Abstract
Background
Preclinical evidence has demonstrated that common intratumor bacteria metabolize the chemotherapeutic drug gemcitabine. The significance of this bacterial metabolism pathway, relative to the known metabolic pathways by host enzymes, is not known. We hypothesized that bacterial metabolism is clinically significant and that “knockdown” by antibacterial therapy has the unintended effect of increasing the effective dose of gemcitabine, thereby increasing the risk for gemcitabine‐associated toxicities.
Materials and Methods
We reanalyzed the comparator arm of the MPACT trial (NCT01442974), made available through Project Data Sphere, LLC (CEO Roundtable on Cancer's Life Sciences Consortium, Cary, NC; www.projectdatasphere.org). In this arm, 430 patients with metastatic pancreatic adenocarcinoma were treated with gemcitabine. We used the Anderson‐Gill survival model to compare the risk of developing an adverse event after antibacterial prescription with time unexposed to antibacterials. Adverse events of grade 3 and greater were considered at three levels of granularity: all aggregated into one endpoint, aggregated by class, and taken individually. Antibiotic exposures were analyzed in aggregate as well as by class.
Results
Antibacterial exposure was associated with an increased risk of adverse events (hazard ratio [HR]: 1.77; confidence interval [CI]: 1.46–2.14), any hematologic adverse event (HR: 1.64; CI: 1.26–2.13), and any gastrointestinal adverse event (HR: 2.14; CI: 1.12–4.10) but not a constitutional (HR: 1.33; CI: 0.611–2.90) or hepatologic adverse event (HR: 0.99; CI: 0.363–2.71). Among specific adverse events, antibacterial exposure was associated with an increased risk of anemia (HR: 3.16; CI: 1.59–6.27), thrombocytopenia (HR: 2.52; CI: 1.31–4.85), leukopenia (HR: 3.91; CI: 1.46–10.5), and neutropenia (HR: 1.53; CI: 1.07–2.17) but not any other specific adverse events.
Conclusion
Antibacterial exposure was associated with an increased risk of gemcitabine‐associated, dose‐limiting adverse events, including aggregate hematologic and gastrointestinal events, as well as four specific hematologic adverse events, suggesting that intratumor bacteria may be responsible for a clinically significant portion of gemcitabine metabolism. Alternative avenues of evidence will be necessary to confirm this preliminary finding and assess its generalizability. There is plentiful opportunity for similar analyses on other clinical trial data sets, where gemcitabine or other biomimetic small molecules were used.
Implications for Practice
Patients treated with gemcitabine for metastatic pancreatic ductal adenocarcinoma have an increased rate of gemcitabine‐associated toxicity during and after antibiotic therapy. This observation is consistent with preclinical evidence that intratumor bacteria metabolize gemcitabine to an inactive form. Further research is needed to determine whether this observation merits any changes in clinical practice.
Keywords: Adverse events, Tumor microbiome, Chemotherapy, Toxicity, Gemcitabine
Short abstract
Previous reports have suggested a potential interaction between gemcitabine and the host microbiome. By reanalyzing an arm of a clinical trial in which patients were treated with gemcitabine, this article provides observational clinical evidence consistent with this potential interaction.
Introduction
Gemcitabine is a nucleoside analogue used in the treatment of advanced pancreatic, bladder, breast, ovarian, and non‐small cell lung cancer 1. Geller et al. recently found that bacteria expressing the long isoform of cytidine deaminase (CDDL) metabolize gemcitabine to its inactive form in vitro. Furthermore, they found that a mouse model of colon carcinoma injected with CDDL‐expressing bacteria could be successfully treated with gemcitabine only when cotreated with an antibacterial to wipe out the bacterial colonization 2. Notably, the bacterial family Enterobacteriaceae expresses CDDL 2, is commonly found in the gut microbiome 3, and was found in appreciable quantities in 30 of 65 pancreatic tumor samples sequenced 2, providing a plausible source for the intratumor bacterial colonization via retrograde migration up the pancreatic duct.
These results suggest a potential interaction between gemcitabine and the host microbiome and provide a plausible biological mechanism for a clinically significant interaction between nucleoside analogues and antibacterials that have activity against CDDL‐expressing bacteria. To date, however, no such clinical evidence has been reported.
