Abstract
A reliable diagnostic assay is crucial to early detect new COVID-19 cases and limit severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) transmission. Since the onset of the COVID-19 pandemic, the World Health Organization has published several diagnostic molecular approaches developed by referral laboratories, including Charité (Germany), HKU (Hong Kong), China CDC (China), US CDC (United States), and Institut Pasteur, Paris (France). We aimed to compare the sensitivity and specificity of these different RT-PCR assays using SARS-CoV-2 cell culture supernatants and clinical respiratory samples. Overall, the different RT-PCR assays performed well for SARS-CoV-2 detection and were all specific except the N Charité (Germany), and N2 US CDC (United States) assays. RdRp Institut Pasteur (IP2, IP4), N China CDC, and N1 US CDC were found to be the most sensitive assays. The data presented herein are of prime importance to facilitate the equipment choice of diagnostic laboratories, as well as for the development of marketed tests.
Keywords: SARS-CoV-2, COVID-19, RT-PCR, diagnostics, sensitivity
1. Introduction
A new human coronavirus called severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) emerged in China in December 2019 [1]. SARS-CoV-2 is responsible for coronavirus disease 2019 (COVID-19), which was declared a pandemic on 12 March 2020 by the World Health Organization (WHO) [2]. As of 26 May 2020, 5,404,512 cases have been reported, including 343,514 deaths [3]. A reliable diagnostic assay is crucial to limit the spread of SARS-CoV-2 as early detection of new cases leads to patient isolation and contact tracing. The first SARS-CoV-2 genome was published on 10 January 2020 [4], enabling the rapid design of a real-time reverse-transcriptase polymerase chain reaction (RT-PCR) assay by Charité (Germany) [5,6]. This test was the first to be dispatched by WHO [7] and was widely implemented in clinical virology laboratories worldwide [8]. Since then, WHO has published [9] other approaches developed by referral laboratories, including HKU (Hong Kong) [10,11], China CDC (China) [12], US CDC (United States) [13], and Institut Pasteur, Paris (France) [14]. These assays included several RT-PCRs targeting two or three different SARS-CoV-2 gene regions, including RdRp (RNA-dependent RNA polymerase), N (nucleocapsid protein), E (envelope protein), ORF1ab nsp10 (non-structural protein 10), and ORF1b nsp14 (non-structural protein 14). In the present study, we aimed to compare the sensitivity and specificity of these different RT-PCR assays. Overall, the different RT-PCR assays performed well for SARS-CoV-2 detection and were all specific except N Charité, (Germany) and N2 US CDC (United States) assays. RdRp Institut Pasteur (IP2, IP4), N China CDC, and N1 US CDC were found to be the most sensitive assays.
2. Materials and Methods
2.1. Study Design
To assess sensitivity, different RNA concentrations of SARS-CoV-2 cell culture supernatants, as well as clinical samples with different viral loads (n = 4), were tested using all RT-PCR assays. For the three most sensitive assays, limit of detection (LoD) was assessed, and additional clinical samples (n = 16) with low viral loads were also tested. To assess specificity, clinical samples negative for SARS-CoV-2 (n = 50) were tested using all RT-PCR assays. All clinical samples (nasopharyngeal aspirates) were provided by the Hospices Civils de Lyon—University Hospital, France, and frozen at 80 °C before extraction.
2.2. Sensitivity
The sensitivity for each RT-PCR assay was first assessed using ten-fold serial dilutions from 10−3 to 10−9 of SARS-CoV-2 cell culture supernatants (one replicate for 10−3 and 10−4, three replicates for 10−5 and 10−6, and five replicates for 10−7 to 10−9). Four positive clinical samples were then tested using all RT-PCR assays to confirm these results.
For the three most sensitive assays, we estimated the LoD using a probit analysis by including five additional replicates of each dilution of the cell culture supernatants. Probit analysis consists of describing the relationship between the probability of detection and concentration using a cumulative probability curve. For each dilution, the ratio (the hit rate) is computed as the number of replicates with a detected outcome per the total number of replicates tested. These hit rates are converted mathematically into cumulative normal probability units (probits) and fitted using a regression model vs. their respective concentrations. The LoD is defined as the lowest amount of viral genome that can be detected with a 95% hit rate. For these three most sensitive assays, the results obtained with probit analysis were confirmed by additional testing of sixteen clinical samples with low viral concentration.
