Abstract
Study Objective
Identify small molecule biomarkers of insufficient sleep using untargeted plasma metabolomics in humans undergoing experimental insufficient sleep.
Methods
We conducted a crossover laboratory study where 16 normal-weight participants (eight men; age 22 ± 5 years; body mass index < 25 kg/m2) completed three baseline days (9 hours sleep opportunity per night) followed by 5-day insufficient (5 hours sleep opportunity per night) and adequate (9 hours sleep opportunity per night) sleep conditions. Energy balanced diets were provided during baseline, with ad libitum energy intake provided during the insufficient and adequate sleep conditions. Untargeted plasma metabolomics analyses were performed using blood samples collected every 4 hours across the final 24 hours of each condition. Biomarker models were developed using logistic regression and linear support vector machine (SVM) algorithms.
Results
The top-performing biomarker model was developed by linear SVM modeling, consisted of 65 compounds, and discriminated insufficient versus adequate sleep with 74% overall accuracy and a Matthew’s Correlation Coefficient of 0.39. The compounds in the top-performing biomarker model were associated with ATP Binding Cassette Transporters in Lipid Homeostasis, Phospholipid Metabolic Process, Plasma Lipoprotein Remodeling, and sphingolipid metabolism.
Conclusion
We identified potential metabolomics-based biomarkers of insufficient sleep in humans. Although our current biomarkers require further development and validation using independent cohorts, they have potential to advance our understanding of the negative consequences of insufficient sleep, improve diagnosis of poor sleep health, and could eventually help identify targets for countermeasures designed to mitigate the negative health consequences of insufficient sleep.
Keywords: sleep loss, sleep restriction, biomarker, metabolomics, sleep deprivation, personalized medicine, lipid metabolism, sex differences
Statement of Significance.
Insufficient sleep is associated with negative health outcomes, including cardiometabolic and neurodegenerative disorders, impaired cognition, and risk of accidents. Clinical biomarkers that can distinguish individuals between adequate versus insufficient sleep have not been identified. Such biomarkers could improve the diagnosis of poor sleep health and inform targets for countermeasures and personalized medicine approaches designed to mitigate the negative health consequences associated with insufficient sleep. Our findings demonstrate that mass spectrometry is a viable approach for identifying candidate biomarkers of insufficient sleep in humans using untargeted plasma metabolomics.
Introduction
The Sleep Research Society and American Academy of Sleep Medicine jointly recommend adults aged 18–60 years regularly obtain 7 hours or more of sleep per night to promote optimal health [1, 2]. However, insufficient sleep is widespread with estimates indicating that ~35% of American adults sleep less than the recommended 7 hours per night and 30% sleep less than 6 hours per night [3–5]. Furthermore, estimates show active military personnel sleep on average 5.5–6.5 hours per night [6, 7]. Findings from cross-sectional, case-control, prospective cohort, and laboratory-controlled clinical translational studies consistently show insufficient sleep is associated with dysregulated physiology and risk of cardiometabolic and neurodegenerative disorders including obesity, diabetes, coronary heart disease, stroke, metabolic syndrome, and Alzheimer’s disease [8–20]. Furthermore, insufficient sleep results in impaired cognition and increased risk of accidents [21–24].
There is a high prevalence of sleep disorders, including insufficient sleep syndrome [25], that are often undiagnosed, in part due to the lack of established clinical biomarkers to discriminate adequate versus insufficient sleep within an individual. Thus, the Sleep Research Society and the National Institutes of Health have recognized an immediate need and potential long-term benefits of identifying objective, point-of-care biomarkers of insufficient sleep [26, 27]. Identifying biomarkers of insufficient sleep would provide a tool that has potential to improve the ability of primary care providers to diagnose insufficient sleep, provide insight into mechanisms underlying the adverse health consequences associated with insufficient sleep, inform novel sleep countermeasures, and support the development of personalized sleep medicine by identifying individuals most likely to benefit from sleep health-based countermeasures.
One approach to identify biomarkers of insufficient sleep is to use omics-based analyses such as transcriptomics, proteomics, or metabolomics. Findings from previous studies have characterized changes in the human blood transcriptome, proteome, and metabolome during insufficient sleep, total sleep deprivation, early versus late sleep timing, and circadian misalignment [28–38]. More specific to developing diagnostic biomarkers, Weljie et al. used metabolomics in rats and humans to identify oxalic acid and diacylglycerol (DAG)-[36:3] as potential single compound biomarkers of sleep debt [39], and Laing et al. used transcriptomics to identify and test the performance of biomarkers of insufficient sleep in humans [40]. Additionally, activity and mRNA levels of salivary amylase increase in response to 28 hours of total sleep deprivation in humans, indicating these amylase metrics are potential biomarkers of extended wakefulness [41]. Still, the field is in the discovery phase of identifying clinically useful biomarkers of insufficient sleep in humans. An optimal biomarker of insufficient sleep should have high sensitivity and specificity, low intra-individual variability, be easily interpreted by clinicians and patients, and be robust to physiological variation such as age, sex, and body mass index. We used untargeted metabolomics to analyze plasma samples from a crossover 14- to 15-day counterbalanced in-laboratory protocol with the goal of identifying and testing biomarkers of insufficient sleep.
Methods
Participants
All procedures were approved by the scientific and advisory review committee of the Colorado Clinical and Translational Sciences Institute, the Colorado Multiple Institutional Review Board (IRB), and the University of Colorado Boulder IRB, and all participants provided written informed consent prior to study procedures. Data were collected in a previously described study (Figure 1) [14, 17]. Sixteen healthy participants (eight men) aged 22 ± 5 years (mean ± SD), with normal body mass index (BMI) 22.9 ± 2.4 kg/m2 completed the study. Each participant completed medical, psychological, and sleep history screening procedures, and polysomnographic clinical sleep disorders screening as previously described [17]. Participants were considered free of medical and psychological disorders based on these screening procedures. Breath alcohol testing and urine toxicology verified drug-free status upon laboratory admission.
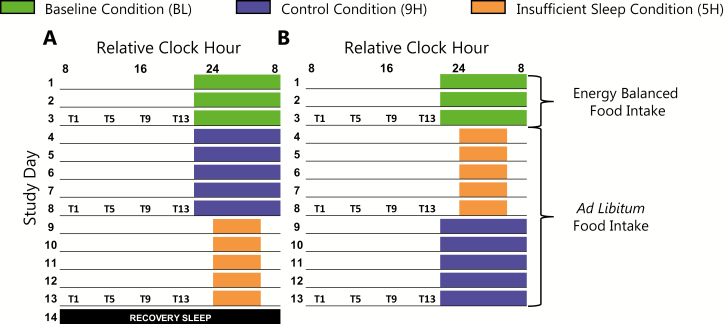
Figure 1.
Experimental protocol. (A; n = 8) condition order A. (B; n = 8) condition order B. Underlines represent scheduled wakefulness and colored boxes represent scheduled sleep. A recovery sleep opportunity (black box) was provided on study day 14 prior to discharge from the laboratory for participants in condition order A. Time of day is plotted as relative clock hour with scheduled waketime arbitrarily assigned to 08:00 am for the baseline and 9H conditions. All other times and protocol events are referenced to this arbitrarily assigned value (e.g. blood draws occurring 1 hours after scheduled waketime during the baseline condition are reported as occurring at 09:00 am). Actual sleep timing and protocol events were based on habitual sleep schedules of each individual participant, thus all participants were studied at their habitual sleep and circadian phase for baseline. Blood collection for untargeted metabolomics occurred on study days 3, 8, and 13, represented by T1, T5, T9, and T13 (note that T17 and T21 blood collections are not represented on the figure as they were not used for biomarker analyses).
Protocol
Participants completed a 7-day home ambulatory monitoring segment with ~9 hours nightly sleep schedules immediately prior to the 14- to 15-day in-laboratory protocol [17]. In-laboratory baseline (BL) assessments consisted of 3 days with 9 hours sleep opportunities with sleep and wake times aligned to individual habitual sleep schedules. Participants then completed two conditions (9H; 9 hours per night sleep opportunities, and 5H; 5 hours per night sleep opportunities) in a crossover counterbalanced design (Figure 1). Each condition lasted 5 days. Men and women were randomized to condition order separately to ensure equal sex allocation to each condition order. All protocol events were scheduled relative to individual participant’s habitual sleep and waketimes. Participants were provided energy-balanced diets during BL, but energy intake was ad libitum during the 9H and 5H conditions [17]. During ad libitum energy intake, registered dietitians weighed all food provided to participants and then weighed-back all uneaten food to facilitate precise measurement of energy intake. Blood was collected every 4 hours across 24 hours (T1, T5, T9, T13, T17, and T21), starting 1 hour after scheduled waketime on study days 3, 8, and 13 (the final day of each condition; Figure 1). T1 blood samples were collected after an overnight fast, whereas the T5, T9, and T13 samples were collected in a non-fasted state. Samples obtained during scheduled sleep (T17 and T21) were excluded from biomarker model analyses.
