Abstract
The CRISPR (clustered regularly interspaced short palindromic repeat)/Cas9 (CRISPR-associated nuclease 9) technology enables rapid, targeted, and efficient changes in the genomes of various model organisms. The short guide RNAs (gRNAs) of the CRISPR/Cas9 system can be designed to recognize target DNA within coding regions for functional gene knockouts. Several studies have demonstrated that the CRISPR/Cas9 system efficiently and specifically targets sea urchin genes and results in expected mutant phenotypes. In addition to disrupting gene functions, modifications and additions to the Cas9 protein enable alternative activities targeted to specific sites within the genome. This includes a fusion of cytidine deaminase to Cas9 (Cas9-DA) for single nucleotide conversion in targeted sites. In this chapter, we describe detailed methods for the CRISPR/Cas9 application in sea urchin embryos, including gRNA design, in vitro synthesis of single guide RNA (sgRNA), and the usages of the CRISPR/Cas9 technology for gene knockout and single nucleotide editing. Methods for genotyping the resultant embryos are also provided for assessing efficiencies of gene editing.
Keywords: CRISPR/Cas9, guide RNA, Cas9-deaminase, knockout, gene editing
Introduction
Targeted genome editing technologies enable site-specific modifications of the genomes of various model organisms. Three major genome editing approaches, ZFNs (zinc-finger nucleases), TALENs (transcription activator-like effector-based nucleases), and CRISPR/Cas9 have been applied to target sea urchin genes. Both ZFNs and TALENs employ modular proteins to recognize target DNA sequences. On the other hand, the CRISPR/Cas9 system uses RNA-based DNA recognition via complementary sequence between a guide RNA (gRNA) and its target DNA (Fig. 1). Cases of gene-targeting studies in sea urchins and comparison of these three editing tools have been reviewed recently (Cui, Lin & Su, 2017). Generally, the CRISPR/Cas9 system is easier to prepare and it offers higher efficiency for mutagenesis.
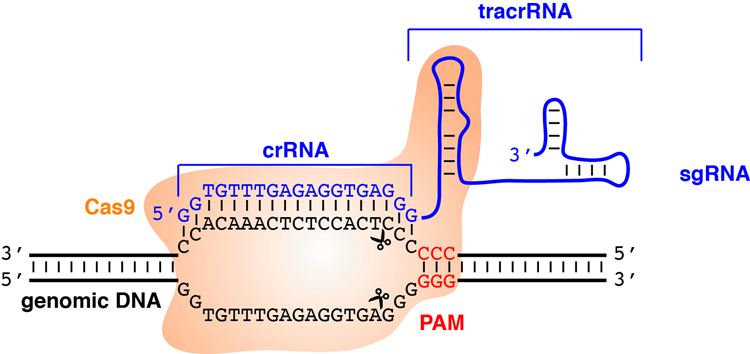
Fig. 1.
Schematic of the CRISPR/Cas9 system. A single guide RNA (sgRNA) is composed of a CRISPR RNA (crRNA) and a trans-activating crRNA (tracrRNA). Cas9 endonuclease recognizes the protospacer adjacent motif (PAM) and the sgRNA is annealed to the complementary region of genomic DNA adjacent to the PAM site. Cas9 cleaves the DNA at about 3-bp upstream of the PAM site and causes a double-stranded break.
The Cas9 endonuclease from the microbial adaptive immune system is brought to the targets by gRNAs that contain two components, a CRISPR RNA (crRNA) and a trans-activating crRNA (tracrRNA). The crRNA possesses an 18~20-nucleotide guide sequence recognizing target sites, while the tracrRNA interacts with the Cas9 endonuclease. These two RNA components can be in vitro-synthesized into a single guide RNA (sgRNA) (Jinek, Chylinski, Fonfara, Hauer, Doudna & Charpentier, 2012). sgRNAs and Cas9 protein form a ribonucleoprotein complex that scans the genome for NGG, the protospacer adjacent motif (PAM) recognized by Cas9. When the adjacent DNA sequence is complementary to the sgRNA sequence, the Cas9 endonuclease creates double-stranded breaks (DSBs). In the absence of a DNA template, DSBs are repaired through non-homologous end joining (NHEJ) pathway, which often results in random insertions or deletions (indels). Modifications of Cas9 have been developed for various applications. For example, two point mutations in the two nuclease domains of Cas9 (D10A and H840A) result in a catalytically dead Cas9 (dCas9) (Larson, Gilbert, Wang, Lim, Weissman & Qi, 2013). The CRISPR interference (CRISPRi) system utilizing a sgRNA-dCas9 complex is unable to create DSBs but remains its targeting ability to silence gene transcription. When only one of the two nuclease domains is mutated (D10A), the mutated Cas9 becomes a nickase (Cas9n) that introduces gRNA targeted single-stranded breaks (Ran, Hsu, Wright, Agarwala, Scott & Zhang, 2013). These activity-impaired Cas9 mutants can be further fused with other proteins to elicit specific responses. For example, tethering a transcription activation domain or a repressive domain to dCas9 converts it to a transcriptional activator or repressor, respectively (Gilbert, Larson, Morsut, Liu, Brar, Torres et al., 2013; Hilton, D’Ippolito, Vockley, Thakore, Crawford, Reddy et al., 2015; Maeder, Linder, Cascio, Fu, Ho & Joung, 2013). Fusions of dCas9 or Cas9n with a cytidine deaminase enzyme retain the ability to interact with gRNA and mediate single nucleotide conversion of cytidine to uridine, causing a C to T (or G to A) substitution (Komor, Kim, Packer, Zuris & Liu, 2016; Nishida, Arazoe, Yachie, Banno, Kakimoto, Tabata et al., 2016).
