Abstract
In this study, fresh seafood in retail markets was investigated for the antibiotic susceptibility patterns of the faecal indicator Escherichia coli and distribution of important β-lactamase encoding genes. E. coli were isolated from 50 (37 fish and 13 shellfish) fresh seafood samples and studied with respect to the phenotypic and genotypic characters of antibiotic resistance. Of 475 E. coli isolates from fresh seafood, 71.58% exhibited extended-spectrum β-lactamase (ESBL)-positive phenotypes. A high percentage of isolates were resistant to indicator cephalosporins cefotaxime (95%), cefpodoxime (90.88%) and ceftazidime (90.29%). Relatively higher susceptibilities were recorded against imipenem (74.41%), cefoxitin (66.76%) and meropenem (51.18%). The multiple antibiotic resistance (MAR) index of 97.35% of the isolates was above 0.18. The ESBL genes blaCTX-M, blaSHV and blaTEM were detected in 62.37%, 23.35% and 2.6% of E. coli isolates, respectively. The ESBL-producing isolates also harboured the metallo-β-lactamase-encoding genes blaOXA (7.06%), blaNDM (4.42%) and blaVIM (0.88%). This study highlights the risk of dissemination of multidrug resistant E. coli in seafood consumer communities and also the need to improve the hygiene of the coastal waters, landing centres and the retail markets.
Keywords: MAR index, India, beta lactamase, MBL, disc diffusion, seafood, PCR
1. Introduction
Escherichia coli is a common inhabitant of the gastrointestinal tract of both humans and animals. Although E. coli is popular as an indicator of faecal contamination of water and foods, it is a significant food-borne human pathogen [1]. Contamination of seafood with E. coli as a consequence of contamination of coastal waters with domestic sewage is common in densely populated countries like India [2,3]. Such pathogenic E. coli can cause a range of infections, such as gastroenteritis, wound infections, septicaemia, urinary tract infections, etc. [4].
Resistance to multiple, clinically relevant antibiotics in E. coli has severely compromised the efficacy of antibiotic chemotherapy. Multidrug-resistant (MDR) E. coli is not restricted to clinical settings alone. Since humans and animals are sources of contamination of aquatic environments, MDR E. coli can be often found in seafood. Seafood as a vehicle of MDR E. coli is a serious concern, since such bacteria can be introduced via seafood to distant geographical regions, where such resistance patterns were unknown before [5]. Globally, beta-lactam antibiotics are among the most commonly prescribed drugs, which include penicillins, cephalosporins, cephamycins and carbapenems. In E. coli, as well as in several other members of the Enterobacteriaceae, production of one or more types of β-lactamases that hydrolyse β-lactam antibiotics is a key mechanism of resistance [6,7]. Among these, the extended-spectrum β-lactamases (ESBLs) can hydrolyse various types of β-lactam antibiotics, including third generation cephalosporins and monobactams, and confer resistance to bacteria against these antibiotics [7,8,9]. ESBLs have evolved from TEM-1, TEM-2 and SHV-1 β-lactamases by point mutation, and are generally susceptible to β-lactamase inhibitors (e.g., clavulanates and sulbactams) [10]. The second group of ESBLs, which are also derived from the classical TEM and SHV enzymes, are not susceptible to antibiotic-inhibitor combinations, except the piperacillin–tazobactam combination [10,11]. A relatively new category of ESBLs represented by CTX-M has a high affinity for cefotaxime and has been divided into five groups with more than 40 variants widely distributed throughout the world [12,13]. The CTX-M enzymes have independently evolved from chromosomally encoded β-lactamases of Kluyvera spp. and are susceptible to clavulanic acid [13].
Carbapenem-resistant Enterobacteriaceae (CRE) produce one or more carbapenemases which have varying abilities to hydrolyse carbapenems and differential susceptibilities to inhibition by clavulanic acid. These belong to the Ambler class A (KPC), class B lactamases, such as New Delhi metallo-β-lactamase (NDM), imipenemase (IMP), Verona integron-encoded metallo-β-lactamase (VIM) and the Class D oxacillinases (OXA-48/-181) [14,15]. CRE are resistant to most β-lactam antibiotics and often exhibit resistance to other antimicrobial classes, thus critically narrowing down the treatment options against them. Since these resistance markers are commonly located on mobile genetic elements, CRE are also important reservoirs of resistance genes that can be transmitted to clinical and environmental bacteria [16].
Seafood contamination of E. coli is primarily due to the faecal contamination of coastal waters. Post-harvest contamination of seafood with E. coli occurs in fish landing centres and markets. Past studies from India have reported the isolation of various pathogroups of E. coli from seafood [3,17,18,19] and also the presence of highly antibiotic resistant E. coli, such as the blaNDM-harbouring strains [20,21,22]. In the study reported here, we investigated the prevalence of antibiotic resistance in Escherichia coli isolated from seafood and the genetic factors responsible for resistance. The study highlights extensive occurrence of multidrug-resistant E. coli in seafood, which can be a potential health risk to seafood consumers and handlers as well.
2. Materials and Methods
2.1. Sample Collection, Isolation and Identification of Escherichia coli From Seafood
Fresh seafood samples (n = 50) comprising of fish (37) and shrimps (13) were collected from retail markets in north-western Mumbai, India, during October 2016 to February 2018. The samples were transported immediately to the laboratory in a chilled condition in sterile containers and processed for the isolation of E. coli. Equal portions from the head, belly and tail regions of finfish were pooled and 25 g of this pooled sample was used for analysis. Clams were opened using a sterile scalpel and the meat was collected from 2–3 specimens and pooled. Similarly, in the case of shrimps, the meat was collected and pooled. Twenty-five grams of the pooled meat was homogenized in 225 mL of tryptone broth (Hi-Media, Mumbai, India) in a stomacher (Seward Stomacher 80, Lab system, London, UK), serially diluted in tryptone water and spread plated on MacConkey agar (Hi-Media, Mumbai, India). Typical pink colonies, 3–5 from the useful dilution, were purified on Luria Bertani (LB) agar. Oxidase-negative isolates were subjected to biochemical tests for the identification of E. coli [23].
