Abstract.
The microbial communities residing in the child gut are thought to play an important role in child growth, although the relationship is not well understood. We examined a cohort of young children from Mirzapur, Bangladesh, prospectively over 18 months. Four fecal markers of environmental enteropathy (EE) (high levels of alpha-1-antitrypsin, calprotectin, myeloperoxidase, and neopterin) were examined and anthropometric measures obtained from a cohort of 68 children. The 16S rRNA gene of bacterial DNA was sequenced from stool samples and used to estimate amplicon sequence variants (ASVs). We age-matched children with poor growth to children with normal growth within 1 month and compared the change in abundance and diversity of ASVs over time. Elevated EE markers and poor linear growth in children were associated with changes in microbial communities in the gut. There were increased amounts of Escherichia/Shigella and Proteobacteria and decreased amounts of Prevotella associated with poorly growing children consistent with the mounting evidence supporting the relationship between intestinal inflammation, child growth, and changes in gut microbiota composition. Future research is needed to investigate this association among young children in low- and middle-income countries.
INTRODUCTION
Worldwide, 159 million young children are estimated to be stunted.1 Stunting has been associated with poor cognitive outcomes and a higher risk for mortality.2,3 There is a growing literature demonstrating an association between environmental enteropathy (EE) and stunting among susceptible pediatric populations.4–8 Environmental enteropathy is defined by abnormal intestinal morphology, reduced intestinal barrier function, and increased inflammation.9–14 This disorder occurs from unhygienic environments leading to repeated pathogen exposure, enteric infections, inflammation, and reduced nutrient absorption.4,5,15 Fecal markers of EE, including higher levels of alpha-1-antitrypsin, calprotectin, myeloperoxidase, and neopterin, are easier to assess than the 5-hour period of urine collection often required for dual sugar permeability tests.13 These four markers have been associated with impaired growth in children.5,9,16
Previous studies have found that the gut microbiota is an important determinant of diarrheal disease and impaired growth among pediatric populations.17,18 Pop et al.19 demonstrated major differences in the gut microbiota composition between those with diarrheal and non-diarrheal stool and identified Escherichia/Shigella and Streptococcus to be associated with diarrhea, whereas Prevotella was negatively associated with diarrhea. Growth faltering has been associated with reduced number and evenness of bacterial taxonomic units (i.e., alpha diversity).20,21 In Bangladesh, malnourished children were found to have a lower alpha diversity and a decreased number of taxonomic units in their gut compared to healthy children and had higher Proteobacteria and Bacteroides.22 Another study found that microbiota immaturity (marked by increased Escherichia/Shigella and Streptococcus), measured using a microbiota-for-age Z-score, was associated with severe acute malnutrition.23 However, the relationship between EE, the gut microbiota, and impaired growth18 must be better understood to develop therapeutic interventions to reduce EE and impaired growth among susceptible pediatric populations. Research in this area has been complicated by measurement issues around how children who are growing poorly can be identified because growth cannot necessarily be measured at a single point in time or with any single specific biomarker.24 Measuring growth is further complicated by different underlying conditions that are thought to drive poor growth, which may include, but are not limited to, gut microbiome composition, nutritional intake, genetic differences between individuals, and chronic infection.25 We aimed to describe the gut microbiota in young children with abnormally poor growth or EE identified by seven common criteria within a cohort of children in rural Bangladesh, and to compare their gut microbiota with similar children with normal or robust growth.
MATERIALS AND METHODS
A cohort of 68 children aged 6–31 months were followed up for a period of 18 months in Mirzapur, Bangladesh, at the site of the Global Enteric Multicenter Study demographic and surveillance system from 2014 to 2015.26 Mirzapur is a primarily agricultural area roughly 60 km northwest of the capital city of Dhaka. Anthropometry of enrolled children was recorded at baseline and 18 months later by trained research associates, where each child’s weight was measured once and their height (or length) was taken three times and averaged at each time point.
Stool samples were collected from children on enrollment and at 18 months after enrollment. After collection, the samples were placed in a cooler and transported to the International Center for Diarrheal Disease Research, Bangladesh (icddr,b) in Dhaka, Bangladesh, where they were stored at −80°C until analysis. Stool samples from baseline were analyzed for four fecal markers of EE: calprotectin, alpha-1-antitrypsin, myeloperoxidase, and neopterin using ELISA kits from ALPCO (Salem, NH), BioVendor (Asheville, NC), ALPCO, and GenWay (San Diego, CA), respectively. All assays were run according to the manufacturer’s specifications. All dilutions were also performed according to the package inserts except for a 1:500 dilution used for initial runs for myeloperoxidase.
