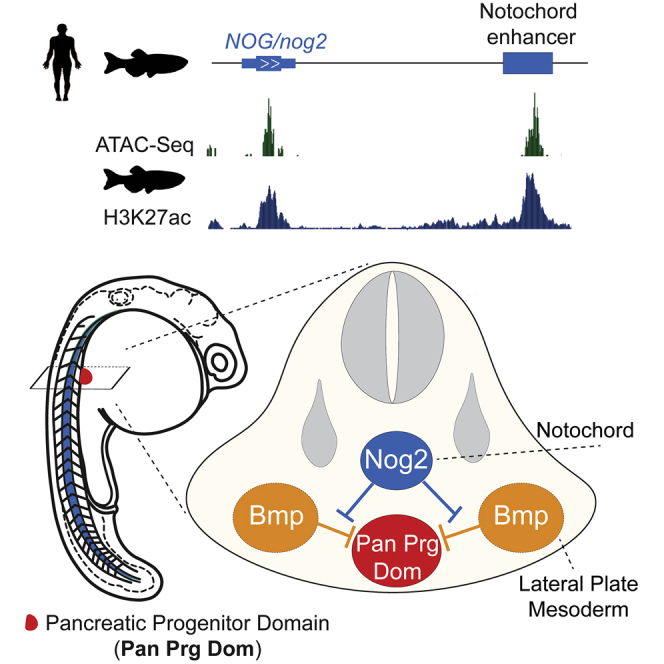
Summary
The notochord is an evolutionary novelty in vertebrates that functions as an important signaling center during development. Notochord ablation in chicken has demonstrated that it is crucial for pancreas development; however, the molecular mechanism has not been fully described. Here, we show that in zebrafish, the loss of function of nog2, a Bmp antagonist expressed in the notochord, impairs β cell differentiation, compatible with the antagonistic role of Bmp in β cell differentiation. In addition, we show that nog2 expression in the notochord is induced by at least one notochord enhancer and its loss of function reduces the number of pancreatic progenitors and impairs β cell differentiation. Tracing Nog2 diffusion, we show that Nog2 emanates from the notochord to the pancreas progenitor domain. Finally, we find a notochord enhancer in human and mice Nog genomic landscapes, suggesting that the acquisition of a Nog notochord enhancer occurred early in the vertebrate phylogeny and contributes to the development of complex organs like the pancreas.
Keywords: pancreas development, transcriptional regulation, signaling diffusion, notochord, zebrafish
Graphical Abstract
Highlights
-
•
nog2 loss of function leads to a reduced number of pancreatic progenitor and β cells
-
•
A tissue-specific enhancer controls the expression of nog2 in the notochord
-
•
The nog2 notochord enhancer is required for establishing a correctly sized pancreas
-
•
Regulation of notochord expression of Nog might be similar in many vertebrate lineages
Amorim et al. find that Nog2 is expressed in the zebrafish notochord by the action of a tissue-specific enhancer, and it diffuses to the pancreatic domain and controls its size. The identification of Nog enhancers in other vertebrate lineages suggests a conserved mechanism for pancreas development in vertebrates.
Introduction
The pancreas is an endoderm-derived organ that has several crucial roles, among them, controlling levels of glucose in the blood by producing insulin from β cells and aiding digestion by secreting digestive enzymes to the digestive system. The pancreas develops from a group of progenitor cells, originating from the endoderm developing gut, under the control of signaling pathways that determine its correct position and size (Prince et al., 2017; Sakhneny et al., 2019). One of these determinant signaling molecules is Bmp, which has various roles in pancreas development. These roles include the regulation of pancreatic size by controlling liver and pancreas fates, partly by restricting the expression of pdx1, an important top hierarchical gene present in pancreatic progenitor cells (Prince et al., 2017; Sakhneny et al., 2019). In zebrafish, and according to this model, bmp2b expressed and secreted from the lateral plate mesoderm generates a gradient at which high levels of Bmp2b restrict pancreatic size, favoring liver fate and establishing the development of the pancreas in a region aligned with the midline of the embryo (Chung et al., 2008, 2010). In this context, Bmp signaling works as negative regulator of pancreas development. Interestingly, it has been long postulated that the notochord, located at the midline of the embryo, dorsal to the pancreas, has a pro-pancreatic function (Prince et al., 2017; Sakhneny et al., 2019). Part of these observations derive from studies in which the notochord was surgically removed from chicken embryos, showing that in its absence, pancreatic markers such as insulin were defectively expressed, as well as early progenitor markers such as Pdx1 (Kim et al., 1997). These results suggest that a non-autonomous signal must emanate from the notochord to induce pancreatic fate. One possibility is that activin βB and fibroblast growth factor 2 (FGF2), expressed in the notochord, might have a role in pancreatic induction via repression of SHH expression in the endoderm (Hebrok et al., 1998). Nevertheless, ectopic pancreatic tissue is not observed in the endoderm of Shh mutant embryos (Hebrok et al., 2000), suggesting that inhibition of Shh signaling is permissive, rather than instructive, for pancreas fate (McCracken and Wells, 2012). A complementary hypothesis to be addressed is the presence of a Bmp antagonist expressed in the notochord that might be required for proper pancreatic induction; however, little is known about how Bmp signaling is modulated in the endoderm to effectively determine pancreatic size and position.
In zebrafish, one of the most notochord-specific Bmp antagonists is Noggin2 (Nog2) (Fürthauer et al., 1999). nog2 starts to be expressed by the end of gastrulation (10 h post-fertilization [hpf]) in the zebrafish axial mesoderm, which shortly gives rise to the notochord (Fürthauer et al., 1999). This expression is maintained up to the 18-somite stage (16 hpf), when the transcript slowly starts to disappear, and persists until 30 hpf in the posterior tip of the notochord. In this work, we disrupted the function of nog2, observed consistent impairment in the number of pancreatic progenitor cells and insulin-expressing cells, and showed nog2 to be an important Bmp antagonist required for pancreatic development. The introduction of an enhancer blocking insulator (Bessa et al., 2014) downstream of nog2 also impaired β cell differentiation and reduced the number of pancreatic progenitor cells, suggesting the existence of an enhancer required for the nog2 pancreatic function. By analyzing histone modifications associated with enhancer activity (histone H3 lysine 27 acetylation [H3K27ac] and histone H3 lysine 4 monomethylation [H3K4me1]) (Bogdanovic et al., 2012; Marlétaz et al., 2018; Rada-Iglesias et al., 2011) and performing circular chromosome conformation capture coupled with next-generation sequencing (4C-seq) (Zhao et al., 2006), we identified a nog2 notochord enhancer, among others. To further demonstrate that the notochord enhancer is crucial for the nog2 pancreatic function, we targeted it using CRISPR-Cas9, inducing genomic deletions (Letelier et al., 2018); complementing these assays, we used Cas9 endonuclease dead (dCas9) fused to a KRAB repressor domain (dCas9KRAB) (Thakore et al., 2015) and observed β cell differentiation impairment. Moreover, in heteroallelic mutant embryos for partial deletion of the notochord enhancer, we observed a reduced number of pancreatic progenitor cells and β cells when performing immunostaining for Nkx6.1 and insulin, respectively. In addition, we traced Nog2 in vivo and observed its diffusion from the notochord to the pancreatic progenitor domain, supporting its non-autonomous function in pancreas development. To understand whether this mechanism could also be present in mammals, we explored the human and mouse Nog regulatory landscapes, identifying notochord enhancers. Overall, these results suggest that Nog has acquired a notochord enhancer, most likely in the basis of the vertebrate phylogeny, that contributes to the crucial function of the notochord as an important signaling center for the development of complex organs such as the pancreas.
