SUMMARY
Better understanding of the progression of neural stem cells (NSCs) in the developing cerebral cortex is important for modeling neurogenesis and defining the pathogenesis of neuropsychiatric disorders. Here, we use RNA sequencing, cell imaging, and lineage tracing of mouse and human in vitro NSCs and monkey brain sections to model the generation of cortical neuronal fates. We show that conserved signaling mechanisms regulate the acute transition from proliferative NSCs to committed glutamatergic excitatory neurons. As human telencephalic NSCs develop from pluripotency in vitro, they transition through organizer states that spatially pattern the cortex before generating glutamatergic precursor fates. NSCs derived from multiple human pluripotent lines vary in these early patterning states, leading differentially to dorsal or ventral telencephalic fates. This work furthers systematic analyses of the earliest patterning events that generate the major neuronal trajectories of the human telencephalon.
In Brief
Micali et al. report that human telencephalic NSCs in vitro transition through the organizer states that pattern the neocortex. Human pluripotent lines vary in organizer formation, generating divergent neuronal differentiation trajectories biased toward dorsal or ventral telencephalic fates and opening further analysis of the earliest cortical specification events.
Graphical Abstract
INTRODUCTION
Defining how cell types emerge in the forebrain is central to understanding the origins of normal and pathological function in the cerebral cortex (Geschwind and Rakic, 2013; Kwan et al., 2012b; Lein et al., 2017; Nowakowski et al., 2017; Sandberg et al., 2016; Wamsley and Fishell, 2017). The neocortex in mammals, including rodents and humans, is the product of fate transitions of radial glial cells (RGCs), which function as neural stem cells (NSCs), sequentially generating waves of post-mitotic neurons that migrate superficially from the ventricular germinal zones (VZs) to form the ontogenic columns of the cortical layers (Angevine and Sidman, 1961; Malatesta et al., 2000; Noctor et al., 2001; Rakic, 1974, 1988). This evidence has led to a sustained interest in defining how the commitment and transition from proliferative RGCs to excitatory cortical neuronal fate are controlled.
In the developing mammalian telencephalon, organizer centers secreting morphogenic signals emerge to pattern the cortical field before neuron specification (Geschwind and Rakic, 2013; Grove and Fukuchi-Shimogori, 2003; O’Leary et al., 2007; Sur and Rubenstein, 2005). Moreover, the excitatory and inhibitory neurons of the cortex emerge in two different zones, the dorsal and the ventral telencephalon (Kwan et al., 2012b; Sandberg et al., 2016; Wonders and Anderson, 2006). In spite of the central importance of this very early period, many features of it, when telencephalic regional identities are first acquired, are not well understood, particularly in humans. Recent reports of species-specific differences in corticogenesis are often focused on relatively late neurogenic stages in which there is an enhanced genesis in humans of superficial neurons from the outer subventricular zone (oSVZ) (Hansen et al., 2010; Namba and Huttner, 2017; Nowakowski et al., 2016; Zhu et al., 2018). However, the evolutionary expansion of the human cerebral primordium is evident from the earliest stages and is already prominent when RGCs produce the first glutamatergic neurons (Bystron et al., 2008; Geschwind and Rakic, 2013). Thus, there is a clear interest in defining how the early patterning mechanisms are coordinated to achieve discrete waves of neurogenesis.
Evidence of the genetic risk for neuropsychiatric disorders has been found in the patterns of genes expressed in the neurogenic fetal cortex (de la Torre-Ubieta et al., 2018; Gulsuner et al., 2013; Parikshak et al., 2013; State and Sestan, 2012; Willsey et al., 2013; Xu et al., 2014). Moreover, risk-associated genes have been identified in the in vitro functional phenotypes of NSCs derived from patient-specific induced pluripotent stem cells (iPSCs) (Brennand et al., 2015; HD iPSC Consortium, 2017; Fujimori et al., 2018; Lang et al., 2019; Madison et al., 2015; Marchetto et al., 2017; Mariani et al., 2015; Schafer et al., 2019). These studies, which define the molecular and developmental origins of risk for brain disorders, point to the importance of early telencephalic fate transitions in the onset of pathogenic mechanisms.
In vitro neural systems are central in modeling these early events in neurogenesis. The growth factors FGF2, insulin, and other extracellular ligands, acting through the mitogen-activated protein kinase-extracellular signal-regulated kinase (MAPK-ERK) and phosphatidylinositol 3-kinase-protein kinase B (PI3K-AKT) pathways on the expression of cell-cycle regulators, control the critical transition when proliferating cortical NSCs initiate neurogenesis, both during brain development and in cell culture (Adepoju et al., 2014; Androutsellis- Theotokis et al., 2006; Cattaneo and McKay, 1990; Johe et al., 1996; Lehtinen et al., 2011; Qi et al., 2017; Rash et al., 2011; Ravin et al., 2008; Vaccarino et al., 1999). Lineage analysis of rodent NSCs differentiating in vitro directly demonstrated a rapid commitment of multipotent cells to neuronal or glial fates (Ravin et al., 2008). However, we still lack a comprehensive view of the molecular events regulating human NSC (hNSC) progression to post-mitotic cortical glutamatergic excitatory neurons.
Here, we modulated FGF2-MAPK signaling to control the developmental progression of mouse and hNSCs toward neurogenesis in vitro. We strictly define the acute molecular events as NSCs commit to excitatory glutamatergic fates. Then, building on previous work (Edri et al., 2015; Sakaguchi et al., 2015), we show that when hNSCs derived from PSCs were serially passaged in vitro, they self-organized to transition through a sequence of dorsal telencephalic developmental stages. At early passages, hNSCs expressed genes that are characteristic of the cortical hem, the dorso-caudal organizer zone of the telencephalon (Caronia-Brown et al., 2014), progressing later to cortical glutamatergic neuronal fates. We then derived multiple human iPSC (hiPSC) lines that showed intrinsic variability in this early organizer state formation and in the consequent differentiation bias to dorsal or ventral telencephalic fates. This work shows that patterning mechanisms and commitment events that generate dorsal or ventral telencephalic neuronal fates are coordinated features that emerge under precise control in hNSCs in vitro. This 2-dimensional (2D) experimental system opens to further systematic analysis the early non-linear fate transitions that specify human cortical fates.
RESULTS
Waves of Transcriptional Dynamics during Mouse In Vitro Neurogenesis Are Regulated by FGF2 Signaling
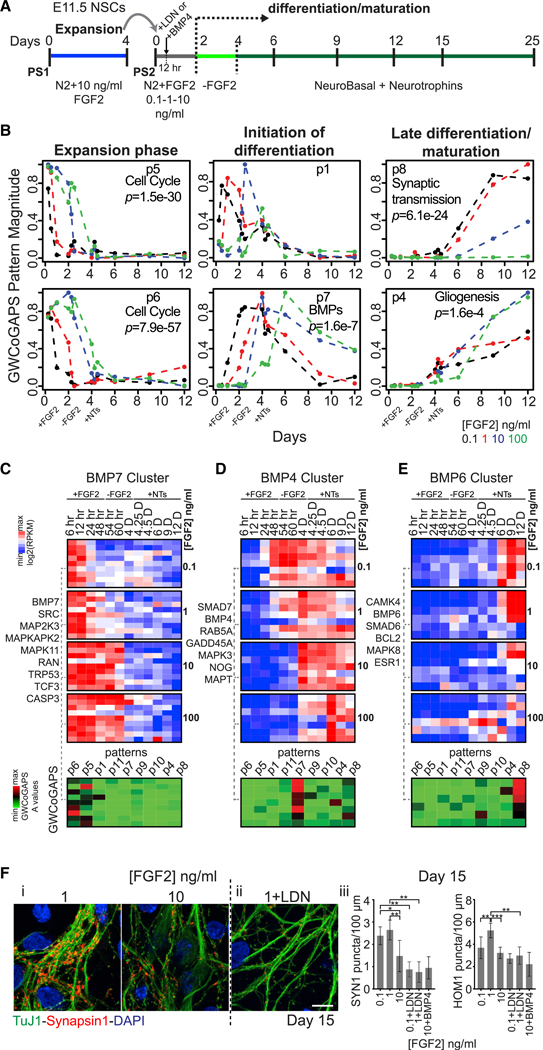
To define the events of cortical neuron commitment, we used primary NSC cultures derived from the mouse dorsal telencephalon at the beginning of neurogenesis, embryonic day 11.5 (E11.5). In vitro, rodent cortical NSCs proliferate maximally in the presence of 10 ng/mL FGF2 and abruptly differentiate when this growth factor is withdrawn (Adepoju et al., 2014; Cattaneo and McKay, 1990; Ravin et al., 2008). Here, we modulated FGF2 signaling to assess the differentiation trajectory of NSCs. NSCs were exposed to a range of FGF2 doses (0.1, 1, or 10 ng/mL) for 48 h after passage, and their later differentiation was monitored on day in vitro (DIV) 15 (Figure 1A). Consistent with a known role for FGF2 in antagonizing neurogenesis (Rash et al., 2011), the cultures exposed to the lower (0.1 or 1.0 ng/mL) rather than the higher (10 ng/mL) dose of FGF2 generated more glutamatergic excitatory neurons of every subtype, as defined using established cortical layer-specific markers (Kwan et al., 2012b; Molyneaux et al., 2007; Shen et al., 2006) (Figures S1A and S1B), and expressed significantly higher levels of the pre- and post-synaptic proteins Synapsin 1 (SYN1) and Homer1 (HOM1) (Figures 1F, i and iii, and S1C). Electrophysiological data also show the regulation of the neuronal activity induced by early FGF2 treatment (Figure S1D). These results confirm that the FGF2 signaling status of NSCs exiting the cell cycle regulates the production and the functionality of glutamatergic neurons many days later. This in vitro experimental system is used here to explore cellular progression during neurogenesis, from proliferation to the post-mitotic state.
Figure 1. Transcriptional Dynamics and BMP Signaling across Mouse In Vitro Neurogenesis.
(A) Experimental design. Passage 1 (PS1) NSCs were passaged into N2 + different FGF2 doses (PS2). Differentiation was induced by FGF2 withdrawal at DIV2. Neurotrophins (NTs) were added from DIV4.
(B) Six of 11 GWCoGAPS patterns shown in Figure S1. p values indicate the enrichment of GO categories in each pattern.
(C–E) Top: expression dynamics of selected gene clusters annotated by “BMP receptor signaling” from NCI (full dendrogram in Figure S1), correlating with BMP7 (C), BMP4 (D), and BMP6 (E). BMP7-related genes were rapidly repressed in low FGF2. BMP4-related gene levels increased at the initiation of differentiation. BMP6-related gene levels increased at late time points. Bottom: gene weights in each GWCoGAPS pattern for the same upper genes.
(F) Immunofluorescence images of SYN1 puncta in neurons cultured over astrocytes (i) at different FGF2 doses, or (ii) at 1 ng/mL FGF2 + LDN (100 nM). Scale bar: 10 μm. (iii) SYN1 and HOM1 puncta counts per 100 mm of neurite length for each condition, assessed by high-throughput image analysis. Means ± SDs, t test, n > 5 fields per measurement. LDN or BMP4 were added 12 h after NSC plating and withdrawn with FGF2 at DIV2 (see A).
To gain insight into the transcriptional dynamics of NSCs progressing to excitatory neuron fates, mouse cortical NSCs were exposed to varying doses of FGF2, and total RNA was collected for RNA sequencing (RNA-seq), from 6 h to 12 days of differentiation (Figure 1A). Principal-component analysis (PCA), which identifies major axes of variation among the samples, defined progressive differentiation across the time in PC1 (Figure S1E), indicating that NSCs exposed to lower FGF2 concentrations moved more rapidly through this transition in gene expression. To explore these transcriptional dynamics at higher resolutions, we used the genome-wide Coordinated Gene Activity in Pattern Sets (GWCoGAPS) non-negative matrix factorization (NMF) method. This method assigns weights for the contribution of every gene to a set number of patterns representing dominant changes in gene expression across all of the in vitro samples (Fertig et al., 2014; Stein-O’Brien et al., 2017). GWCoGAPS analysis, identifying 11 patterns (p1–p11), defined sequential waves of transcriptional extinction and induction as NSCs differentiated. Hierarchical clustering ordered these patterns into 3 groups associated with (1) NSC expansion (p5 and p6), (2) initiation of neural differentiation (p1, p11, p7, p9, and p10), and (3) further maturation (p9, p10, p4, and p8) (Figures 1B, S1F, and S1G). The early patterns (p5 and p6) were maintained in high FGF2 doses and enriched in cell-cycle regulator genes, defined by Gene Ontology (GO) analysis. The initial FGF2 condition controlled sequential waves of gene expression in the first 6 days of differentiation (patterns p1, p11, p7, and p9). GO analysis defined distinct endpoints for low and high FGF2 conditions that reflect either terminal neurogenesis (p8; low FGF2) or gliogenesis- (p4; high FGF2) (Figure 1B). Hence, NSCs passaged into low FGF2 downregulated cell-cycle genes and traversed later steps of neuron differentiation more efficiently than in high FGF2 (Figures S1H–S1K). This high-resolution analysis defines dynamic waves of transcription initiated by variation in FGF2 signaling as NSCs exit the cell cycle, distinguishing differentiation trajectories that from the earliest times were biased toward either cortical neurons or glia.
Early Endogenous BMP Signaling Is Required for Mouse Cortical Neurogenesis
To address the events initiating the differentiation of dorsal telencephalic neurons, we focused on bone morphogenetic protein (BMP) signaling that is known to oppose FGFs both in patterning the telencephalon in vivo and in regulating NSC differentiation in vitro (Lehtinen et al., 2011; Lillien and Raphael, 2000; Mabie et al., 1999; Tiberi et al., 2012). When hierarchical clustering was used to analyze the dynamics of BMP signaling during in vitro neurogenesis, distinct waves of expression of BMP-responsive genes correlated with particular BMP ligands were initiated by the early exposure to FGF2 (Figures 1B, p7, 1C–1E, S1L, and S1M). Nuclear phosphorylated SMAD1/5 (pSMAD1/5) signal monitored a rapid transient induction of endogenous BMP in low FGF2 doses (Figure S1N), which paralleled the induction of mRNA expression for BMP4 and the BMP-induced antagonist Noggin (Figures 1D and S1M).
To determine whether this early BMP signaling had a significant effect on neurogenesis and neuronal maturation, we perturbed it 12 h after plating NSCs, before BMP4 transcription (Figures 1A and S1M). The early inhibition of BMP signaling by the BMP type I receptor (BMPR1) inhibitor LDN193189, applied to NSCs exposed to low FGF2 doses, blocked both pSMAD1/5 induction at DIV 2 and the subsequent neurogenesis and synaptic maturation seen at DIV 15 (Figures 1F, S1O, and S1P). Exposing NSCs to high FGF2 doses plus exogenous BMP4 boosted pSMAD1/5 levels but did not rescue the compromised synaptogenesis, suggesting that a specific BMP-responsive cell state must be induced before the neuronal differentiation program (Figures 1F, iii, and S1O). We previously demonstrated that rodent telencephalic NSCs consist of a heterogeneous and dynamic population with rapidly varying lineage potential (Ravin et al., 2008). These data indicate that low FGF2 induces response to an endogenous and transient wave of BMP signaling that is required for cortical excitatory neuron commitment of NSCs in vitro.
