Abstract
Background:
Primary neuronal cell cultures are useful for studying mechanisms that influence dendritic morphology during normal development and in response to various stressors. However, analyzing dendritic morphology is challenging, particularly in cultures with high cell density, and manual methods of selecting neurons and tracing dendritic arbors can introduce significant bias, and are labor-intensive. To overcome these challenges, semi-automated and automated methods are being developed, with most software solutions requiring computer-assisted dendrite tracing with subsequent quantification of various parameters of dendritic morphology, such as Sholl analysis. However fully automated approaches for classic Sholl analysis of dendritic complexity are not currently available.
New Method:
The previously described Omnisphero software, was extended by adding new functions to automatically assess dendritic mass, total length of the dendritic arbor and the number of primary dendrites, branch points, and terminal tips, and to perform Sholl analysis.
Results:
The new functions for assessing dendritic morphology were validated using primary mouse hippocampal and rat cortical neurons transfected with a fluorescently tagged MAP2 cDNA construct. These functions allow users to select specific populations of neurons as a training set for subsequent automated selection of labeled neurons in high-density cultures.
Comparison with Existing Semi-Automated Methods:
Compared to manual or semi-automated analyses of dendritic arborization, the new functions increase throughput while significantly decreasing researcher bias associated with neuron selection, tracing, and thresholding.
Conclusion:
These results demonstrate the importance of using unbiased automated methods to mitigate experimenter-dependent bias in analyzing dendritic morphology.
Keywords: Dendritic arborization, high-content image analysis, neurite outgrowth, quantitative morphometric analyses, Sholl analysis
1. Introduction
Dendritic morphology is a critical determinant of neuronal connectivity that influences the processing and distribution of information within neural circuits (Libersat and Duch, 2004; Menon and Gupton, 2016; Scott and Luo, 2001). Aberrant dendritic morphology is strongly associated with neurodevelopmental disorders (Bourgeron, 2009; Fukuda et al., 2005; Garey, 2010; Keown et al., 2013; Supekar et al., 2013) and neurodegenerative diseases (Cochran et al., 2014; Kweon et al., 2017). Thus, there is great scientific interest in analyzing dendritic morphology.
Currently, a wide variety of tools exist for morphometric analysis of dendrites. Open source software, including Neurite–Tracer (Pool et al., 2008), CellProfiler (Kamentsky et al., 2011), NeurphologyJ (Ho et al., 2011), and NeuronCyto (Yu et al., 2009) provide semi-automated approaches for analyzing parameters of dendritic morphology such as the number of primary dendrites, total length of the dendritic arbor, number of dendritic branch points and the number of terminal dendritic tips. However, only a few of the currently available analytical approaches automatically extract morphological features like Omnisphero (Schmuck et al., 2017) or the GAIN algorithm (Long et al., 2017). Here, the definition of automatic means that only the input image is required and all following steps, including isolation of single cells, segmentation from the background, transformation into a center line representation (skeletonization of the cell), and location of the center point (middle of the cell somata) for placement of the Sholl rings are performed by the software. This is in contrast to existing semi-automatic methods, such as the Bonfire program (Kutzing et al., 2010) and the Sholl Fiji plugin, in which these steps must be performed manually. This information is required to perform Sholl analysis (Sholl, 1953), a classic approach for quantifying dendritic complexity both in vitro and in vivo that counts the number of dendritic intersections with concentric rings superimposed on the soma as a function of distance from the cell soma.
The Sholl analysis ImageJ Plugin (Ferreira et al., 2014) is an example of semi-automatic Sholl Analysis. In this case, individual neurons are chosen manually, a center point defined and thresholds set manually to generate a binary image. Next, the Sholl Analysis ImageJ Plugin superimposes concentric rings around the center point, determines the number of dendritic intersections with each ring, and generates a distribution plot. With this data, the researcher can obtain important information, such as the number of dendritic intersections at increasing radii from the soma (referred to as area under the curve [AUC]), the radius at which the largest number of intersections occurs (Peak X), and the greatest number of intersections that occurs at any radius (Peak Y). However, neuron selection, manual thresholding and selection of the center points within a neuron can result in significant inter-researcher differences that critically affect data reproducibility (Nobis and Hunziker, 2005), and this approach is time-consuming, which limits the number of cells that can be analyzed due to time and labor constraints.
A method that automates the pre-processing steps for analyzing dendritic morphology by Sholl analysis would be highly desirable in that it would minimize investigator bias and enable medium-to-high throughput screening. While common annotation data can be used for deep learning approaches, such as automatically identifying specific neuronal subtypes (Shen et al., 2017), self-learning approaches demand a high number of annotated objects, usually several thousand, to allow for a successful prediction (Shen et al., 2017). Obtaining these sample sizes can be challenging, and manual annotation by a few researchers as well as archiving these annotations is challenging.
Here, we present a new algorithm that addresses some of these challenges using an extension of the Omnisphero software (Schmuck et al., 2017). The algorithm automates identification of neuronal cells, extraction of image data from the background, selection of center points for the Sholl rings and quantitative analysis of these structures for standard morphological features, such as total neurite length, number of primary processes, and number of terminal tips. In addition, this algorithm performs automated Sholl analysis. Omnisphero’s graphical user interface (GUI) allows the user to display binary images, cell centroids, skeleton structures, terminal tips, and branch points, as well as Sholl masks (rings with dendritic intersections) and overlays obtained from the new extension. The GUI gives the user the power to judge the quality of structure isolation and the accuracy of structures. The new extension is the first algorithm to automatically perform all pre- and post-processing for Sholl analysis and can be run in two different modes: manual or automated object selection, with the later based on a training set of images. If objects are manually selected, the algorithm automatically extracts neuronal structures and determines their centroid. Consequently, the researcher can annotate objects of interest or delete unwanted structures. The extraction of the named features described above is completely automatic.
In this study, various parameters of dendritic morphology were analyzed from images of primary mouse hippocampal and rat cortical neurons grown in high cell density cultures that had been transfected at low efficiency with a fluorescently tagged MAP2B cDNA construct to visualize the dendritic arbors of individual neurons. Data obtained using manual annotation or the new Omnisphero extension were compared. Two researchers manually labeled structures of interest, and objects labeled by both were used as a training set. This allowed for a completely unbiased automated approach to thresholding and selection. The overall goal of this study was to establish a fully automated approach for classic analyses of dendrite morphology similar to other algorithms while eliminating researcher bias in neuron selection and thresholding. We used two experimental sets of neurons to recapitulate common challenges to analysis of dendritic arborization in high-density cell cultures: high signal-to-noise ratios, which make thresholding difficult (mouse cultures), and selection of individual neurons with overlapping dendritic arbors (rat cultures).
2. Materials and Methods
2.1. Primary mouse or rat cortical cell culture
All procedures involving animals were conducted in accordance with the NIH Guide for the Care and Use of Laboratory Animals and were approved by the University of California, Davis, Institutional Animal Care and Use Committee. Timed-pregnant Sprague Dawley rats were purchased from Charles River Laboratories (Hollister, CA, USA). Timed-pregnant C57BL/6J wild-type mice were purchased from Jackson Labs (Bar Harbor, ME, USA). All animals were housed in clear plastic cages containing corn cob bedding in a room with a constant temperature (22 ± 2 °C) and a 12 h light-dark cycle. Food and water were provided ad libitum.
Primary hippocampal or cortical neuron-glia co-cultures were prepared from postnatal day 0 mouse or rat pups, respectively, as previously described (Sethi et al., 2017). Dissociated cells were plated on glass coverslips (BellCo, Vineland, NJ, USA) pre-coated with 500 μg/ml poly-L-lysine (Sigma-Aldrich, St. Louis, MO, USA) and maintained at 37 °C in NeuralQ Basal Medium (MTI-GlobalStem, Gaithersburg, MD, USA) supplemented with 2% GS21 (MTI-GlobalStem) and GlutaMAX (ThermoScientific, Waltham, MA, USA) under 5% CO2. Neurons were plated at 83,000 cells/cm2 in 24-well tissue culture plates for analyses of dendritic morphology. At 4 days in vitro (DIV), cultures were treated with cytosine β-D-arabinofuranoside (Sigma-Aldrich) at 2.5 μM to limit glial growth.
