Abstract.
Orthohantaviruses are negative-sense, single-stranded RNA viruses harbored by multiple small mammals. Dobrava-Belgrade virus (DOBV) and Puumala virus (PUUV) cause hemorrhagic fever with renal syndrome (HFRS) in Europe. In Poland, serological surveys have demonstrated antibodies against DOBV and PUUV in patients with HFRS. Molecular evidence of DOBV and PUUV has been found in Apodemus flavicollis and Myodes glareolus, respectively, in southeastern Poland, and Seewis virus (SWSV) has been reported in Sorex araneus in central Poland. However, data on the geographic distribution and phylogeny of orthohantaviruses are unavailable for other regions in Poland. To ascertain the prevalence and genetic diversity of orthohantaviruses in western and northern Poland, lung tissues from 106 small mammals were analyzed for the presence of orthohantavirus RNA. DOBV and SWSV were detected in two of 42 (4.8%) Apodemus agrarius and in three of 10 (30%) S. araneus, respectively. Phylogenetic analyses of partial L- and S-segment sequences of DOBV indicated a shared genetic lineage with the Kurkino genotype from Slovakia, Russia, and Hungary, whereas the partial M segment of DOBV clustered with the Kurkino genotype from Germany. Phylogenetic relationships of the SWSV L and S segments showed a geographic lineage with SWSV strains from central Poland, Czech Republic, and Germany. In conclusion, the study provides insights into the molecular prevalence, phylogenetic diversity, and evolutionary relationship of DOBV in A. agrarius and SWSV in S. araneus. This report increases awareness among physicians for HFRS outbreaks in western Poland.
INTRODUCTION
Orthohantaviruses (family Hantaviridae, order Bunyavirales) are enveloped, negative-sense, and single-stranded RNA viruses, which pose a critical public health threat owing to endemic outbreaks and lack of effective therapeutics.1–3 Small mammals, such as rodents (order Rodentia) and shrews and moles (order Eulipotyphla), serve as reservoir hosts.4–6 In humans, some rodent-borne orthohantaviruses cause hemorrhagic fever with renal syndrome (HFRS) and hantavirus cardiopulmonary syndrome (HCPS).7 Transmission to humans occurs by inhalation of aerosolized excreta or rarely by a bite from an infected rodent.
Approximately 10,000 clinical cases of orthohantavirus diseases are reported annually in Europe, with case fatality rates (CFRs) ranging from < 1% to more than 10%.8 Dobrava-Belgrade virus (DOBV) causes HFRS of varying severity, whereas Puumala virus (PUUV) causes a milder form of HFRS, known as nephropathia epidemica.9 Dobrava and Belgrade viruses individually isolated from the lung tissue of Apodemus flavicollis in Slovenia and from the blood and urine of patients with HFRS in Serbia were found to be genetically identical.10–12 There are four distinct genotypes of DOBV: the Dobrava genotype is harbored by the yellow-necked mouse (A. flavicollis) in Albania, Croatia, Czech Republic, Germany, Greece, Hungary, Poland, Slovakia, Slovenia, and Turkey; the Kurkino and Saaremaa genotypes are found in the striped field mouse (Apodemus agrarius) in Croatia, Denmark, Estonia, Germany, Hungary, Russia, Serbia, Slovakia, and Slovenia; and the Sochi genotype is carried by the Black Sea field mouse (A. ponticus) in Russia.13 The Dobrava and Sochi genotypes cause moderate to severe HFRS with CFRs of 10–12% and up to 14.5%, respectively, whereas the Kurkino genotype is responsible for mild HFRS with CFRs of 0.3–0.9%.14,15
A serologically confirmed clinical case of HFRS was first reported in Poland in 2005,16 and the first HFRS outbreak of 13 clinical cases occurred in 2007,17 with 10 caused by DOBV and three by PUUV. Antibodies against DOBV and PUUV have been found among forestry workers in distinct areas of Poland.18,19 PUUV strains were detected in the bank vole (Myodes glareolus) in northeastern and southern Poland.20,21 Recent surveillance of small mammals revealed co-circulation of DOBV and PUUV in natural reservoirs in Poland.22,23 Molecular and serological evidence of DOBV infection were observed in A. flavicollis and M. glareolus, whereas PUUV was detected in A. agrarius and M. glareolus. However, the presence of DOBV in A. agrarius in Poland remains unknown.
A shrew-borne hantavirus, Thottapalayam virus, was first discovered in the Asian house shrew (Suncus murinus) in India.24,25 Genetically distinct shrew-borne orthohantaviruses have since been reported in Eurasia, including Asikkala virus in the Eurasian pygmy shrew (Sorex minutus),26 Boginia virus in the Eurasian water shrew (Neomys fodiens),27 Cao Bằng virus in the Chinese mole shrew (Anourosorex squamipes),28 Jeju virus in the Asian lesser white-toothed shrew (Crocidura shantungensis),29 and Kenkeme virus in the flat-skulled shrew (Sorex roboratus).30 Seewis virus (SWSV) was first identified in the Eurasian common shrew (Sorex araneus) in Switzerland31 and is now known to be widespread across the distribution of S. araneus in Austria,32 Czech Republic,32 Finland,33–35 Germany,32 Hungary,33 Poland,27,36 Russia,37 Slovakia,32 and Slovenia.38,39 In addition, SWSV has been detected in S. minutus,26 Neomys anomalus,27 S. daphaenodon,37 and S. tundrensis.37 Polish shrew-borne hantaviruses showed phylogenetic diversity and co-circulation within S. araneus and N. fodiens.27,36
In this report, a small-scale molecular epidemiological investigation demonstrated the prevalence and phylogenetic relationships of DOBV Kurkino genotype in A. agrarius and SWSV in S. araneus collected in western Poland. This study emphasizes the need for continued surveillance of small mammals to better understand the geographic distribution and genetic diversity of orthohantaviruses for assessing the risk of disease.
MATERIALS AND METHODS
Rodent trapping and sampling.
