Abstract
The genus Brassica contains important vegetable crops, which serve as a source of oil seed, condiments, and forages. However, their production is hampered by various diseases such as clubroot and Fusarium wilt, especially in Brassica vegetables. Soil-borne diseases are difficult to manage by traditional methods. Host resistance is an important tool for minimizing disease and many types of resistance (R) genes have been identified. More than 20 major clubroot (CR) disease-related loci have been identified in Brassica vegetables and several CR-resistant genes have been isolated by map-based cloning. Fusarium wilt resistant genes in Brassica vegetables have also been isolated. These isolated R genes encode the toll-interleukin-1 receptor/nucleotide-binding site/leucine-rice-repeat (TIR-NBS-LRR) protein. DNA markers that are linked with disease resistance allele have been successfully applied to improve disease resistance through marker-assisted selection (MAS). In this review, we focused on the recent status of identifying clubroot and Fusarium wilt R genes and the feasibility of using MAS for developing disease resistance cultivars in Brassica vegetables.
Keywords: clubroot, Fusarium wilt, R gene, quantitative trait locus, marker-assisted selection, Brassica
1. Introduction
The genus Brassica belongs to the family Brassicaceae (Cruciferae) containing 37 different species (http://www.theplantlist.org) and has great economic importance [1]. Three species, Brassica rapa L. (2n = 20, AA) and Brassica oleracea L. (2n = 18, CC) and its allotetraploid species, Brassica napus L. (2n = 38, AACC) are included in the genus Brassica and comprise commercially important vegetable and oilseed crops. B. rapa includes leafy vegetables such as Chinese cabbage (var. pekinensis), pak choi (var. chinensis), and komatsuna (var. perviridis), root vegetables such as turnip (var. rapa), and oilseed (var. oleifera). B. oleracea comprises commercially important vegetable crops with morphological variations such as cabbage (var. capitata), broccoli (var. italica), kale (var. acephala), kohlrabi (var. gongylodes), and cauliflower (var. botrytis). B. napus includes the oilseed crop, canola/rapeseed.
Various pathogens such as clubroot, Fusarium wilt, black rot, Sclerotinia stem rot, blackleg, white rust, downy mildew, white leaf spot, and turnip mosaic virus can infect Brassica crops [2,3]. Cultural, physical, biological, or chemical controls, or a combination of these controls, integrated pest management, are used for disease control. If plants have natural resistance against these pathogens, the dependence on these controls is reduced and is cost-effective. Thus, disease resistance is an important trait in plant breeding to prevent quality and yield losses.
The first tier of plant immunity is called pathogen-associated molecular pattern (PAMP)-triggered immunity (PTI) [4,5]. Plants recognize pathogens through the PAMPs by pattern recognition receptors (PRRs) [6] and this recognition leads to the activation of PTI. PTI induces the expression of defense genes such as the mitogen-associated protein kinase (MAPK) cascade or WRKY transcription factors [7,8]. In contrast, pathogens deliver virulence molecules called as effectors to suppress PTI [4]. The failure of PTI defense helps to activate an immune response called effector-triggered immunity (ETI), when plants recognize the effectors (Avr proteins) through disease resistance (R) proteins, an ETI is activated [5]. This recognition between R and Avr is termed ‘gene-for-gene resistance’ [9]. ETI is stronger against newly adapted pathogens in host plants than PTI [10]. R proteins contain nucleotide-binding (NB) and leucine rich repeat (LRR) domains, which are called NBS-LRR (nucleotide-binding site leucine-rich repeat) protein. NBS-LRR proteins are separated into two types by their N-terminus domain, either having a toll interleukin-1 receptor (TIR) domain (TIR-NBS-LRR protein) or coiled-coil (CC) domains (CC-NBS-LRR protein) [11,12,13]. In general, the LRR domain provides recognition specificity, the NB domain regulates activation, and the TIR domain regulates downstream signaling [5]. Besides this, some R genes also encode transmembrane receptor-like protein (RLPs), transmembrane receptor-like kinases (RLKs), cytoplasmic kinases (CKs), and proteins with atypical molecular motifs [4]. The constitution of R genes is different between monocotyledonous and dicotyledonous genomes. TIR-NBS-LRR genes are mostly absent in monocotyledons, while TIR-NBS-LRR genes are present in dicotyledons and usually more abundant than CC-NBS-LRR genes [13]. The R genes have been comprehensively identified in several species of the genus Brassica [14,15,16].
In a practical sense, the successful deployment of a novel R gene into a crop depends on the identification of a positive phenotype, the identification of genetic markers for marker-assisted selection (MAS) breeding, and understanding of how the novel resistance will behave under different genetic backgrounds and pathogenic pressures in the field. Clubroot and Fusarium wilt are considered as devastating diseases, and they cause a significant yield loss of Brassica vegetables for many years over the world. Some clubroot-resistant lines are susceptible to the Fusarium wilt and vice versa. In this review, we focus on recent knowledge about R genes of clubroot and Fusarium wilt as several important R genes/quantitative trait loci (QTL) against these pathogens have been identified in Brassica vegetables. In addition, MAS has been used to improve the disease resistance, and several cultivars with higher resistance in Brassica vegetables have recently been developed. We will introduce recent information about R genes and the prospect of their possible utilization for Brassica breeding.
2. Infection Process of the Pathogens
2.1. Infection Process of Clubroot Pathogen P. brassicae
Clubroot is caused by the obligate parasite Plasmodiophora brassicae Woronin and is recognized as a major devastating disease in Brassicaceae that poses an emerging threat to Brassica crop production [17]. Clubroot disease was first reported in Russia in 1878 by Woronin and rapidly expanded to other countries like Europe, Brazil, South Africa, Australia, New Zealand, and China [17]. The infection of plants by P. brassicae is a two-phase process (Figure 1). The resting spores in soil germinate and the resultant zoospores then attack the plant’s root hairs. The zoospores then grow into multi-nucleate plasmodia (primary plasmodia) within the root hairs. The plasmodia cleave the root tissues and form secondary zoospores. The secondary zoospores penetrate into the root cortical tissues in a process known as cortical infection [18,19]. This cortical infection induces abnormal growth by the development of secondary plasmodia inside the affected cell, and proliferation of the secondary plasmodia leads to the formation of distorted massive gall known as club [18,19,20]. During the development of the pathogen in the plant, these secondary zoospores are capable of infecting the same plant or adjacent plants, thus repeating the cycle. Secondary plasmodia develop into multinuclear plasmodia by a number of nuclear divisions, and further meiosis may appear before the formation of numerous resting spores within the diseased plant tissue [19,20]. Resting spores are released into soil by the decay of clubs and survive for many years in soil. The spores are spread field-to-field via drainage water and infected root debris. Clubroot inhibits nutrient and water transport, resulting in wilting and ultimately the death of the infected plant. It is difficult to control clubroot infection by any means except genetic resistance cultivars due to the longevity of the resting spores. Crop rotation by clubroot resistant cultivars can reduce 100% of the clubroot severity compared with the susceptible cultivars [21]. Practicing two or more years of crop rotation by clubroot resistant cultivars with clubroot host significantly reduces the resting spores in soil, which is near to complete eradication of clubroot [22]. The effective and sustainable clubroot management by clubroot resistant cultivars is now disclosed, and hints at the importance of resistant cultivars for clubroot management. On the other hand, the host-range of the pathogen is mostly restricted within Brassicaceae species [19,23,24].
Figure 1.
Infection process of clubroot disease caused by Plasmodiophora brassicae.
2.2. Infection Process of Fusarium Wilt Pathogen Foc/For
Yellowing or Fusarium wilt is caused by Fusarium oxysporum f. sp. conglutinans/rapae (Foc/For). Fusarium wilt disease was first reported in the USA, then in Japan and China, and has now been found almost all over the world [25,26]. The pathogen (Foc, Fusarium oxysporum f. sp. conglutinans)/For, Fusarium oxysporum f. sp. rapae) usually invades plants through their young roots, but can also invade through wounds in older roots [27,28]. This pathogen moves into and colonizes the xylem tissues, blocking vascular transport, leading to leaf yellowing, wilting, and defoliation, and in older plants, stunting and plant death [29,30]. The browning of vascular tissues can be observed in the stem and petiole of late-stage infected plants. It is a warm-weather disease and is active between 16 °C and 35 °C. The disease is more severe in warm conditions (above 24 °C) and not a problem in cool conditions [28,29]. The pathogen can survive in soil, seeds, and seedlings and can spread through water such as rain and flood [27,28] and remain for several years as resting spores in the soil. Two forma specialis (f. sp.) of F. oxysporum can cause disease in Brassicaceae. Foc causes disease in B. oleracea and B. rapa and For is specific to B. rapa [31]. Only two races in the Foc, race 1 and race 2, have been reported in the genus Brassica to date: race1 has been found worldwide and race 2 has only been found in USA and Russia [32].
