonetheless, important differences exist between iMGLs and ex vivo microglia, even in the presence of human neurons and glia—indeed 1,967 differentially expressed genes (DEGs) remain, although this is a significant improvement on the 4,461 DEGs between ex vivo microglia and iMGL_abud (adj. p < 0.05; Table S1). As ReN cells are derived from fetal ventral mesencephalon, it is possible that co-culture with adult neurons and astrocytes or glia may provide additional maturation signals. Moreover, microglial transcriptomes are regionally heterogeneous, at least within the mouse (Grabert et al., 2016), which may be an epigenetically controlled function of microenvironment and neuronal turnover rates (Ayata et al., 2018); thus, co-culture with cells from a particular region of interest may also yield increased molecular resemblance to bona fide microglia. Together, our data provide a framework as well as laboratory and transcriptional tools. These tools allow the comparison and integration of existing and newly generated datasets, and a pluripotent microglia reporter line that can be used to track iMGLs and their media, alone or in co-culture systems, for a variety of molecular and functional assays or drug screening approaches.
Experimental Procedures
Resource Availability
Materials Availability
The H9.CX3CR1-tdTomato cell line is registered in hPSCReg as WAe009-A-24 (synonym MIPSe010-A-24).
Data and Code Availability
All sequencing data have been deposited to GEO under accession number GEO: GSE125872. The accession number for the raw data reported in this paper is GEO: GSE125872.
Differentiation of iPSCs/ESCs to Microglia-like Cells
Eight iPSC lines, as described previously (Oksanen et al., 2017), were used for validation of the protocol and the data are presented in Figures 1 and S1. The generation of H9 CX3CR1-tdTomato ESCs is described later in the Gene Editing section and was approved by the Monash University Human Research Ethics Committee (Project Number: 8826) and the Monash University Institutional Biosafety Committee (Reference: 23066).
The protocol for iMGL derivation was adapted from Abud et al. with modifications from the StemDiff Hematopoietic Kit (05310, STEMCELL Technologies) similar to (McQuade et al., 2018). H9 CX3CR1-tdTomato cells were cultured on vitronectin (A14700, Thermo Fisher Scientific)-coated T25 flasks in E8 medium (A1517001, Thermo Fisher Scientific). Two days before differentiation, cells were detached in 0.5 mM EDTA and 40–80 colonies/well were seeded in a 12-well plate coated with Matrigel (1:40, 354277, hESC-qualified matrix, LDEV-free, Falcon) in E8 medium. On day 0 (D0), E8 was exchanged for 1 mL of supplemented iHPC differentiation base medium (Supplemental Experimental Procedures). iHPC differentiation base medium supplemented with fibroblast growth factor 2 (FGF2) (50 ng/mL, 130-093-564, Miltenyi Biotec), BMP4 (50 ng/mL, 130-111-165, Miltenyi Biotec), Activin A (12.5 ng/mL, 130-115-010, Miltenyi Biotec), ROCKi (1 μM, 130-103-922, Miltenyi Biotec), and LiCl (2 mM, L7026, Sigma), and incubated in a hypoxic incubator (5% oxygen). On D2, medium was changed to 1 mL of iHPC differentiation base medium supplemented with FGF2 (50 ng/mL) and vascular endothelial growth factor (VEGF) (50 ng/mL, 130-109-385) and incubated in a hypoxic incubator. On D4, medium was changed to 1 mL iHPC differentiation base medium containing human FGF2 (50 ng/mL), VEGF (50 ng/mL), thrombopoietin (TPO) (50 ng/mL, 130-095-752, Miltenyi Biotec), stem cell factor (10 ng/mL, 130-096-695, Miltenyi Biotec), IL-6 (50 ng/mL, PHC0061, Thermo Fisher Scientific), and IL-3 (10 ng/mL, PHC0031, Thermo Fisher Scientific) and incubated under normoxia. Half the medium was replaced on D5 and D7 as on D4. On D10, the supernatant containing the HPCs was collected, centrifuged (300 × g for 5 min at room temperature [RT]), then 0.5 mL cell-containing medium was replaced and supplemented with 0.5 mL fresh medium. We note that hypoxia, TPO, and IL-6 are not required for 2D primitive myelopoiesis as assessed by the percentage of CD43+ cells at D12. On D12, supernatant containing HPCs was collected and plated onto Matrigel-coated 12-well plates at 1 × 105 cells/well in iMGL complete differentiation medium (Supplemental Experimental Procedures). The yield of iMGLs varies from 1:80 to 1:200 (i.e., 1 × 103 ESCs yields 8 × 104 to 2 × 105 HPCs). Every 2 days, each well was supplemented with 0.5 mL of complete differentiation medium, and at D22 cells were replated and a 50% media change was performed every 2 days. From D35-38, iMGLs were cultured in complete iMGL differentiation medium supplemented with human CD200 (100 ng/mL, C311, Novoprotein) and CX3CL1 (100 ng/mL, 300-31, PeproTech). iMGLs were stimulated with LPS for 24 h before RNA isolation or collection of culture supernatant.