In addition to EE fecal marker analysis, DNA was extracted from each stool sample with a QIAGEN kit (Hilden, Germany) according to the manufacturer’s instructions at baseline and 18-month follow-up.27 This process included homogenization with iterative bead-beating steps for optimal disruption of cellular material, and the resulting concentration of DNA was measured by NanoDrop spectrophotometer (Thermo Scientific, Waltham, MA). Extracted DNAs were transferred to the University of Maryland for amplification of bacterial DNA with universal primers specific for the V1–V3 region of the 16S rRNA gene common to all bacteria. Amplified products were sequenced using Illumina® MiSeq® at the Institute for Genome Sciences (San Diego, CA). Raw reads were custom filtered to identify those with the best quality as described by Pop et al.19 The resulting high-quality sequences were used with DADA2 (v. 1.8.0, Stanford, CA) employing an algorithm to resolve sequencing errors for sample-specific resolution of amplicon sequence variants (ASVs).28 Briefly, the sequences were dereplicated, sorted by abundance, and aggregated while accounting for the sequence-specific likelihood that the sequence was read from sample DNA or an artifact of the Illumina processing. The taxonomical annotation was performed on the resulting ASVs using the naïve Bayes classifier described by Wang et al.29 based on the k-mer length 8nd the reference database SILVA (v. 123), Bremen, Germany.30
Ethical approval.
Written informed consent was received from the primary caregivers of participating children. This study was approved by the Institutional Review Board of icddr,b, and an exemption was granted from the Institutional Review Board of the Johns Hopkins Bloomberg School of Public Health.
Statistical methods.
We defined growth by three anthropometric measures: height, weight, and weight for height standardized to Z-scores by age and gender for each child based on the WHO criteria (height for age Z-score (HAZ), weight for age Z-score (WAZ), and weight for height or length Z-score (WFLZ)),31 where a negative change in the Z-score over the 18-month follow-up period was assumed to indicate poor growth. We assumed children in the top tertile (largest increases in Z-score, defined as “control”) to be healthy children who were growing well and that those with the largest decreases in Z-score in the lowest or bottom tertile (defined as a “case”) were growing poorly. These criteria were applied separately, so that a child with a low change in HAZ would be defined as a “case” for HAZ but may have a normal change in WAZ and hence be grouped as a “control” for WAZ. We defined EE by four fecal EE markers, where high levels in the top tertile among all 68 children of each fecal marker (calprotectin, alpha-1-antitrypsin, myeloperoxidase, and neopterin) were defined as “case” and low levels in bottom tertile for each fecal marker were defined as “control.”
For each measure, child pairs were matched by age within one month in a retrospective case–control design, with potentially overlapping definitions of “case” as defined by HAZ, WAZ, WFLZ, and the four fecal markers alpha-1-antitrypsin, calprotectin, myeloperoxidase, and neopterin. Age matching was performed given the known large variability in gut microbiota composition by age.19 Because we did not expect exact agreement between these different growth classifications, we also examined the Pearson correlation between measures. We included children whose stool samples had at least 2,000 reads of 16S RNA at baseline and follow-up in statistical analyses, discarding those with fewer than 2,000 reads at either time. We compared the proportional abundance of phyla and genera between cases and controls with nonparametric comparisons using the Wilcoxon rank sum test. We also compared the change over time in relative abundance with the rank sum test. For comparison, we also compared the relative abundance at baseline between age-matched children using the DESeq method and comparing the change in relative abundance over time, assuming a negative binomial distribution of sequence counts for each taxa.32 We did not adjust for multiple comparisons. In addition, we examined the Bray–Curtis distance between bacterial communities identified from stool samples for pairwise comparisons.33 This distance was clustered hierarchically to identify similar subgroups among samples.34
Diversity.
In addition to comparing proportional abundance at baseline and the change in proportional abundance, the diversity of bacterial communities at each observation time was also compared using alpha diversity described by the Shannon index, with 0 corresponding to total dominance from one ASV and larger values indicating more diversity.20,35 We also examined the turnover of bacterial ASVs over time between children with poor or normal growth and children with low or high EE markers with beta diversity36 using the metric βw as defined by Whittaker,37 ranging from 0 (no change) to 1 (complete turnover). Diversity was estimated using the vegan statistical package in R.38 We compared these measures of diversity and change in bacterial diversity between cases and controls using Student’s t-test.
RESULTS
Of the 68 children included in this analysis, 50% were male (34 of 68), with an average age of 17 months at baseline. Baseline anthropometry and fecal EE markers are shown in Table 1. Relative to the standards defined by the WHO for child growth, children in this cohort were below average height (average Z-score −1.4), weight (average −1.1), and weight for height (average −0.6). In addition, on average, standardized height and weight were declining over the 18-month follow-up period (average decline of 0.5 SD in HAZ and 0.5 SD in WAZ). Further details for this study cohort are available in George et al.5 Levels of fecal markers and change in anthropometric measures that were used to define child pairs for classification of cases and controls are shown in Table 1. Complete distributions of anthropometric factors are available in Supplemental Appendix S1.
Table 1.
