Abstract
MYB proteins are involved in diverse important biological processes in plants. Herein, we obtained the MYB superfamily from the allotetraploid Brassica napus, which contains 227 MYB-related (BnMYBR/Bn1R-MYB), 429 R2R3-MYB (Bn2R-MYB), 22 R1R2R3-MYB (Bn3R-MYB), and two R1R2R2R1/2-MYB (Bn4R-MYB) genes. Phylogenetic analysis classified the Bn2R-MYBs into 43 subfamilies, and the BnMYBRs into five subfamilies. Sequence characteristics and exon/intron structures within each subfamily of the Bn2R-MYBs and BnMYBRs were highly conserved. The whole superfamily was unevenly distributed on 19 chromosomes and underwent unbalanced expansion in B. napus. Allopolyploidy between B. oleracea and B. rapa mainly contributed to the expansion in their descendent B. napus, in which B. rapa-derived genes were more retained. Comparative phylogenetic analysis of 2R-MYB proteins from nine Brassicaceae and seven non-Brassicaceae species identified five Brassicaceae-specific subfamilies and five subfamilies that are lacking from the examined Brassicaceae species, which provided an example for the adaptive evolution of the 2R-MYB gene family alongside angiosperm diversification. Ectopic expression of four Bn2R-MYBs under the control of the viral CaMV35S and/or native promoters could rescue the lesser root hair phenotype of the Arabidopsis thaliana wer mutant plants, proving the conserved negative roles of the 2R-MYBs of the S15 subfamily in root hair development. RNA-sequencing data revealed that the Bn2R-MYBs and BnMYBRs had diverse transcript profiles in roots in response to the treatments with various hormones. Our findings provide valuable information for further functional characterizations of B. napus MYB genes.
Keywords: Brassica napus, MYB transcription factor, complementation, evolution, expression, root hair development
1. Introduction
MYB proteins are characterized by a conserved DNA-binding domain comprising 1–4 conserved MYB repeats (R1–R3) [1,2,3]. MYB proteins are widely distributed in eukaryotes with many more homologs in plants, which comprise a transcription factor (TF) gene superfamily in plant genomes [4,5]. This superfamily has been generally categorized into four major MYB-type families: 2R-MYB (R2R3-MYB), 3R-MYB (R1R2R3-MYB), 4R-MYB (R1R2R2R1/2-MYB), and MYB-related (MYBR/1R-MYB) families, based on the number of their MYB repeats [3]. The MYBR and 2R-MYB proteins form the largest families that usually have from dozens to hundreds of members in higher plants, whereas the other two families have much lower numbers of members and generally have only several members in some species [4,5].
The 2R-MYB genes encode the most typical plant MYB proteins. Since the first 2R-MYB gene was found to be implicated in the maize anthocyanin biosynthesis [1], the 2R-MYB family has been detected in a much wider range of plant species, and represents a large TF family in plant genomes [5,6]. Thus, a great number of recent research has focused more intensively on the roles of 2R-MYB genes in evolution [3,5,6], and the regulation of different biological processes including secondary metabolism [3,7,8], plant development (e.g., root hair development) [9,10] as well as plant responses to environmental stresses [11,12] and different types of hormones [13,14]. The MYBR proteins have also been demonstrated to play key roles in many important biochemical, developmental, and physiological processes in plants such as regulating circadian rhythms [15], anthocyanin biosynthesis [16], trichome and root hair development [17], and triacylglycerol accumulation [18]. The plant 3R-MYB proteins are homologs of animal MYB proteins [19], which play a conserved role in cell cycle regulation [19,20] as well as plant abiotic stress responses [21]. However, the functions of 4R-MYB genes remain unclear.
Genome-wide analysis of the MYB superfamily has been carried out in many plant species including Arabidopsis thaliana [22], Oryza sativa (rice) [23], and Glycine max (soybean) [6], etc. Most of these studies focused on the typical 2R-MYB family, and only a few works have addressed the whole superfamily [6,24,25]. Previously, we carried out a global analysis of the 2R-MYB families across 50 representative eukaryotes and provided a systemic classification for this family [5]. Similarly, we also assessed the MYBR families in 16 major land plants [4]. We found that these two families rapidly expanded alongside the diversification of higher plants, forming a range of new functions, with the 2R-MYBs forming many ’orphan’ genes and/or species-specific subfamilies or clades during evolution [4,5]. However, a detailed evolutionary history of the MYB superfamily in many plant genomes has not yet been elucidated by any study, since the MYB superfamily has a huge number, from hundreds to thousands, of members. Furthermore, detailed mechanisms underlying gene expansion and the subsequent evolution of plant MYB duplicates also remain unknown. Gaining deeper insights into the evolutionary history of all the MYB members in a specific evolutionary lineage of plants may have implications for an in-depth understanding of the evolution and classification of the MYB superfamily.
Brassicaceae is a large eudicot family that includes A. thaliana and many important crops for human food. This family represents one of the largest lineages with a mass of available sequenced genomes; and thus, may have potential for further research on genome duplication and evolution. In the present study, we identified and classified the MYB superfamily in allotetraploid Brassica napus (genome AnAnCnCn), and its ancestors B. rapa (genome ArAr) and B. oleracea (genome CoCo) at a genome-wide level. We focused on exploring the evolutionary history of this superfamily after the recent allopolyploidy event between B. oleracea and B. rapa. Furthermore, given an evident boosting trend of new subfamilies in the 2R-MYB family, to gain further insights into the evolutionary mechanisms of this gene family, we conducted a phylogenetic analysis of all putative 2R-MYB genes identified in 16 species (including nine Brassicaceae and seven closely related non-Brassicaceae species). These data allowed us to provide a more detailed classification and assessment of the evolutionary patterns of differentiation and proliferation of the 2R-MYB family in higher plants. As a means to study the function of several representative members of the Bn2R-MYB subfamily, on the basis of homology with the A. thaliana MYB066 (WER) gene, we selected four 2R-MYB genes for their functional analysis. Our results revealed that these four selected genes could rescue the aberrance in root hair development of the A. thaliana wer mutant plants, indicating a positive correlation between sequence homology and biological function. Finally, to identify the BnMYB genes, which act in the roots in a hormone-dependent manner, we analyzed the RNA-sequencing (RNA-seq) data of the roots of B. napus MYB plants exposed to various hormone treatments.
2. Materials and Methods
2.1. Sequence Retrieval of MYB Genes
To identify the MYB proteins in Brassica napus, BLASTP was applied to search against the proteome of B. napus (Darmor–bzh ecotype) available in GENOSCOPE [26]. To ensure that no MYB proteins were eliminated by lack of correspondence to the consensus, a representative sequence from each subfamily of the MYBR, 2R-MYB, 3R-MYB, and 4R-MYB proteins [4,5,22] was used as a query in the BLASTP search with a low-stringency stringent criterion (cutoff P < 0.1). Each matching sequence was subsequently used to search in the B. napus genome database until no new sequence was found. After removing the incomplete or redundant sequences, putative MYB protein sequences were further confirmed using the PROSITE database [27] and the SMART database [28] to confirm that the candidates possessed the typical MYB domain. The identified sequences were then considered as candidates based on the following three additional criteria: (i) the MYB domain had a significant sequence similarity to that of typical plant MYB proteins; (ii) each MYB repeat contained at least two of the three highly conserved tryptophan (W) residues; and (iii) MYB repeats were adjacent to each other [4,5]. To ensure the integrity of the data, among the sequences that possess incomplete open reading frames (ORFs), only those that have a long deletion in the MYB domain, which will impact the phylogenetic analyses (no common sites for the sequence pair), were excluded from further analyses. Finally, the positive hits were classified into four distinct MYB-types (i.e., MYBR-, 2R-MYB-, 3R-MYB- and 4R-MYB-types according to the detection of one, two, three, and four MYB repeats, respectively). Meanwhile, we also identified the MYB genes from another sequenced B. napus (cv. ZS11) genome found in the NCBI database [29], and compared the sequences of candidates identified from these two B. napus cultivars using the MEGA v5.2 [30]. All the newly identified sequences were then named according to their chromosomal location. The corresponding cDNA and genomic sequences of identified candidates were also acquired from these two genomes.
To further explore the expansion of 2R-MYB genes across the Brassicaceae family or closely related species, the 2R-MYBs in B. rapa FPsc v1.3, B. oleracea v1.0, Boechera stricta v1.2, Capsella grandiflora v1.1, Eutrema salsugineum v1.0, Citrus clementina v1.0, Citrus sinensis v1.1, Theobroma cacao v1.1, Gossypium raimondii v2.1, Carica papaya ASGPB v0.4, Capsella rubella v1.0, A. lyrata v1.0, Manihot esculenta v6.1, and Eucalyptus grandis v2.0 were identified from Phytozome v12.1 [31] using the same method. The newly identified candidate sequences for each Brassicaceae species along with the previously reported A. thaliana sequences [22] and the candidates identified in B. napus (Darmor–bzh ecotype) (this work) were collected into a preliminary dataset for further analyses.