Small mammals were live-trapped using Sherman and wooden traps between 2009–2012 and 2014–2015, in Lower Silesian (Wrocław-Osobowice, 51°9′N, 16°59′E; Milicz Ponds Nature Reserve, 51°32′N, 17°19′E; and Ślęża Landscape Park, 50°51′N, 16°46′E) and Pomeranian (Gdańsk-Sobieszewo Island, 54°21′N, 18°47′E) in Poland (Figure 1). Approximately 100 traps were set daily at 3-m intervals along grid lines at each trap site, for 2–3 days. Captured animals were marked, weighed, and tentatively speciated by morphological features. They were then anesthetized with isoflurane and euthanized by cervical dislocation. Lung tissues were dissected and placed in an RNAlater (Qiagen, Valencia, CA), and then shipped to Korea University, Republic of Korea. All animal procedures were approved by the Local Ethics Committee (Nos. 48/2008 and 48/2012).
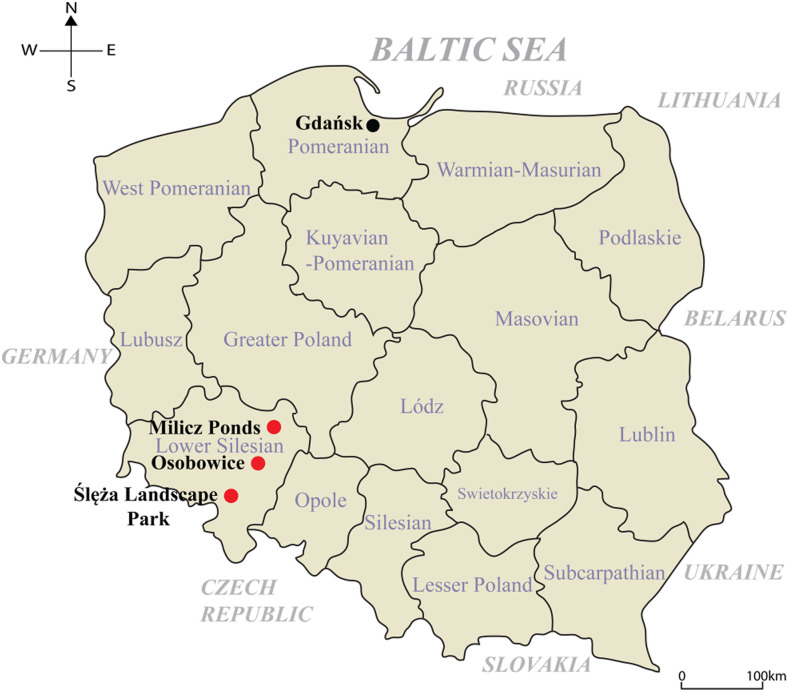
Figure 1.
Geographic location of small mammal trapping sites in Poland. Trapping sites for small mammals are indicated as circles. Red circles represent the location where the genomic sequence of Dobrava-Belgrade virus and Seewis virus was obtained. Dobrava-Belgrade virus was identified in the Milicz Ponds and Ślęża Landscape Park. Seewis virus was discovered in Osobowice. (The map was generated using Adobe illustrator CS6.)
RNA extraction and reverse transcription polymerase chain reaction (RT-PCR).
Total RNA was extracted from lung tissues using a Hybrid R kit (Gene All, Seoul, Republic of Korea), according to the manufacturer’s instructions. cDNA was synthesized using a high-capacity RNA-to-cDNA kit (Applied Biosystems, Foster City, CA). First and nested RT-PCR were performed in 25-µL reaction volumes, containing 0.1 mM dNTP mix, 0.625 units TaKaRa Ex Taq polymerase (Takara, Shiga, Japan), 0.4 µM each primer, and 1.5 µL cDNA. The first and nested PCR conditions were as follows: a cycle of 5 minutes at 94°C; six cycles of 30 seconds at 94°C, 30 seconds at 37°C, and 1 minute at 72°C; 32 cycles of 30 seconds at 94°C, 30 seconds at 42°C, and 1 minute at 72°C; and a final extension at 72°C for 5 minutes. The oligonucleotide primers are shown in Supplemental Table 1. The genomic sequences of DOBV and SWSV were deposited in GenBank (accession numbers: DOBV, MH598462–MH598465; SWSV, MK402015–MK402022).
Mitochondrial DNA (mtDNA) analysis.
Total DNA was extracted from lung tissues using TRIzol reagent solution (AMBION Inc., Austin, TX). The taxonomic identities of small mammals were confirmed by mtDNA cytochrome b gene-specific PCR.40
Phylogenetic analysis.
Partial genome sequences were aligned by the ClustalW method with Lasergene program, version 5.0 (DNASTAR, Madison, WI), and phylogenetic trees were generated by the maximum likelihood method, according to the best-fit substitution model by MEGA 7.41 Topologies were assessed by bootstrap analysis of 1,000 iterations.
RESULTS
Epidemiological survey of small mammals.
A total of 106 small mammals (42 A. agrarius, 25 A. flavicollis, one Crocidura suaveolens, 27 M. glareolus, one N. anomalus, and 10 S. araneus) were captured in northern and western Poland in 2009–2012 and 2014–2015. Using L-segment primers specific to the genus Orthohantavirus, DOBV and SWSV RNA were detected in the lung tissue of two (4.8%) A. agrarius and three (30%) S. araneus, respectively (Table 1). DOBV was found in two (8%) of 25 female and none of 17 male A. agrarius, whereas SWSV RNA was detected in three of five male (60%) and in none of five female S. araneus. Orthohantavirus RNA was not detected in A. flavicollis, M. glareolus, C. suaveolens, and N. anomalus.
Table 1.