Description of cohort of 68 children from Mirzapur, Bangladesh, followed up for 18 months
| Factor | Mean (SD) or N (%) | Control classification | Case classification |
|---|---|---|---|
| Gender | |||
| Male | 34 (50%) | – | – |
| Female | 34 (50%) | – | – |
| Age at baseline (months) | 16.8 (3.6) | – | – |
| Baseline | |||
| Alpha-1-antitrypsin (g/L)* | 0.4 (0.4) | < 0.20 | > 0.36 |
| Calprotectin (µg/g)* | 897.1 (928.6) | < 230.4 | > 1,041 |
| Myeloperoxidase (ng/mL)* | 6,380 (6,224) | < 2,643 | > 6,003 |
| Neopterin (mmol/mL)* | 2,002 (2,395) | < 590.0 | > 1,999 |
| HAZ | −1.4 (1.1) | – | – |
| WAZ | −1.1 (1.1) | – | – |
| WFLZ | −0.6 (1.2) | – | – |
| 18-month follow-up | |||
| HAZ | −1.8 (1.0) | – | – |
| WAZ | −1.4 (1.1) | – | – |
| WFLZ | −0.5 (1.1) | – | – |
| Change from baseline to 18 months | |||
| HAZ** | −0.5 (0.7) | > −0.32 | < −0.73 |
| WAZ** | −0.3 (0.8) | > −0.05 | < −0.65 |
| WFLZ** | 0.1 (1.0) | > 0.23 | < −0.38 |
HAZ = height for age Z-score; WAZ = weight for age Z-score; WFLZ = weight for length or height Z-score.
Controls defined by the tertile of highest growth defined by low fecal markers, and cases were defined in the lowest tertile of growth (highest fecal markers).
Controls defined by the tertile of highest growth, and cases were defined in the lowest tertile of growth.
The correlations between the four EE and three growth measures are shown in Table 2. Baseline fecal markers were not significantly correlated with each other, with the exception of calprotectin and myeloperoxidase, which have a moderate association at 0.41 (P < 0.001). Baseline myeloperoxidase was positively correlated with changes in WFLZ (correlation 0.29, P = 0.021), unexpectedly indicating higher myeloperoxidase being related to a positive change in WFLZ. Among changes in anthropometry, change in HAZ was correlated with change in WAZ (0.35, P = 0.005).
Table 2.
Correlation among different measures defining criteria for poor growth among 68 children from the Mirzapur cohort for measurements at baseline and 18-month follow-up
| AA | CAL | MPO | NEO | Delta HAZ | Delta WAZ | Delta WFLZ | |
|---|---|---|---|---|---|---|---|
| Baseline | |||||||
| AA | 1.00 | – | – | – | – | – | – |
| CAL | −0.03 | 1.00 | – | – | – | – | – |
| MPO | 0.10 | 0.41* | 1.00 | – | – | – | – |
| NEO | −0.07 | 0.06 | 0.18 | 1.00 | – | – | – |
| Change from baseline to 18 months | |||||||
| HAZ | 0.18 | −0.12 | −0.21 | −0.11 | 1.00 | – | – |
| WAZ | −0.02 | −0.01 | 0.16 | −0.10 | 0.35† | 1.00 | – |
| WFLZ | −0.15 | 0.07 | 0.29** | −0.03 | −0.18 | 0.84* | 1.00 |
AA = alpha-1-antitrypsin; CAL = calprotectin; MPO = myeloperoxidase; NEO = neopterin; HAZ = height for age Z-score; WAZ = weight for age Z-score; WFLZ = weight for length or height Z-score.
†P = 0.005.
*P < 0.001.
**P = 0.021.
Each child contributing to case–control pairs had at least 2,000 reads of DNA at each observation time for analysis. For some EE and growth measures, children classified as having poor growth were dissimilar in age (not within 1 month) to all children classified as having normal growth, and 26 stool samples were excluded because of an insufficient number of reads. The resulting number of pairs for growth status defined by high/low alpha-1-antitrypsin, high/low calprotectin, high/low myeloperoxidase, high/low neopterin, short/tall height for age, light/heavy weight for age, and small/large weight for height/length was 10, 8, 4, 8, 7, 8, and 9, respectively.
Using DADA2 to incorporate quality scores from the Illumina MiSeq sequencer for resolving errors in the corresponding read of 16S RNA, we identified 10,676 unique bacterial taxa among all 68 children in this cohort over the observation period. Study children contributed a total of 136 stool samples; however, there were 29 samples with fewer than 2,000 reads of DNA, leaving 107 samples for analysis from 56 children. On average, 26,000 sequences of DNA were read from these 107 samples, with a median of 14,000 sequences.
A summary of common phyla is shown in Figure 1 both for cases and controls at each observation time, excluding phyla with 1% or lower average proportional abundance. Actinobacteria, Bacteroidetes, Cyanobacteria, Firmicutes, Fusobacteria, and Proteobacteria are included. The proportional abundance of bacterial phylum did not appear different at baseline between case and control children for any case/control definition; however, sensitivity analysis with DESeq identified lower Cyanobacteria at baseline among the case children defined by the HAZ as well as the alpha-1-antitrypsin criteria (see Supplemental Appendix S2). Comparing the change over time between child pairs, Proteobacteria increased more among those with high calprotectin than low calprotectin (10% increase versus 15% decrease, P = 0.005).
