Abstract
Long-term care facilities (LTCFs) are an important reservoir of multidrug-resistant organisms (MDROs). Colonization of LTCF residents by MDROs is generally higher in Italy compared to other European countries. The present review by the working group for the study of infections in LTCFs (GLISTer) of the Italian Association of Clinical Microbiologists (AMCLI) aims to propose criteria for a laboratory-based surveillance of MDROs in Italian LTCFs.
We recommend the adhesion to three levels of laboratory-based MDROs surveillance in LTCFs: i) mandatory MDRO surveillance by cumulative retrospective analysis of antimicrobial susceptibility data, obtained as part of routine care of clinical specimens. ii) strongly recommended surveillance by active rectal swab cultures or molecular screening to determine colonization with carbapenemase-producing Enterobacterales, should a resident be proven infected. iii) voluntary surveillance by prospective MDRO surveys, mainly based on point prevalence colonization studies, allowing to determine the MDROs baseline prevalence in the facility.
Laboratory-based surveillance of MDROs in LTCFs is aimed at providing useful epidemiological information to healthcare providers operating in the facility, but it is only effective if the collected data are used for infection prevention and control purposes, targeting the peculiar aspects of LTCFs.
Keywords: Italy, LTCF, MDRO, Surveillance, Screening, Infection control, Prevention
Introduction
Long-term care facilities (LTCFs) are institutions that provide skilled nursing care to residents in need of assistance with activities of daily living. LTCFs encompass nursing homes, residential care centers, chronic disease hospitals, rehabilitation centers and institutions for mentally handicapped persons [1]. These facilities are functionally the home for residents, usually elderly and in declining health status, often staying for months to years. Due to the ageing population in Italy (present life expectancy 80.1 years for males and 84.7 years for females) LTCFs play an important role in the Italian healthcare system; in December 2013, 12.200 LTCFs were counted, hosting 384.000 people, and these figures are intended to increase during the next decades [http://www.istat.it/].
The frequently older, sicker residents cared for in LTCFs, have a variety of risk factors for colonization and infection by multidrug-resistant organisms (MDROs). Pathogen cross-transmission within LTCFs is a significant issue and residents of these facilities are frequently moved between the acute and the long-term care settings [2]. For these reasons, LTCFs are an important reservoir for MDROs, such as methicillin-resistant Staphylococcus aureus (MRSA), vancomycin-resistant enterococci (VRE), Pseudomonas aeruginosa or Acinetobacter baumannii with MDR phenotypes (expressing resistance to ≥3 classes of antibiotics) and Enterobacterales producing extended-spectrum β-lactamases (ESBLs), high-level cephalosporinases (AmpCs) and/or carbapenemases [3, 4]; these MDROs have been included in the present surveillance recommendations.
Surveillance of MDROs in a healthcare institution is of pivotal importance for guiding prevention, therapy and control of infectious diseases caused by these organisms [5]. The urgent need for MDRO surveillance recommendations in Italian LTCFs has been highlighted during the National Congress of AMCLI in Rimini, 2017 [6], and by the national plan for contrasting antimicrobial resistance 2017–20, approved by the Italian Presidency of the Council of Ministers (PNCAR) [7]. Therefore, the Italian working group for the study of infections in LTCFs (GLISTer) of the Association of Italian Clinical Microbiologists (AMCLI), has been prompted to define and recommend general indications for the surveillance of MDROs in Italian LTCFs [8]. For literature search we adopted a search strategy in the Medline/Pubmed database including the following search terms: (nursing home* OR long term care facility*) AND (colonization OR multi drug resistan* OR ESBL OR MRSA OR VRE OR carbapenemase*) AND Italy. We restricted the search to the dates range 01.01.2000–30.08.2019 but did not impose any language restriction. Applying this search strategy, we identified 30 articles; from the reference lists of the above articles we retrieved further studies. Moreover, we included a Poster Abstract from the National Congress AMCLI 2017, the “Piano Nazionale per il Contrasto dell’antimicrobico-resistenza 2017-20 (PNCAR)”, Clinical and Laboratory Standards Institute (CLSI) guidelines for analysis and presentation of cumulative antimicrobial susceptibility test data, and international and national guidelines for infection prevention and control in LTCFs. At least two authors screened every abstract and full-text article or guideline, the results have been collegially discussed before the inclusion in the review article.
The recommendations are intended to be used by local and regional bodies, comprising LTCF physicians, infectious disease specialists and nurses, infection preventionists, LTCF administrations, referral hospital physicians, clinical microbiologists and administrations.
MDR bacteria in Italian LTCFs
Dwelling in a LTCF is an independent risk factor for bloodstream infections by MDROs [9], and Italy has one of the highest prevalence of MDROs isolated from blood cultures in European countries, especially for MRSA, carbapenemase-producing A. baumannii and ESBL- or carbapenemase-producing Enterobacterales (CPE) [10]. A systematic review of studies on colonization by MDROs from Italian LTCFs, on risk factors for colonization and on molecular characteristics of clinical isolates also in comparison with other European countries, has been published recently [11]. Italian review data show an MRSA colonization prevalence of 7.8–38% for residents and 5.2–7.0% for staff members, an ESBL-prevalence of 50–64% for residents and 5.2–14% for staff and a CPE-prevalence of 1.0–6.3% for residents and 0.0–1.5% for staff. These data show that colonization of residents in Italian LTCFs is generally significantly higher than in other European countries. A point prevalence survey undertaken in 2008 in an Italian LTCF [12], repeated in 2012 [13] and 2016 [14], showed high resident colonization rates for ESBL-producing Enterobacterales (49.0–64.0%), a low prevalence for CPE (1.7–6.3%) and a variable prevalence for MRSA-colonization (13.2–38.7%). Two recent multicenter studies in Italian LTCFs, confirmed a high colonization prevalence (ranging from 32.8 to 81.5%) for ESBL-producing Enterobacterales, especially Escherichia coli, whereas lower prevalence was found for CPE (0.0–5.8%) and carbapenemase-producing A. baumannii (0.0–5.8%); highly variable prevalence was found for MRSA (5.0–30.6%) and VRE (0.8–20.2%) [15, 16].
An Italian multicenter study, investigating the frequency of ESBL-producing Enterobacterales in urine specimens from LTCF residents, found a median ESBL-rate of 32.1%, with E. coli as the most frequent ESBL-producing pathogen [17].
