Highlights
-
•
PLP1 promotes PEDV replication and inhibits expression of TNF-α induced IFN-β.
-
•
PLP1 interacts with cellular PCBP2.
-
•
PCBP2 expression affects PEDV replication.
-
•
The interaction of PCBP2 and PLP1 supports PEDV replication.
Keywords: PEDV, Coronavirus, Papain-like proteases, Poly(C) binding protein 2, Virus replication, Interaction
Abstract
Porcine epidemic diarrhea virus (PEDV) belongs to the Alphacoronavirus genus in the Coronaviridae family. Similar to other coronaviruses, PEDV encodes two papain-like proteases. Papain-like protease (PLP)2 has been proposed to play a key role in antagonizing host innate immunity. However, the function of PLP1 remains unclear. In this study, we found that overexpression of PLP1 significantly promoted PEDV replication and inhibited production of interferon-β. Immunoprecipitation and mass spectrometry were used to identify cellular interaction partners of PLP1. Host cell poly(C) binding protein 2 (PCBP2) was determined to bind and interact with PLP1. Both endogenous and overexpressed PCBP2 co-localized with PLP1 in the cytoplasm. Overexpression of PLP1 upregulated expression of PCBP2. Furthermore, overexpression of PCBP2 promoted PEDV replication. Silencing of endogenous PCBP2 using small interfering RNAs attenuated PEDV replication. Taken together, these data demonstrated that PLP1 negatively regulated the production of type 1 interferon by interacting with PCBP2 and promoted PEDV replication.
1. Introduction
Porcine epidemic diarrhea virus (PEDV) was identified as the causative agent of porcine epidemic diarrhea (PED) in 1978 (Pensaert and de Bouck, 1978) and has had catastrophic impacts on the global pig industry. The clinical signs and symptoms of PED include severe enteritis, vomiting, watery diarrhea, and high mortality. In 2010 China experienced and outbreak of a mutant PEDV, leading to huge economic losses (Sun et al., 2012a). PEDV has a positive-sense single stranded RNA genome that is approximately 28 kb in size and contains six open reading frames (ORFs). ORF1ab encodes polyprotein (pp)1a and pp1ab, which are further cleaved into non-structural protein (NSP)1–16. The structural spike (S), envelope (E), membrane (M), and nucleocapsid (N) proteins are encoded by ORF2, ORF4, ORF5 and ORF6, respectively. ORF3 encodes the accessory protein ORF3 (Kocherhans et al., 2001). Nsp3 is the largest Nsp, and comprises papain-like protease (PLP)1 and PLP2 domains (Lei et al., 2018).
The innate immune response is the first line of host defense against viral infection (O'Neill and Bowie, 2010). Type 1 interferons (IFNs) play a key role in host resistance to viral infections. RNA viruses induce the production of IFNs through Toll-like receptor 3 (TLR3) and retinoic acid-inducible gene (RIG)-I-like receptor-dependent pathways (Kawai and Akira, 2007). The virus can evade host innate immune response in two major ways: by modifying or hiding pathogen-associated molecular patterns (PAMPs), and by encoding specific proteins to block immune responses. Coronaviruses have evolved specific mechanisms to evade or inhibit host antiviral innate immune responses (G Devaraj et al., 2007; Zhou and Perlman, 2007). Many PEDV proteins are involved in escaping the innate immune response. Nsp1, Nsp3, Nsp7, Nsp14, Nsp15, and Nsp16, as well as the structural E, M and N proteins, demonstrated IFN antagonism (Zhang et al., 2016). The mechanisms underlying the IFN antagonism of Nsp1, PLP2, Nsp5, and N protein have been elucidated (Wang et al., 2016; Xing et al., 2013; Zhang et al., 2016). PEDV Nsp1 inhibits production of type I IFNs by degrading cyclic adenosine monophosphate responsive element-binding protein-binding protein and inhibiting immune stress granule expression (Dragan et al., 2007; Zhang et al., 2016). PEDV Nsp5 is a 3C-like protease that proteolytically cleaves the nuclear transcription factor kappa B (NF-κB) essential modulator (NEMO) at glutamine 231, impairing the ability of NEMO to activate IFN production (Wang et al., 2016). The PEDV N protein interacts with the TANK-binding kinase, blocking its association with interferon regulatory factor 3 (IRF3) and thus inhibiting IRF3 activation and type I IFN production (Ding et al., 2014; Hu et al., 2018).
PEDV can also resist immune responses by co-opting host cell proteins. Poly(C) binding protein 2 (PCBP2) belongs to a class of proteins that bind poly(C) sequences in both RNA and DNA and are involved in maintaining mRNA stability, regulating translation and cellular antiviral responses (Makeyev and Liebhaber, 2002). PCBP2 expression is induced following viral infection and acts as a negative regulator of mitochondrial antiviral signaling protein (MAVS), triggering its degradation (You et al., 2009). It was previously reported that PCBP2 interacted with porcine reproductive and respiratory syndrome virus (PRRSV) Nsp1β and supported viral replication (Beura et al., 2011; Wang et al., 2012). PCBP2 also antagonized vesicular stomatitis virus growth by affecting viral gene expression (Dinh et al., 2011). It is unclear whether PCBP2 is also involved in PEDV replication. Although significant progress has been made in understanding PEDV evasion of innate immune responses, the mechanism of interaction between viral and host cell proteins remains unclear.
In the present study, we demonstrated that PLP1 interacted with PCBP2 to inhibit IFN-β production and promote PEDV replication.
2. Materials and methods
2.1. Cells and viruses
Human embryonic kidney (HEK) 293 T cells, porcine intestinal epithelial IPEC-J2 cells and African green monkey Vero E6 kidney cells were maintained in Dulbecco’s modified Eagle's Medium supplemented with 10 % heat-inactivated fetal bovine serum (FBS), 100 units/mL penicillin and 100 μg/mL streptomycin. PEDV strain GDgh (Genbank accession number: MG983755) was isolated and stored in our laboratory.
2.2. Plasmid construction
A DNA sequence encoding PEDV PLP1 protein was cloned into pCMV-HA as previously described to yield the pCMV-PLP1-HA expression vector (Yu et al., 2019). PCBP2 and PCBP1 were amplified by PCR and cloned into the pECMV-3×FLAG-N vector using the primers listed in Table 1 . All plasmids were verified by DNA sequencing.
Table 1.
