Abstract.
The eradication of yaws caused by Treponema pallidum subsp. pertenue is constrained by the lack of rapid, accurate diagnosis. We sought to develop a molecular point-of-care test for the diagnosis of yaws. A loop-mediated isothermal amplification (LAMP) assay with primers targeting the conserved gene, tp0967, with visual detection by lateral flow test strip was developed and optimized. The limit of detection was evaluated while 63 samples from clinical cases of yaws and five samples with polymerase chain reaction (PCR)–confirmed syphilis were used to determine the sensitivity and specificity of the assay compared with the current molecular testing protocol. The developed LAMP assay was found to be optimal when run at 65°C for 30 minutes. The limit of detection from extracted DNA was 2.7 × 104 DNA copies/mL. The sensitivity of the LAMP assay using unextracted and DNA extracted samples were 0.67 and 1.00, respectively. None of the syphilis samples tested positive in any of the assays. We show the development of a fast and sensitive LAMP assay for yaws detected by lateral flow test strip. Using extracted DNA, the assay sensitivity is at par with real-time PCR-based detection. The assay can be adapted to minimal sample processing required for infield detection without DNA extraction.
INTRODUCTION
Yaws is a debilitating childhood infectious disease caused by the bacterium Treponema pallidum subsp. pertenue (TPE).1 It is currently known to be endemic in 13 countries, but accurate reporting data are lacking.2,3 Worldwide, the highest number of cases are reported from Ghana, Papua New Guinea, and the Solomon Island.4 Until 2011 and 2013, when Knauf et al.5 reported TPE infection in primates in several West and Central African countries, humans were thought to be the main reservoir of TPE, with children being most affected. Yaws presents clinically as chronic lesions of the skin, bone, and cartilage,6 and without treatment, it can cause gross deformities and lifetime disabilities.7
In 2012, a single dose of oral azithromycin was shown to be as effective as the standard injection of benzathine penicillin.8 This prompted the WHO to launch a road map, called the Morges strategy, to eradicate yaws by 2020.9 Among other things, the Morges strategy stipulates a total community treatment strategy, where treatment of an entire endemic community irrespective of the number of active clinical cases are found.10 Healthcare workers in yaws-endemic countries, however, are faced with two major challenges when it comes to the diagnosis of yaws. First, the genomic structure of TPE which causes yaws has nearly 99.8% similarity with T. pallidum subsp. pallidum (TPA) which causes syphilis.11 As such, all of the current serological diagnostic methods that test positive for yaws also test positive for syphilis. Second, clinical diagnosis which is the main method of diagnosis of several yaws-endemic communities is unreliable.12 Clinical diagnosis relies on identifying lesions that are consistent with yaws. However, studies have shown that lesions consistent with yaws are often caused by Haemophilus ducreyi and not TPE.13 To compound this, TPE and other pathogenic treponemes are unable to be cultured in vitro, thus hindering the ability to properly study the pathogen.14,15 Molecular methods, such as polymerase chain reaction (PCR), although reliable, are not readily available in the field.12 To help with the yaws eradication efforts, there remains a critical need for an accurate, cost-effective, and easy-to-use diagnostic assay for use especially in endemic countries.16
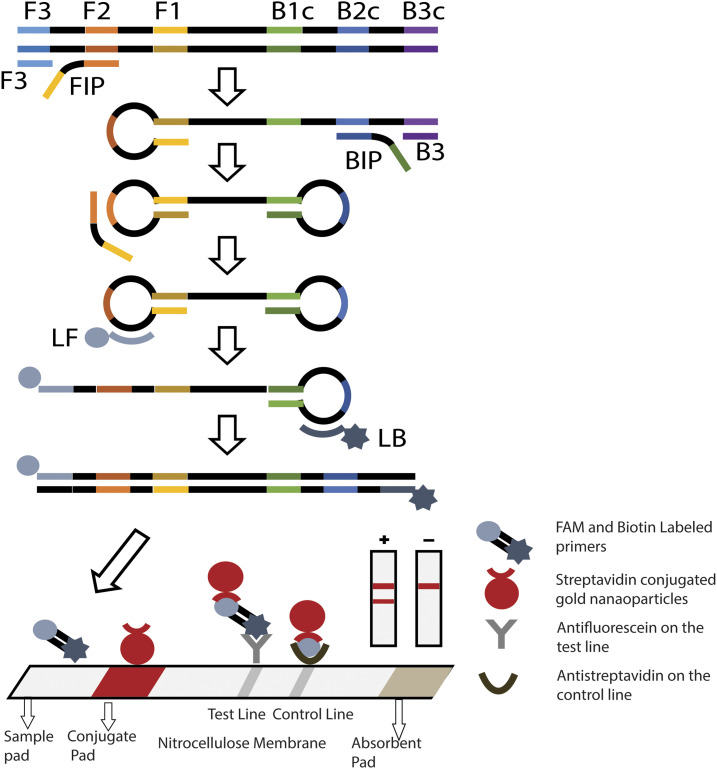
Unlike PCR, loop-mediated isothermal amplification (LAMP) (Figure 1) does not require thermal cycling, hence overcoming the logistical challenge of using complex equipment.17 A simple incubator or water bath which is readily available in most endemic communities could be used to perform LAMP. Furthermore, LAMP produces more DNA in a shorter time than PCR, and, most importantly, the enzymes and reagents used in LAMP are more robust and can be stored and transported in less ideal conditions than PCR.18 An important detection method of LAMP is via nucleic acid lateral flow immunoassay test strips (lateral flow strips), where biotinylated DNA is detected using streptavidin-bound gold nanoparticles. As the nucleic acid amplification product from primers labeled with carboxyfluorescein (FAM) and biotin flow downstream, the DNA–nanoparticle complex, if formed, are then captured by immobilized anti-FAM antibodies at the test line. The remaining nanoparticles are captured by anti-streptavidin antibodies at the control line. Aggregation of the nanoparticles results in a visible reaction at the test and control lines, indicating either a positive (2 lines) or negative (1 line) result (Figure 1).
