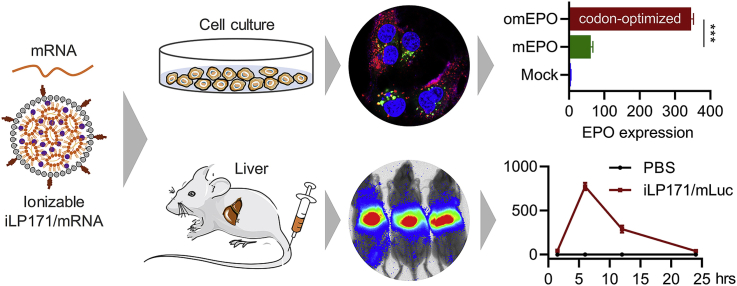
Abstract
mRNA is a novel class of therapeutic modality that holds great promise in vaccination, protein replacement therapy, cancer immunotherapy, immune cell engineering etc. However, optimization of mRNA molecules and efficient in vivo delivery are quite important but challenging for its broad application. Here we present an ionizable lipid nanoparticle (iLNP) based on iBL0713 lipid for in vitro and in vivo expression of desired proteins using codon-optimized mRNAs. mRNAs encoding luciferase or erythropoietin (EPO) were prepared by in vitro transcription and formulated with proposed iLNP, to form iLP171/mRNA formulations. It was revealed that both luciferase and EPO proteins were successfully expressed by human hepatocellular carcinoma cells and hepatocytes. The maximum amount of protein expression was found at 6 h post-administration. The expression efficiency of EPO with codon-optimized mRNA was significantly higher than that of unoptimized mRNA. Moreover, no toxicity or immunogenicity was observed for these mRNA formulations. Therefore, our study provides a useful and promising platform for mRNA therapeutic development.
Keywords: mRNA therapy, Lipid nanoparticle, mRNA delivery, Erythropoietin, Codon optimization
Graphical abstract
Highlights
-
•
mRNA therapy represents a promising revolutionary treatment strategy.
-
•
Sequence optimization and efficient delivery are critical to its wide application.
-
•
Ionizable lipid nanoparticle (iLP171) was developed to deliver codon-optimized mRNA.
-
•
Robust protein expressions of luciferase and erythropoietin (EPO) were achieved both in vitro and in vivo.
-
•
ILP171/mRNA exhibited excellent safety profiles in vivo.
1. Introduction
mRNA is the bridge of the central dogma of molecular biology, which means mRNA is neither the permanent genetic information nor a functional end product, but a transient transition [1]. This transient nature gives mRNA great flexibility, thereby expanding its therapeutic area over most of other known drugs. Although instability and immunogenicity hindered the development of mRNA drugs in previous years, these key problems are gradually overcome with the progresses of chemical modification technologies. Compared with small molecules and antibody drugs, the production of mRNA is faster, cost-effective and more flexible, because mRNA can be easily produced by in vitro transcription (IVT) in large-scale [2]. Compared with plasmid DNA, mRNA also has some advantages, such as: (1) mRNA does not need to enter the nucleus but functions in the cytoplasm; (2) mRNA will not insert into the host genome because it is delivered by non-viral carriers instead of virus vectors; (3) mRNA is biocompatible, non-toxic and immunologically inert. Besides, mRNA theoretically is able to express any protein, so it possesses wide application prospect in vaccination, protein replacement therapy, cancer immunotherapy, immune cell engineering and so on [2,3]. Currently, plenty of mRNA therapeutic pipelines have been established for curing various cancers, infectious diseases, heart disease, fibrosis, etc. [3,4].
Recently, a mRNA vaccine (mRNA-1273) against coronavirus disease 2019 (COVID-19) had advanced into clinical trials (NCT04283461). It took only 25 days and 63 days respectively, from sequence selection to vaccine manufacture for the first clinical batch and the first human dosing. The bright prospect of mRNA is attracting the attentions of scientists, investors, patients and even common people. However, mRNA therapeutic is still facing the challenges of lacking safe and effective delivery system, and requirement of optimizing the sequences and nucleotide compositions [5,6]. Due to its large size (300–5000 kDa, 1–15 kb), negative charge, and degradability, native mRNA cannot readily pass through the cell membrane and efficiently accumulate in the cytoplasm. Therefore, the development of appropriate delivery systems, e.g., lipid nanoparticle (LNP), liposome, lipoplex or polyplex, is urgently required. Among them, lipids are the most frequently used nucleic acid delivery materials [3,7].
Lipids or lipid-like materials (lipidoids) are able to form various vesicles, e.g., LNP, liposome, lipid emulsion, lipid implant, with nuclei acids [3,[7], [8], [9], [10], [11]]. A series of cationic lipids and ionizable lipids such as 1,2-dioleoyloxy-3-trimethylammonium propane chloride (DOTAP), N-[1-(2,3-dioleoyloxy) propyl]-N,N,N-trimethylammo- nium chloride (DOTMA), 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE), dilinoleylmethyl-4-dimethylaminobutyrate (Dlin-MC3-DMA) have been investigated for siRNA or mRNA delivery [5,10,[12], [13], [14], [15], [16], [17]]. Ionizable LNP (iLNP) is more clinically advanced than other deliver systems in the context of RNA delivery. For example, the first siRNA drug Onpattro approved by U.S. Food and Drug Administration's (FDA), and the first COVID-19 mRNA vaccine under clinical investigation, are both iLNP formulations. Because of the core component of ionizable lipid, iLNP is neutral in the physiological condition but builds charges in acidic endosomes, which enable potent intracellular mRNA delivery [10,18]. iLNP has been validated across a variety of cell types, including hepatocytes, immune cells, cancer cells, and so on [[19], [20], [21], [22]]. It is reported that iLNP delivered mRNA into lymphocytes more effectively than commercially available Lipofectamine reagents [19]. In addition, iLNP is easy to be modified and very flexible. By adjusting different components, their ratio and iLNP preparation process, the type of loaded RNA can be changed and encapsulation efficiency can be improved [23,24]. These characteristics make iLNP a powerful platform for nucleic acid delivery. In addition, nucleotide modifications and codon optimization by using pseudouridine (ψ) and synonymous codons, respectively, are of importance for efficient and durable mRNA expression [3].
In this study, we developed an iLNP formulation (iLP171) based on iBL0713 to thoroughly evaluated the mRNA delivery efficiency and protein expression both in vitro and in vivo. Two mRNAs expressing luciferase and erythropoietin (EPO) were prepared by in vitro transcription with decorations of anti-reverse cap analog (ARCA) and poly(A) tail. The codon of EPO mRNA was optimized with synonymous codons to enhance mRNA's expression efficiency and reduce their immunogenicity. The physicochemical properties of mRNA-encapsulated iLNP (iLP171/mRNA) were characterized. The cellular uptake efficiency, subcellular localization and in vivo biodistribution were analyzed with FACS, Confocal microscopy and in vivo imaging, respectively. More importantly, profiles of protein expression and safety of iLP171/mRNA were carefully investigated both in cell line and in animals. As a result, an effective platform potentially can be used to develop mRNA-based therapeutics was established.