Gene Editing
For gene targeting, H9 wild-type hESCs were co-cultured on 6-cm2 dishes with mouse embryonic fibroblasts (MEFs) until 80% confluent in hESC media (20% v/v KnockOut Serum Replacement, 1% non-essential amino acids, 0.5% GlutaMAX I, 1% v/v P/S, 0.625% v/v β-mercaptoethanol in DMEM/F12 + GlutaMAX I (all Thermo Fisher Scientific), replaced daily. For targeting, confluent cells were dissociated in 1 mL Accutase, collected into a 15-mL Falcon, centrifuged (160 × g, 4 min, RT), and depleted of MEFs for 1 h in an uncoated 6-cm2 dish with hESC media. Nucleofection was achieved by following the manufacturer's instructions for the Lonza Amaxa Primary P3 Kit (V4XP-3024, Lonza). On the day of targeting, 12,000/cm2 MEFs were dissociated as described above and one million cells/1 μg vector DNA/1 μg small guide RNA were resuspended in 100 μL of Lonza Amaxa Primary P3 nucleofection solution. Nucleofection was performed using the Lonza Nucleofection cuvette and the CB-156 setting. Cells were immediately replated (12,000/cm2) onto a 6-cm2 dish in hESC media supplemented with 10 μM ROCKi, and 20 ng/mL FGF2. Seventy two hours after nucleofection, cells were incubated in hESC medium containing G418 at 50 ng/mL (Thermo Fisher Scientific). Medium containing G418 was replaced daily for 10 days. Surviving colonies were manually picked using a dissecting microscope, and seeded into individual wells of a 12-well plate (12,000/cm2). Clones were grown to confluency, expanded into 6-well plates, and pellets frozen at −20°C for gDNA extraction and PCR screening, or cells frozen in Freeze Mix (10% v/v DMSO, 50% v/v fetal bovine serum, 40% v/v hESC media).
Cytometric Bead Array
CBA was carried out using the BD CBA human flexi kit using a protocol modified from that of the manufacturer. Five microliters of each standard (highest concentration at 5,000 pg/mL in assay diluent) and samples were incubated with 5 μL of Capture Bead mix (containing 0.1 μL of each cytokine Capture Bead diluted in Capture Bead Diluent) for 1 h in a 96-well V-bottom assay plate. Wells were incubated with 5 μL PE detection reagent mix (containing 0.1 μL of each cytokine PE reagent) diluted in Detection Reagent Diluent for 1 h in the dark. Wells were then washed once with 200 μL of wash buffer, and beads resuspended in 80 μL of wash buffer for analysis by FACS using the LSRFortessa X-20 (BD Biosciences). At least 200 single bead events from each cytokine population were collected. Results obtained were analyzed using the FCAP Array software v.3.0 (BD). Differences between LPS-stimulated and non-stimulated groups for secretion of individual cytokines were calculated using two-tailed student's t test.
