Abstract
Motivation
Accurate morphological models of brain vasculature are key to modeling and simulating cerebral blood flow in realistic vascular networks. This in silico approach is fundamental to revealing the principles of neurovascular coupling. Validating those vascular morphologies entails performing certain visual analysis tasks that cannot be accomplished with generic visualization frameworks. This limitation has a substantial impact on the accuracy of the vascular models employed in the simulation.
Results
We present VessMorphoVis, an integrated suite of toolboxes for interactive visualization and analysis of vast brain vascular networks represented by morphological graphs segmented originally from imaging or microscopy stacks. Our workflow leverages the outstanding potentials of Blender, aiming to establish an integrated, extensible and domain-specific framework capable of interactive visualization, analysis, repair, high-fidelity meshing and high-quality rendering of vascular morphologies. Based on the initial feedback of the users, we anticipate that our framework will be an essential component in vascular modeling and simulation in the future, filling a gap that is at present largely unfulfilled.
Availability and implementation
VessMorphoVis is freely available under the GNU public license on Github at https://github.com/BlueBrain/VessMorphoVis. The morphology analysis, visualization, meshing and rendering modules are implemented as an add-on for Blender 2.8 based on its Python API (application programming interface). The add-on functionality is made available to users through an intuitive graphical user interface, as well as through exhaustive configuration files calling the API via a feature-rich command line interface running Blender in background mode.
Supplementary information
Supplementary data are available at Bioinformatics online.
1. Introduction
The brain requires an adequate supply of energy to drive billions of neurons to fire properly. To ensure its normal functioning, essential energy substrates including oxygen, glucose and lactate are provided, on-demand, through a vast network of cerebral blood vessels via neurovascular coupling (NVC; Iadecola, 2017). The length of this network in human brains is estimated to be ∼700 km and the failure to deliver the proper amount of blood at the right time and location will be accompanied by catastrophic neurodegenerative disorders (Sweeney et al., 2018). While most of the focus in neuroscience research covers the structural and functional aspects of brain cells such as neurons (Markram et al., 2015) and glia (von Bartheld et al., 2016), the dense network of vascular channels that course through our brains has received comparatively less attention. This oversight is unfortunate considering the disproportionately high metabolic demands of neural tissues, the function of which must surely be tied to the availability of energy and other nutrients. There is a large body of work on gross-scale hemodynamics with the use of brain imaging technologies such as functional magnetic resonance (MR) imaging, with which the blood-oxygen level-dependent activity is measured (Logothetis et al., 2001; Raichle and Mintun, 2006). But this and other imaging methods do not resolve the functions of fine-scale structures that form the points of contact between the microvasculature and glial cells or within the effective range of individual neurons. Not only does an understanding of normal brain function require improving our concept of neurovascular structure, but there are numerous disease states associated with blood vessel damage that are broadly referred to as vascular cognitive impairments or dementias. Although all are generally due to cerebral hemodynamic insufficiencies of one sort or another, there is still little etiological or diagnostic agreement about causes or precise definitions, let alone treatments (Frantellizzi et al., 2020). The neuro-glia-vasculature (NGV) ensemble is often considered as a functional unit for the study of interactions among these critical components of brain energy metabolism at an oligocellular scale (Coggan et al., 2018b; Jolivet et al., 2015). Within this unit, detailed vascular maps revealing fine anatomical features of vessels and their interaction with neurons and glia are therefore critical to understanding overall brain function (Calcinaghi et al., 2013; Calì et al., 2019; Coggan et al., 2018a; Marín-Padilla, 2012; Schmid et al., 2019). These maps are essential for developing blood flow models to simulate cerebral blood flow (CBF) in realistic vascular networks (Damseh et al., 2019; Reichold et al., 2009), analyzing the proximity of cell bodies to vasculature elements (Blinder et al., 2013) and also for classifying vascular components into different types of segments (Zeng et al., 2017). Accurate digital reconstructions of vascular ultrastructure down to capillary level (Di Giovanna et al., 2018) are central to this goal, which requires innovative and rapid software visualization tools to investigate the topology of the vascular network and to model its behavior (Smith et al., 2019). Unfortunately, the existence of convenient domain-specific software frameworks capable of visualizing those vascular models in real time is currently lacking. We therefore remedy this limitation and present an integrated tool to address this problem. This tool is primarily devoted to neuroscience researchers studying NVC using computational modeling and simulation.
1.1. Relevant work
Vasculature datasets are originally obtained on multiple scales from two principal sources: imaging scanners such as computed tomography or MR (Preim and Oeltze, 2008; Wright et al., 2013) and microscopy, mainly from optical (Lugo-Hernandez et al., 2017) and electron microscopes (Januszewski et al., 2018). Certain optical techniques have bridged the gap between macroscopic and microscopic imaging, such as ultramicroscopy which can reconstruct cm-sized vascular networks with micrometer resolution (Jährling et al., 2009). A recent study proposed an advanced approach improving vascular demarcation, making it possible to reconstruct a whole brain vasculature with extremely high resolution that can capture the details of a single capillary (Di Giovanna et al., 2018). Nevertheless, exploring those vascular reconstructions and analyzing their fine structures with which we can test our scientific hypotheses, mainly in computational modeling, remains challenging.