In Italian LTCFs, as well as in other European countries, the most prevalent ESBLs from E. coli isolates were found CTX-M-type enzymes, particularly CTX-M-15, mainly belonging to the pandemic ST131 clonal group [11, 15, 18]. On the other hand, carbapenemases in Enterobacterales from Italian LTCFs, generally are of KPC- or VIM-type [11, 15, 16]. Recently, other authors reported a high colonization rate of residents with KPC-type -producing Klebsiella pneumoniae in a LTCF from central Italy: 11.6% of patients were colonized at admission, 9.9% subsequently, with very high rates of carriage and cross-transmission in severe brain injury patients [19].
It is important to emphasize that published surveillance studies from Italian LTCFs are scarce. Moreover, large urban facilities are overrepresented, and there have been no systematic, culture-based or molecular surveys at national or regional level until now.
Surveillance recommendations for MDROs in Italian LTCFs
Recommended laboratory based surveillance strategies for MDROs range from passive surveillance of routine clinical microbiology laboratory-results (1st level) [20], up to retrospective (2nd level) [2] or prospective surveillance screening (3rd level) [21].
| Surveillance level | Strategies | Organisms included | Recommendation | Limitations |
| 1st level laboratory based surveillance | Surveillance of routine clinical microbiology laboratory-results | Various MDROs | Mandatory | Low number of collected samples, identification of specimens as coming from LTCF patients, exclusion of colonization data |
| 2nd level laboratory based surveillance | Active surveillance cultures | Generally CPE, possible extension to other MDROs | Strongly recommended | Generally limitation to colonization by CPE, exclusion of routine isolates from infections |
| 3rd level laboratory based surveillance | Prospective facility screening of LTCF residents for MDROs | Various MDROs | On a voluntary basis, and recommended | Generally point prevalence study, exclusion of routine isolates from infections, costly and time-consuming |
Variation in MDRO surveillance includes facility wide vs. targeted units, patient based vs. laboratory based, colonization vs. infection, passive vs. active, retrospective vs. prospective, and optional vs. required strategies. The significant heterogeneity of Italian LTCFs, as referred to bed number, resident comorbidities and staffing, and the heterogeneity of local, regional and national regulations, complicate the recommendation of generally acceptable surveillance measures for MDROs [7]. Laboratory-based surveillance of MDROs should be associated to a careful infection monitoring -e.g. of incidence of urinary tract infections or central line associated bacteremia-, by healthcare providers within the LTCF, and should also be paralleled by evaluation of the consumption of antibiotics in the institution, as key prerequisites for the implementation of an infection control and antimicrobial stewardship programs [22, 23]. In any case, routine environmental cultures or samples from asymptomatic personnel are not recommended, except as target for an epidemiologic investigation.
Surveillance of routine clinical microbiology laboratory-results (first level laboratory-based surveillance, mandatory)
We define first level surveillance of MDROs in LTCFs as the cumulative retrospective analysis of antimicrobial susceptibility data, obtained as part of routine care from diagnostic cultures of clinical specimens in the referring clinical microbiology laboratory, in accordance with the Clinical Laboratory Standards Institute (CLSI) guideline for recording and analysis of antimicrobial susceptibility data [5]. Local susceptibility data are mainly used to provide useful information to enable laboratories to assist the clinician in the selection of appropriate therapy for infections, but they can also be used as a basis for infection prevention and control procedures.
Microbiology laboratories chosen by LTCFs for analysis of microbiological specimens must comply with the following criteria:
accreditation/certification for the analysis of microbiology samples, including implementation of the required internal and external quality control programs
documentation of an adequate volume of sample processing activity
capability of cryopreservation of MDR organisms
ability to collect, elaborate and present cumulative antibiotic susceptibility data
Therefore, specimen processing and data elaboration in a few regional reference laboratories is recommended: it facilitates standardized cumulative retrospective analysis of antimicrobial susceptibility data and their comparison among different LTCFs.
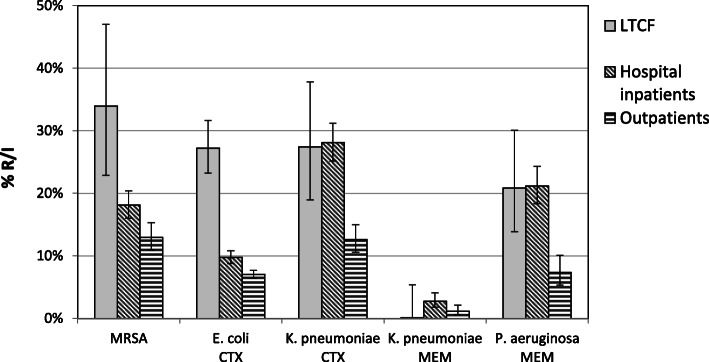
A major disadvantage of using such retrospective susceptibility data is the generally low number of specimens sent to the clinical microbiology laboratory, limiting the number of isolates available for susceptibility testing and cumulative analysis. Therefore, combining of data from consecutive years (e.g. 3 years) and from several comparable LTCFs in a healthcare district area may be required [5]. Elaboration of such combined data is useful for indication of general trends, which can be compared with outpatient or acute-care hospital (ACH) inpatient data from the same geographic region [5]; however, if data are pooled from consecutive years, changes in resistance profiles might be difficult to detect. For example, selective cumulative susceptibility data on routine clinical isolates from LTCFs, inpatients and outpatients from the Bolzano healthcare district, highlighted significantly higher rates of MRSA and cefotaxime-resistance in E. coli (mainly ESBL-producers) from LTCF residents, compared with isolates from hospital inpatients; this was not the case for cefotaxime- or meropenem-resistance in K. pneumoniae or for meropenem-resistance in P. aeruginosa (Fig. 1).
Fig. 1.
Antibiotic resistance in routine clinical isolates from LTCFs, hospital inpatients and outpatients in the Bolzano healthcare district. Resistance rates were calculated using episode based duplicate exclusion within 28 days (Virtuoso Plus, Dedalus Healthcare Systems Group, Florence, Italy). Screening isolates have been excluded. MRSA: methicillin-resistant S. aureus, CTX: cefotaxime, MEM: meropenem. Error bars: 95% confidence interval (https://www.graphpad.com/). Number of isolates: S. aureus (LTCFs: 56; inpatients: 1187; outpatients: 918), E. coli (LTCFs: 430; inpatients: 3202; outpatients: 6202), K. pneumoniae (LTCFs: 84; inpatients: 844; outpatients: 874), P. aeruginosa (LTCFs: 96; inpatients: 723; outpatients: 477)
Therefore, an essential requirement for an efficient laboratory-based passive MDROs surveillance system is the mandatory collection of specimens for diagnostic microbiological testing during infectious episodes. Moreover, because of the generally low number of isolates, we recommend to always calculate the 95% confidence interval for antimicrobial resistance rates. Together with the cumulative retrospective analysis of antimicrobial resistance data, a line listing of residents infected/colonized with MDROs should be provided to the infection control practitioners operating in the LTCFs; the line listing can also be prepared for low isolate numbers of single MDRO species (< 30 isolates).
