Abstract
Next-Generation Risk Assessment is defined as an exposure-led, hypothesis-driven risk assessment approach that integrates new approach methodologies (NAMs) to assure safety without the use of animal testing. These principles were applied to a hypothetical safety assessment of 0.1% coumarin in face cream and body lotion. For the purpose of evaluating the use of NAMs, existing animal and human data on coumarin were excluded. Internal concentrations (plasma Cmax) were estimated using a physiologically based kinetic model for dermally applied coumarin. Systemic toxicity was assessed using a battery of in vitro NAMs to identify points of departure (PoDs) for a variety of biological effects such as receptor-mediated and immunomodulatory effects (Eurofins SafetyScreen44 and BioMap Diversity 8 Panel, respectively), and general bioactivity (ToxCast data, an in vitro cell stress panel and high-throughput transcriptomics). In addition, in silico alerts for genotoxicity were followed up with the ToxTracker tool. The PoDs from the in vitro assays were plotted against the calculated in vivo exposure to calculate a margin of safety with associated uncertainty. The predicted Cmax values for face cream and body lotion were lower than all PoDs with margin of safety higher than 100. Furthermore, coumarin was not genotoxic, did not bind to any of the 44 receptors tested and did not show any immunomodulatory effects at consumer-relevant exposures. In conclusion, this case study demonstrated the value of integrating exposure science, computational modeling and in vitro bioactivity data, to reach a safety decision without animal data.
Keywords: Next-Generation Risk Assessment, new approach methodologies, exposure science
The ambition to conduct human health risk assessments without generating new animal data has resulted in intense efforts over the past few decades from industry, academia, and regulatory bodies to develop and apply new approach methodologies (NAMs) that can form the basis of integrated testing and assessment strategies designed to prevent harm to human health (Carmichael et al., 2009; Council, 2007; Desprez et al., 2018; Thomas et al., 2019; Westmoreland et al., 2010). The momentum created by these efforts has led to a number of studies that employed 1 or more NAMs in risk assessment. The studies that focused on specific pathways, such as DNA damage for quercetin (Adeleye et al., 2015), estrogenic (Becker et al., 2015), and antiandrogenic activity (Dent et al., 2018b) using the concept of Dietary Comparator Ratio have provided a valuable insight into how adverse outcome pathways (AOPs) can be put into the context of safety assessment (Carusi et al., 2018; Villeneuve et al., 2014). However, it may not always be necessary or possible to identify a specific pathway or AOP for a chemical to carry out a risk assessment. In some cases, risk assessments can be completed using low tier approaches such as exposure-based waiving (Yang et al., 2017), history of safe use (Constable et al., 2007; Neely et al., 2011), read-across (Ball et al., 2016), or in vitro OECD test methods that require minimal data generation (eg, genotoxicity, skin, and eye irritation) (OECD, 2017, 2018a,b, 2019a,b).
Despite the rapid advances in biotechnology and computational modeling, regulatory safety assessment of new chemicals continues to rely heavily on in vivo testing in animals, particularly for higher tier hazard endpoints, such as systemic toxicity. The rapid evolution in technology might be part of the challenge; numerous different cell-based assays (eg, high-content screening, omics, and reporter cell lines) and a variety of computational models are available, but guidance on how to combine NAMs in a weight of evidence approach to ensure the robustness and transparency of future risk assessments is still required. Toward that end, the International Cooperation on Cosmetics Regulation (ICCR), a voluntary international group of cosmetic regulatory authorities, has recently outlined the major overarching principles for incorporating NAMs into an integrated strategy for “Next-Generation’ Risk Assessment” (NGRA) (Dent et al., 2018a). A subsequent report provided supplementary guidance on the types of NAMs that may be used in risk assessment of cosmetic ingredients, aligning them to the different tiers of the SEURAT-1 ab initio workflow for systemic repeat-dose toxicity (Berggren et al., 2017; ICCR, 2018).
In this work, we set out to integrate currently available NAMs in an ab initio risk assessment of a cosmetic ingredient to make a safety decision without using any in vivo animal data. To our knowledge, this is the first comprehensive study that brings together available NAMs to fully address more complex health effects associated with systemic exposures. Coumarin, present as an ingredient in hypothetical face cream and body lotion products, was selected as a case study chemical based on the availability of the existing exposure data and its relevance as a cosmetic ingredient (EFSA, 2008). The NGRA carried out was exposure-led and hypothesis-driven guided by the ICCR principles and based solely on in silico, in chemico, and in vitro data.
The overall strategy of the presented NGRA involved collecting and generating a broad suite of bioactivity data to provide a comprehensive set of biomarkers which were used to measure the bioactivity of the ingredient at consumer-relevant concentrations. Relevant internal exposures were estimated using a physiologically based kinetic (PBK) model for coumarin for exposure scenarios based on the habits and practices of the European demographic (Hall et al., 2007; SCCS, 2018). Biomarkers were selected to provide evidence of whether coumarin may cause specific cellular effects (eg, due to G protein-coupled receptors [GPCR] receptor binding) or nonspecific effects (eg, changes reflective of cellular stress). The presented strategy is closely aligned with the recently published NexGen blueprint for toxicology from the U.S. EPA (Thomas et al., 2019), a tiered guidance on how to characterize the mode of action (MoA) of a chemical at consumer-relevant concentrations. In cases when a chemical elicits nonspecific effects, which is particularly relevant to cosmetic ingredients, a point of departure (PoD) is derived using the most sensitive pathway or phenotypic effect. Such a derived PoD does not aim to identify a specific adverse outcome or pathology but rather aims to be protective of human health by estimating an exposure at which no biological response is expected (Friedman et al., 2020; Wetmore et al., 2015).
MATERIALS AND METHODS
Scope of the Risk Assessment
Because the purpose of this study was to perform an NGRA for systemic toxicity using novel safety assessment tools (excluding read-across and exposure-based waiving), local endpoints such as skin sensitization and phototoxicity were excluded from the study, and coumarin was treated as a novel chemical, despite it having been extensively studied in the literature. Furthermore, these endpoints can be addressed with established nonanimal approaches (OECD, 2018a,b, 2019a,b).
Overview of the Risk Assessment Approach
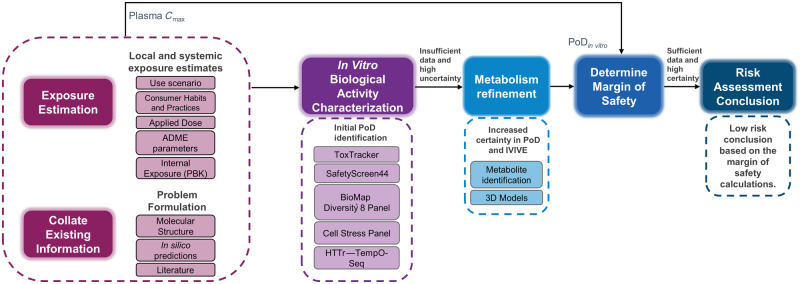
The workflow for the coumarin case study, presented in Figure 1, was shaped based on the principles underpinning the use of NAMs in the safety assessment of cosmetic ingredients (Berggren et al., 2017; Dent et al., 2018a). It uses an iterative, hypothesis-driven decision-making process to guide the risk assessment from problem formulation to safety decision. The first steps consisted of estimation of exposure levels based on the use-case scenario and consumer habits (“Exposure Estimation” step), together with other existing information on the chemical structure, in silico tool predictions (physicochemical properties, structural-based hazard alerts) and pre-existent (ToxCast and PubChem only) bioactivity data (“Collate existing information” step). Systemic exposure levels were estimated using a PBK model (Moxon et al., 2020). Next, to identify or develop mechanistic hypotheses, or to derive PoDs based on biological pathway or cellular phenotype perturbation (Thomas et al., 2019), new in chemico and in vitro bioactivity data were generated. This comprised high-throughput assays based on single and short-term exposures using 2D cell line models. A review of the in silico predicted metabolism and results from ToxTracker revealed that there was insufficient confidence in the PoDs generated in these 2D cell models due to their poor metabolic competence, and therefore a safety decision could not be reached using these data alone. Consequently, in the “Metabolism refinement” step a subset of the in vitro assays were repeated in 3D HepaRG models. In addition, to further refine the in silico predicted metabolism, a metabolite identification study was carried out in primary hepatocytes and a metabolic pathway proposed. Finally, all PoDs were compared with exposure estimates (plasma Cmax) to calculate a margin of safety (MoS) distribution which is used in the risk assessment decision.
Figure 1.
Next-Generation Risk Assessment case study workflow for 0.1% coumarin in consumer products. Initial steps involved collating existing data, generating in silico predictions, and problem formulation. In parallel, applied and systemic consumer exposure estimates were calculated based on the use scenario, habits and practices information, and chemical parameters. A battery of in vitro assays was then conducted to characterize the cellular response to coumarin. From these data, point of departure (PoD) values with associated uncertainties were determined, however, the lack of metabolic capacity of the cell line models used, and the potential toxicity of reactive metabolites led to the generation of additional data (metabolism refinement). All PoDs were compared with exposure estimates (plasma Cmax) to calculate a margin of safety, which was used for the risk assessment decision. Abbreviations: HTTr, high-throughput transcriptomics; IVIVE, in vitro to in vivo extrapolation.
Exposure Estimation
Use Scenario and Consumer Practices and Habits Information
Inclusion of 0.1% coumarin in 2 cosmetic product types, face cream and body lotion were selected as hypothetical use-case scenarios. Corresponding consumer habits and practices information (Hall et al., 2007) are summarized in Table 1.
Table1.
Summary of Habits and Practices Data and Applied Dose Estimates for Face Cream and Body Lotion for the European Consumer
| Product Types | Face Cream | Body Lotion |
|---|---|---|
| Amount of product used per day (g/day) using 90th percentilea | 1.54 | 7.82 |
| Frequency of useb | 2 times/dayc | 2 times/dayd |
| Amount of product in contact with skin per occasion (mg) | 770 | 3910 |
| Ingredient inclusion level | 0.1% | 0.1% |
| Skin surface area (cm2)b | 565 | 15670e |
| Leave on or rinse off | Leave on | Leave on |
| Exposure duration per occasion | 12 h | 12 h |
| Amount of ingredient in contact with skin per occasion (mg)f | 0.77 | 3.91 |
Hall et al. (2007).
Rounded from 2.14 times/day.
Rounded from 2.28 times/day.
Specified as Leg region in GastroPlus.
Based on 100% skin penetration and a body weight of 66.7 kg.
Source: Adapted from Moxon et al. (2020).
Applied Dose and PBK
Under normal use conditions dermal exposure was the only anticipated route of exposure expected for the 2 product types. The applied dose, which was calculated from consumer habits and practices information presented in Table 1, was used as an input to a PBK model of coumarin to estimate the systemic exposure levels. The methodologies used to produce the PBK models are published elsewhere (Moxon et al., 2020). In brief, models were parameterized using in silico generated values, and a sensitivity analysis performed to understand which of these parameters will have the greatest impact on the model output (Cmax). For the most sensitive parameters, experimental data were generated and then the model was reparametrized, and Monte Carlo simulations were conducted using GastroPlus v9.6 (Simulations Plus, Inc) to quantify the uncertainty of model outputs based on population variability and input parameter uncertainties. The results were expressed as the distribution of plasma Cmax.