Metabolomics
Plasma sample preparation and untargeted metabolomics was conducted as previously described [42–44]. Prior to biomarker model analyses, metabolomics data were extracted and processed, including batch correction (Supplementary Figure S1), per standard procedures in the Reisdorph laboratory [42–44], as detailed in the Supplementary Material. In total, 4114 compounds were detected and analyzed. Prior to tandem mass spectrometry (MS/MS), the metabolomics software Mass Profiler Professional (v.B.14.5; Agilent Technologies, Inc.) [45–47] was used to putatively annotate compounds with common chemical names using isotope ratios and an error window of <10 ppm. This software utilizes an in-house database comprising data from METLIN, Human Metabolome Database (HMDB), Kyoto Encyclopedia of Genes and Genomes (KEGG), Lipid Maps, and data from an authentic standards database to match masses from unnamed compounds to database entries.
Biomarker models
During ad libitum energy intake in the 9H and 5H conditions, participants consumed more energy than needed to maintain body weight [17], consistent with findings from other studies of insufficient sleep with ad libitum energy intake [13, 48–50]. To increase the specificity of our biomarker models for insufficient sleep, the compounds most impacted by this increased energy intake were excluded from all biomarker models. To identify the compounds most impacted by increased energy intake we exclusively analyzed samples in 9H versus BL, within condition order A, to remove the potential confounding effects of increased energy intake combined with insufficient sleep that occurred in 5H. Within the 9H versus BL comparison, compounds with an area under the receiver operator characteristic plot (AUROC) ≥ 0.70, detected using the biomarker module in Metaboanalyst [51], were excluded from all biomarker models—95 compounds met this criterion. Of these 95 compounds, three (propionylcarnitine, phosphatidylcholine(PC)-[36:3], and sphingomyelin(SM)-[d34:1]) were previously found to be impacted by insufficient sleep or total sleep deprivation [30, 33, 39].
Because findings from untargeted metabolomics studies show ~15%–20% of the human plasma metabolome is circadian regulated [32, 34, 35], and nearly half of the metabolites in a targeted analysis showed 24-hour time-of-day patterns [29], we trained biomarker models using two planned datasets, one that included and one that a priori excluded compounds with 24-hour time-of-day patterns. We also performed an exploratory analysis using only the compounds with 24-hour time-of-day patterns (Supplementary Table S1). MetaCycle [52] (v1.1.0) for R [53] (v3.5.3) was used to identify compounds with 24-hour time-of-day patterns in at least one condition. LOG2 normalized metabolomics data from the BL, 9H, and 5H conditions, using the T1, T5, T9, T13, T17, and T21 samples, were loaded separately into MetaCycle and the “meta2d” function was used with “cycMethod” set to “JTK” and “LS.” The resulting p-values for each compound for each condition were combined using the minP method [54]. Statistical significance for detecting a 24 hours time-of-day pattern was set at the 10% False Discovery Rate corrected level, similar to our previous proteomics findings [28]. Based on these analyses, we identified 985 compounds with 24-hour time-of-day patterns in at least one condition.
For all biomarker models, plasma samples were divided into training and validation datasets. After filtering and QC analyses, 153 plasma samples were included in the final biomarker model analyses. Within the BL, 9H, and 5H conditions, the plasma samples were randomly divided into training (2/3 samples) and validation (1/3 samples) datasets (Supplementary Figures S2 and S3), balanced by time-points, using the caret [55] package (v6.0) for R (v3.5.3). We assigned 2/3 samples to the training dataset, as opposed to dividing the samples evenly between the training and validation datasets, to maintain higher statistical power in the training dataset during the discovery phase of our biomarker analyses. The training dataset consisted of 104 samples with 57 from men and 47 from women. The validation dataset consisted of 49 plasma samples with 20 from men and 29 from women. Because participants had 9 hours per night sleep opportunities in the BL and 9H conditions, samples from BL and 9H were defined as adequate sleep and 5H samples defined as insufficient sleep.
Compound selection for biomarker models was conducted using least absolute shrinkage and selection operator (LASSO) in the GLMNET [56] package (v2.0–16) for R (v3.5.3), and included annotated and unannotated compounds. Biomarker models were trained individually on datasets that included and excluded the 985 compounds with 24-hour time-of-day patterns using the “cv.glmnet” function with “nfolds” set to the number of samples for leave one sample out cross-validation (LOOCV), “type.measure” set to “mae,” “family” set to “binomial” for logistic regression, “alpha” set to “1.0” for LASSO, and “nlambda” set to “500.” Lambda requires tuning; thus, the mean value of lambda across 500 searches that provided the minimum cross-validated error within the training set was defined as the optimal lambda value for each final model. The predicted class (e.g. adequate sleep) in the validation datasets was determined using the “predict” function with “type” set to “class” and “s” set to the optimal lambda, additionally, the logistic regression linear predictor was determined using the “predict” function with “type” set to “link” and “s” set to the optimal lambda.
We also developed linear support vector machine (SVM) biomarker models using the e1071 package (v1.7-0.1) [27] for R (v3.5.3). SVM models were developed using subsets of the compounds identified by LASSO. Specifically, compound subsets for training the SVM models were created using the “LASSO frequency” function within the Biomarker module in MetaboAnalyst [51] that provides the frequency of LASSO compound selection using Monte Carlo cross-validation. SVM models were trained on the resulting subsets consisting of compounds with ≥100%, 90%, and 80% LASSO selection frequencies. The optimal compound subset for each SVM model was identified based on the compound subset that produced the highest Matthew’s Correlation Coefficient [57, 58] (MCC) in the validation dataset using the insufficient and adequate sleep classifications. MCC is invariant to the balancing of samples between classes [57] and ranges from −1 to 1, with −1 representing perfect negative correlation (all samples misclassified), and 1 representing perfect positive correlation (all samples correctly classified). Thus, MCC was a priori selected as the primary performance metric of each biomarker model. Biomarker models were trained individually on datasets that included and excluded compounds with 24-hour time-of-day patterns using the “svm” function with “cross” set to the number of samples in the training set for LOOCV, “kernel” set to “linear,” “scale” set to “FALSE,” and “type” set to “C-classification.” Additionally, the argument “class.weights” was set to the inverse of the number of samples in the insufficient and adequate sleep classifications in the training dataset to account for the larger number of adequate sleep samples. The “cost” function for SVM requires tuning. The “tune.svm” function was used to identify the optimal “cost” value for each SVM model with “scale” set to “FALSE,” “cost” set to the range 2–10 to 22, “cross” set to “10” for 10-fold cross-validation, “class.weights” set to the inverse of the number of samples for insufficient and adequate sleep classifications in the training dataset as described above, and “kernel” set to “linear.” Ten-fold cross-validation was used to tune the “cost” parameter, and the tuning procedure was repeated 25 times for each SVM model separately. The mode of the optimal “cost” value from the 25 tuning repetitions was selected as the final “cost” value for each SVM model. The predicted class and SVM decision values for the validation datasets were determined using the “predict” function with “decision.values” set to “TRUE” and “scale” set to “FALSE.”
Biomarker model assessments
Performance of the final biomarker models was assessed using LOOCV in the training dataset and by internal validation (IV) in the validation dataset, similar to Laing et al. [40]. For LOOCV and IV the positive classification was insufficient sleep and the negative classification was adequate sleep. For all classification comparisons for each model, we calculated the number of true positive samples (TP), true negative samples (TN), false-positive samples (FP), and false-negative samples (FN). Overall accuracy was calculated as [TP + TN/(TP + TN + FP + FN)], sensitivity as [TP/(TP + FN)], specificity as [TN/(TN + FP)], and MCC as [(TP·TN) – (FP·FN)]/√[(TP + FP)·(TP + FN)·(TN + FP)·(TN + FN)]. We calculated AUROC for each classification comparison for all models using OriginPro (OriginLab Corporation). To test the previously published metabolomics-based biomarkers of insufficient sleep [39], a one-tailed dependent t-test was used to test the targeted statistical analysis of DAG-[36:3]. Oxalic acid was not annotated in our dataset and thus was not tested.
Pathway and functional analyses
Compound annotations for each biomarker model were converted to Human Metabolome Database [59] (HMDB, v4.0) IDs and searched using IMPaLA [60] to match compounds to relevant biochemical pathways in the KEGG [61], Reactome [62], and Small Molecule Pathway Database [63]. Within IMPaLA, over-representation pathway analysis was conducted with statistical significance set at the 10% FDR corrected level for individual pathways. We also developed a gene-metabolite network to further assess potential biochemical functions associated with the top-performing biomarker model, using the Network Explorer module in Metaboanalyst [51], as detailed in the Supplementary Material.