Results
nog2 Controls Non-autonomously the Embryonic Endocrine Pancreas Size
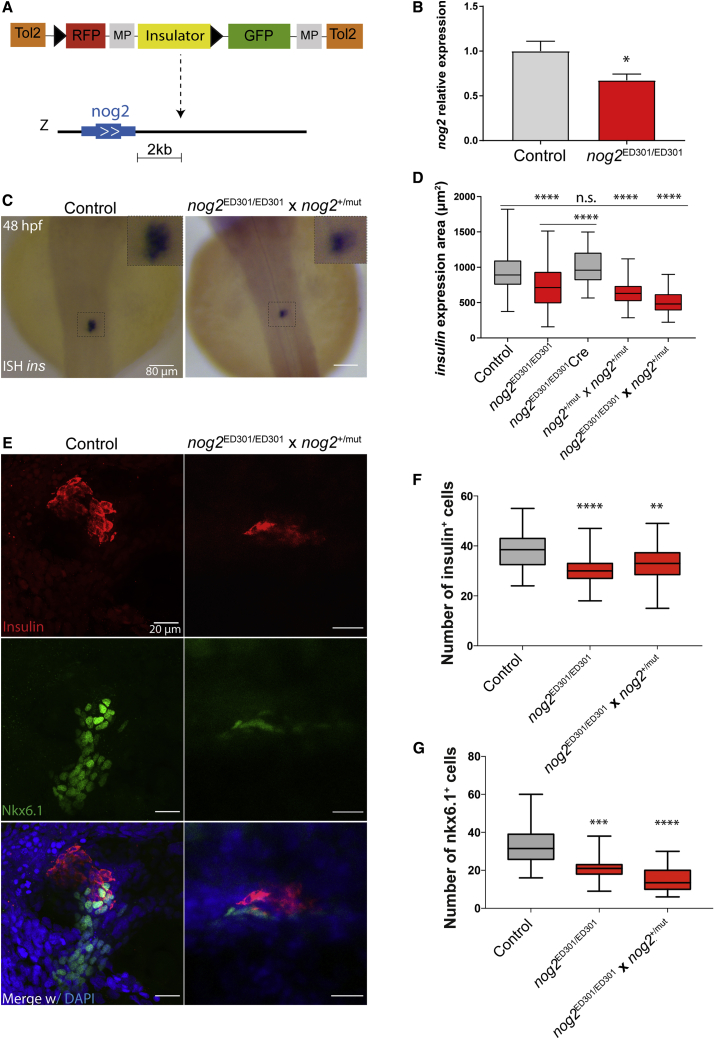
nog2 encodes a zebrafish Bmp antagonist that starts to be expressed at 10 hpf in the primordium of the notochord (Fürthauer et al., 1999). To identify possible non-coding genetic modifications affecting pancreas development, we mobilized a transposable element containing a potent enhancer blocking insulator (Bessa et al., 2014). We found a novel integration of this construct (ED301) located 2 kb downstream of nog2 (nog2ED301) (Figure 1A; Figure S1). Homozygous embryos for this integration showed a decrease of nog2 expression at 10 hpf that was detected by qPCR (Figure 1B). To determine whether the downregulation of nog2 could affect pancreas development, we performed in situ hybridization for insulin and determined that its area of expression was reduced in nog2ED301 homozygous embryos (Figures 1C and 1D) compared with control embryos. Next, to determine whether the phenotype observed in pancreas development was caused by the action of the insulator, we injected Cre recombinase in homozygous embryos for the nog2ED301 integration and performed in situ hybridization for insulin. Cre recombinase targeted two loxP cassettes flanking the insulator in the ED301 construct (Figure 1A). In this experimental condition, we observed a rescue of the decreased area of insulin expression (Figure 1D). These results suggest that the ectopic insulator might be blocking a cis-regulatory element of nog2 required for proper pancreas development. To validate nog2 as an important gene controlling pancreas development, we generated a nog2 mutant line (nog2mut) using the CRISPR-Cas9 targeting system. This mutant line shows a frameshift mutation in the nog2 gene (Figure S2A). Although nog2 is not expressed at the shield developmental stage, when the dorsal/ventral axis of the zebrafish embryo is established by a ventral gradient of Bmp, it is known that ectopic expression of nog2 interferes with Bmp signaling, causing dorsalization phenotypes (Fürthauer et al., 1999). Therefore, to test whether the nog2mut frameshift mutation is a loss of function of the nog2 gene, we injected mRNA of the frameshift version of nog2 in zebrafish embryos. We did not observe a dorsalization phenotype, contrasting the wild-type (WT) version of nog2 mRNA (Figures S2C and S2D). These results show that nog2mut is most likely a null mutant for nog2. We incrossed nog2+/mut, generating a mixture of WT, heterozygous, and homozygous embryos, and outcrossed nog2+/mut with nog2ED301/ED301. In both cases, the resulting embryos had a significant decrease in the area of expression of insulin, demonstrating that nog2 is required for the differentiation of a correctly sized pancreas (Figure 1D). The decrease of the area of expression of insulin could be caused by a decrease in the number of β cells or by a decrease in their size. To discriminate between these two possible scenarios, we injected a morpholino against nog2 in a transgenic reporter line for insulin expression (diIorio et al., 2002) and observed a reduced number of β cells in the nog2 morpholino-injected animals (Figures S2E and S2F). In addition, we performed immunostaining for Nkx6.1, to label pancreatic progenitor cells, and for insulin, to label differentiated β cells, in homozygous embryos for the nog2ED301 integration. We observed that the numbers of both Nkx6.1 and insulin cells decreased compared with controls. A similar decrease in the average number of Nkx6.1- and insulin-expressing cells was also detected in a 50% mixed population of embryos carrying the heteroallelic combination of nog2ED301/mut and nog2+/ED301 (Figures 1E to 1G). These results suggest that the reduced number of insulin-expressing cells might be the consequence of a smaller pancreatic progenitor domain, indicating that nog2 is required for the development of a properly sized pancreas.
Figure 1.
nog2 Is Required for Proper Pancreas Development
(A) ED301 zebrafish line corresponds to an ED transposon integration containing a potent enhancer blocking insulator (yellow) and mapped 2 kb downstream of nog2. The loxP sequences are depicted as black triangles (see also Figure S1). Scale bar represents 2 kilobases.
(B) In the ED301 zebrafish line, nog2 expression is downregulated, as detected by qPCR performed in three biological replicates of 30 embryos each (error bars represent SD; ∗p < 0.05).
(C) In situ hybridization of insulin in 48 hpf representative embryos from an outcross of nog2ED301/ED301 and nog2+/mut lines compared with control embryos. Scale bar represents 80 μm.
(D) Quantification of the insulin expression area detected by in situ hybridization in control (n = 194), nog2 ED301/ED301 (n = 93), nog2ED301/ED301 injected with Cre recombinase (n = 32), nog2+/mut incross (n = 115), and nog2ED301/ED301, nog2+/mut outcross embryos (n = 36) at 48 hpf. Error bars represent SD; ∗∗∗∗p < 0.0001.
(E) Representative confocal images of 48 hpf zebrafish embryos counterstained with a DAPI nuclear marker (blue), an anti-insulin antibody marking β cells (red), and an anti-Nkx6.1 antibody marking pancreatic progenitor cells (green). Images represent the maximum-intensity z projection of several focal planes obtained in a Leica Sp5 confocal microscope using a 40× objective. Scale bars represent 20 μm.
(F) Quantification of the number of insulin-expressing cells in nog2ED301/ED301 and nog2ED301/ED301, nog2+/mut outcross embryos compared with controls (n ≥ 30). Error bars represent SD; ∗∗∗∗p < 0.0001, ∗∗p < 0.01.
(G) Quantification of the number of nkx6.1-expressing cells in nog2 ED301/ED301 and nog2ED301/ED301, nog2+/mut outcross embryos compared with controls (n ≥ 18). Error bars represent SD; ∗∗∗∗p < 0.0001, ∗∗∗p < 0.001.
nog2 Genomic Regulatory Landscape Contains a Notochord-Specific Enhancer
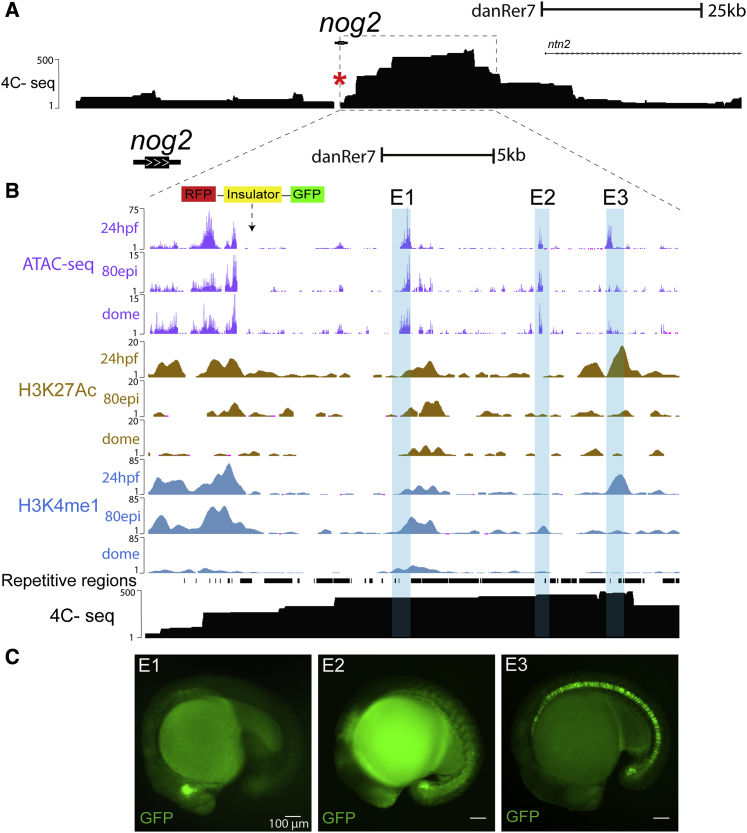
Nog proteins are well-known diffusible molecules that act as Bmp antagonists (Inomata et al., 2013). nog2 function in pancreas development might be explained by its expression in the notochord and subsequent diffusion to the endoderm, which has a non-autonomous, pro-pancreatic role. The nog2ED301 zebrafish line contains an insulator integration in the genomic landscape of nog2 that causes its transcriptional downregulation. This leads us to postulate that an essential enhancer for the nog2 pancreatic function should be present in the nog2 regulatory landscape, downstream of the insulator integration. To identify the possible cis-regulatory element that might be triggering the expression of nog2 in the notochord, we analyzed the landscape of nog2, looking for regions of open chromatin, as determined by Assay for Transposase-Accessible Chromatin using sequencing (ATAC-seq), and enriched for H3K27ac and H3K4me1, two histone marks associated with enhancer activity (Figures 2A and 2B). Because these histone modifications probably occur before the onset of nog2 expression, starting in the notochord at 10 hpf, we looked at histone marks at the developmental stages of dome, 80% epiboly, and 24 hpf (Bogdanovic et al., 2012; Marlétaz et al., 2018). In addition, we performed 4C-seq using the nog2 promoter as the point of view to identify the subset of putative cis-regulatory sequences that interact with the promoter of nog2 and therefore belong specifically to the regulatory landscape of this gene (Figure S3A). Using these strategies, we defined 3 putative nog2 enhancers (Figure 2B, E1 to E3). We performed enhancer transgenesis assays for these sequences and verified that all 3 sequences are enhancers, together recapitulating the early expression pattern of nog2 (Figure 2C; Figure S1E). Of the 3 sequences, E3 was shown to be a strong notochord enhancer; E2 was shown to be is a forebrain, hindbrain, and somite enhancer; and E1 was shown to be a strong ventral forebrain enhancer (Figure 2C).
Figure 2.
nog2 Genomic Regulatory Landscape Contains a Notochord-Specific Enhancer
(A) Genomic landscape of nog2, represented in the University of California Santa Cruz Genomics Institute (UCSC) genome browser (Kent et al., 2002), comprising more than 100 kb. The black track represents chromatin interaction points detected by 4C-seq using the nog2 promoter as the viewpoint (red asterisk). Scale bar represents 25 kilobases.
(B) Zoom-in in the locus of nog2 comprising approximately 25 kb. Representation of the ED transposon integration 2 kb downstream of nog2. Purple (top), brown (middle), and blue (bottom) tracks represent the ATAC-seq and chromatin immunoprecipitation sequencing (ChIP-seq) signal for H3K27ac and H3K4me1, respectively, at three developmental times: 24 hpf, 80% epiboly (80 epi), and dome. The black track represents the 4C-seq signal at 24 hpf using the nog2 promoter as the viewpoint. Pale blue boxes highlight selected sequences for enhancer activity assays (E1 to E3). Two replicates of 4C data are shown in Figure S3. Scale bar represents 5 kilobases.
