Abstract
Background
Chronic thromboembolic pulmonary hypertension (CTEPH) is characterized by elevated pressure in pulmonary arteries. This study was performed to explore the critical miRNAs and genes affecting the pathogenesis of CTEPH.
Methods
GSE56914 dataset (10 CTEPH whole blood samples and 10 control samples) was downloaded from the Gene Expression Omnibus database. Using limma package, the differentially expressed miRNAs (DE-miRNAs) were acquired. After miRNA-target pairs were obtained using miRWalk2.0 tool, a miRNA-target regulatory network was built by Cytoscape software. Using DAVID tool, significantly enriched pathways involving the target genes were identified. Moreover, the protein–protein interaction network and transcription factor-target regulatory network were built by the Cytoscape software. Additionally, quantitative real-time PCR (qRT-PCR) experiments and luciferase assay were conducted to validate miRNA/gene expression and miRNA–target regulatory relationship, respectively.
Results
There were 25 DE-miRNAs (8 up-regulated and 17 down-regulated) between CTEPH and control groups. The target genes of has-let-7b-3p, has-miR-17-5p, has-miR-3202, has-miR-106b-5p, and has-miR-665 were enriched in multiple pathways such as “Insulin secretion”. qRT-PCR analysis confirmed upregulation of hsa-miR-3202, hsa-miR-665, and matrix metalloproteinase 2 (MMP2) as well as downregulation of hsa-let-7b-3p, hsa-miR-17-5p, and hsa-miR-106b-5p. Luciferase assay indicated that MMP2 was negatively mediated by hsa-miR-106b-5p.
Conclusions
These miRNAs and genes were associated with the pathogenesis of CTEPH. Besides, hsa-miR-106b-5p was involved in the development of CTEPH via targeting MMP2.
Keywords: chronic thromboembolic pulmonary hypertension, differentially expressed miRNAs, target genes, regulatory network, protein–protein interaction network, luciferase assay
Introduction
Chronic thromboembolic pulmonary hypertension (CTEPH), as a fourth category of pulmonary hypertension (PH), refers to the elevated pressure in pulmonary arteries, which is induced by stenosis or persistent occlusion of pulmonary arteries.1,2 CTEPH is the result of incomplete thrombolysis of an acute pulmonary embolism that progressively develops into fibrosis.3 The symptoms of early CTEPH are usually absent or non-specific, and advanced CTEPH is only correlated with right heart failure.4 Therefore, CTEPH is hard to be diagnosed at an early stage. Early and accurate diagnosis is important for successful treatment since the outcome of CTEPH patients without timely treatment is usually unfavorable, with a lower than 40% five-year survival rate.5 Thus, the underlying mechanisms of CTEPH should be strongly investigated to offer effective biomarkers or targets for CTEPH diagnosis or treatment.
In recent years, genes or miRNAs involved in CTEPH have been gradually investigated. Satoh et al. have revealed that chronic hypoxia can cause a significant increase in thrombin-activatable fibrinolysis inhibitor and thrombin and thrombomodulin plasma levels, which are associated with thrombus formation in patients with CTEPH.6 In addition, down-regulated let-7d in CTEPH patients can enhance proliferation of pulmonary artery smooth muscle cells (PASMCs), which may be correlated with the development and progression of CTEPH.7,8 Moreover, miR-664a, miR-382, miR-150, miR-337, miR-127, and miR-376c have been found to be abnormally expressed in serum microvesicles of CTEPH patients compared to normal controls.9 Nevertheless, there are still many miRNAs and genes implicated in the pathogenesis of CTEPH that have not been reported.
In 2014, Guo et al. analyzed the dysregulation of miRNAs in plasma samples from healthy controls and CTEPH patients and explored their underlying functions. They discovered an abnormal miRNA signature in CTEPH patients and that decreased levels of let-7b may play a role in CTEPH via affecting endothelin-1 expression, and the function of PASMCs and pulmonary arterial endothelial cells.10 However, they have not explored the key miRNAs involved in CTEPH using comprehensive bioinformatics methods. In this study, multiple bioinformatics analyses were conducted to screen key miRNAs and genes affecting the pathogenesis of CTEPH. Quantitative real-time PCR (qRT-PCR) experiments and luciferase assay were performed to confirm the results of the bioinformatics analysis. Our findings might provide novel targets for the therapy or diagnosis of CTEPH.
Methods
Data source
The microarray dataset GSE56914 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE56914; species: Homo sapiens; platform: GPL18587 miRCURY LNA™ microRNA Arrays (v14.0) was downloaded from the Gene Expression Omnibus (GEO) database. In GSE56914, there were 10 whole blood samples from CTEPH patients and 10 whole blood samples from healthy controls. CTEPH was diagnosed using a previously described protocol.11 Patients with other cardiovascular diseases and malignancies who had undergone treatment for PH except for anticoagulation were excluded from the study. Whole blood samples were collected and centrifuged at 1000 g for 10 min to isolate plasma.10 This study analyzed the microarray dataset uploaded by Guo et al.,10 therefore, ethical review and informed consent were not needed.
Data treatment and differential expression analysis
For the matrix data, expression background correction and standardization pretreatment were performed using the R package limma12 (http://www.bioconductor.org/packages/2.9/bioc/html/limma.html, version 3.10.3).
The miRNA expression matrixes were divided into CTEPH and control groups to screen differentially expressed miRNAs (DE-miRNAs) between the two. Differences were analyzed for significance by the unpaired T test method in limma package.12 The DE-miRNAs were required to have a |log fold change (FC)| > 1 and p value < 0.05.
Prediction of miRNA-target pairs
The top 10 DE-miRNAs (up-regulated or down-regulated) with higher |log FC| values were considered as key DE-miRNAs for further analysis. Using miRWalk2.0 tool (p value < 0.05, seed length ≥ 7, input parameter: 3’UTR, http://zmf.umm.uni-heidelberg.de/mirwalk2),13 target genes of the key DE-miRNAs were predicted. The miRNA-target pairs included in six or more databases of miRWalk, miRanda, mirbridge, miRDB, miRMap, RNA22, Targetscan databases were selected. Using Cytoscape software14 (http://www.cytoscape.org/, version: 3.2.0), a regulatory network was constructed.