Primers used for genes amplication.
| Name | Sequences (5′to3′) | Use |
|---|---|---|
| PEDV N-F | CGCAAAGACTGAACCCACTAA | quantitative RT-PCR |
| PEDV N-R | TTGCCTCTGTTGTTACTTGGAGAT | |
| qPCBP2 F | TCAGGACAGGTACAGCACAG | quantitative RT-PCR |
| qPCBP2 R | CTGGTGCAGCTTGGTCAAAT | |
| PCBP2 siRNA (3) | CCGGAUUCAGUGCAGGUUUTT | Silence PCBP2 gene |
| AAACCUGCACUGAAECCGGTT | ||
| PCBP2 siRNA (2) | CCACUAAUGCCAUCUUCMTT | |
| UUGAAGAUGGCAUUAGUGGTT | ||
| PCBP2 siRNA (1) | CCACUAAUGCCAUCUUCMTT | |
| UUGAAGAUGGCAUUAGUGGTT | ||
| PLP1F | CCGGAATTCGGGAAGTTG TTACTGATGCAC (EcoRI) | PLP1gene amplification and clone |
| PLP1R | CGGGGTACCTCATTTAACACAAATAGTGTTCA (KpnI) | |
| PCBP1F | GGGGTACCATGGATGCCGGTGTGAC(KpnI) | PCBP1gene amplification and clone |
| PCBP1R | CGGAATTCCTAGCTGCACCCCATGC(EcoRI) | |
| PCBP2 F | GGGGTACCATGGACACCGGTGTGAT(KpnI) | PCBP2gene amplification and clone |
| PCBP2 R | CGGAATTCCTAGCTGCTCCCCATGC(EcoRI) | |
| β-actin-F | CTCCGATCTGTGCAGGGTAT | RT-PCR |
| β-actin-R | GAGGCGCGATGATCTTGATC |
2.3. Dual luciferase reporter assay
HEK293 T cells were seeded on coverslips in 24-well plates and cotransfected with 100 ng of IFN-Luc, 5 ng of TK-Luc and 500 ng of pCMV-PLP1-HA or empty pCMV-HA vector. Twenty-four hours post-transfection (hpt), cells were stimulated with 10 ng/mL tumor necrosis factor (TNF)-α for 6 h. Cell lysates were prepared for analysis of luciferase activity using a luciferase enzyme assay system in accordance with the manufacturer's instructions (Promega, Beijing, China).
2.4. RNA isolation and quantitative reverse transcription polymerase chain reaction (RT-PCR)
Total cellular RNA was extracted using the MiniBEST Universal RNA Extraction Kit (Takara, Japan) in accordance with the manufacturer’s protocol. Total RNA was reverse transcribed to cDNA using Prime Script™ RT Master Mix (Takara, Japan) and relative gene expression levels were quantified by quantitative RT-PCR using SYBR® Green real-time PCR master mix (Toyobo, Japan) using the cycle threshold method. β-actin was used as the internal control. All primers were designed using Primer Premier 6.0 software and are listed in Table 1.
2.5. Co-immunoprecipitation
HEK293 T cells were grown to 80 % confluence in a 100 mm dish and transfected with empty pCMV-HA vector or pCMV-PLP1-HA using Lipofectamine LTX Reagent (Thermo Scientific, China) and following the manufacturer’s instructions. At 24 hpt, cells were washed with phosphate-buffered saline (PBS) and lysed with lysis buffer (20 mM Tris, pH 7.5, containing 150 mM NaCl, 1% Triton X-100, sodium pyrophosphate, β-glycerophosphate, ethylenediaminetetraacetic acid, Na3VO4, and leupeptin) supplemented with 1 mM phenylmethylsulfonyl fluoride. Lysates were centrifuged at 12,000 g for 10 min at 4 °C. The supernatant was collected and incubated with 20 μL of anti-HA agarose (Pierce® HA Tag IP/Co-IP Kit, Thermo Scientific) at 4 °C with shaking overnight. Immunoprecipitates were washed with TBST buffer (25 mM Tris-HCl, pH 7.2, containing 0.15 M NaCl and 0.05 % Tween 20) and eluted in non-reducing sample buffer. The samples were separated by SDS-PAGE and either transferred to a nitrocellulose membrane for western blotting or silver stained for mass spectrometry (Pierce TM, Thermo Scientific).
2.6. Liquid chromatography-mass spectrometry analysis
The silver stained gel was analyzed, and one differentially expressed protein spot was selected for liquid chromatography-mass spectrometry (LC—MS/MS) analysis. Briefly, the excised band was cut into small cubes approximately 0.5–1 mm3 in size, decolorized in a 37 °C water bath, further decolorized in decolorizing solution (30 mM potassium ferricyanide and 100 mM sodium thiosulfate) in a 37 °C water bath, reduced with 50 mM dithiothreitol at 56 °C for 1 h, then alkylated with 100 mM iodoacetamide for 40 min at room temperature in the dark. The samples were digested with trypsin overnight.
Tryptic peptides were analyzed using EASY-nL C1200 and Q-Exactive (Thermo Scientific, Waltham, MA). Trapped peptides were separated on an analytical C18 column (75 μm ×25 cm, Thermo Scientific). The mobile phases consisted of 2% acetonitrile (ACN) A and 80 % ACN B, both containing 0.1 % formic acid. A gradient of 5%–100 % solvent B was used to elute the peptides at a constant flow rate of 300 nL/min for 90 min. Data were acquired using a MS scan range (m/z) of 350–1300, acquisition mode DDA and a resolution of 70,000. Thermo Xcalibur 4.0 software (Thermo) was used for data acquisition. Protein identifications were assigned using the NCBI nr databank and SwissProt/UniProt databank.
2.7. Western blotting
Cells were harvested and lysed with lysis buffer. Protein samples were mixed with SDS sample loading buffer, electrophoresed on 10 % or 12 % polyacrylamide gels and transferred to a nitrocellulose membrane. The membrane was incubated with primary antibodies for 2 h at 37 °C. PLP1 was detected using a primary anti-HA-Tag rabbit monoclonal antibody (Cell Signaling Technology, Danvers, MA). An anti-FLAG mouse antibody (Sigma-Aldrich, St. Louis, MO) was used to detect FLAG-tagged proteins and an anti-tubulin antibody (Cell Signaling Technology) was used to detect tubulin. Rabbit polyclonal PCBP2 antibody (Proteintech, Chicago, IL) was used to detect endogenous PCBP2 protein. Horseradish peroxidase-conjugated anti-mouse IgG and goat anti-rabbit IgG antibodies were used as secondary antibodies. Chemiluminescence was detected using the Gel Imaging System Tanon-5200Multi (Tanon, Shanghai, China).