CRISPR/Cas9 technology is highly effective in the sea urchin Strongylocentrotus purpuratus. Single or multiple sgRNAs have been used to target to the nodal (Lin & Su, 2016), the polyketide synthase 1 (pks1), the glial cell missing (gcm) (Oulhen & Wessel, 2016), the nanos2 (Oulhen, Swartz, Laird, Mascaro & Wessel, 2017), and the delta gene (Mellott, Thisdelle & Burke, 2017). A modified CRISPR/Cas9 system fused to cytidine deaminase (Cas9-DA) is also able to edit the alx1, dsh, and pks genes (Shevidi, Uchida, Schudrowitz, Wessel & Yajima, 2017). It should be noted that as with all gene targeting approaches, off target sites must be considered in the experimental interpretation. Fortunately, many of the algorithms for calculating gRNA sites now have the genome of the sea urchin S. purpuratus included in the search analysis, e.g. CRISPRScan.org (Moreno-Mateos, Vejnar, Beaudoin, Fernandez, Mis, Khokha et al., 2015). Perhaps a gold standard in such targeting analysis is to have multiple different targeting mechanisms tested for gene function. In the gene targeting examples listed above, for example, the CRISPR/Cas9 result phenocopies the results using morpholino antisense oligonucletoides (MASO). It should be noted that several recent studies have revealed phenotypic differences between knockouts (i.e., mutants) and knockdowns (e.g., MASO-injected), and genetic compensation by functionally equivalent genes or unpredicted effects on mRNA processing in mutants may be attributable to these differences (Anderson, Mulligan, Shen, Wang, Scahill, Tan et al., 2017; El-Brolosy & Stainier, 2017; Rossi, Kontarakis, Gerri, Nolte, Holper, Kruger et al., 2015). Nevertheless, confidence in MASO and CRISPR/Cas9 applications are high as long as experimental prudence is employed (Angerer & Angerer, 2004; Stainier, Raz, Lawson, Ekker, Burdine, Eisen et al., 2017). In this chapter, we provide a list of materials and detailed methods for using the CRISPR/Cas9 system in sea urchin embryos. The methods include protocols for designing gRNA, preparing sgRNA and Cas9 capped mRNA, performing gene knockout and single nucleotide editing, and isolation of genomic DNA for genotyping.
Materials
- Reagents
-
1.1Addgene (www.addgene.org) has a large variety of plasmids encoding Cas9, and more are added at an increasing pace. Plasmids used in this chapter include pCS2-nCas9n (plasmid #47929), pT7-gRNA (plasmid #46759), and pCS2–3xFLAG-NLS-SpCas9-NLS (plasmid #51307).
-
1.2restriction enzymes and the corresponding reaction buffers (New England BioLabs), including BsmBI for sgRNA cloning into the pT7-gRNA vector and NotI for linearization of the pCS2-nCas9n vector
-
1.3MEGAscript SP6 Transcription Kit (Thermo Fisher Scientific)
-
1.4MEGAshortscript T7 Transcription Kit (Thermo Fisher Scientific)
-
1.5MEGAclear Transcription Clean-Up Kit (Thermo Fisher Scientific)
-
1.6Taq DNA Polymerase
-
1.710x NEBuffer™ 2 (New England BioLabs)
-
1.8proteinase K
-
1.9pGEM®-T Easy Vector Systems (Promega)
-
1.10T7E1 (NEB)
-
1.11KCl
-
1.12M13 Forward primer (5’-TGTAAAACGACGGCCAGT-3’), M13 Reverse primer (5’-CAGGAAACAGCTATGAC-3’), and pT7-gRNA-reverse primer (5’-GATCCGCACCGACTCGGTG-3’)
-
1.13QIAquick Gel Extraction Kit (Qiagen)
-
1.14QIAquick PCR Purification Kit (Qiagen)
-
1.15QIAprep Spin Miniprep Kit (Qiagen)
-
1.1
- Software
-
2.1CRISPRScan (www.crisprscan.org)
-
2.2CHOPCHOP v.2 (chopchop.cbu.uib.no)
-
2.3Primer-BLAST (www.ncbi.nlm.nih.gov/tools/primer-blast)
-
2.4OligoAnalyzer 3.1 (sg.idtdna.com/calc/analyzer)
-
2.5ImageJ (imagej.nih.gov/ij)
-
2.1
- Equipment
-
3.1LabChip (PerkinElmer)
-
3.2Microinjection apparatus
-
3.1
Methods
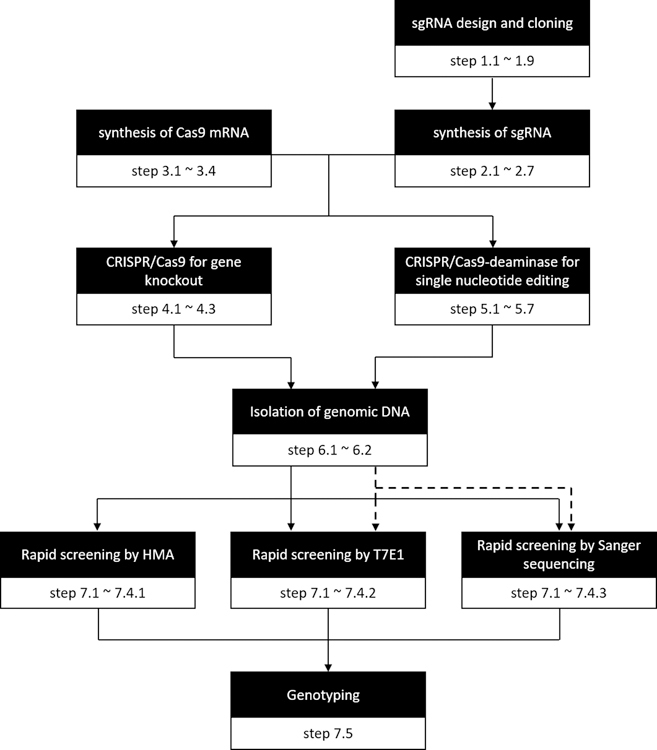
The methods are divided into 7 sections and the flowchart (Fig. 2) serves as a quick guide to specific steps or methods.
