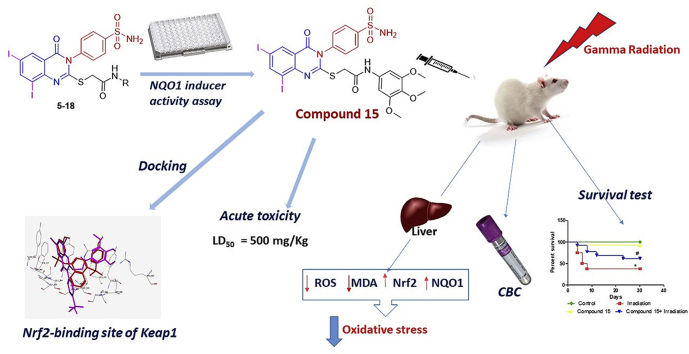
Abstract
Fifteen new quinazolinone derivatives bearing benzenesulfonamide moiety with variable acetamide tail were synthesized. The structures assigned to the products were concordant with the microanalytical and spectral data. Compounds 4–18 were screened for their ability to induce the antioxidant enzyme NAD(P)H: quinone oxidoreductase 1 (NQO1) in cells, a classical target for transcription factor nuclear factor erythroid 2-related factor 2 (Nrf2). The 2-((6,8-diiodo-4-oxo-3-(4-sulfamoylphenyl)-3,4-dihydroquinazolin-2-yl)thio)-N-(3,4,5-trimethoxyphenyl) acetamide 15 showed the most potent NQO1 inducer activity in vitro. Compound 15 had low toxicity in mice (LD50 = 500 mg/kg). It also reduced the damaging effects of gamma radiation, as assessed by the levels of Nrf2, NQO1, reactive oxygen species (ROS) and malondialdehyde (MDA) in liver tissues. In addition, compound 15 showed amelioration in the complete blood count of irradiated mice and enhanced survival over a period of 30 days following irradiation. Molecular docking of 15 inside the Nrf2-binding site of Kelch-like ECH associated protein 1 (Keap1), the main negative regulator of Nrf2, showed the same binding interactions as that of the co-crystallized ligand considering the binding possibilities and energy scores. These findings suggest that compound 15 could be considered as a promising antioxidant and radiomodulatory agent.
Keywords: Diiodoquinazolinone, Sulfonamide, Radiomodulatory, NQO1, Nrf2, Oxidative stress, Docking
Graphical abstract
Highlights
-
•
Novel 6,8-diiodoquinazolinone derivatives carrying sulfonamide moiety were synthetized.
-
•
Compounds 4-18 were assayed in vitro for their NQO1 inducer activity.
-
•
Acute toxicity of the most potent NQO1 inducer 15 was low in vivo (LD50 = 500 mg/kg).
-
•
Compound 15 inhibited oxidative stress and improved survival in irradiated mice.
-
•
Molecular docking showed interactions of 15 with the Nrf2-binding site of Keap1.
1. Introduction
The extensive use of radiotherapy and the damage caused to the surrounding normal organs have provoked researchers to find new strategies to protect normal tissues from radiation hazards [1,2]. The risk of injury from radiation can diminish the value of radiotherapy and contribute to complications for long-term cancer survivors [3]. Ionizing radiation interrupts cell functions through radiolysis of water and the production of reactive oxygen species (ROS) or reactive nitrogen species (RNS) [4,5]. Excessive production of ROS and RNS promotes oxidative stress, which can affect all cellular components, including single or double DNA strand breaks [6]. This ROS-mediated toxicity can lead to mutations and consequently cause cardiovascular, neurological toxicities and sexual dysfunction as well as cancer [[7], [8], [9], [10]]. In order to reduce these radiation-induced side effects, radioprotective drugs are used [11]. Also, the use of multi-target antioxidants that act as radioprotectors can help limit normal tissue damage caused by ionizing radiation [[12], [13], [14]].
Nuclear factor erythroid 2-related factor 2 (Nrf2) is a transcription factor that regulates the expression of various antioxidant proteins to protect against oxidative damage in the cell [15]. The abundance of Nrf2 is negatively regulated by Kelch-like ECH associated protein 1 (Keap1), a substrate adaptor for a Cullin3/Rbx1 ubiquitin ligase that binds and continuously targets Nrf2 for ubiquitination and proteasomal degradation [[16], [17], [18]]. Under conditions of oxidative stress, redox-sensitive cysteine sensors of Keap1 are modified leading to loss of, its ability to target Nrf2 for degradation; consequently, Nrf2 transports into the nucleus where it initiates the transcription of its downstream target genes, such as NAD(P)H: quinone oxidoreductase1 (NQO1) [19].
Quinazolinone is a strategic scaffold that has a wide range of pharmacological activities such as antioxidant, anti-inflammatory and anticancer activities [[20], [21], [22], [23]]. Sulfonamides, in addition to their use as antibiotics [[24], [25], [26], [27]], have many pharmacological activities and can be used as antiviral [28], anti-inflammatory [29], antioxidant [30,31], and anticancer agents [[32], [33], [34], [35]]. These versatile pharmacological activities make the two chemical classes excellent candidates for developing new multi-target agents through a slight alteration in the structure that might lead to diversity in the biological activity [20,36,37]. In addition, numerous studies have revealed iodine to be a potent antioxidant with higher potency than that of ascorbic acid [38,39]. Iodine can act as an electron donor that quenches ROS, such as OH• and H2O2 [40], or decreases the damaging effects of ROS, thus increasing the total antioxidant status in human serum [41].
In this context, it seemed of interest to search for new compounds with the ability to scavenge ROS and protect cells. A series of new 6,8-diiodoquinazolin-4(3H)-one conjugated to benzenesulfonamide was synthesized by the introduction of the sulfonamide group at the N-3 of quinazolinone with the incorporation of varying acetamide terminal aimed at exploring the potential antioxidant and radioprotective activity. The antioxidant potential of the target compounds was first measured using a quantitative and robust NQO1 inducer activity bioassay in cells. Acute toxicity study for the most active compound was then performed in vivo. A non-toxic dose was subsequently selected to investigate the potential protective effect against whole-body gamma irradiation-induced oxidative stress in experimental mice. All groups were observed 30 days after irradiation for survival and weight changes. Additionally, molecular docking was performed inside the Nrf2-binding site of Keap1 to gain insights into the molecular interactions and possible mode of action.
2. Results and discussion
2.1. Chemistry
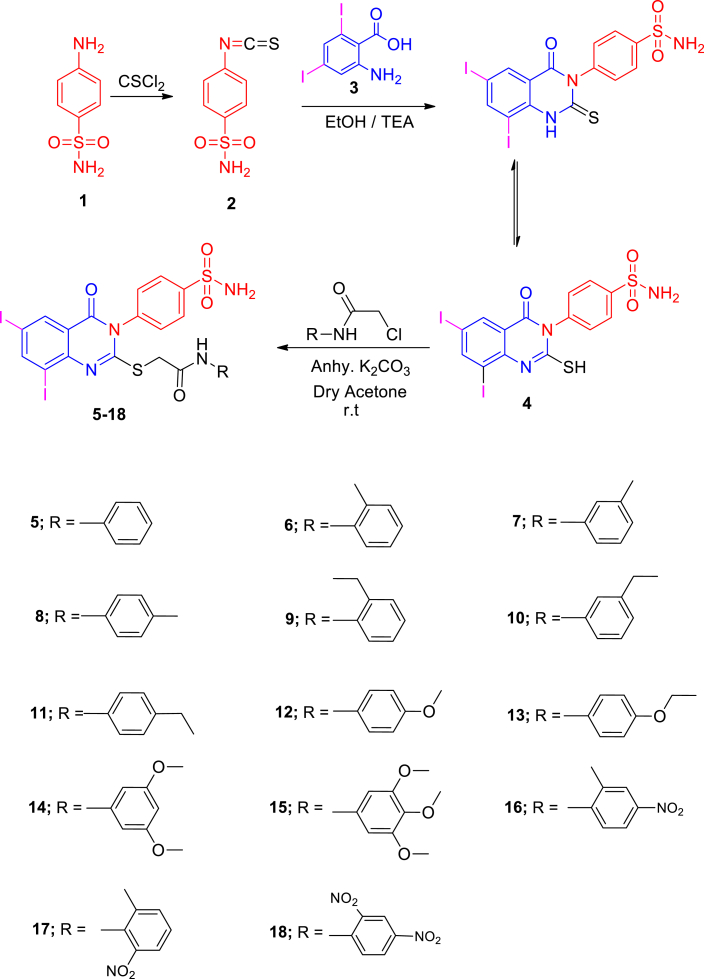
Scheme 1 shows the synthesis of thioacetamide quinazolinone benzenesulfonamide derivatives 5–18. The starting material 4-(6,8-diiodo-2-mercapto-4- oxoquinazolin-3(4H)-yl) benzenesulfonamide 4 was prepared from the reaction of 4-isothiocyanatobenzenesulfonamide 2 [42] and 2-amino-3,5-diiodobenzoic acid 3. The coupling of 4 with the 2-chloro-N-substituted acetamide in dry acetone and anhydrous K2CO3 yielded the corresponding 2-((6,8-diiodo-4-oxo-3-(4-sulfamoylphenyl)-3,4-dihydroquinazolin-2-yl)thio)-N-substituted acetamide 5–18. IR spectra of 5–18 displayed additional NH, CH2 aliphatic and CO bands at their specified regions. 1H NMR spectra of 5–18 revealed the acetamide group through the presence of two singlets, one at 4.17–4.31 ppm referring to the CH2 and the other at 9.66–11.21 ppm attributed to the NH protons with the disappearance of SH singlet of 4 at 1.97 ppm. 13C NMR of 5–18 exhibited two signals peculiar to the CH2 and CO carbons. 1H NMR spectra of 6–8 displayed singlets at 2.21, 2.28 and 2.30 ppm assigned to the CH3 group at the ortho, meta and para-positions of the phenyl group. 13C NMR of 6–8 showed signals at 16.32, 24.13 and 19.21 ppm for the CH3 group. 1H NMR spectra of 9–11 revealed triplets at 1.31, 1.20 and 1.15 ppm attributed to the CH3 ethyl, and quartet at 2.54, 2.58 and 2.55 ppm referring to the CH2 ethyl at the ortho, meta and para-positions. 13C NMR of 9–11 showed two signals at 14.67, 15.21, 17.12 due to CH3 ethyl and 24.23, 24.10, 29.40 due to the CH2 ethyl groups, respectively. 1H NMR spectra of 12 revealed singlet at 3.75 ppm attributed to the OCH3 protons, while 13C NMR of 12 showed a signal at 54.26 ppm due to the OCH3 carbon. 1H NMR spectra of 13 revealed triplet at 1.27 ppm and quartet at 3.97 ppm due to the ethoxy group. 1H NMR spectra of 14 revealed a singlet at 3.74 ppm due to the 2OCH3 protons, while 15 revealed two singlets at 3.70 and 3.81 ppm due to the 3OCH3 protons. IR of 16–18 showed NO2 bands.
Scheme 1.
The synthetic pathways for the development of the diiodoquinazolinone derivatives 4–18.
