Abstract
Genomic data has opened new possibilities to understand how organisms change over time, and could enable the discovery of previously undescribed species. Although taxonomy used to be based on phenotypes, molecular data has frequently revealed that morphological traits are insufficient to describe biodiversity. Genomics holds the promise of revealing even more genetic discontinuities, but the parameters on how to describe species from genomic data remain unclear. Fungi have been a successful case in which the use of molecular markers has uncovered the existence of genetic boundaries where no crosses are possible. In this minireview, we highlight recent advances, propose a set of standards to use genomic sequences to uncover species boundaries, point out potential pitfalls, and present possible future research directions.
Keywords: Speciation, gene flow, fungi
Introduction
Species are defined as genetic clusters of organisms that are isolated from other clusters (Coyne and Orr, 2004; Nosil, 2012). This isolation can be assessed with phenotypic and genetic data alike. In spite of the simplicity of this idea, recognizing species is anything but simple (Figure 1). Gene exchange, phenotypic plasticity, and the infeasibility of culturing (let alone performing experiments) might impede species identification. Nonetheless identifying species boundaries is crucial to understanding how and why genetic variation is portioned in nature.
FIGURE 1. Simplified representation of the process of speciation continuum and the different signatures of speciation.
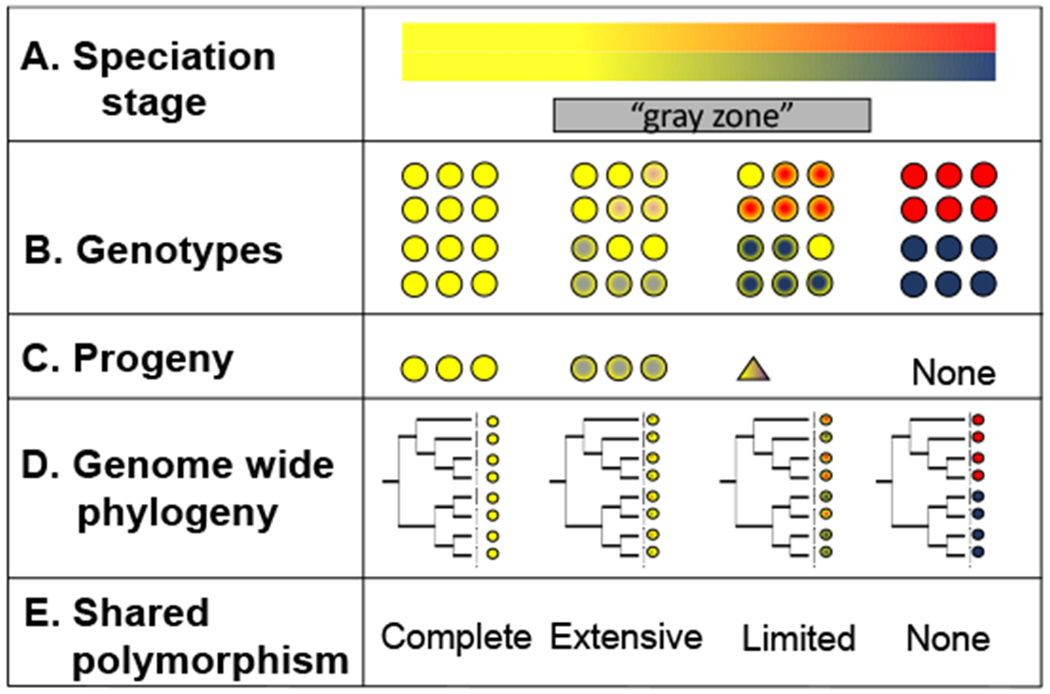
A. Speciation stage: genetic differences accrue in the nascent species to the extent that they become fully differentiated. Detecting speciation can be difficult in cases where divergence is recent (i.e., the gray zone). B. Genotypes: genetic differentiation will manifest in differences in the allele frequencies of the resulting populations (middle panel). Genetic differentiation might also lead to phenotypic differences but that is not always the case. C. Progeny: as divergence accumulates, the resulting species are less likely to interbreed and produce fit progeny to the extent that interbreeding will cease completely (bottom panel). D. Genome wide phylogeny: before speciation occurs (left), populations will exchange genes and polymorphic sites across the genome will be present in both species. As divergence accrues, the likelihood of reciprocal monophyly across the genome increases, and the likelihood of shared polymorphism decreases. Speciation is complete when genomes become reciprocally monophyletic, the magnitude of shared polymorphism is negligible, and reproductive isolation precludes the possibility of admixture. E. Level of shared polymorphism.
At least ten distinct conceptual frameworks and associated metrics for species delimitation have been proposed (listed and reviewed in(Coyne and Orr, 2004; De Queiroz, 2007; Taylor et al., 2000)). Arguably, the most influential concept for recognizing species boundaries is the biological species concept (BSC; (Huxley, 1943; Dobzhansky, 1937)). Under this framework, groups of individuals that are reproductively isolated from each other are considered species. This approach constitutes the basis of diversity studies across domains of life (Bobay and Ochman, 2017). Nevertheless, in spite of its wide application, the BSC is impractical to apply for many types of organisms (De Queiroz, 2007; Sites and Marshall, 2003; Sokal and Crovello, 1970). A nontrivial proportion of species are either asexual or cannot be crossed in an experimental setting and thus are not amenable to the BSC. In other cases, there is evidence that sexual reproduction takes place in nature, but it is infrequent enough or is triggered by such rare conditions that the sexual stages of many species remain unknown. This is a particularly notorious issue for fungi (Cai et al., 2011; Crespo and Lumbsch 2010, Dettman et al., 2003a; Giraud et al., 2010; Grube and Kroken 2000, Harrington and Rizzo 1999, Taylor et al., 2000). Clearly, an alternative that is not based on experimental crosses would be advantageous for organisms where the BSC cannot be implemented.
Genetic data also reveal the signature of speciation (Bobay and Ochman, 2017; Taylor, 2006; Taylor et al., 2000). Several species concepts have been proposed based on population cohesion. All these concepts (henceforth referred to as ‘discontinuity concepts’) and the BSC have a common ground. According to the BSC, speciation has occurred in instances where reproductive isolation (RI) exists. According to the discontinuity concepts, speciation has occurred in instances where there are major genetic gaps between groups (i.e., discontinuities). If RI exists and speciation has proceeded, then there should be genetic differentiation between the putative species which should leave a signature of genetic discontinuity across the whole genome (Avise and Wollenberg, 1997; Bobay and Ochman, 2017; Coyne and Orr, 2004; Sobel et al., 2010). To date, there are few proposals on how to implement species concepts using genome-wide genetic data, and this is particularly evident for fungi. We aim to bridge this gap. In this piece, we review the main approaches to delimiting species, present an overview of the precedents on species boundaries delimitation in fungi to date, and propose four criteria to identify species boundaries using genome-wide data in fungi. We argue that a unified approach to study species boundaries is crucial for our understanding of fungal diversity but also has practical applications for plant pathology and medical mycology.
Current approaches to detect species boundaries in fungi
All discontinuity concepts are based on the detection of genetic clusters that are sufficiently differentiated from one another. Of all the discontinuity concepts, the phylogenetic species concept (PSC) is by far the most widely applied in fungi (Taylor et al., 2000). According to the PSC, speciation has occurred in instances where there are major genetic discontinuities. Under the PSC, species are diagnosed as a cluster of individuals that are sufficiently differentiated from other clusters as revealed by DNA sequences. Gene genealogies are frequently used to determine the clusters used in the PSC, although other approaches with a similar premise have been proposed (reviewed in (Coyne and Orr, 2004; Nixon and Wheeler, 1990; Renard et al., 2003; Wheeler and Platnick, 1989)). The PSC was originally proposed in the 1980s (Baum and Donoghue, 1995; Donoghue, 1985) but did not gain momentum in fungi until multilocus sequence typing was coopted from bacterial studies (Taylor and Fisher, 2003). The PSC in fungi has two broadly-defined forms, the strict genealogic concordance (SGC) and the coalescent-based species delimitation (CBD) approaches (revised in (Taylor et al., 2000)). These approaches are not mutually exclusive as they both use phylogenetic trees but their scope is different. We summarize each of these groups as follows.
SGC approaches assess the extent of genetic concordance across loci in a semi-quantitative framework. Two seminal pieces, Dettman et al. (Dettman et al., 2003a, 2003b) and Liti et al. (Liti et al., 2006), studied what tree metrics might signal discontinuities in genetic variation that likely resulted from reproductive isolation and could thus differentiate between structured populations and true species. First, if all gene genealogies from unlinked loci in the genome show genealogies that are congruent with each other, then speciation might have occurred (i.e., genealogical concordance, or ‘concordance rule’). Second, if a clade or group of putative species forms reciprocally monophyletic groups with Bayesian support of at least 90% and of bootstrap above 70%, then they are likely to be reproductively isolated (i.e., ‘support rule’—based on (Hillis and Bull, 1993)). The SGC approach as proposed by Dettman et al. (Dettman et al., 2003a) is one of the unified approaches for species delimitation in fungi.
The mycology community has extensively used the two rules of the SGC to detect the signature of speciation, especially to identify cryptic species. The focus on this instance of divergence is due to the fact that this is the stage at which species boundaries are the most challenging to detect (reviewed in (Bickford et al., 2007; Cai et al., 2011; Restrepo et al., 2014; Roux et al., 2016)). At least 55 clades (64 studies) used SGC criteria to study potential cryptic speciation in fungi. The application of these two rules has led to the report of over 200 previously-unknown cryptic species, leading to an increase of almost three times in fungal diversity in the studied groups (from 116 to 343 species). Even though this list is certainly not complete, and should be taken as a sample from the complete literature, a pattern emerges from this body of work; most studies used either the support rule (described above; 24 out of 55) or both the support and the concordance rule (30/55; Figure 2A). Only one study used the concordance rule exclusively (1/55)—a study on species boundaries in Histoplasma using whole genome sequences ((Sepúlveda et al., 2017); discussed below). The reason for why the support rule is more commonly used than the concordance rule might be technical. Once a topology is inferred, assessing the level of support of the branches is almost automatic, while assessing the level of concordance requires different approaches.
FIGURE 2. Summary of the approaches used to delimit species in fungi.
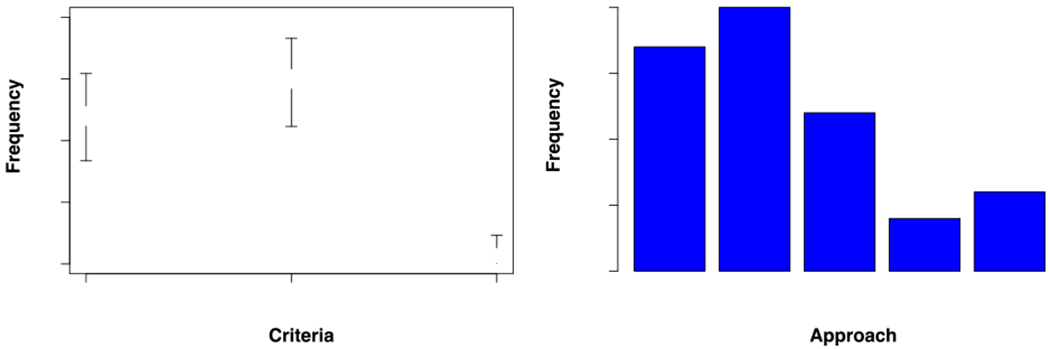
A. Proportion of studies that have used either one or both rules of the SGC to detect species boundaries in fungi. Error bars show the Bayesian 95% confidence intervals of the proportions. B. Proportion of studies that have used different CBD approaches to detect species boundaries in fungi. Please note that we reviewed 41 studies but the total for this panel is 60 as 12 studies used more than one approach.
The application of the SGC rules is not devoid of limitations. First, the ‘rules’ were proposed for a set of clades (Neurospora (Dettman et al., 2003b); Saccharomyces (Liti et al., 2006)), but it is unclear to what extent they can be generalized to other taxa. The evolution of reproductive isolation is particular to each clade and metrics to detect species boundaries might be required for each individual clade. A second caveat stems from the way that alignments are regularly analyzed. Notably, 31 out of the 5 5 studies that used the support rule used concatenated loci instead of individual genealogies. Gene concatenation has been shown to fail to recover true tree topologies when there are high levels of incomplete lineage sorting (as occurs in recently diverged species) and might thus obscure the relationships between sibling species (Edwards et al., 2007; Kubatko and Degnan, 2007; Liu and Edwards, 2009; Mendes and Hahn, 2018).
