Abstract.
Dengue fever and other febrile mosquito-borne diseases place considerable health and economic burdens on small island nations in the Caribbean. Here, we used two methods of cluster detection to find potential hotspots of transmission of dengue and chikungunya in Barbados, and to assess the impact of input surveillance data and methodology on observed patterns of risk. Using Moran’s I and spatial scan statistics, we analyzed the geospatial and temporal distribution of disease cases and rates across Barbados for dengue fever in 2013–2016, and a chikungunya outbreak in 2014. During years with high numbers of dengue cases, hotspots for cases were found with Moran’s I in the south and central regions in 2013 and 2016, respectively. Using smoothed disease rates, clustering was detected in all years for dengue. Hotspots suggesting higher rates were not detected via spatial scan statistics, but coldspots suggesting lower than expected rates of disease activity were found in southwestern Barbados during high case years of dengue. No significant spatiotemporal structure was found in cases during the chikungunya outbreak. Spatial analysis of surveillance data is useful in identifying outbreak hotspots, potentially complementing existing early warning systems. We caution that these methods should be used in a manner appropriate to available data and reflecting explicit public health goals—managing for overall case numbers or targeting anomalous rates for further investigation.
INTRODUCTION
Dengue fever threatens the health of communities throughout Latin America and the Caribbean, where all four serotypes of dengue virus (DENV 1–4) are in circulation following a regional resurgence of the pathogen in the 1980s.1,2 The Caribbean island of Barbados is challenged with managing endemic dengue fever and other febrile mosquito-borne diseases including emerging chikungunya and Zika viral diseases.3–6 In small island nations such as Barbados, outbreaks translate into increased morbidity and mortality, high costs to healthcare systems, and lost economic productivity.7–10 With approximately 40% of employment and gross domestic product linked to the tourism industry, Barbados is particularly vulnerable to the economic impacts of arbovirus outbreaks.11 In addition to lost domestic productivity, travel-related cases and negative health perceptions associated with outbreaks deter potential visitors, further impacting the livelihoods of island residents.12,13 The emergence and subsequent establishment of novel arboviruses in the Caribbean exacerbates matters by complicating disease management while further impacting sources of income.3,14 In response to these social and economic burdens, the Ministry of Health (MoH) and Wellness of Barbados has a long history of engaging in public mosquito control and active disease surveillance, where suspected human cases are laboratory confirmed, and vector control interventions are conducted in response to both laboratory results and mosquito surveillance. Interagency collaborations are part of a comprehensive effort to mitigate the toll of endemic dengue.15 Previous studies performed in Barbados have described climatological and seasonal drivers of dengue transmission, vital components of early warning systems, and forecasting models.16,17 Although large-scale climatological factors undoubtedly play a dominant role in driving outbreaks of mosquito-borne illness, this plays out at the local scale as a function of the human landscape.18,19 Therefore, understanding the local distribution of human cases is also necessary for understanding patterns of exposure risk and guiding vector abatement strategies.
Aedes aegypti is the primary mosquito vector of medical concern throughout the Caribbean. Public health vector control programs are widely acknowledged as cost-effective in controlling arboviruses transmitted by Ae. aegypti, relative to costs associated with the delivery of health services and supportive care.20,21 Nevertheless, public health resources are finite, calling for efficient intervention strategies to target mosquito populations and suppress transmission pathways. Ae. aegypti is a container-breeding mosquito and successfully exploits anthropogenic environments for oviposition and larval rearing. The role of household-level characteristics, such as housing condition and water storage habits, in promoting mosquito production has been repeatedly demonstrated.18,19 In some instances, favorable microhabitats enable mosquitoes, and subsequently disease transmission, to persist despite general unfavorable environmental conditions.22 Thus, identifying spatial clusters of high disease activity, or “hotspots,” can prove invaluable when prioritizing the delivery of abatement and outreach services. Further additional challenges, although not unique in the context of integrated vector control, are essential to address for management of mosquito-borne diseases in Caribbean islands. Vector-borne disease risk can shift rapidly on small islands like Barbados because of many factors, including insecticide resistance, climate variability, climate change, high disease prevalence, and variable mosquito control efforts in response to herd immunity dynamics. Small island developing states in the Caribbean also face challenges to the elimination of Ae. aegypti, as reintroductions of vectors are frequent because of interregional travel, unplanned urbanization, and limited resources for vector control.23 Frequent reintroductions of pathogens are also problematic for elimination campaigns, particularly in locations with high volumes of regional and international travel, as humans are the reservoir host for both epidemic dengue and chikungunya.24,25 These management challenges demand strategies that incorporate spatially and temporally sensitive methods of detecting transmission activity.