A large variety of rendering techniques has been developed to visualize different structural aspects of vasculature networks (Preim and Oeltze, 2008). Complex networks can be visualized relying on direct volume rendering methods, such as slice-based viewing or ray-marching, employing original angiography stacks prior to their segmentation (Kubisch et al., 2012). This approach is substantial to classify and reveal vessel abnormalities more faithfully using multi-dimensional transfer functions, allowing diagnosis of vascular diseases, such as atherosclerosis or stenosis. Visualization of large-scale volumes is not as trivial however. It requires scalable volume rendering workflows that either use out-of-core algorithms, distributed solutions with sort-last rendering or combine the two approaches to load terabyte-sized datasets (Eilemann et al., 2012). These workflows remain unavailable to the public, which drives scientists to use standard visualization packages such as Paraview (Henderson et al., 2004), or even build their custom workflows based on visualization toolkit (VTK; Schroeder et al., 2004; Taka and Srinivasan, 2011). The vascular modeling toolkit is a prominent example, designed based on VTK and insight toolkit specifically to visualize and model vascular data (Antiga and Steinman, 2006). It has several components for (i) segmenting vessels from imaging stacks, (ii) reconstructing surface meshes, (iii) computing centerlines of vascular segments from polygonal surfaces and (iv) creating tetrahedral meshes. Nevertheless, its capability to perform these tasks for complex or large networks is doubtful. Obviously, volume rendering can be useful for segmentation and quantification purposes (Bühler et al., 2004), but it cannot reveal spatial relationships between different structures in the volume. Several limitations of volume rendering can be addressed by relying on polygonal surface meshes instead. Visualization of vascular meshes reconstructed from those stacks can be accomplished with either model-free or model-based techniques (Kretschmer et al., 2013). Conventional surface rendering applications use marching cubes algorithms to visualize a polygonal surface reconstructed during the rendering stage based on an appropriate iso-value. Although the quality of this category of algorithms is relatively poor, it could be enhanced if the surface is explicitly segmented a priori, but it might be subject to other inaccuracies due to user interaction. Unfortunately, even for relatively small-scale networks, the size of corresponding vascular meshes might be large for interactive visualization and analysis.
Skeletal representation of vascular networks has been proposed fundamentally for morphometric analysis purposes (Wright et al., 2013), but it has resolved implicitly the size limitation issue. Storing a complex graph of a vascular network is comparatively more efficient than a corresponding mesh. Although skeletons must be reconstructed either from volumes or meshes, there are advanced skeletonization methods that can perform this task efficiently (Saha et al., 2016). Nevertheless, visualizing those skeletons was not similarly investigated, in particular for brain vasculature. We only found a few studies that have used input vascular trees for visualization or analysis purposes based on convolution surfaces (Oeltze and Preim, 2004), abstract—yet spatially contextualized—approaches (Pandey et al., 2020), illustrative rendering approaches (Ritter et al., 2006) and signed distance fields (Lichtenberg et al., 2019). Nevertheless, these research trials were not accompanied with open source implementations that can be adapted. The presence of a software package that is mainly domain-specific, extensible, cross-platform and more significantly free, capable of interactive visualization and analysis of large-scale morphologies of vascular networks integrated in a single package is still largely missing.
1.2. Contribution
We present VessMorphoVis, a multi-functional, domain-specific and user-friendly add-on leveraging the core functionality provided by Blender. This add-on is primarily developed for visual analysis of large-scale morphological skeletons of vascular networks, allowing to build accurate vasculature structures for modeling and simulating CBF to understand the mechanisms of NVC. The add-on comes with the following features:
Interactive visualization, analysis and automated repair of large-scale vasculature morphology skeletons.
Creation of high-fidelity polygonal mesh models of vascular structures from their morphology skeletons using metaballs.
Automated creation of high-quality multimedia content using artistic shader nodes for debugging, discovery and dissemination.
Extensible python-based application programming interface (API) for analysis and visualization. This API can be called from the user interface of Blender or invoked via feature-rich command line interface (CLI) that runs Blender in background mode.
2 System architecture and results
VessMorphoVis is composed of five modules: (i) data handling, (ii) vascular network analysis and repair, (iii) morphology building for interactive visualization, (iv) polygonal mesh reconstruction and (v) high-quality rendering. In the following part, we present the functionality of each module and then demonstrate the features of the add-on with multiple datasets of varying structure and complexity. A high-level overview of the framework is illustrated in Supplementary Figure S1. The integration of the add-on in the graphical user interface (GUI) of Blender is shown in Supplementary Figures S2 and S3. The corresponding panels of each module as seen in the GUI are shown in Supplementary Material (Section 4).
2.1 Vasculature morphology structure
A vascular morphology skeleton, or a vascular graph (VC), consists of a set of ordered samples or points, where each sample is specified by an index, a three-dimensional Cartesian position vector and a diameter reflecting the cross-sectional extent of the skeleton at this point. Each two consecutive samples account for a segment, where a list of connected segments between two branching points represents a section. The segments are implicitly connected based on the order of their constituent samples in the section. A loop is composed of two independent sections having the same parents and children. For efficient data storage and convenient representation, the sections are logically connected at their respective branching points, i.e. any two connected sections have, at least, a shared terminal sample with the exact same Cartesian position, but with different indices and possibly varying radii for each respective section. In certain cases, the connectivity information is not explicitly encoded in the morphology tree, and therefore, it will be computed on-the-fly if needed. Figure 1 illustrates a schematic view of a small chunk of vasculature morphology skeleton extracted from a larger vascular network.
Fig. 1.

A schematic view of a vascular morphology network
2.2 Data format
VessMorphoVis supports two unique data formats: an internal binary file format based on the HDF5 library (with extension.h5) that targets efficient storage of data to guarantee scalability, and another ASCII format (with extension.vmv) that is mainly provided to external collaborators and the neuroscientific community. The ASCII format is designed to be extensible, allowing users to add further optional parameters that serve their needs. The structure of the file formats is detailed in Supplementary Material (Section 1).
2.3 Skeleton analysis and repair
To complement the subjective visual analysis of the skeleton, it was crucial to integrate another module for automated morphological analysis as well. This module applies a list of predefined kernels on the input network and produces a set of measurements and statistical distributions. This analysis is essential to detect and reveal any structural artifacts in the skeleton. Users can then apply another set of kernels to repair any artifact that might affect the visual quality of the reconstructed morphology, e.g. having a sample with zero diameter. Afterwards, a fact sheet containing the computed measures of the VC is displayed in the analysis panel, allowing users to link those values to the morphology skeleton appearing on the rendering widget. The measurements include the total number of samples, segments and sections in the morphology before and after resampling the skeleton, statistical distributions of the lengths of sections and their corresponding segments and the entire network in addition to the total number of fragmented components and loops. Supplementary Figure S4 shows a screenshot demonstrating a large vascular network loaded in the viewport and its analysis results.