2.2. Antibiotic Susceptibility Tests
The production of extended-spectrum β-lactamase (ESBL) by E. coli isolates was detected by spotting on the chromogenic medium Hi-Chrome ESBL agar (Hi-Media, Mumbai, India), on which ESBL-positive isolates appear as blue/bluish-green colonies. Presumptive ESBL+ E. coli isolates were screened for resistance against the indicator cephalosporins cefotaxime (CTX; 30 µg), ceftazidime (CAZ; 30 µg) and cefpodoxime (CPD; 10 µg) by disk diffusion test. Resistant isolates were further tested against cefoxitin (CX; 30 µg), imipenem (IPM; 10 µg), meropenem (MRP; 10 µg), ertapenem (ETP; 10 µg), aztreonam (AT; 30 µg), amoxicillin/clavulanic acid (AMC; 30 µg), piperacillin/tazobactam (PIT; 100/10 µg) and ciprofloxacin (CIP; 5 µg). The zones of inhibitions were measured and interpreted as resistant or susceptible according to Clinical and Laboratory Standards Institute guidelines [24]. The multiple antibiotic resistance (MAR) index was determined as the ratio of the total antibiotics used to the number of antibiotics to which the bacterium was resistant [25].
2.3. Detection of ESBL and MBL Phenotypes
For phenotypic detection of ESBL production, the double disc synergy test was followed [26]. The test bacterium was grown in Mueller–Hinton (MH) broth to 0.5 McFarland units and a 0.1 mL of the broth culture was spread plated on MH agar. An amoxicillin/clavulanic acid (30/10 µg) disc was placed at the centre of the plate and ceftazidime (30 µg), cefotaxime (30 µg) and cefpodoxime (10 µg) discs were placed 20–30 mm away from the central disc. An extension in the zone of inhibition around the peripheral disc towards the centrally placed amoxicillin/clavulanic acid disc by at least 5 mm was considered as due to ESBL production. For further confirmation of an ESBL+ phenotype, the Triple ESBL detection Ezy MICTM Strip (Hi-Media, Mumbai, India) was used according to the manufacturer’s instructions. The upper half of the strip designated as “mixture+” is coated with ceftazidime, cefotaxime and cefepime plus clavulanic acid, while the lower half of the strip designated as “mixture” is coated with the same antibiotics in a reverse concentration gradient, but without an inhibitor. The strip was placed on the lawn culture of each test strain on MH agar and minimum inhibitory concentrations (MIC)s were read from the mixture and mixture+ ends of the strip. The isolate was considered ESBL-positive if the mix/mix+ value was ≥8. AmpC β-lactamase was inhibited by adding 200 µg/mL cloxacillin in the agar medium.
For detection of metallo-β-lactamase (MBL) production, the double disc diffusion synergy test was used [27]. The test bacterium was grown in Muller–Hinton (MH) broth as described above and spread onto MH agar plates. Two imipenem discs (10 μg), one of which contained 10 μL of 0.1M EDTA (292 μg), were placed 25 mm apart on the inoculated MH plates. MBL production was defined by a zone of inhibition of ≥ 4 mm around the imipenem–EDTA disc compared to the disc containing imipenem alone.
2.4. Detection of Antibiotic Resistance Genes
For detection of known antibiotic resistance genes, primers and PCR protocols previously described for blaCTX-M, [28], blaSHV and blaTEM [29], blaNDM [30,31], blaOXA [32] and blaVIM [33] were used (Table S1). Bacterial DNA was extracted using the Wizard DNA kit (Promega, Madison, WI, USA) and PCR amplifications were done in a SimpliAmp thermal cycler (Thermo Fisher Scientific, Waltham, MA, USA).
3. Results and Discussion
Antibiotic-resistant bacteria are frequently encountered in raw seafood in India [20,34,35,36] and the incidence is alarmingly high in seafood harvested off the Mumbai coast [22,37]. The coastal city of Mumbai is densely populated, and the numerous creeks that lead to the sea act as conduits for draining untreated sewage. Every day, an estimated 2000 million litres of sewage is released into the Arabian sea. Direct contamination occurs from the population that live on the beaches without proper sanitation facilities. Retail markets too lack basic facilities to prevent contamination of fish and maintain hygiene. As a result, contamination of seafood with pathogenic and antibiotic-resistant bacteria is expected to occur right from the harvesting stage, till it reaches the consumer.
In this study, we investigated the occurrence of antibiotic-resistant E. coli in seafood, which directly represents the human cause of seafood contamination. The details of 50 seafood samples (37 fish and 13 shellfish) analysed for the presence of E. coli are shown in the Table 1. Overall, 896 bacterial colonies were isolated from MacConkey agar, of which 475 were identified as E. coli.
Table 1.
Sample-wise distribution of Escherichia coli and ESBL+ isolates.
| Sl. No. | Samples | No. Analysed | No. of E. coli Isolated | No. (%) ESBL+ E. coli |
|---|---|---|---|---|
| Finfish | ||||
| 1. | Sardinella longiceps | 5 | 32 | 24 (75) |
| 2. | Terapon jarbua | 5 | 29 | 29 (100) |
| 3. | Otolithes cuvieri | 3 | 19 | 19 (100) |
| 4. | Epinephelus diacanthus | 2 | 15 | 15 (100) |
| 5. | Nemipterus randalli | 2 | 22 | 14 (64.28) |
| 6. | Mene maculata | 2 | 24 | 13 (53.84) |
| 7. | Coilia dussumieri | 3 | 45 | 32 (71.87) |
| 8. | Harpadon nehereus | 4 | 42 | 35 (82.85) |
| 9. | Trichiurus lepturus | 2 | 21 | 12 (58.33) |
| 10. | Priacanthus hamrur | 2 | 19 | 13 (69.23) |
| 11. | Megalaspis cordyla | 2 | 25 | 25 (100) |
| 12. | Anodontostoma chacunda | 3 | 13 | 13 (100) |
| 13. | Pampus argenteus | 2 | 19 | 17 (88.23) |
| Shellfish | ||||
| 14. | Acetes indicus | 5 | 56 | 27 (48.21) |
| 15. | Metapenaeus dobsoni | 3 | 41 | 19 (73.68) |
| 16. | Meretrix meretrix | 3 | 26 | 18 (44.44) |
| 17. | Loligo duvauceli | 2 | 27 | 15 (26.66) |
| Total | 50 | 475 | 340 (71.58) |
3.1. Distribution of ESBL+ E. coli in Seafood
Out of a total of 475 E. coli isolates, 340 (71.58%) were ESBL+ on the chromogenic medium (Table 1). Among 325 finfish isolates of E. coli, 261(80.30%) were ESBL+ and 79 (52.66%) out of 150 E. coli isolates from shellfish were also ESBL+. All the samples yielded at least one ESBL+ isolate, and all E. coli isolates from fish samples Terapon jarbua, Otolithes cuvieri, Epinephelus diacanthus, Megalaspis cordyla and Anodontostoma chacunda were ESBL+ (Table 1).