Geographic information systems (GISs) offer a powerful tool in the visualization and incorporation of spatial epidemiological data into public health programs.26 Although many health departments and ministries have readily adopted GIS methods into their surveillance and reporting activities, fewer have extended these methodologies to incorporate statistical tests of spatial dependency in human case data. Local indicators of spatial association (LISA) statistics are routinely used in an exploratory framework to quantitatively describe patterns of spatial dependence and clustering, or dispersion, of disease cases within a defined area of study.27 Identifying spatially discrete areas of significantly high (i.e., hotspots) or low (i.e., coldspots) disease activity within functional administrative boundaries is a useful framework for crafting responses to outbreak events and future interventions, enabling agencies to focus their efforts more efficiently. Global and local Moran’s I tests have been applied in public health contexts to describe spatial distributions of mosquito-borne disease outbreaks, including dengue fever, and to detect the location of disease clusters.19,28,29
Although LISA methods give us insight into the spatial structure of disease activity within a given time period, these analyses are temporally static. In instances where georeferenced disease surveillance data are available at regular time intervals, spatial scan statistics can be used to identify local areas of clustering in multiple dimensions (i.e., space, time, or space–time). Spatial scan statistics are capable of detecting possible disease clustering in surveillance data, using a series of variable search windows to evaluate spatial and temporal trends in the dataset.30,31 Application of space–time scan statistics can be a powerful tool in disease surveillance and outbreak detection, where we are interested in describing not only where but also when clusters of events occur over a continuous period of time.
To our knowledge, no previous efforts have described the spatial and temporal distribution of dengue or chikungunya outbreaks in Barbados. Using epidemiological case data collected by the MoH in Barbados from 2013–2016, we used exploratory LISA and space–time scan statistics to test for spatial and temporal autocorrelation of dengue and chikungunya cases within operational health districts. The objectives of this study were to perform the following: 1) detect global spatial dependency, or clustering, of surveillance arbovirus cases reported in Barbados within each year of the study period; 2) when global spatial dependency is detected, identify the locations of hotspots (i.e., clustered) and coldspots (i.e., dispersed) of disease activity; 3) assess the effect of different input data on observed spatial patterns; and 4) detect spatiotemporal patterns in disease rates at finer temporal resolutions via spatial scan statistics. This also provides an important opportunity to discuss and showcase the implications of how these methods are implemented in situations where data are limited, simply as a function of small populations, as seen in small island nations.
METHODS
Study area and epidemiological data.
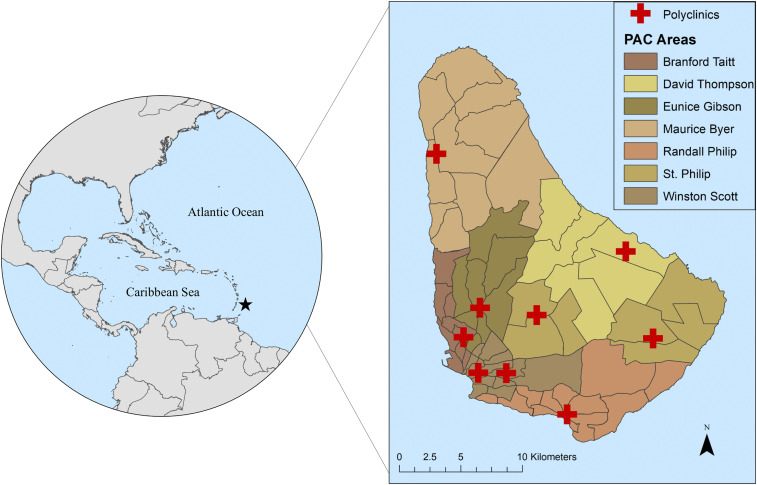
Barbados, situated in the Caribbean, has an estimated residential population of more than 277,000.32 The most densely populated areas are found on the southern side of the island, with the highest population density found around Bridgetown, the capital city.32 Transmission of mosquito-borne diseases in Barbados is seasonal, with peak transmission typically associated with high numbers of mosquitoes during the rainy season (June–November) and fewer disease cases reported during the dry season (December–May).16,33 The MoH of Barbados performs active and passive surveillance for dengue and other mosquito-borne diseases via nine polyclinics. These serve seven polyclinic administrative catchment (PAC) areas (Branford Taitt, David Thompson, Eunice Gibson, Maurice Byer, Randall Philip, St. Philip, and Winston Scott), which are further divided into 63 health districts (Figure 1). Public vector control and health services are delivered at the level of health districts, which range in size from 0.40 km2 to 26.62 km2. The MoH oversees arbovirus sureveillance activities, where suspected human cases of dengue and chikungunya are recorded by the ministry and confirmed in the National Reference Laboratory by reverse transcriptase polymerase chain reaction (RT-PCR) or ELISA. De-identified, monthly case totals for dengue fever in each health district were provided by the MoH for the years 2013–2016, and we defined the location of cases as the centroid of a given district. Georeferenced data on laboratory-confirmed chikungunya cases, aggregated to health districts, were also made available for this study, but were only available for the 2014 outbreak. Additional GIS data were provided for this study by the MoH, including shapefiles of the administrative boundaries for health districts in Barbados.