Pathway enrichment analysis and building of miRNA-target regulatory network
Since there were too many miRNA-target pairs, the targets corresponding to the miRNAs with the highest degrees were selected for Kyoto Encyclopedia of Genes and Genomes15 (KEGG) analysis. Under the significant threshold of p value < 0.05, KEGG analysis was executed using DAVID tool16 (https://david.ncifcrf.gov/, version 6.8).
To better demonstrate the structure of the regulatory network, the target genes with degree = 1 in the above regulatory network were removed. For the remaining target genes, a regulatory network was built by the Cytoscape software.14
Construction of protein–protein interaction (PPI) network and network module
Combined with STRING database (http://string-db.org/, version 10.0),17 PPI pairs were predicted for the target genes in the regulatory network, and PPI score was defined as 0.7. Afterwards, the PPI network for the target genes was built based on Cytoscape software.14 The degree centrality was used to analyze the scores of the network nodes. Higher node score indicated that the location of the network node was more important. Moreover, the MCODE plug-in (http://apps.cytoscape.org/apps/MCODE, version 1.4.2) in Cytoscape software14 was used to analyze the significant network modules, with a score ≥5 as a threshold. In addition, the module nodes were constructed with the Gene Ontology (GO)_biological process (BP)18 enrichment analysis using DAVID tool.16
Prediction of transcription factor (TF)-target pairs
Based on WebGestalt GAST tool19 (http://www.webgestalt.org/option.php), TFs targeting the genes implicated in the PPI network were predicted through overrepresentation enrichment analysis. The enrichment parameter was set as number of enriched genes ≥2.
qRT-PCR analysis
qRT-PCR experiments were performed in a total of eight whole blood samples from CTEPH patients and eight whole blood samples from healthy controls were obtained from Beijing Chao-Yang Hospital, Capital Medical University. This study’s protocol was approved by the Ethics Committee of Beijing Chao-Yang Hospital, Capital Medical University. The requirement to obtain informed written consent was waived. Using Primer Premier 6.0 software (Premier Software Inc., Cherry Hill, NJ), the primer sequences of hsa-let-7b-3p, hsa-miR-17-5p, hsa-miR-3202, hsa-miR-106b-5p, and hsa-miR-665, as well as hsa-miR-106b-5p target genes bone morphogenetic protein receptor 2 (BMPR2), forkhead box J2 (FOXJ2), and matrix metalloproteinase 2 (MMP2) were designed (Table S1). The primer sequences were produced by Sangon Biotech Co., Ltd (Shanghai, CHN). After performing a reverse transcription reaction, qRT-PCR experiments were conducted using SYBR Green master mix kit (Applied Biosystems, Foster City, CA). The 20 µl amplification reaction included: 1 µl forward primer, and 1 µl reverse primer, 8 µl cDNA template (at a constant concentration), and 10 µl SYBR Premix Ex Taq (2×). The reaction conditions were: 50℃ for 3 min; 95℃ for 3 min; 95℃ for 10 s, and 40 cycles at 60℃ for 30 s; melt curve 60–95℃: increment 0.5℃ for 10 s plate read. All experiments were conducted three times to ensure accuracy.
Luciferase assay
Based on the results of qRT-PCR experiments, hsa-miR-106b-5p/MMP2 pair was selected for performing luciferase assays. The MMP2 3’UTR fragments containing the putative or mutated binding site of hsa-miR-106b-5p were synthesized and inserted into the downstream of luciferase coding sequence in pUC57-Amp vector. Then, the recombinant product was transformed into TOP10 competent cells (Invitrogen, Carlsbad, CA). Single colonies were cultured and plasmids were extracted using Plasmid Extraction Kit (Tiangen, Shanghai, China) based on the manufacturer’s instructions. pUC57-MMP2 3’UTR plasmids and pmir-GLO vector were cut with NheI and XbaI restriction enzymes and then ligated. Subsequently, the recombinant product was transformed into stbl3 competent cells (Invitrogen). After plasmid extraction, identification by enzyme digestion, and sequencing were conducted, positive clones were cultured and plasmids were extracted in large quantities.
The 293T cells were obtained from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). The cells were cultured in Dulbecco’s modified eagle medium (DMEM; Gibco, Grand Island, NY) containing 1% penicillin–streptomycin (Gibco), and 10% fetal bovine serum (Gibco). At the logarithmic growth phase, 293T cells were washed with phosphate buffer solution (PBS) once, digested by 0.05% pancreatin at 37℃ for 1 min, and centrifuged at 300 g for 3 min. The cells were then counted and cultured in a 12-well plate (1.5 × 105 cells/well) overnight at 37℃. The cells were randomly divided into Luc-NC + miR-106b-5p mimics, Luc-MMP2 3’UTR (WT) + miR-106b-5p mimics, Luc-MMP2 3’UTR (WT) + miR-NC, Luc-MMP2 3’UTR (MUT) + miR-106b-5p mimics, Luc-MMP2 3’UTR (MUT) + miR-NC, Luc-NC + miR-NC. The plasmids and miR-106b-5p mimic were added into 300 μl opt-MEM medium (Invitrogen) in a 1:1 ratio (1 µg plasmids: 20 pM siRNA, tube A). Meanwhile, 2 μl lipofectamine 2000 was dissolved in 300 μl opt-MEM medium (tube B). A and B tubes were incubated for 5 min at room temperature, then mixed (tube C) and incubated for another 20 min. The media in the 12-well plate were replaced by opt-MEM medium (800 μl/well), and the mixture in tube C was added into the corresponding well. After performing transfection for 4–6 h, FAM-NC fluorescence intensity was observed under fluorescence microscope, and the medium was replaced with DMEM.
Fluorescence intensity of the overexpressed GFP was observed and photographed under a fluorescence microscope 48 h after transfection. Then, the cells were washed with PBS, lysed with lysis buffer (200 μl/well) for 30 min, and centrifuged at 12,000 g at 4℃ for 10 min. The dual-luciferase assay kit (Beyotime, Nantong, China) and a multimode reader (BioTek, Winooski, VT) were used to detect firefly and Renilla luciferases.