2.8. Confocal microscopy
Vero E6 cells were grown in glass slides and cotransfected with pCMV-PLP1-HA, pECMV-3×FLAG-PCBP2 or empty vector (pECMV-3×FLAG). At 36 hpt, the cells were washed three times with PBS, fixed with 0.4 % paraformaldehyde for 1 h at room temperature, and blocked with 1 % bovine serum albumin for 1 h. Subsequently, the cells were permeabilized with 0.5 % Triton X-100 for 5 min, and then incubated at 37 °C for 1 h with the primary antibody. The antibodies used were as follows: rabbit anti-HA monoclonal antibody (Cell Signaling), mouse anti-HA monoclonal antibody (Cell Signaling), mouse anti-FLAG antibody (Sigma-Aldrich) or rabbit anti-PCBP2 polyclonal antibody (Proteintech). All antibodies were diluted 1:100 in 10 % FBS. Goat anti-rabbit IgG conjugated to Alexa Fluor 555 and anti-mouse IgG conjugated to Alexa Fluor 488 (Abcam, Shanghai, China) diluted 1:2000 in 10 % FBS were used as secondary antibodies. The cells were stained with 4′,6-diamidino-2-phenylindole and visualized using a Zeiss LSM 710 confocal microscope (Oberkochen, Germany).
2.9. Small interfering (si)RNA silencing
siRNAs targeting the PCBP2 gene were designed and synthesized by GenePharma (Shanghai, China) (Table 1). IPEC-J2 cells were transfected with 20 nM of siRNAs using Lipofectamine RNAiMax (Thermo Fisher). At 24 hpt, cells were infected with PEDV at a multiplicity of infection (MOI) of 0.1. At 24 h or 48 h post infection (hpi), the cells were collected for viral quantitation. The effects of siRNA were evaluated by quantitative RT-PCR and western blotting to assess the expression level of PCBP2. Primers for PCBP2 quantitative RT-PCR are shown in Table 1. Anti-tubulin antibody and rabbit polyclonal PCBP2 antibody (Proteintech) were used to assess expression levels of tubulin and PCBP2 proteins.
2.10. Statistical analysis
All results were presented as the means ± standard errors of the means of three independent experiments. Data were analyzed using GraphPad Prism 6.0 (GraphPad Software, Inc., La Jolla, CA). Differences between and among groups were assessed using the student’s t-test and two-way analysis of variance, respectively. Values of p < 0.05 were considered statistically significant and were indicated as follows: *p < 0.05, **p < 0.01, and ***p < 0.005.
3. Results
3.1. PLP1 promotes PEDV replication and inhibits TNF-α-induced IFN-β production
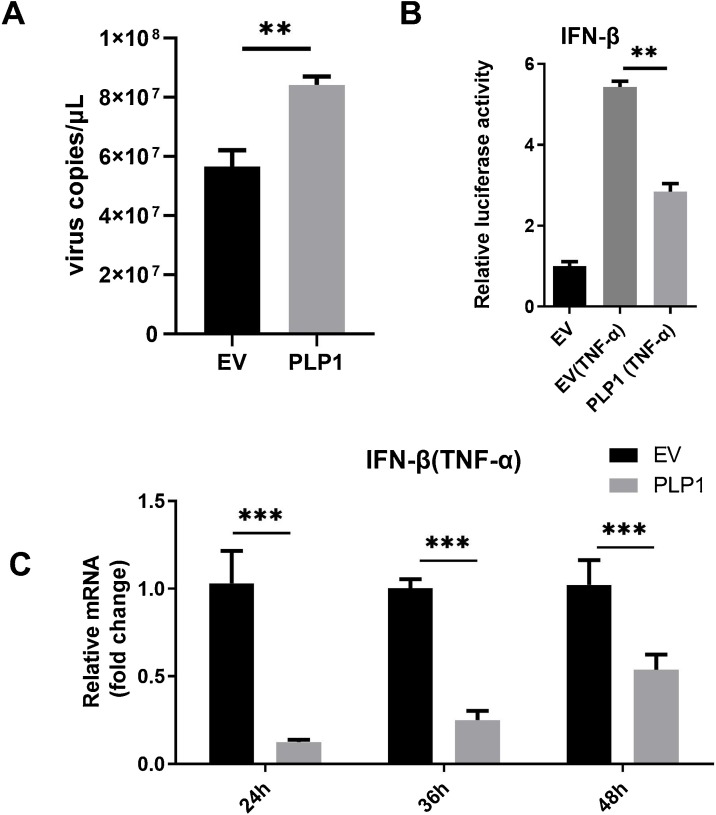
IFN-α/β is a key component of the host i2nnate immune response to viral infection. PEDV PLP2 has been proposed to play a key role in antagonizing innate immunity (Xing et al., 2013). To investigate the effect of PLP1 on PEDV replication, Vero E6 cells were transfected with pCMV-PLP1-HA or empty pCMV-HA vector. At 24 hpt, cells were infected with PEDV at a MOI of 0.1. At 24 hpi, PEDV RNA levels were assessed by quantitative RT-PCR. As shown in Fig. 1 A, PLP1 overexpression increased the levels of PEDV RNA compared with the empty vector control. This result indicated that PLP1 may play an important role in PEDV infection.
Fig. 1.
Overexpression of PLP1 promotes PEDV replication and inhibits IFN-β promoter activation induced by TNF-α.
(A) Vero E6 cells were transfected with pCMV-PLP1-HA or empty pCMV-HA vector. At 24 hpt, cells were infected with PEDV at MOI of 0.1. At 24 hpi, viral RNA levels were determined. (B) HEK293 T cells were seeded in a 24-well plate and cotransfected with IFN-Luc, TK-Luc, and pCMV-PLP1-HA or empty pCMV-HA vector. At 24 hpt, the cells were stimulated with 10 ng/mL TNF-α for 6 h and cell lysates were prepared for luciferase analysis. (C) HEK293 T were stimulated with TNF-α (10 ng/mL), then transfected with pCMV-PLP1-HA or empty pCMV-HA vector. Levels of IFN mRNA were determined by quantitative RT-PCR at 12, 24, 36 and 48 hpi.
To investigate the effect of PLP1 on type I IFN promoter activation, HEK293 T cells were cotransfected with IFN-Luc and pCMV-PLP1-HA or empty pCMV-HA vector. TK-Luc was cotransfected as an internal control. At 24 hpt, the cells were stimulated with 10 ng/mL TNF-α for 6 h and prepared for luciferase analysis. As shown in Fig. 1B, TNF-α significantly activated the IFN promoter and PLP1 overexpression significantly inhibited TNF-α induced IFN-Luc activity.
