Abstract
The HIV intasome is a large nucleoprotein assembly that mediates the integration of a DNA copy of the viral genome into host chromatin. Intasomes are targeted by the latest generation of antiretrovirals, integrase (IN) strand transfer inhibitors (INSTIs). Challenges associated with lentiviral intasome biochemistry have hindered high-resolution structural studies of how INSTIs bind to their native drug target. Here, we present high-resolution cryo-electron microscopy (cryo-EM) structures of HIV intasomes bound to the latest generation INSTIs. These structures highlight how small changes in the IN active site can have significant implications for drug binding and design and provide mechanistic insights into why a leading INSTI retains efficacy against a broad spectrum of drug resistant variants. The data have implications for expanding effective treatments available for HIV-infected individuals.
One Sentence Summary
High-resolution cryo-EM structures of the latest generation antiretroviral drugs bound to HIV intasomes have implications for drug design.
The human immunodeficiency virus (HIV) currently infects ~40 million people worldwide. The virus’s ability to integrate a viral DNA (vDNA) copy of its RNA genome into host chromatin, leading to the establishment of a permanent and irreversible infection of the target cell (and any progeny cells), is the central challenge in developing a cure (1). Integration, catalyzed by the viral integrase (IN) protein, is essential for retroviral replication and results in the covalent linkage of vDNA to the host genome (2, 3). Proper integration depends on the formation of a large oligomeric nucleoprotein complex containing viral IN assembled on the ends of vDNA, commonly referred to as an intasome (4–9). All intasomes contain multimeric IN bound to vDNA ends, but they are characterized by distinct oligomeric configurations and domain arrangements.
Intasome assembly and catalysis proceeds through a multi-step process that involves several distinct intermediates (fig. S1). The catalytically competent cleaved synaptic complex (CSC) intasome, which contains free 3′-OH ends, is the specific target of the IN strand transfer inhibitors (INSTIs), a group of drugs that bind to both the active site of HIV IN and the ends of vDNA, blocking catalysis. Treatment with INSTIs, which are a key component of combined antiretroviral therapy, leads to a rapid decrease in viral load in patients. INSTIs are generally well tolerated, and the second generation drugs do not readily select for resistance (10–13). They are used in the recommended first line combination therapies for treating HIV-infected patients and are prime candidates for future development (14, 15).
The prototype foamy virus (PFV) intasome has been used as a model system to understand INSTI binding (6, 16–19). However, this system has limitations. PFV and HIV INs share only ~25% sequence identity in the catalytic core domain (CCD) (6), and many of the sites where drug resistance mutations occur in HIV IN are not conserved in PFV IN. Moreover, minor changes in the structure of an INSTI can profoundly impact its ability to inhibit mutant forms of HIV (19, 20). Thus, understanding how INSTIs interact with HIV intasomes – their natural target – at a molecular level is needed to overcome drug resistance and to guide development of improved inhibitors.
We established conditions for assembling, purifying, and structurally characterizing HIV CSC intasomes. Previously, we showed that fusion of the small protein Sso7d to the N-terminal domain (NTD) of HIV IN improves its solubility and facilitates assembly and purification of strand transfer complex intasomes (4, 21). We further optimized conditions required for CSC formation and purification and showed that these complexes are biochemically active for concerted integration (fig. S2). We used a tilted cryo-EM data collection strategy to alleviate the effects of preferential specimen orientation on cryo-EM grids (22), allowing us to collect data on the apo form of the HIV CSC intasome. The cryo-EM reconstruction of the HIV CSC intasome reveals a two-fold symmetric dodecameric molecular assembly of IN. The highest resolution (~2.7 Å) resides within the core containing the two catalytic sites and the ends of vDNA (fig. S3 and Table S1).