Statistical analysis
Combined with the 2−ΔΔCt method,20 the expression values of the key miRNAs and genes were calculated. Statistical analysis of the experimental data was conducted using SPSS 16.0 software (SPSS Inc., Chicago, IL). All data were presented as mean ± standard deviation. The one-way analysis of variance21 was utilized for comparisons between groups. The p < 0.05 and p < 0.01 indicated a significant difference and a highly significant difference, respectively.
Results
Differential expression analysis
Following initial data analysis, 898 miRNAs were identified from the raw data. According to the selection criteria, 25 DE-miRNAs between CTEPH and control groups were analyzed, including eight up-regulated miRNAs (such as hsa-miR-3202 and hsa-miR-665) and 17 down-regulated miRNAs (such as hsa-miR-106b, hsa-let-7b, hsa-miR-17, hsa-miR-93, and hsa-miR-20a; Table S2).
Pathway enrichment analysis and miRNA-target regulatory network analysis
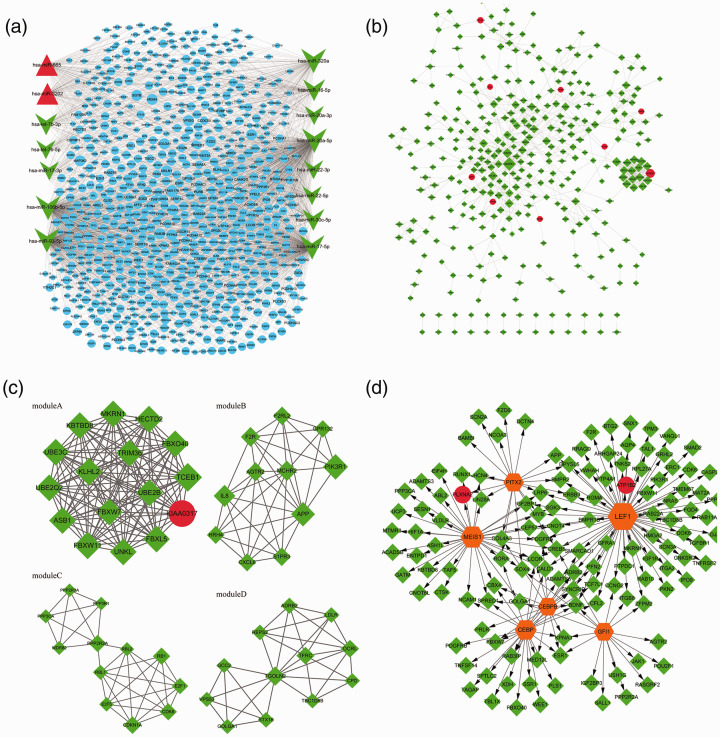
Through limiting miRNAs and the number of target genes, 2269 miRNA-target pairs (involving 2 up-regulated miRNAs, 13 down-regulated miRNAs, and 756 target genes) were finally obtained. Based on the 2269 miRNA-target pairs (including hsa-miR-106b-5p → BMPR2, hsa-miR-106b-5p → FOXJ2, and hsa-miR-106b-5p → MMP2), the miRNA-target regulatory network was constructed (Fig. 1a).
Fig. 1.
The miRNA-target regulatory network. Red triangles, green arrows, and blue circles represent up-regulated miRNAs, down-regulated miRNAs, and target genes, respectively (a); the protein–protein interaction network. Red circles and green diamonds represent the target genes of up-regulated miRNAs and down-regulated miRNAs, respectively. The higher the degree of a node, the larger the node is (b). The modules (a), (b), (c), and (d) identified from the protein–protein interaction network. Red circles and green diamonds represent the target genes of up-regulated miRNAs and down-regulated miRNAs, respectively. The higher the degree of a node, the larger the node is (c). The transcription factor (TF)-target regulatory network. Red circles and green diamonds represent the target genes of up-regulated miRNAs and down-regulated miRNAs, respectively. Orange hexagons stand for TFs. The higher the degree of a node, the larger the node is (d).
Pathway enrichment analysis was conducted for the target genes of the top 15 miRNAs (listed by degree values) in the miRNA-target regulatory network. As a result, the top 10 pathways for the targets of the up-regulated has-miR-665 (such as Hippo signaling pathway) and has-miR-3202 (such as Insulin secretion) are listed in Table 1a. Meanwhile, the top five pathways for the targets of the down-regulated has-miR-17-5p (such as chronic myeloid leukemia), has-miR-93-5p (such as chronic myeloid leukemia), has-miR-20a-5p (such as TGF-beta signaling pathway), has-miR-106b-5p (such as MAPK signaling pathway and pathways in cancer), has-miR-320a (such as signaling pathways regulating pluripotency of stem cells), has-miR-16-5p (such as ubiquitin-mediated proteolysis), has-miR-30c-5p (such as axon guidance), has-let-7b-3p (such as adherens junction), has-miR-17-3p (such as oxytocin signaling pathway), has-miR-22-3p (such as insulin resistance), has-miR-22-5p (such as Hippo signaling pathway), has-let-7b-5p (such as signaling pathways regulating pluripotency of stem cells), and has-miR-20a-3p (such as adherens junction) are listed in Table 1b.
Table 1.
The pathways enriched for the target genes of the top 15 miRNAs in the miRNA-target regulatory network. (a) The pathways enriched for the target genes of the up-regulated miRNAs (top 10 listed); (b) the pathways enriched for the target genes of the down-regulated miRNAs (top 5 listed).
