Abstract
Background
We aimed to systematically evaluate telomere dynamics across a spectrum of pediatric cancers, search for underlying molecular mechanisms, and assess potential prognostic value.
Methods
The fraction of telomeric reads was determined from whole-genome sequencing data for paired tumor and normal samples from 653 patients with 23 cancer types from the Pediatric Cancer Genome Project. Telomere dynamics were characterized as the ratio of telomere fractions between tumor and normal samples. Somatic mutations were gathered, RNA sequencing data for 330 patients were analyzed for gene expression, and Cox regression was used to assess the telomere dynamics on patient survival.
Results
Telomere lengthening was observed in 28.7% of solid tumors, 10.5% of brain tumors, and 4.3% of hematological cancers. Among 81 samples with telomere lengthening, 26 had somatic mutations in alpha thalassemia/mental retardation syndrome X-linked gene, corroborated by a low level of the gene expression in the subset of tumors with RNA sequencing. Telomerase reverse transcriptase gene amplification and/or activation was observed in 10 tumors with telomere lengthening, including two leukemias of the E2A-PBX1 subtype. Among hematological cancers, pathway analysis for genes with expressions most negatively correlated with telomere fractions suggests the implication of a gene ontology process of antigen presentation by Major histocompatibility complex class II. A higher ratio of telomere fractions was statistically significantly associated with poorer survival for patients with brain tumors (hazard ratio = 2.18, 95% confidence interval = 1.37 to 3.46).
Conclusion
Because telomerase inhibitors are currently being explored as potential agents to treat pediatric cancer, these data are valuable because they identify a subpopulation of patients with reactivation of telomerase who are most likely to benefit from this novel therapeutic option.
Telomeres are ribonucleoprotein structures that cap the ends of chromosomes and protect them from exposure to DNA damage to maintain genomic integrity. Human telomeric DNAs are usually 9–15 kilobases long and composed of thousands of tandem repeats of (TTAGGG)n, which are shortened by 100–200 bp per cell division because of incomplete replication (1). It is well known that dysfunctional telomeres can lead to genome instability and disease (2). In cancer cells, telomere shortening can be reversed by activation of telomerase (TERT) (3–5) and/or the alternative lengthening of telomere (ALT) mechanism (6,7), thus maintaining telomere length and permitting cells to divide indefinitely. TERT expression can be deregulated by various modalities including point mutation, amplification, promoter rearrangement, and fusions (4,8–11). Alterations in alpha thalassemia/mental retardation syndrome X-linked (ATRX), a chromatin remodeler gene, contribute to telomere lengthening via the ALT mechanism (12–14).
A recent study analyzed telomere lengths using sequencing data from The Cancer Genome Atlas (TCGA), including tumors and nonneoplastic samples, across 31 primarily adult cancer types (15). Thirty percent of tumor samples showed telomere lengthening (telomere length greater in tumor than in nonneoplastic sample) based on the analysis of nearly 9000 pairs of tumor and matched nonneoplastic samples. The authors systematically surveyed TERT–activating somatic mutations and the spectrum of ATRX–truncating alterations to search for the underlying molecular mechanisms. The data also suggest the existence of other molecular mechanisms potentially affecting telomere length in tumors.
Our group previously analyzed 235 pediatric cancer patients, representing the subset of samples from the Pediatric Cancer Genome Project (PCGP), which had sequencing data available at the time (16). Telomere lengthening was found in 32% of solid tumors and 4% of brain tumors; telomere lengthening was not found among hematological cancers, and the estimated telomere length was experimentally validated by three independent approaches. In the present study, the analyses were expanded to 653 samples with tumor and normal (germline) whole-genome sequencing (WGS) data available and included additional tumor subtypes not covered in the previous study. An integrative analysis using both DNA and RNA sequencing data was performed to search for genetic lesions underlying the observed telomere dynamics, and a survival analysis was conducted to assess the prognostic value of the ratio of telomere fractions.
Methods
Study Samples
A subset of 653 PCGP cohort samples with WGS data available for DNA from both tumor and matched normal samples (17) was analyzed for tumor and normal (T/N) ratio of telomere fractions. Among these, 330 samples with tumor RNA sequencing data available were analyzed for abundance of messenger RNA transcripts (18). The study was approved by the institutional review board at St. Jude Children’s Research Hospital. Written informed consent was provided by a parent or guardian of each child or by a patient who was 18 years or older.
Telomere Fraction Measurement
TelSeq (19), recently developed by Ding et al., was used to calculate telomere length based on next-generation sequencing data. TelSeq classifies a read as telomeric if it contains at least k occurrences (default seven) of the telomere motif (TTAGGG); occurrences are not required to be consecutive. The number of telomeric reads is divided by the number of reads having GC content between 48% and 52%. To obtain an estimate of average telomere length, this fraction is multiplied by a constant equal to the number of base pairs of a reference genome (GRCh37) with GC content between 48% and 52% and divided by the number of chromosome ends (n = 46). We developed a new program called teltale (https://github.com/stjude/teltale) with the following modifications: We kept the k = 7 threshold but excluded reads marked as duplicates or those failing QC and required occurrences of the telomere hexamer motif to be consecutive. We reported the number of telomeric reads per million reads as the telomere fraction instead of estimating absolute length. For each tumor sample, the telomere fraction was adjusted by estimated tumor purity (20). Telomere change between tumor vs normal was assessed by using log2 of the T/N ratio of telomere fractions. We followed a similar approach in classifying the telomere changes as previously reported (16). In brief, Gaussian mixture modeling was applied to the log2(T/N) ratio of telomere fractions using the mclust package (version 5.4.5) in R-3.6.1. The optimal model according to Bayesian information criterion yielded three clusters corresponding to those samples with excessive lengthening, excessive shortening, and moderate changes of telomeres.