2.3. Specificity
The specificity for each RT-PCR assay was assessed using clinical samples (n = 50) tested negative for SARS-CoV-2, including clinical samples (n = 30) tested positive for other respiratory viruses: human coronaviruses 229E, OC43, HKU1, and NL63, human influenza A and B viruses, rhinovirus, respiratory syncytial virus, parainfluenzavirus, adenovirus, metapneumovirus, and picornavirus.
Exploration of false-positive results was performed with additional negative clinical samples, water, and one additional clinical sample tested positive for each target. Amplicon size was analyzed using Agilent DNA 1000 kit (Agilent Technologies, Santa Clara, CA, USA).
2.4. SARS-CoV-2 Cell Culture Supernatants
Cell culture supernatants were obtained from a positive clinical sample cultivated in a biosafety level 3 laboratory on buffalo green monkey cells (cell line provided by the Université Louis Pasteur, Strasbourg, France) [15]. The SARS-CoV-2 culture had an infectious titer of 8.27 log10TCID50/mL as assessed by the Reed and Muench statistical method [16].
2.5. Extraction and RT-PCR
RNA extraction was performed using the EMAG® platform (bioMérieux, Marcy-l’Étoile, France), according to manufacturer’s instructions. RT-PCR was performed following published instructions [5,6,10,11,12,13,14], which are summarized in Table 1; Table 2. Since the China CDC protocol does not specify polymerase, thermocycler, volume of nucleic acid extract, and amplification cycles, the same instructions as those for the HKU assay were applied. RdRp IP2 and IP4 assays from Institut Pasteur, Paris (France) can be multiplexed or used in simplex [14]. Preliminary comparison on SARS-CoV-2 cell culture supernatants found that RdRp IP4 performed better when used in multiplex, whereas IP2 was not significantly impacted (Supplementary Table S1). The CFX 96 Touch™ Real-Time PCR (Bio-Rad, Hercules, CA, USA) was used for all RT-PCR assays.
Table 1.
Summary of the five RT-PCR assays targeting severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2).
| Country (Institute) | Target | Oligonucleotide | Sequence | Amplicon Size a | Polymerase | Thermocycler Used in the Reference Publication | Volume of RNA Extract |
|---|---|---|---|---|---|---|---|
| Charité (Germany) [5,6] | RdRp b | Charité_RdRp_F | GTGARATGGTCATGTGTGGCGG | 100 bp | SuperScript™ III Platinum® One-Step Quantitative RT-PCR System | Light Cycler ® 480II (Roche) or Applied Biosystems ViiA™7 (Therom Fisher) | 5 µL |
| Charité_S_RdRp_P c | FAM-CAGGTGGAACCTCATCAGGAGATGC-BBQ | ||||||
| Charité_NS_RdRp_P d | FAM-CCAGGTGGWACRTCATCMGGTGATGC-BBQ | ||||||
| Charité_RdRp_R | CARATGTTAAASACACTATTAGCATA | ||||||