Fig. 2.
Flowchart of CRISPR/Cas9-mediated genome editing in sea urchin embryos. Specific steps for each method are indicated. The dashed arrows apply to the methods for verifying the results of single nucleotide editing.
-
gRNA design and cloning
Previously, we have used the ZiFiT online tool (zifit.partners.org/ZiFiT) to design sgRNA and used BLAST functions provided by Echinobase (www.echinobase.org) (Cameron, Samanta, Yuan, He & Davidson, 2009; Kudtarkar & Cameron, 2017) to predict potential off-target sites (Lin & Su, 2016). Other online tools without the sea urchin genome information, such as CRISPRko (portals.broadinstitute.org/gpp/public/analysis-tools/sgrna-design), can also be used to design sgRNAs in a similar way. Recently, several new web-based tools have included the sea urchin genome for sgRNA design, allowing us to predict the potential off-target sites. These include CRISPRScan (www.crisprscan.org), CHOPCHOP v2 (chopchop.cbu.uib.no), and CRISPRdirect (http://crispr.dbcls.jp) (Labun, Montague, Gagnon, Thyme & Valen, 2016; Moreno-Mateos et al., 2015), and the former two programs also provide scores of potential cleavage efficiency of CRISPR/Cas9. Here, we recommend using CRISPRScan to design the target sgRNAs. The sgRNAs are then cloned into the pT7-gRNA vector.
-
1.1Go to the CRISPRScan website (www.crisprscan.org).
-
1.1.1To find target sites located in the coding sequences (CDSs) of existing sea urchin gene models, click “By Gene” tab on the website. From the species name dropdown list, select “Sea urchin - Strongylocentrotus purpuratus” and enter a gene name or ensemble ID of a gene of interest. Set other parameters as follows: select “Cas9 – NGG”, the PAM sequence recognized by Cas9, and “In vitro T7 promoter” for finding target sites containing two guanine nucleotides at the 5’end for initial transcription of sgRNAs using the T7 RNA polymerase. Click “Get sgRNAs”.
-
1.1.2To identify target sites located in the non-coding region, click “Submit sequence” tab and enter your DNA sequence. Select the same parameters as above and click “Get sgRNAs”.
-
1.1.1
-
1.2
The result page displays a list of candidate sgRNAs targeting to either forward (+) or reverse (−) strand of genomic DNA in order of priority. For the initial assessment, the sgRNAs labeled in bright green should be considered first due to their higher estimated cleavage efficiencies and lack of potential off-target sites. Positions of sgRNA target sites also need to be taken into consideration (e.g. located at, or upstream of an important functional domain). The second priorities are the sgRNAs labeled in light green. These sgRNAs also lack potential off-target sites, although have lower estimated cleavage efficiencies. The least favored sgRNAs are labeled in gray that have potential off-target sites. In some cases, although only one of the first two nucleotides in the target sequences is guanosine, CRISPRScan still selects those and adds a second guanine nucleotides for transcription initiation of sgRNAs using the T7 RNA polymerase. One mismatch in the first two nucleotides between sgRNAs and their targets does not affect cleavage efficiencies (Thomas, Percival, Yoder & Parant, 2014). Nevertheless, because the sea urchin genome exhibits high degree of polymorphism, resequencing the genomic DNA around sgRNA target sites are recommended before preparing sgRNAs.
-
1.3Prepare sense and antisense oligonucleotides of the selected sgRNAs.
-
1.3.1For the sense oligonucleotide, copy the sequence shown in uppercase of the selected gRNA from step 1.2 and add “TA” to the 5’ end of the sequence. For the antisense oligonucleotide, reverse and complement the same sequence shown in uppercase, add “AAAC” to the 5’ end, and remove the “CC” at the 3’ end. Commercially synthesize the sense and antisense oligonucleotides followed by at least OPC (oligonucleotide purification cartridge) purification method. Continue to step 1.4~1.9 for cloning into the pT7-gRNA vector.
-
1.3.2Alternatively, click on the Help tab of CRISPRScan and construct two overlapping oligonucleotides – one being the unique targeting sequence that targets the Cas9 along with a T7 polymerase promoter on its 5’ end, the other “tail” sequence common to all of the sgRNAs. The two oligos have a common, complementary region for dimerization and extension, and one simply uses the PCR extension reaction given for making double stranded DNA templates for sgRNA synthesis (Vejnar, Moreno-Mateos, Cifuentes, Bazzini & Giraldez, 2016). No modification of sequence is needed with this protocol. Although the sgRNA template is not cloned into a vector for future cellular amplification, the amount of DNA template generated for sgRNA synthesis by the CRISPRscan protocol is extraordinary and will last for many, many experiments. If this protocol is used to synthesize the DNA template sgRNA, proceed to step 2.4 for in vitro synthesis of the sgRNA.
-
1.3.1
-
1.4
Resuspend the oligonucleotides with TE buffer to 100 μM. Make a mixture containing 10 μl of 100 μM sense oligonucleotide, 10 μl of 100 μM antisense oligonucleotide, 5 μl of 10x NEBuffer™ 2, and 25 μl of nuclease-free water. Mix well, spin down, and incubate the mixture at a PCR machine with the following program to anneal the sense and antisense oligonucleotides: 95°C for 5 min, decrease the temperature from 95°C to 85°C within 4 steps (−2.5°C/step and 10 s/step), and decrease the temperature from 85°C to 25°C within 12 steps (−5°C/step and 10 min/step).