Figure 1.
Schematic representation of the principle of loop-mediated isothermal amplification using six primers incorporating biotin and FAM labels into the loop primers. Amplification is followed by detection of the products via lateral flow strip. Amplified samples result in two visible lines (positive detection), whereas those without amplification result in only one line on the lateral flow strip (negative detection). This figure appears in color at www.ajtmh.org.
Yaws is known to affect people living in very rural communities with poor sanitation and a lack of access to healthcare services.19 In Ghana, in particular, children with yaws are often found living in communities and farmlands that are hard to reach.20 Case searches for yaws are often carried out at central locations where a high number of children with yaws-like lesions can be found. These are mostly Community-based Health Planning and Services (CHPS) compounds in a particular locality. Samples of children who are on farms, hard to reach areas, and cottages nearby are then picked up by a community health worker on a motorbike. We therefore sought to design a simple assay that could be run with dried reagents in a water bath at a CHPS compound and detected on a lateral flow test strip.
METHODS
Primer design.
Based on the 1,566-bp gene sequence of the conserved hypothetical protein, tp0967, of the Gauthier strain of TPE (GenBank: HM151371.1),11 a primer set was designed using Primer Explorer V5 software (http://primerexplorer.jp/e/v5_manual/index.html). The primer set targeted six distinct regions on the gene via a forward inner primer (FIP), a backward inner primer (BIP), two outer primers (F3 and B3), and two loop primers (loop F and loop B). A basic local alignment search tool (BLAST) search with primer sequences in GenBank was performed before LAMP to ensure their specificity for TPE.
Loop-mediated isothermal amplification assay.
The 25-μL LAMP reaction included an isothermal master mix with 1.6 μmol each of FIP and BIP, 0.2 μmol each of F3 and B3 primers, and 1.6 μmol each of the loop primers, loops B and F. The isothermal master mix contained 10× isothermal buffer (New England Biolabs, Ipswitch, MA), 1.4 mM of 100 mM dNTPs, 400 mM of 5 M Betaine, and 0.2× and 1× of the fluorescent dyes Evagreen and ROX, respectively. The final constituents of the reaction mix, 2 μL of DNA sample, and 8U of Bst polymerase were added to the reaction mix at last. The LAMP assay was conducted at 65°C for 60 minutes in an ABI 7500 real-time PCR system (Applied Biosystems, Thermofisher Scientific, Waltham, MA). The amplicons were run on 2.0% ethidium bromide in 2.0% agarose gel at 100 V for 60 minutes, and the gels were imaged on an Azure c400 imager (Azure Biosystems, Dublin, CA) using a UV wavelength of 302 nm and an exposure time of 15 seconds.
Optimization of the LAMP reaction.
Concentrations of additional betaine and MgSO4, the optimal temperature for the reaction, and time were optimized for the LAMP assay. In the study, 25 μL of amplified DNA fragments were analyzed using 2.0% agarose gel electrophoresis. For determining the best reaction temperature, the reaction mixture was incubated for 60 min at 61°C, 63°C, 65°C, 67°C, 69°C, and 71°C, respectively, and then terminated at a range from 95 to 60 °C (0.05 °C per sec). The concentrations of betaine (800 mM, 400 mM, and 200 mM) and those of MgSO4 (6 mM, 2 mM, and 0 mM) were tested. Moreover, to investigate the minimum reaction time required in a LAMP run, the reaction was performed at 65°C and assessed with gel electrophoresis and lateral flow at 20 minutes, 25 minutes, 30 minutes, 35 minutes, 40 minutes, 45 minutes, 50 minutes, and 60 minutes, respectively.
Limit of detection determination.