Mouse Cortical NSC Subtypes Show Selective FGF2-Induced BMP Signaling Activation and Distinct Fate Bias
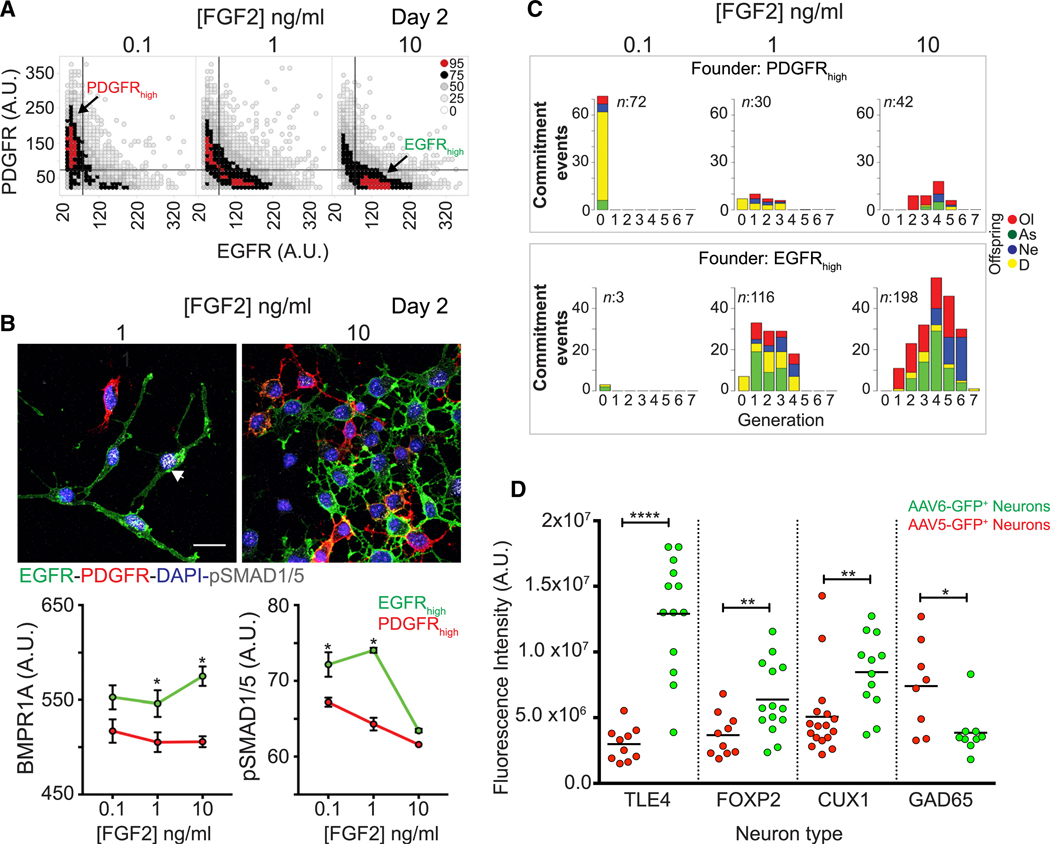
To further explore NSC diversity during cortical neurogenesis, we interrogated the surface expression of the tyrosine kinase receptors platelet-derived growth factor receptor α (PDGFRα) and epidermal growth factor receptor (EGFR), which identify subsets of embryonic dorsal telencephalic NSCs generating neurons and glia in vivo and in vitro (Andrae et al., 2001; Lillien and Raphael, 2000; Park et al., 1999; Sun et al., 2005). The proportion of PDGFRα high and EGFRhigh cells was regulated by FGF2 dose in the first 2 days in vitro, and moreover, the expression of the receptors was mutually exclusive before decreasing at DIV 6 (Figures 2A and S2A). These data indicate that transient waves of EGFR+ and PDGFR+ cell states occurred during the differentiation progression of NSCs in vitro.
Figure 2. Mouse Cortical NSC Subtypes Show Selective BMP Signaling Activation and Distinct Fate Bias.
(A) Density plot of PDGFRα and EGFR expression in individually segmented cells from high-throughput image analysis (images in Figure S2A). Color key: percentile cell counts per bin.
(B) Top: immunofluorescence images of pSMAD1/5 in EGFRhigh and PDGFRαhigh cells. Arrow shows pSMAD1/5 signals. Scale bar: 20 μm. Bottom: mean signal intensity of BMPR1A or pSMAD1/5 in EGFRhigh and PDGFRαhigh cells at DIV 2 from high-throughput image analysis ± SD, t test. Significance is indicated as * for every p < 0.05.
(C) Lineage analysis (see Videos S1, S2, and S3). Commitment events plot across cell generations from PDGFRαhigh (top) or EGFRhigh (bottom) founder cells. An event is given by an initial progenitor generating offspring with the same fate (see Method Details). As, astrocytes; D, death (apoptosis); Ne, neurons; Ol, oligodendrocytes. n = total commitment events.
(D) Fluorescence intensity of each marker in individual AAV5-GFP- or AAV6-GFP-TuJ1+ neurons at DIV15. Mean values (lines), t test.
Time-lapse recording and lineage analysis showed that FGF2 signaling controlled the cell-cycle duration of EGFRhigh cells in a dose-dependent manner (Figure S2B), inducing in these cells higher levels of phosphorylated FGF receptor (pFGFR) and ERK1/2 (pERK1/2) across all of the doses (Figure S2C). However, no differences in FGFR2, FGFR1, and FGFR3 expression were seen between PDGFRα high and EGFRhigh cells (Figure S2C; data not shown). These data suggest that FGF2 induced asymmetric signaling in the two subtypes. Higher expression of the neural fate transcriptional regulators HES1 and PAX6 was seen in EGFRhigh cells, while the levels of the precursor marker SOX2 were higher in PDGFRαhigh cells (Figure S2D). Extending the above data showing that early induction of endogenous BMP signaling was required for cortical neurons to efficiently differentiate, higher BMPR1A expression and pSMAD1/5 signal was found in EGFRhigh cells in low FGF2 (Figure 2B). These results indicate that BMP signaling was initiated in a transient EGFRhigh NSC state.
The fate bias of these NSC subtypes was assessed by time-lapse recording to create lineage dendrograms linking EGFRhigh or PDGFRαhigh founder cells at DIV 1 to the derived neurons, oligodendrocytes, and astrocytes identified at DIV 6 by the expression of TuJ1, O4, or glial fibrillary acidic protein (GFAP) (Figure S2E, i; Videos S1, S2, and S3). PDGFRαhigh progenitors committed with similar proportions to oligodendrocytes or neurons in low FGF2, and predominantly to oligodendrocytes at the higher FGF2 dose. EGFRhigh cells were tripotent, both in 1 and 10 ng/mL FGF2 (Figure 2C). Consistent with the transcriptomic data (Figures 1 and S1), the lineage analysis showed that faster cell-cycle exit correlated with early neuron specification of the EGFRhigh cells (Figures 2C and S2E, ii). These results suggest that efficient neurogenesis was associated with an early wave of differentiating EGFRhigh cells becoming acutely postmitotic in low FGF2 conditions.
To lineage trace neurons at more mature stages of differentiation (DIV 15), we used adeno-associated viruses (AAVs) encoding green fluorescent protein (GFP) with preferential tropism for cells expressing either PDGFRα (AAV5) or EGFR (AAV6) (Di Pasquale et al., 2003; Jackson et al., 2006; Weller et al., 2010) (Figures S2F and S2G). AAV6-infected NSCs preferentially generated CUX1+, FOXP2+, or TLE4+ glutamatergic neurons, while AAV5-infected NSCs were more prone to produce GAD65 expressing putative GABAergic neurons with less complex morphologies (Figures 2D and S2H). These results define an EGFRhigh BMP responsive cell state that efficiently produces cortical glutamatergic neuronal fates.
Cortical Neuron Differentiation Bias Varies with the Passage of hNSCs
Having defined the events of the transition to glutamatergic neurons from mouse cortical NSCs, we investigated neuronal commitment mechanisms in human dorsal telencephalic NSCs derived from PSCs in vitro (Edri et al., 2015; Mariani et al., 2012; Shi et al., 2012). Using standard neural induction protocols (Chambers et al., 2009; Edri et al., 2015), telencephalic NSCs were generated from the widely used human embryonic stem cell (ESC) line H9, which were then serially passaged in high FGF2 (Figure 3A). From passage (PS) 2 to 5, hNSCs were analyzed for the expression of the fate regulators SOX2, HES1, HES5, OTX2, PAX6, and SOX21 (Figures S3A and S3B). The spontaneous emergence of the neural rosette-forming state at PS3 and PS4 and its subsequent decline provided a system to define the dynamics of neurogenic fate mechanisms in humans.
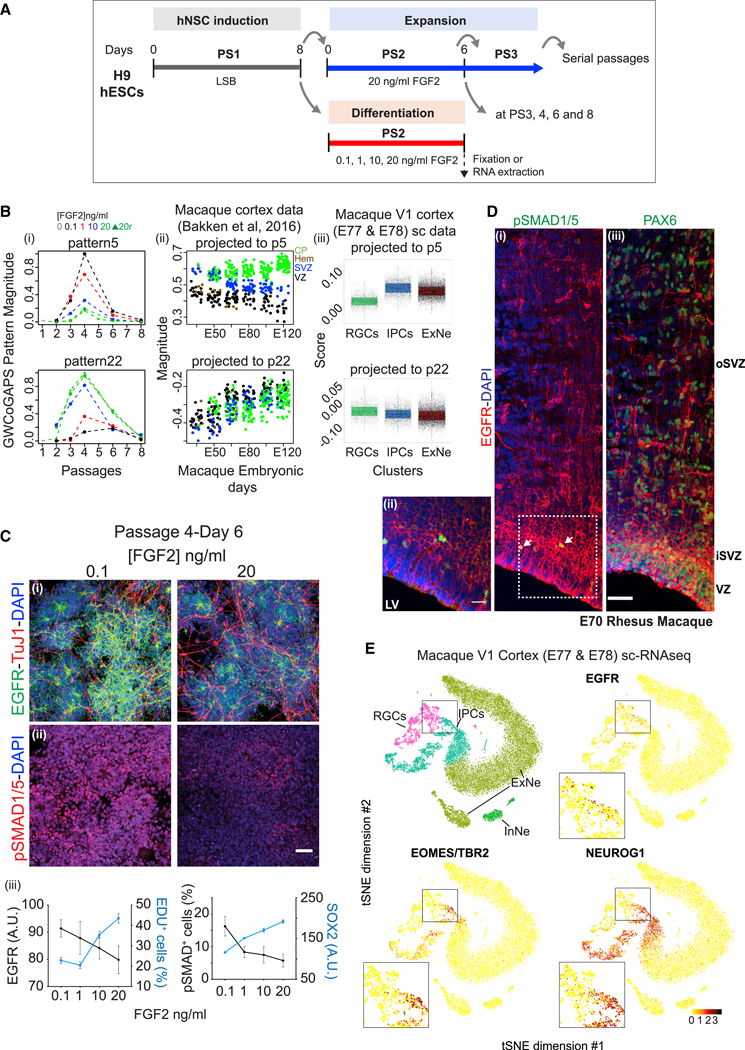
Figure 3. Cortical Excitatory Neuron Fate Bias at a Specific Passage of Human NSCs.
(A) Scheme of H9 hESC differentiation into NSCs. N2 + LSB (LDN193189 + SB431542) was applied at PS1, then hNSCs were serially passaged in N2 + 20 ng/mLFGF2. FGF2 modulation was applied at specific passages for 6 days before RNA collection.
(B) GWCoGAPS patterns p5 and p22 (i). FGF2 doses indicated. 0 refers to PS1; 20r are replicates for 20 ng/mL FGF2. (ii) Projections of macaque cortex microarraydata from (Bakken et al., 2016). CP, cortical plate; Hem, cortical hem; SVZ, subventricular zone; VZ, ventricular zone. (iii) Projection of macaque V1 scRNA-seq data.
(C) Immunofluorescence images of EGFR and TuJ1 (i), or pSMAD1/5 expression (ii). Scale bar: 50 μm. (iii) Mean fluorescence intensity of EGFR or SOX2 from high-throughput image analysis. Proportion of EDU+ or pSMAD1/5+ cells over total cells at DIV 6 for different FGF2 doses. Mean values ± SD.
(D) Immunohistochemistry images of E70 macaque cortex sections for EGFR (red; all panels) with pSMAD1/5 (i) or PAX6 (iii). Scale bar: 50 mm. Arrows indicate some positive cells. (ii) Higher magnification from the dashed area in (i). Scale bar: 20 μm. Nuclei stained with DAPI. LV, lateral ventricle; oSVZ/iSVZ, outer/inner subventricular zone; VZ, ventricular zone.
(E) t-Distributed stochastic neighbor embedding (t-SNE) plots colored by annotated cells and indicated markers of macaque V1 scRNA-seq data. All majorclusters are in Figure S4. EGFR+, TBR2+, and NEUROG1+ IPCs transitioning from RGCs are evident in the insets.
The neurogenic potential of the hNSCs was explored using FGF2 signaling modulation, as with the mouse system. Following expansion, hNSCs from PS2 to PS8 were exposed to varying concentrations of FGF2 for 6 days before RNA collection for RNA-seq (Figure 3A). PC1 revealed transcriptional progression across passages that was independent of the FGF2 dose (Figure S3C, i). Gene expression data can be visualized or “projected” into a low-dimensional space defined by another dataset, allowing the exploration of the transcriptional modules defined in one dataset as they change in the other. To relate the dynamics of hNSCs in vitro to in vivo cortical development, we projected gene expression data from the developing human (Jaffe et al., 2018) or macaque neocortex (Bakken et al., 2016) into the transcriptional space defined by the individual gene weights from PC1, using projectR (Figure S3C, ii and iii) (Stein-O’Brien et al., 2019; see Method Details). The projection analysis indicated that the transcriptional dynamics identified by PC1 in vitro parallel human and macaque development as neurogenesis peaks in vivo.
To characterize these transcriptional dynamics at a more granular level, we explored a set of 24 patterns (p1–p24) defined by the GWCoGAPS algorithm in this in vitro hNSC RNA-seq data (Figure S3D). Three of these patterns (p5, p6, and p22) showed transcriptional differences across the FGF2 dose that peaked at PS4, when neural rosettes were most abundant (Figures 3B, i and S3D). Projection of the developing macaque cortex gene expression data (Bakken et al., 2016) into these patterns shows that p5 specifically identifies a gene signature more highly expressed in the forming cortical plate (CP) than in the germinal SVZ and VZ domains throughout this neurogenic period (Figures 3B, ii and S4A).
To more precisely relate these in vitro transcriptional dynamics to discrete cell types of the developing primate neocortex, we generated a single-cell mRNA-seq (scRNA-seq) dataset from 2 macaque fetal visual cortex (V1) samples collected at E77 and E78, using the 103 Genomics platform. Data from 17,161 single cells passed quality control measures and were included in the present study. Using unsupervised clustering, we identified major cell clusters including RGCs, intermediate neuronal precursor cells (IPCs), excitatory neurons (ExNe), and interneurons (InNe) (Figures S4D and S4E). Projection of this monkey fetal V1 or human developing cortex scRNA-seq data (Nowakowski et al., 2017) into the hNSC GWCoGAPS patterns confirmed that p5 correlates with post-mitotic IPC and excitatory neuron signatures in vivo (Figures 3B, iii and S4F). Pattern p22 induced by high FGF2 was not associated with an excitatory neurogenic signature in either projection (Figures 3B, S4A, and S4F). These results indicate that the in vitro appearance of newborn cortical neurons was favored specifically at PS4 by the acute reduction of FGF2 and, more important, that gene expression dynamics occurring during in vitro neurogenesis parallel those in the neurogenic primate neocortex.
Immunofluorescence was used to further explore the differentiation of PS4 hNSCs into post-mitotic neurons. The data show that low FGF2 favored a switch from proliferating (EDU+) SOX2+ hNSCs to SOX2, PAX6, TuJ1+, EGFRhigh, and DCX+ post-mitotic (EDU) newborn neurons (Figures 3C, i and iii, S4B, iii–v, and S4C). Elevated pSMAD1/5 immunoreactivity showed that this transition from dividing precursors to young neuroblasts induced by low FGF2 was associated with a burst of endogenous BMP signaling occurring most efficiently at passage PS4 (Figures 3C, ii and S4B, vi). These results suggest that post-mitotic neurons with cortical glutamatergic features were specified by a conserved mechanism in human and mouse NSCs and that hNSCs differ across the passages in the generation of pertinent intermediates as excitatory cortical neuronal fates were specified (Figure S4B, i–iv).