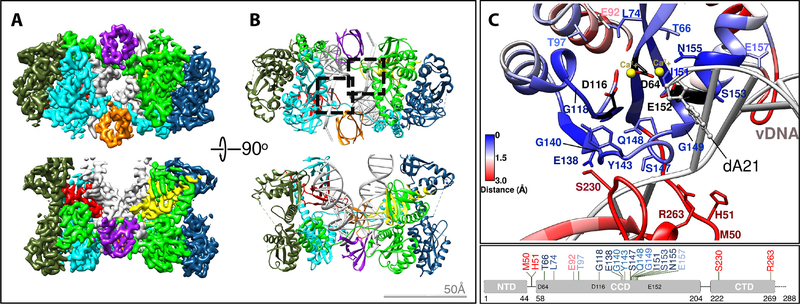
Lentiviral intasomes have a large degree of heterogeneity and vary in size depending on the protein and biochemical conditions, forming tetramers, dodecamers, hexadecamers, and proto-intasome stacks (fig. S4–5). The basic underlying unit, the conserved intasome core (CIC), resembles, but is not identical to, the tetrameric PFV intasome. The CIC is composed of two IN dimers, each of which binds one vDNA end and a C-terminal domain (CTD) from a neighboring protomer (23). In the cryo-EM reconstruction, four fully defined IN protomers, two CTDs from flanking protomers, and two additional CTDs from distal subunits are clearly resolved (Fig. 1A); these were used to build an atomic model (Fig. 1B). With the exception of the additional CTDs from distal subunits, which are not conserved in other retroviral species, the resolved regions comprise the intasome CIC.
Fig. 1. Cryo-EM structure of the HIV intasome core.
(A) Cryo-EM reconstruction and (B) corresponding atomic model of the HIV CIC colored by protomer (red and yellow CTDs from distal protomers do not comprise the CIC, but are conserved among lentiviral intasomes). The two catalytic sites are indicated by dashed squares. (C) Close-up of the HIV intasome active site, colored by root-mean-square deviation from the corresponding region in the PFV intasome (PDB 3L2Q). IN residues frequently mutated in patient-derived clinical samples in response to 2nd generation INSTI treatment are indicated (11, 12).
Each of the two active sites in an HIV intasome contains the catalytic residues Asp64, Asp116, and Glu152, forming the prototypical DDE motif present in many nucleases, transposases, and other INs (24). The regions near the active sites of the PFV and HIV intasomes are similar because many of the residues participate in substrate binding and catalysis. However, further from the active sites, the structures diverge (Figure 1C and fig. S6–7). The largest differences reside in the synaptic CTD from the flanking protomer, specifically the region around the loop spanning HIV IN Arg228-Lys236. The corresponding loop in PFV IN has four additional residues, and assumes a distinct configuration. Clinically relevant drug resistance mutations occur within regions of HIV IN where the amino acid sequences between the two orthologs diverge (11, 12).
To better understand how INSTIs interact with HIV intasomes, we assembled the complex with bictegravir (BIC), a leading 2nd generation INSTI and the most broadly potent of all clinically approved INSTIs (25). We also examined the binding of additional compounds, named 4f, 4d and 4c, which contain a distinct chelating core (Fig. 2A), whose development was motivated by the need to further improve potency against drug resistant variants (19, 20). Currently, 4d is a leading drug candidate that shows improved efficacy over all clinically used and developmental compounds against the known drug resistant variants (25, 26) (fig. S8). Intasomes were co-assembled and co-purified with INSTIs, and we verified their inhibitory activity (fig. S9). The cryo-EM structures of INSTI-bound CSCs extend to a comparable ~2.6–2.7 Å resolution near the active site, allowing derivation of atomic models (fig. S10–12, Table S1).