Ten-fold dilutions of the stock DNA (concentrations ranging from 2.77 × 107 to 2.77 × 100 copies/mL) were prepared using molecular grade water and tested using labeled primers (LF-Biotin and LB-FAM) and the same conditions as described earlier for LAMP. In all, 10 μL of each of the reaction product was pipetted onto a lateral flow test strip (catalog no. D003-03, USTAR Biotechnologies, Hangzhou, China), and 40 μL of each of commercial lateral flow test wash buffer was added. All the test strips were scanned with an Epson V850 Pro scanner (Long Beach, CA), and, then, the intensities of the test lines were analyzed using a custom MATLAB script that quantifies the average of the gray-scale pixel intensities (I) of the lines and subtracts the background noise. This “background-subtracted test line intensity” was then calculated as follows21:
where Pw represents the lowest possible pixel intensity (pure white).
The limit of detection of TPE was determined using test strips from three replicate reactions. The test line intensities were compared with the averaged lateral flow strip experimental negative control test line intensities via GraphPad Prism version 8.0. (GraphPad Software, La Jolla, CA) using a Dunnett’s test with an alpha of 0.05.
Drying LAMP reagents.
The reagents, as well as the conditions for drying, were adapted from Phillips et al.22; briefly, the yaws LAMP reagents were streaked on polyethylene terephthalate film (Apollo, Lake Zurich, IL) and were dried and stored at room temperature. The primer mix which contained 0.2 µM of F3 and B3; 1.6 µm of FIP, BIP, LB, and LF primers; and 2 M sucrose, 50% glycerol, and 0.1% Triton X-100 were deposited by hand. This was then dried for 60 minutes in a biosafety cabinet at room temperature under airflow. The enzyme mix contained 2 M sucrose, 1.4 mM dNTPs, and 8 U Bst 3.0 polymerase. This was pipetted directly on top of the dried primer mix and dried for another 60 minutes. A 25-µL reaction corresponded to a 0.5-cm × 0.5-cm piece of the polyethylene terephthalate film. This was cut and deposited in a clean microcentrifuge tube and stored with silica gel desiccant (Uline, Pleasant Prairie, WI) at room temperature.
Determining stability of the dried reagents.
The dried reagents which had been stored at room temperature were tested on days 0, 1, 7, and 14 to determine the stability of the dried reagents for up to 14 days. The dried reagents were rehydrated with a rehydrating mix comprising isothermal amplification buffer, betaine, and MilliQ water. 20 µL of the sample was added to the reconstituted reagents and run on a simple water bath at 65°C for 30 minutes. 20 µL of the sample and water were added in triplicates to the reconstituted reagents and run on a simple water bath at 65°C for 30 minutes. The amplification products were then run on a lateral flow test strip.
Clinical samples.
Sixty-three suspected cases of TPE from patients with lesions consistent with yaws were obtained from the Noguchi Memorial Institute for Medical Research and from the Asamankese District Hospital Laboratory, both in Ghana. At the time of collection, these cases were presumptively diagnosed in the field by clinically evaluating lesions that were consistent with yaws and the Dual Path Pathway rapid kit (Chembio Diagnostic Systems Inc., Medford, NY), and, then, the communities were treated accordingly with azithromycin. Swab samples from suspected yaws lesions were collected and stored in AssayAssure transport media (Sierra Molecular Corporation, Incline Village, NV) at −20°C. Extracted DNA from five separate PCR-confirmed serologically positive samples for TPA were obtained from the Komfo Anokye Teaching Hospital in Ghana and stored at −20°C.
Sample preparation.
Each of the 63 suspected TPE samples were divided into two equal parts. DNA was extracted from one part, whereas the other part was analyzed without DNA extraction.
DNA extraction.
DNA was extracted using a QIAGEN brand DNA extraction kit. Briefly, cell lysis was achieved by adding 20 μL proteinase K (20 mg/mL) to 200 μL AssayAssure containing sample, incubated at 55°C in a thermomixer for 4 hours. After lysis, the DNA was processed according to the manufacturer’s instructions and eluted into 200 μL elution buffer.
Real-time PCR.
Real-time PCR, developed by the United States CDC, was run as described by Chi et al.,23 using an ABI 7500 Fast Real-Time PCR System (Applied Biosystems, Inc., Foster City, CA). Briefly, a 10-µL sample of DNA was analyzed in a 25-µL reaction volume containing 12.5 µL of PerfeCTa Multiplex qPCR SuperMix (Quanta Biosciences, Gaithersburg, MD). The amplification parameters were as follows: 95°C for 4 minutes, followed by 50 cycles of 95°C for 20 seconds, and 65°C for 1 minute with FAM, Cy5, and Calred dyes as detectors as per the manufacturer’s procedure. This method has previously been shown to be sensitive and specific to TPE without cross-reactivity with TPA.
Sensitivity of LAMP assay.
To determine the sensitivity of the lamp assay, DNA templates of the 63 TPE-suspected samples from Ghana were tested by LAMP with the CDC real-time PCR assay as the reference standard. This CDC-PCR testing method using Cy5-tagged DNA is the diagnostic reference standard for TPE used throughout Ghana.3,24–26
Specificity of LAMP assay.
A primer blast was initially performed to determine the specificity of the primers selected for the assay. Five previously PCR-confirmed serologically positive samples of TPA were run using both LAMP and the CDC real-time PCR assay to rule out cross-reactivity.