Collation of Existing Information
Molecular Structure and In Silico Predictions
Coumarin (CAS No. 91-64-5) is an aromatic organic chemical compound classified as a member of the benzopyrone family. The in silico tools ToxTree, OECD Toolbox, Derek Nexus, Meteor Nexus, TIMES, and molecular initiating events (MIE) ATLAS (see Supplementary material) were run to predict the potential biological activity of coumarin, identify the active groups and predict its metabolic fate.
Free concentration was predicted by a steady-state mass balance partitioning model based on published models (Armitage et al., 2014; Fischer et al., 2017; Kramer et al., 2012). The model is parameterized to describe a specific in vitro assay setup and uses coumarin’s physicochemical properties to predict the partitioning of the chemical into each compartment (media, cells, plastic, etc.) (Supplementary material).
Literature Review
ToxCast data were extracted from the current version of the in vitro database using the CompTox Chemicals Dashboard (https://comptox.epa.gov/dashboard/dsstoxdb/results?search=DTXSID7020348; accessed August 8, 2019). All dose-response curve fits were manually inspected, and graphs exported for the assays considered active (Supplementary material). PubChem data were extracted from the bioassays database (https://pubchem.ncbi.nlm.nih.gov/compound/323; accessed August 8, 2019).
In Vitro Biological Activity Characterization
Genotoxicity Assessment: ToxTracker
The ToxTracker assay and data analysis were performed as previously described (Hendriks et al., 2016). Cells were treated in 3 independent replicates with 5 concentrations of coumarin (62.5, 125, 250, 500, 1000 µM) for 24 h, in the absence or presence of 0.25% rat S9 liver extract and required cofactors (RegenSysA+B, Moltox). ToxTracker is considered positive when a compound induces a minimum 2-fold increase in green fluorescent protein (GFP) induction in any of the reporters.
In Vitro Binding and Enzymatic Assays: Eurofins SafetyScreen44
The panel consists of 24 GPCRs, 7 enzymes, 2 nuclear receptors, 8 ion channels, and 3 transporters (Supplementary material). The experiments were carried out at Eurofins Cerep SA using coumarin sourced from Sigma-Aldrich (C4261, purity > 99%) at 10 µM using 2 replicates. Compound binding from the assay was calculated as a percentage inhibition of the binding of a radioactively labeled ligand specific for each target (Supplementary material). Compound enzyme inhibition effect was calculated as a percentage inhibition of control enzyme activity.
Immunomodulatory Screening Assay: BioMap Diversity 8 Panel
The BioMap Diversity 8 Panel consists of 8 cell-based human primary cell cultures (or cocultures) that have been activated using various stimulants (Supplementary material). Two independent datasets were generated using this panel for coumarin, 1 through the ToxCast program (see above) and 1 as part of this study. Coumarin was added to each system (n = 1) for 24 h at 4 concentrations (18.5, 55.6, 166.7, 500 µM). Detailed methods, including preparation and culture of cells, biomarker readout measurements, and statistical analyses, have been described previously (Bergamini et al., 2012). Biomarkers are annotated if: (1) 2 or more consecutive concentrations are changed in the same direction relative to vehicle controls, (2) at least 1 readout is outside of the significance envelope, (3) and at least 1 concentration has an effect size > 20% versus vehicle controls. A lowest observed effect level (LOEL) was defined for each cell system as the lowest concentration at which a biomarker met the criteria mentioned above.
In Vitro Cell Stress Panel and Bayesian Concentration-response Analysis
Coumarin was tested in 2D HepG2 cells using a recently developed cell stress panel (Hatherell et al., forthcoming). The panel comprises biomarkers that cover 8 key stress pathways (Simmons et al., 2009), mitochondrial toxicity and general cell health (Supplementary Table 3). Measurement timepoints were 1, 6, and 24 h in HepG2 cells at 8 concentrations (0.0128, 0.064, 0.32, 1.6, 8, 40, 200, 1000 µM). This was subsequently extended to 2D normal human epidermal keratinocytes (NHEK) using a reduced panel (Supplementary material).
Concentration-response analysis of the stress panel data was performed using a Bayesian statistical model as described in Hatherell et al. (forthcoming). Briefly, the model is fitted to the data for each biomarker and timepoint separately. From the fit we calculate a concentration dependency score (CDS), which is a measure of the confidence that the test chemical induces a concentration-dependent effect on the biomarker. CDS values range between 0 and 1, with 1 indicating strong evidence of an effect and 0 strong evidence against. Only concentration-response data sets for which CDS > 0.5 were used to calculate a PoD, this being defined as the lowest concentration inducing a response more than 2 standard deviations from the baseline.
High-throughput Transcriptomics
Cell treatment and lysate generation for transcriptomics
High-throughput transcriptomics (HTTr) method used was TempO-Seq (BioSpyder Technologies). HepG2, MCF7, and HepaRG 2D (3 biological replicates each) were treated for 24 h with 6 concentrations of coumarin (0.001, 0.01, 0.1, 1, 10, and 100 µM) using 0.5% DMSO as a vehicle control (Supplementary material). Following treatment, cells were washed in calcium and magnesium-free 1× PBS. After removal of all residual PBS, 2× TempO-Seq lysis buffer (BioSpyder Technologies, proprietary kit) was diluted to 1× with PBS and added at a volume of 1 µl per 1000 cells with a minimum of 10 µl per well and incubated for 10 min at room temperature. Following lysis, the samples were frozen at −80°C prior to sequencing.
RNA-seq and statistical analysis workflow
A consistent workflow was used for determination of a transcriptional point of departure (PoDT) from gene expression data using regular established methods available from the literature, broken down into 3 key steps.
(1) Sample Generation and QC
TempO-Seq analysis was performed as described previously (Yeakley et al., 2017), with a targeted sequence depth of 200 mapped read counts per transcript including the use of the general attenuation panel. The data analysis pipeline followed a similar QC and selection criteria from (Ramaiahgari et al., 2019) with the following modifications. Samples below a read depth of approximately 200 reads per gene were removed from further analysis. Normalization was performed using the rlog transformation from DESeq2 (Love et al., 2014) with outliers removed where replicates of matched samples had a correlation of < 85% and identified within a principal component analysis.
(2) Differential Expression Analysis
Differential expression analysis was performed using DESeq2 analysis (Love et al., 2014) combining either 3 or 5 biological replicates at each treatment concentration against the vehicle control as the primary comparison. Genes were considered differentially expressed using either a p-adj value of < 0.05 or a p value of 0.05 combined with a 1.5-fold change response.
(3) Pathway Analysis and PoD Determination
For the concentration-response analysis samples were filtered in BMDexpress2 (Phillips et al., 2019), using a Williams t test with a threshold of p < 0.05 and a minimum required fold change of 1.5. Data were fitted to 6 different models: Poly2, Exp3, Exp4, Exp5, Power, and Hill. Model selection was based on Akaike Information Criteria. A Benchmark Response factor of 1.349-fold (10%) was used to determine the PoD at a gene level, with a subsequent threshold fit p value filter was used of greater or equal to 0.1. At a functional level a mean pathway Benchmark Dose (BMD) was generated based on the Reactome database with pathways defined as altered that contained at least 3 input genes passing the following thresholds of Fishers Exact test of p < 0.1 with and a lower bound (BMDL)/the upper bound (BMDU) ratio of < 40. Final PoDT was determined based on a subset of methods (1, 3, 4, 5, 9) outlined in Farmahin et al. (2017) (Table 5).
Table 5.
PoDT Values (µM) for Coumarin Treated Across 4 Cell Models for 24 h Using a Subset of Proposed Approaches for Gene Selection Based on Those Proposed by Farmahin et al. (2017)
| Cell Model | HepG2 | MCF7 | HepaRG 2D | HepaRG 3D |
|---|---|---|---|---|
| Pathway-level tests PoDT (µM) | (308 pathways) | (0 pathways) | (17 pathways) | (2 pathways) |
| 20 pathways with the lowest p value Reactome | 70 | NA | 58* | 46* |
| 20 pathways with the lowest BMD Reactome | 44 | NA | 58* | 46* |
| BMD of Reactome pathway with lowest BMD that meets significance threshold criteria | 31 | NA | 38 | 41 |
| Gene-level tests PoDT (µM) | (1570 genes) | (47 genes) | (87 genes) | (9 genes) |
| Mean BMD of 20 genes with largest fold change | 6 | 3 | 54 | 55 |
| Mean BMD of genes between 25th and 75th percentile | 17 | 1 | 59 | 46* |
Highlighted (*) are values where the number of pathways or genes was below the recommended number (ie, 20) for grouping. Abbreviation: NA, not applicable.
Metabolism Refinement
Coumarin Metabolism in Primary Hepatocytes Using CYP2A6 Inhibition
Coumarin metabolic stability screening in expressed human CYP and UGT isoforms (CYP1A2, CYP2A6, CYP2C8, CYP2C9, CYP2C19, CYP2D6, CYP3A4 and UGT1A1, UGT1A3, UGT1A6, UGT1A9, UGT2B7) identified CYP2A6 as the only enzyme metabolizing coumarin (Supplementary material). Pooled human cryopreserved primary hepatocytes (human tissues are only obtained from donors with valid, written consent following full ethics approval prior to tissue collection; 50 multidonors, BioIVT, lot no. YQV) in suspension at a cell density of 0.5 million cells per ml, were incubated at 37°C up to 90 min with 10 µM coumarin (final concentration of 0.25% DMSO) without and with tranylcypromine (0.5, 2 µM) to inhibit the CYP2A6-specific reaction. A second experiment was conducted at a higher concentration of coumarin (1 mM) without the inhibitor, to saturate the 7-hydroxycoumarin pathway; all experiments were performed in duplicate. Full scan liquid chromatography-mass spectrometry data were acquired using multienergy time-of-flight acquisition (Waters Xevo G2 Q-ToF in MSE mode). The data were interrogated for the masses associated with the metabolites predicted by Meteor Nexus (Supplementary material). All possible metabolites were reported based on the Meteor Nexus output and observed fragments. Metabolites were reported as “% total peak area” and “% parent peak area at T = 0.”
Short- and Long-term Exposure in 3D Tissues
Additional cell stress panel assays were conducted in HepaRG 3D cultures (Supplementary material) using a reduced version of the panel at the same concentrations (single dose) as performed in 2D cells, however, longer exposure durations were used (24, 72, and 168 h). HTTr was performed in additional HepaRG 3D cultures (5 biological replicates) at the same concentrations and duration as previously described for the 2D models.