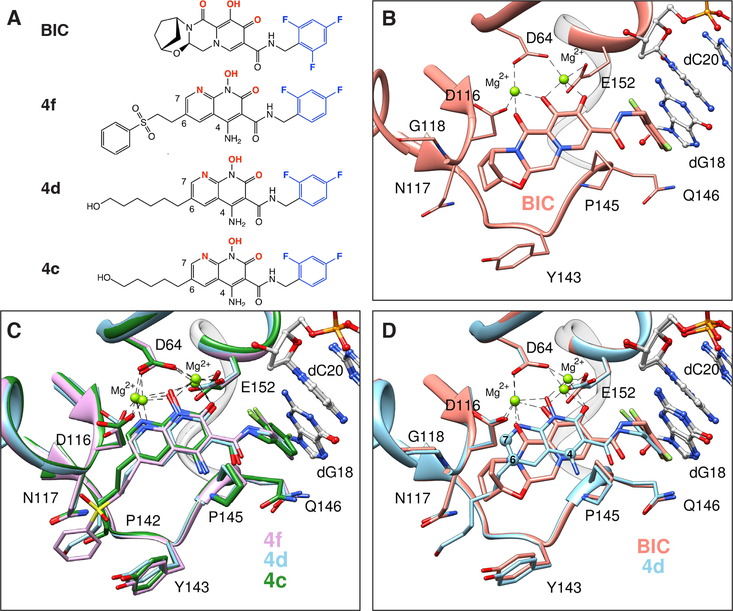
Fig. 2. Structural basis of INSTI binding to HIV intasomes.
(A) Chemical structures of the compounds used in this study, including the leading clinical drug Bictegravir (BIC) and developmental inhibitors 4f, 4d, and 4c [nomenclature based on previously reported work (19)]. Halogenated phenyl groups are shown in blue and the metal chelating heteroatoms are in red. (B-C) Binding modes are depicted for (B) BIC or (C) 4f (pink), 4d (light blue), and 4c (green) in the HIV intasome active site. (D) Superimposed binding modes of BIC and 4d. The terminal adenine base of viral DNA, and all water molecules, are omitted for clarity.
INSTIs bind HIV CSCs within a well-defined pocket formed by the interface between two IN protomers and vDNA. Several important pharmacophores characterize the binding of all INSTIs (Fig. 2B-C). First, three central electronegative heteroatoms chelate two Mg2+ cofactors within the active site of IN. A halogenated benzyl moiety appended to the core by a short linker displaces and substitutes for the 3′ terminal adenosine of processed vDNA, making a π-stacking interaction with the base of the penultimate cytosine. The displaced adenosine can adopt multiple rotameric conformations (17), only one of which contributes to INSTI binding by stacking on the central ring of the INSTI core (fig. S13). Removing the adenosine from the end of vDNA increases INSTI dissociation (27). The nature of the INSTI core and its substituents modulate its binding and help to determine its spatial orientation within the active site. For example, the core naphthyridine ring of the 4c/4d/4f compounds binds closer to the Mg2+ ions than the chelating core of BIC (Fig. 2D). These naphthyridine compounds position their 6-substituents within a constriction formed by the sidechain of Y143 and the backbone of N117. Fifteen of the most commonly found mutations that cause resistance in HIV IN are located within 10 Å of an INSTI core; however, only six are conserved between HIV IN and PFV IN (Table S2). Small chemical modifications can dramatically affect drug potency, as demonstrated previously for compounds targeting reverse transcriptase (28) or protease (29, 30). Thus, it is important to understand all interactions at the molecular level.