2.4 Interactive skeleton visualization
Visualizing complex networks of brain vasculature is relatively challenging with respect to other NGV structures (Coggan et al., 2018b), such as neurons or glial cells. Neuronal and glial morphologies are represented by directed acyclic graphs, which makes storing and traversing them simpler than vasculature which are represented by cyclic ones. In terms of data complexity, neuronal morphologies have—on average—fewer numbers of samples than a small piece of vasculature network. They range from several hundred for simple morphologies and up to a few thousand for a neuron with highly complex arborizations in its dendritic or axonal trees. Subsequently, building interactive visualizers to render individual neuronal graphs in real time relying on Blender (Abdellah et al., 2018) is comparatively trivial. Blender is a powerful application; it comes with a rich high-level API and an intuitive GUI that can be exploited jointly for sketching, animation, mesh reconstruction and even progressive rendering using third party plug-ins such as Cycles. However, it is limited in terms of the number of objects that can be drawn in the scene. For instance, spheres can only be visualized based on explicit geometry (polygonal meshes) using UV spheres or icospheres; they cannot be represented by implicit geometry like signed distance functions (Karlsson et al., 2019; Lichtenberg et al., 2019). This representation impacts the total number of independent sphere objects that can be drawn in the scene.
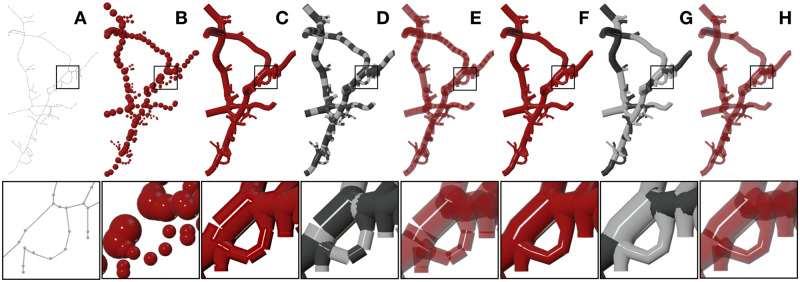
The fundamental objective of VessMorphoVis is to allow neuroscientists and modelers to load, optimize and interactively visualize large-scale vascular networks that can have several millions of traced samples. Visualizing the connectivity of these networks using the most straightforward approach requires drawing each element in the morphology (segment or section) using a unique polyline object. This object can be referenced later to assign a different property to it, e.g. a color code, that maps its radius or any other vascular property given by the user. This entails creating a geometric primitive per element and then linking it to the scene. This approach is not scalable; it can only be used to draw hundreds of polylines at most. Trying to go beyond this limit results in a non-responsive application which ultimately reduces its usability. The vascular network graph is highly interconnected and can contain up to tens of millions of samples if reconstructed at single capillary level (Di Giovanna et al., 2018). Therefore, we had to investigate a radically different approach capable of reconstructing the same polyline geometry and drawing millions of samples in few seconds. We then developed several skeleton builders to visualize vasculature morphologies in various styles, making it possible to visually analyze their structure (e.g. how each section is sampled and whether there is an overlapping between or within the sections or not) and the connectivity between its different components (segments or sections—refer to Fig. 1). Figure 2 shows an exemplar vascular morphology visualized with the different builders.
Fig. 2.
VessMorphoVis implements different algorithms for visualizing vascular networks. The outline of the morphology is sketched in (A) using thin polylines and tiny spheres to represent the sections and samples of the morphology, respectively. In (B), the morphology is illustrated by a list of points showing only the individual samples without any connectivity. The morphology is visualized as a disconnected set of segments and sections using the same color in (C) and (F), with alternating colors in (D) and (G) and also using transparent shaders in (E) and (H), respectively
2.4.1 Disconnected sections builder
This builder converts each section in the morphology into a virtual polyline structure whose segments correspond to those composed by the section itself. The final morphology is constructed by creating all the polylines and combining them into a single object with which it gets linked to the scene to be drawn in the viewport only once. This approach allows to efficiently pack a giant number of polylines into a single object, which in turn makes it possible to visualize large and dense vascular networks as shown in Supplementary Figures S3 and S4. This builder does not require any section-to-section connectivity information; the segment-to-segment connectivity is sufficient, which is already granted by having ordered set of samples per section in the loaded skeleton.
2.4.2 Disconnected segments builder
This builder converts each segment in the morphology into a line. To build up on the implementation of the previous builder, this line is merely a polyline composed of two points. This builder is used to visualize how the segments are distributed along every section in the morphology. It is mainly exploited to debug morphologies containing oversampled sections where we can clearly remove unnecessary samples and preserve the spatial topology of the morphology.
2.4.3 Connected sections builder
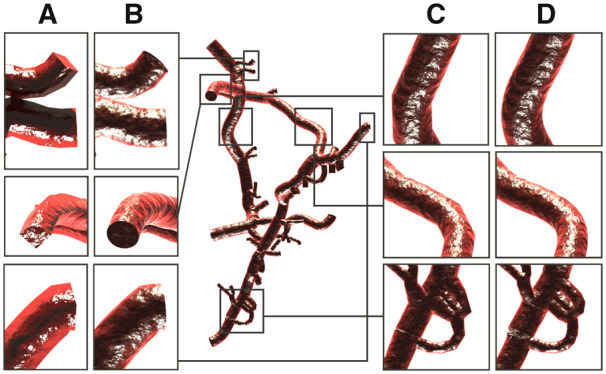
In some cases, connected sections might have certain configurations leading to reconstructed objects with artifacts such as having clear gaps between its different polylines at their branching points. These issues are not visually pleasant when users are interested in creating closeup images of their skeletons. Consequently, we have extended the disconnected sections builder to resolve this issue and create morphologies with connected sections that exhibit natural branching. Nevertheless, this extension entails the presence of valid connectivity list between the different sections in the morphology. As explained, this list might be missing in some morphology files as it requires an extra step after segmenting the skeleton from its microscopy stack to be built. However, it could be easily constructed on-the-fly before building the polylines. If the branching samples have the same indices in the different sections they belong to, this connectivity list can be built by comparing the indices of the terminal samples on a per-section-basis. Otherwise, connectivity can be obtained by evaluating the Euclidean distance between the terminal samples. Once this connectivity list is computed, a long polyline is constructed per section starting from a parent section, passing through the section itself and ending at a child. Therefore, this building strategy will create multiple polylines for every section within the network. If a section is connected between two parent and two child sections, four polylines will be created. Figure 3 shows the same morphology reconstructed with the disconnected and connected builders and how the central gap between two sections at the branching point is filled using the connected sections builder.