2. Materials and methods
2.1. Materials
Luciferase Assay System was purchased from Promega Co. Ltd. (Madison, USA). TRIzol Reagent, cholesterol, and RNAlater were bought from Sigma-Aldrich (St Louis, MO). Lipofectamine 2000, Dulbecco's modified Eagle's medium (DMEM), Opti-MEM, fetal bovine serum, penicillin-streptomycin, trypsin and Lipofectamine 2000 were purchased from Thermo Fisher. ApoB-against siRNA (siApoB) and Cy5-labeled siRNA were provided by Suzhou Ribo Life Science Co. Ltd. (Jiangsu, China). pcDNA3.0-Luc (luciferase) and pcDNA3.0-EPO (erythropoietin) plasmids used as the transcription templates were constructed in-house. BstZ17I–HF, HiScribe T7 ARCA mRNA Kit (with tailing) were provided by New England Biolabs Inc. All of the primers were provided by BioSune Co. (Shanghai, China). 16:0 PEG2000 PE was purchased by Avanti Polar Lipids, Inc (USA, Alabama). Cholesterol and iBL0713 are provided by Suzhou Ribo Life Science (Kunshan, China). iBL0713 is a proprietary ionizable lipid that has been studied for siRNA delivery previously [8].
2.2. Cell culture
Huh7, a human hepatocellular carcinoma cell line, was cultured in Dulbecco's modified Eagle's medium (DMEM) with 10% fetal bovine serum, 100 unit/mL penicillin, 100 μg/mL streptomycin and were cultured in an incubator at 37 °C with 5% CO2.
2.3. In vitro transcription (IVT) of mRNA
In order to generate mRNA with improved expression of recombinant proteins, we firstly performed codon optimization of the erythropoietin (EPO) encoding mRNA sequence. We replaced rare codons with synonymous codons according to the principles of codon usage preference and frequency in different species, the requirement of avoiding specific restriction enzyme cutting site, the computationally predicted secondary structure properties, the GC content, etc. Detailed sequences of EPO coding region before optimization and after optimization were provided in supporting information. The optimized codons were highlighted in bold. Then the codon-optimized coding sequence were cloned into pcDNA3.0 vector (Beijing Aoke Life Science Co. Ltd). The vectors were linearized with BstZ17I digestion and were used as an IVT template. IVT was conducted under the manufacturer's protocol. During the IVT, mRNA products were ARCA capped and enzymatically polyadenylated (New England Biolabs Inc). The final transcribed mRNAs were purified using LiCl Solution (New England Biolabs Inc), and mRNA concentrations were determined by Nanodrop.
2.4. Preparation and characterization of iLP171/mRNA
LP171/mRNA formulation was prepared according to a modified protocol of reference [8]. Briefly, total lipids with compositions of iBL0713, cholesterol and C16-PEG (16:0 PEG2000 PE) in an optimized molar ratio were dissolved in ethanol to prepare a stock solution. The lipid solution was added into sodium acetate solution (200 nM, pH 5.2) under stirring for 2–3 min to form iLNPs preliposome. The volume of aqueous sodium acetate solution is three times over organic lipid solution. Then, the aqueous mRNA solution (mRNA dissolved in sodium acetate solution that containing 25% (v/v) ethanol) was mixed with iLNPs solution at a 1:1 vol ratio. After gently stirring for 2–3 min, the mixture was incubated at 50 °C for 10–20 min, and then dialyzed with a dialysis tube (molecular weight cut-off of 100 kDs, Millipore) for 2 h in PBS buffer at room temperature, to remove the ethanol and saline ions, and adjust the pH of the solution to a neutral pH. The volume ratio of PBS buffer to mRNA solution are higher than 200. The final iLP171/mRNA formulation was used freshly or kept at 4 °C for less than one month.
The size, polydispersity index (PDI) and zeta potential of iLP171/mRNA were measured by dynamic light scattering (DLS) (Malvern, Zetasizer Nano ZS). The morphology was identified by transmission electron microscopy (TEM) (Hitachi, HT7700). The encapsulation efficiency was analyzed with RiboGreen assay. Briefly, the iLP171/mRNA was firstly treated with 2% Triton X-100 to release mRNA. Then, both Triton X-100 treated and untreated iLP171/mRNA were incubated with RiboGreen reagent (Thermo Fisher Scientific, Cat No. R11491). The fluorescence intensity was recorded by a microplate reader to reflect the amount of total mRNA and free mRNA. The encapsulation efficiency is calculated according to the following formula:
Encapsulation efficiency (%) = [(total mRNA - free mRNA)/total mRNA] × 100%
2.5. pKa determination
The pKa value of iLNPs was determined using the fluorescent probe 2-(p-toluidino)-6-napthalene sulfonic acid (TNS). First, prepare a 10 mM iLNP liquid in PBS solution. Then, prepare a series of iLNP solution with pH ranged from 3.00 to 10.00 containing 1 μM TNS, 10 mM HEPES, 10 mM 4-morpholineethanesulfonic acid, 10 mM ammonium acetate and 130 mM NaCl. The fluorescence intensity of each solution was recorded by a spectrophotometer with an excitation and emission wavelengths of 321 nm and 445 nm, respectively. The fluorescence data was analyzed using the sigmoidal best fit analysis. The pKa value was defined as the pH giving rise to the half-maximal fluorescence intensity [18].
2.6. Animals
All animals were maintained in Peking University Laboratory Animal Center (an AAALAC accredited experimental animal facility). All animal experiments were performed in accordance with the principles of care and use of laboratory animals of Peking University (approval number, IMM-LiangZC-4).
2.7. Validation of nucleic acid delivery efficiency of iLP171 with siRNA
Female C57BL/6 mice, 5–7 weeks old, weighting 18–22 g, and ApoB-against siRNA (siApoB) were used to validate the in vivo nucleic acid delivery efficiency of iLP171. The iLP171/siApoB was prepared according to the same protocol of iLP171/mRNA. Then it was diluted to appropriate concentrations in sterile PBS and intravenously injected to the mice through the lateral tail vein at increasing doses ranging from 0.001 mg/kg to 1 mg/kg. After 24 h, mice were sacrificed and their livers were collected. The mRNA expression levels of ApoB in the livers were determined using the quantitative real-time PCR (qRT-PCR) assay. Briefly, total mRNA was extracted using the Trizol method and transcribed into cDNA. qRT-PCR was performed by using SYBR Green PCR Mix. The β-actin primer was used as the internal control. All the primers used in this study are listed in Table S1. ED50 value was calculated with GraphPad Prism 8.0 software.
2.8. Cellular uptake and intracellular distribution of iLNP171/mRNA
In order to investigate the cellular uptake and intracellular distribution of iLP171/mRNA in cells, a hepatoma carcinoma cell line, Huh7, was seeded in 6-well (2 × 105 per well) plate and cultured at 37 °C for 24 h. Then, the cells were transfected with iLP171/siRNA or iLP171/mRNA. iLP171/siRNA was prepared by formulating Cy5-labeled siRNA according to the same protocol with mRNA. iLP171/mRNA used for fluorescent detection was prepared by encapsulating a nucleic acid mixture that containing mRNA and Cy5-labeled siRNA at a weight ratio of 9:1. Because completely identical lipid compositions and protocol are employed to load siRNA and mRNA, it is impossible for iLP171 nanoparticle exhibiting the capability to recognize and select siRNA or mRNA in a mixture, both siRNA and mRNA molecules will be loaded into the same LNP at the same time. Recording the fluorescence signal of LNP containing a mixture of mRNA and siRNA will reflect the behavior of iLP171/mRNA, because 90% of the nucleic acids in the particle is mRNA. The iLP171/siRNA and iLP171/mRNA were transfected at the same siRNA concentration (125 ng per well). Lipofectamine 2000 carrying siRNA, or mixture of mRNA and siRNA at a weight ratio of 9:1, were used as controls. After transfection for 4 h, cell nuclei and lysosomes were stained with Hoechst 33342 and Lysotracker Green, respectively. Then the cells were observed using confocal laser scanning microscope. For cellular uptake quantification, the transfected cells were digested with 0.25% trypsin, washed with PBS three times, and then were analyzed with flow cytometry (FACS).