Figure 1.
Health districts and polyclinic administrative catchment (PAC) areas in Barbados. This figure was produced in ArcMap 10.4 (ESRI) using shapefiles from the GADM database of Global Administrative Areas, v. 2.8 (gadm.org), and shapefiles provided by the Ministry of Health, Barbados. This figure appears in color at www.ajtmh.org.
Global and LISA.
Annual case totals for dengue and chikungunya in each health district were aggregated from monthly case data provided by the MoH for each year of the study. Annual per capita disease rates were derived from annual totals and population data from the most recent national census, conducted in 2010.32 The population of each health district ranged from 68 to 12,743, according to census data. Because of low population in some health districts, raw disease rates may be susceptible to instability due to high variance associated with small numerators or denominators (i.e., the “small number problem”).34 Performing spatial analyses on raw rates with high instability can result in incorrectly identifying artefacts of the small number problem as statistically significant outliers. We performed empirical Bayes (EB) smoothing, where the variance of rate estimates is globally reduced via a priori probability functions on raw disease rates in Geoda (v. 1.12.0) to compensate for high variability in rates due to low health district population. Empirical Bayes smoothed rates were compared with raw disease rates, verifying the overall reduction of variance from smoothing.
Global Moran’s I with inverse distance weighting (ArcMap, v. 10.4) was used to test for spatial autocorrelation in both case counts and smoothed disease rates for dengue and chikungunya in Barbados for each year of the study. A global indicator of spatial dependence, the Moran’s I statistic, provides a measure of the degree of statistically significant clustering or dispersion in disease measures for the entire island. Local Moran’s I is a LISA statistic for identifying locations (e.g., health districts within the study area) with statistically similar spatial patterns (e.g., clustering or dispersion) of high and low values (i.e., hotspots or coldspots) on the island.35 This statistic is also useful for the detection of spatial outliers, locations with significantly extreme values compared with neighboring areas.35 Local Moran’s I with inverse distance weighting was performed for each reported year in ArcMap (v. 10.4) to identify health districts that were hotspots, or coldspots, of dengue or chikungunya activity.
Spatial scan statistics.
We compared the spatial distribution of dengue and chikungunya found via LISA statistical analyses, calculated for each year of the study, to patterns of clustering and dispersion in continuous aggregated cases over the study period. Patterns of spatiotemporal clustering in monthly case totals within each year were tested using the univariate Kulldorff’s spatial scan statistic in SaTScan (v. 9.4.4), where we performed retrospective analyses under the space–time permutation model, scanning for both clustering and dispersion.36 A circular search window was specified to test for spatiotemporal clustering, comparing cases at each location (i.e., centroids of health districts) with neighboring areas within a variable window, using a time precision of 1 month. Clusters were constrained to a maximum cluster size of 50% of case data and a maximum temporal window of 50% of the study period, as recommended in Kulldorff.36,37 Geographic overlap with other clusters was allowed, provided that no neighboring cluster centers were included in a given cluster. Likelihood ratios and associated P-values were reported for each identified cluster, where maximum likelihood values were calculated via Monte Carlo simulation (999 replications). Statistically significant clusters (α = 0.05) from the SaTScan analyses were mapped with LISA results for each year in ArcMap (v. 10.4) for visual comparison.
RESULTS
The number of dengue cases in Barbados reported by the MoH fluctuated greatly during the study period, with large outbreaks occurring in 2013 (n = 526) and 2016 (n = 386), and lower case numbers in 2014 (n = 147) and 2015 (n = 58). Georeferenced cases of chikungunya (n = 57) were only available for 2014. We detected statistically significant (α = 0.05) global clustering (i.e., Moran’s I values > 0) in aggregated case counts during the years of large dengue outbreaks, 2013 and 2016 (Table 1), whereas significant global clustering of EB smoothed rates was found in every year for dengue (Table 2). No significant clustering was detected during the 2014 chikungunya outbreak.
Table 1.
Global Moran’s I values for dengue and chikungunya case totals, aggregated to health district, in each year in the study
| Arbovirus | Year | Total cases | Moran’s I* | Z-score | P-value |
|---|---|---|---|---|---|
| Dengue | 2013 | 526 | 0.23 | 4.40 | < 0.001 |
| Dengue | 2014 | 147 | −0.04 | −0.51 | 0.610 |
| Dengue | 2015 | 58 | 0.04 | 1.06 | 0.291 |
| Dengue | 2016 | 386 | 0.11 | 2.29 | 0.022 |
| Chikungunya | 2014 | 57 | 0.06 | 0.84 | 0.400 |
P-values in bold indicate statistically significant results.