2.2. Sequence and Phylogenetic Analyses
The intron/exon structures and intron phases of B. napus MYB genes (BnMYBs) were analyzed by aligning the genomic and coding sequence (CDS) of each gene using the MUSCLE software in MEGA v5.2 [30] with the default parameters. The gene structures of BnMYBs were then confirmed and visualized with Gene Structure Display Server 2.0 with the default parameters [32]. Subsequently, the intron patterns (including their distribution, positions and phases) of each gene were determined based on the presence of intron(s) in the genomic region spanning the MYB domain encoded by each MYB gene. This region was only used in intron pattern analysis because the length and sequence homology of the other regions were quite divergent. The physical and chemical characteristics (e.g., number of amino acids, theoretical isoelectric points (pI) and molecular weight) of each candidate protein were predicted using the Protparam tool [33] with the default parameters. Subcellular localization prediction was performed by the Plant-mPLoc online tool with the default parameters [34].
Multiple sequence alignment of the protein sequences of the MYB domains was carried out using the online MAFFT v7 under the default parameters [35], and the aligned sequences were then manually checked using the MEGA v5.2 [30]. An unrooted phylogenetic tree was constructed using the neighbor-joining (NJ) method in MEGA v5.2 with 1000 replicates based on proportion distance (p-distance) and pairwise deletion. The tree file was viewed and edited using the FigTree v1.3.1 (http://tree.bio.ed.ac.uk/ software/figtree).
2.3. Chromosomal Locations and Collinearity of MYB Genes in B. napus
The chromosomal distribution of putative MYB genes in the B. napus genome was viewed in MapChart. Collinearity analysis of A. thaliana, B. oleracea, B. rapa, and B. napus genomes was carried out in CoGe (https://genomevolution.org/CoGe) [36]. The colinear relationships of putative MYB genes were identified based on the cross-genome collinearity analysis, and the duplication events were accordingly defined as follows: (i) homeologous exchanges (HE): transfer of genetic information between homeologous sequences of different subgenomes; (ii) segmental exchanges (SE): transfer of genetic information between different chromosomal sequences; (iii) segmental duplications (SD): duplicated copies of chromosomal sequences; (iv) tandem duplications (TD): closely related MYB genes in a single cluster in the phylogenetic tree, which were physically located close to each other on a given chromosome without intervening sequences. The HE, SE, and SD events were distinguished from each other based on the chromosomal homology and the colinear relationship (orthologous gene pairs in orthologous blocks) of the An (derived from B. rapa) and Cn (derived from B. oleracea) subgenomes and their respective progenitor genomes (B. rapa and B. oleracea) in all possible combination pairs. The intron/exon organization and chromosomal information of putative MYB genes in other species were acquired from the Phytozome v12.1 or BRAD database [37].
2.4. Spatial and Hormone-Induced Expression of B. napus MYB Genes
The expression profiles of MYB candidates in the roots of B. napus seedlings (five-leaf stage) exposed to auxin (IAA), gibberellin (GA3), cytokinin (6-BA), abscisic acid (ABA), and ethylene (ACC) treatments, respectively, were evaluated using the RNA-seq data recently created by our lab (BioProject ID PRJNA608211). Briefly, seeds of the ZS11 cultivar were obtained from the College of Agriculture and Biotechnology, Southwest University (Chongqing, China) and grown in soil (nutrition soil : rock = 2:1, v:v) in a plant incubator under the long-day conditions (day/night = 16/8 h). Seedlings with four expanded leaves (four-leaf stage) were transferred to Hoagland liquid medium, and grown in an artificial climate chamber at 25 °C under a 16/8 h (day/night) photoperiod. The seedlings at the five-leaf stage were then treated in Hoagland solution containing indicated concentrations of phytohormones (50 µM ABA, 120 µM GA3, 75 µM 6-BA, 60 µM ACC, and 10 µM IAA, respectively). Root samples were then harvested at 0, 1, 3, 6, 12, and 24 h after the treatments, and were immediately frozen in liquid nitrogen and stored at −80 °C until RNA isolation. RNA-Seq was performed using the Illumina sequencing platform (HiSeq 2000) at Biomarker Technologies Co. Ltd. (Beijing, China) (http://www.biomarker.com.cn).
Genes with no or low expression levels (fragments per kilobase of transcript per million mapped reads (FPKM) < 1) in all of the 60 samples might represent pseudogenes or might be expressed only under special condition(s) or at specific developmental stage(s); thus, they were removed from further analyses. The RNA-Seq data of MYB candidate genes were log2-transformed and median-centered. Then, hierarchical clustering of log2-transformed RNA-Seq data was analyzed using Cluster 3.0 [38]. The heatmaps were viewed in Java Treeview [39].
2.5. Cloning of B. napus WER-Homologous Genes
The full-length CDSs of four B. napus 2R-MYB genes (BnMYB019, BnMYB189, BnMYB231, and BnMYB388) that showed high homology to the A. thaliana MYB066 (WER) [10] were polymerase chain reaction (PCR)-amplified using the cDNA sample prepared from roots with gene-specific primers (Table S1). The PCR reaction was carried out using kodakarensis (KOD) plus DNA polymerase (ToYoBo, Shanghai, China) in a total volume of 50 μL. The procedure was as follows: an initial denaturing step at 94 °C for 5 min; 34 cycles of 94 °C for 30 s, 55 °C for 30 s, and 68 °C for 50 s; and a final extension step at 68 °C for 10 min. Similarly, the 1.5-kb promoter sequences of the BnMYB019 and BnMYB231 genes were amplified from the B. napus genome by PCR using the PrimeSTAR HS DNA Polymerase (TakaRa, Dalian, China).
2.6. Plasmid Construction and Transformation
The corresponding CDSs of the four selected B. napus 2R-MYB genes were first cloned into the binary vector pRI201 (TakaRa), and then subcloned into the pCAMBIA1301 vector with the PstI and SmaI sites for ectopic expression in A. thaliana. The resulting vectors (35S::BnMYB019, 35S::BnMYB189, 35S::BnMYB231, and 35S::BnMYB388) contained the selected genes under the control of the cauliflower mosaic virus (CaMV) 35S promoter, and the phosphinothricin gene as a plant-selectable marker conferring Basta resistance. Similarly, the native promoters of BnMYB019 and BnMYB231 were digested with PstI and XbaI, and were used to replace the 35S promoter in the 35S::BnMYB019 and 35S::BnMYB231 plasmids, resulting in the BnMYB019::BnMYB019 and BnMYB231::BnMYB231 constructs, respectively, for the functional complementation of the A. thaliana wer mutant plants. All of the constructs were sequenced to check for PCR-induced errors.
Gene constructs were transferred into Agrobacterium tumefaciens GV3101 by the freeze–thaw method. A. thaliana plants (wild-type (WT) and wer mutant; both are Col-0 ecotype) were transformed with the GV3101 transformants by the floral dipping method, and transgenic plants were then screened on 0.8% agar plates containing diluted (50%, v/v) Basta 50 mg/L. Transgenic T3 lines were isolated and selected based on their segregation ratios for Basta resistance.
2.7. Growth and Phenotypic Evaluation of Transgenic Plants
For A. thaliana transformation and phenotypic analysis of adult transgenic plants, seeds were sown directly in soil and incubated in an artificial climatic chamber at 22 °C with a 16/8 h (day/night) photoperiod. PCR analysis using gene-specific primers (Table S1) was employed to confirm the presence of the transgenes in the transformed plants. For phenotypic analysis of the seedlings, seeds of A. thaliana wild type (WT) and transgenic T3 lines (homozygous with single copy of transgene) were surface-sterilized and grown on half Murashige & Skoog Medium (MS) solid plates. Seeded plates were kept at 4 °C for two days, and were then vertically incubated at 22 °C with a 16/8 h (day/night) photoperiod for the indicated periods. At least ten individual seven-day-old seedlings of each transgenic line were applied for root hair phenotype survey (root hair number per 2 nm near the root elongation zone) using the OLYMPUS MVX10 (Olympus, Tokyo, Japan). The assays were repeated thrice (n = 3, 10 plants/genotype/experiment) with similar results. One-way Analysis of Variance (ANOVA) test was performed to assess the statistical differences among the mean values (least significant difference test; p < 0.05) using SPSS v.20.
3. Results and Discussion
3.1. Identification and Phylogenetic Analysis of BnMYB Proteins
A total of 680 non-redundant putative BnMYBs were identified in B. napus Darmor–bzh [24] including 227 BnMYBR (33.4%), 429 Bn2R-MYB (63.1%), 22 Bn3R-MYB (3.2% including 17 typical Bn3R-MYB and five Bn3R-MYB-like genes) and two Bn4R-MYB (0.3%) genes. These B. napus candidates represent ~3.4-found of the total number of AtMYBs in A. thaliana [22]. Moreover, to ensure the integrity of our data, we further searched in the B. napus ZS11 genome [29], and compared the sequence similarities and identities of the candidates identified in the B. napus Darmor–bzh and ZS11 cultivars. Results revealed that most of the MYB proteins of the two ecotypes had >90% similarity levels, and the sequence annotation information from Darmor–bzh was more complete (Table S2). Therefore, the candidates of each MYB-type family annotated from Darmor–bzh were used in the present study for further analyses, which were named according to their genome locus (Table S2). The physicochemical property of the 680 candidates is shown in Table S2.