(C) Representative images of GFP reporter lines for enhancers E1 to E3 (see also Figure S1). Scale bars represent 100 μm.
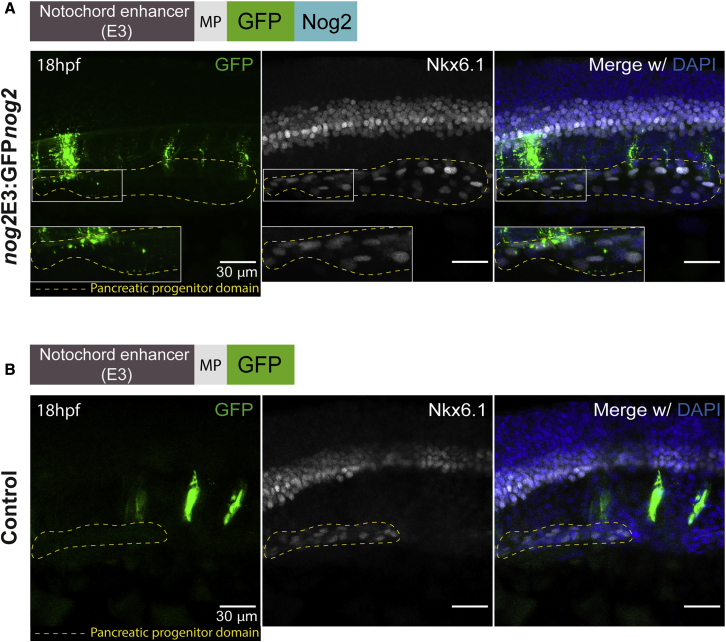
Next, we wanted to address whether nog2 can diffuse from the notochord to the pancreatic progenitor domain. To test this, first we built a construct containing a Nog2-encoded protein fused to GFP. To test whether the GFPNog2 fusion protein maintained its functionality, we injected GFPnog2 mRNA in 1-cell-stage embryos and observed the presence of dorsalization phenotypes, as in the case of WT nog2 mRNA (Figures S2C and S2D). Then, we built a transposable element containing the nog2E3 notochord enhancer and a minimal promoter upstream of GFPnog2 (nog2E3:GFPnog2) to be able to track Nog2 diffusion (Figure 3A; Figure S2B). Upon injection of zebrafish embryos with this construct and Tol2 mRNA, we performed staining using an anti-Nkx6.1 antibody that labels pancreatic progenitor cells (Binot et al., 2010; Hesselson et al., 2011). We observed clear colocalization of GFP aggregates in the pancreatic progenitor domain (Figure 3A) in 16 of 20 embryos (80%), as analyzed by confocal microscopy, showing that Nog2 can diffuse from the notochord to the pancreatic field in the endoderm. These results contrast those of controls expressing only GFP (Figure 3B), in which GFP aggregates were not seen outside of the notochord in 18 analyzed embryos. These results help to explain the non-autonomous effect observed in the pancreas in the nog2 loss-of-function assays.
Figure 3.
Nog2 Diffuses from the Notochord to the Pancreatic Progenitor Domain
(A) Representative confocal image of a 18 hpf zebrafish embryo injected with the nog2E3:GFPnog2 construct. Of 20 embryos, 16 (80%) showed colocalization of GFPNog2 aggregates (green) with pancreatic progenitor cells, labeled with anti-Nkx6.1 antibody (white). Details of the colocalization can be observed in the inset (white box).
(B) Representative confocal image of a 18 hpf zebrafish embryo injected with nog2E3:GFP (control), in which the pattern of GFP expression (green) is restricted to the notochord. None of 18 analyzed embryos displayed colocalization of GFP aggregates with Nkx6.1-labeled cells. Embryos were counterstained with the nuclear marker DAPI (blue), and the endocrine progenitor domain is delimited by a yellow dashed line (see also Figure S2). Images represent the maximum-intensity z projection of several focal planes obtained in a Leica Sp5 confocal microscope using a 40× objective. Scale bars represent 30 μm.
The nog2 Notochord Enhancer Is Required for Proper Pancreas Development
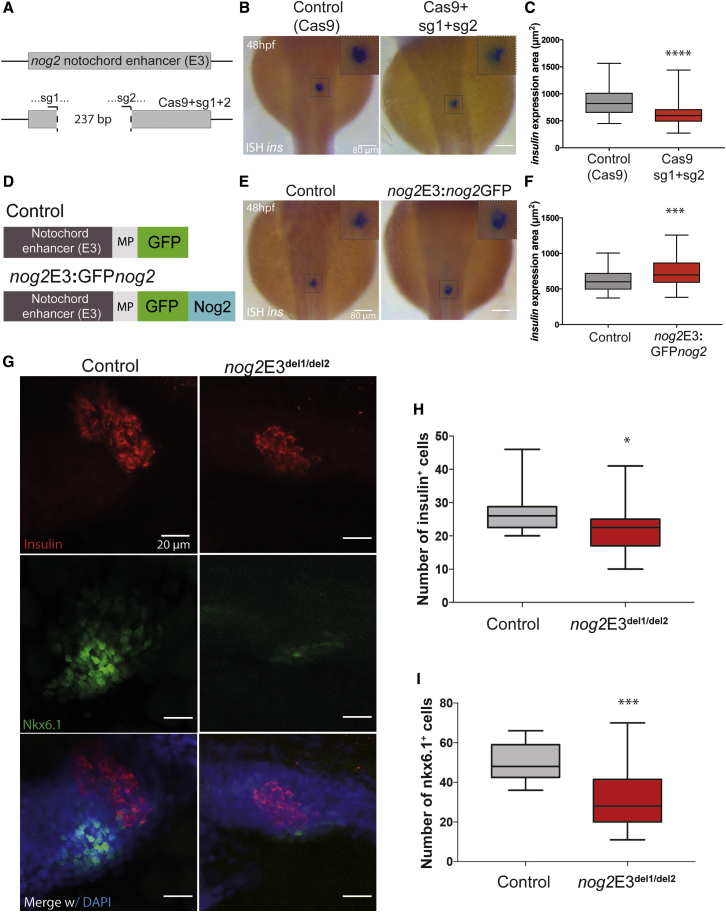
The nog2E3 notochord enhancer most likely controls the expression of nog2 in the notochord and is required for pancreatic development. To address this hypothesis, we synthetized a pair of single-guide RNAs (sgRNAs) 237 bp apart (nog2E3 sg1 and sg2) to target the nog2E3 notochord enhancer and generate genomic deletions for this cis-regulatory sequence. Embryos injected with Cas9 and the combination of nog2E3 sg1 and sg2 showed somatic deletions (Figure 4A; Figures S3E–S3G). In situ hybridization for insulin was performed in these injected embryos, together with controls injected with Cas9 alone, and the area of insulin expression was measured. We observed a decreased area of insulin expression for nog2E3 sg1 and sg2 (Figures 4A–4C). To validate these results, we injected a combination of nog2E3 sg1 and sg2 with dCas9-KRAB (Thakore et al., 2015). When compared with control embryos injected with dCas9-KRAB alone, we observed a reduction in the area of insulin expression (Figures S3B–S3D), suggesting that the nog2E3 notochord enhancer is modulating the expression of nog2 in this tissue and is required for proper pancreas development. Indeed, partial deletion of the nog2E3 notochord enhancer shows a tendency for decreased nog2 expression (Figure S4). Furthermore, we observed a decrease in the average number of Nkx6.1- and insulin-expressing cells in homozygous embryos for this deletion (Figures 4G–4I), recapitulating the phenotype seen in the nog2ED301 integration mutants. Accordingly, ectopic expression of nog2 in the notochord should result in increased pancreas size. To test this, we injected the nog2E3:GFPnog2 construct and measured the size of the area of expression of insulin, noticing an incremental change in the pancreatic field area (Figures 4D–4F). These results demonstrate that the nog2E3 notochord enhancer is required for the proper function of nog2 in pancreatic development and to establish a correctly sized pancreas.
Figure 4.
The nog2 Notochord Enhancer Is Required for Proper Pancreas Development
(A) Representation of the WT nog2E3 enhancer (above) and somatic deletions (below) generated by the injection of the Cas9 protein, together with sg1 and sg2 targeting two regions of the sequence 237 bp apart (see also Figure S3).
(B) In situ hybridization of insulin in 48 hpf representative embryos, injected with the Cas9 protein, and two sgRNAs compared with control embryos, injected with only Cas9. Scale bars represent 80 μm.
(C) Quantification of the insulin expression area detected by in situ hybridization in injected embryos at 48 hpf compared with controls. Error bars represent SD; ∗∗∗∗p < 0.0001. In all cases, n ≥ 97.
(D) Diagram of the nog2E3:GFPnog2 construct used to achieve notochord-specific overexpression of GFPnog2 and the respective control.
(E) In situ hybridization of insulin in 48 hpf representative embryos injected with nog2E3:GFPnog2 compared with control embryos. Scale bars represent 80 μm.
(F) Quantification of the insulin expression area detected by in situ hybridization in injected embryos at 48 hpf compared with controls. Error bars represent SD; ∗∗∗p < 0.001. In all cases, n ≥ 38.
(G) Representative confocal images of 48 hpf zebrafish embryos counterstained with a DAPI nuclear marker (blue), an anti-insulin antibody marking β cells (red), and an anti-Nkx6.1 antibody marking pancreatic progenitor cells (green). Images represent the maximum-intensity z projection of several focal planes obtained in a Leica Sp5 confocal microscope using a 40× objective. Scale bars represent 20 μm.
(H) Quantification of the number of insulin-expressing cells in nog2E3del1/del2 embryos compared with controls (n ≥ 19). Error bars represent SD; ∗p < 0.05.
(I) Quantification of the number of nkx6.1-expressing cells in nog2E3del1/del2 embryos compared with controls (n ≥ 13). Error bars represent SD; ∗∗∗p < 0.001.