Prevalence of orthohantavirus infection in small mammals captured in southwestern and northern Poland, 2009–2012 and 2014–2015
| Animal species | Prevalence of orthohantaviral RNA (%) | Total | |||
|---|---|---|---|---|---|
| Pomeranian | Lower Silesian | ||||
| Gdańsk | Osobowice | Milicz Ponds | Ślęża Landscape Park | ||
| Apodemus agrarius | 0/0 | 0/7 (0.0) | 1/29 (3.4) | 1/6 (16.7) | 2/42 (4.8) |
| Apodemus flavicollis | 0/7 (0.0) | 0/8 (0.0) | 0/10 (0.0) | 0/0 | 0/25 (0.0) |
| Crocidura suaveolens | 0/0 | 0/1 (0.0) | 0/0 | 0/0 | 0/1 (0.0) |
| Myodes glareolus | 0/2 (0.0) | 0/9 (0.0) | 0/3 (0.0) | 0/13 (0.0) | 0/27 (0.0) |
| Neomys anomalus | 0/1 (0.0) | 0/0 | 0/0 | 0/0 | 0/1 (0.0) |
| Sorex araneus | 0/2 (0.0) | 3/8 (37.5) | 0/0 | 0/0 | 3/10 (30) |
| Total | 0/12 (0.0) | 3/33 (9.1) | 1/42 (2.4) | 1/19 (5.3) | 5/106 (4.8) |
Phylogenetic and genetic analyses of DOBV.
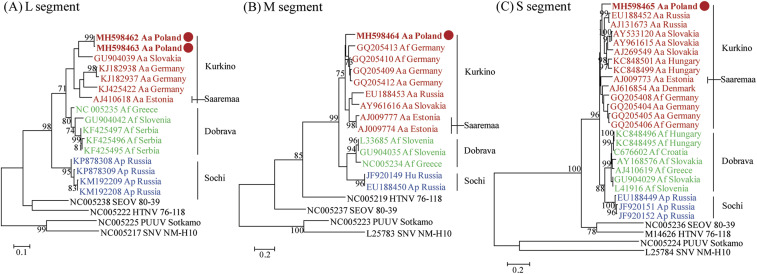
Partial genomic sequences of the DOBV L segment were obtained from Aa15-14 and Aa15-42, whereas partial M and S segments were recovered from Aa15-14 (Table 2). The comparative L, M, and S genome analyses revealed nucleotide sequence similarities of 79.1–87.1%, 81.4–86.2%, and 86.7–88.9%, respectively, with other DOBV strains (Supplemental Table 2). In western Poland, amino acid similarities of L, M, and S segments of DOBV were 93.5–97.2%, 95.7–98.6%, and 97.2–99.1%, respectively. For the partial L segments, the Polish DOBV strains (Aa15-14 and Aa15-42) formed a close genetic lineage with the Kurkino genotype in Slovakia (Figure 2A). The partial M segment of Polish DOBV strain Aa15-14 was phylogenetically clustered with other DOBV Kurkino genotypes in Germany (Figure 2B). The entire S-segment coding sequence of Aa15-14 shared a common ancestor with the Kurkino genotype in Russia, Slovakia, and Hungary (Figure 2C).
Table 2.
Characteristics of orthohantavirus-infected small mammals and the nucleotide sequence of DOBV and SWSV tripartite RNA acquired in the study
| Virus species | Animal species | Sample | Location | Trapping date | Gender | Age | Nucleotide (nt) position of orthohantavirus RNA | ||
|---|---|---|---|---|---|---|---|---|---|
| L segment | M segment | S segment | |||||||
| DOBV | Apodemus agrarius | Aa15-14 | Milicz Ponds | November 2015 | Female | Adult | 2,997–3,312 | 2,046–2,464 | 36–1,325* |
| Aa 15-42 | Sleza Landscape Park | November 2015 | Female | Adult | 2,997–3,312 | –† | –† | ||
| SWSV | Sorex araneus | Sa10-1 | Wrocław Osobowice | October 2010 | Male | Adult | 2,235–3,313 | –† | 14–964 |
| Sa10-5 | Wrocław Osobowice | October 2010 | Male | Adult | 2,235–3,313 | 415–1,219 | 14–1,002 | ||
| Sa10-6 | Wrocław Osobowice | October 2010 | Male | Adult | 2,235–3,313 | 396–534 | 14–1,121 | ||
DOBV = Dobrava-Belgrade virus;PUUV = Puumala virus.
Entire open reading frame sequences were completely obtained.
Not obtained sequence.
Figure 2.
Phylogenetic trees of Dobrava-Belgrade virus (DOBV) L, M, and S segments. Phylogenetic trees were generated by the maximum likelihood method using the T92+G, T92+G, and GTR+G model, based on 326 nt (position 2,997-3,312) of the L segment (A), 419 nt (position 2,046-2,464) of the M segment (B), and 1,290 nt (position 36-1,325) of the entire coding region of the S segment (C), respectively. The numbers at each node are bootstrap probabilities (expressed as percentages), as determined for 1,000 iterations. Dobrava-Belgrade virus strains Aa15-14 (L: MH598462, M: MH598464, and S: MH598465), Aa15-42 (L: MH59846), SK/Aa (L: GU904039, M: AY961616, and S: AY961615), Mu08_0118 (L: KJ182938), Mu07_0293 (L: KJ182937), Mu05_0239 (L: KJ425422), Saaremaa/160V (L: AJ410618, M: AJ009774, and S: AJ009773), Ano-Poroia/Af19 (L: NC005235 and M: NC005234), Slo/Af-BER (L: GU904042, M: GU904035, and S: GU904029), LT-RS1 (L KF425495), LT-RS2 (L: KF425496), LT-RS3 (L: KF425497), 10636/Ap (L: KP878308), 10645/Ap (L: KP878309), Ap/Sochi/43 (L: KM192209 and S: JF920151), Ap/Sochi/79 (L: KM192208 and S: JF920152), GER/08/131/Af (M: GQ205413 and S: GQ205408), GER/07/607/Af (M: GQ205410), GER/07/293/Aa (M: GQ205409), GER/08/118/Aa (M: GQ205412), Aa1854/Lipetsk-02 (M: EU188453 and S: EU188452), Saaremaa/90/Aa/97 (M: AJ009777), Dobrava 3970/87 (M: L33685 and S: L41916), Ap/Sochi/hu (M: JF920149), Ap1584/Sochi-01 (M: EU188450 and S: EU188449), Kurkino/53Aa/98 (S: AJ131673), Esl/81Aa/01 (S: AY533120), Esl/856/Aa (S: AJ269549), Sarmellek/77Aa06 (S: KC848501), Pecs/27Aa/06 (S: KC848499), Lolland/Aa1403 (S: AJ009773), GER/07/1064/Aa (S: GQ205404), GER/05/239/Aa (S: GQ205405), GER/05/477/Af (S: GQ205406), Gola/242Af/07 (S: KC848496), Gola/235Af/07 (S: KC848495), Croatia_Zutica/Af813 (S: KC676602), Esl/400Af/98 (S: AY168576), and Ano-Poroia/13Af/99 (S: AJ410619). Seoul virus strain 80-39 (L: NC005238, M: NC005237, and S: NC005236), Hantaan virus strain 76-118 (L: NC005222, M: NC005219, and S: M14626), PUUV strain Sotkamo (L: NC005225, M: NC005223, and S: NC005224), Sin Nombre virus strain NM-H10 (L: NC005217, M: L25783, and S: L25784). The newly detected DOBV strains in Poland are shown in bold red letters and designated by red circles. The scale bar indicates the number of nucleotide substitutions per site. The genomic sequences of other DOBV genotypes (Kurkino, Saaremaa, Dobrava, and Sochi) and other hantaviruses were analyzed together as out-groups.