| E e | Charité_E_F | ACAGGTACGTTAATAGTTAATAGCGT | 113 bp | ||||
| Charité_E_P | FAM-ACACTAGCCATCCTTACTGCGCTTCG-BBQ | ||||||
| Charité_E_R | ATATTGCAGCAGTACGCACACA | ||||||
| N | Charité_N_F | CACATTGGCACCCGCAATC | 128 bp | ||||
| Charité_N_P | FAM-ACTTCCTCAAGGAACAACATTGCCA-BBQ | ||||||
| Charité_N_R | GAGGAACGAGAAGAGGCTTG | ||||||
| HKU (Hong Kong) [10,11] | ORF1b-nsp14 f | HKU_ORF_F | TGGGGYTTTACRGGTAACCT | 132 bp | TaqMan Fast Virus Master mix | Applied Biosystems ViiA™7 (Therom Fisher) | 4 µL |
| HKU_ORF_P | FAM-TAGTTGTGATGCWATCATGACTAG-TAMRA | ||||||
| HKU_ORF_R | AACRCGCTTAACAAAGCACTC | ||||||
| N e | HKU_N_F | TAATCAGACAAGGAACTGATTA | 110 bp | ||||
| HKU_N_P | FAM-GCAAATTGTGCAATTTGCGG-TAMRA | ||||||
| HKU_N_R | CGAAGGTGTGACTTCCATG | ||||||
| China CDC (China) [12] | N | ChinaCDC_N_F | GGGGAACTTCTCCTGCTAGAAT | 99 bp | Unspecified | Unspecified | Unspecified |
| ChinaCDC_N_P | FAM-TTGCTGCTGCTTGACAGATT-TAMRA | ||||||
| ChinaCDC_N_R | CAGACATTTTGCTCTCAAGCTG | ||||||
| ORF1ab-nsp10 | ChinaCDC_ORF_F | CCCTGTGGGTTTTACACTTAA | 119 bp | ||||
| ChinaCDC_ORF_P | FAM-CCGTCTGCGGTATGTGGAAAGGTTATGG-BHQ1 | ||||||
| ChinaCDC_ORF_R | ACGATTGTGCATCAGCTGA | ||||||
| US CDC (United States) [13] | N1 c | USCDC_N1_F | GACCCCAAAATCAGCGAAAT | 72 bp | TaqPath™ 1-Step RT-qPCR Master Mix, CG (Thermo Fisher) | Applied Biosystems™ 7500 Fast (Thermo Fisher) | 5 µL |
| USCDC_N1_P | FAM-ACCCCGCATTACGTTTGGTGGACC-BHQ1 | ||||||
| USCDC_N1_R | TCTGGTTACTGCCAGTTGAATCTG | ||||||
| N2 c | USCDC_N2_F | TTACAAACATTGGCCGCAAA | 67 bp | ||||
| USCDC_N2_P | FAM-ACAATTTGCCCCCAGCGCTTCAG-BHQ1 | ||||||
| USCDC_N2_R | GCGCGACATTCCGAAGAA | ||||||
| N3 d | USCDC_N3_F | GGGAGCCTTGAATACACCAAAA | 72 bp | ||||
| USCDC_N3_P | FAM-AYCACATTGGCACCCGCAATCCTG-BHQ1 | ||||||
| USCDC_N3_R | TGTAGCACGATTGCAGCATTG | ||||||
| Institut Pasteur, Paris (France) [14] | RdRp IP2 (Flo2) | Pasteur_IP2_F | ATGAGCTTAGTCCTGTTG | 108 bp | SuperScript™ III Platinum® One-Step Quantitative RT-PCR System | LightCycler ® 480 (Roche) | 5 µL |
| Pasteur_IP2_P | HEX-AGATGTCTTGTGCTGCCGGTA-BHQ1 | ||||||
| Pasteur_IP2_R | CTCCCTTTGTTGTGTTGT | ||||||
| RdRp IP4 (Flo4) | Pasteur_IP4_F | GGTAACTGGTATGATTTCG | 107 bp | ||||
| Pasteur_IP4_P | FAM-TCATACAAACCACGCCAGG-BHQ1 | ||||||
| Pasteur_IP4_R | CTGGTCAAGGTTAATATAGG |
a Amplicon size in base-pairs (bp) deduced from BetaCoV/Wuhan-Hu-1/2019 sequence (GISAID |EPI ISL 402125). b Target used for confirmation and discrimination of SARS-CoV-2 and SARS-CoV. c Probe specific for SARS-CoV-2. d Probe detecting SARS-CoV-2, SARS-CoV, and bat-SARS-related CoVs. e Target used for screening. f Target used for confirmation.
Table 2.