Figure 1.
Average proportional abundance of phyla in stool samples at baseline and 18-month follow-up among Mirzapur, Bangladesh, age-matched growth pairs, for seven separate growth criteria. Growth pairs were constructed so that those in the tertile of highest growth were matched to a child in the lowest growth tertile whose age at baseline was within 1 month. This figure appears in color at www.ajtmh.org.
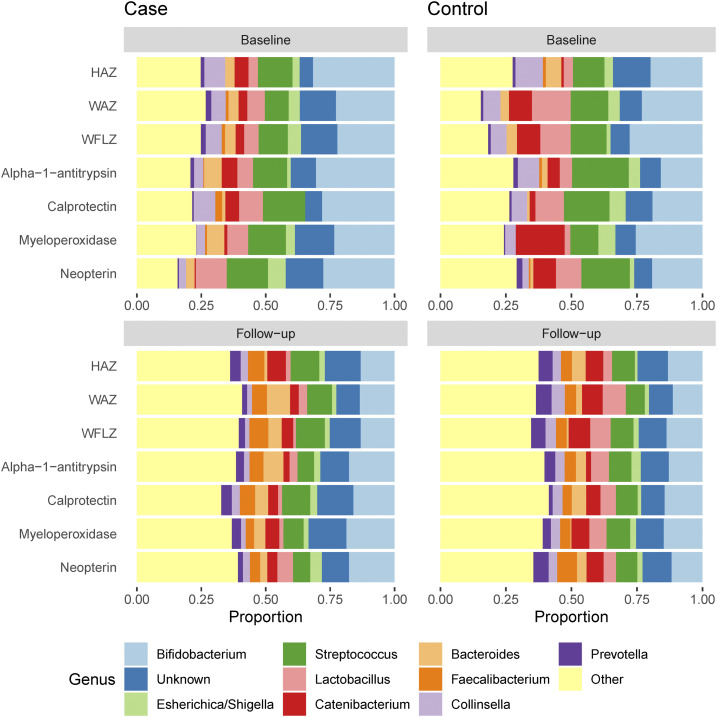
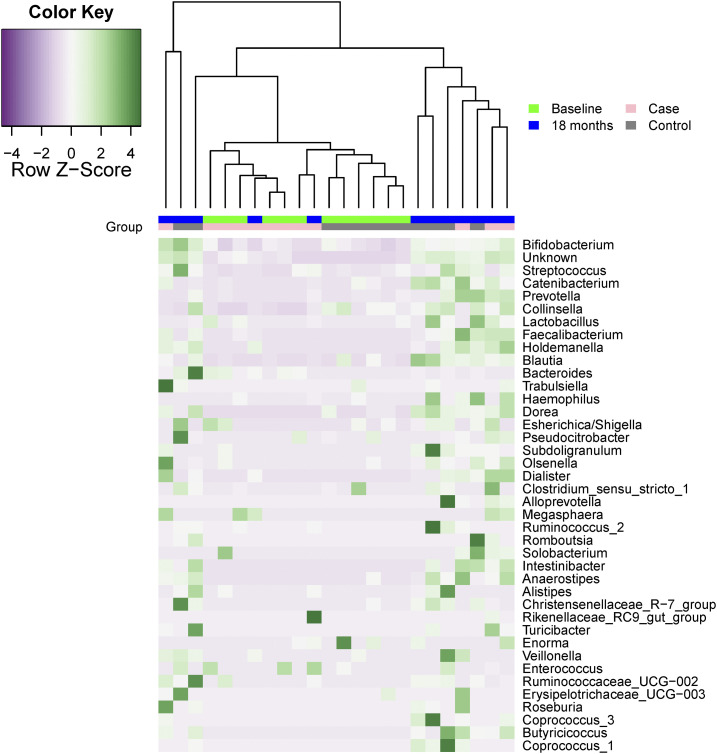
The 10 most commonly observed genera are shown in Figure 2, including Bifidobacterium, Escherichia/Shigella, Streptococcus, Lactobacillus, Catenibacterium, Bacteroides, Faecalibacterium, Collinsella, and Prevotella. At baseline, the proportional abundance of genera was different among Bifidobacterium (25% among those with high alpha-1-antitrypsin versus 17% among those with low alpha-1-antitrypsin, P = 0.035). There were also changes in the microbiota from baseline to 18 months, Escherichia/Shigella increased more for those with high calprotectin than among those with low calprotectin (3% increase versus no change, P = 0.016). Prevotella declined more among those with large decline in WAZ (5% increase versus no change, P = 0.014). The proportion of unidentifiable bacteria increased more among those with large declines in HAZ (8% increase versus no change, P = 0.017). The normalized abundance of the 40 most common genera in those with low change in HAZ and their respective age-matched children with normal or robust change over time in HAZ is shown in Figure 3. Hierarchical clustering does not appear to identify the HAZ growth classification, although many of the ASVs measured at a similar follow-up time are grouped together, either at baseline or at 18 months. Heterogeneity across age may be better identified, not unexpectedly given the strong relationship between age and gut microbiota.19
Figure 2.