Phylogenetic and genetic analyses of SWSV.
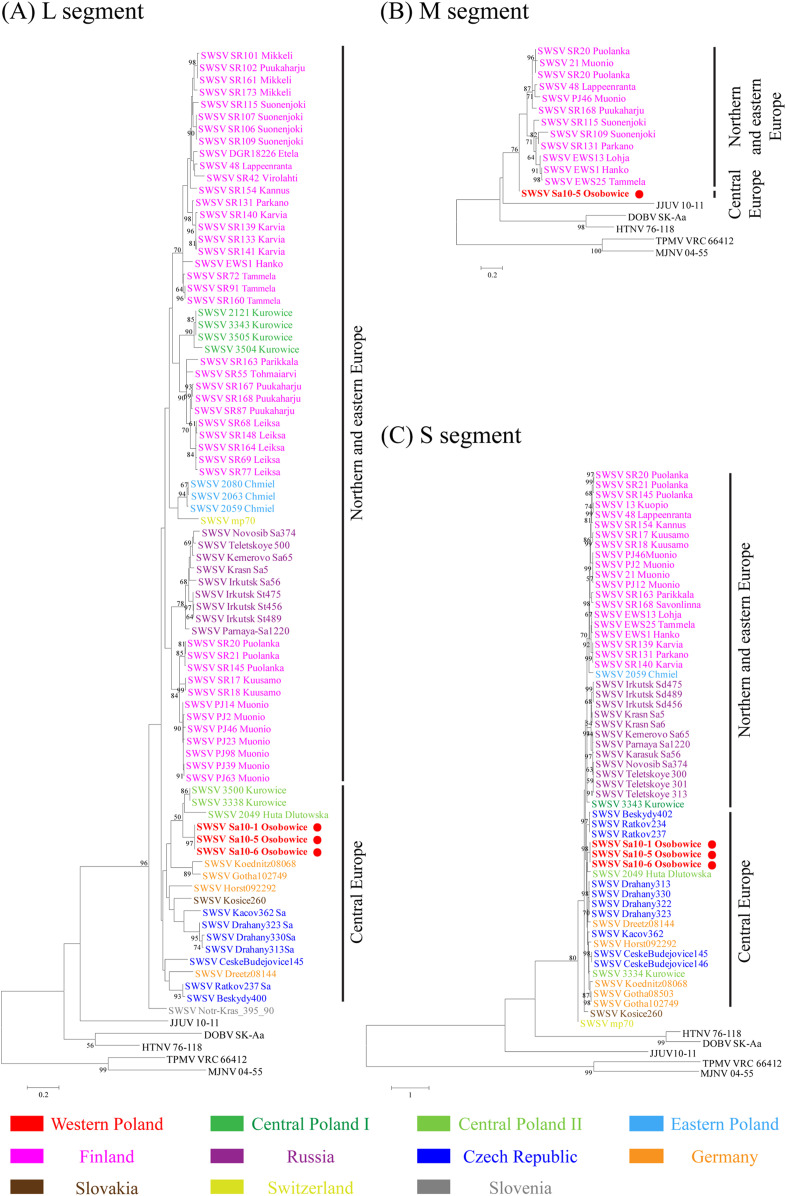
Partial L- and S-genomic sequences of SWSV were obtained in Sa10-1, Sa10-5, and Sa10-6 and the partial M segment in Sa10-5 (Table 2). The nucleotide and amino acid sequence similarities of the SWSV L segment showed 80.3% and 98.1%, respectively, compared with the prototype SWSV mp70 strain (Supplemental Table 3). The sequence homologies of the SWSV S segment were 88.3% and 99.2% at the nucleotide and amino acid levels. The homology analysis of SWSV L and S segments demonstrated highly divergent genomic sequences of shrew-borne orthohantaviruses in Poland, ranging from 78.2–100% to 80.1–97.7%, respectively. Based on the L segment, SWSV strains Sa10-1, Sa10-5, and Sa10-6 from western Poland clustered with SWSV strains 3338 and 3500 from Kurowice and strain 2049 from Huta Dłutowska in central Poland (Figure 3A). This genetic group shared a common ancestor with SWSV strains in central Europe, including the Czech Republic, Germany, and Slovakia. The partial M segment of SWSV Sa10-5 formed a segregated genetic lineage from SWSV strains in Finland, northeastern Europe (Figure 3B). The partial S segments phylogenetically grouped with SWSV strain 2049 from Huta Dłutowska in central Poland (Figure 3C). The genetic lineage belonged to SWSV strains in the Czech Republic and Germany.
Figure 3.