3.2. Antibiotic Resistance Profiles of E. coli
The antibiotic resistance patterns of 340 ESBL+ isolates are shown in Table 2. Studies reporting isolation of ESBL-producing E. coli from seafood in India are rare. We recently reported that 78.6% of the enterobacteria isolates from fresh seafood in Mumbai, India, were ESBL-positive [22]. ESBL+ Gram-negative bacteria were also isolated from effluents of a fish processing plant in Mangalore, India [38]. The latter study suggests that the contaminated fish can again be a source of environmental contamination with antibiotic-resistant bacteria, highlighting the need to treat effluents from fish processing plants. In Germany, ESBL- and AmpC-producing Enterobacteriaceae were detected in 21.3% of seafood samples, with Klebsiella pneumoniae and E. coli being the predominant species [39]. Sellera and colleagues [40] reported isolation of ESBL (CTX-M)-producing E. coli from wild fishes of a polluted area in the South Atlantic coast of Brazil. ESBL-producing E. coli of identical genotypes have been reported from fish, chicken and infected patients in Cambodia [41].
Table 2.
Antibiotic susceptibility profiles of ESBL+ E. coli (n = 340) isolated in this study.
| Antibiotic Tested | No. (%) Resistant | No. (%) Intermediate Resistant | No. (%) Susceptible |
|---|---|---|---|
| Cefoxitin (CX) 30 mcg | 66 (19.41%) | 47 (13.82%) | 227 (66.76%) |
| Cefotaxime (CTX) 30 mcg | 323 (95%) | 12 (3.53%) | 5 (1.47%) |
| Ceftazidime (CAZ) 30mcg | 307 (90.29%) | 26 (7.65%) | 7 (2.05%) |
| Cefpodoxime (CPD) 10 mcg | 309 (90.88%) | 17 (5.0%) | 14 (4.11%) |
| Imipenem (IPM) 10mcg | 31 (9.11%) | 56 (16.47%) | 253 (74.41%) |
| Ertapenem (ETP) 10 mcg | 107 (31.47%) | 81 (23.82%) | 152 (44.71%) |
| Meropenem (MRP) 10 mcg | 87 (25.58%) | 79 (23.24%) | 174 (51.18%) |
| Ciprofloxacin (CIP) 5mcg | 152 (44.71%) | 43 (12.65%) | 145 (42.65%) |
| Aztreonam (AT) 30 mcg | 219 (64.41%) | 57 (16.76%) | 64 (18.82%) |
| Amoxyclav (AMC) 30 mcg | 127 (37.35%) | 83 (24.41%) | 130 (38.24%) |
| Piperacillin/Taz 100/10mcg | 114 (33.53%) | 95 (27.94%) | 131 (38.53%) |
3.3. Molecular Characterization of ESBL+ E. coli
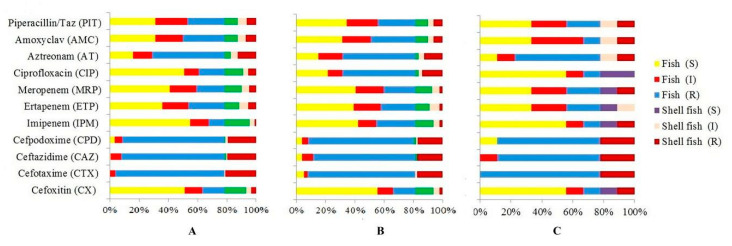
PCR investigation was carried out to identify the molecular mechanisms of antibiotic resistance in E. coli isolates of this study. The results are shown in Table 3. Among the ESBL genes tested in this study, blaCTX-M was detected in the highest number of isolates (212, 62.35%). Sample-wise isolation of blaCTX-M+ E. coli is shown in Table 3. Finfish and shellfish yielded 166 (78.30%) and 46 (21.70%) of blaCTX-M+ isolates, respectively. The highest percentage of blaCTX-M+ E. coli was isolated from Sardinella longiceps (91.66%), followed by Otolithes cuvieri (89.47%), while the least percentage of such isolates was found in squid Loligo duvauceli (20%) (Table 3). The isolates exhibited highest resistance to cefotaxime (158 isolates, 74.53%). A high percentage of isolates (116 isolates, 69.88%) were susceptible to imipenem (Figure 1A). The isolates were largely susceptible to carbapenems and ciprofloxacin. The blaSHV gene was detected in 79 (23.35%) isolates, 64 of which were from finfish and 15 from shellfish (Table 3). The isolates were commonly resistant to cefotaxime and ceftazidime (Figure 1B). Fifty-four (44%) isolates were susceptible to cefoxitin (Figure 1B). The blaTEM gene was detected in the least number of isolates (9, 2.64%) (Table 3). The antibiotic susceptibility patterns of blaTEM-positive E. coli are shown in Figure 1C.
Table 3.
Distribution of β-lactamase genes in Escherichia coli isolated in this study.