Moran’s I values range between -1 and 1, where negative values indicate dispersion and positive values indicate clustering.
Table 2.
Global Moran’s I values for dengue and chikungunya empirical Bayes (EB) smoothed disease rates, aggregated to health district, in each year in the study
| Arbovirus | Year | Total cases | Moran’s I* | Z-score | P-value |
|---|---|---|---|---|---|
| Dengue | 2013 | 526 | 0.290 | 5.77 | < 0.001 |
| Dengue | 2014 | 147 | 0.103 | 2.22 | 0.026 |
| Dengue | 2015 | 58 | 0.166 | 3.35 | < 0.001 |
| Dengue | 2016 | 386 | 0.330 | 6.43 | < 0.001 |
| Chikungunya | 2014 | 57 | 0.06 | 1.30 | 0.192 |
P-values in bold indicate statistically significant results.
Moran’s I values range between -1 and 1, where negative values indicate dispersion and positive values indicate clustering.
Local Moran’s I revealed shifting the locations of dengue hotspots and coldspots at the health district level between years in both case totals (Figure 2) and EB smoothed disease rates (Figure 3). Localized spatial autocorrelation in dengue case counts was found during large outbreak years, whereas significant patterns of clustering in EB smoothed rates of dengue were found in every year. The locations of hot and coldspots differed for case counts versus rates (Figures 3). In years where both cases and rates had significant spatial autocorrelation (i.e., the 2013 and 2016 outbreaks), the highest disease rates were clustered in health districts in the north of the island, whereas dengue case counts had hotspots and clustered outliers (i.e., health districts with a high number of cases relative to neighboring districts with low counts) in central and southern health districts. Statistically significant spatial autocorrelation was only detected in smoothed rates in years of lower dengue burden (i.e., 2014 and 2015). During low burden years, hotspots were generally identified in northern health districts, whereas coldspots were detected in southern health districts (Figure 3).
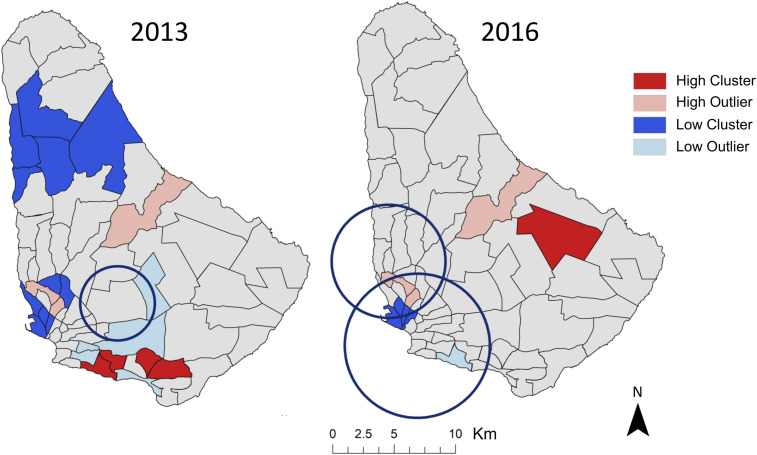
Figure 2.
Patterns of clustering (red) and dispersion (blue) of annual dengue case totals were found at the level of health district in Barbados in 2013 and 2016, as determined by Local Moran’s I. Spatiotemporal areas of low disease risk in monthly cases (blue circles), found via Kulldorff’s spatial scan statistic, were found in both years. This figure was produced in ArcMap 10.4 (ESRI). This figure appears in color at www.ajtmh.org.
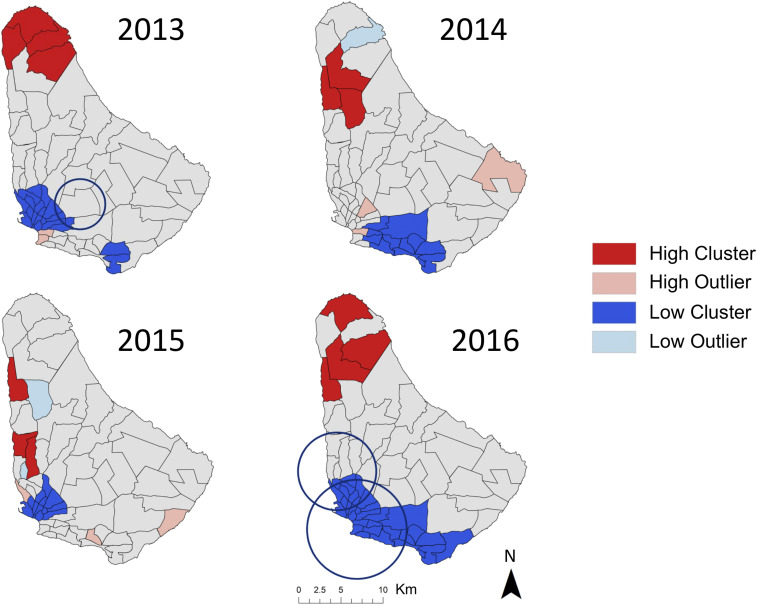
Figure 3.