2.2. Manual morphometric analyses of dendritic arborization
To visualize dendrites, cultures were transfected with a Map2B-pCAG-FusRed or a Map2B-pCAG-GFP plasmid (Wayman et al., 2006) at DIV6 using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s protocol. Neurons were fixed with 4% paraformaldehyde (Sigma-Aldrich) in 0.2 M phosphate buffered saline (PBS) at DIV9. 16bit tiff images were acquired in an unbiased manner using an ImageXpress Micro XL high content imaging system (Molecular Devices, Sunnyvale, CA, USA) with a plan fluor 10x objective (numerical aperture 0.3) and a 4.66 megapixel CMOS camera. Approximately 50 to 75 images were acquired from each well and exported as 16-bit tiff stacks. Neurons were chosen for analysis using previously described criteria (Keil et al., 2017). Manual quantification of dendritic arborization was performed by counting the number of primary dendrites for each neuron and the number of corresponding dendritic tips using ImageJ software (Schneider et al., 2012) with Sholl analysis plug-in v3.4.2 (Ferreira et al., 2014) and with NeuronJ plug-in (Meijering et al., 2004). Researchers were blinded to experimental groups prior to performing all morphometric analyses. Neurons from two different sets of mouse cultures and two different sets of rat cultures, referred to as experimental groups A vs. B and C vs. D, respectively, were examined.
2.3. Image pre-processing using the new Omnisphero extension
Omnisphero, was originally designed to analyze neurons in neurosphere cultures. In addition to automatic identification of neurons, it provides a graphical user interface that allows the user to overlay image analysis results with the original image. We describe here a new function we added to the GUI of Omnisphero to analyze cortical neurons and to show an overlay of the segmentation as well as Sholl-Analysis rings.
Image pre-processing (Fig. 1) started with 16-bit tiff image stacks of rat or mouse neurons imported into the new extension, split into individual images, converted to 8-bit and saved as individual images in ‘portable network graphics’ (png) format (Fig. 1e). Binary images of the original grey scale images were generated using the imopen function with a non-flat structuring element (ball) to estimate the uneven background, which was subtracted from the original image. The resulting image was thresholded using the isodata algorithm (Fig. 1f), and binary particles were analyzed for their ratio of major and minor axes. Round small structures were considered artifacts and removed from further analysis. In addition, different edge detection algorithms (Canny, prewitt, sobel, Roberts and log) were applied to the background-corrected grey scale image and filtered for small objects considered originating from artifacts (Fig. 1g).
Figure 1. Semi- and fully-automated morphological features and Sholl analysis.
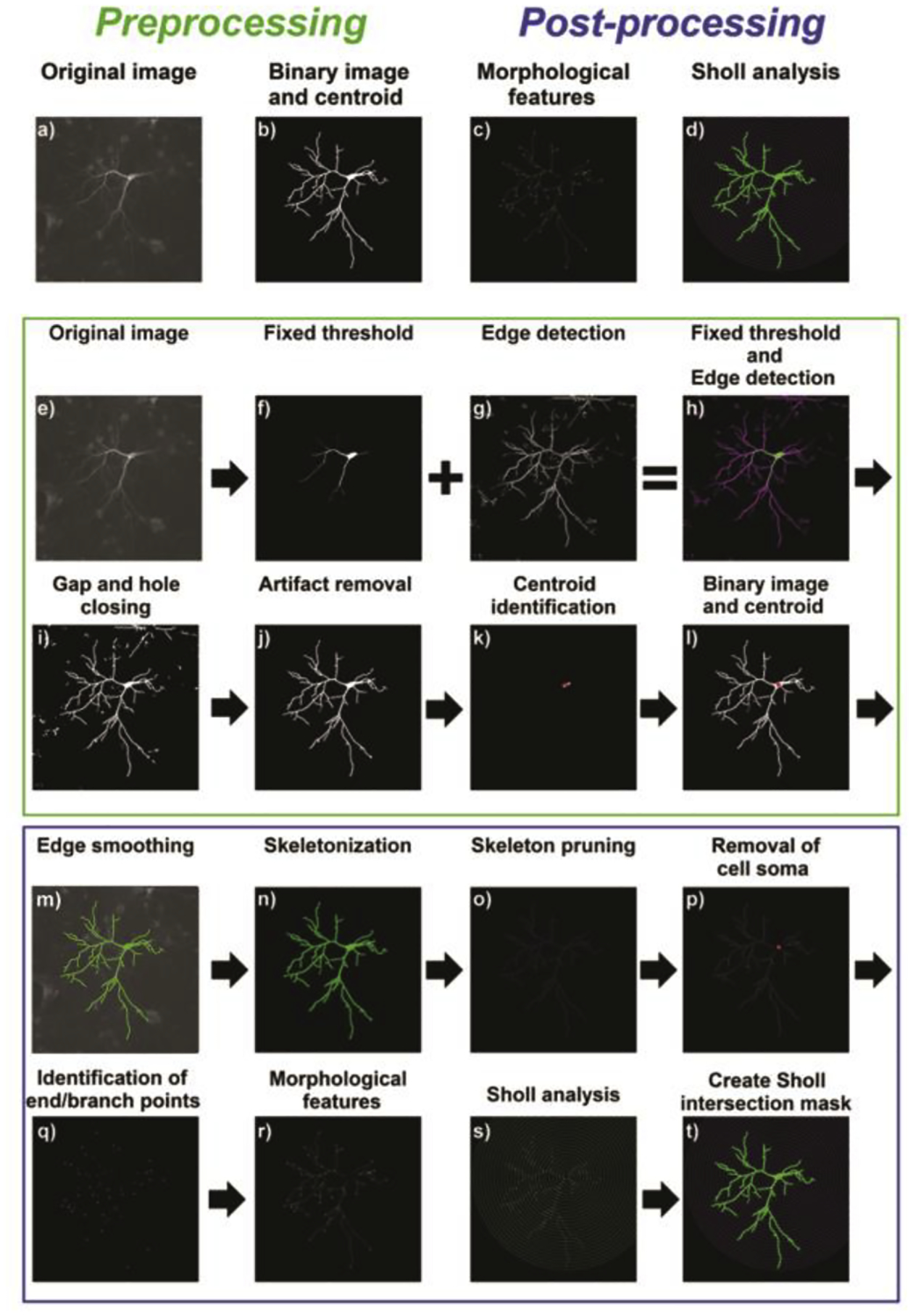
(a, b) Overall preprocessing of raw images involves crucial thresholding prior to post-processing examination of (c) morphological features and (d) Sholl analysis. (e, f) 8-bit raw images were thresholded. (g, h) Artifacts were removed by using different combinations of edge detection algorithms that were applied and combined with the thresholded image. (i) Gaps and holes were closed, and (j) non-dendritic structures were removed. (k, l) To obtain the centroid of the cell soma, an erosion was performed, and the centroid (red) of the resulting particle computed. Post-processing was performed in a semi-automated way by manually selecting neurons of interest, or in a fully automated approach in which the software calculates similarities towards a training set of selected neurons. (m, n) Selected neuronal structures were smoothed and skeletonized. (o) Resulting skeletons were pruned to remove artificial branching points and small branches. (p-s) For analysis, the cell soma was removed and endpoints like the number of primary processes, total dendritic length, number of terminal tips, number of branching points were computed and Sholl analysis was performed. (t) Results can be visualized in the graphical user interface of Omnisphero and data are stored as .xlsx files.
Resulting binary images were added to the binary image acquired using the isodata algorithm. The resulting image (Fig. 1h) was further processed by morphological closing of gaps in fragmented dendrites using the bridge function and consequent filling of the structures with the imfill holes function (Fig. 1i). In order to circumvent filling of spaces enclosed by cellular processes, only small holes not exceeding 50 pixels (pixel size can be defined by the user and is dependent on the optical setup, see Supplementary Information) were filled. The resulting binary image was filtered for objects larger than 2000 pixels (can be parametrized by the user) to maintain neuronal structures (Fig. 1j). Next, neuronal structures were masked out from the original image using the generated binary image. The median pixel value multiplied by 1.5 was calculated, and used as a threshold value. Only pixels equal or brighter than these values were masked, resulting in masks of only the brightest structures. Resulting structures were filled with the imfill function and eroded until only the thickest part, the cell soma, remained. Soma centroids were identified (Fig. 1k), and their coordinates were saved in a matrix. Particles identified as cell somata were used to mask the respective region from the original grey scale image. Binary images and cell soma images were saved and centroids returned to the GUI of Omnisphero (Fig. 1l).
Since images of mouse neurons tend to be noisy, an additional run of the same edge detection algorithms was performed without filtering small artifacts, and an image dilation of the binary image was done to check whether dilation was able to close gaps in broken neurites. Lastly, the binary image obtained from filtered edge detection was subtracted from the unfiltered approach. Remaining structures were validated to be an extension of the neuron by analyzing the weight factor, which was calculated as the ratio of the area of the binary particle to a spanned rectangle (obtained by identifying the smallest surrounding rectangle) multiplied by the number of endpoints of the structure, divided by the particle skeleton length and normalized to the area of the particle.