Results
Planned analyses focused on four biomarker models of insufficient sleep, two that include compounds with and without 24-hour time-of-day patterns and two that exclude compounds with 24-hour time-of-day patterns (Table 1). Collectively, there are 88 compounds in these four biomarker models, with 33 compounds overlapping across the four models (Supplementary Table S2). LOOCV accuracy from the training dataset was higher than IV accuracy from the validation dataset for all four models (Table 1). The logistic regression and SVM models that excluded compounds with 24-hour time-of-day patterns had the highest IV accuracy at 74% (Table 1). The SVM model that excluded compounds with 24-hour time-of-day patterns had the highest IV sensitivity at 60%, whereas all other models had IV sensitivity of 47% (Table 1). The logistic regression model that excluded compounds with 24-hour time-of-day patterns had the highest IV specificity at 85%, whereas all other models ranged from 77% to 82% (Table 1). The SVM model that excluded compounds with 24-hour time-of-day patterns had the highest IV MCC at 0.39, whereas all other models ranged from 0.23 to 0.34 (Table 1).
Table 1.
Overall performance metrics of biomarker models
| LOOCV ACC | IV ACC | IV Sensitivity | IV Specificity | IV MCC | |
|---|---|---|---|---|---|
| Compounds with and without 24-hour time-of-day patterns included | |||||
| Logistic Regression (66) | 100% | 71% | 47% | 82% | 0.30 |
| Linear SVM (47) | 97% | 67% | 47% | 77% | 0.23 |
| Compounds with 24-hour time-of-day patterns excluded | |||||
| Logistic Regression (65) | 100% | 74% | 47% | 85% | 0.34 |
| Linear SVM (65) | 95% | 74% | 60% | 79% | 0.39 |
Numbers in parenthesis indicate total number of compounds in each biomarker model. ACC, accuracy.
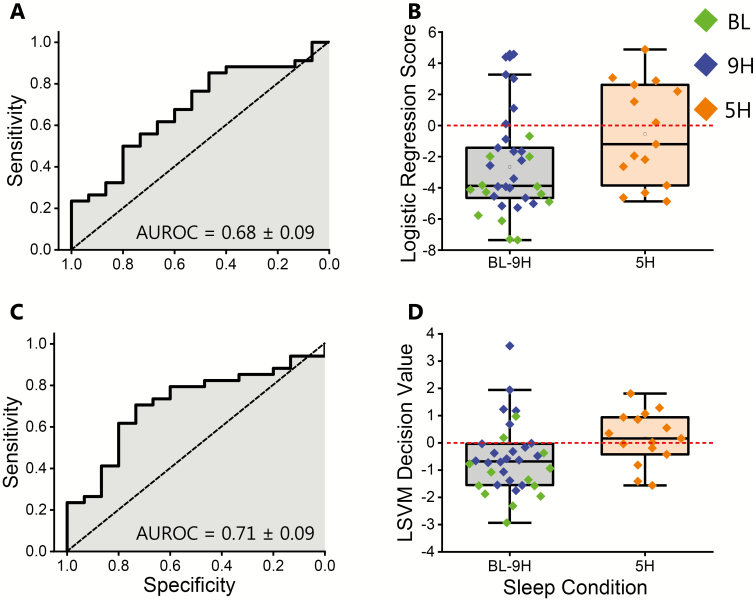
We compared the best performing model that included compounds with and without 24-hour time-of-day patterns (logistic regression model) versus the best performing model that excluded compounds with 24-hour time-of-day patterns (SVM model). The SVM model that excluded compounds with 24-hour time-of-day patterns had higher MCC (0.39), IV sensitivity (60%), and AUROC (0.71 ± 0.09; Figure 2), but slightly lower IV specificity (79%) compared to the logistic regression model that included compounds with and without 24-hour time-of-day patterns (82%). Thus, the SVM that excluded compounds with 24-hour time-of-day patterns correctly classified more of the insufficient sleep samples, but less of the adequate sleep samples, compared to this logistic regression model (Figure 2, B and D), and was, therefore, the overall top-performing biomarker model. Additionally, as exploratory analyses, we generated two biomarker models that only included compounds with 24-hour time-of-day patterns (Supplementary Table S1). Both the logistic regression and SVM models that only included compounds with 24-hour time-of-day patterns had lower IV MCCs versus the planned logistic regression and SVM models that excluded compounds with 24-hour time-of-day patterns. Because these exploratory biomarker models had lower MCCs, the following in-depth analyses focus on the four planned biomarker models.
Figure 2.
Top-performing biomarker models that include and exclude compounds with 24-hour time-of-day patterns. (A) Receiver operator characteristic plot for the top-performing biomarker model that included compounds with 24-hour time-of-day patterns (logistic regression model). (B) Box plot of the logistic regression scores for all samples in the validation dataset for the logistic regression biomarker model that included compounds with 24-hour time-of-day patterns. Note that the logistic regression scores were multiplied by −1 so that positive values were classified as insufficient sleep. (C) Receiver operator characteristic plot for the top-performing biomarker model that excluded compounds with 24-hour time-of-day patterns (support vector machine model). (D) Box plot of the decision values for all samples in the validation dataset for the support vector machine biomarker model that excluded compounds with 24-hour time-of-day patterns. Positive values were classified as insufficient sleep. BL, baseline condition; 9H, control condition; 5H, insufficient sleep condition; AUROC, area under the receiver operator characteristic plot.
We further analyzed the SVM model that excluded compounds with 24-hour time-of-day patterns, by sex and by condition order (Table 2). MCC for men (0.42) and women (0.40) were similar. However, IV sensitivity was 20% higher for men versus women, whereas IV specificity was 8% lower for men versus women. MCC for condition order A (0.34) and condition order B (0.38) were similar. Furthermore, IV sensitivity was 6% lower for condition order A versus condition order B, whereas IV specificity was similar for condition order A (77%) and condition order B (75%).
Table 2.
Performance metrics of linear SVM biomarker model that excluded compounds with 24-hour time-of-day patterns, by sex and condition order (top overall performing biomarker)
| IV ACC | IV sensitivity | IV specificity | IV MCC | |
|---|---|---|---|---|
| Performance by sex | ||||
| Men | 75% | 75% | 75% | 0.42 |
| Women | 72% | 55% | 83% | 0.40 |
| Performance by condition order (excluding BL samples) | ||||
| Condition order A (9H first) | 70% | 57% | 77% | 0.34 |
| Condition order B (5H first) | 69% | 63% | 75% | 0.38 |
ACC, accuracy. The BL condition is excluded from all analyses by condition order as all participants completed the BL condition prior to the 9H or 5H conditions, regardless of condition order.
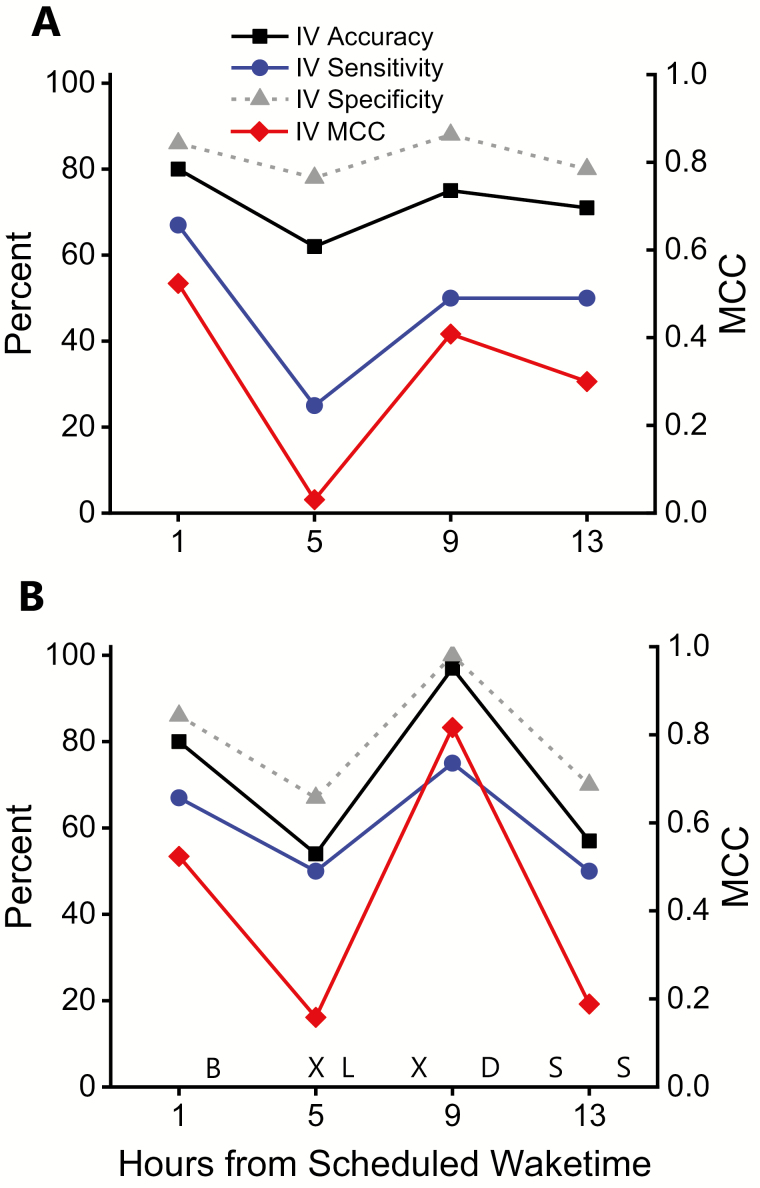
We also analyzed by time-points across the day the top-performing biomarker model that included compounds with and without 24-hour time-of-day patterns, and that excluded compounds with 24-hour time-of-day patterns, in part, to assess potential differences between fasting and non-fasting time-points (Figure 3). Using IV MCC, the T1 fasting time-point had the highest performance for the logistic regression model that included compounds with and without 24-hour time-of-day patterns (Figure 3A), whereas the T9 non-fasting time-point had the highest performance for the SVM model that excluded compounds with 24-hour time-of-day patterns (Figure 3B). Note that the general pattern of T1 and T9 having higher IV MCC compared to the T5 and T13 time-points was consistent in the logistic regression and SVM models, with IV accuracy, IV sensitivity, and IV specificity following the same general pattern.