A second set of approaches, the coalescent-based delimitation (CBD) involves understanding how several species are related by modeling the genealogical history of individuals back to a common ancestor (i.e., coalescent theory; reviewed in (Fujita et al., 2012)). Similar to the SGC approaches, the original coalescent-based methods to detect species boundaries leverage the fact that speciation might be inferred in cases where multiple loci show coincident splits for a sample of individuals. CBD can be divided into at least three families of methods. The first family of methods estimates the likelihood of obtaining a given set of gene genealogies given a species tree. This in turn allows for the estimation of the probability of a species tree and the probability of a particular species delimitation given multilocus data (e.g., BP&P, (Yang, 2015a, 2015b; Yang and Rannala, 2010)). These approaches sequentially collapse nodes to identify the potential species until all descendants are assigned to one species using a prespecified topology. A second family explores the full space of possible species tree topologies which allows them to delimit species and infer the species tree simultaneously (e.g., STEM (Kubatko et al., 2009), SpedeSTEM (Ence and Carstens, 2011), DISSECT (Jones et al., 2015), STACEY (Jones, 2017), and recent iterations of BP&P (Yang, 2015a)). A third family uses the distinct branching patterns between divergence (using either Yule or Poisson models) and intraspecific diversification (using coalescent models) to distinguish between species and populations. The transition between these different branching patterns is inferred to predict species boundaries. The main assumption of these methods is that within species branching events will be substantially more frequent than between species. GMYC (Generalized Mixed Yule Coalescent; (Fontaneto et al., 2007; Pons et al., 2006)) and PTP (Poisson Tree Processes; (Zhang et al., 2013)) are examples of this framework.
Each of these method families has been used in studies of fungal species boundaries. In a similar way to the compilation we did for SGC criteria, we focused on obtaining a sample from cases of cryptic speciation. We compiled a sample of 41 cases which studied potential cryptic speciation using coalescent-based species delimitation in fungi. Three general patterns emerged. First, the most common approach to delimit species boundaries in fungi under the CBD approach is modeling branching patterns across species following a Yule model, i.e., GMYC (Figure 2B, Table S2). A second pattern suggests that studies that delimit cryptic species in fungi using the CBD often use methods from more than one family. Ten (out of 41) studies used a combination of methods from different families, while two used more than one method from the same family (GMYC and PTP in both cases, Table S3). The species boundaries inferred by different approaches are usually similar but not always identical (see below). This discordance usually leads to authors choosing the most conservative number of species (e.g., (Pino-Bodas et al., 2018; Singh et al., 2015)) and highlights the subjectivity of current species boundaries delimitation.
It is worth noting that the effectiveness of the multispecies coalescent to detect species boundaries has been called into consideration as it cannot disentangle strong population structure from incipient speciation, especially in cases where the distribution of potential species is allopatric (Leaché et al., 2019; Sukumaran and Knowles, 2017). This is an important distinction as not all populations become species, and most species show population structure usually associated with geography. The putative species identified using the multispecies coalescent (and the concordance-based PSC) must be considered as tentative hypotheses and must be confirmed through the collection of other sources of information.
Some general patterns also emerge for both the use of the SGC and CBD. First, a large proportion of cases used a set of common markers and most of them were nuclear loci (e.g., ITS, TEF1-α, β-TUB, RPB2; Figure 3A, B). This is certainly not a coincidence as these loci were originally chosen for being single copy genes, harboring polymorphism (suggestive of not being completely constrained by purifying selection), and being amenable to PCR-amplification using universal primers. Some of these markers (e.g., ITS) have come under scrutiny as they might underestimate the magnitude of differentiation (Gazis et al., 2011). Second, the mean number of loci studied to define species boundaries is 4.178 for SGC and 3.897 for CBD (with medians of 3 and 4, respectively). The ranges are broad and goes from one locus to fifteen unlinked loci (not including studies using whole genome sequencing, Figure 3C, D). This pattern can be concerning as individual loci (or a small group of loci) are more likely to be affected by selection, recombination, or even drift than large sets of unlinked loci (Dupuis et al., 2012; Edwards et al., 2007; Rokas et al., 2003). Simulations indicate that the accuracy of species delimitation is much higher when five or more loci are sampled for divergences larger than 2N generations (Avise and Ball, 1990; Baum and Shaw, 1995; Hudson and Coyne, 2002).
FIGURE 3. Characteristics of the SGC and CBD approaches in fungi.
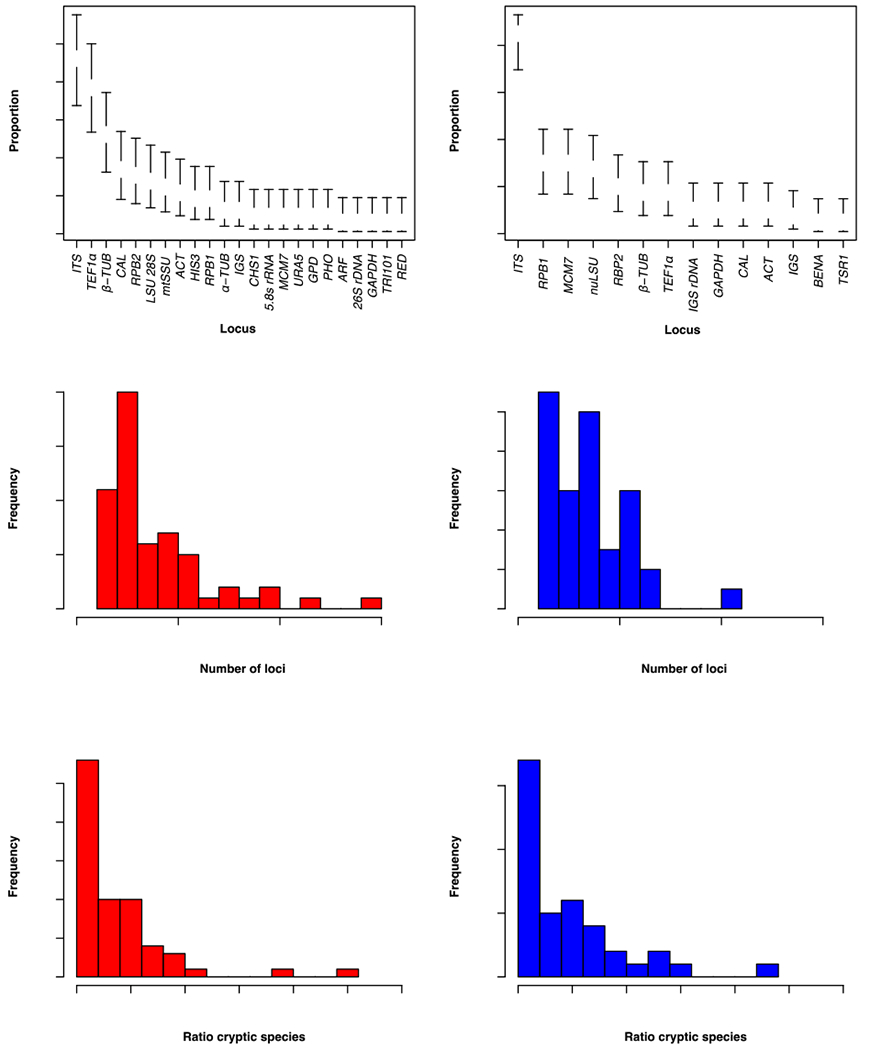
A. Histogram showing the most used loci in SGC studies in fungi. B. Histogram showing the most used loci in CBD studies in fungi. C. Distribution of the number of loci used in SGC studies in fungi. Error bars show the Bayesian 95% confidence intervals of the proportions. D. Distribution of the number of loci used in CBD studies in fungi. E. Ratio of resulting species to initially reported species for studies using the SGC criteria. When this index is higher than one, the study found cryptic species; when it is lower, the study collapsed species (i.e., found no support for previously names species). F. Ratio of resulting species to initially reported species for studies using the CBD criteria. Note that we excluded a study (Lucking et al. 2014), that found 126 potentially cryptic species from the histogram but not from the mean calculations.
The range of reported cryptic species is large when using either the SGC or the CBD delimitation (Figure 3E, F), and in some notable cases a single species has been proposed to be split into dozens of species (e.g., (Lucking et al., 2014)). In the majority of cases, the number of proposed cryptic species is much lower (Figure 3E, F) and is remarkably similar for both approaches (mean ratio of resulting species to previously known species = 2.96 and 3.00 for SGC and CBD, respectively). Notably, the vast majority of studies that contemplated the possibility of cryptic speciation found a signature of it (Figure 3E, F). A handful of studies found no support for previously proposed species boundaries (Boluda et al., 2018; Chen et al., 2011; Douanla-Meli et al., 2018; Liu et al., 2016; Pino-Bodas et al., 2018; Saag et al., 2014; Zhao et al., 2015, 2017). This apparent prevalence of cryptic speciation begs the question whether the relative abundance of unidentified species is real or whether it is driven by the ascertainment bias of the published studies (i.e., ‘the drawer effect’—negative results do not get published). Few systematic studies have been published on the matter. The most influential, perhaps, studied the possibility of cryptic speciation in Coccidioides and its closest relatives, Auxarthron zuffianum and Uncinocarpus reesii (Koufopanou et al., 2001) . Using common criteria (i.e., the concordance and support rules) and genetic markers, the study found that the three genera harbored cryptic species, lending support to the hypothesis that unidentified species are common. Similar studies across all fungal groups are sorely needed.
A set of studies used both the SGC and CBD and the results were largely concordant (Table 1). This is not surprising given that the two lines of eviden ce aim to detect similar patterns across the genome which in turn signal deep splits. Yet, there are some exceptions. Studies within the lichen-forming fungal genus Protoparmellia are representative of this situation (Singh et al., 2015). Using the concordance and support rules (SGC), Protoparmellia seemed to be composed by 25 species. CBD approaches did not fully agree: spedeSTEM suggested 19 species, while BP&P suggested 23. Not all species supported by BPP were supported by spedeSTEM; 16 received high support from all methods. This reflects the difficulties of applying cutoffs to species boundaries even when the premise of approaches is similar. There is no guarantee that the application of any PSC-related approach will give an unambiguous delimitation of species boundaries in any given case.
TABLE 1. A sample of studies that have used both the SGC and the CBD criteria in fungi.
Table S2 shows a complete list of studies using SGC; Table S3 shows a complete list of studies using CBD.
| Taxon | SGC | Species reported with SGC | CBD | Species reported with CBD |
|---|---|---|---|---|
| Protoparmelia s. str. | Concordance + Support | 25 | BP&P, spedeSTEM | 19,23 |
| Trebouxia | Concordance + Support | 6 | GMYC | 4,6 |
| Geopyxis | Concordance + Support | 6 | BP&P | 8 |
| Diploschistes | Support | 8 | BP&P | 8 |
| Serendipita vermifera | Support | 8 | BP&P | 8 |
| Colletotrichum cliviae | Support | 3 | PTP | 3 |
| Cryptococcus gattii/Cryptococcus neoformans | Support | 7 | GMYC | 6-8 |
| Cladonia mediterranea | Concordance + Support | 2 | spedeSTEM | 2 |
| Peltigera | Support | 5 | bGMYC, bPTP, spedeSTEM, BP&P | 14* |
| Montanelia | Support | at least 8 | GMYC, PTP | 6-8 |
| Oropogon | Support | at least 10 | GMYC | at least 10 |
| Collemopsidium | Support | 26 | GMYC | 26-28 |
| Hesperomyces virescens | Concordance + Support | 3 | GMYC, PTP | 3 |
| Macalpinomyces | Support | 1 | GMYC, PTP | 10,11 |
| Graphis scripta | Support | at least 7 | GMYC | 6,7 |
| Fomitopis pinicola | Support | 3 | BP&P | 4 |
authors state that this result might be intraspecific population structure.
Some caveats apply to both the SGC and CBD. Both approaches work under the assumption that speciation is predicted by sequence divergence (Dettman et al., 2003b; Liti et al., 2006); nevertheless, this assumption is often violated. While the number of hybrid incompatibilities increase as genetic distance accrues (Matute et al., 2010; Matute and Gavin-Smyth, 2014; Moyle and Nakazato, 2010; Wang et al., 2015), reproductive isolation might be achieved with just one single gene (Orr, 2006; Richards et al., 2017) which in turn reduces the predictability of the signal of speciation given molecular divergence. For example, a single epistatic interaction can cause complete reproductive isolation. In interspecific hybrids between Saccharomyces bayanus and S. cerevisiae, the S. bayanus Aep2 protein cannot regulate the translation of the S. cerevisiae OLI1 mRNA (Chou et al., 2010; Lee et al., 2008). This interaction is sufficient to cause sterility in hybrids. On the other hand, some fungal species show an exceptionally high level of genetic diversity; individuals from the same species might be more differentiated than species from different genera, without being isolated from each other. Haploid genotypes of a split-gill fungus, Schizophyllum commune, show a staggering diversity at synonymous sites of 20%, the highest of any Eukaryote, without apparent incompatibility (Baranova et al., 2015; Leffler et al., 2012).