| Samples Analysed (No.) | No. of E. coli Isolated | No. (%) ESBL+ | No. (%) CTX-M+ | No. (%) SHV+ | No. (%) TEM+ | No. (%) OXA+ | No. (%) VIM+ | No. (%) NDM+ |
|---|---|---|---|---|---|---|---|---|
| Sardinella longiceps (5) | 32 | 24 (75) | 22 (91.66) | 3 (12.5) | 3 (12.5) | 3 (12.5) | 1 (3.13) | 3 (9.38) |
| Terapon jarbua (5) | 29 | 29 (100) | 18 (62.06) | 1 (3.45) | - | 1 (3.45) | 1 (3.45) | - |
| Otolithes cuvieri (3) | 19 | 19 (100) | 17 (89.47) | 3 (15.79) | 1(5.26) | 1(5.26) | 1 (5.26) | 3 (15.78) |
| Epinephelus diacanthus (2) | 15 | 15 (100) | 13 (86.66) | - | 1 (6.67) | 1(6.67) | - | 1 (6.66) |
| Nemipterus randalli (2) | 22 | 14 (64.28) | 5 (35.71) | 5 (35.71) | - | - | - | 2 (9.09) |
| Mene maculata (2) | 24 | 13 (53.84) | 5 (38.46) | 2 (15.38) | - | - | - | - |
| Coilia dussumieri (3) | 45 | 32 (71.87) | 25 (78.13) | 7 (21.86) | 1 (3.13) | 7 (21.88) | - | 3 (6.66) |
| Harpadon nehereus (4) | 42 | 35 (82.85) | 27 (77.14) | 7 (20) | 1 (2.86) | 4 (11.43) | - | 1 (2.38) |
| Trichiurus lepturus (2) | 21 | 12 (58.33) | 5 (41.67) | 9 (75) | - | - | - | 2 (9.52) |
| Priacanthus hamrur (2) | 19 | 13 (69.23) | 3 (23.08) | 7 (53.85) | - | - | - | - |
| Megalaspis cordyla (2) | 25 | 25 (100) | 9 (36) | 11 (44) | - | 1 (4) | - | 2 (8) |
| Anodontostoma chacunda (3) | 13 | 13 (100) | 4 (30.77) | - | - | 1 (7.69) | - | - |
| Pampus argenteus (2) | 19 | 17 (88.23) | 13 (76.47) | 9 (52.94) | - | 2 (11.76) | - | - |
| Acetes indicus (5) | 27 | 27 (100) | 23 (85.19) | 12 (44.44) | - | 1 (3.70) | - | 2 (7.4) |
| Metapenaeus dobsoni (3) | 26 | 19 (73.68) | 11 (57.89) | 2 (10.53) | 2 (10.53) | 1 (5.26) | - | - |
| Meretrix meretrix (3) | 41 | 18 (44.44) | 9 (50) | 1 (5.56) | - | 1 (5.56) | - | 2 (4.88) |
| Loligo duvauceli (2) | 56 | 15 (26.66) | 3 (20) | - | - | - | - | - |
| Total | 475 | 340 (71.58) | 212 (62.35) | 79 (21.35) | 9 (2.65) | 24 (7.06) | 3 (0.88) | 21 (4.42) |
Figure 1.
Antibiotic resistance profiles of E. coli harbouring blaCTX-M (n = 212; fish 166, shellfish 46) (A), blaSHV (n = 79; fish 64, shellfish 15) (B) and blaTEM (n = 9; fish 7, shellfish 2) (C). R = resistant, I = Intermediate resistant and S = susceptible.
The CTX-M group of enzymes, which are commonly found in E. coli, confer resistance to cefotaxime, ceftriaxone and aztreonam [12,15]. Although CTX-M was originally found to be less active against ceftazidime, its variants, such CTX-M-15, hydrolyse ceftazidime efficiently [42,43,44]. In our study, 141 of 212 (66%) blaCTX-M-positive E. coli were susceptible to cefoxitin (Figure 1A). ESBL-producing E. coli is generally susceptible to cephamycins, thus making cephamycins, such as cefoxitin, viable options for treating ESBL-producing E. coli infections [45]. However, cefoxitin resistance due to loss of a porin has been reported in K. pneumonia and E. coli [46]. TEM, SHV and CTX-M-producing organisms have been reported from a variety of food-producing animals, such as poultry, swine, bovine, horse, rabbit, ostrich, wild boars, etc. [47].
In the present study, the blaCTX-M-harbouring E. coli were predominantly isolated compared to the blaSHV- or blaTEM-harbouring E. coli. The CTX-M group has the highest number of variants, with E. coli being the major producer of this enzyme [48]. The extraordinary mobility of plasmid and/transposon-located blaCTX-M genes within clones of E. coli that are successful as pathogens has made CTX-M β-lactamase the most dominant ESBL currently [48]. The high prevalence of E. coli producing CTX-M β-lactamase in seafood might reflect on the local antibiotic consumption patterns as well as specific resistant clones circulating in the community.
3.4. Detection of Carbapenemase-Encoding Genes
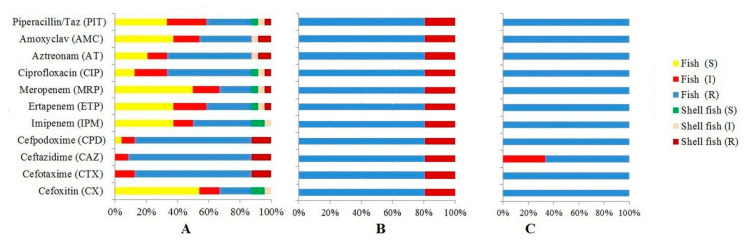
Carbapenem-resistant Enterobacteriaceae (CRE) have become the biggest challenge in healthcare settings, as these bacteria are recalcitrant to most of the common antibiotics used. In our study, 21 (4.42%) isolates, 17 from fish and 4 from shellfish, harboured the blaNDM gene (Table 3). The isolates were resistant to all tested antibiotics except colistin (Figure 2B). The present study strengthens our previous report of an alarmingly high occurrence of blaNDM-harbouring E. coli in fresh seafood [21,22].
Figure 2.
Antibiotic resistance profiles of Escherichia coli isolates harbouring blaOXA (n = 24; fish 21, shellfish 3) (A), blaNDM (n = 21, fish 17, shellfish 4 (B) and blaVIM (n = 3, fish 3, shellfish 0) (C) genes. R = resistant, I = Intermediate resistant and S = susceptible.
The blaOXA and blaVIM genes were detected in 7.06% and 0.88% of the isolates, respectively. The antibiotic resistance profiles of these isolates are shown Figure 2. The blaVIM was first reported in Pseudomonas aeruginosa [49]. Of the three isolates harbouring blaVIM, two isolates were resistant to all 11 antibiotics, while one had intermediate resistant to ceftazidime (Figure 2C). The VIM type metallo-β-lactamases are mostly reported from Pseudomonas aeruginosa, and from several non-related bacterial species like Achromobacter xylosoxidans, E. coli, Klebsiella pneumoniae, etc., which suggests its horizontal transfer ability among non-related species [50]. Worldwide, blaVIM-2 has been reported to be the dominant MBL variant [51]. Studies from India have reported VIM lactamase producing clinical isolates of P. aeruginoasa [52], Acinetobacter baumannii [53] and E. coli [54]. We did not find any reports of food-associated Gram-negative bacteria harbouring VIM and OXA type β-lactamase genes from India. Elsewhere, a recent study has reported the presence of blaVIM in a food-associated Vibrio parahaemolyticus [55]. In our study, we screened E. coli isolates for ESBL production first using indicator cephalosporins, and this might have resulted in underestimation of certain ceftazidime-susceptible carbapenemase-producing E. coli (e.g., OXA-48) that lack ESBL and AmpC enzymes [56].