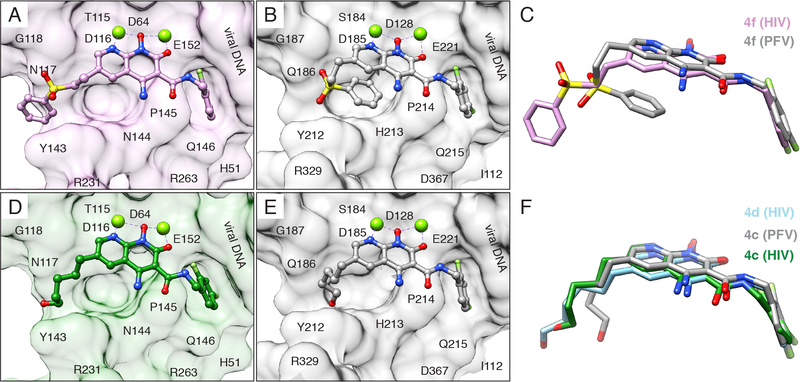
One strategy for developing inhibitors with broad potency against rapidly evolving enzyme targets is based on the concept of filling the “substrate envelope” (29). The rationale is that if inhibitory compounds bind entirely within a conserved consensus volume occupied by an enzyme’s natural substrates, this limits the ability of the virus to evolve changes in the target enzyme that allow it to discriminate between its normal substrates and synthetic inhibitors. The concept was originally used to guide the development of protease inhibitors and resulted in compounds with broad potency against viral resistant variants (31). We extended the substrate envelope hypothesis to the development of INSTIs; however, the structural models initially used were based on PFV intasomes (19). The cryo-EM structures of HIV intasomes with bound INSTIs reveal key differences in the substrate binding region. For example, although the chelating naphthyridine core of 4f binds to PFV and HIV intasomes similarly, the 6-substituted sulfonyl benzyl moiety, which is key to the potency of the compound (19, 20), adopts distinct configurations (Fig. 3A-C). In compound 4c, the 6-substitution is an n-pentanol chain. When bound to the HIV CSC, the pentanol group of 4c adopts an extended configuration and makes contacts with HIV IN that are distinct from interactions that the pentanol substituent of 4c makes with PFV IN (Fig. 3D-F) (19, 26). Compound 4d, which is more potent than 4c (fig. S8), adapts a similar extended configuration (Fig. 3F). Therefore, the differences in INSTI configuration are induced by the nature of the IN to which they bind. The simplest explanation for these differences is that multiple, minor variations in the amino acids that surround the bound INSTI and DNA substrates affect the binding of the compound in the active site. These compounds mimic aspects of bound forms of vDNA and tDNA substrates, residing within the substrate envelope (fig. S14).
Fig. 3. INSTIs can bind differently to PFV and HIV intasomes.
(A) Compound 4f bound to the HIV (pink) and (B) PFV intasome (gray). (C) Overlay of compound 4f binding modes. (D) Compound 4c, containing a 6-pentanol substituent, bound to the HIV (green) and (E) PFV intasome (gray, PDB 5FRN). (F) Overlay of compound 4c binding modes. Compound 4d, containing a 6-hexanol substituent, is also shown in its binding mode to the HIV (light blue) intasome. In panels A, B, D, and E, intasome active sites are shown as surface views, with labeled residues. R231 is modeled as an Ala stub due to flexibility. The terminal adenine is removed, for clarity.
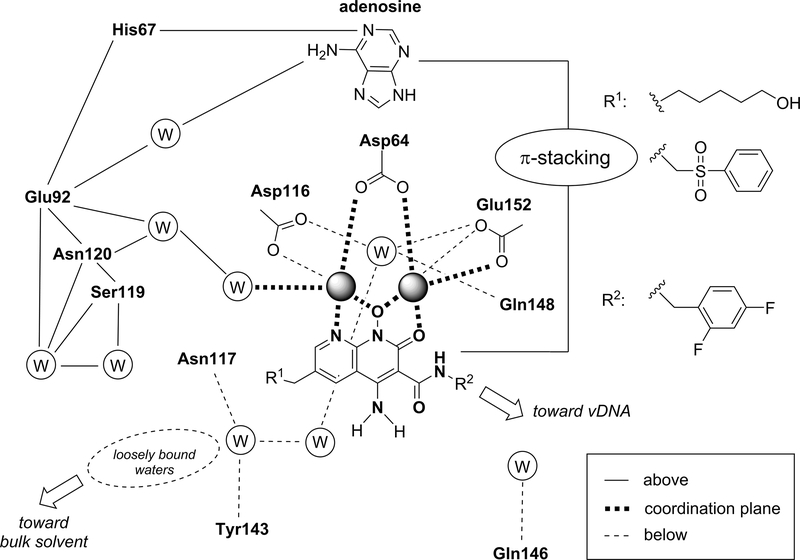
We were particularly interested in understanding why 4d is, in general, more broadly effective against resistant mutants than other INSTIs (fig. S8). The high-resolution maps revealed a complex and dynamic network of water molecules surrounding bound INSTIs (fig. S15). The binding sites of many waters appear to be conserved, occupying similar positions in the unliganded and INSTI-bound CSC structures. However, some waters are displaced or shifted as a consequence of INSTI binding; others are found only when INSTIs are bound, suggesting that the conformational changes induced by the binding stabilize their position. To simplify the analysis, INSTI interactions and waters can be subdivided by their relative position with respect to the plane formed by the Mg2+-coordinating ligand scaffolds, respectively above, in-plane, and below the plane, as depicted in Fig. 4. The naphthyridine cores are engaged from above by the purine ring of the 3′-adenosine via a π-stacking interaction, helping to stabilize a hydrogen bonding network involving the phosphate and N1 nitrogen of the adenine on one end and four waters in the cavity delimited by His67, Glu92, Asn120, and Ser119 on the other