2.9. mRNA-mediated protein expression in vitro
To evaluate the luciferase expression in vitro, Huh7 cells were firstly seeded in 24-well plate for 24 h, then were transfected with iLP171/mLuc at a concentration of 200 ng mRNA per well. At indicated time points, cells were lysed with lysis buffer (Promega Co., Madison) and the lysate was collected. After centrifuging at 12, 000 rpm for 1 min, the supernatant was collected for luminescence measurement with a multi-mode microplate reader (Synergy HT, BioTek).
For quantification of EPO protein expression in vitro, the effect of codon optimization of mEPO was firstly investigated. The Huh7 cells were transfected with iLNP containing unoptimized mEPO (iLP171/mEPO) or codon-optimized mEPO (iLP171/omEPO) for 4 h at a concentration of 400 ng mRNA per well. Then the cell culture supernatant was collected and the EPO concentration was determined with EPO detection ELISA Kit (R&D, DEP00). In addition, iLP171/omEPO was transfected at increasing concentrations of 200 ng, 400 ng, and 600 ng mRNA per well, respectively. At desired transfection time, EPO protein concentration was measured accordingly.
2.10. Biodistribution of iLP171/mRNA in mice
For in vivo biodistribution study, iLP171/siRNA and iLP171/mRNA were prepared according to aforementioned protocols and were intravenously injected into the C57BL/6 mice at the dose of 10 μg Cy5-labeled nucleic acid per mouse. Whole-body imaging was performed at 1 h, 4 h, 6 h, 10 h, and 24 h after injection by using in vivo imaging system (Kodak In-Vivo Imaging System FX Pro, Carestream Health, USA). Meanwhile, one animal was sacrificed at 1 h and 6 h after injection, and the main tissues were collected and re-examined by in vivo imaging. The observations was terminated at 24 h after administration. Two mice were sacrificed, and the main tissues were also isolated and re-analyzed. Finally, the mean fluorescence intensities of the livers from the whole-body images and the isolated tissues of the sacrificed animals at given time points were quantitatively analyzed using a molecular imaging software package (Carestream Health, USA).
2.11. Luciferase and EPO expression in vivo
To examine the delivery and expression efficiency of iLP171/mRNA, both iLP171/mLuc and iLP171/omEPO were employed. For evaluating in vivo luciferase expression, either PBS or iLP171/mLuc was injected into mice intravenously at a dose of 2 mg/kg (milligrams per kilograms, mpk). At given time points, bioluminescence in vivo imaging was performed by treating with D-luciferin potassium salt (SynChem OHG, Kalles, Germany) at the dose of 150 mg/kg (10 μL/g of body weight, 15 mg/mL stock solution, i.p.). One animal in each group was sacrificed at 1.5 h, 6 h, and 24 h post injection, and the main tissues were collected. Fifty micrograms of each tissue were weighed and homogenated. The luciferase expression in the supernatant of the homogenate was further examined. Meanwhile, the signal intensities from the liver position of the whole-body images were also analyzed with a molecular imaging software package (Carestream Health, USA). Fluorescence imaging by recoding the Cy5 signal in iLP171/mLuc complexes was also performed in parallel with the bioluminescence imaging.
In addition, the iLP171/omEPO was intravenously injected into the mice at a dose of 1 mpk for three animals per group. PBS and iLP171/mLuc were included as negative controls. The liver tissues were harvested at 6 h and 24 h after injection, and the EPO concentrations were determined by using ELISA Kit.
2.12. In vivo toxicity evaluation
To evaluate the in vivo toxicity of iLNPs, the PBS, iLP171/mLuc and iLP171/omEPO were intravenously injected into the C57BL/6 mice at the doses of 2 mpk (for iLP171/mLuc) or 1 mpk (for iLP171/omEPO). Blood was drawn retro-orbitally and serum was isolated after 6 h and 24 h injection. Cytokines in serum were measured with immunoassays based on Luminex xMAP (multi-analyte profiling) technology (Beijing 4A Biotech Co., Ltd) according to the manufacturer's protocol. For histological examination, the main organs including the liver, the heart, the spleen, the lung and the kidneys were collected 48 h after administration. Organs were fixed with 4% paraformaldehyde and embedded in paraffin, followed by sectioning and staining with H&E, analyzing with an inverted microscope (Olympus X71, Olympus, Tokyo, Japan).
2.13. Statistical analysis
All data are shown as the mean ± SD, differences with p < 0.05 were considered significant. Student's t-test and One-way ANOVA (GraphPad Software, La Jolla, CA, USA) were used to perform the statistical analysis for comparison between two groups and comparison between multiple groups, respectively. Statistical differences were defined as *p < 0.05, **p < 0.01, ***p < 0.001.
3. Results and discussion
3.1. Characterization of iLP171/mRNA
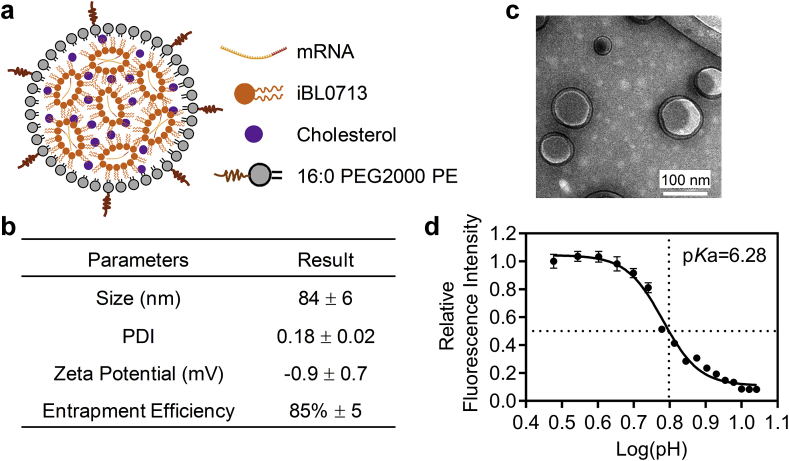
The iLNP formulation (iLP171) studied here is composed of three lipids, iBL0713, cholesterol and C16-PEG (16:0 PEG2000 PE) (Fig. 1a). The molar ratio of three lipids and weight ratio of total lipids to mRNA were optimized based on the formulation employing the same lipid components but used for siRNA delivery [8]. According to the DLS and TEM data, the size of iLP71/mRNA was approximate 84 nm with a polydispersity index (PDI) of 0.18. (Fig. 1b and c). The average surface charge at physiological pH environment was near neutral (−0.9 ± 0.7 mV). The iLP71/mRNA showed spherical shape, and higher than 80% mRNA encapsulation efficiency could be achieved.
Fig. 1.
Characterization of the physicochemical properties of iLP171/mRNA formulation. (a) Schematic diagram and (b) physicochemical parameters of iLP171/mRNA. PDI, polydispersity index. (c) TEM image of iLP171/mRNA nanoparticle. (d) Determination of pKa value of iLP171 by in situ TNS fluorescence titration. The pKa value was calculated with GraphPad Prism 8.0 software. Data are presented as mean ± SD. n = 6.