We hypothesized that intratumor bacteria metabolize a clinically meaningful fraction of administered gemcitabine. Based on this hypothesis, antibacterial therapy that reduces the population of CDDL‐expressing bacteria was expected to increase the effective dose of gemcitabine. We sought to test this hypothesis by reanalyzing a clinical trial data set in which patients were treated with gemcitabine and incidentally cotreated with antibacterials. The possible effects of an increased dose of gemcitabine include an increase in clinical efficacy (survival, an increase in tumor response) and an increase in adverse events. We elected to focus our study on adverse events because of their perceived greater sensitivity to gemcitabine dose. Furthermore, the development of adverse events is of immediate clinical relevance in this population because adverse events are often dose‐limiting and it has been shown that the patients who discontinued treatment owing to adverse events had shorter overall survival and shorter disease‐free survival than patients who continued treatment until disease progression 4.
Materials and Methods
We identified the comparator arm of the MPACT trial (NCT01442974), freely available through Project Data Sphere, LLS (CEO Roundtable on Cancer's Life Sciences Consortium, Cary, NC; www.projectdatasphere.org) 5, 6, as an appropriate data source to investigate the possible interaction between antibacterial exposure and gemcitabine toxicity.
Patients with metastatic pancreatic cancer were randomized to receive either gemcitabine or gemcitabine plus nab‐paclitaxel. The gemcitabine comparator arm contained 430 patients who were treated with gemcitabine at a dose of 1,000 mg per square meter of body surface area weekly for 7 of the first 8 weeks after randomization (cycle one) and 3 of each subsequent 4 weeks (cycles two and subsequent) until disease progression 7. Baseline and demographic characteristics of this population are shown in Table 1.
Table 1.
Baseline characteristics of patients in the comparator arm of the MPACT trial
| Characteristic | Patient statistics, n (%) |
|---|---|
| Age, years | |
| Median | 63 |
| Range | 32–88 |
| Sex | |
| Female | 173 (40) |
| Male | 257 (60) |
| Race (self‐reported) | |
| White | 401 (93) |
| Black | 16 (4) |
| Asian | 9 (2) |
| Other | 4 (1) |
| Region | |
| North America | 271 (63) |
| Eastern Europe | 62 (14) |
| Australia | 59 (14) |
| Western Europe | 38 (9) |
| Karnofsky Performance Status | |
| 100 | 70 (16) |
| 90 | 191 (45) |
| 80 | 135 (31) |
| 70 | 33 (8) |
| 60 | 0 (0) |
| Pancreatic tumor location | |
| Head | 180 (42) |
| Body | 136 (32) |
| Tail | 110 (26) |
| Unknown | 4 (1) |
| Number of organs with metastasis | |
| 1 | 21 (5) |
| 2 | 206 (48) |
| 3 | 140 (33) |
| 4+ | 63 (15) |
| Metastasis in: | |
| Liver | 360 (84) |
| Lung | 184 (43) |
| Bone | 18 (4) |
| Peritoneum | 10 (2) |
Over the course of the MPACT trial, physicians prescribed antibacterials as clinically indicated. We considered patients “exposed” from the first day of their first antibiotic prescription until they left the trial. As a sensitivity analysis, we also considered a “temporary exposure” framework, where patients were considered exposed only during the time period they were prescribed antibiotics plus an optional lag period of 1 or 2 weeks (supplemental online Table 2). In both frameworks, adverse events that started the same day as an antibiotic exposure were considered to have started during an unexposed time period.
We considered antibacterial exposure at two levels of granularity. First, we considered an aggregate “any antibacterial” exposure. Second, we considered antibacterials separated into the most common classes used in the MPACT trial: quinolones, penicillins, nonpenicillin beta‐lactams, and other.
Over the course of the MPACT trial, adverse events were noted at clinic visits. We considered only adverse events of grade 3 or greater. We considered adverse events at three levels of granularity: (1) an aggregate of “any adverse event,” (2) any one of four subgroups of adverse events: hematologic, gastrointestinal, constitutional, and hepatologic, and (3) each of the 20 most common adverse events individually.