The Expression of Nog in the Notochord Is Conserved in Vertebrates and Is Mediated by Equivalent Notochord Enhancers
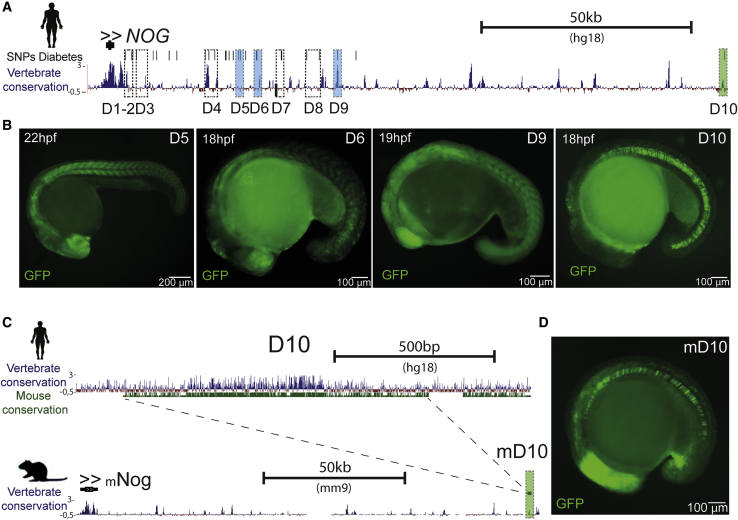
Next, we wanted to understand whether NOG expression in humans could be controlled by a notochord enhancer. To clarify this, we screened the human NOG genomic landscape for putative enhancers, based on sequence conservation and SNPs associated with type 2 diabetes and hyperglycemic traits (Figure 5A) (Morris et al., 2012). We selected 9 candidate human sequences (D12 to D10) (Figure 5A) to perform enhancer reporter assays in zebrafish and determined whether any of these sequences is a notochord enhancer. Preliminary results obtained by the generation of mosaic zebrafish transgenic embryos showed that sequence D10 was able to drive expression of GFP in the notochord (Figure S5D). We confirmed this result by generating stable zebrafish transgenic reporter lines for the selected sequences (Figure 5A). Apart from the D10 notochord enhancer, we found that D5 is a somite, forebrain, and hindbrain enhancer; D6 is a forebrain and somite enhancer; and D9 has broad enhancer activity and does not show high tissue specificity (Figure 5B; Figures S5A–S5D). Enhancers D5, D6, D9, and D10 were confirmed and obtained concordant expression patterns in at least two independent genomic integrations of the reporter constructs (Figures S5A–S5D). Sequences D7, D11, and D12 showed unreproducible patterns, and D12 and D8 showed no expression of the reporter gene. The expression patterns of the different human enhancers correlate with the known expression of Nog in mice, chicken, and Xenopus (Figure S5E), including the notochord and nervous system, suggesting that the Nog vertebrate genomic landscape has functionally similar cis-regulatory elements.
Figure 5.
The Expression of Nog in the Notochord Is Conserved in Vertebrates and Is Mediated by Equivalent Notochord Enhancers
(A) Genomic landscape of human NOG, comprising approximately 150 kb. Black vertical lines represent SNPs associated with diabetes from the Diabetes Genetics Replication and Meta-analysis (DIAGRAM) Consortium (p < 0.02) (Morris et al., 2012; Pasquali et al., 2014). The dark blue track represents vertebrate conservation and shows multiple alignments of 100 vertebrate species and measurements of evolutionary conservation (phyloP) (Pollard et al., 2010). Dashed line boxes are putative enhancer sequences selected for enhancer assays. Sequences in pale blue shaded boxes showed reproducible expression patterns. D10 (green shaded box) showed enhancer activity in the notochord. Scale bar represents 50 kilobases.
(B) Representative images of GFP reporter lines for enhancers D5, D6, D9, and D10 (see also Figure S5). Scale bars represent 100 or 200 μm.
(C) Genomic landscape of the human D10 sequence, showing vertebrate (blue track) and mouse conservation (green track) (above). Genomic landscape of mouse Nog, including the conserved mouse D10 (mD10) sequence, located approximately 150 kb downstream of the promoter (see also Figures S5 and S6). Scale bars represent 500 basepairs or 50 kilobases.
(D) Representative image of the GFP reporter line for the mouse enhancer mD10. Scale bar represents 100 μm.
We were able to align the human D10 enhancer with the genome of other mammal species, suggesting that this cis-regulatory element is conserved. The most distantly related species with which D10 can be aligned are two representatives of marsupials, opossum (Monodelphis domestica) and Tasmanian devil (Sarcophilus harrisii), that diverged from humans around 180 million years ago (Figure S5F). Interestingly, in four analyzed cases—mouse, dog, armadillo, and opossum—the D10 ortholog sequences are always found in synteny with the respective Nog ortholog genes (Figures S6A and S6B), suggesting that these conserved sequences belong to the regulatory landscape of Nog. Furthermore, looking at HiC data from human and mouse tissues, it is possible to appreciate that NOG and D10 enhancer, as well as the corresponding mouse ortholog sequences, are within the same topologically associating domain (TAD) (Dixon et al., 2012) in a genomic region enriched for chromatin interactions (Figures S6C–S6F). Next, we wanted to understand whether the conserved D10 orthologs correspond to notochord enhancers. In mice, the D10 ortholog sequence is located 157 kb downstream of Nog (Figure 5C). We isolated this sequence, tested it for enhancer activity using reporter assays, and demonstrated that this sequence is also a notochord enhancer (Figure 5D). Overall, these results suggest that the regulatory mechanism for the expression of Nog in the notochord might be equivalent in many vertebrate lineages.
Discussion
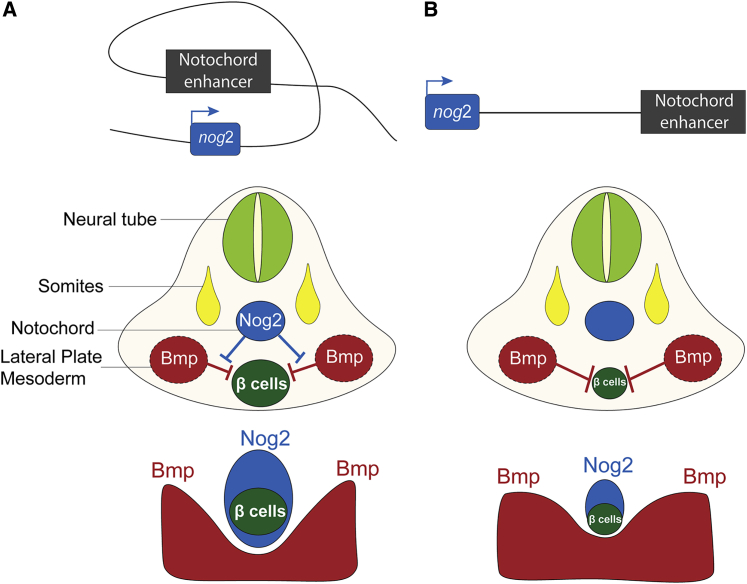
Non-autonomous cell signaling is crucial for proper organogenesis. It has been demonstrated that several signaling pathways regulate pancreatic development in vertebrates. Among them, Hedgehog signaling represses the expression of the pancreatic multipotent progenitor marker Pdx1 during late gastrulation (Tehrani and Lin, 2011). In mice, activin βB and Fgf2 expressed in the notochord might repress Shh expression in the endoderm (Hebrok et al., 1998). However, ectopic pancreatic tissue is not observed in Shh mutant embryos (Hebrok et al., 2000), suggesting that Shh inhibition is permissive, rather than instructive, for pancreas fate (McCracken and Wells, 2012). Experiments of notochord ablation in chicken embryos showed that pancreatic development is impaired (Kim et al., 1997). These results highlight the notochord as an important signaling center for pancreatic development. In subsequent experiments using coculture of mice tissues, it has been shown that the notochord is sufficient to trigger expression of Pdx1 in the endoderm, but not Insulin, whereas cells from the dorsal aorta are able to trigger the expression of Pdx1 and Insulin. These results further suggest that the notochord and the dorsal aorta have a role in the control of pancreas progenitor cells, whereas aorta endothelial cells have an extra role in the differentiation of β cells (Lammert et al., 2001). Here we show that Nog is expressed in the notochord, controlled transversely in humans, mice, and zebrafish by at least one notochord enhancer, and that nog2 gain and loss of function in zebrafish correlate with an increase and a decrease of pancreatic differentiation, respectively. In addition, we show that Nog2 diffuses from the notochord to the pancreatic domain, as detected indirectly by an ectopic expression system (Figure 3). These results indicate that Nog is one of the crucial pro-pancreatic signaling molecules emanating from the notochord. In agreement with this, Nog is a well-known Bmp signaling inhibitor (Fürthauer et al., 1999), and this signaling pathway is known to antagonize pancreas differentiation (Chung et al., 2008, 2010) while promoting liver differentiation. In this model, the Bmp2b gradient emanating from the lateral plate mesoderm is not enough for the establishment of a proper pancreatic progenitor domain by control of the expression of pdx1. Nog2 from the notochord, located in the midline, counteracts the lateral signaling of Bmp2b, allowing the proper placement and proper size of the pancreatic progenitor domain in the midline (Figure 6). In absence of Nog2 from the notochord, Bmp2b signaling is most likely expanded, resulting in a reduced pancreatic progenitor domain that will cause partial impairment of β cell development (Figure 6). These phenotypes are recapitulated by impairing a nog2 notochord enhancer, either by the action of an insulator or by the deletion of the enhancer. Nevertheless, in these genetic backgrounds, the downregulation of nog2 is more robustly detected in the insulator background, suggesting the existence of several notochord enhancers or that partial deletion of the enhancer results in partial loss of expression of nog2. In both cases, knockdown of nog2 is sufficient to induce a pancreatic phenotype.
Figure 6.
Proper Pancreas Development Requires the Expression of Nog2 in the Notochord
(A) Diagram of the nog2 notochord enhancer driving expression of nog2. Nog2 diffusion from the notochord, located in the midline of the embryo, counteracts the signaling of Bmp2b from the lateral plate mesoderm, allowing the proper placement and proper size of the endocrine pancreas.