Somatic Mutations
Somatic mutations, including single nucleotide variants, insertion/deletions, copy number, and structural variations in gene coding regions, were obtained from the St. Jude PCGP mutation database (18).
Gene Expression From RNA Sequencing
RNA sequencing data were mapped using our internal StrongArm pipeline as previously described (21). Gene-level read count was generated using HTseq-count (22). Fragments per kilobase of transcript per million mapped reads (FPKM) were calculated based on the transcript models in GENCODE v19, and then normalized (23) using the limma R package (version 3.30.13) in R-3.3.1 separately for three cancer categories (hematological, solid tumors, and brain tumors). The PANTHER classification system (24,25) was used to analyze the overrepresentation of genes in biological pathways.
Molecular Mechanisms for Telomere Lengthening
Tumors were assigned to an ATRX group based on the following scenarios: ATRX truncation mutation; ATRX copy loss; ATRX fusion; or ATRX low expression (with normalized FPKM < 5). If expression data and ATRX mutation both existed, the classifications were considered cross-validated. Tumors were assigned to a TERT expression group based on the following scenarios: when TERT expression data were available and normalized FPKM for TERT expression exceed 2; and a focal copy gain of TERT exists in the absence of both TERT expression and ATRX alteration/expression data.
Statistical Analysis
To explore other possible molecular mechanisms beyond TERT and ATRX, we conducted agnostic correlation analyses based on the samples for which telomere lengthening could not be attributed to dysregulation of TERT expression or somatic mutations in ATRX. Thirty-seven samples with these known mechanisms were thus excluded from the analyses described below. A Wilcoxon rank sum test was performed for each gene to compare the log2 of the ratio of telomere fractions (tumor vs normal) for samples with and without somatic mutations in the coding region, for brain, solid, and hematological cancers. Only genes (n = 160) with mutations in at least seven samples (∼1.1% of the cohort of 653 samples) were tested. A Spearman rank correlation analysis was performed for samples with RNA sequencing data between the tumor telomere fractions and normalized FPKM values in brain, solid, and hematological cancers. Bonferroni correction (26) was applied to control for multiple testing.
The association between ratio of telomere fractions and survival status was assessed by Cox regression using age as the time scale, adjusting for age at diagnosis, sex, and race. Follow-up of survivors started at primary diagnosis and was censored at their last date of contact. The same model was fit using subgroups of patients stratified by three major categories (hematologic, brain, and solid) or subtypes (medulloblastoma [MB], osteosarcoma [OS], and high-grade glioma [HGG], each with a total number of patients of 30 or greater and a total number of deaths of five or greater). The proportionality assumption of Cox models was assessed graphically using Schoenfeld residuals.
Tests of statistical significance are two-sided, and P less than .05 is regarded as statistical significance.
Results
Tumor and Normal Ratio of Telomere Fraction Across Pediatric Cancer Subtypes
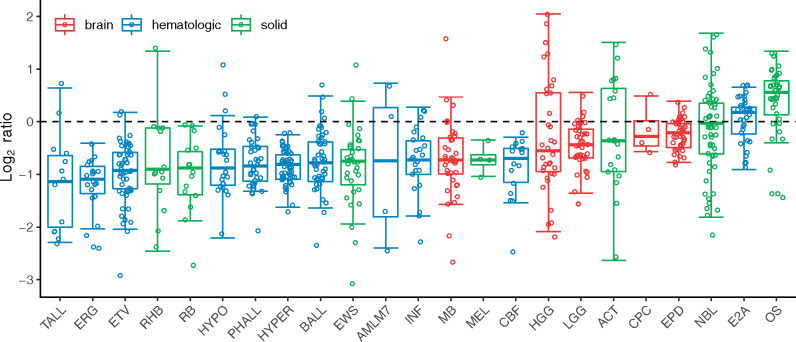
WGS data were available for 664 paired tumor and normal samples from the PCGP cohort (Table 1). Ten of the 23 pediatric cancer subtypes including choroid plexus brain tumor, six acute lymphoblastic leukemia (ALL) subtypes (27) (B-lineage, E2A-PBX1, ERG, ETV-RUNX1, hyperdiploid, Philadelphia, PA), M7 acute myeloid leukemia, Ewing sarcoma, and melanoma, were not included in the previous telomere analysis (16). Eleven samples were excluded because tumor purity cannot be estimated (mostly because of their quiet tumor genomes). The T/N ratio of telomere fractions for the remaining 653 samples was analyzed, and it differed by pediatric cancer subtype (Figure 1). T-lineage ALL samples showed the most telomere shortening, and OS samples showed the most telomere lengthening. In general, telomere shortening was observed in hematological cancers except for the E2A-PBX1 subtype, where 24.2% (8 of 33) showed telomere lengthening. Telomere fractions for paired tumor and normal samples and the T/N ratio of telomere fractions are listed for each patient in Supplementary Table 1 (available online). The distribution of log2(T/N) ratio is shown in Supplementary Figure 1 (available online). Overall, telomere lengthening was found in 28.7% (51 in 178) of solid tumors, 10.5% (16 in 153) of brain tumors, and 4.3% (14 in 322) of hematological cancers.