Average proportional abundance of genera in stool samples at baseline and 18-month follow-up among Mirzapur, Bangladesh, age-matched growth pairs, for seven separate growth criteria. Pairs were constructed so that those in the tertile of highest growth were matched to a child in the lowest growth tertile whose age at baseline was within 1 month. This figure appears in color at www.ajtmh.org.
Figure 3.
Abundance of 40 most common genera in stool samples at baseline and 18-month follow-up among Mirzapur, Bangladesh, age-matched pairs, for the height/length for age growth criteria. Pairs were constructed so that those in the tertile of highest growth were matched to a child in the lowest growth tertile whose age at baseline was within one month. Genera are shown in the decreasing order of overall average abundance. This figure appears in color at www.ajtmh.org.
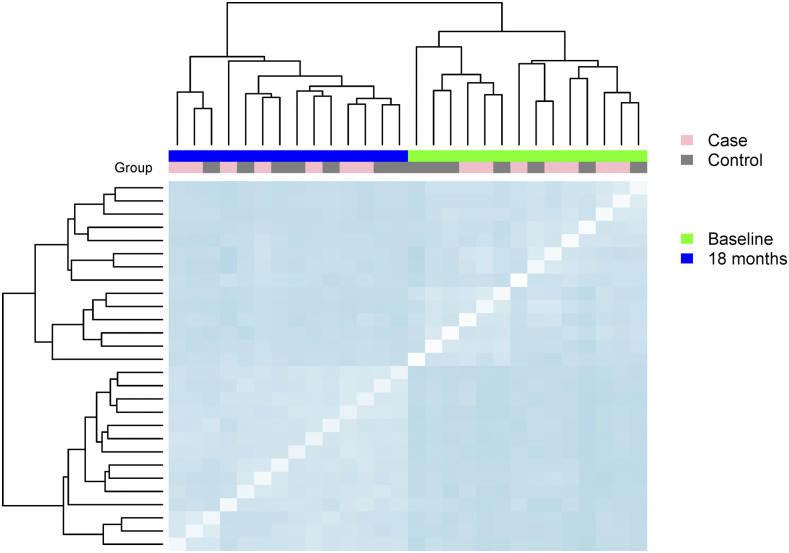
The Bray–Curtis distance among sample bacterial communities based on genus for child pairs with large decline versus small changes in HAZ is shown in Figure 4. The Bray–Curtis distance between sample communities was similar between those with low change in HAZ and high change in HAZ (average 0.66) at baseline to that within cases (0.69) and within controls (0.64). At 18-month follow-up, the distance between cases and controls was also similar (0.52) and maintained similarity with the average distance within cases (0.54) and within controls (0.54).
Figure 4.
Dissimilarity among and between cases and controls for seven pairs matched by age and for differential growth based on the change in height for age Z-score over an 18-month follow-up period. Dissimilarity estimated with Bray–Curtis distance, where darker blue indicates more difference in the community structure defined by genera. This figure appears in color at www.ajtmh.org.
Diversity.
Diversity at each observation time for 56 children with samples having at least 2,000 sequences at both observation times was quantified. Both Simpson and Shannon diversity, as well as richness and evenness are all increasing over time, as expected for children in this period of rapid gut microbiota maturation, having P < 0.001 when compared with a paired t-test. Diversity at each observation time and for turnover in ASVs during the study period are shown in Table 3. Follow-up ASV Shannon diversity was significantly higher in children identified in the top tertile of change in WAZ than those with poor growth for change in WAZ (5.47 versus 5.34, P = 0.036). Baseline Shannon diversity was also higher among controls defined by low neopterin concentration than in children with high neopterin (2.48 versus 2.11, P = 0.016).
Table 3.