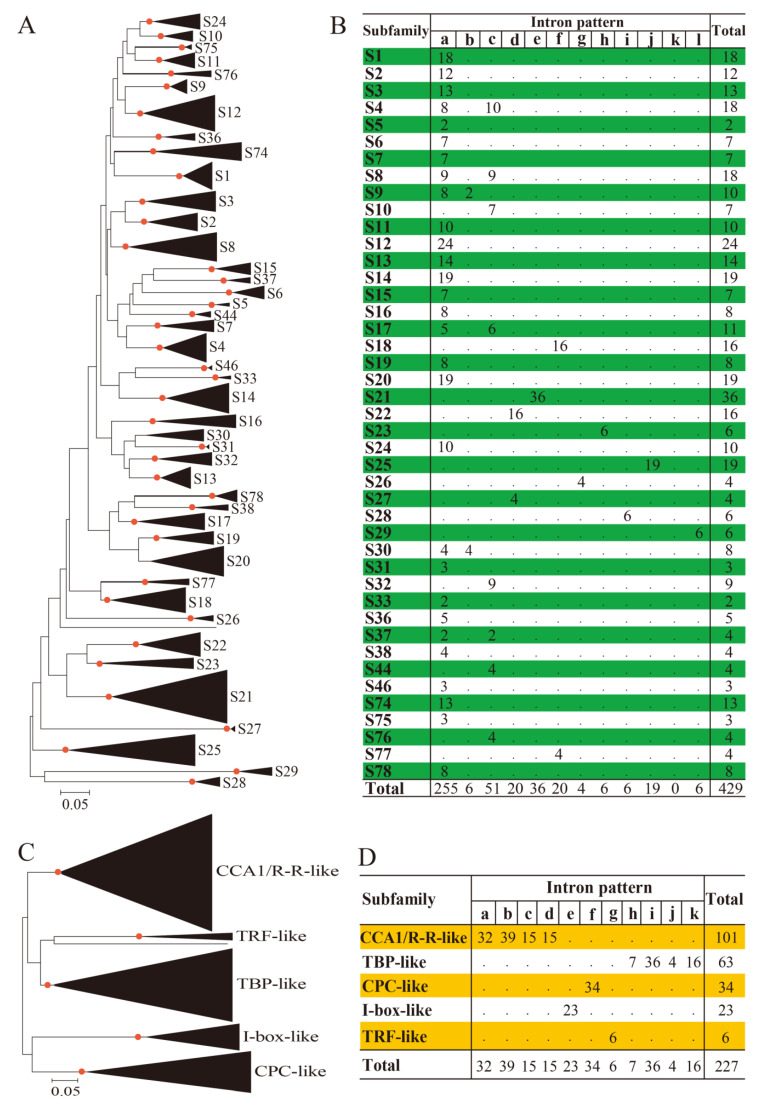
Given the limited number (generally one to three members in many plant species) and high sequence homology of the 3R-MYB and 4R-MYB proteins, in this study, we focused on the evolutionary relationships of the BnMYBR and Bn2R-MYB families by constructing separate unrooted NJ trees for the members of these two families, based on the multiple alignments of their MYB domains. We also included AtMYBRs and At2R-MYBs from A. thaliana into the respective phylogenetic analysis (Figure S1). For subfamily classification, we considered the results received from A. thaliana [22] and our previous studies in 50 eukaryotes [4,5]. With respect to the Bn2R-MYBs, based on the bootstrap values and topology of the phylogenetic tree, the candidates were classified into 43 subfamilies (Figure 1A and Figure S1), 38 of which were consistent with our previous studies [5], strengthening that the 2R-MYBs are distributed in a conserved way in many plant species. Some of the Bn2R-MYBs and previously reported At2R-MYBs encoded by ‘orphan’ genes were clustered into five new subfamilies (S74–S78) with strong bootstrap support (=100%) (Figure 1A and Figure S1), which indicates the presence of some previously neglected Brassicaceae-specific subfamilies. The numbers of Bn2R-MYBs in each subfamily were different, ranging from two to 36 members (Figure 1B and Figure S1). For example, there are 36 and 24 members in subfamilies S21 and S12, respectively, whereas there are only two members in S5 and S33 (Figure 1B and Table S2). This result shows that there was a clear expansion bias of Bn2R-MYBs in different subfamilies, implying that the gene retention and loss rates of different Bn2R-MYB subfamilies after duplications were diverse during the evolution.
Figure 1.
Phylogenetic relationships and intron patterns of MYBR and 2R-MYB proteins from Brassica napus (Bn) and Arabidopsis thaliana (At). The unrooted phylogenetic trees were constructed using the neighbor-joining (NJ) method based on the alignment of the MYB domains of the 429 B. napus 2R-MYBs and 127 A. thaliana 2R-MYBs (including the CDC5 like proteins), and the MYB-like domains of the 227 B. napus and 68 A. thaliana MYBR proteins, respectively. Nodes with bootstrap values ≥70% are dotted in red. The scale shows the relative differences in the examined sequences. Subfamilies are represented in compressed subtree by black triangles with both depth and width proportional to sequence divergence and size, respectively. (A) The NJ tree of 556 2R-MYB proteins from B. napus (429 proteins) and A. thaliana (127 proteins), which were divided into 43 subfamilies (S1–S33, S36–S38, S44, S46, and S74-S78). (B) Summary of intron patterns identified for the 43 B. napus 2R-MYB subfamilies. Intron patterns ‘d–h’ and ‘l’ were designated by referring to Du et al. 2015 [5]. (C) The NJ tree of 295 MYBR proteins from B. napus (227 proteins) and A. thaliana (68 proteins), which were clustered into five subfamilies, namely CCA1/R-R-like, TBP-like, CPC-like, I-box-like and TRF-like. (D) Summary of intron patterns identified for the five B. napus MYBR subfamilies. Intron pattern designation (‘a–k’) was referred to Du et al. 2013 [4].
Next, we analyzed the intron patterns including the intron distribution, positions, and phases over the genomic regions encoding the MYB domains of the Bn2R-MYBs, which revealed conserved intron patterns. Furthermore, according to the data on the absolute conserved intron insertion positions and phases, the gene structures of the MYB domains of Bn2R-MYBs were divided into 11 conserved intron patterns (‘a–l’, while pattern ‘k’ is lacking) that were highly conserved in each subfamily (Figure 1B and Table S2). This result was consistent with the result of our previous study of 2R-MYBs identified in 50 major eukaryotic species [5]. Thus, in this study, we followed the intron pattern designation previously reported [5]. As a result, 34 of the 43 subfamilies contained patterns ‘a–c’ accounting for ~73% of Bn2R-MYBs (a = 59.4%, b = 1.4%, c = 11.9%), whereas the remaining nine subfamilies contained patterns ‘d–h’ and ‘l’ (~27% of Bn2R-MYBs). Four of the five newly identified subfamilies (S74–S76 and S78) had patterns ‘a’ or ‘c’, whereas S77 shared pattern ‘f’ with S18 (Figure 1B and Table S2). These results demonstrated that the intron patterns of Bn2R-MYBs were highly conserved in each subfamily. The gene numbers and expansion trends of different subfamilies were quite diverse, where the rapid expansion and preferential retention of this gene family mainly occurred in genes with patterns ‘a–c’, especially pattern ‘a’.
The BnMYBRs were classified into five major subfamilies: CCA1/R-R-like, I-box-like, CPC-like, TRF-like, and TBP-like (Figure 1C and Figure S2). Unlike the Bn2R-MYBs, no species-specific subfamilies and/or clades were observed, implying that the BnMYBR family was more conserved than the Bn2R-MYB family during evolution. CCA1-like/R-R (101 members) and TBP-like (63 members) were the largest subfamilies, and could be further divided into several clades with different intron patterns (Figure S2 and Table S2). All members of the CCA1-like/R-R subfamily contained the common motif SHAQK(Y/F)F. Similar to the Bn2R-MYBs, intron patterns in the regions spanning MYB domains encoded by the BnMYBRs of this subfamily differed, but were highly conserved in each of the four major clades (patterns ‘a–d’) (Figure 1D). Similarly, the reliability of the TBP-like subfamily was well supported by the presence of the consensus motif LKDKW(R/K)(N/T), and the intron patterns ‘h–k’ were conserved in each clade (Table S2). The CPC-like subfamily (34 members) was characterized by only having a MYB domain without a transcriptional activation domain, and all members shared intron pattern ‘f’. The I-box-like subfamily consisted of 23 members sharing intron pattern ‘e’, whereas the TRF-like subfamily was the smallest (six members) with intron pattern ‘g’. It is worth mentioning that no BnMYBR homologs of the ’orphan‘ genes AtMYBR23 and AtMYBR48 were identified in B. napus. The diverse intron patterns in CCA1-like/R-R and TBP-like BnMYBR subfamilies were consistent with the large numbers of their members and multiple separated clades, which indicates the expansion and functional diversification trend in the BnMYBR family. In contrast, members in the last three BnMYBR subfamilies were relatively conserved during evolution in terms of conserved gene number and gene structure (intron patterns).
