Abstract
Many substances for which consumer safety risk assessments need to be conducted are not associated with specific toxicity modes of action, but rather exhibit nonspecific toxicity leading to cell stress. In this work, a cellular stress panel is described, consisting of 36 biomarkers representing mitochondrial toxicity, cell stress, and cell health, measured predominantly using high content imaging. To evaluate the panel, data were generated for 13 substances at exposures consistent with typical use-case scenarios. These included some that have been shown to cause adverse effects in a proportion of exposed humans and have a toxicological mode-of-action associated with cellular stress (eg, doxorubicin, troglitazone, and diclofenac), and some that are not associated with adverse effects due to cellular stress at human-relevant exposures (eg, caffeine, niacinamide, and phenoxyethanol). For each substance, concentration response data were generated for each biomarker at 3 timepoints. A Bayesian model was then developed to quantify the evidence for a biological response, and if present, a credibility range for the estimated point of departure (PoD) was determined. PoDs were compared with the plasma Cmax associated with the typical substance exposures, and indicated a clear differentiation between “low” risk and “high” risk chemical exposure scenarios. Developing robust methods to characterize the in vitro bioactivity of xenobiotics is an important part of non-animal safety assessment. The results presented in this work show that the cellular stress panel can be used, together with other new approach methodologies, to identify chemical exposures that are protective of consumer health.
Keywords: alternatives to animal testing, risk assessment, systems biology, redox signaling, inflammation, oxidative injury, glutathione, dose-response, cytotoxicity, computational modeling
Historically, safety assessments of ingredients in consumer products, such as cosmetics and food have relied on apical endpoints derived from animal testing. However, ethical and regulatory considerations on animal use, in addition to the scientific need to use more human-relevant data, have led to the emergence of next-generation risk assessment (NGRA) (U.S. EPA, 2014). NGRA is an exposure-led and hypothesis-driven approach, wherein safety assessments are conducted in a tiered manner using detailed information on levels of consumer exposure to the ingredient together with appropriate new approach methodologies (NAMs), including in silico, in chemico, and in vitro approaches (Dent et al., 2018).
For many substance exposures, low tier approaches such as exposure-based waiving will provide sufficient information to make a decision on safety (Yang et al., 2017, 668). However, when this is not the case, higher tier approaches can be deployed. For exposures where systemic toxicity is predicted to be significant, physiologically based kinetic (PBK) models can be used to simulate the distribution of the substance throughout the body (ie, the bioavailability) (Campbell et al., 2012; Moxon et al., 2020). The output from such models can be combined with high-throughput in vitro cell assays where toxicity biomarkers of concern and the concentrations at which they are perturbed (ie, the point of departure [PoD]) are identified (Thomas et al., 2019; Wetmore et al., 2015).
Developing suitable high-throughput assays for different toxicity outcomes remain a major challenge within NGRA (Middleton et al., 2017). In particular, studies have suggested that many substances for which a chemical risk assessment needs to be conducted are associated with nonspecific toxicity modes of action (Sipes et al., 2013; Thomas et al., 2013), leading to cellular stress or mitochondrial toxicity, which in turn are associated with various organ toxicities (Fromenty, 2019; Ipsen et al., 2018; Ramachandran et al., 2018; Rana et al., 2019). Perhaps, the most comprehensive datasets looking at the general bioactivity of substances have been produced as part of the U.S. EPA Toxcast (Dix et al., 2007; Judson et al., 2010; Kavlock et al., 2012) and the U.S. federal cross agency Tox21 programs (Attene-Ramos et al., 2013; Collins et al., 2008; Shukla et al., 2010; Tice et al., 2013). Analysis of these datasets revealed that there is a disproportionate increase in positive assay responses at concentrations that coincide with cytotoxicity and cell stress (Judson et al., 2016). However, in that analysis it was generally not possible to distinguish between specific stress responses triggered by a chemical at subcytotoxic concentrations (that may have subsequently lead to cytotoxicity at higher concentrations or later timepoints), and cell stress events that coincided with, and potentially occurred as a consequence of cytotoxicity (referred to in this work as a “cytotoxic burst”). This limits the degree to which the data could be used to develop a hypothesis on a potential mechanism of toxicity. As such, developing a suitable set of assays and analysis approaches to unravel these events is an on-going challenge in NGRA.
The objective of this work was to develop and evaluate a cellular stress response panel that could form part of an early tier screen for identifying substances that could, at relevant exposure levels, be associated with causing adverse effects in humans. The panel consisted of biomarkers covering the key cellular stress pathways already identified (Simmons et al., 2009), together with mitochondrial toxicity and various cell health effects. To evaluate the suitability of the panel for chemical risk assessment, data were generated using two sets of benchmark chemicals. The first set included chemicals that at defined human exposures are known to cause adverse systemic effects due to cellular stress in a proportion of exposed individuals. The second set included chemicals that at relevant human exposures have not been associated with adverse systemic effects related to cellular stress.
A key principle of NGRA is that the various sources of uncertainty, such as identifying positive biomarkers and estimating the associated PoDs, should be robustly characterized (Dent et al., 2018). To this end, a novel concentration-response model was developed. The approach used Bayesian statistics, which allowed for uncertainties in the model outputs (ie, PoD estimates) to be quantified in a probabilistic manner (Gelman et al., 2013; Reynolds et al., 2019; Shao and Shapiro, 2018). Using the stress panel, together with the statistical approach described here, it was possible to largely distinguish between chemical exposures that are associated with adverse health outcomes and chemical exposures that pose a low risk for the consumer.
MATERIALS AND METHODS
Composition of the cellular stress panel
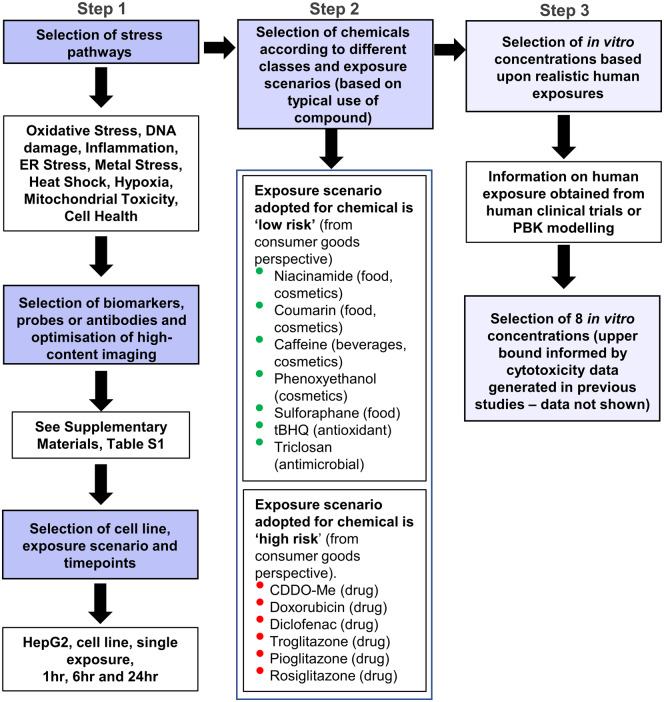
Development of the cellular stress response panel followed three stages, depicted in Figure 1. First, a comprehensive literature review was performed to identify biomarkers representative of key stress pathways already identified based on Simmons et al. (2009), together with mitochondrial toxicity and various cell health effects (Step 1, Figure 1). A complete list of these biomarkers, together with a detailed description of their mechanistic interpretation and associated references is provided in Table 1. Where possible, measurements for the same pathway were multiplexed on the same plate, as indicated in Supplementary Table 1 (making a total of 15 assays). However, this was not always feasible due to not being able to multiplex antibodies from the same species or fluorescent antibodies/probes of the same wavelength in the same well. For all chemicals, each assay was run once with 3 technical replicates per concentration tested. The positive controls used for each biomarker can also be found in Supplementary Table 1. In order to generate reliable PoD estimates for each biomarker and detect potentially transient responses (Shah et al., 2016), data were generated for 8 concentration points per test chemical at 1, 6, and 24 h post-treatment, except for phospholipidosis and steatosis (6 and 24 h only). Test substances were selected (Step 2, Figure 1) such that there exists evidence either that (at defined levels of human exposure) the substance was known to cause adverse effects in humans due to cellular stress or that there was a history of safe use. Substance exposures in the former group were considered “high” risk and those in the latter group “low” risk, in the context of safety decisions made in the consumer goods industry. The only exception to this was diethyl maleate (DEM), which was the only test substance used in the panel for which a typical systemic exposure level in humans could not be defined. A summary of the chemicals and corresponding exposure scenarios is provided in Table 2, and the rationale for whether a chemical exposure is low or high risk is provided in Supplementary Material (Supplementary Substance Information). Where available, total Cmax estimates (see Table 2) were obtained from the literature, otherwise they were calculated using PBK models (see Exposure assessment section). These were then used to establish the concentration ranges for the in vitro assays of each substance, together with cytotoxicity data from previous studies (Step 3, Figure 1; data not shown). In addition, in vitro nominal concentrations were selected in order to cover the predicted/observed free in vivo plasma concentrations. To achieve this, the free concentration was calculated for both plasma Cmax concentrations and in vitro concentrations by applying a steady-state mass balance partitioning model based on published models (Armitage et al., 2014; Fischer et al., 2017; Kramer et al., 2012). For all chemicals tested, the quantitative in vitro to in vivo extrapolation factor was higher than 1 (Supplementary Table 2), which indicated that for the same total plasma Cmax and in vitro concentration, the resulting free concentration that was available for uptake into the cells was higher in vitro than in vivo. This meant that an initial comparison between plasma Cmax and in vitro PoD was conservative and therefore nominal concentrations were applied throughout. The 8 concentration points for each substance based on these ranges are provided in Supplementary Table 3.
Figure 1.
Overview of composition of the stress panel and experimental design for benchmark data generation. Diethyl maleate (DEM) was also included as a test chemical in the panel, but could not be designated as high or low risk due to lack of exposure information.
Table 1.