3.5. Multidrug Resistance in Seafood E. coli
Of the 340 isolates, 331 (97.35%) were resistant to two or more antibiotics with a MAR index of >0.18 (Table 4). A MAR index of more than 0.2 suggests contamination from high-risk sources where antibiotics are extensively used [25], and the majority of our isolates belonged to this category. All (100%) of the E. coli isolates recovered from nine samples were multidrug-resistant. These samples consisted of fish (Sardinella longiceps, Otolithes cuvieri, Epinephelus diacanthus, Mene maculata, Priacanthus hamrur and Pampus argenteus) and shellfish, such as crustaceans (Acetes indicus and Metapenaeus dobsoni) and clams (Meretrix meretrix). Twenty-one E. coli isolates were resistant to the entire range of 11 antibiotics tested.
Table 4.
Multiple antibiotic resistance (MAR) indices of Escherichia coli.
| No. of Antibiotic | MAR Index | No. (%) of Isolates |
|---|---|---|
| 1 | 0.09 | 9 (2.64) |
| 2 | 0.18 | 7 (2.06) |
| 3 | 0.27 | 71 (20.88) |
| 4 | 0.36 | 11 (3.24) |
| 5 | 0.45 | 102 (30) |
| 6 | 0.55 | 13 (3.82) |
| 7 | 0.64 | 53 (15.59) |
| 9 | 0.82 | 41 (12.06) |
| 11 | 1.00 | 21 (6.18) |
In this study, we analysed 50 samples of fish and shellfish that belonged to 17 different species. Since we isolated multiple isolates of E. coli from a single sample, we selected one E. coli isolate from each species of fish or shellfish that was most resistant, 17 isolates in total. The antibiotic resistance profiles and the genotypes are given in the Table 5. Ten isolates were resistant to 11 antibiotics and susceptible to only colistin. Two isolates harboured all six β-lactamase genes tested. Other isolates showed high but varying levels of resistance to the tested antibiotics, and also harboured multiple β-lactamase genes.
Table 5.
Characteristics of the E. coli isolates exhibiting the highest resistance phenotypes from each sample type.
| Isolate No. | Source | Resistance Genotype | Antibiotic Resistance Profile | Antibiotic to which Susceptible |
|---|---|---|---|---|
| EC21 | Sardinella longiceps | blaCTX-M, blaSHV, blaTEM, blaNDM, blaOXA, blaVIM | CX, CTX, CAZ, CPD, IPM, ETP, MRP, CIP, AT, AMC, PIT | CL |
| EC123 | Terapon jarbua | blaCTX-M, blaSHV, blaOXA,blaVIM | CX, CTX, CAZ, CPD, MRP, AT | IPM, CIP, ETP, AMC, PIT |
| EC31 | Otolithes cuvieri | blaCTX-M, blaSHV, blaTEM, blaNDM, blaOXA, blaVIM | CX, CTX, CAZ, CPD, IPM, ETP, MRP, CIP, AT, AMC, PIT | CL |
| EC91 | Epinephelus diacanthus | blaCTX-M, blaTEM, blaNDM, blaOXA | CX, CTX, CAZ, CPD, IPM, ETP, MRP, CIP, AT, AMC, PIT | CL |
| EC81 | Nemipterus randalli | blaCTX-M, blaSHV, blaNDM | CX, CTX, CAZ, CPD, IPM, ETP, MRP, CIP, AT, AMC, PIT | CL |
| EC13 | Mene maculata | blaCTX, blaSHV | CX, CTX, CAZ, CPD, CIP, AT | IPM, MRP, ETP, AMC, PIT |
| EC221 | Coilia dussumieri | blaCTX-M, blaSHV, blaTEM, blaNDM, blaOXA | CX, CTX, CAZ, CPD, IPM, ETP, MRP, CIP, AT, AMC, PIT | CL |
| EC201 | Harpadon nehereus | blaCTX-M, blaSHV, blaTEM, blaNDM, blaOXA | CX, CTX, CAZ, CPD, IPM, ETP, MRP, CIP, AT, AMC, PIT | CL |
| EC303 | Trichiurus lepturus | blaCTX-M, blaSHV, blaNDM | CX, CTX, CAZ, CPD, IPM, ETP, MRP, CIP, AT, AMC, PIT | CL |
| EC48 | Priacanthus hamrur | blaCTX, blaSHV | CX, CTX, CAZ, CPD, CIP, AT, AMC, PIT | IPM, ETP, MRP |
| EC51 | Megalaspis cordyla | blaCTX-M, blaSHV, blaNDM, blaOXA | CX, CTX, CAZ, CPD, IPM, ETP, MRP, CIP, AT, AMC, PIT | CL |
| EC253 | Anodontostoma chacunda | blaCTX, blaOXA | CX, CTX, CAZ, CPD, CIP, AT, ETP | IPM, MRP, AMC, PIT |
| EC271 | Pampus argenteus | blaCTX-M, blaSHV, blaOXA | CX, CTX, CAZ, CPD, CIP, AT, PIT | IPM, ETP, MRP, AMC |
| EC281 | Acetes indicus | blaCTX-M, blaSHV, blaNDM, blaOXA | CX, CTX, CAZ, CPD, IPM, ETP, MRP, CIP, AT, AMC, PIT | CL |
| EC253 | Metapenaeus dobsoni | blaCTX-M, blaSHV, blaTEM, blaOXA | CX, CTX, CAZ, CPD, IPM, CIP, AT, AMC, PIT | IPM, AMC, PIT |
| EC305 | Meretrix meretrix | blaCTX-M, blaSHV, blaNDM, blaOXA | CX, CTX, CAZ, CPD, IPM, ETP, MRP, CIP, AT, AMC, PIT | CL |
| EC131 | Loligo duvauceli | bla CTX-M | CX, CTX, CAZ, CPD, CIP | IPM, ETP, MRP, AMC, PIT |
4. Conclusions
The resistance patterns of E. coli observed in this study suggests anthropogenic sources of contamination of seafood. Release of raw or partially treated sewage into fishing areas is a major concern that requires the attention of the local regulatory bodies. Seafood can be a source of dissemination of antibiotic-resistant bacteria into the community. Incidence of MDR bacteria can also potentially jeopardise the international fish trade. In this regard, urgent measures are necessary to implement suitable policy to monitor the quality of coastal waters, ensure hygiene in retail fish markets and establish suitable testing methods to detect MDR bacteria in seafood.