Fig. 3.

A close up showing how the connected sections builder is applied to visualize the morphology skeleton (right) to resolve any visual artifacts (gaps between sections) revealed by the disconnected sections builder (left)
2.4.4 Samples builder
We also implemented another builder to construct the skeleton directly from its raw samples as a group of spheres, each of them has a center and radius matching a corresponding sample in the morphology. Blender has no implicit sphere representation; therefore, we based our implementation on an isotropic simplicial polyhedron approximating a sphere called icosphere. To maximally reduce the number of vertices, a subdivision of two is used to construct the icosphere. To accelerate building the entire skeleton, each icosphere is created relying on an internal mesh editing library in Blender called bmesh, i.e. all the icospheres are created and joined together into a single object before being linked to the scene (Conlan, 2017). This visualization mode is selected to visualize the distribution of samples along each section in the morphology. Nevertheless, it is not advised to visualize dense skeletons as it becomes less efficient and cluttered as shown in Supplementary Figure S5.
2.4.5 General features
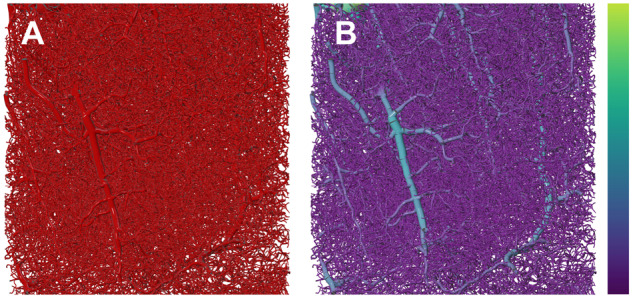
The morphology visualization module was integrated with a set of features that are generic and transparent to all the builders. These features include resampling the morphology skeleton, changing the tubular quality of the polylines or using spline interpolations to plot the polylines and several other features that are detailed in the documentation. Skeleton resampling is encouraged for two reasons. First of all, it removes any visual artifacts introduced from overlapping segments having different radii. Moreover, it eliminates all unnecessary samples along every section in the morphology, and therefore, optimizes the building process and makes the visualization further interactive. Figure 4 shows the substantial differences between an original highly oversampled vascular morphology and an optimized one after the resampling process. A combined rendering of a small morphology before and after resampling is shown in Supplementary Figure S6 to demonstrate how the resampling process affects the morphology.
Fig. 4.

(A) A highly oversampled (∼1.5 million samples) vascular network extracted from the cortex. Removing the unnecessary samples (B and C) without changing the connectivity of the morphology evidently improves the visual quality of the reconstructed morphology (D and E) and increases the rendering interactivity
The tubular surface of the polylines is reconstructed from interpolating a bevel object whose sides determine its cross-sectional shape and quality. We added an external parameter that allows users to control the number of sides of this bevel object, with minimum of 4 and maximum of 32 sides. Increasing the number of sides increases the geometry of the reconstructed polygons and, in turn, reduces the rendering performance. By default, we set this parameter to eight to balance between quality and interactivity, but we also expose this parameter to be controlled by users. This allows to load extremely large networks, where they can use 4 sides for interactive rendering and 16 sides for creating high-quality images. For the same reason, we added an option allowing the users to use spline interpolation to improve the visual quality of the reconstructed polylines. These features are illustrated on the exemplar morphology in Figure 5. In summary, Supplementary Videos S1–S3 demonstrate how to use the morphology visualization module interactively.
Fig. 5.
Users can control the visual quality of the skeleton, choosing between highly optimized geometry (A and C) for global far views or high-quality reconstructions (B and D) for close up views. Morphology polylines are rendered using bevel objects with 4 and 16 sides in A and B, respectively. The piecewise segments of the polylines (C) might limit the visual quality in case of close ups; therefore, we added another parameter to use spline interpolation to smooth their curvature (D)
Certain CBF modeling experiments mandate visualizing some vascular properties per segment such as its length, diameter, pressure or flow (Reichold et al., 2009; Smith et al., 2019). Therefore, it was essential to implement different coloring schemes allowing the users to select between using a single color to visualize the entire morphology, or using alternating colors to see some structural aspects of the morphology—Figure 4C and E, or even use a high dynamic range colormap to reveal other functional aspects. We added another option that allows users to color every segment or section in the morphology according to a specific property defined by the user. This feature is demonstrated in Figure 6.
Fig. 6.
Users can assign a color map to reveal the spatial distribution of the radii of all the segments in the morphology
2.5. Polygonal mesh reconstruction
Packing the vascular morphology as a list of polylines into a single object was essential for interactive visual analysis of dense vascular networks. Nevertheless, this polyline-based structure is limited in two aspects. First of all, it is merely convenient for visualization and structural validation purposes; it cannot be used to perform or even visualize stochastic reaction-diffusion simulations that entail highly accurate and optimized polygonal meshes associated with a color mapping scheme on a per-vertex basis. Furthermore, it can be only loaded in Blender which ultimately limits the usability of datasets beyond their visualization or analysis. Therefore, it was significant to integrate a mesh reconstruction module in VessMorphoVis, making it possible to build vascular surface meshes from their morphological skeletons, with which they can be used by other applications. In general, mesh generation of graph-based structures is known to be an offline process that might even take several hours based on the graph complexity, so the performance of this module was not a concern.