Patterns of clustering (red) and dispersion (blue) of EB smoothed annual dengue rates were found at the level of health district in Barbados for all years of the study, as determined by Local Moran’s I. Spatiotemporal areas of low disease risk in monthly cases (blue circles), found via Kulldorff’s spatial scan statistic, were found in 2013 and 2016 for dengue. This figure was produced in ArcMap 10.4 (ESRI). This figure appears in color at www.ajtmh.org.
In years with large outbreaks of dengue, health districts in the southernmost Randall Philip PAC area were identified as hotspots of cases in 2013 (n = 4) and in the centrally located St. Philip PAC area in 2016 (n = 1) (Figure 2). Coldspots for dengue cases were detected in health districts located in the Maurice Byer (n = 5), Branford Taitt (n = 4), and Eunice Gibson (n = 2) PAC areas in 2013. In 2016, only three districts, in the Branford Taitt (n = 2) and Winston Scott (n = 1) administrative regions, were significant coldspots of cases (Figure 2). Three health districts, located in Branford Taitt, Eunice Gibson, and David Thompson catchment areas, were found to be clustered outliers in both 2013 and 2016 (Figure 2). When performing LISA analyses on smoothed dengue rates, the northern Maurice Byer PAC area contained all health districts that were hotspots of disease rates in 2013 (n = 4) and 2016 (n = 4) (Figure 3). Significantly, low rates of dengue were consistently found in southern health districts throughout the study period (Figure 3).
Analysis of monthly case data in Barbados via spatial scan statistics did not identify statistically significant clustering of high rates for either dengue or chikungunya. However, spatiotemporal locations with low rates of dengue were found in years with high case counts, indicating the duration and location of low disease activity during outbreak years (Table 3). In 2013, an area with low relative risk (RR = 0.08) spanning nine health districts across three PAC areas persisted from January to March (Table 3, Figure 2). A large area with low relative risk (RR = 0.44) was also identified in 2016 from August to October, comprising 29 health districts across five PAC areas, with a smaller overlapping area of low risk (RR = 0.39) found in June to September of the same year.
Table 3.
Statistically significant coldspots in monthly disease counts, calculated using the space–time permutation spatial scan statistic in SaTScan
| Arbovirus | Year | PAC areas | Health districts | Duration | Radius (km) | Observed cases | Expected cases | Relative risk | P-value |
|---|---|---|---|---|---|---|---|---|---|
| Dengue | 2013 | SP, WS, and EG | 9 | January 2013–March 2013 | 3.03 | 1 | 11.89 | 0.08 | 0.034 |
| Dengue | 2016 | WS, RP, SP, and BT | 29 | August 2016–October 2016 | 5.90 | 24 | 54.18 | 0.44 | < 0.001 |
| Dengue | 2016 | BT, EG, and WS | 17 | June 2016–September 2016 | 4.63 | 12 | 30.40 | 0.39 | 0.055 |
BT = Branford Taitt; EG = Eunice Gibson; PAC = polyclinic administrative catchment; RP = Randall Philip; SP = St. Philip; WS = Winston Scott. P-values in bold indicate statistically significant results.
DISCUSSION
In this study, we found that cases of dengue fever in Barbados detected via surveillance in 2013–2016 exhibit both spatial and temporal structure. Dengue cases clustered in the central and southwestern health districts in years with elevated case counts. By contrast, smoothed rates of population-derived incidence revealed clustering in all years for dengue, with many hotspots found in northern health districts. The identification of spatial dependence in disease cases is highly relevant for public health professionals working to suppress arbovirus transmission in Barbados, where there is a call to allocate public health resources efficiently.
Spatial discrepancies in data inputs (i.e., case numbers vs. population-derived rates) were driven in part by the small and spatially heterogeneous population density of Barbados. Consequently, any analyses performed on this system are susceptible to the “small numbers problem,” where estimates of disease prevalence and incidence rates can dramatically fluctuate as an artefact of either low-density underlying populations or relatively low case detection in high density populations.34 Procedures to reduce variance in rates, such as EB smoothing, are recommended to reduce the effect of unstable rates in disease mapping and tests for spatial autocorrelation.38 However, broad geospatial prescriptive remedies for the small number problem may unintentionally subvert public health agency management priorities, particularly in small island systems with extreme spatial population heterogeneity. Even after smoothing, we observed consistent hotspots of disease activity in northern health districts, where population densities are very low. Health districts with significantly high disease rates in low populations may not represent pragmatic management targets, especially in years where resources are limited or outbreaks are focused in urban centers. Although statistically sound, practical application of such analyses should be tempered by the expectations and priorities of public health agencies. In this context, raw case counts may give us a better understanding of operational disease burden on Barbados despite the problems typically associated with disregarding underlying population in morbidity metrics, where we would expect to detect more cases in densely populated areas regardless of true risk.