Figure 3.
Top-performing biomarker models that included and excluded compounds with 24-hour time-of-day patterns, plotted by time of day. IV accuracy (black line), IV sensitivity (blue line), IV specificity (gray line), and IV MCC (red line and right y-axis) for (A) the top-performing biomarker model that included compounds with 24-hour time-of-day patterns (logistic regression model); and the (B) the top-performing biomarker model that excluded compounds with 24-hour time-of-day patterns (support vector machine model), plotted by hours from scheduled waketime. B, breakfast; L, lunch; D, dinner; S, scheduled snack; X, 20-minute stair-stepping session.
During ad libitum energy intake in the 9H and 5H conditions, participants consumed more energy than needed to maintain energy balance, leading to weight gain [17]. Thus, comparing biomarker model performance using the condition-specific classifications exclusively from 5H and BL versus exclusively from 5H and 9H allowed us to assess how positive energy balance and weight-gain during adequate sleep impacted biomarker model performance. We analyzed the performance of the biomarker models by condition-specific comparisons using only the 5H and BL conditions, or only the 5H and 9H conditions (Table 3). Based on IV MCC, both models that included compounds with and without 24-hour time-of-day patterns and the logistic regression model that excluded compounds with 24-hour time-of-day patterns had the highest performance for classifying samples from the 5H and BL conditions, each with an IV MCC of 0.54 (Table 3). Furthermore, each of these models had 71% IV accuracy, 47% IV sensitivity, and 100% IV specificity, but the SVM model that included compounds with and without 24-hour time-of-day patterns had the highest IV AUROC at 0.91 ± 0.06 (Table 3). Alternatively, the SVM model that excluded compounds with 24-hour time-of-day patterns had the highest performance for classifying samples from the 5H and 9H conditions with an IV MCC of 0.37. Furthermore, this model had the highest IV accuracy, IV sensitivity, IV specificity, and IV AUROC for classifying samples from the 5H and 9H conditions (Table 3).
Table 3.
Between condition performance metrics
| IV ACC | IV sensitivity | IV specificity | IV MCC | IV AUROC | |
|---|---|---|---|---|---|
| Compounds with and without 24-hour time-of-day patterns included | |||||
| Logistic Regression: 5H and BL (66) | 71% | 47% | 100% | 0.54 | 0.82 ± 0.08 |
| Logistic Regression: 5H and 9H (66) | 61% | 47% | 71% | 0.19 | 0.60 ± 0.10 |
| Linear SVM: 5H and BL (47) | 71% | 47% | 100% | 0.54 | 0.91 ± 0.06 |
| Linear SVM: 5H and 9H (47) | 56% | 47% | 62% | 0.09 | 0.63 ± 0.10 |
| Compounds with 24-hour time-of-day patterns excluded | |||||
| Logistic Regression: 5H and BL (65) | 71% | 47% | 100% | 0.54 | 0.83 ± 0.08 |
| Logistic Regression: 5H and 9H (65) | 64% | 47% | 76% | 0.24 | 0.63 ± 0.10 |
| Linear SVM: 5H and BL (65) | 71% | 60% | 85% | 0.46 | 0.83 ± 0.08 |
| Linear SVM: 5H and 9H (65) | 69% | 60% | 76% | 0.37 | 0.64 ± 0.10 |
5H and BL indicates samples from the 9H condition were excluded from the calculation of performance metrics. 5H and 9H indicates samples from the BL condition were excluded from the calculation of performance metrics. Numbers in parenthesis indicate total number of compounds in each biomarker model. BL, baseline condition; 9H, control condition; 5H, insufficient sleep condition; ACC, accuracy.
Pathway analyses identified 24 pathways associated with the logistic regression model that included compounds with and without 24-hour time-of-day patterns, 15 pathways associated with the SVM model that included compounds with and without 24-hour time-of-day patterns, and 13 pathways associated with the logistic regression and SVM models that excluded compounds with 24-hour time-of-day patterns (Table 4). Of these pathways, five were commonly associated with all four planned biomarker models, ATP Binding Cassette (ABC) Transporters in Lipid Homeostasis, High-Density Lipoprotein (HDL) Remodeling, Plasma Lipoprotein Remodeling, Golgi-to-endoplasmic reticulum (ER) retrograde transport, and Intra-Golgi and Retrograde Golgi-to-ER Traffic (Table 4). Alternatively, five of the 13 pathways associated with the overall top-performing biomarker model based on MCC (SVM model that excluded compounds with 24-hour time-of-day patterns) were uniquely associated with the models that excluded compounds with 24-hour time-of-day patterns (Table 4). We also generated a gene-metabolite interaction network derived from the compounds in the overall top-performing biomarker model based on MCC (SVM model that excluded compounds with 24-hour time-of-day patterns). The top 10 gene ontology biological processes associated with this gene-metabolite interaction network are presented in Supplementary Table S3. Consistently, the biochemical pathways and gene ontology biological processes associated with the overall top-performing biomarker model overlap in phospholipid metabolism.
Table 4.