EGFR-BMP Signaling Interaction Defines a Neurogenic Transition State in RGCs of the Developing Primate Cortex
Next, we investigated whether the progression of the NSC states seen in vitro recapitulates fundamental events of RGC development in vivo. In primates, the expression of PAX6 defines the neurogenic potential of RGCs in the VZ and marks committed neurons in the oSVZ (Hansen et al., 2010; Mo and Zecevic, 2008). In the E70 macaque dorsal parietal cortex, we detected EGFR and PAX6 co-expression in RGCs of the VZ/iSVZ (inner SVZ) and in committed neurons delaminating through the oSVZ (Figures 3D and S4G, i). The induction of nuclear pSMAD1/5 detected in EGFRhigh expressing RGCs indicates the BMP signaling response, suggesting the transition of these cells to neurogenic precursors (Lehtinen et al., 2011; Li et al., 1998; Saxena et al., 2018) (Figure 3D). Moreover, we found EGFRhigh precursors expressing the neurogenic transcription factor EOMES/TBR2 in iSVZ, confirming the neuronal commitment of these cells (Figure S4G, ii and iii) (Arnold et al., 2008; Englund et al., 2005; Pollen et al., 2015). EGFR staining was not detected in CP cells expressing later neuronal markers such as TBR1 (Figure S4G, i, ii, and iv).
We explored this transition further in the monkey fetal V1 scRNA-seq data and identified a population of early neuronal IPCs progressing from RGCs, expressing EGFR, TBR2, SOX2, and NEUROG1 (Figures 3E and S4E). These results are consistent with the hypothesis that hNSCs in vitro model a mid-neurogenic phase of primate cortex development, when an EGFRhigh expressing the RGC population uses BMP signaling in a transition state from proliferative cells to committed glutamergic cortical neurons.
Early Passage hNSCs Show Cortical Organizer Identities
To understand the origins of the neurogenic peak, we focused on NSC states before PS4. Five GWCoGAPS patterns from the in vitro hNSC data showed the highest gene expression at PS2 and PS3 (p8, p11, p2, p4, and p19) (Figures 4A, i and S5A, i). Of these, p2, p11, p4, and p19 but not p8 were dependent on the initial FGF2 status of the cells. These transcriptional dynamics suggest that these early stages of hNSCs also undergo an FGF2-regulated developmental progression that we explore further here.
Figure 4. Cortical Organizer Identities of Early Passage hNSCs.
(A) GWCoGAPS patterns p8, p11, and p2 (i). FGF2 doses indicated. 0 refers to PS1; 20r are replicates for 20 ng/mL FGF2. (ii) Projections of macaque cortex datafrom Bakken et al. (2016).
(B) Expression of hem-related genes. Gene list derived from (Aii) contrasting hem with other cortical regions from Bakken et al. (2016). BMP2, −4, and −7, not derived from the gene list of (Aii), are in a separate heatmap at the bottom.
(C) Immunofluorescence images of (i) LMX1A and LHX2, (ii) FOXG1, and (iii) EGFR and OTX2, in cells cultured with 0.1 ng/mL FGF2 for 6 days from PS2 to PS4. FOXG1 (ii) and EGFR/OTX2 (iii) are from the same field. Dashed lines enclose OTX2− FOXG1− EGFRhigh cells. Scale bar: 50 μm. (iv and v) From high-throughput image analysis: (iv) proportion of LMX1A and LHX2; (v) scatterplot of EGFR versus OTX2 expressing cells, colored by FOXG1 level.
(D) Model of the hNSC state progression in vitro. (Left) View of a coronal section of the developing mammalian telencephalon. CP, cortical plate; DM/DL, dorsomedial/-lateral; GE, ganglionic eminence; LV, lateral ventricle; MP, medial pallium; SVZ, subventricular zone; VZ, ventricular zone. (Right) Distinct states of in vitro hNSCs indicated by markers. State transition of hNSCs induced by low FGF2, revealing lineage progression from hem state at PS2 to cortical neurogenic identity at PS4. The transition from NSCs to newborn post-mitotic neurons is defined by EGFRhigh cells responsive to BMP signaling.
Projection of the developing macaque cortex gene expression data (Bakken et al., 2016) into these early passage hNSC GWCoGAPS patterns showed that p8 and the low FGF2 patterns p11 and p2 expressed genes characteristic of the cortical hem domain (Figures 4A, ii and S5B). Neither the early high FGF2 patterns p4 and p19 nor the later patterns p5 and p22 (described above) showed an enrichment of genes of the cortical hem domain (Figures 3B, ii and S5A, ii). This analysis indicates that low FGF2 conditions at early passages induced elements of the hem transcriptional signature. The cortical hem is a source of BMP and WNT morphogens that pattern the dorso-caudal domain of the telencephalon (Caronia-Brown et al., 2014; Grove et al., 1998). To monitor the emergence of this cellular organizer identity across the hNSC passages, we used the gene expression data from the developing macaque cortex (Bakken et al., 2016) to generate a list of hem-specific genes of the primate telencephalon (Figure 4B; see Method Details). PS2 and PS3 were enriched in these genes, including LMX1A, WNT8B, WNT3A, BMP2/4/6/7, RSPO2/3, and BAMBI, indicating coordinated transient expression of the cortical hem transcriptional signature, sensitive to FGF2 dose, at early passages of in vitro hNSCs. The expression patterns of genes such as FGF8, TTR, and SFRP2 at these early passages suggest that other organizer states, including rostral patterning center (RPC), choroid plexus, and antihem, were also represented (Assimacopoulos et al., 2003; Sakaguchi et al., 2015; Storm et al., 2006) (Figure S5C).
Consistent with the known function of LMX1A in the specification of neural organizers, including the cortical hem in the mouse (Chizhikov et al., 2010), the expression of this transcription factor was highest at PS2 in low FGF2 and subsequently decreased (Figure 4C, i and iv). In contrast, FOXG1 and LHX2, which are required in the mouse to form the medial pallium and lateral cortex and to repress the formation of the cortical hem (Bulchand et al., 2001; Hanashima et al., 2007; Mangale et al., 2008; Molyneaux et al., 2007; Monuki et al., 2001), were expressed more prominently at later passages (Figure 4C, i, ii, iv, and v). At PS3, the majority of cells expressed LMX1A or LHX2, suggesting co-emergence of the caudal hem organizer and precursor cells of the cortical field, which then become dominant in PS4 (Figure 4C, iv). The progressive appearance of cells co-expressing OTX2, a transcriptional regulator of the choroid plexus and cortical hem (Sakaguchi et al., 2015), and FOXG1 at PS3, followed by OTX2− FOXG1− EGFRhigh cells at PS4 (Figure 4C, ii, iii, and v), is consistent with the progression of hNSCs from a hem to a cortical neurogenic identity (Figures 4C and 4D). This coordinated change indicates that cortical patterning and dorsal excitatory neuronal specification mechanisms are efficiently executed across early passages of hNSCs generated in vitro.
hNSC Line Variation in Organizer States Results in Divergent Neuronal Fate Trajectories
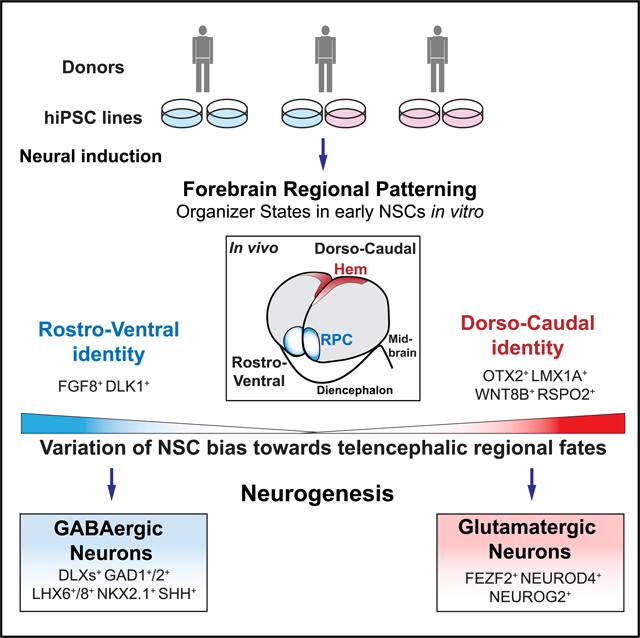
To address the stability of these early telencephalic differentiation steps, we analyzed multiple hiPSC lines. Six hiPSC lines (2063–1, −2; 2053–2, −6; 2075–1, −3) were generated from the scalp fibroblasts of 3 donors and differentiated toward forebrain fates. Aiming to probe intrinsic developmental bias in the early steps of telencephalic commitment in different hiPSC lines, we used a 32-day differentiation protocol that included the DKK1 mimetic XAV939 (X), which inhibits BMP and WNT signaling promoting anterior fates (Glinka et al., 1998). To limit FGF2-mediated effects on the regional identity of the NSCs and neurogenesis (Hendrickx et al., 2009; Korada et al., 2002; Raballo et al., 2000; Rash et al., 2013), the neuronal differentiating conditions X + LSB (XLSB) were applied directly to the iPSCs, without intervening passages of the NSCs in the presence of FGF2 (Figure 5A).
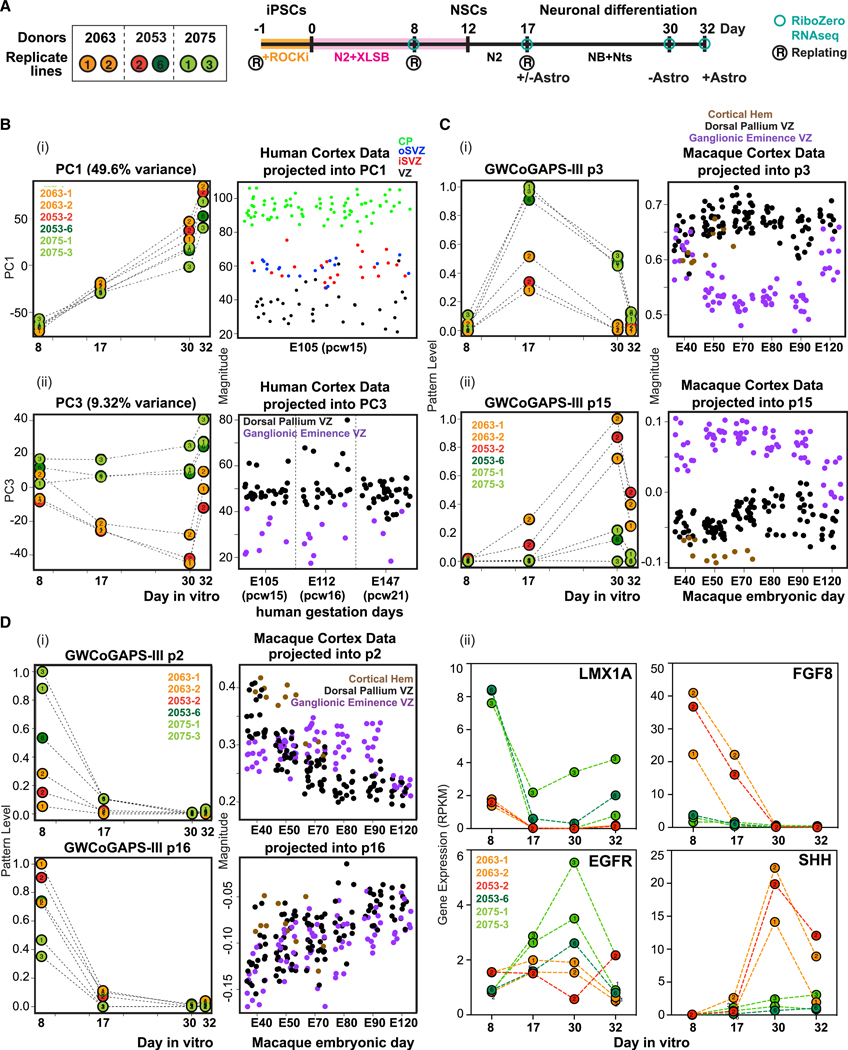
Figure 5. Human NSC Line Variation in Organizer States Results in Divergent Neuronal Fate Trajectories.
(A)Left: six hiPSC lines from 3 donors. Colors indicate divergent neuronal trajectory bias (see below). (Right) Experimental design. hiPSC lines were passaged inmTesR + Rock inhibitor, then differentiated into NSCs in N2-B27 + XLSB medium for 12 days (Maroof et al., 2013). Sample collection and passages are indicated. On day 17, NSCs were passaged and terminally differentiated with or without astrocytes. RNA was collected on day 30 (−Astro) or 32 (+Astro).
(B) Distinct forebrain trajectories within 6 lines. (Left) Progression of neural differentiation represented by PC1 (i) and emergence of divergent trajectories represented by PC3 (ii). (Right) Projection of human cortex data (Miller et al., 2014). CP, cortical plate; oSVZ/iSVZ, outer/inner subventricular zone; VZ, ventricular zone.
(C) Left: Dorsal or ventral trajectories revealed by GWCoGAPS-III p3 (i) and p15 (ii). Right: Projection of macaque cortex data (Bakken et al., 2016).
(D) Hem genes are highly expressed in lines with dorsal lineage bias. (i) Left: divergent bias of lines at day 8 revealed by GWCoGAPS-III p2 and p16. Right: projection of macaque cortex data. (ii) Expression of the indicated genes across neural differentiation of the 6 hiPSC lines.
To analyze the differentiation states induced by this treatment, RNA was collected at different time points for sequencing analysis. PC1 of the gene expression data showed that all of the samples progressed with similar kinetics through a common landscape of differentiation, promoted with similar efficiency in all six lines by growing NSCs either with or without rat astrocytes, which are known to facilitate neuronal maturation (Figure 5B, i). NSC fate regulators (SOX21, OTX2, and HES3) had high expression at early time points, in contrast to the regulators of neuronal function (NEFL, SYP, SYT4, and SNAP25) expressed at later times (Figure S6A). The projection of gene expression data from micro-dissected regions of fetal human cortex (Miller et al., 2014) into PC1 demonstrated that the in vivo spatiotemporal progression that generates post-mitotic neurons of the CP maps onto the time course delineated in this PC1 (Figure 5B, i).
In contrast to PC1, in which all 6 iPSC lines progressed equivalently, PC3 identified distinct patterns of gene expression on days 17 and 30 in 2 groups of cell lines: (2063–1, −2 and 2053–2) and (2053–6 and 2075–1, −3) (Figure 5B, ii). Projection of the fetal human cortex gene expression data (Miller et al., 2014) into PC3 distinguished cell lines with differential expression of dorsal pallium (2053–6 and 2075–1, −3) or ventral ganglionic eminence genes (2063–1, −2 and 2053–2] (Figure 5B, ii). Dorsal telencephalic regulators such as PAX6, FEZF2, NEUROD4, and NEUROG2 were highly expressed in 1 group of cells (2053–6, 2075–1 and −3), while regulators of ventral telencephalic fates such as NKX2–1, FOXG1, LHX6, LHX8, SHH, and DLX genes (Sandberg et al., 2016) were highly expressed in the other (2063–1, −2 and 2053–2) (Figure S6B). This analysis suggests that by day 17, these 2 groups of hiPSC lines have accessed dorsal or ventral telencephalic fates with different efficiencies.
To gain a higher-resolution view of transcriptional change in this system, we decomposed the RNA-seq data of the 6 hiPSC lines with the informatic tool GWCoGAPS, identifying a set of 30 gene expression patterns (GWCoGAPS-III p1–p30) (Figure S6C). Among these, patterns of genes restricted to the 2 groups of cell lines were identified. GWCoGAPS-III patterns p3, p14, and p15 showed differential expression in the 2 groups of cell lines at days 17 and 30 (Figures 5C and S6C), again involving dorsal versus ventral telencephalic genes, as indicated by the projection of the macaque developing cortex gene expression dataset (Bakken et al., 2016) into GWCoGAPS-III patterns using projectR (Figure 5C, i and ii). The GWCoGAPS-III p3 revealed enhanced expression of genes promoting dorsal telencephalic fates, including FEZF2 and PAX6 (p = 5.0e11 and p = 9.0e13 in DESeq2 on day 17). In contrast, the top weighted genes in the GWCoGAPS-III p15 included regulators of inhibitory neuron differentiation and function (ASCL1*, NKX2–1*, LHX6, LHX8, GAD1, DLX1*, DLX2*, DLX5*, and DLX6*; *p = 1.0e5 in DESeq2 analysis of differential expression on day 30) (Figure S6D).