We propose an offline, but highly accurate mesh generator based on implicit structures called metaobjects (Oeltze and Preim, 2004; Zoppè et al., 2008) allowing to create high-fidelity vascular meshes from raw morphologies even without the necessity to have any section connectivity information in the morphology. Metaobjects are implicit surfaces defined procedurally. Unlike meshes that are composed of vertices or surfaces that are defined by control points, these objects are merely represented by mathematical functions that are computed on-the-fly. The fundamental feature that makes metaobjects significant is their ability to blend, i.e. when two independent metaobjects are getting closer to each other, they start to interact and blend together into a single object. We, based on metaobjects and in particular metaballs, propose an algorithm to reconstruct vasculature meshes—refer to Supplementary Figure S7. This algorithm is capable of handling highly complex branching scenarios avoiding to create intersecting geometry that is common in other meshing algorithms, e.g. skin modifiers (Abdellah et al., 2019).
Generating a vasculature mesh from its morphological skeleton using metaballs requires three principal stages: (i) initializing the metaobject, (ii) building the metaobject structure on a per-section-basis and (iii) converting the metaobject into a target surface mesh. During the initialization stage, an empty metaobject of type bpy.data.metaball is created and linked to the scene. The initial resolution of this object is set to 1; however, this resolution will be modified in the finalization stage based on the radius of the smallest sample along the morphology skeleton to avoid reconstructing a fragmented mesh as shown in Figure 7 and Supplementary Video S4.
Fig. 7.
A vasculature mesh created with the proposed metaballs implementation. The resolution of the metaobject is automatically set to 0.5 to create a highly tessellated mesh in (A). The meshes in (B), (C) and (D) are created with metaobject resolutions of 1.0, 2.0 and 4.0, respectively. These meshes have much fewer polygons than the original mesh in A, but their quality is degrading correspondingly. The small vessels and loops are not reconstructed in C and the large vessels are completely fragmented in D
Afterwards, the metaobject body is constructed on a per-section-basis, where each step converts a raw list of ordered samples that belong to the section to what is called a meta-section. During this step, each pair of connected samples along the section is used to create a meta-segment and append it to the metaobject using sphere marching. The segment length and direction are initially computed and then the spatial extent of the segment is filled with multiple interpolated points based on the radii and position vectors of the two samples of the segment until the traveled distance along the ray exceeds the segment length. The radius of the smallest sample in the morphology skeleton is computed during the building of each meta-section. At the finalization stage, this radius is used to set the actual resolution of the metaobject and then the metaobject is converted to a manifold surface mesh. The reconstructed mesh is tessellated or decimated to result in a smooth and non-bumpy surface and then linked to the rendering widget.
It has to be noted that our implementation does not require the connectivity information between the sections. However, it assumes that the corresponding branching points of the connected sections are logically located at the same Cartesian positions even if they have different radii. The segment connectivity information per section is mandatory to be able to construct correct structures. Supplementary Figure S8 shows a combined rendering of a reconstructed vascular mesh and its corresponding morphology for qualitative validation.
The mesh generation time depends mainly on the size of the vascular morphology and the metaballs resolution used to build the mesh. Building meshes for morphologies with less than few thousands of samples at full resolution takes several hundreds of milliseconds to a few seconds. For a relatively larger dataset with 55 807 samples, we performed the meshing at three different resolutions. The mesh reconstruction process took 107.45, 6.37 and 2.49 s corresponding to metaball resolutions of 1, 2 and 3, respectively.
2.6. Mesh optimization
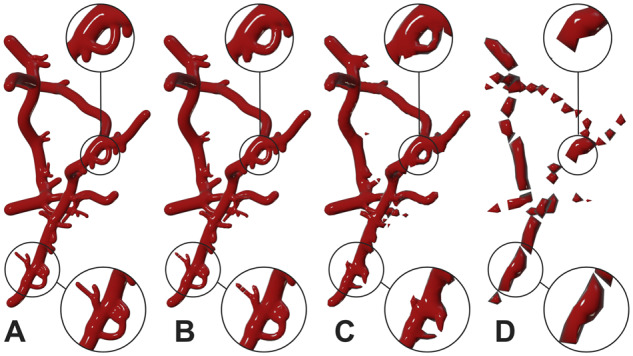
The tessellation of the reconstructed meshes from the metaballs implementation is primarily based on the radius of the smallest sample along the skeleton. Consequently, the presence of relatively small samples would significantly impact the geometry of the mesh by creating very small polygons that are not necessarily essential to preserve the structure of the vascular network. By default, the resolution of the metaobject is obtained during the metaobject building process, and set at the finalization stage before converting the metaobject into a mesh. But, for convenience, we added another option that allows users to manually set the metaobject resolution. Although very useful for testing, inappropriate values of this parameter can result in highly fragmented mesh as obvious in Figure 7.
To overcome this issue, we added another option to decimate the geometry allowing to create optimized meshes which could be used for several applications, e.g. real-time rendering or stochastic simulations. To avoid destructing the mesh, the decimation range exposed to the user is limited between 0.99 and 0.125. This range has been set based on trial-and-error. This allows users to create a mesh with decent size without sacrificing its quality by reducing the metaobject resolution. Figure 8 demonstrates how a highly tessellated mesh with ∼68 000 polygons can be optimized to ∼17 000 polygons without any loss in visual quality. We can also notice that decimation factors beyond the 0.125 can introduce visual artifacts due to loss of topological detail.
Fig. 8.
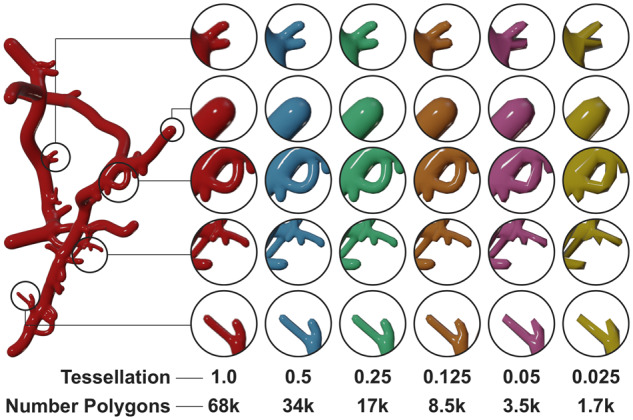
Decimation is essential to reduce the tessellation of meshes with large polygon counts. At certain point, it can reduce the visual quality of the reconstructed mesh, but it does not fragment the mesh into several pieces. The mesh complexity (number of polygons) has been reduced 40 times and the branching is still preserved
There are different metrics that define mesh quality including its minimum and maximum dihedral angles, edge and radius ratios (Hu et al., 2018). Due to the nature of the metaobject algorithm itself, the topology of the resulting meshes is not optimized according to these metrics. We therefore have added another utility (implemented in the C code of Blender) to optimize any generated mesh according to these metrics based on an open source mesh optimization library (Yu et al., 2008). The resulting meshes from this optimization process are shown in Figure 9.