We conducted a survival analysis on pairs of (a) antibacterial exposure and (b) adverse event when two criteria were met: at least 15 adverse events were observed and at least 5 adverse events were observed in exposed patients. These criteria were set to ensure stability of the effect estimates.
For the 12 pairs that met these criteria, we used the Andersen‐Gill survival model 8 to assess the association between antibacterial exposure and the risk of developing each adverse event. This statistical model extends the Cox proportional hazards model 9 by accommodating recurrent events. We included age, sex, race, and baseline performance status as covariates to control for their effects on outcomes.
Reproducibility
All data analyzed here are freely available on Project DataSphere, and all analysis scripts are available at https://github.com/rcorty/gemcitabine.
Results
After collapsing overlapping prescriptions, 179 patients experienced 271 antibacterial exposure periods covering 2,588 patient‐days. After collapsing overlapping adverse events, there were 537 adverse event periods of grade 3+, of which 268 were hematologic, 48 were gastrointestinal, 33 were constitutional, and 21 were hepatologic. The frequency of common specific adverse events is detailed in Table 2.
Table 2.
For each of the adverse events of grade 3+ for which results are presented, the number of patients who experienced that adverse event, the total number of experiences, and the total duration for which that adverse event was experienced
| Adverse eventa | Patients, n (%) | No. of events | Total duration, days | Incidence rate, events per 1,000 patient‐days |
|---|---|---|---|---|
| Any adverse event | 264 (61) | 537 | 5108 | 11.5 |
| Any hematologic | 131 (30) | 268 | 2547 | 5.8 |
| Neutropenia | 81 (19) | 155 | 1403 | 3.3 |
| Thrombocytopenia | 29 (7) | 45 | 354 | 1 |
| Anemia | 30 (7) | 44 | 459 | 0.9 |
| Neutropenia | 16 (4) | 30 | 270 | 0.6 |
| Leukopenia | 13 (3) | 21 | 198 | 0.5 |
| Any gastrointestinal | 39 (9) | 48 | 279 | 1 |
| Abdominal pain | 20 (5) | 26 | 131 | 0.6 |
| Any constitutional | 28 (7) | 33 | 354 | 0.7 |
| Fatigue | 19 (4) | 23 | 271 | 0.5 |
| Any hepatologic | 18 (4) | 21 | 251 | 0.5 |
| AST elevated | 12 (3) | 14 | 160 | 0.3 |
A longer version of this table that includes the many adverse events for which insufficient data were available to stably estimate the effect of antibacterial exposure is included in supplemental online Table 1.
Adverse events were defined based on patient report and physician recording, rather than by lab values.
Abbreviation: AST, aspartate aminotransferase.
In the top‐level analysis, the hazard ratio of antibacterial exposure on “any adverse event” was at 1.77 with 95% confidence interval (CI) of 1.46–2.14 and a t test p value of 1.7 × 10−9, as shown in Figure 1.
Figure 1.
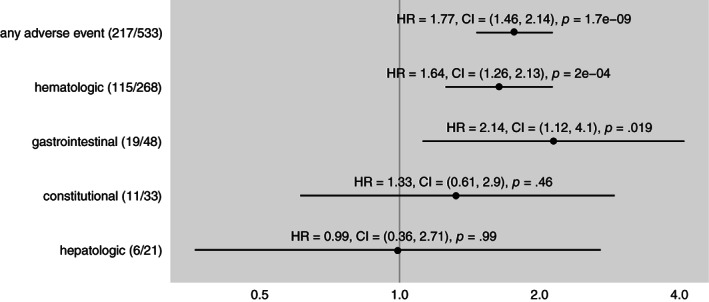
Hazard ratio of antibacterial exposure for the development any adverse event of grade 3 or greater and each of the four most common categories of adverse events in the comparator (gemcitabine alone) arm of the MPACT trial. Dot indicates point estimate of hazard ratio and line indicates 95% confidence interval. In parentheses after the adverse event name, numerator is the number of occurrences of the adverse event during antibacterial exposure and denominator is the number of occurrences of the adverse event overall. p values from t test. All adverse events are derived from the adverse events recorded by the investigators, not by lab values. Abbreviations: CI, confidence interval; HR, hazard ratio.