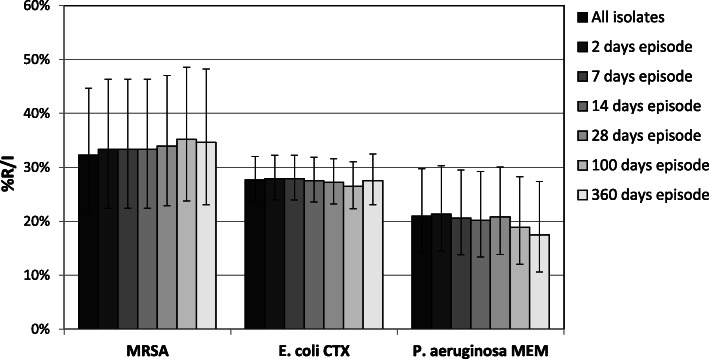
Various criteria for exclusion of multiple isolates of the same species from single patients have been proposed [5]. The frequently used first isolate per episode based calculation, includes the first isolate of a given species recovered from each episode of infection, and an episode is defined as the set of all isolates from a patient in which the interval between consecutive isolates is less or equal to a defined time interval, e.g. 28 days. In order to analyze the effect of removal of duplicate isolates by an episode based strategy, we considered all diagnostic isolates (active surveillance isolates excluded) recovered during 2015–17 from LTCF residents in the Bolzano healthcare district, and generated cumulative susceptibility reports for MRSA, cefotaxime-resistant E. coli and meropenem-resistant P. aeruginosa, based on the all isolates strategy and on different episode-based strategies (minimal interval of time: 2, 7, 14, 28, 100 or 360 days) (Fig. 2). Of note, though a considerable percentage of isolates was removed from calculation in the episode based strategy, with a minimal time interval of 360 days, compared to the all isolates approach (MRSA: 16.1%, E. coli: 23.5%, P. aeruginosa: 16.4%), the multiple isolates removal did not significantly change resistance rates. Similar studies on the effect of duplicate isolates removal may lead to varying results, depending not only on the method used for the exclusion of multiple isolates, but also on local epidemiology, sampling practice and patient demographics [24]. Recommendations for the application of various episode based duplicate exclusion strategies have been published; it was found that cumulative antimicrobial susceptibility reports based on a 28 days’ episode strategy, reflect resistance rates among hospital-acquired infections [25]. A similar duplicate exclusion strategy has therefore been proposed by an Italian guideline [26]; other authors consider a 10 days episode strategy as first choice for the hospital inpatient setting [24]. LTCF residents stay in the facility for months or years and, due to their underlying chronic diseases, MDROs can give repeatedly rise to infections, requiring frequent antimicrobial treatments. For these reasons, we recommend a 28 days’ episode based strategy for multiple isolates from LTCF residents, but it should be emphasized that the time interval chosen does not significantly change the obtained susceptibility rates; moreover, a 28 days’ episode based exclusion strategy for duplicate isolates has recently been adopted for blood culture isolates by the Italian PNCAR [8].
Fig. 2.
Antibiotic resistance dependent on the method of duplicate isolate removal. Resistance rates were calculated using different methods of duplicate isolate removal (Virtuoso Plus, Dedalus Healthcare Systems Group, Florence, Italy). Screening isolates have been excluded. MRSA: methicillin-resistant S. aureus, CTX: cefotaxime, MEM: meropenem. Error bars: 95% confidence interval (https://www.graphpad.com/). Number of isolates (from left to right): S. aureus (62–57–57-57-56-54-52), E. coli (451–444–444-443-430-393-345), P. aeruginosa (105–103–102-99-96-90-80)
It is essential that clinical specimens from LTCF residents are clearly associated to a specific LTCF, but this is not always the case in Italian LTCFs; mixing LTCF data with those of ambulatory patients is a major mistake when it comes to tracing the epidemiology of antimicrobial resistance [20].
Patients hospitalized in an LTCF are sometimes transferred to the referral ACH for treatment of serious infections and then they undergo diagnostic cultures of clinical specimens; it is generally a difficult data management problem to include these isolates in the 1st level LTCF surveillance, but results of these cultures should not be missed, as they represent the most serious infectious episodes. A proposed solution to this limitation is to integrate the 1st level LTCF surveillance into the general cumulative antimicrobial susceptibility report, stratifying data according to outpatients, patients hospitalized in the referral ACH and LTCF residents; isolates from residents in LTCFs and from residents admitted to the referral ACH (within 48 h) should, if possible, be pooled together in the drafting of the report.
Based on published international and national guidelines [5, 20, 27], and applying various modifications for LTCFs, for the mandatory first level laboratory-based MDROs surveillance in LTCFs we suggest the following recommendations and strategies.