| miRNA | ID | Name | Count | p value |
|---|---|---|---|---|
| (a) | ||||
| has-miR-665 | hsa04390 | Hippo signaling pathway | 10 | 2.31E − 03 |
| hsa04022 | cGMP-PKG signaling pathway | 9 | 1.39E − 02 | |
| hsa04020 | Calcium signaling pathway | 9 | 2.09E − 02 | |
| hsa04014 | Ras signaling pathway | 10 | 2.86E − 02 | |
| hsa05212 | Pancreatic cancer | 5 | 3.52E − 02 | |
| hsa05200 | Pathways in cancer | 14 | 3.67E − 02 | |
| hsa05205 | Proteoglycans in cancer | 9 | 3.72E − 02 | |
| hsa04115 | p53 signaling pathway | 5 | 3.87E − 02 | |
| hsa04917 | Prolactin signaling pathway | 5 | 4.63E − 02 | |
| hsa04520 | Adherens junction | 5 | 4.63E − 02 | |
| has-miR-3202 | hsa05214 | Glioma | 5 | 5.01E − 03 |
| hsa05206 | MicroRNAs in cancer | 9 | 1.08E − 02 | |
| hsa04151 | PI3K-Akt signaling pathway | 10 | 1.09E − 02 | |
| hsa04911 | Insulin secretion | 5 | 1.28E − 02 | |
| hsa05215 | Prostate cancer | 5 | 1.44E − 02 | |
| hsa04024 | cAMP signaling pathway | 7 | 1.89E − 02 | |
| hsa05161 | Hepatitis B | 6 | 1.92E − 02 | |
| hsa04261 | Adrenergic signaling in cardiomyocytes | 6 | 1.97E − 02 | |
| hsa05223 | Non-small cell lung cancer | 4 | 2.18E − 02 | |
| hsa04022 | cGMP-PKG signaling pathway | 6 | 3.21E − 02 | |
| (b) | ||||
| has-miR-17-5p | hsa05220 | Chronic myeloid leukemia | 8 | 1.73E − 03 |
| hsa05214 | Glioma | 7 | 4.77E − 03 | |
| hsa05160 | Hepatitis C | 10 | 4.95E − 03 | |
| hsa04012 | ErbB signaling pathway | 8 | 5.06E − 03 | |
| hsa05218 | Melanoma | 7 | 7.34E − 03 | |
| has-miR-93-5p | hsa05220 | Chronic myeloid leukemia | 8 | 1.26E − 03 |
| hsa05160 | Hepatitis C | 10 | 3.44E − 03 | |
| hsa05200 | Pathways in cancer | 19 | 3.85E − 03 | |
| hsa05166 | HTLV-I infection | 14 | 5.93E − 03 | |
| hsa04360 | Axon guidance | 9 | 8.74E − 03 | |
| has-miR-20a-5p | hsa04350 | TGF-beta signaling pathway | 7 | 1.15E − 02 |
| hsa04310 | Wnt signaling pathway | 9 | 1.26E − 02 | |
| hsa04360 | Axon guidance | 8 | 2.45E − 02 | |
| hsa05166 | HTLV-I infection | 12 | 2.98E − 02 | |
| hsa05200 | Pathways in cancer | 16 | 3.14E − 02 | |
| has-miR-106b-5p | hsa04010 | MAPK signaling pathway | 13 | 8.35E − 03 |
| hsa05200 | Pathways in cancer | 16 | 2.07E − 02 | |
| hsa04310 | Wnt signaling pathway | 8 | 2.83E − 02 | |
| has-miR-320a | hsa04550 | Signaling pathways regulating pluripotency of stem cells | 7 | 1.01E − 02 |
| hsa05211 | Renal cell carcinoma | 5 | 1.03E − 02 | |
| hsa05203 | Viral carcinogenesis | 8 | 1.81E − 02 | |
| hsa05215 | Prostate cancer | 5 | 2.84E − 02 | |
| has-miR-16-5p | hsa04120 | Ubiquitin-mediated proteolysis | 7 | 3.34E − 03 |
| hsa04070 | Phosphatidylinositol signaling system | 6 | 3.85E − 03 | |
| hsa04115 | p53 signaling pathway | 4 | 3.47E − 02 | |
| has-miR-30c-5p | hsa04360 | Axon guidance | 5 | 2.54E − 02 |
| hsa04710 | Circadian rhythm | 3 | 3.03E − 02 | |
| has-let-7b-3p | hsa04520 | Adherens junction | 4 | 1.02E − 02 |
| hsa05200 | Pathways in cancer | 8 | 1.13E − 02 | |
| hsa04024 | cAMP signaling pathway | 5 | 3.64E − 02 | |
| has-miR-17-3p | hsa04921 | Oxytocin signaling pathway | 5 | 9.66E − 03 |
| has-miR-22-3p | hsa04931 | Insulin resistance | 5 | 6.97E − 03 |
| hsa04144 | Endocytosis | 6 | 3.49E − 02 | |
| hsa00510 | N-Glycan biosynthesis | 3 | 4.69E − 02 | |
| has-miR-22-5p | hsa04390 | Hippo signaling pathway | 4 | 4.63E − 02 |
| hsa05205 | Proteoglycans in cancer | 7 | 2.43E − 03 | |
| hsa04916 | Melanogenesis | 5 | 4.93E − 03 | |
| hsa05206 | MicroRNAs in cancer | 7 | 1.33E − 02 | |
| hsa04390 | Hippo signaling pathway | 5 | 2.03E − 02 | |
| has-let-7b-5p | hsa04550 | Signaling pathways regulating pluripotency of stem cells | 7 | 3.79E − 04 |
| hsa05205 | Proteoglycans in cancer | 7 | 2.43E − 03 | |
| hsa04916 | Melanogenesis | 5 | 4.93E − 03 | |
| hsa05206 | MicroRNAs in cancer | 7 | 1.33E − 02 | |
| hsa04390 | Hippo signaling pathway | 5 | 2.03E − 02 | |
| has-miR-20a-3p | hsa04520 | Adherens junction | 4 | 8.38E − 03 |
Building of PPI network and network module
Using targets in the miRNA-target network, a PPI network (involving 349 nodes and 751 edges) was built (Fig. 1b). From the PPI network, four significant modules (module A, score = 16, involving 120 edges and 16 nodes; module B, score = 7.6, involving 38 edges and 11 nodes; module C, score = 5.818, involving 32 edges and 12 nodes; module D, score = 5.818, involving 32 edges and 12 nodes) were screened (Fig. 1c).
For the genes included in each module, GO_BP enrichment analysis was conducted. The top 5 GO_BP terms for the nodes in module A (such as protein polyubiquitination), B (such as inflammatory response), C (such as regulation of lipid kinase activity), and D (such as retrograde transport, endosome to Golgi) are listed in Table 2.
Table 2.