Diversity of bacterial ASVs at baseline and 18-month follow-up among age-matched cases and controls for seven separate growth criteria in the Mirzapur, Bangladesh cohort with at least 2,000 reads
| Case/Control Classification | Number of children | Mean among cases | Mean among controls | P-value* |
|---|---|---|---|---|
| Shannon (alpha) diversity at baseline | ||||
| ΔHAZ | 14 | 2.31 | 2.24 | 0.710 |
| ΔWAZ | 16 | 2.46 | 2.27 | 0.257 |
| ΔWFLZ | 18 | 2.42 | 2.26 | 0.328 |
| Shannon (alpha) diversity at 18 months | ||||
| ΔHAZ | 14 | 5.34 | 5.47 | 0.314 |
| ΔWAZ | 16 | 5.45 | 5.59 | 0.036 |
| ΔWFLZ | 18 | 5.41 | 5.48 | 0.473 |
| Beta diversity for ASV turnover from baseline to 18 months | ||||
| ΔHAZ | 14 | 1.00 | 1.00 | 0.172 |
| ΔWAZ | 16 | 1.00 | 1.00 | 0.430 |
| ΔWFLZ | 18 | 1.00 | 1.00 | 0.632 |
| Shannon (alpha) diversity at baseline | ||||
| Alpha-1-antitrypsin | 20 | 2.30 | 2.32 | 0.901 |
| Calprotectin | 16 | 2.28 | 2.12 | 0.340 |
| Myeloperoxidase | 8 | 2.26 | 2.30 | 0.885 |
| Neopterin | 16 | 2.11 | 2.48 | 0.016 |
| Shannon (alpha) diversity at 18 months | ||||
| Alpha-1-antitrypsin | 20 | 5.41 | 5.50 | 0.442 |
| Calprotectin | 16 | 5.33 | 5.40 | 0.521 |
| Myeloperoxidase | 8 | 5.35 | 5.37 | 0.918 |
| Neopterin | 16 | 5.36 | 5.44 | 0.618 |
| Beta diversity for ASV turnover from baseline to 18 months | ||||
| Alpha-1-antitrypsin | 20 | 1.00 | 1.00 | 0.162 |
| Calprotectin | 16 | 1.00 | 1.00 | 0.447 |
| Myeloperoxidase | 8 | 1.00 | 0.99 | 0.289 |
| Neopterin | 16 | 1.00 | 1.00 | 0.692 |
ASV = amplicon sequence variant; HAZ = height for age Z-score; WAZ = weight for age Z score; WFLZ = weight length or age Z-score.
Difference in mean diversity determined by Student’s t-test.
DISCUSSION
We examined the association between markers of EE, growth, and gut microbial communities among a cohort of children from a demographic surveillance area in rural Bangladesh over an 18-month period. Elevated EE markers and poor linear growth in children were associated with changes in microbial communities in the gut. There were increased amounts of Escherichia/Shigella and Proteobacteria and decreased amounts of Prevotella consistent with the growing evidence demonstrating the relationship between intestinal inflammation, child growth, and changes in gut microbiota composition.
Our findings are consistent with previous observations. Segata et al.39 found higher bacteria in the Proteobacteria phylum in children who were stunted. In the study by Dinh et al.,40 children who were not stunted had a gut microbiome with higher Bifidobacterium longum than children who were stunted. In addition, we previously found that in this cohort, children with enterotoxigenic Escherichia. coli infections had elevated fecal calprotectin concentrations, and children with Shigella infections were at significantly higher odds of being stunted.41
We observed low diversity in ASVs among children with higher markers for EE at study enrollment. This is consistent with previous studies that have found an association between elevated EE markers and low diversity in gut microbial communities in young children.18,21,42 In in the study by Subramanian et al.23 in Bangladesh, severe acute malnutrition was also associated with reduced bacterial diversity. Low ASV diversity in children with poor growth may indicate dominance by a few bacterial species, when more species are thought to be beneficial.16
Our findings are also consistent with the theory that younger aged microbiota containing more Escherichia/Shigella are associated with poor growth. Escherichia/Shigella may be contributing to EE as part of a vicious cycle. This genera contains numerous strains known to be associated with inflammation and have enzymatic pathways that detoxify nitric oxide (NO).43–45 Nitric oxide is released by human colonic epithelial cells as part of the normal inflammatory response. Although NO can kill most bacteria without a pathway that can detoxify it, when NO is produced Escherichia/Shigella can actually thrive and potentially gain a selective advantage. By producing inflammatory biomolecules that cause human cells to produce NO, Escherichia/Shigella enters into a cycle in the human intestine that inhibits other bacteria and allows Escherichia/Shigella to proliferate, reinforcing the pattern of enteric infection, inflammation, and poor growth.41
Our study has limitations. First, we only measured fecal markers of EE at study enrollment, so we were unable to quantify their changes or consistency over time. We did not observe strong agreement between fecal markers and change in anthropometry. We also observed a tendency for children in our sample with high myeloperoxidase to have higher change in WFLZ over time, indicating more weight gain, although the expectation is that high myeloperoxidase would correspond to less weight gain. This counterintuitive result may also indicate poorer gains in height for those with high myeloperoxidase, which would similarly result in more increases in WFLZ. Second, we did not measure the nutritional intake for children in this study and, therefore, were unable to examine how child diet was directly related to gut microbiota composition in this population or to account for diet when comparing ASVs across groups of children. We also did not measure micronutrient deficiencies nor whether children were receiving supplementary nutrition. Third, we quantified ASVs from DNA isolated from stool samples, not in biopsy samples from endoscopy directly from colon ileum. Endoscopy is challenging to perform in low-resource settings such as rural Bangladesh. Fourth, we were constrained by our sample size, increasing the likelihood of differences due to sequencing depth or other technical artifacts. We did not rarefy sampled sequences as rarefication may result in the loss of useful sample data.46,47 Fifth, although gut bacteria populations are highly variable, we were not able to measure all variability in gut microbiota over the study period, since we only measured gut microbiota composition at two time points for each child.