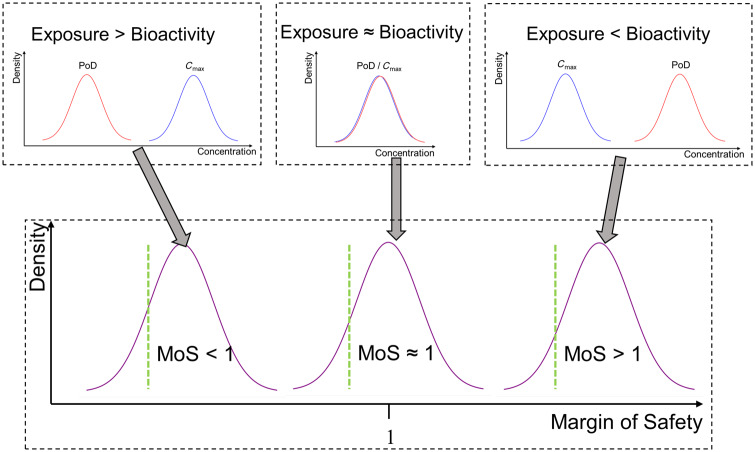
Determination of the MoS
For a given PoD, the MoS was defined as the ratio between the nominal concentration at which the PoD is defined, and the relevant plasma Cmax estimate (Figure 2). Distributions were obtained for plasma Cmax and stress panel PoDs as described above; only point estimates were available for PoDs obtained from the remaining sources (transcriptomics data, ToxCast, PubChem). To calculate the corresponding MoS distributions (which propagates uncertainty in the PoD and Cmax estimates), a Monte Carlo approach was used. This entails generating multiple MoS samples, whereby a single sample is calculated by resampling the Cmax distribution and then resampling either the PoD distribution (in case of cell stress panel PoDs) or the PoD point estimate (for all other PoDs). Statistics on the MoS (e.g. percentiles, mean, median) were then calculated from these samples using standard approaches (Gelman et al., 2013).
Figure 2.
C max and points of departure (PoDs) were inferred as probability distributions encompassing the uncertainty in their estimates. The margin of safety (MoS) was defined as the PoD/Cmax ratio. The uncertainty in the Cmax and PoD estimates is propagated through this calculation such that MoS estimate is also a distribution. When the distribution for the PoD is predominantly lower than the distribution for the Cmax (exposure > bioactivity), this produces a distribution for the MoS in which almost all mass is less than 1. Conversely, if the distribution for the PoD was predominantly greater than the Cmax distribution (exposure < bioactivity), almost all the mass of corresponding MoS distribution is greater than 1. When the distributions for the Cmax and PoD strongly overlap (exposure ≈ bioactivity), these results in an MoS distribution centered around 1. The location of the 5th percentile for the MoS is as illustrated on the graph (green line). For this value to be greater than 1 the PoD must, on average, be greater than the Cmax.
RESULTS
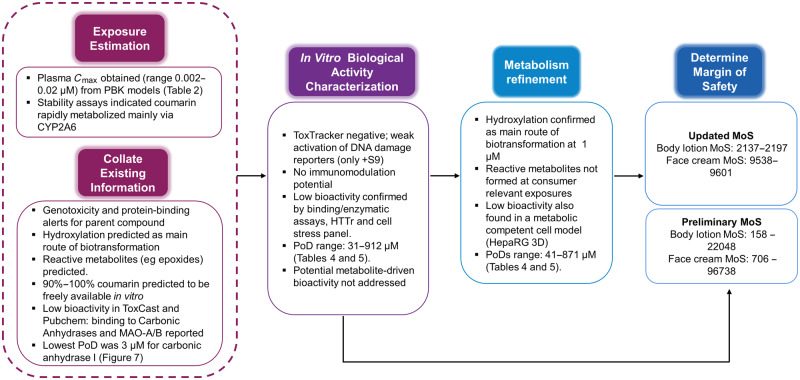
A schematic overview of the key results and how they lead to the risk assessment conclusion is provided in Figure 3.
Figure 3.
Summary of the key results from each step on the Next-Generation Risk Assessment case study workflow (see Figure 1) for 0.1% coumarin in face cream and body lotion. Abbreviations: HTTr, high-throughput transcriptomics; MoS, margin of safety; PBK, physiologically based kinetic; PoD, point of departure.
Exposure Estimation
Systemic exposure (mean plasma Cmax) to coumarin from daily application use of a face cream or a body lotion was estimated to be 0.01 and 0.0022 µM, respectively (Table 2). Coumarin was predicted to be cleared mainly via metabolism by the Extended Clearance Classification System (Varma et al., 2015); CYP450 isoforms stability data showed to be exclusively by CYP2A6 with a half-life of < 5 min in microsomes, and 13 min in human cryopreserved hepatocyte stability assays (Supplementary material). Nevertheless, for this exposure scenario, concentration is never completely cleared from the plasma due to the prolonged application time (12 h) and slow absorption/clearance into and from the plasma. Steady-state plasma concentrations were reached within 5 days for face cream and 20 days for body lotion.
Table 2.
Internal Exposures From Use of 0.1% Coumarin in Face Cream and Body Lotion Following the Exposure Scenario Outlined in Table 1
| Total Plasma Cmax (µM) | Mean | Median | 90th Percentile | 95th Percentile | 97.5th Percentile | 99th Percentile |
|---|---|---|---|---|---|---|
| Body lotion | 0.01 | 0.01 | 0.018 | 0.019 | 0.02 | 0.022 |
| Face cream | 0.0022 | 0.0021 | 0.004 | 0.0043 | 0.0046 | 0.005 |
Collation of Existing Information
In silico predictions
ToxTree predicted that coumarin belongs to the Cramer class III (Cramer et al., 1976; Patlewicz et al., 2008) and can bind to proteins and DNA via Michael addition and acyl transfer mechanism. Similarly, the OECD toolbox predicted binding to DNA and proteins via SN2 mechanisms after oxidation to epoxide. No positive results were obtained from the MIE ATLAS tool (Table 3).
Table 3.
Summary of the In Silico Alerts for Coumarin and 22 of Its Predicted Metabolites (Number of Metabolites Triggering the Alert per Category)
| OECD Toolbox |
ToxTree |
DEREK NEXUS |
MIE Atlas |
|||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mutagenicity | Endocrine activity | Chromosome damage | Protein binding | DNA binding | Mutagenicity | Genotoxicity | Protein binding | DNA binding | Mutagenicity | Genotoxicity | Endocrine activity | Chromosome damage | Skin/eye irritation | MAO-A 2a | MAO-A 2b | COX 2 | MAR (1,2,3) | PDE 3A 4 | A1a ADR | |
| Coumarin | Y | — | — | Y | Y | — | Y | Y | Y | — | — | — | — | — | — | — | — | — | — | — |
| Primary metabolites (n = 6) | 3 | 5 | 1 | 6 | 6 | 4 | 2 | 6 | 6 | 2 | 1 | 1 | 2 | 1 | — | — | — | — | — | — |
| Secondary metabolites (n = 13) | — | 7 | — | 13 | 10 | — | — | 13 | 13 | — | 2 | 1 | 5 | 3 | — | — | — | — | — | — |
| Tertiary metabolites (n = 3) | — | 3 | 3 | 3 | 2 | — | — | 3 | 3 | 1 | — | — | — | — | — | — | — | — | — | — |
Based on the predicted rapid metabolism and the in silico predictions of toxic metabolite formation, Meteor Nexus was run to simulate the potential metabolic pathway of coumarin. Meteor Nexus identified hydroxylation as the main route of biotransformation followed by glucuronidation and sulfation with a total of 22 possible metabolites. Meteor Nexus also predicted the formation of epoxides (primary and secondary metabolites) (Supplementary material). Table 3 shows the summary of in silico alerts for coumarin and its predicted metabolites. Most primary, secondary, and tertiary metabolites were predicted to bind to proteins and DNA. Protein binding is a flag for skin sensitization potential; however, this endpoint was considered out of scope of this study. In summary, these in silico alerts indicated a need to investigate the genotoxicity potential of coumarin and its metabolites using an appropriate in vitro cell assay (namely ToxTracker, see below).
The free fraction of coumarin in the culture media was predicted to be between 90% and 100%, based on coumarin’s relatively low log P and low affinity to plasma (serum) proteins (Supplementary material). For all evaluated in vitro setups, the quantitative in vitro to in vivo extrapolation (QIVIVE) factor was approximately 3 based on a free fraction in vitro of approximately 90% and a free fraction in vivo of approximately 30%. A QIVIVE factor greater than 1 indicates that for the same total medium and plasma concentration the resulting free concentration, which is available for uptake into the cells is higher in vitro compared with in vivo. This means that for coumarin an initial risk assessment based on in vitro nominal and total plasma concentrations is conservative and therefore nominal concentrations were applied throughout.
Literature review
Coumarin was tested in 642 assays of the ToxCast panel, 18 of which were considered active. It has been previously noted that not all responses that the data processing pipeline labels as “active” appear to be truly positive results, therefore graphs were exported from the dashboard and curve fits were manually curated (Filer et al., 2016). The most credible hits in terms of quality of concentration-response and putative MoA were the 3 concordant positive hit-calls in the cell-free enzymatic assays for inhibition of monoamine oxidases (MAO): NVS_ENZ_rMAOBC (rat brain), NVS_ENZ_rMAOAP, and NVS_ENZ_rMAOBP (both rat liver mitochondrial membranes) (Sipes et al., 2013) occurring at similar concentrations (15–19 µM).
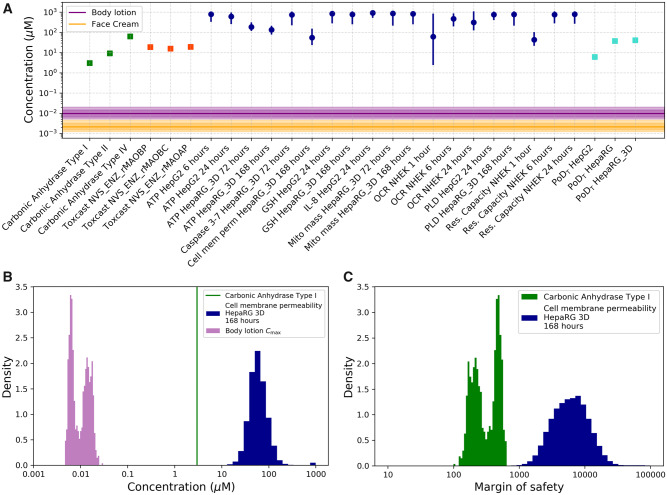
PubChem contains 4254 records of biological tests carried out using coumarin, of which only carbonic anhydrase (EC 4.2.1.1) and MAO inhibition assays were positive. Specifically, coumarin was found to interact with the carbonic anhydrase type I with inhibitory constant (Ki) = 3.1 µM, type II with Ki = 9.2 µM, and type IV with Ki = 62.3 µM (Maresca and Supuran, 2010; Maresca et al., 2009). Coumarin showed very low inhibition toward the other isoforms of carbonic anhydrase with Ki > 500 µM. In addition, coumarin was reported to inhibit MAO-A and MAO-B with IC50 values of 12 µM and 40.7 µM, respectively (Gnerre et al., 2000). For the MoS calculation, the following PoDs were used: IC50s for MAO-A and MAO-B of 18.5, 15.8, and 19 µM and Ki values for carbonic anhydrase I, II, and IV Ki of 3, 9.1, and 62.3 µM.