Many factors need to be considered in rational design of lipids or lipidoids for mRNA delivery, such as the pKa value, lipid saturation, the distance and flexibility of the charged group relative to the lipid caliper interface, the length of hydrophobic chain, etc. [3,18]. Among them, pKa value is of great importance for efficient nucleic acid delivery [9,25]. Ionizable lipid nanoparticle (iLNP) is significantly superior to cationic lipid nanoparticle (cLNP) in delivering nucleic acid. Firstly, iLNP with proper pKa value (normally between 6.0 and 6.5) can entrap nucleic acid at acidic pH, and exhibit neutral zeta potential at physiological pH (pH 7.4), which may dramatically enhance the safety performance by reducing nonspecific protein binding in circulation. cLNP may interact with various proteins, which potentially will trigger immune response, or accumulate in the capillaries of the lung and thereby influence the blood flow. Secondly, it is reported that iLNP can deliver nucleic acid to hepatocytes by binding with Apolipoprotein E (ApoE) and thereby interacting with LDLR (low-density lipoprotein receptor), which is highly expressed by hepatocytes. This is an active liver-targeting delivery mechanism. In contrast, cLNP delivers nucleic acid to hepatocytes in an ApoE and LDLR dually-independent manner, which reduces its transportation efficiency. Thirdly, ionizable lipids will be protonated and positive-charged in acidic environment, e.g., in endosome and lysosome, which may significantly enhance the endosomal escape efficiency of nucleic acids, and facilitate their accumulation in cytoplasm. Therefore, we determined the pKa value of iLP171 formulation by titrating with 2-(p-toluidino)-6-napthalene sulfonic acid (TNS). It was revealed that the pKa value is 6.28 (Fig. 1d), which perfectly matched with the best pKa range for nucleic acid delivery (between 6.0 and 6.5) [9,10,26].
To validate the capability of iLP171 in delivering nucleic acid to hepatocytes, siRNA targeting Apolipoprotein B (ApoB) is firstly employed to evaluate the gene silencing efficiency and determine the ED50 of iLP171. iLP171/siRNA nanoparticles were prepared according to the same protocol with mRNA, and intravenously injected into mice. Data manifested that the expression of ApoB mRNA was dose-dependently inhibited by this formulation. The ED50 is as low as 0.018 mg/kg (Fig. S1a and S1b), which is comparable to Dlin-MC3-DMA-formulated iLNP, whose ED50 is 0.03 mg/kg before optimizing the compositions when using Factor VII-against siRNA [10]. After composition optimization, the ED50 of Dlin-MC3-DMA-based siRNA liposome in mice is 0.005 mg/kg [10]. The ED50 determined with Factor VII-against siRNA normally is lower than that determined with ApoB-against siRNA. This assay demonstrated that the proposed iL171 could effectively entrap nucleic acids and deliver them to hepatocytes.
3.2. Internalization and protein expression of iLP171/mRNA in vitro
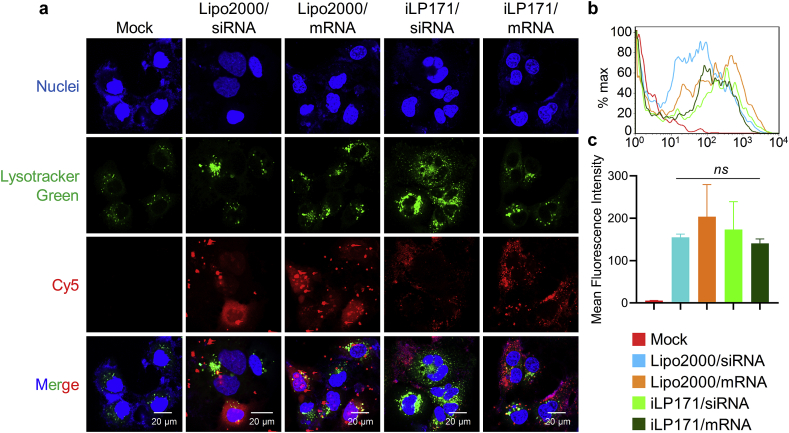
Confocal laser scanning microscopy (CLSM) and fluorescence-activated cell sorting (FACS) were employed to examine the delivery efficiency and subcellular localization of iLP171/mRNA in Huh7 cell, a hepatocellular carcinoma cell line. iLP171/siRNA and lipofectamine 2000 were included as controls. Fluorescence imaging illustrated that nucleic acid was successfully delivered into the cells. The Cy5 signal from iLP171/siRNA and iLP171/mRNA was dispersedly distributed in cytoplasm, around the nuclei (Fig. 2a). While many granular Cy5 signal was observed in lipofectamine 2000-treated cells, although dispersed signal was also recorded. Usually, it is the dispersedly-distributed nucleic acids finally accomplish their mission in the cell [27]. As we all know, lipofectamine 2000 is a representative and widely-used cationic LNP. These data revealed some detailed difference between lipofectamine 2000 and iLP171, and iLP171 exhibited more efficient endosomal escape than lipofectamine 2000. In addition, FACS data showed that nucleic acid was effectively transfected into the cells, no significant difference in percentage of fluorescent cells and mean fluorescence intensity was observed between lipofectamine 2000 and iLP171 (Fig. 2b and c).
Fig. 2.
Cellular uptake and intracellular distribution of iLP171/mRNA in Huh7 cells. (a) Confocal images of Huh7 cells after transfected with Lipofectamine2000/siRNA, Lipofectamine2000/mRNA, iLP171/siRNA or iLP171/mRNA for 4 h. (b and c) The cellular uptake of nucleic acid recorded by FACS. (c) Quantitative analysis of (b). Data were shown as the mean ± SD. ns: no significant difference. Scale bar: 20 μm.
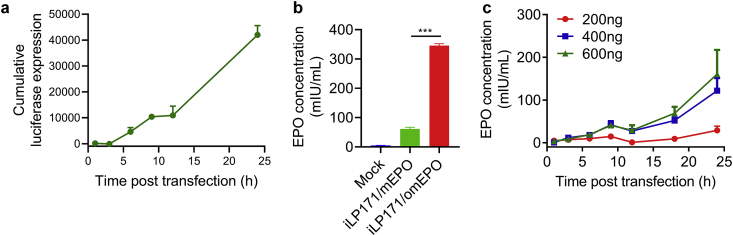
In addition, to explore the in vitro mRNA delivery efficiency of iLNP, we examined the protein expression of two different iLP171/mRNAs in Huh7. Firstly, mRNA-encoding firefly luciferase (mLuc) was loaded by iLP171 and transfected into Huh7. The cells were harvested at indicated time points and luciferase expression was determined by adding enzyme substrate. It is clearly suggested that luciferase was successfully and effectively expressed in Huh7. The cumulative expression increased along with the extension of expression time (Fig. 3a). Secondly, erythropoietin (EPO) expressing mRNA was also encapsulated by iLP171 and tested in Huh7. Two EPO mRNA sequences were in vitro transcribed from two different plasmids, one of which was codon-optimized (omEPO) and another was not (mEPO) (Supporting Materials). Current approaches to codon optimization are based on sequence characteristics considered to influence protein expression efficiency. The coding sequences of many endogenous genes contain frequent codons and rare codons. Frequent codons usually show higher protein expression efficiency than the rare codons, thus increasing the codon frequency is considered to improve the desired protein expression. Besides, mRNA may has many complicated secondary structures, which will inhibit its translation. Introducing of synonymous substitution codons is also considered to change the mRNA's secondary structure, which may improve translation efficiency. In addition, avoiding specific restriction enzyme cutting site, regulating the GC content, adding anti-reverse-cap analog (ARCA) and poly A tail are also common methods to improve translation and stability. By comprehensively considering these issues, the codons of EPO coding region was optimized, as shown in supporting materials. As a result, the codon-optimized EPO mRNA was generated by an IVT process with codon optimization, ARCA capping and enzymatically polyadenylation.