Statistical analysis.
All reactions in the study were run in triplicates with triplicate lateral flow tests per sample except the clinical sample which had one reaction per sample and one lateral flow test per sample. All data were first entered into Microsoft Excel (2016) and then transferred onto GraphPad Prism version 8.00 (GraphPad Software). To determine the limit of quantification, one-way ANOVA followed by Dunnett’s multiple comparison test was used to compare the mean of each of the samples with the mean of no template control. The positive predictive value (PPV) and negative predictive value (NPV) of the LAMP assay, the sensitivity, and specificity were calculated as described by Parikh et al.27
Ethical approval for the study.
Written informed consent was obtained from the parent or guardian of each participant. The study protocol was approved by the Ethical Review Committee of the Ghana Health Service (Accra, Ghana; November 13, 2014) and the Institutional Review Board of Purdue University (West Lafayette, IN; 1804020526).
RESULTS
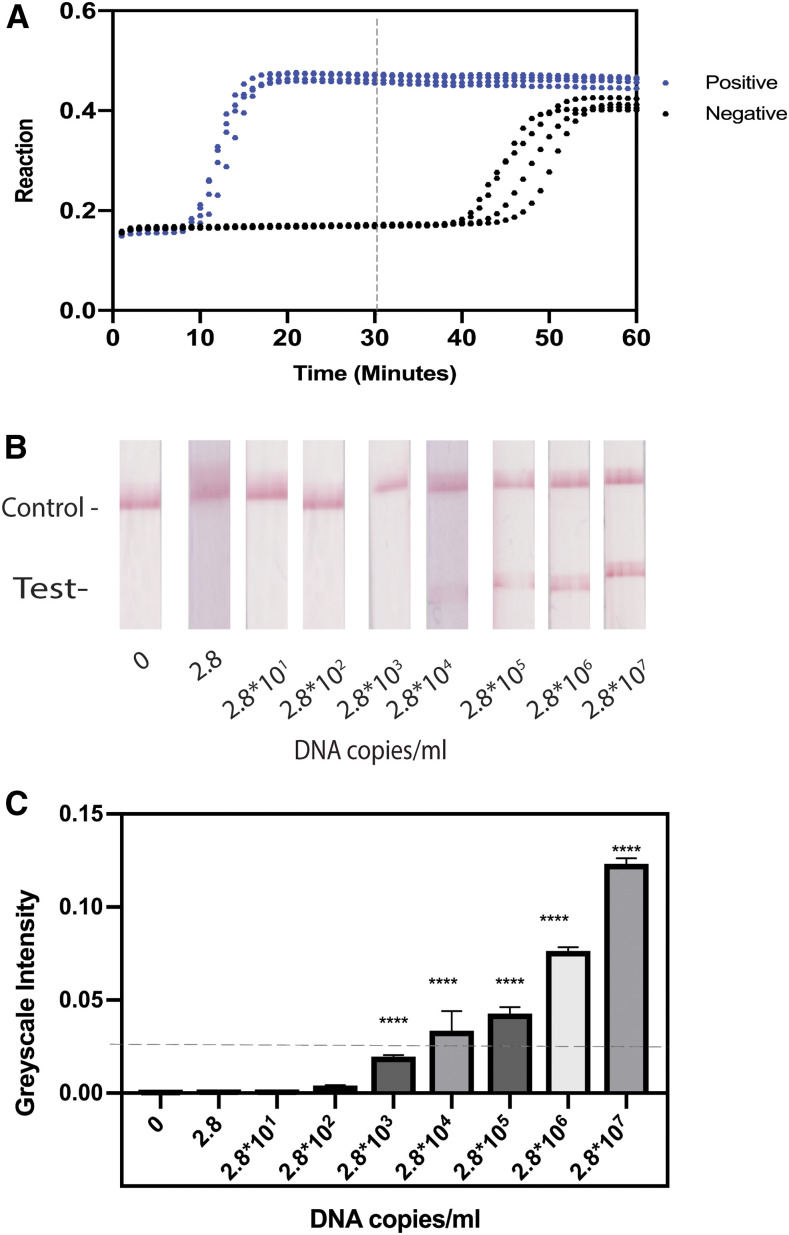
The primer blast showed that the primers were specific to all the strains of TPE. The assay was found to be optimal at 65°C for 30 minutes. The concentration of betaine and MgSO4 was found to be optimal at 400 mM and 0 mM, respectively (Supplemental data). Under the optimized conditions, 2.8 × 107 copies/mL of starting DNA amplified within 15 minutes, whereas negative controls required more than 40 minutes to amplify (Figure 2A).
Figure 2.
Amplification profile and the limit of detection of the loop-mediated isothermal amplification assay. (A) LAMP profile for both positive samples and negative samples. Whereas the positive samples for the primer set amplified with a Ct of 10 minutes, the negative samples had a Ct of 40 minutes or greater. Therefore, further reactions were terminated at 30 minutes. (B) Results of the limit of detection performed on lateral flow test strips. The reaction of 2.8 × 104 DNA copies/mL was visible to the naked eye. C) Background-subtracted line intensities (A.U) of each of the lateral flow test strips in B at each concentration of DNA. **** indicates P < −0.005 compared with 0 copies/mL. This figure appears in color at www.ajtmh.org.