The Gene Ontology (GO)_biological process (BP) terms enriched for the nodes in module A, B, C, and D (top 5 listed).
| Module | ID | Name | Count | p value | Genes |
|---|---|---|---|---|---|
| Module A | GO:0000209 | Protein polyubiquitination | 8 | 5.44E − 11 | MKRN1, FBXW7, TRIM36, HECTD2, UBE3C, UBE2B, FBXW11, UNKL |
| GO:0016567 | Protein ubiquitination | 8 | 5.81E − 09 | FBXW7, FBXO40, KBTBD8, FBXL5, ASB1, UBE2B, FBXW11, KLHL2 | |
| GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process | 3 | 1.76E − 04 | FBXW7, FBXL5, FBXW11 | |
| GO:0042787 | Protein ubiquitination involved in ubiquitin-dependent protein catabolic process | 3 | 6.99E − 03 | HECTD2, UBE3C, TCEB1 | |
| GO:0070936 | Protein K48-linked ubiquitination | 2 | 3.85E − 02 | UBE2Q2, UBE2B | |
| Module B | GO:0006954 | Inflammatory response | 5 | 2.94E − 05 | S1PR3, AGTR2, HRH4, CXCL6, F2R |
| GO:0007186 | G-protein coupled receptor signaling pathway | 5 | 8.28E − 04 | S1PR3, AGTR2, GPR132, CXCL6, F2R | |
| GO:0032651 | Regulation of interleukin-1 beta production | 2 | 1.61E − 03 | S1PR3, F2R | |
| GO:0030168 | Platelet activation | 3 | 1.62E − 03 | F2RL2, PIK3R1, F2R | |
| GO:0007204 | Positive regulation of cytosolic calcium ion concentration | 3 | 2.19E − 03 | S1PR3, HRH4, F2R | |
| Module C | GO:0043550 | Regulation of lipid kinase activity | 3 | 3.90E − 06 | RBL2, RBL1, RB1 |
| GO:0000082 | G1/S transition of mitotic cell cycle | 4 | 3.47E − 05 | CDKN1A, CDK6, PPP3CA, RB1 | |
| GO:0051726 | Regulation of cell cycle | 4 | 6.21E − 05 | E2F5, RBL2, RBL1, RB1 | |
| GO:0006470 | Protein dephosphorylation | 4 | 6.51E − 05 | PPP2R3A, PPP3R1, PPP3CA, PPP2R2A | |
| GO:0010629 | Negative regulation of gene expression | 4 | 8.36E − 05 | CDKN1A, RBL2, RBL1, RB1 | |
| Module D | GO:0042147 | Retrograde transport, endosome to Golgi | 3 | 8.94E − 04 | STX16, VPS53, GCC2 |
| GO:0090161 | Golgi ribbon formation | 2 | 7.18E − 03 | STX16, GCC2 | |
| GO:0000042 | Protein targeting to Golgi | 2 | 1.30E − 02 | GOLGA1, GCC2 |
Construction of TF-target regulatory network
Among the top 10 predicted results, six known TFs predicted for the genes in the PPI network. The TF-target regulatory network was comprised of 187 TF-target pairs, and 134 nodes (involving 6 TFs, 2 targets of up-regulated miRNAs, and 126 targets of down-regulated miRNAs; Fig. 1d).
qRT-PCR analysis
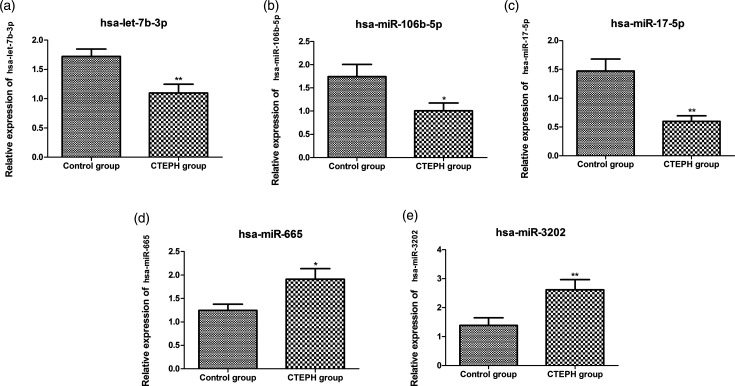
In comparison to control group, hsa-let-7b-3p, hsa-miR-106b-5p, and hsa-miR-17-5p were significantly down-regulated, and hsa-miR-665 and hsa-miR-3202 were significantly up-regulated in the CTEPH group (p < 0.05, Fig. 2). Besides, the expression levels of hsa-miR-106b-5p targets BMPR2, FOXJ2 and MMP2 were also measured. The results showed that the expression levels of BMPR2 and FOXJ2 were not significantly changed, while the expression of MMP2 was significantly down-regulated in the CTEPH group compared to the control group (p < 0.05, Fig. S1).
Fig. 2.
The relative expression of hsa-let-7b-3p (a), hsa-miR-106b-5p (b), hsa-miR-17-5p (c), hsa-miR-665 (d), and hsa-miR-3202 (e). CTEPH: chronic thromboembolic pulmonary hypertension. *Indicates p < 0.05 and **indicates p < 0.01.
Luciferase assay
The luciferase ratio in wide-type Luc-MMP2 3’UTR + miR-106b-5p mimics group was significantly reduced in comparison to other five groups (Luc-NC + miR-106b-5p mimics, wide-type Luc-MMP2 3’UTR + miR-NC, Luc-NC + miR-NC, mutant MMP2 3’UTR + miR-NC and mutant MMP2 3’UTR + miR-106b-5p mimics groups (p < 0.01, Fig. S2). However, no significant changes of luciferase activity were identified in these five groups. The inhibitory effect of luciferase ratio was recovered in mutant MMP2 3’UTR + miR-NC and mutant MMP2 3’UTR + miR-106b-5p mimics groups than wide-type Luc-MMP2 3’UTR + miR-106b-5p mimics group (Fig. S2).
Discussion
A total of 25 DE-miRNAs (8 up-regulated (e.g., hsa-miR-665 and hsa-miR-3202) and 17 down-regulated (e.g., hsa-let-7b-3p, hsa-miR-106b-5p, and hsa-miR-17-5p)) between CTEPH and control groups were identified by differential expression analysis. In agreement with the bioinformatic prediction, the upregulation of hsa-miR-665 and hsa-miR-3202, and downregulation of hsa-let-7b-3p, hsa-miR-106b-5p, and hsa-miR-17-5p were also detected in eight CTEPH samples through qRT-PCR analysis. Several pathways were enriched for the genes targeted by has-miR-3202 (such as insulin resistance). In addition, the result of the luciferase assay showed that hsa-miR-106b-5p negatively regulated MMP2 expression.