(B) When the activity of the nog2 notochord enhancer is disrupted, nog2 expression in the notochord is reduced. In this case, Bmp2b signaling is expanded, impairing β cell development and reducing pancreas size.
Although nog2 has a clear pro-pancreatic role during zebrafish development, null mutants for nog2 do not show complete loss of pancreas differentiation. Chicken notochord ablation has been shown to induce complete loss of pancreatic differentiation (Kim et al., 1997). Therefore, it is possible that apart from nog2, there might be synergistic action with other pro-pancreatic signaling molecules diffusing from the notochord.
In this work, we postulate that the function of NOG is conserved in humans. Supporting this, the genomic landscape of NOG contains a notochord enhancer, clearly showing the potential of this gene to be expressed in these cells. A study of the human notochordal transcriptome during embryogenesis was able to detect NOG expression during development (Rodrigues-Pinto et al., 2018), specifically at a late developmental time in human pancreatic formation. This interpretation is strengthened by advances in in vitro differentiation of human β cells from human embryonic stem cells. Optimization protocols have empirically determined that the addition of NOG was able to improve the efficiency of such protocols, clarifying the importance of BMP inhibition in the proper development of the human pancreas (Kroon et al., 2008; Kumar et al., 2014; Mfopou et al., 2010; Zhang et al., 2009). Cai and colleagues explored the role of NOG in human β cell differentiation, showing that the inclusion of NOG in the media resulted in inhibition of phosphorylation of Smad1/5/8, a BMP signaling transducer (Cai et al., 2010). From an evolutionary point of view, it is known that the vertebrate phylum contains at least three distinct Nog genes: Nog1, Nog2, and Nog3 (Eroshkin et al., 2006). In mammals, only Nog1 was kept, most likely retaining all functions and expression patterns of the three ancestral genes from the vertebrate phylum. An interesting open question relates to the emergence of the cis-regulatory module of the notochord that in humans and mice is present in the landscape of Nog, the ortholog of the zebrafish nog1, whereas in zebrafish, the notochord enhancer is present in the landscape of nog2. This could be explained by the independent recruitment of recently evolved notochord enhancers, as observed in the rapid evolution of cis-regulatory sequences of the mammal liver (Villar et al., 2015). An alternative explanation could be the existence of an ancestral regulatory module driving the expression of Nog in the notochord that upon gene duplication degenerated in particular Nog genes. In the tetrapod lineage, the notochord cis-regulatory elements were most likely kept in the landscape of Nog1, a scenario supported by (1) the high conservation of the enhancer from humans to marsupials (Figure S6), (2) the expression of Nog1 in the notochord from humans to Xenopus (Figure 5; Figure S5), and (3) the functional characterization of the mice and human Nog notochord enhancers. In contrast, the existence of the notochord enhancer in the landscape of zebrafish nog2 suggests that this enhancer was kept in this locus in the ray-finned fish lineage. Most likely, the acquisition of the Nog notochord enhancer occurred after the divergence of urochordates, because expression of Nog is not detected in the sea squirt Ciona intestinalis or in a cephalochordate, the amphioxus Branchiostoma lanceolatum. Interestingly, insulin-producing cells in basal cephalochordates are not organized in a distinct organ and are instead disseminated in the gut (Arntfield and van der Kooy, 2011). Finally, and in light of the axochord hypothesis (Brunet et al., 2015), the Nog notochord enhancer might have an even more ancient origin. Supporting this hypothesis, the annelids Terebratalia transversa and Platymeris dumerilii show expression of Nog in the axochord, a structure reminiscent of the notochord that has a mixture of muscle and notochord double properties (Lauri et al., 2014; Passamaneck et al., 2015).
In summary, in this work, we demonstrate the transversal existence of a notochord enhancer in humans, mice, and zebrafish in the landscape of Nog, a known Bmp antagonist. We show that this enhancer is required for proper pancreas differentiation in the zebrafish, contributing to understanding of the notochord as an important signaling center required for the complex development of the vertebrate pancreas.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal anti-Nkx6.1 | Developmental Studies Hybridoma Bank | Cat# F55A12; RRID: AB_532379 |
| Rabbit polyclonal anti-insulin | abcam | Cat# ab210560 |
| Deposited Data | ||
| 4C-seq data | This paper | ENA: PRJEB36984 |
| UCSC session with 4C-seq processed bedgraph tracks (smoothened to windows containing 30 fragments) | This paper | http://genome-euro.ucsc.edu/cgi-bin/hgTracks?db=danRer7&lastVirtModeType=default&lastVirtModeExtraState=&virtModeType=default&virtMode=0&nonVirtPosition=&position=chr24%3A40302659%2D40356334&hgsid=239502130_A8rG2FyY9qsTK3VDD7dRiuUMjRi5 |
| Experimental Models: Organisms/Strains | ||
| Zebrafish: WT AB | This paper | N/A |
| Zebrafish: WT TU | This paper | N/A |
| Zebrafish: nog2ED301 | This paper | N/A |
| Zebrafish: nog2mut | This paper | N/A |
| Zebrafish: nog2E3del1/del2 | This paper | N/A |
| Zebrafish: Tg (Ins:GFP) | This paper | N/A |
| Zebrafish: nog2E1:GFP | This paper | N/A |
| Zebrafish: nog2E2:GFP | This paper | N/A |
| Zebrafish: nog2E3:GFP | This paper | N/A |
| Zebrafish: NOGD10:GFP | This paper | N/A |
| Zebrafish: NOGD5:GFP | This paper | N/A |
| Zebrafish: NOGD6:GFP | This paper | N/A |
| Zebrafish: NOGD9:GFP | This paper | N/A |
| Zebrafish: mD10:GFP | This paper | N/A |
| Oligonucleotides | ||
| See Table S1 for primers and oligonucleotides used in this paper | ||
| Recombinant DNA | ||
| Zebrafish Enhancer Detection (ZED) Vector | Bessa et al., 2009 | N/A |
| pminiTol2- Z48-Gw-CARGFP | Bessa et al., 2009 | N/A |
| pDR274 | Hwang et al., 2013 | Addgene #42250 |
| nog2E3:GFPnog2 | This paper | N/A |
| Software and Algorithms | ||
| ImageJ | Schneider et al., 2012 | https://imagej.nih.gov/ij/ |
| Graphpad Prism | N/A | https://www.graphpad.com/ |
| bowtie | Langmead et al., 2009 | http://bowtie-bio.sourceforge.net/index.shtml |
| Bedtools | Quinlan and Hall., 2010 | https://github.com/arq5x/bedtools2 |
| 4C-seq processing perl script | This paper | Available upon request to Juan Tena (jjtenagu@upo.es) |
| Other | ||
| Fastqc reports from the raw 4C-seq fastq files from this study | This paper | Data S1 |
Resource Availability
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, José Carlos Ribeiro Bessa (jose.bessa@ibmc.up.pt)
Materials Availability
All unique/stable reagents generated in this study are available from the Lead Contact with a completed Materials Transfer Agreement.
Data and Code Availability
Raw reads from 4C experiments (fastq files of the 2 replicates) are accessible in the European Nucleotide Archive (ENA, https://www.ebi.ac.uk/ena, project PRJEB36984, study ERP120260, runs ERR3964955 and ERR3964956). The processed signal is available for visualization in the following UCSC genome browser session http://genomeeuro.ucsc.edu/s/vdr/danRer7_4Cseq_nog2. Perl script for processing 4C seq data is available upon request.
Experimental Model and Subject Details
Zebrafish
Zebrafish (Danio rerio) fish were handled according to European animal welfare regulations and standard protocols. The Animal Facility of the i3S is licensed by the Portuguese official veterinary department (DGAV) and accredited by the AAALAC (June, 2018), complies with Portuguese law (Portaria 1005/02 and Portaria 1131/97), the 2010/63/EU Directive, and follows the FELASA (Federation of European Laboratory Animal Science Associations) guidelines and recommendations concerning laboratory animal welfare. The current project is licensed by DGAV (reference: 0421/000/000/2016). Wild-Type (WT) zebrafish animals from AB or TU strain were used in this study. The following genotypes were used, as described in the text and Key Resources Table: nog2ED301, nog2mut, nog2E3del1/del2, Tg (Ins:GFP), nog2E1:GFP, nog2E2:GFP, nog2E3:GFP, NOGD10:GFP, NOGD5:GFP, NOGD9:GFP and mD10:GFP. Adult animals are kept at a controlled temperature (25°C) and under a 14h light-8h dark photoperiod. Recirculated water is continuously monitored regarding pH (7), salinity (900 μs) and temperature (28°C) values. In order to obtain zebrafish embryos, adult zebrafish were bred in ratios of 3 females to 2 males or 1 male to 1 female, depending on the purpose of the experiment. Embryos ranging from 16hpf to 48hpf were used.
Method Details
Zebrafish transgenesis
ED301 line was generated as described by Bessa et al. (2014). Enhancer reporter assays were performed by cloning the selected genomic fragments into the ZED vector (Bessa et al., 2009). For the mouse sequence mD10 only, the pminiTol2- Z48-Gw-CARGFP vector was used (Bessa et al., 2009). Briefly, genomic regions of interest were PCR amplified using iMAXII polymerase and specific primers (Table S1). TA cloning was used to introduce PCR products in the pCR8/GW/TOPO vector, as described in the pCR8/GW/TOPO TA Cloning Kit (ThermoFisher Scientific). To clone these sequences in the destination vectors, the Gateway LR Clonase II Enzyme mix (ThermoFisher Scientific) and respective protocol was used. Vectors where purified using phenol:cloroform and injected in one-cell-stage zebrafish embryos (AB strain), together with mRNA encoding Tol2 protein, in a 1:1 ratio and a final concentration of 25 ng/ul each. Injected embryos showing reporter expression were grown to adulthood and germinal line transmission was determined by outcrossing injected fish with wild-type animals and isolating F1 embryos with GFP expression. Embryos were kept in E3 medium with 0.003% 1-phenyl-2-thiourea (SigmaAldrich) to prevent pigmentation and were staged according to Kimmel et al. (1995). Embryos showing GFP expression were anesthetized by adding tricaine (ethyl -3-aminobenzoate; SigmaAldrich) to the E3 medium, and expression patterns were documented at 18-24 hpf, using the Leica M205 stereomicroscope. Post-imaging analysis was done in Fiji-ImageJ (Schneider et al., 2012). Stable transgenic lines were selected to grow and maintained by outcrossing with wild-type animals.