Table 1.
Samples included in the telomere analysis
| Group | Tumor type | Tumor subtype | No. of patients* | Patients with RNA sequencing† | Tumor type included in Parker et al (16) |
|---|---|---|---|---|---|
| Brain | Choroid plexus | – | 4 | 1 | No |
| Brain | Ependymoma | – | 40 | 34 | Yes |
| Brain | High-grade glioma | – | 35 (34) | 26 (25) | Yes |
| Brain | Low-grade glioma | – | 38 | 27 | Yes |
| Brain | Medulloblastoma | – | 37 | 5 | Yes |
| Hematological | Acute lymphoblastic leukemia | B-lineage | 47 | 36 (32) | No |
| Hematological | Acute lymphoblastic leukemia | B-lineage E2A-PBX1 | 33 | 8 | No |
| Hematological | Acute lymphoblastic leukemia | ERG | 22 | 22 | No |
| Hematological | Acute lymphoblastic leukemia | ETV6-RUNX1 | 50 | 48 | No |
| Hematological | Acute lymphoblastic leukemia | Hyperdiploid | 53 | 5 | No |
| Hematological | Acute lymphoblastic leukemia | Hypodiploid | 24 | 4 | Yes |
| Hematological | Acute lymphoblastic leukemia | Infant | 23 | 20 | Yes |
| Hematological | Acute lymphoblastic leukemia | Ph+ (Philadelphia) | 37 | 25 | No |
| Hematological | Acute lymphoblastic leukemia | T-lineage | 13 | 2 | Yes |
| Hematological | Acute myeloid leukemia | Core binding factor | 16 | 9 | Yes |
| Hematological | Acute myeloid leukemia | M7 | 4 | 4 | No |
| Solid | Adrenocortical tumor | – | 20 | 20 | Yes |
| Solid | Ewing sarcoma | – | 33 | 0 | No |
| Solid | Melanoma | – | 4 | 4 (3) | No |
| Solid | Neuroblastoma | – | 55 | 0 | Yes |
| Solid | Osteosarcoma | – | 37 | 17 | Yes |
| Solid | Retinoblastoma | – | 15 | 5 | Yes |
| Solid | Rhabdomyosarcoma | – | 14 | 14 | Yes |
| Total | NA | NA | 664 (653) | 336 (330) | NA |
Number in parentheses is the final number of samples included in tumor and normal telomere analysis. NA = not applicable.
Number in parentheses is the final number of samples included in correlation analyses.
Figure 1.
Boxplot of log2 of tumor and normal (T/N) ratio of telomere fractions across pediatric cancer subtypes. Boxes colored in red, blue, and green are for brain tumors, hematological cancers, and solid tumors, respectively. Samples with log2 of T/N ratio of telomere fractions greater than 0 (above the dotted line) had telomere lengthening, otherwise, telomere shortening. ACT = adrenocortical tumor; AMLM7 = M7 acute myeloid leukemia; BALL = B-lineage acute lymphoblastic leukemia; CBF = core binding factor acute myeloid leukemia; CPC = choroid plexus carcinoma; E2A = E2A-PBX1 acute lymphoblastic lymphoma; EPD = ependymoma; ERG = ERG acute lymphoblastic leukemia; ETV = ETV6-RUNX1 acute lymphoblastic leukemia; EWS = Ewing sarcoma; HGG = high-grade glioma; HYPO = hypodiploid acute lymphoblastic leukemia; INF = infant acute lymphoblastic leukemia; LGG = low-grade glioma; MB = medulloblastoma; MEL = melanoma; NBL = neuroblastoma; OS = osteosarcoma; PHALL = Philadelphia acute lymphoblastic leukemia; RB = retinoblastoma; RHB = rhabdomyosarcoma; TALL = T-lineage acute lymphoblastic leukemia; HYPER = hyperdiploid acute lymphoblastic leukemia.