To investigate the effects of PLP1 on type I IFN production, HEK293 T cells were stimulated with TNF-α (10 ng/mL), then cells were cotransfected with pCMV-PLP1-HA or empty pCMV-HA vector. At 24, 36 and 48 hpt, cells were harvested for total RNA extraction and IFN-β mRNA abundance was assessed by quantitative RT-PCR. As shown in Fig. 1C, TNF-α stimulated IFN-β mRNA transcription, whereas overexpression of PLP1 significantly inhibited TNF-α-induced transcription of IFN-β mRNA. These results indicated that PLP1 acted as a negative regulator of IFN-β, thus augmenting PEDV infection.
3.2. Identification of cellular interaction partners of PLP1
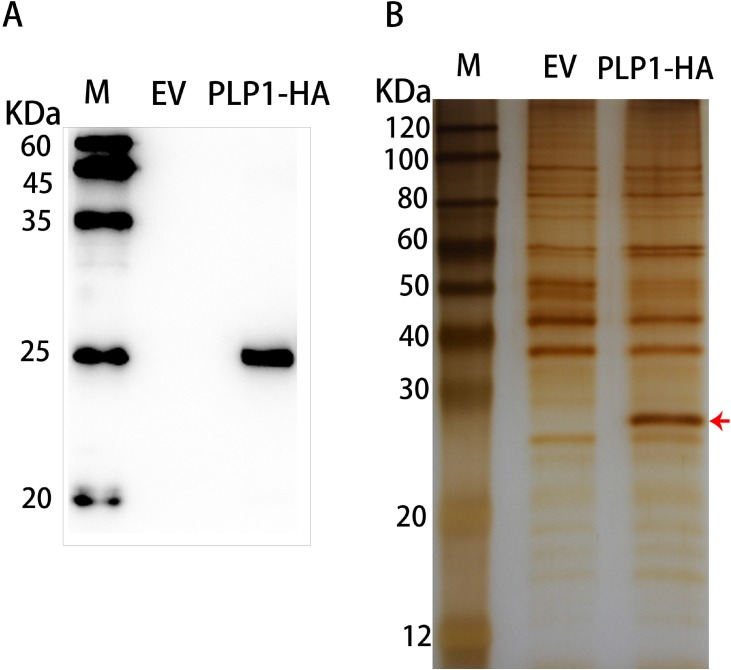
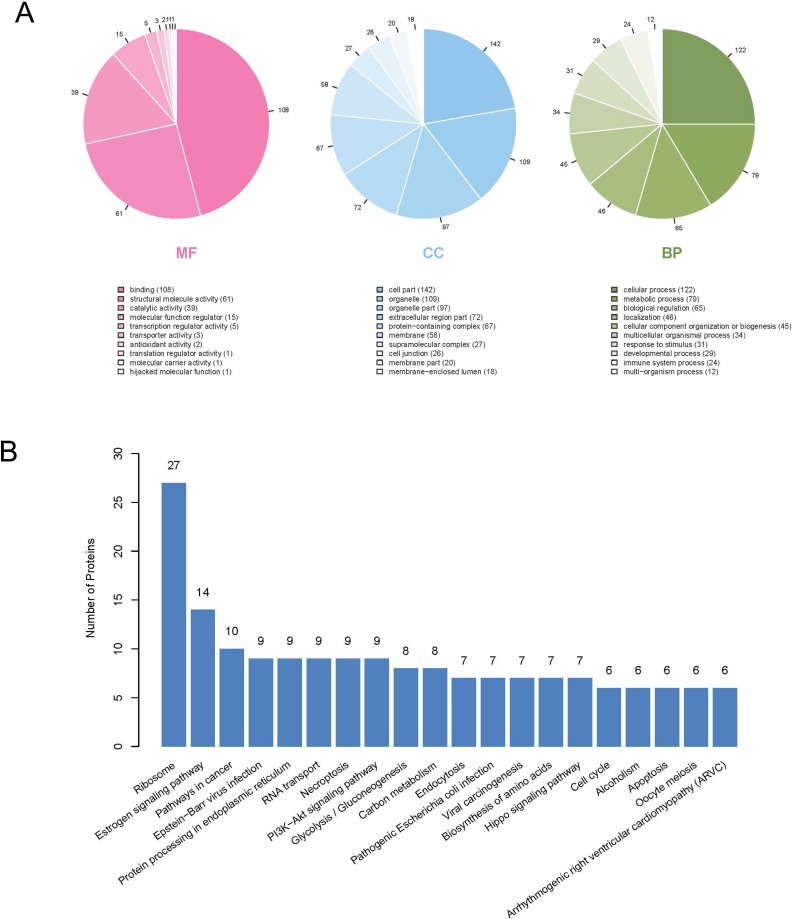
To further investigate the mechanism through which PEDV PLP1 inhibited expression of IFN-β, we identified cellular interaction partners of PLP1 using immunoprecipitation and mass spectrometry. HEK293 T cells were transfected with pCMV-PLP1-HA or pCMV-HA empty vector. At 24 hpt, the cells were lysed and proteins were immunoprecipitated using the Pierce® HA Tag IP/Co-IP Kit (Thermo Scientific, China). The eluent was collected and analyzed by SDS-PAGE. Silver staining showed multiple and distinct protein bands immunoprecipitated in PLP1 overexpressing cells but not in cells transfected with empty vector. We chose the band with the largest expression difference for mass spectrometry analysis (Fig. 2 B). Gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis was performed to analyze the functions of target proteins (Fig. 3 ). The identified host proteins were classified based on molecular function (MF), cellular component (CC) and biological process (BC) (Fig. 3A). The proteins were involved in cellular processes, metabolic processes and biological regulation. The majority of proteins pulled down with PLP1 are listed in Table 2 . Analysis of the top 20 pathways showed that the majority of proteins played a role in organismal systems, human diseases and viral infection (Fig. 3B). Identification of cellular interaction partners showed that PCBPs were involved in the immune response against PEDV infection.
Fig. 2.
Western blotting and silver staining of immunoprecipitated PLP1 samples.
(A and B) HEK293 T cells were transfected with pCMV-PLP1-HA or empty pCMV-HA vector. At 24 hpt, the cells were lysed and the proteins were immunoprecipitated using the Pierce® HA Tag IP/Co-IP Kit. The immunoprecipitated proteins were analyzed by western blotting (A) and by silver staining (B).
Fig. 3.