3. Identification and Molecular Mechanism of Clubroot Resistant (CR) Genes
3.1. CR Loci in B. rapa
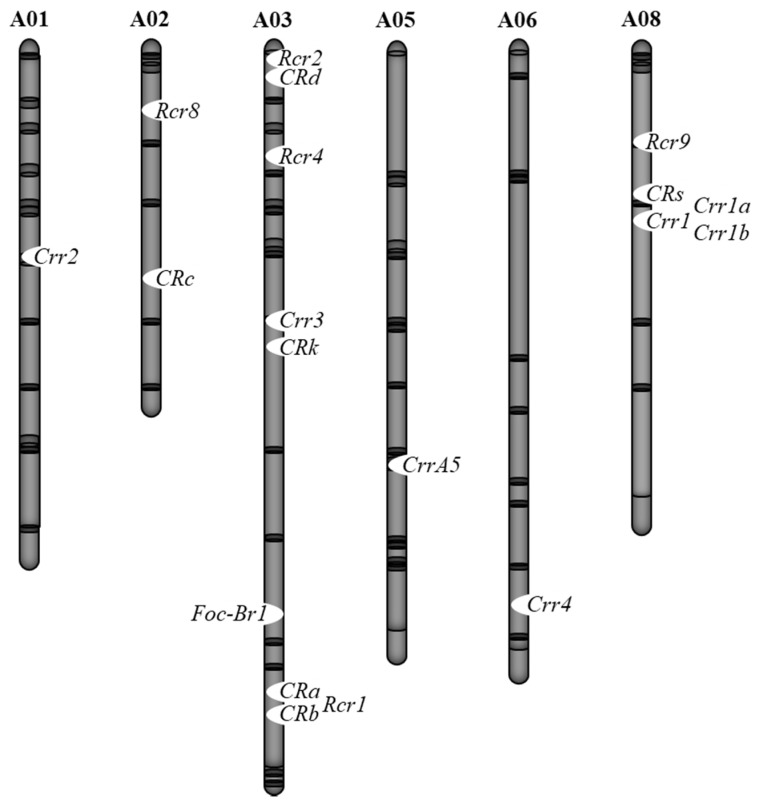
Clubroot disease resistance has been extensively studies in the genus Brassica. Several CR genes have been identified and mapped in B. rapa, B. oleracea, and other Brassica species [33]. In B. rapa, 18 major CR loci have been identified (Figure 2, Table 1); Crr2 mapped on chromosome 1 [34], CRc and CR QTL, designated as Rcr8, on chromosome 2 [35,36], Crr3, CRa, CRb, CRd, CRk, Rcr1, Rcr2, and Rcr4 on chromosome 3 [35,36,37,38,39,40,41,42,43,44,45,46,47,48,49,50], CrrA5 on chromosome 5 [51], Crr4 on chromosome 6 [52], Crr1 (Crr1a, Crr1b), Rcr9, and CRs on chromosome 8 [36,44,53,54]. Most of the CR genes were identified through QTL mapping using a range of resistant sources based on molecular markers, genotyping-by-sequencing (GBS), or bulked segregant RNA sequencing (BSR-seq) strategies.
Figure 2.
Chromosomal locations of clubroot resistant (CR) and Fusarium wilt resistant loci in B. rapa.
Table 1.
CR loci reported on B. rapa.
| QTLs | P/PR | Position | Linked Marker | Gene Source | References |
|---|---|---|---|---|---|
| Crr1 | PR4 | A08 | BRMS-088 | Turnip (G004-Siloga derived) | [34] |
| Crr1a | PR3,4 | A08 | BSA7 | Turnip (G004-DH line) | [53] |
| Crr1b | A08 | AT27 | |||
| Crr2 | PR4 | A01 | BRMS-096 | Turnip (G004-Siloga derived) | [34] |
| Crr3 | PR3 | A03 | OPC11-2S | Turnip (Milan white) | [40,42] |
| Crr4 | PR2,4 | A06 | WE24-1 | Turnip (G004-Siloga derived) | [52] |
| CrrA5 | A05 | RAPD 1, SSR 2 | Chinese cabbage (Inbreed line 20-2ccl) | [51] | |
| CRa | PR2 | A03 | HC352b-SCAR 3 | Chinese cabbage (DH line T136-8) | [39] |
| PR2, P3 | Chinese cabbage (CR Shinki) | [37,38] | |||
| CRb | PR2,4,8 | A03 | TCR09 | Chinese cabbage (CR Shinki DH line, Akiriso) | [41] |
| P3 | KBrH059N21F | Chinese cabbage (CR Shinki) | [43] | ||
| P3 | B0902 | [38,50] | |||
| P4 | KBrB085J21 | Chinese cabbage (CR Shinki DH line) | [46] | ||
| CRc | PR2,4 | A02 | m6R | C9 (DH line of Debra) | [35] |
| CRd | PR4 | A03 | Chinese cabbage (Line 85–74) | [49] | |
| CRk | PR2,4 | A03 | OPC11-2S | K 10 (DH line of CR Kanko) | [35] |
| CRs | P4 | A08 | SNP 4 | Chinese cabbage (cv. Akimeki) | [54] |
| Rcr1 | P3 | A03 | SSR 2 | Flower Nabana (Pak choy) | [45] |
| P2,5,6 | Flower Nabana | [47] | |||
| Rcr2 | P3 | A03 | Chinese cabbage (Jazz) | [48] | |
| Rcr4 | P2,3,5,6,8 | A03 | SNP 4 | Chinese cabbage (T19) | [36] |
| Rcr8 | P5X | A02 | |||
| Rcr9 | P5X | A08 |
P, pathotypes; PR, physiological race of P. brassicae. 1 RAPD-Random Amplification of Polymorphic DNA, 2 SSR-Simple Sequence Repeat, 3 SCAR-Sequence Characterized Amplified Region, 4 SNP-Single Nucleotide Polymorphism.
The first CR gene was identified in the turnip cultivar Siloga using a doubled haploid (DH) population [55] and a dominant major gene CRa was mapped on chromosome 3. A candidate gene of CRa has been identified, and it encodes a TIR-NBS-LRR protein [37]. Crr1a and CRb genes have also been identified by map-based cloning [38,46,53]. CRb was isolated independently of CRa, but they were identical genes [37,38]. Crr1a encodes TIR-NBS-LRR [38].
Recently, proteomics in Chinese cabbage during response to P. brassicae infection identified differentially expressed proteins (DEPs) between the susceptible and resistant lines [56]. Gene ontology analysis using DEPs showed that the category of ‘Glutathione transferase activity’ was overrepresented, suggesting that glutathione transferase is responsible for protecting plants from disease [56].
3.2. CR Loci in B. oleracea
In contrast to B. rapa, no major CR genes or lines with strong resistance have been identified in B. oleracea [57]; only a few completely resistant accessions have been identified in B. oleracea. Genetic analysis of CR in B. oleracea was studied using diallel crossing methods or segregating populations. Only one major resistance gene, Rcr7, has been identified, and it might be located on chromosome 7 (LG 7) in cultivars, Tekila and Kilaherb of cabbage [57]. About fifty QTLs have been identified in the studies using different populations of B. oleracea (Table 2): three QTLs in broccoli [58], two in kale [59], two in cabbage [60], one in kale [61], three in kale [62], nine in kale [63], five in cabbage [64], three in cabbage using the GBS technique [65], and twenty-three QTLs in cabbage using single-nucleotide polymorphism (SNP) microarray technique [66]. The identification of several CR loci indicates that clubroot resistance in B. oleracea is controlled in a polygenic manner, confirming the complex genetic basis of the resistance, where a single resistance locus is not enough to confer sufficient resistance [67]. The comparison of these QTLs is currently impossible due to a lack of common molecular markers among different researchers and the use of different CR sources and races of pathogen [64].
Table 2.