These data raise the question of whether the differential bias toward dorsal or ventral telencephalon shown by these hiPSC lines emerges from an earlier fate segregation process. GWCoGAPS-III p2 and p16 distinguished the 2 groups of cell lines already at day 8 (Figure 5D, i). The projection of macaque developing cortex gene expression data (Bakken et al., 2016) into the GWCoGAPS-III patterns indicates that cortical hem genes, including OTX2, WNT8B, RSPO2, and WLS were highly weighted in p2 more than in p16 (Figures 5D, i and S6E). These dynamics were confirmed by the high expression of LMX1A at day 8 followed by the induction of EGFR at later time points in the lines with dorsal bias, which is consistent with the appearance of cortical excitatory neuronal precursors following patterning states (Figure 5D, ii). These data indicate that the three lines that most efficiently generate cortical excitatory neurons were biased toward dorso-caudal organizer fates at earlier steps. The generality of the emergence of early organizer states in vitro was confirmed in the public sequencing data of differentiating cortical neurons derived from hiPSCs (Edri et al., 2015; van de Leemput et al., 2014; Ziller et al., 2015) by projection into the hem-associated GWCoGAPS-III p2 (Figure S6F). NKX2.1 and SHH, well-known ventral fate regulators (Sandberg et al., 2016), were maximally expressed in the cell lines with ventral bias on days 17 and 30 (Figures 5D, ii and S6G). Before this, on day 8, FGF8, which is known to pattern the antero-ventral telencephalon (Storm et al., 2006), was maximally expressed in the cell lines with ventral bias (Figure 5D, ii). These data indicate that the divergent neuronal trajectory bias of hNSCs was coordinated with early mechanisms regulating the emergence of distinct organizer signals in the generation of forebrain NSCs from pluripotent states. The molecular and cellular bases for this variation may be readily probed further across many human pluripotent lines, progressing in vitro, as we describe here.
DISCUSSION
In the present study, we defined the transcriptional progression of mouse and human cortical NSCs transitioning to glutamatergic excitatory neuronal fates in vitro. Our understanding of the cellular dynamics across dorsal telencephalic neuronal commitment was extended by the analysis of mouse NSC populations transiently expressing distinct levels of EGFR or PDGFR. We showed that FGF2 signaling generates asymmetry within NSC progenitors instructing a wave of EGFRhigh cells that take on cortical excitatory neuronal fates. Mouse and human EGFRhigh NSCs in low doses of FGF2 initiated an endogenous BMP signaling cascade as cortical glutamatergic neurons first differentiated. In higher FGF2 concentrations, we observed compromised neurogenesis that we attribute to a delayed BMP activity and differentiation of EGFRhigh cells, preferentially generating glia fates in these conditions. The different neuronal trajectories traversed by in vitro NSCs exposed to varying FGF2 doses are consistent with previous in vivo results, indicating that FGF2 signaling perturbation during mouse embryogenesis affects neurogenesis in the dorsal telencephalon specifically (Korada et al., 2002; Raballo et al., 2000; Rash et al., 2011, 2013). It has been shown in the mouse that PDGFRα marks dorsal telencephalic NSCs that differentiate into neurons in vivo (Andrae et al., 2001) and in vitro after exposure to PDGF ligands (Johe et al., 1996; Park et al., 1999). We showed that PDGFRαhigh NSCs do not respond to FGF2-induced BMP and generate GABAergic inhibitory more efficiently than cortical excitatory neurons in our in vitro system. A subpallial origin of these NSCs seems unlikely, as fluorescence-activated cell sorting data in our lab (not shown in this work) indicate that the discrete PDGFRαhigh and EGFRhigh subtypes become one population that is double positive for both receptors in suspension, suggesting that these are two states of the same precursor that probably reset potency with passage, as previously reported for NSCs (Ravin et al., 2008). It has been shown in the mouse that olfactory bulb (OB) interneurons generated postnatally share common embryonic progenitors with cortical projection neurons in vivo (Fuentealba et al., 2015) and in vitro (Cai et al., 2013). It will be interesting to determine whether the embryonic PDGFRαhigh NSCs defined in this work share features with other previously identified adult mouse telencephalic precursors expressing PDGFRα, including a subset of B type NSCs in the SVZ generating OB interneurons and oligodendrocytes (Jackson et al., 2006), or others defining the oligodendrocyte lineage (Marques et al., 2016). Extending previous work identifying EGFR-expressing cells as astro-glial precursors of the late SVZ in rodents and monkeys (Burrows et al., 1997; Lillien and Raphael, 2000; Rash et al., 2019; Sun et al., 2005), our work defines a transient population of neurogenic EGFRhigh RGCs responsive to BMP signaling undergoing acute neuronal commitment during cortical development in vitro.
From the first generation of neurons from mouse pluripotent sources in vitro (Kim et al., 2002; Okabe et al., 1996) to the many recent studies of the transcriptomic and epigenetic landscapes of the developing human cerebral cortex (de la Torre-Ubieta et al., 2018; Nowakowski et al., 2017; Zhu et al., 2018), it has been shown that neuronal differentiation from NSCs requires transitions through a series of cellular intermediates. Here, after defining the acute transition events leading to post-mitotic excitatory neuron fates, we explored in vitro the intrinsic patterning mechanisms that coordinate corticogenesis before the ingrowth of thalamic afferents in vivo (Armentano et al., 2007; Bishop et al., 2000; Cholfin and Rubenstein, 2007; Fukuchi-Shimogori and Grove, 2001; Miyashita-Lin et al., 1999; Nakagawa et al., 1999; Shimogori and Grove, 2005). Hence, we demonstrate the sequential appearance of organizer and cortical neuron precursor domains in vitro. Previous work has demonstrated the presence of organizer structures in cerebral organoids derived from human iPSCs (Renner et al., 2017). We show that the earliest events in human telencephalic patterning and neurogenesis may be systematically analyzed with PSCs using this 2D in vitro system, which gives different advantages compared to 3D cultures (Pasxca, 2018), and is where we can observe the emergence of competing cortical patterning signals that determine regional neuronal fates.
The variation in the organizer states and subsequent divergent telencephalic trajectories revealed in the newly generated iPSC lines validates this concept, providing an opportunity to define how morphogenetic spatial patterning is coordinated with the specification of excitatory or inhibitory neurons in the human telencephalon. Consistent with these data, another recent study shows variation in dorso-ventral telencephalic fate bias across cerebral organoids derived from multiple donor iPSCs (Kanton et al., 2019). We also observed that replicate lines from same individual (2053–2, −6) can traverse divergent telencephalic regional fates under the same differentiation conditions. Previous reports have shown that transcriptional heterogeneity or differentiation capacity variability of multiple donor iPSC lines are under both genetic and epigenetic control (Carcamo-Orive et al., 2017; Nishizawa et al., 2016). Future studies will link genetic and epigenetic mechanisms to the divergent transcriptional phenotypes and fate bias we observed here.
The developmental mechanisms controlling these early telencephalic fates are of great interest as they are proximal to genetic risk for many neurodevelopmental disorders, including autism spectrum disorders (ASD) (HD iPSC Consortium, 2017; Kwan et al., 2012a; Madison et al., 2015; Marchetto et al., 2017; Mariani et al., 2015; Schafer et al., 2019) and brain cancers (Crawley et al., 2016; Ernst, 2016). Progress continues in defining disease-relevant in vitro phenotypes using genetically distinct hPSC lines as a central tool in the development of novel therapeutic interventions (Fujimori et al., 2018; Hubler et al., 2018; Lang et al., 2019). Our study suggests that to achieve accurate models of risk for neuropsychiatric disease, it will be necessary to more powerfully assess the extent and origin of the developmental variation in patient-specific iPSCs, as they progress through the non-linear transitions described here. Defining, as we described, cell state transitions and subsequent distinct neuronal differentiation trajectories will be central in selecting optimal lines for specific fates and designing cell assays that more efficiently reveal phenotypes of interest across a more precisely controlled neurogenic landscape in vitro. When integrated with the unprecedented transcriptomic and epigenetic mapping of the human forebrain (Amiri et al., 2018; de la Torre-Ubieta et al., 2018; Li et al., 2018; Wang et al., 2018; Zhu et al., 2018), the neural cell state transitions defined in this study will yield functional insights into the origin of developmental risk for neuropsychiatric disorders.
STAR★METHODS
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Ronald D. McKay (ronald.mckay@libd.org) or (ronaldmckay@me.com).
Materials Availability
All unique/stable reagents generated in this study (6 hiPSCs lines) are available from the Lead Contact with a completed Materials Transfer Agreement.
Data and Code Availability
The RNA-sequencing data generated in this study are available at GEO Accession #: GSE144158; GSE144156; GSE144157; GSE144508.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mouse NSC culture and differentiation
Embryonic day (E)11.5 embryos were isolated from pregnant C57BL/6 mice (Charles River) following the LIBD/Johns Hopkins University School of Medicine’s Institutional Animal Care and Use Committee (IACUC) guidelines. Dorsal cortices were collected in DMEM/F12 medium with N2 supplement (described below) and by mechanical trituration separated into single cells, as previously described in (Androutsellis-Theotokis et al., 2008; Ravin et al., 2008). 1 × 106 cells were plated over 10 cm culture plates (Falcon, 35–3003), previously coated with poly-L-ornithine (PLO) (Sigma, P3655) and fibronectin (FN) (R&D Systems, 1030FN), and incubated at 37°C, 5% O2 and 5% CO2 for 5 days in DMEM/F12 medium (Mediatech 16–405-CV) plus N2 supplement, containing 25 μg/ml bovine insulin (Sigma, I6634), 100 μg/mL apotransferrin (Sigma, T2036), 20 nM progesterone (Sigma, P8783), 100 mM putrescine (Sigma, P5780), 30 nM sodium selenite (Sigma, S5261), penicillin/streptomycin (Life Technology, 15140–122). We refer to DMEM/F12 medium with N2 supplement as N2 medium, hereafter. 10 ng/ml bFGF (R&D Systems, 4114-TC) was added daily and the medium was changed every other day. NSCs were lifted with HBSS (Thermo Fisher Scientific, 14185052) and aliquots of 1 × 106 cells/ 100 μl of N2 were frozen at −80°C. Thawed NSCs were expanded in presence of 10 ng/ml FGF2 for 5 days until confluence (we indicated this as passage 1, PS1). Then NSCs were dissociated with HBSS and passaged (PS2) into PLO/ fibronectin-coated 24 well plates (IBIDI, 82406), at a density of 50.000/well in N2 medium + FGF2. For FGF2 modulation experiment, NSCs were exposed to varying doses of FGF2 (0.1, 1, 10 or 100 ng/ml) on DIV 1 and 2. FGF2 was added daily. For differentiation experiments, NSCs were plated at PS2 over primary E18 rat astrocytes (see below) or, as indicated in the experiment description, without astrocytes. Differentiation of NSCs was induced 2 days after plating, by FGF2 withdrawal. At DIV 4, N2 was replaced with NeuroBasal medium (NB) (Life Technology, 12348–017), containing 25 μg/ml insulin, 30 nM sodium selenite, Glutamax (Life Technology, 35050061), 1x B27 (Life Technologies, 17504–044), 10 ng/ml BDNF (R&D Systems, 248-BD) and 10 ng/ml NT-3 (R&D Systems, 267-N3) until the end of experiment.
Treatment with LDN193189 (100 nM) (Stemgent, 04–0074) or BMP4 (10 ng/ml) (R&D System, 5020-BP-010/CF) was performed 12 hours after NSCs were plated with different FGF2 doses and repeated 24 hours later. LDN193189, BMP4 and FGF2 were withdrawn 48 hours after cell plating (DIV 2), and differentiation was induced as described above.
hPSC culture and neural differentiation
For maintenance of H9 human pluripotent stem cells (hPSCs) in feeder-free condition, cells were dissociated to single cells with Accutase (Life Technologies, A11105), plated at a density of 1 × 105 cells/cm2 in a Matrigel (BD, 354277)-coated 6 well plates (Falcon, 35–3046) with mTeSR1 (Stem Cell Technology, 05850) containing 5 μM Y27632, ROCK inhibitor (Sigma-Aldrich, Y0503) at 37°C, 5% CO2. ROCK inhibitor was removed from the medium 24 hours after plating and cells were cultured for another 4 days before the next passage (Chen et al., 2012). Neural differentiation from hPSCs was induced using dual SMAD inhibition protocol (Chambers et al., 2009). Cells were plated at a density of 6×104 cells in Matrigel-coated 24 well plates (IBIDI, 82406) or (Corning, 3526), with mTeSR1 plus ROCK inhibitor at 37°C, 5% CO2. Cells were switched to Aggrewell medium (Stem Cell Technology, 05893) for 2 days and then to N2+ B27. 100 nM LDN193189 (Stemgent, 04–0074) and 2 μM SB431542 (Sigma-Aldrich, S4317) were added in the medium after ROCK inhibitor withdrawal for 8 days (passage 1). Passage (PS) 1 hNSCs were passaged using Accutase. For expansion, hNSCs were plated at a density of 4×105 cells (from PS1 to PS2) or 2×105 cells (from PS3 to PS8) in a PLO/Fibronectin-coated 24 well plates, cultured in N2 medium with 20 ng/ml FGF2 at 37°C, 5% O2 and 5% CO2 and serially passaged every 6 days, after dissociation with HBSS. NSCs were PFA-fixed or RNA was collected at the end of every passage from PS1 to PS8. For FGF2 modulation experiments, at each passage from PS2, parallel hNSC cultures were exposed to different FGF2 doses (0.1, 1, 10 and 20 ng/ml) for 6 days and then PFA-fixed or processed for RNA extraction.
The 6 human iPSC lines (2063–1, −2; 2053–2, −6, 2075–1, −3) were differentiated into forebrain NSCs as previously described (Maroof et al., 2013). hNSCs were cultured in N2-B27 medium supplemented with 2 μM XAV939 (Stemgent, 04–0046), LDN193189 (100 nM) and SB431542 (10 μM) (XLSB) for 12 days. mTesR medium was gradually switched to N2-B27 in the first 7 days as following: day 0 100% mTesR + ROCK inhibitor, day 1 75% mTesR + 25% N2-B27, day 3 50% mTesR and N2-B27, day 5 25% mTesR + 75% N2-B27, day 7 100% N2-B27. NSCs were passaged on day 8 in N2 + B27 +XLSB; XLSB was withdrawn on day 12. On day 17, NSCs were passaged over astrocytes or without astrocytes and terminally differentiated in Neurobasal medium (NB) + B27 until day 32. RNA was collected on day 30 from neurons without astrocytes, on day 32 from neurons co-cultured with astrocytes.
Generation of human induced pluripotent stem cell (hiPSC) lines
Human fibroblasts (Donor 2075, 2053, and 2063) were seeded at 5 × 103 cells/cm2 in a Matrigel-coated plate and cultured with DMEM medium supplemented with 10% FBS (Life Technologies, 16000–044) and 2 mM L-glutamine. After 24 hours (day 1), the medium was changed to Pluriton human NUFF conditioned media with 300 ng/ml B18R protein. On day 1 and 5, the microRNA booster kit (Stemgent, 00–0073) was used with the StemFect RNA transfection reagent kit (Stemgent, 00–0069) to enhance reprogramming. On day 2–12, the OSKML RNAs were transfected. The mRNA reprogramming process was performed at 37°C in 5% O2 and CO2 incubator.
Rat astrocyte culture
Sprague-Dawley rat embryos hippocampi were dissected on E18 and subjected to papain dissociation (Worthington Biochemical, Ls 3126). Astrocytes were plated at a density of 80.000 cells/ml on glass coverslips coated with Laminin (Life Technologies, 23017–015) and poly-D-lysine (Sigma-Aldrich, P6407) in 24 well plates with 0.5 mL DMEM (Life Technologies, 11960069) with 10% FBS (Life Technologies, 16000–044), 6 mM Glutamax, at 37°C, 5% O2 and 5% CO2, for 2–3 weeks before over-seeding with mouse or human NSCs. After astrocytes seeding, medium was replaced every 7 days. The Johns Hopkins University School of Medicine’s Institutional Animal Care and Use Committee approved all the experimental protocols involving rats.