The need for revised standards for species delimitation in fungi
Dettman et al.’s (Dettman et al., 2003b, 2003a) criteria (SGC) and the CBD of species are milestones in the quest to identify species boundaries. Nonetheless, these rules were proposed for species delimitation using a set of unlinked loci. However, the application of these rules can be misinformative (Phillips et al., 2004; Rokas et al.,2003). First, bootstrap measures the sampling effects in an alignment but not if a tree topology is actually correct (Hillis and Bull, 1993; Holmes, 2003; Phillips et al., 2004; Sanderson, 1995; Susko, 2009). In large datasets (i.e., genomic data) of closely related species, sampling error should disappear and bootstrap values will almost invariably approach 100%. In cases of truly deep divergence, bootstrap might underestimate the support of branches because difficulties in detecting homology will increase the sampling error (Lemoine et al., 2018). New methods to assess branch support incorporating the challenges of genomic data have been proposed (Carstens and Knowles, 2007; Mirarab et al., 2014; Sayyari and Mirarab, 2016), but thus far no method has tried to determine to what extent different magnitudes of support (if any) signal reproductive isolation in different taxa.
Second, genome congruence cannot be assessed in the same way as multilocus gene genealogies. As the number of studied loci increases, the likelihood of complete reciprocal monophyly decreases precipitously (Knowles and Carstens, 2007). The genome of fungi contains on average 5,000 to 10,000 genes ((Desjardins et al., 2011; Martinez et al., 2008; Seixas et al., 2019) reviewed in (Mohanta and Bae, 2015)) and some of these genes might show gene genealogies that are different from the species tree (i.e., are incongruent) just by chance (Knowles and Carstens, 2007). Sister species might retain ancestral alleles (Gao et al., 2015; Hudson and Coyne, 2002) that precede speciation or might exchange genes through hybridization (reviewed in (Schardl and Craven, 2003)). These two causes of shared genetic ancestry might lead to gene genealogies that could contradict the species tree. The increasing ease of obtaining genome-wide data uncovers the need to redefine the approaches to establish species boundaries. In light of these limitations, we propose a unified dataset and four criteria which incorporate the uncertainty associated with large datasets generated by genome sequencing but also leverage the power of such datasets.
Proposed standards for using genome sequences to delimit species boundaries
We propose that the use of whole genome sequences will allow for a systematic and comparable metric of species differentiation in fungi. This level of data not only reveals diagnostic markers, but allows for direct comparisons across studies (a major issue in reduced representation sequencing, such as RAD-Seq; (Burns et al., 2017)). We expect that whole genome sequences will become more ubiquitous over time and will end up largely replacing alternative markers. Here we propose four criteria that we consider sufficient and necessary to leverage genome sequences in identifying bona fide species boundaries in fungi.
1. Mostly reciprocal monophyly.
Reciprocal monophyly along the genome is one of the advanced stages of the speciation continuum. Groups that are reciprocally monophyletic are more likely to represent species. The proportion of loci across the genome that are reciprocally monophyletic is proportional to the the time since species divergence and represents an advanced stage in the speciation continuum (Figure 1; Hudson and Coyne 2002, Rosenberg et al. 2005, Mehta et al. 2016).
One way to identify reciprocally monophyletic groups is to generate a rooted phylogeny using the whole genome as a concatenated dataset. This approach is discouraged because when the gene trees of loci are discordant (as usually occurs in recently diverged species), concatenating data across loci can result in misleading inferences about the history of divergence (Kubatko and Degnan, 2007; Liu and Edwards, 2009). In particular, concatenated markers regularly fail at revealing the true species tree, especially in instances where there is incomplete lineage sorting ((Mendes and Hahn, 2018; Roch and Steel, 2015); discussed above). A different possibility is to find blocks of SNPs (single nucleotide polymorphisms) that are under linkage equilibrium and generate phylogenies for each block. In fungi with high levels of clonality, this genetic unit can be a chromosome (or a supercontig in unfinished genome assemblies). This procedure then produces several phylogenies that can be inspected for genome-wide support and concordance (see immediately below). Reciprocal monophyly is expected to occur after 4×Ne generations ((average number of generations until fixation where Ne is the effective population size) (Hudson and Coyne, 2002)) . Enforcing reciprocal monophyly will also ensure that all individuals in the population are placed within a phylogenetic species (exhaustive subdivision, (Dettman et al. 2003a)). Note that enforcing this criterion will prevent identification of species that are early in the speciation continuum as not all species are reciprocally monophyletic and some of them are nested within each other (Knowles and Carstens, 2007). Early stages of divergence should be treated with care as genetic data evidence might be insufficient to determine whether these groups are species or structured populations.
2. High concordance among genomic partitions.
The use of genome-wide markers will allow for species trees based on thousands of markers. In cases where speciation has truly occurred, the vast majority of the genome should show the signal that reflects the proposed species boundary. In essence, this is an extension of the genealogical concordance criterion (Dettman et al., 2003a). There are several methods to infer species trees and estimate the magnitude of genome-wide concordance (Chung and Ane, 2011; Jackson et al., 2017; Larget et al., 2010; Liu, 2008; Mirarab et al., 2014; Roch and Steel, 2015; Yang, 2015a; Zhang et al., 2018) and all of them have common elements. The main premise of concordance analyses is to generate species trees from individual gene trees and calculate the magnitude of discordance. Concordance trees and Bayesian Concordance Factors (CFs; range: 0-1) allow for this test. Each locus is assumed to have a unique genealogy, and different loci might have different genealogies. The estimated concordance tree (i.e., the dominant phylogenetic history for a group of organisms) is then built from the signal revealed from individual gene genealogies only for clades with the highest estimated CFs. The concordance tree reveals what is the most likely species tree while the CFs reveal the level of discordance among loci and reveal what proportion of genes are discordant with the rest of the genome. Formal concordance analyses account for biological processes like hybridization, incomplete lineage sorting, and lateral gene transfer, which may result in different loci having different genealogies (Ané et al., 2006; Baum, 2007).
What magnitude of CFs might be sufficient to detect species boundaries? This question was preemptively responded since the proposal of CFs: ‘the search for a particular CF threshold that denotes the boundary between reticulation and divergence is doomed’ (Baum, 2007). One of the difficulties with the question is that, although the CF increases until it reaches 1 as speciation proceeds, truly isolated species might show CFs lower than 1 because of shared ancestry (see below). Similarly, the value of the metric depends on the number of loci included in a study. We propose that the solution to this difficulty is to implement CFs not in isolation but with all the other guidelines here proposed and to report the associated CF of any proposed species.
3. Lower interspecies differentiation than intraspecific differentiation.
Most species concepts use genetic discontinuities to identify species boundaries. This general premise is common to all the species concepts. Under this framework, individuals from the same species should be more closely related to each other than to individuals from other species. In other words, in cases where speciation has really occurred, the mean distance between individuals from the two species (Dxy) should be larger than the mean distance between individuals within each of the species (π). Dxy and π can then be compared using permutation tests. This test reveals whether genetic variation in a putative species pair is partitioned across species or not, either at the individual locus level or genome wide (Hughes et al., 2009; Matute et al., 2006; Sepúlveda et al., 2017). It is worth noting that these comparisons require extensive sampling across the whole geographic range of a species to avoid missing clinal variation which with incomplete sampling might look like genetic discontinuities (Piedra-Malagón et al., 2011).
An alternative possibility is to use the magnitude of neutral differentiation to detect well-formed species. A seminal study found that the intermediate “gray zone” of speciation (i.e., cases in which population structure and incipient speciation cannot be differentiated thus making taxonomy controversial, Figure 1), usually happens in animals when synonymous divergence between the diverging species occurs between 0.5% to 2% (Roux et al., 2016). (A similar rule and rationale was proposed for single loci (Fujisawa and Barraclough, 2013)). Diverging populations in which divergence is lower than 0.5% are likely to have pervasive gene exchange and merge into a single lineage (Roux et al., 2016). The magnitude of gene exchange does fall precipitously after 2%. This allows us to formulate a tentative metric; if the magnitude of genome-wide synonymous divergence is above 2% and is significantly lower than the magnitude of polymorphism within either species, then the two lineages can be described as different species. Notably, no studies of this type have been done in fungi and the application of metrics devised for animals can be misinformative. As mentioned before, there is no perfect metric neither for Dxy/π comparisons nor for the magnitude of synonymous divergence to detect species boundaries (but see the 4× rule; (Barraclough et al., 2003; Birky, 2013)).
4. Low shared polymorphism.
As speciation progresses, genomes differentiate, and the likelihood of shared ancestry decreases between species (Figure 1). This occurs because the two events that lead to shared ancestry, retention of ancestral polymorphism (that pre-dates speciation) and gene exchange, are less likely as genetic divergence accrues. In the case of ancestral polymorphism, the magnitude of neutral polymorphism that is shared in sister species (i.e., variants that come from the ancestral species) decays precipitously as divergence increases (Gao et al., 2015; Hudson and Coyne, 2002). As the number of incompatible alleles increases rapidly with divergence time (Matute et al., 2010; Matute and Gavin-Smyth, 2014; Moyle and Nakazato, 2010; Wang et al., 2015), the likelihood of a neutral (or even advantageous) introgression being linked to a hybrid incompatibility increases and the permeability of the genome to introgression decreases (Carneiro et al., 2014, 2010; Muirhead and Presgraves, 2016; Payseur et al., 2004; Turissini and Matute, 2017). Several methods have been proposed to quantify shared ancestry and differentiate introgression and incomplete lineage sorting (Guerrero and Hahn, 2018), some of which have been specifically applied to fungi. These approaches fall into two types: the use of summary statistics, such as D (i.e., ABBA/BABA tests, (Green et al., 2010)) and fD (Martin et al., 2013), and the identification of shared haplotypes (e.g., (Guan, 2014; Hellenthal et al., 2014; Price et al., 2009; Turissini and Matute, 2017)). The former is amenable to all genomic analysis (including reduced representation approaches, reviewed in (Davey et al., 2011)), while the latter is better suited for whole genome analyses but can be applied to reduced representation data in biological systems with extensive linkage disequilibrium and a reference genome (as is the case of humans; e.g., (Price et al., 2009)). Both approaches have been used in fungi, but summary statistics seem to be slightly more common than assessments of local ancestry (Table S3). A formal test of the magnitude of introgression across multiple species of fungi is sorely needed, but analyses of shared ancestry suggest that hybridization and introgression are of common occurrence in fungi. Similar to observations in animals, introgression seems to be less prevalent between diverged species (Maxwell et al., 2018b) than between recently diverged ones (Desjardins et al., 2017; Gladieux et al., 2015; Maxwell et al., 2018a) which follows expectations regarding the evolution of hybrid incompatibilities (Muirhead and Presgraves, 2016).
Why is it important to define species?
The ultimate goal of identifying species boundaries is not merely describing species by itself. Properly defining species boundaries is just the beginning of a robust research program in evolutionary biology. To understand diversification patterns in fungi, we first need to quantify the magnitude of extant diversity in the group. If cryptic species are particularly prevalent in fungi, then our understanding of macroevolutionary patterns—in fungi or any other taxon— will be warped by faulty species definitions. Similarly, in order to understand how fungal genomes evolve and how divergence unfolds, we need to have reliable species definitions that can be applied across taxa. Even our understanding of how genetic diversity is portioned across populations and species will be misinformed if species boundaries are not properly defined.
Properly defining species boundaries also has strong implications for other areas of biology. Cryptic species tend to be phenotypically similar, but this is not always the case. Moreover, since it is impossible to exhaustively measure the phenotype of an organism, it is difficult to rule out the possibility that they may differ in some important aspect. In fact, in hindsight some previously cryptic species differ in phenotypes that are medically or economically significant. Fungal pathogens provide an example of the correct delimitation of species boundaries providing insight into pathogenesis. For example, the application of the SGC approach in Magnaporthe grisea revealed the existence of two species, M. oryzae and M. grisea sensu stricto (Couch and Kohn, 2002). While the former infects rice cultivars, the latter is restricted to crabgrass (Digitaria). Notably, M. oryzae might contain multiple lineages, each of which is preferentially associated with different hosts, which in turns suggests incipient speciation following host shifts (Gladieux et al., 2018). Similarly, species of Histoplasma differ in their virulence, resistance to antifungals, and in some instances genome size (Kasuga et al., 2002; Goughenour et al., 2015; Sepúlveda et al., 2017, 2014). In contrast, the sibling species of the genus Coccidioides, C. immitis and C. posadasii, are deeply genetically differentiated but few phenotypic differences have been identified (Fisher et al., 2002; Neafsey et al., 2010; Ramani and Chaturvedi, 2007; Sharpton et al., 2009). Despite overlapping geographic ranges, which could facilitate introgression, these species share few alleles, indicating rare hybridization (Maxwell et al., 2018b; Neafsey et al., 2010).