Composition of the Cellular Stress Panel
| Pathway | Biomarker | Description | Interpretation | References |
|---|---|---|---|---|
| Cell health and physiology | Cell count (nuclei)a/b | Number of cells calculated by counting stained nuclei. | A decreasing number of cells per well indicates toxicity due to necrosis, apoptosis or a reduction in cellular proliferation. | Bauch et al. (2015), Nikoletopoulou et al. (2013) |
| Cell health and physiology | Nuclear sizeb | Nuclear area measured using DNA stain. | An increase in nuclear area can indicate necrosis or G2 cell cycle arrest and a decrease can indicate apoptosis. | |
| Cell health and physiology | DNA structureb | DNA structure measured using DNA stain. | An increase in DNA structure can indicate chromosomal instability and DNA fragmentation. | |
| Cell health and physiology | Cell cycle arrest | Determined as the ratio of G0/G1(2N) to G2/M(4N) | An increase is linked to G0/G1 arrest and a decrease is linked to G2/M arrest. | |
| Cell health and physiology | Cell membrane permeability (necrosis)a | Detected using a cell-impermeant nucleic acid stain. | An increase in cell membrane permeability is a general indicator of cell death via necrosis. | |
| Cell health and physiology | Caspase 3/7 intensity (apoptosis)a | Following activation of Caspase-3/7 in apoptotic cells, the detection reagent is cleaved, enabling the dye to bind to DNA & generate fluorescence. | An increase in Caspase 3/7 activity indicates the onset of the cell signaling cascade leading to cell signaled cell death (apoptosis). | |
| Cell health and physiology | LDH releasea | Determined by detecting the level of LDH released from cells measured by the conversion of resazurin into resorfin. | An increase in LDH is due to the release of LDH from cells which have damaged membranes. | |
| Cell health and physiology | Intracellular pH | Determined by measuring the intensity of a fluorogenic probe that increases as the pH drops. | Changes in intracellular pH can indicate the interference of the compound with either the regulation of intracellular pH or the protonation of the compound itself. Specific intracellular pH is required for optimum cellular processes, distribution or target binding of the compound. | |
| Cell health and physiology | Phospholipidosis (PLD) | Detected following conjugation of a fluorescent dye to phospholipids within cells. | An increase in phospholipidosis (PLD) indicates an accumulation of phospholipids and/or compounds within lysosomes. Lysosomes are organelles essential in cellular biogenesis and if compromised can lead to cellular toxicity. PLD can also occur indirectly by altering synthesis and/or degradation of phospholipids. | |
| Cell health and physiology | Steatosis | Detected using a fluorescent neutral lipid stain with a high affinity for neutral lipid droplets (mainly consisting of triglycerides). | An increase in steatosis indicates an accumulation of triglycerides within the cytoplasm of treated cells, often triggered by compounds that affect the metabolism of fatty acids and/or neutral lipids. Large accumulations can disrupt cell constituents, and in severe cases the cell may burst. | |
|
| ||||
| Mitochondrial toxicity | PGC1alpha | PGC1alpha is a transcription factor coactivator involved in mitochondrial biogenesis and metabolic homeostasis. | An increase in the protein expression of PGC1alpha indicates mitochondrial toxicity. | Attene-Ramos et al. (2013, 2015), Eakins et al. (2016), Nadanaciva and Will (2011), Yuan et al. (2016) |
| Mitochondrial toxicity | Mitochondrial ROS (MitoROS) | Detected following oxidation by superoxide of a fluorogenic dye specifically targeted to mitochondria. | An increase in mitochondrial superoxide production indicates mitochondrial toxicity and oxidative damage. | |
| Mitochondrial toxicity | Mitochondrial mass (mito-mass) | Mitochondrial mass is measured by a fluorescent mitochondrial specific stain and is an indicator of both the size and number of mitochondria present in a cell. | A decrease in mitochondrial mass indicates loss of total mitochondria and an increase implies mitochondrial swelling or an adaptive response to cellular energy demands. | |
| Mitochondrial toxicity | Mitochondrial membrane potential (MMP) | The mitochondrial membrane potential (MMP) plays a key role in ATP production and mitochondrial homeostasis, and is measured by staining the cells with a fluorescent dye specific for active mitochondria prior to compound treatment. | A decrease indicates a loss of mitochondrial membrane potential and mitochondrial toxicity, as well as a potential role in apoptosis signaling, an increase in mitochondrial membrane potential indicates an adaptive response to cellular energy demands. | |
| Mitochondrial toxicity | Cellular ATP | Cellular ATP levels are detected using a luminescence-based assay. After cell lysis the endogenous enzymes are released from the cell. Cells which are not metabolically active will not release any ATP. | A decrease in metabolically active cells will result in a decrease in the level of ATP detected indicating mitochondrial toxicity and loss of cell viability. An increase in cellular ATP levels could also indicate an effect on cellular metabolism. | |
| Mitochondrial toxicity | Oxygen consumption rate (OCR) | OCR is a measurement of oxygen content in extracellular media using an XFe96 Extracellular Flux Analyzer. | Changes in OCR indicate effects on mitochondrial function and can be bidirectional. A decrease is due to an inhibition of mitochondrial respiration, while an increase may indicate an uncoupler, in which respiration is not linked to energy production. | |
| Mitochondrial toxicity | Reserve capacity | The reserve capacity is the measured ability of cells to respond to an increase in energy demand. Detected using an XFe96 Extracellular Flux Analyzer following addition of the protonophoric uncoupler FCCP. | A reduction indicates mitochondrial dysfunction. This measurement demonstrates how close to the bioenergetic limit the cell is. | |
|
| ||||
| Oxidative stress | NRF2 | Nrf2 is a transcription factor that is key for regulation of cellular redox balance and adaptive responses to oxidative stress. | An increase in translocation of Nrf2 into the nucleus indicates oxidative stress and results in the expression of a wide range of antioxidant-response genes. | Hiemstra et al. (2017), Loboda et al. (2016), Ramesh et al. (2014), Wink et al. (2014) |
| Oxidative stress | Heme oxygenase 1 (HMOX1) | Heme oxygenase 1 is one of the many genes that has its expression induced by Nrf2 activation and has several antioxidant roles including the removal of toxic heme. | An increase in the protein level of heme oxygenase 1 indicates induction of the Nrf2/oxidative stress-response pathway. | |
| Oxidative stress | Oxidative stress (ROS) | Reactive oxygen species (ROS) are free radicals that cause damage to a range of macromolecules including DNA, RNA, and protein. Detected using a probe that fluoresces following reaction with superoxide or hydrogen peroxide. | An increase in ROS indicates the formation of toxic superoxide intermediates, an early cytotoxic response and indicator of oxidative stress. | |
| Oxidative stress | Glutathione content (GSH) | Glutathione is one of the most abundant cellular antioxidants and helps to maintain cysteine-thiol groups of proteins in the reduced state. An increased GSSG (oxidized glutathione) to GSH (reduced glutathione) ratio is indicative of oxidative stress. | A decrease in glutathione content can result from production of reactive oxygen species or from direct binding of electrophiles. An increase in glutathione content represents an adaptive cellular response to oxidative stress. | |
| Inflammation | NFkB | NFkB is a transcription factor, which resides in the cytoplasm bound to IkB. Upon cellular stress the complex dissociates and NFkB translocates into the nucleus, where it triggers the expression of cytokines, enzymes and growth factors. | An increase in NFkB signal indicates the activation of the NFkB pathway and its translocation to the nucleus to initiate downstream gene expression. | Ben-Neriah and Karin (2011), Kany et al. (2019), Verstrepen et al. (2010), Xu et al. (2017) |
| Inflammation | IL-8 | IL-8 is a chemokine involved in inflammation and stimulation of the innate immune system. | An increase in IL-8 secretion may suggest an inflammatory response. | |
| Inflammation | TNFAIP3 (A20) | TNFAIP3 (A20) is a cytoplasmic protein that plays a key role in the negative regulation of inflammation and immunity. | An increase in TNFAIP3 (A20) is likely to be seen if a compound induces an inflammatory response and can lead to inhibition of NFkB activation. | |
|
| ||||
| ER Stress | Endoplasmic reticulum (ER) | The ER plays a crucial role in the synthesis of cellular proteins. The level of ER in a cell was detected using a fluorescent dye selective for ER in live cells. | Cells increase biogenesis of components of the ER in order to increase protein-folding capacity. Therefore, an increase in the size of the endoplasmic reticulum is an indicator of ER stress. | Foufelle and Fromenty (2016), Oslowski and Urano (2011), Teske et al. (2011), Wang and Kaufman (2016), Wink et al. (2014) |
| ER stress | BiP | BiP is an ER chaperone with a high affinity for misfolded proteins. | An increase in the protein levels of BiP indicates ER stress. | |
| ER stress | XBP1 | XBP1 is a transcription factor activated by the ER stress sensor IRE1 and induces transcription of genes involved in ER size and function. | An increase in the protein levels of XBP1 indicates ER stress. | |
| ER stress | PERK | The kinase PERK is an ER stress sensor that plays a key role in inhibiting the synthesis of new proteins and activation of the transcription factors ATF4 and CHOP. | An increase in the protein levels of PERK indicates ER stress. | |
| ER stress | ATF4 | ATF4 is a transcription factor activated via the PERK branch of the ER stress pathway that transcriptionally activates CHOP. | An increase in the protein levels of ATF4 indicates ER stress. | |
| ER stress | CHOP | CHOP is a transcription factor activated via the PERK branch of the ER stress pathway. Low levels of CHOP results in the transcription of pro-survival proteins including chaperones. High levels of CHOP lead to the initiation of apoptosis. | An increase in the protein levels of CHOP indicates ER stress. | |
|
| ||||
| Metal stress | MTF1 | Metal-responsive transcription factor 1 (MTF-1) is a transcription factor that regulates the expression of genes involved in metal homeostasis. | An increase in protein levels of MTF-1 indicates metal stress. | Gunther et al. (2012), Park and Jeong (2018), Wink et al. (2014) |
| Metal stress | Metallothionein (MT) | Metallothionein expression is induced by the transcription factor MTF-1. Metallothioneins bind and sequester toxic heavy metal ions. | An increase in protein levels of MT indicates metal stress. | |
| DNA damage | DNA damage (p-H2AX) | DNA double-strand breaks (DSBs) cause the phosphorylation of histone H2AX at Ser139. DSBs are an indication of genotoxicity and can lead to apoptosis. | An increase in p-H2AX indicates a rise in the number of DSBs and therefore DNA damage induction. | Ando et al. (2014), Banerjee and Chakravarti (2011), Khoury et al. (2013) |
|
| ||||
| Heat shock response | Heat shock response (Hsp70) | Hsp70 protects against cellular stress particularly through its key role in protein folding and inhibition of apoptosis. | An increase in Hsp70 indicates a general cellular stress response which could include thermal, metal, oxidative and ER stress. | Boudesco et al. (2018), Wang et al. (2014a), Westerheide and Morimoto (2005), Wink et al. (2014) |
|
| ||||
| Hypoxia | HIF1alpha | Hypoxia-inducible factor-1 alpha (HIF1alpha) is a transcription factor that plays a key role in the cellular response to hypoxia (low oxygen levels) and also responds to changes in the redox state of the cell. | An increase in the level of HIF1alpha indicates hypoxia. | Dengler et al. (2014), Wenger et al. (2005), Wink et al. (2014) |
|
| ||||
| Aryl hydrocarbon receptor (AhR) | AhR translocation | AhR is a multifunctional transcription factor that cross-talks with other transcription factors including Nrf2 and NFkB, and cytochrome P450 enzymes. | An increase in AhR translocation indicates a general cellular stress response which could include oxidative stress, inflammation, and other chemical defense mechanisms. | Bock (2019), Furue et al. (2017), Nebert et al. (2000) |
Cytotoxicity biomarker.
Biomarker measured in every assay.
Table 2.
Exposure Data Used for the Estimation of Internal Concentration Expressed as total Plasma Cmax (µM)
| Chemical | Chemical-Exposure Classification a | Exposure Data Description | Cmax (µM) b | Reference |
|---|---|---|---|---|
| Niacinamide | Low risk | PBK model predicting niacinamide plasma exposure (Cmax)c based on the tolerable upper daily intake level of 12.5 mg/kg bw/day established by the European Food Safety Authority (EFSA) | 163 | See Supplementary Material (Supplementary Substance Information) |
| Coumarin | Low risk | PBK model predicting coumarin plasma exposure (Cmax)c based on tolerable daily intake of 0.1 mg/kg bw/day established by the European Food Safety Authority (EFSA) | 0.01 | See Supplementary Material (Supplementary Substance Information) |
| Caffeine | Low risk | Human plasma exposure for caffeine was estimated based on a pharmacokinetic study following single oral consumption of 315–530 mg/day | 52 | Blanchard and Sawers (1983) |
| Phenoxyethanol | Low risk | PBK model predicting phenoxyethanol plasma exposure (Cmax) based on cosmetic aggregate exposure of 2.69 mg/kg/day for adults using the conservative assumption that all of the products contain phenoxyethanol at a maximum of 1% and that the total daily aggregate exposure would be given as a single dose (once daily) | 4 | Troutman et al. (2015) |
| Sulforaphane | Low risk | Human pharmacokinetic data describing Cmaxc following repeated daily oral consumption of liquidized broccoli containing 3.9 mg of sulforaphane | 0.07 | Hanlon et al. (2009) |
| tertiary-Butylhydroquinone (tBHQ) | Low risk | PBK model predicting t-BHQ plasma exposure (Cmax)c based on the acceptable daily intake of 0.7 mg/kg bw/day established by the European Food Safety Authority (EFSA) | 1.4 | See Supplementary Material (Supplementary Substance Information) |
| Triclosan | Low risk | Predicted human exposure levels corresponding to the reference dose or MoS targets from the U.S. FDA and the Scientific Committee on Consumer Safety (SCCS), respectively | 2 | Krishnan et al. (2010) |
| CDDO-Me | High risk | Human pharmacokinetic data describing Cmaxd following repeated oral exposure of 900 mg/day for 21 days. Exposure corresponds to maximum tolerated dose | 0.05 | Hong et al. (2012) |
| Doxorubicin | High risk | Human pharmacokinetic data describing Cmax following an i.v. 40 min infusion of 60 mg/m2 of body surface area | 1 | Speth et al. (1987) |
| Diclofenac | High risk | Human pharmacokinetic data describing Cmax following exposure to a single oral dose (50 mg) of diclofenac | 4 | Davies and Anderson (1997), Marzo et al. (2000) |
| Diethyl maleate | No defined exposure scenario | |||
| Troglitazone | High risk | Human pharmacokinetic data describing Cmax following a single oral dose of 400 mg | 3 | Loi et al. (1999), Plosker and Faulds (1999), FDA submission dossierd |
| Pioglitazone | High risk | Human pharmacokinetic data describing Cmax following a single oral dose of 45 mg | 4.5 | Christensen et al. (2005), FDA submission dossierf |
| Rosiglitazone | High risk | Human pharmacokinetic data describing Cmax following a single oral dose of 8 mg | 1.5 | Avandia prescribing informationd |
Exposure scenario adopted for chemical is either “low risk” or “high risk” (from consumer goods perspective).
Mean plasma Cmax values were calculated from clinical trials or PBK models. Further details can be found in the Supplementary Material (Supplementary Substance Information).
C max values were calculated when compound reached steady state after repeat dosing.
https://www.fda.gov/media/75754/download. Accessed June 20, 2019.
https://www.accessdata.fda.gov/drugsatfda_docs/label/1999/20720s12lbl.pdf. Accessed June 20, 2019.
https://www.accessdata.fda.gov/drugsatfda_docs/nda/99/021073A_Actos.cfm, Clinical Pharmacology Biopharmaceutics Review(s), Parts 1 to 4. Accessed June 20, 2019.
Materials
Cell culture media, supplements, and buffers were purchased from ThermoFisher Scientific (Loughborough, UK). All test compounds were purchased from Sigma-Aldrich (Dorset, UK). Supplier information for antibodies, cellular dyes, and assay kits can be found in Supplementary Table 1.
Cell culture
The cell line HepG2 (human hepatoblastoma) was obtained from Public Health England European Collection of Cell Cultures (ECACC, Salisbury, UK). Cells were cultured in complete minimal essential medium (MEM) supplemented with 10% fetal bovine serum (FBS), 2 mM L-GlutaMAX, 1% nonessential amino acids (NEAA), 53 U/ml penicillin, and 53 µg/ml streptomycin in 75 cm2 cell culture flasks. Cells were maintained in a humidified atmosphere with 5% CO2 at 37°C. Cells were kept at a confluence below 85% and not maintained in culture more than 4 weeks (8 passages).
Compound treatment
Compounds were prepared as stock solutions at a 200-fold higher concentration than the desired top concentration in appropriate vehicle (100% DMSO). Compounds were serially diluted in appropriate vehicle to give an 8-point concentration curve using custom dilution series (see Supplementary Table 3). Dosing solutions were prepared by diluting the compound stocks 1:40 in the appropriate assay media and the cells were exposed by adding 25 µl of the dosing solution to the appropriate wells (resulting in a total volume of 125 µl/well of a 96-well plate and 0.5% DMSO v/v). Compound treatment was performed for the appropriate time (1, 6, or 24 h) in a humidified atmosphere with 5% CO2 at 37°C.