CR loci reported on B. oleracea.
| QTLs | P/PR | Position | Linked Marker | Gene Source | References |
|---|---|---|---|---|---|
| Rcr7 | P3,5X | LG7 | Cabbage cv. Tekila and Kilaherb | [57] | |
| 3 QTLs | PR7 | LG1 LG4 LG9 |
14a 48 177b |
Broccoli (CR-7) | [58] |
| 2 QTLs | ECD 16/31/31 | - | OPL6-780, OPB11-740, OPA18-14900, OPA4-700, OPE20-1250, OPA1-1880, OPA16-510 | Kale (C10) | [59] |
| Pb-3 Pb-4 |
ECD 16/3/30 | LG3 LG1 |
4NE11a 2NA8c |
Cabbage (Bindsachsener) | [60] |
| 1 QTL | PR2 | LG3 | WG6A1, WG1G5 | Kale (K269) | [61] |
| QTL1 QTL3 QTL9 |
PR2 | LG1 LG3 LG9 |
SCA02a2 SCB50b, SCB74c SOPT15a, SCA25 |
Kale (K269) | [62] |
| Pb-Bo1 Pb-Bo2 Pb-Bo3 Pb-Bo4 Pb-Bo5a Pb-Bo5b Pb-Bo8 Pb-Bo9a Pb-Bo9b |
P1,2,4,7 | LG1 LG2 LG3 LG4 LG5 LG5 LG8 LG9 LG9 |
Ae05.8800, T2 PBB38a, r10.1200 Ae15.100, RGA8.450 ELI3.983, aa9.983 PBB7b, ae05.135 ELI3.115, a18.1400 C01.980, t16.500 Aj16.570, W22B.400 A04.1900, ae03.136 |
Kale (C10) | [63] |
| Pb-Bo(Anju)1 Pb-Bo(Anju)2 Pb-Bo(Anju)3 Pb-Bo(Anju)4 Pb-Bo(GC)1 |
PR4 | LG2 LG2 LG3 LG7 LG5 |
KBrHo59L13 CB10026 KBrB068C04 KBrB089H07 CB10065 |
Cabbage (cv. Anju) |
[64,67] |
| 2 QTLs | PR2 | LG2 | Cabbage (C1220) | [65] | |
| 1 QTL | PR9 | LG3 | |||
| 23 QTLs | PR4 | - | Cabbage (GZ87) | [66] |
P, pathotypes; PR, physiological race of P. brassicae; ECD, European Clubroot Differential set pathotype.
3.3. CR Loci other Brassica Species
In B. napus, the majority of CR identified genes are derived from B. rapa var. rapifera [57]. In B. napus, one dominant gene and more than 30 QTLs were identified (Table 3). Two QTLs, CR2a and CR2b, were identified using Rutabaga (cv. Wilhelmsburger) showing resistance to race 2 of P. brassicae [68]. A major gene, Pb-Bn1, mapped on chromosome A03 was reported first and two minor QTLs were mapped on linkage groups C02 and C09 [69]. Nineteen race-specific resistance QTLs were mapped on eight different chromosomes, including the A genome (A02, A03, A08, A09) and C genome (C03, C05, C06, C09) [70]. Besides this, five QTLs using a DH line of canola against pathotype 3 [71], and nine QTLs from different accession of oilseed rape were identified, seven of which were novel through integrative analysis [8]. They first applied genome-wide association study (GWAS) based on whole-genome SNP data to detect that nine QTLs and reported that these QTLs cover genes encoding TIR-NBS gene family [8]. Some resistance loci with one dominant and two recessive loci were identified [72], and one locus linked to CRa gene [73] and a genomic region on chromosome A08 carrying resistance to all five pathotypes, namely pathotypes 2, 3, 5, 6, and 8, were also identified [74]. This suggests that a single gene or a cluster of genes located in this genomic region is involved in the control of resistance to these pathotypes [74]. Recently, two major loci on chromosome A02 and A03 controlling resistance, and seven minor loci, were identified by a SNP association analysis [75].
Table 3.
CR loci reported on B. napus and B. nigra.
| QTLs | PG/PR | Position | Process | Gene Source | References |
|---|---|---|---|---|---|
| B. napus | |||||
|
CR2a
CR2b |
PR2 | LG6 LG1 |
RFLP 2 | Rutabage (cv. Wilhelmsburger) | [68] |
|
Pb-Bn1 1 QTL 1 QTL |
P4,7 | A03 C02 C09 |
RAPD 3 | Oilseed rape (cv. Darmor-bzh) | [69] |
| 3 QTLs | SRSI | LG6 1 | AFLP 4, SSR 5 | Canola (cv. Mendel) | [72] |
| 19 QTLs | 7 isolates with dissimilar P | A02, A03, A08, A09, C03, C05, C06, C09 | AFLP 4, SSR 5 | Oilseed rape (cv. Boohmerwaldkohl and ECD04) | [70] |
| 5 QTLs | P3,5,6,8 | A03 | SSR 5/InDel 6 | Canola (cv. Mendel) | [71] |
| 1 QTL | P3 | A03 | PCR-based marker | Canola (DH line 12-3, ECD04 derived) | [73] |
| 1 QTL | P2,3,5,6,8 | A08 | SSR 5 | Rutabage (BF) | [74] |
| 9 QTLs | P4 | - | Oilseed rape (different accession) | [8] | |
| 2 QTLs | ECD 17/31/31 | A02, A03 | SNP 7 | Oilseed rape | [75] |
| B. nigra | |||||
| Rcr6 | P3 | B03 | Accession PI 219,576 (parental line) | [33] |
P, pathotypes; PR, physiological race of P. brassicae; SRSI, Single Resting Spore Isolate of P. brassicae; ECD, European Clubroot Differential set pathotypes. 1 Dominant locus (with two recessive loci), 2 Restriction Fragment Length Polymorphism, 3 Random Amplification of Polymorphic DNA, 4 Amplified Fragment Length Polymorphism, 5 Simple Sequence Repeat, 6 Insertion-Deletion, 7 Single Nucleotide Polymorphism.
A single dominant gene Rcr6 was also identified on chromosome 3 of the B genome (B03) through BSR-Seq and further mapped with Kompetitive Allele Specific PCR (KASP) analysis in Brassica nigra lines PI 219,576 [33]. The authors declared that Rcr6 was the first gene identified and mapped in the B genome of Brassica species. All of the CR genes found in the genus Brassica encode TIR-NBS-LRR proteins [57].
4. Identification and Molecular Mechanism of Fusarium Wilt Resistance Genes
Two types of resistance (Type A and Type B) in B. oleracea have been reported against Fusarium wilt [76]. Type A resistance is controlled by a single dominant gene and is stable at temperatures higher than 24 °C where Type B is polygenic and becomes unstable at temperatures above 24 °C [27,76,77]. Type A resistance is controlled by a single dominant gene against race 1 in B. rapa and B. oleracea and has been studied extensively in recent years (Figure 3, Table 4) [29,30,32,78,79,80].
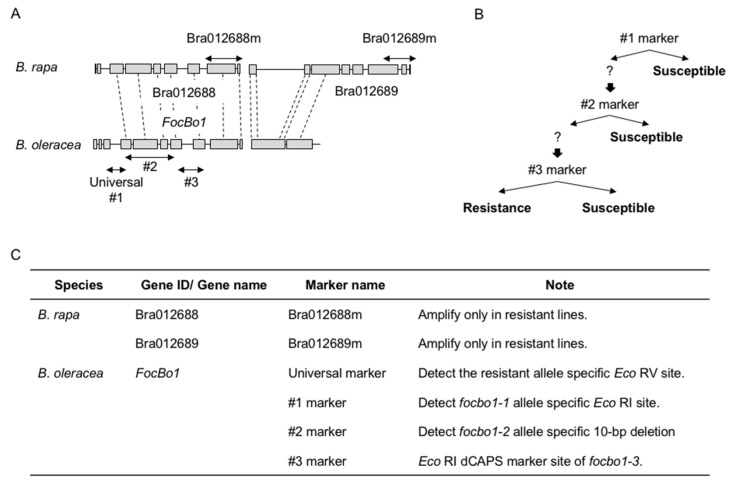
Figure 3.
Schematic view of the alignment of resistance genes of Fusarium wilt disease. (A). DNA marker positions of resistance genes in B. rapa and B. oleracea. Arrows indicate marker positions. (B). Scheme of marker assisted selection in B. oleracea. (C). DNA marker list for marker assisted selection in B. rapa and B. oleracea.