Taxonomy is a hypothesis and if proposed species cannot be verified or are not used they can simply be archived. This is perhaps the ultimate ‘species concept’: if a species classification is evidence-based and turns out to be useful to researchers and others, then it will persist and spread. By infusing taxonomy with genetics and genomics, we can make the concept of ‘species’ in fungi more useful by making it more robust, testable, comparable to other taxa, and devoid of the personal attachments that researchers might feel to particular species names.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Colin S. Maxwell, Juan G. McEwen and William Goldman for their comments on the manuscript. Three anonymous reviewers helped tremendously improve every aspect of this manuscript. This work was funded by grant R01GM121750 to DRM.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Alamouti SM, Wang V, Diguistini S, Six DL, Bohlmann J, Hamelin RC, Feau N, Breuil C, 2011. Gene genealogies reveal cryptic species and host preferences for the pine fungal pathogen Grosmannia clavigera. Mol. Ecol 20(12): 2581–2602. 10.1111/j.1365-294X.2011.05109.x [DOI] [PubMed] [Google Scholar]
- Alcoba-Flórez J, Méndez-Álvarez S, Cano J, Guarro J, Pérez-Roth E, Del Pilar Arévalo M, 2005. Phenotypic and molecular characterization of Candida nivariensis sp. nov., a possible new opportunistic fungus. J. Clin. Microbiol 43(8): 4107–4111. 10.1128/JCM.43.8.4107-4111.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida P, Barbosa R, Bensasson D, Gonçalves P, Sampaio JP, 2017. Adaptive divergence in wine yeasts and their wild relatives suggests a prominent role for introgressions and rapid evolution at noncoding sites. Mol. Ecol 26(7): 2167–2182. 10.1111/mec.14071. [DOI] [PubMed] [Google Scholar]
- Alors D, Lumbsch HT, Divakar PK, Leavitt SD, Crespo A, 2016. An integrative approach for understanding diversity in the Punctelia rudecta species complex (Parmeliaceae, Ascomycota). PLoS One. 11(2): e0146537 10.1371/journal.pone.0146537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alves A, Crous PW, Correia A, Phillips a J.L., 2008. Morphological and molecular data reveal cryptic speciation in Lasiodiplodia theobromae. Fungal Diversity. 28: 1–13. [Google Scholar]
- Ané C, Larget B, Baum DA, Smith SD, Rokas A 2006. Bayesian estimation of concordance among gene trees. Mol. Biol. Evol 24(2):412–426. 2007 doi: 10.1093/molbev/msl170 [DOI] [PubMed] [Google Scholar]
- Avise J, Ball RM, 1990. Principles of genealogical concordance in species concepts and biological taxonomy. Oxford Surv. Evol. Biol 7: 45–67 [Google Scholar]
- Avise JC, Wollenberg K, 1997. Phylogenetics and the origin of species. Proc. Natl. Acad. Sci 94 (15) 7748–7755. 10.1073/pnas.94.15.7748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baranova MA, Logacheva MD, Penin AA, Seplyarskiy VB, Safonova YY, Naumenko SA, Klepikova AV, Gerasimov ES, Bazykin GA, James TY, Kondrashov AS, 2015. Extraordinary genetic diversity in a wood decay mushroom. Mol. Biol. Evol 32(10): 2775–2783. 10.1093/molbev/msv153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barraclough TG, Birky CW, Burt A, 2003. Diversification in sexual and asexual to organisms. Evolution. 57(9): 2166–2172 10.1111/j.0014-3820.2003.tb00394.x [DOI] [PubMed] [Google Scholar]
- Baum DA, Shaw KL, 1995. Genealogical perspectives on the species problem. Experimental and Molecular Approaches to Plant Biosystematics. (Hoch PC and Stephenson AG, eds.) Monogr. Syst, Missouri Bot. Gard 53: 289–303. [Google Scholar]
- Baum DA, 2007. Concordance trees, concordance factors, and the exploration of reticulate genealogy. Taxon. 56(2): 417–426. 10.1007/sl0869-007-9037-x [DOI] [Google Scholar]
- Baum DA, Donoghue MJ, 1995. Choosing among alternative “phylogenetic” species concepts. Syst. Bot 20(4): 560–573. [Google Scholar]
- Bazzicalupo AL, Buyck B, Saar I, Vauras J, Carmean D, Berbee ML, 2017. Troubles with mycorrhizal mushroom identification where morphological differentiation lags behind barcode sequence divergence. Taxon. 66(4): 791–810. 10.12705/664.1 [DOI] [Google Scholar]
- Bickford D, Lohman DJ, Sodhi NS, Ng PKL, Meier R, Winker K, Ingram KK, Das I, 2007. Cryptic species as a window on diversity and conservation. Trends Ecol. Evol 22(3): 148–155. 10.1016/j.tree.2006.11.004 [DOI] [PubMed] [Google Scholar]
- Bidochka MJ, Kamp AM, Lavender TM, Dekoning J, De Croos JNA, 2001. Habitat association in two genetic groups of the insect-pathogenic fungus Metarhizium anisopliae: uncovering cryptic species? Appl. Environ. Microbiol 67(3): 1335–1342. 10.1128/AEM.67.3.1335-1342.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bidochka MJ, Small CLN, Spironello M, 2005. Recombination within sympatric cryptic species of the insect pathogenic fungus Metarhizium anisopliae. Environ. Microbiol 7(9): 1361–1368. 10.1111/j.1462-5822.2005.00823.x [DOI] [PubMed] [Google Scholar]
- Birky CW Jr, 2013. Species detection and identification in sexual organisms using population genetic theory and DNA sequences. PLoS One. 8(1): e52544 10.1371/journal.pone.0052544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff JF, Rehner SA, Humber RA, 2006. Metarhizium frigidum sp. nov.: a cryptic species of M. anisopliae and a member of the M. flavoviride complex. Mycologia. 98(5): 737–745. 10.3852/mycologia.98.5.737 [DOI] [PubMed] [Google Scholar]
- Bobay L-M, Ochman H, 2017. Biological Species Are Universal across Life’s Domains. Genome Biol. Evol 9(3): 491–501. 10.1093/gbe/evx026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boluda CG, Rico VJ, Divakar PK, Nadyeina O, Myllys L, McMullin RT, Zamora JC, Scheidegger C, Hawksworth DL, 2018. Evaluating methodologies for species delimitation: the mismatch between phenotypes and genotypes in lichenized fungi (Bryoria sect. Implexae, Parmeliaceae). Persoonia - Mol. Phylogeny Evol. Fungi. 42(2019): 75–100. 10.3767/persoonia.2019.42.04 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown EM, McTaggart LR, Zhang SX, Low DE, Stevens DA, Richardson SE, 2013. Phylogenetic analysis reveals a cryptic species Blastomyces gilchristii, sp. nov. within the human pathogenic fungus Blastomyces dermatitidis. PLoS One. 8(3): e59237 10.1371/journal.pone.0059237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns M, Starrett J, Derkarabetian S, Richart CH, Cabrero A, Hedin M, 2017. Comparative performance of double-digest RAD sequencing across divergent arachnid lineages. Mol. Ecol. Resour 17(3): 418–430. 10.1111/1755-0998.12575 [DOI] [PubMed] [Google Scholar]
- Cai L, Giraud T, Zhang N, Begerow D, Cai G, Shivas RG, 2011. The evolution of species concepts and species recognition criteria in plant pathogenic fungi. Fungal Divers. 5: 121 10.1007/s13225-011-0127-8 [DOI] [Google Scholar]
- Carneiro M, Albert FW, Afonso S, Pereira RJ, Burbano H, Campos R, Melo-Ferreira J, Blanco Aguiar JA, Villafuerte R, Nachman MW, Good JM, Ferrand N, 2014. The genomic architecture of population divergence between subspecies of the European rabbit. PLoS Genet. 10(8), e1003519. doi: 10.1371/journal.pgen.1003519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carneiro M, Blanco Aguiar JA, Villafuerte R, Ferrand N, Nachman MW, 2010. Speciation in the european rabbit (Oryctolagus cuniculus): islands of differentiation on the X chromosome and autosomes. Evolution. 64(12): 3443–3460. doi: 10.1111/j.1558-5646.2010.01092.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carriconde F, Gardes M, Jargeat P, Heilmann-Clausen J, Mouhamadou B, Gryta H, 2008. Population evidence of cryptic species and geographical structure in the cosmopolitan ectomycorrhizal fungus, Tricholoma scalpturatum. Microb. Ecol 56(3): 513–524. 10.1007/s00248-008-9370-2 [DOI] [PubMed] [Google Scholar]
- Carstens BC, Knowles LL, 2007. Estimating species phylogeny from gene-tree probabilities despite incomplete lineage sorting: An example from Melanoplus grasshoppers. Syst. Biol 56(3): 400–411. 10.1080/10635150701405560 [DOI] [PubMed] [Google Scholar]
- Chen J, Guo SX, Liu PG, 2011. Species recognition and cryptic species in the Tuber indicum complex. PLoS One. 6(1): e14625 10.1371/journal.pone.0014625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou J-Y, Hung Y-S, Lin K-H, Lee H-Y, Leu J-Y, 2010. Multiple molecular mechanisms cause reproductive isolation between three yeast species. PLoS Biol. 8(7): e1000432 10.1371/journal.pbio.1000432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung Y, Ané C, 2011. Comparing two bayesian methods for gene tree/species tree reconstruction: Simulations with incomplete lineage sorting and horizontal gene transfer. Syst. Biol 60(3): 261–275. 10.1093/sysbio/syr003 [DOI] [PubMed] [Google Scholar]
- Corcoran P, Anderson JL, Jacobson DJ, Sun Y, Ni P, Lascoux M, Johannesson H, 2016. Introgression maintains the genetic integrity of the mating-type determining chromosome of the fungus Neurospora tetrasperma. Genome Res. 26(4): 486–98. doi: 10.1101/gr.197244.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correia A, Sampaio P, James S, Pais C, 2006. Candida bracarensis sp. nov., a novel anamorphic yeast species phenotypically similar to Candida glabrata. Int. J. Syst. Evol. Microbiol 56: 313–317. 10.1099/ijs.0.64076-0 [DOI] [PubMed] [Google Scholar]
- Couch BC, Kohn LM, 2002. A multilocus gene genealogy concordant with host preference indicates segregation of a new species, Magnaporthe oryzae, from M. grisea. Mycologia. 94:4, 683–693. 10.1080/15572536.2003.11833196 [DOI] [PubMed] [Google Scholar]
- Coyne JA, Orr HA, 2004. Speciation. Sunderland, MA: 1–475. [Google Scholar]
- Crous PW, Groenewald JZ, Pongpanich K, Himaman W, Arzanlou M, Wingfield MJ, 2004. Cryptic speciation and host specificity among Mycosphaerella spp. occurring on Australian Acacia species grown as exotics in the tropics. Studies in Mycology. 50: 457–469. [Google Scholar]
- Crespo A and Lumbsch HT, 2010. Cryptic species in lichen-forming fungi. IMA fungus, 1(2), pp.167–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruse M, Telerant R, Gallagher T, Lee T, Taylor JW, 2002. Cryptic species in Stachybotrys chartarum. Mycologia. 94(5): 814–822. 10.1080/15572536.2003.11833175 [DOI] [PubMed] [Google Scholar]
- Davey JW, Hohenlohe PA, Etter PD, Boone JQ, Catchen JM, Blaxter ML, 2011. Genome-wide genetic marker discovery and genotyping using next-generation sequencing. Nat. Rev. Genet 12: 499–510. 10.1038/nrg3012. [DOI] [PubMed] [Google Scholar]
- Del-Prado R, Divakar PK, Lumbsch HT, Crespo AM, 2016. Hidden genetic diversity in an asexually reproducing lichen forming fungal group. PLoS One. 11(8): e0161031 10.1371/journal.pone.0161031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Queiroz K, 2007. Species concepts and species delimitation. Syst. Biol 56(6):879–886 10.1080/10635150701701083 [DOI] [PubMed] [Google Scholar]