Phylogenetic trees of Seewis virus (SWSV) L, M, and S segments. Phylogenetic trees were generated by the maximum likelihood method using the GTR+G, T92+G, and TN93+G model, based on 325 nt (position 2,968–3,292) of the L segment (A), 794 nt (position 446–1,239) of the M segment (B), and 732 nt (position 407–1,138) of the S segment (C), respectively. The numbers at each node are bootstrap probabilities (expressed as percentages), as determined for 1,000 iterations. The GenBank accession numbers for virus strains in the L, M, and S segment trees are as follows: SWSV strains mp70 (L: EF636026, M: EF636025, and S: EF636024), 10-1 (L: MK402015 and S: MK402016), 10-5 (L: MK402017, M: MK402018, and S: MK402019), 10-6 (L: MK402020, M: MK402021, and S: MK402022), 2121 (L: KC537794), 2049 (L: JX990941 and S: KM394269), 2063 (L: JX990942), 2080 (L: JX990943), 2049 (L: 990945 and S: KM394271), 3334 (S: KM43419 and S: KM434198), 3338 (L: KM394264), 3343 (L: KM394265 and S: KM434199), 3500 (L: KM394266), 3504 (L: KM394267), 3505 (L: KM394268), Beskydy400 (L: JQ425336), Beskydy402 (S: JQ425302), CeskeBudejovice145 (L: JQ425313 and S: JQ425267), CeskeBudejovice146 (S: JQ425268), Drahany313 (L: JQ425318 and S: JQ425273), Drahany322 (S: JQ425274), Drahany323 (L: JQ425320 and S: JQ425275), Drahany330 (L: JQ425322 and S: JQ425277), Kacov362 (L: JQ425329 and S: JQ425287), Ratkov234 (S: JQ425269), Ratkov237 (L: JQ425316 and S: JQ425271), Dreetz08144 (L: JQ425310 and S: JQ425261), Gotha08503 (S: JQ425263), Gotha102749 (L: JQ425312 and S: JQ425266), Horst092292 (L: JQ425311 and S: JQ425265), Koednitz08068 (L: JQ425309 and S: JQ425260), DGR18226 (L: GQ293102), 13 Kuopio (S: KJ136615 ), 21 Muonio (M: KJ136604 and S: KJ136610), 48 Lappeenranta (L: KJ136619, M: KJ136603, and S: KJ136609), EWS1 Hanko (L: KJ136629, M: KJ136607, and S: KJ136612), EWS13 Lohja (M: KJ136605 and S: KJ136613), EWS25 Tammela (M: KJ136608 and S: KJ136614), PJ12 Mounio (L: KY651079) PJ14 Mounio (L: KY651023), PJ23 Mounio (L: KY651024), SR154 Kannus (L: KY651025 and S: KY651077), PJ2 Mounio (L: KY651026 and S: KY651080), PJ39 Mounio (L: KY651027), PJ46 Mounio (L: KY651028, M: KY651067, and S: KY651078), PJ63 Mounio (L: KY651029), PJ98 Mounio (L: KY651030), SR17 Kuusamo (L: KY651052 and S: KY651076), SR18 Kuusamo (L: KY651054 and S: KY651075), SR20 Puolanka (L: KY651055, M: KY651065, and S: KY651073), SR21 Puolanka (L: KY651056, M: KY651069, and S: KY651074), SR42 Virolahti (L: KY651057), SR55 Tohmaiarvi (L: KY651058), SR68 Leiksa (L: KY651059), SR69 Leiksa (L: KY651060), SR72 Tammela (L: KY651061), SR77 Leiksa (L: KY651062), SR87 Puukaharju (L: KY651048), SR91 Tammela (L: KY651044), SR101 Mikkeli (L: KY651032), SR102 Puukaharju (L: KY651033), SR106 Suonenjoki (L: KY651034), SR107 Suonenjoki (L: KY651035), SR109 Suonenjoki (L: KY651036 and M: KY651064), SR115 Suonenjoki (L: KY651037 and M: KY651066), SR131 Parkano (L: KY651038, M: KY651063, and S: KY651082), SR133 Karvia (L: KY651039), SR139 Karvia (L: KY651040 and S: KY651072), SR140 Karvia (L: KY651041 and S: KY651081), SR141 Karvia (L: KY651042), SR145 Puolanka (L: KY651043 and S: KY651083), SR148 Leiksa (L: KY651049), SR160 Tammela (L: KY651045), SR161 Mikkeli (L: KY651031), SR163 Parikkala (L: KY651046 and S: KY651071), SR164 Leiksa (L: KY651047), SR167 Puukaharju (L: KY651050), SR168 Puukaharju (L: KY651051, M: KY651068, and S: KY651070), SR173 Mikkeli (L: KY651053), Novosib-Sa374 (L: GQ267804), Irkutsk St456 (L: GQ267805 and S: GQ284572), Irkutsk St475 (L: GQ267806 and S: GQ284573), Irkutsk St489 (L: GQ267807 and S: GQ284574), Irkutsk Sa56 (L: GQ267809 and S: GQ284586), Parnaya-Sa1220 (L: GQ267810 and S: GQ284580), Krasn-Sa5 (L: GQ267811 and S: GQ284584), Krasn-Sa6 (S: GQ284583), Kemerovo-Sa65 (L: GQ267812 and S: GQ284582), Novosib-Sa374 (S: GQ284581), Telet-Sa300 (S: GQ284578), Telet-Sa301 (S: GQ284577), Telet-Sa313 (S: GQ284579), Teletskoye 500 (L: EU424338), Kosice260 (L: JQ425317 and S: JQ425272), Notr-Kras_395_90 (L: KF060922). Dobrava-Belgrade virus strain DOBV SK-Aa (L: GU904039, M: AY961616, and S: AY961615), Hantaan virus strain 76-118 (L: KT885047, M: Y00386, and S: M14626), Jeju virus strain 10-11 (L: HQ834697, M: HQ834696, and S: HQ834695), Thottapalayam virus strain TPMV VRC66412 (L: EU001330, M: EU001329, and S: AY526097), and Imjin virus strain MJNV 04-55 (L: EF641807, M: EF641799, and S: EF641805). The newly detected SWSV strains in Poland are shown in bold red lettering and designated by red circles. The scale bar indicates the number of nucleotide substitutions per site.