-
1.5
To linearize the pT7-gRNA vector, make a mixture containing 10 μg of plasmid DNA, 2.5 μl of BsmBI restrict enzyme (10 U/μl), 10 μl of 10x NEBuffer™ 3.1, and nuclease-free water to a final volume of 50 μl. Mix well, spin down and incubate the mixture at 55°C for 3h, followed by heat inactivation for 20 min at 80°C. After enzyme digestion, the linearized vector is separated from the residual circular plasmid DNA by running through an 1% agarose gel following by purification using the QIAquick Gel Extraction Kit (Qiagen). At the wash step of the gel extraction procedures, wash the column twice with 740 μl of the PE buffer. For the elution step, elute DNA with 25 μl of nuclease-free water or the elution buffer (EB). Measure the concentration of the purified vector using a spectrophotometer (expected concentration ~ 150 ng/μl).
-
1.6
To ligate the annealed oligonucleotides into the BsmBI digested pT7-gRNA vector, make a mixture containing 100 ng of the digested pT7-gRNA vector, 1 μl of the 1:50 diluted annealed oligonucleotides, 5 μl of the 2x ligase buffer, 1 μl of T4 DNA ligase, and nuclease-free water to a final volume of 10 μl. Mix well, spin down and incubate the mixture overnight at 16°C.
-
1.7
Perform plasmid transformation following a standard protocol. Spread the transformed competent cells onto an agar plate with ampicillin and incubate overnight at 37°C.
-
1.8
Perform colony PCR using the sense oligonucleotide and M13 Forward primer to identify clones with complete sgRNA. Make a PCR mixture containing 1 μl of 10 μM sense oligonucleotide, 1 μl of 10 μM M13 Forward primer, 10 μl of 2x Taq master mix enzyme, and 8 μl of nuclease-free water. Pick a small amount of a single colony into the PCR mixture and run PCR with the following program: 94°C for 5 min, 35 cycles of (94°C for 30 s, 61°C for 30 s, and 72°C for 30 s), and 72°C for 7 min. After PCR amplification, check the size (~150 bp) by running 1 μl of the PCR products in an 1.5% agarose gel.
-
1.9
Pick one clone with correct insert size into 2 ml of LB broth (with ampicillin) and incubate overnight at 37°C. Isolate the plasmid DNA using a miniprep kit following the standard protocol. Sequence the plasmid DNA from both ends of the sgRNA using the M13 Forward primer and the M13 Reverse primer.
-
1.1
- Synthesis of sgRNA
-
2.1To synthesize sgRNAs, the DNA templates are prepared by PCR amplification using the M13 Reverse primer and the pT7-gRNA-reverse primer. Make a PCR mixture containing 10 ng of the sgRNA plasmid, 1 μl of 10 μM forward primer, 1 μl of 10 μM reverse primer, 25 μl of 2x Taq master mix enzyme, and nuclease-free water to a final volume of 50 μl. PCR with 30 cycles is then conducted with the following program: 94°C for 5 min, 30 cycles of (94°C for 30 s, 61°C for 30 s and 72°C for 30 s), and 72°C for 7 min.
-
2.2After PCR amplification, check the size (~160 bp) by running 1 μl of the PCR products in an 1.5% agarose gel.
-
2.3Purify the PCR products by using a PCR Purification Kit following the standard protocol. For the elution step, elute the DNA with 25 μl nuclease-free water or the elution buffer (EB). Measure the concentration of the purified PCR products by using a spectrophotometer.
-
2.4In vitro synthesize the sgRNA by using the MEGAshortscript T7 Transcription Kit following the standard protocol. Thaw the frozen reagents (10X Reaction Buffer and the A/C/G/UTP solution) and vortex until completely dissolved. Make a mixture containing DNA templates (up to 8 μg), 2 μl of the 10X Reaction Buffer, 2 μl of the A/C/G/UTP solution, 2 μl of T7 Enzyme Mix, and nuclease-free water to a final volume of 20 μl. Gently mix well by pipetting, briefly spin down and incubate the mixture at 37°C for 3.5 h. Add 1 μl of TURBO DNase to remove DNA, mix well and incubate at 37°C for 15 min. [Alternatively, the investigator may pool multiple DNA templates (either plasmid or PCR-extended templates) for a combined sgRNA product for initial testing.]
-
2.5Purify the in vitro synthesized sgRNAs by using the MEGAclear Transcription Clean-Up Kit following the standard protocol. To maximize the yield of sgRNAs, elute RNA twice from the column with 50 μl nuclease-free water to a final volume of 100 μl. [Alternatively, the miRNeasy Mini Kit (Qiagen) can be used to purify the in vitro synthesized sgRNAs according to manufacturer’s instructions and the final product eluted in 30 μl nuclease-free water following column purification. This yields 2,000 to 3,000 ng/μl. No EtOH precipitation step is necessary with this protocol.]
-
2.6Perform ethanol precipitation to concentrate the sgRNA by following the manufacture’s protocol (MEGAclear Transcription Clean-Up Kit). Add 10 μl of 5 M Ammonium Acetate to the purified sgRNA. Then add 275 μl of 100% ethanol, mix well, and incubate at −20°C for 30 min. Centrifuge at full speed for 15 min at 4°C. Carefully remove the supernatant and wash the pellets with 750 μl of 75% ethanol. Centrifuge again at full speed for 15 min at 4°C and carefully remove the supernatant. Repeat the ethanol wash and centrifugation steps again. Spin down and remove the residual ethanol by using a p20 pipette tip. Air dry the pellet until no ethanol remains. Resuspend the pellet in 20 μl of nuclease-free water. Check the size of the in vitro synthesized sgRNA by running 1 μl in an 1.5% agarose gel.
-
2.7Measure the concentration of the in vitro synthesized sgRNA by using a spectrophotometer (expected concentration ~2000 ng/μl). Aliquot the sgRNA into a small volume and store it at −80 °C. Prevent repeated freeze-thaw cycles of the sgRNA samples.