Let-7b and let-7g expression in remodeled pulmonary artery are significantly decreased, and the loss of the inhibitory effect of let-7g on c-myc-B lymphoma Mo-MLV insertion region 1 homolog (Bmi-1)-p16 pathway may induce PASMCs proliferation resulting in vascular remodeling in rats with hypoxic pulmonary hypertension.22 miR-17 improves the functions of the lungs and heart in experimental PH through up-regulating p21 and, therefore, may be used for PH treatment.23 miR-17 affects the proliferation and apoptosis of PASMCs via regulating mitofusin 2, and thus may serve as a novel target for treating pulmonary arterial hypertension (PAH).24 These results indicated that hsa-let-7b-3p and hsa-miR-17-5p might be related to the mechanisms of CTEPH.
In this study, increased expression of hsa-miR-3202 and hsa-miR-665 was detected in CTEPH patients, and hsa-miR-3202 was predicted to be associated with the function “Insulin secretion”. It has been reported that miR-3202 overexpression may have a protective effect on epithelial cells by inhibiting Fas-apoptotic inhibitory molecule 2 (FAIM2), and thus miR-3202 may be considered as a promising marker of smoking-related chronic obstructive pulmonary disease (COPD).25 Similarly, miR-3202 is implicated in the proliferation and apoptosis of epithelial cells in high-glucose medium via targeting FAIM2.26 Interestingly, it has been revealed that hyperglycemia and insulin resistance account for the development of PAH via regulating the proliferation of epithelial or smooth muscle cells (SMCs) and fibrosis.27 Thus, we hypothesized that miR-3202 might be related to the development of CTEPH via participating in insulin resistance. In addition, it has been reported that miR-665 overexpression can promote the proliferation and migration of vascular SMCs through regulating myocyte enhancer factor 2D and fibroblast growth factor 9.28 Notably, the proliferation of pulmonary artery SMCs is a key factor for CTEPH development.29 Collectively, hsa-miR-3202 and hsa-miR-665 might play essential roles in the pathogenesis of CTEPH.
MMP2, a Zn+2-dependent endopeptidase, plays a role in the degradation of proteins in the extracellular matrix, which is implicated in many BPs such as oncogenesis, regulation of vascularization, and wound healing.30 MMP2/tissue inhibitor of metalloprotease 4 ratio is found to be an indicator of right ventricular function, disease severity, and survival prognosis, which may contribute to the progression of idiopathic PAH.31,32 Our RT-PCR results showed that MMP2 was up-regulated in CTEPH patients compared with controls. Consistently, increased expression of MMP2 is found in CTEPH patients, showing that its dysregulation is involved in pulmonary vascular remodeling.33 In addition, we found a significant decrease in the expression of miR-106b-5p in CTEPH patients, and the targeting of the 3’UTR of MMP2 was confirmed by luciferase reporter assay. Reportedly, the progressive decline of plasma miR-106b may reflect the changes occurring in the patients with COPD, indicating that miR-106b may be correlated with the development and progression of COPD.34 Moreover, downregulation of miR-106b may promote the invasion and motility of breast cancer cells by mediating upregulation of MMP2.35 Notably, the correlation of miR-106b-5p and MMP2 in CTEPH has not been reported. Thus, we suggested that downregulation of miR-106b-5p might be implicated in the development of CTEPH via upregulation of MMP2. However, our study is limited by lacking of the expression verification of MMP2 in the presence of a miR-106b-5p mimic or inhibitor, and following functional experiments.
In conclusion, several dysregulated miRNAs (including hsa-let-7b-3p, hsa-miR-17-5p, hsa-miR-665, and hsa-miR-3202) might account for the pathogenesis of CTEPH. In addition, hsa-miR-106b-5p may be involved in the development of CTEPH via negatively regulating MMP2 expression. However, more experiments are still necessary to explore and confirm the roles of these miRNAs and genes in CTEPH.
Supplemental Material
Supplemental material, sj-pdf-1-pul-10.1177_2045894020928300 for hsa-miR-106b-5p participates in the development of chronic thromboembolic pulmonary hypertension via targeting matrix metalloproteinase 2 by Ran Miao, Xingbei Dong, Juanni Gong, Ying Wang, Xiaojuan Guo, Yidan Li, Min Liu, Jun Wan, Jifeng Li, Suqiao Yang, Wang Wang, Tuguang Kuang, Jiuchang Zhong, Zhenguo Zhai and Yuanhua Yang in Pulmonary Circulation
Supplemental material, sj-pdf-2-pul-10.1177_2045894020928300 for hsa-miR-106b-5p participates in the development of chronic thromboembolic pulmonary hypertension via targeting matrix metalloproteinase 2 by Ran Miao, Xingbei Dong, Juanni Gong, Ying Wang, Xiaojuan Guo, Yidan Li, Min Liu, Jun Wan, Jifeng Li, Suqiao Yang, Wang Wang, Tuguang Kuang, Jiuchang Zhong, Zhenguo Zhai and Yuanhua Yang in Pulmonary Circulation
Supplemental material, sj-pdf-3-pul-10.1177_2045894020928300 for hsa-miR-106b-5p participates in the development of chronic thromboembolic pulmonary hypertension via targeting matrix metalloproteinase 2 by Ran Miao, Xingbei Dong, Juanni Gong, Ying Wang, Xiaojuan Guo, Yidan Li, Min Liu, Jun Wan, Jifeng Li, Suqiao Yang, Wang Wang, Tuguang Kuang, Jiuchang Zhong, Zhenguo Zhai and Yuanhua Yang in Pulmonary Circulation
Supplemental material, sj-pdf-4-pul-10.1177_2045894020928300 for hsa-miR-106b-5p participates in the development of chronic thromboembolic pulmonary hypertension via targeting matrix metalloproteinase 2 by Ran Miao, Xingbei Dong, Juanni Gong, Ying Wang, Xiaojuan Guo, Yidan Li, Min Liu, Jun Wan, Jifeng Li, Suqiao Yang, Wang Wang, Tuguang Kuang, Jiuchang Zhong, Zhenguo Zhai and Yuanhua Yang in Pulmonary Circulation
Acknowledgments
All authors have read and approved the manuscript, and ensure that this is the case.