Our study contributes to building hypotheses and informing future studies relating to the gastrointestinal microbiota of young children and how to prevent poor growth in this and other similarly vulnerable populations in low- and middle-income countries.
Supplemental Appendices
Note: Supplemental appendices appear at www.ajtmh.org.
REFERENCES
- 1.United Nations Children’s Fund, World Health Organization , 2015. World Bank Group Joint Child Malnutrition Estimates. Levels and Trends in Child Malnutrition: Key Findings of the 2015 Edition. Global Database on Child Growth and Malnutrition Geneva, Switzerland: World Health Organization. [Google Scholar]
- 2.Berkman DS, Lescano AG, Gilman RH, Lopez SL, Black MM, 2002. Effects of stunting, diarrhoeal disease, and parasitic infection during infancy on cognition in late childhood: a follow-up study. Lancet 359: 564–571. [DOI] [PubMed] [Google Scholar]
- 3.Walker SP, Chang SM, Powell CA, Simonoff E, Grantham-McGregor SM, 2007. Early childhood stunting is associated with poor psychological functioning in late adolescence and effects are reduced by psychosocial stimulation. J Nutr 137: 2464–2469. [DOI] [PubMed] [Google Scholar]
- 4.Lin A, et al. 2013. Household environmental conditions are associated with enteropathy and impaired growth in rural Bangladesh. Am J Trop Med Hyg 89: 130–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.George CM, et al. 2015. Geophagy is associated with environmental enteropathy and stunting in children in rural Bangladesh. Am J Trop Med Hyg 92: 1117–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.George CM, et al. 2015. Fecal markers of environmental enteropathy are associated with animal exposure and caregiver hygiene in Bangladesh. Am J Trop Med Hyg 93: 269–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Humphrey JH, 2009. Child undernutrition, tropical enteropathy, toilets, and handwashing. Lancet 374: 1032–1035. [DOI] [PubMed] [Google Scholar]
- 8.Guerrant RL, Oria RB, Moore SR, Oria MO, Lima AA, 2008. Malnutrition as an enteric infectious disease with long-term effects on child development. Nutr Rev 66: 487–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kosek M, et al. 2013. Fecal markers of intestinal inflammation and permeability associated with the subsequent acquisition of linear growth deficits in infants. Am J Trop Med Hyg 88: 390–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu JR, Sheng XY, Hu YQ, Yu XG, Westcott JE, Miller LV, Krebs NF, Hambidge KM, 2012. Fecal calprotectin levels are higher in rural than in urban Chinese infants and negatively associated with growth. BMC Pediatr 12: 129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Campbell DI, Elia M, Lunn PG, 2003. Growth faltering in rural Gambian infants is associated with impaired small intestinal barrier function, leading to endotoxemia and systemic inflammation. J Nutr 133: 1332–1338. [DOI] [PubMed] [Google Scholar]
- 12.Campbell DI, McPhail G, Lunn PG, Elia M, Jeffries DJ, 2004. Intestinal inflammation measured by fecal neopterin in Gambian children with enteropathy: association with growth failure, Giardia lamblia, and intestinal permeability. J Pediatr Gastroenterol Nutr 39: 153–157. [DOI] [PubMed] [Google Scholar]
- 13.Lunn PG, Northrop-Clewes CA, Downes RM, 1991. Intestinal permeability, mucosal injury, and growth faltering in Gambian infants. Lancet 338: 907–910. [DOI] [PubMed] [Google Scholar]
- 14.Weisz AJ, Manary MJ, Stephenson K, Agapova S, Manary FG, Thakwalakwa C, Shulman RJ, Manary MJ, 2012. Abnormal gut integrity is associated with reduced linear growth in rural Malawian children. J Pediatr Gastroenterol Nutr 55: 747–750. [DOI] [PubMed] [Google Scholar]
- 15.George CM, et al. 2016. Unsafe child feces disposal is associated with environmental enteropathy and impaired growth. J Pediatr 176: 43–49. [DOI] [PubMed] [Google Scholar]
- 16.Richard SA, et al. 2019. Enteric dysfunction and other factors associated with attained size at 5 years: MAL-ED birth cohort study findings. Am J Clin Nutr 110: 131–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kane AV, Dinh DM, Ward HD, 2015. Childhood malnutrition and the intestinal microbiome. Pediatr Res 77: 256–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kosek MN, MAL-ED Network Investigators , 2017. Causal pathways from enteropathogens to environmental enteropathy: findings from the MAL-ED birth cohort study. EBioMedicine 18: 109–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pop M, et al. 2014. Diarrhea in young children from low-income countries leads to large-scale alterations in intestinal microbiota composition. Genome Biol 15: R76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hurlbert SH, 1971. The nonconcept of species diversity: a critique and alternative parameters. Ecology 52: 577–586. [DOI] [PubMed] [Google Scholar]