In the case of rat neurons, a particular challenge was separating neurons with overlapping dendritic arbors. This was achieved by using isodata to determine the number of binary structures located within a structure obtained by combination of isodata and edge detection. If more than one structure was found with edge detection, the connecting neurite was split. Additionally, the majority function was used to identify structures connected by one pixel and to separate the structures. If two neuronal structures were connected via multiple neurites, the binary particles were excluded from further analysis because a reliable separation was not possible. These structures were identified with a binary mask, which subtracted only neuronal structures from the original grey level image, and then thresholded via the isodata method. If the area of the resulting binary image exceeded a certain threshold, then the threshold was doubled. If the threshold reduced the area below 20%, the threshold was reduced to 1.5 times the original threshold. The binary image was consequently dilated with a disk element to combine binary particles close together because two neuronal somata should have a certain distance from each other, due to the overall low transfection efficacy. Next, the number of individual particles was determined. In some instances, the intensity of the two overlapping neurons strongly differed, and a second approach was taken in which the image was thresholded with only half of the initial threshold and checked for particles not overlapping with those from the hard threshold. Overlapping particles were considered outside the main neuron soma and to overlap with a dimmer neuron (which is dimmer because of lower transfection efficacy).
To identify air bubbles, which are very bright and can erroneously be identified by both isodata and edge detection, all binary structures were analyzed for their ratio of area to spanned rectangle (obtained by identifying the smallest surrounding rectangle) and the ratio of length (obtained via infinite thinning of the particles) to branches. Structures having a very small ratio of area to rectangle and length to branches are usually linear, very long, and thus, highly likely to originate from air bubbles. These structures were removed from the analysis. In addition, all structures touching the edge of the image were excluded from analysis.
2.4. Morphometric analyses of dendritic arborization using the new Omnisphero extension
The new Omnisphero extension provides a choice of two different modes: semi-automated or automated. In the semi-automated mode, auto-thresholding is performed and centroids of binary structures are identified during image preprocessing but selection of which neurons should be analyzed is determined by the user. The user can select which structures within the GUI of Omnisphero to analyze (illustrated using rat cultures) or to exclude (illustrated using mouse cultures), with the decision usually based on the overlap of neurons. Marked objects are saved in a separate matrix for further analysis.
2.4.1. Semi-automated mode
Following manual annotation of binary structures, all non-annotated objects were deleted, and binary structures were smoothed by applying a canny edge detection algorithm to each particle (Fig. 1m). Remaining particles were further analyzed by generating a raw skeleton via morphological thinning (Fig. 1n). The skeleton was postprocessed by identifying all endpoints and branches and deleting all branching less than one pixel from the endpoints. This commonly occurred when very small processes, less than 10 pixels, originated from imperfections of the binary image. Next, all branches from the skeleton were pruned and checked for small extensions by measuring the length of each resulting skeleton fragment and verifying that endpoints were located on each fragment (Fig. 1o). This was done to prevent the generation of holes within the skeleton. If branching points were located directly next to each other, they were eliminated since this is most likely attributed to imperfections in the skeleton line. The skeleton was checked for gaps by assessing the number of newly introduced endpoints and reconnecting them. Next, the cell soma was subtracted from the skeleton (Fig. 1p). Endpoints and branching points were saved in separate matrices (Fig. 1q), and the corresponding images were created displaying the endpoints as solid dots and the branching points as circles (Fig. 1r). Results were displayed in the Omnisphero GUI.
A Sholl analysis was performed using the preprocessed skeleton (Fig. 1s). The inner ring centered on the cell soma was set to a radius of 10 μm. Within the inner ring, the skeleton was deleted and the numbers of intersections with the ring were counted. Additional rings were added in a nested loop at a distance of 10 μm for quantification of intersections, added length of processes, and added number of branches, all of which were saved in a separate matrix. This process was terminated when there were no new intersections. Resulting ring masks with intersections, visualized as circles, were saved as a png file for visualization in the Omnisphero GUI (Fig 1t). Lastly, matrices of morphological endpoints and Sholl results were written to an xlsx file.
2.4.2. Fully automated mode
In the fully automated mode, a “gold-standard” of selected neuronal structures, obtained by manual annotation of six experiments by two different researchers, was used to automatically identify specific neuronal subtypes. Only those structures selected by both researchers were used, and Sholl plots of these neurons were created to generate a model function, utilizing a polynomial fit. Respective y-values were calculated for each ring distance and compared to the expected y-values for each neuronal structure. The sum of squared errors (SSE) was calculated to estimate how similar the Sholl plot of an individual neuron was compared to the model function. This method was first applied to the gold-standard to estimate the SSE by calculating the median and standard deviation. The median plus one standard deviation served as a boundary for similar neurons. The complete source code as well as sample image stacks are provided in supplementary material.
2.5. Statistical analyses
Statistics were performed using GraphPad Prism 7 (San Diego, CA, USA). Data in each experimental group were obtained from 3–4 independent dissections and were plotted as the mean ± standard error of the mean (SEM). Each data set was tested for normal distribution using the D’Agostino-Pearson omnibus and the Shapiro-Wilk normality test. Normal data were analyzed by comparison of two groups using an unpaired t-test with a post hoc Sidak test (p < 0.05). Data that did not pass normality tests were compared using the Mann-Whitney test with a post hoc Sidak test (p < 0.05). Differences in variance were determined using the F-test
3. Results
3.1. Automated thresholding of images of primary mouse neurons reduced inter-researcher variability
Manual estimations of thresholds using ImageJ by two researchers using images from the same set of primary mouse neuronal cell cultures resulted in very different binary images (Fig. 2a). For example, the average AUC of binary images was significantly higher for researcher 1 compared to researcher 2 (Fig. 2b). Thresholding not only affected obtained areas, but was also crucial for excluding background signal, which can negatively influence morphological endpoints and Sholl analysis. A challenge to thresholding neuronal cells is variability in fluorescence intensity throughout the dendritic arbor of a single cell (i.e., the fluorescent signal is typically very bright in the cell soma and proximal dendritic branches, but very dim in the distal dendritic arbor). An additional challenge is staining imperfections that result in gaps after thresholding. Therefore, thresholding using a fixed value is a trade-off between maintaining dim structures and not introducing too much background signal to the binary image. For Sholl analysis, background structures can lead to artificial intersections (Fig. 2c), resulting in different Sholl plots (Fig. 2d) and a significant difference in AUC, Peak X (the radius at which the largest number of intersections occurs in Sholl analysis) and Y (the highest number of intersections that occurs at any radius in Sholl Analysis) for manual evaluation as occurred between the two researchers for different populations of mouse neurons (experimental groups A and B).
Figure 2. Automated thresholding reduces inter-researcher variability in mouse neuronal cell cultures.