DISCUSSION
The approximately 10,000 cases of HFRS occurring annually in Europe pose a critical public health threat.8 In Poland, a previous study showed specific IgM and IgG antibodies against orthohantaviruses in 3% of forestry workers.42 During 2007–2017, 128 patients with orthohantavirus infection were reported in the central, eastern, and southern areas.43 However, there is a lack of epidemiological surveillance and genetic sequences of orthohantaviruses in other areas of Poland. Here, we investigated the geographic distribution and genetic diversity of orthohantaviruses from small mammals collected in western and northern Poland.
The phylogeny of DOBV L and S segments in western Poland showed a genetic lineage with DOBV in Russia, Hungary, and Slovakia, whereas the DOBV M segment clustered with that in Germany. In addition, the phylogenies of SWSV strains were extended from central and eastern areas to western areas in Poland. The phylogeny of SWSV was highly divergent compared with strains from Finland.27,35 The western Poland SWSV formed a genetic lineage with SWSV strains in central Poland (Kurowice and Huta Dłutowska) and central European countries (Czech Republic and Germany). The northern and eastern European SWSV lineage comprised strains circulating in central (Kurowice) and eastern (Chmiel) Poland and northeastern Europe (Finland and Russia).
The historical biogeographic analysis of S. araneus demonstrates that ancestors of shrews were widely distributed in Eurasia, spanning from northern Asia to western Europe.44 Genetically analyzed chromosome races and karyotypic groups to S. araneus species described well-visible genetic diversity and evolutionary history of the shrews, for which karyotypes of the shrews were differentiated into the West European Karyological Group (WEKG) and the East European Karyological Group (EEKG).45 The WEKG of shrews originated from southern and western Poland, Czech Republic, and Germany, whereas the EEKG shrews showed close genetic relationships in eastern and northeastern Poland. The L and S segments of western Polish SWSV formed phylogenetic groups with the strains in the Czech Republic and Germany, indicating the WEKG shrews might harbor SWSV. By contrast, the phylogenies of SWSV L and S segments from eastern Poland shared common ancestors with SWSV strains from Finland, Russia, and northeastern European countries. These SWSV strains were likely carried by EEKG shrews. The phylogenetic inference suggests the co-circulation of distinct SWSV lineages derived from WEKG and EEKG shrews in central Poland. This observation led us to hypothesize that central Poland might be a hybrid zone for SWSV circulating in Europe. Hybrid zones for S. araneus were described in northern and eastern Poland.46,47 It is plausible that distinct genotypes of SWSV could be present in the hybrid zone due to the host specificity and divergence of shrew-borne hantaviruses. However, to better understand the evolutionary history of SWSV with their natural hosts, future research should focus on 1) the coexistence of different genotypes (karyotypes) of natural hosts in central Poland, 2) whether EEKG shrews include small mammals in Finland and Russia, and 3) the evolutionary link of SWSV with shrews.
The incidence of orthohantavirus diseases is positively associated with sharing of habitats between humans and rodents.48 Recreational activities are well-described risk factors for HFRS and HCPS.49–51 In 2012, an HCPS outbreak comprising 10 cases and three deaths occurred among overnight visitors at Yosemite National Park.52 Moreover, HFRS cases were correlated with human activities in recreation areas in Germany.53 These outbreaks indicate the increased likelihood of human outdoor activities resulting in contact with rodents, thereby causing a higher incidence of orthohantavirus infections. In this study, we collected small mammals from four investigation sites in Poland. Milicz Ponds Nature Reserve and Ślęża Landscape Park are important habitats and breeding grounds for waterfowl. Wrocław-Osobowice comprises irrigation fields outside the city, and Gdańsk-Sobieszewo Island is located on an estuary of the Vistula River to the Baltic Sea and is near the ornithological nature reserve Ptasi Raj. In particular, DOBV RNA–positive rodents were identified in the Milicz Ponds and Ślęża Landscape Park, the protected nature zones in western Poland. Ślęża Landscape Park is a popular recreation area for Wrocław residents. Recently, a serological study demonstrated an anti-DOBV/HTNV IgM/IgG antibody seroprevalence of 3.9% among forestry workers in Wrocław.54 These observations suggest that DOBV may be a public health threat in western Poland. The surveillance and monitoring of circulating DOBV remain to be investigated within the scope of disease-risk assessment and mitigation.
In conclusion, this study provides preliminary insights into the prevalence, phylogeography, and genetic diversity of DOBV and SWSV circulating in western Poland. Larger scale studies and continued surveillance will improve our understanding about the evolutionary relationships and disease risk of rodent- and shrew-borne orthohantaviruses in Poland.
Supplemental table
Note: Supplemental tables appear at www.ajtmh.org.