GO and KEGG analysis of the PLP1 interactome.
The LC–MS/MS results were analyzed by(A) GO and (B) KEGG pathway analysis.
Table 2.
Identified proteins from immunoprecipitation reaction of PLP1 transfected cell lysates.
| protein ID | Coverage (%) | Name |
|---|---|---|
| M0R210 | 12 | 40S ribosomal protein S16 |
| F8W6I7 | 11 | Heterogeneous nuclear ribonucleoprotein A1 |
| Q15365 | 11 | Poly(rC)-binding protein 1 |
| P61981 | 11 | 14-3-3 protein gamma |
| P62937 | 11 | Peptidyl-prolyl cis-trans isomerase A |
| Q5T123 | 11 | SH3 domain-binding glutamic acid-rich-like protein 3 |
| H3BMH2 | 11 | Ras-related protein (Fragment) |
| B8ZZK4 | 11 | 60S ribosomal protein L31 |
| F8WBR5 | 11 | Calmodulin-2 |
| C9J0E4 | 11 | Cystatin-A |
| H3BRU6 | 10 | Poly(rC)-binding protein 2 (Fragment) |
| P06733 | 10 | Alpha-enolase |
| G3V555 | 10 | Heterogeneous nuclear ribonucleoproteins C1/C2 (Fragment) |
3.3. Validation of proteins interacting with PLP1
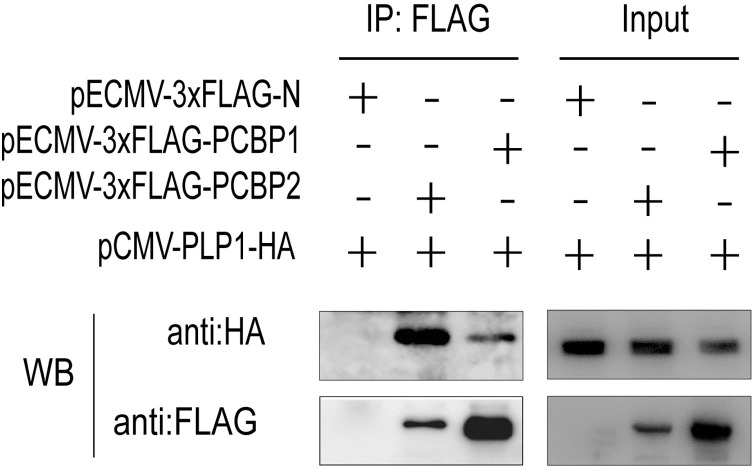
On the basis of the mass spectrometry results, we initially identified two proteins (PCBP1 and PCBP2) related to immune responses for further study. The genes encoding these two proteins were amplified and cloned into pECMV-3×FLAG, and HEK293 T cells were cotransfected with pCMV-PLP1-HA and pECMV-3×FLAG-PCBP1 or pECMV-3×FLAG-PCBP2. At 24 hpt, cells were lysed and immunoprecipitated with mouse anti-FLAG antibody. The results showed that PCBP1 and PCBP2 were both pulled down, and that PCBP2 interacted most strongly with PLP1 (Fig. 4 ). Therefore, PCBP2 was selected for further studies.
Fig. 4.
Identification of proteins that interact with PLP1.
HEK293 T cells were seeded in a six-well plate and cotransfected with pCMV-PLP1-HA, pECMV-3×FLAG-PCBP1, pECMV-3×FLAG-PCBP2 or empty vector. At 24 hpt, cells were lysed and immunoprecipitated with mouse anti-FLAG antibody. Immunoprecipitated samples were analyzed by western blotting.
3.4. Characterization of the PCBP2-PLP1 interaction
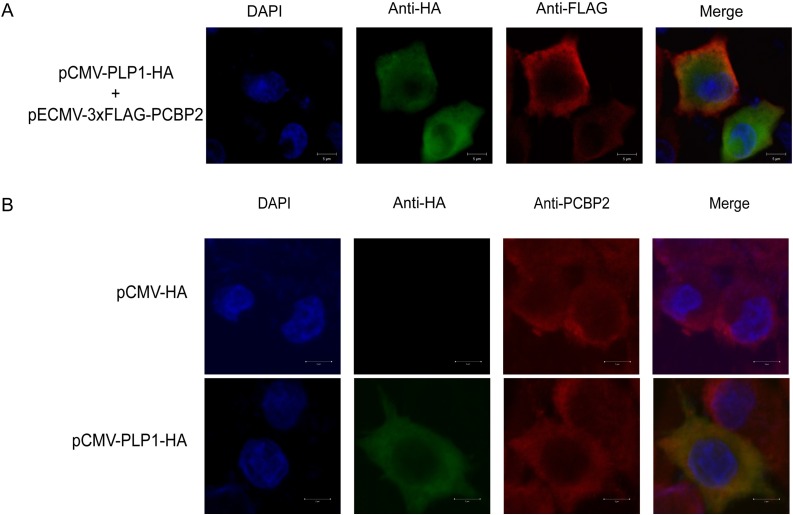
To examine the co-localization of PCBP2 with PLP1, Vero E6 cells were cotransfected with pCMV-PLP1-HA and pECMV-3×FLAG-PCBP2. At 24 hpt, the cells were stained with anti-HA and anti-FLAG antibodies and analyzed by confocal microscopy. Overexpressed PLP1 and PCBP2 were mainly co-located in the cytoplasm (Fig. 5 A).
Fig. 5.
Co-localization of PCBP2 with PLP1.
(A) Vero E6 cells were cotransfected with pCMV-PLP1-HA and pECMV-3×FLAG-PCBP2 or empty pCMV-HA vector. The cells were assessed using an indirect immunofluorescence assay with anti-HA and anti-FLAG antibodies. (B) Co-localization of PLP1 and endogenous PCBP2 in Vero E6 cells. The cells were transfected with pCMV-PLPL-HA or empty pCMV-HA vector and then subjected to the IFA with anti-HA and anti-PCBP2 antibodies.
We further analyzed the co-localization of PLP1 and endogenous PCBP2 in Vero E6 cells. The cells were transfected with pCMV-PLP1-HA or pCMV-HA, then analyzed by confocal microscopy following staining with anti-HA and anti-PCBP2 antibodies. As shown in Fig. 5B, PLP1 was mainly co-localized with endogenous PCBP2 in the cytoplasm.