Amplification cycles of the five RT-PCR assays targeting SARS-CoV-2.
| Institute (Country) | Charité (Germany) [5,6] | HKU (Hong Kong) [10,11] | China CDC (China) [12] | US CDC (United States) [13] | Institut Pasteur, Paris (France) [14] | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Amplification Cycles | T °C | Time (min) | Number of Cycles | T °C | Time (min) | Number of Cycles | T°C | Time (min) | Number of Cycles | T °C | Time (min) | Number of Cycles | T °C | Time (min) | Number of Cycles |
| Uracil-N-glycosylase activation | Unspecified | 25 | 02:00 | 1 | |||||||||||
| Reverse transcription | 55 | 10:00 | 1 | 50 | 05:00 | 1 | 50 | 15:00 | 55 | 20:00 | 1 | ||||
| RT inactivation/Enzyme activation | 95 | 03:00 | 95 | 00:20 | 95 | 02:00 | 95 | 03:00 | |||||||
| Denaturation | 95 | 00:15 | 45 | 95 | 00:05 | 40 | 95 | 00:03 | 45 | 95 | 00:15 | 50 | |||
| Annealing/Extending | 58 | 00:30 | 60 | 00:30 | 55 | 00:30 | 58 | 00:30 | |||||||
| Cooling | 40 | 00:30 | 1 | ||||||||||||
3. Results
3.1. Sensitivity Comparison of the Five RT-PCR Assays
The E Charité and N2 US CDC assays were positive for all specimens, including negative samples and negative controls (water). These false-positive results were explored (details below), but the sensitivity of these assays was not further assessed.
Sensitivity was first assessed using SARS-CoV-2 cell culture supernatants. Using both specific SARS-CoV-2 (S) and non-specific (NS; detecting SARS-CoV-2, SARS-CoV, and bat-SARS-related CoVs) RdRp Charité assays, all replicates of the 10−5 dilution (and inferior); 1/3 (S RdRp) and 3/3 (NS RdRp) of the 10−6 dilution replicates; and none of the 10−7, 10−8, and 10−9 dilutions (0/5) were detected. ORF1b and N HKU, and ORF1ab China CDC assays detected all replicates of dilutions inferior or equal to 10−6 and detected 4/5, 3/5, and 2/5 for 10−7 dilutions, respectively. None of these assays detected replicates of 10−8 (0/5) and 10−9 (0/5) dilutions. In contrast, N Charité, N China CDC, N1 and N3 US CDC, and duplex RdRp IP2/IP4 were positive for most replicates of the 10−7 (5/5, 5/5, 5/5, 4/5, 5/5 and 5/5, respectively) and 10−8 dilutions (3/5, 2/5, 4/5, 5/5, 3/5, 3/5, respectively; Figure 1). At 10−9 dilution, N China, N1 and N3 US CDC, and duplex RdRp IP2/IP4 assays were able to detect replicates (1/5, 1/5, 2/5, 3/5, 1/5, respectively).
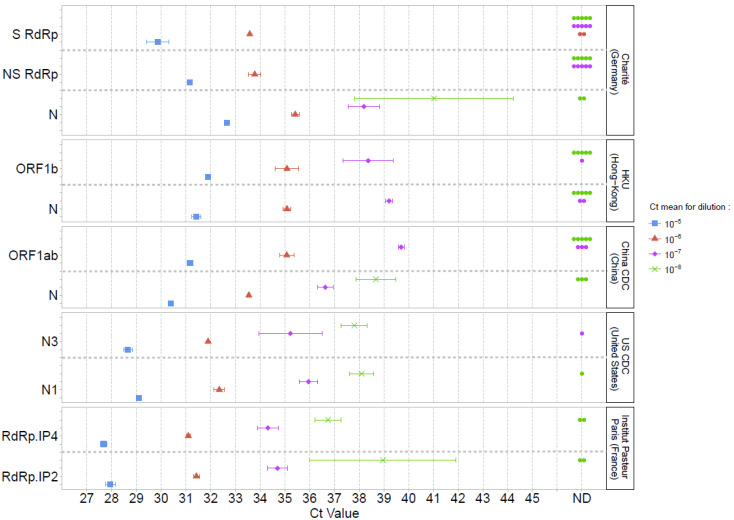
Figure 1.