Fig. 9.
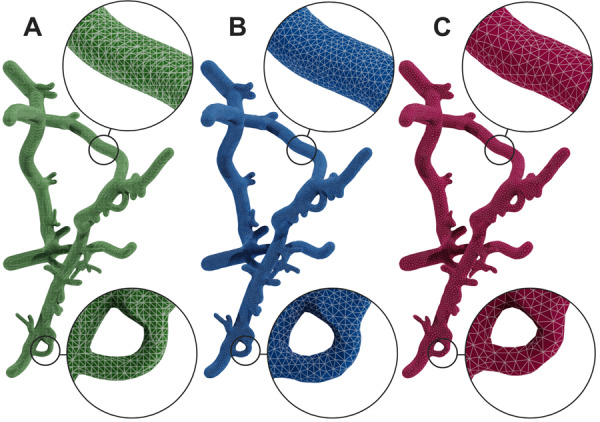
Mesh optimization. A mesh reconstructed from the metaballs implementation in (A) is optimized with two different decimation factors in (B) and (C)
2.7. High-quality rendering and multimedia generation
High-quality scientific illustrations are essential in any research process, but creating these illustrations requires a substantial amount of time and effort, deep knowledge of media design applications in addition to having individual artistic skills to a certain degree. Therefore, we have integrated an extra module in VessMorphoVis specifically designed to seamlessly create high-quality multimedia content that reveals the beauty of vascular networks. The rendering module is transparently connected to the morphology and meshing modules. It consists of a list of artistic shader nodes and light setups that are automatically configured to render scientific illustrations for vascular morphologies and their corresponding surface meshes including high-resolution static images and 360 sequences. These renderings are also helpful to debug the connectivity of the skeletons. Supplementary Videos S5 and S6 show 360 sequences for two meshes of different network complexities. The rendering components including camera, lighting setup and the selection of the rendering engine itself are automatically added to the scene based on three items: (i) the rendering resolution (or scale), (ii) the selected shader (or material) and (iii) the type of the geometry that exists in the scene. The frustum of the camera is defined based on its projection type (whether orthographic or perspective) and the bounding box of the geometry that exist in the scene. Users can create two sets of images, either for debugging and analysis or for scientific articles media. The first set uses glossy shaders with the Workbench renderer, making it possible to create very high-resolution images in only few seconds. The other set uses physically based shaders with Cycles to create cinematic renderings (Eid et al., 2017) even for highly dense vascular networks. Figure 10 shows a vascular mesh rendered with different types of shaders and rendering engines. Supplementary Figure S9 shows other shading styles using a radius-based colormap.
Fig. 10.

The same mesh reconstructed with metaballs algorithm is rendered with four different shaders: glossy, flat, artistic bumby and artistic glossy in A, B, C and D, respectively, using the Workbench and Cycles renderes in Blender
3. Discussion and experts feedback
VessMorphoVis is designed to be an open source and domain-specific software solution. It serves diverse categories of users, primarily neuroscience modeling researchers in addition to visualization specialists and scientific multimedia designers. For this reason, we based our implementation on Blender, exploiting its user-friendly GUI, well-documented Python API to accomplish multiple design goals. Starting from an input vascular morphology, users can analyze its graph using automated kernels and interactive visual inspection, allowing them to repair the morphology and then create an implicit representation that is converted into a surface mesh. This polygonal mesh is further optimized and can be used to reconstruct a tetrahedral mesh for simulating the CBF. Moreover, scientists can seamlessly create visualizations to debug the vascular network and render high-quality multimedia using ray tracing for scientific articles. Figure 11 shows a high-quality rendering of a large mesh reconstructed from a very dense skeleton. The API was also designed taking into consideration extensibility, allowing researchers who have basic Python programming knowledge to implement certain custom features needed for their research purposes with minimal efforts.
Fig. 11.
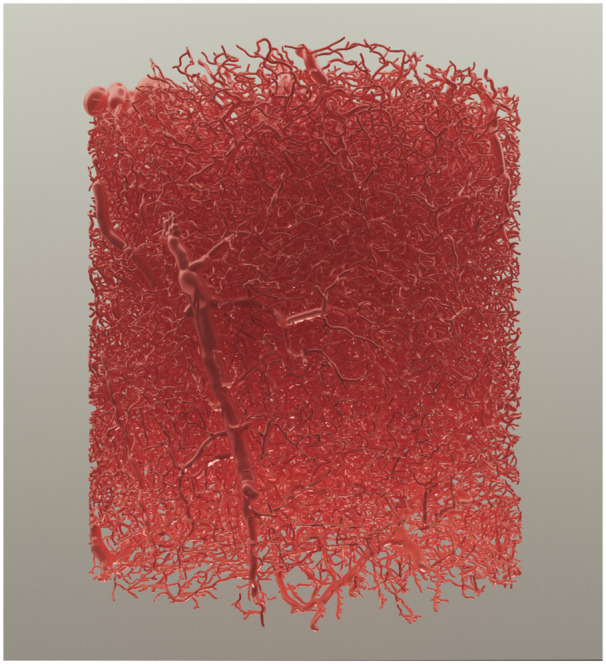
A high-quality rendering of a large vasculature mesh reconstructed from a vascular graph having ∼2.1 million samples based on our metaballs implementation. The mesh is rendered using the artistic glossy shader with Cycles
To assess the overall performance, usability and impact of the add-on, we prepared a collection of vascular morphologies with varying size and complexity. Then, we presented the features of the tool to a group of domain experts. This group included neuroscientists working on computational modeling of brain vasculature, visualization researchers and also multimedia specialists whose mission is creating scientific illustrations of neuroscientific content with Blender. The users were asked to report their feedback indicating (i) points of strength and (ii) current limitations that can be improved, and (iii) other features that can be implemented to be helpful for their specific needs.