Declaration of conflicting interests
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding
The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This study was supported by National Natural Science Foundation of China (Project number: 81300044, 31670928, 81871356, 81871328, 81770253), Beijing Natural Science Foundation (Project number: 7162069, 7182149), Beijing Municipal Administration of Hospitals’ Youth Programme (Project number: QML20160301), the Open Foundation from Beijing Key Laboratory of Hypertension Research (2018GXY-KFKT-02), the National Key Research and Development Program of China (Project number: 2016YFC0905600), the National Major Research Plan Training Program of China (91849111), and Chinese Academy of Medical Sciences Central Public-interest Scientific Institution Basal Research Fund Young Medical Talents Award Project (Project number: 2018RC320013).
Ethical approval
This study was approved by Ethics Committee of Beijing Chao-Yang Hospital, Capital Medical University.
Authors’ contribution
Conception and design of the research: Ran Miao and Yuanhua Yang. Acquisition of data: Xingbei Dong, Juanni Gong, Yidan Li, Suqiao Yang and Zhenguo Zhai. Analysis and interpretation of data: Xingbei Dong, Juanni Gong, Yidan Li, Ying Wang, Jun Wan, Jiuchang Zhong and Wang Wang. Statistical analysis: Xiaojuan Guo, Min Liu, Jifeng Li and Tuguang Kuang. Obtaining funding; Ran Miao, Zhenguo Zhai, Jiuchang Zhong and Yuanhua Yang. Drafting the manuscript: Ran Miao and Yuanhua Yang. Revision of manuscript for important intellectual content: Ran Miao and Yuanhua Yang.
ORCID iDs
Min Liu https://orcid.org/0000-0003-1298-4441
Suqiao Yang https://orcid.org/0000-0001-7369-5999
Yuanhua Yang https://orcid.org/0000-0002-4197-9097
Supplemental Material
Supplemental material for this article is available online.
References
- 1.Simonneau G, Torbicki A, Dorfmüller P, et al. The pathophysiology of chronic thromboembolic pulmonary hypertension. Eur Respir Rev 2017; 26: 160112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lang IM, Raffaele P, Diana B, et al. Risk factors and basic mechanisms of chronic thromboembolic pulmonary hypertension: a current understanding. Eur Respir J 2013; 41: 462–468. [DOI] [PubMed] [Google Scholar]
- 3.Delcroix M, Kerr K, Fedullo P. Chronic thromboembolic pulmonary hypertension. Epidemiology and risk factors. Ann Am Thorac Soc 2016; 13: S201. [DOI] [PubMed] [Google Scholar]
- 4.Takumi I, Masaharu K, Haruhisa I, et al. Percutaneous transluminal pulmonary angioplasty for chronic thromboembolic pulmonary hypertension with severe right heart failure. Am J Respir Crit Care Med 2014; 189: 1437. [DOI] [PubMed] [Google Scholar]
- 5.Rintaro N, Nobuhiro T, Toshihiko S, et al. Improved survival in medically treated chronic thromboembolic pulmonary hypertension. Circ J 2013; 77: 2110–2117. [DOI] [PubMed] [Google Scholar]
- 6.Satoh T, Satoh K, Yaoita N, et al. Activated TAFI promotes the development of chronic thromboembolic pulmonary hypertension: a possible novel therapeutic target. Circul Res 2017; 120: 1246–1262. [DOI] [PubMed] [Google Scholar]
- 7.Wang L, Guo LJ, Liu J, et al. MicroRNA expression profile of pulmonary artery smooth muscle cells and the effect of let-7d in chronic thromboembolic pulmonary hypertension. Pulm Circ 2013; 3: 654–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Izumiya Y, Kimura Y, Onoue Y, et al. Expression of Let-7 family microRNAs in skin correlates negatively with severity of pulmonary hypertension in patients with systemic scleroderma. Int J Cardiol Heart Vasc 2015; 8: 98–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lipps C, Northe P, Figueiredo R, et al. P535 Identification of specific profiles of small non-coding RNAs derived from microvesicles of CTEPH patients. Cardiovasc Res 2018; 114: S131–S131. [Google Scholar]
- 10.Guo L, Yang Y, Liu J, et al. Differentially expressed plasma microRNAs and the potential regulatory function of Let-7b in chronic thromboembolic pulmonary hypertension. PLoS One 2014; 9: e101055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barst RJ, Michael MG, Adam T, et al. Diagnosis and differential assessment of pulmonary arterial hypertension. J Am Coll Cardiol 2004; 43: S40–S47. [DOI] [PubMed] [Google Scholar]
- 12.Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015; 43: e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dweep H, Gretz N. miRWalk2.0: a comprehensive atlas of microRNA-target interactions. Nat Methods 2015; 12: 697. [DOI] [PubMed] [Google Scholar]
- 14.Kohl M, Wiese S, Warscheid B. Cytoscape: software for visualization and analysis of biological networks. Methods Mol Biol 2011; 696: 291–303. [DOI] [PubMed] [Google Scholar]
- 15.Kanehisa M, Sato Y, Kawashima M, et al. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res 2016; 44: D457–D462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009; 4: 44. [DOI] [PubMed] [Google Scholar]
- 17.Szklarczyk D, Morris JH, Cook H, et al. The STRING database in 2017: quality-controlled protein–protein association networks, made broadly accessible. Nucleic Acids Res 2017; 45: D362–D368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Young MD, Wakefield MJ, Smyth GK, et al. Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol 2010; 11: R14–R14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang J, Vasaikar S, Shi Z, et al. WebGestalt 2017: a more comprehensive, powerful, flexible and interactive gene set enrichment analysis toolkit. Nucleic Acids Res 2017; 45: w130–w137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arocho A, Chen B, Ladanyi M, et al. Validation of the 2-DeltaDeltaCt calculation as an alternate method of data analysis for quantitative PCR of BCR-ABL P210 transcripts. Diagn Mol Pathol 2006; 15: 56. [DOI] [PubMed] [Google Scholar]
- 21.Quirk T.J., Quirk M.H., and Horton H.F. One-Way Analysis of Variance (ANOVA). In: Quirk T.J., Quirk M.H., and Horton H.F (eds) Excel 2013 for Physical Sciences Statistics. Springer, Switzerland, 2016, pp. 167–183.