Fig. 3.
In vitro protein expression mediated by iLP171/mRNA. (a) Cumulative luciferase expression in Huh7 cells after being transfected with iLP171/mLuc. (b) EPO protein expression in Huh7 receiving the transfection of iLP171/mEPO (mRNA without codon optimization) or iLP171/omEPO (mRNA with codon optimization). (c) The EPO protein expression of iLP171/omEPO in Huh7 cells was recorded at various time points after cells were transfected at three different concentrations.
Twenty-four hours after transfection, codon-optimized EPO mRNA showed significantly higher protein expression level (345 mIU/mL) than codon-unoptimized EPO mRNA (61 mIU/mL) at the same mRNA transfection concentration (Fig. 3b). Therefore, codon-optimized EPO mRNA was employed in following tests. Moreover, omEPO was transfected with iLP171 at three different concentrations, 200 ng, 400 ng and 600 ng per well, respectively. Because EPO can be secreted from cells, the culture medium was collected at indicated time points, and the expression of EPO was determined using ELISA Kit. Data revealed that EPO was effectively expressed by Huh7 cells in a dose-dependent and time-dependent manner (Fig. 3c).
3.3. In vivo biodistribution
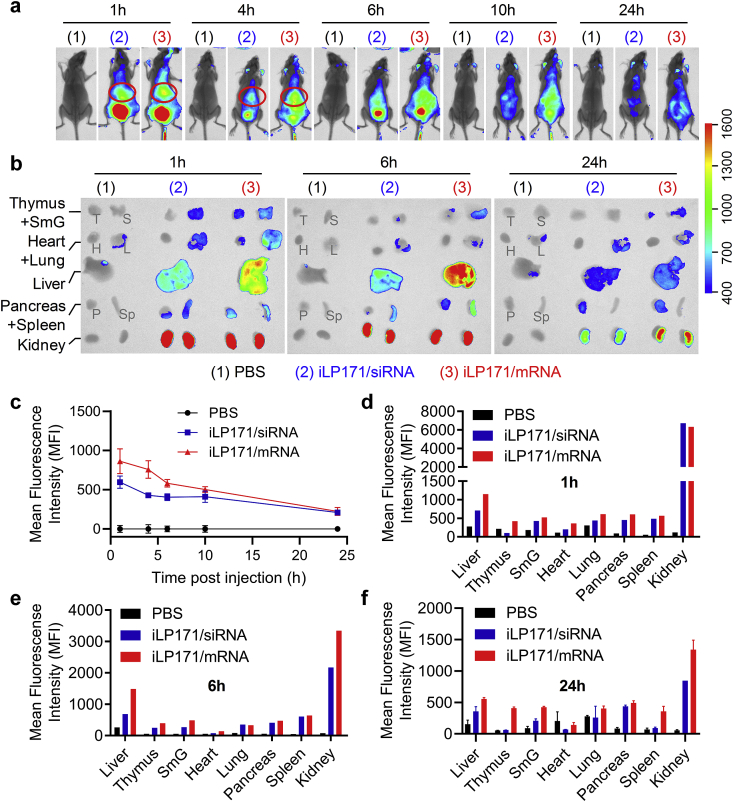
Given the superior mRNA delivery efficiency of iLP171 in vitro, we next evaluated the biodistribution of iLP171/mRNA in mice by intravenous injection. It was observed that iLP171/mRNA accumulated predominantly in the liver (as the red circles indicated) and the kidneys (Fig. 4a and b, and Fig. S2). Kidney is the dominant elimination tissue for most of nucleic acid modalities [28,29]. It is speculated that both passive and active liver-targeting behaviors happened for iLP171/mRNA accumulation in the liver. Most kinds of nanoparticles will be entrapped by the reticuloendothelial system (RES), only the percentages of injection dose of nanoparticles in the RES vary from each other, and 90% of the RES are located in the liver. The size (~80 nm) and morphology (spherical shape) of iLP171/mRNA may also contribute to the accumulation in the liver. More importantly, because iLP171 is an iLNP, ApoE may bind on the surface of iLP171/mRNA and promote its accumulation in the liver and internalization of hepatocytes via LDLR-mediated endocytosis [30]. Quantitative analyses of the whole-body images (Fig. 4c) and isolated organs (Fig. 4d–f) further confirmed these observations. No significant difference was displayed between iLP171/siRNA and iLP171/mRNA.
Fig. 4.
In vivo biodistribution of iLP171/mRNA. The animals were treated with PBS, or iLP171/siRNA, or iLP171/mRNA by intravenous injection. At indicated time points, whole-bodies of the mice (a) or the main organs (b) were fluorescently examined using an in vivo imaging system. Red circles indicate the liver positions in living mice. The mean fluorescence intensities of the liver in living mice (c) and the main organs from sacrificed animals (d–f) were quantitatively analyzed. SmG, submandibular gland; T, thymus; H, heart; L, lung; P, pancreas; Sp, spleen.
3.4. Protein expression mediated by iLP171/mRNA in vivo
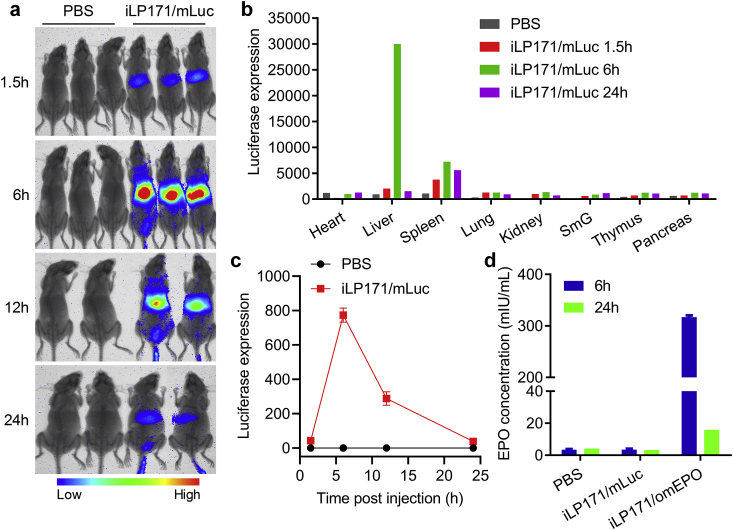
The excellent performances of iLP171/mRNA in in vitro transfection and protein expression, as well as in vivo biodistribution encouraged us to further evaluate the mRNA delivery efficiency and protein expression in mice. Firstly, luciferase-expressing mRNA (mLuc) was loaded with iLP171 and dosed at 2 mg/kg via intravenous injection. Bioluminescence images were acquired at different time points using an in vivo imaging system, which proved that luciferase was robustly expressed in the liver, and the maximum expression reached at 6 h post administration (Fig. 5a). This bioluminescence observations were in line with the Cy5-fluorescence imaging data that were acquired simultaneously (Fig. S2), which proved that iLP171/mRNA dominantly accumulated in the liver. Moreover, one animal was sacrificed at 1.5 h, 6 h, and 24 h after injection, respectively. The luciferase expression analyzed with the homogenates of the major organs manifested that mRNA was mainly delivered to and expressed in the liver, followed by the spleen. Luciferase was barely expressed in other organs. This was in line with the in vivo biodistribution profiles of the iLNP. The highest expression of luciferase in the liver was also observed at 6 h after injection, which is also consistent with the whole-body imaging data (Fig. 5b). Quantitative analysis of the whole-body images further confirmed above observations (Fig. 5c).