- Desjardins CA, Champion MD, Holder JW, Muszewska A, Goldberg J, Bailao AM, Brigido MM, da Silva Ferreira ME, Garcia AM, Grynberg M, Gujja S, Heiman DI, Henn MR, Kodira CD, Leon-Narvaez H, Longo LVG, Ma LJ, Malavazi I, Matsuo AL, Morais FV, Pereira M, Rodriguez-Brito S, Sakthikumar S, Salem-Izacc SM, Sykes SM, Teixeira MM, Vallejo MC, Walter MEMT, Yandava C, Young S, Zeng Q, Zucker J, Felipe MS, Goldman GH, Haas BJ, McEwen JG, Nino-Vega G, Puccia R, San-Blas G, de Soares CMA, Birren BW, Cuomo CA, 2011. Comparative genomic analysis of human fungal pathogens causing paracoccidioidomycosis. PLoS Genet. 7(10): e1002345 10.1371/journal.pgen.1002345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desjardins CA, Giamberardino C, Sykes SM, Yu C-H, Tenor JL, Chen Y, Yang T, Jones AM, Sun S, Haverkamp MR, Heitman J, Litvintseva AP, Perfect JR, Cuomo CA, 2017. Population genomics and the evolution of virulence in the fungal pathogen Cryptococcus neoformans. Genome Res. 27, 1207–1219. 10.1101/gr.218727.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dettman JR, Jacobson DJ, Taylor JW, 2006. Multilocus sequence data reveal extensive phylogenetic species diversity within the Neurospora discreta complex. Mycologia. 98(3): 436–446. 10.1080/15572536.2006.11832678 [DOI] [PubMed] [Google Scholar]
- Dettman JR, Jacobson DJ, Taylor JW, 2003a. A multilocus genealogical approach to phylogenetic species recognition in the model eukaryote Neurospora. Evolution. 57(12): 2703–2720. 10.1111/j.0014-3820.2003.tb01514.x [DOI] [PubMed] [Google Scholar]
- Dettman JR, Jacobson DJ, Turner E, Pringle A, Taylor JW, 2003b. Reproductive isolation and phylogenetic divergence in Neurospora: Comparing methods of species recognition in a model eukaryote. Evolution. 57(12): 2721–2741. 10.1111/j.0014-3820.2003.tb01515. [DOI] [PubMed] [Google Scholar]
- Dobzhansky T 1937. Genetics and the Origin of Species. Columbia University Press. [Google Scholar]
- Donoghue MJ, 1985. A critique of the biological species concept and recommendations for a phylogenetic alternative. Bryologist. 88(3): 172–181. 10.2307/3243026 [DOI] [Google Scholar]
- Dobzhansky T 1937. Genetics and the Origin of Species. Columbia University Press. Donoghue, M.J., 1985. A critique of the biological species concept and recommendations for a phylogenetic alternative. Bryologist. 88(3): 172–181. 10.2307/3243026 [DOI] [Google Scholar]
- Douanla-Meli C, Unger JG, Langer E, 2018. Multi-approach analysis of the diversity in Colletotrichum cliviae sensu lato. Antonie van Leeuwenhoek, Int. J. Gen. Mol. Microbiol. 111(3): 423–435. 10.1007/s10482-017-0965-9 [DOI] [PubMed] [Google Scholar]
- Dupuis JR, Roe AD, Sperling FAH, 2012. Multi-locus species delimitation in closely related animals and fungi: One marker is not enough. Mol. Ecol 21: 4422–4436. 10.1111/j.1365-294X.2012.05642.x [DOI] [PubMed] [Google Scholar]
- Edwards SV, Liu L, Pearl DK, 2007. High-resolution species trees without concatenation. Proc. Natl. Acad. Sci 104(14): 5936–5941. 10.1073/pnas.0607004104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ence DD, Carstens BC, 2011. SpedeSTEM: A rapid and accurate method for species delimitation. Mol. Ecol. Resour 11(3): 473–480. 10.1111/j.1755-0998.2010.02947.x [DOI] [PubMed] [Google Scholar]
- Fisher MC, Koenig GL, White TJ, Taylor JW, 2002. Molecular and phenotypic description of Coccidioides posadasii sp. nov., previously recognized as the non-California population of Coccidioides immitis. Mycologia. 94(1): 73–84. 10.1080/15572536.2003.11833250 [DOI] [PubMed] [Google Scholar]
- Fontaneto D, Herniou EA, Boschetti C, Caprioli M, Melone G, Ricci C, Barraclough TG, 2007. Independently evolving species in asexual bdelloid rotifers. PLoS Biol. 5(4): e87 10.1371/journal.pbio.0050087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujisawa T, Barraclough TG, 2013. Delimiting species using single-locus data and the generalized mixed yule coalescent approach: A revised method and evaluation on simulated data sets. Syst. Biol 62(5): 707–724. 10.1093/sysbio/syt033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita MK, Leaché AD, Burbrink FT, McGuire JA, Moritz C, 2012. Coalescent-based species delimitation in an integrative taxonomy. Trends Ecol. Evol 27(9): 480–488. 10.1016/j.tree.2012.04.012 [DOI] [PubMed] [Google Scholar]
- Gao Z, Przeworski M, Sella G, 2015. Footprints of ancient-balanced polymorphisms in genetic variation data from closely related species. Evolution 69, 431–446. doi: 10.1111/evo.12567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazis R, Rehner S, Chaverri P, 2011. Species delimitation in fungal endophyte diversity studies and its implications in ecological and biogeographic inferences. Mol. Ecol 20(14): 3001–3013. 10.1111/j.1365-294X.2011.05110.x [DOI] [PubMed] [Google Scholar]
- Geiser DM, Pitt JI, Taylor JW, 1998. Cryptic speciation and recombination in the aflatoxin-producing fungus Aspergillus flavus. Proc. Natl. Acad. Sci 95(1): 388–393. 10.1073/pnas.95.1388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlach A da CL, Toprak Z, Naciri Y, Caviro EA., da Silveira RMB, Clerc P, 2019. New insights into the Usnea cornuta aggregate (Parmeliaceae, lichenized Ascomycota): Molecular analysis reveals high genetic diversity correlated with chemistry. Mol. Phylogenet. Evol 131: 125–137. 10.1016/j.ympev.2018.10.035 [DOI] [PubMed] [Google Scholar]
- Gilgado F, Cano J, Gene J, Guarro J, 2005. Molecular phylogeny of the Pseudallescheria boydii species complex: Proposal of two new species. J. Clin. Microbiol 43(10): 4930–4942. doi: 10.1128/JCM.43.10.4930-4942.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraud T, Gladieux P, Hood M, 2010. The origin of species in Fungi. Fungi. 3(4): 23–27. [Google Scholar]
- Giraud T, Gladieux P, Hood M, 2010. The origin of species in Fungi. Fungi. 3(4): 23–27. [Google Scholar]
- Gladieux P, Condon B, Ravel S, Soanes D, Maciel JLN, Nhani A, Chen L, Terauchi R, Lebrun MH, Tharreau D, Mitchell T, Pedley KF, Valent B, Talbot NJ, Farman M, Fournier E, 2018. Gene flow between divergent cereal- and grass-specific lineages of the rice blast fungus Magnaporthe oryzae. mBio. 9:e01219–17. 10.1128/mBio.01219-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gladieux P, Wilson BA, Perraudeau F, Montoya LA, Kowbel D, Hann-Soden C, Fischer M, Sylvain I, Jacobson DJ, Taylor JW, 2015. Genomic sequencing reveals historical, demographic and selective factors associated with the diversification of the fire-associated fungus Neurospora discreta. Mol. Ecol 24, 5657–5675. 10.1111/mec.13417 [DOI] [PubMed] [Google Scholar]
- Goughenour KD, Balada-Llasat JM, Rappleye CA, 2015. Quantitative microplate-based growth assay for determination of antifungal susceptibility of Histoplasma capsulatum yeasts. J Clin Microbiol 53(10):3286–3295. doi: 10.1128/JCM.00795-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green RE, Krause J, Briggs AW, Maricic T, Stenzel U, Kircher M, Patterson N, Li H, Zhai W, Fritz MH-Y, Hansen NF, Durand EY, Malaspinas A-S, Jensen JD, Marques-Bonet T, Alkan C, Prüfer K, Meyer M, Burba no HA, Good JM, Schultz R, Aximu-Petri A, Butthof A, Hober B, Hoffner B, Siegemund M, Weihmann A, Nusbaum C, Lander ES, Russ C, Novod N, Affourtit J, Egholm M, Verna C, Rudan P, Brajkovic D, Kucan Z, Gusic I, Doronichev VB, Golovanova LV, Lalueza-Fox C, de la Rasilla M, Fortea J, Rosas A, Schmitz RW, Johnson PLF, Eichler EE, Falush D, Birney E, Mullikin JC, Slatkin M, Nielsen R, Kelso J, Lachmann M, Reich D, Paabo S, 2010. A Draft Sequence of the Neandertal Genome. Science. 328(5979): 710–722. doi: 10.1126/science.1188021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groenewald M, Groenewald JZ, Crous PW, 2005. Distinct species exist within the Cercospora apii morphotype. Phytopathology. 95(8):951–959. 10.1094/PHYTO-95-0951 [DOI] [PubMed] [Google Scholar]
- Grube M and Kroken S, 2000. Molecular approaches and the concept of species and species complexes in lichenized fungi. Mycological Research, 104(11), pp.1284–1294. [Google Scholar]
- Grünig CR, Duò A, Sieber TN, 2006. Population genetic analysis of Phialocephala fortinii s.l. and Acephala applanata in two undisturbed forests in Switzerland and evidence for new cryptic species. Fungal Genet. Biol 43(6): 410–421. 10.1016/j.fgb.2006.01.007 [DOI] [PubMed] [Google Scholar]
- Grünig CR, McDonald BA, Sieber TN, Rogers SO, Holdenrieder O, 2004. Evidence for subdivision of the root-endophyte Phialocephala fortinii into cryptic species and recombination within species. Fungal Genet. Biol 41(7): 676–687. 10.1016/j.fgb.2004.03.004 [DOI] [PubMed] [Google Scholar]
- Guan Y, 2014. Detecting structure of haplotypes and local ancestry. Genetics. 196(3): 625–642. doi: 10.1534/genetics.113.160697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrero RF, Hahn MW, 2018. Quantifying the risk of hemiplasy in phylogenetic inference. Proc. Natl. Acad. Sci. 115(50): 12787–12792. 10.1073/pnas.1811268115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillin EA, Grijalba PE, Oliveira L.O. de, Gottlieb AM, 2014. Specific boundaries between the causal agents of the soybean stem canker. Trop. Plant Pathol 39(4):316–325. 10.1590/s1982-56762014000400006 [DOI] [Google Scholar]
- Hagen F, Khayhan K, Theelen B, Kolecka A, Polacheck I, Sionov E, Falk R, Parnmen S, Lumbsch HT, Boekhout T, 2015. Recognition of seven species in the Cryptococcus gattii/Cryptococcus neoformans species complex. Fungal Genet. Biol 75: 16–48 10.1016/j.fgb.2015.02.009 [DOI] [PubMed] [Google Scholar]
- Harrington TC and Rizzo DM, 1999. Defining species in the fungi In Structure and dynamics of fungal populations (pp. 43–71). Springer, Dordrecht. [Google Scholar]
- Hellenthal G, Busby GBJ, Band G, Wilson JF, Capelli C, Falush D, Myers S, 2014. A genetic atlas of human admixture history. Science. 343(6172): 747–751. doi: 10.1126/science.1243518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillis DM, Bull JJ, 1993. An empirical test of bootstrapping as a method for assessing confidence in phylogenetic analysis. Syst. Biol 42(2): 182–192. 10.1093/sysbio/42.2.182 [DOI] [Google Scholar]
- Holmes S, 2003. Bootstrapping phylogenetic trees: theory and methods. Stat. Sci 18(2): 241–255. 10.1214/ss/1063994979 [DOI] [Google Scholar]
- Hubka V, Nováková A, Jurjević Z, Sklenář F, Frisvad JC, Houbraken J, Arendrup MC, Jørgensen KM, Siqueira JPZ, Gené J, Kolařík M, 2018. Polyphasic data support the splitting of Aspergillus candidus into two species; proposal of Aspergillus dobrogensis sp. nov. Int. J. Syst. Evol. Microbiol 68: 995–1011. 10.1099/ijsem.0.002583 [DOI] [PubMed] [Google Scholar]
- Hudson RR, Coyne JA, 2002. Mathematical consequences of the genealogical species concept. Evolution. 56(8): 1557–1565. 10.1111/j.0014-3820.2002.tb01467.x. [DOI] [PubMed] [Google Scholar]
- Hughes KW, Petersen RH, Lickey EB, 2009. Using heterozygosity to estimate a percentage DNA sequence similarity for environmental species’ delimitation across basidiomycete fungi. New Phytol. 10.1111/j.1469-8137.2009.02802.x [DOI] [PubMed] [Google Scholar]
- Huxley J, 1943. Systematics and the origin of Species: from the Viewpoint of a Zoologist. Nature. 151, 347–348. 10.1038/151347a0 [DOI] [Google Scholar]
- Jackson ND, Carstens BC, Morales AE, O’Meara BC, 2017. Species delimitation with gene flow. Syst. Biol 66(5): 799–812. 10.1093/sysbio/syw117 [DOI] [PubMed] [Google Scholar]
- Jones G, 2017. Algorithmic improvements to species delimitation and phylogeny estimation under the multispecies coalescent. J. Math. Biol 74: 447–467. 10.1007/S00285-016-1034-0 [DOI] [PubMed] [Google Scholar]