Using the optimized assay conditions, quantification of the background-subtracted LFA test lines (Figure 2B) resulted in a statistically significant difference of 2.8 × 103 copies/mL of starting DNA input into the LAMP assay compared with the controls without DNA (Figure 2C). Although statistically significant at 2.8 × 103 copies/mL, a study by Phillips et al.28 indicates that only the 2.8 × 104 copies/mL would be visible to the human eye; hence, the limit of detection was determined to be 2.8 × 104 copies/mL.
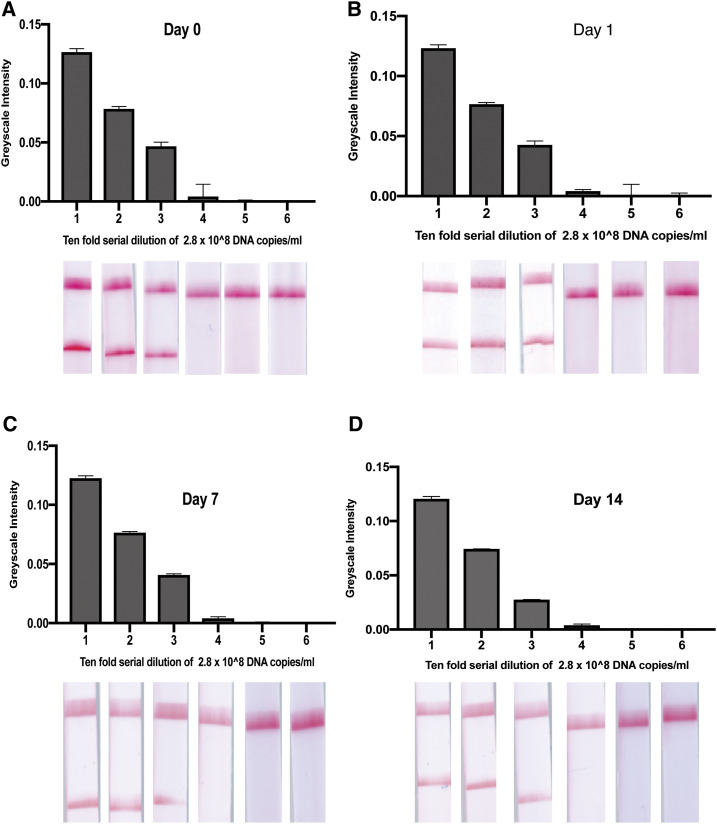
In Figure 3, we show the stability of the dried reagents up to 14 days. The dried reagents were tested fresh (day 0) and on days 1, 7, and 14. The results were consistent on all the days tested, with no reduction in the limit of detection.
Figure 3.
Stability of the dried reagents up to 14 days. The dried reagents were tested fresh on (day 0), day 1, day 7, and day 14. The results were consistent on all the days tested with no reduction in the limit of detection. This figure appears in color at www.ajtmh.org.
Of the 63 samples evaluated with the CDC PCR reference standard, LAMP using extracted DNA samples and LAMP using unextracted samples, 12, 14, and eight samples were found to be positive, respectively. Comparing the CDC PCR reference standard and LAMP using extracted DNA samples (Table 1), the PPV was found to be 0.86, and the NPV was 1.00, with a sensitivity of 1.00 and a specificity of 0.96. Comparing the CDC PCR reference standard with the unextracted DNA samples (Table 2), the PPV was 1.00, and the NPV was 0.927, with a sensitivity of 0.67 and a specificity of 1.00. All of the five PCR-confirmed syphilis samples tested negative using the CDC PCR assay and the LAMP assay specific for yaws (Table 3).
Table 1.
Sensitivity and specificity of the LAMP assay on extracted DNA compared with the reference standard CDC PCR assay
| PCR result | LAMP (extracted) | ||
|---|---|---|---|
| Negative | Positive | Total | |
| Negative | 49 | 2 | 51 |
| Positive | 0 | 12 | 12 |
| Total | 49 | 14 | 63 |
LAMP = loop-mediated isothermal amplification; PCR = polymerase chain reaction.
Table 2.
Sensitivity and specificity of the LAMP assay on unextracted DNA compared with the reference standard CDC PCR assay
| PCR result | LAMP (unextracted) | ||
|---|---|---|---|
| Negative | Positive | Total | |
| Negative | 51 | 0 | 51 |
| Positive | 4 | 8 | 12 |
| Total | 55 | 8 | 63 |
LAMP = loop-mediated isothermal amplification; PCR = polymerase chain reaction.
Table 3.