Table 4.
Loci of resistance gene to Foc reported in Brassica species.
| QTLs | Position | Linked Marker/Process | Gene Source | References |
|---|---|---|---|---|
| B. rapa | ||||
| Foc-Br1a | A03 | Bra012688m | Chinese cabbage (F2 population) | [81] |
| Foc-Br1b | Bra012689m | |||
| B. oleracea | ||||
| FOC | C06 | InDel marker: M10 and A1 | Cabbage (DH lines) | [78] |
| FOC1 1 | C06 | InDel marker: Bol037156 and Bol037158 | Cabbage (DH line and F2 population) | [32] |
| QTL1 | C04 | SSR marker: KBrS003O1N10 | Cabbage (AnjuP01): F2 population | [29] |
| QTL2 (Foc-Bo1)2 | C07 | |||
| Foc-Bo11 (SDG) | C07 | InDel marker: BoInd 2 and BoInd 11 | Cabbage (AnjuP01): Recombinant F2 population | [30] |
1 Single dominant gene, 2 major QTL.
In B. rapa, transcriptome analysis was performed using resistant and susceptible lines. The differentially expressed R genes were identified and seven dominant DNA markers at R genes were developed. Two dominant DNA markers on Bra012688 and Bra012689 were completely linked to the resistance phenotype by an inoculation test, indicating that these two genes are candidates for Fusarium wilt resistance genes in B. rapa (Figure 3, Table 4). These two genes encode TIR-NBS-LRR proteins [81]. Dominant DNA markers, Bra012688m and Bra012689m, were applied to Chinese cabbage inbred lines and confirmed close linkage to the Fusarium wilt resistant phenotype [82]. Furthermore, the transcriptome profiles following Foc inoculation between Fusarium wilt-resistant and -susceptible lines in B. rapa were compared and differentially expressed genes were identified [79]. These genes may be responsible for the resistance mechanism to Foc [79]. Differentially expressed genes between B. rapa and Arabidopsis thaliana after Foc inoculation at the same time point were compared and up-regulated genes related to defense response were identified [79], that may be candidates for conferring resistance against Foc.
Recently, Type A resistance has been mapped and molecular markers have been developed in B. oleracea [29,30,32]. The Fusarium wilt resistance gene, FocBo1, was mapped on chromosome 7 by both segregation testing and QTL analysis, and the closest simple sequence repeat (SSR) marker KBrS003O1N10 was developed [29]. One minor QTL was also detected on chromosome 4. In a previous study, the resistance gene on chromosome 6 of cabbage was linked to two insertion/deletion (InDel) markers: M10 and A1 [78]. Later, it was shown that the resistance of Fusarium wilt was controlled by a single dominant gene based on the segregation ratio of two populations (resistant inbred line, 99–77 and highly susceptible line, 99–91). Two R genes in the target region, re-Bol037156 and re-Bol0371578, were predicted as resistance genes, and re-Bol037156 gene, which encodes a putative TIR-NBS-LRR type R protein, has highly similar sequences among the resistant lines [31]. FocBo1 locus was identified on chromosome 7 and this locus was fine-mapped by using 139 recombinant F2 plants derived from resistant cabbage (AnjuP01) and susceptible broccoli (GCP04) DH lines [30]. The FocBo1 gene was shown by fine mapping to be an orthologous gene of Bra012688 in Chinese cabbage [30].
The proteome of xylem sap of the non-infected and Foc infected plants in both resistant and susceptible cabbage cultivars was also investigated using liquid chromatography-tandem mass spectrometry (LC-MS/MS) after the in-solution digestion of xylem sap proteins [83]. Twenty-five proteins in the infected xylem sap were found and ten of them were cysteine-containing secreted small proteins, suggesting that they are candidates for virulence and/or avirulence effectors. The transcriptome profiling of resistance to Foc in cabbage roots were also analyzed [26], where 885 differentially expressed genes (DEGs) were identified between infected and control samples at 4, 12, 24, and 48 h after inoculation. Some genes involved in Salicylic acid (SA)-dependent systemic acquired resistance (SAR), ethylene (ET)-, jasmonic acid (JA)-mediated, and the lignin biosynthesis pathways showed differential expression; the authors discussed the possibility that DEGs involved in these pathways may play important roles in resistance against Foc inoculation [26].
5. Resistant Breeding, Gene Accumulation, and MAS
MAS is an indirect selection process where a trait of interest is selected based on a marker (morphological, biochemical, or DNA/RNA variation) linked to that trait. Selecting individuals with disease resistance using MAS involves identifying a marker allele that is linked to disease resistance rather than to the level of disease resistance. There are several types of DNA markers that have been used to identify disease resistance genes [32,41,43,46,59,78,82,84].
The complexity of plant–pathogen interaction is a problematic in the case of CR breeding due to the appearance of multiple races of the pathogen [85]. Combinations of different CR genes exhibit higher resistance to the disease [62,67,86]. Though CR cultivars have been used widely for major production areas, field isolates of P. brassicae show variation, and different resistance sources from either B. rapa or B. oleracea vegetables were attained by P. brassicae. This suggests a serious risk that a resistance gene can be overcome by pathogen variants [3]. For example, seven CR canola cultivars were characterized for virulence in 106 P. brassicae population, and 61 of 106 P. brassicae population overcame the resistance in at least one of the seven CR cultivars [87]. There are many reports that CR genes show different reactions against the variable virulence of P. brassicae [34,36,44,48,61,62,63,66,74], but heterozygous CR loci are less resistant than the homozygous state [18]. B. rapa possesses several major CR loci (Table 1), which may confer differential (pathotype-specific) resistance to particular isolates of P. brassicae, and sometimes this may have a large effect on resistance [34,52,85,88]. The NARO Institute of Vegetable and Tea Science (NIVTS) has developed a high CR Chinese cabbage cultivar, ‘Akimeki’, by the accumulation of Crr1, Crr2, and CRb genes. It was proven that the accumulation of CR genes through MAS strengthened resistance and, consequently, it can be resistant to the multiple races of P. brassicae in B. rapa. Three CR genes, CRa, CRk, and CRc, were accumulated in Chinese cabbage through MAS [85] and the homozygous lines for the CR genes exhibited exceedingly high resistance against all six field isolates of P. brassicae. The effect of accumulation of different CR genes could be controlled by the dose-dependent accumulation of CR proteins [53,89]. In B. oleracea, resistance in genotypes has generally been identified less frequently than in the genotypes of B. rapa and the level of resistance is low [90]. This might be due to the polygenic nature of resistance in B. oleracea [67]. B. oleracea progeny were developed by accumulating major and minor QTLs to evaluate its effectiveness to the clubroot disease [64]. Three QTLs in the F2/F3 population from the cross between cabbage and kale line K269 were identified [62]. The accumulation of those three CR genes showed broad resistance to three isolates. It was observed that only one major QTL PbBo(Anju)1 showed moderate resistance, whereas three minor QTLs without the major one showed distinct susceptibility [64]. Later, it was proven that PbBo(Anju)1 and three minor QTLs PbBo(Anju)2, PbBo(Anju)4, and PbBo(GC)1 play a critical role in the acquisition of resistance to clubroot disease [67,86]. Here, PbBo(Anju)1 plays a crucial role in the expression of clubroot resistance, and pyramiding minor CR genes are also essential for achieving higher resistance [67,86]. Their effectiveness was verified for controlling disease involving various isolates of P. brassicae [67]. Recently, two CR genes, CRb and PbBa8.1, were combined through MAS and CR homozygous lines in developed B. napus. The homozygous lines demonstrated a higher resistance than the heterozygous lines [91].
The Type A resistance to Fusarium wilt disease controlled by a single dominant gene has been successfully mapped and molecular markers have been developed: SSR marker KBrS003O1N10 [29], InDel markers M10 and A1 [78], Indel markers Bra012688m and Bra012689m [81,82], and DNA marker sets [30,80,84], which are used to generate a series of resistance cultivars (Figure 3).