- Jones G, Aydin Z, Oxelman B, 2015. DISSECT: An assignment-free Bayesian discovery method for species delimitation under the multispecies coalescent. Bioinformatics. 31(7): 991–998. 10.1093/bioinformatics/btu770 [DOI] [PubMed] [Google Scholar]
- Kasuga T, White TJ, Koenig G, Mcewen J, Restrepo A, Castaneda E, Da Silva Lacaz CDA, Heins-Vaccari EM, De Freitas RS, Zancope-Oliveira RM, Qin Z, Negroni R, Carter DA, Mikami Y, Tamura M, Taylor ML, Miller GF, Poonwan N, Taylor JW, 2003. Phylogeography of the fungal pathogen Histoplasma capsulatum. Mol. Ecol 12: 3383–3401. 10.1046/j.1365-294X.2003.01995.x [DOI] [PubMed] [Google Scholar]
- Kasuga T, White TJ, Taylor JW, 2002. Estimation of nucleotide substitution rates in Eurotiomycete fungi. Mol. Biol. Evol 19(12): 2318–2324. 10.1093/oxfordjournals.molbev.a004056 [DOI] [PubMed] [Google Scholar]
- Knowles LL, Carstens BC, 2007. Delimiting species without monophyletic gene trees. Syst. Biol 56(6): 887–895. 10.1080/10635150701701091 [DOI] [PubMed] [Google Scholar]
- Koufopanou V, Burt A, Szaro T, Taylor JW, 2001. Gene genealogies, cryptic species, and molecular evolution in the human pathogen Coccidioides immitis and relatives (Ascomycota, Onygenales). Mol. Biol. Evol 18(7): 1246–1258. 10.1093/oxfordjournals.molbev.a003910 [DOI] [PubMed] [Google Scholar]
- Kubatko LS, Carstens BC, Knowles LL, 2009. STEM: Species tree estimation using maximum likelihood for gene trees under coalescence. Bioinformatics. 25(7): 971–973. 10.1093/bioinformatics/btp079 [DOI] [PubMed] [Google Scholar]
- Kubatko LS, Degnan JH, 2007. Inconsistency of phylogenetic estimates from concatenated data under coalescence. Syst. Biol 56(1): 17–24. 10.1080/10635150601146041 [DOI] [PubMed] [Google Scholar]
- Larget BR, Kotha SK, Dewey CN, Ane C, 2010. BUCKy: Gene tree/species tree reconciliation with Bayesian concordance analysis. Bioinformatics. 26(22): 2910–2911. 10.1093/bioinformatics/btq539 [DOI] [PubMed] [Google Scholar]
- Laurence MH, Summerell BA, Burgess LW, Liew ECY, 2014. Genealogical concordance phylogenetic species recognition in the Fusarium oxysporum species complex. Fungal Biol. 118(4): 374–384. 10.1016/j.funbio.2014.02.002 [DOI] [PubMed] [Google Scholar]
- Le Gac M, Hood ME, Giraud T, 2007a. Evolution of reproductive isolation within a parasitic fungal species complex. Evolution. 61(7): 1781–1787. 10.1111/j.1558-5646.2007.00144.x [DOI] [PubMed] [Google Scholar]
- Le Gac M, Hood MEME, Fournier E, Giraud T, 2007b. Phylogenetic evidence of host-specific cryptic species in the anther smut fungus. Evolution. 61(1): 15–26. 10.1111/j.1558-5646.2007.00002.x [DOI] [PubMed] [Google Scholar]
- Leaché AD, Zhu T, Rannala B, Yang Z, 2019. The Spectre of too many species. Syst. Biol 68(1): 168–181. 10.1093/sysbio/syy051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leavitt S, Fernández-Mendoza F, Pérez-Ortega S, Sohrabi M, Divakar P, Lumbsch T, St. Clair L, 2013a. DNA barcode identification of lichen-forming fungal species in the Rhizoplaca melanophthalma species-complex (Lecanorales, Lecanoraceae), including five new species. MycoKeys. 7: 1–22. 10.3897/mycokeys.7.4508 [DOI] [Google Scholar]
- Leavitt SD, Esslinger TL, Divakar PK, Crespo A, Lumbsch HT, 2016. Hidden diversity before our eyes: Delimiting and describing cryptic lichen-forming fungal species in camouflage lichens (Parmeliaceae, Ascomycota). Fungal Biol 120(11): 1374–1391. 10.1016/j.funbio.2016.06.001 [DOI] [PubMed] [Google Scholar]
- Leavitt SD, Fankhauser JD, Leavitt DH, Porter LD, Johnson LA, St. Clair LL, 2011. Complex patterns of speciation in cosmopolitan “rock posy” lichens - Discovering and delimiting cryptic fungal species in the lichen-forming Rhizoplaca melanophthalma species-complex (Lecanoraceae, Ascomycota). Mol. Phylogenet. Evol 59(3): 587–602. 10.1016/j.ympev.2011.03.020 [DOI] [PubMed] [Google Scholar]
- Leavitt SD, Fernández-Mendoza F, Pérez-Ortega S, Sohrabi M, Divakar PK, Vondrak J, Thorsten Lumbsch H, Clair LLS, 2013b. Local representation of global diversity in a cosmopolitan lichen-forming fungal species complex (Rhizoplaca, Ascomycota). J. Biogeogr 40(9): 1792–1806. 10.1111/jbi.12118 [DOI] [Google Scholar]
- Leavitt SD, Westberg M, Nelsen MP, Elix JA, Timdal E, Sohrabi M, St. Clair LL, Williams L, Wedin M, Lumbsch HT, 2018. Multiple, distinct intercontinental lineages but isolation of Australian populations in a cosmopolitan lichen-forming Fungal Taxon, Psora decipiens (Psoraceae, Ascomycota). Front. Microbiol 9: 283 10.3389/fmicb.2018.00283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leducq JB, Nielly-Thibault L, Charron G, Eberlein C, Verta JP, Samani P, Sylvester K, Hittinger CT, Bell G, Landry CR, 2016. Speciation driven by hybridization and chromosomal plasticity in a wild yeast. Nat. Microbiol 1:1–10. 10.1038/nmicrobiol.2015.3 [DOI] [PubMed] [Google Scholar]
- Lee H-Y, Chou J-Y, Cheong L, Chang N-H, Yang S-Y, Leu J-Y, 2008. Incompatibility of nuclear and mitochondrial genomes causes hybrid sterility between two yeast species. Cell. 135(6): 1065–1073. 10.1016/j.cell.2008.10.047. [DOI] [PubMed] [Google Scholar]
- Leffler EM, Bullaughey K, Matute DR, Meyer WK, Segurel L, Venkat A, Andolfatto P, Przeworski M, 2012. Revisiting an old riddle: what determines genetic diversity levels within species? PLoS Biol. 10(9): e1001388 10.1371/journal.pbio.1001388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemoine F, Domelevo Entfellner JB, Wilkinson E, Correia D, Davila Felipe M, De Oliveira T, Gascuel O, 2018. Renewing Felsenstein’s phylogenetic bootstrap in the era of big data. Nature. 556: 452–456. 10.1038/s41586-018-0043-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YM, Shivas RG, Cai L, 2017. Cryptic diversity in Tranzscheliella spp. (Ustilaginales) is driven by host switches. Sci. Rep 7: 43549 10.1038/srep43549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liti G, Barton DBH, Louis EJ, 2006. Sequence diversity, reproductive isolation and species concepts in Saccharomyces. Genetics. 174(2): 839–850. 10.1534/genetics.106.062166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Wang M, Damm U, Crous PW, Cai L, 2016. Species boundaries in plant pathogenic fungi: A Colletotrichum case study. BMC Evol. Biol 16(81). 10.1186/s12862-016-0649-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, 2008. BEST: Bayesian estimation of species trees under the coalescent model. Bioinformatics. 24(21): 2542–2543. 10.1093/bioinformatics/btn484 [DOI] [PubMed] [Google Scholar]
- Liu L, Edwards SV, 2009. Phylogenetic analysis in the anomaly zone. Syst. Biol 58(4): 452–460. 10.1093/sysbio/syp034 [DOI] [PubMed] [Google Scholar]
- Lombard L, Crous PW, Wingfield BD, Wingfield MJ, 2010. Multigene phylogeny and mating tests reveal three cryptic species related to Calonectria pauciramosa. Stud. Mycol 66: 15–30. 10.3114/sim.2010.66.02 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucking R, Dal-Forno M, Sikaroodi M, Gillevet PM, Bungartz F, Moncada B, Yanez-Ayabaca A, Chaves JL, Coca LF, Lawrey JD, 2014. A single macrolichen constitutes hundreds of unrecognized species. Proc. Natl. Acad. Sci 111(30). 11091–11096. 10.1073/pnas.1403517111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludlow CL, Cromie GA, Garmendia-Torres C, Sirr A, Hays M, Field C, Jeffery EW, Fay JC, Dudley AM, 2016. Independent origins of yeast associated with coffee and cacao fermentation. Curr. Biol 26(7): 965–971. 10.1016/j.cub.2016.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marimon R, Cano J, Gene J, Sutton DA, Kawasaki M, Guarro J, 2007. Sporothrix brasiliensis, S. globosa, and S. mexicana, three new Sporothrix species of clinical interest. J. Clin. Microbiol 45(10): 3198–3206. 10.1128/JCM.00808-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin SH, Dasmahapatra KK, Nadeau NJ, Salazar C, Walters JR, Simpson F, , Blaxter M, Manica A, Mallet J, Jiggins CD, 2013. Genome-wide evidence for speciation with gene flow in Heliconius butterflies. Genome Res. 23(11): 1817–1828. doi: 10.1101/gr.159426.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez D, Berka RM, Henrissat B, Saloheimo M, Arvas M, Baker SE, Chapman J, Chertkov O, Coutinho PM, Cullen D, Danchin EGJ, Grigoriev IV, Harris P, Jackson M, Kubicek CP, Han CS, Ho I, Larrondo LF, De Leon AL, Magnuson JK, Merino S, Misra M, Nelson B, Putnam N, Robbertse B, Salamov AA, Schmoll M, Terry A, Thayer N, Westerholm-Parvinen A, Schoch CL, Yao J, Barbote R, Nelson MA, Detter C, Bruce D, Kuske CR, Xie G, Richardson P, Rokhsar DS, Lucas SM, Rubin EM, Dunn-Coleman N, Ward M, Brettin TS, 2008. Genome sequencing and analysis of the biomass-degrading fungus Trichoderma reesei (syn. Hypocrea jecorina). Nat. Biotechnol 26(5): 553–560. 10.1038/nbt1403 [DOI] [PubMed] [Google Scholar]
- Matute DR, Butler IA, Turissini DA, Coyne JA, 2010. A test of the snowball theory for the rate of evolution of hybrid incompatibilities. Science. 329(5998): 1518–1521. 10.1126/science.1193440. [DOI] [PubMed] [Google Scholar]
- Matute DR, Gavin-Smyth J, 2014. Fine mapping of dominant X-linked incompatibility alleles in Drosophila hybrids. PLoS Genet. 10(4): e1004270. doi: 10.1371/journal.pgen.1004270 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matute DR, McEwen JG, Puccia R, Montes BA, San-Blas G, Bagagli E, Rauscher JT, Restrepo A, Morais F, Nino-Vega G, Taylor JW, 2006. Cryptic speciation and recombination in the fungus Paracoccidioides brasiliensis as revealed by gene genealogies. Mol. Biol. Evol 23(1): 65–73. 10.1093/molbev/msj008 [DOI] [PubMed] [Google Scholar]
- Maxwell CS, Mattox K, Turissini DA, Teixeira MM, Barker BM, Matute DR, 2018a. Gene exchange between two divergent species of the fungal human pathogen, Coccidioides. Evolution. 73(1): 42–58. 10.1111/evo.13643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell CS, Sepúlveda VE, Turissini DA, Goldman WE, Matute DR, 2018b. Recent admixture between species of the fungal pathogen Histoplasma. Evol. Lett 2(3): 210–220. 10.1002/evl3.59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta RS, Bryant D, Rosenberg NA. 2016. The probability of monophyly of a sample of gene lineages on a species tree. Proc. Natl. Acad. Sci July 19;113(29):8002–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendes FK, Hahn MW, 2018. Why concatenation fails near the anomaly zone. Syst. Biol 67(1): 158–169. 10.1093/sysbio/syx063 [DOI] [PubMed] [Google Scholar]
- Millanes AM, Truong C, Westberg M, Diederich P, Wedin M, 2014. Host switching promotes diversity in host-specialized mycoparasitic fungi: Uncoupled evolution in the biatoropsis-usnea system. Evolution. 68(6): 1576–1593. 10.1111/evo.12374 [DOI] [PubMed] [Google Scholar]
- Mirarab S, Reaz R, Bayzid MS, Zimmermann TS Swenson M, Warnow T., 2014. ASTRAL: Genome-scale coalescent-based species tree estimation. Bioinformatics. 30(17): i541–i548. 10.1093/bioinformatics/btu462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra B, Choi YJ, Thines M, 2018. Phylogenomics of Bartheletia paradoxa reveals its basal position in Agaricomycotina and that the early evolutionary history of basidiomycetes was rapid and probably not strictly bifurcating. Mycol. Prog 17(3): 333–341. 10.1007/s11557-017-1349-2 [DOI] [Google Scholar]