Fig. 5.
In vivo protein expression of iLP171/mRNA in mice. (a) Bioluminescence imaging of the mice receiving intravenous injection of PBS or iL171/mLuc (2 mg/kg). n = 2 or 3. (b) Luciferase expression recorded with the homogenates of 50 μg of tissues collected at 1.5 h, 6 h and 24 h after administration. (c) Quantitative analysis of the livers in (a). (d) iLP171/omEPO was injected into mice at a dose of 1 mg/kg, and EPO protein level in circulation was determined at 6 h and 24 h post injection with ELISA Kit.
Secondly, iLP171/omEPO was administrated into mice at the dose of 1 mg/kg. Because EPO can be secreted into the circulation from the hepatocytes, the serum samples were collected at given time points. It is suggested that EPO concentration was significantly enhanced in circulation, and the highest concentration also reached at 6 h after administration (Fig. 5d), revealing that the process of mRNA delivery and translation, and protein secretion was extremely effective and rapid. In contrast, iLP171/mLuc, a negative control for analyzing EPO expression, showed no EPO expression at all testing time points. Therefore, these results proved that iLP171 is able to deliver mRNA to hepatocytes effectively, achieve therapeutic protein expression in the liver, and can be used for mRNA therapeutic development.
The pKa value (6.28) of iLP171 formulation is considered to play an essential role in determining the efficiency of mRNA delivery to hepatocytes. As an ionizable lipid nanoparticle, it may bind with ApoE, interact with LDLR, and achieve an active liver-targeting transportation. Meanwhile, the surface charge of iLP171 is near neutral at physiological pH, which reduces the nonspecific disruption of plasma membranes in circulation. More importantly, it can become protonated and positively charged in acidic endosomes, leading to destabilization of both the liposome and the membrane of endosome. As a result, mRNA can quickly escape from the endosome and release from the liposome, which is crucial for following protein expression.
In addition, Dlin-MC3-DMA, a clinically-approved ionizable lipid, also showed ideal mRNA (e.g, mRNA encoding luciferase or Factor IX) delivery efficiency in the liver [3,31]. The transportation efficiencies of iBL0713 and Dlin-MC3-DMA-based formulations are comparable. However, iBL0713 is a proprietary lipid that can potentially be used to develop nucleic acid therapeutics with freedom to operate, which is an important starting point for this research.
3.5. Safety evaluation of iLP171/mRNA in vivo
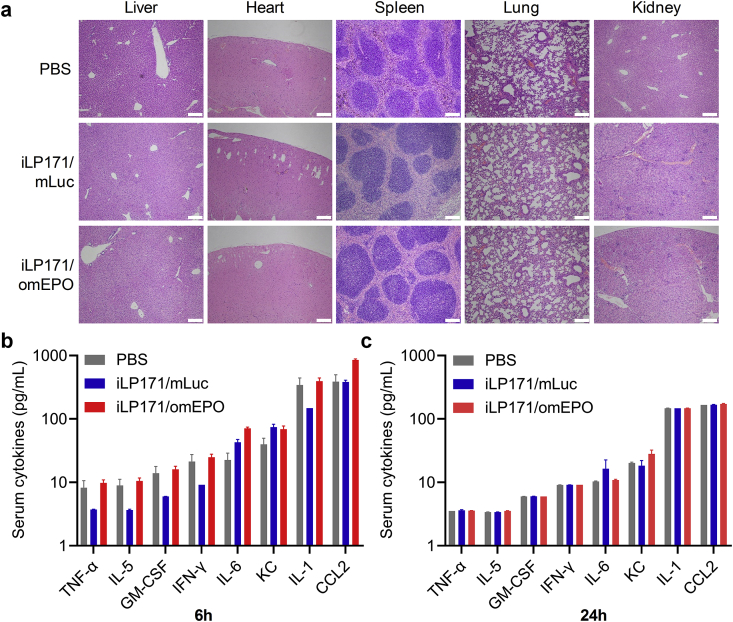
Finally, we evaluated whether iLP171/mRNA formulation is safe in mice. The major organs, including the heart, the liver, the spleen, the lung and the kidneys, were collected from the animals treated with PBS, or iLP171/mLuc, or iLP171/omEPO, and the histopathological sections were prepared. Dara revealed that no pathological change was observed (Fig. 6a). More importantly, serum specimens were also collected at 6 h and 24 h post administration. The levels of several cytokines, including TNF-α (tumor necrosis factor alpha), IL-5 (interleukin 5), GM-CSF (granulocyte-macrophage colony-stimulating factor (GM-CSF), or CSF2, colony stimulating factor 2), IFN-γ (interferon gamma), IL-6 (interleukin 6), KC (keratinocyte-derived cytokine, or CXCL1, chemokine (C-X-C motif) ligand 1), IL-1 (interleukin 1) and CCL2 (chemokine (C–C motif) ligand 2, or monocyte chemoattractant protein-1 (MCP-1)) were measured with Luminex technology. As a result, no significant immune response was observed in iLP171/mRNA-treated mice because no difference was observed in these animals compared to PBS-treated animals (Fig. 6b and 6c). These data further proved that the iLP171/mRNA has no immune stimulation effect and can be used for safe and effective nucleic acid delivery in vivo.
Fig. 6.
In vivo safety evaluation of iLP171/mRNA. The PBS, iLP171/mLuc or iLP171/omEPO were intravenously injected into the mice at the dose of 2 mg/kg (for iLP171/mLuc) or 1 mg/kg (for iLP171/omEPO). (a) H&E staining of the major organ sections were performed 48 h after injection. (b and c) Cytokine stimulation of these formulations recorded at 6 h (b) and 24 h (c) post injection. Scale bar: 200 μm.
4. Conclusion
In this study, we developed an ionizable lipid-based mRNA delivery system termed iLP171. mRNA can be efficiently encapsulated by iLP171 and formed regular spherical nanoparticles. Cellular uptake, intracellular localization and in vivo biodistribution were thoroughly investigated, suggesting mRNA was effectively transfected into the cells or delivered to the liver. Robust protein expression was achieved both in vitro and in vivo without triggering obvious adverse effects. The highest expression was observed at approximate 6 h after transfection or administration. Considering the great potential of mRNA therapy and the lack of systemic studies of in vivo mRNA delivery, our study sheds light on the application of lipid-mediated mRNA delivery for treating liver-relative diseases, such as anemia and hepatocellular carcinoma.
Author contribution
T. Y., X. W, and D. Z. constructed the plasmids and prepared the mRNAs. T. Y. and C. L. performed most of the studies. X. W. and M. Z. involved in animal studies. C. L., Y. W. and T. Y. characterized the iLNPs. Z. L. and X.-J. L. provided critical suggestions and involved in data analysis. Y. H. supervised on the project and analyzed the data. Y. H., Y. W. and T. Y. wrote the manuscript.
CRediT authorship contribution statement
Tongren Yang: Investigation, Methodology, Formal analysis, Data curation, Writing - original draft. Xiaoxia Wang: Investigation, Methodology, Resources, Data curation. Deyao Zhao: Investigation, Methodology. Mengjie Zhang: Investigation. Huiqing Cao: Resources. Zicai Liang: Resources. Haihua Xiao: Formal analysis. Xing-Jie Liang: Formal analysis. Yuhua Weng: Investigation, Formal analysis, Writing - original draft. Yuanyu Huang: Conceptualization, Methodology, Formal analysis, Data curation, Writing - review & editing, Visualization, Supervision, Funding acquisition.