In Vitro Biological Activity Characterization
Genotoxicity assessment: ToxTracker
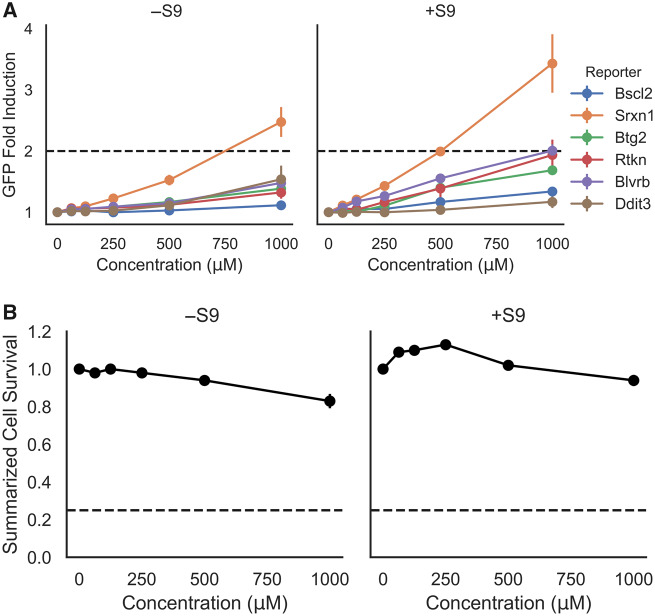
In silico analysis of coumarin using DEREK Nexus, OECD QSAR toolbox, and ToxTree determined several genotoxicity alerts for its primary (n = 7), secondary (n = 12), and tertiary (n = 3) predicted metabolites (Supplementary material). Thus, coumarin’s genotoxic potential was further investigated with the ToxTracker assay. In this assay, a weak activation (between 1.5- and 2.0-fold) of the Rtkn-GFP marker, associated with DNA double-strand breaks, was observed in the presence of rat liver S9 (Figure 4). However, no activation of the genotoxicity reporter Bscl2-GFP, which indicates DNA replication inhibition and induction of promutagenic DNA adducts, was observed in the absence or presence of S9. A weak activation was observed for the p53 response (Btg2-GFP reporter) in the presence of S9 which suggests general cellular stress (Hendriks et al., 2016; Karlsson et al., 2014). Similarly, exposure to coumarin without metabolic activation induced the oxidative stress reporter Srxn1-GFP, whilst in the presence of S9 both oxidative stress markers (Srxn1-GFP and Blvrb-GFP) were activated. All controls caused GFP induction levels consistent with historical data and demonstrated the functionality of the mES reporter cell lines. No significant cytotoxicity occurred up to the maximum tested concentration of 1000 µM in the absence or presence of S9.
Figure 4.
A, The ToxTracker toxicity pathway markers Bscl2-GFP and Rtkn-GFP (DNA damage), Btg2-GFP (p53-associated cellular stress), Srxn1-GFP and Blvrb-GFP (oxidative stress), and Ddit-GFP (unfolded protein response) observed by flow cytometry after 24 h exposure to coumarin (0, 62.5, 125, 250, 500, and 1000 μM) in mES cells. A 2-fold green fluorescent protein (GFP) induction level was defined as the threshold for a positive test result, whilst the cell survival rate determined by cell count was always > 25%. Each data point on the graph represents the mean fold induction of 3 independent experiments ± SD. B, Summarized cell survival is expressed as the average across all cell lines and the 3 independent experiments ± SD per concentration. .
In conclusion, the weak activation of DNA damage reporters in the presence of rat liver S9-mediated metabolism was not sufficient to classify coumarin as genotoxic in the ToxTracker assay. However, these results suggested that reactive coumarin metabolite(s) could induce DNA lesions secondary to oxidative stress, rather than directly interacting with DNA.
In vitro binding and enzymatic assays: Eurofins SafetyScreen44
The SafetyScreen44 panel includes 44 targets associated with in vivo adverse drug reactions (Bowes et al., 2012). Coumarin showed no significant effect in any of the targets. The highest activity was 22% and was recorded for MAO-A (Supplementary material). This was consistent with the in silico predictions of the MIE ATLAS.
Immunomodulatory screening assay: BioMap Diversity 8 Panel
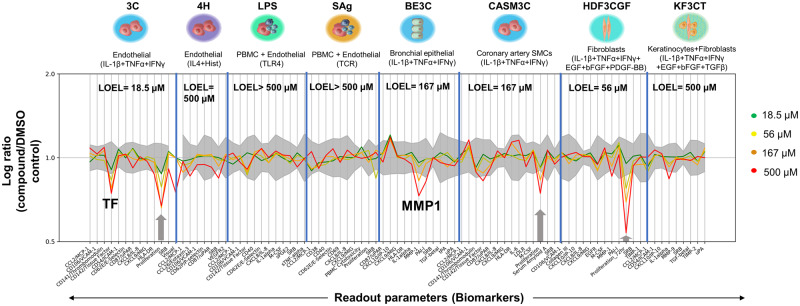
To increase the biological coverage, a screening panel of 8 primary cell systems, that are stimulated to replicate complex cell and pathway interactions of vascular inflammation, immune activation, and tissue remodeling, was conducted. The most significantly affected biological readouts across all cell systems in the BioMap Diversity 8 Panel were associated with antiproliferative and tissue remodeling activities (Figure 5), and the most sensitive cell system was the 3C endothelial cell system where antiproliferation (−33%) was observed at the lowest dose tested (18.5 µM). No biomarkers were affected in the LPS or SAg-stimulated PBMC and endothelial cell coculture systems. No concentration-response was observed in the BioMap data generated during the ToxCast program (see https://comptox.epa.gov/dashboard/dsstoxdb/results?search=DTXSID7020348; accessed August 8, 2019).
Figure 5.
An overview of coumarin activity in the BioMap panel. There was no cytotoxicity observed at any concentrations tested. Antiproliferation is indicated by a gray arrow. Biomarkers are annotated if: (1) 2 or more consecutive concentrations are changed in the same direction relative to vehicle controls, (2) at least 1 readout is outside of the significance envelope, and (3) at least 1 concentration has an effect size > 20%versus vehicle controls. A lowest observed effect level (LOEL) was defined for each cell system as the lowest concentration at which a biomarker was significantly changed outside of the vehicle envelope and a dose-response was observed.
In vitro cell stress panel
Cellular stress-response assays are useful to characterize nonspecific biological activity which is not mediated via a specific protein/receptor interaction. Coumarin data was previously published elsewhere (Hatherell et al., forthcoming) and results are briefly described as follows. In HepG2 cells, a dose-response with a CDS > 0.5 was only observed for 4 biomarkers (ATP, GSH, phospholipidosis, and IL-8) out of the 36 biomarkers across the 10 pathways with PoDs in the 500–800 µM range (Table 4). In NHEK cells, only the mitochondrial respiration parameters (oxygen consumption rate [OCR] and reserve capacity) decreased (Table 4). The lowest PoD observed in the 2D models was 44 µM for reserve capacity at 1 h, however the PoD increased to approximately 700 µM at the later timepoints. It is also worth noting that in this Seahorse assay the PoD was uncertain, partially explained by plate effects in this experiment (Supplementary material). All PoDs outlined in Table 4 were used in the calculation of MoS.
Table 4.
PoDs From Cell Stress Panel After Acute Exposure (24 h) in HepG2 and NHEK and Long-term Exposure (168 h) in HepaRG 3D Spheroids
| Biomarker | Cell Type | Stress Pathway | PoD (µM) | Effect | CDS | |
|---|---|---|---|---|---|---|
|
HepG2 | Cell health |
|
|
|
|
| Phospholipidosis (24 h) | HepG2 | Cell health | 759 (437–977) | Down | 0.93 | |
| GSH (24h) | HepG2 | Oxidative stress | 851 (301–1000) | Up | 0.92 | |
| IL-8 (24h) | HepG2 | Inflammation | 912 (575–1000) | Down | 0.61 | |
|
NHEK | Mitochondrial toxicity |
|
Down |
|
|
|
NHEK | Mitochondrial toxicity |
|
Down |
|
|
| Caspase 3–7 (72 h) | HepaRG 3D | Cell health | 741 (245–977) | Up | 0.95 | |
| Cell membrane permeability (168 h) | HepaRG 3D | Cell health | 55 (26–141) | Up | 0.99 | |
|
HepaRG 3D | Cell health |
|
|
1 | |
| Phospholipidosis (168h) | HepaRG 3D | Cell health | 776 (234–1000) | Up | 0.86 | |
| GSH (168 h) | HepaRG 3D | Oxidative stress | 776 (275–1000) | Down | 0.92 | |
|
HepaRG 3D | Mitochondrial toxicity |
|
|
|
|
Only PoDs from concentration-responses with CDS > 0.5 were considered as true representations of bioactivity. Reported values are the mode (most likely value in bold) and 95% highest-density-interval (in brackets) summarizing the distribution for the PoD as reported in Hatherell et al. (forthcoming).
High-throughput transcriptomics
Transcriptomics was applied as a broad nontargeted biological screen of in vitro cellular perturbation following coumarin treatment, to complement the targeted assays (eg, Eurofins SafetyScreen44, and BioMap). Use of concentration-response modeling of transcriptomics data has been utilized to derive PoDs for chemical risk assessment (Farmahin et al., 2017; Thomas et al., 2013, 2019) and to identify potential MoAs for target organ toxicity (Limonciel et al., 2018; Ramaiahgari et al., 2019). This study used 3 different cell lines (HepG2, MCF7, and 2D HepaRG) to extend biological coverage and address potential cellular variation in response to coumarin including any consequences of metabolism. RNA-seq data have been deposited in the ArrayExpress database at EMBL-EBI (www.ebi.ac.uk/arrayexpress) under accession number E-MTAB-8985 titled RNA-Seq of human cell lines, MCF-7, HepG2 and HepaRG treated with a dose range of Coumarin from 0.001 μM to 100 μM against untreated vehicle controls. No significant cytotoxicity (> 20%) was observed in any of the cell lines up to the maximum tested coumarin concentration (data not shown).