| Theme of the recommendation | Recommended strategies | Comments | Specific responsibilities |
| Microbiological specimen collection | Require from LTCF healthcare providers the mandatory collection of specimens for microbiological testing during infectious episodes in LTCF residents | Essential for empiric therapy and for infection control | LTCF physicians, nurses, LTCF infection preventionists |
| Microbiology laboratory selection | Select laboratories accredited/certified for analysis of microbiological specimens that are also equipped for long-term cryopreservation of MDRO isolates and for the collection, elaboration and presentation of antibiotic susceptibility data | Preferably choose a few regional reference laboratories | LTCF administration, clinical microbiologists |
| Communication | Communicate immediately the isolation of MDROs from LTCF residents to infection control practitioners in the LTCFs (by phone, fax, e-mail, or smartphone message) and by preliminary reporting | Application of the internal protocol for communication of laboratory-results (e.g. MRSA, VRE, P. aeruginosa or A. baumannii with MDR phenotype and Enterobacterales producing ESBLs, high-level AmpCs and/or carbapenemases) | Clinical microbiologists, LTCF physicians, LTCF infection preventionists |
| Data elaboration | Produce cumulative antibiograms annually and separately for each LTCF if a sufficient number of isolates per species and year is available (≥30 isolates per species) |
If the number of isolates per species and year is < 30, extend the time period up to 3 years and/or include several comparable LTCFs in the same geographic area, in agreement with the infection control practitioners in the involved LTCFs. Prepare for isolate numbers < 30 for single bacterial species a line listing of identified MDROs |
Clinical microbiologists and/or epidemiologists |
| Data elaboration | Exclude isolates from active surveillance and screening studies from the cumulative antibiograms and, if available, elaborate this data separately | Elaborate surveillance and screening data separately (see surveillance levels 2 and 3) | Clinical microbiologists and/or epidemiologists |
| Data elaboration | Calculate antimicrobial susceptibility rates at species level, focusing on the most important resistance phenotypes | Include at least S. aureus (MRSA), E. faecalis and E. faecium (VRE), E. coli and K. pneumoniae (ESBL, carbapenemases), P. aeruginosa and A. baumannii (carbapenem-resistance, carbapenemases) | Clinical microbiologists and/or epidemiologists |
| Data elaboration | Adhere to the European Committee on Antimicrobial Susceptibility Testing (EUCAST) recommendations to ensure standardized antimicrobial susceptibility testing | Disclose any change in antimicrobial susceptibility testing methodology and interpretative reading (for example therapeutic correction of cephalosporin interpretation for ESBL producers) | Clinical microbiologists and/or epidemiologists |
| Data elaboration | Use an episode-based strategy for exclusion of multiple isolates per resident, with a minimum default interval of time between their recovery of 28 days | Discuss different other possible removal strategies for duplicate isolates with the LTCF infection control practitioners. Define and disclose the calculation algorithm used for cumulative antimicrobial susceptibility test reports | Clinical microbiologists and/or epidemiologists, LTCF infection control practitioners |
| Data elaboration | Use stratification of data by specimen type (for example urine, blood, others) only if sufficiently high isolate numbers (≥30 isolates per species and specimen subgroup) have been tested | Discuss various data stratification options with the infection control practitioners in the LTCF | Clinical microbiologists and/or epidemiologists, LTCF infection control practitioners |
| Data elaboration | Calculate the percentage of MDRO-isolates/total number of isolates × 100 (applying a 28 days’ episode based multiple isolates exclusion strategy) | Always add 95% confidence intervals to susceptibility rates | Clinical microbiologists and/or epidemiologists |
| Data elaboration | Integrate the LTCF surveillance report into a general cumulative antimicrobial susceptibility data report, comparing isolates from LTCF residents with outpatient isolates and isolates from referral acute care hospital patients | If possible, stratify isolates from LTCF residents admitted within 48 h to the referral ACH together with LTCF isolates | Clinical microbiologists and/or epidemiologists |
| Data reporting | Report antimicrobial susceptibility rates only for antibiotics routinely tested on all isolates | Do not report supplemental antimicrobial agents that are selectively tested on resistant isolates | Clinical microbiologists and/or epidemiologists |
| Data reporting | Report antimicrobial susceptibility data as percentages of susceptible-standard dosing regimen, susceptible-increased exposure and resistant isolates | According to EUCAST criteria | Clinical microbiologists and/or epidemiologists |
| Data reporting | Present the report preferably in a graphic format, easily accessible to the healthcare practitioners in the LTCFs, for example on the institution’s website | Discuss data with healthcare providers in the LTCFs | Clinical microbiologists and/or epidemiologists, healthcare providers in LTCFs |
| Isolate conservation | Cryopreserve MDR-isolates (at −80 °C) | Preserve for further molecular characterization, in collaboration with a reference molecular biology laboratory | Clinical microbiologists |
Surveillance by active surveillance cultures (second level laboratory-based surveillance, strongly recommended)
Active surveillance of LTCF residents for carriage of MDROs can be defined as obtaining isolates from cultures of specimens that are collected for determining if a patient is harboring a particular organism, and are not from cultures that are obtained as part of the clinical evaluation of a resident’s illness. Active surveillance cultures are specifically designed for MDROs, for which usual reservoirs are established and validated screening tests are available, e.g. CPE [28]. Active surveillance of MDROs can also be based on molecular amplification methods. Use of active surveillance has been shown to improve detection of MDROs, compared with reliance on culture of specimens collected for clinical reasons alone [29].
Specimen types used for active MDRO-screening may influence surveillance data. ESBL- and carbapenemase-producing Enterobacterales and VRE are most often recovered from rectal samples (possibly combined with urine samples), whereas we have shown that for MRSA-screening a combination of oropharyngeal (or nasal) and inguinal (or perineal) swabs gives highest yields; testing of only nasal swabs results in substantial underestimation of colonization with MRSA [12–14]. For carbapenem-resistant P. aeruginosa and A. baumannii oropharyngeal and rectal swabs are recommended (possibly combined with urine samples) [16, 30–32].
According to LTCF infection prevention and control guidelines active surveillance of MDROs in LTCFs by culture methods or by using molecular amplification assays should not imply routine screening of residents at the time of admission to the facility, nor should it be repeated on a periodic basis, in the absence of an epidemic of infections by MDROs, because the application of standard precautions, as applied to all residents, is sufficient [2]. Active surveillance screening for various MDROs should be individualized to residents at risk for shedding large numbers of bacteria into the environment, e.g. residents with colonized wounds not covered fully with dressings, incontinent residents with urinary or fecal carriage, or residents with tracheostomies and difficulty in handling respiratory secretions [1].
CPE, especially KPC-producing K. pneumoniae, are epidemically spread in Italy [10, 33] and the emergence of this MDR-phenotype in LTCFs is widely expanding the reservoir of this health-care threat [11, 15, 16, 19]. Based on the current epidemiology and the clinical significance of CPE, we recommend targeting active MDROs surveillance screening in Italian LTCFs on CPE, according to AMCLI indications for the screening of carbapenemases in Enterobacteriales [28]; identification of the main carbapenemase types in Italy (KPC, OXA-48-like, VIM, NDM) by antigenic or molecular methods is essential. Though active surveillance in Italian LTCFs is generally restricted to CPE, in accordance with healthcare providers in the LTCFs and taking into consideration the various levels of complexity of the single LTCF, other MDROs may be considered for screening.