2.2. Biological activity
2.2.1. In vitro screening
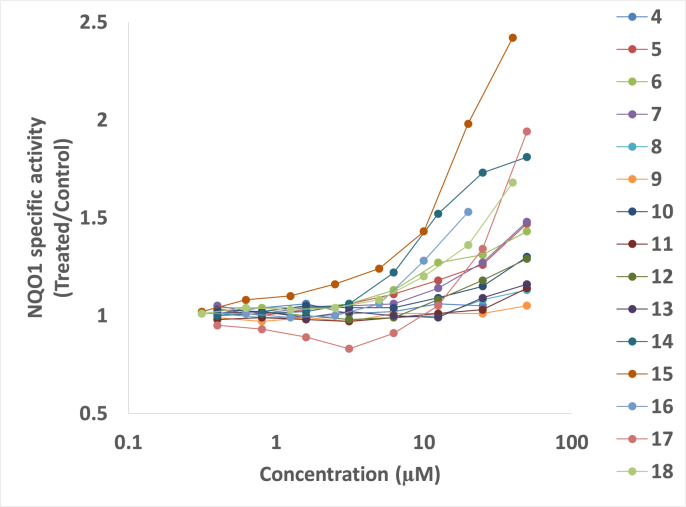
The antioxidant activity of compounds 4–18 was screened using the NQO1 inducer activity assay. The Concentration of the novel compounds to Double the specific enzyme activity of NQO1 (CD value) was used as a measure of inducer potency and results obtained are presented in Fig. 1 & Table 1. Evaluation of the NQO1 inducer activity showed that compounds 4, 8, 9, 11 and 13 were inactive, whereas compounds 5, 6, 7, 10, 12, 14 and 18 had activity; however, CD value was not reached. Compounds 15 (CD = 20 μM), and 17 (CD = 50 μM) showed concentration-dependent inducer activity. These diiodoquinazolinones represent a new chemical class of NQO1 inducers, thus adding to the existing knowledge of the diversity of the many chemical scaffolds that have been reported to induce this antioxidant enzyme. The classical NQO1 inducers are primarily oxidants and electrophiles or other compounds that react (or are metabolized to products that react) and chemically modify cysteine sensors of Keap1 [43]. A new generation of NQO1 inducers is also emerging, that of noncovalent small-molecule modulators of the Keap1–Nrf2 protein-protein interaction [[44], [45], [46]]. Because our diiodoquinazolinones have some common features with the Keap1–Nrf2 protein-protein interaction inhibitors, in this study we tested the potential ability of these compounds to directly disrupt the binding of Keap1 to Nrf2 by molecular modeling (see section 2.3).
Fig. 1.
Concentration dependence of the NQO1 inducer activity of compounds 4–18.
Table 1.
NQO1 inducer activity and CD values of compounds 4–18.
| Conc. (μM) | Compound no. |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | |
| 0.313 | NR | NR | NR | NR | NR | 0.99 | 1.03 | NR | NR | 1.01 | NR | 1.02 | 1.01 | NR | 1.01 |
| 0.4 | 1.01 | 1.03 | 1.04 | 1.05 | 1.01 | NR | NR | 0.98 | 1.01 | NR | 1.00 | NR | NR | 0.95 | NR |
| 0.625 | NR | NR | NR | NR | NR | 0.97 | 1.02 | NR | NR | 1.02 | NR | 1.08 | 1.01 | NR | 1.04 |
| 0.8 | 1.04 | 1.00 | 1.04 | 1.01 | 0.99 | NR | NR | 0.99 | 1.02 | NR | 1.01 | NR | NR | 0.93 | NR |
| 1.25 | NR | NR | NR | NR | NR | 0.99 | 1.05 | NR | NR | 0.99 | NR | 1.10 | 0.99 | NR | 1.03 |
| 1.6 | 1.06 | 1.03 | 1.04 | 1.04 | 0.99 | NR | NR | 0.98 | 1.00 | NR | 1.02 | NR | NR | 0.89 | NR |
| 2.5 | NR | NR | NR | NR | NR | 0.97 | 1.04 | NR | NR | 1.02 | NR | 1.16 | 1.00 | NR | 1.04 |
| 3.125 | 1.01 | 1.06 | 1.06 | 1.05 | 0.97 | NR | NR | 0.97 | 0.98 | NR | 1.06 | NR | NR | 0.83 | NR |
| 5 | NR | NR | NR | NR | NR | 1.01 | 1.04 | NR | NR | 1.00 | NR | 1.24 | 1.06 | NR | 1.08 |
| 6.25 | 1.02 | 1.11 | 1.13 | 1.06 | 0.99 | NR | NR | 0.99 | 0.99 | NR | 1.22 | NR | NR | 0.91 | NR |
| 10 | NR | NR | NR | NR | NR | 1.01 | 1.09 | NR | NR | 0.99 | NR | 1.43 | 1.28 | NR | 1.20 |
| 12.5 | 1.06 | 1.18 | 1.27 | 1.14 | 1.00 | NR | NR | 1.01 | 1.08 | NR | 1.52 | NR | NR | 1.05 | NR |
| 20 | NR | NR | NR | NR | NR | 1.01 | 1.15 | NR | NR | 1.09 | NR | 1.98 | 1.53 | NR | 1.36 |
| 25 | 1.05 | 1.26 | 1.31 | 1.27 | 1.08 | NR | NR | 1.03 | 1.18 | NR | 1.73 | NR | NR | 1.34 | NR |
| 40 | NR | NR | NR | NR | NR | 1.05 | 1.30 | NR | NR | 1.16 | NR | 2.42 | NR | NR | 1.68 |
| 50 | NR | 1.47 | 1.43 | 1.48 | 1.13 | NR | NR | 1.14 | 1.29 | NR | 1.81 | NR | NR | 1.94 | NR |
| CDa | NR | NR | NR | NR | NR | NR | NR | NR | NR | NR | NR | 20 | NR | 50 | NR |
NR means not recorded.
CD values are the averages of three independent experiments, each with eight replicate wells of cells, and SD for each data point was within 5% of the value.
2.2.2. In vivo evaluation
2.2.2.1. Determination of toxicity (lethal dose fifty, LD50) of compound 15
The most promising compound, 15, was investigated in vivo for acute toxicity (LD50) in albino mice, and the value was found to be 500 mg/kg body weight (i.p.). Subsequently, one-tenth of this dose was selected as the therapeutic dose for further evaluation of the potential radioprotective effects of compound 15.
2.2.2.2. Evaluation of the radiomodulatory effect of compound 15 in mice
Four groups of mice were used, the first group served as control, the second group was irradiated at a dose of 7 Gy as a single dose, the third group was injected i.p. with compound 15 only for 5 consecutive days and the last group received compound 15 then exposed to 7 Gy of gamma radiation. After 3 days from irradiation, five mice were checked for liver and hematopoietic system toxicities. The residual mice in all groups were monitored over 30 days to evaluate the survival rate and body weight changes.
2.2.2.2.1. The effect of compound 15 on radiation-induced liver toxicity
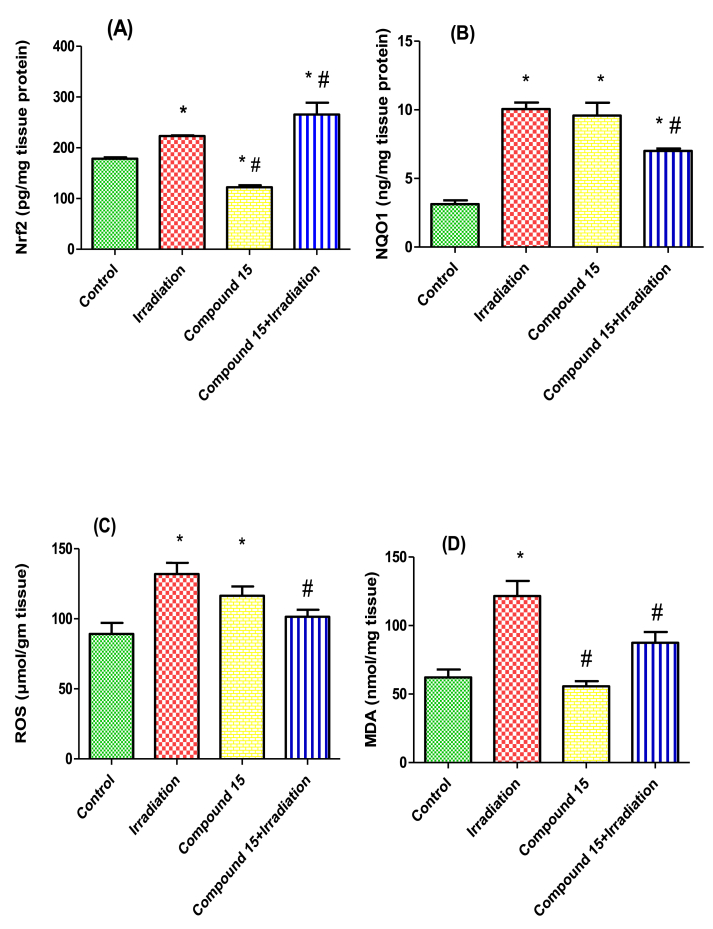
Gamma radiation-induced hepatic oxidative stress as shown by a significant increase in hepatic levels of nuclear Nrf2 (1.3-fold), NQO1 (3.2-fold), ROS (1.5-fold) and the lipid peroxidation product malondialdehyde (MDA) (2-fold) as compared to non-irradiated (control) mice. This was in agreement with other studies [2,47].
Ionizing radiation is believed to induce damage through the generation of ROS, resulting in an imbalance in the oxidant/antioxidant ratio in cells [8,48]. In the current experiment, the presence of ROS-mediated damage was confirmed by the increase in MDA levels in irradiated liver, in addition to the increase in the expression of the enzymatic antioxidant system. Moreover, these results support the notion that Nrf2 is an initial regulator of cellular responses to radiation exposure [49]. Once Nrf2 translocates to the nucleus it induces expression of endogenous antioxidant enzymes, such as NQO1 [50], a flavoprotein involved in cellular protection against oxidative stress [51].
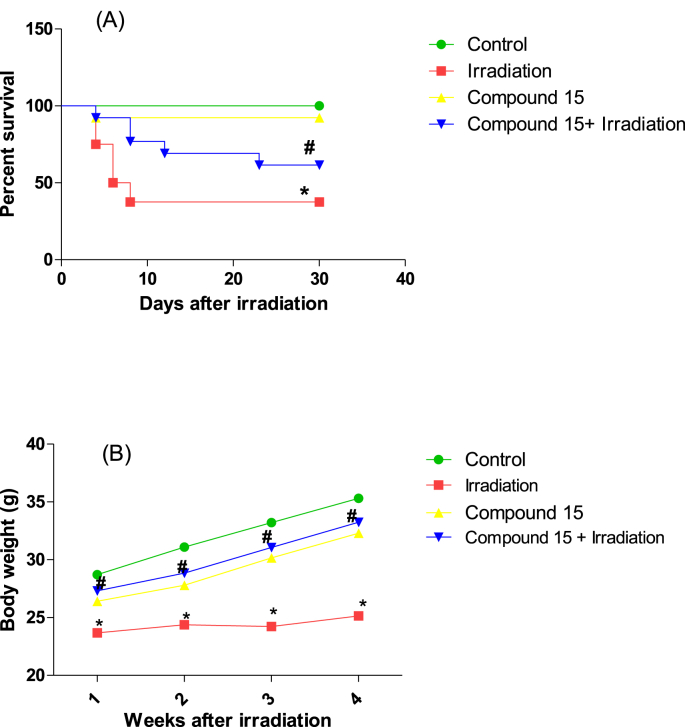
Treatment of non-irradiated mice with compound 15 led to an increase in NQO1 and ROS levels and a decrease in Nrf2, with no significant change in MDA level as compared to normal (non-irradiated) mice (Fig. 2). A significant increase in Nrf2 levels (19%) as well decrease in the levels of NQO1 (30%), ROS (23%) and MDA (28%) was observed in irradiated mice livers treated with compound 15 when compared to the group subjected to radiation alone (Fig. 2). Moreover, treatment with compound 15 improved both survival and body weight of the animals following irradiation (Fig. 3) without affecting the liver weight (Fig. 4) as compared to irradiated mice. The present results indicate that compound 15 has an antioxidant capacity as the treatment of irradiated mice with 15 prevents oxidative stress, reducing the increase in lipid peroxidation markers and maintaining the expression of Nrf2 compared with the irradiated group suggesting improved hepatic antioxidant capacity. Hence, compound 15 validated its radiomodulatory and antioxidant effect through its main structure; quinazolinone and sulfonamide that goes in line with Soliman et al. [52]. Also, this finding was reinforced by Cuadrado and his colleagues, who emphasized the importance of therapeutic targeting for Nrf2 because of its resourceful cytoprotective roles against a plethora of diseases that are associated with oxidative stress [53].
Fig. 2.
Effect of compound 15 on (A) Nrf2, (B) NQO1, (C) ROS and (D) MDA levels in liver of non-irradiated (control) and irradiated mice after 3 days of irradiation. The results were expressed as mean ± S.E. Statistical analysis was carried out by one-way ANOVA followed by Bonferroni’s multiple comparison test. ∗: significantly different from control group, #: significantly different from irradiated group at p < 0.05. (n = 5).
Fig. 3.