Breeding cultivars that have resistance to both clubroot and Fusarium wilt is desired. However, inoculation tests against multiple pathogens or multiple races are difficult to perform on the same individual plant. Thus, DNA marker-based selection is useful for the identification of plants that have one Fusarium wilt resistance gene and multiple clubroot resistance genes. Furthermore, it is necessary to confirm whether these resistance genes are linked. In B. rapa, a Fusarium wilt resistance gene is located on chromosome 3, and CRa/CRb, Crr3, and CRk are located near this Fusarium wilt resistance gene. The CRa/CRb gene is the closest, being approximately 2 Mb in physical distance to the Fusarium wilt resistance gene (Figure 2). Since recombination between these two genes can occur [82], it is possible to inherit both resistance genes. In B. oleracea, a Fusarium wilt resistance gene is located on chromosome 7, and there is a minor QTL for clubroot resistance, PbBo(Anju)4, nearby this Fusarium wilt resistance gene. However, these loci are not completely linked to each other [81,83,84]. Therefore, it is possible to have both resistance genes. In B. napus, the association between susceptibility to Fusarium wilt and clubroot resistance against pathotype 3 was found, and these two resistance genes are located about 10 cM apart [92]. However, recombination between these two genes has been reported [92], suggesting that it is possible to inherit both resistance genes and identify them by DNA marker-based selection.
From the results from various researchers, it has been demonstrated that the DNA markers developed can select for the genes that are required for the acquisition of resistance, and these markers could be a powerful tool for resistance breeding in Brassica species. The novel breeding method developed can reinforce resistance by pyramiding R genes through MAS. For the genetic accumulation of R genes corresponding to wide pathogenicity, MAS is indispensable because it allows a precise identification of how many R genes are involved in a cultivar, and can monitor the accumulation of R genes in the progeny in the breeding program. To increase the durability of resistant cultivars to a broader spectrum of pathogen races, the combination of different R genes into a single line will be indispensable.
6. Conclusions
Brassica production is hampered by various diseases, especially clubroot and Fusarium wilt. Many types of R genes/QTLs have been identified in Brassica against the diseases and are being used for the improvement of resistance in cultivars. In case of clubroot disease, a total of 18 major CR loci have been identified in B. rapa, whereas only one major CR locus (Rcr7) and about 50 QTLs were detected in B. oleracea. Moreover, one locus (Pb-Bn1) on the A genome with more than 30 QTLs in B. napus and one locus (Rcr6) on the B genome in B. nigra were also identified. Several types of DNA markers that are linked with disease resistance allele have been developed, and they have been used for MAS. However, when there are several pathotypes, it is necessary to match effective R genes with a specific pathotypes and develop the DNA markers. The accumulation of CR genes corresponding to a wide pathogenicity will be important for breeding resistant cultivars.
A single type A dominant locus (Foc-1) was identified in B. rapa and B. oleracea, and several DNA markers have been developed.
R genes found from both diseases mostly encode a putative TIR-NBS-LRR. Understanding how plants cope with exposure to multiple pathogens such as P. brassicae and Foc will be important in breeding cultivars with multiple disease resistance.
Acknowledgments
We thank Kenji Osabe and Elizabeth S. Dennis for their helpful comments and manuscript editing.
Author Contributions
Conceptualization, H.M., R.F., and M.A.-u.D.; writing—original draft preparation, H.M., A.A., N.M., J.M., D.J.S., R.F., and M.A.-u.D.; supervision, wrote the manuscript. R.F.; funding acquisition, R.F. and M.A.u.D. All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded by grants from Project of the NARO Bio-oriented Technology Research Advancement Institution (Research program on development of innovation technology) and International Research Fellow of JSPS (Invitation Fellowships for Research in Japan (Long-term)).
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Dixon G.R. Vegetable Brassicas and related crucifers. In: Atherton J., editor. Crop Production Science in Horticulture Series. Volume 14. CABI; Wallingford, UK: 2007. p. 327. [Google Scholar]
- 2.Lv H., Fang Z., Yang L., Zhang Y., Wang Y. An update on the arsenal: Mining resistance genes for disease management of Brassica crops in the genomic era. Hortic. Res. 2020;7:34. doi: 10.1038/s41438-020-0257-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Neik T.X., Barbetti M.J., Batley J. Current status and challenges in identifying disease resistance genes in Brassica napus. Front. Plant Sci. 2017;8:1788. doi: 10.3389/fpls.2017.01788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jones J.D., Dangl J.L. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- 5.Dodds P.N., Rathjen J.P. Plant immunity: Towards an integrated view of plant-pathogen interactions. Nat. Rev. Genet. 2010;11:539–548. doi: 10.1038/nrg2812. [DOI] [PubMed] [Google Scholar]
- 6.Boller T., Felix G. A renaissance of elicitors: Perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu. Rev. Plant Biol. 2009;60:379–406. doi: 10.1146/annurev.arplant.57.032905.105346. [DOI] [PubMed] [Google Scholar]
- 7.Bigeard J., Colcombet J., Hirt H. Signaling mechanism in pattern-triggered immunity (PTI) Mol. Plant. 2015;8:521–539. doi: 10.1016/j.molp.2014.12.022. [DOI] [PubMed] [Google Scholar]
- 8.Li L., Luo Y., Chen B., Xu K., Zhang F., Li H., Huang Q., Xiao X., Zhang T., Hu J., et al. A genome-wide association study reveals new loci for resistance to clubroot disease in Brassica napus. Front. Plant Sci. 2016;7:1483. doi: 10.3389/fpls.2016.01483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dangl J.L., Jones J.D. Plant pathogens and integrated defence responses to infection. Nature. 2001;411:826–833. doi: 10.1038/35081161. [DOI] [PubMed] [Google Scholar]
- 10.Noman A., Aqeel M., Lou Y. PRRs and NB-LRRs: From signal perception to activation of plant innate immunity. Int. J. Mol. Sci. 2019;20:1882. doi: 10.3390/ijms20081882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meyers B.C., Kozik A., Griego A., Kuang H., Michelmore R.W. Genome-wide analysis of NBS-LRR-encoding genes in Arabidopsis. Plant Cell. 2003;15:809–834. doi: 10.1105/tpc.009308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Joshi R.K., Nayak S. Functional characterization and signal transduction ability of nucleotide-binding site-leucine-rich repeat resistance genes in plant. Genet. Mol. Res. 2011;10:2637–2652. doi: 10.4238/2011.October.25.10. [DOI] [PubMed] [Google Scholar]