-
2.1
- Synthesis of Cas9 capped mRNA
-
3.1To synthesize Cas9 capped mRNA, the DNA templates (pCS2-nCas9n vector) are prepared by restriction enzyme digestion. Make a mixture containing 10 μg of pCS2-nCas9n vector, 2.5 μl of Not I restriction enzyme (10 U/μl), 5 μl of 10x NEBuffer™ 3.1, and nuclease-free water to a final volume of 50 μl. Incubate the mixture at 37°C for 2 h, followed by heat inactivation for 20 min at 65°C.
-
3.2After enzyme digestion, purify the linearized DNA by using a PCR Purification Kit following the standard protocol. For the elution step, elute the DNA with 25 μl nuclease-free water or the elution buffer (EB). Measure the concentration of the purified DNA by using a spectrophotometer.
-
3.3In vitro synthesize the Cas9 capped mRNA using the MEGAscript SP6 (or mMessage mMachine; ThermoFisher) Transcription Kit following the standard protocol. Thaw the frozen reagents (10X Reaction Buffer and 2X NTP/CAP) and vortex until completely dissolved. Make a mixture containing 1 μg of DNA templates, 2 μl of 10X Reaction Buffer, 10μl of 2X NTP/CAP, 2 μl of sp6 Enzyme Mix, and nuclease-free water to a final volume of 20 μl. Gently mix well by pipetting, briefly spin down, and incubate the mixture at 37°C for 3.5 h. Add 1 μl of TURBO DNase, mix well and incubate at 37°C for 15 min.
-
3.4Purify the in vitro synthesized Cas9 capped mRNA by using the MEGAclear Transcription Clean-Up Kit. Ethanol precipitate and resuspend the mRNA following the steps 2.6 and 2.7. If the mMessage mMachine kit is used for mRNA synthesis, a LiCl precipitation is suggested and present in the kit. To increase efficiency of Cas9 mRNA translation, the mRNA can be polyadenylated using the Poly(A) Tailing kit (AM1350; ThermoFisher). This does, however, make injection more difficult (stickiness and aggregation of the RNA mix).
-
3.1
- CRISPR/Cas9 for gene knockout
-
4.1Make a microinjection mixture containing 1500 ng of Cas9 capped mRNA, 300 ng of sgRNA, 0.25 μl of 1M KCL, and nuclease-free water to a final volume of 2 μl. The concentrations of Cas9 and sgRNA are 750 and 150 ng/μl, respectively. Keep the mixture on ice before microinjection. One might also consider adding a fluorescent dye e.g. FITC-dextran to identify injected embryos.
-
4.2Inject the solution into unfertilized sea urchin eggs. The diameter of the injected solution is about one third to one quarter of the egg (<5% of the egg volume). Controls for this experiment should include injecting Cas9 mRNA alone, without the sgRNAs, to evaluate the effect of injection and expressing exogenous mRNAs. Incubate the injected embryos and control sibling embryos as a negative control at 14°C. After the embryos reached the desired stage, they are subjected to the isolation of genomic DNA for genotyping.
-
4.3If there is no detectable phenotype in the sgRNA/Cas9 injected embryos, the injected concentration of sgRNA/Cas9 may be increased. However, in our experience, higher dosages of Cas9 mRNA (>1500 ng/μl) are toxic to the sea urchin embryos. On the other hand, if the injected embryos exhibit severe and nonspecific phenotypes, the injected concentration can be decreased. The ratio of the amount of Cas9 mRNA to the amount of sgRNA can also be adjusted (from 30:1 to 1:1). We find that easy visual phenotypes are advantageous to consider when beginning this protocol in the lab for the first time. For example, targeting the polyketide synthase gene (Calestani, Rast & Davidson, 2003) is advantageous because pigmentation presence or absence in the larva is clear and direct, whereas all other cells of the larvae are otherwise normal. Thus, one can easily interpret whether the experiment has off target, or Cas9 toxicity effects (unusual larva phenotype), or is highly selective (normal looking larva lacking pigment), all in the same larva.
-
4.1
-
CRISPR/Cas9-deaminase for single nucleotide editing
The Cas9 protein can be modified to contain a variety of DNA modifying activities. One can mutate the two nickase sites within Cas9 so that it still targets DNA, but does not cut the targeted site (dCas9), or only cuts one strand of the DNA (Cas9n or sCas9). Severed DNA in a cell may induce repair mechanisms that could alter the cell cycle and yield results independent of the actual gene. One could envisage adding sequences to a dead Cas9 (double nickase mutation, dCas9) such that targeted dCas9 now mutates DNA independent of cutting the DNA. This may include modifying chromatin structures, recruiting transcriptional regulators, or labeling sites within the genome in situ. Here we document how single nucleotide changes can be made, without severing genomic DNA, by adding a cytidine deaminase sequence to dCas9. Details of this protocol can be found in Shevidi et al., 2017.-
5.1The Shevidi et al. study used a Cas9 (plasmid #51307), which had been previously reported to function in Xenopus tropicalis (Nakayama, Fish, Fisher, Oomen-Hajagos, Thomsen & Grainger, 2013). This Cas9 construct demonstrated high translational efficiency in sea urchins, as well as nuclear localization from an added nuclear localization signal in the DNA construct. This Cas9 construct was used for the majority of functions, including mutations of the first nickase site (D10A; to yield sCas9) and the second site (H840A; dCas9) to completely inactivate Cas9’s cleaving activities. Mutagenesis of the nickase sites was performed using QuikChange II Site-Directed Mutagenesis Kit (cat. #200523–5) by following the manufacturer’s protocol.
-
5.2Cytosine deaminase 1 (PmCDA; GenBank accession #A5H718) from Petromyzon marinus (Nishida et al., 2016) was added to Cas9 at its C-terminus prior to the single or double mutagenesis protocol.