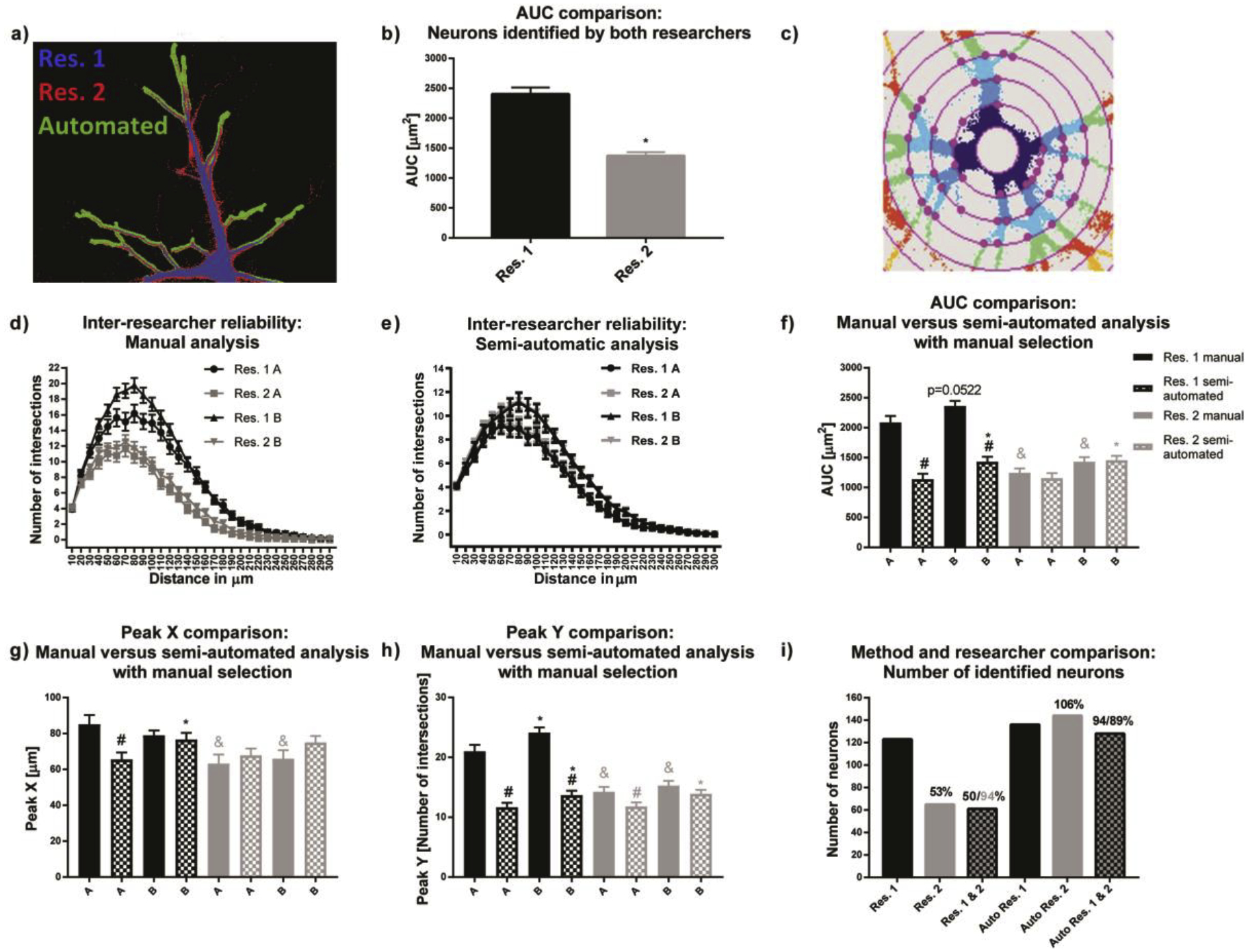
(a) Overlay of the same neuron manually thresholded by two researchers (blue and red) and the automated thresholding (green). b) respective average neuronal areas of all neurons of the experiment. (c) Imperfect thresholding introduces artificial intersections when background structures are located on the ring mask in the ImageJ Sholl Analysis plugin (Ferreira et al., 2014, Nature Methods 11(10): 982–984). (d) As a result, there is a high inter-researcher variability in the Sholl plots and (e) significant inter-researcher variability for the area under the curve (AUC) for the same set of images. (e) Automated thresholding significantly reduced inter-researcher variability and (f-h) delivered results comparable to those obtained using manual evaluation by researcher two (Res. 2). (i) The reason for reduced inter-researcher variability was that with automated thresholding, the overlap between selected neurons was 89–94% compared to 50–94% overlap between the two researchers using the manual approach. Data are shown as mean ± SEM (n=4 independent dissections). *Significantly different (p<0.05) between experimental groups for the same researcher, # between evaluation methods for the same researcher and & for the same experimental group and evaluation method between different researchers as determined using unpaired student’s t-test.
Using the semi-automated approach (manual neuron selection, but automated thresholding) significantly reduced inter-researcher differences in Sholl plots (Fig. 2e). Comparison of AUC, Peak X and Y (Fig. 2f–h, respectively) revealed identical statistical differences between both experimental groups, but no statistical significances between both researchers. Furthermore, results from the semi-automated analysis were highly comparable to those of the manual analysis of researcher 2, as indicated by the absence of statistical difference between methods except for Peak Y (Fig. 2h). Only researcher 1 found significant differences between group A and B and only for Peak Y manual evaluation.
Researcher results with the semi-automated analysis are most likely more similar than with the manual approach because of a high overlap in neuron selection (94%/89%) compared to only a 50% overlap with manual analysis (Fig. 2i), and due to the fact that automated thresholding estimates thresholding biases. Because inter-researcher variance is a known pitfall of morphological analyses, each experiment was manually analyzed by the same person to minimize user-introduced thresholding variability.
3.2. Automated neuronal thresholding of neuronal images from primary rat neuronal cell cultures introduced inter-method variation
Using ImageJ to manually threshold neuronal images from primary rat neuronal cell cultures resulted in smaller differences between the results of the two researchers compared to manual thresholding of neuronal images from primary mouse neuronal cell cultures (see overlays in Fig. 2a vs. Fig. 3a). Still, a significant difference was observed for AUC values between the two researchers (Fig. 3b, c). Similarity of results with the rat data may be due to a higher signal-to-noise ratio in images of transfected rat neurons. Both researchers detected significant differences between experimental groups C and D, as shown by Sholl analysis (Fig. 3d); however, the absolute peak height for the respective conditions was higher for researcher 1 compared to researcher 2 (Fig. 3f). However, since the trend of differences in Sholl curve heights between experimental conditions was preserved for both researchers, significant differences for AUC, Peak X and Peak Y were found between experimental groups C and D by both researchers (Fig. 3f, g, h). Due to the differences in the height of the Sholl curves, there was a statistically significant difference between researchers for AUC, Peak X, and Peak (Fig. 3f, g, h).
Figure 3. Automated thresholding did not reduce inter-researcher variability in rat neuronal cultures.
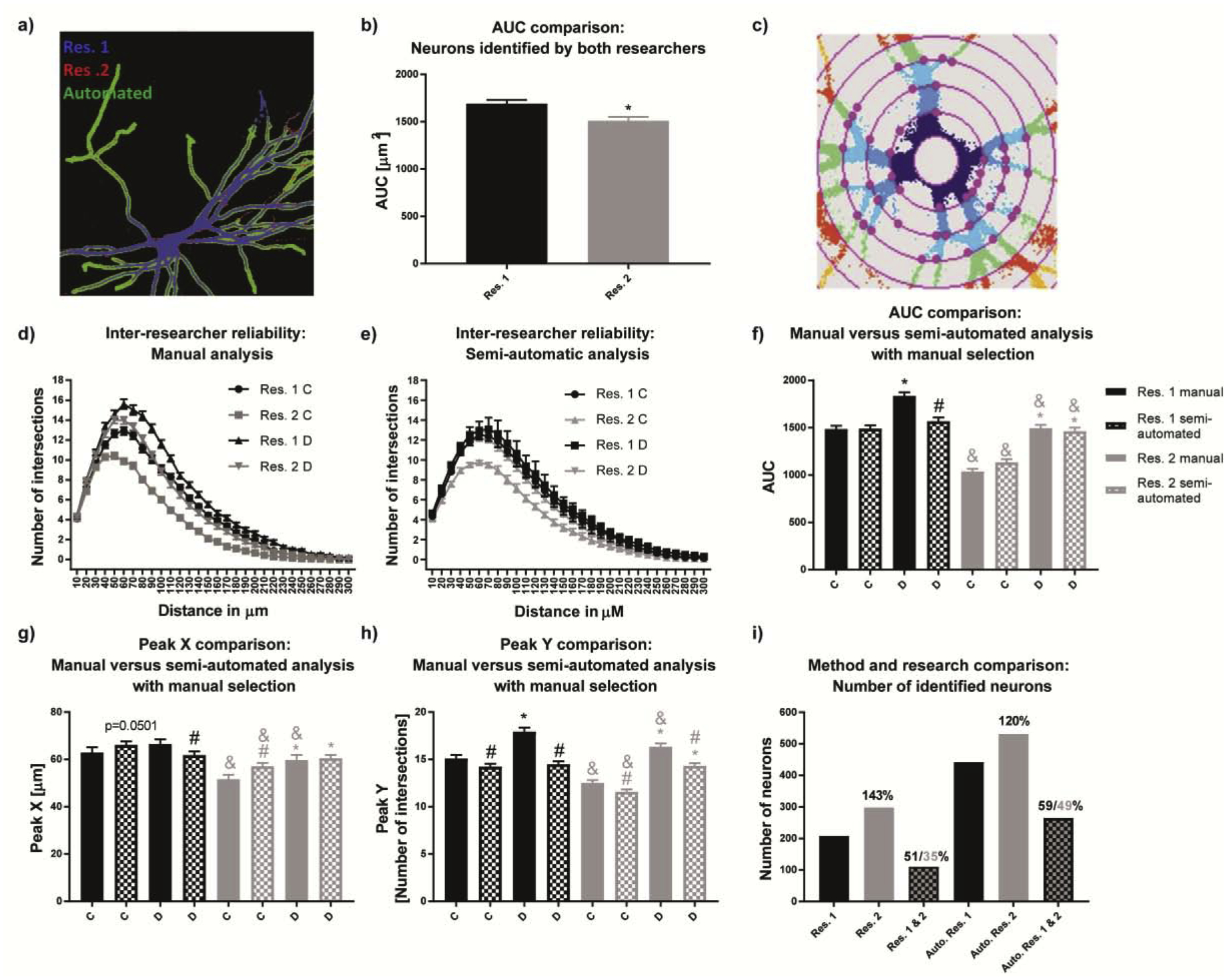
(a) The overlay of two manually thresholded rat neurons (blue and red) and the automated thresholding (green). b) the inter-researcher variability in neuronal area, and (c) artificial intersections from imperfect thresholding. (d) Sholl plots of primary rat neurons manually thresholded and (e) semi-automated analysis (manual selection but automated thresholding) reveal inter-researcher and inter-methodological differences. These differences are reflected in the (f) AUC, (g) Peak X and (h) Peak Y. (i) Inter-researcher variance originated from low overlap of manually selected neurons of 35–59%. Data are shown as mean ± SEM (n=3 independent dissections). *Significantly different (p<0.05) between experimental groups for the same researcher, # between evaluation methods for the same researcher, & for the same experimental group and evaluation method between different researchers as determined using unpaired student’s t-test.