- Mohanta TK, Bae H, 2015. The diversity of fungal genome. Biol. Proced. Online. 17:8 10.1186/s12575-015-0020-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina MC, Del-Prado R, Divakar PK, Sanchez-Mata D, Crespo A, 2011. Another example of cryptic diversity in lichen-forming fungi: The new species Parmelia mayi (Ascomycota: Parmeliaceae). Org. Divers. Evol 11(5): 331–342. 10.1007/s13127-011-0060-4 [DOI] [Google Scholar]
- Moyle LC, Nakazato T, 2010. Hybrid incompatibility “Snowballs” between Solanum species. Science. 329(5998): 1521–1523. 10.1126/science.1193063 [DOI] [PubMed] [Google Scholar]
- Muirhead CA, Presgraves DC, 2016. Hybrid incompatibilities, local adaptation, and the genomic distribution of natural introgression between species. Am. Nat 187(2): 249–261. 10.1086/684583 [DOI] [PubMed] [Google Scholar]
- Neafsey DE, Barker BM, Sharpton TJ, Stajich JE, Park DJ, Whiston E, Hung C-Y, McMahan C, White J, Sykes S, Heiman D, Young S, Zeng Q, Abouelleil A, Aftuck L, Bessette D, Brown A, FitzGerald M, Lui A, Macdonald JP, Priest M, Orbach MJ, Galgiani JN, Kirkland TN, Cole GT, Birren BW, Henn MR, Taylor JW, Rounsley SD, 2010. Populatio n genomic sequencing of Coccidioides fungi reveals recent hybridization and transposon control. Genome Res. 20(7): 938–946. 10.1101/gr.103911.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen NH, Landeros F, Garibay-Orijel R, Hansen K, Vellinga EC, 2013. The Helvella lacunosa species complex in western North America: cryptic species, misapplied names and parasites. Mycologia. 105(5): 1275–1286. 10.3852/12-391 [DOI] [PubMed] [Google Scholar]
- Nixon KC, Wheeler QD, 1990. An amplification of the phylogenetic species concept. Cladistics. 6(3): 221–223. 10.1111/j.1096-0031.1990.tb00541.x [DOI] [Google Scholar]
- Nosil P, 2012. Ecological speciation, Ecological Speciation. 10.1093/acprof:osobl/9780199587100.001.0001 [DOI] [Google Scholar]
- O’Donnell K, Kistler HC, Tacke BK, Casper HH, 2000. Gene genealogies reveal global phylogeographic structure and reproductive isolation among lineages of Fusarium graminearum, the fungus causing wheat scab. Proc. Natl. Acad. Sci 97(14): 7905–7910. 10.1073/pnas.130193297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell K, Sutton DA, Fothergill A, McCarthy D, Rinaldi MG, Brandt ME, Zhang N, Geiser DM, 2008. Molecular phylogenetic diversity, multilocus haplotype nomenclature, and in vitro antifungal resistance within the Fusarium solani Species complex. J. Clin. Microbiol 46(8): 2477–2490. 10.1128/JCM.02371-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oono R, Lutzoni F, Arnold AE, Kaye L, U’Ren JM, May G, Carbone I, 2014. Genetic variation in horizontally transmitted fungal endophytes of pine needles reveals population structure in cryptic species. Am. J. Bot 101(8): 1362–1374. 10.3732/ajb.1400141 [DOI] [PubMed] [Google Scholar]
- Orr HA, 2006. Is single-eene speciation possible? Evolution. 45(3): 764–769. 10.2307/2409927 [DOI] [PubMed] [Google Scholar]
- Parnmen S, Rangsiruji A, Mongkolsuk P, Boonpragob K, Nutakki A, Lumbsch HT, 2012. Using phylogenetic and coalescent methods to understand the species diversity in the Cladia aggregata complex (Ascomycota, Lecanorales). PLoS One. 7(12): e52245 10.1371/journal.pone.0052245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payseur BA, Krenz JG, Nachman MW, 2004. Differential patterns of introgression across the X chromosome in a hybrid zone between two species of house mice. Evolution 58(9): 2064–2078. 10.1554/03-738 [DOI] [PubMed] [Google Scholar]
- Pelin A, Selman M, Aris-Brosou S, Farinelli L, Corradi N, 2015. Genome analyses suggest the presence of polyploidy and recent human-driven expansions in eight global populations of the honeybee pathogen Nosema ceranae. Environ. Microbiol 15(11): 4443–4458. 10.1111/1462-2920.12883 [DOI] [PubMed] [Google Scholar]
- Peris D, Langdon QK, Moriarty RV, Sylvester K, Bontrager M, Charron G, Leducq JB, Landry CR, Libkind D, Hittinger CT, 2016. complex ancestries of lager-brewing hybrids were shaped by standing variation in the wild yeast Saccharomyces eubayanus. PLoS Genet. 12(7): e1006155 10.1371/journal.pgen.1006155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrone G, Stea G, Epifani F, Varga J, Frisvad JC, Samson RA, 2011. Aspergillus niger contains the cryptic phylogenetic species A. awamori. Fungal Biol. 115(11): 1138–1150. 10.1016/j.funbio.2011.07.008 [DOI] [PubMed] [Google Scholar]
- Phillips MJ, Delsuc F, Penny D, 2004. Genome-scale phylogeny and the detection of systematic biases. Mol. Biol. Evol 21(7): 1455–1458. 10.1093/molbev/msh137 [DOI] [PubMed] [Google Scholar]
- Piedra-Malagón EM, Sosa V, Ibarra-Manríquez G, 2011. Clinal variation and species boundaries in the Ficus petiolaris Complex (Moraceae). Syst. Bot 31(1): 80–87. 10.1600/036364411X553153 [DOI] [Google Scholar]
- Pino-Bodas R, Burgaz AR, Ahti T, Stenroos S, 2018. Taxonomy of Cladonia angustiloba and related species. Lichenologist. 50(3): 267–282. 10.1017/S002428291800018X [DOI] [Google Scholar]
- Pons J, Barraclough TG, Gomez-Zurita J, Cardoso A, Duran DP, Hazell S, Kamoun S, Sumlin WD, Vogler AP, 2006. Sequence-based species delimitation for the DNA taxonomy of undescribed insects. Syst. Biol 55(4): 595–609. 10.1080/10635150600852011 [DOI] [PubMed] [Google Scholar]
- Powell JR, Monaghan MT, Opik M, Rillig MC, 2011. Evolutionary criteria outperform operational approaches in producing ecologically relevant fungal species inventories. Mol. Ecol 10.1111/j.1365-294X.2010.04964.x [DOI] [PubMed] [Google Scholar]
- Price AL, Tandon A, Patterson N, Barnes KC, Rafaels N, Ruczinski I, Beaty TH, Mathias R, Reich D, Myers S, 2009. Sensitive detection of chromosomal segments of distinct ancestry in admixed populations. PLoS Genet. 5(6): e1000519. doi: 10.1371/journal.pgen.1000519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pringle A, Baker DM, Platt JL, Wares JP, Latge JP, Taylor JW, 2005. Cryptic speciation in the cosmopolitan and clonal human pathogenic fungus Aspergillus fumigatus. Evolution. 59(9): 1886–1899. 10.1111/j.0014-3820.2005.tb01059.x [DOI] [PubMed] [Google Scholar]
- Ramani R, Chaturvedi V, 2007. Antifungal susceptibility profiles of Coccidioides immitis and Coccidioides posadasii from endemic and non-endemic areas. Mycopathologia 163: 315–319. 10.1007/s11046-007-9018-7. [DOI] [PubMed] [Google Scholar]
- Rehner SA, Buckley E, 2005. A Beauveria phylogeny inferred from nuclear ITS and EF1-a sequences: evidence for cryptic diversification and links to Cordyceps teleomorphs. Mycologia. 97(1): 84–98. 10.1080/15572536.2006.11832842 [DOI] [PubMed] [Google Scholar]
- Renard D, Valle G Della, Eds, Y P., Royal T., 2003. Species concepts and phylogenetic theory: a debate. Edited by Wheeler Q, and Meier R Columbia University Press. [Google Scholar]
- Restrepo S, Tabima JF, Mideros MF, GrOnwald NJ, Matute DR, 2014. Speciation in fungal and Oomycete plant pathogens. Annu. Rev. Phytopathol. 52(1): 289–316. 10.1146/annurev-phyto-102313-050056 [DOI] [PubMed] [Google Scholar]
- Richards PM, Morii Y, Kimura K, Hirano T, Chiba S, Davison A, 2017. Single-gene speciation: Mating and gene flow between mirror-image snails. Evol. Lett 1(6): 282–291. 10.1002/evl3.31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roch S, Steel M, 2015. Likelihood-based tree reconstruction on a concatenation of aligned sequence data sets can be statistically inconsistent. Theor. Popul. Biol 100: 56–62. 10.1016/j.tpb.2014.12.005 [DOI] [PubMed] [Google Scholar]
- Rokas A, Williams BI, King N, Carroll SB, 2003. Genome-scale approaches to resolving incongruence in molecular phylogenies. Nature. 425: 798–804. 10.1038/nature02053 [DOI] [PubMed] [Google Scholar]
- Rosenberg NA 2003. The shapes of neutral gene genealogies in two species: Probabilities of monophyly, paraphyly, and polyphyly in a coalescent model. Evolution 57(7): 1465–1477. [DOI] [PubMed] [Google Scholar]
- Roux C, Fra’isse C, Romiguier J, Anciaux Y, Galtier N, Bierne N, 2016. Shedding light on the grey zone of speciation along a continuum of genomic divergence. PLoS Biol. 14(12): e2000234 10.1371/journal.pbio.2000234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saag L, Mark K, Saa A, Randlane T, 2014. Species delimitation in the lichenized fungal genus Vulpicida (Parmeliaceae, Ascomycota) using gene concatenation and coalescent-based species tree approaches. Am. J. Bot 101(12): 2169–2182. 10.3732/ajb.1400439 [DOI] [PubMed] [Google Scholar]
- Sadowska-Deś AD, Dal Grande F, Lumbsch HT, Beck A, Otte J, Hur JS, Kim JA, Schmitt I, 2014. Integrating coalescent and phylogenetic approaches to delimit species in the lichen photobiont Trebouxia. Mol. Phylogenet. Evol 76: 202–210. 10.1016/j.ympev.2014.03.020 [DOI] [PubMed] [Google Scholar]
- Salgado-Salazar C, Rossman AY, Chaverri P, 2013. Not as ubiquitous as we thought: taxonomic crypsis, hidden diversity and cryptic speciation in the cosmopolitan fungus Thelonectria discophora (Nectriaceae, Hypocreales, Ascomycota). PLoS One. 8(10): e76737 10.1371/journal.pone.0076737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuels GJ, Ismaiel A, Bon M-C, De Respinis S, Petrini O, 2010. Trichoderma asperellum sensu lato consists of two cryptic species. Mycologia. 102(4): 944–966. 10.3852/09-243 [DOI] [PubMed] [Google Scholar]
- Sanchez-Ramírez S, Tulloss RE, Guzmán-Dávalos L, Cifuentes-Blanco J, Valenzuela R, Estrada-Torres A, Ruan-Soto F, Diaz-Moreno R, Hernandez-Rico N, Torres-Gómez M, León H, Moncalvo JM, 2015. In and out of refugia: Historical patterns of diversity and demography in the North American Caesar’s mushroom species complex. Mol. Ecol 24(23): 5938–5956. 10.1111/mec.13413 [DOI] [PubMed] [Google Scholar]
- Sanderson MJ, 1995. Objections to bootstrapping phylogenies: A critique. Syst. Biol 10.1093/sysbio/44.3.299 [DOI] [Google Scholar]
- Sato H, Murakami N, 2008. Reproductive isolation among cryptic species in the ectomycorrhizal genus Strobilomyces: Population-level CAPS marker-based genetic analysis. Mol. Phylogenet. Evol 48: 326–334. 10.1016/j.ympev.2008.01.033 [DOI] [PubMed] [Google Scholar]
- Sato H, Yumoto T, Murakami N, 2007. Cryptic species and host specificity in the ectomycorrhizal genus Strobilomyces (Strobilomycetaceae). Am. J. Bot 94(10): 1630–1641. 10.3732/ajb.94.10.1630. [DOI] [PubMed] [Google Scholar]
- Savary R, Masclaux FG, Wyss T, Droh G, Cruz Corella J, Machado AP,Morton JB, Sanders IR, 2017. A population genomics approach shows widespread geographical distribution of cryptic genomic forms of the symbiotic fungus Rhizophagus irregularis. ISME J. 12(1): 17–30. doi: 10.1038/ismej.2017.153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayyari E, Mirarab S, 2016. Fast coalescent-based computation of local branch support from quartet frequencies. Mol. Biol. Evol 33(7): 1654–1668. 10.1093/molbev/msw079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schardl CL, Craven KD, 2003. Interspecific hybridization in plant-associated fungi and oomycetes: A review. Mol. Ecol 12(11): 2861–2873. 10.1046/j.1365-294X.2003.01965.x [DOI] [PubMed] [Google Scholar]