Biochemical pathways associated with each biomarker model
| Pathway | Pathway source | Compound hits | Total compounds in pathway | p-value | FDR |
|---|---|---|---|---|---|
| Logistic regression—compounds with and without 24-hour time-of-day patterns included (66) | |||||
| Sphingolipid de novo biosynthesis | Reactome | 4 | 29 | <0.0001 | 0.0104 |
| Sphingolipid metabolism | Reactome | 4 | 52 | <0.0001 | 0.0288 |
| Immune System | Reactome | 4 | 56 | <0.0001 | 0.0288 |
| Synthesis of PC | Reactome | 3 | 22 | <0.0001 | 0.0372 |
| Synthesis of PS | Reactome | 2 | 5 | 0.0001 | 0.0609 |
| Acyl chain remodeling of PE | Reactome | 2 | 5 | 0.0001 | 0.0609 |
| Adaptive Immune System | Reactome | 3 | 29 | 0.0001 | 0.0609 |
| Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell | Reactome | 2 | 6 | 0.0002 | 0.0661 |
| Acyl chain remodeling of CL | Reactome | 2 | 6 | 0.0002 | 0.0661 |
| ABC transporters in lipid homeostasisa | Reactome | 2 | 7 | 0.0003 | 0.0767 |
| Sphingolipid Metabolism | SMPDB | 3 | 40 | 0.0004 | 0.0767 |
| Gaucher Disease | SMPDB | 3 | 40 | 0.0004 | 0.0767 |
| Globoid Cell Leukodystrophy | SMPDB | 3 | 40 | 0.0004 | 0.0767 |
| Metachromatic Leukodystrophy (MLD) | SMPDB | 3 | 40 | 0.0004 | 0.0767 |
| Fabry disease | SMPDB | 3 | 40 | 0.0004 | 0.0767 |
| Krabbe disease | SMPDB | 3 | 40 | 0.0004 | 0.0767 |
| Golgi-to-ER retrograde transport a | Reactome | 2 | 9 | 0.0005 | 0.0891 |
| Glycerophospholipid biosynthesis | Reactome | 3 | 46 | 0.0006 | 0.0913 |
| HDL remodelinga | Reactome | 2 | 10 | 0.0006 | 0.0913 |
| Plasma lipoprotein remodelinga | Reactome | 2 | 10 | 0.0006 | 0.0913 |
| Synthesis of PG | Reactome | 2 | 10 | 0.0006 | 0.0913 |
| Metabolism of proteins | Reactome | 4 | 117 | 0.0007 | 0.0949 |
| Intra-Golgi and retrograde Golgi-to-ER traffica | Reactome | 2 | 11 | 0.0007 | 0.0949 |
| Phospholipid metabolism | Reactome | 3 | 50 | 0.0007 | 0.0949 |
| Linear SVM—Compounds with and without 24-hour time-of-day patterns included (47) | |||||
| ABC transporters in lipid homeostasisa | Reactome | 2 | 7 | <0.0001 | 0.0826 |
| Golgi-to-ER retrograde transporta | Reactome | 2 | 9 | 0.0001 | 0.0826 |
| HDL remodelinga | Reactome | 2 | 10 | 0.0002 | 0.0826 |
| Plasma lipoprotein remodelinga | Reactome | 2 | 10 | 0.0002 | 0.0826 |
| Synthesis of PG | Reactome | 2 | 10 | 0.0002 | 0.0826 |
| Intra-Golgi and retrograde Golgi-to-ER traffica | Reactome | 2 | 11 | 0.0002 | 0.0826 |
| Role of phospholipids in phagocytosis | Reactome | 2 | 12 | 0.0003 | 0.0826 |
| Surfactant metabolism | Reactome | 2 | 13 | 0.0003 | 0.0826 |
| ABC-family proteins mediated transport | Reactome | 2 | 13 | 0.0003 | 0.0826 |
| Plasma lipoprotein assembly_ remodeling_ and clearance | Reactome | 2 | 13 | 0.0003 | 0.0826 |
| Fcgamma receptor (FCGR) dependent phagocytosis | Reactome | 2 | 14 | 0.0003 | 0.0903 |
| Synthesis of PA | Reactome | 2 | 15 | 0.0004 | 0.0936 |
| PLC beta mediated events | Reactome | 2 | 17 | 0.0005 | 0.0936 |
| G-protein mediated events | Reactome | 2 | 17 | 0.0005 | 0.0936 |
| PI Metabolism | Reactome | 2 | 17 | 0.0005 | 0.0936 |
| Logistic regression and linear SVM—Compounds with 24-hour time-of-day patterns excluded (65) | |||||
| Acyl chain remodeling of PCb | Reactome | 2 | 6 | <0.0001 | 0.0671 |
| Acyl chain remodeling of CL | Reactome | 2 | 6 | <0.0001 | 0.0671 |
| ABC transporters in lipid homeostasisa | Reactome | 2 | 7 | <0.0001 | 0.0671 |
| COPI-independent Golgi-to-ER retrograde trafficb | Reactome | 2 | 7 | <0.0001 | 0.0671 |
| Phospho-PLA2 pathwayb | Reactome | 2 | 7 | <0.0001 | 0.0671 |
| Golgi-to-ER retrograde transporta | Reactome | 2 | 9 | 0.0001 | 0.0673 |
| HDL remodelinga | Reactome | 2 | 10 | 0.0002 | 0.0673 |
| Plasma lipoprotein remodelinga | Reactome | 2 | 10 | 0.0002 | 0.0673 |
| Choline metabolism in cancer—Homo sapiens (human)b | KEGG | 2 | 10 | 0.0002 | 0.0673 |
| Ca-dependent eventsb | Reactome | 2 | 11 | 0.0002 | 0.0673 |
| Intra-Golgi and retrograde Golgi-to-ER traffica | Reactome | 2 | 11 | 0.0002 | 0.0673 |
| ABC-family proteins mediated transport | Reactome | 2 | 13 | 0.0003 | 0.0729 |
| Plasma lipoprotein assembly_ remodeling_ and clearance | Reactome | 2 | 13 | 0.0003 | 0.0729 |
Pathways within each biomarker model are ordered by p-value. Number of compounds indicates the number of compounds from the given biomarker model that appear in the pathway. Total compounds in the pathway indicates the number of compounds in the pathway that are in the background list. Numbers in parenthesis indicate total number of compounds in each biomarker model. FDR, false discovery rate.
aPathways that overlap between all four biomarker models.
bPathways that are unique to the models that exclude compounds with 24-hour time-of-day patterns.
Finally, based on the findings of Weije et al. [39], we conducted a targeted data analysis on fasting oxalic acid and DAG-[36:3]. Oxalic acid was not annotated in our dataset; however, we did identify a compound annotated as DAG-[36:3] at the Metabolomics Standards Initiative level 3 [64]. When classifying fasting (T1) samples from the validation dataset as insufficient sleep or adequate sleep (BL-9H), DAG-[36:3] has an AUROC of 0.69 ± 0.10, 73% accuracy, 64% sensitivity, 77% specificity, and MCC of 0.40 based on the optimal relative abundance cutoff value determined using the ROC. Fasting DAG-[36:3] was reduced (p < 0.05) in 5H versus BL, but not statistically different (p = 0.12) in 5H versus 9H (Supplementary Figure S4). We also assessed the individual AUROC for all compounds in each of our four planned biomarker models. Including all time-points, individual compound AUROCs in the validation dataset ranged from 0.44 to 0.60 for 69 compounds and from >0.60 to 0.69 for 19 compounds (Supplementary Table S2).
Discussion
We used untargeted plasma metabolomics to identify potential biomarkers of insufficient sleep in humans. The SVM biomarker model that excluded compounds with 24-hour time-of-day patterns produced the best overall performance with 74% accuracy and MCC = 0.39. The logistic regression biomarker model that also excluded compounds with 24-hour time-of-day patterns produced a similar overall performance with 74% accuracy and MCC = 0.34. Using whole blood transcriptomics, Laing et al. identified biomarker models that predicted total sleep deprivation with 92% accuracy and MCC = 0.83, and insufficient sleep (1 week of 6 hours sleep opportunities) with 57% accuracy and MCC = 0.14 [40]. Our findings in combination with the findings from Laing et al., suggest omics-based approaches are viable for identifying biomarkers of insufficient sleep and total sleep deprivation. A key strength of the findings from Laing et al. and us is the use of independent training and validation datasets since performance assessments on training data often result in overly optimistic findings. Our findings of higher LOOCV accuracy in the training dataset compared to IV accuracy in the validation dataset for all our biomarker models is consistent with this concept and supports the need for true independent validation. Ultimately, the candidate omics-based biomarkers identified to date will require further work to refine and validate for clinical and research uses. Such efforts will help advance the biomarker goals of the Sleep Research Society and the National Institutes of Health [26, 27].
The five biochemical pathways associated with all four planned biomarker models were ABC Transporters in Lipid Homeostasis, High-Density Lipoprotein (HDL) Remodeling, Plasma Lipoprotein Remodeling, Golgi-to-ER retrograde transport, and Intra-Golgi and Retrograde Golgi-to-ER Traffic. The ABC transporters are highly conserved cellular transmembrane transport proteins, have high expression levels in monocytes and macrophages, are regulated in part by sterol flux, and have been linked to chronic inflammatory conditions including atherosclerosis [65]. Specifically, the ABC family A and G transporters facilitate reverse cholesterol transport from macrophage-foam cells to HDL particles, and are reported to be down-regulated during insufficient sleep [66]. Accordingly, potential changes in the ABC transporters during insufficient sleep may be linked to an inflammatory response and risk of atherosclerosis associated with insufficient sleep, although follow-up targeted studies are needed to confirm our findings. The associations with other pathways related to lipid metabolism consist of HDL Remodeling and Plasma Lipoprotein Remodeling, which may also be linked to changes in cholesterol homeostasis through the ABC transporters, further suggesting insufficient sleep can impact cholesterol metabolism. In general, Golgi-to-ER retrograde transport must be tightly regulated with anterograde transport from the ER to the Golgi to maintain functionality of these organelles, especially in relation to transporting proteins and lipids [67]. However, much is still unknown about the regulation and function of Golgi-to-ER retrograde transport. Thus, in the context of our current findings, the functional significance of Golgi-to-ER retrograde transport during insufficient sleep is not clear.
Of the five unique pathways associated with our top-performing biomarker model, two are linked to phospholipid metabolism (Acyl Chain Remodeling of Phosphatidylcholine and Phospho-PLA2 Pathway), implicating altered phospholipid metabolism with insufficient sleep. The two altered compounds in both of these pathways are lysoPC-[18:3] and PC-[40:5]. Phospholipids are key structural and functional components of cell membranes [58]. Changes in skeletal muscle phospholipid metabolism have been linked to altered insulin sensitivity [68, 69]. We previously reported that insulin sensitivity was reduced during insufficient sleep in the current study [14]. It is possible that such changes in insulin sensitivity may be linked to some of the altered compounds involved in phospholipid metabolism observed here, including lysoPC-[18:3], lysophosphatidylethanolamine (PE)-[22:5], PC-[38:5], PC-[40:5], PE-[35:3], PE-[39:2], and phosphatidylinositol (PI)-[38:3] from our top-performing biomarker model.