Phagocytosis Assay
Phagocytosis assays were performed using pHrodo-red or pHrodo-green E. coli bio-particles (P35361 or P35366, Thermo Fisher Scientific) as per the manufacturer's instructions. In brief, particles were resuspended in 2 mL of iMGL differentiation medium, sonicated for 50 s, then vortexed for at least 30 s. Microglia were grown on 12-well plates at 1 × 105 cells per well in complete iMGL medium. Twenty four hours before phagocytosis assay, half the wells were treated with LPS (100 ng/mL, L4391, Sigma). One hour before assay, half of the LPS and non-LPS wells were treated with cytochalasin D (C8273, Sigma) at 10 μM to inhibit phagocytosis, and incubated for 1 h at 37°C. Immediately before the phagocytosis assay, particles were vortexed for at least 30 s, and diluted at a 1:60 concentration. Particles were added to wells, and incubated for 1 h at 37°C in the dark. Following incubation with bio-particles, iMGLs were collected in a 15-mL tube, washed in PBS 2×, and centrifuged at 300 × g for 5 min between washes. Microglia were resuspended in 100 μL of FACS buffer (PBS with 0.1% w/v BSA, 2.5 mM EDTA) before FACS analysis.
Synaptosome Isolation, Labeling, and Phagocytosis
Synaptosomes were isolated from human brain tissue (obtained from Victorian Brain Bank, Ethics Approval: Monash University MUHREC, 2016-0554) according to the Syn-PER Synaptic Protein Extraction Reagent (87793, Thermo Fisher Scientific) protocol. Protein concentrations were measured by nanodrop, and synaptosomes labeled with blue fluorescent 2.0-μm FluoSpheres (carboxylate-modified microspheres; F8824, Life Technologies) according to the manufacturer’s instructions. In brief, 5.5 mg synaptosomes was resuspended at 5 mg/mL in MES buffer (1.1 mL) for 15 min at RT and labeled by addition of 7.6 mg EDAC (E2247, Thermo Fisher Scientific) for 2 h at RT, then overnight (O/N) at 4°C. Conjugates were sonicated for 2 × 10 cycles (20 s on, 30 s off), then 16.2 mg glycine was added. After washing, conjugates were resuspended in 1 mL 1% (w/v) BSA with 2 mM sodium azide, and stored at 4°C before addition to cells. After 22.5 h LPS treatment, iMGLs were incubated with conjugated synaptosomes (3.44 μg/mL) for a further 1.5 h. iMGLs were collected as above and analyzed by FACS for internalization of synaptosome labeled particles.
Nanoluciferase Assay
Nanoluciferase activity in iMGL supernatants was assessed using the Nano-Glo Luciferase Assay System (N1110, Promega) according to the manufacturer's instructions. In brief, Nanoluciferase Assay Reagent was prepared immediately before the assay with Nano-Glo Luciferase Assay Substrate diluted 1:50 in Nano-Glo Luciferase Assay Buffer. Assay reagent (50 μL) was mixed with 50 μL cell-conditioned culture medium for 3 min and nanoluciferase activity was measured using the luminescence detection mode on a FLUOstar Omega microplate reader (BMG Labtech). The average signal per well over 10 min, beginning 3 min after substrate addition, was used as the final reading.
Culture and Differentiation of ReN Neural Progenitor Cells
ReN cells (SCC008, Millipore) were maintained and differentiated as described previously (Choi et al., 2014). In brief, cells were maintained on Matrigel-coated flasks (BD Biosciences) in DMEM/F12 (11320033, Thermo Fisher Scientific) supplemented with 2 μg/mL heparin (07980, STEMCELL Technologies), 2% (v/v) B27, 20 μg/mL EGF (130-097-749, Miltenyi Biotec), 20 μg/mL basic FGF (130-093-843, Miltenyi Biotec), and 1% (v/v) P/S. Neuronal and glial differentiation was achieved by growth factor withdrawal, with twice-weekly half media changes. We note that ReN cells do not differentiate to convincing GLAST+/S100+ astrocytes, although they do stain for GFAP. For co-culture assays, iMGLs were added to D21-differentiated ReN cells for a further 21 days at a ratio of (1:10), either in 6- or 48-well plate on coverslips, for FACS and immunofluorescence, respectively.