METHOD DETAILS
Immuno-fluorescence
For immuno-fluorescence (IF) cells were fixed with 4% paraformaldehyde for 15 min and permeabilized 1 hour using 0.1% Triton X-100 (Sigma-Aldrich)- PBS plus 10% donkey serum (Sigma-Aldrich, D9663) or 3% BSA, then incubated with primary antibodies overnight 4C. The following primary antibodies were used at the concentration indicated by manufacturer’s protocol: CaM Kinase II alpha (6G9) (NB100–1983), LMX1A (NBP1–81303) Novusbio; SYNAPSIN (106 001), HOMER (160 003) Synaptic System; EGFR (Ab231), FGFR1 phosphoY654 (Ab59194), TBR1 (Ab31940), REELIN (Ab18570), CYCLIN D1 (Ab10540), FGFR2 (Ab10648), BMPR1A (Ab38560) Abcam; HES1 (11988), p-SMAD1/5 (9516), CYCLIN D1 (2926), pERK1/2 (4370), FGFR1 (9740) Cell Signaling Technology; PAX6 (PRB-278P) BioLegend; NESTIN (MAB1259), OTX2 (AF1979), PDGFR alpha (AF1062; AF307), SOX2 (AF2018; MAB2018), SOX21 (AF3538), TuJ1 (MAB1195), EGFR (AF1280), O4 (MAB 1326) R&D Systems; GFAP (Z 0334) DAKO; HES5 (sc-13859), CUX1 (sc-13024), TLE4 (sc-9125), FGFR3 (sc-9007), LHX2 (sc-19344) Santa Cruz Biotechnology; FOXP2 (ABE73), TBR1 (AB2261), REELIN (MAB5366) Millipore; FOXG1 (M227) Takara/Clontech; anti GAD65/67 was kindly gifted by Dr. Christian Geis, Hans Berger Department of Neurology, Jena University Hospital, Germany (Hansen et al., 2013). Secondary antibody incubation was performed 1 hour RT with Alexa conjugated antibodies (Life Technologies). Nuclei were counterstained with DAPI (Sigma, D8417). Detection of EGFR and PDGFRα in mouse NSCs was performed without permeabilization, using fluorochrome-conjugated anti EGFR (AF1280) (1:100) and anti PDGFRα (AF1062) (1:500) primary antibodies (R&D System), prepared with Alexa Fluor antibody labeling kit (Life Technologies, A20181; A20184; A20186). Detection of a third marker was performed after fluorochrome-conjugated EGFR and PDGFRα antibodies incubation. When cell permebilization was required, cells were post-fixed, permeabilized and incubated with a primary antibody and Alexa conjugated secondary antibody, as described above. Edu incorporation assay was performed with hNSCs at PS4, 6 days after plating, using Click-iT EdU Cell Proliferation Kit (Invitrogen), as previously reported (Adepoju et al., 2014). Images were captured with Zeiss LSM780 microscope or Image M1 microscope fitted with Apotome2 confocal (Zeiss) and finally assembled with ImageJ.
High-content imaging analysis
Images were acquired with the Operetta automatic microscope (Perkin Elmer). Quantification of the cell fluorescence signals were performed by analysis of the images in batch mode with custom building blocks on a Columbus server (Perkin Elmer). Data were visualized with Spotfire (TIBCO). To facilitate cell-object identification, in addition to DAPI nuclei counterstaining, HSC-Cell Mask blue (Life Technology, H32720) or DRAQ5 (Cell Signaling Technology, 4084) labeling was performed according to manufacturer’s protocol.
Electrophysiological recordings
Coverslips were transferred to a recording chamber perfused at a rate of 2 ml/min at 32°C with ACSF containing 140 mM NaCl, 5 mM KCl, 2 mM CaCl2, 2 mM MgCl2, 10 mM HEPES, and 10 mM glucose (pH 7.3 adjusted with NaOH). Recordings were performed with an Axopatch 700b amplifier equipped with Digidata 1440 digitizer and pClamp10 software (Molecular Devices). Whole cell voltage and current clamp recordings were performed with borosilicate glass pipettes (4–6 MΩ) backfilled with a solution containing 135 mM K-gluconate, 20 mM KCl, 10 mM HEPES, 4 mM Mg-ATP, 0.3 mM Na2GTP, and 0.5 mM EGTA. Traces were analyzed offline using Clampfit software (Molecular Devices). Whole-cell voltage clamp recording were performed at a holding potential of −70 mV in neurons cultured over E18 rat astrocytes. Action potentials were induced by a series of 100 mS, 10 pA depolarizing current injections under current clamp configuration. Data are presented as means ± the standard error of the mean. For statistical analysis, a Student’s t test was performed using GraphPad Prism 6 unless otherwise noticed.
Time-lapse micro-recording
Passage 1 mouse NSCs were passaged onto coverslip-bottom, PLO-fibronectin-coated, dishes (Advanced TC 35 mm, Greiner Bio-one, 627965) for imaging, in N2 medium at the density of 100.000 cells/well and the indicated FGF2 dose, and incubated as above. 16–24 hours later, plates were etched with a custom etching device, to create an array of restricted growth areas of 300 × 300 um, as described in Ravin et al. (2008). Imaging was performed on a Zeiss 780 LSM microscope with scanning stage and 20× 0.8 NA objective inside an environmental enclosure (PECON) set to 37°C, 5% CO2 and 5% O2. Each of 20 restricted growth areas was imaged every 2 minutes using the 633 nm laser, DIC optics, and T-PMT detector. PDGFRα and EGFR expression was measured 1 hour after plate etching, by live labeling with fluorochrome-conjugated anti EGFR and PDGFRα antibodies, in N2 + FGF2, for 10 minutes at 37°C, 5% CO2 and 5% O2. Unbound antibody/fluorophore was removed with a N2 + FGF2 wash and labeled cells were imaged. Cells were continuously imaged with DIC optics for 48 hours (short-term lineage analysis) for cell cycle length assessment, or 120 hours (long-term lineage analysis) when differentiated. At the end of the time series, cells were fixed with 4% paraformaldehyde for 15 min, stained and imaged. Neurons, oligodendrocytes and astrocytes were identified by immuno-fluorescence using antibodies specific for TuJ1, O4 and GFAP. Movies were exported and assembled using ImageJ.
Lineage analysis
From each time-lapse micro-recording, complete lineage dendrograms were derived using BioCell (SIMI Reality Motion Systems), linking EGFR and PDGFRα expression in founder cells to terminal fates.
48 hours (2 days) lineage analysis: individual cell cycle lengths were determined by measurement of the time difference between the 1st and 2nd mitoses (generation 1) during the 48-hour observation window. When the cycle of a founder cell lasted more than 24 hours from the beginning of the recording and before the first mitosis, this time was considered as generation 1 and plotted. Aggregate statistics (mean, SD, p value Mann-Whitney U test) were calculated with GraphPad (Prism).
120 hours (5 days) lineage analysis: individual cells in the dendrograms are colored to reflect terminal fates. Cell lineage specification was defined by labeling retrospectively the ancestors of every generation as in Ravin et al. (2008). Briefly: if siblings share the same fate, the parent cell takes their same color; if they have different commitment, or one of them is not determined or lost, then the parent cell is marked as not specified; if one cell dies, the parent cell is defined with the fate of the other daughter. Commitment was considered as the point beyond which all the progeny of a cell belonged to the same determined lineage. These events were tallied and displayed as histograms across generation in Figure 2C, and as density plots across time in Figure S2E. Number of cells traced: 1106 in 10 ng/ml FGF2; 646 in 1 ng/ml FGF2; 202 in 0.1 ng/ml FGF2.
Adeno-associated virus (AAV) infection
AAV5-CMV-eGFP (Lot Number RVC0041; titer: 2.62E+13 vg/ml) and AAV6-CMV-eGFP (Lot Number RVC0065; titer: 1.98E+12vg/ml) were provided by Children’s Hospital of Philadelphia, Viral Vector Core. 5.2 ×1010 vg AAV5 or 1.98 × 109 vg AAV6 were used to infect NSCs. NSCs were cultured in 1 ng/ml FGF2 on 24 wells plates (IBIDI, 82406) with or without rat astrocytes, as indicated in the experiments, and infected in distinct wells with AAV5 or AAV6, 24 hours after seeding. Then, NSCs were fixed at DIV 2, or at DIV 15 after differentiation, and stained. Images were captured with the 40× 0.8NA objective-Zeiss LSM780 microscope. Fluorescence signal intensity was calculated as pixel intensity in each nucleus. High-content analysis was performed using custom blocks on a Columbus server (Perkin Elmer) and t tests were performed using GraphPad (Prism 6).
Neuro-morphology
AAV5- and AAV6-GFP+ neurons were imaged with 10× 0.45 NA objective using an Image M1 microscope fitted with Apotome2 confocal (Zeiss). Structural analysis was performed on single neurons using Neurolucida software (MBF Bioscience). GFP signal was used to guide the morphological tracing of the neurons randomly picked in the field. For the statistical analysis, Student’s t test was performed using GraphPad (Prism 6).
Bioinformatic and Statistical Analysis
Genome-wide CoGAPS (GWCoGAPS) analysis in parallel sets was run using default parameters as previously described (Fertig et al., 2010, 2012), for a range of k patterns (k = 11, 24 selected for mouse and human, respectively) and uncertainty as 10% of the data. Briefly, whole transcriptomic data was parallelized into seven sets. GWCoGAPS decomposes a matrix of experimental observations, D—here, log2 RNaseq RPKMs—with genes as rows and samples as columns, into two matrices, by the following equation:
Where, A is the amplitude matrix indicating the strength of involvement of a given gene in each pattern, P is the pattern matrix defining relationships (i.e., patterns) between samples. N and S are both functions of each element of AP and represent the Normal distribution and the standard deviation, respectively. Principle component analysis of gene-level RPKM from RNaseq data was done using the prcomp() function in R. Projection of GWCoGAPS and principal components gene weights defines patterns of relationships between samples in a new data associated with the gene expression signatures of the patterns from the primary data. These were achieved using the default projectR function in the projectR package available at https://github.com/genesofeve/projectR and (Sharma et al., 2020; Stein-O’Brien et al., 2019). Enrichment was calculated via either the calcCoGAPSStat function in the CoGAPS Bioconductor package or the geneSetTest function in the limma Bioconductor package in R. Agglomerative hierarchical clustering of genes using gene-level RPKM from RNaseq data or GWCoGAPS genes weights was performed using hclust() and cutree() with correlational distance (dist = 1-r) in the R statistical language.
To generate the list of genes specific to the cortical hem we used in vivo data from laser microdissected tissue from the developing macaque cortex (Bakken et al., 2016). Note that in Bakken et al. (2016) the designation of a sample as coming from the hem or other cortical regions is given in the primary data associated with this reference. In order to distinguish the hem from other regions of the developing cortex, we used t tests to contrast the microdissected hem tissue versus the ventricular zone, and hem versus all other developing cortical samples. For each gene we selected the maximum p value of these two contrasts. Then, we ranked all genes by this maximum p value, selecting the top genes from this list to create the heatmap in Figure 4B.
RNA-sequencing library preparation
2.5 × 105 PS2 mouse NSCs were plated over PLO/ fibronectin-coated 6 wells culture plates, as described above, with no astrocytes, and exposed to different doses of FGF2 (0.1, 1, 10 or 100 ng/ml). Human NSCs were plated over 24 well plates (IBIDI, 82406), as described above, and after PS1 serially passaged in 20 ng/ml FGF2 up to PS8, or exposed to different doses of FGF2 (0.1, 1, 10 or 20 ng/ml) for 6 days. At the indicated time points, total RNA was extracted from at least 3 different wells/condition using RNeasy Mini Kit (QIAGEN, 74106), according to manufacturer’s protocol. RNA quality control was performed using the Agilent 2100 Bio-analyzer System. RNaseq libraries were constructed using Illumina TruSeq Stranded Total RNA Ribo-Zero sample Prep Kit (for strand-specific libraries) following the manufacturer’s protocol. Briefly, ribosomal RNAs were removed using Ribo-Zero beads from ~800 ng DNase treated total RNA. Following purification, the resulting RNA was fragmented into small pieces using divalent cations under elevated temperature at 94°C for 2 min. Under this condition, the range of the fragment length obtained was 130–290 bp, with a median length of 185 bp. Reverse transcriptase and random primers were used to copy the cleaved RNA fragments into first strand cDNA. The second strand cDNA was synthesized using DNA Polymerase I and RNase H. These cDNA fragments then went through an end repair process using T4 DNA polymerase, T4 PNK and Klenow DNA polymerase, the addition of a single ‘A’ base using Klenow exo (3’ to 5’ exo minus) and the ligation of Illumina PE adapters using T4 DNA Ligase. An index was inserted into Illumina adapters so that multiple samples can be sequenced in one lane of 8-lane flow cell if necessary. The concentration of RNA libraries was measured by Qubit (Life Technologies). The quality of RNA-seq library was measured by LabChipGX (Caliper) using HT DNA 1K/ 12K/HiSens Labchip. The final cDNA libraries were sequenced using HiSeq 3000 for high throughput DNA sequencing.
RNA-sequencing data processing
After sequencing run the Illumina Real Time Analysis (RTA) module was used to perform image analysis, base calling, and the BCL Converter (CASAVA v1.8.2) were followed to generate FASTQ files which contain the sequence reads. The sequencing depth was over 80 million (40 million paired-end) mappable sequencing reads. Read-level Q/C was performed by FastQC (v0.10.1). Pair-end reads of cDNA sequences were aligned back to the human genome (UCSC hg19 from Illumina iGenome) or to the mouse genome (UCSC mm10 from Illumina iGenome) by the spliced read mapper TopHat (v2.0.4) with default option with “–mate-innder-dist 160” and “–library-type fr-firststrand” options. The alignment statistics and Q/C was achieved by samtools (v0.1.18) and RSeQC (v2.3.5) to calculate quality control metrics on the resulting aligned reads, which provided useful information on mappability, uniformity of gene body coverage, insert length distributions and junction annotation, respectively. To achieve gene-level expression profile, the properly paired and mapped reads were achieved by “samtools sort –n” option, and these reads were counted by htseq-count v0.5.3 (with intersection-strict mode and stranded option for RiboZero samples) according to gene annotation (Illumina iGenome), and RPKM was calculated. This provided 23,368 gene-level expression profiles.
Monkeys tissue processing
All animal procedures were performed in accordance with the policies of the Yale Institutional Animal Care and Use Committee. Rhesus macaque monkeys were bred in Rakic primate breeding colony at Yale and timed pregnant females underwent caesarian section at the required embryonic age. Macaque fetal brains were dissected, divided into coronal blocks, and immerse fixed in 4% paraformaldehyde overnight. Fixed brain blocks were cryoprotected in step-gradients of up to 30% sucrose/PBS for several days and then frozen. Sections were prepared at 30 μm on a Leica CM3050S cryostat. For single cell RNA-sequencing, fetal Visual cortices (V1) were isolated from one E77 and one E78 monkeys and mechanically triturated in single cell suspension as described below. The Yale University IACUC approved all experimental protocols involving monkeys.
Monkey single cell dissociation
One E77 and one E78 fetal rhesus macaque brain were sectioned. V1 cortex was placed in a tube containing HBSS. The sample was incubated with HBSS-Papain (2 mg/ml) (BrainBits)-DNase I 0.1mg/ml (StemCells) solution 25 min at 37Co. The solution was removed and the tissue was mechanically triturated in HBSS and filtered using a 30 μm cell strainer. Samples were diluted to 1000 cells/micro-liter and processed for single cell RNA-seq analysis within 20 min at Yale Center for Genome Analysis (YCGA) core facility.
Single cell RNA-sequencing
Single cell RNA-sequencing was performed using the 10X Genomics Single Cell 3’ RNA-Seq V2 libraries and sequencing protocol.
GEM Generation and Barcoding
Single cell suspension in RT Master Mix was loaded on the Single Cell A Chip and partition with a pool of about 750,000 barcoded gel beads to form nanoliter-scale Gel Beads-In-Emulsions (GEMs). Gel beads have primers containing (i) an Illumina R1 sequence (read 1 sequencing primer), (ii) a 16 nt 10x Barcode, (iii) a 10 nt Unique Molecular Identifier (UMI), and (iv) a poly-dT primer sequence. Upon dissolution of the Gel Beads in a GEM, the primers are released and mixed with cell lysate and Master Mix. Incubation of the GEMs then produced barcoded, full-length cDNA from poly-adenylated mRNA.