3.2. Diversification of 2R-MYB Genes in Higher Plants
As shown in Figure 1A, some of the 429 Bn2R-MYBs were clustered into five new subfamilies (S74–S78) alongside several A. thaliana ‘orphan’ genes (Figure S1), indicating the expansion trend of the Bn2R-MYB family, and perhaps functional diversification of its members. Genome-wide analyses of the 2R-MYB gene family have been carried out in many plant species such as A. thaliana [22], rice [23], Populus trichocarpa [40], Ananas comosus [41], and potato (Solanum tuberosum) [42]. The classification of this gene family in these studies commonly refers to the results obtained from A. thaliana [22], which includes 25 main subfamilies. Based on the analysis of 50 major eukaryotic lineages, we found many species- or lineage-specific subfamilies in land plants [5]. These results imply that the distribution and classification of this gene family are much wider than we have ever expected.
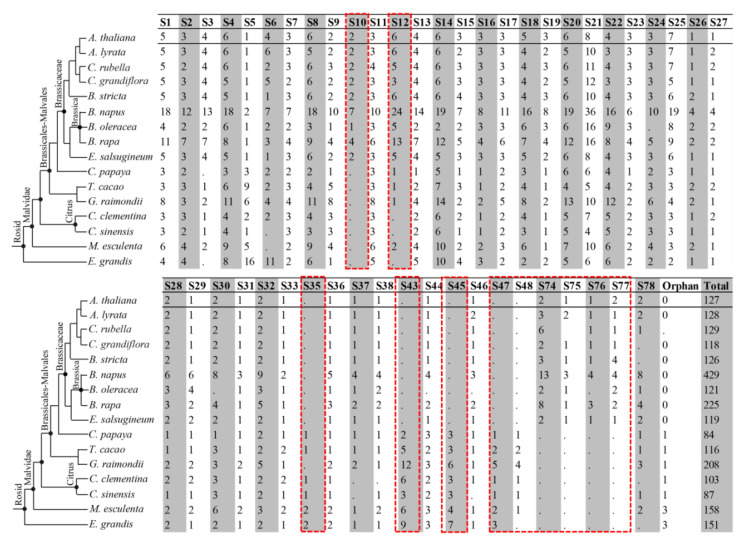
To gain further insights into the evolutionary mechanisms of the 2R-MYBs in Brassicaceae, we identified putative 2R-MYB genes in eight other Brassicaceae and seven out-lineages in Phytozome including E. grandis v2.0, M. esculenta v6.1, C. clementina v1.0, C. papaya ASGPB v0.4, G. raimondii v2.1, T. cacao v1.1, A. lyrata v1.0, A. thaliana TAIR10, C. grandiflora v1.1, C. rubella v1.0, E. salsugineum v1.0, B. stricta v1.2, B. rapa FPsc v1.3, and B. oleracea for a phylogenetic analysis. The non-redundant set of 2R-MYBs from C. sinensis (87 genes) was taken from our previous report [5]. Finally, a total of 2429 typical 2R-MYBs were identified in these 16 species including B. napus (Figure 2 and Table S3), and a NJ tree was constructed based on the multiple alignment of their MYB domains (Figure S3). The topology of this NJ tree was mostly the same as that obtained in our previous study for 50 selected species [5]. Therefore, to avoid confusion in terms of subfamily names, in this study, we retained the same nomenclature, which was used for A. thaliana [22] and in our previous study [5], for classification of the 2R-MYB members.
Figure 2.
Presence/absence of the 2R-MYB genes in 16 representative plant genomes by phylogenetic position. Numbers of 2R-MYB genes in each subfamily (S1–S78) are shown. Red boxes indicate the lineage-specific subfamilies present or absent in nine Brassicaceae species.
Phylogenetic analysis revealed that many 2R-MYBs representing various classes showed very high homology within or across species that were grouped as sister pairs (Figure S3). Generally, the 2429 putative 2R-MYBs from different species were clustered in many compact clades with high support values. Most subfamilies contained members from all of the 16 species investigated, and 38 of the 73 subfamilies defined in eukaryotes in our previous study [5] were well supported in the new tree, demonstrating the conservation of these subfamilies during the evolution. Meanwhile, some newly identified 2R-MYBs and the previously defined A. thaliana ’orphan‘ genes (AtMYB039, AtMYB047, and AtMYB049) were clustered into seven new subfamilies that had been previously neglected. Furthermore, two subfamilies containing candidates from Malvidae, except Brassica, were classified as S77 and S78 (Figure 2). In addition, homologs of AtMYB048 and AtMYB104, previously classified into S38 and S18, were reclassified into S77 and S78, respectively, based on topology and bootstrap values. Four of the newly identified subfamilies (S74, S75, S76, and S77) and S10 were likely to be Brassica-specific because no counterparts were identified in other species (Figure 2 and Figure S3). Similarly, the previously defined A. thaliana-specific subfamily S12 [22,42,43], which is specifically involved in the glucosinolate biosynthesis pathway [42], was demonstrated to be distributed only in the nine Brassicaceae species and M. esculenta among the 16 plant species investigated in this study. Conversely, five subfamilies (S35, S43, S45, S47, and S48) were absent from the nine investigated Brassicaceae species. Our results suggested that there are many more lineage-specific subfamilies of 2R-MYBs in plants that exhibited lineage-specific expansion or deficiency (Figure 2 and Figure S3).
Recently, based on the analyses in 35 plant species, 18 rosid-specific subfamilies/clusters/clades (e.g., S1, S3–5, S12, etc.), four asterid-specific subfamilies (S39, S40, S43, and S80), and nine subfamilies (S29–36 and S69) specific to one of the 35 investigated species were identified [43]. Similarly, many subfamilies/clusters/clades specific to different lineages and species were found in numerous studies [42,43,44,45,46,47]. For example, subfamily S29 was proven to harbor only potato genes [42], seven subfamilies (e.g., S1, S5, S11, etc.) contained only 2R-MYB genes of Sesamum indicum [44], S26 harbored only 2R-MYB genes from Ginkgo biloba [45], 12 subfamilies (e.g., S5, S25, S32, etc.) contained only grass-species 2R-MYB genes, and S15 and S18 subfamilies possessed only Phyllostachys edulis 2R-MYB genes [46]. These results suggest that lineage- or species-specific 2R-MYBs may be genomic relics that have evolved independently or may have unique function(s) in particular lineage or species, implying that expansion and diversification of 2R-MYBs occurred during the evolution. Although the functions of these subfamilies remain unknown, our analysis provided evidence to support that some members of the 2R-MYB gene family might have evolved to fulfill lineage-specific functions [47,48].
3.3. Chromosomal Distribution and Expansion of the MYB Members in B. napus
To explore the duplication events of each type of the B. napus MYB superfamily, we analyzed their chromosomal distribution and colinear relationships with their respective counterparts in B. rapa, B. oleracea, and A. thaliana. Chromosomal distribution analysis showed that 429 Bn2R-MYBs were distributed on all 19 B. napus chromosomes (Figure S4A). There were 213 and 216 Bn2R-MYBs on An and Cn subgenomes, respectively. In subgenome An, 16, 21, 37, 9, 14, 26, 22, 14, 21, and 12 Bn2R-MYBs were located on chromosomes A01–A10, respectively, while the chromosomal locations of the remaining 21 genes remain to be determined. In subgenome Cn, 14, 21, 31, 15, 16, 15, 23, 19, and 25 Bn2R-MYBs were distributed on chromosomes C01–C09, respectively, while the chromosomal locations of the remaining 37 genes remain unclear. High densities of the Bn2R-MYBs were observed on the top and/or bottom end(s) of each chromosome such as C03 and C07–C09 (Figure S4A). Thus, the distributions of Bn2R-MYBs across the 19 chromosomes were uneven. A similar trend was observed in the BnMYBR family. The 227 BnMYBRs were also distributed on all 19 chromosomes, with 114 and 113 on subgenomes An and Cn, respectively (Figure S4B). In both the An and Cn subgenomes, chromosome 3 contained the largest number of candidates (19 and 22 genes, respectively). Furthermore, the numbers of BnMYBRs were also higher on the top and/or bottom end(s) of each chromosome. As for the 22 Bn3R-MYB/Bn3R-MYB-like genes, 11 and 11 genes were located on the An and Cn subgenomes, respectively (Table S2). With respect to the two Bn4R-MYBs, one is located on chromosome A05 (Bn4R-MYB1) and another on chromosome C01 (Bn4R-MYB2) (Table S2).
Collinearity analysis showed that the B. napus An and Cn subgenomes were widely colinear to the corresponding diploid B. rapa and B. oleracea genomes, respectively. Accordingly, many orthologous gene pairs between the An and Cn subgenomes and their respective progenitor genomes were observed (Table S4). A total of 362 of the 429 (~84%) Bn2R-MYBs were involved in colinear relationships in B. napus (Figure S4A and Table S4). Among the 362 Bn2R-MYBs, 126 (35%) had colinear relationships with 2R-MYBs of B. rapa (96 genes, ~76%) and/or B. oleracea (30 genes, ~24%). Thus, many Bn2R-MYBs were derived from the two progenitor genomes, with more genes from B. rapa. After the allopolyploidy event, 68 (~19%), 109 (~30%), and 59 (~16%) of the 362 Bn2R-MYBs underwent SE, HE, and SD events, respectively, while no TD events were identified. These results demonstrated that the small-scale duplication events (including SE, HE, and SD) might also play a major role for the large gene expansion of the Bn2R-MYB family in the B. napus genome.