The semi-automated approach (manual neuron selection but automated thresholding) did not negate differences between researchers. However, the effects between experimental groups completely disappeared in the Sholl plot for researcher 1 (Fig. 3e). Although this was not observed for researcher 2, the overall peak height of experimental condition was comparable between the two researchers. As a result, significant differences between the two researchers for AUC, Peak X and Peak Y were only found for experimental group C (Fig. 3f, g, h, respectively).
AUC and Peak Y manual ImageJ analyses of both researchers (Fig 3f, h) revealed significant differences between experimental groups, but only for researcher 2 using the semi-automated approach. For Peak X, only researcher 2 found significant differences using either manual or semi-automated analysis (Fig. 3g). The differences observed in the numbers of selected neurons between manual and semi-automatic method were much more pronounced in rats than in mice (Fig. 2i), which is attributed to a higher transfection efficacy in rat than in mouse. While in mouse the total number of neurons is usually around 80–100, there can be found almost 10-fold more rat neurons. For both researchers, the number of primary rat neurons chosen using the semi-automated approach was roughly 2-fold higher compared to the manual assessment (Fig. 3i). The overlap between the two researchers was 51% towards researcher 1 and 35% towards researcher 2 for manual assessment, and 59% towards researcher 1 and 49% towards researcher 2 for the semi-automated analysis (Fig. 3i). Collectively, inter-researcher differences using the semi-automated analysis can be attributed to different selection of neurons by the researchers, with researcher 2 choosing larger and more branched neurons.
3.3. Automated neuronal selection reduces inter-researcher variability in analysis of neuronal images from primary rat neuronal cell cultures
To overcome selection bias, a training set of rat neuronal structures from the same image set were manually selected by both researchers. Sholl curves were generated (Fig. 4a), and the mean and standard deviation of the sum of squared errors (SSE) of each neuron in the image set was used to calculate a Fit curve. Neurons with an SSE smaller than the mean plus one standard deviation of the training data were considered similar to the training set Fit curve and included in the analysis (Fig. 4a: blue circles represent neurons with high similarity to the fitted curve, red squares represent neurons with low similarity). When the same criteria were applied to the entire image set, researcher 1’s semi-automated analysis was similar to the fully automated process (Sholl plots, Fig. 4b). Unlike the semi-automated approach, the fully automated approach resulted in no significant differences between the experimental groups for AUC, Peak X and Y (Fig. 4c, d, e, respectively). Differences for AUC, Peak X and Peak Y were more pronounced with the semi-automated approach of researcher 2 than 1, suggesting a difference in neuron selection.
Figure 4. Automated neuron selection decreases researcher bias in rat neuronal cultures.
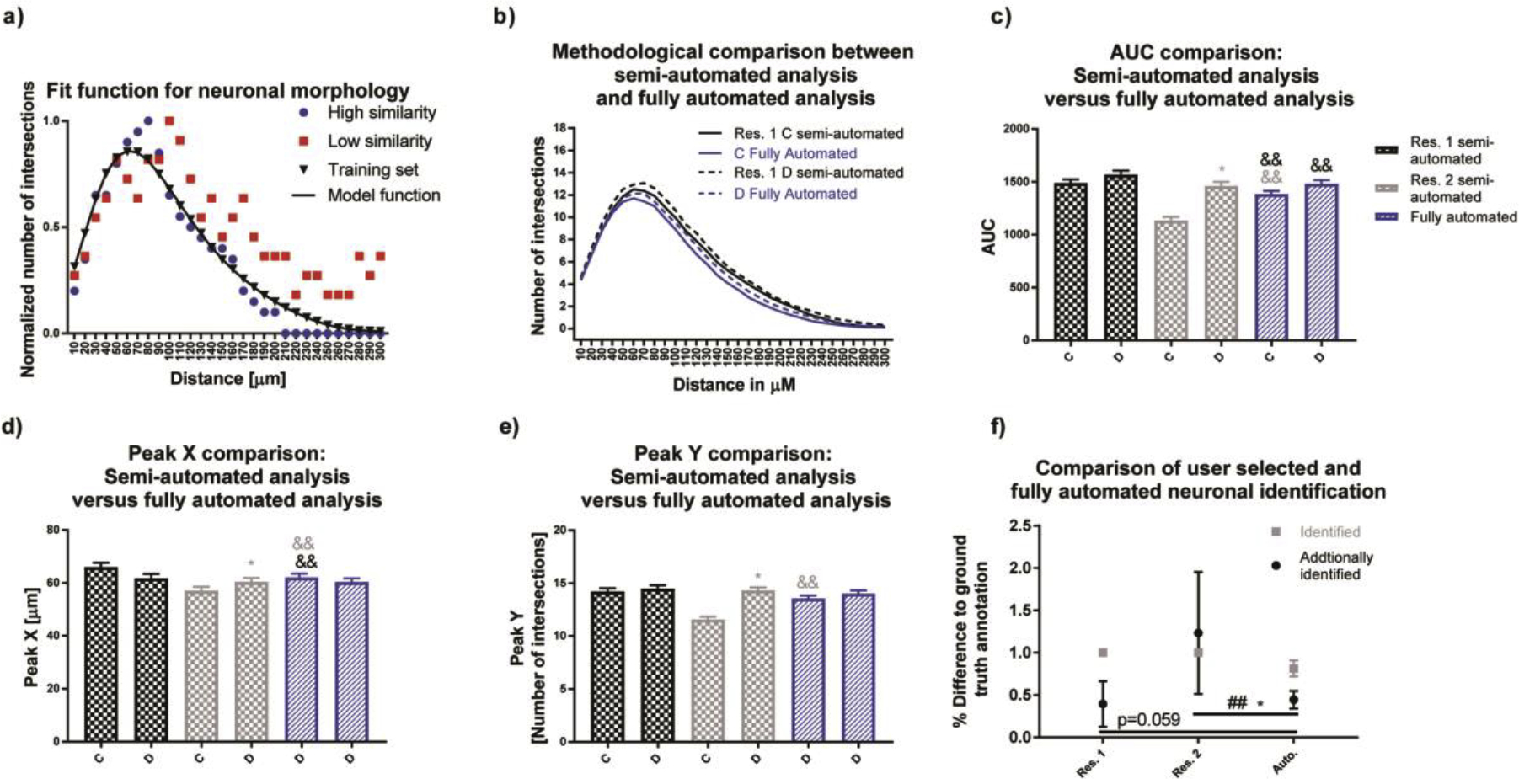
(a) Using neurons identified by both researchers, a Fit curve was generated and used to calculate the sum of squared errors (SSE) for individual neurons contained in this data set. We considered neurons if their SSE was smaller than the mean plus one standard deviation of our training data set. (b-e) Data generated using this fully automated method were more similar to researcher 1 than researcher 2. (f) The automated method was able to identify 81% of neurons from the ground truth annotation and additional identified neurons percentages were comparable to researcher one with a significantly lower variance than researcher 2. Data are shown as mean ± SEM (n=3 independent dissections). *Significantly different (p<0.05) between experimental groups for the same researcher, && significant differences between the fully automated method versus semi-automated method as determined using unpaired student’s t-test. ## Significant differences between variations as determined using the F-test.
The percentage of overlap for semi-automated and fully automated methods was determined using the training set. Since the training set was derived from the combined neuron selection of the two researchers, both researchers identified 100% of the training set (Fig. 4f, gray squares for researchers). The fully automated method identified 81% of the neurons (gray square for Auto), selected by both researchers. Both researchers and the fully automated method identified additional neurons (Fig. 4f, black circles), with researcher 2 having the highest additional identification. The fully automated method possessed the lowest variance towards the training set, with significantly lower variability than researcher 2 (Fig. 4f).