Post GEM-RT Cleanup, cDNA Amplification and library construction
Full-length, barcoded cDNA was amplified by PCR to generate sufficient mass for library construction. R1 (read 1 primer sequence) were added to the molecules during GEM incubation. P5, P7, a sample index, and R2 (read 2 primer sequence) were added during library construction via End Repair, A-tailing, Adaptor Ligation, and PCR. The final libraries contained the P5 and P7 primers used in Illumina bridge amplification.
Sequencing libraries
A Single Cell 3’ Library comprises standard Illumina paired-end constructs which begin and end with P5 and P7. The Single Cell 3’ 16 bp 10x Barcode and 10 bp UMI were encoded in Read 1, while Read 2 was used to sequence the cDNA fragment. Sequencing a Single Cell 3’ Library produces a standard Illumina BCL data output folder. The BCL data include the paired-end Read 1 (containing the 16 bp 10x Barcode and 10 bp UMI) and Read 2 and the sample index in the i7 index read.
Single Cell RNA-sequencing analysis
The Cell Ranger pipeline was used to perform the reads alignment, barcode counting and UMI counting. Macaca mulatta genome (Mmul_10) and annotation (release 103) were downloaded from https://www.ncbi.nlm.nih.gov/genome/215 and were provided for the reads alignment to gain better mapping quality (Pruitt et al., 2014). The filtered cells from Cell Ranger were not used, instead, real cells were recovered by EmptyDrops (Lun et al., 2019). Furthermore, iteratively clustering was used to remove low-quality clusters or clusters with high doublet scores, which were calculated via Scrublet package (Wolock et al., 2019). After quality control, 17161 cells with average 2216 detected UMIs from two samples were obtained for downstream analysis. The R Seurat package was applied to integrate the two datasets (Butler et al., 2018; Stuart et al., 2019). More specifically, 25 CCA dimensions were used for anchor finding and top 2500 highly variable genes were used for PCA and clustering analysis after data integration. For projection of single cell RNA-seq data into human H9 ESC derived NSC GWCoGAPS patterns (Figure 3), homolog genes expressed in at least 10 cells were included for the projection analysis.
Immuno-histochemistry
Immuno-histochemistry (IHC) was performed using methanol permeabilization (incubation in 100% methanol for 10 min at −20°C) after initial rehydration in PBS, followed by antigen retrieval via incubation in 0.01M Na citrate pH 6.0 at 95°C for 7 min. Blocking in 10% normal donkey serum (NDS)/PBS included 0.1% tween-20 and 0.2% Triton X-100, as did all subsequent steps. Primary and secondary antibody incubations included 5% NDS. Primary antibodies and dilutions for monkey tissue were: chicken anti-TBR2 (Millipore, AB15894, 1:200), rabbit anti-TBR1 (Abcam, AB31940, 1:750), rabbit anti-PAX6 (Millipore, AB2237, 1:750), goat anti-EGFR (R&D Systems, AF1280, 10 μg/mL), rabbit anti-pSMAD1/5 (Cell Signaling, 9510, 1:800). Secondary antibodies were Jackson DyLight donkey anti-(species) 488, 543, 647, and DAPI nuclear staining was provided by Vector mounting medium H-1200. Confocal imaging was performed on a Zeiss LSM 510 coupled with a Chameleon titanium sapphire 2-photon laser by Coherent. Large strips of macaque tissue were imaged with automated z stack tile scanning. Slight tile stitching defects or misalignments, and DAPI intensity artifacts were manually corrected in Adobe Photoshop or ImageJ. In Figure S4G i, two panels, each of 3 sequential images, were acquired. One from VZ to intermediate zone, and another from intermediate zone to CP. Then, the 2 panels were stitched. Slight low DAPI intensity artifact in CP tile was corrected.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical analysis and significance levels can be found in the figure legends. High-throughput image quantification of individual cell fluorescence was performed acquiring 50–100 fields per cell culture well of at least 3 replicates followed by analysis with Columbus server (Perkin Elmer). After a first analysis and visualization with Spotfire (TIBCO), the data were exported and re-elaborated with GraphPad Software Prism (6.0 or 7.0) for statistical analysis. Statistical analysis of the percentage of DIV 15 TuJ1 neurons/field, synaptic puncta/neurite length and EPSC frequency among the different FGF2 doses, shown in Figures 1 and S1, was performed employing unpaired nonparametric Mann-Whitney test with 95% confidence. Significance is indicated with *, **, or ***, meaning p-value ≤ 0.05, ≤ 0.01, or ≤ 0.001, respectively. Statistical analysis of marker fluorescence intensity across time and FGF2 doses (CUX1, TLE4, pERK, CycD1, CAMK2α, pSMAD1/5), shown in Figure S1, was performed using one-way ANOVA and Tukey test. Notice that only significance between FGF2 doses at a specific day is shown, not between each dose across the days. Significance is indicated for every p < 0.05 as as *: 0.1 versus 10; #: 1 versus 10; +: 0.1 versus 1 ng/ml FGF2. For statistical analysis of the quantification of the markers expressed in EGFRhigh versus PDGFRhigh cells shown in Figures 2 and S2 (BMPR1A, pSMAD1/5, FGFR2, pFGFR1, pERK1/2, SOX2, PAX6, HES1), thousands of cells from each well were individually segmented using Columbus. EGFRhigh and PDGFRhigh cells were identified and the fluorescence intensity of a given marker was quantified in every cell, obtaining mean value and SD of its expression in each population and condition. The analysis was done for 3 replicates. Then, unpaired multiple t-test was used. Significance is indicated as * for every p < 0.05. For statistical analysis of EGFRhigh versus PDGFRhigh cell cycle length, neuromorphology and layer marker fluorescence intensity in AAV5-GFP+ versus AAV6-GFP+ neurons, shown in Figures 2 and S2, unpaired nonparametric Mann-Whitney Test was employed. Significance is indicated with *, **, ***, or ****, meaning p-value ≤ 0.05, ≤ 0.01, ≤ 0.001, or ≤ 0.0001, respectively.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| CaM Kinase II alpha (6G9) | Novusbio | # NB100–1983; RRID:AB_10001339 |
| LMX1A | Novusbio | # NBP1–81303; RRID:AB_11008396 |
| SYNAPSIN | Synaptic System | # 106 001; RRID:AB_887805 |
| HOMER | Synaptic System | # 160 003; RRID:AB_887730 |
| EGFR | Abcam | # Ab231; RRID:AB_2293306 |
| FGFR1 phosphoY654 | Abcam | # Ab59194; RRID:AB_941585 |
| TBR1 | Abcam | # Ab31940; RRID:AB_2200219 |
| REELIN | Abcam | # Ab18570; RRID:AB_444539 |
| CYCLIN D1 | Abcam | # Ab10540; RRID:AB_297280 |
| FGFR2 | Abcam | # Ab10648; RRID:AB_297369 |
| BMPR1A | Abcam | # Ab38560; RRID:AB_722713 |
| HES1 | Cell Signaling Technology | # 11988; RRID:AB_2728766 |
| p-SMAD1/5 | Cell Signaling Technology | # 9516; RRID:AB_491015 |
| CYCLIN D1 | Cell Signaling Technology | # 2926; RRID:AB_2070400 |
| pERK1/2 | Cell Signaling Technology | # 4370; RRID:AB_2315112 |
| FGFR1 | Cell Signaling Technology | # 9740; RRID:AB_11178519 |
| PAX6 | BioLegend | # PRB-278P; RRID:AB_291612 |
| NESTIN | R&D Systems | # MAB1259; RRID:AB_2251304 |
| OTX2 | R&D Systems | # AF1979; RRID:AB_2157172 |
| PDGFR alpha | R&D Systems | # AF1062; # AF307; RRID:AB_2236897; RRID:AB_354459 |
| SOX2 | R&D Systems | # AF2018; # MAB2018; RRID:AB_355110; RRID:AB_358009 |
| SOX21 | R&D Systems | # AF3538; RRID:AB_2195947 |
| TuJ1 | R&D Systems | # MAB1195; RRID:AB_357520 |
| EGFR | R&D Systems | # AF1280; RRID:AB_354717 |
| O4 | R&D Systems | # MAB 1326;RRID:AB_357617 |
| GFAP | DAKO | # Z 0334; RRID:AB_10013382 |
| HES5 | Santa Cruz Biotechnology | # sc-13859; RRID:AB_2233041 |
| CUX1 | Santa Cruz Biotechnology | # sc-13024; RRID:AB_2261231 |
| TLE4 | Santa Cruz Biotechnology | # sc-9125; RRID:AB_793141 |
| FGFR3 | Santa Cruz Biotechnology | # sc-9007; RRID:AB_640487 |
| LHX2 | Santa Cruz Biotechnology | # sc-19344; RRID:AB_2135660 |
| FOXP2 | Millipore | # ABE73; RRID:AB_11214319 |
| TBR1 | Millipore | # AB2261; RRID:AB_10615497 |
| REELIN | Millipore | # MAB5366; RRID:AB_2285132 |
| FOXG1 | Takara/Clontech | # M227; RRID:AB_2827749 |
| GAD65/67 | Kindly gifted by Dr. Christian Geis, Hans Berger Department of Neurology, Jena University Hospital, Germany | (Hansen et al., 2013) |
| TBR2 | Millipore | # AB15894; RRID:AB_10615604 |
| PAX6 | Millipore | # AB2237; RRID:AB_1587367 |
| Alexa 488, 568, 647 conjugated antibodies | Life Technologies | N/A |
| Secondary antibodies donkey anti- (species) 488, 543, 647 | Jackson DyLight | N/A |
| Bacterial and Virus Strains | ||
| AAV5-CMV-eGFP | Children’s Hospital of Philadelphia, Viral Vector Core | Lot # RVC0041; titer:2.62E+13 vg/ml |
| AAV6-CMV-eGFP | Children’s Hospital of Philadelphia, Viral Vector Core | Lot # RVC0065; titer: 1.98E+12vg/ml |
| Biological Samples | ||
| Rhesus macaque monkey embryonic brain cells | Primate breeding colony at Yale | N/A |
| C57BL/6 mouse embryonic neural stem cells | Charles River | N/A |
| Sprague-Dawley rat embryonic astrocytes | Charles River | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Poly-L-ornithine (PLO) | Sigma | # P3655 |
| Fibronectin (FN) | R&D Systems | # 1030FN |
| DMEM/F12 medium | Mediatech | # 16–405-CV |
| Insulin | Sigma | # I6634 |
| Apotransferrin | Sigma | # T2036 |
| Progesterone | Sigma | # P8783 |
| Putrescine | Sigma | # P5780 |
| Sodium Selenite | Sigma | # S5261 |
| bFGF | R&D Systems | # 4114-TC |
| NeuroBasal medium | Life Technology | # 12348–017 |
| Glutamax | Life Technology | # 35050061 |
| B27 | Life Technology | # 17504–044 |
| BDNF | R&D Systems | # 248-BD |
| NT-3 | R&D Systems | # 267-N3 |
| LDN193189 | Stemgent | # 04–0074 |
| BMP4 | R&D System | # 5020-BP-010/CF |
| mTeSR1 | Stem Cell Technology | # 05850 |
| ROCK inhibitor | Sigma-Aldrich | # Y0503 |
| Matrigel | BD | # 354277 |
| Aggrewell medium | Stem Cell Technology | # 05893 |
| SB431542 | Sigma-Aldrich | # S4317 |
| XAV939 | Stemgent | # 04–0046 |
| FBS | Life Technologies | # 16000–044 |
| Poly-D-lysine | Sigma-Aldrich | # P6407 |
| DMEM | Life Technologies | # 11960069 |
| Laminin | Life Technologies | # 23017–015 |
| Papain | BrainBits | PAP |
| Critical Commercial Assays | ||
| microRNA booster kit | Stemgent | # 00–0073 |
| StemFect RNA transfection reagent kit | Stemgent | # 00–0069 |
| Alexa Fluor antibody labeling kit (488, 568, 647) | Life Technologies | # A20181; # A20184; # A20186 |
| RNeasy Mini Kit | QIAGEN | # 74106 |
| 10X Genomics Single Cell 3“ RNA-Seq V2 | 10X Genomics | N/A |
| Deposited Data | ||
| mouse RNaseq: in vitro E11.5 mouse cortical NSCs across neural differentiation Series GSE144158 |
This paper | https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE144158 |
| human RNaseq #1: H9 hESC-derived NSCs serially passaged Series GSE144156 | This paper | https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE144156 |
| human RNaseq #2: 6 hiPSC lines [2063–1,−2; 2053–2,−6; 2075–1,−3] across neural differentiation Series GSE144157 | This paper | https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE144157 |
| single-cell RNA-sequencing of E77 and E78 Rhesus macaque Visual cortex (V1) Series GSE144508 | This paper | https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE144508 |
| Developing human DLPF cortex RNaseq dataset | (Jaffe et al., 2018) | http://eqtl.brainseq.org/phase1/ |
| Laser micro-dissected macaque developing cortex microarray data | (Bakken et al., 2016) | http://www.blueprintnhpatlas.org/static/download |
| Human developing cortex single cell RNAseq data | (Nowakowski et al., 2017) | https://cells.ucsc.edu/?ds=cortex-dev |
| Fetal human cortex gene expression microarray data | (Miller et al., 2014) | http://www.brainspan.org/static/download.html |
| Cortecon | (van de Leemput et al., 2014) | http://cortecon.neuralsci.org/ |
| Differentiating cortical neurons derived from hiPSCs | (Ziller et al., 2015) | https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE62193 (supplemental file “GSE62193_ AllSamples_ geneExpression.fpkm_tracking.gz”) |
| Experimental Models: Cell Lines | ||
| H9 human pluripotent stem cell line | LIBD | N/A |
| human induced pluripotent stem cell lines | LIBD | N/A |
| Experimental Models: Organisms/Strains | ||
| C57BL/6 mouse embryos | Charles River | N/A |
| Rhesus macaque monkey embryos | Primate breeding colony at Yale | N/A |
| Software and Algorithms | ||
| Columbus server | Perkin Elmer | N/A |
| Spotfire | TIBCO | N/A |
| pClamp10 software | Molecular Devices | N/A |
| Clampfit software | Molecular Devices | N/A |
| GraphPad Prism | GraphPad Software | N/A |
| BioCell | SIMI Reality Motion Systems | (Ravin et al., 2008) |
| Neurolucida software | MBF Bioscience | N/A |
| GWCoGAPS | (Fertig et al., 2014; Stein-O’Brien et al., 2017) | In the R statistical language: if (!requireNamespace(“BiocManager,” quietly = TRUE)) install. packages (“BiocManager”) BiocManager:: install(“CoGAPS”) browseVignettes (“CoGAPS”) |
| ProjectR | (Sharma et al., 2020; Stein-O’Brien et al., 2019) | In the R statistical language: if (!requireNamespace(“BiocManager,” quietly = TRUE)) install.packages (“BiocManager”) BiocManager:: install(“projectR”) browseVignettes (“projectR”) |
Highlights.
EGFR and BMP signaling define the transition of cortical NSCs into neurons
In vitro hNSCs differ between passages in generating cortical neuronal fates
Cortical patterning and neurogenesis are executed across passages of in vitro hNSCs
Variation in organizer states precedes divergent neuronal bias across human pluripotent lines
ACKNOWLEDGMENTS
This work was supported by the Lieber Institute for Brain Development (LIBD), AstraZeneca, and NIH grants DA02399 and EY002593 (to P.R.) and MH106934, MH106874, MH116488, MH110926, and MH109904 (to N.S.). E.J.F. and G.S.-O. were supported by the NIH (CA177669, CA212007, and CA006973), the Johns Hopkins University Discovery Award, and the Johns Hopkins School of Medicine Synergy Award. We thank many members of LIBD and the Yale University Department of Neuroscience (Rakic and Sestan labs) for their helpful comments on this work and manuscript. We thank Dr. Alvaro Duque (Rakic lab) and MacBrainResource (https://medicine.yale.edu/neuroscience/macbrain/) for providing non-human primate (NHP) tissue (National Institute of Mental Health (NIMH) R01-MH113257 to Alvaro Duque).