GLISTer recommends active routine surveillance screening of MDROs in LTCFs as follows.
| Theme of the recommendation | Recommended strategies | Comments | Specific responsibilities |
| Selection of MDRO types | Generally restrict active MDRO surveillance screening to CPE | In accordance with healthcare providers in the LTCFs, other MDROs may be considered for screening | LTCF physicians, LTCF infection preventionists, clinical microbiologists |
| Selection of LTCF residents | Perform active surveillance cultures (or molecular screening) in the presence of infection by CPE (index case), excluding colonization such as asymptomatic bacteriuria | Screening is especially recommended if the index case is at risk for shedding large numbers of bacteria into the environment, e.g. residents with colonized wounds not fully covered with dressings, incontinent residents with urinary or fecal carriage, or residents with tracheostomies and difficulty in handling respiratory secretions | LTCF physicians, LTCF infection preventionists, Clinical microbiologists |
| Selection of LTCF residents | Perform active CPE surveillance cultures (or molecular methods) on all residents hosted in the same LTCF unit as the index case | Possible extension of surveillance cultures to other close contacts of the index case in other LTCF units; extension of CPE-screening to all residents of the LTCF can be considered, in agreement with the infection control practitioners in the involved LTCFs | LTCF physicians, LTCF infection preventionists, clinical microbiologists |
| Specimen types | Use rectal or fecal swabs for CPE-screening | Add other specific specimen types for possible screening of other MDROs | LTCF physicians, LTCF infection preventionists, clinical microbiologists |
| Frequency of surveillance | Repeat CPE surveillance cultures (or molecular screening) as recommended by the infection control professionals of the LTCF | Discuss frequency of possible surveillance for other MDROs with LTCF infection preventionists | LTCF infection preventionists, clinical microbiologists |
| Environmental or staff screening | Do not perform routine environmental cultures or screening cultures from asymptomatic personnel | Discuss possible extension of screening to environmental samples or asymptomatic personnel (generally not recommended) with infection preventionists, clinical microbiologists, epidemiologists and infectious disease specialists |
LTCF infection preventionists, infectious disease specialists, clinical microbiologists, epidemiologists, |
| Data elaboration | Integrate active MDRO cultural or molecular screening data from LTCF residents into a general antimicrobial susceptibility data report, comparing screening data from LTCF residents with those from referral ACH patients | If possible, stratify screening data from LTCF residents, admitted within 48 h to the referral ACH, together with LTCF screening data | Clinical microbiologists, epidemiologists, |
| Isolate conservation | Cryopreserve CPE-isolates (at − 80 °C) | Preserve for further molecular characterization, in collaboration with a reference molecular biology laboratory | Clinical microbiologists |
Prospective MDRO surveillance surveys (third level laboratory-based surveillance, on a voluntary basis)
Analysis of routine isolates from clinical diagnostic or active screening cultures obtained from LTCF residents is easy and cheap to perform, but the generally low number of routine clinical samples sent to the microbiology laboratory for analysis limits the value of this strategy; nevertheless, it is generally difficult to include isolates from LTCF residents admitted to the acute care referral hospital in the LTCF surveillance report. Moreover, with respect to the current epidemiological situation of CPE in Italy, the present recommendations for active surveillance screening are focused on CPE. Therefore, prospective surveillance surveys, mainly based on point prevalence colonization studies are recommended, allowing to determine the baseline prevalence of various MDROs in the facility; repeating of the active surveillance can then be used to evaluate the success of an intervention that was implemented in response to high MDRO-rates. We recommend selecting highest prevalence LTCFs for inclusion in the prospective surveillance study, allowing to obtain a “worst case scenario” for a specific healthcare region. Prospective surveillance can be limited to a resident subgroup or may be facility-wide. We recommend performing local, regional and national surveys to delineate the extent of MDRO colonization and infection in residents of LTCFs. For culture based or molecular screening of MDROs, microbiology methods routinely used in the referring microbiology laboratory can be applied. Prospective surveys for MDROs based on culturing specimens from clinically suspected infections can also give significant laboratory-based surveillance results [17].
Molecular typing of the associated isolates is important for identification of resistance plasmids and of hyper-epidemic clonal groups [34], and may be helpful in assessing whether resident-to-resident transmission has occurred. Genotyping can utilize various methods, such as pulsed-field gel electrophoresis (PFGE) [12, 13, 35], polymerase chain reaction and sequence based methods [12, 15, 36], multi locus sequence typing (MLST) [15], up to whole-genome sequencing (WGS) [37], which is rapidly becoming the new standard for the molecular characterization of isolates.
We recommend prospective third level laboratory-based surveillance as follows.
| Theme of the recommendation | Recommended strategies | Comments | Specific responsibilities |
| Preparation of surveillance project | Prepare a surveillance survey project, in collaboration with the LCTF physicians and the infection control practitioners, to the LTCF administration and the referral ethics committee, that includes institutional review board approval for the survey | Consent from residents (or their legal representatives) to participate is required | LTCF administration, LTCF physicians, LTCF infection preventionists, clinical microbiologists, epidemiologists, local ethics committee |
| LTCF selection | Select at least one representative LTCF in a healthcare district for performing a point prevalence screening survey, to obtain baseline MDRO colonization data | Selection should be done by the body oversighting the surveillance. Preferably select LTCFs with high prevalence of MDROs (as derived from routine clinical and/or screening data) | LTCF physicians, infection control practitioners, clinical microbiologists, epidemiologists |
| MDRO types | Include at least ESBL- and carbapenemase-producing Enterobacterales, carbapenemase-producing A. baumannii, carbapenem-resistant P. aeruginosa, MRSA and VRE | Possible extension to other MDROs | Infection control practitioners, clinical microbiologists, epidemiologists |
| Specimen types | Collect from each LTCF resident at least a rectal (or fecal), an oropharyngeal (or nasal) and inguinal (or perineal) swab | Specimen types recommended for screening of ESBL- and carbapenemase-producing Enterobacterales and for screening of VRE are rectal or fecal swabs (possibly combined with urine samples), for carbapenem-resistant P. aeruginosa and A. baumannii we recommend rectal and oropharyngeal swabs (possibly combined with urine samples), and for MRSA we recommend a combination of oropharyngeal (or nasal) and inguinal (or perineal) swabs (together with swabs from wounds, if present) | Infection control practitioners, clinical microbiologists, epidemiologists |
| Frequency of surveillance project | Repeat surveillance cultures in the same LTCF at least in a four-year interval | Shorter intervals are preferred | Infection control practitioners, clinical microbiologists, epidemiologists |
| Staff screening | Perform screening cultures from asymptomatic personnel only if staff members agree to participate in the screening study | Anonymous and only for epidemiologic data collection | Infection control practitioners, LTCF personnel, clinical microbiologists, epidemiologists |
| Isolate conservation | Cryopreserve MDR-isolates (at −80 °C) | Preserve for further molecular characterization, in collaboration with a reference molecular biology laboratory | Clinical microbiologists |
Infection control measures
Laboratory-based surveillance of MDROs is only effective if the collected data are used by healthcare professionals [38]:
to better address empiric therapy of infectious diseases
to guide infection control activities (e.g. isolation precautions for residents to prevent transmission)
to plan educational programs
to identify trends in resistance
to detect outbreaks requiring prompt therapeutic and infection control actions
Exhaustive guidelines providing basic information for infection prevention and control in LTCFs have been published by the Society for Healthcare Epidemiology of America (SHEA) in collaboration with the Association for Professionals in Infection Control (APIC) and by the Royal College of Physicians Clinical Advisory Group on Healthcare Associated Infections [1, 2, 39]. Guidelines agree that for the control of MDROs in LTCFs, besides hygiene measures, the implementation of antimicrobial stewardship programs is pivotal for the improvement of antibiotic use [3].