Nine out of the 10 gene ontology biological processes associated with the gene-metabolite network derived from our top-performing biomarker model are linked to either phospholipid metabolism or sphingolipid metabolism. Specifically, the sphingolipids ceramide (CER)-[40:2], CER-[d41:2], sphingomyelin(SM)-[43:2], and SM-[d33:2] were altered during insufficient sleep in our top-performing biomarker model. In addition to phospholipids, changes in sphingolipids, and notably ceramides have also been linked to inflammation and risk of metabolic disorders [68, 70, 71], further suggesting that our biomarker model may in part be reflecting changes in inflammation and insulin sensitivity associated with insufficient sleep. Furthermore, altered phospholipid and sphingolipid metabolism have been linked to mild cognitive impairment and Alzheimer’s Disease [72]. As insufficient sleep is believed to contribute to risk of Alzheimer’s Disease [18, 20], altered phospholipid and sphingolipid metabolism may represent a common pathway linking insufficient sleep and risk of Alzheimer’s Disease. While it is possible that changes in phospholipid and sphingolipid metabolism contribute to risk of metabolic and neurodegenerative disorders associated with insufficient sleep, causality cannot be determined in our current study. Ultimately, if our findings are validated in larger prospective trials it will strengthen the links between insufficient sleep, altered lipid metabolism, increased inflammation, and risk of disease.
Consistent with Weljie et al. [39], we show decreased fasting DAG-36:3 during insufficient sleep versus adequate sleep in the BL condition. However, fasting DAG-36:3 was not statistically different in 5H versus 9H, suggesting DAG-36:3 may also be impacted by factors other than sleep duration. Fasting DAG-36:3 produced an MCC of 0.40 whereas our top-performing biomarker model produced an MCC of 0.53 at the fasting time-point, supporting the hypothesis that biomarkers of insufficient sleep with multiple compounds are likely to produce better overall performance compared to any single compound. Although DAG-36:3 was not in any of our current biomarker models, as a single compound, the MCC of 0.40 shows promise and fasting DAG-36:3 should be considered in future biomarker studies as a potential candidate, especially for samples collected after an overnight fast.
There are known biochemical links between the changes in DAG, phospholipid, and sphingolipid metabolism we observed during insufficient sleep. Specifically, the enzyme sphingomyelin synthase catalyzes the reaction CER + PC ↔ SM + DAG [68], and CER can increase the activity of phospholipase A2 [73], that hydrolyzes phospholipids at the SN-2 position resulting in a fatty acid and a lysophospholipid. Furthermore, the collective changes in DAG, phospholipid, and sphingolipid metabolism are hypothesized to be linked to reduced insulin sensitivity through mitochondrial dysfunction, reactive oxygen species, and inflammation, ultimately forming a negative feedback loop that contributes to metabolic dysregulation [68]. Since altered DAG-36:3, PCs, and SMs have now been observed in humans and rats during insufficient sleep [39], a more thorough and targeted investigation of lipid metabolism including gene expression and lipidomic analyses during insufficient sleep is warranted. Such analyses will provide further insight into how lipid metabolism is altered during sleep loss and its potential impacts on insulin sensitivity and risk of disease.
Because we collected samples across the 24-hour day, we trained biomarker models on datasets that included and excluded compounds with 24-hour time-of-day patterns. Consistently, a priori exclusion of compounds with 24-hour time-of-day patterns resulted in equal or better overall performance compared to the biomarker models that included compounds with and without 24-hour time-of-day patterns, and compared to the exploratory biomarker models that included only compounds with 24-hour time-of-day patterns. Multiple factors such as the fasting/feeding cycle, sleep-wake cycle, and the circadian clock [29, 32, 34, 74], likely contributed to the 24-hour time-of-day patterns in many of the compounds. Laing et al. also showed a priori variable selection improved some of their reported transcriptomics-based biomarker models [40]. Thus, in the context of biomarker development it is critical to test a range of potential biomarkers, and when possible, explore a priori variable selection to minimize known biological variation such as 24-hour time-of-day patterns. Future metabolomics-based biomarker trials should include compounds with and without 24-hour time-of-day patterns to confirm our current findings. Furthermore, studies with ideally larger sample sizes and more frequent sampling, and that utilize established circadian protocols such as the constant routine or forced desynchrony are required to identify 24-hour time-of-day patterns regulated by the behavioral (e.g. sleep-wake or fasting/feeding) versus circadian cycles [75].
When analyzing the performance of our biomarker models at individual time-points, the T1 fasting time-point and T9 non-fasting time-point consistently produced the best performance. The T1 fasting time-point is the most clinically relevant as fasting blood collection is most common in clinical settings. Since consuming a single meal or a small snack acutely impacts the human plasma metabolome [76–79], energy intake throughout the day likely increased the variability in non-fasted samples. Notably, during ad libitum energy intake during insufficient sleep, more calories were consumed as post-dinner snacks compared to any other single meal [17], likely contributing to the lower performance of our biomarker models in the T13 samples that were collected during this post-dinner snack timeframe. Furthermore, participants completed two 20-minute stair-stepping sessions per day to simulate activity outside the laboratory setting [17] (Figure 3). The T5 samples were collected immediately before a stair-stepping session, whereas, the T9 samples were collected ~40 minutes following a stair-stepping session. The different timing of the stair-stepping sessions relative to the T5 and T9 sample collections may have contributed to the differences in performance metrics between time-points. Our findings show normal activities of daily living can impact the human plasma metabolome and the performance of omics-based biomarkers must be carefully tested and validated using the specific conditions of their anticipated clinical and research uses.
Our top-performing biomarker model, the SVM model that excluded compounds with 24-hour time-of-day patterns, had higher IV sensitivity in men versus women indicating this biomarker model correctly classified insufficient sleep more frequently in men versus women. Alternatively, IV specificity was lower in men versus women indicating this biomarker model misclassified adequate sleep more frequently in men versus women. As previously described [17], women participants had a history of regular menstrual cycles, and men were in greater positive energy balance and gained more weight versus women during the 5H and 9H conditions with ad libitum energy intake. Because blood was collected across 13 days, and condition order was balanced by sex, we did not have statistical power to analyze model performance by different menstrual cycle phases. However, it is likely that samples collected across different menstrual cycle phases, and sex differences in energy intake and weight-gain contributed to some of the differences in biomarker model performance between men and women. Optimally, as omics-based biomarkers of insufficient sleep progress, biomarker models will be trained and validated on men and women independently to determine the influence of menstrual cycle phase, optimize sensitivity and specificity, and determine if there is a core biochemical signature of insufficient sleep common to men and women.
For the four planned biomarker models, the IV accuracy, IV specificity, IV MCC, and IV AUROC were higher when exclusively classifying samples in the 5H and BL conditions compared to exclusively classifying samples in the 5H and 9H conditions, suggesting ad libitum energy intake in the 9H condition reduced the performance of our models. It is also possible that condition order contributed to these findings. Specifically, during condition order B, where participants completed the 5H condition followed by the 9H condition, 5 days of 9 hours sleep opportunities during 9H may not have fully recovered our biomarker models back to BL levels. However, in our top-performing biomarker model, MCC and IV specificity were similar between condition order A and B suggesting that our biomarker model recovered after the 9H condition in condition order B. Another consideration is the regularity of sleep timing and duration. Under free-living conditions, we expect the regularity of insufficient sleep and any attempts at recovery sleep to be more variable than our laboratory protocol since we controlled all sleep opportunities, keeping the mid-point of sleep centered across conditions. How the regularity of insufficient and recovery sleep may impact metabolomics-based biomarkers of insufficient sleep remains to be determined, especially since insufficient and recovery sleep can alter energy intake [8, 13, 17]. As the field progresses, it is essential to validate our preliminary findings using independent cohorts studied under controlled laboratory conditions as well as free-living conditions ranging from days to years to quantify the timescale of changes in our biomarker model with different sleep patterns.
Our findings are derived from a laboratory-controlled study of scheduled insufficient sleep based on habitual bed and wake times in otherwise healthy adults. As others have suggested [26, 80, 81], this is a preferred experimental design during the biomarker discovery phase. The primary limiting factor of our biomarkers is the high rate of false negatives (i.e. misclassifying insufficient sleep). Moving forward, large-scale independent validation studies conducted in laboratory and “real-world” or field settings using completely independent cohorts are necessary to refine and validate our metabolomics-based biomarkers of insufficient sleep. Such independent validation studies could also facilitate direct comparison of the performance of different omics-based biomarkers such as transcriptomics and metabolomics. Factors including age, sex, body mass index, physical activity, and chronic disease status have the potential to influence biomarker performance and these factors must be considered at the population level in validation trials. Beyond our findings, there are opportunities to identify omics-based biomarkers that link insufficient sleep with behavior and physiology such as cognitive performance, sleep staging, slow-wave activity, insulin sensitivity, and blood pressure. Sleep health is multi-dimensional [82] including aspects of regularity, satisfaction, alertness, timing, efficiency, and duration. Integrating omics-based biomarkers into the larger construct of overall sleep health will facilitate translation of such biomarkers, and if further developed and validated using independent cohorts, could eventually help identify novel countermeasures designed to mitigate the negative health consequences of insufficient sleep.