Flow Cytometry and Cell Sorting
Cells were stained with antibodies to microglial and macrophage cell surface markers (CX3CR1-BV786, 1:20, 744489, BD Biosciences; CD11b-BV650, 1:100, 101259, BioLegend; CCR2-BV421, 1:20, 564067, BD Biosciences; TREM2-APC, 1:10, FAB17291A R&D Systems or human TREM2 antibody 1:15, AF1828, R&D; and donkey anti-goat Alexa Fluor 647 1:2,000, A-21447, Thermo Fisher Scientific; CD45-APC.Cy7, 1:200, 25-0459-T100, Tonbo Biosciences; CD43-APC, 1:200, 343206, BioLegend, TRA-1-60 BUV395, 1:100, 563878, BD Biosciences; CD235a, PE-cy7, 1:200, 349112, BioLegend). Zombie violet (1:200) or propidium iodide (1:500) were used to discriminate live/dead cells, as appropriate for the antibody panel. For co-culture RNA-seq experiments, the CX3CR1-tdTomato reporter was used for isolation of iMGLs from glia-neuron co-cultures using the FACSAria III cell sorter.
Immunofluorescence
iMGLs were grown alone or in co-culture with ReN cells, on 8-mm glass coverslips (72296-08 PD25, Emgrid Australia) coated with Matrigel. Cells were fixed with 4% (v/v) paraformaldehyde (PFA) for 1 min, added directly to medium. Medium and PFA were replaced with pre-warmed 4% (v/v) PFA for 10 min at RT. After three PBS washes, cells were blocked for 1 h with 10% (v/v) donkey serum or normal goat serum, then stained with the primary antibodies O/N at 4°C: P2RY12 (1:400, HPA013796, Sigma), TREM2 (1:200, AF1828-SP, R&D), IBA1 (1:500, 019–19741, Novachem), MAP2 (1:500, MAB3418, Millipore), GFAP (1:500, Z0334, Dako), Synapsin I (1:1,500, 574777, Millipore), PSD95 (1:500, ab12093 Abcam). After 3× with PBST washes, cells were incubated rocking for 2 h at RT with either Alexa Fluor 488 donkey anti-goat (1:800, A-11055, Thermo Fisher Scientific), Alexa Fluor 488 goat anti-rabbit (1:800, A-11008, Thermo Fisher Scientific), Alexa Fluor 488 goat anti-mouse (1:800, A-11001, Thermo Fisher Scientific), or Alexa Fluor 647 donkey anti-rabbit (1:800, A31573, Thermo Fisher Scientific), Alexa Fluor 647 donkey anti-goat (1:800, A21447, Thermo Fisher Scientific), followed by DAPI (1 μg/mL, D1306, Thermo Fisher Scientific). Coverslips were washed 2× with PBST, then mounted with Dako Fluorescence Mounting Medium (S3023, Dako). Slides from ReN-iMGL co-cultures were imaged on a Nikon C1 confocal microscope using a 40× oil 1.4 NA objective and 1× zoom with 1,024 × 1,024 resolution, resulting in a pixel size of 90 nm. Slides for iMGL monocultures were imaged on a Leica SP8 confocal microscope using a 40× oil 1.24 NA objective and 0.75× zoom with 2,048 × 2,048 resolution, resulting in a pixel size of 188 nm.
RNA-Seq
RNA-Seq Library Construction and Sequencing
RNA extraction from 1 to 10 × 104 FACS-sorted CX3CR1+ iMGLs or from two wells of iMGLs harvested directly from 12-well plates, was performed on QIAcube (QIAGEN) using an RNeasy Micro Kit (74004, QIAGEN). RNA quality was assessed on the Bioanalyser (Agilent RNA 6000 Pico Kit). Libraries were prepared with 0.5–2 ng RNA, RNA integrity number ≥ 8. An 8-bp sample index (list below) and a 10-bp unique molecular identifier (UMI) were added during initial poly(A) priming and pooled samples were amplified using a template switching oligonucleotide. The Illumina P5 (5′ AAT GAT ACG GCG ACC GA 3') and P7 (5′ CAA GCA GAA GAC GGC ATA CGA GAT 3') sequences were added by PCR and Nextera transposase, respectively. The library was designed so that the forward read (R1) uses a custom primer (5′ GCC TGT CCG CGG AAG CAG TGG TAT CAA CGC AGA GTA C 3') to sequence directly into the index and then the 10-bp UMI. The reverse read (R2) uses the standard R2 primer to sequence cDNA in the sense direction for transcript identification. Sequencing was on the NextSeq 550 (Illumina), using the V2 High-Output Kit (Illumina) and the Illumina Protocol 15046563 v.02, generating two reads per cluster composed of a 19-bp R1 and a 72-bp R2.