The differences observed in the spatial distribution of cases versus rates have critical implications with regard to intervention strategies and management goals. Although we accounted for inflated variance in rates by performing EB smoothing, hotspots in northern districts still reflect lower absolute caseloads than those found in densely populated areas in the south, especially in the vicinity of Bridgetown, the capital city. It is therefore imperative that management objectives are clearly specified before using spatial analyses on health surveillance data for planning purposes. Prioritization of goals is particularly important in a small island with high heterogeneity in population density, where making management decisions based on unstable rates could drive misallocation of resources. When responding to endemic transmission or emerging pathogens, such as chikungunya, targeting areas with the highest transmission rates (i.e., high numbers of cases relative to the underlying population) should be prioritized to prevent further spread. Conversely, when considering large outbreaks of endemic diseases, such as dengue, the management focus may be instead on reducing the total number of infections, regardless of population density, to mitigate hyperendemic peak years and reduce costs associated with the delivery of health services. In Barbados, these fundamental management distinctions may be subtle, but as our analyses demonstrate, it can require vastly different spatial representations of disease clustering in the study area. This would directly translate to choices of where to allocate resources in particular health districts.
Although we observed shifts in the clustering and dispersion of disease activity in Barbados between years, there were nevertheless consistencies in the location of health districts with clustered dengue cases or rates, especially in outbreak years. In particular, health districts identified as high clustering outliers during peak years were identical in 2013 and 2016, suggesting that some areas may have an underlying susceptibility to localized outbreaks when transmission is high (Figure 2). Although the analyses presented here represent a reactive management approach, in which there is lagged decision-making in response to previously reported case data, spatial methodologies can also be incorporated into proactive strategies as part of an early warning system framework. Predictive climatological models of dengue risk have been developed for Barbados, enabling the anticipation of large outbreak events driven by environmental factors.16 Although useful in terms of triggering agency response ahead of major island wide outbreak events, current probabilistic forecast models do not provide information on where to intervene. Here, our spatial analyses point to consistent areas of transmission peaks, providing complementary analyses to predictive climate modeling efforts, which can be incorporated into MoH decision-making, targeting discrete locations for mosquito control ahead of anticipated outbreak events.
We did not identify significant spatiotemporal clustering of dengue or chikungunya with spatial scan statistics within surveillance years, although locations with low dengue activity were found during some months in years with higher case totals. Previous studies have indicated that spatial scan statistics are often more sensitive to the detection of hotspots, particularly when relative risk is low, than other exploratory methods of spatial analysis.39,40 Spatial scan statistics have also been used to successfully identify hotspots of mosquito-borne diseases at fine temporal resolutions in systems where diseases are endemic.41,42 Our inability to detect disease clustering at higher temporal resolutions, even in years with high case counts, perhaps points to a lack of within-season localized clustering. The areas of low risk detected in 2016 for dengue coincide with the rainy season in Barbados, when we would expect to see increased transmission (Table 3). Areas with significantly low disease rates arise as a result of spatial uniformity in risk outside these locations of unexpectedly low transmission. Our ability to detect spatial clustering at subseason scales may alternatively be hindered by human movements, reflecting the difficulty of performing local disease surveillance in transient populations (e.g., commuters and international travelers).43 Although the inability to detect monthly clustering of arbovirus cases may limit the utility of spatial scan statistics to direct mosquito control activities at fine temporal scales in Barbados, our identification of locations with lower risk during active transmission seasons warrants investigation and future research into potential drivers.
Aggregated surveillance data are routinely collected in a public health context, but are not free from limitations. The MoH verifies dengue and chikungunya cases in the laboratory, but underreporting of cases is a commonly encountered issue with passive surveillance data in Latin America and the Caribbean, resulting in an underestimation of true disease risk in some areas.44 This is particularly true on islands with large transient populations, where travelers with mild or asymptomatic cases may not be detected by local surveillance networks, yet still serve to maintain local transmission cycles. The availability of current population data for calculating disease rates is also a limitation of this work. Although the projected population growth for Barbados is quite low, the most recent census data were collected in 2010, nearly a decade ago.32 In a spatial analysis context, the aggregation of cases to health districts prevents us from drawing conclusions at finer scales. Although this limits our ability to inform household-level interventions within disease clusters, identifying health districts with high level of disease transmission is nevertheless relevant to the operational scale of health services delivered by the MoH in Barbados. Despite these limitations, the data used in this study represent the most accurate and up-to-date estimates of population and disease risk in Barbados.
Public health implications.