Funding
This work was supported by NIH-HL109706, HL132150, HL085705, DK111161, DK048520, NIH-TR002535, Sleep Research Society Foundation-011-JP-16, and the Howard Hughes Medical Institute with Biological Sciences Initiative/Undergraduate Research Opportunities Program at University of Colorado Boulder.
Conflict of interest statement. C.M.D. has received research support from the National Institutes of Health and the Sleep Research Society Foundation. Any opinions, findings, and conclusions or recommendations expressed in this publication are those of the author(s), and do not necessarily reflect the views of the Sleep Research Society Foundation. K.P.W. has received research support from the National Institutes of Health, Office of Naval Research, Pac-12, and SomaLogic, Inc., consulting fees from or served as a paid member of scientific advisory boards for the Sleep Disorders Research Advisory Board - National Heart, Lung and Blood Institute, CurAegis Technologies, Circadian Therapeutics, LTD. and has received speaker/educational/travel consultant honorarium fees from the American Academy of Sleep Medicine, American College of Chest Physicians, American College of Sports Medicine, American Diabetes Association, Associated Professional Sleep Societies, Kellogg Company, Obesity Medicine Association, and The European Association for the Study of Obesity. E.L.M. is supported by resources from the Geriatric Research, Education, and the Clinical Center at the Denver VA Medical Center. The contents do not represent the views of the US Department of Veterans Affairs or the United States Government.
Author Contributions
R.R.M., E.L.M, and K.P.W designed the experiments and collected data; all authors analyzed data and edited the paper; C.M.D. and K.P.W. wrote the manuscript; C.M.D., R.R.M., E.L.M, N.R., and K.P.W. obtained funding and supervised the research.
Deposit of Material in a Data Repository
The mass spectrometry data from this publication is available at MetabolomicsWorkbench database: http://www.metabolomicsworkbench.org/ (Study ID: ST001808).
Supplementary Material
Acknowledgments
We thank the research participants and the staff and students of the Sleep and Chronobiology Laboratory for their contributions. We also thank the Clinical Translational Research Center physicians, nurses, registered dieticians, and technicians for their support; and B. Griffin, B. Ball, U. Mohapatra, S. Peralta, B. Perry, A. W. McHill, and G. Wright for their assistance.
References Cited
- 1. Watson NF, et al. Recommended amount of sleep for a healthy adult: a joint consensus statement of the american academy of sleep medicine and sleep research society. Sleep. 2015;38(6):843–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Watson NF, et al. Recommended amount of sleep for a healthy adult: a joint consensus statement of the American Academy of Sleep Medicine and Sleep Research Society. J Clin Sleep Med. 2015;11(6):591–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Centers for Disease Control and Prevention. Effect of short sleep duration on daily activities—United States, 2005–2008. Morb Mortal Wkly Rep. 2011;60(8):239–242. [PubMed] [Google Scholar]
- 4. Centers for Disease Control and Prevention. Short Sleep Duration Among US Adults.2014. https://www.cdc.gov/sleep/data_statistics.html. Accessed September 24, 2018.
- 5. Ford ES, et al. Trends in self-reported sleep duration among US Adults from 1985 to 2012. Sleep. 2015;38(5):829–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Luxton DD, et al. Prevalence and impact of short sleep duration in redeployed OIF soldiers. Sleep. 2011;34(9):1189–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Seelig AD, et al. ; Millennium Cohort Study Team. Sleep patterns before, during, and after deployment to Iraq and Afghanistan. Sleep. 2010;33(12):1615–1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Depner CM, et al. Metabolic consequences of sleep and circadian disorders. Curr Diab Rep. 2014;14(7):507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. St-Onge MP, et al. ; American Heart Association Obesity, Behavior Change, Diabetes, and Nutrition Committees of the Council on Lifestyle and Cardiometabolic Health; Council on Cardiovascular Disease in the Young; Council on Clinical Cardiology; and Stroke Council. Sleep duration and quality: impact on lifestyle behaviors and cardiometabolic health: a scientific statement from the American Heart Association. Circulation. 2016;134(18):e367–e386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cappuccio FP, et al. Sleep duration predicts cardiovascular outcomes: a systematic review and meta-analysis of prospective studies. Eur Heart J. 2011;32(12):1484–1492. [DOI] [PubMed] [Google Scholar]
- 11. Cappuccio FP, et al. Quantity and quality of sleep and incidence of type 2 diabetes: a systematic review and meta-analysis. Diabetes Care. 2010;33(2):414–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cappuccio FP, et al. Meta-analysis of short sleep duration and obesity in children and adults. Sleep. 2008;31(5):619–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Depner CM, et al. Ad libitum weekend recovery sleep fails to prevent metabolic dysregulation during a repeating pattern of insufficient sleep and weekend recovery sleep. Curr Biol. 2019;29(6):957–967.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Eckel RH, et al. Morning circadian misalignment during short sleep duration impacts insulin sensitivity. Curr Biol. 2015;25(22):3004–3010. [DOI] [PubMed] [Google Scholar]
- 15. Spiegel K, et al. Impact of sleep debt on metabolic and endocrine function. Lancet. 1999;354(9188):1435–1439. [DOI] [PubMed] [Google Scholar]
- 16. Broussard JL, et al. Impaired insulin signaling in human adipocytes after experimental sleep restriction: a randomized, crossover study. Ann Intern Med. 2012;157(8):549–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Markwald RR, et al. Impact of insufficient sleep on total daily energy expenditure, food intake, and weight gain. Proc Natl Acad Sci U S A. 2013;110(14):5695–5700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lucey BP, et al. How amyloid, sleep and memory connect. Nat Neurosci. 2015;18(7):933–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bokenberger K, et al. Association between sleep characteristics and incident dementia accounting for baseline cognitive status: a prospective population-based study. J Gerontol A Biol Sci Med Sci. 2017;72(1):134–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Winer JR, et al. Sleep as a potential biomarker of tau and β-Amyloid burden in the human brain. J Neurosci. 2019;39(32):6315–6324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Dinges DF, et al. Cumulative sleepiness, mood disturbance, and psychomotor vigilance performance decrements during a week of sleep restricted to 4-5 hours per night. Sleep. 1997;20(4):267–277. [PubMed] [Google Scholar]
- 22. Carskadon MA, et al. Cumulative effects of sleep restriction on daytime sleepiness. Psychophysiology. 1981;18(2):107–113. [DOI] [PubMed] [Google Scholar]
- 23. Barger LK, et al. ; Harvard Work Hours, Health, and Safety Group. Extended work shifts and the risk of motor vehicle crashes among interns. N Engl J Med. 2005;352(2):125–134. [DOI] [PubMed] [Google Scholar]
- 24. Åkerstedt T, et al. Accidents and sleepiness: a consensus statement from the International Conference on Work Hours, Sleep and Accidents, Stockholm, 8–10. J Sleep Res. 1994;3:195. [Google Scholar]
- 25.Academy of Sleep Medicine. International Classification of Sleep Disorders: Diagnostic and Coding Manual (ICSD-3), 3rd ed. Darien, IL: American Academy of Sleep Medicine; 2014. [Google Scholar]
- 26. Mullington JM, et al. Developing biomarker arrays predicting sleep and circadian-coupled risks to health. Sleep. 2016;39(4):727–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Depner CM, et al. Wearable technologies for developing sleep and circadian biomarkers: a summary of workshop discussions. Sleep. 2019;43(2). doi: 10.1093/sleep/zsz254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Depner CM, et al. Mistimed food intake and sleep alters 24-hour time-of-day patterns of the human plasma proteome. Proc Natl Acad Sci U S A. 2018;115(23):E5390– E5399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Skene DJ, et al. Separation of circadian- and behavior-driven metabolite rhythms in humans provides a window on peripheral oscillators and metabolism. Proc Natl Acad Sci U S A. 2018;115(30):7825–7830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Davies SK, et al. Effect of sleep deprivation on the human metabolome. Proc Natl Acad Sci U S A. 2014;111(29):10761–10766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bell LN, et al. Effects of sleep restriction on the human plasma metabolome. Physiol Behav. 2013;122:25–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Dallmann R, et al. The human circadian metabolome. Proc Natl Acad Sci U S A. 2012;109(7):2625–2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chua EC, et al. Changes in Plasma Lipids during Exposure to Total Sleep Deprivation. Sleep. 