An 8-bp sample index sequences used for sample multiplexing for RNA-seq:
TAAGGCGA; CGTACTAG; AGGCAGAA; TCCTGAGC; GGACTCCT; TAGGCATG; CTCTCTAC; CGAGGCTG; AAGAGGCA; GTAGAGGA; GCTCATGA; ATCTCAGG; ACTCGCTA; GGAGCTAC; GCGTAGTA; CGGAGCCT; TACGCTGC; ATGCGCAG; TAGCGCTC; ACTGAGCG; CCTAAGAC; CGATCAGT; TGCAGCTA; TCGACGTC.
Demultiplexing and Mapping
Sequencing reads were processed using in-house pipelines consisting of sabre tools (https://github.com/serine/sabre) and RNAsik (Tsyganov et al., 2018). Samples were demultiplexed with a fork of sabre tools with the commands below. Raw data were processed with an RNAsik pipeline to generate QC metrics, including percentage of reads mapped and assigned to the reference genome and duplication rates, and raw read counts for differential expression analysis. Demultiplexed UMI-tagged sequencing reads were aligned to the human genome (Ensembl GRCh38 primary assembly) using RNAsik. Read deduplication based on UMIs was performed with Je markdupes in RNAsik and transcript read counts calculated with featureCounts (Liao et al., 2014).
sabre pe -f ${RAW_DATA}/MultiplexRNASeq_S1_R1_001.fastq.gz∖
-r ${RAW_DATA}/MultiplexRNASeq_S1_R2_001.fastq.gz∖
-b ${BARCODE}∖
--combine∖
--umi∖
--max-mismatch 1∖
--min-umi-len 9∖
--max-5prime-crop 2∖
--stats demultiplex.stats∖
--no-comment
`combine`—merges R1 and R2 since R1 only holds "metadata," i.e., sample identity
`umi`—append umi into FASTQ header
`min-umi-len`—trim longer umis to 9 bases, discard umi (reads) that are shorter than 9
`max-5prime-crop`—if matching barcode cannot be found at 5ʹ of R1, remove 1 base, with maximum bases allowed to be removed set to 2
Normalization and Integration of Existing Microglia RNA-Seq Datasets
To compare different microglia-like cells and effect of in vitro culture we integrated the publicly available datasets (below).
| Set Number | Authors | GEO Accession | No. of Samples (Used for This Comparison) |
|---|---|---|---|
| 1 | Muffat et al. | GEO: GSE85839 | 16 |
| 2 | Douvaras et al. | GEO: GSE97744. | 24 (22) |
| 3 | Abud et al. | GEO: GSE89189. | 43 (40) |
| 4 | Gosselin et al. | GEO: GSE89960 | 64 |
| 5 | Galatro et al. | GEO: GSE99074 | 65 |
| 6 | Ormel et al. | GEO: GSE102335 | 16 (13) |
| 7 | Grubman et al. | GEO: GSE125872 | 16 (14) |
| Combined | 244 (234) |
Processed RNA-seq data were used where possible. Datasets 1–3 are available as fragments per kilobase of transcript per million mapped reads (FPKM) tables; datasets 4, 5, and 6 as counts. Read counts tables were transformed to FPKM with the edgeR (Robinson et al., 2010) (v.3.22.3) rpkm function and using average transcript length as gene length. Data were normalized with the removeBatchEffect function specifying the dataset numbers as batch and common groups where possible. The shared groups include brain samples, ex vivo monocytes, ex vivo microglia, and iPSCs. Normalized data were log transformed and used as input for plotMDS to generate the MDS analysis. For the heatmaps to visualize clustering of samples and genes, normalized data were used as described above. Samples pertaining to the ex vivo and in vitro microglia cells were selected (a total of 98 samples). The genes of interest (63 TFs from Gosselin et al., 2017) are selected from the expression data. Genes that were not widely expressed with an RPKM value of >0.5 in more than 10 samples, including ex vivo microglia, were filtered out. For Figure 4C, Pearson correlation was computed for all samples and visualized with heatmap.2 (using "ward.D2" clustering method) from the gplots package (v.3.0.1). For Figure S4B the RPKM values are shown and heatmap.2 was used to cluster samples using ward.D2.