REFERENCES
- 1.Vaheri A, Strandin T, Hepojoki J, Sironen T, Henttonen H, Makela S, Mustonen J, 2013. Uncovering the mysteries of hantavirus infections. Nat Rev Micro 11: 539–550. [DOI] [PubMed] [Google Scholar]
- 2.Fill MA, et al. 2017. Notes from the field: multiple cases of Seoul virus infection in a household with infected pet rats-Tennessee, December 2016-April 2017. Morb Mortal Wkly Rep 66: 1081–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thangavel RR, Bouvier NM, 2014. Animal models for influenza virus pathogenesis, transmission, and immunology. J Immunol Methods 410: 60–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee HW, Lee PW, Johnson KM, 1978. Isolation of the etiologic agent of Korean hemorrhagic fever. J Infect Dis 190: 1711–1721. [DOI] [PubMed] [Google Scholar]
- 5.Guo WP, et al. 2013. Phylogeny and origins of hantaviruses harbored by bats, insectivores, and rodents. PLoS Pathog 9: e1003159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yanagihara R, Gu SH, Arai S, Kang HJ, Song JW, 2014. Hantaviruses: rediscovery and new beginnings. Virus Res 187: 6–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Krüger DH, Figueiredo LT, Song JW, Klempa B, 2015. Hantaviruses – globally emerging pathogens. J Clin Virol 64: 128–136. [DOI] [PubMed] [Google Scholar]
- 8.Vaheri A, Henttonen H, Voutilainen L, Mustonen J, Sironen T, Vapalahti O, 2013. Hantavirus infections in Europe and their impact on public health. Rev Med Virol 23: 35–49. [DOI] [PubMed] [Google Scholar]
- 9.Papa A, 2012. Dobrava-Belgrade virus: phylogeny, epidemiology, disease. Antiviral Res 95: 104–117. [DOI] [PubMed] [Google Scholar]
- 10.Xiao SY, Diglisic G, Avšič-Županc T, LeDuc JW, 1993. Dobrava virus as a new Hantavirus: evidenced by comparative sequence analysis. J Med Virol 39: 152–155. [DOI] [PubMed] [Google Scholar]
- 11.Avšič-Županc T, Xiao SY, Stojanovic R, Gligić A, van der Groen G, LeDuc JW, 1992. Characterization of Dobrava virus: a hantavirus from Slovenia, Yugoslavia. J Med Virol 38: 132–137. [DOI] [PubMed] [Google Scholar]
- 12.Gligić A, et al. 1992. Belgrade virus: a new hantavirus causing severe hemorrhagic fever with renal syndrome in Yugoslavia. J Infect Dis 166: 113–120. [DOI] [PubMed] [Google Scholar]
- 13.Klempa B, et al. 2013. Complex evolution and epidemiology of Dobrava-Belgrade hantavirus: definition of genotypes and their characteristics. Arch Virol 158: 521–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Klempa B, Schutt M, Auste B, Labuda M, Ulrich R, Meisel H, Krüger DH, 2004. First molecular identification of human Dobrava virus infection in central Europe. J Clin Microbiol 42: 1322–1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klempa B, Tkachenko EA, Dzagurova TK, Yunicheva YV, Morozov VG, Okulova NM, Slyusareva GP, Smirnov A, Krüger DH, 2008. Hemorrhagic fever with renal syndrome caused by 2 lineages of Dobrava hantavirus, Russia. Emerg Infect Dis 14: 617–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Knap JP, Trybusz A, 2006. Haemorrhagic fever with renal syndrome (HFRS)-hantavirus infection disease appearing in Poland. Pol Merkur Lekarski 21: 411–417. [PubMed] [Google Scholar]
- 17.Nowakowska A, Heyman P, Knap JP, Burzynski W, Witas M, 2009. The first established focus of hantavirus infection in Poland, 2007. Ann Agric Environ Med 16: 79–85. [PubMed] [Google Scholar]
- 18.Grygorczuk S, Pancewicz S, Zajkowska J, Kondrusik M, Swierzbinska R, Moniuszko A, Pawlak-Zalewska W, 2008. Detection of anti-hantavirus antibodies in forest workers in the north-east of Poland. Przegl Epidemiol 62: 531–537. [PubMed] [Google Scholar]
- 19.Zukiewicz-Sobczak W, Zwolinski J, Chmielewska-Badora J, Galinska EM, Cholewa G, Krasowska E, Zagorski J, Wojtyla A, Tomasiewicz K, Klapec T, 2014. Prevalence of antibodies against selected zoonotic agents in forestry workers from eastern and southern Poland. Ann Agric Environ Med 21: 767–770. [DOI] [PubMed] [Google Scholar]
- 20.Ali HS, Drewes S, Sadowska ET, Mikowska M, Groschup MH, Heckel G, Koteja P, Ulrich RG, 2014. First molecular evidence for Puumala hantavirus in Poland. Viruses 6: 340–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rosenfeld UM, Drewes S, Ali HS, Sadowska ET, Mikowska M, Heckel G, Koteja P, Ulrich RG, 2017. A highly divergent Puumala virus lineage in southern Poland. Arch Virol 162: 1177–1185. [DOI] [PubMed] [Google Scholar]
- 22.Michalski A, Niemcewicz M, Bielawska-Drozd A, Nowakowska A, Gawel J, Pitucha G, Joniec J, Zielonka K, Marciniak-Niemcewicz A, Kocik J, 2014. Surveillance of hantaviruses in Poland: a study of animal reservoirs and human hantavirus disease in Subcarpathia. Vector Borne Zoonotic Dis 14: 514–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kolodziej M, Melgies A, Joniec-Wiechetek J, Michalski A, Nowakowska A, Pitucha G, Niemcewicz MN, 2018. First molecular characterization of Dobrava-Belgrade virus found in Apodemus flavicollis in Poland. Ann Agric Environ Med 25: 368–373. [DOI] [PubMed] [Google Scholar]
- 24.Carey DE, Reuben R, Panicker KN, Shope RE, Myers RM, 1971. Thottapalayam virus: a presumptive arbovirus isolated from a shrew in India. Indian J Med Res 59: 1758–1760. [PubMed] [Google Scholar]
- 25.Song JW, Baek LJ, Schmaljohn CS, Yanagihara R, 2007. Thottapalayam virus, a prototype shrewborne hantavirus. Emerg Infect Dis 13: 980–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Radosa L, et al. 2013. Detection of shrew-borne hantavirus in Eurasian pygmy shrew (Sorex minutus) in Central Europe. Infect Genet Evol 19: 403–410. [DOI] [PubMed] [Google Scholar]