A) Survival percent and B) Body weight changes of control, irradiated, compound 15 and compound 15 + irradiated mice through 30 days after irradiation. The results were expressed as mean ± S.E. (n = 15). Statistical analysis was carried out by Kaplan-Meier method followed by the Mantel–Cox test for survival analysis. Body weight changes between groups were analyzed by two-way ANOVA followed by Bonferroni’s post test. ∗: significantly different from control group, #: significantly different from irradiated group at p < 0.05.
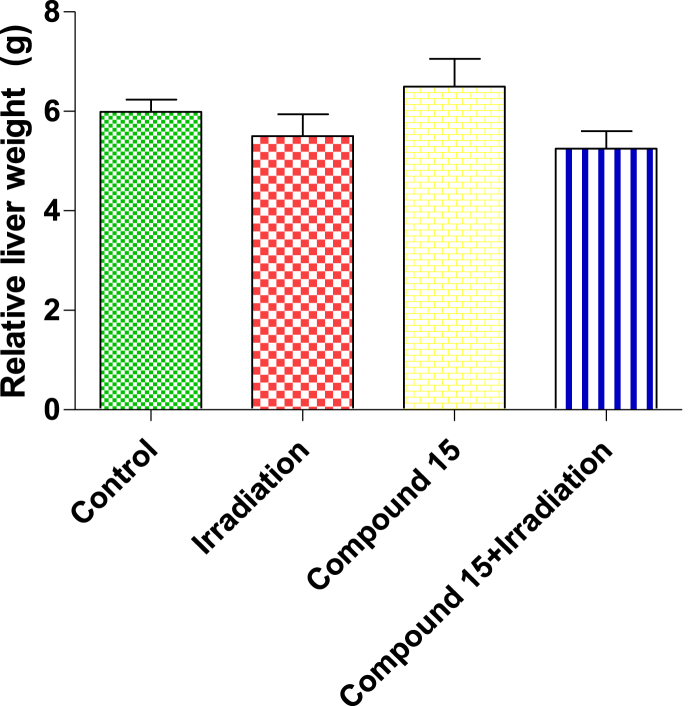
Fig. 4.
Effect of compound 15 on relative liver weight in non-irradiated (control) and irradiated mice after 3 days of irradiation. The results were expressed as mean ± S.E. (n = 5). Statistical analysis was carried out by one-way ANOVA followed by Bonferroni’s multiple comparison test. There were no significant differences between groups.
At the same time, it was found that NQO1 expression levels of irradiated mice treated with 15 were significantly lower as compared to vehicle-treated irradiated ones, but still significantly higher than normal levels. Interestingly, the levels of NQO1 in all experimental groups correlate with the levels of ROS, suggesting ROS involvement in the NQO1 induction. The lower levels of NQO1 and ROS in the irradiated group that also received 15 could be the results of increased antioxidant capacity due to Nrf2 activation [54]. Additionally, it has been reported that Nrf2 modifies ROS production partly by regulating NQO1 expression [55]. On the other hand, the NQO1 levels were significantly higher than the non-irradiated controls, in agreement with the cell culture results (this study). Notably, the increased levels of ROS in non-irradiated mice treated with compound 15 are consistent with the increased levels of ROS following genetic Nrf2 activation by Keap1 knockdown [54]. Importantly however, the increased ROS production that accompanies NQO1 induction does not lead to damage, as evidenced by the lack of increase in the levels of MDA (this study).
2.2.2.2.2. The effect of compound 15 on the hematopoietic system
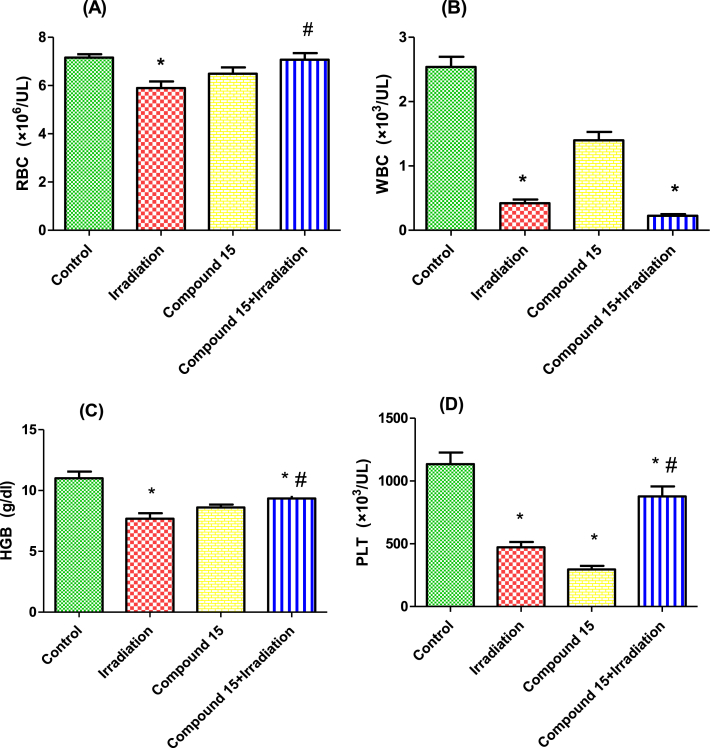
To examine the possible role of compound 15 in protecting the hematopoietic system against irradiation, we measured the peripheral blood cell counts of red blood cells (RBCs), white blood cells (WBCs), hemoglobin (HGB) and platelets (PLT). The irradiated mice exhibited a significant decrease in RBCs, WBCs, HGB and PLT compared with the control group (Fig. 5). These results are mainly attributed to the fact that irradiation causes the formation of free radicals which initiate a chain of events leading to the decline in the levels of hematological parameters [56]. Indeed, it has been well established that gamma irradiation induces RBC injury, including morphological and quantitative changes of RBCs. These alternations may be partly attributed to radiation-induced oxidative stress in RBCs. Exposure to radiation results in the formation of reactive oxygen species (ROS) and reactive nitrogen species (RNS), as well as DNA damage, which can then lead to severe injury to the hematopoietic system [57]. This is in harmony with Wang et al. [58] who stated that, injury to the hematopoietic system is the most common injury induced by irradiation. This was attributed to the effect of ionizing radiation on hematopoietic stem cells and hematopoietic progenitor cells, which are principally responsible for hematopoietic recovery. Treatment of irradiated mice with compound 15 ameliorated the decrease in peripheral blood cells, particularly RBCs, HGB and PLT. Hence, the antioxidant properties of compound 15 may contribute to the amelioration of RBC counts and HGB in irradiated mice. This is consistent with other studies for antioxidants effects on the hematopoietic system [57,59]. This might be explained through the promotion effect of radioprotectors to proliferate hematopoietic stem cells and they also could increase the levels of leukocyte growth factors [60,61]. Besides, several potent radioprotectors protect various membrane systems, as well as hematopoietic stem cells from peroxidative damages, that happened after irradiation so it could protect blood components against irradiation [62]. Taken all together, these results demonstrate the protective effect of compound 15 against gamma radiation.
Fig. 5.
Effect of compound 15 on: (A) RBCs, (B) WBCs, (C) HGB concentration and (D) PLT counts in non-irradiated (control) and irradiated mice after 3 days of irradiation. The results were expressed as mean ± S.E. (n = 5). Statistical analysis was carried out by one-way ANOVA followed by Bonferroni’s multiple comparison test. ∗: significantly different from control group, #: significantly different from irradiated group.
2.3. Molecular docking
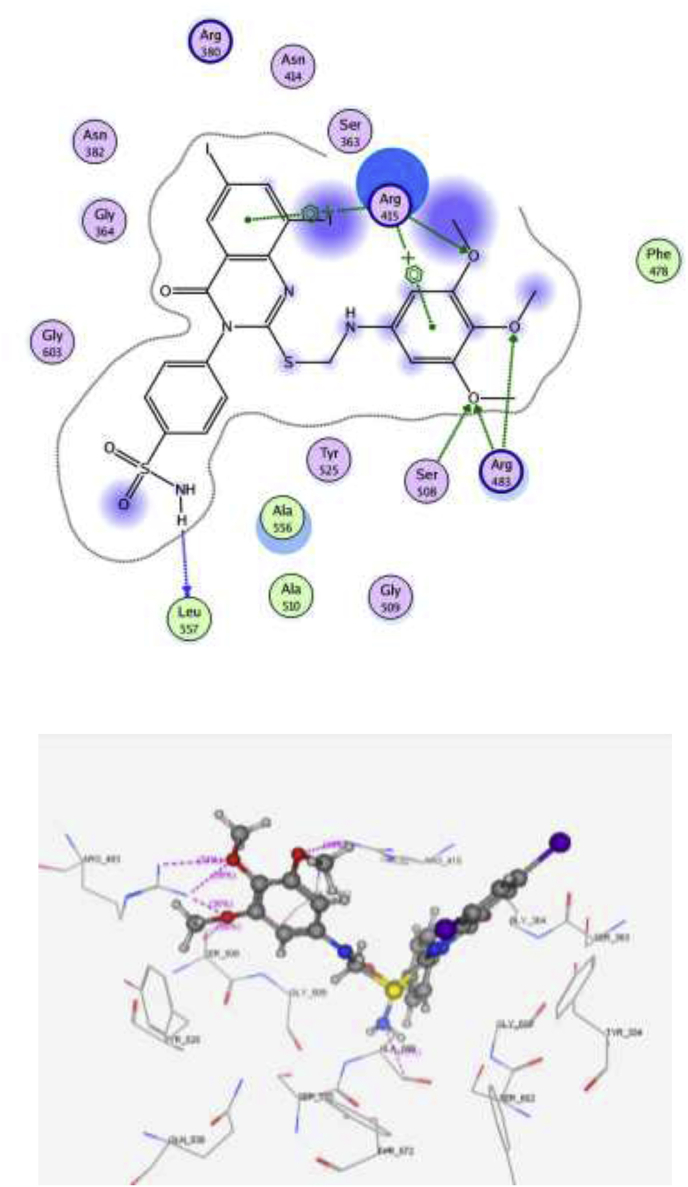
Molecular docking was performed to assess the ability of compound 15 to block the Kelch domain of Keap1. Through its Kelch domain, Keap1 binds to Nrf2, promoting its degradation, resulting in low cytoprotective gene levels [63]. The PDB file: 4IQK was obtained from the Protein Data Bank. The binding site of Kelch domain has been reported to have five subpockets: P1, P2, P3, P4 and P5 [64]. P1 and P2 are positively charged pockets that contain the arginine triad (Arg 415, Arg 483 and Arg 380). This triad is crucial for the selectivity of the molecular recognition, together with a group of hydrophobic residues contributes to the stability of the complex. P1 is formed by residues Arg 415, Ile 461, Gly 462, Phe 478, Arg 483 and Ser 508. P2 is formed by Ser 363, Arg 380, Asn 382 and Asn 414. P3 is a neutrally charged pocket composed of Gly 509, Ser 555, Ala 556, Gly 571, Ser 602 and Gly 603. P4 is formed by Tyr 525, Gln 530 and Tyr 572, whereas P5 is formed by Tyr 334 and Phe 577.