Mean Ct values and standard deviations obtained using five PCR-based methods for SARS-CoV-2 detection. Serial dilutions of SARS-CoV-2 cell culture supernatants were used and are represented by a single color (10−5 blue, 10−6 red, 10−7 pink, 10−8 green). A point in the ND (non-detected) column (Ct value axis) indicates a negative result for one replicate.
The mean cycle threshold (Ct) values obtained for each assay were then compared for dilutions 10−5 to 10−8 (Figure 1, Supplementary Table S2). Since the accepted technical variability of RT-PCR is below 0.5 log10, we considered a difference of 2 Ct as significant [17]. At 10−5 dilution, the lowest Ct value was 27.7 for RdRp IP4. No significant difference in Ct values (Ct ranged from 28.0 to 29.1) was reported with N1 and N3 US CDC, and RdRp IP2. A similar Ct profile was observed for these assays at 10−6 and 10−7 dilutions. At 10−8 dilution, only N Charité had significantly higher Ct values (41.0 vs. 36.7 to 39.0 for N China CDC, N1 and N3 US CDC, and duplex RdRp IP2/IP4).
Clinical samples (n = 4) were then tested using all RT-PCR assays to confirm the results obtained on SARS-CoV-2 cell culture supernatants (Supplementary Table S3). ORF1b and N HKU, ORF1ab and N China CDC, N1 and N3 US CDC, and RdRp IP2 and RdRp IP4 assays detected all four positive samples. S and NS RdRp, and N Charité assays did not detect the positive sample with the lowest viral concentration.
Taken together, N China CDC, N1 and N3 US CDC, as well as RdRp IP2 and IP4 were the most sensitive assays.
3.2. Limit of Detection for the Most Sensitive Assays
Due to a limited quantity of clinical sample available, only one target from each referral laboratory providing the most sensitive assays was tested: N China CDC, N1 US CDC, and RdRp IP2. The LoD of N3 US CDC was not determined as this assay is not specific for SARS-CoV-2 detection and was removed from the new version of the US CDC assay [13]. We chose to determine the LoD for RdRp IP2 and not IP4 as IP2 detected more replicates at the 10−9 dilution on cell culture supernatants.
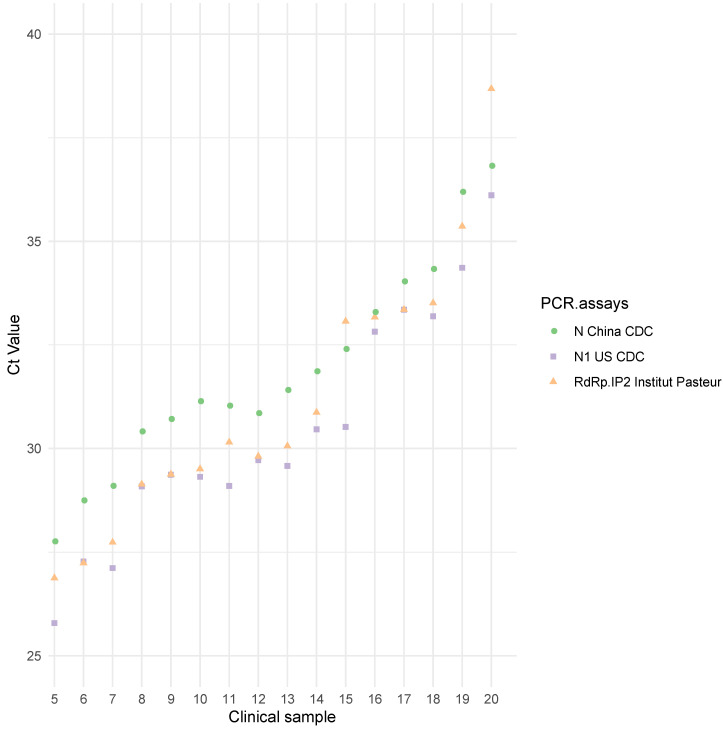
The 95% hit rate obtained was 1.36 log10TCID50/mL [0.8; 3.09] for N China CDC, 0.44 log10TCID50/mL [0.05; 1.83] for N1 US CDC, and 0.63 log10TCID50/mL [0.25; 1.9] for RdRp IP2. The differences observed were not statistically significant. For these three assays, the results were confirmed by additional testing of clinical samples (Figure 2, Supplementary Table S4). N1 US CDC and RdRp IP2 had lower Ct values than N China CDC (Figure 2), but no significant differences (Ct difference < 2) were observed.