In the level two analysis, antibacterial exposure was associated with a statistically significant increased risk of developing hematologic (hazard ratio [HR]: 1.64; CI: 1.26–2.13; p = 2 × 10−4) and gastrointestinal (HR: 2.14; CI: 1.12–4.1; p = .019) adverse events, but not constitutional (HR: 1.33; CI: 0.61–2.9; p = .46) or hepatologic (HR: 0.99; CI: 0.36–2.71; p = .99) adverse events, as shown in Figure 1.
In the level three analysis, antibacterial exposure was associated with a statistically significant increased risk of developing all hematologic adverse for which hazard ratios were estimable: neutropenia (HR: 1.53; CI: 1.07–2.17; p = .017), thrombocytopenia (HR: 2.52; CI: 1.31–4.85; p = .005), anemia (HR: 3.16; CI: 1.59–6.27; p < .0001), and leukopenia (HR: 3.91; CI: 1.46–10.46; p = .0056), as shown in Figure 2. Among gastrointestinal, constitutional, and hepatologic adverse events, antibacterial exposure was not associated with a statistically significant increased risk of any specific adverse event: abdominal pain (HR: 1.83; CI: 0.74–4.52; p = .18), fatigue (HR: 0.98; CI: 0.36–2.68; p = .97), and aspartate aminotransferase elevation (HR: 1.43; CI: 0.45–4.57; p = .54).
Figure 2.
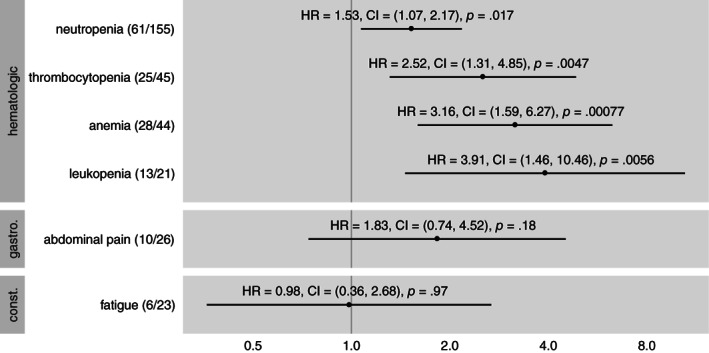
Hazard ratio of antibacterial exposure for the development of specific adverse events of grade 3 or greater, grouped by category. Dot indicates point estimate of hazard ratio and line indicates 95% confidence interval. In parentheses after the adverse event name, numerator is the number of occurrences of the adverse event during antibacterial exposure and denominator is the number of occurrences of the adverse event overall. p values from t test. All adverse events are derived from the adverse events recorded by the investigators, not by lab values. Abbreviations: CI, confidence interval; HR, hazard ratio.
In the antibacterial class–specific analyses, the only statistically significant associations identified were between the quinolone class and risk of “any adverse event” (HR: 1.53; CI: 1.22–1.91; p = .001) and between the beta‐lactam class and “any hematologic event” (HR: 1.76; CI: 1.21–2.55; p = .01; results not shown graphically).
Discussion
Preclinical evidence has demonstrated that bacteria commonly found in the gut flora and in pancreatic ductal adenocarcinoma samples can metabolize gemcitabine 2. Whether this metabolic pathway holds any clinical significance, however, is unknown. We addressed this question with reanalysis of the MPACT clinical trial 7, where, in the comparator arm, patients with advanced pancreatic cancer were treated with gemcitabine. We reasoned that if the microbial metabolic pathway consumes a significant fraction of administered gemcitabine, patients would be expected to experience a higher rate of gemcitabine‐associated toxicities after taking antibacterials, owing to the decreased microbial population.