Author Contributions
A.G., T.H.V., C.W.P., and J.M.P. conceived and designed the experiments. M.O. and S.L. generated iPSC lines. J.E.K. conceived and supervised generation of iPSC lines. T.H.V. and C.H. generated the reporter line. J.H.-H. and J.C. performed teratoma assays. A.G., T.H.V., and G.S. performed iMGL differentiation experiments. A.G. and G.S. performed validation experiments. J.S. integrated datasets and performed RNA-seq analysis. A.G., T.H.V., J.S., J.M.P., and C.W.P. analyzed and interpreted the data. A.G., T.H.V., J.M.P., J.S., and C.W.P. wrote the manuscript. J.M.H., G.S., C.H., S.K.N., and J.E.K., edited and approved the manuscript.
Acknowledgments
A.G. is supported by an NHMRC-ARC Dementia Research Development Fellowship (GNT1097461). A.G., J.M.P., C.W.P., and J.M.H. are supported by a Monash Joint Medicine-Pharmacy Grant (JMP17-0669), a Monash University International Network of Excellence Grant (NOE170089), and an NHMRC Grant APP1105786, and C.W.P. and J.M.H. by NHMRC grant APP1099372. A.G. was supported by the Dementia Australia Research Foundation and Yulgilbar Foundation Innovation Grant. G.S. and J C. were supported by the Yulgilbar Foundation. J.M.P. was supported by Sylvia and Charles Viertel Senior Medical Research Fellowship and ARC Future Fellowship (FT180100674). J.E.K. was supported by the Sigrid Jusélius Foundation, Finland. S.K.N. is supported by an OCE CSIRO Science Leader Fellowship. The Australian Regenerative Medicine Institute is supported by grants from the State Government of Victoria and the Australian Government. We thank the following Monash Platforms for the provision of instrumentation, training and technical support: Flowcore, Monash Micro Imaging, Monash Health Translation Precinct Medical Genomics Facility, Monash Bioinformatics Platform.
Published: May 21, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.stemcr.2020.04.007.
Contributor Information
Colin W. Pouton, Email: colin.pouton@monash.edu.
Jose M. Polo, Email: jose.polo@monash.edu.
Supplemental Information
References
- Abud E.M., Ramirez R.N., Martinez E.S., Healy L.M., Nguyen C.H.H., Newman S.A., Yeromin A.V., Scarfone V.M., Marsh S.E., Fimbres C. iPSC-derived human microglia-like cells to study neurological diseases. Neuron. 2017;94:278–293.e9. doi: 10.1016/j.neuron.2017.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amit I., Winter D.R., Jung S. The role of the local environment and epigenetics in shaping macrophage identity and their effect on tissue homeostasis. Nat. Immunol. 2016;17:18–25. doi: 10.1038/ni.3325. [DOI] [PubMed] [Google Scholar]
- Ayata P., Badimon A., Strasburger H.J., Duff M.K., Montgomery S.E., Loh Y.E., Ebert A., Pimenova A.A., Ramirez B.R., Chan A.T. Epigenetic regulation of brain region-specific microglia clearance activity. Nat. Neurosci. 2018;21:1049–1060. doi: 10.1038/s41593-018-0192-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett F.C., Bennett M.L., Yaqoob F., Mulinyawe S.B., Grant G.A., Hayden Gephart M., Plowey E.D., Barres B.A. A combination of ontogeny and CNS environment establishes microglial identity. Neuron. 2018;98:1170–1183.e8. doi: 10.1016/j.neuron.2018.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohlen C.J., Bennett F.C., Tucker A.F., Collins H.Y., Mulinyawe S.B., Barres B.A. Diverse requirements for microglial survival, specification, and function revealed by defined-medium cultures. Neuron. 2017;94:759–773 e758. doi: 10.1016/j.neuron.2017.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownjohn P.W., Smith J., Solanki R., Lohmann E., Houlden H., Hardy J., Dietmann S., Livesey F.J. Functional studies of missense TREM2 mutations in human stem cell-derived microglia. Stem Cell Reports. 