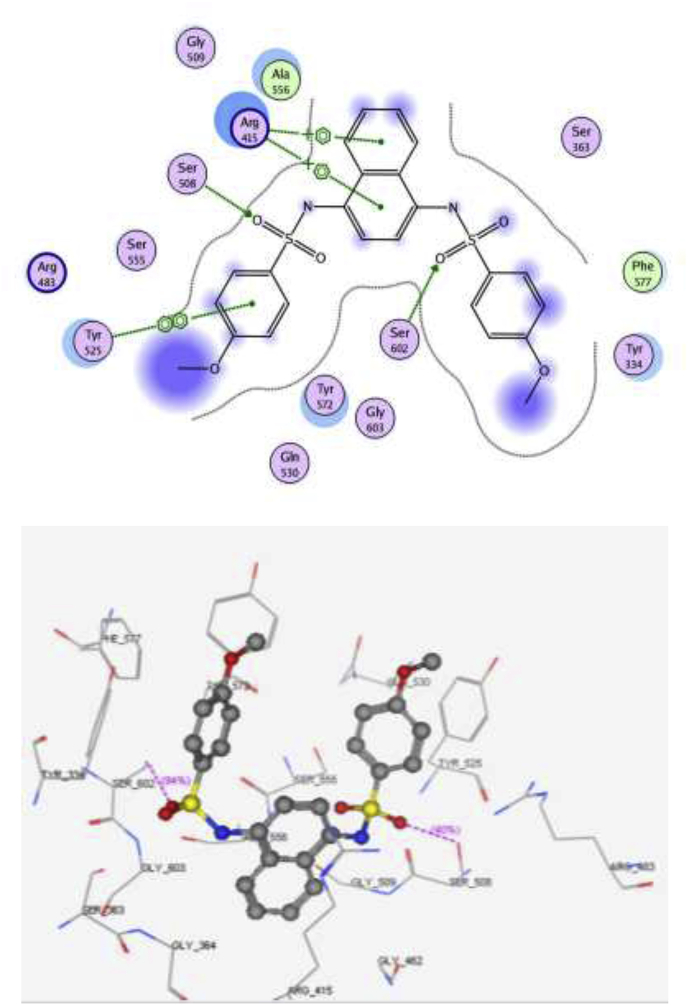
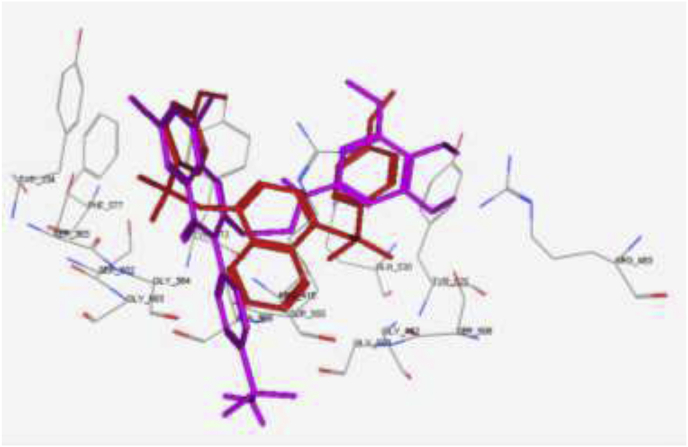
The main interactions observed by the co-crystallized ligand (N, N′-naphthalene-1,4-diylbis(4-methoxybenzenesulfonamide) are two cation-pi interaction with Arg 415, pi–pi interaction with Tyr 525 and two hydrogen bonds with Ser 602 and Ser 508, with S = -10.11 kcal/mol (Fig. 6). Compound 15 showed the same key interactions exhibited by the co-crystallized ligand. Compound 15 (S = -9.61 kcal/mol, RMSD = 1.34 Å) has adopted a conformation allowing the presence of two cation-pi interaction between Arg 415 and the aromatic rings in addition to a hydrogen bond with the methoxy group (Fig. 7), three hydrogen bonds made by ser 508 and Arg 483 towards the methoxy groups, and another hydrogen bond between Leu 557 and NH2 group of the sulfonamide. Superimposition between compound 15 and the co-crystallized ligand showed that they adopt the same orientation inside the binding site (Fig. 8). Finally, compound 15 possessing the highest NQO1 inducer activity (CD = 20 μM) in this series showed the same interactions and the same orientation of the native ligand inside the receptor, indicating a possible correlation between those multiple interactions and the noted higher potency. Based on the above-mentioned results, compound 15 could possibly bind to Keap1 and disrupt its interaction with Nrf2.
Fig. 6.
2D and 3D interaction poses of the N,N′-naphthalene-1,4-diylbis(4-methoxybenzenesulfonamide showing cation-π, π-π interaction and hydrogen bonds with the key amino acids inside the binding pocket.
Fig. 7.
2D and 3D interaction pose of compound 15 showing cation-π, π-π interactions inside the binding pocket of 4IQK.
Fig. 8.
Superimposition of compound 15 (magenta) and the co-crystallized ligand (red) showed that they adopt the same orientation inside the receptor. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
The results from this study complement previous reports showing that the classical electrophilic Nrf2 activator sulforaphane protects cells, including human retinal pigment epithelial cells, keratinocytes, and mouse leukemia cells against oxidative damage caused by oxidative stressors of four different types, namely menadione, tert-butyl hydroperoxide, 4-hydroxynonenal, and peroxynitrite, as well as by exposure to ultraviolet radiation [65,66]. Furthermore, unlike the effects of most direct antioxidants, the indirect antioxidant effect of sulforaphane, which results from Nrf2 activation, persists for several days after sulforaphane is no longer present in the cell culture medium. This is because direct antioxidants, such as ascorbic acid, tocopherols, carotenoids, and polyphenols, which neutralize ROS and other chemical oxidants are consumed in these reactions, whereas Nrf2 activation results in transcriptional upregulation of antioxidant defences, which are mediated by proteins with long half-lives, often several days. The new compounds generated in the current study have an additional advantage in that they are non-electrophilic and are therefore expected to have a broader therapeutic window compared to electrophilic Nrf2 activators. This is supported by the very low toxicity of compound 15 in mice. Taken together, these results demonstrate the powerful effect of Nrf2 activation and induction of NQO1 in protecting cells and animals against high levels of ROS and preventing ROS-mediated damage. This is of particular relevance to protecting the hematopoietic system, which is highly sensitive to ROS.
3. Conclusion
In summary, a hybridization strategy was adopted using the iodinated quinazolinone scaffold and sulfonamide moiety to produce the 2-((6,8-diiodo-4-oxo-3-(4-sulfamoylphenyl)-3, 4-dihydroquinazolin-2-yl)thio)-N-(substituted) acetamide derivatives 5–18. Different substitutions were introduced to the acetamide group to study the structure-activity relationship. All the compounds were screened for their antioxidant potential using the NQO1 inducer activity assay. The 3,4,5-trimethoxyphenyl derivative 15 showed the highest inducer activity in this series (CD = 20 μM) and had low toxicity (LD50 = 500 mg/kg). Treatment of gamma-irradiated mice with compound 15 lowered oxidative stress as evidenced by the lower levels of MDA, ROS and NQO1 in liver. Furthermore, compound 15 ameliorated the complete blood picture of irradiated mice, as well as enhanced the survival of mice over a period of 30 days post-irradiation. Molecular docking of 15 inside the active site of Keap1 confirmed that it binds in the same manner as that of the co-crystallized ligands. The inducer activity of compound 15 in upregulating NQO1 strongly suggests that it could be used as a lead antioxidant and radiomodulatory agent for further optimization of the quinazolinone scaffold.
4. Materials and methods
4.1. Chemistry
All chemicals were purchased from Sigma-Aldrich and are of AR grade. Melting points were determined in open capillary on a Gallen Kamp melting point apparatus (Sanyo Gallen Kamp, UK). Thin layer chromatography using precoated silica gel plates (Kieselgel 0.25 mm, 60 F254, Merck, Germany) was performed with a solvent system of chloroform/methanol (8:2) to detect the spots by UV light. IR spectra (KBr disc) were recorded using an FT-IR spectrophotometer (Perkin Elmer, USA). NMR spectra were scanned on NMR spectrophotometer (Bruker AXS Inc., Switzerland), operating at 500 MHz for 1H and 125.76 MHz for 13C. Mass spectra were recorded on the ISQ LT Thermo Scientific GCMS model (Massachusetts, USA). Chemical shifts are expressed in δ-values (ppm) relative to TMS as an internal standard, using DMSO‑d6 as a solvent. Elemental analyses were done on a model 2400 CHNSO analyser (Perkin Elmer, USA). All the values were within ±0.4% of the theoretical values.
4.1.1. 4-(6, 8-diiodo-2-mercapto-4- oxoquinazolin-3(4H)-yl) benzenesulfonamide(4)
A mixture of 2-amino-3,5-diiodobenzoic acid 3 (3.88 g, 0.01 mol) and 4- isothiocyanatobenzenesulfonamide 2 (2.14 g, 0.01 mol) in absolute ethanol (30 mL) containing 3 drops of triethylamine, was refluxed for 3 h. The solid product formed was collected by filtration and crystallized from ethanol to give 4.
4: Yield, 88%; m.p. > 300 °C. IR (KBr, ʋ cm−1): 3311, 3210 (NH2), 3098 (arom.), 1701 (CO), 1618 (CN), 1379, 1160 (SO2). 1H NMR (DMSO‑d6, δ, ppm): 1.97 (s, 1H), 7.86 (d, 2H, J = 7 Hz, AB), 8.06 (d, 2H, J = 7 Hz, AB), 8.23 (d, 1H, J = 2 Hz), 8.58 (d, 1H, J = 2 Hz), 10.05 (s, 2H). 13C NMR (DMSO‑d6, δ, ppm): 86.84, 89.74, 118.49, 118.84, 122.99, 127.24 (2), 133.61, 134.21, 136.20, 144.48, 158.31, 165.76, 176.14. Anal. Calcd. for C14H9I2N3O3S2 (584.82): C, 28.73; H, 1.55; N, 7.18. Found: C, 29.02; H, 1.82; N, 7.41.
4.1.2. 3,4-Dihydroquinazolin-sulfonamide derivatives(5-18)
4.1.2.1. General procedure
A mixture of 4 (5.85 g, 0.01 mol) and 2-chloro-N-substituted acetamide derivatives (0.01 mol) in dry acetone (30 mL) and anhydrous K2CO3 (1.38 g, 0.01 mol) was stirred at room temperature for 18 h, filtered and the solid product formed was crystallized from dioxane to give 5–18.
4.1.2.2. 2-((6,8-Diiodo-4-oxo-3-(4-sulfamoylphenyl)-3,4-dihydroquinazolin-2-yl)thio)-N-phenylacetamide(5)
5: Yield, 79%; m.p. > 300 °C. IR (KBr, ʋ cm−1): 3408, 3310, 3231 (NH, NH2), 3079 (arom.), 2945, 2881 (aliph.), 1702, 1679 (2CO), 1620 (CN), 1349, 1170 (SO2). 1H NMR (DMSO‑d6, δ, ppm): 4.17 (s, 2H), 7.03–7.30 (m, 3H), 7.60–7.83 (m, 4H), 8.02 (s, 2H), 8.09 (d, 2H, J = 10 Hz, AB), 8.35 (d, 1H, J = 2 Hz), 8.60 (d, 1H, J = 2 Hz), 10.12 (s, 1H). 13C NMR (DMSO‑d6, δ, ppm): 29.73, 81.27, 89.61, 118.79 (2), 123.62 (2), 124.33, 127.61, 128.05 (2), 128.80 (2), 133.12, 137.64, 137.90, 139.42, 145.13, 158.92, 163.31, 164.02, 169.81. MS m/z [%]: 718 [M+, 29.82], 719 [M+1, 19.85], 454 [100]. Anal. Calcd. for C22H16I2N4O4S2 (718.30): C, 36.78; H, 2.25; N, 7.80. Found: C, 37.01; H, 2.51; N, 8.12.
4.1.2.3. 2-((6,8-Diiodo-4-oxo-3-(4-sulfamoylphenyl)-3,4-dihydroquinazolin-2-yl)thio)-N-o-tolylacetamide(6)
6: Yield, 81%; m.p. > 300 °C. IR (KBr, ʋ cm−1): 3403, 3321, 3216 (NH, NH2), 3098 (arom.), 2956, 2891 (aliph.), 1711, 1681 (2CO), 1631 (CN), 1355, 1188 (SO2). 1H NMR (DMSO‑d6, δ, ppm): 2.21 (s, 3H), 4.30 (s, 2H), 7.10 (ddd, 1H, J = 8 & 2.5 Hz), 7.30–7.55 (m, 3H), 7.87 (d, 2H, J = 8.5 Hz, AB), 8.01 (s, 2H), 8.05 (d, 2H, J = 8.5 Hz, AB), 8.28 (d, 1H, J = 1.5 Hz), 8.55 (d, 1H, J = 1.5 Hz), 9.66 (s, 1H). 13C NMR (DMSO‑d6, δ, ppm): 16.32, 26.61, 82.13, 91.41, 120.80 (2), 122.81, 123.90, 124.63, 128.72, 128.91 (2), 129.66, 130.81, 133.54, 134.02, 135.25, 135.87, 147.31, 153.82, 158.10, 159.62, 167.53. Anal. Calcd. for C23H18I2N4O4S2 (732.35): C, 37.72; H, 2.48; N, 7.65. Found: C, 38.04; H, 2.68; N, 7.96.