Measurement of mitochondrial toxicity using the extracellular flux assay
The extracellular flux assay was used to assess mitochondrial toxicity in HepG2 cells by determining the oxygen consumption rate (OCR), reserve capacity, and extracellular acidification rate (ECAR) using the XFe96 flux analyzer (Agilent), as described previously (Brand and Nicholls, 2011). In brief HepG2 cells were seeded at 16 000 cells/well onto XFe96 plates (Agilent) in complete MEM supplemented with 10% FBS, 2 mM L-GlutaMAX, 1% NEAA, 53 U/ml penicillin, and 53 µg/ml streptomycin and left overnight to attach in a humidified atmosphere with 5% CO2 at 37°C. Dosing solutions were prepared as described above. Compound treatment of the HepG2 cells was performed for 1, 6, or 24 h (see Composition of the cellular stress panel section). Due to some components of the complete MEM media being known to interfere with the assay readout (Brand and Nicholls, 2011), the last 60 min of each treatment (or the entire period of the 1 h treatment) was conducted in appropriately dosed unbuffered DMEM assay medium (Sigma-Aldrich) supplemented with 10 mM glucose, 30 mM NaCl, 1 mM pyruvate, and 2 mM L-alanyl-glutamine (medium pH7.4, 37°C). The XFe 96 microplate cartridges (Agilent) were loaded with 20 µl of dosing solution. Four initial baseline OCR and ECAR measurements prior to the addition of test compound were determined. Each measurement consisted of a 3-min mix and 4-min read cycle. Following treatment with the test compound a further six measurements of OCR and ECAR were taken. Subsequently a mitochondrial stress test was performed by consecutive addition of the inhibitors oligomycin (1 μM), carbonyl cyanide 4-(trifluoromethoxy) phenylhydrazone (FCCP, 0.5 μM), and rotenone (1 μM) plus antimycin A (1 μM) (Rot/AA). Two subsequent OCR measurements were taken following each inhibitor addition. Basal OCR (the sixth OCR measurement following compound/vehicle addition) were normalized to the baseline OCR measurements, and all measurements were corrected for the nonmitochondrial OCR (the final OCR measurement following the addition of Rot/AA). The reserve capacity is a measurement of the maximal OCR (following FCCP addition), and was determined as change from the baseline OCR, and corrected for the nonmitochondrial OCR. ECAR measurements were taken after the addition of vehicle or test compound and normalized to baseline ECAR. On each plate no cells were seeded into A1, H1, or column 12, the 4 corner wells were used as the temperature control wells. Control cell free wells were used to identify compounds, which interfered with either OCR or ECAR as a result of compound induced pH changes or interference due to compound color.
High content imaging assays
Cell imaging with fluorescence analysis was performed with a Cellomics ArrayScan VTI or Cellomics ArrayScan XTI Infinity High Content Screening platform (ThermoFisher, UK), which used HCS Studio 2.0 software (ThermoFisher, UK) and the compartmental analysis bioapplication for image analysis. HepG2 cells were seeded in complete EMEM supplemented with 10% FBS, 2 mM L-GlutaMAX, 1% NEAA, 53 U/ml penicillin, and 53 µg/ml streptomycin at cell densities of 10 000, 12 500 or 12 500 cells/well, for the exposure times of 24, 6 and 1 h, respectively. The cells were seeded in 96-well black-walled clear-bottom Greiner micro plates (Sigma-Aldrich) and were allowed to adhere overnight. Test compounds were prepared as described above. Cells were treated in triplicates at 8 different concentrations of each test compound. Following compound treatment, the culture media were removed and if appropriate cells were stained with the specific dye/antibody for each HCS endpoint (Supplementary Table 1). If required cells were fixed with 100 μl/well fixation solution (4% formaldehyde in phosphate-buffered saline [PBS] containing 20 μg/ml Hoechst 33342) for 30 min at room temperature protected from light. Cells were washed 3 times with PBS and fluorescence image acquisition was performed. Per well 8 fields of view were imaged using a 10× wide-field objective. Cell nuclei were detected by analyzing the Hoechst 33342 (Sigma) fluorescence signal using a 360–400-nm excitation filter and 410- to 480-nm emission filter or SYTO 11 Green Fluorescent Nucleic Acid Stain (ThermoFisher Scientific) using 505-nm excitation filter and 525-nm emission filter depending upon the endpoint combination used per plate. Endpoints detailed in Supplementary Table 1 were multiplexed where possible based upon emission spectra, each endpoint was analyzed for changes in fluorescent intensity signal in either the cytoplasmic or nuclear regions of each cell and compared against the vehicle control wells.
Cellular ATP and lactate dehydrogenase release
Cellular ATP was determined in HepG2 cells following dosing procedures as outlined above using luminescence following the manufacturers guidelines (CellTiterGlo, Promega), luminescence was determined using a BioTek Synergy 2 (BioTek). Raw fluorescence intensity values (RFU) were normalized to vehicle control wells in all cases and expressed as fold changes in assay signal. The presence of lactate dehydrogenase (LDH) was determined from 50 µl supernatant samples of HepG2-treated cells using the CytoTox 96 Non-Radioactive Cytotoxicity Assay (Promega), following the manufacturers guidelines, absorbance was determined at 490 nm using a SpectraMax (Molecular Devices).
ELISA tests in the HepG2 cell culture supernatants
HepG2 cells were cultured and exposed as detailed above. Supernatants were collected from treated HepG2 cells following the appropriate incubation period (see Table 1). Fifty microliters of supernatant was analyzed for levels of IL-8 using an ELISA kit, according to the manufacturer’s instructions.
Exposure assessment
Physiologically based kinetic models were constructed with the simulation software GastroPlus 9.6 (Simulation Plus, Lancaster, California). Briefly, the PBK models developed represent the whole body as a series of compartments parameterized based on physiology of tissues and organs (eg, blood flow, volume, etc.). PBK models integrate this physiological description with compound-specific data to predict the pharmacokinetics of compounds (ie, concentration over time in plasma and tissues). In Table 2, a summary of the exposure scenarios and total Cmax for each chemical is provided. The full details regarding the choice of the exposure scenario, the literature clinical data underpinning the plasma Cmax values selected, and the PBK model assumptions are described in the Supplementary Material (Supplementary Substance Information). When available, the performance of the PBK model was checked against clinical data.
Concentration-response analysis using Gaussian processes
Concentration-response analysis was performed using Gaussian processes (Gelman et al., 2013) within a Bayesian statistical framework. A brief overview of the approach is provided below; a more detailed description is given in the Supplementary Material (supplementary information on concentration-response modeling). The approach differs from other more commonly used methods such as benchmark dose (BMD) as follows. Typically, concentration-response analysis involves fitting several different curves (mathematical functions, such as exponential, Hill, polynomial) to a single concentration-response dataset. These functions (or curves) are dependent on one or more parameters. This process of fitting a curve to the data by optimizing over the parameters is known as a parametric approach. Model selection criteria then need to be established in order to select the “best” curve that describes the data, from which the PoD can be calculated. However, finding this “best” curve can be regarded as source of uncertainty in itself, as it can impact the estimate of the PoD (Watt and Judson, 2018). Gaussian processes are, in contrast, an example of a nonparametric modeling approach, whereby the Gaussian process defines effectively a probability distribution of random functions. Using this approach, the data are able to dictate the shape of the fitted curves rather than have the curves be of a pre-defined shape, as in the parametric approach. This allows for a single (nonparameteric) model to be flexible enough to describe a wide range of concentration-response shapes.
The model was constructed as follows. Raw measurements for each plate were divided by the corresponding median control measurement, which was then log2 transformed. Nominal concentrations were used for calculating the PoDs; these were log10 transformed.
Let be the concentration of a test compound with corresponding mean response denoted by . The change in the mean response with respect to is modeled using a Gaussian processes (Gelman et al., 2013). Thus are distributed according to a multivariate Gaussian distribution with constant mean vector and covariance matrix such that where
| (1) |
and is the identity matrix. This covariance function has been constructed so that the sampled curves will have a constant mean up to some threshold , and that above the response may change in any direction provided that it does so smoothly with respect to the concentration . The quadratic terms preceding the exponential ensure that samples of are continuous and smooth over the whole test concentration range. The scale parameter is used to model variability in the mean response as a function of the well location along a plate, thereby allowing the model to account for potential biases arising from plate effects (Malo et al., 2006), see Supplementary Material (accounting for biases in well location and reducing sensitivity to outliers) for further details.
The distribution of replicate measurements at the same test concentration, , is modeled using a Student’s t distribution, with 5 degrees of freedom, centered on the mean response and having scale parameter ,
| (2) |
The mean response in each group of control wells was modeled using the variable and was assumed to be normally distributed with mean and scale ,
The replicates, , within each group of control wells for each plate were assumed to be distributed as in equation 2 such that
Independent prior distributions were assigned to the parameters , and (see Supplementary Material).
The model was fitted using Markov-chain Monte Carlo (MCMC) approaches (see Software details for further information). For each concentration-response dataset, 10 000 model parameter sample sets () were drawn from the posterior distribution. For each set, the concentration-dependent mean response , was calculated at 100 uniformly spaced concentrations between the minimum and maximum tested concentrations using the standard equation for the marginal distribution of a multivariate normal distribution (Gelman et al., 2013).
PoDs were defined as the lowest concentration at which the mean response deviated more than . This threshold was based on the heuristic that the 95% credibility range of the combined variability emanating from the plate bias and replicate fluctuations is given (approximately) by this term, when both effects are normally distributed (which is not strictly true in the mathematical sense, because Student’s t distributions were used in the model formulation above). The PoD for sample is therefore defined as the maximum value of such that
| (3) |
Because is only defined at discrete concentrations , linear interpolation is used to solve the above equation. If the first lowest value of which satisfied the above was the maximum tested concentration, then no PoD was defined for that sample.
For a given concentration response, the above procedure can result in anywhere between 0 and 10 000 samples for the PoD. The proportion of samples for which a PoD could be detected (according to equation 3) was used as a confidence score that the response was concentration dependent (termed the Concentration Dependency Score or CDS). A default choice of 50% of the samples (corresponding to a CDS of 0.5) was required for the concentration-response dataset to be classified as concentration dependent. In other words, at least 5000 of the sampled curves attain a value (at some point along the curve) that exceeded the threshold defined in equation 3 in order for the response to be considered concentration dependent.
Modes and a 95% highest density interval (HDI) were estimated from the posterior samples to summarize the distribution (Kruschke, 2014). For datasets in which two or more modes were inferred, the HDI is split into subintervals, one for each mode. A representative mode was chosen by inspection of the curve fit, and the corresponding subinterval is used as the credibility range for the POD. Otherwise, in cases where there is only one mode, the credibility range is based on the entire HDI. In most cases, the representative mode corresponded to the mode of highest density. However, in a small number of cases a lower density mode was chosen for an alternative reason, such as consistency with other timepoints. Choices and selection rationale are provided as Supplementary Material, mode selection file.
A summary file indicating chemical, assay, timepoint, CDS, and PoD distribution modes for each profile, together with an image for every concentration-response fit, is provided via the Dryad repository (https://doi.org/10.5061/dryad.cnp5hqc20). These images include an overlay of the maximal fold change (up or down) attained by the corresponding positive control (see Supplementary Table 1). Here, note that almost all positive controls caused an increased response, except cellular ATP, OCR, reserve capacity, glutathione content, and intracellular pH which were decreased.
Software details for the Gaussian process model
Data processing was performed using the Anaconda bundle for Python (version 2019.07, Python 3.7.3). MCMC sampling was performed using Pystan 2.19.0 (Carpenter et al., 2017). Python code for running the model and reproducing the fits, together with a brief tutorial, is provided via the Dryad repository (see above).
Benchmark dose analysis
Benchmark dose and benchmark dose lower confidence limit (BMDL) maximum likelihood estimates were obtained using BMD Software (BMDS) version 3.1 (available from https://www.epa.gov/bmds/benchmark-dose-software-bmds-version-30). For a single concentrations-response dataset, the software fits multiple parametric models and calculates corresponding estimates of the BMD, the BMDL, and the Akaike Information Criteria (AIC) for each fit. The AIC provides a quantitative estimate of the quality of the fit, offset by the complexity of the models, and can be used to guide model selection; a generally accepted heuristic is that the model with the lowest AIC is likely to be the “best” one among those that were considered, although this does not guarantee that the model in itself provides a good description of the data. The following continuous models were used: exponential, polynomial, power, Hill function, and linear, using both restricted frequentist and unrestricted frequentist settings (where possible [U.S. EPA, 2019]). BMDs were calculated using the “standard deviation” to set the benchmark response (BMR), where the BMR was (by definition) the baseline level of the response plus or minus the standard deviation of the control samples, times the BMR Factor (BMFR); the BMFR was set equal to 2 to ensure that the BMD definition would be approximately equivalent to the PoD definition provided in equation 3. Before fitting the models, raw measurements were processed using the same settings the Gaussian process concentration-response model described above, but the nominal concentrations of the test compound were not log10 transformed as BMDS was not designed to accept concentrations in log scale. Otherwise, default settings were used. All output files from the analysis are available via the Dryad repository (see above).
RESULTS
Generation and Analysis of Concentration-Response Cell Stress Data
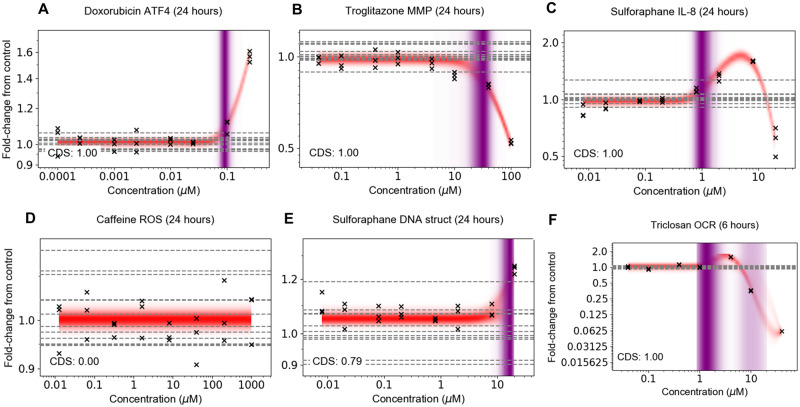
Concentration response data were generated for each of the 36 biomarkers representing mitochondrial toxicity, cell stress, and cell health in Table 1 at 1, 6 and 24 h (unless otherwise stated) using the 13 benchmark chemicals (listed in Table 2) and DEM (a model substance for oxidative stress), making a total of 2965 concentration-response datasets. In this work, cytotoxicity is represented by biomarkers of cell death (indicated in Table 1). The stress panel datasets comprised a wide variety of different concentration-dependent trends, depending on the substance, biomarker, and timepoint, representative examples of which are provided in Figure 2. This included response profiles that were either monotonic (ie, the response only increased or decreased with concentration; Figs. 2A and 2B) or nonmonotonic (ie, hormesis: increased at low concentrations before reaching a plateau and then decreasing at higher concentrations, or vice-versa; Figs. 2C and 2F) (Hill et al., 2018; O’Brien et al., 2006), profiles where no discernable concentration-dependent response could be observed (Figure 2D), or cases where there was only a very weak response (Figure 2E), which typically occurred at the top concentration tested.