These initial results serve as the foundation for incorporating spatial analyses into the existing arbovirus surveillance network in Barbados. Moving forward, these methodologies provide us not only with a means of guiding ministry responses to outbreaks of mosquito-borne diseases but also with the impetus for future geospatial analytical health studies in Barbados. Exploratory spatial analyses allow us to test hypotheses related to dominant social-ecological drivers of spatial clustering in health districts. Understanding the human characteristics that underlie observed spatial patterns can contribute to the development of better intervention methods.
REFERENCES
- 1.Knudsen AB, 1983. Aedes aegypti and dengue in the Caribbean. AMCA Mosq News 43: 269–275. [Google Scholar]
- 2.Brathwaite Dick O, San Martin JL, Montoya RH, del Diego J, Zambrano B, Dayan GH, 2012. The history of dengue outbreaks in the Americas. Am J Trop Med Hyg 87: 584–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ryan SJ, et al. 2018. Zika virus outbreak, Barbados, 2015–2016. Am J Trop Med Hyg 98: 1857–1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gittens-St Hilaire M, Clarke-Greenidge N, 2008. An analysis of the subtypes of dengue fever infections in Barbados 2003–2007 by reverse transcriptase polymerase chain reaction. Virol J 5: 152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kumar A, Nielsen AL, 2015. Trends in the patterns of IgM and IgG antibodies in febrile persons with suspected dengue in Barbados, an English-speaking Caribbean country, 2006–2013. J Infect Public Health 8: 583–592. [DOI] [PubMed] [Google Scholar]
- 6.Kumar A, Best C, Benskin G, 2017. Epidemiology, clinical and laboratory features and course of chikungunya among a cohort of children during the first Caribbean epidemic. J Trop Pediatr 63: 43–49. [DOI] [PubMed] [Google Scholar]
- 7.Bhatt S, et al. 2013. The global distribution and burden of dengue. Nature 496: 504–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gubler DJ, 2012. The economic burden of dengue. Am J Trop Med Hyg 86: 743–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Halasa YA, Zambrano B, Shepard DS, Dayan GH, Coudeville L, 2011. Economic impact of dengue illness in the Americas. Am J Trop Med Hyg 84: 200–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shepard DS, Undurraga EA, Halasa YA, Stanaway JD, 2016. The global economic burden of dengue: a systematic analysis. Lancet Infect Dis 16: 935–941. [DOI] [PubMed] [Google Scholar]
- 11.WTTC , 2018. Travel and Tourism Economic Impact 2018 Barbados. London, United Kingdom: World Travel and Tourism Council. [Google Scholar]
- 12.WTTC , 2017. Travel and Tourism Economic Impact 2017 Barbados. London, United Kingdom: World Travel and Tourism Council. [Google Scholar]
- 13.Shirtcliffe P, Cameron E, Nicholson KG, Wiselka MJ, 1998. Don’t forget dengue! Clinical features of dengue fever in returning travellers. J R Coll Physicians Lond 32: 235–237. [PMC free article] [PubMed] [Google Scholar]
- 14.Jamil Z, Waheed Y, Durrani TZ, 2016. Zika virus, a pathway to new challenges. Asian Pac J Trop Med 9: 626–629. [DOI] [PubMed] [Google Scholar]
- 15.Trotman A, Mahon R, Shumake-Guillemot J, Lowe R, Stewart-Ibarra AM, 2018. Strenghthening climate services for the health sector in the Caribbean. World Meteorol Organ 67: 14–19. [Google Scholar]
- 16.Lowe R, Gasparrini A, Van Meerbeeck CJ, Lippi CA, Mahon R, Trotman AR, Rollock L, Hinds AQJ, Ryan SJ, Stewart-Ibarra AM, 2018. Nonlinear and delayed impacts of climate on dengue risk in Barbados: a modelling study. PLoS Med 15: e1002613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Depradine CA, Lovell EH, 2004. Climatological variables and the incidence of Dengue fever in Barbados. Int J Environ Health Res 14: 429–441. [DOI] [PubMed] [Google Scholar]
- 18.Stewart-Ibarra AM, Muñoz ÁG, Ryan SJ, Ayala EB, Borbor-Cordova MJ, Finkelstein JL, Mejía R, Ordoñez T, Recalde-Coronel GC, Rivero K, 2014. Spatiotemporal clustering, climate periodicity, and social-ecological risk factors for dengue during an outbreak in Machala, Ecuador, in 2010. BMC Infect Dis 14, 610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lippi CA, Stewart-Ibarra AM, Muñoz ÁG, Borbor-Cordova MJ, Mejía R, Rivero K, Castillo K, Cárdenas WB, Ryan SJ, 2018. The social and spatial ecology of dengue presence and burden during an outbreak in Guayaquil, Ecuador, 2012. Int J Environ Res Public Health 15: E827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pepin KM, Marques-Toledo C, Scherer L, Morais MM, Ellis B, Eiras AE, 2013. Cost-effectiveness of novel system of mosquito surveillance and control, Brazil. Emerg Infect Dis 19: 542–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Momoh AA, Fügenschuh A, 2017. Optimal control of intervention strategies and cost effectiveness analysis for a Zika virus model. Oper Res Health Care 18: 99–111. [Google Scholar]