Finally, any discussion of MDRO control issues in LTCFs must be made in the context of these facilities as a home for residents, in which they usually reside for months or years, and therefore the resident’s living comfort must be addressed together with the control of MDROs. In the absence of risk factors for shedding large numbers of MDROs, LTCF residents should not be restricted from participation in social or therapeutic group activities within the facility, unless there is reason to think that they have been implicated in the development of infections in other residents [2].
Summary and conclusions
Routine surveillance of MDROs in LTCFs needs to be simple, pragmatic and sustainable. Laboratory-based routine testing of isolates from cultures taken for “clinical” purposes is the most cost-effective and less labor-intensive method to track MDROs. Therefore, we propose this method as mandatory first level laboratory-based MDRO surveillance strategy in Italian LTCFs.
On the other hand, active surveillance cultures (or PCR based methods) report asymptomatic colonization by MDROs (e.g., rectal swabs for CPE). Laboratory-based active screening results may complement routine diagnostic data, and are strongly recommended when an LTCF reports a case of infection by CPE (second level laboratory-based MDRO-screening).
Prospective surveillance surveys for various MDROs (third level laboratory-based MDRO-screening), based on prospective culturing of specimens from clinically suspected infections, and especially on point prevalence studies, is costly and time-consuming, but yields the most significant laboratory-based surveillance results; especially if isolates are molecularly characterized and genotyped.
We therefore recommend a stepwise application of the three MDROs surveillance levels, initially focusing on the elaboration of data from diagnostic bacterial isolates, in a second step extending this procedure to active surveillance screening for CPE, and finally performing prospective point prevalence studies in selected LTCFs. Finally, it is mandatory that the collected and elaborated MDROs surveillance data be used by healthcare professionals to inform decisions for infection prevention and control strategies, targeting the unique aspects of LTCFs.
Acknowledgements
These recommendations were prepared and approved by the GLISTer Working Group of AMCLI and do not necessarily reflect the opinions of Antimicrobial Resistance and Infection Control or its Editors.
Members of the GLISTer working group: Massimo Confalonieri (Piacenza), Claudio Farina (Bergamo), Paolo Fazii (Pescara), Francesco Luzzaro (Lecco), Roberto Rigoli (Treviso), Melissa Spalla (Pavia).
Abbreviations
- GLISTer
Gruppo di Lavoro per lo studio delle Infezioni nelle Residenze Sanitarie Assistite e Strutture Territoriali Assimilabili (Italian working group for the study of infections in LTCFs)
- AMCLI
Associazione Microbiologi Clinici Italiani (Italian Association of Clinical Microbiologists)
- LTCF
Long-term care facility: encompass nursing homes, residential care centers, chronic disease hospitals, rehabilitation centers and institutions for mentally handicapped persons
- MDRO
Multidrug-resistant organism: methicillin-resistant Staphylococcus aureus (MRSA), vancomycin-resistant enterococci (VRE), Pseudomonas aeruginosa or Acinetobacter baumannii with MDR phenotype (resistance to ≥3 classes of antibiotics), Enterobacterales producing extended-spectrum β-lactamases (ESBL), high-level cephalosporinases (AmpCs) and/or carbapenemases
- PNCAR 2017–20
Piano Nazionale di Contrasto dell’Antimicrobico-Resistenza 2017–20 (Italian plan for contrasting of antimicrobial resistance 2017–20)
Authors’ contributions
RA conceived the study, participated in its design and coordination and drafted the manuscript; LP participated in the study design, provided technical improvement, and drafted the manuscript; RM participated in the study design and coordination and drafted the manuscript; LP conceived the study, coordinated and led its different parts and reviewed the manuscript. The authors read and approved the final manuscript.
Funding
No source of funding to declare.
Availability of data and materials
Data sharing not applicable to this article as no datasets were generated or analyzed during the current study.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Leonardo Pagani, Email: lpagani.id@gmail.com.
the GLISTer (Gruppo di Lavoro per lo Studio delle Infezioni nelle Residenze Sanitarie Assistite e Strutture Assimilabili) working group:
Massimo Confalonieri, Claudio Farina, Paolo Fazii, Francesco Luzzaro, Roberto Rigoli, and Melissa Spalla
References
- 1.Strausbaugh LJ, Crossley KB, Nurse BA, Thrupp LD. Antimicrobial resistance in long-term-care facilities. Infect Control Hosp Epidemiol. 1996;17:129–140. doi: 10.2307/30141013. [DOI] [PubMed] [Google Scholar]
- 2.Smith PW, Bennett G, Bradley S, Drinka P, Lautenbach E, Marx J, et al. SHEA/APIC guideline: infection prevention and control in the long-term care facility. Infect Control Hosp Epidemiol. 2008;29:785–814. doi: 10.1086/592416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dyar OJ, Pagani L, Pulcini C. Strategies and challenges of antimicrobial stewardship in long-term care facilities. Clin Microbiol Infect. 2015;21:10–19. doi: 10.1016/j.cmi.2014.09.005. [DOI] [PubMed] [Google Scholar]
- 4.Moro ML, Gagliotti C. Antimicrobial resistance and stewardship in long-term care settings. Future Med. 2013;8:1011–1025. doi: 10.2217/fmb.13.75. [DOI] [PubMed] [Google Scholar]
- 5.CLSI . Analysis and presentation of cumulative antimicrobial susceptibility test data; approved guideline - fourth edition. CLSI document M39-A4. Wayne: Clinical and Laboratory Standards Institute; 2014. [Google Scholar]
- 6.Confalonieri M. Patogeni MDR tra territorio e strutture sanitarie: è possibile contrastare l’emergenza? La governance delle residenze sanitarie assistite e strutture territoriali assimilabili: il quadro italiano. Abstract from the National Congress AMCLI 2017, Rimini, https://www.amcli.it/eventi/congressi-nazionali/. Last access on 30/03/2019.