Of the 227 BnMYBRs, 182 (~80%) genes exhibited colinear relationships in B. napus. Out of these 182 BnMYBRs, 107 (~59%) and 55 (~30%) genes were involved in colinear relationships with MYBR genes of B. rapa and B. oleracea, respectively (Figure S4B and Table S4). This result indicates that the majority of BnMYBRs were obtained through the allopolyploidy event, and the MYBR genes derived from B. rapa tended to be retained in B. napus. Furthermore, 32 (~18%), 32 (~18%), and 20 (~11%) of the 182 BnMYBRs (~18%) underwent SE, HE, and SD events, respectively, while no TD events were identified. Of the 22 Bn3R-MYB/Bn3R-MYB-like genes, 19 (~86%) exhibited colinear relationships in B. napus, with 11 and five genes also showing colinear relationships with 3R-MYB genes in B. rapa (50%) and B. oleracea (23%), respectively. This finding indicates that the majority of Bn3R-MYBs were also obtained from the allopolyploidy event. Additionally, one (5%), eight (36%), and three (14%) of the 19 3R-MYB genes were derived from the SE, HE, and SD events, respectively, while no 3R-MYB genes were involved in any TD event (Table S4).
Overall, our results collectively demonstrated that large-scale duplication event (allopolyploidy) and small-scale duplication events (SD, SE, and HE) were the major forces driving the large expansion of the MYB superfamily in B. napus. After allopolyploidy, many SE and HE events occurred across the An and Cn subgenomes, resulting in a similar number of genes for all MYB-type genes in both subgenomes of B. napus with higher preference for the retention of MYB genes from B. rapa. On the contrary, TD events likely had less effect on the expansion of BnMYBs in B. napus.
3.4. Conserved Functions of B. napus WER Homologs in Root Hair Development in A. thaliana
To date, many MYBs, especially 2R-MYBs, have been experimentally demonstrated to play key roles in root development. For example, AtMYB30 and AtMYB60 of S1 have been shown to regulate root elongation [49,50], AtMYB36 of the S14 subfamily is involved in lateral root primordium development [51], AtMYB066/WER homologs of S15 are well-known in regulating root hair patterning [10,52], AtMYB33, AtMYB65, and AtMYB101 of S18 have been reported to regulate primary root growth [53], AtMYB77 of S22 interacted with auxin response factors (ARFs) to regulate lateral root formation [54], AtMYB93 of S24 acted as a negative regulator of lateral root development [55], AtMYB88 of S28 regulated root gravitropism [56], and AtMYB59 of S38 could inhibit root growth by extending the metaphase of mitotic cells [57].
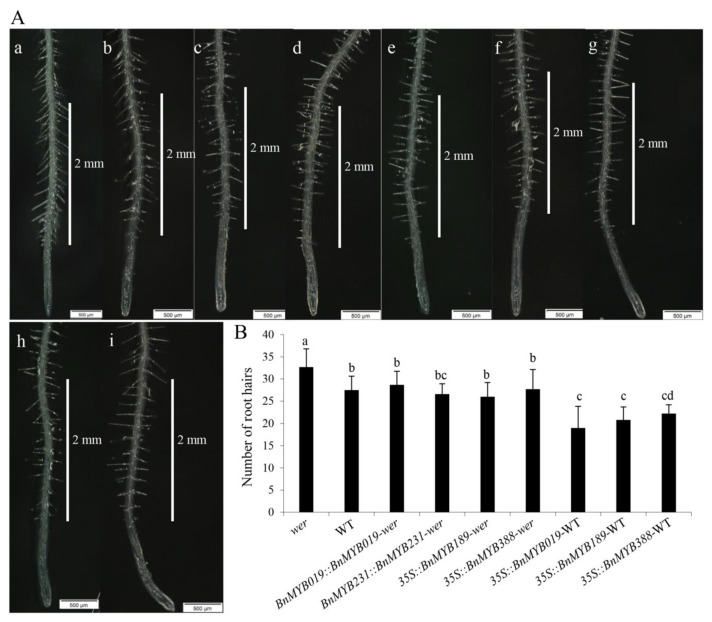
Root hair patterning in A. thaliana is controlled by the interplay of several TFs including the 2R-MYB-type WER gene [10]. Our phylogenetic analysis showed that seven Bn2R-MYBs (BnMYB019, BnMYB042, BnMYB077, BnMYB118, BnMYB189, BnMYB231, and BnMYB388) and three A. thaliana WER/AtMYB066 homologs (AtMYB066, AtMYB023, and AtMYB000) were clustered in the S15 subfamily (Figure S1). Among them, BnMYB019, BnMYB189, BnMYB231, and BnMYB388 are the homologs of WER; thus, they were named BnWERs. To further explore the roles of these four BnWERs in root hair development, we introduced them into A. thaliana WT and/or wer mutant plants under the control of the 35S promoter. At the same time, BnMYB019 and BnMYB231 genes directed by their native promoter were also used to transform A. thaliana wer mutant plants.
The wer mutant had more root hairs than WT as reported earlier [10] (Figure 3A,B). The T3 homozygous lines ectopically expressing BnMYB189 or BnMYB388 in wer background using the 35S promoter showed reduced root hair numbers that were comparable to that of WT plants (Figure 3A,B). Likewise, all homozygous transgenic lines harboring the BnMYB019::BnMYB019 or BnMYB231::BnMYB231 construct in the wer background completely rescued the aberrant root hair density of the wer mutant (Figure 3A,B). These results indicated that all four of the examined BnMYB019, BnMYB189, BnMYB231, and BnMYB388 genes have conserved functions with the A. thaliana WER gene in regulating root hair formation. Furthermore, the transgenic lines carrying the 35S::BnMYB019, 35S::BnMYB189 or 35S::BnMYB388 construct in the WT background displayed lower root hair numbers than the WT plants (Figure 3A,B), strengthening their negative regulatory roles in root hair development.
Figure 3.
Complementation assay of root hair density in the Arabidopsis thaliana wer mutant background using the four Brassica napus WER-homologous genes BnMYB019, BnMYB189, BnMYB231, and BnMYB388. (A) Representative picture showing root hair morphology and density (2 nm) in 7-day-old A. thaliana seedlings of wer mutant (a), wild-type (WT) (b), and wer mutant plants harboring BnMYB019::BnMYB019 (c), wer mutant plants harboring BnMYB0231::BnMYB231 (d), wer mutant plants harboring 35S::BnMYB189 (e), wer mutant plants harboring 35S::BnMYB388 (f), WT plants harboring 35S::BnMYB019 (g), WT plants harboring 35S::BnMYB189, and (h) WT plants harboring 35S::BnMYB388 (i). (B) Root hair density in 7-day-old A. thaliana seedlings of the corresponding lines (n = 3, 10 plants/genotype/experiment). Different letters indicate significant differences (least significant difference test; p < 0.05).
Taken together, our results demonstrated that the Bn2R-MYB homologs in the S15 subfamily shared a conserved function in root hair development, and could complement the loss-of-function of the A. thaliana WER gene. This finding supports that in most cases, genes in a same subfamily with high similarity in sequence and expression patterns tend to have functional redundancy, indicating a positive correlation among expression patterns, sequence features, and gene functions across different species such as B. napus and A. thaliana.
3.5. Hormone-Induced Expression Profiling of B. napus MYB Genes by RNA-Seq
It was demonstrated that the functions of MYB genes in plant development, stress responses, and metabolism were generally mediated by phytohormones. For example, homologs of S18 (e.g., A. thaliana AtMYB33 and AtMYB65, G. hirsutum GhMYB24, and H. vulgare HvGAMYB) were shown to regulate anther/pollen development by the GA signal [58,59,60]. AtMYB21, AtMYB24, and AtMYB57 of S19 play roles in stamen development in a jasmonate-dependent manner [61,62], while AtMYB77 of S22 regulates lateral root growth through modulating auxin signal transduction [54]. In the present study, we performed a global expression profiling of the B. napus MYB superfamily under ABA, IAA, GA3, 6-BA, and ACC treatments in a time-course manner to identify those BnMYBs whose functions in B. napus might be mediated by hormones (Figure 4).