end. In-plane, the presence of the amino group at the 4-position of the naphthyridine core was previously shown to impart a >10-fold increase in potency (20). This improved efficacy appears to be due to: 1) formation of an intramolecular hydrogen bond with the halobenzylamide oxygen, which stabilizes its planar conformation; 2) electronic/inductive effects on the aromatic core increasing the metal coordination strength and electrostatic potential over the ring (i.e., stronger π-stacking) (Fig. S16 and Supplementary Note 1). Below the plane, the R1 substituent points toward the bulk solvent and the positioning of its long chain displaces loosely-bound waters. Displacement of solvent should be entropically advantageous. In turn, the location of one of the displaced waters closely matches the location of the hydroxyl moiety of 4d, providing additional enthalpic gain. This observation helps to explain why the 6-hexanol sidechain of 4d imparts this derivative with superior potency against resistant viral variants (sometimes up to ~10-fold) compared to very similar compounds in which the lengths of the side chain are shorter (propanol or pentanol) or longer (octanol) (19, 26). Finally, there are three tightly bound water molecules underneath the DDE motif, reaching toward the backbone of Asn117 and Tyr143 and projecting toward the bulk solvent. These bound waters can be exploited for the development of improved compounds.
Fig. 4. Interactions of naphthyridine-based INSTIs and HIV intasomes.
Schematic representation that recapitulates the receptor molecular environment and the water networks with which the naphthyridine scaffold ligands interact when coordinating the Mg2+ ions. The scheme summarizes interactions by their location with respect to the metal coordination plane of the naphthyridine scaffold (above, in-plane, and below). For clarity, the two waters coordinating the Mg2+ ions from above are not shown.
Within the substrate envelope, differences in geometry of the catalytic pockets, their overall volume, the locations of bound waters, among other features, all matter for understanding INSTI interactions. The current work highlights how small changes in the active site modulate drug binding and have implications for drug design. Structures of WT and mutant HIV intasomes bound to INSTIs should improve our understanding of resistance mechanisms and lead to the development of better drugs to be used in combination antiretroviral therapy for targeting viral escape mutants.
Supplementary Material
Acknowledgements
The authors acknowledge Bill Anderson at The Scripps Research Institute for help with EM data collection, Phil Baldwin at Salk for assistance with the local computational infrastructure, and Tim Grant at Janelia for providing the beam-tilt refinement program. Some of the early work identifying conditions for sample vitrification was performed by Venkata Dandey at the National Resource for Automated Molecular Microscopy, which is supported by a grant from the National Institute of General Medical Sciences (9 P41 GM103310) from the National Institutes of Health. Molecular graphics and analyses were performed with the USCF Chimera package (supported by NIH P41 GM103331). This work was supported by NIH grants R01 AI136680 and R01 AI146017 (to DL), R01 GM069832 (to SF), U54 AI150472 (to DL and SF) and by the Intramural Programs of the National Institute of Diabetes and Digestive Diseases (RC) and the National Cancer Institute (XZ, TB, SS and SH) and the Intramural AIDS Targeted Anti-Viral Program (IATAP) of the National Institutes of Health.
Footnotes
Accession Codes
The cryo-EM maps and atomic models have been deposited into the EMDB and PDB under the following accession codes: CSCAPO (EMD-20481 and 6PUT); CSCBIC (EMD-20483 and 6PUW); CSC4d (EMD-20484 and 6PUY); CSC4f (EMD-20485 and 6PUZ); CSC4c (EMD-21038 and 6V3K).