- 7.Piano Nazionale per il Contrasto dell’antimicrobico-resistenza 2017–20 (PNCAR), ottobre 2017, Presidenza del Consiglio dei Ministri. http://www.salute.gov.it/imgs/C_17_pubblicazioni_2660_allegato.pdf. Last access on 30/03/2019.
- 8.Pagani L, Confalonieri M, Aschbacher R, Farina C, Fazii P, Luzzaro F et al. Linee di indirizzo per la sorveglianza dei batteri multi-resistenti nelle Long-Term Care Facilities (LTCFs) italiane, proposte dal Gruppo di Lavoro per lo Studio delle Infezioni nelle Residenze Sanitarie Assistenziali e Strutture assimilabili (GLISTer) dell’Associazione Microbiologi Clinici Italiani (AMCLI). 2018. http://www.amcli.it/wp-content/uploads/2015/09/Linee-di-indirizzo-GLISTer-versione-definitiva04lug2018-1.pdf. Last access on 30/03/2019.
- 9.Venkatachalam I, Yang HL, Fisher D, Lye DC, Moi Lin L, Tambyah P, et al. Multidrug-resistant gram-negative bloodstream infections among residents of long-term care facilities. Infect Control Hosp Epidemiol. 2014;35:519–526. doi: 10.1086/675823. [DOI] [PubMed] [Google Scholar]
- 10.Annual report of the European Antimicrobial Resistance Surveillance Network (EARS-Net) 2018. https://www.ecdc.europa.eu/sites/default/files/documents/surveillance-antimicrobial-resistance-Europe-2018.pdf. Last access on 28/11/2019.
- 11.Aschbacher R, Pagani E, Confalonieri M, Farina C, Fazii P, Luzzaro F, et al. Review on colonization of residents and staff in Italian long-term care facilities by multidrug-resistant bacteria compared with other European countries. Antimicrob Resist Infect Control. 2016;5:33. doi: 10.1186/s13756-016-0136-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.March A, Aschbacher R, Dhanji H, Livermore DM, Böttcher A, Sleghel F, et al. Colonization of residents and staff of a long-term-care facility and adjacent acute-care hospital geriatric unit by multiresistant bacteria. Clin Microbiol Infect. 2010;16:934–944. doi: 10.1111/j.1469-0691.2009.03024.x. [DOI] [PubMed] [Google Scholar]
- 13.March A, Aschbacher R, Pagani E, Sleghel F, Soelva G, Hopkins KL, et al. Changes in colonization of residents and staff of a long-term care facility and an adjacent acute-care hospital geriatric unit by multidrug-resistant bacteria over a four-year period. Scand J Infect Dis. 2014;46:114–122. doi: 10.3109/00365548.2013.859392. [DOI] [PubMed] [Google Scholar]
- 14.March A, Aschbacher R, Sleghel F, Soelva S, Kaczor M, Migliavacca R, et al. Colonization of residents and staff of an Italian long-term care facility and an adjacent acute-care hospital geriatric unit by multidrug-resistant bacteria. 2017. New Microbiol. 2017;40:258–263. [PubMed] [Google Scholar]
- 15.Giufrè M, Ricchizzi E, Accogli M, Barbanti F, Monaco M, Pimentel de Araujo F, et al. Colonization by multidrug-resistant organisms in long-term care facilities in Italy: a point-prevalence study. Clin Microbiol Infect. 2017;23:961–967. doi: 10.1016/j.cmi.2017.04.006. [DOI] [PubMed] [Google Scholar]
- 16.Nucleo E, Caltagirone M, Marchetti VM, D’Angelo R, Fogato E, Confalonieri M, et al. Colonization of long-term care facility residents in three Italian provinces by multidrug-resistant bacteria. Antimicrob Resist Infect Control. 2018;7:33. doi: 10.1186/s13756-018-0326-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Giufrè M, Accogli M, Ricchizzi E, Barbanti F, Farina C, Fazii P, et al. Multidrug-resistant infections in long-term care facilities: extended-spectrum β-lactamase-producing Enterobacteriaceae and hypervirulent antibiotic resistant Clostridium difficile. Diagn Microbiol Infect Dis. 2018;91:275–281. doi: 10.1016/j.diagmicrobio.2018.02.018. [DOI] [PubMed] [Google Scholar]
- 18.Tinelli M, Cataldo MA, Mantengoli E, Cadeddu C, Cunietti E, Luzzaro F, et al. Epidemiology and genetic characteristics of extended-spectrum β-lactamase-producing gram-negative bacteria causing urinary tract infections in long-term care facilities. J Antimicrob Chemother. 2012;67:2982–2987. doi: 10.1093/jac/dks300. [DOI] [PubMed] [Google Scholar]
- 19.Arena F, Vannetti F, Di Pilato V, Fabbri L, Colavecchio OL, Giani T, et al. Diversity of the epidemiology of carbapenemase-producing Enterobacteriaceae in long-term acute care rehabilitation settings from an area of hyperendemicity, and evaluation of an intervention bundle. J Hosp Infect. 2018;100:29–34. doi: 10.1016/j.jhin.2018.05.025. [DOI] [PubMed] [Google Scholar]
- 20.Cornaglia G, Hryniewicz W, Jarlier V, Kahlmeter G, Mittermayer H, Stratchounski L, et al. European recommendations for antimicrobial resistance surveillance. Clin Microbiol Infect. 2004;10:349–383. doi: 10.1111/j.1198-743X.2004.00887.x. [DOI] [PubMed] [Google Scholar]
- 21.Orsi GB, Falcone M, Venditti M. Surveillance and Management of Multidrug-resistant Microorganisms. Expert Rev Anti-Infect Ther. 2011;9:653–679. doi: 10.1586/eri.11.77. [DOI] [PubMed] [Google Scholar]
- 22.Ricchizzi E, Gagliotti G, Buttazzi R, Moro ML. La prevalenza di infezioni correlate all’assistenza e di uso di antibiotici nelle strutture residenziali per anziani in Emilia-Romagna, I risultati del Progetto europeo HALT3 2017. Rapporto Regionale. http://assr.regione.emilia-romagna.it/it/notizie/home/ica-risultati-2016-2017. Last access on 30/03/2019.