4.1.2.4. 2-((6,8-Diiodo- 4-oxo-3-(4-sulfamoylphenyl)-3,4-dihydroquinazolin-2-yl)thio)-N-(m-tolyl)acetamide(7)
7: Yield, 86%; m.p. > 300 °C. IR (KBr, ʋ cm−1): 3421, 3318, 3207 (NH, NH2), 3095 (arom.), 2978, 2842 (aliph.), 1707, 1675 (2CO), 1625 (CN), 1378, 1145 (SO2). 1H NMR (DMSO‑d6, δ, ppm): 2.28 (s, 3H), 4.25 (s, 2H), 6.87 (m, 1H), 7.31–7.56 (m, 3H), 7.82 (d, 2H, J = 6.5 Hz, AB), 8.03 (d, 2H, J = 6.5 Hz, AB), 8.05 (s, 2H), 8.30 (d, 1H, J = 2 Hz), 8.50 (d, 1H, J = 2 Hz), 10.28 (s, 1H). 13C NMR (DMSO‑d6, δ, ppm): 24.13, 27.62, 84.64, 93.71, 117.10, 119.21, 120.63 (2), 122.81, 123.74, 127.62, 128.31 (2), 133.04, 134.22, 135.41, 137.15, 137.90, 146.33, 153.91, 158.03, 161.85, 169.63. Anal. Calcd. for C23H18I2N4O4S2 (732.35): C, 37.72; H, 2.48; N, 7.65. Found: C, 38.07; H, 2.72; N, 7.98.
4.1.2.5. 2-((6,8-Diiodo-4-oxo-3-(4-sulfamoylphenyl)-3,4-dihydroquinazolin-2-yl)thio)-N-(p-tolyl)acetamide(8)
8: Yield, 85%; m.p. > 300 °C. IR (KBr, ʋ cm−1): 3419, 3331, 3197 (NH, NH2), 3089 (arom.), 2995, 2852 (aliph.), 1710, 1682 (2CO), 1623 (CN), 1391, 1161 (SO2). 1H NMR (DMSO‑d6, δ, ppm): 2.30 (s, 3H), 4.24 (s, 2H), 7.11 (d, 2H, J = 6 Hz, AB), 7.61 (m, 2H), 7.90 (d, 2H, J = 8 Hz, AB), 8.04 (d, 2H, J = 7 Hz, AB), 8.05 (s, 2H), 8.37 (d, 1H, J = 2 Hz), 8.58 (d, 1H, J = 2 Hz), 10.43 (s, 1H). 13C NMR (DMSO‑d6, δ, ppm): 19.21, 25.64, 80.92, 94.07, 119.21 (2), 120.16 (2), 123.72, 128.35 (2), 129.12 (2), 132.73, 134.61, 134.90, 135.11, 135.85, 145.62, 154.57, 160.61, 161.43, 165.90. MS m/z [%]: 733 [M+, 68.30], 681 [100]. Anal. Calcd. for C23H18I2N4O4S2 (732.35): C, 37.72; H, 2.48; N, 7.65. Found: C, 38.11; H, 2.80; N, 8.02.
4.1.2.6. 2-((6,8-Diiodo-4-oxo-3-(4-sulfamoylphenyl)-3,4-dihydroquinazolin-2-yl)thio)-N-(2-ethylphenyl) acetamide(9)
9: Yield, 79%; m.p. > 300 °C. IR (KBr, ʋ cm−1): 3402, 3321, 3181 (NH, NH2), 3092 (arom.), 2982, 2860 (aliph.), 1720, 1685 (2CO), 1619 (CN), 1387, 1142 (SO2). 1H NMR (DMSO‑d6, δ, ppm): 1.31 (t, 3H, J = 15 Hz), 2.54 (q, 2H, J = 12 Hz), 4.25 (s, 2H), 7.03 (dd, 1H, J = 8 & 2 Hz), 7.15 (m, 1H), 7.25 (ddd, 1H, J = 7 & 6 Hz), 7.30 (dd, 1H, J = 7 & 2 Hz), 7.76 (d, 2H, J = 9 Hz, AB), 8.03 (d, 2H, J = 7.5 Hz, AB), 8.04 (m, 2H), 8.32 (d, 1H, J = 2.5 Hz), 8.61 (d, 1H, J = 2.5 Hz), 9.79 (s, 1H). 13C NMR (DMSO‑d6, δ, ppm): 14.67, 24.23, 37.14, 91.81, 101.47, 121.87 (2), 126.36, 126.47, 126.67, 127.59, 128.88, 130.59 (2), 135.59, 135.74, 138.65, 139.69 (2), 146.65, 151.48, 158.80, 159.67, 165.71. MS m/z [%]: 746 [M+, 26.98], 747 [M+1, 5.94], 368 [100]. Anal. Calcd. for C24H20I2N4O4S2 (746.38): C, 38.62; H, 2.70; N, 7.51. Found: C, 39.01; H, 3.01; N, 7.78.
4.1.2.7. 2-((6,8-Diiodo-4-oxo-3-(4-sulfamoylphenyl)-3,4-dihydroquinazolin-2-yl)thio)-N-(3-ethylphenyl) acetamide(10)
10: Yield, 75%; m.p. > 300 °C. IR (KBr, ʋ cm−1): 3412, 3313, 3192 (NH, NH2), 3085 (arom.), 2994, 2853 (aliph.), 1714, 1679 (2CO), 1622 (CN), 1393, 1157 (SO2). 1H NMR (DMSO‑d6, δ, ppm): 1.20 (t, 3H, J = 15 Hz), 2.58 (q, 2H, J = 12 Hz), 4.21 (s, 2H), 6.98 (ddd, 1H, J = 8 & 2 Hz), 7.21–7.48 (m, 2H), 7.70–8.04 (m, 5H), 8.05 (s, 2H), 8.23 (d, 1H, J = 3 Hz), 8.54 (d, 1H, J = 3 Hz), 10.32 (s, 1H). 13C NMR (DMSO‑d6, δ, ppm): 15.21, 24.10, 29.82, 82.60, 92.01, 120.43 (2), 122.71, 128.58, 125.73, 126.02, 126.68, 128.81, 129.12 (2), 133.65, 134.01, 135.28, 135.91, 144.80, 156.02, 160.51, 160.95, 168.12. Anal. Calcd. for C24H20I2N4O4S2 (746.38): C, 38.62; H, 2.70; N, 7.51. Found: C, 38.31; H, 2.44; N, 7.20.
4.1.2.8. 2-((6,8-Diiodo-4-oxo-3-(4-sulfamoylphenyl)-3,4-dihydroquinazolin-2-yl)thio)-N-(4-ethylphenyl) acetamide(11)
11: Yield, 73%; m.p. > 300 °C. IR (KBr, ʋ cm−1): 3422, 3330, 3201 (NH, NH2), 3094 (arom.), 2986, 2847 (aliph.), 1710, 1678 (2CO), 1631 (CN), 1384, 1156 (SO2). 1H NMR (DMSO‑d6, δ, ppm): 1.15 (t, 3H, J = 10 Hz), 2.55 (q, 2H, J = 8 Hz), 4.23 (s, 2H), 7.12 (d, 2H, J = 9 Hz, AB), 7.61 (d, 2H, J = 9 Hz, AB), 7.79 (d, 2H, J = 8.5 Hz), 7.80–8.05 (m, 4H), 8.39 (d, 1H, J = 2 Hz), 8.58 (d, 1H, J = 2 Hz), 10.31 (s, 1H). 13C NMR (DMSO‑d6, δ, ppm): 17.12, 29.40, 29.62, 83.25, 93.04, 120.61 (2), 120.83 (2), 123.10, 127.65 (2), 128.02 (2), 133.71, 135.07, 135.63, 135.80, 140.92, 150.13, 153.81, 160.72, 160.97, 166.32. Anal. Calcd. for C24H20I2N4O4S2 (746.38): C, 38.62; H, 2.70; N, 7.51. Found: C, 38.97; H, 2.96; N, 7.78.
4.1.2.9. 2-((6,8-Diiodo- 4-oxo-3-(4-sulfamoylphenyl)-3, 4-dihydroquinazolin-2-yl)thio)-N -(4-methoxyphenyl) acetamide(12)
12: Yield, 79%; m.p. > 300 °C. IR (KBr, ʋ cm−1): 3397, 3325, 3194 (NH, NH2), 3087 (arom.), 2990, 2852 (aliph.), 1716, 1684 (2CO), 1627 (CN), 1396, 1140 (SO2). 1H NMR (DMSO‑d6, δ, ppm): 3.75 (s, 3H), 4.29 (s, 2H), 6.87 (d, 2H, J = 9 Hz, AB), 7.56 (d, 2H, J = 9 Hz, AB), 7.78 (d, 2H, J = 7 Hz, AB), 7.95 (d, 2H, J = 7 Hz), 8.04 (s, 2H), 8.27 (d, 1H, J = 2.5 Hz), 8.61 (d, 1H, J = 2.5 Hz), 10.20 (s, 1H). 13C NMR (DMSO‑d6, δ, ppm): 29.43, 54.26, 83.03, 91.67, 112.01 (2), 118.33 (2), 123.14 (2), 123.85, 127.82 (2), 129.64, 134.12, 134.83, 135.90, 150.23, 156.71, 157.42, 160.04, 161.62, 169.31. Anal. Calcd. for C23H18I2N4O5S2 (748.35): C, 36.91; H, 2.42; N, 7.49. Found: C, 37.27; H, 2.70; N, 7.82.
4.1.2.10. 2-((6,8-Diiodo-4-oxo-3-(4-sulfamoylphenyl)-3,4-dihydroquinazolin-2-yl)thio)-N-(4-ethoxyphenyl) acetamide(13)
13: Yield, 78%; m.p. > 300 °C. IR (KBr, ʋ cm−1): 3385, 3295, 3188 (NH, NH2), 3096 (arom.), 2979, 2844 (aliph.), 1705, 1680 (2CO), 1608 (CN), 1387, 1148 (SO2). 1H NMR (DMSO‑d6, δ, ppm): 1.27 (t, 3H, J = 12 Hz), 3.97 (q, 2H, J = 10 Hz), 4.21 (s, 2H), 6.89 (d, 2H, J = 10 Hz, AB), 7.39 (d, 2H, J = 10 Hz, AB), 7.76 (d, 2H, J = 8 Hz, AB), 8.03–8.10 (m, 4H), 8.29 (d, 1H, J = 2 Hz), 8.62 (d, 1H, J = 2 Hz), 10.29 (s, 1H). 13C NMR (DMSO‑d6, δ, ppm): 15.21, 29.63, 62.04, 83.72, 92.57, 115.03 (2), 120.61 (2), 121.40 (2), 122.74, 127.66 (2), 129.52, 134.67, 134.91, 135.78, 146.83, 154.02, 155.25, 160.12, 162.46, 169.60. Anal. Calcd. for C24H20I2N4O5S2 (762.38): C, 37.81; H, 2.64; N, 7.35. Found: C, 38.20; H, 3.00; N, 7.65.
4.1.2.11. 2-((6,8-Diiodo-4-oxo-3-(4-sulfamoylphenyl)-3,4-dihydroquinazolin-2-yl)thio)-N-(3,5-dimethoxyphenyl) acetamide(14)
14: Yield, 67%; m.p. > 300 °C. IR (KBr, ʋ cm−1): 3256, 3209, 3142 (NH, NH2), 3065 (arom.), 2912, 2844 (aliph.), 1676, 1663 (2CO), 1608 (CN), 1385, 1161 (SO2). 1H NMR (DMSO‑d6, δ, ppm): 3.74 (s, 6H), 4.21 (s, 2H), 6.25 (dd, 1H, J = 3 & 1.5 Hz), 6.87 (dd, 2H, J = 3 & 1.5 Hz), 7.77 (d, 2H, J = 9 Hz, AB), 8.04 (d, 2H, J = 9 Hz, AB), 8.05 (s, 2H), 8.34 (d, 1H, J = 2.5 Hz), 8.63 (d, 1H, J = 2.5 Hz), 10.38 (s, 1H). 13C NMR (DMSO‑d6, δ, ppm): 31.32, 55.56 (2), 91.66, 95.96, 98.12, 101.11 (2), 121.92 (2), 127.61, 130.64 (2), 135.64, 138.77 (2), 140.93, 146.14, 151.43, 158.86, 159.59 (2), 160.90, 165.20. Anal. Calcd. for C24H20I2N4O6S2 (778.38): C, 37.03; H, 2.59; N, 7.20. Found: C, 37.34; H, 2.83; N, 7.43.