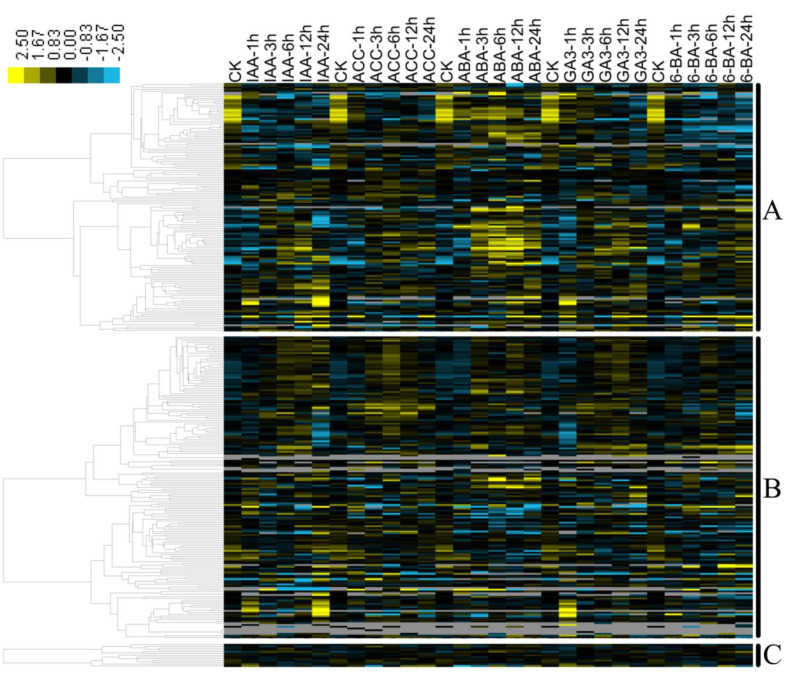
Figure 4.
Transcript profiles of members of the B. napus MYB superfamily in seedling roots under individual treatments with auxin (IAA), gibberellin (GA3), cytokinin (6-BA), abscisic acid (ABA) and ethylene (ACC) in a time-course manner. (A–C) Expression patterns of 141 Bn2R-MYB (A), 171 BnMYBR (B) and 12 Bn3R-MYB (C) genes in roots of B. napus seedlings under the IAA, ACC, ABA, GA3 and 6-BA treatments. Transcript data were obtained by RNA-Seq. Log2 (FPKM ≥ 1) values were displayed according to the color bar (top left). BnMYBs with no or weak expression (FPKM < 1) in all samples were excluded from this figure. Expression values of all BnMYBs under hormone treatments are supplied in Table S5. CK, control (non-treatment at 0 h).
Regarding the Bn2R-MYB family (429 members), 141 genes (~33%) showed detectable transcript levels (FPKM ≥ 1) in the roots under hormone treatments, whereas the remaining genes had weak (FPKM < 1) or no transcript accumulation (Figure 4A and Table S5). The expression profiles of these 141 Bn2R-MYBs, which are distributed into 25 of the 43 Bn2R-MYB subfamilies, were classified into three major patterns. The first group (73 genes) was evidently upregulated (≥1.5-fold) by all five hormones and under most of the treatment conditions, especially ABA (e.g., Bn2R-MYB126 and Bn2R-MYB170 in S4, Bn2R-MYB189 and Bn2R-MYB252 in S9, Bn2R-MYB148, Bn2R-MYB341 in S11, etc.). The second group (51 genes) was evidently downregulated (≥1.5-fold) by all five hormones and in most of the treatment conditions, especially the S22 members (e.g., Bn2R-MYB010, Bn2R-MYB127, and Bn2R-MYB330). The members of the third group (20 genes) had their expression altered by a few hormones, for example, Bn2R-MYB209 in S78, Bn2R-MYB331 in S28, and Bn2R-MYB322 in S1 were upregulated (≥1.5 fold change) by IAA, ABA and/or GA3 treatments, while Bn2R-MYB334 in S1, Bn2R-MYB299 in S12, and Bn2R-MYB001 and Bn2R-MYB215 in S22 were downregulated by GA3 and/or 6-BA treatments (Figure 4A and Table S5). The different expression patterns of the Bn2R-MYBs suggest their diverse roles in responses to hormones. Accordingly, consistent with the diverse hormone-induced expression profile, homologs of S1 were demonstrated to be involved in ABA-mediated defense (e.g., against drought) and development (e.g., roots) [14,50,63], members of S22 that regulate plant responses to abiotic stresses (e.g., drought, salt, and cold stresses) [13] and lateral root development [64] were also ABA-mediated, while AtMYB88 in S28, which was involved in stomatal formation and root gravitropism, was auxin-mediated [56]. Although only a few members of the MYB superfamily have been functionally characterized in plants to date, our results suggest that their potential roles in root-related processes may be commonly hormone-mediated.
With respect to the BnMYBR family, 171 of the 227 genes (75%) exhibited detectable transcript levels (FPKM ≥ 1) in roots under hormone treatments (Figure 4B and Table S5). This ratio was much higher than that of the Bn2R-MYB family, suggesting that BnMYBRs may be more sensitive to exogenous hormone treatments. Among the five subfamilies, members of the CCA1/R-R-like, TBP-like, and TRF-like subfamilies tended to be expressed in roots under various hormone treatments. Except for a few genes (e.g., BnMYBR099, BnMYBR128, and BnMYBR077), the majority of BnMYBRs (130 genes) were upregulated (≥1.5-fold) at several time points by at least one of the five hormones, and similar expression patterns were often observed for homologs in the same subfamily or clade (Figure 4B and Table S5). On the other hand, 41 BnMYBRs were downregulated (≥1.5-fold) by at least one of the five hormones (Figure 4B and Table S5), many of which, mainly the CCA1/R-R-like subfamily genes such as BnMYBR103, BnMYBR132, and BnMYB179 were downregulated by all five hormones under all treatment conditions.
As for the 22 Bn3R-MYB and Bn3R-MYB-like genes, 12 of the 17 Bn3R-MYBs (71%), and one of the five Bn3R-MYB-like genes (20%) exhibited detectable transcript levels (FPKM ≥ 1) in roots upon hormone treatments (Figure 4C and Table S5). Out of these 13 Bn3R-MYB and Bn3R-MYB-like genes, six and seven genes were found to be upregulated and/or downregulated, respectively, by hormone treatments (Figure 4C and Table S5). For example, Bn3R-MYB08 was upregulated by both ACC and ABA treatments, Bn3R-MYB07 was induced by IAA, ABA, and 6-BA treatments, while Bn3R-MYB10 was upregulated by IAA, ABA, and 6-BA treatments (Table S5). None of the two identified Bn4R-MYBs had detectable expression levels in any hormone-treated samples (Table S5). These data together suggest that the 3R-MYB, Bn3R-MYB-like, and Bn4R-MYB genes may be less affected by hormones.
4. Conclusions
In this study, we systematically identified and classified the BnMYB superfamily. Exon–intron structure analyses of this superfamily confirmed the conserved exon–intron structures within the conserved MYB domain. Collinearity analyses revealed that allopolyploidy between B. rapa and B. oleracea mainly contributed to the large BnMYB gene expansion in B. napus. Comparative analysis of the 2R-MYB families in 16 Brassicaceae and non-Brassicaceae species identified five Brassicaceae-specific subfamilies (S10, S74-77) and five subfamilies (S35, S43, S45, S47, and S48) that are absent from the nine examined Brassicaceae species. In addition, one subfamily (S12) was found to be merely distributed in the nine Brassicaceae species and M. esculenta. Functional analysis of the four WER-homologous Bn2R-MYBs in a complementary assay indicated their conserved roles in root hair patterning. Global expression profiling demonstrated that the Bn2R-MYBs and BnMYBRs were widely responsive to hormone treatments in roots, suggesting that the hormone-responsive BnMYBs may regulate biological processes in B. napus in a hormone-dependent manner.
Supplementary Materials
The following are available online at https://www.mdpi.com/2218-273X/10/6/875/s1. Figure S1: Phylogenetic tree of the R2R3-MYB proteins (2R-MYBs) in Brassica napus and Arabidopsis thaliana; Figure S2: Phylogenetic tree of the MYB-related proteins (MYBRs) from Brassica napus and Arabidopsis thaliana; Figure S3: Phylogenetic tree of 2429 plant 2R-MYB proteins from 16 plant species; Figure S4: Physical positions and duplications of 429 Bn2R-MYB and 227 BnMYBR genes in 19 Brassica napus chromosomes; Table S1: Primers used in this study; Table S2: The MYB genes identified the Brassica napus genome; Table S3: The 2R-MYB genes identified in 15 plant species (excepting Brassica napus) used in this study; Table S4: Duplications of the MYB genes in the Brassica napus, B. rapa, and B. oleracea genomes; Table S5: Expression value (FPKM) of the Brassica napus MYB genes under five hormone treatments in roots.