Figure 2.
Representative concentration response data and model fits. (A) Concentration response that strictly increased, (B) strictly decreased, and (C) increased but then decreased at a higher concentration. (D) Example where the substance had negligible influence on the measured biomarker at all tested concentrations. (E) Example in which there was a high uncertainty regarding whether the observed change was due to the test chemical, a chance fluctuation in the replicates and/or a bias in the response due to well location or due to well position. F) Example in which the PoD distribution is bimodal, resulting in more than one plausible PoD found by the model to be consistent with the data. Crosses correspond to individual data points; horizontal dashed lines indicate control values.
To analyze the data, a nonparametric (Gaussian process) Bayesian concentration response model was applied. Fits between the model and data are represented in Figure 2 using the mean concentration response distribution, indicated by the red shaded bands. The nonparametric nature of the approach ensured that the model provided a standalone method that was flexible enough to describe all possible concentration-response shapes. In the model, a change in concentration-response measurement could be caused through a combination of 3 possible effects: (1) chance fluctuations in the response arising from biological or technical variability; (2) biases in the location of wells used to generate the measurement samples (see Supplementary Material, “Accounting for biases in well location and reducing sensitivity to outliers” for further details); and (3) the test chemical induced a concentration-dependent effect. To quantify the uncertainty in whether a change in the response was due to the latter (a concentration-dependent effect), rather than either of the former explanations, the model generated a concentration-dependency score (CDS; see Materials and Methods section). A CDS value >0.5 indicated that, conditional on the model, it was statistically more likely than not that the test chemical was inducing a change in the response measurement (conversely, a CDS value <0.5 indicated it was more likely that the chemical did not induce such a change, and the effect was due to a statistical fluctuation or a bias in well location). In Figures 2A–C and F, the corresponding CDS values were all close to 1.00, reflecting the strong evidence that the observed change in the biomarker could be attributed to the test chemical, rather than say, well location on the plate. Equally, the CDS in Figure 2D was close to 0.0, providing strong evidence that there was no response. Figure 2E provides an example where the CDS was slightly above 0.5, and there was high uncertainty in whether the response at the maximum tested concentration was due to a weak response by the test substance, or another factor such as a plate bias or a chance fluctuation.
In cases where the CDS was above 0.5, the plausible regions for the PoD are reflected via the purple-shaded regions in Figure 2, where the intensity of the shading reflects its underlying distribution, and gives an indication of how likely the different PoD locations were. For most datasets, there was a solitary mode for the PoD (as the case in Figs. 2A–C and E). However, in some instances the model suggested two distinct modes, as is the case, for example, in Figure 2F. This arose because the model suggested two possible sets of response curves which would be consistent with the data. In the first set (corresponding to the lower mode), the mean response increased from around 1 µM to reach a peak at around 5 µM before decreasing well beyond the baseline. In the second set of trajectories (upper mode), the increase in the response observed at 5 µM was attributed to either a plate bias or statistical chance and the associated fits were constant up to ∼7 µM and decreased thereafter. The depth of purple shading reflects the fact that the lower POD mode was considerably more likely than the upper PoD mode.
To compare the nonparametric Bayesian model developed in this work with a more commonly used approach, the 6 datasets in Figure 2 were also analyzed using the U.S. EPA BMD Software implementation of BMD (Haber et al., 2018) (see Materials and Methods section). Details of this analysis can be found in the Supplementary Material (Benchmark dose analysis section). In summary, for each concentration-response dataset in Figure 2, multiple BMD estimates were generated using different parametric models. According to the default model selection process in BMDS, none of the fits were reported as “recommended” (all were “questionable” or “unviable”), and in some cases (depending on the dataset and model fit), BMDS was observed to overfit the data. However, in general the “questionable” BMD estimates were found to be contained within the corresponding PoD credibility ranges inferred using the Bayesian approach, illustrating how the latter was able to effectively quantify the uncertainty in deciding “which” curve to use for PoD estimation, this uncertainty being quantified in terms of a PoD distribution. Furthermore, multiple BMDs were also reported for the data in Figure 2D, even though there is clearly no concentration-dependent response, because the BMDS implementation of BMD implicitly assumed that there was a concentration response prior to fitting any curves (other implementations of BMD address this through additional computational steps, eg, by using appropriate statistical tests [Phillips et al., 2019]). Overall, the Bayesian concentration response model reported here provided a robust statistical analysis of concentration-response datasets without the need for model selection or additional steps to manage datasets where there is likely no concentration-dependent effect by the test chemical (instead, providing a measure of uncertainty in this, the CDS). Furthermore, based on these results the nonparametric nature of the Bayesian model means it is less susceptible to overfitting (see the Supplementary Materials, Benchmark dose analysis section for further details).
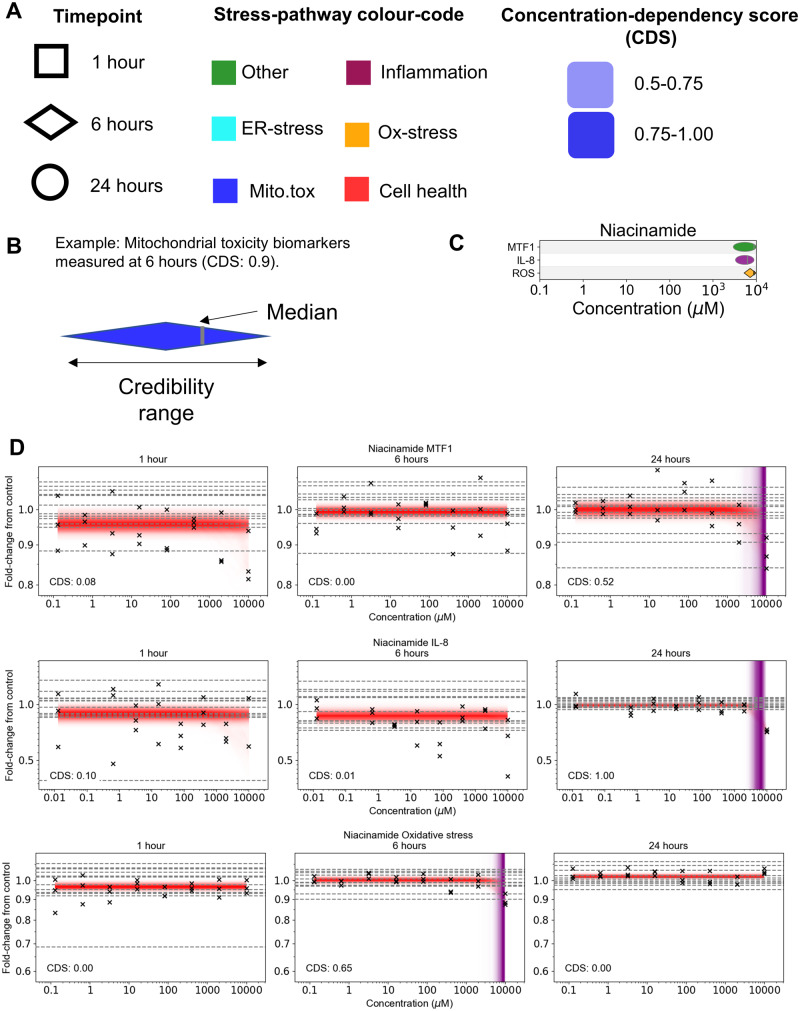
Generation of PoD Summary Plots
For most of the substances tested in this study (listed in Table 2), multiple biomarkers were triggered at varying concentrations and timepoints, spanning different stress pathways and cell health effects. In order to visualize these data effectively, separate plots of the PoDs for each chemical were constructed as follows (Figure 3): all biomarkers (regardless of timepoint) likely to be perturbed by the substance (ie, have a CDS > 0.5) are listed along the y-axis, grouped by pathway, and the corresponding PoDs are plotted along the x-axis. For biomarkers in which multiple biological replicates were available (namely cell count, nuclear size, and DNA structure, see Table 1), the PoD samples were pooled in order to compute statistics (eg, CDS and PoD credibility range) for the summary plots.
Figure 3.
Overview of PoD summary plots. A, Information on the PoD timepoint, stress pathway, and CDS are indicated using shape, color and depth of shading. B, The credibility range for the representative PoD is indicated using the width of the symbol, the median is given by a vertical gray line. C, PoD summary plot for niacinamide. D, Corresponding niacinamide concentration-response data. Crosses correspond to individual data points; horizontal dashed lines indicate control values.
Different symbols are used to indicate the measurement timepoint (Figure 3A), and width of the PoD symbol reflects the credibility range of the PoD (Figure 3B). Finally, the color of the PoD symbols corresponds to the pathway that the biomarker represents (Figure 3A). An example summary PoD plot for niacinamide, which is used in many cosmetic formulations (see Supplementary Materials) is shown in Figure 3C. Here, there were only three PoDs for which the CDS was above 0.5, corresponding to a small but appreciable decrease in IL-8 at 24 h and relatively weak changes in MTF1 and ROS at 6 and 24 h respectively, all at the top concentration tested (10 000 µM; Figure 3D). All other concentration-response data generated for niacinamide (including those associated with MTF1 and IL-8 at 1 and 6 h for, and ROS at 1 or 24 h) had CDS values below 0.5 and so were excluded from the summary plots.
Using the Cell Stress Panel to Distinguish Chemicals Based on Bioactivity
Analysis of the data revealed that the different benchmark substances could be divided broadly into 3 groups based on their bioactivity, these being: (1) substances with very low levels of bioactivity, corresponding to none or few of the 36 stress panel biomarkers being triggered at the concentrations tested; (2) substances for which cellular stress responses largely coincided with cytotoxicity, reflecting a nonspecific burst of activity (ie, the “cytotoxic burst”; (3) substances for which, at subcytotoxic concentrations, bioactivity was limited to specific effects on one or more cellular stress pathways.
Compounds in group 1 were phenoxyethanol, niacinamide, coumarin, and caffeine. The least bioactive was phenoxyethanol, a substance that is commonly used as a preservative in personal care products (see Supplementary Material): no concentration-responses could be detected among all 36 biomarkers (the maximum CDS from all the assays tested was 0.29). For caffeine only, a single concentration-response could be detected, which had a CDS of 0.54 (barely above the 0.5 threshold), corresponding to phospho-gamma-H2AX at 1 h; this observation is unlikely to have biological significance as a concentration response was not detected at 6 or 24 h. For coumarin, all PoDs also occurred around the top concentration tested (1000 µM). The lowest PoD was due to a decrease in ATP at both 6 and 24 h, which was accompanied by a decrease in IL-8, an increase in glutathione levels and a decrease in phospholipidosis at 24 h. It is possible that the increase in glutathione levels was a protective response of the cells to the decrease in ATP that occurred at the earlier timepoint of 6 h and that the decrease in IL-8 was due to the anti-inflammatory properties of coumarin (Hadjipavlou-Litina et al., 2007).
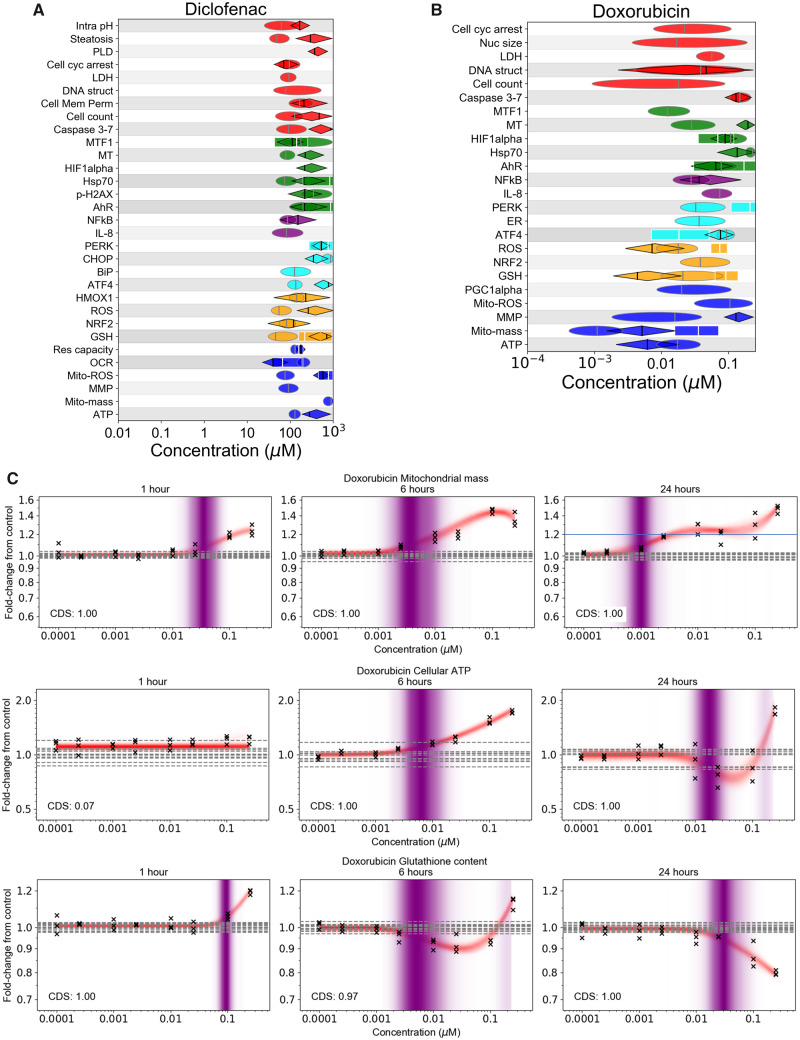
These results are in stark contrast to the bioactivity observed for substances in group 2, which were diclofenac (an anti-inflammatory drug) and doxorubicin (a chemotherapy drug), both of which are known to cause adverse effects in humans through mitochondrial toxicity (Burridge et al., 2016; Damiani et al., 2016; Eakins et al., 2016; Ramachandran et al., 2018). In the case of diclofenac (Figure 4A), multiple cellular stress effects were detected, all coinciding with cell injury. For doxorubicin (Figure 4B), the only subcytotoxic PoD was associated with increases in mitochondrial mass that occurred at 1, 6 and 24 h (Figure 4C). This was accompanied by increases in cellular ATP at 6 h, potentially indicating an increase in mitochondrial biogenesis, a mechanism used by cells to protect against various forms of cellular stress. Increases in glutathione could also be observed at 1 h, likely reflecting a homeostatic cellular response to increased oxidative stress (Lushchak, 2012). However, both apparent mechanisms were overwhelmed by 24 h, leading to a loss of glutathione and cellular ATP. In both cases, a specific mode of action (such as mitochondrial toxicity) could not be determined, as changes in most biomarkers coincided with concentrations causing cell death.