4.1.2.12. 2-((6,8-Diiodo-4-oxo-3-(4-sulfamoylphenyl)-3,4-dihydroquinazolin-2-yl)thio)-N-(3,4,5-trimethoxyphenyl) acetamide(15)
15: Yield, 80%; m.p. > 300 °C. IR (KBr, ʋ cm−1): 3416, 3319, 3200 (NH, NH2), 3093 (arom.), 2982, 2822 (aliph.), 1689, 1666 (2CO), 1616 (CN), 1370, 1151 (SO2). 1H NMR (DMSO‑d6, δ, ppm): 3.70 (s, 6H), 3.81 (s, 3H), 4.31 (s, 2H), 6.90 (d, 2H, J = 3 Hz), 7.78 (d, 2H, J = 9 Hz, AB), 8.04 (s, 2H), 8.05 (d, 2H, J = 9 Hz), 8.28 (d, 1H, J = 2.5 Hz, AB), 8.54 (d, 1H, J = 2.5 Hz), 10.36 (s, 1H). 13C NMR (DMSO‑d6, δ, ppm): 27.23, 55.60 (2), 61.32, 82.74, 95.07, 101.41 (2), 119.60 (2), 121.82, 130.74 (2), 131.90, 133.81, 134.15, 134.67, 134.92, 146.04, 151.32 (2), 153.50, 160.01, 160.73, 166.37. MS m/z [%]: 809 [M+, 15.75], 456 [100]. Anal. Calcd. for C25H22I2N4O7S2 (808.40): C, 37.14; H, 2.74; N, 6.93. Found: C, 37.44; H, 3.03; N, 7.29.
4.1.2.13. 2-((6,8-Diiodo-4-oxo-3-(4-sulfamoylphenyl)-3,4-dihydroquinazolin-2-yl)thio)-N-(2-methyl-4-nitrophenyl) acetamide(16)
16: Yield, 74%; m.p. > 300 °C. IR (KBr, ʋ cm−1): 3313, 3265, 3203 (NH, NH2), 3055 (arom.), 2943, 2811 (aliph.), 1683, 1660 (2CO), 1621 (CN), 1530, 1370 (NO2), 1397, 1155 (SO2). 1H NMR (DMSO‑d6, δ, ppm): 2.39 (s, 3H), 4.30 (s, 2H), 7.72 (d, 1H, J = 9.5 Hz), 7.88 (d, 2H, J = 8 Hz, AB), 7.90–8.05 (m, 6H), 8.26 (d, 1H, J = 3 Hz), 8.59 (d, 1H, J = 3 Hz), 10.27 (s, 1H). 13C NMR (DMSO‑d6, δ, ppm): 18.05, 27.34, 83.69, 92.03, 106.32, 120.85, 121.71 (2), 125.83, 127.61, 130.64 (2), 133.72, 134.80, 134.92, 135.01, 141.42, 141.94, 147.56, 154.02, 160.82, 161.04, 167.42. Anal. Calcd. for C23H17I2N5O6S2 (777.35): C, 35.54; H, 2.20; N, 9.01. Found: C, 35.78; H, 2.57; N, 9.32.
4.1.2.14. 2-((6, 8-Diiodo-4-oxo-3-(4-sulfamoylphenyl)-3, 4-dihydroquinazolin-2-yl)thio)- N-(2-methyl-6-nitrophenyl) acetamide(17)
17: Yield, 70%; m.p. > 300 °C. IR (KBr, ʋ cm−1): 3329, 3283, 3174 (NH, NH2), 3087 (arom.), 2952, 2828 (aliph.), 1680, 1662 (2CO), 1615 (CN), 1545, 1340 (NO2), 1376, 1155 (SO2). 1H NMR (DMSO‑d6, δ, ppm): 2.21 (s, 3H), 4.27 (s, 2H), 7.34 (dd, 1H, J = 9 & 6 Hz), 7.76 (dd, 1H, J = 9 & 2.5 Hz), 7.82 (d, 2H, J = 10 Hz), 8.01–8.10 (m, 5H), 8.27 (d, 1H, J = 2.5 Hz), 8.67 (d, 1H, J = 2.5 Hz), 10.02 (s, 1H). 13C NMR (DMSO‑d6, δ, ppm): 18.17, 36.90, 91.83, 101.40, 121.87 (2), 122.70, 127.43, 127.63, 128.79, 130.67 (2), 135.49, 135.62 (2), 137.85 (2), 138.68, 146.93, 151.50, 158.34, 159.67, 165.87. MS m/z [%]: 777 [M+, 4.30], 64 [100]. Anal. Calcd. for C23H17I2N5O6S2 (777.35): C, 35.54; H, 2.20; N, 9.01. Found: C, 35.80; H, 2.47; N, 9.33.
4.1.2.15. 2-((6, 8-Diiodo-4-oxo-3-(4-sulfamoylphenyl)-3, 4-dihydroquinazolin-2-yl)thio)- N-(2,4-dinitrophenyl) acetamide(18)
18: Yield, 66%; m.p. > 300 °C. IR (KBr, ʋ cm−1): 3432, 3303, 3165 (NH, NH2), 3058 (arom.), 2961, 2850 (aliph.), 1690, 1683 (2CO), 1608 (CN), 1537, 1361 (NO2), 1388, 1161 (SO2). 1H NMR (DMSO‑d6, δ, ppm): 4.31 (s, 2H), 7.75 (d, 2H, J = 8 Hz, AB), 7.90 (d, 1H, J = 10 Hz), 8.00–8.04 (m, 2H), 8.04 (d, 2H, J = 8 Hz, AB), 8.29 (d, 1H, J = 2.5 Hz), 8.30–8.34 (m, 3H), 11.21 (s, 1H). 13C NMR (DMSO‑d6, δ, ppm): 29.21, 86.23, 102.13, 117.69, 121.03 (2), 122.12, 124.25, 127.43 (2), 131.16, 134.64 (2), 139.50 (2), 142.62, 144.12, 149.04, 154.91, 160.24, 165.65, 167.45. Anal. Calcd. for C22H14I2N6O8S2 (808.32): C, 32.69; H, 1.75; N, 10.40. Found: C, 32.36; H, 1.47; N, 10.18.
4.2. Biological evaluation
4.2.1. NQO1 in vitro inducer activity
Hepa1c1c7 murine hepatoma cells were grown in a humidified atmosphere at 37 °C, 5% CO2. The cells were tested routinely to ensure that they were mycoplasma-free. The α-minimum essential medium (α-MEM) supplemented with 10% (v/v) heat- and charcoal-inactivated (1 g/100 mL, 90 min at 55 °C) fetal bovine serum was used. For evaluation of the potential NQO1 inducer activity, cells (104/well) were grown in transparent flat-bottom plastic 96-well plates for 24 h, after which the cell culture medium was replaced with fresh medium containing each inducer (dissolved in DMSO and diluted in the medium 1:1000), and the cells were grown for further 48 h. Three replicates of each treatment of eight serial dilutions of inducers were used. The final DMSO concentration in the cell culture medium was maintained at 0.1% (v/v) in all wells. Cell lysates were prepared in digitonin and the specific activity of NQO1 was determined using menadione as a substrate as described [67,68]. Briefly, the cell culture medium was removed from each well, and the cells were washed three times with 200 μL of phosphate buffered saline (PBS), and subsequently lysed in 75 μL of digitonin suspension in the presence of EDTA for 20 min with shaking. Of the cell lysate, 20 μL was transferred to the well of a new plate and used to determine the protein concentration by adding 300 μL of bicinchoninic acid reagent and measuring the reaction product spectrophotometrically in a 96-well plate reader at 550 nm after 30 min incubation at room temperature. The remaining 55 μL of the cell lysate was used to measure the enzyme activity of NQO1. This was done by addition of 200 μL of enzyme assay buffer, containing NADPH-generating system (glucose-6-phosphate, glucose-6-phosphate dehydrogenase, NADP) that maintained a constant NADPH concentration, FAD, menadione (2-methyl-1,4-naphthoquinone, a quinone that is reduced to menadiol by NQO1 in the presence of NADPH), and MTT (3-[4,5-dimethylthiazo-2-yl]-2,5-diphenyltetrazolium bromide; a tetrazolium dye that is reduced non-enzymatically to a formazan dye by menadiol). The reaction was terminated after 5 min by the addition of dicumarol (a potent inhibitor of NQO1), and the reduced formazan dye was measured spectrophotometrically at 610 nm. The Concentration that Doubles the specific activity of NQO1 (CD value) was used as a measure of inducer potency. Mean values for the eight replicate wells are shown for each data point. The standard deviation for each data point was within 5% of the mean value. The classical NQO1 inducer sulforaphane was included as a positive control in each bioassay and was consistently giving a CD value of 0.2 μM.
4.2.2. In vivo evaluation
4.2.2.1. Animals
Eight-week old Swiss albino male mice (20–25 g) were supplied from the breeding unit of the National Center for Radiation Research and Technology (NCRRT), Cairo, Egypt. They were housed in the laboratory room for one week prior to the experiment for acclimatization to the lab environment. Water and food were allowed ad libitum. Mice were kept under controlled conditions: room temperature of 25 ± 5 °C, humidity (60 ± 5%), alternating 12 h dark and 12 h light cycle. Animals were treated gently; squeezing, pressure and tough handling were avoided. All animal procedures were performed in accordance with the Ethics Committee for Animal Experimentations, Faculty of Pharmacy, Cairo University, which complies with the Guide for the Care and Use of Laboratory Animals issued by the US National Institutes of Health (NIH Publication No. 85-23, revised 2011).
4.2.2.2. Irradiation process
Mice were exposed to whole-body gamma radiation as a single dose of 7 Gy using Canadian Gamma Cell-40 biological irradiator (137Cs) located at the NCRRT, Cairo, Egypt and the dose rate was 0.655 rad/s.
4.2.2.3. Acute toxicity study
The median lethal dose (LD50) of the most promising compound, 15 was determined according to Chinedu et al. [69].
4.2.2.4. Experimental design
Eighty mice were randomly classified into four groups. First (control) group was injected i.p. with 10% DMSO, daily, for 5 days. Second (irradiated) group was treated as control, and after 1 h from last DMSO injection, the mice were irradiated at a dose of 7 Gy. Third (Compound 15) group; received 50 mg/kg/day i.p. (1/10 LD50) of compound 15, daily for 5 days. Fourth (Compound 15 + Irradiation) group was treated as third group then on the last day, after 1 h of injection, mice exposed to 7 Gy gamma radiation. On the third day, five mice from each group (n = 5) were weighed and anesthetized using urethane (1.2 mg/kg i.p) [70]. Then the blood samples were collected by cardiac puncture. At that time, they were euthanized by cervical dislocation. Each blood sample was collected into EDTA coated tubes for complete blood picture. Liver tissues were rinsed with ice-cold saline, dried on a filter paper and weighed to calculate the relative liver/body weight ratio. Then, it was homogenized in ice-cold 0.1 M phosphate buffer saline (pH 7.4) and stored at −80 °C till used for subsequent biochemical analysis. The residual of mice in all the groups was monitored on a daily basis for 30 days to check the survival rate, as well as their body weight, were recorded weekly to estimate the changes in body weight.
4.2.2.5. Biochemical parameters investigated in liver homogenate
Liver homogenates were used for measuring the level of Nrf2 using colorimetric cell-based Elisa kit (OKAG00918) Aviva systems biology (San Diego, CA., USA), as well as the level of NAD(P)H: quinone oxidoreductase 1 (NQO1) was measured using an ELISA Kit (OKCD02727) Aviva Systems Biology (San Diego, CA., USA). Liver lipid peroxides were determined by measuring MDA as an indicator according to the method of Yoshioka et al. [71]. The generation of ROS in liver tissues was measured according to a modified technique of Vrablic et al. [72].
4.2.2.6. Hematological analysis
Complete blood count with platelet count was determined using the automated micro-analyzer (BC-2800 Mindray, China).