Differential expression analysis
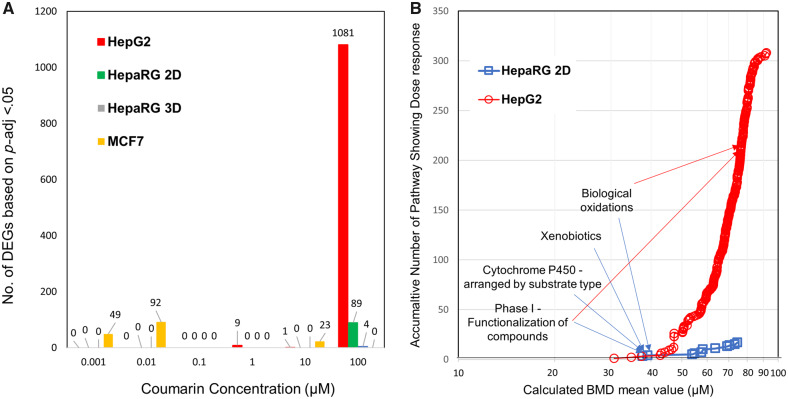
DESeq2 is regarded as one of the leading tools for pairwise differential expression analysis when fewer than 12 replicates are used (Schurch et al., 2016). In general, across the cell lines, treatment with coumarin resulted in limited gene expression changes at concentrations below 100 µM suggesting limited cellular effects at lower concentrations (Figure 6A). Specifically, in HepG2 cells there were no significant gene changes (p-adj < 0.05) at concentrations under 1 µM. By 10 µM, only 10 genes were found to be differentially expressed, which increased to 1081 genes at 100 µM (Figure 6A). In the 2D HepaRG cell model the overall gene expression response to coumarin was even weaker with 89 differentially expressed genes identified at the highest concentration of 100 µM. Cytochrome P450 genes including both CYP3A4 and CYP2A6 that have been reported to be potentially involved in the metabolism of coumarin (Farinola and Piller, 2007) were among those observed to be upregulated. In contrast, differentially expressed genes were detected in MCF7 cells at 0.001, 0.01, and 10 µM, but not at 0.1, 1, or 100 µM, with none of the genes observed to be altered at the lower concentrations continuing to be differentially expressed at the higher concentrations, suggesting this effect was unrelated to the coumarin treatment (Figure 6A).
Figure 6.
Summary of transcriptomic data analysis. A, Total Differentially Expressed Genes (DEGs) identified for each concentration for each cell line following DESeq2 analysis highlighting limited responses until the highest dose. B, Pathway based Benchmark Dose (BMD) mean accumulation plot. Each data point represents the total number of Reactome pathways that met the significance criteria for both HepG2 (o) and HepaRG (□) cell lines plotted against the corresponding calculated BMD mean value across the range of signaling pathways identified. The curves slope indicates the rate that pathways are showing differential regulation as concentration increases. Labeled are the lowest reported Reactome pathways for each HepaRG and where they correspondingly are identified in the HepG2 accumulation curve.
Pathway analysis and PoD determination
There is still significant discussion of what approaches to use for deriving a PoDT at both gene and pathway level, and therefore multiple PODTs were derived using several published methods (Farmahin et al., 2017) that have been shown to correlate closely to BMDL derived from equivalent treated samples using standard pathology studies. These included the mean of the 20 pathways with the lowest p value, or the 20 pathways with the lowest transcriptional BMDs and finally the lowest pathway BMD that meets the significant enrichment criteria. At the gene level this included both the mean BMD of 20 genes with largest fold change and the mean BMD of genes between 25th and 75th percentile. Only HepG2 met the recommendation (Farmahin et al., 2017) that at least 20 pathways be detected to apply the pathway-level tests (Table 5); no pathways were detected for MCF7, reflecting the DESeq2 results, and 17 pathways were detected using the 2D HepaRG cell model. Analysis of these changes using the Reactome pathway database (Fabregat et al., 2018) corroborates a shift in metabolic responses for HepaRG 2D with pathways such as Metabolism, Biological oxidations, and Phase 1-Functionalization of compounds amongst the lowest concentration related pathway responses (Figure 6B, Supplementary material). Where the number of pathways fell below 20, all relevant pathways were included in the analysis. Using this selection, the observed pathway-level PoDT ranged from 44 µM to 58 µM across cell lines. Taking the lowest pathway identified as the PoDT this value reduces to between 31 and 38 µM (Table 5).
In contrast, using the aggregated gene-level tests, the lowest PoDT was 1 µM derived using the MCF7 cell line (Table 5). However, the low number of genes that pass the gene-level test filtering criteria, lack of detectable pathway responses, and results from the DESeq2 analysis (described above) indicate that evidence supporting this PoDT was weak. Using the more metabolically competent and physiological-relevant 2D HepaRG cell line resulted in a PODTs between 54 and 59 µM, where again the cytochrome P450 genes were amongst the most sensitive, similar to the pathway-level test results. The gene-level test PoDTs for HepG2s ranged between 6 and 17 µM and were slightly lower than the values derived for the pathway-level tests.
In conclusion, the MCF7 PoDT was not considered to be sufficiently robust to derive an MoS. Because there is still significant discussion of which approach is the most appropriate to derive a PoDT, the lowest PoDT for each cell model was selected (regardless of method) for the MoS calculation, as this represented the most conservative PoDT.
Preliminary MoS
All PoDs estimated from the cell stress panel and HTTr in the 2D systems provided an MoS of 706-96738 for face cream and an MoS of 158-22048 for body lotion (Figure 3, Supplementary material). The lowest MoS (158) across all assays generated at the “In vitro bioactivity characterization” step was derived using the PoD (represented by Ki) for the inhibition of carbonic anhydrase I for the body lotion exposure (Figure 8). At this stage, sufficient data had been generated to derive an MoS for coumarin itself, however the weak activation of DNA damage reporters observed in ToxTracker in the present of S9 increased our uncertainty around the impact of toxic metabolites in the overall toxicity of coumarin which might not be covered by the MoS obtained above. Therefore, next steps included the generation of data in more complex models with longer exposure duration, and the investigation of coumarin’s metabolic fate in primary hepatocytes (Figure 3).
Figure 8.
A, Margin of safety (MoS) plot for face cream (orange band) and body lotion (purple band). Plasma Cmax (total, μM) expressed as distribution, purple or orange line (median, 50th percentile), inner dark band (25–75th percentile), outer light band (2.5th–97.5th percentile [95th credible interval]). Points of departure (PoDs) expressed as nominal concentration (μM) as single values for carbonic anhydrase enzymatic assays (green squares), monoamine oxidases (MAO) enzymatic assays (red squares), and transcriptional point of departure (PoDT) from high-throughput transcriptomics assays (light blue squares). PoDs from cell stress panel (dark blue circles) are reported in terms of the mode (circle) and the 95% credibility interval (solid lines). B, Histogram representations of the distribution for the predicted Cmax (purple), example PoD distribution from the stress panel (blue) and PoD summarized as point-estimate from PubChem (green). C, Distributions for the MoS calculated using both examples. Abbreviations: NHEK, normal human epidermal keratinocytes; OCR, oxygen consumption rate.
Metabolism Refinement
Coumarin metabolism in primary human hepatocytes
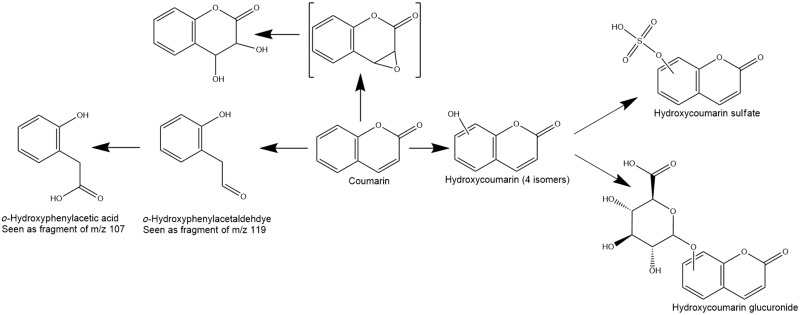
Human CYP2A6 was identified as the enzyme responsible for the major route of biotransformation of coumarin in human hepatocytes (Supplementary material ). To further investigate other pathways of coumarin metabolism in humans, 2 high-resolution mass spectrometry-based approaches for metabolite-ID were used. In the first, a high (1 mM) concentration of coumarin was used to saturate the CYP2A6 pathway. In the second, a lower concentration of coumarin (10 µM) was used, both with and without inhibition of CYP2A6 (using either 0.5 or 2 µM tranylcypromine, based on the method of Taavitsainen et al. [2001]). Across all experimental conditions the metabolites detected were different forms of hydroxylated coumarin including 7-hydroxycoumarin and its glucuronide or sulfated conjugates (Figure 7). These hydroxylated metabolites were detected in lower amounts in the presence of tranylcyropamine but did not differ significantly between the 10 μM and 1 mM coumarin concentration groups, indicating that the pathway had become saturated. Also observed in all concentration groups (and therefore not impacted by the inhibitor) were fragments of 2 metabolites (m/z 107 and 119 in negative ion MS); the lower mass fragment coeluted with a signal which was consistent with hydroxyphenylacetic acid (o-HPAA), and the higher mass fragment could arise from o-hydroxyphenylacetaldehyde (o-HPA), which has been reported to be the major metabolite in other studies (Fentem and Fry, 1992). These fragments were minor metabolites at 10 μM but were the most intense signals at 1 mM, indicating a pathway that becomes significant only at concentrations significantly above expected exposure levels. A further metabolite, corresponding to the diol arising from hydrolysis of an epoxide, was detected at low levels only in the 1 mM concentration group. The overall proposed coumarin metabolism pathway, using both experimental conditions, is summarized in Figure 7.
Figure 7.
Coumarin’s proposed metabolic pathway based on the in vitro experiments.
Short- and long-term exposure in 3D tissues
To increase our confidence in the initial PoDs from the 2D cell models using the cell stress panel, coumarin was tested for longer exposure durations in a 3D HepaRG model with potentially higher metabolic capacity and in vivo-like physiology than HepG2 cells (Ramaiahgari et al., 2017). Even though spheroid size was unchanged throughout the duration of the experiment, early signs of cell damage were observed at low concentrations with a dose-dependent increase in cell permeability at 168 h (PoD = 56 µM), and ATP decrease at 72 and 168 h (PoD = 190 and 144 µM, respectively) (Table 4). At higher concentrations (approximately 700 µM) a mixture of biomarkers related to mitochondrial toxicity, oxidative stress, and cell health were affected (Table 4). It is worth noting that in this system there were no changes at 24 h in contrast to biomarker changes in HepG2 and NHEK at earlier timepoints.
Similarly, HTTr was repeated in a HepaRG 3D model where cells were exposed to coumarin for 24 h. Using the same analysis approaches as previously described, the response observed was very limited for DESeq2 with only 4 genes meeting the p-adj value of .05, all seen at the top dose (200 µM) (Supplementary material). Similarly using BMDexpress2 only 9 genes passed the Williams trend test for dose-response with a greater than 1.5-fold change. Functional enrichment indicated that 2 pathways met the significance threshold resulting in the summarized PoDs after 24 h exposure (41–55 µM). The lowest PoDT of 41 µM was used for the calculation of MoS (Table 5).
Updated MoS and Risk Assessment Conclusion
The metabolism refinement results demonstrated that coumarin is primarily detoxified to the respective glucuronides via hydroxylation, and epoxide formation only occurred at high concentrations (1 mM) when the CYP2A6 is saturated. PoDs derived from the cell stress panel and HTTr assays in more metabolic competent cells (HepaRG 2D and HepaRG 3D) were not significantly different from PoDs generated in HepG2 and NHEK (Tables 4 and 5). From both these results we can conclude that at consumer-relevant exposures, metabolite formation does not impact the MoS (Figs. 3 and 8, Supplementary material).