Figure 2.
Ct values for 16 positive clinical samples using the three most sensitive assays. N China CDC is represented by green circles, N1 US CDC by purple squares, and RdRp IP2 Institut Pasteur, Paris, by orange triangles.
3.3. Specificity
No false-positive results were obtained on clinical samples that tested negative for SARS-CoV-2 and/or positive for other viruses than SARS-CoV-2, except for E Charité and N2 US CDC, which were positive for all specimens (Supplementary Table S5).
3.4. Exploration of E Charité and N2 US CDC False-Positive Results
Since E Charité and N2 US CDC assays were positive for all specimens and replicates, including negative samples and controls, false-positive results were further explored (Supplementary Figure S1).
For E Charité, negative samples showed two amplicons, one at 84 base pairs (bp) and one at 121 bp, whereas the positive sample only had one amplicon at 121 bp, which is close to the expected size of a specific amplification (Table 1). Thus, the false-positive amplification obtained using E Charité might be derived from a contamination (amplicon size at 121 bp) but could also be associated with an aspecific amplification (amplicon size at 84 bp). Using the N2 US CDC assay, negative samples showed one amplicon at 73 bp, which is close to the expected size of a specific amplification (Table 1). Thus, the false-positive amplification obtained using N2 US CDC might be due to a contamination. Sequencing of these amplicon products should be performed for further investigation.
4. Discussion
The present study compared the performance of five RT-PCR-based methods developed by referral laboratories. N China CDC, N1 US CDC, and RdRp IP2 and IP4 were found to be the most sensitive assays on SARS-CoV-2 cell culture supernatants and clinical respiratory samples. Vogels et al. compared the performance of SARS-CoV-2 PCR assays developed by the same referral laboratories, except those from Institut Pasteur. Using RNA-spiked mock samples, they found that ORF HKU was one of the most sensitive assays [18]. Herein, ORF HKU was more sensitive than RdRp Charité but slightly less sensitive than other assays, such as N1 US CDC or N China. Although RdRp Charité performed well for the lowest dilutions, it was nevertheless found to be less sensitive than others, a result in line with those of Vogels et al. [18]. It is worth noting that the Charité assay was the first to be published at the early stage of the pandemic [9] and has been widely used worldwide [8]. This assay was initially designed for the diagnosis of SARS-related CoVs and then optimized for SARS-CoV-2 detection [5]. Thanks to this assay, an important number of COVID-19 diagnoses were made, which contributed to limiting the spread of the outbreak. In line with the present results, it was reported that RdRp IP2 and RdRp IP4 sensitivity was similar when used in multiplex [14], suggesting that the Institut Pasteur assay should preferentially be used in multiplex. Of note, we did not apply the Ct cut-off values above, in which a sample would be considered negative, since such values were not provided in the protocols made available by the referral laboratories.
As previously reported [19], we identified probable primer contamination using N2 US CDC and E Charité, which prevented us from further evaluating their sensitivity and specificity. Except for these two assays, no false-positive results were observed when testing a wide range of respiratory viruses, a result consistent with previous studies [5,6,10,11,13,14]. Although not observed herein, the amplification of non-specific products for ORF1 and N China CDC, and N2 and N3 US CDC has also been reported [18].
The performance of other RT-PCR tests recently developed [7] should be explored in further studies. In addition, the quantification of SARS-CoV-2 could be performed to assess the effectiveness of potential treatments. The data presented herein are of prime importance to facilitate the equipment choice of all diagnostic laboratories, as well as for the development of marketed tests. Sensitive tests should be widely implemented to limit the spread of the current outbreak and prepare for the post-epidemic phase and future seasonal epidemics.
Acknowledgments
We would like to thank all the patients, clinicians, laboratory technicians, and informatics department who contributed to this investigation. We are also grateful to Véréna Landel and Philip Robinson (DRCI, Hospices Civils de Lyon) for help in the manuscript preparation. We also gratefully acknowledge the authors and laboratories that originated and submitted the sequence from GISAID’s used to calculate amplicon sizes.