2015;38(11):1683–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chua EC, et al. Extensive diversity in circadian regulation of plasma lipids and evidence for different circadian metabolic phenotypes in humans. Proc Natl Acad Sci U S A. 2013;110(35):14468–14473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Grant LK, et al. Circadian and wake-dependent changes in human plasma polar metabolites during prolonged wakefulness: A preliminary analysis. Sci Rep. 2019;9(1):4428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Archer SN, et al. Mistimed sleep disrupts circadian regulation of the human transcriptome. Proc Natl Acad Sci U S A. 2014;111(6):E682–E691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Möller-Levet CS, et al. Effects of insufficient sleep on circadian rhythmicity and expression amplitude of the human blood transcriptome. Proc Natl Acad Sci U S A. 2013;110(12):E1132–E1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Xiao Q, et al. Habitual Sleep and human plasma metabolomics. Metabolomics. 2017;13(5):1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Weljie AM, et al. Oxalic acid and diacylglycerol 36:3 are cross-species markers of sleep debt. Proc Natl Acad Sci U S A. 2015;112(8):2569–2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Laing EE, et al. Identifying and validating blood mRNA biomarkers for acute and chronic insufficient sleep in humans: a machine learning approach. Sleep. 2019;42(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Seugnet L, et al. Identification of a biomarker for sleep drive in flies and humans. Proc Natl Acad Sci U S A. 2006;103(52):19913–19918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Cruickshank-Quinn C, et al. Multi-step preparation technique to recover multiple metabolite compound classes for in-depth and informative metabolomic analysis. J Vis Exp. 2014;89:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hughes G, et al. MSPrep–summarization, normalization and diagnostics for processing of mass spectrometry-based metabolomic data. Bioinformatics. 2014;30(1):133–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yang Y, et al. New sample preparation approach for mass spectrometry-based profiling of plasma results in improved coverage of metabolome. J Chromatogr A. 2013;1300:217– 226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Reisdorph NA, et al. Application of metabolomics in lung research. Methods Mol Biol. 2018;1809:263–288. [DOI] [PubMed] [Google Scholar]
- 46. Cruickshank-Quinn C, et al. Impact of blood collection tubes and sample handling time on serum and plasma metabolome and lipidome. Metabolites. 2018;8(4):1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Cruickshank-Quinn CI, et al. Metabolomics and transcriptomics pathway approach reveals outcome-specific perturbations in COPD. Sci Rep. 2018;8(1):17132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Brondel L, et al. Acute partial sleep deprivation increases food intake in healthy men. Am J Clin Nutr. 2010;91(6):1550– 1559. [DOI] [PubMed] [Google Scholar]
- 49. Spaeth AM, et al. Effects of experimental sleep restriction on weight gain, caloric intake, and meal timing in healthy adults. Sleep. 2013;36(7):981–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Spaeth AM, et al. Phenotypic vulnerability of energy balance responses to sleep loss in healthy adults. Sci Rep. 2015;5:14920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Chong J, et al. MetaboAnalyst 4.0: towards more transparent and integrative metabolomics analysis. Nucleic Acids Res. 2018;46(W1):W486–W494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wu G, et al. MetaCycle: an integrated R package to evaluate periodicity in large scale data. Bioinformatics. 2016;32(21):3351–3353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. R Development Core Team. R: A Language and Enviornment for Statistical Computing.Vienna, Austria: R Foundation for Statistical Computing; https://www.R-project.org/. [Google Scholar]
- 54. Tseng GC, et al. Comprehensive literature review and statistical considerations for microarray meta-analysis. Nucleic Acids Res. 2012;40(9):3785–3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kuhn M. Building predictive models in R Using the caret Package. J Stat Softw. 2008;28(5). [Google Scholar]
- 56. Friedman J, et al. Regularization paths for generalized linear models via coordinate descent. J Stat Softw. 2010;33(1):1–26. [PMC free article] [PubMed] [Google Scholar]
- 57. Vihinen M. How to evaluate performance of prediction methods? Measures and their interpretation in variation effect analysis. BMC Genomics. 2012;13 Suppl 4:S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Matthews BW. Comparison of the predicted and observed secondary structure of T4 phage lysozyme. Biochim Biophys Acta. 1975;405(2):442–451. [DOI] [PubMed] [Google Scholar]
- 59. Wishart DS, et al. HMDB 4.0: the human metabolome database for 2018. Nucleic Acids Res. 2018;46(D1):D608–D617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Kamburov A, et al. Integrated pathway-level analysis of transcriptomics and metabolomics data with IMPaLA. Bioinformatics. 2011;27(20):2917–2918. [DOI] [PubMed] [Google Scholar]
- 61. Kanehisa M, et al. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28(1):27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Fabregat A, et al. The reactome pathway knowledgebase. Nucleic Acids Res. 2018;46(D1):D649–D655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Jewison T, et al. SMPDB 2.0: big improvements to the Small Molecule Pathway Database. Nucleic Acids Res. 2014;42(Database issue):D478–D484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Sumner LW, et al. Proposed minimum reporting standards for chemical analysis Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI). Metabolomics. 2007;3(3):211–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Schmitz G, et al. ABC transporters in cellular lipid trafficking. Curr Opin Lipidol. 2000;11(5):493–501. [DOI] [PubMed] [Google Scholar]
- 66. Aho V, et al. Prolonged sleep restriction induces changes in pathways involved in cholesterol metabolism and inflammatory responses. Sci Rep. 2016;6:24828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Heffernan LF, et al. The trials and tubule-ations of Rab6 involvement in Golgi-to-ER retrograde transport. Biochem Soc Trans. 2014;42(5):1453–1459. [DOI] [PubMed] [Google Scholar]
- 68. Meikle PJ, et al. Sphingolipids and phospholipids in insulin resistance and related metabolic disorders. Nat Rev Endocrinol. 2017;13(2):79–91. [DOI] [PubMed] [Google Scholar]
- 69. Funai K, et al. Skeletal muscle phospholipid metabolism regulates insulin sensitivity and contractile function. Diabetes. 2016;65(2):358–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Turpin-Nolan SM, et al. CerS1-Derived C18:0 ceramide in skeletal muscle promotes obesity-induced insulin resistance. Cell Rep. 2019;26(1):1–10.e7. [DOI] [PubMed] [Google Scholar]
- 71. Chaurasia B, et al. Targeting a ceramide double bond improves insulin resistance and hepatic steatosis. Science. 2019;365(6451):386–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Mapstone M, et al. Plasma phospholipids identify antecedent memory impairment in older adults. Nat Med. 2014;20(4):415–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Koumanov KS, et al. Ceramides increase the activity of the secretory phospholipase A2 and alter its fatty acid specificity. Biochem J. 2002;363(Pt 1):45–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Ang JE, et al. Identification of human plasma metabolites exhibiting time-of-day variation using an untargeted liquid chromatography-mass spectrometry metabolomic approach. Chronobiol Int. 2012;29(7):868–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Broussard JL, et al. Circadian rhythms versus daily patterns in human physiology and behavior. In: Kumar V, ed. Biological Timekeeping: Clocks, Rhythms and Behaviour. New Delhi, India: Springer India; 2017:279–295. [Google Scholar]
- 76. Pellis L, et al. Plasma metabolomics and proteomics profiling after a postprandial challenge reveal subtle diet effects on human metabolic status. Metabolomics. 2012;8(2):347–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Krug S, et al. The dynamic range of the human metabolome revealed by challenges. FASEB J. 2012;26(6):2607–2619. [DOI] [PubMed] [Google Scholar]
- 78. Ho JE, et al. Metabolite profiles during oral glucose challenge. Diabetes. 2013;62(8):2689–2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Wahl S, et al. Comparative analysis of plasma metabolomics response to metabolic challenge tests in healthy subjects and influence of the FTO obesity risk allele. Metabolomics. 2014;10(3):386–401. [Google Scholar]
- 80. Mullington JM. Please forgive our appearance while under biomarker construction. Sleep. 2019;42(1). [DOI] [PubMed] [Google Scholar]
- 81. Mullington J, et al. In pursuit of sleep-circadian biomarkers. Sleep. 2015;38(11):1665–1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Buysse DJ. Sleep health: can we define it? Does it matter? Sleep. 2014;37(1):9–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Broadhurst D, et al. Guidelines and considerations for the use of system suitability and quality control samples in mass spectrometry assays applied in untargeted clinical metabolomic studies. Metabolomics. 2018;14(6):72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Stein S. Nist MS Search Program.https://chemdata.nist.gov/dokuwiki/doku.php?id=chemdata:nist17. Accessed 30 October, 2018.
- 85. Stacklies W, et al. pcaMethods–a bioconductor package providing PCA methods for incomplete data. Bioinformatics. 2007;23(9):1164–1167. [DOI] [PubMed] [Google Scholar]
- 86. Johnson WE, et al. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8(1):118–127. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.