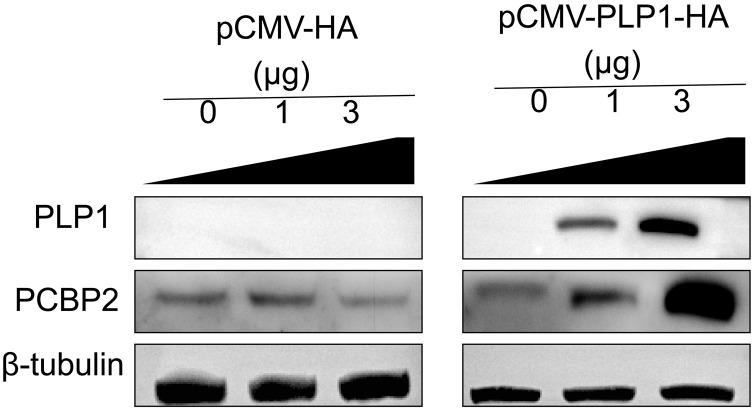
To study whether PLP1 promoted PCBP2 upregulation, Vero E6 cells were transfected with different concentrations of pCMV-PLP1-HA or empty pCMV-HA vector and western blotting was performed using anti-HA, anti-PCBP2, and anti-t3.4ulin antibodies. PLP1 overexpression enhanced endogenous accumulation of PCBP2 protein in a dose-dependent manner (Fig. 6 ).
Fig. 6.
PLP1 up-regulates PCBP2 expression.
Vero 6 cells were transfected with different concentrations of pCMV-PLP1-HA or empty pCMV-HA vector and western blotting was performed using anti-HA, anti-PCBP2 and anti-tubulin antibodies.
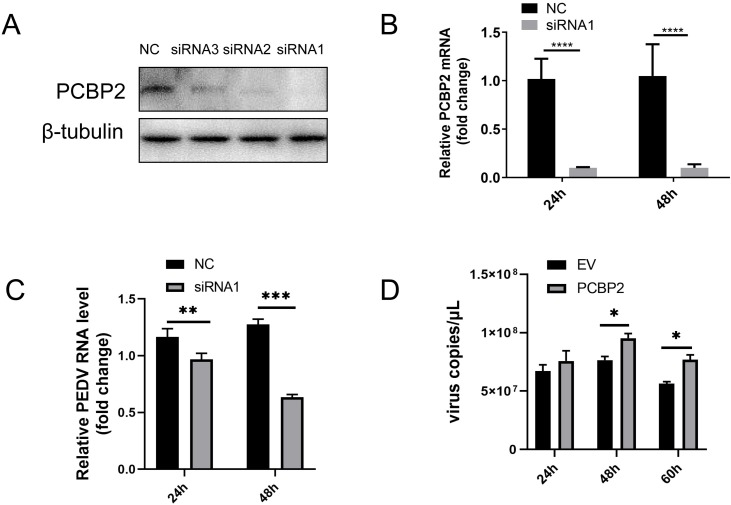
3.5. PCBP2 affects PEDV infection
To investigate the effect of endogenous PCBP2 on viral replication, expression of PCBP2 was silenced using siRNAs. Three siRNAs (1–3) were designed. Following transfection of IPEC-J2 cells, the effect of knockdown was assessed by western blotting. Levels of PCBP2 protein were significantly decreased in all siRNA-treated groups compared with the control group, and siRNA1 showed the most efficient PCBP2 silencing (Fig. 7 A). Next, IPEC-J2 cells were treated with siRNA1 or a negative control (NC) siRNA for 24 h and then infected with PEDV at a MOI of 0.1. Levels of PCBP2 mRNA and PEDV RNA were determined at 24 and 48 hpi by quantitative RT-PCR. As shown in Fig. 7B and 7C, PCBP2 mRNA levels in siRNA-treated IPEC-J2 cells were significantly reduced compared with NC-siRNA-treated cells, and PEDV replication was significantly suppressed (Fig. 7B and C).
Fig. 7.
PCBP2 affects PEDV infection.
IPEC-J2 cells were transfected with three different PCBP2-specific siRNAs. After 24 h, PCBP2 protein expression was assessed by western blotting with anti-PCBP2 and anti-tubulin antibodies. (B to C) IPEC-J2 cells were transfected with siRNA1, then 24 h later infected with PEDV for 24 h and 48 h. PCBP2 mRNA levels and PEDV RNA levels were determined by quantitative RT-PCR. (D) IPEC-J2 cells were transfected with pECMV-3×FLAG-PCBP2 or empty vector. At 24 hpt, the cells were infected with PEDV at a MOI of 0.1. PEDV viral loads were determined at 24, 48 and 60 hpi by quantitative RT-PCR.
To further investigate whether overexpression of PCBP2 would affect PEDV replication, IPEC-J2 cells were transfected with pECMV-3×FLAG-PCBP2 or empty vector. At 24 hpt, the cells were infected with PEDV at a MOI of 0.1 and PEDV viral loads were determined at 24, 48 and 60 hpi by quantitative RT-PCR. We found that PCBP2 overexpression enhanced PEDV replication (Fig. 7D). These results indicated that PLP1 acted as a negative regulator of IFN-β, and PLP1 functions to augment PEDV infection by interacting with PCBP2.
4. Discussion
Nsp3 is the largest protein encoded in the coronavirus genome and comprises two subdomains (PLP1 and PLP2), which are mainly responsible for cleavage of Nsp1/Nsp2 and Nsp2/Nsp3, respectively (Barretto et al., 2005; Harcourt et al., 2004). Not all coronaviruses have a PLP1 domain, while the PLP2 domain is conserved in all coronaviruses. PLP proteins encoded by various viruses have been shown to inhibit innate immunity (Zheng et al., 2008). Severe acute respiratory syndrome coronavirus PLP inhibits IRF3 activation by blocking ubiquitination of RIG-I, TNF-receptor associated factor 3, and stimulator of interferon genes (A Lindner et al., 2007). Transmissible gastroenteritis virus PLP1 and PRRSV PLP2 rely on deubiquitination activity to antagonize IFN production (Chen et al., 2007; Hu et al., 2017; Sun et al., 2012b). Similar to other coronaviruses, PEDV can also inhibit the production of type 1 IFNs (Zheng et al., 2008). PEDV PLP2 has been shown to have deubiquitination activity and is an IFN antagonist, but PLP1 has no deubiquitination activity (Xing et al., 2013). Our results showed that PLP1 is also an IFN antagonist. Overexpression of PLP1 enhanced PEDV infection (Fig. 1A). Using a dual fluorescent reporter, we found that PLP1 could inhibit TNF-α-induced production of IFN-β. In HEK293 T cells transfected with PLP1 expression vectors, induction of IFN-β by TNF-α was inhibited (Fig. 1C). These results suggested that PLP1 could enhance PEDV infection by inhibiting IFN-β production. Studies have shown that the PLP1 of PEDV CV777 strain has no IFN antagonism (Zheng et al., 2008). There are 10 amino acid substitutions distinguishing the PLP1s of PEDV GDgh and CV777 strains (data not shown). We suspect that these substitutions may lead to differences in PLP1 function. Future studies should investigate the effects of amino acid mutations at these sites on PLP1 function.