4.2.2.7. Statistical analysis
Data were analyzed using Prism 5.03 (GraphPad, San Diego, CA, USA) and expressed as means ± standard error. Comparisons between groups were analyzed by one-way analysis of variance (ANOVA) followed by Bonferroni’s multiple comparison test. Survival was analyzed by the Kaplan–Meier method followed by the Mantel–Cox (log rank) test. Body weight changes between groups through 30 days were analyzed by two-way ANOVA followed by Bonferroni’s post test. P < 0.05 was considered to represent statistically significant differences.
4.3. Molecular docking
The molecular modeling studies were fulfilled by the Molecular Operating Environment software (MOE, 10.2008). The receptor was chosen from the protein data bank; 4IQK that represents Keap1 co-crystallized with N,N′-naphthalene-1,4-diylbis(4-methoxybenzenesulfonamide. The protein was prepared for docking by ignoring water in the receptor. Hydrogen atoms were added to the structure with their standard geometry. The co-crystallized ligand was used to determine the binding site. Triangle Matcher placement method and dG scoring function were used for docking. Energy minimizations were performed with an RMSD gradient of 0.1 kcal mol−1Å−1with the MMFF94X force field and the partial charges were automatically calculated. Validation of the docking protocol was performed by re-docking of the co-crystallized ligands into the active site of Keap1 protein followed by docking of compound 15. The obtained data were used to interpret the ligand-protein interactions at the Nrf2-binding site.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgment
The authors appreciate the staff members of the gamma irradiation unit at the National Center for Radiation Research and Technology (NCRRT) for carrying out the irradiation process. Maureen Higgins and Albena T Dinkova-Kostova are grateful to Cancer Research UK (C20953/A18644) for financial support.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ejmech.2020.112467.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Citrin D., Cotrim A.P., Hyodo F., Baum B.J., Krishna M.C., Mitchell J.B. Radioprotectors and mitigators of radiation-induced normal tissue injury. Oncol. 2010;15:360–371. doi: 10.1634/theoncologist.2009-S104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Karam H.M., Gharib O.A. The therapeutic role of ziziphus extract on liver injury induced by electromagnetic waves and ionizing radiation as environmental pollutants. J. Nucl. Tech. Appl. Sci. 2018;6:207–219. [Google Scholar]
- 3.Radwan R.R., Karam H.M. Resveratrol attenuates intestinal injury in irradiated rats via PI3K/Akt/mTOR signaling pathway. Environ. Toxicol. 2020;35:223–230. doi: 10.1002/tox.22859. [DOI] [PubMed] [Google Scholar]
- 4.Haffor A., Al-Sadoon M. Increased antioxidant potential and decreased free radical production in response to mild injection of crude venom, Cerastes cerastes gasperetti. Toxicol. Mech. Methods. 2008;18:11–16. doi: 10.1080/15376510701728455. [DOI] [PubMed] [Google Scholar]
- 5.Shimura T., Kunugita N. Mitochondrial reactive oxygen species-mediated genomic instability in low-dose irradiated human cells through nuclear retention of cyclin D1. Cell Cycle. 2016;15:1410–1414. doi: 10.1080/15384101.2016.1170271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Selim M., Saha A., Mukherjea K.K. Protection of radiation induced DNA damage by a newly developed molybdenum complex. J. Radioanal. Nucl. Chem. 2017;311:189–193. [Google Scholar]
- 7.Abdel Fattah S.M., Mohmed H.K., Mohamed M.A.E.H. The potential protective effect of ferulic acid against gamma irradiation induced ovarian failure in rats, Egypt. J. Radit. Sci. Appl. 2019;32:1–12. [Google Scholar]
- 8.Ibrahim A.A., Karam H.M., Shaaban E.A., Safar M.M., El-Yamany M.F. MitoQ ameliorates testicular damage induced by gamma irradiation in rats: modulation of mitochondrial apoptosis and steroidogenesis. Life Sci. 2019;232:116655. doi: 10.1016/j.lfs.2019.116655. [DOI] [PubMed] [Google Scholar]
- 9.Karam H.M., Radwan R.R. Metformin modulates cardiac endothelial dysfunction, oxidative stress and inflammation in irradiated rats: a new perspective of an antidiabetic drug. Clin. Exp. Pharmacol. Physiol. 2019;46:1124–1132. doi: 10.1111/1440-1681.13148. [DOI] [PubMed] [Google Scholar]
- 10.Valko M., Leibfritz D., Moncol J., Cronin M.T., Mazur M., Telser J. Free radicals and antioxidants in normal physiological functions and human disease. Int. J. Biochem. Cell Biol. 2007;39:44–84. doi: 10.1016/j.biocel.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 11.Kamran M.Z., Ranjan A., Kaur N., Sur S., Tandon V. Radioprotective agents: strategies and translational advances. Med. Res. Rev. 2016;36:461–493. doi: 10.1002/med.21386. [DOI] [PubMed] [Google Scholar]
- 12.Greenberger J.S. Radioprotection. In Vivo. 2009;23:323–336. [PMC free article] [PubMed] [Google Scholar]
- 13.Tsutomu N., Munetaka Y., Toshihiko O., Shunro K. Suppression of active oxygen-induced cytotoricity by flavonoids. Biochem. Pharmacol. 1993;45:265–267. doi: 10.1016/0006-2952(93)90402-i. [DOI] [PubMed] [Google Scholar]
- 14.Devasagayam T., Tilak J., Boloor K., Sane K.S., Ghaskadbi S.S., Lele R. Free radicals and antioxidants in human health: current status and future prospects. J. Assoc. Phys. India. 2004;52:794–804. [PubMed] [Google Scholar]
- 15.Kensler T.W., Wakabayashi N., Biswal S. Cell survival responses to environmental stresses via the Keap1-Nrf2-ARE pathway. Annu. Rev. Pharmacol. Toxicol. 2007;47:89–116. doi: 10.1146/annurev.pharmtox.46.120604.141046. [DOI] [PubMed] [Google Scholar]
- 16.Cullinan S.B., Gordan J.D., Jin J., Harper J.W., Diehl J.A. The Keap1-BTB protein is an adaptor that bridges Nrf2 to a Cul3-based E3 ligase: oxidative stress sensing by a Cul3-Keap1 ligase. Mol. Cell Biol. 2004;24:8477–8486. doi: 10.1128/MCB.24.19.8477-8486.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kobayashi A., Kang M.-I., Okawa H., Ohtsuji M., Zenke Y., Chiba T., Igarashi K., Yamamoto M. Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2. Mol. Cell Biol. 2004;24:7130–7139. doi: 10.1128/MCB.24.16.7130-7139.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang B., Schmoyer D., Kirov S., Snoddy J. GOTree Machine (GOTM): a web-based platform for interpreting sets of interesting genes using Gene Ontology hierarchies. BMC Bioinf. 2004;5:16–24. doi: 10.1186/1471-2105-5-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dinkova-Kostova A.T., Talalay P. NAD (P) H: quinone acceptor oxidoreductase 1 (NQO1), a multifunctional antioxidant enzyme and exceptionally versatile cytoprotector. Arch. Biochem. Biophys. 2010;501:116–123. doi: 10.1016/j.abb.2010.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alsaid M.S., Al-Mishari A.A., Soliman A.M., Ragab F.A., Ghorab M.M. Discovery of Benzo [g] quinazolin benzenesulfonamide derivatives as dual EGFR/HER2 inhibitors. Eur. J. Med. Chem. 2017;141:84–91. doi: 10.1016/j.ejmech.2017.09.061. [DOI] [PubMed] [Google Scholar]
- 21.Ghorab M.M., Alsaid M.S., Soliman A.M., Al-Mishari A.A. Benzo [g] quinazolin-based scaffold derivatives as dual EGFR/HER2 inhibitors. J. Enzym. Inhib. Med. Chem. 2018;33:67–73. doi: 10.1080/14756366.2017.1389922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kumar A., Sharma P., Kumari P., Kalal B.L. Exploration of antimicrobial and antioxidant potential of newly synthesized 2, 3-disubstituted quinazoline-4 (3H)-ones. Bioorg. Med. Chem. Lett. 2011;21:4353–4357. doi: 10.1016/j.bmcl.2011.05.031. [DOI] [PubMed] [Google Scholar]
- 23.Soliman A.M., Ghorab M.M. Exploration of N-alkyl-2-[(4-oxo-3-(4-sulfamoylphenyl)-3, 4-dihydroquinazolin-2-yl) thio] acetamide derivatives as anticancer and radiosensitizing agents. Bioorg. Chem. 2019;88:102956. doi: 10.1016/j.bioorg.2019.102956. [DOI] [PubMed] [Google Scholar]
- 24.Genç Y., Özkanca R., Bekdemir Y. Antimicrobial activity of some sulfonamide derivatives on clinical isolates of Staphylococus aureus. Ann. Clin. Microbiol. Antimicrob. 2008;7:7–17. doi: 10.1186/1476-0711-7-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ghorab M.M., Alsaid M.S., El-Gaby M.S., Safwat N.A., Elaasser M.M., Soliman A.M. Biological evaluation of some new N-(2, 6-dimethoxypyrimidinyl) thioureido benzenesulfonamide derivatives as potential antimicrobial and anticancer agents. Eur. J. Med. Chem. 2016;124:299–310. doi: 10.1016/j.ejmech.2016.08.060. [DOI] [PubMed] [Google Scholar]
- 26.Ghorab M.M., Soliman A.M., Alsaid M.S., Askar A.A. Synthesis, antimicrobial activity and docking study of some novel 4-(4, 4-dimethyl-2, 6-dioxocyclohexylidene) methylamino derivatives carrying biologically active sulfonamide moiety. Arab. J. Chem. 2020;13:545–556. [Google Scholar]
- 27.Ghorab M.M., Alqahtani A.S., Soliman A.M., Askar A.A. Novel N-(Substituted) thioacetamide quinazolinone benzenesulfonamides as antimicrobial agents. Int. J. Nanomed. 2020;15:3161–3180. doi: 10.2147/IJN.S241433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.De Clercq E. Hamao Umezawa memorial award Lecture:‘An odyssey in the viral chemotherapy field’. Int. J. Antimicrob. Agents. 2001;18:309–328. doi: 10.1016/s0924-8579(01)00411-3. [DOI] [PubMed] [Google Scholar]
- 29.Borne R.F., Peden R.L., Waters I., Weiner M., Jordan R., Coats E.A. Anti-inflammatory activity of para-substituted N-benzenesulfonyl derivatives of anthranilic acid. J. Pharm. Sci. 1974;63:615–617. doi: 10.1002/jps.2600630428. [DOI] [PubMed] [Google Scholar]
- 30.Doungsoongnuen S., Worachartcheewan A., Pingaew R., Suksrichavalit T., Prachayasittikul S., Ruchirawat S., Prachayasittikul V. Investigation on biological activities of anthranilic acid sulfonamide analogs. EXCLI J. 2011;10:155–161. [PMC free article] [PubMed] [Google Scholar]
- 31.Gouda M.A., Hussein B.H. Synthesis and anti-oxidant evaluation of some novel sulfa drugs. Lett. Drug Des. Discov. 2017;14:1425–1432. [Google Scholar]
- 32.Soliman A.M., Alqahtani A.S., Ghorab M.M. Novel sulphonamide benzoquinazolinones as dual EGFR/HER2 inhibitors, apoptosis inducers and radiosensitizers. J. Enzym. Inhib. Med. Chem. 2019;34:1030–1040. doi: 10.1080/14756366.2019.1609469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ghorab M.M., Ragab F.A., Heiba H.I., Soliman A.M. Design and synthesis of some novel 4-Chloro-N-(4-(1-(2-(2-cyanoacetyl) hydrazono) ethyl) phenyl) benzenesulfonamide derivatives as anticancer and radiosensitizing agents. Eur. J. Med. Chem. 2016;117:8–18. doi: 10.1016/j.ejmech.2016.04.009. [DOI] [PubMed] [Google Scholar]
- 34.Ghorab M.M., Alsaid M.S., Samir N., Abdel-Latif G.A., Soliman A.M., Ragab F.A., El Ella D.A.A. Aromatase inhibitors and apoptotic inducers: design, synthesis, anticancer activity and molecular modeling studies of novel phenothiazine derivatives carrying sulfonamide moiety as hybrid molecules. Eur. J. Med. Chem. 2017;134:304–315. doi: 10.1016/j.ejmech.2017.04.028. [DOI] [PubMed] [Google Scholar]