Genomic Lesion and/or Dysregulation of Gene Expression Driving the Telomere Dynamics
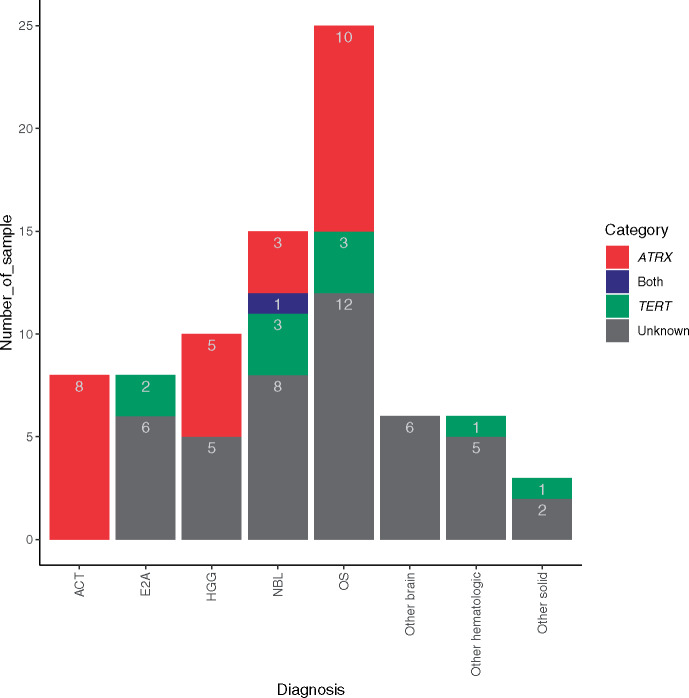
We surveyed genomic and transcriptomic lesions for 81 pediatric cancer patients with observed telomere lengthening in their tumors (Supplementary Table 2, available online). Based on available mutation and expression data, 36 pediatric patients were grouped into TERT-expressing (n = 10) or ATRX–altered (n = 26) categories. Although these mechanisms were mostly mutually exclusive, one neuroblastoma (NBL) sample (SJNBL044) had focal copy gain for TERT and copy loss for ATRX because of a deletion of the entire non-PAR region of the X chromosome. For tumors (n = 36) where telomere lengthening was classified as associated with one of these two mechanisms, ATRX-altered was the only mechanism observed in adrenocortical tumor (ACT) and HGG samples, and TERT-expressing was the only mechanism observed in hematological cancers and Ewing sarcoma. Both mechanisms were observed in NBL and OS samples (Figure 2). To summarize, the ATRX mechanism was observed only in solid or brain tumors, and the TERT mechanism was found in hematological cancers or solid tumors, but not in brain tumors with telomere lengthening. The mechanisms for the remaining 44 (54.3%) tumors with telomere lengthening remain unknown (Supplementary Table 2, available online).
Figure 2.
Molecular mechanisms for telomere lengthening in tumors. Number of samples with telomere lengthening had ATRX, TERT, or both or unknown mechanisms plotted for each subtype. Subtypes with fewer than five samples were pooled into one of the “other” groups. Note, one NBL patient (SJNBL044) had both ATRX alteration and TERT-expressing (depicted as a blue-colored bar). ACT = adrenocortical tumor; E2A = acute lymphoblastic leukemia B-lineage E2A-PBX1 subtype; HGG = high-grade glioma; NBL = neuroblastoma; OS = osteosarcoma.
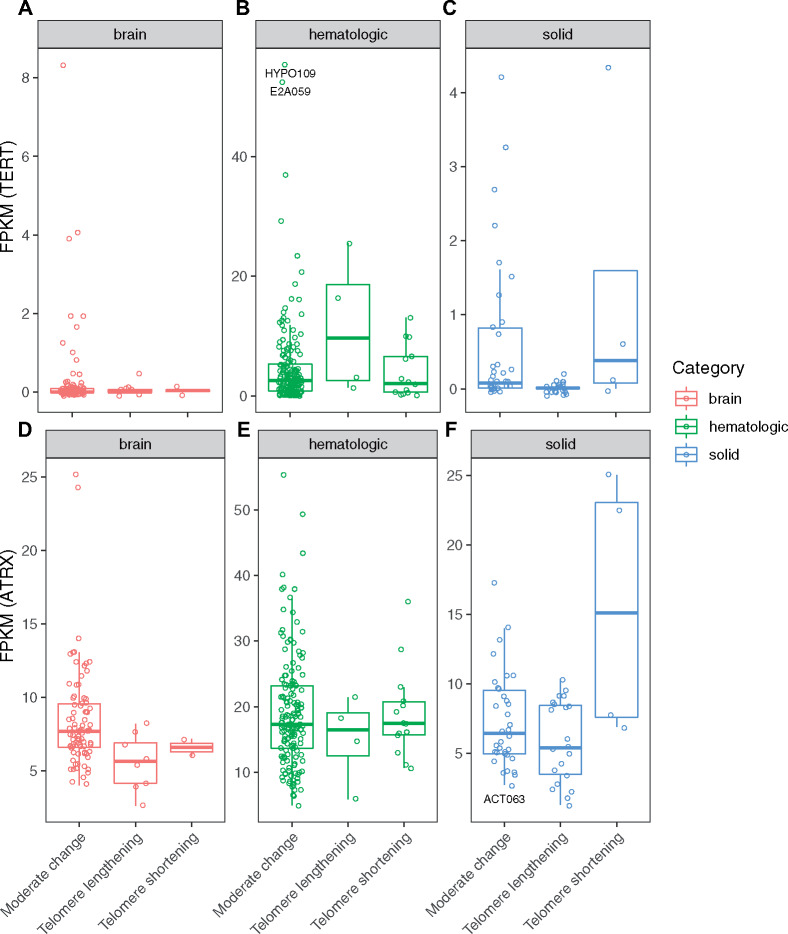
Tumors with telomere lengthening tended to have higher expression of TERT (more evident in hematological cancers) or lower expression of ATRX (more evident in solid or brain tumors) (Figure 3). However, high expression of TERT (> 2 FPKM) or low expression of ATRX (< 5 FPKM) in tumor samples did not guarantee telomere lengthening. For example, sample SJHYPO109 had the highest expression of TERT (normalized FPKM = 55.4), and sample SJE2A059 had the second highest expression of TERT (normalized FPKM = 52.4), yet neither showed telomere lengthening (Figure 3B and Supplementary Table 3, available online). On the other hand, sample SJACT063 had low ATRX expression (normalized FPKM = 2.73) but with telomere shortening (Figure 3F and Supplementary Table 3 available online).