The experimental feedback was largely positive and satisfactory. The scientists provided the following comments. The morphology builders are extremely insightful to verify several structural aspects of the vascular morphology and reveal certain inaccuracies of its graph including false loops, disconnected fragments, misaligned sections, incorrect branching and overlapping components. The automated analysis module is very helpful and efficient. A single user click creates an inclusive fact sheet summarizing different analysis results of the whole network. Further kernels can be effortlessly implemented to enhance the results. To them, resampling the skeleton without altering its spatial structure was substantial; several datasets are excessively oversampled and using those datasets without resampling in computational modeling increases the simulation time significantly. The design of the add-on is intuitive and even with no previous Blender experience, it can be used seamlessly. Every item on the interface comes with a clear tooltip documentation displayed upon mouse hover over the element. Moreover, designing a rich CLI to the add-on was as crucial as its GUI to allow its integration into other software pipelines, which would potentially magnify the impact of VessMorphoVis. The visualization and media specialists have mainly liked the integrity of the tool. The meshing and rendering modules were of significant value to them. It would be very helpful to create Blender scenes where they can apply further shading nodes to create outstanding multimedia.
We then received several feature requests to further enhance the add-on functionality including (i) adding other coloring schemes to visualize more vascular properties, (ii) visualizing dynamic graphs to reveal changing structures over time to help in silico experimentalists to explore the impact of varying modeling parameters on the network, (iii) integrating another module allowing scientists to manually reconnect disconnected or fragmented pieces that were poorly segmented, (iv) adding support to visualize a region of interest selected from the GUI to focus on specific parts of the morphology and (v) finally, making the API callable from a web interface. Surprisingly, we were excited to see that other collaborators are willing and deeply interested to implement some of these requests by themselves to get to know more about the API.
4. Conclusion
Vascular modeling efforts in the neuroscientific community are lacking domain-specific frameworks enabling interactive visual analysis of vast morphological networks of brain vasculature. VessMorphoVis is presented to fill this gap. Integrated in a single open source toolbox, this Blender add-on is capable of interactive visual analysis of vascular networks, synthesizing high-fidelity polygonal meshes from their morphologies and also creating high-quality scientific illustrations and sequences of these networks. The capabilities of the add-on were demonstrated on several vascular morphologies with varying sizes and complexity. We performed a set of user experiments to evaluate different aspects of the add-on including its performance, functionality and interface intuitiveness. We concluded from their feedback that our tool will be an essential component in vascular modeling and simulation in the future.
Add-on Requirements
VessMorphoVis is designed as a python add-on for Blender 2.80. The add-on depends only on the following software components: Blender and the python bindings of HDF5 and MorphIO.
Data sources
The vascular morphologies used in this paper are kindly provided by Bruno Weber, Institute of Pharmacologyand Toxicology –Experimental Imaging and Neuroenergetics, University of Zürich (UZH), Switzerland, Pablo Blinder, Department of Neurobiology, George S. Wise Faculty of Life Sciences, Tel Aviv University (TAU), Israel and David Kleinfeld, University of California at San Diego (UCSD), USA.
Supplementary Material
Acknowledgements
We thank Stéphanie Battini, Alexis Arnaudon, Eleftherios Zisis and Pablo Blinder for their user feedback. We also thank Karin Holm, Alessandro Foni and Judit Planas for their valuable comments and suggestions.
Funding
This study was supported by funding to the Blue Brain Project, a research center of the École Polytechnique Fédérale de Lausanne (EPFL), from the Swiss Government’s ETH Board of the Swiss Federal Institutes of Technology.
Conflict of Interest: none declared.
References
- Abdellah M. et al. (2018) Neuromorphovis: a collaborative framework for analysis and visualization of neuronal morphology skeletons reconstructed from microscopy stacks. Bioinformatics, 34, i574–i582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdellah M. et al. (2019) Generating high fidelity surface meshes of neocortical neurons using skin modifiers. In: Vidal,F.P. et al. (eds), Computer Graphics and Visual Computing (CGVC), The Eurographics Association.
- Antiga L., Steinman D.A. (2006) VMTK: Vascular Modeling Toolkit. VMTK, San Francisco, CA, USA. [Google Scholar]
- Blinder P. et al. (2013) The cortical angiome: an interconnected vascular network with noncolumnar patterns of blood flow. Nat. Neurosci., 16, 889–897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bühler K. et al. (2004) Geometric methods for vessel visualization and quantification—a survey In Geometric Modeling for Scientific Visualization , ; pp. 399–419. Springer, Berlin, Heidelberg. [Google Scholar]
- Calcinaghi N. et al. (2013) Multimodal imaging in rats reveals impaired neurovascular coupling in sustained hypertension. Stroke, 44, 1957–1964. [DOI] [PubMed] [Google Scholar]
- Calì C. et al. (2019) Sparse reconstruction of neurons and glial cells of layer vi somatosensory cortex of a juvenile rat. IBRO Rep., 6, S383–S384. [Google Scholar]
- Coggan J.S. et al. (2018. a) Norepinephrine stimulates glycogenolysis in astrocytes to fuel neurons with lactate. PLoS Comput. Biol., 14, e1006392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coggan J.S. et al. (2018. b) A process for digitizing and simulating biologically realistic oligocellular networks demonstrated for the neuro-glio-vascular ensemble. Front. Neurosci., 12, 664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conlan C. (2017) The bmesh module In The Blender Python API, pp. 27–42. Apress, Berkeley, CA. [Google Scholar]
- Damseh R. et al. (2019) Automatic graph-based modeling of brain microvessels captured with two-photon microscopy. IEEE J. Biomed. Health Inf., 23, 2551–2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Giovanna A.P. et al. (2018) Whole-brain vasculature reconstruction at the single capillary level. Sci. Rep., 8, 12573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eid M. et al. (2017) Cinematic rendering in CT: a novel, lifelike 3d visualization technique. Am. J. Roentgenol., 209, 370–379. [DOI] [PubMed] [Google Scholar]
- Eilemann S. et al. (2012) Parallel rendering on hybrid multi-GPU clusters. In Eurographics Symposium on Parallel Graphics and Visualization, Number EPFL-CONF-216016, pp. 109–117. The Eurographics Association, Cagliari, Italy.