- 35.El Ella D.A.A., Ghorab M.M., Heiba H.I., Soliman A.M. Synthesis of some new thiazolopyrane and thiazolopyranopyrimidine derivatives bearing a sulfonamide moiety for evaluation as anticancer and radiosensitizing agents. Med. Chem. Res. 2012;21:2395–2407. [Google Scholar]
- 36.Chinthakindi P.K., Naicker T., Thota N., Govender T., Kruger H.G., Arvidsson P.I. Sulfonimidamides in medicinal and agricultural chemistry. Angew. Chem. Int. Ed. 2017;56:4100–4109. doi: 10.1002/anie.201610456. [DOI] [PubMed] [Google Scholar]
- 37.Winum J.-Y., Maresca A., Carta F., Scozzafava A., Supuran C.T. Polypharmacology of sulfonamides: pazopanib, a multitargeted receptor tyrosine kinase inhibitor in clinical use, potently inhibits several mammalian carbonic anhydrases. Chem. Commun. 2012;48:8177–8179. doi: 10.1039/c2cc33415a. [DOI] [PubMed] [Google Scholar]
- 38.Venturi S. Evolutionary significance of iodine. Curr. Chem. Biol. 2011;5:155–162. [Google Scholar]
- 39.Aceves C., Anguiano B., Delgado G. The extrathyronine actions of iodine as antioxidant, apoptotic, and differentiation factor in various tissues. Thyroid. 2013;23:938–946. doi: 10.1089/thy.2012.0579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smyth P.P. Role of iodine in antioxidant defence in thyroid and breast disease. Biofactors. 2003;19:121–130. doi: 10.1002/biof.5520190304. [DOI] [PubMed] [Google Scholar]
- 41.Winkler R., Griebenow S., Wonisch W. Effect of iodide on total antioxidant status of human serum. Cell Biochem. Funct. 2000;18:143–146. doi: 10.1002/(SICI)1099-0844(200006)18:2<143::AID-CBF857>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 42.Ghorab M.M., El Ella D.A.A., Heiba H.I., Soliman A.M. Synthesis of certain new thiazole derivatives bearing a sulfonamide moiety with expected anticancer and radiosensitizing activities. J. Mater. Sci. Eng. 2011;1:684–691. [Google Scholar]
- 43.Dinkova-Kostova A.T., Kostov R.V., Canning P. Keap1, the cysteine-based mammalian intracellular sensor for electrophiles and oxidants. Arch. Biochem. Biophys. 2017;617:84–93. doi: 10.1016/j.abb.2016.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bertrand K.A., Baer H.J., Orav E.J., Klifa C., Shepherd J.A., Van Horn L., Snetselaar L., Stevens V.J., Hylton N.M., Dorgan J.F. Body fatness during childhood and adolescence and breast density in young women: a prospective analysis. Breast Cancer Res. 2015;17:95–105. doi: 10.1186/s13058-015-0601-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hu L., Magesh S., Chen L., Wang L., Lewis T.A., Chen Y., Khodier C., Inoyama D., Beamer L.J., Emge T.J. Discovery of a small-molecule inhibitor and cellular probe of Keap1–Nrf2 protein–protein interaction. Bioorg. Med. Chem. Lett. 2013;23:3039–3043. doi: 10.1016/j.bmcl.2013.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lazzara P.R., David B.P., Ankireddy A., Richardson B.G., Dye K., Ratia K.M., Reddy S.P., Moore T.W. Isoquinoline Kelch-like ECH-associated protein 1-nuclear factor (Erythroid-Derived 2)-like 2 (KEAP1-NRF2) inhibitors with high metabolic stability. J. Med. Chem. 2019 doi: 10.1021/acs.jmedchem.9b01074. under press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Marina R., González P., Ferreras M.C., Costilla S., Barrio J.P. Hepatic Nrf2 expression is altered by quercetin supplementation in X-irradiated rats. Mol. Med. Rep. 2015;11:539–546. doi: 10.3892/mmr.2014.2741. [DOI] [PubMed] [Google Scholar]
- 48.Karam H.M., Radwan R.R. Metformin modulates cardiac endothelial dysfunction, oxidative stress and inflammation in irradiated rats: a new perspective of an antidiabetic drug. Clin. Exp. Pharmacol. Physiol. 2019;46:1124–1132. doi: 10.1111/1440-1681.13148. [DOI] [PubMed] [Google Scholar]
- 49.McDonald J.T., Kim K., Norris A.J., Vlashi E., Phillips T.M., Lagadec C., Della Donna L., Ratikan J., Szelag H., Hlatky L. Ionizing radiation activates the Nrf2 antioxidant response. Canc. Res. 2010;70:8886–8895. doi: 10.1158/0008-5472.CAN-10-0171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Aleksunes L.M., Manautou J.E. Emerging role of Nrf2 in protecting against hepatic and gastrointestinal disease. Toxicol. Pathol. 2007;35:459–473. doi: 10.1080/01926230701311344. [DOI] [PubMed] [Google Scholar]
- 51.Xu J., Jaiswal A.K. NAD (P) H: quinone oxidoreductase 1 (NQO1) competes with 20S proteasome for binding with C/EBPα leading to its stabilization and protection against radiation-induced myeloproliferative disease. J. Biol. Chem. 2012;287:41608–41618. doi: 10.1074/jbc.M112.387738. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 52.Soliman A.M., Karam H.M., Mekkawy M.H., Ghorab M.M. Antioxidant activity of novel quinazolinones bearing sulfonamide: potential radiomodulatory effects on liver tissues via NF-κB/PON1 pathway. Eur. J. Med. Chem. 2020;197:112333. doi: 10.1016/j.ejmech.2020.112333. [DOI] [PubMed] [Google Scholar]
- 53.Cuadrado A., Rojo A.I., Wells G., Hayes J.D., Cousin S.P., Rumsey W.L., Attucks O.C., Franklin S., Levonen A.-L., Kensler T.W. Therapeutic targeting of the NRF2 and KEAP1 partnership in chronic diseases. Nat. Rev. Drug Discov. 2019;18:295–317. doi: 10.1038/s41573-018-0008-x. [DOI] [PubMed] [Google Scholar]
- 54.Kovac S., Angelova P.R., Holmström K.M., Zhang Y., Dinkova-Kostova A.T., Abramov A.Y. Nrf2 regulates ROS production by mitochondria and NADPH oxidase. Biochim. Biophys. 2015;1850:794–801. doi: 10.1016/j.bbagen.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Han X., Zhang J., Xue X., Zhao Y., Lu L., Cui M., Miao W., Fan S. Theaflavin ameliorates ionizing radiation-induced hematopoietic injury via the NRF2 pathway. Free Radic. Biol. Med. 2017;113:59–70. doi: 10.1016/j.freeradbiomed.2017.09.014. [DOI] [PubMed] [Google Scholar]
- 56.Begum N., Prasad N.R., Thayalan K. Apigenin protects gamma-radiation induced oxidative stress, hematological changes and animal survival in whole body irradiated Swiss albino mice. Int. J. Nutr. Pharmacol. Neurol. Dis. 2012;2:45–52. [Google Scholar]
- 57.Yi L., Tian M., Piao C., Gao G., Wu L., Pan Y., Liu J. The protective effects of 1, 2-propanediol against radiation-induced hematopoietic injury in mice. Biomed. Pharmacother. 2019;114:108806. doi: 10.1016/j.biopha.2019.108806. [DOI] [PubMed] [Google Scholar]
- 58.Wang Y., Liu L., Pazhanisamy S.K., Li H., Meng A., Zhou D. Total body irradiation causes residual bone marrow injury by induction of persistent oxidative stress in murine hematopoietic stem cells. Free Rad. Biol. Med. 2010;48:348–356. doi: 10.1016/j.freeradbiomed.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Thimmaiah N.M., Joshi C.G., Patil R.K., Khandagale A.S., Somashekarappa H., Ananda D., Manjunath H. Mitigation of radiation-induced oxidative stress by methanolic extract of Tragia involucrata in swiss albino mice. Pharmacogn. Res. 2019;11:236–243. [Google Scholar]
- 60.Lu L., Dong J., Li D., Zhang J., Fan S. 3, 3′-diindolylmethane mitigates total body irradiation-induced hematopoietic injury in mice. Free Rad. Biol. Med. 2016;99:463–471. doi: 10.1016/j.freeradbiomed.2016.09.007. [DOI] [PubMed] [Google Scholar]
- 61.Singh V.K., Newman V.L., Seed T.M. Colony-stimulating factors for the treatment of the hematopoietic component of the acute radiation syndrome (H-ARS): a review. Cytokine. 2015;71:22–37. doi: 10.1016/j.cyto.2014.08.003. [DOI] [PubMed] [Google Scholar]
- 62.Li Z.-T., Wang L.-M., Yi L.-R., Jia C., Bai F., Peng R.-J., Yu Z.-Y., Xiong G.-L., Xing S., Shan Y.-J. Succinate ester derivative of δ-tocopherol enhances the protective effects against 60 Co γ-ray-induced hematopoietic injury through granulocyte colony-stimulating factor induction in mice. Sci. Rep. 2017;7:40380. doi: 10.1038/srep40380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Marcotte D., Zeng W., Hus J.-C., McKenzie A., Hession C., Jin P., Bergeron C., Lugovskoy A., Enyedy I., Cuervo H. Small molecules inhibit the interaction of Nrf2 and the Keap1 Kelch domain through a non-covalent mechanism. Bioorg. Med. Chem. 2013;21:4011–4019. doi: 10.1016/j.bmc.2013.04.019. [DOI] [PubMed] [Google Scholar]
- 64.Lo S.C., Li X., Henzl M.T., Beamer L.J., Hannink M. Structure of the Keap1: Nrf2 interface provides mechanistic insight into Nrf2 signaling. EMBO J. 2006;25:3605–3617. doi: 10.1038/sj.emboj.7601243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gao X., Dinkova-Kostova A.T., Talalay P. Powerful and prolonged protection of human retinal pigment epithelial cells, keratinocytes, and mouse leukemia cells against oxidative damage: the indirect antioxidant effects of sulforaphane. Proc. Natl. Acad. Sci. Unit. States Am. 2001;98:15221–15226. doi: 10.1073/pnas.261572998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gao X., Talalay P. Induction of phase 2 genes by sulforaphane protects retinal pigment epithelial cells against photooxidative damage. Proc. Natl. Acad. Sci. Unit. States Am. 2004;101:10446–10451. doi: 10.1073/pnas.0403886101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fahey J.W., Dinkova-Kostova A.T., Stephenson K.K., Talalay P. The “Prochaska” microtiter plate bioassay for inducers of NQO1. Methods Enzymol. 2004;384:243–258. doi: 10.1016/S0076-6879(04)82014-7. [DOI] [PubMed] [Google Scholar]
- 68.Prochaska H.J., Santamaria A.B.J.A.b. Direct measurement of NAD (P) H: quinone reductase from cells cultured in microtiter wells: a screening assay for anticarcinogenic enzyme inducers. Anal. Biochem. 1988;169:328–336. doi: 10.1016/0003-2697(88)90292-8. [DOI] [PubMed] [Google Scholar]
- 69.Chinedu E., Arome D., Ameh F.S. A new method for determining acute toxicity in animal models. Toxicol. Int. 2013;20:224–226. doi: 10.4103/0971-6580.121674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Flecknell P. Anaesthesia of animals for biomedical research. Br. J. Anaesth. 1993;71:885–894. doi: 10.1093/bja/71.6.885. [DOI] [PubMed] [Google Scholar]
- 71.Yoshioka T., Kawada K., Shimada T., Mori M. Lipid peroxidation in maternal and cord blood and protective mechanism against activated-oxygen toxicity in the blood. Am. J. Obstet. Gynecol. 1979;135:372–376. doi: 10.1016/0002-9378(79)90708-7. [DOI] [PubMed] [Google Scholar]
- 72.Vrablic A.S., Albright C.D., Craciunescu C.N., Salganik R.I., Zeisel S.H. Altered mitochondrial function and overgeneration of reactive oxygen species precede the induction of apoptosis by 1-O-octadecyl-2-methyl-rac-glycero-3-phosphocholine in p53-defective hepatocytes. Faseb. J. 2001;15:1739–1744. doi: 10.1096/fj.00-0300com. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.