Genomic DNA extraction
Batches of embryos were incubated with Chelex 100 sodium form resin (Sigma #C7901 25 gr) 5% in TE buffer and with proteinase K 1 mg/mL (ThermoFisher Scientific), at 56°C for 3 hours. After that, proteinase K activity was inactivated by incubating the samples at 100°C for 10 minutes.
Cas9 targeting, sgRNA synthesis, mutant generation
The sgRNAs targeting the coding regions of nog2 and the regions flanking nog2E3 (Table S1) were designed using the Crisprscan software (MorenoMateos et al., 2015). These oligonucleotides were annealed in vitro by denaturation at 95°C for 5 min followed by slow cooling at RT. Annealed oligonucleotides (1:10) were further inserted into 100ng of pDR274 vector (Hwang et al., 2013; Addgene plasmid #42250) previously cut with BsaI (Anza, ThermoScientific). pDR274 vectors carrying sgRNA sequences were linearized with HindIII, purified with phenol:chloroform and transcribed with T7 RNA polymerase (ThermoFisher Scientific). sgRNAs were purified following the mRNA purification protocol previously described (Bessa et al., 2009). To achieve nog2 knockdown, one- cell-stage zebrafish embrycos were injected with Cas9 protein (PNA Bio Inc) (150 250 ng/ul) and 1 sgRNA (80125 ng/ul), targeting the coding region of nog2 (sg1nog2, Table S1). To perform the nog2E3 enhancer knockdown, embryos were coinjected with a mixture containing a combination of either Cas9 protein or dCas9-KRAB mRNA and 2 sgRNAs (80125 ng/ul each) (sg1E3, sg2E3, Table S1). In both cases, zebrafish mutants for nog2E3 were generated using the combination of sgRNA1 and sgRNA2. Enhancer deletion in zebrafish was detected by PCR with HOT FIREPol® DNA Polymerase (Solis BioDyne) using primers flanking the region of interest (see Figure S3 and Table S1). PCR product was analyzed by electrophoresis in 2% agarose gel, and further confirmed by Sanger sequencing. Injected embryos were grown and adult fish fins were cut to perform genomic DNA extraction and genotyping as described above (see Figure S4 and Table S1). One heterozygous male and one female having similar deletions were selected and crossed. Embryos were genotyped by fin clipping and genomic DNA extraction, followed by PCR amplification using the primers depicted in Table S1 (see also Figure S4).
Whole-mount in situ hybridization
Whole-mount in situ hybridization was performed on zebrafish embryos staged according to Kimmel et al. (1995), using antisense riboprobes targeting nog2 or insulin. nog2 probe was amplified from zebrafish cDNA, using the primers depicted in Table S1, and cloned into pCR2.1-TOPO TA vector (ThermoFisher Scientific). A vector for generating an insulin-targeting probe was kindly sent to us by Francesco Argenton (Argenton et al., 1999). Vectors were linearized using HindIII restriction enzyme (ThermoFisher Scientific). Antisense RNA probes were synthesized, using T7 RNA polymerase (ThermoFisher Scientific) and digoxigenin-labeled nucleotides (Roche) and kept in ISH hybridization buffer (HYB+; Thisse et al., 1993). Zebrafish embryos were collected at 18 or 48hpf and fixed overnight in 4% paraformaldehyde (PFA) in phosphatebuffered saline (PBS). Fixed embryos were washed in PBS-Tween 0.1% (PBT) and dehydrated in methanol (MeOH), in a series of MeOH/PBT solutions. Dehydrated embryos were kept at −20°C in 100% MeOH. After this, embryos were rehydrated through a reverse MeOH/PBT series and then treated with proteinase K 1 mg/mL (ThermoFisher Scientific), for 1–10 min. Embryos were washed in PBT, post-fixed in 4% PFA for 20 min and washed again in PBT. Pre-hybridization was performed by incubating embryos in HYB+ buffer for 3 h, at 70°C. After that, hybridization buffer was replaced with solution containing riboprobes at the concentration of 2 ng/uL and an overnight incubation was performed, at 70°C. In the next day, embryos were washed with a series of solutions containing washing buffer (HYB-) and sodium saline citrate buffer (SSC), as described by Thisse et al. (1993). Embryos were incubated for 1 h in blocking buffer, containing 2% normal goat serum (ThermoFisher Scientific) and 2 mg/mL bovine serum albumin (BSA; Sigma-Aldrich). Following this, embryos were incubated for 2 h with anti-digoxigenin-AP antibody (Sigma-Aldrich) 1:5000, dissolved in blocking buffer. Several rinses in PBT were performed. In the third day, embryos were washed with alkaline-phosphatase (AP) buffer (Thisse et al., 1993) and AP activity was revealed through incubation in NBT/BCIP substrate (Sigma-Aldrich), in the dark, for at least 30 min. Embryos were rinsed in PBT to stop the reaction and kept in 100% glycerol at 4°C. Coloration associated to the presence of mRNA was observed and photographed, using the Leica M205 stereomicroscope. Post-imaging analysis was done in Fiji-ImageJ.
Immunohistochemistry
Whole-mount immunohistochemistry was performed using mouse anti-rat nkx6.1 (Developmental Studies Hybridoma Bank) and rabbit anti-insulin (abcam) antisera. Goat antimouse IgG AlexaFluor 488 (Invitrogen), goat anti-rabbit IgG AlexaFluor 568 (Invitrogen) and goat anti-mouse IgG AlexaFluor 647 (Invitrogen) were used as secondary antisera. Briefly, embryos were fixed overnight in 4% paraformaldehyde (PFA) in phosphatebuffered saline (PBS). Fixed embryos were sequentially washed with PBS Triton 0.1%, then treated for 30 min to 2 h with PBS-Triton 1%, washed again with PBS-Triton 0.1% and incubated for 1h with PBS-T+BSA 5% before the incubation with the primary antisera, diluted 1:50 in PBS-T+BSA, overnight at 4°C. After extensive wash, embryos were incubated with the secondary antisera, overnight at 4°C, washed extensively and mounted in 50% glycerol in PBS. Embryos were stained with DAPI and 1,5 μm Z-series stacks were acquired on a Leica SP5 confocal microscope, using a 40x objective. Post-imaging analysis and quantifications were done in Fiji-ImageJ.
Nog2 overexpression vector
The sequence encoding the signaling domain of the Nog2 protein was PCR amplified from zebrafish genomic DNA, using primers carrying adaptor ends for BamHI and NcoI restriction enzymes. The mature encoding nog2 domain was amplified using 5′EcoRI and 3′ XhoI restriction sites. The PCR products were sequentially cloned into pCS2GFP (Bessa et al., 2008) by restriction enzyme cloning and the vector was sequenced. The nog2 signaling_GFP_mature nog2 sequence was introduced into the #237 pMEMCS vector of the tol2 kit (Kwan et al., 2007), by BamHI/ XhoI cloning. The minimal promoter–E3 enhancer cassette was obtained by BamHI/ SmaI digestion from the ZEDnog2E3 vector and cloned into the #228 p5EMCS vector of the tol2 kit. The generated #237 and #228 vectors were recombined with the #302 into the #394 destination vector using recombination driven by clonase II.
Morpholino injections
For the nog2 knockdown, 25 nL of 1 mM solution of morpholino targeting nog2 5′UTR or Danio rerio standard control morpholino (Table S1) (Gene tools, LLC) were injected in one-cell-stage Tg(ins:GFP) zebrafish embryos. Prior to the injection, morpholinos were incubated at 65°C for 10 min, vortexed, and diluted to 1mM in HyPure H20. Injected embryos were grown to 48 hpf and fixed overnight at 4°C in 4% paraformaldehyde in PBS. Fixed embryos were stained with DAPI and 1,5 μm Z-series stacks were acquired on a Leica SP5 confocal microscope, using a 40x objective. GFP + cells were counted using Fij-ImageJ. Embryos from 3 independent injections were analyzed for each condition.
Nog2 mRNA injections
25 nL of an 8 ng/uL solution of Nog2 WT or Nog2mut mRNAs were injected in one-cell-stage zebrafish embryos. Dorsalization phenotypes were observed and photographed around 24hpf, using the Leica M205 stereomicroscope. Post-imaging analysis was done in Fiji-ImageJ.
4C-seq
4C-seq was performed according with Fernández-Miñán et al. (2016), with minor alterations. For each replicate, at least 200 24hpf zebrafish embryos were collected. Embryos were fixed in 2% formaldehyde for 10 min, and stored at 80°C. Cell lysis was performed on ice, with a 15 mL Tenbroeck Homogenizer, not exceeding 10 min. Ligation was performed by using 60U T4 DNA Ligase (#EL0012, ThermoFisher Scientific). Restriction enzymes used were DpnII (#R0543M, NEB) and Csp6I (#ER0211, ThermoScientific) for the first and second cuts, respectively. Chromatin was purified by Amicon Ultra15 Centrifugal Filter Device (Milipore). 4C libraries were prepared for Illumina sequencing by using Expand Long Template Polymerase (Roche) with primers targeting the TSSs of nog2 (Table S1) and including Illumina adapters. Final PCR product was purified by High Pure PCR Product Purification Kit (Roche) and AMPure XP PCR purification kit (Agencourt AMPure XP). 4C-seq libraries were first inspected for quality control using FASTQC v.0.11.5 (Data S1) and demultiplexed using the script “demultiplex.py” from the FourCSeq package (Klein et al., 2015), allowing for 1 mismatch in the primer sequence. A custom perl script was used for subsequent processing, as previously described (Noordermeer et al., 2011, Splinter et al., 2012). Shortly, reads were aligned to the zebrafish genome (Zv9/danRer7) using Bowtie (Langmead et al., 2009), keeping only uniquely mapping reads (v1.1.2, m 1). Reads within fragments flanked by restriction sites of the same enzyme, checked with BedTools (Quinlan and Hall, 2010), or fragments smaller than 40 bp were filtered out. Mapped reads were then converted to readsperfirstenzymefragmentend units, and smoothed using a 30 fragment mean running window algorithm.