There is not yet agreement on how large an MoS derived in an NGRA needs to be to assure human safety. However, the predicted Cmax values for face cream and body lotion were lower than all PoDs with an MoS (the 5th percentile) higher than 100. Furthermore, we can conclude that coumarin is not genotoxic, does not bind to any of the 44 targets and does not show any immunomodulatory effects at consumer-relevant exposures. In conclusion, the weight of evidence suggests that the inclusion of 0.1% coumarin in these products is safe for the consumer.
DISCUSSION
Several theoretical frameworks describing a tiered approach for NGRA have been published over the past few years (Andersen et al., 2019; Berggren et al., 2017; Thomas et al., 2019), but concrete NGRA examples of how to analyze, integrate and interpret all the data obtained from NAMs to inform a safety decision are still lacking. Therefore, this work represents a milestone in the development and application of nonanimal approaches to assess human safety, showing for the first time that in chemico, in silico, and in vitro approaches can be integrated to arrive at a consumer safety decision for systemic effects. Furthermore, this work demonstrates several key principles of NGRA (Dent et al., 2018a). The overall goal was to perform an exposure-led human safety assessment designed to prevent harm by applying robust and relevant methods in a hypothesis-driven way. The philosophy behind this type of risk assessment aimed at preventing harm is based on the premise of “Protection not Prediction” (Kavlock et al., 2018; Thomas et al., 2019). Such a safety assessment approach is possible because it does not attempt to replicate the results of the animal tests historically used in safety assessment. Instead, the hypothesis underpinning this type of NGRA is that if there is no bioactivity observed at consumer-relevant concentrations, there can be no adverse health effects.
One of the strengths of this case study was the conservative and rigorous consumer exposure assessment by assuming a worst case scenario for the estimation of applied dose and by building a refined PBK model parametrized with in vitro ADME parameters (Moxon et al., 2020), respectively. Furthermore, systemic exposure was expressed as distribution of plasma Cmax across a population, taking into account inter-individual variability. Even though PBK is a well-established tool in pharmaceutical development (Jones et al., 2015), there are still issues around the confidence in predictions, especially when in vivo data is unavailable for model validation. In this work, we attempted to mitigate some of these issues by following good practices outlined in Paini et al. (2017), and applying the PBK framework developed in Moxon et al. (2020). This framework utilizes sensitivity and uncertainty analysis to guide parameter generation and attempts to increase confidence in the model output without the explicit need for in vivo studies. Following completion of the work reported here, coumarin clinical data (Ritschel et al., 1977) was compared with the simulated area under the curve, Cmax and total clearance, and the results were generally within 2-fold of the measured clinical data (Moxon et al., 2020). Therefore, our confidence in the estimation of exposure to coumarin using this PBK framework is high.
Another strength of this work is the wide range of NAMs incorporated in the testing strategy, covering multiple biological pathways. The biomarkers were selected to provide evidence of whether coumarin may cause specific cellular effects (eg, due to nuclear receptor binding, ion channels, and enzymes inhibition) or nonspecific effects (eg, changes reflective of cellular stress and inflammation) across different cell models and exposure durations. The lack of predicted binding activity from both MIE ATLAS (Allen et al., 2018) and Eurofins SafetyScreen44 suggested that coumarin does not activate specific targets. Furthermore, biological coverage was increased by information provided by the immunomodulatory screening assay tested in 8 primary systems, by HTTr and cell stress panel in multiple cell models. HTTr was primarily used as a nontargeted approach for characterizing biological responses potentially not covered by the other tools. In this paper, we have applied different approaches for aggregating gene and pathway-level BMDs from HTTr based on previous work by Farmahin and colleagues. The results are combined in a weight of evidence to provide an overall understanding of transcriptional responses. The limited gene expression changes seen at concentrations below the maximal used, suggest the compound is having a minimal biological effect on the cells. Farmahin et al. (2017) indicated that summarizing BMD modeling in different ways has a comparatively small impact on the PoDT (varying up to 1 order of magnitude); a similar result was seen in this study across all cell lines. Although such analysis was not performed for this study, the use of 3D spheroid cultures of HepaRG could have provided a further refinement of the PoDT by extending the exposure duration or performing repeat dosing. However, further work is required to understand the advantages of this for human health protection due to the added uncertainty in modeling the compound’s concentration in vitro following repeat dosing. Recent findings showed that in vitro PoDs estimates from single 24 h in vitro exposure studies are as conservative as in vivo studies for most chemicals tested (Paul Friedman et al., 2020).
The reduced number of active assays in large assay panels such as ToxCast and PubChem further contributed to the low bioactivity weight of evidence gained from the other assays. However, it is worth noting that one of the lowest PoDs was derived from the carbonic anhydrase assay from PubChem which was not covered in any of the other assays. Ensuring all relevant biological endpoints is an ongoing challenge in NGRA, and future research is needed to identify which additional molecular targets or cell models would increase our confidence that no biological activity will occur at relevant exposures.
A key limitation in the development and execution of ToxCast and Tox21 was the reduced xenobiotic metabolism in vitro in comparison with in vivo (Thomas et al., 2019). Likewise, the role of metabolism in driving the effects of coumarin represented a major source of uncertainty. However, our approach exemplifies how a hypothesis-driven risk assessment can guide the selection of the appropriate assays. In this case study, evidence from in silico tools highlighted the potential formation of reactive metabolites (ie, epoxides) with alerts for genotoxicity and protein binding. Furthermore, information on coumarin metabolic clearance (in vitro half-life 5–20 min) underlined the potential for a considerable high exposure to metabolites in the liver. The ToxTracker assays and metabolism identification assays were critical to understand 2 aspects: (1) reactive coumarin metabolite(s) and not coumarin are able to induce DNA lesions, possibly secondary to oxidative stress; and (2) coumarin is preferentially detoxified to hydroxycoumarins and respective glucuronides. Moreover, reactive metabolites such as the epoxide, o-HPAA, and o-HPA were detected at concentrations below the expected plasma concentration, and therefore genotoxicity or other adverse health effects associated with these compounds (Born et al., 1997; Fentem and Fry, 1992; Fentem et al., 1991; Vassallo et al., 2004) are not expected to occur for this exposure scenario. Lastly, PoDs and MoS were not significantly impacted by the generation of cell stress panel and transcriptomics data in cells with higher metabolic competence (Jackson et al., 2016; Yokoyama et al., 2018).
Analogous to traditional approaches, in NGRA the MoS (SCCS, 2018) is used to characterize chemical exposure risks, except instead of using PoDs from animal studies and applied dose (both expressed as external dose in mg/kg), the MoS was calculated using in vitro PoDs and the blood plasma Cmax (both expressed as internal concentration in µM). However, traditionally, a point estimate of the MoS has been used, whereas in this work the MoS was expressed as a distribution, reflecting the combined uncertainty in the in vitro PoDs (where possible) and the Cmax estimates. The risk assessment conclusion was based on the 5th percentile of the distribution, which represented a lower limit for the MoS. This is in agreement with one of the key principles of NGRA, which is that sources of uncertainty are characterized and documented appropriately (Dent et al., 2018a). A critical question is how large should this lower limit be for use in NGRA? Using traditional approaches, a default value of 100 based on the MoS point estimate has been generally accepted to account for the uncertainty in the extrapolation from animal studies to the human population and be adequately protective (SCCS, 2018). However, PoDs derived from NAMs (ToxCast, Tox21, HTTr, and high-throughput phenotypic profiling) have been shown to be more conservative than in vivo PoDs for most tested substances (Paul Friedman et al., 2020), and these findings corroborate previous studies that have proposed that for most industrial chemicals, a PoD based on the most sensitive pathway or biological response provides a conservative estimate of the PoD in vivo (Thomas et al., 2013; Wetmore et al., 2013). Thus, because the PoDs in this study were based on activity rather than adverse effects, and robust probabilistic approaches were applied to characterize uncertainty in both the exposure and PoD determinations, a decision based on the 5th percentile of the MoS distribution being over 100 could be sufficiently conservative. This was the case for both face cream and body lotion suggesting that the inclusion of 0.1% coumarin in these products is safe for the consumer.
Using historical benchmark chemicals with well-characterized human exposures and known human outcomes could help define a protective exposure level and an evidence-based MoS values. This benchmarking approach has not yet been developed for overall systemic toxicity, but examples applied to drug-induced liver injury (Albrecht et al., 2019; Williams et al., 2020), reproductive (Becker et al., 2015; Dent et al., 2018b), and cardiac toxicity (Lazic et al., 2018) have been previously published.
For this case study, we selected a well-known chemical, and therefore the NGRA based on NAMs can be benchmarked against the current risk assessment approach (Abraham et al., 2010; EFSA, 2008). The tolerable daily intake (TDI) established by EFSA for coumarin in foods is 0–0.1 mg/kg/day (EFSA, 2008), based on applying an overall assessment factor of 100 to the animal NOAEL. Using the PBK model developed, plasma Cmax at the upper end of the TDI (7 mg per day, assuming a body weight of 70 kg) would be 0.1 µM. Using the in vitro PoDs derived in this study, this would result in an MoS of 30–600 which means that a safety assessment based on NAMs is at least as protective as the risk assessment based on traditional approaches. More examples of safety assessments, especially for high-risk exposures and a variety of MoAs are needed to test whether the panels used are protective. The final area of uncertainty that warrants additional work is the extrapolation of PoDs from static, short-term in vitro systems to longer term systems able to mimic in vivo exposures (Beilmann et al., 2019; Ewart et al., 2018).
In conclusion, this case study has demonstrated that NAMs can provide robust insights to support exposure estimation and mechanistic in vitro bioactivity data to inform nonanimal safety assessments. The continued development and application of NAMs in a decision-making context will play an increasing role in fulfilling the ambition to assure safety of novel ingredients without the need for any animal testing, but confidence in NAMs will only come with learning by doing and sharing more case studies.
SUPPLEMENTARY DATA
Supplementary data are available at Toxicological Sciences online.
Supplementary Material
ACKNOWLEDGMENTS
The authors would like to thank Katarzyna Przybylak, Katie Paul-Friedman, Gavin Maxwell, Paul Russell, and Mabel Cotter for their helpful technical advice and helpful comments during the preparation of this manuscript. The authors would also like to acknowledge the laboratories, where the data were generated, namely Charles River Research Services UK Limited (Discovery UK), Eurofins Discovery, Cyprotex Discovery Ltd, and BioSpyder Technologies.
FUNDING
Unilever as part of Unilever’s ongoing effort to develop new ways of assuring consumer safety.