Figure 4.
Summary of group 2 substances, diclofenac and doxorubicin. (A) PoD summary plot for diclofenac and (B) doxorubicin (see Figure 3A for corresponding legend). C, Concentration responses to doxorubicin for mitochondrial mass, cellular ATP, and glutathione content at 1, 6, and 24 h. The PoD distributions (indicated by purple shading) for cellular ATP at 24 h and glutathione at 6 h were both bimodal (ie, two district shaded bands); the lower mode was selected as the representative mode in both cases. Crosses correspond to individual data points; horizontal dashed lines indicate control values.
Compounds in group 3, namely triclosan, troglitazone, pioglitazone, rosiglitazone, CDDO-Me, DEM, tBHQ, and sulforaphane, were in general either found to trigger specific mitochondrial toxicity effects or oxidative stress at subcytotoxic concentrations. Overall, these responses reflected the substances’ known bioactivity, showing the utility of the cellular stress panel for developing mechanistic hypotheses for NGRA, and are described in greater detail below.
Cell Stress Panel Identifies a Specific Mode of Toxicity for a Subset of Substances: Mitochondrial Toxicants and Nrf2 Activators
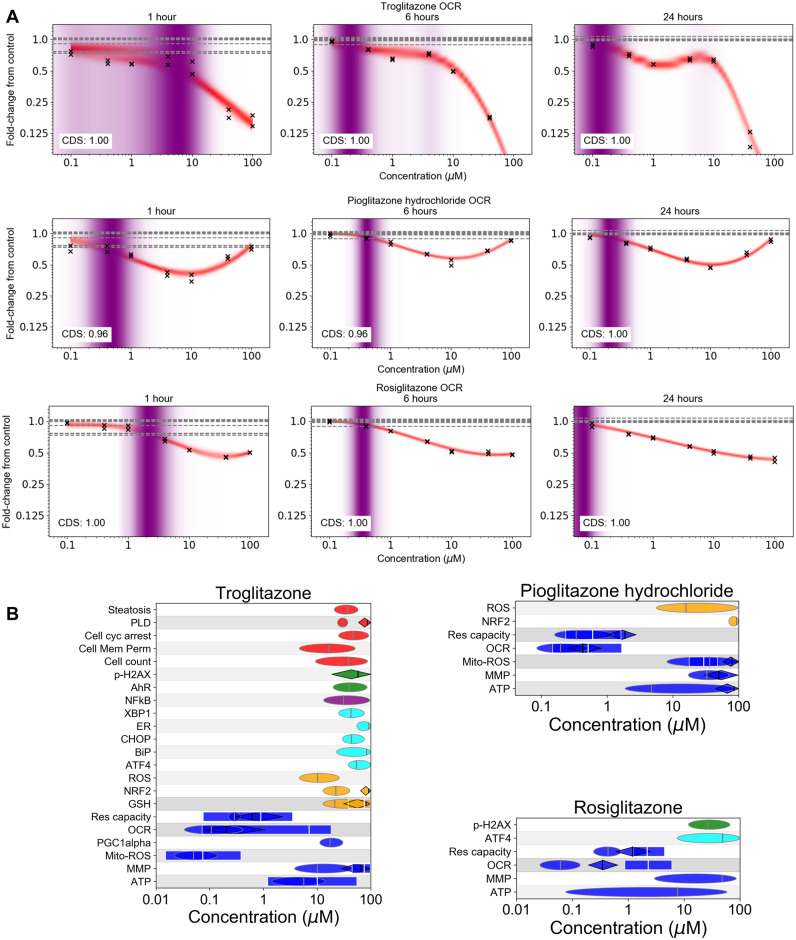
A slightly reduced panel of biomarkers (exceptions were hypoxia and metal stress, see Supplementary Table 1) was tested using 3 members of the thiazolidinedione family of drugs (troglitazone, pioglitazone, and rosiglitazone), used in the treatment of diabetes (Hauner, 2002). These are widely used by researchers as model chemicals for cell systems and assays designed to predict either liver or mitochondrial toxicity (Bell et al., 2018; Hu et al., 2015; Masubuchi et al., 2006; Nadanaciva et al., 2007). All 3 thiazolidinediones exhibited a significant concentration-dependent inhibitory effect on cellular respiration measured by monitoring the OCR and reserve capacity (see Figure 5A and Supplementary Figure 1) at subcytotoxic concentrations. These results showed that the maximum inhibition of oxidative phosphorylation occurred as early as 1 h with no further significant changes by 24 h. Overall, the extracellular flux assays were the most sensitive for all 3 chemicals: the 24 h PoDs for OCR and reserve capacity were in the range of 0.05–0.5 µM, occurring at concentrations at least 10-fold lower than those which caused changes in MMP, ATP, and cell count (Figure 5B). However, changes to mitochondrial ROS could be detected at even lower concentrations for troglitazone. This is consistent with the observations of Liao et al. (2010), where this effect was attributed to troglitazone (but not rosiglitazone or pioglitazone) stimulating PGC-1α protein degradation, reducing superoxide dismutase 1 and 2 expression.
Figure 5.
Summary of stress panel responses for 3 group 3 substances (troglitazone, rosiglitazone, and pioglitazone). A, Oxygen consumption rate data for all 3 substances at 1, 6, and 24 h measured using the extracellular flux assay. Crosses correspond to individual data points; horizontal dashed lines indicate control values. B, Summary PoD plots (see Figure 3A for corresponding legend).
Despite the OCR PoDs occurring at similar concentrations, the fold change from control was significantly different between the three substances. Exposure to troglitazone at the two highest concentrations (40 and 100 µM) reduced the OCR near to zero which was not observed for the other chemicals. Pioglitazone on the other hand showed a completely different concentration-response curve, causing a decrease in both OCR and reserve capacity at concentrations up to 10 µM, and an increase at concentrations between 40 and 100 µM (Figure 5A and Supplementary Figure 1).
The magnitude of the response observed in these extracellular flux assays implied that troglitazone is a more potent chemical than pioglitazone and rosiglitazone as previously reported in the literature (Hu et al., 2015). This was further confirmed by observations in the other assays where pioglitazone and rosiglitazone only triggered mitochondria-related biomarkers (MMP, MitoROS, ATP, and ROS) at concentrations around 10–100 µM (Figure 5B). At similar concentrations, troglitazone not only induced mitochondrial stress but also perturbed the oxidative stress, ER and AhR stress pathways, and cell health-related biomarkers which seemed to coincide with a cytotoxic burst (Figure 5B).
Triclosan is used as antimicrobial in many consumer products. Using the full stress panel, the lowest PoDs for this chemical were obtained using the OCR and reserve capacity assays (similarly to the thiazolidinediones, see Supplementary Figs. 2A and 2B). These results were consistent with previous reports that triclosan has been shown to inhibit mitochondrial complex II, superoxide release, and uncoupling of oxidative phosphorylation (Teplova et al., 2017). At higher concentrations, other mitochondrial biomarkers were also affected by triclosan with a strong increase (2- to 4-fold) in mitochondrial superoxide (MitoROS) across all timepoints at the highest concentration, an increase in ATP at 6 h followed by a reduction in ATP at 24 h, but minor changes in MMP, mitochondrial mass, and PGC1α (see Supplementary Figure 2B).
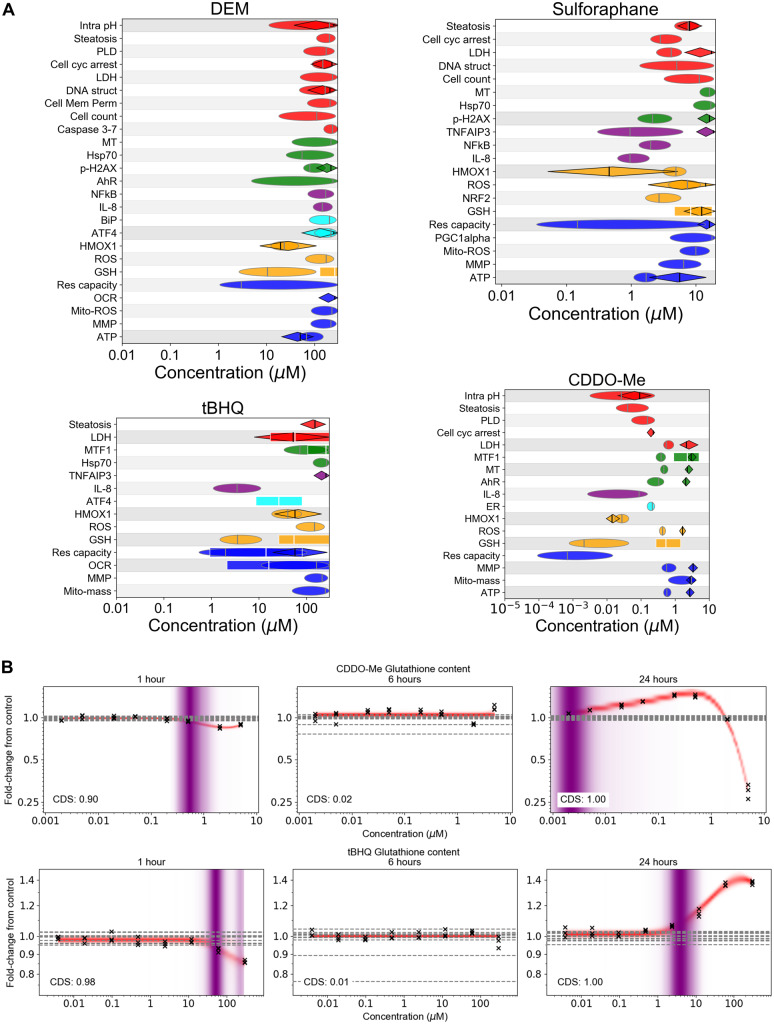
The full stress panel was also tested using 4 soft electrophiles: CDDO-Me, sulforaphane, DEM, and tBHQ. Soft electrophiles are thought to react predominantly with soft nucleophiles, including glutathione and the sulfhydryl groups of cysteines in Keap1 (LoPachin and Gavin, 2016). The former can cause depletion of glutathione, which is counteracted by the latter by disrupting interactions between Keap1 and Nrf2, leading Nrf2 nuclear translocation and the consequent upregulation of various antioxidant response mechanisms, including increased expression and activity of glutathione synthesis and ROS-detoxification enzymes (Magesh et al., 2012).
Across all 4 soft electrophiles, the lowest PoDs triggered below cytotoxicity were associated with changes in glutathione content, Heme Oxygenase 1 (HMOX1), and reserve capacity via the extracellular flux assay (see Figure 6A). However, unlike the 3 thiazolidinediones and triclosan described above, changes in reserve capacity were not accompanied by changes in OCR or other mitochondrial toxicity biomarkers (with the exception of transient effects on OCR by tBHQ at 1 h). Responses by Nrf2 to treatment appeared relatively weak and difficult to detect for all substances, possibly due to its transient nature, and the fact that only small increases in nuclear Nrf2 are needed to induce downstream effects (Wink et al., 2017). However, HMOX1, which is downstream of Nrf2 (Loboda et al., 2016), responded at subcytotoxic concentrations with the levels increasing from ∼75% to 200% of control at both 6 and 24 h for all substances (see Supplementary Figure 3).
Figure 6.
A, Summary of PODs for group 3 substances tested in the panel that are soft electrophiles (CDDO-Me, sulforaphane, tBHQ, and DEM). See Figure 3A for legend. B, Representative glutathione concentration-response plots for CDDO-Me and tBHQ measured at 1, 6, and 24 h. The glutathione response to tBHQ at 1 h results in a bimodal PoD distribution; the lower mode was chosen to be the representative the PoD. Crosses correspond to individual data points; horizontal dashed lines indicate control values.
Changes in glutathione abundance were similar across all 4 soft electrophiles: short exposures (1 h) to high concentrations resulted in depletion of glutathione, whereas longer-term exposures (6–24 h) caused glutathione levels to increase and accumulate back to, and in some cases exceed, baseline levels (see Figure 6B and Supplementary Figure 4). This resulted in nonmonotonic response profiles at 6 and 24 h that at low concentrations increased in a concentration dependent manner, but then began to decrease at higher concentrations. In all the cases, the concentration that gave rise to the 1 h glutathione PoD that overlapped with multiple other cellular stress, cell health, and physiology biomarker PoDs at 24 h, including the accumulation of ROS. Thus, although glutathione levels were typically replenished for concentrations close to the high-concentration PoD by 24 h, consistent with an adaptive response by cells to electrophilic or oxidative stress, the insult was severe enough at these concentrations to cause cell injury by 24 h; the nonmonotonic nature of the observed glutathione responses were consistent with those made by Wang et al. (2014b) for CDDO-Me.