All of the five PCR-confirmed syphilis samples tested negative using the CDC PCR assay and the LAMP assay specific for yaws
| PCR result | LAMP (extracted) | ||
|---|---|---|---|
| Negative | Positive | Total | |
| Negative | 5 | 0 | 5 |
| Positive | 0 | 0 | 0 |
| Total | 5 | 0 | 5 |
LAMP = loop-mediated isothermal amplification; PCR = polymerase chain reaction.
DISCUSSION
This study shows the feasibility of using a LAMP assay coupled with a lateral flow test strip for yaws eradication purposes in the remote parts of yaws-endemic communities. The result of the study by Mitja et al.8 in 2012, showing that a single dose of azithromycin could treat yaws, was the first sign of significant hope that yaws could be eradicated. Azithromycin requires no cold chain and can be delivered orally. Thus, the drug provides a convenient and fast way of ensuring that all children are treated to eradicate the disease. Treponema pallidum subsp. pallidum (causing syphilis), TPE (causing yaws), and T. pallidum subsp. endemicum (causing bejel) differ from each other by less than 0.2% of the entire genome29; thus, it is difficult to differentiate between them using microscopy and serological methods. As can be seen from the results in Table 1, not all yaws-suspected lesions are positive for TPE. Several studies have shown that H. ducreyi is responsible for several of the yaws-like lesions found in endemic communities. Indeed, another study by Mitja et al.13 in 2014 showed that many of the lesions thought to be yaws were indeed H. ducreyi infection.
In 2015, researchers at the CDC developed a real-time PCR assay which targeted a predicted outer membrane protein gene, tp0858,23 which is currently being used by health ministries in several yaws-endemic countries including Ghana. In our study, we targeted the hypothetical protein gene, tp0967, of the Gautheir strain of TPE which had been shown to have variable genomic regions when compared with TPA.11 By performing a primer BLAST, we confirmed that our designed LAMP primers were specific to all strains of TPE but not TPA. We further tested PCR-confirmed TPA DNA to ensure that no cross-reactivity occurred.
The limit of detection of the yaws LAMP assay was found to be from 2.8 × 103 to 2.8 × 104 copies/mL or about 2–20 copies/reaction involving 2 μL of the sample. This limit of detection was found to be lower than the CDC PCR assay which had an analytical sensitivity of 10–100 copies per reaction involving a 10-µL sample or 1.0 × 105 to 1.0 × 106 copies/mL.23 This result is expected as lamp assays are known to be more sensitive than conventional PCR.18
To be able to detect the amplified products in yaws-endemic district health facilities, we explored the possibility of using dried reagents that could be stored at room temperature. Being able to dry the LAMP reagents and being able to store these reagents provide a solution to a very major logistical challenge when it comes to molecular point-of-care diagnostics in resource-limited countries. Because of cold chain requirements for reagents, most of these technologies are not deployed because most of those who need it do not really have the resources to keep these reagents refrigerated. As can be seen in Figure 3, we had consistent results between day 0 and day 14 (n = 3), indicating that the dried reagents could provide consistent results in endemic communities up to 7 days on the field. Because most endemic countries have reference laboratories in the big cities, the reagents can be dried and packaged and then deployed on the field when there are yaws case search activities.
Twelve of 63 deidentified samples tested with the CDC PCR assay were positive for TPE DNA. This is consistent with molecular data from studies conducted in Ghana showing prevalence between 15% and 27.6%.24–26 These results were also high because these samples were taken from patients with lesions, which had a high suspicion of yaws. When these same DNA-extracted samples were run using the Lamp assay, 14 of the 63 were found to be positive. This is possibly because the limit of detection of the CDC PCR assay is higher than that of the LAMP assay. It is possible that two discordant samples may have been below the limit of detection of the CDC assay. Furthermore, a study by Marks et al.30 in 2017 showed that 22% of samples previously shown to be negative by the CDC PCR assay turned out to be positive when whole genome sequencing was used because of mutations in the PCR primer-binding sites targeted by the CDC assay. The CDC assay does not detect all the isolates of TPE; therefore, other molecular-based tests, such as the LAMP assay described herein, may be necessary in the fight against yaws.
To increase the convenience of performing the assay in low-resource health facilities in yaws-endemic districts, we explored the possibility of using the LAMP assay on samples that had been taken directly from swab samples in transport medium. Although other studies31–36 integrated the extraction step into their devices, we explored the possibility of bypassing the extraction step altogether, thereby speeding the detection process and removing the need for a bulky device and eliminating the multiple hands-on steps associated with an integrated extraction. In most yaws-endemic districts, DNA extraction kits and reagents are largely unavailable. Bypassing the DNA extraction step is essential in making a LAMP-based rapid test that is applicable to yaws eradication programs in endemic communities. When these unextracted samples were tested with LAMP, eight of 63 samples were found to be positive. Reasons why the other previously amplified samples did not amplify in this experiment remain unknown. However, it is suspected that because the DNA extraction process gives rise to highly concentrated elution of DNA and high DNA yields, it was easier to detect DNA in the extracted samples, whereas it was more difficult to detect DNA in unextracted samples with low DNA concentrations.