Author Contributions
Conceptualization, H. D.; Methodology, P.L. and P.C.; Software, J.W.; Validation, P.L. and J.W.; Formal analysis, Y.K.; Resources, P.G., M.W., and M.L.; Data curation, J.W., P.L., and L.-S.P.T.; Writing—original draft preparation, H.D. and J.W.; Writing—review and editing, H.D. and L.-S.P.T.; Supervision, J.L.; Project administration, H.D.; Funding acquisition, H.D. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Natural Science Foundation of China, grant number 31671727.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- 1.Martin C., Paz-Ares J. MYB transcription factors in plants. Trends Genet. 1997;13:67–73. doi: 10.1016/S0168-9525(96)10049-4. [DOI] [PubMed] [Google Scholar]
- 2.Lipsick J.S. One billion years of Myb. Oncogene. 1996;13:223–235. [PubMed] [Google Scholar]
- 3.Dubos C., Stracke R., Grotewold E., Weisshaar B., Martin C., Lepiniec L. MYB transcription factors in Arabidopsis. Trends Plant Sci. 2010;15:573–581. doi: 10.1016/j.tplants.2010.06.005. [DOI] [PubMed] [Google Scholar]
- 4.Du H., Wang Y.B., Xie Y., Liang Z., Jiang S.J., Zhang S.S., Huang Y.B., Tang Y.X. Genome-wide identification and evolutionary and expression analyses of MYB-related genes in land plants. DNA Res. 2013;20:437–448. doi: 10.1093/dnares/dst021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Du H., Liang Z., Zhao S., Nan M.G., Tran L.S., Lu K., Huang Y.B., Li J.N. The Evolutionary History of R2R3-MYB Proteins across 50 Eukaryotes: New Insights into Subfamily Classification and Expansion. Sci. Rep. 2015;5:11037. doi: 10.1038/srep11037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Du H., Yang S.S., Liang Z., Feng B.R., Liu L., Huang Y.B., Tang Y.X. Genome-wide analysis of the MYB transcription factor superfamily in soybean. BMC Plant Biol. 2012;12:106. doi: 10.1186/1471-2229-12-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li S., Wang W., Gao J., Yin K., Wang R., Wang C., Petersen M., Mundy J., Qiu J.L. MYB75 Phosphorylation by MPK4 Is Required for Light-Induced Anthocyanin Accumulation in Arabidopsis. Plant Cell. 2016;28:2866–2883. doi: 10.1105/tpc.16.00130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stracke R., Ishihara H., Huep G., Barsch A., Mehrtens F., Niehaus K., Weisshaar B. Differential regulation of closely related R2R3-MYB transcription factors controls flavonol accumulation in different parts of the Arabidopsis thaliana seedling. Plant J. 2007;50:660–677. doi: 10.1111/j.1365-313X.2007.03078.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Albert N.W., Lewis D.H., Zhang H., Schwinn K.E., Jameson P.E., Davies K.M. Members of an R2R3-MYB transcription factor family in Petunia are developmentally and environmentally regulated to control complex floral and vegetative pigmentation patterning. Plant J. 2011;65:771–784. doi: 10.1111/j.1365-313X.2010.04465.x. [DOI] [PubMed] [Google Scholar]
- 10.Lee M.M., Schiefelbein J. WEREWOLF, a MYB-Related Protein in Arabidopsis, Is a Position-Dependent Regulator of Epidermal Cell Patterning. Cell. 1999;99:473–483. doi: 10.1016/S0092-8674(00)81536-6. [DOI] [PubMed] [Google Scholar]
- 11.Kim J.H., Nguyen N.H., Jeong C.Y., Nguyen N.T., Hong S.W., Lee H. Loss of the R2R3 MYB, AtMyb73, causes hyper-induction of the SOS1 and SOS3 genes in response to high salinity in Arabidopsis. J. Plant Physiol. 2013;170:1461–1465. doi: 10.1016/j.jplph.2013.05.011. [DOI] [PubMed] [Google Scholar]
- 12.Zhang P., Wang R., Ju Q., Li W., Tran L.P., Xu J. The R2R3-MYB Transcription Factor MYB49 Regulates Cadmium Accumulation. Plant Physiol. 2019;180:529–542. doi: 10.1104/pp.18.01380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jung C., Seo J.S., Han S.W., Koo Y.J., Kim C.H., Song S.I., Nahm B.H., Choi Y.D., Cheong J.J. Overexpression of AtMYB44 enhances stomatal closure to confer abiotic stress tolerance in transgenic Arabidopsis. Plant Physiol. 2008;146:623–635. doi: 10.1104/pp.107.110981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liao C., Zheng Y., Guo Y. MYB30 transcription factor regulates oxidative and heat stress responses through ANNEXIN-mediated cytosolic calcium signaling in Arabidopsis. New Phytol. 2017;216:163–177. doi: 10.1111/nph.14679. [DOI] [PubMed] [Google Scholar]
- 15.Nagel D.H., Doherty C.J., Pruneda-Paz J.L., Schmitz R.J., Ecker J.R., Kay S.A. Genome-wide identification of CCA1 targets uncovers an expanded clock network in Arabidopsis. Proc. Natl. Acad. Sci. USA. 2015;112:E4802–E4810. doi: 10.1073/pnas.1513609112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nguyen N.H., Lee H. MYB-related transcription factors function as regulators of the circadian clock and anthocyanin biosynthesis in Arabidopsis. Plant Signal. Behav. 2016;11:e1139278. doi: 10.1080/15592324.2016.1139278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wada T., Tachibana T., Shimura Y., Okada K. Epidermal cell differentiation in Arabidopsis determined by a Myb homolog, CPC. Science. 1997;277:1113–1116. doi: 10.1126/science.277.5329.1113. [DOI] [PubMed] [Google Scholar]
- 18.Pant B.D., Burgos A., Pant P., Cuadros-Inostroza A., Willmitzer L., Scheible W.R. The transcription factor PHR1 regulates lipid remodeling and triacylglycerol accumulation in Arabidopsis thaliana during phosphorus starvation. J. Exp. Bot. 2015;66:1907–1918. doi: 10.1093/jxb/eru535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ito M. Conservation and diversification of three-repeat Myb transcription factors in plants. J. Plant Res. 2005;118:61–69. doi: 10.1007/s10265-005-0192-8. [DOI] [PubMed] [Google Scholar]
- 20.Haga N., Kobayashi K., Suzuki T., Maeo K., Kubo M., Ohtani M., Mitsuda N., Demura T., Nakamura K., Jurgens G., et al. Mutations in MYB3R1 and MYB3R4 cause pleiotropic developmental defects and preferential down-regulation of multiple G2/M-specific genes in Arabidopsis. Plant Physiol. 2011;157:706–717. doi: 10.1104/pp.111.180836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dai X., Xu Y., Ma Q., Xu W., Wang T., Xue Y., Chong K. Overexpression of an R1R2R3 MYB gene, OsMYB3R-2, increases tolerance to freezing, drought, and salt stress in transgenic Arabidopsis. Plant Physiol. 2007;143:1739–1751. doi: 10.1104/pp.106.094532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stracke R., Werber M., Weisshaar B. The R2R3-MYB gene family in Arabidopsis thaliana. Curr. Opin. Plant Biol. 2001;4:447–456. doi: 10.1016/S1369-5266(00)00199-0. [DOI] [PubMed] [Google Scholar]
- 23.Katiyar A., Smita S., Lenka S.K., Rajwanshi R., Chinnusamy V., Bansal K.C. Genome-wide classification and expression analysis of MYB transcription factor families in rice and Arabidopsis. BMC Genom. 2012;13:544. doi: 10.1186/1471-2164-13-544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang C., Ma R., Xu J., Yan J., Guo L., Song J., Feng R., Yu M. Genome-wide identification and classification of MYB superfamily genes in peach. PLoS ONE. 2018;13:e0199192. doi: 10.1371/journal.pone.0199192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qing J., Dawei W., Jun Z., Yulan X., Bingqi S., Fan Z. Genome-wide characterization and expression analyses of the MYB superfamily genes during developmental stages in Chinese jujube. PeerJ. 2019;7:e6353. doi: 10.7717/peerj.6353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chalhoub B., Denoeud F., Liu S., Parkin I.A., Tang H., Wang X., Chiquet J., Belcram H., Tong C., Samans B., et al. Plant genetics. Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science. 2014;345:950–953. doi: 10.1126/science.1253435. [DOI] [PubMed] [Google Scholar]
- 27.Mitchell A., Chang H.Y., Daugherty L., Fraser M., Hunter S., Lopez R., McAnulla C., McMenamin C., Nuka G., Pesseat S., et al. The InterPro Protein Families Database: The Classification Resource After 15 Years. Nucleic. Acids. Res. 2015;43:D213–D221. doi: 10.1093/nar/gku1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Letunic I., Copley R.R., Schmidt S., Ciccarelli F.D., Doerks T., Schultz J., Ponting C.P., Bork P. SMART 4.0: Towards genomic data integration. Nucleic. Acids. Res. 2004;32:D142–D144. doi: 10.1093/nar/gkh088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun F., Fan G., Hu Q., Zhou Y., Guan M., Tong C., Li J., Du D., Qi C., Jiang L., et al. The high-quality genome of Brassica napus cultivar ‘ZS11’ reveals the introgression history in semi-winter morphotype. Plant J. 2017;92:452–468. doi: 10.1111/tpj.13669. [DOI] [PubMed] [Google Scholar]