Figure 3.
Distribution of TERT expression and ATRX expression in groups with excessive telomere lengthening, moderate change of telomeres, and excessive telomere shortening. A) TERT in brain tumors; B) TERT in hematological cancers; C) TERT in solid tumors; D) ATRX in brain tumors; E) ATRX in hematological cancers; and F) ATRX in solid tumors.
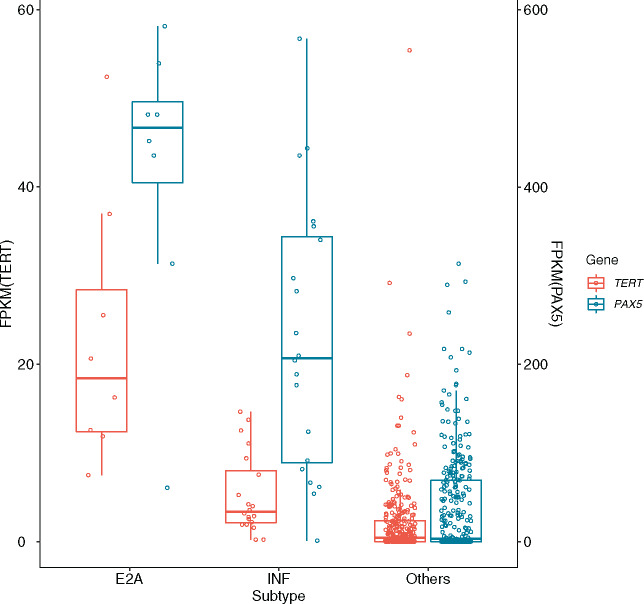
Among hematological cancers, leukemia samples of the E2A-PBX1 subtype had the highest expressions of PAX5, a well-known B cell–specific transcription factor, together with the highest expressions of TERT, followed by the ALL Infant subtype (Figure 4 and Supplementary Table 3 available online).
Figure 4.
Coexpression of PAX5 and TERT in E2A ALL (E2A), infant ALL (INF), and other subtypes of hematological cancers. Expression of TERT measured in FPKM (brown-colored bar and Y-axis on the left); expression of PAX5 measured in FPKM (blue-colored bar and Y-axis on the right).
Search for Other Potential Molecular Mechanisms to Explain Telomere Lengthening
The T/N ratio of telomere fractions showed a statistically significant difference between somatic mutation carriers vs noncarriers for genes including TTN (P = .01) among patients with brain tumors, MUC12 (P = .03) among patients with solid tumors, and TPRXL (P = .02) among patients with hematological cancers (Supplementary Table 4, available online), but nothing remained statistically significant after Bonferroni correction for 160 genes.
In the evaluation of associations between telomere fraction and RNA expression in tumor, after Bonferroni correction for 19 521 genes, nothing was correlated in brain tumor samples or in solid tumor samples (Supplementary Table 5, available online); however, four genes were positively and 10 genes negatively correlated in hematological samples (Supplementary Table 5, available online). Further pathway analysis of the 30 most negatively correlated genes using the PANTHER program (24,25) showed greater than 100-fold (P = 2.1 × 10-7) enrichment of genes pertaining to antigen processing and presentation of exogenous peptide, highlighting four genes belonging to the MHC class II family: HLA-DPB1, HLA-DPA1, HLA-DMA, and HL-DQA1.
Association Between Ratio of Telomere Fraction and Survival Status
Cox regression, using age as the time scale and adjusting for age at diagnosis, as well as for sex and race, showed a statistically significant association of telomere lengthening with higher mortality (hazard ratio [HR] = 2.18; 95% CI = 1.37 to 3.46) for patients with brain tumors, but not those with hematological cancers or solid tumors (Table 2). The same model fit for each tumor subtype, with the total number of patients of 30 or greater and total number of deaths of five or greater, showed a statistically significant association between telomere lengthening and higher mortality (HR = 5.48; 95% CI = 1.24 to 24.29) for MB patients but for neither OS nor HGG patients (Supplementary Table 6, available online).
Table 2.
Cox regression analysis for tumor and normal (T/N) ratio of telomere fractions and survival status for patients with hematologic cancers and brain and solid tumors
| Tumor type | Hematologic (n = 239) |
Brain (n = 148) |
Solid (n = 101) |
||||||
|---|---|---|---|---|---|---|---|---|---|
| HR | 95% CI | P * | HR | 95% CI | P * | HR | 95% CI | P * | |
| Age at diagnosis, y | |||||||||
| < 5 | 0.02 | 0.00 to 0.14 | <.001 | 0.02 | 0.00 to 0.14 | <.001 | 0.11 | 0.02 to 0.63 | .01 |
| 5–9 | 0.15 | 0.03 to 0.77 | .02 | 0.04 | 0.01 to 0.24 | <.001 | 0.16 | 0.04 to 0.65 | .01 |
| 10–14 | 0.45 | 0.10 to 2.01 | .29 | 0.30 | 0.07 to 1.31 | 0.11 | 0.30 | 0.11 to 0.80 | .02 |
| 15+ | 1.00 | (Referent) | 1.00 | (Referent) | 1.00 | (Referent) | |||
| Female vs male | 0.95 | 0.49 to 1.82 | .87 | 1.24 | 0.66 to 2.31 | 0.50 | 0.83 | 0.40 to 1.71 | .62 |
| Race | |||||||||
| White | 1.00 | (Referent) | 1.00 | (Referent) | 1.00 | (Referent) | |||
| Black | 1.09 | 0.49 to 2.42 | .83 | 1.44 | 0.70 2.98 | 0.32 | 1.86 | 0.81 to 4.28 | .14 |
| Other | 0.68 | 0.16 to 3.00 | .61 | 0.96 | 0.28 to 3.32 | 0.95 | 0.99 | 0.22 to 4.58 | .99 |
| Telomere ratio | 0.81 | 0.27 to 2.39 | .70 | 2.18 | 1.37 to 3.46 | 0.001 | 1.07 | 0.61 to 1.87 | .82 |
P values are from two-sided Wald tests. HR = hazard ratio; CI = confidence interval; Referent = reference category.