Quantitative PCR
Total mRNA was extracted from batches of 30 embryos or single embryos at 12 hpf with TRIzol (Invitrogen) and subsequent phenol:chloroform purification. Total mRNA was used for reverse transcription with random primers (SuperScript II, Invitrogen). Quantitative PCR was performed on a BioRad CFX96 realtime system using Sybr (iSybr Green Supermix, BioRad), using the primers depicted in Table S1. Each biological replicate was quantified in triplicates. eef1a and beta-actin or beta-actin alone were used as internal controls, and expression levels were compared using the DDCT method.
Quantification and Statistical Analysis
Assuming that the data are normally distributed, an unpaired t-test with Welch’s correction was performed to test the significance of differences among sample averages. For the comparison of nog2 relative expression between single embryos, a Mann-Whitney test to compare ranks was used. Asterisks in each figure represent significance level: ∗p < 0.05, ∗∗p < 0,01, ∗∗∗p < 0,001, n.s. = non-significant. Statistics and plots were done using GraphPad Prism 6.
Acknowledgments
We thank José Luís Gómez-Skarmeta and Paulo Pereira for helpful suggestions and critical reading of the manuscript, Vitor Silva for support in the phylogenetic tree design, and Marta Duque for help in designing the graphical abstract. This study was supported by the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation program (grant ERC2015StG680156ZPR). J.B. acknowledges Fundação para a Ciência e a Tecnologia (FCT) for an FCT Investigator position (grant IF/00654/2013). J.T. and J.P.A. are PhD fellows from FCT (grant SFRH/BD/126467/2016 to J.T. and grant SFRH/BD/145110/2019 to J.P.A.). M.G. was supported by the EnvMetaGen project via the European Union’s Horizon 2020 research and innovation program (grant 668981). The authors acknowledge the support of i3S Scientific Platform Advanced Light Microscopy, a member of the national infrastructure PPBI (Portuguese Platform of BioImaging) (supported by POCI010145FEDER022122). We acknowledge the I3S hpc facility, used for processing the 4C-seq data, and André Torres for the useful support. We also thank Francesco Argenton for providing us with a vector for generating an insulin-targeting probe. Finally, we thank Yolanda Roncero and Isabel Guedes for helping to screen and maintain the nog2ED301 zebrafish line and Tania Medeiros for preliminary work on the function of the notochord in pancreas development.
Author Contributions
J.B. conceived and supervised the project. J.P.A., A.G.-M., H.M., and J.B. conceived, designed, and analyzed the data. R.B.C. performed 4C-seq experiments. J.T. and M.G. performed computational analyses. S.N., S.R.-G., and J.M. contributed with reagents and materials. J.P.A., A.G.-M., and J.B. wrote the paper.
Declaration of Interests
The authors declare no competing interests.
Published: July 7, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2020.107862.
Supporting Citations
The following references appear in the Supplemental Information: Blanchette et al. (2004); Schmitt et al. (2016); Wang et al. (2018).
Supplemental Information
References
- Argenton F., Zecchin E., Bortolussi M. Early appearance of pancreatic hormone-expressing cells in the zebrafish embryo. Mech. Dev. 1999;87:217–221. doi: 10.1016/s0925-4773(99)00151-3. [DOI] [PubMed] [Google Scholar]
- Arntfield M.E., van der Kooy D. β-Cell evolution: How the pancreas borrowed from the brain: The shared toolbox of genes expressed by neural and pancreatic endocrine cells may reflect their evolutionary relationship. BioEssays. 2011;33:582–587. doi: 10.1002/bies.201100015. [DOI] [PubMed] [Google Scholar]
- Bessa J., Tavares M.J., Santos J., Kikuta H., Laplante M., Becker T.S., Gómez-Skarmeta J.L., Casares F. meis1 regulates cyclin D1 and c-myc expression, and controls the proliferation of the multipotent cells in the early developing zebrafish eye. Development. 2008;135:799–803. doi: 10.1242/dev.011932. [DOI] [PubMed] [Google Scholar]
- Bessa J., Tena J.J., de la Calle-Mustienes E., Fernández-Miñán A., Naranjo S., Fernández A., Montoliu L., Akalin A., Lenhard B., Casares F., Gómez-Skarmeta J.L. Zebrafish enhancer detection (ZED) vector: a new tool to facilitate transgenesis and the functional analysis of cis-regulatory regions in zebrafish. Dev. Dyn. 2009;238:2409–2417. doi: 10.1002/dvdy.22051. [DOI] [PubMed] [Google Scholar]
- Bessa J., Luengo M., Rivero-Gil S., Ariza-Cosano A., Maia A.H., Ruiz-Ruano F.J., Caballero P., Naranjo S., Carvajal J.J., Gómez-Skarmeta J.L. A mobile insulator system to detect and disrupt cis-regulatory landscapes in vertebrates. Genome Res. 2014;24:487–495. doi: 10.1101/gr.165654.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binot A.C., Manfroid I., Flasse L., Winandy M., Motte P., Martial J.A., Peers B., Voz M.L. Nkx6.1 and nkx6.2 regulate α- and β-cell formation in zebrafish by acting on pancreatic endocrine progenitor cells. Dev. Biol. 2010;340:397–407. doi: 10.1016/j.ydbio.2010.01.025. [DOI] [PubMed] [Google Scholar]
- Blanchette M., Kent W.J., Riemer C., Elnitski L., Smit A.F.A., Roskin K.M., Baertsch R., Rosenbloom K., Clawson H., Green E.D. Aligning multiple genomic sequences with the threaded blockset aligner. Genome Res. 2004;14:708–715. doi: 10.1101/gr.1933104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogdanovic O., Fernandez-Miñán A., Tena J.J., de la Calle-Mustienes E., Hidalgo C., van Kruysbergen I., van Heeringen S.J., Veenstra G.J., Gómez-Skarmeta J.L. Dynamics of enhancer chromatin signatures mark the transition from pluripotency to cell specification during embryogenesis. Genome Res. 2012;22:2043–2053. doi: 10.1101/gr.134833.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunet T., Lauri A., Arendt D. Did the notochord evolve from an ancient axial muscle? The axochord hypothesis. BioEssays. 2015;37:836–850. doi: 10.1002/bies.201500027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai J., Yu C., Liu Y., Chen S., Guo Y., Yong J., Lu W., Ding M., Deng H. Generation of homogeneous PDX1(+) pancreatic progenitors from human ES cell-derived endoderm cells. J. Mol. Cell Biol. 2010;2:50–60. doi: 10.1093/jmcb/mjp037. [DOI] [PubMed] [Google Scholar]
- Chung W.S., Shin C.H., Stainier D.Y.R. Bmp2 signaling regulates the hepatic versus pancreatic fate decision. Dev. Cell. 2008;15:738–748. doi: 10.1016/j.devcel.2008.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung W.S., Andersson O., Row R., Kimelman D., Stainier D.Y.R. Suppression of Alk8-mediated Bmp signaling cell-autonomously induces pancreatic beta-cells in zebrafish. Proc. Natl. Acad. Sci. USA. 2010;107:1142–1147. doi: 10.1073/pnas.0910205107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- diIorio P.J., Moss J.B., Sbrogna J.L., Karlstrom R.O., Moss L.G. Sonic hedgehog is required early in pancreatic islet development. Dev. Biol. 2002;244:75–84. doi: 10.1006/dbio.2002.0573. [DOI] [PubMed] [Google Scholar]
- Dixon J.R., Selvaraj S., Yue F., Kim A., Li Y., Shen Y., Hu M., Liu J.S., Ren B. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eroshkin F.M., Ermakova G.V., Bayramov A.V., Zaraisky A.G. Multiple noggins in vertebrate genome: cloning and expression of noggin2 and noggin4 in Xenopus laevis. Gene Expr. Patterns. 2006;6:180–186. doi: 10.1016/j.modgep.2005.06.007. [DOI] [PubMed] [Google Scholar]
- Fernández-Miñán A., Bessa J., Tena J.J., Gómez-Skarmeta J.L. Assay for transposase-accessible chromatin and circularized chromosome conformation capture, two methods to explore the regulatory landscapes of genes in zebrafish. Methods Cell Biol. 2016;135:413–430. doi: 10.1016/bs.mcb.2016.02.008. [DOI] [PubMed] [Google Scholar]
- Fürthauer M., Thisse B., Thisse C. Three different noggin genes antagonize the activity of bone morphogenetic proteins in the zebrafish embryo. Dev. Biol. 1999;214:181–196. doi: 10.1006/dbio.1999.9401. [DOI] [PubMed] [Google Scholar]
- Hebrok M., Kim S.K., Melton D.A. Notochord repression of endodermal Sonic hedgehog permits pancreas development. Genes Dev. 1998;12:1705–1713. doi: 10.1101/gad.12.11.1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebrok M., Kim S.K., St Jacques B., McMahon A.P., Melton D.A. Regulation of pancreas development by hedgehog signaling. Development. 2000;127:4905–4913. doi: 10.1242/dev.127.22.4905. [DOI] [PubMed] [Google Scholar]
- Hesselson D., Anderson R.M., Stainier D.Y.R. Suppression of Ptf1a activity induces acinar-to-endocrine conversion. Curr. Biol. 2011;21:712–717. doi: 10.1016/j.cub.2011.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang W.Y., Fu Y., Reyon D., Maeder M.L., Tsai S.Q., Sander J.D., Peterson R.T., Yeh J.R., Joung J.K. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat. Biotechnol. 2013;31:227–229. doi: 10.1038/nbt.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inomata H., Shibata T., Haraguchi T., Sasai Y. Scaling of dorsal-ventral patterning by embryo size-dependent degradation of Spemann’s organizer signals. Cell. 2013;153:1296–1311. doi: 10.1016/j.cell.2013.05.004. [DOI] [PubMed] [Google Scholar]
- Kent W.J., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.K., Hebrok M., Melton D.A. Notochord to endoderm signaling is required for pancreas development. Development. 1997;124:4243–4252. doi: 10.1242/dev.124.21.4243. [DOI] [PubMed] [Google Scholar]