-
5.3For in vitro transcription (IVT), Cas9 was linearized with XhoI (Cas9) or NotI (sCas9, dCas9-DA) and transcribed using the mMESSAGE mMACHINE® SP6 Transcription Kit (Thermo Fisher; catalog #AM1340) by following the manufacturer’s protocol, with incubation for 5 hours at 37°C, followed by DNaseI treatment for 15 minutes at 37°C.
-
5.4RNA was then purified using the RNeasy Micro Kit from Qiagen (Cat. #74004) following the manufacturer’s protocol.
-
5.5The sgRNAs as constructed by CRISPRscan.org as described in section 1.4 above were used both for conventional CRISPR and CRISPR-DA analyses.
-
5.6By virtue of the single nucleotide changes in uncut DNA that result from such a modified Cas9, sequencing of the DNA was prudent to assess mutation sites and rates. See section 7.4.3 below.
-
5.7Sequencing results revealed that the CRISPR/Cas9-DA system favorably edits C-to-T changes between 8~20 bases upstream of the PAM sequence to the gRNA. The mutations may form a non-synonymous change in coding of the protein causing increased or decreased activity, or may form a stop codon prematurely in the coding region. Adjusting linker sequences between Cas9 and the deaminase sequence may enable increased targeting, or more flexibility in sites altered, depending on the goals of the experiment.
-
5.1
- Isolation of genomic DNA
-
6.1To rapidly screen the efficiencies of sgRNAs on DNA mutations, embryos collected in bulk (five or twenty embryos) are used for genomic DNA isolation. Wash the embryos with filtered sea water. Transfer the embryos with a minimum volume of sea water (0.5~2 μl) into a 0.2 ml PCR tube containing 5 μl of 1x NEBuffer™ 2. Incubate the samples at 94°C for 10 min, and then cool down to 4°C for 10 min. Add 1 μl of proteinase K (20 mg/ml) and 1x NEBuffer™ 2 to a final volume of 10 μl and then incubate at 55°C for 2 h. Boil the samples at 94°C for 10 min to inactivate proteinase K. Store the DNA solution at −20°C and the solution can be diluted 2–5 fold for use in PCR.
-
6.2To precisely link the phenotype to genotype, genomic DNA also can be isolated from a single embryo. Wash the embryos with filtered seawater. Transfer a single embryo in a volume of 0.5 μl of sea water into 0.2 ml PCR tube containing 1 μl of 1x NEBuffer™ 2. Incubate the samples at 94°C for 10 min, and then cool down to 4°C for 10 min. Add 0.5 μl of proteinase K (5 mg/ml) and then incubate at 55°C for 2 h. Boil the samples at 94°C for 10 min and the solution is diluted 2–5 fold for PCR.
-
6.1
-
Genotyping
The indels resulted from the NHEJ pathway following site-specific DSBs by Cas9 can be screened and examined. Here we provide three methods for rapid screening and one method used to examine indels for genotyping CRISPR/Cas9-injected sea urchin embryos. If an automated electrophoresis system is available, an initial high throughput rapid screening using the heteroduplex mobility assay (HMA) is recommended for identifying effective sgRNAs, although only indels of more than 2 base pairs can be detected using this method. T7 endonuclease I assay can also be performed to reveal non-perfectly matched DNAs resulted from the presence of indels. Sequencing genomic DNA around the target sites is ultimately required to examine the outcomes of editing and the possible gene products produced.-
7.1Design primer pairs encompassing the sgRNA target region using the Primer-BLAST online tool (www.ncbi.nlm.nih.gov/tools/primer-blast) or other available tools. Check the free energy (ΔG) of the selected primer pairs using the OligoAnalyzer 3.1 online tool (sg.idtdna.com/calc/analyzer). The recommended parameters are: ΔG of 3’ end hairpin >−2 kcal/mol; ΔG of 3’ end self/cross dimer >−5 kcal/mol.
-
7.2Amplify the DNA fragments (from both injected and un-injected embryos) by PCR with a total volume of 25 μl. After PCR amplification, perform electrophoresis with 1 μl of PCR products in an 1.5% agarose gel.
-
7.3Reanneal the PCR products by using the following program: 95°C for 5 min, decrease the temperature from 95°C to 85°C within 4 steps (−2.5°C/step and 10 s/step), and decrease the temperature from 85°C to 25°C within 12 steps (−5°C/step and 10 min/step).
-
7.4For the initial identification of effective sgRNAs, we recommend the high throughput rapid screening method (>24 samples) by using HMA in a commercial automated electrophoresis system, LabChip (7.4.1). For low throughput rapid screening (<12 samples), we perform the T7 endonuclease I (T7E1) assay (7.4.2) or Sanger sequencing of the purified PCR products (7.4.3).
-
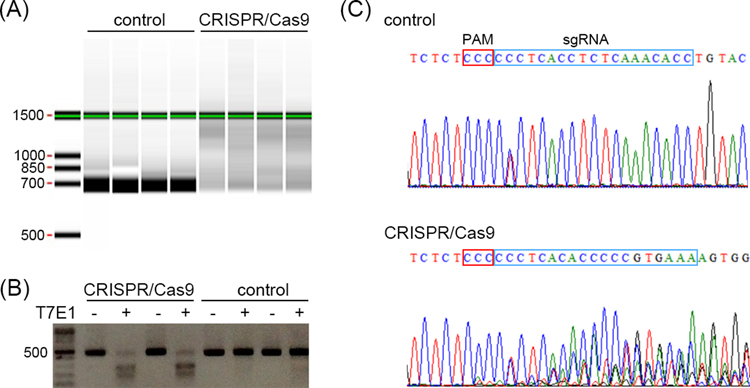
7.4.1Perform rapid screening by HMA on a LabChip following the standard protocol. In the uninjected, wild type embryos, the homoduplex DNA fragments can be detected as a clear, single band in the Gel View. When target genomic sites are cleaved by Cas9 endonuclease in the sgRNA/Cas9 injected embryos, the heteroduplex DNA fragments can be detected as smeared bands (Fig. 3A).