Acknowledgments
The authors thank Director, ICAR-CIFE, Mumbai for help in carrying out this work. Thanks to K.V. Rajendran and A.K. Balange for their advice during the course of this research.
Supplementary Materials
The following are available online at https://www.mdpi.com/2306-7381/7/2/46/s1, Table S1: Oligonucleotide primers used in this study for the detection of antibiotic resistance genes.
Author Contributions
Conceptualization, S.H.K. and B.B.N.; supervision, S.H.K. and B.B.N.; methodology, S.H.K., B.B.N. and A.S.S.; bench work, A.S.S.; manuscript writing and editing, S.H.K. and B.B.N. All authors have read and agreed to the published version of the manuscript.
Funding
No external funding received for this work. The work was done under student (AS) research programme funded by the university (ICAR-CIFE, Mumbai).
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Jang J., Hur H.-G., Sadowsky M.J., Byappanahalli M.N., Yan T., Ishii S. Environmental Escherichia coli: Ecology and public health implications-a review. J. Appl. Microbiol. 2017;123:570–581. doi: 10.1111/jam.13468. [DOI] [PubMed] [Google Scholar]
- 2.Kumar H.S., Karunasagar I., Karunasagar I., Teizou T., Shima K., Yamasaki S. Characterisation of Shiga toxin-producing Escherichia coli (STEC) isolated from seafood and beef. FEMS Microbiol. Lett. 2004;233:173–178. doi: 10.1016/j.femsle.2004.02.008. [DOI] [PubMed] [Google Scholar]
- 3.Prakasan S., Prabhakar P., Lekshmi M., Kumar S., Nayak B.B. Isolation of Shiga toxin-producing Escherichia coli harboring variant Shiga toxin genes from seafood. Vet. World. 2018;11:379–385. doi: 10.14202/vetworld.2018.379-385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gomes T.A.T., Elias W.P., Scaletsky I.C.A., Guth B.E.C., Rodrigues J.F., Piazza R.M.F., Ferreira L.C.S., Martinez M.B. Diarrheagenic Escherichia coli. Braz. J. Microbiol. 2016;47(Suppl. 1):3–30. doi: 10.1016/j.bjm.2016.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lekshmi M., Ammini P., Kumar S., Varela M.F. The food production environment and the development of antimicrobial resistance in human pathogens of animal origin. Microorganisms. 2017;5:11. doi: 10.3390/microorganisms5010011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pitout J.D.D. Multiresistant Enterobacteriaceae: New threat of an old problem. Expert Rev. Anti-Infect. Ther. 2008;6:657–669. doi: 10.1586/14787210.6.5.657. [DOI] [PubMed] [Google Scholar]
- 7.Sheng W.-H., Badal R.E., Hsueh P.-R. SMART Program Distribution of extended-spectrum β-lactamases, AmpC β-lactamases, and carbapenemases among Enterobacteriaceae isolates causing intra-abdominal infections in the Asia-Pacific region: Results of the study for Monitoring Antimicrobial Resistance Trends (SMART) Antimicrob. Agents Chemother. 2013;57:2981–2988. doi: 10.1128/AAC.00971-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hawkey P.M. Prevalence and clonality of extended-spectrum beta-lactamases in Asia. Clin. Microbiol. Infect. 2008;14(Suppl. 1):159–165. doi: 10.1111/j.1469-0691.2007.01855.x. [DOI] [PubMed] [Google Scholar]
- 9.McDanel J., Schweizer M., Crabb V., Nelson R., Samore M., Khader K., Blevins A.E., Diekema D., Chiang H.-Y., Nair R., et al. Incidence of extended-spectrum β-lactamase (ESBL)-producing Escherichia coli and Klebsiella Infections in the United States: A systematic literature review. Infect. Control Hosp. Epidemiol. 2017;38:1209–1215. doi: 10.1017/ice.2017.156. [DOI] [PubMed] [Google Scholar]
- 10.Paterson D.L., Bonomo R.A. Extended-spectrum beta-lactamases: A clinical update. Clin. Microbiol. Rev. 2005;18:657–686. doi: 10.1128/CMR.18.4.657-686.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Drieux L., Brossier F., Sougakoff W., Jarlier V. Phenotypic detection of extended-spectrum beta-lactamase production in Enterobacteriaceae: Review and bench guide. Clin. Microbiol. Infect. 2008;14(Suppl. 1):90–103. doi: 10.1111/j.1469-0691.2007.01846.x. [DOI] [PubMed] [Google Scholar]
- 12.Bradford P.A. Extended-spectrum β-lactamases in the 21st century: Characterization, epidemiology, and detection of this important resistance threat. Clin. Microbiol. Rev. 2001;14:933–951. doi: 10.1128/CMR.14.4.933-951.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bush K. Past and Present Perspectives on β-Lactamases. Antimicrob. Agents Chemother. 2018;62 doi: 10.1128/AAC.01076-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nordmann P., Dortet L., Poirel L. Carbapenem resistance in Enterobacteriaceae: Here is the storm! Trends Mol. Med. 2012;18:263–272. doi: 10.1016/j.molmed.2012.03.003. [DOI] [PubMed] [Google Scholar]
- 15.Tzouvelekis L.S., Tzelepi E., Tassios P.T., Legakis N.J. CTX-M-type beta-lactamases: An emerging group of extended-spectrum enzymes. Int. J. Antimicrob. Agents. 2000;14:137–142. doi: 10.1016/S0924-8579(99)00165-X. [DOI] [PubMed] [Google Scholar]
- 16.Lutgring J.D., Limbago B.M. The Problem of Carbapenemase-Producing-Carbapenem-Resistant-Enterobacteriaceae Detection. J. Clin. Microbiol. 2016;54:529–534. doi: 10.1128/JCM.02771-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sanath Kumar H., Otta S.K., Karunasagar I., Karunasagar I. Detection of Shiga-toxigenic Escherichia coli (STEC) in fresh seafood and meat marketed in Mangalore, India by PCR. Lett. Appl. Microbiol. 2001;33:334–338. doi: 10.1046/j.1472-765X.2001.01007.x. [DOI] [PubMed] [Google Scholar]