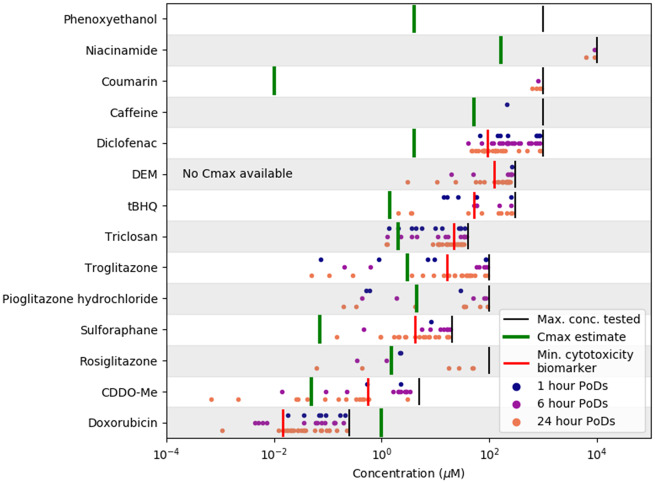
Comparison Between POD and Cmax Values for Typical Exposure Scenarios
To obtain a broad summary of cell stress panel results, the representative mode for all concentration-response datasets is plotted separately for all chemicals in Figure 7. To compare the PoDs with the typical exposure scenarios associated with each substance (summarized in Table 2), estimates of the blood plasma Cmax are also plotted, along with the maximum concentration tested. The lowest PoD associated with cytotoxicity is also highlighted to indicate the onset of cell death (see Table 1 for a list of relevant biomarkers).
Figure 7.
Overview of PoD modes (corresponding to concentration-response datasets where the CDS is larger 0.5) and associated mean Cmax estimates for each substance. The ordering of the chemicals along the y-axis is determined by ranking chemicals based on the mean of all displayed PoDs.
For all the chemical exposures categorized as low risk (see Table 2) except triclosan, the estimated Cmax was below the minimum PoD detected for that substance (or, in the case of phenoxyethanol, the maximum tested concentration, as no bioactivity was detected). In contrast, for chemical-exposure scenarios identified as high risk from a consumer safety perspective (except diclofenac), the estimated Cmax values were above the minimum PoD, and were generally found to be at concentrations where the substance would be expected to be causing significant cellular stress. Although the estimated Cmax was approximately one order of magnitude below the lowest PoD for diclofenac, the adverse cellular stress effects are known to be caused by a reactive metabolite (Ponsoda et al., 1995), and these may not have been present at human relevant levels in these assays because the HepG2s that were used lack many phase I enzymes required for the metabolism of diclofenac (Bort et al., 1999).
DISCUSSION
Within NGRA there is an ongoing need to develop robust and relevant assays that can be used to characterize bioactivity of chemicals at human-relevant exposures (Dent et al., 2018). To that end, the cell stress panel presented here was developed to serve as a low-tier broad-spectrum tool for determining whether a chemical is likely to cause cell stress (Thomas et al., 2019), and if so provide an estimate of the concentrations at which this perturbation occurs. The intention is that information from the cellular stress panel can be combined with appropriate human exposure estimates (eg, the Cmax) to provide an indication of how likely this bioactivity is to occur in humans under the relevant exposure scenario (ie, the risk). This is in contrast to other panels that have been developed for the purposes of hazard identification (eg, drug-induced liver injury or genotoxicity) and focus on one or two stress pathways at a time, or mitochondrial toxicity (Clewell et al., 2014; Eakins et al., 2016; van der Linden et al., 2014; Wink et al., 2017). Although cellular stress responses have been studied extensively in the literature, a major challenge of implementing such panels for NGRA is not only in their development (ie, selecting biomarkers, timepoints, etc.), but also ensuring that the panel is fit for purpose (Middleton et al., 2017). In other words, can the cell panel be used to distinguish between high- and low-risk chemical-exposure scenarios that are driven by cellular stress? To this end, various substances (pharmaceuticals, foods, cosmetics) were used as benchmarks.
This research builds on work conducted as part of the ToxCast and Tox21 U.S. governmental programs (Judson et al., 2016). The added value of the present study is the ability to distinguish the “trigger” stress pathways from those that coincide with cytotoxicity. For example, it was observed that the lowest PoDs for troglitazone, pioglitazone, rosiglitazone, and triclosan were all specifically associated with mitochondrial toxicity at subcytotoxic concentrations, consistent with their known mechanisms of toxicity. At higher (ie, cytotoxic) concentrations, however, a burst of activity could be observed, where multiple cellular stress pathways are triggered, notably ER stress, but also other pathways including DNA damage and hypoxia. In contrast, the soft electrophiles triggered a distinctly different set of responses at low concentrations, notably causing potentially adaptive effects such as glutathione upregulation; at higher cytotoxic concentrations nonspecific stress responses could again be observed. The potential consequences of the 2 types of effect, mitochondrial disruption and glutathione upregulation, are stark: depending on the dose used, the former can be associated with various forms of organ toxicity, including liver failure (Ramachandran et al., 2018; Rana et al., 2019), whereas the latter can be associated with beneficial health effects (Wu and Batist, 2013). Thus, being able to disentangle the “trigger” stress pathways from those caused by nonspecific cytotoxicity is highly beneficial from a risk assessment perspective.
One of the strengths of the work presented here is that the uncertainties associated with the data analysis are quantified probabilistically, a key principle of NGRA (Dent et al., 2018). This was made possible through the novel modeling approach developed as part of the work. First, the model can be used to generate a quantitative measure of the plausibility that a response of a biomarker to the test chemical can be detected, namely the CDS. This approach has the advantage of providing a measure of confidence attributed to when there is no response in a set of assays, or flagging when a response is particularly weak (ie, when the CDS is slightly above 0.5). This is very relevant for consumer safety assessments, because the PoDs represent the lowest concentration at which a response can be detected, rather than an arbitrary threshold on the response, such as an AC50. Calculating the PoD in this way may provide a more sensitive metric for the analysis of high-throughput assay data, and allow differentiation between low concentration and high concentration events. This could explain why, with our analysis, we can disentangle the subcytotoxic and cytotoxic cell stress responses as discussed above. Second, using a nonparametric approach removes the issue over which set of parametric models (ie, Hill function, exponential, linear, etc.) one must choose to fit the data, which represents a source of uncertainty in itself and is frequently a challenge in concentration-response modeling (Watt and Judson, 2018).
To illustrate how the stress panel might be deployed in future, we provided a comparison between the PoDs and the estimated exposure levels based on typical use-case scenarios listed in Table 2 (see Figure 7). For this, Cmax was used as a surrogate metric for systemic exposure levels. It should be noted that adverse effects caused by cellular stress may not always be driven by Cmax, and in certain cases the duration of exposure may also need to be considered. Nevertheless, the results obtained from this preliminary analysis were promising: generally, of the low-risk substance-exposure scenarios (except triclosan), the PoDs occurred at higher concentrations than the corresponding Cmax values, whereas for all of the high-risk substance-exposure scenarios (except diclofenac), there was a clear overlap between the PoDs and Cmax values (Figure 7). The ability to distinguish low from high risk is influenced heavily by the degree of the “biological coverage” of the panel. The biomarkers that were associated with low sub-cytotoxic PoDs that could be considered to be especially protective from a risk assessment perspective for the compounds tested were glutathione content, HMOX1, IL-8 or MTF1, OCR and reserve capacity, whereas many of the other stress biomarkers were only responsive at cytotoxic concentrations and were rather “nonspecific” in their response. This was likely a consequence of the benchmark substances that were selected for testing. Running more chemical-exposure scenarios will allow us to determine whether the degree of coverage is sufficient, and if there is a “core” set of biomarkers that can be identified. However, in reality it is expected that this panel would be used alongside other in vitro cell assays, such as high-throughput transcriptomics, receptor binding screens, and other bespoke assays designed to investigate specific modes of action (Daston et al., 2015; Dent et al., 2018). In particular, some substances relevant to consumer safety may act via specific targets and mechanisms, such as interfering with the function of key receptors, enzymes, or ion channels (Bowes et al., 2012). Going forward, it will be important to understand how different NAMs covering both specific and nonspecific toxicity endpoints can be combined to define substance exposures that are protective of consumer safety. How this might work in practice for the NGRA of a consumer product has recently been demonstrated in Baltazar et al. (2020).
The discrepancy between PoD and Cmax for diclofenac in terms of its risk category from a consumer safety perspective highlights the importance of incorporating metabolism within in vitro assays, which is an on-going challenge in NGRA and may be addressed either by using more organotypic models (Ramaiahgari et al., 2017, 2019) that are metabolically competent, artificially increasing the expression of specific CYP enzymes (DeGroot et al., 2018), or directly testing predicted metabolites.
One further consideration is that in the cellular stress panel, 3 timepoints were selected for data generation (1, 6, and 24 h). Depending on the biomarker and chemical being tested, PoD values vary over time, and one key challenge is determining how to incorporate this time-dependence into the risk assessment approach. Based on our observations, the lowest PoDs (ie, most conservative) typically occurred at 24 h. However, in some cases the value of the PoDs could continue to change, and in particular decrease, over longer time periods (eg, several days) (Zhang et al., 2018). This issue is intricately linked with the tipping point between adaptive and adverse responses (Middleton et al., 2017; Shah et al., 2016; Wink et al., 2017). In particular, although the data in this work were generated using a single dose of the test chemicals, most exposure scenarios relevant to consumer safety involve long-term repeated exposures, and the nature of the exposure scenario itself (eg, whether cells or tissues are left with a period of recovery following insult) may also affect the PoD values (see, eg, Bischoff et al., 2019). Together, these considerations lead to a key question: is the 24 h timepoint sufficiently conservative for a low-tier screen? Answering this will require generating data under long-term exposures, which would most likely require using more sophisticated organotypic cell models, potentially within a microfluidic-based system (Trietsch et al., 2017; Wevers et al., 2018). Such data would then need to be complimented with appropriate computational approaches that take into consideration the biodynamic changes cellular stress responses (Zhang et al., 2018), in order to quantify the uncertainties associated with the time-dependent effects on PoDs and tipping points (eg, using the approach introduced by Shah et al. [2016]). However, given the possible approaches and technologies available, these challenges appear to be surmountable.
CONCLUSION
Using the in vitro cellular stress panel and statistical approach described here it was possible to identify substance exposures that may be associated with adverse health effects due to cellular stress. In combination with other cellular assays and in silico approaches, this panel could provide a powerful tool to use in nonanimal safety decision making. The preliminary analysis presented here will be built upon to ensure the approach described is sufficiently protective of consumer safety for a larger group of substances, including those that require metabolic activation.
SUPPLEMENTARY DATA
Supplementary data are available at Toxicological Sciences online.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank Beate Nicol, Cameron Mackay, Carl Westmoreland, and Paul Russell for their helpful comments in preparing the manuscript.
FUNDING
Unilever Plc.