-
7.4.2Perform rapid screening using the T7E1 assay. Purify the reannealed PCR products using a PCR purification kit following the standard protocol. Measure concentrations of the purified DNA fragments using a spectrophotometer. Add ~1 μg of PCR products, 1 μl of 10x NEBuffer™ 2, 0.5 μl of T7E1 enzyme, and nuclease-free water to a final volume of 10 μl. Prepare a negative control with PCR products and the buffer, but without the T7E1 enzyme. Incubate at 37 °C for 30 min. Perform electrophoresis in an 1.5% agarose gel. In the uninjected, wild type embryos, the homoduplex DNA fragments are shown as a single band. In embryos injected with sgRNA/Cas9, T7E1 enzyme cleaves non-perfectly matched DNA resulting in two smaller fragments (Fig. 3B). Using the ImageJ software (https://imagej.nih.gov/ij/) to calculate the ratio of the undigested band to the two smaller bands for estimating the efficiency of gene editing.
-
7.4.3Perform rapid screening using Sanger sequencing. Purify the PCR products using a gel extraction kit or a PCR purification kit. For the wash step, wash the column twice with 740 μl of the PE buffer. Sequence the purified DNA fragments using the same forward or reverse primer used for PCR. Analyze the sequencing results. If Cas9 endonuclease cleaves the target genomic site, the sequencing chromatogram will reveal multiple peaks near the cutting site as compared to the distinctive, single peaks from the unedited embryos (Fig. 3C).
-
7.4.1
-
7.5To examine indels, we clone the PCR amplicons into the pGEM-T Easy Vector for subsequent sequencing. PCR amplification is performed with a final volume of 50 μl. Do not reanneal the DNA fragments after PCR amplification. Purify the PCR products using a gel extraction kit or PCR purification kit. For the wash step, wash the column twice with 740 μl of the PE buffer. Ligate the purified DNA fragments into the pGEM-T Easy vector using a T4 DNA ligase. Transform the plasmids into bacterial host cells and perform colony PCR using T7 and sp6 primers to check the insert size. Pick positive clones into 2 ml of LB broth and incubate overnight at 37°C. Isolate plasmid DNAs from ~10 positive clones and subject to sequencing using T7 or sp6 primer. Check indel events surrounding the sgRNA target site as compared to the sequences of the control embryos.
-
7.1
- General comments on strategies to consider in experimental design
-
8.1If beginning CRISPR/Cas9 in the lab for the first time, or for an animal not yet proven effective by this technology, consider targeting genes with easily visible outcomes. Pigment, unique cilia functions, a gene function in a specific cell type not otherwise needed for development, all allow easy assessment visually without needing to evaluate DNA immediately. Analysis of the genetic locus for mutations is far more time consuming and costly than performing the mutations themselves.
-
8.2Use of a MASO in combination with CRISPR/Cas9 targeting of the same gene function is a powerful approach to gene analysis. MASOs can block translation of maternal mRNAs, that CRISPR/Cas9 would not interfere with, whereas CRISPR/Cas9-induced mutations will last throughout the lifetime of the animal injected.
-
8.3We have noted that by using mRNA for Cas9 expression, and sgRNA injection for targeting that induced mutations can often be complete in all cells in both copies by as early as the four cell stage. We have also used recombinant expressed Cas9 protein complexed with sgRNAs is less successful. One, it costs more to purchase or make the Cas9 protein, but the protein and sgRNA form complexes that clog the injection needles of an opening needed for injecting sea urchin eggs and embryos.
-
8.4We routinely design and purchase 4–6 non-overlapping oligos per gene for constructing the sgRNA-template DNA. We will then initially inject a composite of these synthesized sgRNAs for initial phenotype analysis. If the phenotype proves sufficiently interesting for further study, one should consider then injecting individual sgRNAs and assessing the phenotype individually. When considering how to assess off target effects, multiple different sgRNAs yielding the same phenotype is a powerful dataset.
-
8.5In designing the sgRNA primers from cDNA or from transcriptome sequences, it is prudent to exclude sites at intron/exon boundaries. If the gene anatomy is not known, one should design additional sgRNA primers in the coding sequence with the realization that some of them will be hybrid sequences from two different exons and not be a legitimate genomic targeting sequence.
-
8.1
Fig. 3.
Three rapid screening methods for detecting genome editing in sea urchin embryos. (A) Rapid screening using the heteroduplex mobility assay (HMA) on a LabChip. DNA fragments from the CRISPR/Cas9 injected embryos are detected as smeared bands. (B) Rapid screening using the T7E1 assay. DNA fragments from the CRISPR/Cas9 injected embryos are cleaved into two smaller fragments. (C) Rapid screening using the Sanger sequencing. Sequencing chromatogram from the control embryo shows single peaks representing single PCR product. The only base showing two peaks may be a polymorphic site. In CRISPR/Cas9 injected embryos, multiple peaks near the cutting site are observed, indicating multiple PCR products due to indels introduced by NHEJ. The PAM site and the sgRNA target site are indicated with the red and blue box, respectively.
Summary
The CRISPR/Cas9 system is a game changer for mechanistic studies of gene function in echinoderms. Within a week, using the protocol documented at CRISPRScan.org mentioned above, and for about $20US, one can go from ordering sgRNA primers to having a tested phenotype in embryos. This approach is highly efficient and cost effective, but must also be conducted and interpreted carefully, with a plethora of controls. The CRISPR/Cas9 system is also in its infancy and investigators should consider creative approaches to modifying the Cas9 protein to target new functions, with greater control, to specific sites within the genome. The CRISPRScan.org site also includes other CRISPR-associated enzymes e.g. Cpf1, that may have additional applications. Such creative approaches, in concert with gene knock-outs, MASO comparisons, and the optical clarity of many echinoderms may make this experimental system of genetic analysis unique.