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2020.107599.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Adepoju A, Micali N, Ogawa K, Hoeppner DJ, and McKay RD (2014). FGF2 and insulin signaling converge to regulate cyclin D expression in multipotent neural stem cells. Stem Cells 32, 770–778. [DOI] [PubMed] [Google Scholar]
- Amiri A, Coppola G, Scuderi S, Wu F, Roychowdhury T, Liu F, Pochareddy S, Shin Y, Safi A, Song L, et al. ; PsychENCODE Consortium (2018). Transcriptome and epigenome landscape of human cortical development modeled in organoids. Science 362, eaat6720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrae J, Hansson I, Afink GB, and Niste r, M. (2001). Platelet-derived growth factor receptor-alpha in ventricular zone cells and in developing neurons. Mol. Cell. Neurosci. 17, 1001–1013. [DOI] [PubMed] [Google Scholar]
- Androutsellis-Theotokis A, Leker RR, Soldner F, Hoeppner DJ, Ravin R, Poser SW, Rueger MA, Bae SK, Kittappa R, and McKay RD (2006). Notch signalling regulates stem cell numbers in vitro and in vivo. Nature 442, 823–826. [DOI] [PubMed] [Google Scholar]
- Androutsellis-Theotokis A, Murase S, Boyd JD, Park DM, Hoeppner DJ, Ravin R, and McKay RD (2008). Generating neurons from stem cells. Methods Mol. Biol. 438, 31–38. [DOI] [PubMed] [Google Scholar]
- Angevine JB Jr., and Sidman RL (1961). Autoradiographic study of cell migration during histogenesis of cerebral cortex in the mouse. Nature 192, 766–768. [DOI] [PubMed] [Google Scholar]
- Armentano M, Chou SJ, Tomassy GS, Leinga rtner, A., O’Leary DD, and Studer M (2007). COUP-TFI regulates the balance of cortical patterning between frontal/motor and sensory areas. Nat. Neurosci. 10, 1277–1286. [DOI] [PubMed] [Google Scholar]
- Arnold SJ, Huang GJ, Cheung AF, Era T, Nishikawa S, Bikoff EK, Molnár Z, Robertson EJ, and Groszer M (2008). The T-box transcription factor Eomes/Tbr2 regulates neurogenesis in the cortical subventricular zone. Genes Dev. 22, 2479–2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assimacopoulos S, Grove EA, and Ragsdale CW (2003). Identification of a Pax6-dependent epidermal growth factor family signaling source at the lateral edge of the embryonic cerebral cortex. J. Neurosci. 23, 6399–6403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakken TE, Miller JA, Ding SL, Sunkin SM, Smith KA, Ng L, Szafer A, Dalley RA, Royall JJ, Lemon T, et al. (2016). A comprehensive transcriptional map of primate brain development. Nature 535, 367–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop KM, Goudreau G, and O’Leary DD (2000). Regulation of area identity in the mammalian neocortex by Emx2 and Pax6. Science 288, 344–349. [DOI] [PubMed] [Google Scholar]
- Brennand K, Savas JN, Kim Y, Tran N, Simone A, Hashimoto-Torii K, Beaumont KG, Kim HJ, Topol A, Ladran I, et al. (2015). Phenotypic differences in hiPSC NPCs derived from patients with schizophrenia. Mol. Psychiatry 20, 361–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulchand S, Grove EA, Porter FD, and Tole S (2001). LIM-homeodomain gene Lhx2 regulates the formation of the cortical hem. Mech. Dev. 100, 165–175. [DOI] [PubMed] [Google Scholar]
- Burrows RC, Wancio D, Levitt P, and Lillien L (1997). Response diversity and the timing of progenitor cell maturation are regulated by developmental changes in EGFR expression in the cortex. Neuron 19, 251–267. [DOI] [PubMed] [Google Scholar]
- Butler A, Hoffman P, Smibert P, Papalexi E, and Satija R (2018). Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bystron I, Blakemore C, and Rakic P (2008). Development of the human cerebral cortex: Boulder Committee revisited. Nat. Rev. Neurosci. 9, 110–122. [DOI] [PubMed] [Google Scholar]
- Cai Y, Zhang Y, Shen Q, Rubenstein JL, and Yang Z (2013). A subpopulation of individual neural progenitors in the mammalian dorsal pallium generates both projection neurons and interneurons in vitro. Stem Cells 31, 1193–1201. [DOI] [PubMed] [Google Scholar]
- Carcamo-Orive I, Hoffman GE, Cundiff P, Beckmann ND, D’Souza SL, Knowles JW, Patel A, Papatsenko D, Abbasi F, Reaven GM, et al. (2017). Analysis of Transcriptional Variability in a Large Human iPSC Library Reveals Genetic and Non-genetic Determinants of Heterogeneity. Cell Stem Cell 20, 518–532.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caronia-Brown G, Yoshida M, Gulden F, Assimacopoulos S, and Grove EA (2014). The cortical hem regulates the size and patterning of neocortex. Development 141, 2855–2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cattaneo E, and McKay R (1990). Proliferation and differentiation of neuronal stem cells regulated by nerve growth factor. Nature 347, 762–765. [DOI] [PubMed] [Google Scholar]
- Chambers SM, Fasano CA, Papapetrou EP, Tomishima M, Sadelain M, and Studer L (2009). Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat. Biotechnol. 27, 275–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen KG, Mallon BS, Hamilton RS, Kozhich OA, Park K, Hoeppner DJ, Robey PG, and McKay RD (2012). Non-colony type monolayer culture of human embryonic stem cells. Stem Cell Res. (Amst.) 9, 237–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chizhikov VV, Lindgren AG, Mishima Y, Roberts RW, Aldinger KA, Miesegaes GR, Currle DS, Monuki ES, and Millen KJ (2010). Lmx1a regulates fates and location of cells originating from the cerebellar rhombic lip and telencephalic cortical hem. Proc. Natl. Acad. Sci. USA 107, 10725–10730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cholfin JA, and Rubenstein JL (2007). Patterning of frontal cortex subdivisions by Fgf17. Proc. Natl. Acad. Sci. USA 104, 7652–7657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawley JN, Heyer WD, and LaSalle JM (2016). Autism and Cancer Share Risk Genes, Pathways, and Drug Targets. Trends Genet. 32, 139–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Torre-Ubieta L, Stein JL, Won H, Opland CK, Liang D, Lu D, and Geschwind DH (2018). The Dynamic Landscape of Open Chromatin during Human Cortical Neurogenesis. Cell 172, 289–304.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Pasquale G, Davidson BL, Stein CS, Martins I, Scudiero D, Monks A, and Chiorini JA (2003). Identification of PDGFR as a receptor for AAV-5 transduction. Nat. Med. 9, 1306–1312. [DOI] [PubMed] [Google Scholar]
- Edri R, Yaffe Y, Ziller MJ, Mutukula N, Volkman R, David E, JacobHirsch J, Malcov H, Levy C, Rechavi G, et al. (2015). Analysing human neural stem cell ontogeny by consecutive isolation of Notch active neural progenitors. Nat. Commun. 6, 6500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Englund C, Fink A, Lau C, Pham D, Daza RA, Bulfone A, Kowalczyk T, and Hevner RF (2005). Pax6, Tbr2, and Tbr1 are expressed sequentially by radial glia, intermediate progenitor cells, and postmitotic neurons in developing neocortex. J. Neurosci. 25, 247–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst C (2016). Proliferation and Differentiation Deficits are a Major Convergence Point for Neurodevelopmental Disorders. Trends Neurosci. 39, 290–299. [DOI] [PubMed] [Google Scholar]
- Fertig EJ, Ding J, Favorov AV, Parmigiani G, and Ochs MF (2010). CoGAPS: an R/C++ package to identify patterns and biological process activity in transcriptomic data. Bioinformatics 26, 2792–2793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fertig EJ, Ren Q, Cheng H, Hatakeyama H, Dicker AP, Rodeck U, Considine M, Ochs MF, and Chung CH (2012). Gene expression signatures modulated by epidermal growth factor receptor activation and their relationship to cetuximab resistance in head and neck squamous cell carcinoma.BMC Genomics 13, 160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fertig EJ, Stein-O’Brien G, Jaffe A, and Colantuoni C (2014). Pattern identification in time-course gene expression data with the CoGAPS matrix factorization. Methods Mol. Biol. 1101, 87–112. [DOI] [PubMed] [Google Scholar]
- Fuentealba LC, Rompani SB, Parraguez JI, Obernier K, Romero R, Cepko CL, and Alvarez-Buylla A (2015). Embryonic Origin of Postnatal Neural Stem Cells. Cell 161, 1644–1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimori K, Ishikawa M, Otomo A, Atsuta N, Nakamura R, Akiyama T, Hadano S, Aoki M, Saya H, Sobue G, and Okano H (2018). Modeling sporadic ALS in iPSC-derived motor neurons identifies a potential therapeutic agent. Nat. Med. 24, 1579–1589. [DOI] [PubMed] [Google Scholar]
- Fukuchi-Shimogori T, and Grove EA (2001). Neocortex patterning by the secreted signaling molecule FGF8. Science 294, 1071–1074. [DOI] [PubMed] [Google Scholar]
- Geschwind DH, and Rakic P (2013). Cortical evolution: judge the brain by its cover. Neuron 80, 633–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glinka A, Wu W, Delius H, Monaghan AP, Blumenstock C, and Niehrs C (1998). Dickkopf-1 is a member of a new family of secreted proteins and functions in head induction. Nature 391, 357–362. [DOI] [PubMed] [Google Scholar]
- Grove EA, and Fukuchi-Shimogori T (2003). Generating the cerebral cortical area map. Annu. Rev. Neurosci. 26, 355–380. [DOI] [PubMed] [Google Scholar]
- Grove EA, Tole S, Limon J, Yip L, and Ragsdale CW (1998). The hem of the embryonic cerebral cortex is defined by the expression of multiple Wnt genes and is compromised in Gli3-deficient mice. Development 125, 2315–2325. [DOI] [PubMed] [Google Scholar]
- Gulsuner S, Walsh T, Watts AC, Lee MK, Thornton AM, Casadei S, Rippey C, Shahin H, Nimgaonkar VL, Go RC, et al. ; Consortium on the Genetics of Schizophrenia (COGS); PAARTNERS Study Group (2013). Spatial and temporal mapping of de novo mutations in schizophrenia to a fetal prefrontal cortical network. Cell 154, 518–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanashima C, Fernandes M, Hebert JM, and Fishell G (2007). The role of Foxg1 and dorsal midline signaling in the generation of Cajal-Retzius subtypes. J. Neurosci. 27, 11103–11111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen DV, Lui JH, Parker PR, and Kriegstein AR (2010). Neurogenic radial glia in the outer subventricular zone of human neocortex. Nature 464, 554–561. [DOI] [PubMed] [Google Scholar]
- Hansen N, Grünewald B, Weishaupt A, Colaç o, M.N., Toyka, K.V., Sommer, C., and Geis, C. (2013). Human Stiff person syndrome IgG-containing high-titer anti-GAD65 autoantibodies induce motor dysfunction in rats. Exp.Neurol. 239, 202–209. [DOI] [PubMed] [Google Scholar]
- HD iPSC Consortium (2017). Developmental alterations in Huntington’s disease neural cells and pharmacological rescue in cells and mice. Nat. Neurosci.20, 648–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickx M, Van XH, and Leyns L (2009). Anterior-posterior patterning of neural differentiated embryonic stem cells by canonical Wnts, Fgfs, Bmp4 and their respective antagonists. Dev. Growth Differ. 51, 687–698. [DOI] [PubMed] [Google Scholar]
- Hubler Z, Allimuthu D, Bederman I, Elitt MS, Madhavan M, Allan KC, Shick HE, Garrison E, T Karl M, Factor DC, et al. (2018). Accumulation of 8,9-unsaturated sterols drives oligodendrocyte formation and remyelination. Nature 560, 372–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson EL, Garcia-Verdugo JM, Gil-Perotin S, Roy M, Quinones-Hinojosa A, VandenBerg S, and Alvarez-Buylla A (2006). PDGFR alpha-positive B cells are neural stem cells in the adult SVZ that form glioma-like growths in response to increased PDGF signaling. Neuron 51, 187–199. [DOI] [PubMed] [Google Scholar]
- Jaffe AE, Straub RE, Shin JH, Tao R, Gao Y, Collado-Torres L, Kam-Thong T, Xi HS, Quan J, Chen Q, et al. ; BrainSeq Consortium (2018). Developmental and genetic regulation of the human cortex transcriptome illuminate schizophrenia pathogenesis. Nat. Neurosci. 21, 1117–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johe KK, Hazel TG, Muller T, Dugich-Djordjevic MM, and McKay RD (1996). Single factors direct the differentiation of stem cells from the fetal and adult central nervous system. Genes Dev. 10, 3129–3140. [DOI] [PubMed] [Google Scholar]
- Kanton S, Boyle MJ, He Z, Santel M, Weigert A, Sanchís-Calleja F, Guijarro P, Sidow L, Fleck JS, Han D, et al. (2019). Organoid single-cell genomic atlas uncovers human-specific features of brain development. Nature 574, 418–422. [DOI] [PubMed] [Google Scholar]
- Kim JH, Auerbach JM, Rodríguez-Gómez JA, Velasco I, Gavin D, Lumelsky N, Lee SH, Nguyen J, Sànchez-Pernaute R, Bankiewicz K, and McKay R (2002). Dopamine neurons derived from embryonic stem cells function in an animal model of Parkinson’s disease. Nature 418, 50–56. [DOI] [PubMed] [Google Scholar]
- Korada S, Zheng W, Basilico C, Schwartz ML, and Vaccarino FM (2002). Fibroblast growth factor 2 is necessary for the growth of glutamate projection neurons in the anterior neocortex. J. Neurosci. 22, 863–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwan KY, Lam MM, Johnson MB, Dube U, Shim S, Rašin MR, Sousa AM, Fertuzinhos S, Chen JG, Arellano JI, et al. (2012a). Species-dependent posttranscriptional regulation of NOS1 by FMRP in the developing cerebral cortex. Cell 149, 899–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwan KY, Sestan N, and Anton ES (2012b). Transcriptional co-regulation of neuronal migration and laminar identity in the neocortex. Development 139, 1535–1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang C, Campbell KR, Ryan BJ, Carling P, Attar M, Vowles J, Pere-stenko OV, Bowden R, Baig F, Kasten M, et al. (2019). Single-Cell Sequencing of iPSC-Dopamine Neurons Reconstructs Disease Progression and Identifies HDAC4 as a Regulator of Parkinson Cell Phenotypes. Cell Stem Cell 24, 93–106.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehtinen MK, Zappaterra MW, Chen X, Yang YJ, Hill AD, Lun M, Maynard T, Gonzalez D, Kim S, Ye P, et al. (2011). The cerebrospinal fluid provides a proliferative niche for neural progenitor cells. Neuron 69, 893–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lein ES, Belgard TG, Hawrylycz M, and Molnár Z (2017). Transcriptomic Perspectives on Neocortical Structure, Development, Evolution, and Disease. Annu. Rev. Neurosci. 40, 629–652. [DOI] [PubMed] [Google Scholar]
- Li W, Cogswell CA, and LoTurco JJ (1998). Neuronal differentiation of precursors in the neocortical ventricular zone is triggered by BMP. J. Neurosci. 18, 8853–8862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Santpere G, Imamura Kawasawa Y, Evgrafov OV, Gulden FO, Pochareddy S, Sunkin SM, Li Z, Shin Y, Zhu Y, et al. ; BrainSpan Consortium; PsychENCODE Consortium; PsychENCODE Developmental Subgroup (2018). Integrative functional genomic analysis of human brain development and neuropsychiatric risks. Science 362, eaat7615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lillien L, and Raphael H (2000). BMP and FGF regulate the development of EGF-responsive neural progenitor cells. Development 127, 4993–5005. [DOI] [PubMed] [Google Scholar]