3.4. Morphometric analysis of mouse and rat cortical neurons reflects inter-researcher differences observed Sholl analysis and are comparable using semi-automated analysis for mouse and fully automated analysis for rat
Morphological parameters of neurite mass, number of primary dendrites, total dendritic length, number of dendritic branch points and number of terminal dendritic tips were obtained from the skeleton binary image. Inter-researcher variability occurred with manual evaluation of both experimental groups of mouse neurons, as demonstrated by researcher 1’s finding of a significantly higher neurite mass than researcher 2 (Fig. 5a).
Figure 5. Morphological endpoints analyzed by the semi-automatic approach.
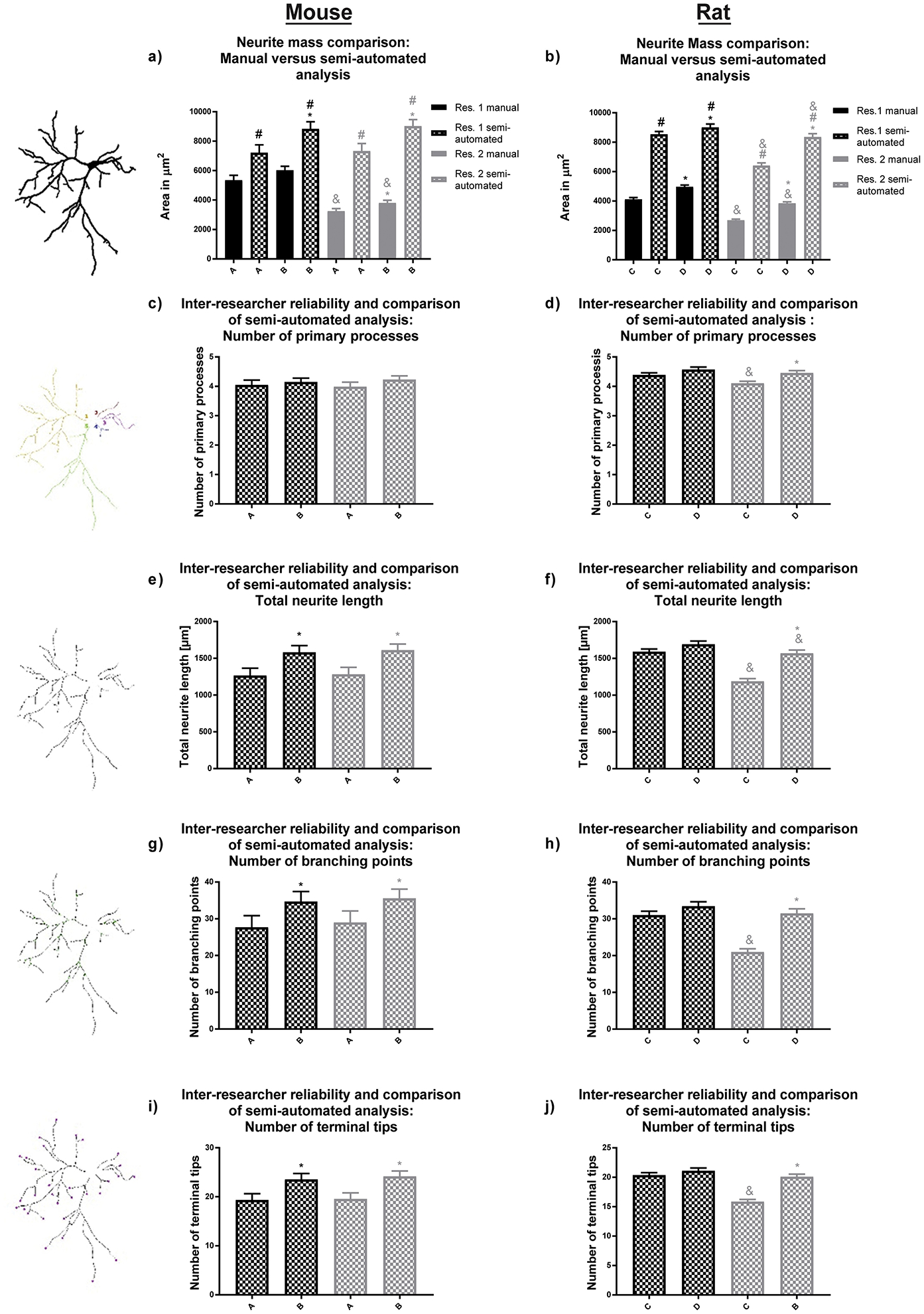
(a, b) Our software is capable of analyzing neural mass (area of the entire neuronal structure), (c, d) the number of primary processes, (e, f) total dendritic length, (g, h) number of dendritic branch points, and (i, j) number of terminal dendritic tips for both primary mouse hippocampal and rat cortical neurons. Experimental groups A and B for mouse and groups C and D for rat neuronal cultures were analyzed by two independent researchers. Data are shown as mean ± SEM (n=3 (rat) or 4 (mouse) independent dissections per condition). *Significantly different (p<0.05) between experimental groups for the same researcher; # between evaluation methods for the same researcher, & for the same experimental group and evaluation method between different researchers, using unpaired student’s t-test.
Both researchers found a significant difference between the two experimental groups for neurite mass, total dendritic length, number of branches and terminal tips using the semi-automated approach (Fig. 5a, e, g, i, respectively). No statistically significant difference was observed by the researchers when they evaluated the number of mouse primary processes using the semi-automated approach (Fig. 5c). This reassembles the results shown in Fig. 2, since the differences in Sholl plots were found to be attributed to the difference in thresholding and thus to obtained cell skeletons, which are used to calculate the morphometric attributes neurite length, branching and number of terminal tips.
Analysis of total dendritic mass in rat neurons revealed statistically significant higher values for semi-automated analysis compared to the manual analysis for both researchers and experimental groups (Fig. 5b). Comparable to the endpoints obtained by Sholl analysis, researcher 2 alone found significant differences for the number of primary processes, total neurite length, number of branching points and number of terminal tips between the experimental conditions (Fig. 5f, h, j). It is important to note that those endpoints were only evaluated utilizing the semi-automated approach. These variances can therefore be explained by the selection of neurons.
The fully automated analysis showed no differences between the experimental groups for any of the analyzed morphological endpoints (Fig. 6a–e). Neither the semi-automated approach of researcher 1 nor the fully automated approach revealed significant differences among experimental groups. Finally, we determined the accuracy of the training set’s produced by researchers 1 and 2 by examining the difference of the fully automated approach to the direct observations made by the researchers (ground truth annotation) (Cardoso et al., 2014; Wiesmann et al., 2017). Significant differences in neuron selection were observed between researcher 2 and the fully automated approach. Similarly, data variance was found significant between the fully automated approach and researcher 2, but not researcher 1. The fully automated method was able to identify approximately 80% of the neurons most comparable with researcher 1.
Figure 6. Morphological endpoints analyzed by the fully automated approach.
