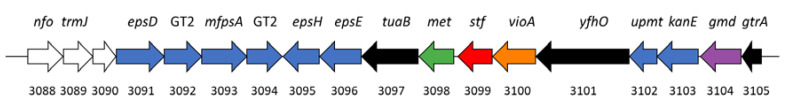
Figure 5.
Putative sulfated glycosaminoglycan biosynthesis gene cluster in strain F21T. Locus tag numbers without the “SCARR_” prefix are indicated below the genes. Genes encoding glycosyltransferases are marked blue, auxiliary proteins marked black, methyltransferases marked green, sulfotransferases marked red, transaminases marked orange, dehydratases marked purple and genes of other or unknown function are marked white. Genes with the prefix eps are homologs with putative EPS biosynthesis glycosyltransferases from Bacillus subtilis strain 168. Abbreviations from left to right: nfo, apurinic endonuclease; trmJ, tRNA methyltransferase; epsD, GT4; mfpsA, mannosylfructose-phosphate synthase (GT4); epsH, GT2; epsE, GT2; tuaB, teichuronic acid biosynthesis protein; met, methyltransferase; stf, sulfotransferase; vioA, dTDP-4-amino-4,6-dideoxy-D-glucose transaminase; yfhO, bacterial membrane protein; upmt, undecaprenyl-phosphate mannosyltransferase (GT2); kanE, α-D-kanosaminyltransferase (GT4); gmd, GDP-mannose 4,6-dehydratase; gtrA, GtrA-like protein.