2018;10:1294–1307. doi: 10.1016/j.stemcr.2018.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi S.H., Kim Y.H., Hebisch M., Sliwinski C., Lee S., D'Avanzo C., Chen H., Hooli B., Asselin C., Muffat J. A three-dimensional human neural cell culture model of Alzheimer's disease. Nature. 2014;515:274–278. doi: 10.1038/nature13800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douvaras P., Sun B., Wang M., Kruglikov I., Lallos G., Zimmer M., Terrenoire C., Zhang B., Gandy S., Schadt E. Directed differentiation of human pluripotent stem cells to microglia. Stem Cell Reports. 2017;8:1516–1524. doi: 10.1016/j.stemcr.2017.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galatro T.F., Holtman I.R., Lerario A.M., Vainchtein I.D., Brouwer N., Sola P.R., Veras M.M., Pereira T.F., Leite R.E.P., Möller T. Transcriptomic analysis of purified human cortical microglia reveals age-associated changes. Nat. Neurosci. 2017;20:1162–1171. doi: 10.1038/nn.4597. [DOI] [PubMed] [Google Scholar]
- Garcia-Reitboeck P., Phillips A., Piers T.M., Villegas-Llerena C., Butler M., Mallach A., Rodrigues C., Arber C.E., Heslegrave A., Zetterberg H. Human induced pluripotent stem cell-derived microglia-like cells harboring TREM2 missense mutations show specific deficits in phagocytosis. Cell Rep. 2018;24:2300–2311. doi: 10.1016/j.celrep.2018.07.094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gjoneska E., Pfenning A.R., Mathys H., Quon G., Kundaje A., Tsai L.H., Kellis M. Conserved epigenomic signals in mice and humans reveal immune basis of Alzheimer's disease. Nature. 2015;518:365–369. doi: 10.1038/nature14252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosselin D., Link V.M., Romanoski C.E., Fonseca G.J., Eichenfield D.Z., Spann N.J., Stender J.D., Chun H.B., Garner H., Geissmann F. Environment drives selection and function of enhancers controlling tissue-specific macrophage identities. Cell. 2014;159:1327–1340. doi: 10.1016/j.cell.2014.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosselin D., Skola D., Coufal N.G., Holtman I.R., Schlachetzki J.C.M., Sajti E., Jaeger B.N., O'Connor C., Fitzpatrick C., Pasillas M.P. An environment-dependent transcriptional network specifies human microglia identity. Science. 2017;356:eaal3222. doi: 10.1126/science.aal3222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabert K., Michoel T., Karavolos M.H., Clohisey S., Baillie J.K., Stevens M.P., Freeman T.C., Summers K.M., McColl B.W. Microglial brain region-dependent diversity and selective regional sensitivities to aging. Nat. Neurosci. 2016;19:504–516. doi: 10.1038/nn.4222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haenseler W., Sansom S.N., Buchrieser J., Newey S.E., Moore C.S., Nicholls F.J., Chintawar S., Schnell C., Antel J.P., Allen N.D. A highly efficient human pluripotent stem cell microglia model displays a neuronal-co-culture-specific expression profile and inflammatory response. Stem Cell Reports. 2017;8:1727–1742. doi: 10.1016/j.stemcr.2017.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konttinen H., Cabral-da-Silva M.E.C., Ohtonen S., Wojciechowski S., Shakirzyanova A., Caligola S., Giugno R., Ishchenko Y., Hernández D., Fazaludeen M.F. PSEN1DE9, APPswe, and APOE4 confer disparate phenotypes in human iPSC-derived microglia. Stem Cell Reports. 2019;13:669–683. doi: 10.1016/j.stemcr.2019.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y., Smyth G.K., Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- Mali P., Aach J., Stranges P.B., Esvelt K.M., Moosburner M., Kosuri S., Yang L., Church G.M. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat. Biotechnol. 