- 13.Marone D., Russo M.A., Laido G., De Leonardis A.M., Mastrangelo A.M. Plant nucleotide binding site-leucine-rich repeat (NBS-LRR) gene: Active guardians in host defense responses. Int. J. Mol. Sci. 2013;14:7302–7326. doi: 10.3390/ijms14047302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yu J., Tehrim S., Zhang F., Tong C., Huang J., Cheng X., Dong C., Zhou Y., Qin R., Hua W., et al. Genome-wide comparative analysis of NBS-encoding genes between Brassica species and Arabidopsis thaliana. BMC Genom. 2014;15:3. doi: 10.1186/1471-2164-15-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bayer P.E., Golicz A.A., Tirnaz S., Chan C.K., Edwards D., Batley J. Variation in abundance of predicted resistance genes in the Brassica oleracea pangenome. Plant Biotechnol. J. 2019;17:789–800. doi: 10.1111/pbi.13015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dolatabadian A., Bayer P.E., Tirnaz S., Hurgobin B., Edwards D., Batley J. Characterization of disease resistance genes in the Brassica napus pangenome reveals significant structural variation. Plant Biotechnol. J. 2020;18:969–982. doi: 10.1111/pbi.13262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Perez-Lopez E., Waldner M., Hossain M., Kusalik A.J., Wei Y., Bonham-Smith P.C., Todd C.D. Identification of Plasmodiophora brassicae effectors—A challenging goal. Virulence. 2018;9:1344–1353. doi: 10.1080/21505594.2018.1504560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hirai M. Genetic analysis of clubroot resistance in Brassica crops. Breed. Sci. 2006;56:223–229. doi: 10.1270/jsbbs.56.223. [DOI] [Google Scholar]
- 19.Kageyama K., Asano T. Life cycle of Plasmodiophora brassicae. J. Plant Growth Regul. 2009;28:203–211. doi: 10.1007/s00344-009-9101-z. [DOI] [Google Scholar]
- 20.Schwelm A., Fogelqvist J., Knaust A., Jülke S., Lilja T., Bonilla-Rosso G., Karlsson M., Shevchenko A., Dhandapani V., Choi S.R., et al. The Plasmodiophora brassicae genome reveals insights in its life cycle and ancestry of chitin synthases. Sci. Rep. 2015;5:11153. doi: 10.1038/srep11153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hwang S.F., Ahmed H.U., Zhou Q., Fu H., Turnbull G.D., Fredua-Agyeman R., Strelkov S.E., Gossen B.D., Peng G. Influence of resistant cultivars and crop intervals on clubroot of canola. Can. J. Plant Sci. 2019;99:862–872. doi: 10.1139/cjps-2019-0018. [DOI] [Google Scholar]
- 22.Ernst T.W., Kher S., Stanton D., Rennie D.C., Hwang S.F., Strelkov S.E. Plasmodiophora brassicae resting spore dynamics in clubroot resistant canola (Brassica napus) cropping systems. Plant Pathol. 2019;68:399–408. doi: 10.1111/ppa.12949. [DOI] [Google Scholar]
- 23.Rolfe S.A., Strelkov S.E., Links M.G., Clarke W.E., Robinson S.J., Djavaheri M., Malinowski R., Haddadi P., Kagale S., Parkin I.A.P., et al. The compact genome of the plant pathogen Plasmodiophora brassicae is adapted to intracellular interactions with host Brassica spp. BMC Genom. 2016;17:272. doi: 10.1186/s12864-016-2597-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wagner G., Laperche A., Lariagon C., Marnet N., Renault D., Guitton Y., Bouchereau A., Delourme R., Manzanares-Dauleux M.J., Gravot A. Quantitative resistance to clubroot deconvoluted into QTL-specific metabolic modules. J. Exp. Bot. 2019;70:5375–5390. doi: 10.1093/jxb/erz265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smith E.F. The fungus infection of agricultural soils in the United States. Sci. Am. 1899;48:19981–19982. [Google Scholar]
- 26.Xing M., Lv H., Ma J., Xu D., Li H., Yang L., Kang J., Wang X., Fang Z. Transcriptome profiling of resistance to Fusarium oxysporum f. sp. conglutinans in cabbage (Brassica oleracea) roots. PLoS ONE. 2016;11:e0148048. doi: 10.1371/journal.pone.0148048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sherf A.F., MacNab A.A. Vegetable Diseases and Their Control. 2nd ed. John Wiley & Sons; New York, NY, USA: 1986. pp. 1–22. [Google Scholar]
- 28.Daly P., Tomkins B., Rural Industries Research and Development Corporation (Canbera, Australia) Production and postharvest handling of Chinese cabbage (Brassica rapa var. pekinensis): A review of literature. Rural Ind. Res. Dev. Corp. Barton ACT. 1997;97:32–35. [Google Scholar]
- 29.Pu Z., Shimizu M., Zhang Y., Nagaoka T., Hayashi T., Hori H., Matsumoto S., Fujimoto R., Okazai K. Genetic mapping of a Fusarium wilt resistance gene in Brassica oleracea. Mol. Breed. 2012;30:809–818. doi: 10.1007/s11032-011-9665-8. [DOI] [Google Scholar]
- 30.Shimizu M., Pu Z., Kawanabe T., Kitashiba H., Matsumoto S., Ebe Y., Sano M., Funaki E., Fujimoto R., Okazai K. Map-based cloning of a candidate gene conferring Fusarium yellows resistance in Brassica oleracea. Theor. Appl. Genet. 2015;128:119–130. doi: 10.1007/s00122-014-2416-6. [DOI] [PubMed] [Google Scholar]
- 31.Enya J., Togawa M., Takeuchi T., Yoshida S., Tsushima S., Arie T., Sakai T. Biological and phylogenetic characterization of Fusarium oxysporum complex, which causes yellows on Brassica spp. and proposal of F. oxysporum f. sp. rapae, a novel forma specialis pathogenic on B. rapa in Japan. Phytopathology. 2008;98:475–483. doi: 10.1094/PHYTO-98-4-0475. [DOI] [PubMed] [Google Scholar]
- 32.Lv H., Fang Z., Yang L., Zhang Y., Wang Q., Liu Y., Zhuang M., Yang Y., Xie B., Liu B., et al. Mapping and analysis of a novel candidate Fusarium wilt resistance gene FOC1 in Brassica oleracea. BMC Genom. 2014;15:1094. doi: 10.1186/1471-2164-15-1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chang A., Lamara M., Wei Y., Hu H., Parkin I.A.P., Gossen B.D., Peng G., Yu F. Clubroot resistance gene Rcr6 in Brassica nigra resides in a genomic region homologous to chromosome A08 in B. rapa. BMC Plant Biol. 2019;19:224. doi: 10.1186/s12870-019-1844-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Suwabe K., Tsukazaki H., Iketani H., Hatakeyama K., Fujimura M., Nunome T., Fukuoka H., Matsumoto S., Hirai M. Identification of two loci for resistance to clubroot (Plasmodiophora brassicae Woronin) in Brassica rapa L. Theor. Appl. Genet. 2003;107:997–1002. doi: 10.1007/s00122-003-1309-x. [DOI] [PubMed] [Google Scholar]
- 35.Sakamoto K., Saito A., Hayashida N., Taguchi G., Matsumoto E. Mapping of isolate-specific QTLs for clubroot resistance in Chinese cabbage (Brassica rapa L. spp. pelkinensis) Theor. Appl. Genet. 2008;117:759–767. doi: 10.1007/s00122-008-0817-0. [DOI] [PubMed] [Google Scholar]
- 36.Yu F., Zhang X., Peng G., Falk K.C., Strelkov S.E., Gossen B.D. Genotyping-by-sequencing reveals three QTL for clubroot resistance to six pathotypes of Plasmodiophora brassicae in Brassica rapa. Sci. Rep. 2017;7:4516. doi: 10.1038/s41598-017-04903-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ueno H., Matsumoto E., Aruga D., Kitagawa S., Matumura H., Hayashida N. Molecular characterization of the CRa gene conferring clubroot resistance in Brassica rapa. Plant Mol. Biol. 2012;80:621–629. doi: 10.1007/s11103-012-9971-5. [DOI] [PubMed] [Google Scholar]
- 38.Hatakeyama K., Niwa T., Kato T., Ohara T., Kakizaki T., Matsumoto S. The tandem repeated organization of NB-LRR genes in the clubroot-resistant CRb locus in Brassica rapa L. Mol. Genet. Genom. 2017;292:397–405. doi: 10.1007/s00438-016-1281-1. [DOI] [PubMed] [Google Scholar]
- 39.Matsumoto E., Yasui C., Ohi M., Tsukada M. Linkage analysis of RFLP markers for clubroot resistance and pigmentation in Chinese cabbage (Brassica rapa ssp. pekinensis) Euphytica. 1998;104:79–86. doi: 10.1023/A:1018370418201. [DOI] [Google Scholar]
- 40.Hirai M., Harada T., Kubo N., Tsukada M., Suwabe K., Matsumoto S. A novel locus for clubroot resistance in Brassica rapa and its linkage markers. Theor. Appl. Genet. 2004;108:639–643. doi: 10.1007/s00122-003-1475-x. [DOI] [PubMed] [Google Scholar]
- 41.Piao Z.Y., Deng Y.Q., Choi S.R., Park Y.J., Lim Y.P. SCAR and CAPS mapping of CRb, a gene conferring resistance to Plasmodiophora brassicae in Chinese cabbage (Brassica rapa ssp. pekinensis) Theor. Appl. Genet. 2004;108:1458–1465. doi: 10.1007/s00122-003-1577-5. [DOI] [PubMed] [Google Scholar]