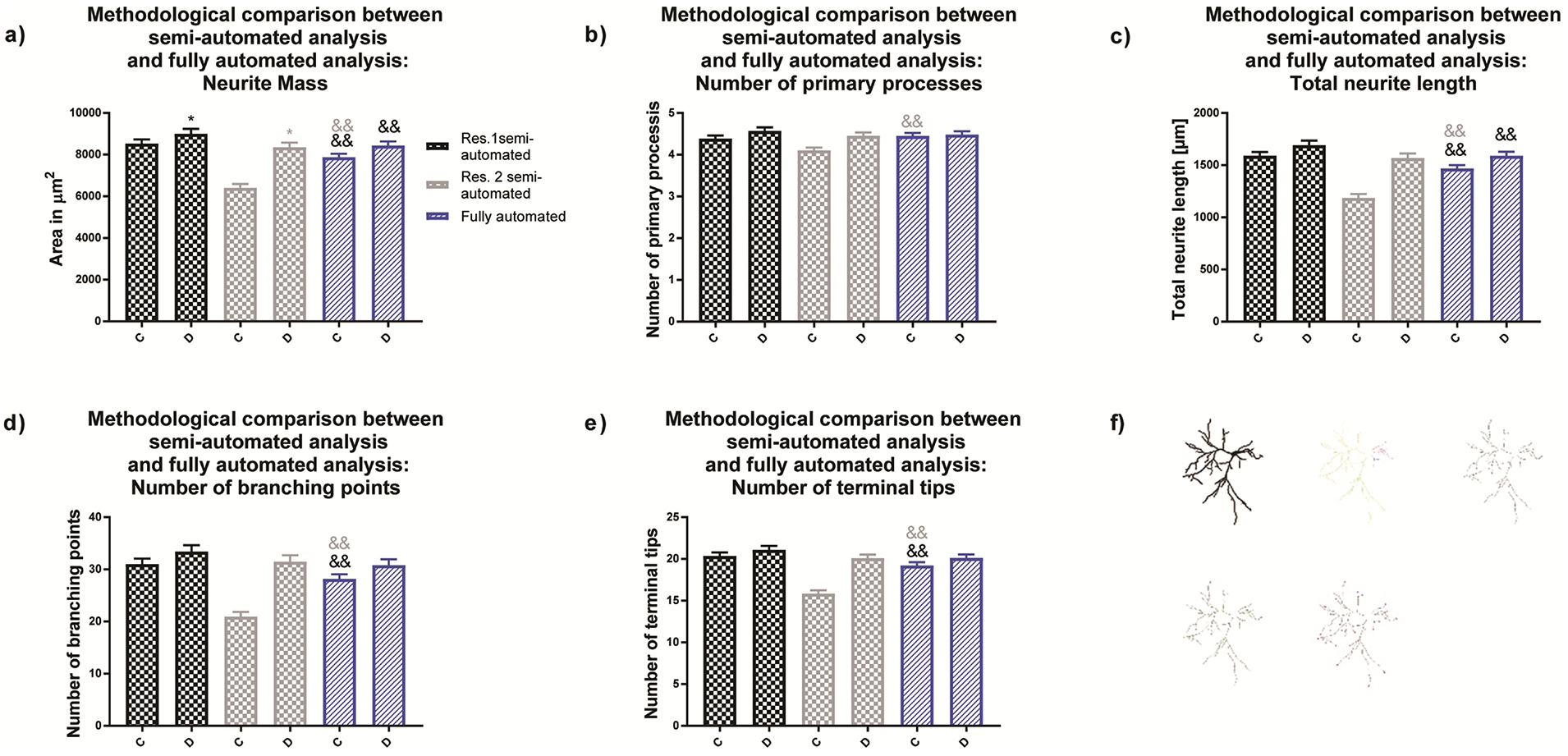
(a) Comparison of neural mass (area of the entire neuronal structure, (b) number of primary processes, (c) total dendritic length, (d) number of dendritic branch points and (e) number of terminal dendritic tips for primary rat cortical neurons obtained by (f) semi-automated and fully automated analysis. Data are shown as mean ± SEM (n=3 independent dissections per condition). *Significantly different (p<0.05) between experimental groups for the same researcher and && semi-automated method vs. fully automated method as determined using unpaired student’s t-test.
4. Discussion
Immunocytochemical approaches for visualizing the dendritic arbor of neurons in cell culture create challenges for analyzing dendritic morphology. When plated at high densities to better reflect the in vivo situation, the dendritic arbors of individual neurons can be more readily visualized by transfection of neurons with cDNA encoding fluorescently-tagged MAP2B. Because lipid-based transfection efficiencies range from 5–30% in primary cultured neurons (Ohki et al., 2001), only a small percentage of the neuronal cell population is labeled. While entire dendritic arbors of individual cells can be seen in fluorescent images (Wayman et al., 2012a; Wayman et al., 2006; Wayman et al., 2012b), overlapping dendritic arbors are a major issue and isolated neurons still need to be manually extracted. Thresholding dendritic arborization images is quite difficult and requires manual annotation until now.
Modern image analysis tools have increased the number of features that can be rapidly extracted from cells, allowing for a more comprehensive evaluation of cell morphology (Ho et al., 2011; Kamentsky et al., 2011; Long et al., 2017; Pool et al., 2008; Schmuck et al., 2017; Yu et al., 2009). However, for dendritic tracings, almost all of the available algorithms rely on manual annotations or parameter settings (Ferreira et al., 2014; Ho et al., 2011; Kamentsky et al., 2011; Pool et al., 2008; Yu et al., 2009), which can introduce unintentional variability or bias. Traditional endpoints like dendritic length, number of primary dendrites, number of dendritic branch points and number of terminal dendritic tips are assessed by first removing the cell soma followed by skeletonizing of dendritic structures. Pixels of the resulting dendritic skeleton can be analyzed in reference to neighboring cells to determine whether they are starting points or terminal tips, branching points, crossing points or line points (Ferreira et al., 2014). Fully automated skeletonizing algorithms rely on a good signal-to-noise ratio and often lose distal dendrites that are dimly fluorescent, and introduce gaps due to staining imperfections (Long et al., 2017). Therefore, computer-assisted manual tracing tools, such as NeuronJ and NeuronCyto, are often used (Pool et al., 2008; Yu et al., 2009), but they are extremely time-consuming. Thus, the number of cells that can be analyzed is limited within typical time constraints.
Another widely used method for analyzing dendritic complexity is Sholl analysis, for which a completely automated method has been lacking to date. While the Sholl plugin in ImageJ is frequently used for Sholl analysis (Ferreira et al., 2014), it requires multiple user interactions. First, thresholding has to be performed by the user, and the center of the cell soma has to be marked. Subsequently, the plugin fits concentric rings around neuron soma and counts the number of intersections of dendritic structures with each ring. The outcome is highly dependent on the applied threshold. For example, the lower the threshold, the higher the numbers of intersections, since more parts of the cell remain intact. At the same time background signal will be added to the binary image (Fig. 2a, c) due to a lower signal-to-noise ratio (Fig. 2b). Since this method is also time consuming, only a subset of cells acquired with a slide scanner can be analyzed, which increases the possibility of researcher bias in cell selection. Bias occurred for both species (mouse and rat) investigated here (Fig. 2i, Fig. 3i). Therefore, a software solution that can identify neurons, analyze morphological characteristics and perform Sholl analysis is highly desirable to eliminate potential user bias and to minimize manual annotation time.
The newly introduced extension to the Omnisphero software eliminates user interaction, automatically assesses morphological features, and performs Sholl analysis, after labeling an initial data set for similarity. This step has to be performed only once or when image acquisition, staining or cell type changes. Here, we described two experimental sets of neurons used to recapitulate common challenges to neuron analysis such as high signal-to-noise ratios (i.e., difficulty in thresholding), and selection of neurons (i.e., dealing with overlapping dendritic arbors of neighboring neurons).
With the mouse neurons, a significant difference was observed between manual researcher results for average dendritic area and Sholl plots. The differences are most likely due to the low signal-to-noise ratio of the images and dissimilar thresholding between researchers. High thresholds eliminate dim structures, and low thresholds capture background information. Due to the low signal-to-noise ratio, it is not surprising that the researchers’ results varied with regards to the obtained dendritic area because small changes in thresholding values result in large increases in area. Automated analysis completely removed inter-experimenter differences in Sholl plots of the mouse neurons (Fig. 2e), and led to the same significant difference between experimental groups A and B for AUC (Fig. 2f) and Peak Y (Fig. 2h) for both researchers.
With the rat cortical neurons, the two researchers still produced significantly different results for AUC even though the signal-to-noise ratio was better than with the mouse neurons. Semi-automated thresholding did not deliver comparable Sholl plots, as was observed for mouse neurons, but led to a loss of effect for researcher 2 compared to researcher 1 (Fig. 3e). Most likely, the two researchers selected different objects because thresholding of the same cells with the automated approach delivered the same values. Rat neurons are transfected with a much higher efficacy compared to mouse neurons (10-fold), which leads to strong selection bias for both the Fiji App as well as Omnisphero.
Automated selection of neurons based on a training set derived by both researchers revealed that thresholding (in the case of mouse) and selection criteria (in the case of rat) of different researchers can highly influence conclusions. Therefore, a fully unbiased method is desirable to obtain higher reproducibility among different researchers (Versi, 1992). The training set used in this study was derived from three experiments, but can be continuously enlarged with new manual annotations of neurons. Upon collecting 2000 to 5000 annotated neurons, the SSE approach could be supplemented with artificial intelligence solutions (U_Net (Ronneberger et al., 2015) for segmentation and transfer learning from pretrained networks such as resnets for classification (He et al., 2015) to get even more robust automated selection criterions (Shen et al., 2017).
While Sholl analysis offers a graphical representation of altered dendritic arborization, the reason for the alteration is not easily extractable. For example, the number of intersections is dependent on the number of primary processes, the number of branching points or a combination of both. Therefore, analysis of morphological characteristics, such as total dendritic length, number of primary processes, number of branch points and number of terminal tips, is essential to further understand the effect of experimental manipulations on dendritic arborization. The new extension of Omnisphero allows a fast and unbiased analysis of dendritic morphology. Novel aspects compared to existing methods are complete automated identification of neuronal subtypes, automated thresholding, identification of cell somata and skeletonization, allowing or an implementation into a fully automated screening cascade. This method will increase throughput, the number of cells analyzed, and the robustness of observed effects. To date, this automated method has markedly decreased time associated with data analysis and improved experimental quality in ongoing neurotoxicity screens in our laboratory.