Discussion
Based on a systematic survey of telomere dynamics performed on 653 pediatric cancer patients for 23 subtypes from the PCGP study, we observed that 10.5% of brain tumors and 28.7% of solid tumors had telomere lengthening, as compared with 4% of brain tumors and 32% of solid tumors reported in our earlier study of 235 pediatric cancer patients, a subset of the samples analyzed here (16). In contrast, most hematological cancers displayed excessive telomere shortening or moderate changes except for E2A-PBX1 leukemia, where eight out of 33 (24.2%) showed telomere lengthening. To the best of our knowledge, this finding has not been previously reported in the literature. Even within the same cancer subtype there were substantial variations, highlighting the complexity of telomere dynamics among pediatric cancers.
Additionally, integrative analysis using both WGS and RNA sequencing data identified potential mechanisms for 37 (45.7%) of 81 samples with telomere lengthening. The finding of higher coexpression of PAX5 and TERT in E2A leukemia is consistent with experimental results showing that PAX5 activates the transcription of TERT in B cells (28). Interestingly, we identified an NBL case where telomere lengthening can likely be attributed to both ATRX and TERT mechanisms. This was corroborated by our previous publication, which highlighted the upregulation of TERT that occurred in a high-risk subgroup with poor outcomes (4). Notably, we found no samples with telomere lengthening attributable to the ALT mechanism among hematological cancers, which is consistent with previous reports (29). Clinically, telomere-based diagnostic/treatment strategies should be augmented by mutation and expression characterizations of ATRX and TERT genes (at a minimum) to choose an effective therapeutic option, for example, telomerase inhibitors (5).
Recently, two hot-spot mutations in the promoter region of TERT have been reported in different tumors (8,11,30,31) and were associated with a poor prognosis (32,33). The most common mutations were a single cytosine exchange to thymine at chromosome 5 base position 1 295 228 (hg19) (C228T), and less frequently at base position 1 295 250 (hg19) (C250T) (−124 and −146 base pair from the ATG start site, respectively). In our analysis, we found one occurrence of C228T in an NBL sample but without telomere lengthening. We also noted that high expression of TERT or low expression of ATRX does not guarantee telomere lengthening.
Although both TERT–activating and ATRX–truncating alterations exist in adult and pediatric tumors, the prevalence varies across different tumor types. It is important to note that only 10.9% of tumors had WGS–derived telomere length in the previous TCGA telomere study (15), which may invalidate the following comparisons. Approximately 27.1% of adult sarcomas underwent an ALT mechanism through somatic ATRX and DAXX alterations (15). Similarly, we found 40.0% (10 in 25) of osteosarcoma patients with telomere lengthening had ATRX alterations. However, we observed no evidence suggestive of an ALT mechanism among patients with Ewing sarcoma or rhabdomyosarcoma. Additionally, TERT was expressed in most adult glioblastoma (121 out of 132) with telomere shortening, though a substantial proportion of adult low-grade glioma (197 out of 450) had ATRX alterations and telomere lengthening (15). In contrast, only 1 of 38 pediatric low-grade glioma cases in our study had telomere lengthening but with no ATRX alteration.
Our work represents the largest study of the telomere dynamics in pediatric cancers, but the sample sizes are still limited for individual cancer subtypes, and this consequently led to limited power for comprehensive evaluation of prognostic values of telomere dynamics. For the same reason, some findings were inconsistent with the literature. For example, we did not observe any HGG sample with telomere lengthening with activated TERT expression, contradicting previous reports that 18–21% had both ATRX alteration and TERT activation (34). Another limitation of our study is the estimated ratios of telomere fractions may be confounded by possible age mismatch between tumor and normal sample; however, this did not appear to be a major issue based on our evaluation of age-dependent telomere attrition, which suggests an average telomere shortening of approximately 1.1% per year as compared with a substantially higher magnitude of changes (lengthening or shortening) in a tumor vs its paired normal sample (Supplementary Figure 2, available online). A further limitation of our study is that the RNA sequencing data were acquired from prior studies, each of which had performed RNA sequencing for either a subset or all samples of the study (we did not have RNA sequencing data available with neuroblastoma or Ewing sarcoma patients); therefore, selection bias potentially affects the interpretation of telomere and gene-expression association results for solid tumors. It is intriguing that the pathway analysis revealed that telomere length in hematological cancers were negatively correlated with the expression level of MHC class II genes; further validation studies and mechanistic investigations using a system biology approach are warranted. With an even larger number of patient samples, other mechanisms for telomere dynamics may be further explored, and risk estimates, such as hazard ratios for telomere dynamics on patient survival, can be more reliable.