- 30.Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Goodstein D.M., Shu S., Howson R., Neupane R., Hayes R.D., Fazo J., Mitros T., Dirks W., Hellsten U., Putnam N., et al. Phytozome: A comparative platform for green plant genomics. Nucleic. Acids. Res. 2012;40:D1178–D1186. doi: 10.1093/nar/gkr944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hu B., Jin J., Guo A.Y., Zhang H., Luo J., Gao G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics. 2015;31:1296–1297. doi: 10.1093/bioinformatics/btu817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bjellqvist B., Hughes G.J., Pasquali C., Paquet N., Ravier F., Sanchez J.C., Frutiger S., Hochstrasser D. The Focusing Positions of Polypeptides in Immobilized Ph Gradients Can Be Predicted from Their Amino-Acid-Sequences. Electrophoresis. 1993;14:1023–1031. doi: 10.1002/elps.11501401163. [DOI] [PubMed] [Google Scholar]
- 34.Chou K.C., Shen H.B. Cell-PLoc: A package of Web servers for predicting subcellular localization of proteins in various organisms. Nat. Protoc. 2008;3:153–162. doi: 10.1038/nprot.2007.494. [DOI] [PubMed] [Google Scholar]
- 35.Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lyons E., Pedersen B., Kane J., Alam M., Ming R., Tang H., Wang X., Bowers J., Paterson A., Lisch D. Finding and comparing syntenic regions among Arabidopsis and the outgroups papaya, poplar and grape: CoGe with rosids. Plant Physiol. 2008;148:1772–1781. doi: 10.1104/pp.108.124867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cheng F., Liu S., Wu J., Fang L., Sun S., Liu B., Li P., Hua W., Wang X. BRAD, the genetics and genomics database for Brassica plants. BMC Plant Biol. 2011;11:136. doi: 10.1186/1471-2229-11-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.De Hoon M.J., Imoto S., Nolan J., Miyano S. Open source clustering software. Bioinformatics. 2004;20:1453–1454. doi: 10.1093/bioinformatics/bth078. [DOI] [PubMed] [Google Scholar]
- 39.Saldanha A.J. Java Treeview--extensible visualization of microarray data. Bioinformatics. 2004;20:3246–3248. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- 40.Wilkins O., Nahal H., Foong J., Provart N.J., Campbell M.M. Expansion and diversification of the Populus R2R3-MYB family of transcription factors. Plant Physiol. 2009;149:981–993. doi: 10.1104/pp.108.132795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu C., Xie T., Chen C., Luan A., Long J., Li C., Ding Y., He Y. Genome-wide organization and expression profiling of the R2R3-MYB transcription factor family in pineapple (Ananas comosus) BMC Genom. 2017;18:503. doi: 10.1186/s12864-017-3896-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li X., Guo C., Ahmad S., Wang Q., Yu J., Liu C., Guo Y. Systematic Analysis of MYB Family Genes in Potato and Their Multiple Roles in Development and Stress Responses. Biomolecules. 2019;9:317. doi: 10.3390/biom9080317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Han Y., Yu J., Zhao T., Cheng T., Wang J., Yang W., Pan H., Zhang Q. Dissecting the Genome-Wide Evolution and Function of R2R3-MYB Transcription Factor Family in Rosa chinensis. Genes. 2019;10:823. doi: 10.3390/genes10100823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mmadi M.A., Dossa K., Wang L., Zhou R., Wang Y., Cisse N., Sy M.O., Zhang X. Functional Characterization of the Versatile MYB Gene Family Uncovered Their Important Roles in Plant Development and Responses to Drought and Waterlogging in Sesame. Genes. 2017;8:362. doi: 10.3390/genes8120362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu X., Yu W., Zhang X., Wang G., Cao F., Cheng H. Identification and expression analysis under abiotic stress of the R2R3-MYB genes in Ginkgo biloba L. Physiol. Mol. Biol. Plants. 2017;23:503–516. doi: 10.1007/s12298-017-0436-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hou D., Cheng Z., Xie L., Li X., Li J., Mu S., Gao J. The R2R3 MYB Gene Family in Phyllostachys edulis: Genome-Wide Analysis and Identification of Stress or Development-Related R2R3MYBs. Front. Plant Sci. 2018;9:738. doi: 10.3389/fpls.2018.00738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Soler M., Camargo E.L., Carocha V., Cassan-Wang H., Clemente H.S., Savelli B., Hefer C.A., Paiva J.A., Myburg A.A., Grima-Pettenati J. The Eucalyptus grandis R2R3-MYB transcription factor family: Evidence for woody growth-related evolution and function. New Phytol. 2015;206:1364–1377. doi: 10.1111/nph.13039. [DOI] [PubMed] [Google Scholar]
- 48.Bailey P.C., Dicks J., Wang T.L., Martin C. IT3F: A web-based tool for functional analysis of transcription factors in plants. Phytochemistry. 2008;69:2417–2425. doi: 10.1016/j.phytochem.2008.08.003. [DOI] [PubMed] [Google Scholar]
- 49.Sakaoka S., Mabuchi K., Morikami A., Tsukagoshi H. MYB30 regulates root cell elongation under abscisic acid signaling. Commun. Integr. Biol. 2018;11:e1526604. doi: 10.1080/19420889.2018.1526604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Oh J.E., Kwon Y., Kim J.H., Noh H., Hong S.W., Lee H. A dual role for MYB60 in stomatal regulation and root growth of Arabidopsis thaliana under drought stress. Plant Mol. Biol. 2011;77:91–103. doi: 10.1007/s11103-011-9796-7. [DOI] [PubMed] [Google Scholar]
- 51.Liberman L.M., Sparks E.E., Moreno-Risueno M.A., Petricka J.J., Benfey P.N. MYB36 regulates the transition from proliferation to differentiation in the Arabidopsis root. Proc. Natl. Acad. Sci. USA. 2015;29:12099–12104. doi: 10.1073/pnas.1515576112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ryu K.H., Kang Y.H., Park Y.H., Hwang I., Schiefelbein J., Lee M.M. The WEREWOLF MYB protein directly regulates CAPRICE transcription during cell fate specification in the Arabidopsis root epidermis. Development. 2005;132:4765–4775. doi: 10.1242/dev.02055. [DOI] [PubMed] [Google Scholar]
- 53.Xue T., Liu Z., Dai X., Xiang F. Primary root growth in Arabidopsis thaliana is inhibited by the miR159 mediated repression of MYB33, MYB65 and MYB101. Plant Sci. 2017;262:182–189. doi: 10.1016/j.plantsci.2017.06.008. [DOI] [PubMed] [Google Scholar]
- 54.Shin R., Burch A.Y., Huppert K.A., Tiwari S.B., Murphy A.S., Guilfoyle T.J., Schachtman D.P. The Arabidopsis transcription factor MYB77 modulates auxin signal transduction. Plant Cell. 2007;19:2440–2453. doi: 10.1105/tpc.107.050963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gibbs D.J., Voss U., Harding S.A., Fannon J., Moody L.A., Yamada E., Swarup K., Nibau C., Bassel G.W., Choudhary A., et al. AtMYB93 is a novel negative regulator of lateral root development in Arabidopsis. New Phytol. 2014;203:1194–1207. doi: 10.1111/nph.12879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang H.Z., Yang K.Z., Zou J.J., Zhu L.L., Xie Z.D., Morita M.T., Tasaka M., Friml J., Grotewold E., Beeckman T., et al. Transcriptional regulation of PIN genes by FOUR LIPS and MYB88 during Arabidopsis root gravitropism. Nat. Commun. 2015;6:8822. doi: 10.1038/ncomms9822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mu R.L., Cao Y.R., Liu Y.F., Lei G., Zou H.F., Liao Y., Wang H.W., Zhang W.K., Ma B., Du J.Z., et al. An R2R3-type transcription factor gene AtMYB59 regulates root growth and cell cycle progression in Arabidopsis. Cell Res. 2009;19:1291–1304. doi: 10.1038/cr.2009.83. [DOI] [PubMed] [Google Scholar]
- 58.Millar A.A., Gubler F. The Arabidopsis GAMYB-like genes, MYB33 and MYB65, are microRNA-regulated genes that redundantly facilitate anther development. Plant Cell. 2005;17:705–721. doi: 10.1105/tpc.104.027920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li Y., Jiang J., Du M.L., Li L., Wang X.L., Li X.B. A cotton gene encoding MYB-like transcription factor is specifically expressed in pollen and is involved in regulation of late anther/pollen development. Plant Cell Physiol. 2013;54:893–906. doi: 10.1093/pcp/pct038. [DOI] [PubMed] [Google Scholar]
- 60.Murray F., Kalla R., Jacobsen J., Gubler F. A role for HvGAMYB in another development. Plant J. 2003;33:481–491. doi: 10.1046/j.1365-313X.2003.01641.x. [DOI] [PubMed] [Google Scholar]
- 61.Song S., Qi T., Huang H., Ren Q., Wu D., Chang C., Peng W., Liu Y., Peng J., Xie D. The Jasmonate-ZIM domain proteins interact with the R2R3-MYB transcription factors MYB21 and MYB24 to affect Jasmonate-regulated stamen development in Arabidopsis. Plant Cell. 2011;23:1000–1013. doi: 10.1105/tpc.111.083089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mandaokar A., Browse J. MYB108 acts together with MYB24 to regulate jasmonate-mediated stamen maturation in Arabidopsis. Plant Physiol. 2009;149:851–862. doi: 10.1104/pp.108.132597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lee H.G., Mas P., Seo P.J. MYB96 shapes the circadian gating of ABA signaling in Arabidopsis. Sci. Rep. 2016;6:17754. doi: 10.1038/srep17754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yang Y., Zhang L., Chen P., Liang T., Li X., Liu H. UV-B photoreceptor UVR8 interacts with MYB73/MYB77 to regulate auxin responses and lateral root development. EMBO J. 2020;39:e101928. doi: 10.15252/embj.2019101928. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.