To be able to detect the amplified products in yaws-endemic district health facilities, we tested these amplified DNA products on lateral flow test strips. Although we ran the samples on agarose gels as controls, this may not be applicable to yaws-endemic communities and greatly increases both the infrastructure and the assay time to result. Running these samples on a lateral flow strips, therefore, provides a convenient and effective way of detecting TPE in the resource-limited settings of yaws-endemic communities. The downside to a field-based test, however, is the risk of contamination, which can be mitigated by properly training the users.
Although using LAMP on extracted DNA samples may have a sensitivity of 100%, it can only be performed in settings with equipment to adequately extract DNA, therefore introducing a lag time between sample collection and treatment. Many yaws eradication programs suffer from loss to follow-up from sample collection to treatment.5,26,37 Many people living in yaws-endemic communities move around for agricultural purposes and may have left communities that were previously screened. Although the unextracted samples have a sensitivity of only 67%, this method offers the convenience of same day sample-to-answer testing and treatment. Because of the minimal sample handling requirements and rapid time to result (30 minutes for the lamp assay and 15 minutes for the lateral flow test strips), it is recommended that this LAMP assay be used on unextracted DNA samples on the field in endemic communities. However, negative samples with a high index of suspicion can then be sent to reference laboratories for extraction and retesting.
CONCLUSION
Herein, we have shown the development of a fast and sensitive LAMP assay for T. pallidum pertenue as detected by lateral flow test strips. This new assay provides a sensitive and convenient same day sample-to-answer testing and provides an important tool for the global yaws eradication effort.
Supplemental information, table, and figures
Acknowledgments:
We would like to thank Frank Biney of the Asamankese District Hospital for his help in acquiring the yaws-suspected clinical samples and Eliot Eli Dogbe for providing the syphilis samples.
Note: Supplemental information, table, and figures appear at www.ajtmh.org.
REFERENCES
- 1.Marks M, Mitjà O, Solomon AW, Asiedu KB, Mabey DC, 2015. Yaws. Br Med Bull 113: 91–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marks M, et al. 2015. Challenges and key research questions for yaws eradication. Lancet Infect Dis 15: 1220–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ghinai R, et al. 2015. A cross-sectional study of ‘yaws’ in districts of Ghana which have previously undertaken azithromycin mass drug administration for trachoma control. PLoS Negl Trop Dis 9: e0003496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mitjà O, et al. 2015. Global epidemiology of yaws: a systematic review. Lancet Glob Health 3: e324–e331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marks M, Sokana O, Nachamkin E, Puiahi E, Kilua G, Pillay A, Bottomley C, Solomon AW, Mabey DC, 2016. Prevalence of active and latent yaws in the Solomon Islands 18 months after azithromycin mass drug administration for trachoma. PLoS Negl Trop Dis 10: e0004927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mitjà O, Asiedu K, Mabey D, 2013. Yaws. Lancet 381: 763–773. [DOI] [PubMed] [Google Scholar]
- 7.Mitjà O, Hays R, Ipai A, Wau B, Bassat Q, 2011. Osteoperiostitis in early yaws: case series and literature review. Clin Infect Dis 52: 771–774. [DOI] [PubMed] [Google Scholar]
- 8.Mitjà O, Hays R, Ipai A, Penias M, Paru R, Fagaho D, De Lazzari E, Bassat Q, 2012. Single-dose azithromycin versus benzathine benzylpenicillin for treatment of yaws in children in Papua New Guinea: an open-label, non-inferiority, randomised trial. Lancet 379: 342–347. [DOI] [PubMed] [Google Scholar]
- 9.WHO , 2012. Eradication of yaws–the Morges strategy. Relev Epidemiol Hebd 87: 189–194. [PubMed] [Google Scholar]
- 10.Marks M, 2016. Yaws: towards the WHO eradication target. Trans R Soc Trop Med Hyg 110: 319–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mikalová L, Strouhal M, Čejková D, Zobaníková M, Pospíšilová P, Norris SJ, Sodergren E, Weinstock GM, Šmajs D, 2010. Genome analysis of Treponema pallidum subsp. pallidum and subsp. pertenue strains: most of the genetic differences are localized in six regions. PLoS One 5: e15713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marks M, Goncalves A, Vahi V, Sokana O, Puiahi E, Zhang Z, Dalipanda T, Bottomley C, Mabey D, Solomon AW, 2014. Evaluation of a rapid diagnostic test for yaws infection in a community surveillance setting. PLoS Negl Trop Dis 8: e3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mitjà O, et al. 2014. Haemophilus ducreyi as a cause of skin ulcers in children from a yaws-endemic area of Papua New Guinea: a prospective cohort study. Lancet Glob Health 2: e235–e241. [DOI] [PubMed] [Google Scholar]
- 14.Lewis DA, Mitjà O, 2016. Haemophilus ducreyi: from sexually transmitted infection to skin ulcer pathogen. Curr Opin Infect Dis 29: 52–57. [DOI] [PubMed] [Google Scholar]