DECLARATION OF CONFLICTING INTERESTS
The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
REFERENCES
- Ando M., Yoshikawa K., Iwase Y., Ishiura S. (2014). Usefulness of monitoring gamma-H2AX and cell cycle arrest in HepG2 cells for estimating genotoxicity using a high-content analysis system. J. Biomol. Screen. 19, 1246–1254. [DOI] [PubMed] [Google Scholar]
- Armitage J. M., Wania F., Arnot J. A. (2014). Application of mass balance models and the chemical activity concept to facilitate the use of in vitro toxicity data for risk assessment. Environ. Sci. Technol. 48, 9770–9779. [DOI] [PubMed] [Google Scholar]
- Attene-Ramos M. S., Huang R., Michael S., Witt K. L., Richard A., Tice R. R., Simeonov A., Austin C. P., Xia M. (2015). Profiling of the Tox21 chemical collection for mitochondrial function to identify compounds that acutely decrease mitochondrial membrane potential. Environ. Health Perspect. 123, 49–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attene-Ramos M. S., Huang R., Sakamuru S., Witt K. L., Beeson G. C., Shou L., Schnellmann R. G., Beeson C. C., Tice R. R., Austin C. P., et al. (2013). Systematic study of mitochondrial toxicity of environmental chemicals using quantitative high throughput screening. Chem. Res. Toxicol. 26, 1323–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baltazar M. T., Cable S., Carmichael P. L., Cubberley R., Cull T. A., Delagrange M., Dent M. P., Houghton J., Kukic P., Li H., Lee M.-Y., et al. (2020). A next generation risk assessment case study for coumarin in hypothetical cosmetic products. Toxicol. Sci. Available at: https://doi.org/10.1093/toxsci/kfaa048 [DOI] [PMC free article] [PubMed]
- Banerjee T., Chakravarti D. (2011). A peek into the complex realm of histone phosphorylation. Mol. Cell Biol. 31, 4858–4873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauch C., Bevan S., Woodhouse H., Dilworth C., Walker P. (2015). Predicting in vivo phospholipidosis-inducing potential of drugs by a combined high content screening and in silico modelling approach. Toxicol. In Vitro 29, 621–630. [DOI] [PubMed] [Google Scholar]
- Bell C. C., Dankers A. C. A., Lauschke V. M., Sison-Young R., Jenkins R., Rowe C., Goldring C. E., Park K., Regan S. L., Walker T., et al. (2018). Comparison of hepatic 2D sandwich cultures and 3D spheroids for long-term toxicity applications: A multicenter study. Toxicol. Sci. 162, 655–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Neriah Y., Karin M. (2011). Inflammation meets cancer, with NF-kappaB as the matchmaker. Nat. Immunol. 12, 715–723. [DOI] [PubMed] [Google Scholar]
- Bischoff L. J. M., Kuijper I. A., Schimming J. P., Wolters L., Braak B. T., Langenberg J. P., Noort D., Beltman J. B., van de Water B. (2019). A systematic analysis of Nrf2 pathway activation dynamics during repeated xenobiotic exposure. Arch. Toxicol. 93, 435–451. [DOI] [PubMed] [Google Scholar]
- Blanchard J., Sawers S. J. A. (1983). The absolute bioavailability of caffeine in man. E. J. Clin. Pharmacol. 24, 93–98. [DOI] [PubMed] [Google Scholar]
- Bock K. W. (2019). Aryl hydrocarbon receptor (AHR): From selected human target genes and crosstalk with transcription factors to multiple AHR functions. Biochem. Pharmacol. 168, 65–70. [DOI] [PubMed] [Google Scholar]
- Bort R., Ponsoda X., Jover R., Gomez-Lechon M. J., Castell J. V. (1999). Diclofenac toxicity to hepatocytes: A role for drug metabolism in cell toxicity. J. Pharmacol. Exp. Ther. 288, 65–72. [PubMed] [Google Scholar]
- Boudesco C., Cause S., Jego G., Garrido C. (2018). Hsp70: A cancer target inside and outside the cell. Methods Mol. Biol. 1709, 371–396. [DOI] [PubMed] [Google Scholar]
- Bowes J., Brown A. J., Hamon J., Jarolimek W., Sridhar A., Waldron G., Whitebread S. (2012). Reducing safety-related drug attrition: The use of in vitro pharmacological profiling. Nat. Rev. Drug Discov. 11, 909–922. [DOI] [PubMed] [Google Scholar]
- Brand M. D., Nicholls D. G. (2011). Assessing mitochondrial dysfunction in cells. Biochem. J. 435, 297–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burridge P. W., Li Y. F., Matsa E., Wu H., Ong S. G., Sharma A., Holmstrom A., Chang A. C., Coronado M. J., Ebert A. D., et al. (2016). Human induced pluripotent stem cell-derived cardiomyocytes recapitulate the predilection of breast cancer patients to doxorubicin-induced cardiotoxicity. Nat. Med. 22, 547–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell J. L. Jr, Clewell R. A., Gentry P. R., Andersen M. E., Clewell H. J. 3rd (2012). Physiologically based pharmacokinetic/toxicokinetic modeling. Methods Mol. Biol. 929, 439–499. [DOI] [PubMed] [Google Scholar]
- Carpenter B., Gelman A., Hoffman M. D., Lee D., Goodrich B., Betancourt M., Brubaker M., Guo J., Li P., Riddell A. (2017). Stan: A probabilistic programming language. J. Stat. Softw.76, 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen M. L., Meibohm B., Capparelli E. V., Velasquez-Mieyer P., Burghen G. A., Tamborlane W. V. (2005). Single- and multiple-dose pharmacokinetics of pioglitazone in adolescents with type 2 diabetes. J. Clin. Pharmacol. 45, 1137–1144. [DOI] [PubMed] [Google Scholar]
- Clewell R. A., Sun B., Adeleye Y., Carmichael P., Efremenko A., McMullen P. D., Pendse S., Trask O. J., White A., Andersen M. E. (2014). Profiling dose-dependent activation of p53-mediated signaling pathways by chemicals with distinct mechanisms of DNA damage. Toxicol. Sci. 142, 56–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins F. S., Gray G. M., Bucher J. R. (2008). Transforming environmental health protection. Science 319, 906–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damiani R. M., Moura D. J., Viau C. M., Caceres R. A., Henriques J. A. P., Saffi J. (2016). Pathways of cardiac toxicity: Comparison between chemotherapeutic drugs doxorubicin and mitoxantrone. Arch. Toxicol. 90, 2063–2076. [DOI] [PubMed] [Google Scholar]
- Daston G., Knight D. J., Schwarz M., Gocht T., Thomas R. S., Mahony C., Whelan M. (2015). SEURAT: Safety evaluation ultimately replacing animal testing–recommendations for future research in the field of predictive toxicology. Arch. Toxicol. 89, 15–23. [DOI] [PubMed] [Google Scholar]
- Davies N. M., Anderson K. E. (1997). Clinical pharmacokinetics of diclofenac. Therapeutic insights and pitfalls. Clin. Pharmacokinet. 33, 184–213. [DOI] [PubMed] [Google Scholar]
- DeGroot D. E., Swank A., Thomas R. S., Strynar M., Lee M. Y., Carmichael P. L., Simmons S. O. (2018). mRNA transfection retrofits cell-based assays with xenobiotic metabolism. J. Pharmacol. Toxicol. Methods 92, 77–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dengler V. L., Galbraith M., Espinosa J. M. (2014). Transcriptional regulation by hypoxia inducible factors. Crit. Rev. Biochem. Mol. Biol. 49, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dent M., Amaral R. T., Da Silva P. A., Ansell J., Boisleve F., Hatao M., Hirose A., Kasai Y., Kern P., Kreiling R., et al. (2018). Principles underpinning the use of new methodologies in the risk assessment of cosmetic ingredients. Comput. Toxicol. 7, 20–26. [Google Scholar]
- Dix D. J., Houck K. A., Martin M. T., Richard A. M., Setzer R. W., Kavlock R. J. (2007). The ToxCast program for prioritizing toxicity testing of environmental chemicals. Toxicol. Sci. 95, 5–12. [DOI] [PubMed] [Google Scholar]
- Eakins J., Bauch C., Woodhouse H., Park B., Bevan S., Dilworth C., Walker P. (2016). A combined in vitro approach to improve the prediction of mitochondrial toxicants. Toxicol. In Vitro 34, 161–170. [DOI] [PubMed] [Google Scholar]
- Fischer F. C., Henneberger L., Konig M., Bittermann K., Linden L., Goss K. U., Escher B. I. (2017). Modeling exposure in the Tox21 in Vitro bioassays. Chem. Res. Toxicol. 30, 1197–1208. [DOI] [PubMed] [Google Scholar]
- Foufelle F., Fromenty B. (2016). Role of endoplasmic reticulum stress in drug-induced toxicity. Pharmacol. Res. Perspect. 4, e00211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromenty B. (2019). Inhibition of mitochondrial fatty acid oxidation in drug-induced hepatic steatosis. Liver Res. 3, 157–169. [Google Scholar]
- Furue M., Uchi H., Mitoma C., Hashimoto-Hachiya A., Chiba T., Ito T., Nakahara T., Tsuji G. (2017). Antioxidants for healthy skin: The emerging role of aryl hydrocarbon receptors and nuclear factor-erythroid 2-related factor-2. Nutrients 9, 223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelman A., Carlin J. B., Stern H. S., Dunson D. B., Vehtari A., Rubin D. B. (2013). Bayesian Data Analysis. Chapman and Hall/CRC, Boca Raton, FL. [Google Scholar]
- Gunther V., Lindert U., Schaffner W. (2012). The taste of heavy metals: Gene regulation by MTF-1. Biochim. Biophys. Acta. 1823, 1416–1425. [DOI] [PubMed] [Google Scholar]
- Haber L. T., Dourson M. L., Allen B. C., Hertzberg R. C., Parker A., Vincent M. J., Maier A., Boobis A. R. (2018). Benchmark dose (BMD) modeling: Current practice, issues, and challenges. Crit. Rev. Toxicol. 48, 387–415. [DOI] [PubMed] [Google Scholar]
- Hadjipavlou-Litina D., Kontogiorgis C., Pontiki E., Dakanali M., Akoumianaki A., Katerinopoulos H. E. (2007). Anti-inflammatory and antioxidant activity of coumarins designed as potential fluorescent zinc sensors. J. Enzym. Inhib. Med. Chem. 22, 287–292. [DOI] [PubMed] [Google Scholar]
- Hanlon N., Coldham N., Gielbert A., Sauer M. J., Ioannides C. (2009). Repeated intake of broccoli does not lead to higher plasma levels of sulforaphane in human volunteers. Cancer Lett. 284, 15–20. [DOI] [PubMed] [Google Scholar]
- Hauner H. (2002). The mode of action of thiazolidinediones. Diabetes Metab. Res. Rev. 18, S10–S15. [DOI] [PubMed] [Google Scholar]
- Hiemstra S., Niemeijer M., Koedoot E., Wink S., Klip J. E., Vlasveld M., de Zeeuw E., van Os B., White A., Water B. v d. (2017). Comprehensive landscape of Nrf2 and p53 pathway activation dynamics by oxidative stress and DNA damage. Chem. Res. Toxicol. 30, 923–933. [DOI] [PubMed] [Google Scholar]
- Hill C. E., Myers J. P., Vandenberg L. N. (2018). Nonmonotonic dose-response curves occur in dose ranges that are relevant to regulatory decision-making. Dose Res. 16, 155932581879828. 1559325818798282–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong D. S., Kurzrock R., Supko J. G., He X., Naing A., Wheler J., Lawrence D., Eder J. P., Meyer C. J., Ferguson D. A., et al. (2012). A phase I first-in-human trial of bardoxolone methyl in patients with advanced solid tumors and lymphomas. Clin. Cancer Res. 18, 3396–3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu D., Wu C. Q., Li Z. J., Liu Y., Fan X., Wang Q. J., Ding R. G. (2015). Characterizing the mechanism of thiazolidinedione-induced hepatotoxicity: An in vitro model in mitochondria. Toxicol. Appl. Pharmacol. 284, 134–141. [DOI] [PubMed] [Google Scholar]
- Ipsen D. H., Lykkesfeldt J., Tveden-Nyborg P. (2018). Molecular mechanisms of hepatic lipid accumulation in non-alcoholic fatty liver disease. Cell. Mol. Life Sci. 75, 3313–3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judson R., Houck K., Martin M., Richard A. M., Knudsen T. B., Shah I., Little S., Wambaugh J., Woodrow Setzer R., Kothya P., et al. (2016). Editor’s highlight: Analysis of the effects of cell stress and cytotoxicity on in vitro assay activity across a diverse chemical and assay space. Toxicol. Sci. 152, 323–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judson R. S., Houck K. A., Kavlock R. J., Knudsen T. B., Martin M. T., Mortensen H. M., Reif D. M., Rotroff D. M., Shah I., Richard A. M., et al. (2010). In vitro screening of environmental chemicals for targeted testing prioritization: The ToxCast project. Environ. Health Perspect. 118, 485–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kany S., Vollrath J. T., Relja B. (2019). Cytokines in inflammatory disease. Int. J. Mol. Sci. 20, 6008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavlock R., Chandler K., Houck K., Hunter S., Judson R., Kleinstreuer N., Knudsen T., Martin M., Padilla S., Reif D., et al. (2012). Update on EPA’s ToxCast program: Providing high throughput decision support tools for chemical risk management. Chem. Res. Toxicol. 25, 1287–1302. [DOI] [PubMed] [Google Scholar]
- Khoury L., Zalko D., Audebert M. (2013). Validation of high-throughput genotoxicity assay screening using gammaH2AX in-cell western assay on HepG2 cells. Environ. Mol. Mutagen. 54, 737–746. [DOI] [PubMed] [Google Scholar]
- Kramer N. I., Krismartina M., Rico-Rico A., Blaauboer B. J., Hermens J. L. (2012). Quantifying processes determining the free concentration of phenanthrene in Basal cytotoxicity assays. Chem. Res. Toxicol. 25, 436–445. [DOI] [PubMed] [Google Scholar]
- Krishnan K., Gagne M., Nong A., Aylward L. L., Hays S. M. (2010). Biomonitoring Equivalents for triclosan. Regul. Toxicol. Pharmacol. 58, 10–17. [DOI] [PubMed] [Google Scholar]
- Kruschke J. (2014). Doing Bayesian Data Analysis: A Tutorial with R, JAGS, and Stan. Academic Press, Cambridge, MA. [Google Scholar]
- Liao X., Wang Y., Wong C.-W. (2010). Troglitazone induces cytotoxicity in part by promoting the degradation of peroxisome proliferator-activated receptor γ co-activator-1α protein. Br. J. Pharmacol. 161, 771–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loboda A., Damulewicz M., Pyza E., Jozkowicz A., Dulak J. (2016). Role of Nrf2/HO-1 system in development, oxidative stress response and diseases: An evolutionarily conserved mechanism. Cell. Mol. Life Sci. 73, 3221–3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loi C. M., Young M., Randinitis E., Vassos A., Koup J. R. (1999). Clinical pharmacokinetics of troglitazone. Clin. Pharmacokinet. 37, 91–104. [DOI] [PubMed] [Google Scholar]