Supplementary Material
Highlights.
OmniBrain performs manual, semi- and fully automated Sholl analysis
Manual cell selection and tracing increases inter-researcher variability
OmniBrain eliminated researcher bias from neuron selection and image thresholding
OmniBrain provides medium to high throughput analysis of dendritic morphology
Acknowledgements
This research was supported by the National Institutes of Health [grants R01 ES014901 to PJL, K99 ES029537 to KPK, F32 HD088016 to KPK, T32 ES007059 predoctoral fellowship to SS, P30 ES023513, P01 ES011269, and U54 HD079125].
Abbreviations
- AUC
area under the curve in Sholl Analysis
- GUI
graphical user interface
- MAP2
microtubule associated protein 2
- Peak X
the radius at which the largest number of intersections occurs in Sholl analysis
- Peak Y
the highest number of intersections that occurs at any radius in Sholl Analysis
- SEM
standard error of the mean
- SSE
sum of squared errors
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bourgeron T A synaptic trek to autism. Curr Opin Neurobiol, 2009; 19: 231–4. [DOI] [PubMed] [Google Scholar]
- Cardoso JR, Pereira LM, Iversen MD, Ramos AL. What is gold standard and what is ground truth? Dental press journal of orthodontics, 2014; 19: 27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cochran JN, Hall AM, Roberson ED. The dendritic hypothesis for Alzheimer’s disease pathophysiology. Brain Res Bull, 2014; 103: 18–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira TA, Blackman AV, Oyrer J, Jayabal S, Chung AJ, Watt AJ, Sjostrom PJ, van Meyel DJ. Neuronal morphometry directly from bitmap images. Nat Methods, 2014; 11: 982–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda T, Itoh M, Ichikawa T, Washiyama K, Goto Y. Delayed maturation of neuronal architecture and synaptogenesis in cerebral cortex of Mecp2-deficient mice. J Neuropathol Exp Neurol, 2005; 64: 537–44. [DOI] [PubMed] [Google Scholar]
- Garey L When cortical development goes wrong: schizophrenia as a neurodevelopmental disease of microcircuits. J Anat, 2010; 217: 324–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He K, Zhang X, Ren S, Sun J. Deep Residual Learning for Image Recognition. 2015; arXiv:1512.03385. [Google Scholar]
- Ho SY, Chao CY, Huang HL, Chiu TW, Charoenkwan P, Hwang E. NeurphologyJ: an automatic neuronal morphology quantification method and its application in pharmacological discovery. BMC bioinformatics, 2011; 12: 230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamentsky L, Jones TR, Fraser A, Bray MA, Logan DJ, Madden KL, Ljosa V, Rueden C, Eliceiri KW, Carpenter AE. Improved structure, function and compatibility for CellProfiler: modular high-throughput image analysis software. Bioinformatics, 2011; 27: 1179–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keil KP, Sethi S, Wilson MD, Chen H, Lein PJ. In vivo and in vitro sex differences in the dendritic morphology of developing murine hippocampal and cortical neurons. Sci Rep, 2017; 7: 8486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keown CL, Shih P, Nair A, Peterson N, Mulvey ME, Muller RA. Local functional overconnectivity in posterior brain regions is associated with symptom severity in autism spectrum disorders. Cell Rep, 2013; 5: 567–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutzing MK, Langhammer CG, Luo V, Lakdawala H, Firestein BL. Automated Sholl analysis of digitized neuronal morphology at multiple scales. J Vis Exp, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kweon JH, Kim S, Lee SB. The cellular basis of dendrite pathology in neurodegenerative diseases. BMB Rep, 2017; 50: 5–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libersat F, Duch C. Mechanisms of dendritic maturation. Mol Neurobiol, 2004; 29: 303–20. [DOI] [PubMed] [Google Scholar]
- Long BL, Li H, Mahadevan A, Tang T, Balotin K, Grandel N, Soto J, Wong SY, Abrego A, Li S, Qutub AA. GAIN: A graphical method to automatically analyze individual neurite outgrowth. Journal of neuroscience methods, 2017; 283: 62–71. [DOI] [PubMed] [Google Scholar]
- Meijering E, Jacob M, Sarria JC, Steiner P, Hirling H, Unser M. Design and validation of a tool for neurite tracing and analysis in fluorescence microscopy images. Cytometry A, 2004; 58: 167–76. [DOI] [PubMed] [Google Scholar]
- Menon S, Gupton SL. Building Blocks of Functioning Brain: Cytoskeletal Dynamics in Neuronal Development. Int Rev Cell Mol Biol, 2016; 322: 183–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobis M, Hunziker U. Automatic thresholding for hemispherical canopy-photographs based on edge detection. Agricultural and Forestry Meterology, 2005; 128: 243–50. [Google Scholar]
- Ohki EC, Tilkins ML, Ciccarone VC, Price PJ. Improving the transfection efficiency of post-mitotic neurons. Journal of neuroscience methods, 2001; 112: 95–9. [DOI] [PubMed] [Google Scholar]
- Pool M, Thiemann J, Bar-Or A, Fournier AE. NeuriteTracer: a novel ImageJ plugin for automated quantification of neurite outgrowth. Journal of neuroscience methods, 2008; 168: 134–9. [DOI] [PubMed] [Google Scholar]
- Ronneberger O, Fischer P, Brox T. U-Net: Convolutional Networks for Biomedical Image Segmentation. 2015; arXiv: 1505.04597.[cs.CV]. [Google Scholar]
- Schmuck MR, Temme T, Dach K, de Boer D, Barenys M, Bendt F, Mosig A, Fritsche E. Omnisphero: a high-content image analysis (HCA) approach for phenotypic developmental neurotoxicity (DNT) screenings of organoid neurosphere cultures in vitro. Arch Toxicol, 2017; 91: 2017–28. [DOI] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods, 2012; 9: 671–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott EK, Luo L. How do dendrites take their shape? Nat Neurosci, 2001; 4: 359–65. [DOI] [PubMed] [Google Scholar]
- Sethi S, Keil KP, Chen H, Hayakawa K, Li X, Lin Y, Lehmler HJ, Puschner B, Lein PJ. Detection of 3,3’-Dichlorobiphenyl in Human Maternal Plasma and Its Effects on Axonal and Dendritic Growth in Primary Rat Neurons. Toxicol Sci, 2017; 158: 401–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen D, Wu G, Suk HI. Deep Learning in Medical Image Analysis. Annu Rev Biomed Eng, 2017; 19: 221–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sholl DA. Dendritic organization in the neurons of the visual and motor cortices of the cat. J Anat, 1953; 87: 387–406. [PMC free article] [PubMed] [Google Scholar]
- Supekar K, Uddin LQ, Khouzam A, Phillips J, Gaillard WD, Kenworthy LE, Yerys BE, Vaidya CJ, Menon V. Brain hyperconnectivity in children with autism and its links to social deficits. Cell Rep, 2013; 5: 738–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Versi E. “Gold standard” is an appropriate term. BMJ, 1992; 305: 187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wayman GA, Bose DD, Yang D, Lesiak A, Bruun D, Impey S, Ledoux V, Pessah IN, Lein PJ. PCB-95 modulates the calcium-dependent signaling pathway responsible for activity-dependent dendritic growth. Environmental health perspectives, 2012a; 120: 1003–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wayman GA, Impey S, Marks D, Saneyoshi T, Grant WF, Derkach V, Soderling TR. Activity-dependent dendritic arborization mediated by CaM-kinase I activation and enhanced CREB-dependent transcription of Wnt-2. Neuron, 2006; 50: 897–909. [DOI] [PubMed] [Google Scholar]
- Wayman GA, Yang D, Bose DD, Lesiak A, Ledoux V, Bruun D, Pessah IN, Lein PJ. PCB-95 promotes dendritic growth via ryanodine receptor-dependent mechanisms. Environmental health perspectives, 2012b; 120: 997–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiesmann V, Bergler M, Palmisano R, Prinzen M, Franz D, Wittenberg T. Using simulated fluorescence cell micrographs for the evaluation of cell image segmentation algorithms. BMC bioinformatics, 2017; 18: 176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W, Lee HK, Hariharan S, Bu W, Ahmed S. Quantitative neurite outgrowth measurement based on image segmentation with topological dependence. Cytometry A, 2009; 75: 289–97. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