- 22.Paaijmans KP, Thomas MB, 2011. The influence of mosquito resting behaviour and associated microclimate for malaria risk. Malar J 10: 183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nathan MB, 1993. Critical review of Aedes aegypti control programs in the Caribbean and selected neighborhing countries. J Am Mosq Control Assoc 9: 1–7. [PubMed] [Google Scholar]
- 24.Moncayo AC, et al. 2004. Dengue emergence and adaptation to peridomestic mosquitoes. Emerg Infect Dis 10: 1790–1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Staples JE, Hills SL, Powers AM, 2020. Chikungunya. CDC Yellow Book, Vol. 4 New York, NY: Oxford University Press, 78. [Google Scholar]
- 26.McLafferty SL, 2003. GIS and health care. Annu Rev Public Health 24: 25–42. [DOI] [PubMed] [Google Scholar]
- 27.Oliveira MA, Ribeiro H, Castillo-Salgado C, 2013. Geospatial analysis applied to epidemiological studies of dengue: a systematic review. Rev Bras Epidemiol 16: 907–917. [DOI] [PubMed] [Google Scholar]
- 28.Castillo KC, Körbl B, Stewart A, Gonzalez JF, Ponce F, 2011. Application of spatial analysis to the examination of dengue fever in Guayaquil, Ecuador. Proc Environ Sci 7: 188–193. [Google Scholar]
- 29.Sugumaran R, Larson SR, DeGroote JP, 2009. Spatio-temporal cluster analysis of county-based human West Nile virus incidence in the continental United States. Int J Health Geogr 8: 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kulldorff M, 1997. A spatial scan statistic. Commun Stat Theor Methods 26: 1481–1496. [Google Scholar]
- 31.Kulldorff M. SaTScanTM v7.0.3: Software for the Spatial and Space-Time Scan Statistics. Information Management Services, Inc. [Google Scholar]
- 32.BSS , 2010. Barbados Population and Housing Census. Available at: http://www.barstats.gov.bb/census/. Accessed January 11, 2018. [Google Scholar]
- 33.WMO Caribbean Regional Climate Centre, 2017. Available at: http://rcc.cimh.edu.bb/. Accessed December 29, 2017. [Google Scholar]
- 34.Kennedy S, 1989. The small number problem and the accuracy of spatial databases. The Accuracy of Spatial Databases. London, United Kingdom: CRC Press, 308. [Google Scholar]
- 35.Anselin L, 1995. Local indicators of spatial association—LISA. Geogr Anal 27: 93–115. [Google Scholar]
- 36.Kulldorff M, Heffernan R, Hartman J, Assunção R, Mostashari F, 2005. A space–time permutation scan statistic for disease outbreak detection. PLoS Med 2: e59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kulldorff M, 2018. SaTScanTM User Guide. Available at: www.satscan.org. Accessed July 24, 2018.
- 38.Olsen SF, Martuzzi M, Elliott P, 1996. Cluster analysis and disease mapping–why, when, and how? A step by step guide. BMJ 313: 863–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barro AS, Kracalik IT, Malania L, Tsertsvadze N, Manvelyan J, Imnadze P, Blackburn JK, 2015. Identifying hotspots of human anthrax transmission using three local clustering techniques. Appl Geogr 60: 29–36. [Google Scholar]
- 40.Aamodt G, Samuelsen SO, Skrondal A, 2006. A simulation study of three methods for detecting disease clusters. Int J Health Geogr 5: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Desjardins MR, Whiteman A, Casas I, Delmelle E, 2018. Space-time clusters and co-occurrence of chikungunya and dengue fever in Colombia from 2015 to 2016. Acta Trop 185: 77–85. [DOI] [PubMed] [Google Scholar]
- 42.Coleman M, Coleman M, Mabuza AM, Kok G, Coetzee M, Durrheim DN, 2009. Using the SaTScan method to detect local malaria clusters for guiding malaria control programmes. Malar J 8: 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Walker AT, LaRocque RC, Sotir MJ, 2019. Travel epidemiology. CDC Yellow Book. New York, NY: Oxford University Press, 720. [Google Scholar]
- 44.Torres JR, Orduna TA, Piña-Pozas M, Vázquez-Vega D, Sarti E, 2017. Epidemiological characteristics of dengue disease in Latin America and in the Caribbean: a systematic review of the literature. J Trop Med 2017: 8045435. [DOI] [PMC free article] [PubMed] [Google Scholar]