- Frantellizzi V. et al. (2020) Neuroimaging in vascular cognitive impairment and dementia: a systematic review. J. Alzheimers Dis., 73, 1279–1294. [DOI] [PubMed] [Google Scholar]
- Henderson A. et al. (2004) The ParaView Guide. Kitware, Clifton Park, NY, USA. [Google Scholar]
- Hu Y. et al. (2018) Tetrahedral meshing in the wild. ACM Trans. Graph., 37, 60. [Google Scholar]
- Iadecola C. (2017) The neurovascular unit coming of age: a journey through neurovascular coupling in health and disease. Neuron, 96, 17–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jährling N. et al. (2009) 3D-reconstruction of blood vessels by ultramicroscopy. Organogenesis, 5, 227–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Januszewski M. et al. (2018) High-precision automated reconstruction of neurons with flood-filling networks. Nat. Methods, 15, 605–610. [DOI] [PubMed] [Google Scholar]
- Jolivet R. et al. (2015) Multi-timescale modeling of activity-dependent metabolic coupling in the neuron-glia-vasculature ensemble. PLoS Comput. Biol., 11, e1004036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson J. et al. (2019) High fidelity visualization of large scale digitally reconstructed brain circuitry with signed distance functions. In: 2019 IEEE Visualization Conference (VIS), pp. 176–180. IEEE, Vancouver, Canada.
- Kretschmer J. et al. (2013) Interactive patient-specific vascular modeling with sweep surfaces. IEEE Trans. Vis. Comput. Graph., 19, 2828–2837. [DOI] [PubMed] [Google Scholar]
- Kubisch C. et al. (2012) Vessel visualization with volume rendering In Visualization in Medicine and Life Sciences II, pp. 109–132. Springer, Berlin, Heidelberg. [Google Scholar]
- Lichtenberg N. et al. (2019) Distance field visualization and 2d abstraction of vessel tree structures with on-the-fly parameterization.
- Logothetis N.K. et al. (2001) Neurophysiological investigation of the basis of the fMRI signal. Nature, 412, 150–157. [DOI] [PubMed] [Google Scholar]
- Lugo-Hernandez E. et al. (2017) 3D visualization and quantification of microvessels in the whole ischemic mouse brain using solvent-based clearing and light sheet microscopy. J. Cereb. Blood Flow Metab., 37, 3355–3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marín-Padilla M. (2012) The human brain intracerebral microvascular system: development and structure. Front. Neuroanat., 6, 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markram H. et al. (2015) Reconstruction and simulation of neocortical microcircuitry. Cell, 163, 456–492. [DOI] [PubMed] [Google Scholar]
- Oeltze S., Preim B. (2004) Visualization of anatomic tree structures with convolution surfaces. In: Joint EUROGRAPHICS - IEEE TCVG Symposium on Visualization (2004), Eurographics. pp. 311–320.
- Pandey A. et al. (2020) Cerebrovis: designing an abstract yet spatially contextualized cerebral artery network visualization. IEEE Trans. Vis. Comput. Graph., 26, 938–948. [DOI] [PubMed] [Google Scholar]
- Preim B., Oeltze S. (2008) 3D visualization of vasculature: an overview In Visualization in Medicine and Life Sciences, pp. 39–59. Springer, Berlin, Heidelberg. [Google Scholar]
- Raichle M.E., Mintun M.A. (2006) Brain work and brain imaging. Annu. Rev. Neurosci., 29, 449–476. [DOI] [PubMed] [Google Scholar]
- Reichold J. et al. (2009) Vascular graph model to simulate the cerebral blood flow in realistic vascular networks. J. Cereb. Blood Flow Metab., 29, 1429–1443. [DOI] [PubMed] [Google Scholar]
- Ritter F. et al. (2006) Real-time illustration of vascular structures. IEEE Trans. Vis. Comput. Graph., 12, 877–884. [DOI] [PubMed] [Google Scholar]
- Saha P.K. et al. (2016) A survey on skeletonization algorithms and their applications. Pattern Recognit. Lett., 76, 3–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid F. et al. (2019) Vascular density and distribution in neocortex. Neuroimage, 197, 792–805. [DOI] [PubMed] [Google Scholar]
- Schroeder W.J. et al. (2004) The Visualization Toolkit: An Object-Oriented Approach to 3D Graphics. Kitware, Clifton Park, NY, USA. [Google Scholar]
- Smith A.F. et al. (2019) Brain capillary networks across species: a few simple organizational requirements are sufficient to reproduce both structure and function. Front. Physiol., 10, 233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeney M.D. et al. (2018) The role of brain vasculature in neurodegenerative disorders. Nat. Neurosci., 21, 1318–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taka S.J., Srinivasan S. (2011) Nirviz: 3D visualization software for multimodality optical imaging using visualization toolkit (VTK) and insight segmentation toolkit (ITK). J. Digit. Imaging, 24, 1103–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Bartheld C.S. et al. (2016) The search for true numbers of neurons and glial cells in the human brain: a review of 150 years of cell counting. J. Comp. Neurol., 524, 3865–3895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright S.N. et al. (2013) Digital reconstruction and morphometric analysis of human brain arterial vasculature from magnetic resonance angiography. Neuroimage, 82, 170–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z. et al. (2008) Feature-preserving adaptive mesh generation for molecular shape modeling and simulation. J. Mol. Graph. Model., 26, 1370–1380. [DOI] [PubMed] [Google Scholar]
- Zeng Y-z. et al. (2017) Liver vessel segmentation and identification based on oriented flux symmetry and graph cuts. Comput. Methods Prog. Biomed., 150, 31–39. [DOI] [PubMed] [Google Scholar]
- Zoppè M. et al. (2008) Using blender for molecular animation and scientific representation. In Blender Conference. Amsterdam.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.