Supplementary Materials
The following are available online at https://www.mdpi.com/2077-0383/9/6/1871/s1. Figure S1: Electropherograms of amplicon sizes obtained using Agilent DNA 1000 kit (Agilent Technologies) for one positive sample and one negative sample for E Charité (Germany) and N2 US CDC (United States), respectively. Table S1: Cycle threshold values and mean ± standard deviation for serial dilutions of SARS-CoV-2 cell culture supernatants obtained using IP2 and IP4 Institut Pasteur, Paris (France) when used in simplex and multiplex. Table S2: Cycle threshold values and mean ± standard deviation for serial dilutions of SARS-CoV-2 cell culture supernatants obtained using five RT-PCR assays for SARS-CoV-2 detection made available by WHO, Table S3: Cycle thresholds values obtained on clinical samples (n = 4) using five RT-PCR assays for SARS-CoV-2 detection made available by WHO, Table S4: Cycle threshold values obtained on clinical samples (n = 16) using the three most sensitive methods: N China CDC, N1 US CDC, and IP2 Institut Pasteur, Paris (France). Table S5: Specificity assessment of five RT-PCR assays made available by WHO.
Author Contributions
Conceptualization, F.M. and A.G.; Methodology, S.E. and A.B.; Validation, S.E., A.B., and A.G.; Formal Analysis, S.E. and A.B.; Investigation, K.B.-P., V.C., and L.G.; Resources, S.E., A.B., K.B.-P., M.B., V.C., L.G., and A.G.; Data Curation, K.B.-P., V.C., and L.G.; Writing—Original Draft Preparation, S.E. and A.B.; Writing—Review and Editing, S.E., A.B., V.E., K.B.-P., M.B., G.D., G.B., L.J., E.F., F.M., and A.G.; Visualization, S.E., A.B., K.B.-P., G.O., and A.G.; Supervision, F.M. and A.G. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding. Reagents were provided by Laboratoire Commun de Recherche Hospices Civils de Lyon-bioMérieux, Centre Hospitalier Lyon Sud, Pierre-Bénite, France.
Conflicts of Interest
S.E., V.E., M.B., G.D., G.B., L.J., E.F., F.M., and A.G. declare no conflict of interest. A.B has received a research grant from bioMérieux. K.B.-P., V.C., L.G., and G.O. are employees at bioMérieux. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- 1.Coronaviridae Study Group of the International Committee on Taxonomy of Viruses The species Severe acute respiratory syndrome related coronavirus: Classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020;5:536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.World Health Organization WHO Announces COVID-19 Outbreak a Pandemic. [(accessed on 6 April 2020)]; Available online: http://www.euro.who.int/en/health-topics/health-emergencies/coronavirus-covid-19/news/news/2020/3/who-announces-covid-19-outbreak-a-pandemic.
- 3.World Health Organization Coronavirus Disease 2019 (COVID-19)—Situation Report—127. [(accessed on 26 May 2020)]; Available online: https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200416-sitrep-87-covid-19.pdf?sfvrsn=9523115a_2.
- 4.Holmes E.C. Novel 2019 Coronavirus Genome. [(accessed on 24 March 2020)]; Available online: http://virological.org/t/novel-2019-coronavirus-genome/319.
- 5.Corman V., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D., Bleicker T., Brünink S., Schneider J., Schmidt M.L., et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eurosurveillance. 2020;25:2000045. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Corman V., Bleicker T., Brünink S., Drosten C. Diagnostic Detection of 2019-nCoV by Real-Time RT-PCR; Charité Virology, Berlin, Germany. [(accessed on 24 March 2020)]; Available online: https://www.who.int/docs/default-source/coronaviruse/protocol-v2-1.pdf?sfvrsn=a9ef618c_2.