References
- 1.Martin AR, Siliciano RF, Progress Toward HIV Eradication: Case Reports, Current Efforts, and the Challenges Associated with Cure. Annu. Rev. Med. 67, 215–228 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Lesbats P, Engelman AN, Cherepanov P, Retroviral DNA Integration. Chem. Rev. 116, acs.chemrev.6b00125–12757 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Craigie R, Bushman FD, HIV DNA integration. Cold Spring Harb Perspect Med. 2, a006890–a006890 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Passos DO et al. , Cryo-EM structures and atomic model of the HIV-1 strand transfer complex intasome. Science. 355, 89–92 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maertens GN, Hare S, Cherepanov P, The mechanism of retroviral integration from X-ray structures of its key intermediates. Nature. 468, 326–329 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hare S, Gupta SS, Valkov E, Engelman A, Cherepanov P, Retroviral intasome assembly and inhibition of DNA strand transfer. Nature. 464, 232–236 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ballandras-Colas A et al. , A supramolecular assembly mediates lentiviral DNA integration. Science. 355, 93–95 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ballandras-Colas A et al. , Cryo-EM reveals a novel octameric integrase structure for betaretroviral intasome function. Nature. 530, 358–361 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yin Z et al. , Crystal structure of the Rous sarcoma virus intasome. Nature. 530, 362–366 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hazuda DJ, HIV integrase as a target for antiretroviral therapy. Current Opinion in HIV and AIDS. 7, 383–389 (2012). [DOI] [PubMed] [Google Scholar]
- 11.Grobler JA, Hazuda DJ, Resistance to HIV integrase strand transfer inhibitors: in vitro findings and clinical consequences. Current Opinion in Virology. 8, 98–103 (2014). [DOI] [PubMed] [Google Scholar]
- 12.Anstett K, Brenner B, Mesplede T, Wainberg MA, HIV drug resistance against strand transfer integrase inhibitors. Retrovirology. 14, 36 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arts EJ, Hazuda DJ, HIV-1 Antiretroviral Drug Therapy. Cold Spring Harb Perspect Med. 2, a007161–a007161 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Riddell J, 2018 IAS-USA Recommendations for the Use of Antiretroviral Therapy for HIV: Building on Decades of Progress. JAMA. 320, 347–349 (2018). [DOI] [PubMed] [Google Scholar]
- 15.Volberding PA, HIV Treatment and Prevention: An Overview of Recommendations From the IAS-USA Antiretroviral Guidelines Panel. Top Antivir Med. 25, 17–24 (2017). [PMC free article] [PubMed] [Google Scholar]
- 16.Hare S et al. , Molecular mechanisms of retroviral integrase inhibition and the evolution of viral resistance. Proc. Natl. Acad. Sci. U.S.A. 107, 20057–20062 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hare S et al. , Structural and functional analyses of the second-generation integrase strand transfer inhibitor dolutegravir (S/GSK1349572). Mol. Pharmacol. 80, 565–572 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao XZ et al. , Structure-Guided Optimization of HIV Integrase Strand Transfer Inhibitors. J. Med. Chem. 60, 7315–7332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao XZ et al. , HIV-1 Integrase Strand Transfer Inhibitors with Reduced Susceptibility to Drug Resistant Mutant Integrases. ACS Chem. Biol. 11, 1074–1081 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao XZ et al. , 4-Amino-1-hydroxy-2-oxo-1,8-naphthyridine-Containing Compounds Having High Potency against Raltegravir-Resistant Integrase Mutants of HIV-1. J. Med. Chem. 57, 5190–5202 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li M, Jurado KA, Lin S, Engelman A, Craigie R, Engineered hyperactive integrase for concerted HIV-1 DNA integration. PLoS ONE. 9, e105078 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tan YZ et al. , Addressing preferred specimen orientation in single-particle cryo-EM through tilting. Nature Methods. 14, 793–796 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Engelman AN, Cherepanov P, Retroviral intasomes arising. Curr. Opin. Struct. Biol. 47, 23–29 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rice P, Craigie R, Davies DR, Retroviral integrases and their cousins. Curr. Opin. Struct. Biol. 6, 76–83 (1996). [DOI] [PubMed] [Google Scholar]