- Lun ATL, Riesenfeld S, Andrews T, Dao TP, Gomes T, and Marioni JC; Participants in the 1st Human Cell Atlas Jamboree (2019). EmptyDrops: distinguishing cells from empty droplets in droplet-based single-cell RNA sequencing data. Genome Biol. 20, 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mabie PC, Mehler MF, and Kessler JA (1999). Multiple roles of bone morphogenetic protein signaling in the regulation of cortical cell number and phenotype. J. Neurosci. 19, 7077–7088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madison JM, Zhou F, Nigam A, Hussain A, Barker DD, Nehme R, van der Ven K, Hsu J, Wolf P, Fleishman M, et al. (2015). Characterization of bipolar disorder patient-specific induced pluripotent stem cells from a family reveals neurodevelopmental and mRNA expression abnormalities. Mol. Psychiatry 20, 703–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malatesta P, Hartfuss E, and Go tz, M. (2000). Isolation of radial glial cells by fluorescent-activated cell sorting reveals a neuronal lineage. Development 127, 5253–5263. [DOI] [PubMed] [Google Scholar]
- Mangale VS, Hirokawa KE, Satyaki PR, Gokulchandran N, Chikbire S, Subramanian L, Shetty AS, Martynoga B, Paul J, Mai MV, et al. (2008). Lhx2 selector activity specifies cortical identity and suppresses hippocampal organizer fate. Science 319, 304–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchetto MC, Belinson H, Tian Y, Freitas BC, Fu C, Vadodaria KC, Beltrao-Braga PC, Trujillo CA, Mendes AP, Padmanabhan K, et al. (2017). Altered proliferation and networks in neural cells derived from idiopathic autistic individuals. Mol. Psychiatry 22, 820–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariani J, Simonini MV, Palejev D, Tomasini L, Coppola G, Szekely AM, Horvath TL, and Vaccarino FM (2012). Modeling human cortical development in vitro using induced pluripotent stem cells. Proc. Natl. Acad. Sci. USA 109, 12770–12775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariani J, Coppola G, Zhang P, Abyzov A, Provini L, Tomasini L, Amenduni M, Szekely A, Palejev D, Wilson M, et al. (2015). FOXG1-Dependent Dysregulation of GABA/Glutamate Neuron Differentiation in Autism Spectrum Disorders. Cell 162, 375–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maroof AM, Keros S, Tyson JA, Ying SW, Ganat YM, Merkle FT, Liu B, Goulburn A, Stanley EG, Elefanty AG, et al. (2013). Directed differentiation and functional maturation of cortical interneurons from human embryonic stem cells. Cell Stem Cell 12, 559–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques S, Zeisel A, Codeluppi S, van Bruggen D, Mendanha Falcão A, Xiao L, Li H, Häring M, Hochgerner H, Romanov RA, et al. (2016). Oligodendrocyte heterogeneity in the mouse juvenile and adult central nervous system. Science 352, 1326–1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JA, Ding SL, Sunkin SM, Smith KA, Ng L, Szafer A, Ebbert A, Riley ZL, Royall JJ, Aiona K, et al. (2014). Transcriptional landscape of the prenatal human brain. Nature 508, 199–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyashita-Lin EM, Hevner R, Wassarman KM, Martinez S, and Rubenstein JL (1999). Early neocortical regionalization in the absence of thalamic innervation. Science 285, 906–909. [DOI] [PubMed] [Google Scholar]
- Mo Z, and Zecevic N (2008). Is Pax6 critical for neurogenesis in the human fetal brain? Cereb. Cortex 18, 1455–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molyneaux BJ, Arlotta P, Menezes JR, and Macklis JD (2007). Neuronal subtype specification in the cerebral cortex. Nat. Rev. Neurosci. 8, 427–437. [DOI] [PubMed] [Google Scholar]
- Monuki ES, Porter FD, and Walsh CA (2001). Patterning of the dorsal telencephalon and cerebral cortex by a roof plate-Lhx2 pathway. Neuron 32, 591–604. [DOI] [PubMed] [Google Scholar]
- Nakagawa Y, Johnson JE, and O’Leary DD (1999). Graded and areal expression patterns of regulatory genes and cadherins in embryonic neocortex independent of thalamocortical input. J. Neurosci. 19, 10877–10885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namba T, and Huttner WB (2017). Neural progenitor cells and their role in the development and evolutionary expansion of the neocortex. Wiley Interdis-cip. Rev. Dev. Biol. 6 10.1002/wdev.256. [DOI] [PubMed] [Google Scholar]
- Nishizawa M, Chonabayashi K, Nomura M, Tanaka A, Nakamura M, Inagaki A, Nishikawa M, Takei I, Oishi A, Tanabe K, et al. (2016). Epigenetic Variation between Human Induced Pluripotent Stem Cell Lines Is an Indicator of Differentiation Capacity. Cell Stem Cell 19, 341–354. [DOI] [PubMed] [Google Scholar]
- Noctor SC, Flint AC, Weissman TA, Dammerman RS, and Kriegstein AR (2001). Neurons derived from radial glial cells establish radial units in neocortex. Nature 409, 714–720. [DOI] [PubMed] [Google Scholar]
- Nowakowski TJ, Pollen AA, Sandoval-Espinosa C, and Kriegstein AR (2016). Transformation of the Radial Glia Scaffold Demarcates Two Stages of Human Cerebral Cortex Development. Neuron 91, 1219–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowakowski TJ, Bhaduri A, Pollen AA, Alvarado B, Mostajo-Radji MA, Di Lullo E, Haeussler M, Sandoval-Espinosa C, Liu SJ, Velmeshev D, et al. (2017). Spatiotemporal gene expression trajectories reveal developmental hierarchies of the human cortex. Science 358, 1318–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Leary DD, Chou SJ, and Sahara S (2007). Area patterning of the mammalian cortex. Neuron 56, 252–269. [DOI] [PubMed] [Google Scholar]
- Okabe S, Forsberg-Nilsson K, Spiro AC, Segal M, and McKay RD (1996). Development of neuronal precursor cells and functional postmitotic neurons from embryonic stem cells in vitro. Mech. Dev. 59, 89–102. [DOI] [PubMed] [Google Scholar]
- Parikshak NN, Luo R, Zhang A, Won H, Lowe JK, Chandran V, Horvath S, and Geschwind DH (2013). Integrative functional genomic analyses implicate specific molecular pathways and circuits in autism. Cell 155, 1008–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JK, Williams BP, Alberta JA, and Stiles CD (1999). Bipotent cortical progenitor cells process conflicting cues for neurons and glia in a hierarchical manner. J. Neurosci. 19, 10383–10389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pașca SP (2018). The rise of three-dimensional human brain cultures. Nature 553, 437–445. [DOI] [PubMed] [Google Scholar]
- Pollen AA, Nowakowski TJ, Chen J, Retallack H, Sandoval-Espinosa C, Nicholas CR, Shuga J, Liu SJ, Oldham MC, Diaz A, et al. (2015). Molecular identity of human outer radial glia during cortical development. Cell 163, 55–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt KD, Brown GR, Hiatt SM, Thibaud-Nissen F, Astashyn A, Ermo-laeva O, Farrell CM, Hart J, Landrum MJ, McGarvey KM, et al. (2014). RefSeq: an update on mammalian reference sequences. Nucleic Acids Res.42, D756–D763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Y, Zhang XJ, Renier N, Wu Z, Atkin T, Sun Z, Ozair MZ, Tchieu J, Zimmer B, Fattahi F, et al. (2017). Combined small-molecule inhibition accelerates the derivation of functional cortical neurons from human pluripotent stem cells. Nat. Biotechnol. 35, 154–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raballo R, Rhee J, Lyn-Cook R, Leckman JF, Schwartz ML, and Vaccarino FM (2000). Basic fibroblast growth factor (Fgf2) is necessary for cell proliferation and neurogenesis in the developing cerebral cortex. J. Neurosci. 20, 5012–5023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakic P (1974). Neurons in rhesus monkey visual cortex: systematic relation between time of origin and eventual disposition. Science 183, 425–427. [DOI] [PubMed] [Google Scholar]
- Rakic P (1988). Specification of cerebral cortical areas. Science 241, 170–176. [DOI] [PubMed] [Google Scholar]
- Rash BG, Lim HD, Breunig JJ, and Vaccarino FM (2011). FGF signaling expands embryonic cortical surface area by regulating Notch-dependent neurogenesis. J. Neurosci. 31, 15604–15617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rash BG, Tomasi S, Lim HD, Suh CY, and Vaccarino FM (2013). Cortical gyrification induced by fibroblast growth factor 2 in the mouse brain. J. Neurosci. 33, 10802–10814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rash BG, Duque A, Morozov YM, Arellano JI, Micali N, and Rakic P (2019). Gliogenesis in the outer subventricular zone promotes enlargement and gyrification of the primate cerebrum. Proc. Natl. Acad. Sci. USA 116, 7089–7094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravin R, Hoeppner DJ, Munno DM, Carmel L, Sullivan J, Levitt DL, Miller JL, Athaide C, Panchision DM, and McKay RD (2008). Potency and fate specification in CNS stem cell populations in vitro. Cell Stem Cell 3, 670–680. [DOI] [PubMed] [Google Scholar]
- Renner M, Lancaster MA, Bian S, Choi H, Ku T, Peer A, Chung K, and Knoblich JA (2017). Self-organized developmental patterning and differentiation in cerebral organoids. EMBO J. 36, 1316–1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaguchi H, Kadoshima T, Soen M, Narii N, Ishida Y, Ohgushi M, Takahashi J, Eiraku M, and Sasai Y (2015). Generation of functional hippocampal neurons from self-organizing human embryonic stem cell-derived dorsomedial telencephalic tissue. Nat. Commun. 6, 8896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandberg M, Flandin P, Silberberg S, Su-Feher L, Price JD, Hu JS, Kim C, Visel A, Nord AS, and Rubenstein JLR (2016). Transcriptional Networks Controlled by NKX2–1 in the Development of Forebrain GABAergic Neurons. Neuron 91, 1260–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxena M, Agnihotri N, and Sen J (2018). Perturbation of canonical and non-canonical BMP signaling affects migration, polarity and dendritogenesis of mouse cortical neurons. Development 145, dev147157. [DOI] [PubMed] [Google Scholar]
- Schafer ST, Paquola ACM, Stern S, Gosselin D, Ku M, Pena M, Kuret TJM, Liyanage M, Mansour AA, Jaeger BN, et al. (2019). Pathological priming causes developmental gene network heterochronicity in autistic subject-derived neurons. Nat. Neurosci. 22, 243–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma G, Colantuoni C, Goff LA, Fertig EJ, and Stein-O’Brien G (2020). projectR: An R/Bioconductor package for transfer learning via PCA, NMF, correlation, and clustering. Bioinformatics. 10.1093/bio-informatics/btaa183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Q, Wang Y, Dimos JT, Fasano CA, Phoenix TN, Lemischka IR, Ivanova NB, Stifani S, Morrisey EE, and Temple S (2006). The timing of cortical neurogenesis is encoded within lineages of individual progenitor cells.Nat. Neurosci. 9, 743–751. [DOI] [PubMed] [Google Scholar]
- Shi Y, Kirwan P, Smith J, Robinson HP, and Livesey FJ (2012). Human cerebral cortex development from pluripotent stem cells to functional excitatory synapses. Nat. Neurosci. 15, 477–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimogori T, and Grove EA (2005). Fibroblast growth factor 8 regulates neocortical guidance of area-specific thalamic innervation. J. Neurosci. 25, 6550–6560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- State MW, and Sestan N (2012). The emerging biology of autism spectrum disorders. Science 337, 1301–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein-O’Brien GL, Carey JL, Lee WS, Considine M, Favorov AV, Flam E, Guo T, Li S, Marchionni L, Sherman T, et al. (2017). PatternMarkers & GWCoGAPS for novel data-driven biomarkers via whole transcriptome NMF.Bioinformatics 33, 1892–1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein-O’Brien GL, Clark BS, Sherman T, Zibetti C, Hu Q, Sealfon R, Liu S, Qian J, Colantuoni C, Blackshaw S, et al. (2019). Decomposing Cell Identity for Transfer Learning across Cellular Measurements, Platforms, Tissues, and Species. Cell Syst. 8, 395–411.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storm EE, Garel S, Borello U, Hebert JM, Martinez S, McConnell SK, Martin GR, and Rubenstein JL (2006). Dose-dependent functions of Fgf8 in regulating telencephalic patterning centers. Development 133, 1831–1844. [DOI] [PubMed] [Google Scholar]
- Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM 3rd, Hao Y, Stoeckius M, Smibert P, and Satija R (2019). Comprehensive Integration of Single-Cell Data. Cell 177, 1888–1902.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Goderie SK, and Temple S (2005). Asymmetric distribution of EGFR receptor during mitosis generates diverse CNS progenitor cells. Neuron 45, 873–886. [DOI] [PubMed] [Google Scholar]
- Sur M, and Rubenstein JL (2005). Patterning and plasticity of the cerebral cortex. Science 310, 805–810. [DOI] [PubMed] [Google Scholar]
- Tiberi L, Vanderhaeghen P, and van den Ameele J (2012). Cortical neurogenesis and morphogens: diversity of cues, sources and functions. Curr. Opin. Cell Biol. 24, 269–276. [DOI] [PubMed] [Google Scholar]
- Vaccarino FM, Schwartz ML, Raballo R, Nilsen J, Rhee J, Zhou M, Doetschman T, Coffin JD, Wyland JJ, and Hung YT (1999). Changes in cerebral cortex size are governed by fibroblast growth factor during embryogenesis. Nat. Neurosci. 2, 848. [DOI] [PubMed] [Google Scholar]
- van de Leemput J, Boles NC, Kiehl TR, Corneo B, Lederman P, Menon V, Lee C, Martinez RA, Levi BP, Thompson CL, et al. (2014). CORTECON: a temporal transcriptome analysis of in vitro human cerebral cortex development from human embryonic stem cells. Neuron 83, 51–68. [DOI] [PubMed] [Google Scholar]
- Wamsley B, and Fishell G (2017). Genetic and activity-dependent mechanisms underlying interneuron diversity. Nat. Rev. Neurosci. 18, 299–309. [DOI] [PubMed] [Google Scholar]
- Wang D, Liu S, Warrell J, Won H, Shi X, Navarro FCP, Clarke D, Gu M, Emani P, Yang YT, et al. ; PsychENCODE Consortium (2018). Comprehensive functional genomic resource and integrative model for the human brain. Science 362, eaat8464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weller ML, Amornphimoltham P, Schmidt M, Wilson PA, Gutkind JS, and Chiorini JA (2010). Epidermal growth factor receptor is a co-receptor for adeno-associated virus serotype 6. Nat. Med. 16, 662–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willsey AJ, Sanders SJ, Li M, Dong S, Tebbenkamp AT, Muhle RA, Reilly SK, Lin L, Fertuzinhos S, Miller JA, et al. (2013). Coexpression networks implicate human midfetal deep cortical projection neurons in the pathogenesis of autism. Cell 155, 997–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolock SL, Lopez R, and Klein AM (2019). Scrublet: Computational Identification of Cell Doublets in Single-Cell Transcriptomic Data. Cell Syst. 8, 281– 291.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wonders CP, and Anderson SA (2006). The origin and specification of cortical interneurons. Nat. Rev. Neurosci. 7, 687–696. [DOI] [PubMed] [Google Scholar]
- Xu X, Wells AB, O’Brien DR, Nehorai A, and Dougherty JD (2014). Cell type-specific expression analysis to identify putative cellular mechanisms for neurogenetic disorders. J. Neurosci. 34, 1420–1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Sousa AMM, Gao T, Skarica M, Li M, Santpere G, Esteller-Cucala P, Juan D, Ferrández-Peral L, Gulden FO, et al. (2018). Spatiotemporal transcriptomic divergence across human and macaque brain development. Science 362, eaat8077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziller MJ, Edri R, Yaffe Y, Donaghey J, Pop R, Mallard W, Issner R, Gifford CA, Goren A, Xing J, et al. (2015). Dissecting neural differentiation regulatory networks through epigenetic footprinting. Nature 518, 355–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-sequencing data generated in this study are available at GEO Accession #: GSE144158; GSE144156; GSE144157; GSE144508.