- 27.Gu SH, Markowski J, Kang HJ, Hejduk J, Sikorska B, Liberski PP, Yanagihara R, 2013. Boginia virus, a newfound hantavirus harbored by the Eurasian water shrew (Neomys fodiens) in Poland. Virol J 10: 160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gu SH, Arai S, Yu HT, Lim BK, Kang HJ, Yanagihara R, 2016. Genetic variants of Cao Bang hantavirus in the Chinese mole shrew (Anourosorex squamipes) and Taiwanese mole shrew (Anourosorex yamashinai). Infect Genet Evol 40: 113–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Arai S, et al. 2012. Divergent ancestral lineages of newfound hantaviruses harbored by phylogenetically related crocidurine shrew species in Korea. Virology 424: 99–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kang HJ, Arai S, Hope AG, Cook JA, Yanagihara R, 2010. Novel hantavirus in the flat-skulled shrew (Sorex roboratus). Vector Borne Zoonotic Dis 10: 593–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Song JW, Gu SH, Bennett SN, Arai S, Puorger M, Hilbe M, Yanagihara R, 2007. Seewis virus, a genetically distinct hantavirus in the Eurasian common shrew (Sorex araneus). Virol J 4: 114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schlegel M, et al. 2012. Broad geographical distribution and high genetic diversity of shrew-borne Seewis hantavirus in Central Europe. Virus Genes 45: 48–55. [DOI] [PubMed] [Google Scholar]
- 33.Kang HJ, Arai S, Hope AG, Song JW, Cook JA, Yanagihara R, 2009. Genetic diversity and phylogeography of Seewis virus in the Eurasian common shrew in Finland and Hungary. Virol J 6: 208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ling J, Sironen T, Voutilainen L, Hepojoki S, Niemimaa J, Isoviita VM, Vaheri A, Henttonen H, Vapalahti O, 2014. Hantaviruses in Finnish soricomorphs: evidence for two distinct hantaviruses carried by Sorex araneus suggesting ancient host-switch. Infect Genet Evol 27: 51–61. [DOI] [PubMed] [Google Scholar]
- 35.Ling J, Smura T, Tamarit D, Huitu O, Voutilainen L, Henttonen H, Vaheri A, Vapalahti O, Sironen T, 2018. Evolution and postglacial colonization of Seewis hantavirus with Sorex araneus in Finland. Infect Genet Evol 57: 88–97. [DOI] [PubMed] [Google Scholar]
- 36.Gu SH, Hejduk J, Markowski J, Kang HJ, Markowski M, Polatynska M, Sikorska B, Liberski PP, Yanagihara R, 2014. Co-circulation of soricid- and talpid-borne hantaviruses in Poland. Infect Genet Evol 28: 296–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yashina LN, et al. 2010. Seewis virus: phylogeography of a shrew-borne hantavirus in Siberia, Russia. Vector Borne Zoonotic Dis 10: 585–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Resman K, Korva M, Fajs L, Zidaric T, Trilar T, Županc TA, 2013. Molecular evidence and high genetic diversity of shrew-borne Seewis virus in Slovenia. Virus Res 177: 113–117. [DOI] [PubMed] [Google Scholar]
- 39.Korva M, Knap N, Rus KR, Fajs L, Grubelnik G, Bremec M, Knapic T, Trilar T, Županc TA, 2013. Phylogeographic diversity of pathogenic and non-pathogenic hantaviruses in Slovenia. Viruses 5: 3071–3087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Irwin DM, Kocher TD, Wilson AC, 1991. Evolution of the cytochrome b gene of mammals. J Mol Evol 32: 128–144. [DOI] [PubMed] [Google Scholar]
- 41.Kumar S, Stecher G, Tamura K, 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33: 1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wroblewska-Luczka P, Chmielewska-Badora J, Zwolinski J, Galinska EM, Adamczuk P, Zukiewicz-Sobczak W, Zagorski J, Wojtyla A, 2017. Seroepidemiologic evaluation of exposure to infection with hantavirus (serotype Puumala) among forestry workers in Poland. Baltic Forestry 23: 612–618. [Google Scholar]
- 43.Stojak J, 2019. Hantavirus infections in humans in Poland-current state of knowledge and perspectives for research. Eur J Public Health. Available at: 10.1093/eurpub/ckz144. [DOI] [PubMed] [Google Scholar]
- 44.Mackiewicz P, Moska M, Wierzbicki H, Gagat P, Mackiewicz D, 2017. Evolutionary history and phylogeographic relationships of shrews from Sorex araneus group. PLoS One 12: e0179760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wójcik JM, 1993. Chromosome races of the common shrew Sorex araneus in Poland: a model of karyotype evolution. Acta Theriologica 38: 315–338. [Google Scholar]
- 46.Fedyk S, Chetnicki W, Banaszek A, 1991. Genetic differentiation of Polish populations of Sorex araneus L. III. Interchromosomal recombination in a hybrid zone. Evolution 45: 1384–1392. [DOI] [PubMed] [Google Scholar]
- 47.Szalaj KA, Fedyk S, Banaszek A, Chetnicki W, Ratkiewicz M, 1996. A hybrid zone between two chromosome races of the common shrew, Sorex araneus, in eastern Poland: preliminary results. Hereditas 125: 169–176. [Google Scholar]
- 48.Tian H, et al. 2018. Urbanization prolongs hantavirus epidemics in cities. Proc Natl Acad Sci U S A 115: 4707–4712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Park KH, Kang YU, Kang SJ, Jung YS, Jang HC, Jung SI, 2011. Experience with extrarenal manifestations of hemorrhagic fever with renal syndrome in a tertiary care hospital in South Korea. Am J Trop Med Hyg 84: 229–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Martinez VP, Bellomo CM, Cacace ML, Suarez P, Bogni L, Padula PJ, 2010. Hantavirus pulmonary syndrome in Argentina, 1995–2008. Emerg Infect Dis 16: 1853–1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.de St Maurice A, et al. 2017. Exposure characteristics of hantavirus pulmonary syndrome patients, United States, 1993–2015. Emerg Infect Dis 23: 733–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nunez JJ, et al. 2014. Hantavirus infections among overnight visitors to Yosemite National Park, California, USA, 2012. Emerg Infect Dis 20: 386–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Essbauer SS, et al. 2007. Nephropathia epidemica in metropolitan area, Germany. Emerg Infect Dis 13: 1271–1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wróblewska-Łuczka P, Chmielewska-Badora J, Zwoliński J, Galińska EM, Adamczuk P, Żukiewicz-Sobczak W, Zagórski J, Tomasiewicz K, Wojtyła A, 2017. Exposure to infection with hantavirus (serotype Dobrava/Hantaan) among forestry workers in Poland. Folia Forestalia Polonica 59: 198–206. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.