DECLARATION OF CONFLICTING INTERESTS
The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
REFERENCES
- Abraham K., Wohrlin F., Lindtner O., Heinemeyer G., Lampen A. (2010). Toxicology and risk assessment of coumarin: Focus on human data. Mol. Nutr. Food Res. 54, 228–239. [DOI] [PubMed] [Google Scholar]
- Adeleye Y., Andersen M., Clewell R., Davies M., Dent M., Edwards S., Fowler P., Malcomber S., Nicol B., Scott A., et al. (2015). Implementing toxicity testing in the 21st century (TT21C): Making safety decisions using toxicity pathways, and progress in a prototype risk assessment. Toxicology 332, 102–111. [DOI] [PubMed] [Google Scholar]
- Albrecht W., Kappenberg F., Brecklinghaus T., Stoeber R., Marchan R., Zhang M., Ebbert K., Kirschner H., Grinberg M., Leist M., et al. (2019). Prediction of human drug-induced liver injury (DILI) in relation to oral doses and blood concentrations. Arch. Toxicol. 93, 1609–1637. [DOI] [PubMed] [Google Scholar]
- Allen T. E. H., Goodman J. M., Gutsell S., Russell P. J. (2018). Using 2D structural alerts to define chemical categories for molecular initiating events. Toxicol. Sci. 165, 213–223. [DOI] [PubMed] [Google Scholar]
- Andersen M., McMullen P., Phillips M., Yoon M., Pendse S., Clewell H., Hartman J., Moreau M., Becker R., Clewell R. (2019). Developing context appropriate toxicity testing approaches using new alternative methods (NAMs). ALTEX 36, 523–534. [DOI] [PubMed] [Google Scholar]
- Armitage J. M., Wania F., Arnot J. A. (2014). Application of mass balance models and the chemical activity concept to facilitate the use of in vitro toxicity data for risk assessment. Environ. Sci. Technol. 48, 9770–9779. [DOI] [PubMed] [Google Scholar]
- Ball N., Cronin M. T., Shen J., Blackburn K., Booth E. D., Bouhifd M., Donley E., Egnash L., Hastings C., Juberg D. R., et al. (2016). Toward good read-across practice (GRAP) guidance. ALTEX 33, 149–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker R. A., Friedman K. P., Simon T. W., Marty M. S., Patlewicz G., Rowlands J. C. (2015). An exposure: Activity profiling method for interpreting high-throughput screening data for estrogenic activity-proof of concept. Regul. Toxicol. Pharmacol. 71, 398–408. [DOI] [PubMed] [Google Scholar]
- Beilmann M., Boonen H., Czich A., Dear G., Hewitt P., Mow T., Newham P., Oinonen T., Pognan F., Roth A., et al. (2019). Optimizing drug discovery by investigative toxicology: Current and future trends. ALTEX 36, 289–313. [DOI] [PubMed] [Google Scholar]
- Bergamini G., Bell K., Shimamura S., Werner T., Cansfield A., Muller K., Perrin J., Rau C., Ellard K., Hopf C., et al. (2012). A selective inhibitor reveals PI3Kgamma dependence of T(H)17 cell differentiation. Nat. Chem. Biol. 8, 576–582. [DOI] [PubMed] [Google Scholar]
- Berggren E., White A., Ouedraogo G., Paini A., Richarz A.-N., Bois F. Y., Exner T., Leite S., van Grunsven L. A, Worth A., et al. (2017). Ab initio chemical safety assessment: A workflow based on exposure considerations and non-animal methods. Comput. Toxicol. 4, 31–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Born S. L., Rodriguez P. A., Eddy C. L., Lehman-McKeeman L. D. (1997). Synthesis and reactivity of coumarin 3,4-epoxide. Drug Metab. Dispos. 25, 1318–1324. [PubMed] [Google Scholar]
- Bowes J., Brown A. J., Hamon J., Jarolimek W., Sridhar A., Waldron G., Whitebread S. (2012). Reducing safety-related drug attrition: The use of in vitro pharmacological profiling. Nat. Rev. Drug Discov. 11, 909–922. [DOI] [PubMed] [Google Scholar]
- Carmichael P., Davies M., Dent M., Fentem J., Fletcher S., Gilmour N., MacKay C., Maxwell G., Merolla L., Pease C., et al. (2009). Non-animal approaches for consumer safety risk assessments: Unilever’s scientific research programme. Altern. Lab. Anim. 37, 595–610. [DOI] [PubMed] [Google Scholar]
- Carusi A., Davies M. R., De Grandis G., Escher B. I., Hodges G., Leung K. M. Y., Whelan M., Willett C., Ankley G. T. (2018). Harvesting the promise of AOPs: An assessment and recommendations. Sci. Total Environ. 628–629, 1542–1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constable A., Jonas D., Cockburn A., Davi A., Edwards G., Hepburn P., Herouet-Guicheney C., Knowles M., Moseley B., Oberdorfer R., et al. (2007). History of safe use as applied to the safety assessment of novel foods and foods derived from genetically modified organisms. Food Chem. Toxicol. 45, 2513–2525. [DOI] [PubMed] [Google Scholar]
- Council N. R. (2007). Toxicity Testing in the 21st Century: A Vision and a Strategy. The National Academies Press, Washington, DC. [Google Scholar]
- Cramer G. M., Ford R. A., Hall R. L. (1976). Estimation of toxic hazard—A decision tree approach. Food Cosmet. Toxicol. 16, 255–276. [DOI] [PubMed] [Google Scholar]
- Dent M., Amaral R. T., Da Silva P. A., Ansell J., Boisleve F., Hatao M., Hirose A., Kasai Y., Kern P., Kreiling R., et al. (2018. a). Principles underpinning the use of new methodologies in the risk assessment of cosmetic ingredients. Comput. Toxicol. 7, 20–26. [Google Scholar]
- Dent M. P., Li H., Carmichael P. L., Martin F. L. (2018. b). Employing dietary comparators to perform risk assessments for anti-androgens without using animal data. Toxicol. Sci. 167, 375–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desprez B., Dent M., Keller D., Klaric M., Ouédraogo G., Cubberley R., Duplan H., Eilstein J., Ellison C., Grégoire S., et al. (2018). A strategy for systemic toxicity assessment based on non-animal approaches: The cosmetics Europe long range science strategy programme. Toxicol. In Vitro 50, 137–146. [DOI] [PubMed] [Google Scholar]
- EFSA. (2008). Coumarin in flavourings and other food ingredients with flavouring properties—Scientific opinion of the panel on food additives, flavourings, processing aids and materials in contact with food (AFC). EFSA J. 6, 793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewart L., Dehne E. M., Fabre K., Gibbs S., Hickman J., Hornberg E., Ingelman-Sundberg M., Jang K. J., Jones D. R., Lauschke V. M., et al. (2018). Application of microphysiological systems to enhance safety assessment in drug discovery. Annu. Rev. Pharmacol. Toxicol. 58, 65–82. [DOI] [PubMed] [Google Scholar]
- Fabregat A., Jupe S., Matthews L., Sidiropoulos K., Gillespie M., Garapati P., Haw R., Jassal B., Korninger F., May B., et al. (2018). The Reactome pathway knowledgebase. Nucleic Acids Res. 46, D649–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farinola N., Piller N. B. (2007). CYP2A6 polymorphisms: Is there a role for pharmacogenomics in preventing coumarin-induced hepatotoxicity in lymphedema patients? Pharmacogenomics 8, 151–158. [DOI] [PubMed] [Google Scholar]
- Farmahin R., Williams A., Kuo B., Chepelev N. L., Thomas R. S., Barton-Maclaren T. S., Curran I. H., Nong A., Wade M. G., Yauk C. L. (2017). Recommended approaches in the application of toxicogenomics to derive points of departure for chemical risk assessment. Arch. Toxicol. 91, 2045–2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fentem J. H., Fry J. R. (1992). Metabolism of coumarin by rat, gerbil and human liver microsomes. Xenobiotica 22, 357–367. [DOI] [PubMed] [Google Scholar]
- Fentem J. H., Fry J. R., Whiting D. A. (1991). o-Hydroxyphenylacetaldehyde: A major novel metabolite of coumarin formed by rat, gerbil and human liver microsomes. Biochem. Biophys. Res. Commun. 179, 197–203. [DOI] [PubMed] [Google Scholar]
- Filer D. L., Kothiya P., Setzer R. W., Judson R. S., Martin M. T. (2016). Tcpl: The ToxCast pipeline for high-throughput screening data. Bioinformatics 33, 618–620. [DOI] [PubMed] [Google Scholar]
- Fischer F. C., Henneberger L., Konig M., Bittermann K., Linden L., Goss K. U., Escher B. I. (2017). Modeling exposure in the tox21 in vitro bioassays. Chem. Res. Toxicol. 30, 1197–1208. [DOI] [PubMed] [Google Scholar]
- Friedman K. P., Gagne M., Loo L. H., Karamertzanis P., Netzeva T., Sobanski T., Franzosa J., Richard A., Lougee R., Gissi A., et al. (2020). Utility of in vitro bioactivity as a lower bound estimate of in vivo adverse effect levels and in risk-based prioritization. Toxicol. Sci. 173, 202–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelman A., Carlin J. B., Stern H. S., Dunson D. B., Vehtari A., Rubin D. B. (2013). Bayesian Data Analysis. Chapman and Hall: / CRC, Boca Raton, FL. [Google Scholar]
- Gnerre C., Catto M., Leonetti F., Weber P., Carrupt P.-A., Altomare C., Carotti A., Testa B. (2000). Inhibition of monoamine oxidases by functionalized coumarin derivatives: Biological activities, QSARs, and 3D-QSARs. J. Med. Chem. 43, 4747–4758. [DOI] [PubMed] [Google Scholar]
- Hall B., Tozer S., Safford B., Coroama M., Steiling W., Leneveu-Duchemin M. C., McNamara C., Gibney M. (2007). European consumer exposure to cosmetic products, a framework for conducting population exposure assessments. Food Chem. Toxicol. 45, 2097–2108. [DOI] [PubMed] [Google Scholar]
- Hatherell, S., Baltazar, M. T., Reynolds, J., Carmichael, P. L., Dent, M., Li, H., Ryder, S., White, A., Walker, P., Middleton, A. M. Identifying and characterizing stress pathways of concern for consumer safety in next generation risk assessment. Toxicol. Sci. Forthcoming. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendriks G., Derr R. S., Misovic B., Morolli B., Calleja F. M., Vrieling H. (2016). The extended ToxTracker assay discriminates between induction of DNA damage, oxidative stress, and protein misfolding. Toxicol. Sci. 150, 190–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ICCR. (2018). Report on integrated strategies for safety assessment of cosmetic ingredients: Part 2. Available at: https://www.iccr-cosmetics.org/files/8315/4322/3079/ICCR_Integrated_Strategies_for_Safety_Assessment_of_Cosmetic_Ingredients_Part_2.pdf. Accessed April 19, 2020.