- 15.Gangaiah D, Spinola SM, 2016. Haemophilus ducreyi cutaneous ulcer strains diverged from both class I and class II genital ulcer strains: implications for epidemiological studies. Small PLC. PLoS Negl Trop Dis 10: e0005259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ayove T, et al. 2014. Sensitivity and specificity of a rapid point-of-care test for active yaws: a comparative study. Lancet Glob Health 2: e415–e421. [DOI] [PubMed] [Google Scholar]
- 17.Li Y, Fan P, Zhou S, Zhang L, 2017. Loop-mediated isothermal amplification (LAMP): a novel rapid detection platform for pathogens. Microb Pathog 107: 54–61. [DOI] [PubMed] [Google Scholar]
- 18.Notomi T, Mori Y, Tomita N, Kanda H, 2015. Loop-mediated isothermal amplification (LAMP): principle, features, and future prospects. J Microbiol 53: 1–5. [DOI] [PubMed] [Google Scholar]
- 19.Mabey D, 2015. Mapping the geographical distribution of yaws. Lancet Glob Health 3: e300–e301. [DOI] [PubMed] [Google Scholar]
- 20.Marks M, et al. 2017. Knowledge, attitudes and practices towards yaws and yaws-like skin disease in Ghana. PLoS Negl Trop Dis 11: e0005820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Holstein CA, 2015. Development of a Novel Paper-Based Flu Test for Improved Diagnosis at the Point of Care. Seattle, WA: University of Washington. [Google Scholar]
- 22.Phillips EA, et al. 2019. Microfluidic rapid and autonomous analytical device (microRAAD) to detect HIV from whole blood samples. Lab Chip 19: 3375–3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chi K-H, et al. 2015. Molecular differentiation of Treponema pallidum subspecies in skin ulceration clinically suspected as yaws in Vanuatu using real-time multiplex PCR and serological methods. Am J Trop Med Hyg 92: 134–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Abdulai AA, et al. 2018. Community-based mass treatment with azithromycin for the elimination of yaws in Ghana—results of a pilot study. PLoS Negl Trop Dis 12: e0006303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kwakye-Maclean C, Agana N, Gyapong J, Nortey P, Adu-Sarkodie Y, Aryee E, Asiedu K, Ballard R, Binka F, 2017. A single dose oral azithromycin versus intramuscular benzathine penicillin for the treatment of yaws-A randomized non inferiority trial in Ghana. PLoS Negl Trop Dis 11: e0005154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marks M, et al. 2018. Comparative efficacy of low-dose versus standard-dose azithromycin for patients with yaws: a randomised non-inferiority trial in Ghana and Papua New Guinea. Lancet Glob Health 6: e401–e410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parikh R, Mathai A, Parikh S, Chandra Sekhar G, Thomas R, 2008. Understanding and using sensitivity, specificity and predictive values. Indian J Ophthalmol 56: 45–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Phillips EA, Moehling TJ, Bhadra S, Ellington AD, Linnes JC, 2018. Strand displacement probes combined with isothermal nucleic acid amplification for instrument-free detection from complex samples. Anal Chem 90: 6580–6586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Šmajs D, Norris SJ, Weinstock GM, 2012. Genetic diversity in Treponema pallidum: implications for pathogenesis, evolution and molecular diagnostics of syphilis and yaws. Infect Genet Evol 12: 191–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marks M, et al. 2018. Diagnostics for yaws eradication: insights from direct next-generation sequencing of cutaneous strains of Treponema pallidum. Clin Infect Dis 66: 818–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Heiniger EK, Buser JR, Mireles L, Zhang X, Ladd PD, Lutz BR, Yager P, 2016. Comparison of point-of-care-compatible lysis methods for bacteria and viruses. J Microbiol Methods 128: 80–87. [DOI] [PubMed] [Google Scholar]
- 32.Lafleur LK, et al. 2016. A rapid, instrument-free, sample-to-result nucleic acid amplification test. Lab Chip 16: 3777–3787. [DOI] [PubMed] [Google Scholar]
- 33.Mahalanabis M, Do J, ALMuayad H, Zhang JY, Klapperich CM, 2010. An integrated disposable device for DNA extraction and helicase dependent amplification. Biomed Microdevices 12: 353–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rodriguez NM, Wong WS, Liu L, Dewar R, Klapperich CM, 2016. A fully integrated paperfluidic molecular diagnostic chip for the extraction, amplification, and detection of nucleic acids from clinical samples. Lab Chip 16: 753–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Horst AL, Rosenbohm JM, Kolluri N, Hardick J, Gaydos CA, Cabodi M, Klapperich CM, Linnes JC, 2018. A paperfluidic platform to detect Neisseria gonorrhoeae in clinical samples. Biomed Microdevices 20: 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Connelly JT, Rolland JP, Whitesides GM, 2015. “Paper machine” for molecular diagnostics. Anal Chem 87: 7595–7601. [DOI] [PubMed] [Google Scholar]
- 37.Mitjà O, et al. 2018. Re-emergence of yaws after single mass azithromycin treatment followed by targeted treatment: a longitudinal study. Lancet 391: 1599–1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.