- 18.Thampuran N., Surendraraj A., Surendran P.K. Prevalence and characterization of typical and atypical Escherichia coli from fish sold at retail in Cochin, India. J. Food Prot. 2005;68:2208–2211. doi: 10.4315/0362-028X-68.10.2208. [DOI] [PubMed] [Google Scholar]
- 19.Sehgal R., Kumar Y., Kumar S. Prevalence and geographical distribution of Escherichia coli O157 in India: A 10-year survey. Trans. R. Soc. Trop. Med. Hyg. 2008;102:380–383. doi: 10.1016/j.trstmh.2008.01.015. [DOI] [PubMed] [Google Scholar]
- 20.Kumar H.S., Parvathi A., Karunasagar I., Karunasagar I. Prevalence and antibiotic resistance of Escherichia coli in tropical seafood. World J. Microbiol. Biotechnol. 2005;21:619–623. doi: 10.1007/s11274-004-3555-8. [DOI] [Google Scholar]
- 21.Singh A.S., Lekshmi M., Nayak B.B., Kumar S.H. Isolation of Escherichia coli harboring blaNDM-5 from fresh fish in India. J. Microbiol. Immunol. Infect. 2016;49:822–823. doi: 10.1016/j.jmii.2014.11.004. [DOI] [PubMed] [Google Scholar]
- 22.Sanjit Singh A., Lekshmi M., Prakasan S., Nayak B.B., Kumar S. Multiple antibiotic-resistant, extended spectrum-β-lactamase (ESBL)-producing enterobacteria in fresh seafood. Microorganisms. 2017;5:53. doi: 10.3390/microorganisms5030053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.MacFaddin J.F. Biochemical Tests for Identification of Medical Bacteria. 3rd ed. Lippincott Williams & Wilkins; Philadelphia, PA, USA: 2000. [Google Scholar]
- 24.CLSI . Performance Standards for Antimicrobial Susceptibility Testing. 28th ed. Clinical and Laboratory Standards Institute; Wayne, PA, USA: 2018. CLSI supplement M100. [Google Scholar]
- 25.Krumperman P.H. Multiple antibiotic resistance indexing of Escherichia coli to identify high-risk sources of fecal contamination of foods. Appl. Environ. Microbiol. 1983;46:165–170. doi: 10.1128/AEM.46.1.165-170.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.CLSI . Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Fourth Informational Supplement Performance Standards for Antimicrobial Susceptibility Testing. Document M100-24. Clinical and Laboratory Standards Institute (CLSI); Wayne, PA, USA: 2014. [Google Scholar]
- 27.Nordmann P., Poirel L., Carrër A., Toleman M.A., Walsh T.R. How to detect NDM-1 producers. J. Clin. Microbiol. 2011;49:718–721. doi: 10.1128/JCM.01773-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bonnet R. Growing group of extended-spectrum beta-lactamases: The CTX-M enzymes. Antimicrob. Agents Chemother. 2004;48:1–14. doi: 10.1128/AAC.48.1.1-14.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sharma J., Sharma M., Ray P. Detection of TEM & SHV genes in Escherichia coli & Klebsiella pneumoniae isolates in a tertiary care hospital from India. Indian J. Med. Res. 2010;132:332–336. [PubMed] [Google Scholar]
- 30.Hornsey M., Phee L., Wareham D.W. A novel variant, NDM-5, of the New Delhi metallo-β-lactamase in a multidrug-resistant Escherichia coli ST648 isolate recovered from a patient in the United Kingdom. Antimicrob. Agents Chemother. 2011;55:5952–5954. doi: 10.1128/AAC.05108-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mushtaq S., Irfan S., Sarma J.B., Doumith M., Pike R., Pitout J., Livermore D.M., Woodford N. Phylogenetic diversity of Escherichia coli strains producing NDM-type carbapenemases. J. Antimicrob. Chemother. 2011;66:2002–2005. doi: 10.1093/jac/dkr226. [DOI] [PubMed] [Google Scholar]
- 32.Siu L.K., Lo J.Y., Yuen K.Y., Chau P.Y., Ng M.H., Ho P.L. beta-lactamases in Shigella flexneri isolates from Hong Kong and Shanghai and a novel OXA-1-like beta-lactamase, OXA-30. Antimicrob. Agents Chemother. 2000;44:2034–2038. doi: 10.1128/AAC.44.8.2034-2038.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shirani K., Ataei B., Roshandel F. Antibiotic resistance pattern and evaluation of metallo-beta lactamase genes (VIM and IMP) in Pseudomonas aeruginosa strains producing MBL enzyme, isolated from patients with secondary immunodeficiency. Adv. Biomed. Res. 2016:5. doi: 10.4103/2277-9175.1869866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Adesiji Y.O., Deekshit V.K., Karunasagar I. Antimicrobial-resistant genes associated with Salmonella spp. isolated from human, poultry, and seafood sources. Food Sci. Nutr. 2014;2:436–442. doi: 10.1002/fsn3.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Deekshit V.K., Kumar B.K., Rai P., Karunasagar I., Karunasagar I. Differential expression of virulence genes and role of gyrA mutations in quinolone resistant and susceptible strains of Salmonella Weltevreden and Newport isolated from seafood. J. Appl. Microbiol. 2015;119:970–980. doi: 10.1111/jam.12924. [DOI] [PubMed] [Google Scholar]
- 36.Deekshit V.K., Kumar B.K., Rai P., Srikumar S., Karunasagar I., Karunasagar I. Detection of class 1 integrons in Salmonella Weltevreden and silent antibiotic resistance genes in some seafood-associated nontyphoidal isolates of Salmonella in south-west coast of India. J. Appl. Microbiol. 2012;112:1113–1122. doi: 10.1111/j.1365-2672.2012.05290.x. [DOI] [PubMed] [Google Scholar]
- 37.Das U.N., Singh A.S., Lekshmi M., Nayak B.B., Kumar S. Characterization of blaNDM-harboring, multidrug-resistant Enterobacteriaceae isolated from seafood. Environ. Sci. Pollut. Res. Int. 2019;26:2455–2463. doi: 10.1007/s11356-018-3759-3. [DOI] [PubMed] [Google Scholar]