The innate immune response is an important line of defense, and type 1 IFNs are essential for host defense against viral infection. The double-stranded RNA (dsRNA) molecules produced by viral RNA replication viral are PAMPs and can be recognized by pattern recognition receptors (PRRs) (Matzinger, 2002). TLRs and RIG-I-like receptors are two crucial PRRs that recognize pathogens and stimulate downstream signaling to activate immune responses (Huang et al., 2009; Kawai and Akira, 2007). MAVS is a downstream adaptor protein of RIG-1. RIG-1 mediates the activation of NF-κB and IRF3 by interacting with the caspase-recruiting-like domain of MAVS to inhibit the production of type I IFNs (Biacchesi et al., 2009; Kawai et al., 2005; Meylan et al., 2005; Seth et al., 2005). PCBP2 is a negative regulator of MAVS whose expression is induced following viral infection and results in MAVS degradation (You et al., 2009). We found that overexpression of PCBP2 promoted PEDV replication (Fig. 7D). PEDV replication could be inhibited by silencing PCBP2 expression (Fig. 7C). PCBP2 has been reported to play a role in a variety of viral infections (Dinh et al., 2011; Luo et al., 2014; Wang et al., 2012). We found that PLP1 interacted with PCBP2 (Fig. 4, Fig. 5), and that overexpression of PLP1 induces the expression of PCBP2 (Fig. 6). These results indicated that PLP plays an important role in the antagonism of IFN responses. Therefore, the study of PLP proteins is important for understanding viral escape from host immune responses. Compared with PLP2, the function of PLP1 has received less attention. The PLP2s of many coronaviruses have deubiquitination activity, and can interfere with host antiviral responses (Barretto et al., 2005). We found that PEDV PLP1 also has IFN antagonism and contributes to viral replication. Our results indicate that PLP1 interacted with PCBP2, thereby helping PEDV escape from host immune responses. Our findings reveal a new mechanism evolved by PEDV to circumvent the host antiviral response, and contribute to our understanding of the role of coronavirus PLP1 in the viral life cycle.
Acknowledgments
This work was supported by the National Key Technologies R&D Program (2015BAD12B02-5), Frontiers and Key Technological Innovations (2015B020230004) and the Guangdong Innovation Team of Modern Agricultural Industry Technology System (2016LM2150), the Key and Cultivation Discipline of Xinyang Agriculture and Forestry University (ZDXK201702) and University-level Science and Technology Innovation Team (CXTD—201801), Science and Technology Innovation Team Construction Project of 2019 Xinyang Agriculture and Forestry University (Pathogenesis and comprehensive prevention and control of major poultry diseases), and Youth Fund Project of Xinyang Agriculture and Forestry University (2019LG012).
Contributor Information
Li Huang, Email: cnvet19@126.com.
Changxu Song, Email: cxsong2004@163.com, cxsong@scau.edu.cn.
References
- Barretto N., Jukneliene D., Ratia K., Chen Z., Mesecar A.D., Baker S.C. The papain-like protease of severe acute respiratory syndrome coronavirus has deubiquitinating activity. J. Virol. 2005;79:15189–15198. doi: 10.1128/JVI.79.24.15189-15198.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beura L.K., Dinh P.X., Osorio F.A., Pattnaik A.K. Cellular poly(c) binding proteins 1 and 2 interact with porcine reproductive and respiratory syndrome virus nonstructural protein 1beta and support viral replication. J. Virol. 2011;85:12939–12949. doi: 10.1128/JVI.05177-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biacchesi S., LeBerre M., Lamoureux A., Louise Y., Lauret E., Boudinot P., Bremont M. Mitochondrial antiviral signaling protein plays a major role in induction of the fish innate immune response against RNA and DNA viruses. J. Virol. 2009;83:7815–7827. doi: 10.1128/JVI.00404-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Wang Y., Ratia K., Mesecar A.D., Wilkinson K.D., Baker S.C. Proteolytic processing and deubiquitinating activity of papain-like proteases of human coronavirus NL63. J. Virol. 2007;81:6007–6018. doi: 10.1128/JVI.02747-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaraj G., S Wang, N Chen, Z Chen, Z Tseng, M Barretto, N Lin, R Peters, C Tseng, C.-T Evans, S Li., K. Vol. 282. 2007. pp. 32208–32221. (Regulation of IRF-3-Dependent Innate Immunity by the Papain-Like Protease Domain of the Severe Acute Respiratory Syndrome Coronavirus). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Z., Fang L., Jing H., Zeng S., Wang D., Liu L., Zhang H., Luo R., Chen H., Xiao S. Porcine epidemic diarrhea virus nucleocapsid protein antagonizes beta interferon production by sequestering the interaction between IRF3 and TBK1. J. Virol. 2014;88:8936–8945. doi: 10.1128/JVI.00700-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinh P.X., Beura L.K., Panda D., Das A., Pattnaik A.K. Antagonistic effects of cellular poly(C) binding proteins on vesicular stomatitis virus gene expression. J. Virol. 2011;85:9459–9471. doi: 10.1128/JVI.05179-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dragan A.I., Hargreaves V.V., Makeyeva E.N., Privalov P.L. Mechanisms of activation of interferon regulator factor 3: the role of C-terminal domain phosphorylation in IRF-3 dimerization and DNA binding. Nucleic Acids Res. 2007;35:3525–3534. doi: 10.1093/nar/gkm142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harcourt B.H., Jukneliene D., Kanjanahaluethai A., Bechill J., Severson K.M., Smith C.M., Rota P.A., Baker S.C. Identification of severe acute respiratory syndrome coronavirus replicase products and characterization of papain-like protease activity. J. Virol. 2004;78:13600–13612. doi: 10.1128/JVI.78.24.13600-13612.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X., Tian J., Kang H., Guo D., Liu J., Liu D., Jiang Q., Li Z., Qu J., Qu L. Transmissible gastroenteritis virus papain-like protease 1 antagonizes production of interferon-beta through its deubiquitinase activity. Biomed. Res. Int. 2017 doi: 10.1155/2017/7089091. 