- 23.Menichetti F, Falcone M, Lopalco P, Tascini C, Pan A, Busani L, et al. The GISA call to action for the appropriate use of antimicrobials and the control of antimicrobial resistance in Italy. Int J Antimicrob Agents. 2018;52:127–134. doi: 10.1016/j.ijantimicag.2018.05.010. [DOI] [PubMed] [Google Scholar]
- 24.Kohlmann R, Gatermann SG. Analysis and presentation of cumulative antimicrobial susceptibility test data – the influence of different parameters in a routine clinical microbiology laboratory. PLoS One. 2016;11:1. doi: 10.1371/journal.pone.0147965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fridkin SK, Edwards JR, Tenover FC, Gaynes RP, McGowan JE., Jr Antimicrobial resistance prevalence rates in hospital antibiograms reflect prevalence rates among pathogens associated with hospital-acquired infections. Clin Infect Dis. 2001;33:324–330. doi: 10.1086/321893. [DOI] [PubMed] [Google Scholar]
- 26.De Paoli P, Goglio A, Nicoletti P. Proposta di linee guida per l’analisi e la presentazione Dei risultati cumulativi degli antibiogrammi. Associazione per la Prevenzione ed il Controllo delle Infezioni (APSI) Giorn Ital Infez Osped. 2002;9:66–73. [Google Scholar]
- 27.Ambretti S, Gagliotti C, Luzzaro F, Malacarne P, Pan A, Pieretti B, et al. Comitato CoSIAS-AMCLI. Reporting epidemiology of antibiotic resistance. Microbiol Med. 2015;30:5308. doi: 10.4081/mm.2015.5308. [DOI] [Google Scholar]
- 28.Luzzaro F, Pagani L, Rossolini GM, Sarti M, Stefani S, Varaldo PE. Indicazioni per lo screening colturale dei pazienti colonizzati da Enterobatteri produttori di carbapenemasi, AMCLI 2012 (http://www.amcli.it/wp-content/uploads/2015/10/Screening_Enterob._prod_di_carbapenemasi.pdf). Last access on 30/03/2019.
- 29.Weber SG, Huang SS, Oriola S, Huskins WC, Noskin GA, Harriman K et al. Legislative mandates for use of active surveillance cultures to screen for methicillin-resistant Staphylococcus aureus and vancomycin-resistant enterococci: position statement from the joint SHEA and APIC task force. SHEA/APIC position statement 2007 (https://www.shea-online.org/images/position-statements/2007_SHEA-APIC_position_paper_Final.pdf). Last access on 30/03/2019. [DOI] [PubMed]
- 30.APIC guideline, 2010. Guide to the Elimination of Multidrug-resistant Acinetobacter baumannii Transmission in Healthcare Settings (https://apic.org/Resource_/EliminationGuideForm/b8b0b11f-1808-4615-890b-f652d116ba56/File/APIC-AB-Guide.pdf). Last access on 30/03/2019.
- 31.ECDC guideline, 2016. Carbapenem-resistant Acinetobacter baumannii in health-care setting. European Center for Disease Prevention and Control (https://ecdc.europa.eu/sites/portal/files/media/en/publications/Publications/8-Dec-2016-RRA-Acinetobacter%20baumannii-Europe.pdf). Last access on 30/03/2019.
- 32.Araoka H, Kimura M, Abe M, Takahashi N, Yoneyama A. Appropriate sampling sites for the surveillance of multidrug-resistant Pseudomonas aeruginosa colonization. Jpn J Infect Dis. 2014;67:118–119. doi: 10.7883/yoken.67.118. [DOI] [PubMed] [Google Scholar]
- 33.Giani T, Antonelli A, Caltagirone M, Mauri C, Nicchi J, Arena F, et al. Evolving beta-lactamase epidemiology in Enterobacteriaceae from Italian nationwide surveillance, October 2013: KPC-carbapenemase spreading among outpatients. Euro Surveill. 2017;22:31. doi: 10.2807/1560-7917.ES.2017.22.31.30583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mathers AJ, Peirano G, Pitout JD. The role of epidemic resistance plasmids and international high-risk clones in the spread of multidrug-resistant Enterobacteriaceae. Clin Microbiol Rev. 2015;28:565–591. doi: 10.1128/CMR.00116-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brugnaro P, Fedeli U, Pellizzer G, Buonfrate D, Rassu M, Boldrin C, et al. Clustering and risk factors of methicillin-resistant Staphylococcus aureus carriage in two Italian long-term care facilities. Infection. 2009;37:216–221. doi: 10.1007/s15010-008-8165-1. [DOI] [PubMed] [Google Scholar]
- 36.Monaco M, Bombana E, Trezzi L, Regattin L, Brusaferro S, Pantosti A, et al. Meticillin-resistant Staphylococcus aureus colonising residents and staff members in a nursing home in northern Italy. J Hosp Infect. 2009;73:182–184. doi: 10.1016/j.jhin.2009.06.026. [DOI] [PubMed] [Google Scholar]
- 37.Brodrick HJ, Rave KE, Kallonen T, Jamrozy D, Blane B, Brown NM, et al. Longitudinal genomic surveillance of multidrug-resistant Escherichia coli carriage in a long-term care facility in the United Kingdom. Genome Med. 2017;9:70. doi: 10.1186/s13073-017-0457-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Critchley IA, Karlowsky JA. Optimal use of antibiotic resistance surveillance systems. Clin Microbiol Infect. 2004;10:502–511. doi: 10.1111/j.1469-0691.2004.00911.x. [DOI] [PubMed] [Google Scholar]
- 39.Royal College of Physicians clinical advisory group on healthcare associated infections . Guidelines for the prevention and control of multi-drug resistant organisms (MDRO) excluding MRSA in the healthcare setting. 2012. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing not applicable to this article as no datasets were generated or analyzed during the current study.