- 22.Zhang WF, Xiong YW, Zhu TT, et al. MicroRNA let-7g inhibited hypoxia-induced proliferation of PASMCs via G 0 /G 1 cell cycle arrest by targeting c-myc. Life Sci 2016; 170: 9. [DOI] [PubMed] [Google Scholar]
- 23.Pullamsetti SS, Carmen D, Ariane F, et al. Inhibition of microRNA-17 improves lung and heart function in experimental pulmonary hypertension. Am J Respir Crit Care Med 2012; 185: 409. [DOI] [PubMed] [Google Scholar]
- 24.Zheng L, Li S, Zhao S, et al. Upregulated miR-17 regulates hypoxia-mediated human pulmonary artery smooth muscle cell proliferation and apoptosis by targeting mitofusin 2. Med Sci Monit 2016; 22: 3301–3308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shen W, Liu J, Fan M, et al. MiR-3202 protects smokers from chronic obstructive pulmonary disease through inhibiting FAIM2: an in vivo and in vitro study. Exp Cell Res 2018; 362: 370–377. [DOI] [PubMed] [Google Scholar]
- 26.Huang X, Xie H, Xue G, et al. MiR-3202 – promoted H5V cell apoptosis by directly targeting Fas apoptotic inhibitory molecule 2 (FAIM2) in high glucose condition. Med Sci Monit 2017; 23: 975–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grinnan D, Farr G, Fox A, et al. The role of hyperglycemia and insulin resistance in the development and progression of pulmonary arterial hypertension. J Diabetes Res 2016; 2016: 2481659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li K, Pan J, Wang J, et al. MiR-665 regulates VSMCs proliferation via targeting FGF9 and MEF2D and modulating activities of Wnt/β-catenin signaling. Am J Transl Res 2017; 9: 4402. [PMC free article] [PubMed] [Google Scholar]
- 29.Opitz I, Kirschner MB. Molecular research in chronic thromboembolic pulmonary hypertension. Int J Mol Sci 2019; 20: 784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kundu GC, Patil DP. MMP2 (matrix metallopeptidase 2 (gelatinase A, 72 kDa gelatinase, 72 kDa type IV collagenase). Atlas Genet Cytogene Oncol Haematol 2006; 10: 88–89. [Google Scholar]
- 31.Wetzl V, Tiede SL, Faerber L, et al. Plasma MMP2/TIMP4 ratio at follow-up assessment predicts disease progression of idiopathic pulmonary arterial hypertension. Lung 2017; 195: 1–8. [DOI] [PubMed] [Google Scholar]
- 32.Wang XM, Shi K, Li JJ, et al. Effects of angiotensin II intervention on MMP-2, MMP-9, TIMP-1, and collagen expression in rats with pulmonary hypertension. Genet Mol Res 2015; 14: 1707–1717. [DOI] [PubMed] [Google Scholar]
- 33.Wilde M, BmedSci MM, Sheth A, et al. Elevated VEGF, TIMP-1 and MMP-2 in chronic thromboembolic pulmonary hypertension – a potential role in pathogenesis. Am J Respir Crit Care Med. Epub ahead of print 2009. 10.1164/ajrccm-conference.2009.179.1_MeetingAbstracts.A3327. [DOI] [Google Scholar]
- 34.Seiko S, Ohyashiki JH, Kazushige O, et al. Clinical relevance of plasma miR-106b levels in patients with chronic obstructive pulmonary disease. Int J Mol Med 2013; 31: 533–539. [DOI] [PubMed] [Google Scholar]
- 35.Xiaojian N, Tiansong X, Yingchun Z, et al. Downregulation of miR-106b induced breast cancer cell invasion and motility in association with overexpression of matrix metalloproteinase 2. Cancer Sci 2014; 105: 18–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-pdf-1-pul-10.1177_2045894020928300 for hsa-miR-106b-5p participates in the development of chronic thromboembolic pulmonary hypertension via targeting matrix metalloproteinase 2 by Ran Miao, Xingbei Dong, Juanni Gong, Ying Wang, Xiaojuan Guo, Yidan Li, Min Liu, Jun Wan, Jifeng Li, Suqiao Yang, Wang Wang, Tuguang Kuang, Jiuchang Zhong, Zhenguo Zhai and Yuanhua Yang in Pulmonary Circulation
Supplemental material, sj-pdf-2-pul-10.1177_2045894020928300 for hsa-miR-106b-5p participates in the development of chronic thromboembolic pulmonary hypertension via targeting matrix metalloproteinase 2 by Ran Miao, Xingbei Dong, Juanni Gong, Ying Wang, Xiaojuan Guo, Yidan Li, Min Liu, Jun Wan, Jifeng Li, Suqiao Yang, Wang Wang, Tuguang Kuang, Jiuchang Zhong, Zhenguo Zhai and Yuanhua Yang in Pulmonary Circulation
Supplemental material, sj-pdf-3-pul-10.1177_2045894020928300 for hsa-miR-106b-5p participates in the development of chronic thromboembolic pulmonary hypertension via targeting matrix metalloproteinase 2 by Ran Miao, Xingbei Dong, Juanni Gong, Ying Wang, Xiaojuan Guo, Yidan Li, Min Liu, Jun Wan, Jifeng Li, Suqiao Yang, Wang Wang, Tuguang Kuang, Jiuchang Zhong, Zhenguo Zhai and Yuanhua Yang in Pulmonary Circulation
Supplemental material, sj-pdf-4-pul-10.1177_2045894020928300 for hsa-miR-106b-5p participates in the development of chronic thromboembolic pulmonary hypertension via targeting matrix metalloproteinase 2 by Ran Miao, Xingbei Dong, Juanni Gong, Ying Wang, Xiaojuan Guo, Yidan Li, Min Liu, Jun Wan, Jifeng Li, Suqiao Yang, Wang Wang, Tuguang Kuang, Jiuchang Zhong, Zhenguo Zhai and Yuanhua Yang in Pulmonary Circulation