- 7.Cormac S. Coronavirus and the Race to Distribute Reliable Diagnostics. [(accessed on 24 March 2020)]; doi: 10.1038/d41587-020-00002-2. Available online: https://www.nature.com/articles/d41587-020-00002-2. [DOI] [PubMed]
- 8.Reusken C., Broberg E., Haagmans B., Meijer A., Corman V., Papa A., Charrel R., Drosten C., Koopmans M., Leitmeyer K., et al. Laboratory readiness and response for novel coronavirus (2019-nCoV) in expert laboratories in 30 EU/EEA countries, January 2020. Eurosurveillance. 2020;25:2000082. doi: 10.2807/1560-7917.ES.2020.25.6.2000082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.World Health Organization Coronavirus Disease (COVID-19) Technical Guidance: Laboratory Testing for 2019-nCoV in Humans. [(accessed on 30 March 2020)]; Available online: https://www.who.int/emergencies/diseases/novel-coronavirus-2019/technical-guidance/laboratory-guidance.
- 10.Chu D., Pan Y., Cheng S., Hui K., Krishnan P., Liu Y., Ng D., Wan C., Yang P., Wang Q., et al. Molecular diagnosis of a novel Coronavirus (2019-nCoV) causing an outbreak of pneumonia. Clin. Chem. 2020;66:1–7. doi: 10.1093/clinchem/hvaa029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.HKU Med Detection of 2019 Novel Coronavirus (2019-nCoV) in Suspected Human Cases by RT-PCR. [(accessed on 24 March 2020)]; Available online: https://www.who.int/docs/default-source/coronaviruse/peiris-protocol-16-1-20.pdf?sfvrsn=af1aac73_4.
- 12.China CDC China CDC Primers and Probes for Detection 2019-nCoV. [(accessed on 24 March 2020)]; Available online: http://ivdc.chinacdc.cn/kyjz/202001/t20200121_211337.html.
- 13.Centers for Disease Control and Prevention A CDC 2019-Novel Coronavirus (2019-nCoV) Real-Time RT-PCR Diagnostic Panel. [(accessed on 24 March 2020)]; Available online: https://www.fda.gov/media/134922/download.
- 14.Institut Pasteur, Paris Protocol: Real-Time RT-PCR Assays for the Detection of SARS-CoV-2. [(accessed on 24 March 2020)]; Available online: https://www.who.int/docs/default-source/coronaviruse/real-time-rt-pcr-assays-for-the-detection-of-sars-cov-2-institut-pasteur-paris.pdf?sfvrsn=3662fcb6_2.
- 15.Barron A., Olshevsky C., Cohen M. Characteristics of the BGM line of cells from African green monkey kidney. Brief report. Arch. Gesamte Virusforsch. 1970;32:389–392. doi: 10.1007/BF01250067. [DOI] [PubMed] [Google Scholar]
- 16.Reed L., Muench H. A Simple Method of Estimating Fifty Per Cent Endpoints. Am. J. Epidemiol. 1938;27:493–497. doi: 10.1093/oxfordjournals.aje.a118408. [DOI] [Google Scholar]
- 17.Baraduc M., Baume A., Bouvet F., Canis C., Cattoen S., Charachon V., Cocquerelle R., Courcol C., de Champs M., Ferroni N., et al. Comite Qualite (QUAMIC) de la Société Francaise de Microbiologie—Recommandations 2014. Société Francaise de Microbiologie; Paris, France: 2014. SFM comparaison de methodes; p. 146. [Google Scholar]
- 18.Vogels C., Brito A., Wyllie A., Fauver J., Ott I., Kalinich C., Petrone M., Landry M., Foxman E., Grubaugh N. Analytical sensitivity and efficiency comparisons of SARS-COV-2 qRT-PCR assays (Preprint) [(accessed on 2 April 2020)];Prepr. MedRxiv. Available online: https://www.medrxiv.org/content/10.1101/2020.03.30.20048108v1.full.pdf.
- 19.Mögling R., Meijer A., Berginc N., Bruisten S., Charrel R., Coutard B., Eckerle I., Enouf V., Hungnes O., Korukluoglu G., et al. Early release—Delayed laboratory response to COVID-19 caused by molecular diagnostic contamination. Emerg. Infect. Dis. J. CDC. 2020;26:1843. doi: 10.3201/eid2608.201843. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.