Some earlier studies showed that inhibition of telomerase in leukemia cell lines induces progressive telomere shortening and eventual proliferative arrest or cell apoptosis (35–38). The observed functional requirement of telomerase in established hematologic malignancies provides a rationale to target telomerase therapeutically. In addition, telomere inhibition was shown to prevent tumorigenicity of pediatric ependymoma tumor-initiating cells (39). With the further development of telomerase inhibitors (40–42), our work provides the first line of evidence in classifying a subpopulation of pediatric cancer patients with reactivation of telomerase who are most likely to benefit from these therapeutic options.
Funding
This research was supported by funding from the American Lebanese Syrian Associated Charities to St. Jude Children’s Research Hospital and by grants (CA021765) from the National Institutes of Health to St. Jude Children’s Research Hospital.
Notes
Affiliations of authors: Department of Epidemiology and Cancer Control (ZW, NQ, KS, JQL, CLW, KKN, YY, LLR), Department of Computational Biology (ZW, SVR, TCC, YL, DKP, MCR, MNE, GW, JE, XC, JZ), Department of Pathology (JRD), and Department of Oncology (CAK, KEN), St. Jude Children’s Research Hospital, Memphis, TN; School of Public Health, University of Alberta, Edmonton, Alberta, Canada (QL).
The funders of the study had no role in the design and conduct of the study and were not involved in collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; or the decision to submit the manuscript for publication. The authors have no conflicts of interest to disclose.
Authors’ contributions: conception and design: ZW, SVR, JZ; development of methodology: ZW, SVR, JZ; acquisition of data (genetic and clinical data): ZW, SVR, YL, KS, JQL, CLW, KKN, MCR, MNE, GW, JE, JZ; analysis and interpretation of data (eg, statistical analysis, biostatistics, and computational analysis): ZW, SVR, T-CC, YL, QL, NQ, DKP, XC, YY, JZ; writing, review, and/or revision of the manuscript: all coauthors; administrative, technical, or material support (ie, reporting or organizing data and constructing databases): JRD, JZ; study supervision: ZW, JZ.
The authors thank all the individuals who participated in this study.
Aligned BAM files and the mutation call set are accessible through the St. Jude Cloud (https://stjude.cloud). The teltale program is available for download at github (https://github.com/stjude/teltale).
Supplementary Material
References
- 1. O’Sullivan RJ, Karlseder J.. Telomeres: protecting chromosomes against genome instability. Nat Rev Mol Cell Biol. 2010;11(3):171–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Shammas MA. Telomeres, lifestyle, cancer, and aging. Curr Opin Clin Nutr Metab Care. 2011;14(1):28–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Greider CW, Blackburn EH.. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell. 1985;43(2):405–413. [DOI] [PubMed] [Google Scholar]
- 4. Peifer M, Hertwig F, Roels F, et al. Telomerase activation by genomic rearrangements in high-risk neuroblastoma. Nature. 2015;526(7575):700–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jafri MA, Ansari SA, Alqahtani MH, et al. Roles of telomeres and telomerase in cancer, and advances in telomerase-targeted therapies. Genome Med. 2016;8(1):69.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bryan TM, Englezou A, Dalla-Pozza L, et al. Evidence for an alternative mechanism for maintaining telomere length in human tumors and tumor-derived cell lines. Nat Med. 1997;3(11):1271–1274. [DOI] [PubMed] [Google Scholar]
- 7. Dilley RL, Verma P, Cho NW, et al. Break-induced telomere synthesis underlies alternative telomere maintenance. Nature. 2016;539(7627):54–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Huang FW, Hodis E, Xu MJ, et al. Highly recurrent TERT promoter mutations in human melanoma. Science. 2013;339(6122):957–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kyo S, Takakura M, Fujiwara T, et al. Understanding and exploiting hTERT promoter regulation for diagnosis and treatment of human cancers. Cancer Sci. 2008;99(8):1528–1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhang A, Zheng C, Lindvall C, et al. Frequent amplification of the telomerase reverse transcriptase gene in human tumors. Cancer Res. 2000;60(22):6230–6235. [PubMed] [Google Scholar]
- 11. Horn S, Figl A, Rachakonda PS, et al. TERT promoter mutations in familial and sporadic melanoma. Science. 2013;339(6122):959–961. [DOI] [PubMed] [Google Scholar]
- 12. Jiao Y, Shi C, Edil BH, et al. DAXX/ATRX, MEN1, and mTOR pathway genes are frequently altered in pancreatic neuroendocrine tumors. Science. 2011;331(6021):1199–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Heaphy CM, de Wilde RF, Jiao Y, et al. Altered telomeres in tumors with ATRX and DAXX mutations. Science. 2011;333(6041):425.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ramamoorthy M, Smith S.. Loss of ATRX suppresses resolution of telomere cohesion to control recombination in ALT cancer cells. Cancer Cell. 2015;28(3):357–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Barthel FP, Wei W, Tang M, et al. Systematic analysis of telomere length and somatic alterations in 31 cancer types. Nat Genet. 2017;49(3):349–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Parker M, Chen X, Bahrami A, et al. Assessing telomeric DNA content in pediatric cancers using whole-genome sequencing data. Genome Biol. 2012;13(12):R113.