- Kimmel C.B., Ballard W.W., Kimmel S.R., Ullmann B., Schilling T.F. Stages of embryonic development of the zebrafish. Dev. Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Klein F.A., Pakozdi T., Anders S., Ghavi-Helm Y., Furlong E.E.M., Huber W. FourCSeq: analysis of 4C sequencing data. Bioinformatics. 2015;31:3085–3091. doi: 10.1093/bioinformatics/btv335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroon E., Martinson L.A., Kadoya K., Bang A.G., Kelly O.G., Eliazer S., Young H., Richardson M., Smart N.G., Cunningham J. Pancreatic endoderm derived from human embryonic stem cells generates glucose-responsive insulin-secreting cells in vivo. Nat. Biotechnol. 2008;26:443–452. doi: 10.1038/nbt1393. [DOI] [PubMed] [Google Scholar]
- Kumar S.S., Alarfaj A.A., Munusamy M.A., Singh A.J., Peng I.C., Priya S.P., Hamat R.A., Higuchi A. Recent developments in β-cell differentiation of pluripotent stem cells induced by small and large molecules. Int. J. Mol. Sci. 2014;15:23418–23447. doi: 10.3390/ijms151223418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwan K.M., Fujimoto E., Grabher C., Mangum B.D., Hardy M.E., Campbell D.S., Parant J.M., Yost H.J., Kanki J.P., Chien C.B. The Tol2kit: a multisite gateway-based construction kit for Tol2 transposon transgenesis constructs. Dev. Dyn. 2007;236:3088–3099. doi: 10.1002/dvdy.21343. [DOI] [PubMed] [Google Scholar]
- Lammert E., Cleaver O., Melton D. Induction of pancreatic differentiation by signals from blood vessels. Science. 2001;294:564–567. doi: 10.1126/science.1064344. [DOI] [PubMed] [Google Scholar]
- Langmead B., Trapnell C., Pop M., Salzberg S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauri A., Brunet T., Handberg-Thorsager M., Fischer A.H., Simakov O., Steinmetz P.R., Tomer R., Keller P.J., Arendt D. Development of the annelid axochord: insights into notochord evolution. Science. 2014;345:1365–1368. doi: 10.1126/science.1253396. [DOI] [PubMed] [Google Scholar]
- Letelier J., de la Calle-Mustienes E., Pieretti J., Naranjo S., Maeso I., Nakamura T., Pascual-Anaya J., Shubin N.H., Schneider I., Martinez-Morales J.R., Gómez-Skarmeta J.L. A conserved Shh cis-regulatory module highlights a common developmental origin of unpaired and paired fins. Nat. Genet. 2018;50:504–509. doi: 10.1038/s41588-018-0080-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marlétaz F., Firbas P.N., Maeso I., Tena J.J., Bogdanovic O., Perry M., Wyatt C.D.R., de la Calle-Mustienes E., Bertrand S., Burguera D. Amphioxus functional genomics and the origins of vertebrate gene regulation. Nature. 2018;564:64–70. doi: 10.1038/s41586-018-0734-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCracken K.W., Wells J.M. Molecular pathways controlling pancreas induction. Semin. Cell Dev. Biol. 2012;23:656–662. doi: 10.1016/j.semcdb.2012.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mfopou J.K., Chen B., Mateizel I., Sermon K., Bouwens L. Noggin, retinoids, and fibroblast growth factor regulate hepatic or pancreatic fate of human embryonic stem cells. Gastroenterology. 2010;138:2233–2245. doi: 10.1053/j.gastro.2010.02.056. [DOI] [PubMed] [Google Scholar]
- Moreno-Mateos M.A., Vejnar C.E., Beaudoin J.D., Fernandez J.P., Mis E.K., Khokha M.K., Giraldez A.J. CRISPRscan: designing highly efficient sgRNAs for CRISPR-Cas9 targeting in vivo. Nat. Methods. 2015;12:982–988. doi: 10.1038/nmeth.3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris A.P., Voight B.F., Teslovich T.M., Ferreira T., Segrè A.V., Steinthorsdottir V., Strawbridge R.J., Khan H., Grallert H., Mahajan A., Wellcome Trust Case Control Consortium. Meta-Analyses of Glucose and Insulin-related traits Consortium (MAGIC) Investigators. Genetic Investigation of ANthropometric Traits (GIANT) Consortium. Asian Genetic Epidemiology Network-Type 2 Diabetes (AGEN-T2D) Consortium. South Asian Type 2 Diabetes (SAT2D) Consortium. DIAbetes Genetics Replication And Meta-analysis (DIAGRAM) Consortium Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat. Genet. 2012;44:981–990. doi: 10.1038/ng.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noordermeer D., Leleu M., Splinter E., Rougemont J., Laat W.D., Duboule D. The Dynamic Architecture of Hox Gene Clusters. Science. 2011;334(6053):222–225. doi: 10.1126/science.1207194. [DOI] [PubMed] [Google Scholar]
- Pasquali L., Gaulton K.J., Rodríguez-Seguí S.A., Mularoni L., Miguel-Escalada I., Akerman İ., Tena J.J., Morán I., Gómez-Marín C., van de Bunt M. Pancreatic islet enhancer clusters enriched in type 2 diabetes risk-associated variants. Nat. Genet. 2014;46:136–143. doi: 10.1038/ng.2870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passamaneck Y.J., Hejnol A., Martindale M.Q. Mesodermal gene expression during the embryonic and larval development of the articulate brachiopod Terebratalia transversa. Evodevo. 2015;6:10. doi: 10.1186/s13227-015-0004-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard K.S., Hubisz M.J., Rosenbloom K.R., Siepel A. Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res. 2010;20:110–121. doi: 10.1101/gr.097857.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prince V.E., Anderson R.M., Dalgin G. Zebrafish Pancreas Development and Regeneration: Fishing for Diabetes Therapies. Curr. Top. Dev. Biol. 2017;124:235–276. doi: 10.1016/bs.ctdb.2016.10.005. [DOI] [PubMed] [Google Scholar]
- Quinlan A.R., Hall I.M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rada-Iglesias A., Bajpai R., Swigut T., Brugmann S.A., Flynn R.A., Wysocka J. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011;470:279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues-Pinto R., Ward L., Humphreys M., Zeef L.A.H., Berry A., Hanley K.P., Hanley N., Richardson S.M., Hoyland J.A. Human notochordal cell transcriptome unveils potential regulators of cell function in the developing intervertebral disc. Sci. Rep. 2018;8:12866. doi: 10.1038/s41598-018-31172-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakhneny L., Khalifa-Malka L., Landsman L. Pancreas organogenesis: Approaches to elucidate the role of epithelial-mesenchymal interactions. Semin. Cell Dev. Biol. 2019;92:89–96. doi: 10.1016/j.semcdb.2018.08.012. [DOI] [PubMed] [Google Scholar]
- Schmitt A.D., Hu M., Jung I., Xu Z., Qiu Y., Tan C.L., Li Y., Lin S., Lin Y., Barr C.L., Ren B. A Compendium of Chromatin Contact Maps Reveals Spatially Active Regions in the Human Genome. Cell Rep. 2016;17:2042–2059. doi: 10.1016/j.celrep.2016.10.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C.A., Rasband W.S., Eliceiri K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Splinter E., Wit E.D., Werken H.J., Klous P., Laat W.D. Determining long-range chromatin interactions for selected genomic sites using 4C-seq technology: From fixation to computation. Methods. 2012;58(3):221–230. doi: 10.1016/j.ymeth.2012.04.009. [DOI] [PubMed] [Google Scholar]
- Tehrani Z., Lin S. Antagonistic interactions of hedgehog, Bmp and retinoic acid signals control zebrafish endocrine pancreas development. Development. 2011;138:631–640. doi: 10.1242/dev.050450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakore P.I., D’Ippolito A.M., Song L., Safi A., Shivakumar N.K., Kabadi A.M., Reddy T.E., Crawford G.E., Gersbach C.A. Highly specific epigenome editing by CRISPR-Cas9 repressors for silencing of distal regulatory elements. Nat. Methods. 2015;12:1143–1149. doi: 10.1038/nmeth.3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thisse C., Thisse B., Schilling T.F., Postlethwait J.H. Structure of the zebrafish snail1 gene and its expression in wild-type, spadetail and no tail mutant embryos. Development. 1993;119:1203–1215. doi: 10.1242/dev.119.4.1203. [DOI] [PubMed] [Google Scholar]
- Villar D., Berthelot C., Aldridge S., Rayner T.F., Lukk M., Pignatelli M., Park T.J., Deaville R., Erichsen J.T., Jasinska A.J. Enhancer evolution across 20 mammalian species. Cell. 2015;160:554–566. doi: 10.1016/j.cell.2015.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Song F., Zhang B., Zhang L., Xu J., Kuang D., Li D., Choudhary M.N.K., Li Y., Hu M. The 3D Genome Browser: a web-based browser for visualizing 3D genome organization and long-range chromatin interactions. Genome Biol. 2018;19:151. doi: 10.1186/s13059-018-1519-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D., Jiang W., Liu M., Sui X., Yin X., Chen S., Shi Y., Deng H. Highly efficient differentiation of human ES cells and iPS cells into mature pancreatic insulin-producing cells. Cell Res. 2009;19:429–438. doi: 10.1038/cr.2009.28. [DOI] [PubMed] [Google Scholar]
- Zhao Z., Tavoosidana G., Sjölinder M., Göndör A., Mariano P., Wang S., Kanduri C., Lezcano M., Sandhu K.S., Singh U. Circular chromosome conformation capture (4C) uncovers extensive networks of epigenetically regulated intra- and interchromosomal interactions. Nat. Genet. 2006;38:1341–1347. doi: 10.1038/ng1891. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw reads from 4C experiments (fastq files of the 2 replicates) are accessible in the European Nucleotide Archive (ENA, https://www.ebi.ac.uk/ena, project PRJEB36984, study ERP120260, runs ERR3964955 and ERR3964956). The processed signal is available for visualization in the following UCSC genome browser session http://genomeeuro.ucsc.edu/s/vdr/danRer7_4Cseq_nog2. Perl script for processing 4C seq data is available upon request.