- LoPachin R. M., Gavin T. (2016). Reactions of electrophiles with nucleophilic thiolate sites: Relevance to pathophysiological mechanisms and remediation. Free Radic. Res. 50, 195–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lushchak V. I. (2012). Glutathione homeostasis and functions: Potential targets for medical interventions. J. Amino Acids 2012, 736837–736837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magesh S., Chen Y., Hu L. (2012). Small molecule modulators of Keap1-Nrf2-ARE pathway as potential preventive and therapeutic agents. Med. Res. Rev. 32, 687–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malo N., Hanley J. A., Cerquozzi S., Pelletier J., Nadon R. (2006). Statistical practice in high-throughput screening data analysis. Nat. Biotechnol. 24, 167–175. [DOI] [PubMed] [Google Scholar]
- Marzo A., Bo L., Verga F., Monti N., Abbondati G., Tettamanti R., Crivelli F., Uhr M., Ismaili S. (2000). Pharmacokinetics of diclofenac after oral administration of its potassium salt in sachet and tablet formulations. Arzneimittelforschung 50, 43–47. [DOI] [PubMed] [Google Scholar]
- Masubuchi Y., Kano S., Horie T. (2006). Mitochondrial permeability transition as a potential determinant of hepatotoxicity of antidiabetic thiazolidinediones. Toxicology 222, 233–239. [DOI] [PubMed] [Google Scholar]
- Middleton A., Cooper S., Cull T., Stark R., Adeleye Y., Boekelheide K., Clewell R., Jennings P., Guo J., Liu C., et al. (2017). Case studies in cellular stress: Defining adversity/adaptation tipping points. Appl. In Vitro Toxicol. 3, 199–210. [Google Scholar]
- Moxon T. E., Li H., Lee M. Y., Piechota P., Nicol B., Pickles J., Pendlington R., Sorrell I., Baltazar M. T. (2020). Application of physiologically based kinetic (PBK) modelling in the next generation risk assessment of dermally applied consumer products. Toxicol. In Vitro 63, 104746. [DOI] [PubMed] [Google Scholar]
- Nadanaciva S., Dykens J. A., Bernal A., Capaldi R. A., Will Y. (2007). Mitochondrial impairment by PPAR agonists and statins identified via immunocaptured OXPHOS complex activities and respiration. Toxicol. Appl. Pharmacol. 223, 277–287. [DOI] [PubMed] [Google Scholar]
- Nadanaciva S., Will Y. (2011). New insights in drug-induced mitochondrial toxicity. Curr. Pharm. Des. 17, 2100–2112. [DOI] [PubMed] [Google Scholar]
- Nebert D. W., Roe A. L., Dieter M. Z., Solis W. A., Yang Y., Dalton T. P. (2000). Role of the aromatic hydrocarbon receptor and [Ah] gene battery in the oxidative stress response, cell cycle control, and apoptosis. Biochem. Pharmacol. 59, 65–85. [DOI] [PubMed] [Google Scholar]
- Nikoletopoulou V., Markaki M., Palikaras K., Tavernarakis N. (2013). Crosstalk between apoptosis, necrosis and autophagy. Biochim. Biophys. Acta 1833, 3448–3459. [DOI] [PubMed] [Google Scholar]
- O’Brien P. J., Irwin W., Diaz D., Howard-Cofield E., Krejsa C. M., Slaughter M. R., Gao B., Kaludercic N., Angeline A., Bernardi P., et al. (2006). High concordance of drug-induced human hepatotoxicity with in vitro cytotoxicity measured in a novel cell-based model using high content screening. Arch. Toxicol. 80, 580–604. [DOI] [PubMed] [Google Scholar]
- Oslowski C. M., Urano F. (2011). Measuring ER stress and the unfolded protein response using mammalian tissue culture system. Methods Enzymol. 490, 71–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park C., Jeong J. (2018). Synergistic cellular responses to heavy metal exposure: a minireview. Biochim. Biophys. Acta Gen. Subj. 1862, 1584–1591. [DOI] [PubMed] [Google Scholar]
- Phillips J. R., Svoboda D. L., Tandon A., Patel S., Sedykh A., Mav D., Kuo B., Yauk C. L., Yang L., Thomas R. S., et al. (2019). BMDExpress 2: enhanced transcriptomic dose-response analysis workflow. Bioinformatics 35, 1780–1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plosker G. L., Faulds D. (1999). Troglitazone: a review of its use in the management of type 2 diabetes mellitus. Drugs 57, 409–438. [DOI] [PubMed] [Google Scholar]
- Ponsoda X., Bort R., Jover R., Gomez-Lechon M. J., Castell J. V. (1995). Molecular mechanism of diclofenac hepatotoxicity: Association of cell injury with oxidative metabolism and decrease in ATP levels. Toxicol. In Vitro 9, 439–444. [DOI] [PubMed] [Google Scholar]
- Ramachandran A., Duan L., Akakpo J. Y., Jaeschke H., (2018). Mitochondrial dysfunction as a mechanism of drug-induced hepatotoxicity: Current understanding and future perspectives. J. Clin. Transl. Res. 4, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramaiahgari S. C., Auerbach S. S., Saddler T. O., Rice J. R., Dunlap P. E., Sipes N. S., DeVito M. J., Shah R. R., Bushel P. R., Merrick B. A., et al. (2019). The power of resolution: Contextualized understanding of biological responses to liver injury chemicals using high-throughput transcriptomics and benchmark concentration modeling. Toxicol. Sci. 169, 553–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramaiahgari S. C., Waidyanatha S., Dixon D., DeVito M. J., Paules R. S., Ferguson S. S. (2017). From the cover: Three-dimensional (3D) HepaRG spheroid model with physiologically relevant xenobiotic metabolism competence and hepatocyte functionality for liver toxicity screening. Toxicol. Sci. 159, 124–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramesh A., Varghese S. S., Doraiswamy J., Malaiappan S. (2014). Role of sulfiredoxin in systemic diseases influenced by oxidative stress. Redox Biol. 2, 1023–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rana P., Aleo M. D., Gosink M., Will Y. (2019). Evaluation of in vitro mitochondrial toxicity assays and physicochemical properties for prediction of organ toxicity using 228 pharmaceutical drugs. Chem. Res. Toxicol. 32, 156–167. [DOI] [PubMed] [Google Scholar]
- Reynolds J., MacKay C., Gilmour N., Miguel-Vilumbrales D., Maxwell G. (2019). Probabilistic prediction of human skin sensitiser potency for use in next generation risk assessment. Comput. Toxicol. 9, 36–49. [Google Scholar]
- Shah I., Setzer R. W., Jack J., Houck K. A., Judson R. S., Knudsen T. B., Liu J., Martin M. T., Reif D. M., Richard A. M., et al. (2016). Using ToxCast data to reconstruct dynamic cell state trajectories and estimate toxicological points of departure. Environ. Health Perspect. 124, 910–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao K., Shapiro A. J. (2018). A web-based system for Bayesian Benchmark dose estimation. Environ. Health Perspect. 126, 017002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla S. J., Huang R., Austin C. P., Xia M. (2010). The future of toxicity testing: A focus on in vitro methods using a quantitative high-throughput screening platform. Drug Discov. Today 15, 997–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons S. O., Fan C. Y., Ramabhadran R. (2009). Cellular stress response pathway system as a sentinel ensemble in toxicological screening. Toxicol. Sci. 111, 202–225. [DOI] [PubMed] [Google Scholar]
- Sipes N. S., Martin M. T., Kothiya P., Reif D. M., Judson R. S., Richard A. M., Houck K. A., Dix D. J., Kavlock R. J., Knudsen T. B., (2013). Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem. Res. Toxicol. 26, 878–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speth P. A., Linssen P. C., Boezeman J. B., Wessels H. M., Haanen C. (1987). Cellular and plasma adriamycin concentrations in long-term infusion therapy of leukemia patients. Cancer Chemother Pharmacol. 20, 305–310. [DOI] [PubMed] [Google Scholar]
- Teplova V. V., Belosludtsev K. N., Kruglov A. G. (2017). Mechanism of triclosan toxicity: Mitochondrial dysfunction including complex II inhibition, superoxide release and uncoupling of oxidative phosphorylation. Toxicol. Lett. 275, 108–117. [DOI] [PubMed] [Google Scholar]
- Teske B. F., Wek S. A., Bunpo P., Cundiff J. K., McClintick J. N., Anthony T. G., Wek R. C. (2011). The eIF2 kinase PERK and the integrated stress response facilitate activation of ATF6 during endoplasmic reticulum stress. Mol. Biol. Cell 22, 4390–4405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas R. S., Bahadori T., Buckley T. J., Cowden J., Deisenroth C., Dionisio K. L., Frithsen J. B., Grulke C. M., Gwinn M. R., Harrill J. A., et al. (2019). The next generation blueprint of computational Toxicology at the U.S. Environmental Protection Agency. Toxicol. Sci. 169, 317–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas R. S., Philbert M. A., Auerbach S. S., Wetmore B. A., Devito M. J., Cote I., Rowlands J. C., Whelan M. P., Hays S. M., Andersen M. E., et al. (2013). Incorporating new technologies into toxicity testing and risk assessment: Moving from 21st century vision to a data-driven framework. Toxicol. Sci. 136, 4–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tice R. R., Austin C. P., Kavlock R. J., Bucher J. R. (2013). Improving the human hazard characterization of chemicals: A Tox21 update. Environ. Health Perspect. 121, 756–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trietsch S. J., Naumovska E., Kurek D., Setyawati M. C., Vormann M. K., Wilschut K. J., Lanz H. L., Nicolas A., Ng C. P., Joore J., et al. (2017). Membrane-free culture and real-time barrier integrity assessment of perfused intestinal epithelium tubes. Nat. Commun. 8, 262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troutman J. A., Rick D. L., Stuard S. B., Fisher J., Bartels M. J. (2015). Development of a physiologically-based pharmacokinetic model of 2-phenoxyethanol and its metabolite phenoxyacetic acid in rats and humans to address toxicokinetic uncertainty in risk assessment. Regul. Toxicol. Pharmacol. 73, 530–543. [DOI] [PubMed] [Google Scholar]
- U.S. EPA. (2014). Next generation risk assessment: Incorporation of recent advances in molecular, computational, and systems biology, Final report. U.S. Environmental Protection Agency. Washington, DC, EPA/600/R-14/004.
- U.S. EPA. (2019). Benchmark Dose Software User Guide, Version 3.1. Washington, DC.
- van der Linden S. C., von Bergh A. R., van Vught-Lussenburg B. M., Jonker L. R., Teunis M., Krul C. A., van der Burg B. (2014). Development of a panel of high-throughput reporter-gene assays to detect genotoxicity and oxidative stress. Mutat. Res. Genet. Toxicol. Environ. Mutagen. 760, 23–32. [DOI] [PubMed] [Google Scholar]
- Verstrepen L., Verhelst K., van Loo G., Carpentier I., Ley S. C., Beyaert R. (2010). Expression, biological activities and mechanisms of action of A20 (TNFAIP3). Biochem. Pharmacol. 80, 2009–2020. [DOI] [PubMed] [Google Scholar]
- Wang M., Kaufman R. J. (2016). Protein misfolding in the endoplasmic reticulum as a conduit to human disease. Nature 529, 326–335. [DOI] [PubMed] [Google Scholar]
- Wang X., Chen M., Zhou J., Zhang X. (2014. a). HSP27, 70 and 90, anti-apoptotic proteins, in clinical cancer therapy (Review). Int. J. Oncol. 45, 18–30. [DOI] [PubMed] [Google Scholar]
- Wang Y. Y., Yang Y. X., Zhe H., He Z. X., Zhou S. F. (2014. b). Bardoxolone methyl (CDDO-Me) as a therapeutic agent: An update on its pharmacokinetic and pharmacodynamic properties. Drug Des. Dev. Ther. 8, 2075–2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt E. D., Judson R. S. (2018). Uncertainty quantification in ToxCast high throughput screening. PLoS One 13, e0196963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenger R. H., Stiehl D. P., Camenisch G. (2005). Integration of oxygen signaling at the consensus HRE. Sci STKE 2005, re12. [DOI] [PubMed] [Google Scholar]
- Westerheide S. D., Morimoto R. I. (2005). Heat shock response modulators as therapeutic tools for diseases of protein conformation. J. Biol. Chem. 280, 33097–33100. [DOI] [PubMed] [Google Scholar]
- Wetmore B. A., Wambaugh J. F., Allen B., Ferguson S. S., Sochaski M. A., Setzer R. W., Houck K. A., Strope C. L., Cantwell K., Judson R. S., et al. (2015). Incorporating high-throughput exposure predictions with dosimetry-adjusted in vitro bioactivity to inform chemical Toxicity testing. Toxicol. Sci. 148, 121–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wevers N. R., Kasi D. G., Gray T., Wilschut K. J., Smith B., van Vught R., Shimizu F., Sano Y., Kanda T., Marsh G., et al. (2018). A perfused human blood-brain barrier on-a-chip for high-throughput assessment of barrier function and antibody transport. Fluids Barriers CNS 15, 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wink S., Hiemstra S., Herpers B., van de Water B. (2017). High-content imaging-based BAC-GFP toxicity pathway reporters to assess chemical adversity liabilities. Arch. Toxicol. 91, 1367–1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wink S., Hiemstra S., Huppelschoten S., Danen E., Niemeijer M., Hendriks G., Vrieling H., Herpers B., van de Water B. (2014). Quantitative high content imaging of cellular adaptive stress response pathways in toxicity for chemical safety assessment. Chem. Res. Toxicol. 27, 338–355. [DOI] [PubMed] [Google Scholar]
- Wu J. H., Batist G. (2013). Glutathione and glutathione analogues; therapeutic potentials. Biochim. Biophys. Acta 1830, 3350–3353. [DOI] [PubMed] [Google Scholar]
- Xu H., Wang L., Zheng P., Liu Y., Zhang C., Jiang K., Song H., Ji G. (2017). Elevated serum A20 is associated with severity of chronic hepatitis B and A20 inhibits NF-kappaB-mediated inflammatory response. Oncotarget 8, 38914–38926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C., Barlow S. M., Muldoon Jacobs K. L., Vitcheva V., Boobis A. R., Felter S. P., Arvidson K. B., Keller D., Cronin M. T., Enoch S.,. et al. (2017). Thresholds of toxicological concern for cosmetics-related substances: New database, thresholds, and enrichment of chemical space. Food Chem. Toxicol. 109, 170–193. [DOI] [PubMed] [Google Scholar]
- Yuan H., Zhang Q., Guo J., Zhang T., Zhao J., Li J., White A., Carmichael P. L., Westmoreland C., Peng S. (2016). A PGC-1alpha-mediated transcriptional network maintains mitochondrial redox and bioenergetic homeostasis against doxorubicin-induced Toxicity in human cardiomyocytes: Implementation of TT21C. Toxicol. Sci. 150, 400–417. [DOI] [PubMed] [Google Scholar]
- Zhang Q., Li J., Middleton A., Bhattacharya S., Conolly R. B. (2018). Bridging the data gap from in vitro toxicity testing to chemical safety assessment through computational modeling. Front. Public Health 6, 261. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.