- 21.Gough EK, Stephens DA, Moodie EE, Prendergast AJ, Stoltzfus RJ, Humphrey JH, Manges AR, 2015. Linear growth faltering in infants is associated with Acidaminococcus sp. and community-level changes in the gut microbiota. Microbiome 3: 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Monira S, et al. 2011. Gut microbiota of healthy and malnourished children in Bangladesh. Front Microbiol 2: 228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Subramanian S, et al. 2014. Persistent gut microbiota immaturity in malnourished Bangladeshi children. Nature 510: 417–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Campbell RK, et al. 2017. Biomarkers of environmental enteric dysfunction among children in rural Bangladesh. J Pediatr Gastroenterol Nutr 65: 40–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schaible UE, Kaufmann SH, 2007. Malnutrition and infection: complex mechanisms and global impacts. PLoS Med 4: e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kotloff KL, et al. 2012. The Global Enteric Multicenter Study (GEMS) of diarrheal disease in infants and young children in developing countries: epidemiologic and clinical methods of the case/control study. Clin Infect Dis 55 (Suppl_4): S232–S245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qiagen SAT , 2010. QIAamp DNA Stool Handbook, 2nd edition Available at: http://www.qiagen.com/. Accessed July 1, 2019. [Google Scholar]
- 28.Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJ, Holmes SP, 2016. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods 13: 581–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang Q, Garrity GM, Tiedje JM, Cole JR, 2007. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73: 5261–5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO, 2012. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41: D590–D596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Onis M, 2006. WHO Child Growth Standards based on length/height, weight and age. Acta Paediatr 95: 76–85. [DOI] [PubMed] [Google Scholar]
- 32.Anders S, Huber W, 2012. Differential Expression of RNA-Seq Data at the Gene Level–The DESeq Package. Heidelberg, Germany: European Molecular Biology Laboratory (EMBL). [Google Scholar]
- 33.Clarke KR, Somerfield PJ, Chapman MG, 2006. On resemblance measures for ecological studies, including taxonomic dissimilarities and a zero-adjusted Bray–Curtis coefficient for denuded assemblages. J Exp Mar Biol Ecol 330: 55–80. [Google Scholar]
- 34.Murtagh F, Legendre P, 2014. Ward’s hierarchical agglomerative clustering method: which algorithms imp.lement Ward’s criterion? J Classif 31: 274–295. [Google Scholar]
- 35.Hill MO, 1973. Diversity and evenness: a unifying notation and its consequences. Ecology 54: 427–432. [Google Scholar]
- 36.Koleff P, Gaston KJ, Lennon JJ, 2003. Measuring beta diversity for presence-absence data. J Anim Ecol 72: 367–382. [Google Scholar]
- 37.Whittaker RH, 1960. Vegetation of Siskiyou Mountains, Oregon and California. Ecol Monogr 30: 279–338. [Google Scholar]
- 38.Dixon P, 2003. VEGAN, a package of R functions for community ecology. J Veg Sci 14: 927–930. [Google Scholar]
- 39.Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, Huttenhower C, 2011. Metagenomic biomarker discovery and explanation. Genome Biology 12: R60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dinh DM, et al. 2016. Longitudinal analysis of the intestinal microbiota in persistently stunted young children in south India. PLoS One 11: e0155405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.George CM, et al. 2018. Enteric infections in young children are associated with environmental enteropathy and impaired growth. Trop Med Int Health 23: 26–33. [DOI] [PubMed] [Google Scholar]
- 42.Kristensen KH, Wiese M, Rytter MJ, Özçam M, Hansen LH, Namusoke H, Friis H, Nielsen DS, 2016. Gut microbiota in children hospitalized with oedematous and non-oedematous severe acute malnutrition in Uganda. PLoS Negl Trop Dis 10: e0004369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Spees AM, Lopez CA, Kingsbury DD, Winter SE, Bäumler AJ, 2013. Colonization resistance: battle of the bugs or ménage à trois with the host? PLoS Pathog 9: e1003730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sorbara MT, Pamer EG, 2018. Interbacterial mechanisms of colonization resistance and the strategies pathogens use to overcome them. Mucosal Immunol 12: 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stecher B, et al. 2007. Salmonella enterica serovar typhimurium exploits inflammation to compete with the intestinal microbiota. PLoS Biol 5: 2177–2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McMurdie PJ, Holmes S, 2014. Waste not, want not: why rarefying microbiome data is inadmissible. PLoS Comput Biol 10: e1003531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Willis A, 2016. Extrapolating abundance curves has no predictive power for estimating microbial biodiversity. Proc Natl Acad Sci USA 113: E5096. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.