2013;31:833–838. doi: 10.1038/nbt.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQuade A., Coburn M., Tu C.H., Hasselmann J., Davtyan H., Blurton-Jones M. Development and validation of a simplified method to generate human microglia from pluripotent stem cells. Mol. Neurodegener. 2018;13:67. doi: 10.1186/s13024-018-0297-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muffat J., Li Y., Yuan B., Mitalipova M., Omer A., Corcoran S., Bakiasi G., Tsai L.H., Aubourg P., Ransohoff R.M. Efficient derivation of microglia-like cells from human pluripotent stem cells. Nat. Med. 2016;22:1358–1367. doi: 10.1038/nm.4189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksanen M., Petersen A.J., Naumenko N., Puttonen K., Lehtonen Š., Gubert Olivé M., Shakirzyanova A., Leskelä S., Sarajärvi T. PSEN1 mutant iPSC-derived model reveals severe astrocyte pathology in Alzheimer's disease. Stem Cell Reports. 2017;9:1885–1897. doi: 10.1016/j.stemcr.2017.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ormel P.R., Vieira de Sá R., van Bodegraven E.J., Karst H., Harschnitz O., Sneeboer M.A.M., Johansen L.E., van Dijk R.E., Scheefhals N., Berdenis van Berlekom A. Microglia innately develop within cerebral organoids. Nat. Commun. 2018;9:4167. doi: 10.1038/s41467-018-06684-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandya H., Shen M.J., Ichikawa D.M., Sedlock A.B., Choi Y., Johnson K.R., Kim G., Brown M.A., Elkahloun A.G., Maric D., Sweeney C.L. Differentiation of human and murine induced pluripotent stem cells to microglia-like cells. Nat. Neurosci. 2017;20:753–759. doi: 10.1038/nn.4534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paolicelli R.C., Bolasco G., Pagani F., Maggi L., Scianni M., Panzanelli P., Giustetto M., Ferreira T.A., Guiducci E., Dumas L., Ragozzino D. Synaptic pruning by microglia is necessary for normal brain development. Science. 2011;333:1456–1458. doi: 10.1126/science.1202529. [DOI] [PubMed] [Google Scholar]
- Ran F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skene N.G., Grant S.G. Identification of vulnerable cell types in major brain disorders using single cell transcriptomes and expression weighted cell type enrichment. Front. Neurosci. 2016;10:16. doi: 10.3389/fnins.2016.00016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith A.M., Dragunow M. The human side of microglia. Trends Neurosci. 2014;37:125–135. doi: 10.1016/j.tins.2013.12.001. [DOI] [PubMed] [Google Scholar]
- Takata K., Kozaki T., Lee C.Z.W., Thion M.S., Otsuka M., Lim S., Utami K.H., Fidan K., Park D.S., Malleret B. Induced-pluripotent-stem-cell-derived primitive macrophages provide a platform for modeling tissue-resident macrophage differentiation and function. Immunity. 2017;47:183–198.e6. doi: 10.1016/j.immuni.2017.06.017. [DOI] [PubMed] [Google Scholar]
- Thion M.S., Ginhoux F., Garel S. Microglia and early brain development: an intimate journey. Science. 2018;362:185–189. doi: 10.1126/science.aat0474. [DOI] [PubMed] [Google Scholar]
- Tsyganov K., Perry A., Archer S., Powell D. RNAsik: a Pipeline for complete and reproducible RNA-seq analysis that runs anywhere with speed and ease. J. Open Source Softw. 2018;3(28):583. [Google Scholar]
- Zhang B., Gaiteri C., Bodea L.G., Wang Z., McElwee J., Podtelezhnikov A.A., Zhang C., Xie T., Tran L., Dobrin R. Integrated systems approach identifies genetic nodes and networks in late-onset Alzheimer's disease. Cell. 2013;153:707–720. doi: 10.1016/j.cell.2013.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequencing data have been deposited to GEO under accession number GEO: GSE125872. The accession number for the raw data reported in this paper is GEO: GSE125872.