Declaration of competing interest
Z. L. is the founder of Suzhou Ribo Life Science Co. Ltd. The other authors declare no competing financial interests.
Acknowledgment
This work was supported by the National Natural Science Foundation of China (31871003, 81402863, 31901053), the Beijing Institute of Technology Research Fund Program for Young Scholars and the Fundamental Research Funds for the Central Universities (3052018065, 1870012222004), the Natural Science Foundation of Guangdong Province (2019A1515010776), the Hunan Provincial Natural Science Foundation of China (2018JJ1019, 2019JJ50196) and the Hu-Xiang Young Talent Program (2018RS3094).
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
Supplementary data related to this article can be found at https://doi.org/10.1016/j.bioactmat.2020.07.003.
Contributor Information
Yuhua Weng, Email: wengyh@bit.edu.cn.
Yuanyu Huang, Email: yyhuang@bit.edu.cn.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- 1.Hajj K.A., Whitehead K.A. Tools for translation: non-viral materials for therapeutic mRNA delivery. Nat Rev Mater. 2017;2:17056. [Google Scholar]
- 2.Sahin U., Kariko K., Tureci O. mRNA-based therapeutics--developing a new class of drugs. Nat. Rev. Drug Discov. 2014;13:759–780. doi: 10.1038/nrd4278. [DOI] [PubMed] [Google Scholar]
- 3.Weng Y., Li C., Yang T., Hu B., Zhang M., Guo S., Xiao H., Liang X.J., Huang Y. The challenge and prospect of mRNA therapeutics landscape. Biotechnol. Adv. 2020;40:107534. doi: 10.1016/j.biotechadv.2020.107534. [DOI] [PubMed] [Google Scholar]
- 4.Kowalski P.S., Rudra A., Miao L., Anderson D.G. Delivering the messenger: advances in technologies for therapeutic mRNA delivery. Mol. Ther. 2019;27:710–728. doi: 10.1016/j.ymthe.2019.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Geall A.J., Verma A Fau - Otten G.R., Otten Gr Fau - Shaw C.A., Shaw Ca Fau - Hekele A., Fau - Banerjee K.Hekele A., Banerjee K Fau - Cu Y., Fau - Beard C.W.Cu Y., Beard Cw Fau - Brito L.A., Brito La Fau - Krucker T., Krucker T Fau - O'Hagan D.T., O'Hagan Dt Fau - Singh M., Singh M Fau - Mason P.W., Mason Pw Fau - Valiante N.M., Valiante Nm Fau - Dormitzer P.R., Dormitzer Pr Fau - Barnett S.W., Barnett Sw Fau - Rappuoli R., Rappuoli R Fau - Ulmer J.B., Ulmer Jb Fau - Mandl C.W., Mandl C.W. Nonviral delivery of self-amplifying RNA vaccines. Proc. Natl. Acad. Sci. U. S. A. 2012;109:14604–14609. doi: 10.1073/pnas.1209367109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kauffman K.J., Mir F.F., Jhunjhunwala S., Kaczmarek J.C., Hurtado J.E., Yang J.H., Webber M.J., Kowalski P.S., Heartlein M.W., DeRosa F., Anderson D.G. Efficacy and immunogenicity of unmodified and pseudouridine-modified mRNA delivered systemically with lipid nanoparticles in vivo. Biomaterials. 2016;109:78–87. doi: 10.1016/j.biomaterials.2016.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weng Y., Xiao H., Zhang J., Liang X.J., Huang Y. RNAi therapeutic and its innovative biotechnological evolution. Biotechnol. Adv. 2019;37:801–825. doi: 10.1016/j.biotechadv.2019.04.012. [DOI] [PubMed] [Google Scholar]
- 8.Zheng S., Wang X., Weng Y.H., Jin X., Ji J.L., Guo L., Hu B., Liu N., Cheng Q., Zhang J., Bai H., Yang T., Xia X.H., Zhang H.Y., Gao S., Huang Y. siRNA knockdown of RRM2 effectively suppressed pancreatic tumor growth alone or synergistically with doxorubicin. Mol. Ther. Nucleic Acids. 2018;12:805–816. doi: 10.1016/j.omtn.2018.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Whitehead K.A., Dorkin J.R., Vegas A.J., Chang P.H., Veiseh O., Matthews J., Fenton O.S., Zhang Y., Olejnik K.T., Yesilyurt V., Chen D., Barros S., Klebanov B., Novobrantseva T., Langer R., Anderson D.G. Degradable lipid nanoparticles with predictable in vivo siRNA delivery activity. Nat. Commun. 2014;5:4277. doi: 10.1038/ncomms5277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jayaraman M., Ansell S.M., Mui B.L., Tam Y.K., Chen J., Du X., Butler D., Eltepu L., Matsuda S., Narayanannair J.K., Rajeev K.G., Hafez I.M., Akinc A., Maier M.A., Tracy M.A., Cullis P.R., Madden T.D., Manoharan M., Hope M.J. Maximizing the potency of siRNA lipid nanoparticles for hepatic gene silencing in vivo. Angew Chem. Int. Ed. Engl. 2012;51:8529–8533. doi: 10.1002/anie.201203263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dong Y., Love K.T., Dorkin J.R., Sirirungruang S., Zhang Y., Chen D., Bogorad R.L., Yin H., Chen Y., Vegas A.J., Alabi C.A., Sahay G., Olejnik K.T., Wang W., Schroeder A., Lytton-Jean A.K., Siegwart D.J., Akinc A., Barnes C., Barros S.A., Carioto M., Fitzgerald K., Hettinger J., Kumar V., Novobrantseva T.I., Qin J., Querbes W., Koteliansky V., Langer R., Anderson D.G. Lipopeptide nanoparticles for potent and selective siRNA delivery in rodents and nonhuman primates. Proc. Natl. Acad. Sci. U. S. A. 2014;111:3955–3960. doi: 10.1073/pnas.1322937111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dwarki V.J., Malone R.W., Verma I.M. Cationic liposome-mediated RNA transfection. Methods Enzymol. 1993;217:644–654. doi: 10.1016/0076-6879(93)17093-k. [DOI] [PubMed] [Google Scholar]
- 13.Felgner P.L., Gadek T.R., Holm M., Roman R., Chan H.W., Wenz M., Northrop J.P., Ringold G.M., Danielsen M. Lipofection: a highly efficient, lipid-mediated DNA-transfection procedure. Proc. Natl. Acad. Sci. U. S. A. 1987;84:7413–7417. doi: 10.1073/pnas.84.21.7413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Manunta M.D.I., Tagalakis A.D., Attwood M., Aldossary A.M., Barnes J.L., Munye M.M., Weng A., McAnulty R.J., Hart S.L. Delivery of ENaC siRNA to epithelial cells mediated by a targeted nanocomplex: a therapeutic strategy for cystic fibrosis. Sci. Rep. 2017;7:700. doi: 10.1038/s41598-017-00662-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao P., Hou X., Yan J., Du S., Xue Y., Li W., Xiang G., Dong Y. Long-term storage of lipid-like nanoparticles for mRNA delivery. Bioact Mater. 2020;5:358–363. doi: 10.1016/j.bioactmat.2020.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang Y., Gao F., Jiang X., Zhao X., Wang Y., Kuai Q., Nie G., He M., Pan Y., Shi W., Ren S., Yu Q. Co-delivery of gemcitabine and mcl-1 SiRNA via cationic liposome-based system enhances the efficacy of chemotherapy in pancreatic cancer. J. Biomed. Nanotechnol. 2019;15:966–978. doi: 10.1166/jbn.2019.2762. [DOI] [PubMed] [Google Scholar]