References
- Anderson JL, Mulligan TS, Shen MC, Wang H, Scahill CM, Tan FJ, … Farber SA (2017). mRNA processing in mutant zebrafish lines generated by chemical and CRISPR-mediated mutagenesis produces unexpected transcripts that escape nonsense-mediated decay. PLoS Genetics, 13, e1007105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angerer LM, & Angerer RC (2004). Disruption of gene function using antisense morpholinos. Methods Cell Biol, 74, 699–711. [DOI] [PubMed] [Google Scholar]
- Calestani C, Rast JP, & Davidson EH (2003). Isolation of pigment cell specific genes in the sea urchin embryo by differential macroarray screening. Development, 130, 4587–4596. [DOI] [PubMed] [Google Scholar]
- Cameron RA, Samanta M, Yuan A, He D, & Davidson E (2009). SpBase: the sea urchin genome database and web site. Nucleic Acids Research, 37, D750–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui M, Lin CY, & Su YH (2017). Recent advances in functional perturbation and genome editing techniques in studying sea urchin development. Brief Funct Genomics, 16, 309–318. [DOI] [PubMed] [Google Scholar]
- El-Brolosy MA, & Stainier DYR (2017). Genetic compensation: A phenomenon in search of mechanisms. PLoS Genetics, 13, e1006780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert LA, Larson MH, Morsut L, Liu Z, Brar GA, Torres SE, … Qi LS (2013). CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell, 154, 442–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilton IB, D’Ippolito AM, Vockley CM, Thakore PI, Crawford GE, Reddy TE, & Gersbach CA (2015). Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers. Nature Biotechnology, 33, 510–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, & Charpentier E (2012). A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science, 337, 816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komor AC, Kim YB, Packer MS, Zuris JA, & Liu DR (2016). Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature, 533, 420-+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudtarkar P, & Cameron RA (2017). Echinobase: an expanding resource for echinoderm genomic information. Database (Oxford), 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labun K, Montague TG, Gagnon JA, Thyme SB, & Valen E (2016). CHOPCHOP v2: a web tool for the next generation of CRISPR genome engineering. Nucleic Acids Research, 44, W272–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson MH, Gilbert LA, Wang X, Lim WA, Weissman JS, & Qi LS (2013). CRISPR interference (CRISPRi) for sequence-specific control of gene expression. Nat Protoc, 8, 2180–2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CY, & Su YH (2016). Genome editing in sea urchin embryos by using a CRISPR/Cas9 system. Developmental Biology, 409, 420–428. [DOI] [PubMed] [Google Scholar]
- Maeder ML, Linder SJ, Cascio VM, Fu YF, Ho QH, & Joung JK (2013). CRISPR RNA-guided activation of endogenous human genes. Nature Methods, 10, 977-+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellott DO, Thisdelle J, & Burke RD (2017). Notch signaling patterns neurogenic ectoderm and regulates the asymmetric division of neural progenitors in sea urchin embryos. Development, 144, 3602–3611. [DOI] [PubMed] [Google Scholar]
- Moreno-Mateos MA, Vejnar CE, Beaudoin JD, Fernandez JP, Mis EK, Khokha MK, & Giraldez AJ (2015). CRISPRscan: designing highly efficient sgRNAs for CRISPR-Cas9 targeting in vivo. Nature Methods, 12, 982–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama T, Fish MB, Fisher M, Oomen-Hajagos J, Thomsen GH, & Grainger RM (2013). Simple and Efficient CRISPR/Cas9-Mediated Targeted Mutagenesis in Xenopus tropicalis. Genesis, 51, 835–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida K, Arazoe T, Yachie N, Banno S, Kakimoto M, Tabata M, … Kondo A (2016). Targeted nucleotide editing using hybrid prokaryotic and vertebrate adaptive immune systems. Science, 353. [DOI] [PubMed] [Google Scholar]
- Oulhen N, Swartz SZ, Laird J, Mascaro A, & Wessel G (2017). Transient translational quiescence in primordial germ cells. Development. [DOI] [PMC free article] [PubMed]
- Oulhen N, & Wessel GM (2016). Albinism as a visual, in vivo guide for CRISPR/Cas9 functionality in the sea urchin embryo. Molecular Reproduction and Development, 83, 1046–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, & Zhang F (2013). Genome engineering using the CRISPR-Cas9 system. Nature Protocols, 8, 2281–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi A, Kontarakis Z, Gerri C, Nolte H, Holper S, Kruger M, & Stainier DY (2015). Genetic compensation induced by deleterious mutations but not gene knockdowns. Nature, 524, 230–233. [DOI] [PubMed] [Google Scholar]
- Shevidi S, Uchida A, Schudrowitz N, Wessel GM, & Yajima M (2017). Single Nucleotide Editing Without DNA Cleavage Using CRISPR/Cas9-deaminase in the Sea Urchin Embryo. Developmental Dynamics, 246, 1036–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stainier DYR, Raz E, Lawson ND, Ekker SC, Burdine RD, Eisen JS, … Moens CB (2017). Guidelines for morpholino use in zebrafish. PLoS Genetics, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas HR, Percival SM, Yoder BK, & Parant JM (2014). High-throughput genome editing and phenotyping facilitated by high resolution melting curve analysis. PLoS One, 9, e114632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vejnar CE, Moreno-Mateos MA, Cifuentes D, Bazzini AA, & Giraldez AJ (2016). Optimized CRISPR-Cas9 System for Genome Editing in Zebrafish. Cold Spring Harb Protoc, 2016. [DOI] [PubMed] [Google Scholar]