2017, 7089091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y.W., Zhang J., Wu X.M., Cao L., Nie P., Chang M.X. TANK-Binding Kinase 1 (TBK1) Isoforms Negatively Regulate Type I Interferon Induction by Inhibiting TBK1-IRF3 Interaction and IRF3 Phosphorylation. Front. Immunol. 2018;9 doi: 10.3389/fimmu.2018.00084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y., Walstrom A., Zhang L., Zhao Y., Cui M., Ye L., Zheng J.C. Type I interferons and interferon regulatory factors regulate TNF-Related apoptosis-inducing ligand (TRAIL) in HIV-1-Infected macrophages. PLoS One. 2009;4:e5397. doi: 10.1371/journal.pone.0005397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai T., Akira S. Antiviral signaling through pattern recognition receptors. J. Biochem. 2007;141:137–145. doi: 10.1093/jb/mvm032. [DOI] [PubMed] [Google Scholar]
- Kawai T., Takahashi K., Sato S., Coban C., Kumar H., Kato H., Ishii K.J., Takeuchi O., Akira S. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat. Immunol. 2005;6:981–988. doi: 10.1038/ni1243. [DOI] [PubMed] [Google Scholar]
- Kocherhans R., Bridgen A., Ackermann M., Tobler K. Vol. 23. 2001. pp. 137–144. (Completion of the Porcine Epidemic Diarrhoea Coronavirus (PEDV) Genome Sequence). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei J., Kusov Y., Hilgenfeld R. Nsp3 of coronaviruses: structures and functions of a large multi-domain protein. Antiviral Res. 2018;149:58–74. doi: 10.1016/j.antiviral.2017.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindner H., Lytvyn V., Qi H., Lachance P., Ziomek E., Ménard R. Vol. 466. 2007. pp. 8–14. (Selectivity in ISG15 and Ubiquitin Recognition by the SARS Coronavirus Papain-Like Protease). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Z., Dong X., Li Y., Zhang Q., Kim C., Song Y., Kang L., Liu Y., Wu K., Wu J. PolyC-binding protein 1 interacts with 5’-untranslated region of enterovirus 71 RNA in membrane-associated complex to facilitate viral replication. PLoS One. 2014;9 doi: 10.1371/journal.pone.0087491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makeyev A.V., Liebhaber S.A. The poly(C)-binding proteins: a multiplicity of functions and a search for mechanisms. RNA (New York, N.Y.) 2002;8:265–278. doi: 10.1017/s1355838202024627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzinger P. The danger model: a renewed sense of self. Science. 2002;296:301. doi: 10.1126/science.1071059. [DOI] [PubMed] [Google Scholar]
- Meylan E., Curran J., Hofmann K., Moradpour D., Binder M., Bartenschlager R., Tschopp J. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature. 2005;437:1167–1172. doi: 10.1038/nature04193. [DOI] [PubMed] [Google Scholar]
- O’Neill L., Bowie A. Vol. 20. 2010. pp. R328–333. (Sensing and Signaling in Antiviral Innate Immunity). [DOI] [PubMed] [Google Scholar]
- Pensaert M., de Bouck P. Vol. 58. 1978. pp. 243–247. (A New Coronavirus-Like Particle Associated With Diarrhea in Swine). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seth R.B., Sun L., Ea C.K., Chen Z.J. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell. 2005;122:669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- Sun R.-Q., Cai R.-J., Chen Y.-Q., Liang P.-S., Chen D.-K., Song C.-X. Vol. 18. 2012. pp. 161–163. (Outbreak of Porcine Epidemic Diarrhea in Suckling Piglets, China). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z., Li Y., Ransburgh R., Snijder E.J., Fang Y. Nonstructural protein 2 of porcine reproductive and respiratory syndrome virus inhibits the antiviral function of interferon-stimulated gene 15. J. Virol. 2012;86:3839–3850. doi: 10.1128/JVI.06466-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., He Q., Gao Y., Guo X., Ge X., Zhou L., Yang H. Interaction of cellular poly(C)-binding protein 2 with nonstructural protein 1beta is beneficial to Chinese highly pathogenic porcine reproductive and respiratory syndrome virus replication. Virus Res. 2012;169:222–230. doi: 10.1016/j.virusres.2012.08.002. [DOI] [PubMed] [Google Scholar]
- Wang D., Fang L., Shi Y., Zhang H., Gao L., Peng G., Chen H., Li K., Xiao S., Perlman S. Porcine epidemic diarrhea virus 3C-LiK.e protease regulates its interferon antagonism by cleaving NEMO. J. Virol. 2016;90:2090–2101. doi: 10.1128/JVI.02514-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing Y., Chen J., Tu J., Zhang B., Chen X., Shi H., Baker S.C., Feng L., Chen Z. The papain-like protease of porcine epidemic diarrhea virus negatively regulates type I interferon pathway by acting as a viral deubiquitinase. J. Gen. Virol. 2013;94:1554–1567. doi: 10.1099/vir.0.051169-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You F., Sun H., Zhou X., Sun W., Liang S., Zhai Z., Jiang Z. PCBP2 mediates degradation of the adaptor MAVS via the HECT ubiquitin ligase AIP4. Nat. Immunol. 2009;10:1300–1308. doi: 10.1038/ni.1815. [DOI] [PubMed] [Google Scholar]
- Yu L., Dong J., Wang Y., Zhang P., Liu Y., Zhang L., Liang P., Wang L., Song C. Porcine epidemic diarrhea virus nsp4 induces pro-inflammatory cytokine and chemokine expression inhibiting viral replication in vitro. Arch. Virol. 2019;164:1147–1157. doi: 10.1007/s00705-019-04176-2. [DOI] [PubMed] [Google Scholar]
- Zhang Q., Shi K., Yoo D. Suppression of type I interferon production by porcine epidemic diarrhea virus and degradation of CREB-binding protein by nsp1. Virology. 2016;489:252–268. doi: 10.1016/j.virol.2015.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng D., Chen G., Guo B., Chen G., Tang H. PLP2, a potent deubiquitinase from murine hepatitis virus, strongly inhibits cellular type I interferon production. Cell Res. 2008;18:1105–1113. doi: 10.1038/cr.2008.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H., Perlman S. Vol. 81. 2007. pp. 568–574. (Mouse Hepatitis Virus Does Not Induce Beta Interferon Synthesis and Does Not Inhibit Its Induction by Double-Stranded RNA). [DOI] [PMC free article] [PubMed] [Google Scholar]