- 25.Smith SJ, Zhao XZ, Burke TR, Hughes SH, Efficacies of Cabotegravir and Bictegravir against drug-resistant HIV-1 integrase mutants. Retrovirology. 15, 37 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smith SJ, Zhao XZ, Burke TR Jr, Hughes SH, HIV-1 integrase inhibitors that are broadly effective against drug-resistant mutants. Antimicrob. Agents Chemother., AAC.01035–18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Langley DR et al. , The Terminal (Catalytic) Adenosine of the HIV LTR Controls the Kinetics of Binding and Dissociation of HIV Integrase Strand Transfer Inhibitors. Biochemistry. 47, 13481–13488 (2008). [DOI] [PubMed] [Google Scholar]
- 28.Smith SJ et al. , Rilpivirine analogs potently inhibit drug-resistant HIV-1 mutants. Retrovirology. 13, 11 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.King NM, Prabu-Jeyabalan M, Nalivaika EA, Schiffer CA, Combating susceptibility to drug resistance: lessons from HIV-1 protease. Chemistry & Biology. 11, 1333–1338 (2004). [DOI] [PubMed] [Google Scholar]
- 30.Nalam MNL et al. , Substrate envelope-designed potent HIV-1 protease inhibitors to avoid drug resistance. Chemistry & Biology. 20, 1116–1124 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kurt Yilmaz N, Swanstrom R, Schiffer CA, Improving Viral Protease Inhibitors to Counter Drug Resistance. Trends Microbiol. 24, 547–557 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Suloway C et al. , Automated molecular microscopy: the new Leginon system. J. Struct. Biol. 151, 41–60 (2005). [DOI] [PubMed] [Google Scholar]
- 33.Zheng SQ et al. , MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nature Methods. 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lander GC et al. , Appion: an integrated, database-driven pipeline to facilitate EM image processing. J. Struct. Biol. 166, 95–102 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grant T, Grigorieff N, Measuring the optimal exposure for single particle cryo-EM using a 2.6 Å reconstruction of rotavirus VP6. elife. 4, e06980 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tegunov D, Cramer P, Real-time cryo-electron microscopy data preprocessing with Warp. Nature Methods. 71, 1–7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grant T, Rohou A, Grigorieff N, cisTEM, user-friendly software for single-particle image processing. elife. 7, e14874 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Adams PD et al. , PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hohn M et al. , SPARX, a new environment for Cryo-EM image processing. J. Struct. Biol. 157, 47–55 (2007). [DOI] [PubMed] [Google Scholar]
- 40.Emsley P, Lohkamp B, Scott WG, Cowtan K, Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moriarty NW, Grosse-Kunstleve RW, Adams PD, IUCr, electronic Ligand Builder and Optimization Workbench (eLBOW): a tool for ligand coordinate and restraint generation. Acta Crystallogr. D Biol. Crystallogr. 65, 1074–1080 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schüttelkopf AW, van Aalten DMF, PRODRG: a tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr. D Biol. Crystallogr. 60, 1355–1363 (2004). [DOI] [PubMed] [Google Scholar]
- 43.Pintilie G et al. , Measurement of Atom Resolvability in CryoEM Maps with Q-scores. bioRxiv. 15, 722991 (2019). [Google Scholar]
- 44.Chen VB et al. , MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pettersen EF et al. , UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 46.Krissinel E, Henrick K, IUCr, Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. D Biol. Crystallogr. 60, 2256–2268 (2004). [DOI] [PubMed] [Google Scholar]
- 47.Smith SJ, Hughes SH, Rapid screening of HIV reverse transcriptase and integrase inhibitors. J Vis Exp, e51400 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Baldwin PR, Lyumkis D, Non-Uniformity of Projection Distributions Attenuates Resolution in Cryo-EM. Prog. Biophys. Mol. Biol. (2019), doi:10.1016/j.pbiomolbio.2019.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.