- Jackson J. P., Li L., Chamberlain E. D., Wang H., Ferguson S. S. (2016). Contextualizing hepatocyte functionality of cryopreserved HepaRG cell cultures. Drug Metab. Dispos. 44, 1463–1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones H., Chen Y., Gibson C., Heimbach T., Parrott N., Peters S., Snoeys J., Upreti V., Zheng M., Hall S. (2015). Physiologically based pharmacokinetic modeling in drug discovery and development: A pharmaceutical industry perspective. Clin. Pharmacol. Ther. 97, 247–262. [DOI] [PubMed] [Google Scholar]
- Karlsson H. L., Gliga A. R., Calleja F. M., Goncalves C. S., Wallinder I. O., Vrieling H., Fadeel B., Hendriks G. (2014). Mechanism-based genotoxicity screening of metal oxide nanoparticles using the ToxTracker panel of reporter cell lines. Part. Fibre Toxicol. 11, 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavlock R. J., Bahadori T., Barton-Maclaren T. S., Gwinn M. R., Rasenberg M., Thomas R. S. (2018). Accelerating the pace of chemical risk assessment. Chem. Res. Toxicol. 31, 287–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer N. I., Krismartina M., Rico-Rico Á., Blaauboer B. J., Hermens J. (2012). Quantifying processes determining the free concentration of phenanthrene in basal cytotoxicity assays. Chem. Res. Toxicol. 25, 436–445. [DOI] [PubMed] [Google Scholar]
- Lazic S. E., Edmunds N., Pollard C. E. (2018). Predicting drug safety and communicating risk: Benefits of a Bayesian approach. Toxicol. Sci. 162, 89–98. [DOI] [PubMed] [Google Scholar]
- Limonciel A., Ates G., Carta G., Wilmes A., Watzele M., Shepard P. J., VanSteenhouse H. C., Seligmann B., Yeakley J. M., van de Water B., et al. (2018). Comparison of base-line and chemical-induced transcriptomic responses in HepaRG and RPTEC/TERT1 cells using TempO-Seq. Arch. Toxicol. 92, 2517–2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M. I., Huber W., Anders S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maresca A., Supuran C. T. (2010). Coumarins incorporating hydroxy- and chloro-moieties selectively inhibit the transmembrane, tumor-associated carbonic anhydrase isoforms IX and XII over the cytosolic ones I and II. Bioorg. Med. Chem. Lett. 20, 4511–4514. [DOI] [PubMed] [Google Scholar]
- Maresca A., Temperini C., Vu H., Pham N. B., Poulsen S.-A., Scozzafava A., Quinn R. J., Supuran C. T. (2009). Non-zinc mediated inhibition of carbonic anhydrases: Coumarins are a new class of suicide inhibitors. J. Am. Chem. Soc. 131, 3057–3062. [DOI] [PubMed] [Google Scholar]
- Moxon T. E., Li H., Lee M.-Y., Piechota P., Nicol B., Pickles J., Pendlington R., Sorrell I., Baltazar M. T. (2020). Application of physiologically based kinetic (PBK) modelling in the Next Generation Risk Assessment of dermally applied consumer products. Toxicol. In Vitro 63, 104746. [DOI] [PubMed] [Google Scholar]
- Neely T., Russell P., O′Hagan S., Walsh-Mason B., Horst A. D., Lahorkar P. (2011). A multi-criteria decision analysis model to assess the safety of botanicals utilizing data on history of use. Toxicol. Int. 18, 20–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OECD. (2017). Test no. 405: Acute eye irritation/corrosion.
- OECD. (2018. a). Test no. 442d: In vitro skin sensitisation.
- OECD. (2018. b). Test no. 442e: In vitro skin sensitisation.
- OECD. (2019. a). Test no. 439: In vitro skin irritation: Reconstructed human epidermis test method.
- OECD. (2019. b). Test no. 442c: In chemico skin sensitisation.
- Paini A., Leonard J. A., Kliment T., Tan Y.-M., Worth A. (2017). Investigating the state of physiologically based kinetic modelling practices and challenges associated with gaining regulatory acceptance of model applications. Regul. Toxicol. Pharmacol. 90, 104–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patlewicz G., Jeliazkova N., Safford R. J., Worth A. P., Aleksiev B. (2008). An evaluation of the implementation of the Cramer classification scheme in the ToxTree software. SAR QSAR Environ. Res. 19, 495–524. [DOI] [PubMed] [Google Scholar]
- Paul Friedman K., Gagne M., Loo L.-H., Karamertzanis P., Netzeva T., Sobanski T., Franzosa J. A., Richard A. M., Lougee R. R., Gissi A., et al. (2020). Utility of in vitro bioactivity as a lower bound estimate of in vivo adverse effect levels and in risk-based prioritization. Toxicol. Sci. 173, 202–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips J. R., Svoboda D. L., Tandon A., Patel S., Sedykh A., Mav D., Kuo B., Yauk C. L., Yang L., Thomas R. S., et al. (2019). BMDexpress2: Enhanced transcriptomic dose-response analysis workflow. Bioinformatics 35, 1780–1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramaiahgari S. C., Auerbach S. S., Saddler T. O., Rice J. R., Dunlap P. E., Sipes N. S., DeVito M. J., Shah R. R., Bushel P. R., Merrick B. A., et al. (2019). The power of resolution: Contextualized understanding of biological responses to liver injury chemicals using high-throughput transcriptomics and benchmark concentration modeling. Toxicol. Sci. 169, 553–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramaiahgari S. C., Waidyanatha S., Dixon D., DeVito M. J., Paules R. S., Ferguson S. S. (2017). From the cover: Three-dimensional (3D) HepaRG spheroid model with physiologically relevant xenobiotic metabolism competence and hepatocyte functionality for liver toxicity screening. Toxicol. Sci. 159, 124–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritschel W. A., Brady M. E., Tan H. S., Hoffmann K. A., Yiu I. M., Grummich K. W. (1977). Pharmacokinetics of coumarin and its 7-hydroxy-metabolites upon intravenous and peroral administration of coumarin in man. Eur. J. Clin. Pharmacol. 12, 457–461. [DOI] [PubMed] [Google Scholar]
- SCCS. (2018). The SCCS Note of Guidance for the Testing of Cosmetic Ingredients and Their Safety Evaluation—10th Revision SCCS/1602/18—Final Version.
- Schurch N. J., Schofield P., Gierliński M., Cole C., Sherstnev A., Singh V., Wrobel N., Gharbi K., Simpson G. G., Owen-Hughes T., et al. (2016). How many biological replicates are needed in an RNA-seq experiment and which differential expression tool should you use? RNA (New York, NY). 22, 839–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons S. O., Fan C. Y., Ramabhadran R. (2009). Cellular stress response pathway system as a sentinel ensemble in toxicological screening. Toxicol. Sci. 111, 202–225. [DOI] [PubMed] [Google Scholar]
- Sipes N. S., Martin M. T., Kothiya P., Reif D. M., Judson R. S., Richard A. M., Houck K. A., Dix D. J., Kavlock R. J., Knudsen T. B. (2013). Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem. Res. Toxicol. 26, 878–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taavitsainen P., Juvonen R., Pelkonen O. (2001). In vitro inhibition of cytochrome P450 enzymes in human liver microsomes by a potent CYP2A6 inhibitor, trans-2-phenylcyclopropylamine (tranylcypromine), and its nonamine analog, cyclopropylbenzene. Drug Metab. Dispos. 29, 217–222. [PubMed] [Google Scholar]
- Thomas R. S., Bahadori T., Buckley T. J., Cowden J., Deisenroth C., Dionisio K. L., Frithsen J. B., Grulke C. M., Gwinn M. R., Harrill J. A., et al. (2019). The next generation blueprint of computational toxicology at the U.S. Environmental Protection Agency. Toxicol. Sci. 169, 317–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas R. S., Philbert M. A., Auerbach S. S., Wetmore B. A., DeVito M. J., Cote I., Rowlands J. C., Whelan M. P., Hays S. M., Andersen M. E., et al. (2013). Incorporating new technologies into toxicity testing and risk assessment: Moving from 21st century vision to a data-driven framework. Toxicol. Sci. 136, 4–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varma M. V., Steyn S. J., Allerton C., El-Kattan A. F. (2015). Predicting clearance mechanism in drug discovery: Extended Clearance Classification System (ECCS). Pharm. Res. 32, 3785–3802. [DOI] [PubMed] [Google Scholar]
- Vassallo J. D., Hicks S. M., Born S. L., Daston G. P. (2004). Roles for epoxidation and detoxification of coumarin in determining species differences in clara cell toxicity. Toxicol. Sci. 82, 26–33. [DOI] [PubMed] [Google Scholar]
- Villeneuve D. L., Crump D., Garcia-Reyero N., Hecker M., Hutchinson T. H., LaLone C. A., Landesmann B., Lettieri T., Munn S., Nepelska M., et al. (2014). Adverse outcome pathway (AOP) development I: Strategies and principles. Toxicol. Sci. 142, 312–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westmoreland C., Carmichael P., Dent M., Fentem J., MacKay C., Maxwell G., Pease C., Reynolds F. (2010). Assuring safety without animal testing: Unilever’s ongoing research programme to deliver novel ways to assure consumer safety. ALTEX 27, 207–265. [PubMed] [Google Scholar]
- Wetmore B. A., Wambaugh J. F., Allen B., Ferguson S. S., Sochaski M. A., Setzer R. W., Houck K. A., Strope C. L., Cantwell K., Judson R. S., et al. (2015). Incorporating high-throughput exposure predictions with dosimetry-adjusted in vitro bioactivity to inform chemical toxicity testing. Toxicol. Sci. 148, 121–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wetmore B. A., Wambaugh J. F., Ferguson S. S., Li L., Clewell H. J., Judson R. S., Freeman K., Bao W., Sochaski M. A., Chu T.-M., et al. (2013). Relative impact of incorporating pharmacokinetics on predicting in vivo hazard and mode of action from high-throughput in vitro toxicity assays. Toxicol. Sci. 132, 327–346. [DOI] [PubMed] [Google Scholar]
- Williams D. P., Lazic S. E., Foster A. J., Semenova E., Morgan P. (2020). Predicting drug-induced liver injury with Bayesian machine learning. Chem. Res. Toxicol. 33, 239–248. [DOI] [PubMed] [Google Scholar]
- Yang C., Barlow S. M., Muldoon Jacobs K. L., Vitcheva V., Boobis A. R., Felter S. P., Arvidson K. B., Keller D., Cronin M. T. D., Enoch S., et al. (2017). Thresholds of toxicological concern for cosmetics-related substances: New database, thresholds, and enrichment of chemical space. Food Chem. Toxicol. 109, 170–193. [DOI] [PubMed] [Google Scholar]
- Yeakley J. M., Shepard P. J., Goyena D. E., VanSteenhouse H. C., McComb J. D., Seligmann B. E. (2017). A trichostatin A expression signature identified by TempO-Seq targeted whole transcriptome profiling. PLoS One 12, e0178302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama Y., Sasaki Y., Terasaki N., Kawataki T., Takekawa K., Iwase Y., Shimizu T., Sanoh S., Ohta S. (2018). Comparison of drug metabolism and its related hepatotoxic effects in HepaRG, cryopreserved human hepatocytes, and HepG2 cell cultures. Biol. Pharm. Bull. 41, 722–732. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.