- Seixas I, Barbosa C, Mendes-Faia A, GQldener U, Tenreiro R, Mendes-Ferreira A, Mira NP, 2019. Genome sequence of the non-conventional wine yeast Hanseniaspora guilliermondii UTAD222 unveils relevant traits of this species and of the Hanseniaspora genus in the context of wine fermentation. DNA Res. 26(1): 67–83. 10.1093/dnares/dsy039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sepúlveda VE, Marquez R, Turissini DA, Goldman WE, Matute DR, 2017. Genome sequences reveal cryptic speciation in the human pathogen Histoplasma capsulatum. mBio. 8:e01339–17. 10.1128/mBio.01339-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sepúlveda VE, Williams CL, Goldman WE, 2014. Comparison of phylogenetically distinct Histoplasma strains reveals evolutionarily divergent virulence strategies. mBio. 5(4):e01376–14. doi: 10.1128/mBio.01376-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharpton TJ, Stajich JE, Rounsley SD, Gardner MJ, Wortman JR, Jordar VS, Maiti R, Kodira CD, Neafsey DE, Zeng Q, Hung C-Y, McMahan C, Muszewska A, Grynberg M, Mandel MA, Kellner EM, Barker BM, Galgiani JN, Orbach MJ, Kirkland TN, Cole GT, Henn MR, Birren BW, Taylor JW, 2009. Comparative genomic analyses of the human fungal pathogens Coccidioides and their relatives. Genome Res. 19(10): 1722–1731. doi: 10.1101/gr.087551.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva DN, Varzea V, Paulo OS, Batista D, 2018. Population genomic footprints of host adaptation, introgression and recombination in coffee leaf rust. Mol. Plant Pathol 19(7): 1742–1753. 10.1111/mpp.12657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh G, Aptroot A, Rico VJ, Otte J, Divakar PK, Crespo A, Cáceres ME da 5, Lumbsch HT, Schmitt I, 2018. Neoprotoparmelia gen. nov. and Maronina (Lecanorales, Protoparmelioideae): species description and generic delimitation using DNA barcodes and phenotypical characters. MycoKeys. 10.3897/mycokeys.44.29904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh G, Dal Grande F, Divakar PK, Otte J, Crespo A, Schmitt I, 2017. Fungal-algal association patterns in lichen symbiosis linked to macroclimate. New Phytol.214(1): 317–329. 10.1111/nph.14366. [DOI] [PubMed] [Google Scholar]
- Singh G, Dal Grande F, Divakar PK, Otte J, Leavitt SD, Szczepanska K,Crespo A, Rico VJ, Aptroot A, Da Silva Cáceres ME, Lumbsch HT, Schmitt I., 2015. Coalescent-based species delimitation approach uncovers high cryptic diversity in the cosmopolitan lichen-forming fungal genus Protoparmelia (Lecanorales, Ascomycota). PLoS One. 10(5): e0124625. 10.1371/journal.pone.0124625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sites JW, Marshall JC, 2003. Delimiting species: a Renaissance issue in systematic biology. Trends Ecol. Evol 18(9): 462–470. 10.1016/S0169-5347(03)00184-8 [DOI] [Google Scholar]
- Sklenář F, Jurjević, Zalar P, Frisvad JC, Visagie CM, Kolařík M, Floubraken J, Chen AJ, Yilmaz N, Seifert KA, Coton M, Déniel F, Gunde-Cimerman N, Samson RA, Peterson SW, Flubka V, 2017. Phylogeny of Xerophilic aspergilli (subgenus Aspergillus) and taxonomic revision of section Restrict!. Stud. Mycol 88: 161–236. 10.1016/j.simyco.2017.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobel JM, Chen GF, Watt LR, Schemske DW, 2010. The biology of speciation. Evolution.64(2): 295–315. 10.1111/j.1558-5646.2009.00877.X [DOI] [PubMed] [Google Scholar]
- Sokal RR, Crovello TJ, 1970. The biological species concept: a critical evaluation. Am. Nat 104(936). 10.1086/282646 [DOI] [Google Scholar]
- Soto Medina E, Prieto M, Wedin M, 2018. A new Bunodophoron species (Sphaerophoraceae, Lecanorales) from the Neotropics. Lichenologist. 50(3): 255–266. 10.1017/S0024282917000743 [DOI] [Google Scholar]
- Starkey DE, Ward TJ, Aoki T, Gale LR, Kistler HC, Geiser DM, Suga H, Toth B, Varga J, O’Donnell K, 2007. Global molecular surveillance reveals novel Fusarium head blight species and trichothecene toxin diversity. Fungal Genet. Biol 44(11): 1191–1204. 10.1016/j.fgb.2007.03.001 [DOI] [PubMed] [Google Scholar]
- Steenkamp ET, Wingfield BD, Desjardins AE, Marasas WFO, Wingfield MJ, 2002. Cryptic speciation in Fusarium subglutinans. Mycologia. 94(6): 1032–1043. 10.1080/15572536.2003.11833158. [DOI] [PubMed] [Google Scholar]
- Sukumaran J, Knowles LL, 2017. Multispecies coalescent delimits structure, not species. Proc. Natl. Acad. Sci. U. S. A 114(17): 1607–1612. 10.1073/pnas.1607921114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Corcoran P, Menkis A, Whittle CA, Andersson SGE, Johannesson H, 2012. Large-scale introgression shapes the evolution of the mating-type chromosomes of the filamentous Ascomycete Neurospora tetrasperma. PLoS Genet. 8(7): e1002820. doi: 10.1371/journal.pgen.1002820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Susko E, 2009. Bootstrap support is not first-order correct. Syst. Biol 58(2): 211–223. 10.1093/sysbio/syp016 [DOI] [PubMed] [Google Scholar]
- Svantesson S, Larsson K-H, Kõljalg U, May TW, Cangren P, Nilsson RH, Larsson E, 2019. Solving the taxonomic identity of Pseudotomentella tristis s.l. (Thelephorales, Basidiomycota) - a multi-gene phylogeny and taxonomic review, integrating ecological and geographical data. MycoKeys. 50: 1–77. 10.3897/mycokeys.50.32432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor JW, 2006. Evolution of human-pathogenic fungi: phylogenies and species, in: molecular principles of fungal pathogenesis. American Society of Microbiology, pp. 113–132. [Google Scholar]
- Taylor JW, Fisher MC, 2003. Fungal multilocus sequence typing - It’s not just for bacteria. Curr. Opin. Microbiol 6(4): 351–356. 10.1016/S1369-5274(03)00088-2 [DOI] [PubMed] [Google Scholar]
- Taylor JW, Jacobson DJ, Kroken S, Kasuga T, Geiser DM, Hibbett DS, Fisher MC, 2000. Phylogenetic species recognition and species concepts in fungi. Fungal Genet. Biol, 10.1006/fgbi.2000.1228 [DOI] [PubMed] [Google Scholar]
- Teixeira MM, Theodoro RC, De Oliveira FFM, Machado GC, Hahn RC, Bagagli E, San-Blas G, Felipe MSS, 2015. Paracoccidioides lutzii sp. nov.: biological and clinical implications. J. Music Ther 52(1): 19–28. 10.3109/13693786.2013.794311. [DOI] [PubMed] [Google Scholar]
- Teixeira MM, Theodoro RC, Nino-Vega G, Bagagli E, Felipe MSS, 2014. Paracoccidioides species complex: ecology, phylogeny, sexual reproduction, and virulence. PLoS Pathog. 10(10): e1004397 10.1371/journal.ppat.1004397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turissini DA, Gomez OM, Teixeira MM, McEwen JG, Matute DR, 2017. Species boundaries in the human pathogen Paracoccidioides. Fungal Genet. Biol 106: 925 10.1016/j.fgb.2017.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turissini DA, Matute DR, 2017. Fine scale mapping of genomic introgres sions within the Drosophila yakuba clade. PLoS Genet. 13(9): e1006971 10.1371/journal.pgen.1006971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udayanga D, Castlebury LA, Rossman AY, Chukeatirote E, Hyde KD, 2014. Insights into the genus Diaporthe: phylogenetic species delimitation in the D. eres species complex. Fungal Divers. 67(1): 203–229. 10.1007/s13225-014-0297-2 [DOI] [Google Scholar]
- Vaghefi N, Kikkert JR, Hay FS, Carver GD, Koenick LB, Bolton MD, Hanson LE, Secor GA, Pethybridge SJ, 2018. Cryptic diversity, pathogenicity, and evolutionary species boundaries in Cercospora populations associated with Cercospora leaf spot of Beta vulgaris. Fungal Biol. 122(4): 264–282. 10.1016/j.funbio.2018.01.008 [DOI] [PubMed] [Google Scholar]
- Van Niekerk JM, Crous PW, Groenewald JZ, Fourie PH, Halleen F, 2004. DNA phylogeny, morphology and pathogenicity of Botryosphaeria species on grapevines. Mycologia. 96(4): 781–298. 10.1080/15572536.2005.11832926 [DOI] [PubMed] [Google Scholar]
- Vieira A, Silva DN, Varzea V, Paulo OS, Batista D, 2018. Novel insights on colonization routes and evolutionary potential of Colletotrichum kahawae, a severe pathogen of Coffea arabica. Mol. Plant Pathol 19(11): 2488–2501. 10.1111/mpp.12726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker A-S, Gautier A, Confais J, Martinho D, Viaud M, Le Pecheur P, Dupont J, Fournier E, 2011. Botrytis pseudocinerea, a new cryptic species causing gray mold in french vineyards in sympatry with Botrytis cinerea. Phytopathology. 101(12): 1433–1445. 10.1094/PHYT0-04-11-0104 [DOI] [PubMed] [Google Scholar]
- Wang RJ, White MA, Payseur BA, 2015. The Pace of hybrid incompatibility evolution in house mice. Genetics 201,229-242 201(1): 229–242. 10.1534/genetics.115.179499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X-H, Huhtinen S, Hansen K, 2016. Multilocus phylogenetic and coalescent-based methods reveal dilemma in generic limits, cryptic species, and a prevalent intercontinental disjunct distribution in Geopyxis (Pyronemataceae s. l., Pezizomycetes). Mycologia. 108(6): 1189–1215. 10.3852/16-100 [DOI] [PubMed] [Google Scholar]
- Wheeler QD, Platnick NI, 1989. The phylogenetic species concept and phylogenetic theory: a debate. Columbia University Press. [Google Scholar]
- Whitehead MR, Catullo RA, Ruibal M, Dixon KW, Peakall R, Linde CC, 2017. Evaluating multilocus Bayesian species delimitation for discovery of cryptic mycorrhizal diversity. Fungal Ecol. 26: 74–84. 10.1016/j.funeco.2016.11.009 [DOI] [Google Scholar]
- Yang Z, 2015a. The BPP program for species tree estimation and species delimitation. Curr. Zool 61(5): 854–865. 10.1093/czoolo/61.5.854 [DOI] [Google Scholar]
- Yang Z, 2015b. A tutorial of BPP for species tree estimation and species delimitation. Curr. Zool 10.1093/czoolo/61.5.854 [DOI] [Google Scholar]
- Yang Z, Rannala B, 2010. Bayesian species delimitation using multilocus sequence data. Proc. Natl. Acad. Sci 10.1073/pnas.0913022107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Ogilvie HA, Drummond AJ, Stadler T, 2018. Bayesian inference of species networks from multilocus sequence data. Mol. Biol. Evol 35(2): 504–517. 10.1093/molbev/msx307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Kapli P, Pavlidis P, Stamatakis A, 2013. A general species de limitation method with applications to phylogenetic placements. Bioinformatics. 29(22): 2869–2876. 10.1093/bioinformatics/btt499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Zhang X, Li K, Wang C, Cai L, Zhuang W, Xiang M, Liu X, 2018. Introgression and gene family contraction drive the evolution of lifestyle and host shifts of hypocrealean fungi. Mycology. 9(3): 176–188. 10.1080/21501203.2018.1478333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao P, Liu F, Li YM, Cai L, 2016. Inferring phylogeny and speciation of Gymnosporangium species, and their coevolution with host plants. Sci. Rep 6: 29339 10.1038/srep29339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao P, Wang QH, Tian CM, Kakishima M, 2015. Integrating a numerical taxonomic method and molecular phylogeny for species delimitation of Melampsora species (Melampsoraceae, Pucciniales) on willows in China. PLoS One. 10(12): e0144883 10.1371/journal.pone.0144883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Fernández-Brime S, Wedin M, Locke M, Leavitt SD, Lumbsch HT, 2017. Using multi-locus sequence data for addressing species boundaries in commonly accepted lichen-forming fungal species. Org. Divers. Evol 17(2): 351–363. 10.1007/s13127-016-0320-4 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