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhang J, Walsh MF, Wu G, et al. Germline mutations in predisposition genes in pediatric cancer. N Engl J Med. 2015;373(24):2336–2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Downing JR, Wilson RK, Zhang J, et al. The pediatric cancer genome project. Nat Genet. 2012;44(6):619–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ding Z, Mangino M, Aviv A, et al. Estimating telomere length from whole genome sequence data. Nucleic Acids Res. 2014;42(9):e75.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Putnam DK, Ma X, Rice SV, et al. VCF2CNA: a tool for efficiently detecting copy-number alterations in VCF genotype data and tumor purity. Sci Rep. 2019;9(1):10357.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Parker M, Mohankumar KM, Punchihewa C, et al. C11orf95-RELA fusions drive oncogenic NF-kappaB signalling in ependymoma. Nature. 2014;506(7489):451–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Anders S, Pyl PT, Huber W.. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31(2):166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ritchie ME, Phipson B, Wu D, et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mi H, Muruganujan A, Casagrande JT, et al. Large-scale gene function analysis with the PANTHER classification system. Nat Protoc. 2013;8(8):1551–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mi H, Huang X, Muruganujan A, et al. PANTHER version 11: expanded annotation data from gene ontology and reactome pathways, and data analysis tool enhancements. Nucleic Acids Res. 2017;45(D1):D183–D189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bland JM, Altman DG.. Multiple significance tests: the Bonferroni method. BMJ. 1995;310(6973):170.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hunger SP, Mullighan CG.. Acute lymphoblastic leukemia in children. N Engl J Med. 2015;373(16):1541–1552. [DOI] [PubMed] [Google Scholar]
- 28. Bougel S, Renaud S, Braunschweig R, et al. PAX5 activates the transcription of the human telomerase reverse transcriptase gene in B cells. J Pathol. 2010;220(1):87–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Heaphy CM, Subhawong AP, Hong SM, et al. Prevalence of the alternative lengthening of telomeres telomere maintenance mechanism in human cancer subtypes. Am J Pathol. 2011;179(4):1608–1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Vinagre J, Almeida A, Populo H, et al. Frequency of TERT promoter mutations in human cancers. Nat Commun. 2013;4(1):2185.. [DOI] [PubMed] [Google Scholar]
- 31. Koelsche C, Renner M, Hartmann W, et al. TERT promoter hotspot mutations are recurrent in myxoid liposarcomas but rare in other soft tissue sarcoma entities. J Exp Clin Cancer Res. 2014;33(1):33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Melo M, da Rocha AG, Vinagre J, et al. TERT promoter mutations are a major indicator of poor outcome in differentiated thyroid carcinomas. J Clin Endocrinol Metab. 2014;99(5):E754–E765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Batista R, Cruvinel-Carloni A, Vinagre J, et al. The prognostic impact of TERT promoter mutations in glioblastomas is modified by the rs2853669 single nucleotide polymorphism. Int J Cancer. 2016;139(2):414–423. [DOI] [PubMed] [Google Scholar]
- 34. Dorris K, Sobo M, Onar-Thomas A, et al. Prognostic significance of telomere maintenance mechanisms in pediatric high-grade gliomas. J Neurooncol. 2014;117(1):67–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Tauchi T, Nakajima A, Sashida G, et al. Inhibition of human telomerase enhances the effect of the tyrosine kinase inhibitor, imatinib, in BCR-ABL-positive leukemia cells. Clin Cancer Res. 2002;8(11):3341–3347. [PubMed] [Google Scholar]
- 36. Delhommeau F, Thierry A, Feneux D, et al. Telomere dysfunction and telomerase reactivation in human leukemia cell lines after telomerase inhibition by the expression of a dominant-negative hTERT mutant. Oncogene. 2002;21(54):8262–8271. [DOI] [PubMed] [Google Scholar]
- 37. Roth A, Vercauteren S, Sutherland HJ, et al. Telomerase is limiting the growth of acute myeloid leukemia cells. Leukemia. 2003;17(12):2410–2417. [DOI] [PubMed] [Google Scholar]
- 38. Nakajima A, Tauchi T, Sashida G, et al. Telomerase inhibition enhances apoptosis in human acute leukemia cells: possibility of antitelomerase therapy. Leukemia. 2003;17(3):560–567. [DOI] [PubMed] [Google Scholar]
- 39. Barszczyk M, Buczkowicz P, Castelo-Branco P, et al. Telomerase inhibition abolishes the tumorigenicity of pediatric ependymoma tumor-initiating cells. Acta Neuropathol. 2014;128(6):863–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Thompson PA, Drissi R, Muscal JA, et al. A phase I trial of imetelstat in children with refractory or recurrent solid tumors: A Children’s Oncology Group Phase I Consortium Study (ADVL1112). Clin Cancer Res. 2013;19(23):6578–6584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Baerlocher GM, Oppliger Leibundgut E, Ottmann OG, et al. Telomerase inhibitor imetelstat in patients with essential thrombocythemia. N Engl J Med. 2015;373(10):920–928. [DOI] [PubMed] [Google Scholar]
- 42. Tefferi A, Lasho TL, Begna KH, et al. A pilot study of the telomerase inhibitor imetelstat for myelofibrosis. N Engl J Med. 2015;373(10):908–919. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.