- 38.Divyashree M., Kumar D.V., Ballamoole K.K., Shetty V., Chakraborty A., Karunasagar I. Occurrence of antibiotic resistance among gram-negative bacteria isolated from effluents of fish processing plants in and around Mangalore. Int. J. Environ. Health Res. 2019:1–8. doi: 10.1080/09603123.2019.1618799. [DOI] [PubMed] [Google Scholar]
- 39.Vu T.T.T., Alter T., Roesler U., Roschanski N., Huehn S. Investigation of extended-spectrum and AmpC β-lactamase-producing Enterobacteriaceae from retail seafood in Berlin, Germany. J. Food Prot. 2018;81:1079–1086. doi: 10.4315/0362-028X.JFP-18-029. [DOI] [PubMed] [Google Scholar]
- 40.Sellera F.P., Fernandes M.R., Moura Q., Carvalho M.P.N., Lincopan N. Extended-spectrum-β-lactamase (CTX-M)-producing Escherichia coli in wild fishes from a polluted area in the Atlantic Coast of South America. Mar. Pollut. Bull. 2018;135:183–186. doi: 10.1016/j.marpolbul.2018.07.012. [DOI] [PubMed] [Google Scholar]
- 41.Nadimpalli M., Vuthy Y., de Lauzanne A., Fabre L., Criscuolo A., Gouali M., Huynh B.-T., Naas T., Phe T., Borand L., et al. Meat and fish as sources of extended-spectrum β-lactamase-producing Escherichia coli, Cambodia. Emerg. Infect. Dis. 2019;25 doi: 10.3201/eid2501.180534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Baraniak A., Fiett J., Hryniewicz W., Nordmann P., Gniadkowski M. Ceftazidime-hydrolysing CTX-M-15 extended-spectrum β-lactamase (ESBL) in Poland. J. Antimicrob. Chemother. 2002;50:393–396. doi: 10.1093/jac/dkf151. [DOI] [PubMed] [Google Scholar]
- 43.Naas T., Oxacelay C., Nordmann P. Identification of CTX-M-type extended-spectrum-β-lactamase genes using real-time PCR and pyrosequencing. Antimicrob. Agents Chemother. 2007;51:223–230. doi: 10.1128/AAC.00611-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pitout J.D.D., Nordmann P., Laupland K.B., Poirel L. Emergence of Enterobacteriaceae producing extended-spectrum beta-lactamases (ESBLs) in the community. J. Antimicrob. Chemother. 2005;56:52–59. doi: 10.1093/jac/dki166. [DOI] [PubMed] [Google Scholar]
- 45.Lepeule R., Ruppé E., Le P., Massias L., Chau F., Nucci A., Lefort A., Fantin B. Cefoxitin as an alternative to carbapenems in a murine model of urinary tract infection due to Escherichia coli harboring CTX-M-15-type extended-spectrum β-lactamase. Antimicrob. Agents Chemother. 2012;56:1376–1381. doi: 10.1128/AAC.06233-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ananthan S., Subha A. Cefoxitin resistance mediated by loss of a porin in clinical strains of Klebsiella pneumoniae and Escherichia coli. Indian J. Med. Microbiol. 2005;23:20–23. doi: 10.4103/0255-0857.13867. [DOI] [PubMed] [Google Scholar]
- 47.Mesa R.J., Blanc V., Blanch A.R., Cortés P., González J.J., Lavilla S., Miró E., Muniesa M., Saco M., Tórtola M.T., et al. Extended-spectrum beta-lactamase-producing Enterobacteriaceae in different environments (humans, food, animal farms and sewage) J. Antimicrob. Chemother. 2006;58:211–215. doi: 10.1093/jac/dkl211. [DOI] [PubMed] [Google Scholar]
- 48.Cantón R., González-Alba J.M., Galán J.C. CTX-M enzymes: Origin and diffusion. Front. Microbiol. 2012;3 doi: 10.3389/fmicb.2012.00110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lauretti L., Riccio M.L., Mazzariol A., Cornaglia G., Amicosante G., Fontana R., Rossolini G.M. Cloning and characterization of blaVIM, a new integron-borne metallo-beta-lactamase gene from a Pseudomonas aeruginosa clinical isolate. Antimicrob. Agents Chemother. 1999;43:1584–1590. doi: 10.1128/AAC.43.7.1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Walsh T.R. The emergence and implications of metallo-beta-lactamases in Gram-negative bacteria. Clin. Microbiol. Infect. 2005;11(Suppl. 6):2–9. doi: 10.1111/j.1469-0691.2005.01264.x. [DOI] [PubMed] [Google Scholar]
- 51.Cornaglia G., Giamarellou H., Rossolini G.M. Metallo-β-lactamases: A last frontier for β-lactams? Lancet Infect. Dis. 2011;11:381–393. doi: 10.1016/S1473-3099(11)70056-1. [DOI] [PubMed] [Google Scholar]
- 52.Castanheira M., Bell J.M., Turnidge J.D., Mathai D., Jones R.N. Carbapenem resistance among Pseudomonas aeruginosa strains from India: Evidence for nationwide endemicity of multiple metallo-β-lactamase clones (VIM-2, -5, -6, and -11 and the newly characterized VIM-18) Antimicrob. Agents Chemother. 2009;53:1225–1227. doi: 10.1128/AAC.01011-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Subramaniyan J.S., Sundaram J.M. Occurrence of bla genes encoding carbapenem-resistant Pseudomonas aeruginosa and Acinetobacter baumannii from intensive care unit in a tertiary care hospital. J. Lab. Physicians. 2018;10:208. doi: 10.4103/JLP.JLP_108_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nagaraj S., Chandran S.P., Shamanna P., Macaden R. Carbapenem resistance among Escherichia coli and Klebsiella pneumoniae in a tertiary care hospital in south India. Indian J. Med. Microbiol. 2012;30:93. doi: 10.4103/0255-0857.93054. [DOI] [PubMed] [Google Scholar]
- 55.Zheng Z., Ye L., Chan E.W.-C., Chen S. Identification and characterization of a conjugative blaVIM-1-bearing plasmid in Vibrio alginolyticus of food origin. J. Antimicrob. Chemother. 2019;74:1842–1847. doi: 10.1093/jac/dkz140. [DOI] [PubMed] [Google Scholar]
- 56.Rodríguez-Baño J., Gutiérrez-Gutiérrez B., Machuca I., Pascual A. Treatment of Infections Caused by Extended-Spectrum-Beta-Lactamase-, AmpC-, and Carbapenemase-Producing Enterobacteriaceae. Clin. Microbiol. Rev. 2018:31. doi: 10.1128/CMR.00079-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.