- 17.Yu S., Bi X., Yang L., Wu S., Yu Y., Jiang B., Zhang A., Lan K., Duan S. Co-delivery of paclitaxel and PLK1-targeted siRNA using aptamer-functionalized cationic liposome for synergistic anti-breast cancer effects in vivo. J. Biomed. Nanotechnol. 2019;15:1135–1148. doi: 10.1166/jbn.2019.2751. [DOI] [PubMed] [Google Scholar]
- 18.Semple S.C., Akinc A., Chen J., Sandhu A.P., Mui B.L., Cho C.K., Sah D.W., Stebbing D., Crosley E.J., Yaworski E., Hafez I.M., Dorkin J.R., Qin J., Lam K., Rajeev K.G., Wong K.F., Jeffs L.B., Nechev L., Eisenhardt M.L., Jayaraman M., Kazem M., Maier M.A., Srinivasulu M., Weinstein M.J., Chen Q., Alvarez R., Barros S.A., De S., Klimuk S.K., Borland T., Kosovrasti V., Cantley W.L., Tam Y.K., Manoharan M., Ciufolini M.A., Tracy M.A., de Fougerolles A., MacLachlan I., Cullis P.R., Madden T.D., Hope M.J. Rational design of cationic lipids for siRNA delivery. Nat. Biotechnol. 2010;28:172–176. doi: 10.1038/nbt.1602. [DOI] [PubMed] [Google Scholar]
- 19.Oberli M.A., Reichmuth A.M., Dorkin J.R., Mitchell M.J., Fenton O.S., Jaklenec A., Anderson D.G., Langer R., Blankschtein D. Lipid nanoparticle assisted mRNA delivery for potent cancer immunotherapy. Nano Lett. 2017;17:1326–1335. doi: 10.1021/acs.nanolett.6b03329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Billingsley M.M., Singh N., Ravikumar P., Zhang R., June C.H., Mitchell M.J. Ionizable lipid nanoparticle-mediated mRNA delivery for human CAR T cell engineering. Nano Lett. 2020;20:1578–1589. doi: 10.1021/acs.nanolett.9b04246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Islam M.A., Xu Y., Tao W., Ubellacker J.M., Lim M., Aum D., Lee G.Y., Zhou K., Zope H., Yu M., Cao W., Oswald J.T., Dinarvand M., Mahmoudi M., Langer R., Kantoff P.W., Farokhzad O.C., Zetter B.R., Shi J. Restoration of tumour-growth suppression in vivo via systemic nanoparticle-mediated delivery of PTEN mRNA. Nat Biomed Eng. 2018;2:850–864. doi: 10.1038/s41551-018-0284-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kranz L.M., Diken M., Haas H., Kreiter S., Loquai C., Reuter K.C., Meng M., Fritz D., Vascotto F., Hefesha H., Grunwitz C., Vormehr M., Hüsemann Y., Selmi A., Kuhn A.N., Buck J., Derhovanessian E., Rae R., Attig S., Diekmann J., Jabulowsky R.A., Heesch S., Hassel J., Langguth P., Grabbe S., Huber C., Türeci Ö., Sahin U. Systemic RNA delivery to dendritic cells exploits antiviral defence for cancer immunotherapy. Nature. 2016;534:396–401. doi: 10.1038/nature18300. [DOI] [PubMed] [Google Scholar]
- 23.Ickenstein L.M., Garidel P. Lipid-based nanoparticle formulations for small molecules and RNA drugs. Expet Opin. Drug Deliv. 2019;16:1205–1226. doi: 10.1080/17425247.2019.1669558. [DOI] [PubMed] [Google Scholar]
- 24.Bose R.J.C., Ravikumar R., Karuppagounder V., Bennet D., Rangasamy S., Thandavarayan R.A. Lipid-polymer hybrid nanoparticle-mediated therapeutics delivery: advances and challenges. Drug Discov. Today. 2017;22:1258–1265. doi: 10.1016/j.drudis.2017.05.015. [DOI] [PubMed] [Google Scholar]
- 25.Lee J.B., Zhang K., Tam Y.Y., Tam Y.K., Belliveau N.M., Sung V.Y., Lin P.J., LeBlanc E., Ciufolini M.A., Rennie P.S., Cullis P.R. Lipid nanoparticle siRNA systems for silencing the androgen receptor in human prostate cancer in vivo. Int. J. Canc. 2012;131:E781–E790. doi: 10.1002/ijc.27361. [DOI] [PubMed] [Google Scholar]
- 26.Cheng Q., Wei T., Farbiak L., Johnson L.T., Dilliard S.A., Siegwart D.J. Selective organ targeting (SORT) nanoparticles for tissue-specific mRNA delivery and CRISPR-Cas gene editing. Nat. Nanotechnol. 2020;15:313–320. doi: 10.1038/s41565-020-0669-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou J., Wu Y., Wang C., Cheng Q., Han S., Wang X., Zhang J., Deng L., Zhao D., Du L., Cao H., Liang Z., Huang Y., Dong A. pH-sensitive nanomicelles for high-efficiency siRNA delivery in vitro and in vivo: an insight into the design of polycations with Robust cytosolic release. Nano Lett. 2016;16:6916–6923. doi: 10.1021/acs.nanolett.6b02915. [DOI] [PubMed] [Google Scholar]
- 28.Huang Y., Cheng Q., Ji J.L., Zheng S., Du L., Meng L., Wu Y., Zhao D., Wang X., Lai L., Cao H., Xiao K., Gao S., Liang Z. Pharmacokinetic behaviors of intravenously administered siRNA in glandular tissues. Theranostics. 2016;6:1528–1541. doi: 10.7150/thno.15246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou J., Neff C.P., Liu X., Zhang J., Li H., Smith D.D., Swiderski P., Aboellail T., Huang Y., Du Q., Liang Z., Peng L., Akkina R., Rossi J.J. Systemic administration of combinatorial dsiRNAs via nanoparticles efficiently suppresses HIV-1 infection in humanized mice. Mol. Ther. 2011;19:2228–2238. doi: 10.1038/mt.2011.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Akinc A., Querbes W., De S., Qin J., Frank-Kamenetsky M., Jayaprakash K.N., Jayaraman M., Rajeev K.G., Cantley W.L., Dorkin J.R., Butler J.S., Qin L., Racie T., Sprague A., Fava E., Zeigerer A., Hope M.J., Zerial M., Sah D.W., Fitzgerald K., Tracy M.A., Manoharan M., Koteliansky V., Fougerolles A., Maier M.A. Targeted delivery of RNAi therapeutics with endogenous and exogenous ligand-based mechanisms. Mol. Ther. 2010;18:1357–1364. doi: 10.1038/mt.2010.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ramaswamy S., Tonnu N., Tachikawa K., Limphong P., Vega J.B., Karmali P.P., Chivukula P., Verma I.M. Systemic delivery of factor IX messenger RNA for protein replacement therapy. Proc. Natl. Acad. Sci. U. S. A. 2017;114:E1941–e1950. doi: 10.1073/pnas.1619653114. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.