- 42.Saito M., Kubo N., Matsumoto S., Suwabe K., Tsukada M., Hirai M. Fine mapping of the clubroot resistance gene, Crr3, in Brassica rapa. Theor. Appl. Genet. 2006;114:81–91. doi: 10.1007/s00122-006-0412-1. [DOI] [PubMed] [Google Scholar]
- 43.Kato T., Hatakeyama K., Fukino N., Matsumoto S. Identification of a clubroot resistance locus conferring resistance to a Plasmodiophora brassicae classified into pathotype group 3 in Chinese cabbage (Brassica rapa L.) Breed. Sci. 2012;62:282–287. doi: 10.1270/jsbbs.62.282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen J., Jing J., Zhan Z., Zhang T., Zhang C., Piao Z. Identification of novel QTLs for isolate-specific partial resistance to Plasmodiophora brassicae in Brassica rapa. PLoS ONE. 2013;8:e85307. doi: 10.1371/journal.pone.0085307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chu M., Song T., Falk K.C., Zhang X., Liu X., Chang A., Lahlali R., McGregor L., Gossen B.D., Yu F., et al. Fine mapping of Rcr1 and analysis of its effect on transcriptome patterns during infection by Plasmodiophora brassicae. BMC Genom. 2014;15:1166. doi: 10.1186/1471-2164-15-1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang T., Zhao Z., Zhang C., Pang W., Choi S.R., Lim Y.P., Piao Z. Fine genetic and physical mapping of CRb gene conferring resistance to clubroot disease in Brassica rapa. Mol. Breed. 2014;34:1173–1183. doi: 10.1007/s11032-014-0108-1. [DOI] [Google Scholar]
- 47.Yu F., Zhang X., Huang Z., Chu M., Song T., Falk K.C., Deora A., Chen Q., Zhang Y., McGregor L., et al. Identification of genome-wide variants and discovery of variants associated with Brassica rapa clubroot resistance gene Rcr1 through bulked segregant RNA sequencing. PLoS ONE. 2016;11:e0153218. doi: 10.1371/journal.pone.0153218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huang Z., Peng G., Liu X., Deora A., Falk K.C., Gossen B.D., McDonald M.R., Yu F. Fine mapping of a clubroot resistance gene in Chinese cabbage using SNP markers identified from bulked segregant RNA Sequencing. Front. Plant Sci. 2017;8:1448. doi: 10.3389/fpls.2017.01448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pang W., Fu P., Li X., Zhan Z., Yu S., Piao Z. Identification and Mapping of the clubroot resistance gene CRd in Chinese cabbage (Brassica rapa spp. pekinensis) Front. Plant Sci. 2018;9:653. doi: 10.3389/fpls.2018.00653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kato T., Hatakeyama K., Fukino N., Matsumoto S. Fine mapping of the clubroot resistance gene CRb and development of a useful selectable marker in Brassica rapa. Breed. Sci. 2013;63:116–124. doi: 10.1270/jsbbs.63.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nguyen M.L., Monakhos G.F., Komakhin R.A., Monakhos S.G. The new clubroot resistance locus is located on chromosome A05 in Chinese cabbage (Brassica rapa L.) Russ. J. Genet. 2018;54:296–304. doi: 10.1134/S1022795418030080. [DOI] [Google Scholar]
- 52.Suwabe K., Tsukazaki H., Iketani H., Hatakeyama K., Kondo M., Fujimura M., Nunome T., Fukuoka H., Hirai M., Matsumoto S. Simple sequence repeat-based comparative genomic between Brassica rapa and Arabidopsis thaliana: The genetic origin of clubroot resistance. Genetics. 2006;173:309–319. doi: 10.1534/genetics.104.038968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hatakeyama K., Suwabe K., Tomita R.N., Kato T., Nunome T., Fukuoka H., Matsumoto S. Identification and characterization of Crr1a, a gene for resistance to clubroot disease (Plasmodiophora brassicae Woronin) in Brassica rapa L. PLoS ONE. 2013;8:e54745. doi: 10.1371/journal.pone.0054745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Laila R., Park J.I., Robin A.H.K., Natarajan S., Vijayakumar H., Shirasawa K., Isobe S., Kim H.T., Nou I.S. Mapping of a novel clubroot resistance QTL using ddRAD-seq in Chinese cabbage (Brassica rapa L.) BMC Plant Biol. 2019;19:13. doi: 10.1186/s12870-018-1615-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kuginuki Y., Ajisaka H., Yui M., Yoshikawa H., Hida K., Hirai M. RAPD markers linked to a clubroot-resistance locus in Brassica rapa L. Euphytica. 1997;98:149–154. doi: 10.1023/A:1003147815692. [DOI] [Google Scholar]
- 56.Lan M., Li G., Hu J., Yang H., Zhang L., Xu X., Liu J., He J., Sun R. iTRAQ-based quantitative analysis reveals proteomic changes in Chinese cabbage (Brassica rapa L.) response to Plasmodiophora brassicae infection. Sci. Rep. 2019;9:12058. doi: 10.1038/s41598-019-48608-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dakouri A., Zhang X., Peng G., Falk K.C., Gossen B.D., Strelkov S.E., Yu F. Analysis of genome-wide variants through bulked segregant RNA sequencing reveals a major gene of resistance to Plasmodiophora brassicae in Brassica oleracea. Sci. Rep. 2018;8:17657. doi: 10.1038/s41598-018-36187-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Figdore S.S., Ferreia M.E., Slocum M.K., Williams P.H. Association of RFLP markers with trait loci affecting clubroot resistance and morphological characters in Brassica oleracea L. Euphytica. 1993;69:33–44. doi: 10.1007/BF00021723. [DOI] [Google Scholar]
- 59.Grandclemant C., Laurens F., Thomas G. Genetic analysis of resistance to clubroot (Plasmodiophora brassicae Woron) in two Brassica oleracea groups (spp. acephala and spp. botrytis) through diallel analysis. Plant Breed. 1996;115:152–156. doi: 10.1111/j.1439-0523.1996.tb00892.x. [DOI] [Google Scholar]
- 60.Voorrips R.E., Jongerius M.C., Kanne H.J. Mapping of two genes for resistance to clubroot (Plasmodiophora brassicae) in a population of double haploid lines of Brassica oleracea by means of RFLP and AFLP markers. Theor. Appl. Genet. 1997;94:75–82. doi: 10.1007/s001220050384. [DOI] [PubMed] [Google Scholar]
- 61.Moriguchi K., Kimizuka-Takagi C., Ishii K., Nomura K. A genetic map based on RAPD, RFLP, isozyme, morphological markers and QTL analysis for clubroot resistance in Brassica oleracea. Breed. Sci. 1999;49:257–265. doi: 10.1270/jsbbs.49.257. [DOI] [Google Scholar]
- 62.Nomura K., Minegishi Y., Kimizuka-Takagi C., Fujioka T., Moriguchi K., Shishido R., Ikehashi H. Evaluation of F2 and F3 plants introgressed with QTLs for clubroot resistance in cabbage developed by using SCAR markers. Plant Breed. 2005;124:371–375. doi: 10.1111/j.1439-0523.2005.01105.x. [DOI] [Google Scholar]
- 63.Rocherieux J., Glory P., Giboulot A., Boury S., Barbeyron G., Thomas G., Manzanares-Dauleux M.J. Isolate-specific and broad-spectrum QTLs are involved in the control of clubroot in Brassica oleracea. Theor. Appl. Genet. 2004;108:1555–1563. doi: 10.1007/s00122-003-1580-x. [DOI] [PubMed] [Google Scholar]
- 64.Nagaoka T., Doullah M.A.U., Matsumoto S., Kawasaki S., Ishikawa T., Hori H., Okazaki K. Identification of QTLs that control clubroot resistance in Brassica oleracea and comparative analysis of clubroot resistance genes between B. rapa and B. oleracea. Theor. Appl. Genet. 2010;120:1335–1346. doi: 10.1007/s00122-010-1259-z. [DOI] [PubMed] [Google Scholar]