We found that after antibacterial exposure, patients have an increased risk of developing any adverse event as well as an increased risk of developing two classes of adverse events: hematologic and gastrointestinal. Within the hematologic class, we identified four specific adverse events for which patients exposed to antibacterials had an increased risk: neutropenia, thrombocytopenia, anemia, and leukopenia. This study design and analysis cannot distinguish between pure antibiotic toxicity and antibiotic‐exacerbated gemcitabine toxicity. However, based on known patterns of drug toxicity, the increased risk for hematologic toxicities is most consistent with an antibiotic‐exacerbated gemcitabine toxicity and the increased risk for gastrointestinal toxicities is most consistent with antibiotic toxicity.
Thus, the preclinical findings of Geller et al. may be clinically significant. There remain, however, notable gaps between the two studies.
Geller et al. studied one specific bacterial species (Escherichia coli K‐12) and one specific antimicrobial (ciprofloxacin). This study makes no observation about any specific species of bacteria or any specific antibiotic. These limitations are inherent in the data set analyzed. The sample size provided little power to detect interactions between individual antibiotic classes and adverse events. The limiting factor in most cases was the number of adverse events observed during antibiotic treatment. Because the original study was conducted to assess clinical outcomes, not microbiological mechanisms, no measurement of any microbial load, diversity, or gemcitabine‐metabolizing activity was made.
Another important limitation of this study is its observational nature. The observed effect could be influenced by potential confounding due to the patient's overall health, the specific illness that motivated the antibiotic prescription, or toxicity from the antibiotic itself. We have endeavored to make our analysis as robust as possible against these concerns by adjusting for performance status, conducting the analysis in both a permanent effect and temporary effect framework, considering a variety of “lags” between exposure and effect in the temporary effect framework (supplemental online Table 2), quantifying the frequency of adverse events before antibiotic exposure (supplemental online Table 3), and comparing the timing of patient‐days spent “exposed” and “unexposed” (supplemental online Table 4). But only a randomized trial can provide conclusive evidence and fully guard against these potential threats to study validity.
Conclusion
Additional evidence to assess this hypothesis should be sought through additional observational studies with larger sample size, more elaborate statistical models, or additional, relevant measurements, such as sequenced tumor samples or chemotherapeutic concentration. Finally, whether this effect extends to other nucleoside analogues or other classes of chemotherapeutics remains to be investigated.
This study provides observational clinical evidence that patients cotreated with gemcitabine and antibiotics experience a higher effective dose of gemcitabine than patients treated with gemcitabine alone at the same nominal dose. Based on preclinical evidence, the hypothesized mechanism of this effect is decreased metabolism of gemcitabine by intratumor bacteria, possibly seeded from the gastrointestinal tract.
Recent work on the effects of antibiotic treatment on survival in patients treated with immunotherapy for melanoma 10 and a variety of other cancers 11 points to, on average, an association between antibiotic exposure and decreased survival. In the context of this rich literature of observational studies in a related clinical context, this study suggests the possibility of an effect in the other direction, where a cancer treatment may actually be more effective (and more toxic) when coadministered with antibiotics.
Author Contributions
Conception/design: Robert W. Corty, Benjamin W. Langworthy, Jason P. Fine, John B. Buse, Hanna K. Sanoff, Jennifer L. Lund
Provision of study material or patients: Robert W. Corty
Data analysis and interpretation: Robert W. Corty, Benjamin W. Langworthy
Manuscript writing: Robert W. Corty, Benjamin W. Langworthy, Jason P. Fine, John B. Buse, Hanna K. Sanoff, Jennifer L. Lund
Final approval of manuscript: Robert W. Corty, Benjamin W. Langworthy, Jason P. Fine, John B. Buse, Hanna K. Sanoff, Jennifer L. Lund
Disclosures
John B. Buse: Adocia, AstraZeneca, Dance Biopharm, Dexcom, Eli Lilly, Fractyl, GI Dynamics, Intarcia Therapeutics, Lexicon, MannKind, Metavention, NovaTarg, Novo Nordisk, Orexigen, PhaseBio, Sanofi, Senseonics, vTv Therapeutics, Zafgen (C/A [paid to institution]), AstraZeneca, Eli Lilly, Intarcia Therapeutics, Johnson & Johnson, Lexicon, Medtronic, Novo Nordisk, Sanofi, Theracos, vTv Therapeutics, PCORI, ADA (RF), Cirius Therapeutics Inc, CSL Behring, Neurimmune AG, Whole Biome (C/A), Mellitus Health, PhaseBio, Stability Health, Whole Biome (OI); Hanna K. Sanoff: Bayer, Merck (RF); Jennifer L. Lund: GlaxoSmithKline (E [spouse]). The other authors indicated no financial relationships.