- 65.Lee J., Izzah N.K., Choi B.S., Joh H.J., Lee S.C., Perumal S., Seo J., Ahn K., Jo E.J., Choi G.J., et al. Genotyping-by-sequencing map permits identification of clubroot resistance QTLs and revision of the reference genome assembly in cabbage (Brassica oleracea L.) DNA Res. 2016;23:29–41. doi: 10.1093/dnares/dsv034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Peng L., Zhou L., Li Q., Wei D., Ren X., Song H., Mei J., Si J., Qian W. Identification of quantitative trait loci for clubroot resistance in Brassica oleracea with the use of Brassica SNP microarray. Front. Plant Sci. 2018;9:822. doi: 10.3389/fpls.2018.00822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tomita H., Shimizu M., Doullah M.A.U., Fujimoto R., Okazaki K. Accumulation of quantitative trait loci conferring broad-spectrum clubroot resistance in Brassica oleracea. Mol. Breed. 2013;32:889–900. doi: 10.1007/s11032-013-9918-9. [DOI] [Google Scholar]
- 68.Landry B.S., Hubert N., Crete R., Chang M.S., Lincoln S.E., Etoh T. A genetic map for Brassica oleracea based on RFLP markers detected with expressed DNA sequences and mapping of resistance genes to race 2 of Plasmodiophora brassicae (Woronin) Genome. 1992;35:409–420. doi: 10.1139/g92-061. [DOI] [Google Scholar]
- 69.Manzanares-Dauleux M.J., Delourme R., Baron F., Tomas G. Mapping of one major gene and of QTLs involved in resistance to clubroot in Brassica napus. Theor. Appl. Genet. 2000;101:885–891. doi: 10.1007/s001220051557. [DOI] [Google Scholar]
- 70.Werner S., Diederichsen E., Frauen M., Schondelmaier J., Jung C. Genetic mapping of clubroot resistance genes in oilseed rape. Theor. Appl. Genet. 2008;116:363–372. doi: 10.1007/s00122-007-0674-2. [DOI] [PubMed] [Google Scholar]
- 71.Fredua-Agyeman R., Rahman R. Mapping of the clubroot disease resistance in spring Brassica napus canola introgressed from European winter canola cv. ‘Mendel’. Euphytica. 2016;211:201–213. doi: 10.1007/s10681-016-1730-2. [DOI] [Google Scholar]
- 72.Diederichsen E., Beckmann J., Schondelmeier J., Dreyer F. Genetics of clubroot resistance in Brassica napus ‘Mendel’. Acta. Hort. 2006;706:307–312. doi: 10.17660/ActaHortic.2006.706.35. [DOI] [Google Scholar]
- 73.Zhang X., Feng J., Hwang S.F., Strelkov S.E., Falak I., Huang X., Sun R. Mapping of clubroot (Plasmodiophora brassicae) resistance in canola (Brassica napus) Plant Pathol. 2016;65:435–440. doi: 10.1111/ppa.12422. [DOI] [Google Scholar]
- 74.Hasan M.J., Rahman R. Genetics and molecular mapping of resistance to Plasmodiophora brassicae pathotypes 2, 3, 5, 6 and 8 in rutabaga (Brassica napus var. napobrassica) Genome. 2016;59:805–815. doi: 10.1139/gen-2016-0034. [DOI] [PubMed] [Google Scholar]
- 75.Hejna O., Havlickova L., He Z., Bancroft I., Curn V. Analysing the genetic architecture of clubroot resistance variation in Brassica napus by associative transcriptomics. Mol. Breed. 2019;39:112. doi: 10.1007/s11032-019-1021-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Blank L.M. Fusarium resistance in Wisconsin all seasons cabbage. J. Agric. Res. 1937;55:497–510. [Google Scholar]
- 77.Walker J.C. Plant Disease-Vegetable Crops-Cauliflower, Cabbage, and Others. US Department of Agriculture, US Government Printing Office; Washington, DC, USA: 1953. [Google Scholar]
- 78.Lv H., Yang L., Kang J., Wang Q., Wang X., Fang Z., Liu Y., Zhuang M., Zhang Y., Lin Y., et al. Development of InDel markers linked to Fusarium wilt resistance in cabbage. Mol. Breed. 2013;32:961–967. doi: 10.1007/s11032-013-9925-x. [DOI] [Google Scholar]
- 79.Miyaji N., Shimizu M., Miyazaki J., Osabe K., Sato M., Ebe Y., Takada S., Kaji M., Dennis E.S., Fujimoto R., et al. Comparison of transcriptome profiles by Fusarium oxysporum inoculation between Fusarium yellows resistant and susceptible lines in Brassica rapa L. Plant Cell Rep. 2017;36:1841–1854. doi: 10.1007/s00299-017-2198-9. [DOI] [PubMed] [Google Scholar]
- 80.Sato M., Shimizu M., Shea D.J., Hoque M., Kawanabe T., Miyaji N., Fujimoto R., Fukai E., Okazaki K. Allele specific DNA marker for fusarium resistance gene FocBo1 in Brassica oleracea. Breed. Sci. 2019;69:308–315. doi: 10.1270/jsbbs.18156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Shimizu M., Fujimoto R., Ying H., Pu Z., Ebe Y., Kawanabe T., Saeki N., Taylor J.M., Kaji M., Dennis E.S., et al. Identification of candidate genes for Fusarium yellows resistance in Chinese cabbage by differential expression analysis. Plant Mol. Biol. 2014;85:247–257. doi: 10.1007/s11103-014-0182-0. [DOI] [PubMed] [Google Scholar]
- 82.Kawamura K., Kawanabe T., Shimizu M., Nagano A.J., Saeki N., Okazaki K., Kaji M., Dennis E.S., Osabe K., Fujimoto R. Genetic characterization of inbred lines of Chinese cabbage by DNA markers; towards the application of DNA markers to breeding of F1 hybrid cultivars. Data Brief. 2016;6:229–237. doi: 10.1016/j.dib.2015.11.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Pu Z., Ino Y., Kimura Y., Tago A., Shimizu M., Natsume S., Sano Y., Fujjimoto R., Kaneko K., Shea D.J., et al. Changes in the proteome of xylem sap in Brassica oleracea in response to Fusarium oxysporum stress. Front. Plant Sci. 2016;7:31. doi: 10.3389/fpls.2016.00031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kawamura K., Shimizu M., Kawanabe T., Pu Z., Kodama T., Kaji M., Kenji O., Fujimoto R., Keiichi O. Assessment of DNA markers for seed contamination testing and selection of disease resistance in cabbage. Euphytica. 2017;213:28. doi: 10.1007/s10681-016-1821-0. [DOI] [Google Scholar]
- 85.Matsumoto E., Ueno H., Aruga D., Sakamoto K., Hayashida N. Accumulation of three clubroot resistance genes through marker-assisted selection in Chinese cabbage (Brassica rapa spp. pekinensis) J. Jpn. Soc. Hortic. Sci. 2012;81:184–190. doi: 10.2503/jjshs1.81.184. [DOI] [Google Scholar]
- 86.Doullah M.A.U., Tomita H., Shimizu M., Matsumoto S., Fujimoto R., Okazaki K. Recent progress of clubroot resistance breeding through marker assisted selection in Brassica rapa and Brassica oleracea. J. Sylhet Agric. Univ. 2014;1:139–146. [Google Scholar]
- 87.Strelkov S.E., Hwang S.F., Manolii V.P., Cao T., Fredua-Agyeman R., Harding M.W., Peng G., Gossen B.D., Mcdonald M.R., Feindel D. Virulence and pathotype classification of Plasmodiophora brassicae populations collected from clubroot resistant canola (Brassica napus) in Canada. Can. J. Plant Pathol. 2018;40:284–298. doi: 10.1080/07060661.2018.1459851. [DOI] [Google Scholar]
- 88.Kuginuki Y., Yoshikawa H., Hirai M. Variation in virulence of Plasmodiophora brassicae in Japan tested with clubroot-resistant cultivars of Chinese cabbage (Brassica rapa L. spp. pekinensis) Eur. J. Plant Pathol. 1999;105:327–332. doi: 10.1023/A:1008705413127. [DOI] [Google Scholar]
- 89.Kou Y., Wang S. Broad-spectrum and durability: Understanding of quantitative disease resistance. Curr. Opin. Plant Biol. 2010;13:181–185. doi: 10.1016/j.pbi.2009.12.010. [DOI] [PubMed] [Google Scholar]
- 90.Crisp P., Crute I.R., Sutherland R.A., Angell S.M., Bloor K., Burgess H., Gordon P.L. The exploitation of genetic resources of Brassica oleracea in breeding for resistance to clubroot (Plasmodiophora brassicae) Euphytica. 1889;42:215–226. [Google Scholar]
- 91.Shah N., Sun J., Yu S., Yang Z., Wang Z., Huang F., Dun B., Gong J., Liu Y., Li Y., et al. Genetic variation analysis of field isolates of clubroot and their responses to Brassica napus lines containing resistant genes CRb and PbBa8.1 and their combination in homozygous and heterozygous state. Mol. Breed. 2019;39:153. doi: 10.1007/s11032-019-1075-3. [DOI] [Google Scholar]
- 92.Rahman H., Franke C. Association of fusarium wilt susceptibility with clubroot resistance derived from winter Brassica napus L. ‘Mendel’. Can. J. Plant Pathol. 2019;41:60–64. doi: 10.1080/07060661.2018.1505780. [DOI] [Google Scholar]