(C/A) Consulting/advisory relationship; (RF) Research funding; (E) Employment; (ET) Expert testimony; (H) Honoraria received; (OI) Ownership interests; (IP) Intellectual property rights/inventor/patent holder; (SAB) Scientific advisory board
Supporting information
See http://www.TheOncologist.com for supplemental material available online.
Supplemental Tables
Acknowledgments
J.B.B. was supported by National Institutes of Health grants UL1TR002489, U01DK098246, UC4DK108612, and U54DK118612. R.W.C. was supported by National Institute of General Medical Sciences grant 2T32GM008719.
Disclosures of potential conflicts of interest may be found at the end of this article.
No part of this article may be reproduced, stored, or transmitted in any form or for any means without the prior permission in writing from the copyright holder. For information on purchasing reprints contact Commercialreprints@wiley.com. For permission information contact permissions@wiley.com.
Footnotes
For Further Reading: Nadina Tinsley, Cong Zhou, Grace Tan et al. Cumulative Antibiotic Use Significantly Decreases Efficacy of Checkpoint Inhibitors in Patients with Advanced Cancer. The Oncologist 2020;25:55–63.
Implications for Practice: Antibiotic use is negatively associated with treatment outcomes of immune checkpoint inhibitors (ICI) in advanced cancer. Cumulative antibiotic use is associated with a marked negative survival outcome. Judicious antibiotic prescribing is warranted in patients receiving treatment with ICI for treatment of advanced malignancy.
References
- 1. Hertel LW, Boder GB, Kroin JS et al. Evaluation of the antitumor activity of gemcitabine (2',2'‐difluoro‐2'‐deoxycytidine). Cancer Res 1990;50:4417–4422. [PubMed] [Google Scholar]
- 2. Geller LT, Barzily‐Rokni M, Danino T et al. Potential role of intratumor bacteria in mediating tumor resistance to the chemotherapeutic drug gemcitabine. Science 2017;357:1156–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zou Y, Xue W, Luo G et al. 1,520 reference genomes from cultivated human gut bacteria enable functional microbiome analyses. Nat Biotechnol 2019;37:179–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Vogel A, Römmler‐Zehrer J, Li JS et al. Efficacy and safety profile of nab‐paclitaxel plus gemcitabine in patients with metastatic pancreatic cancer treated to disease progression: A subanalysis from a phase 3 trial (MPACT). BMC Cancer 2016;16:817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hede K. Project data sphere to make cancer clinical trial data publicly available. J Natl Cancer Inst 2013;105:1159–1160. [DOI] [PubMed] [Google Scholar]
- 6. Green AK, Reeder‐Hayes KE, Corty RW et al. The Project Data Sphere Initiative: Accelerating cancer research by sharing data. The Oncologist 2015;20:464–e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Von Hoff DD, Ervin T, Arena FP et al. Increased survival in pancreatic cancer with nab‐paclitaxel plus gemcitabine. N Engl J Med 2013;369:1691–1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Andersen PK, Gill RD. Cox's regression model for counting processes: A large sample study. Ann Stat 1982;10:1100–1120. [Google Scholar]
- 9. Cox DR. Regression models and life‐tables. J R Stat Soc Series B Stat Methodol 1972;34:187–220. [Google Scholar]
- 10. Elkrief A, El Raichani L, Richard C et al. Antibiotics are associated with decreased progression‐free survival of advanced melanoma patients treated with immune checkpoint inhibitors. Oncoimmunology 2019;8:e1568812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Elkrief A, Derosa L, Kroemer G et al. The negative impact of antibiotics on outcomes in cancer patients treated with immunotherapy: A new independent prognostic factor? Ann Oncol 2019;30:1572–1579. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
See http://www.TheOncologist.com for supplemental material available online.
Supplemental Tables


