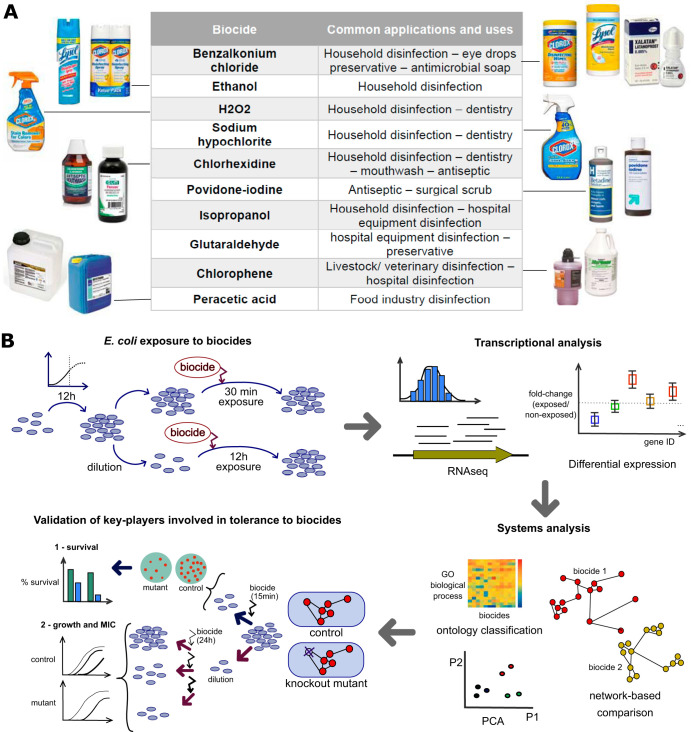
Antiseptics and disinfectant products are of great importance to control and eliminate pathogens, especially in settings such as hospitals and the food industry. Such products are widely distributed and frequently poorly regulated. Occasional outbreaks have been associated with microbes resistant to such compounds, and researchers have indicated potential cross-resistance with antibiotics. Despite that, there are many gaps in knowledge about the bacterial stress response and the mechanisms of microbial resistance to antiseptics and disinfectants. We investigated the stress response of the bacterium Escherichia coli to 10 common disinfectant and antiseptic chemicals to shed light on the potential mechanisms of tolerance to such compounds.
KEYWORDS: antiseptics, biocides, disinfectants, microbial resistance, stress response, transcriptomics
ABSTRACT
The mechanisms of the bacterial response to biocides are poorly understood, despite their broad application. To identify the genetic basis and pathways implicated in the biocide stress response, we exposed Escherichia coli populations to 10 ubiquitous biocides. By comparing the transcriptional responses between a short-term exposure (30 min) and a long-term exposure (8 to 12 h) to biocide stress, we established the common gene and pathway clusters that are implicated in general and biocide-specific stress responses. Our analysis revealed a temporal choreography, starting from the upregulation of chaperones to the subsequent repression of motility and chemotaxis pathways and the induction of an anaerobic pool of enzymes and biofilm regulators. A systematic analysis of the transcriptional data identified a zur-regulated gene cluster to be highly active in the stress response against sodium hypochlorite and peracetic acid, presenting a link between the biocide stress response and zinc homeostasis. Susceptibility assays with knockout mutants further validated our findings and provide clear targets for downstream investigation of the implicated mechanisms of action.
IMPORTANCE Antiseptics and disinfectant products are of great importance to control and eliminate pathogens, especially in settings such as hospitals and the food industry. Such products are widely distributed and frequently poorly regulated. Occasional outbreaks have been associated with microbes resistant to such compounds, and researchers have indicated potential cross-resistance with antibiotics. Despite that, there are many gaps in knowledge about the bacterial stress response and the mechanisms of microbial resistance to antiseptics and disinfectants. We investigated the stress response of the bacterium Escherichia coli to 10 common disinfectant and antiseptic chemicals to shed light on the potential mechanisms of tolerance to such compounds.
INTRODUCTION
Biocides, by the definition of the European Commission, include antiseptics, disinfectants, and preservatives. They are intended to control, eliminate, or reduce the number of undesired organisms, similar to antibiotics, which are used to eradicate infections in humans and animals (1). Several researchers have suggested the possibility of cross-resistance between biocides and antibiotics or other biocides (1–5). In contrast to the microbial mechanistic response to antibiotics, which has been extensively studied, there is a lack of understanding regarding the microbial response to biocides (6, 7), and the Food and Drug Administration (FDA) has recently expressed the need to collect more data related to biocide cross-resistance (8).
Research on biocide resistance is limited, partially due to a general belief that, unlike antibiotics, their multitarget action in the cell does not select for resistance to a specific target (9). In contrast, exposure to a subinhibitory concentration of triclosan, a broad-spectrum bisphenol biocide, was shown to select for Escherichia coli mutants with mutations in the fabI gene (10). fabI is homologous to genes found in other species, such as Staphylococcus aureus, and is also the target of therapeutic drugs against tuberculosis (11). Triclosan had been added to various household products for at least a decade, until very recently, when the FDA took action to remove this compound from most products since neither the safety nor the efficacy of triclosan has been shown (12). As the scientific community adds to our knowledge regarding the safety of biocide utilization, regulatory agencies become able to provide consumers and manufacturers with updated guidelines for the adequate use of these compounds.
Bacterial adaptation to biocides, which may include changes in gene expression as well as selection for mutants, may emerge for various reasons. Those include the irregular use of biocidal products, a gradient distribution around corners and difficult-to-reach areas, and improper disposal in the environment (7, 13). Aside from triclosan’s selection of fabI mutants (9, 10), multidrug efflux pumps have been one of the few well-studied mechanisms implicated in adaptation to biocides (14). Additional mechanisms of resistance and tolerance to biocides, such as changes in biofilm formation (15) and activation of soxRS and oxyR by oxidative agents, have also been proposed (16). Overall modification of the membrane composition has been reported and indirectly associated with resistant and cross-resistant phenotypes (17–19). Additionally, several Pseudomonas spp. are capable of degrading biocides such as quaternary ammonium compounds (7, 20). In contrast, the bacterial response to other biocides, such as povidone-iodine (POV) and glutaraldehyde (GLUTA), has not been studied, even though strains isolated from the environment were tolerant to such chemicals (21).
Transcriptomic data for microbes exposed to biocides can provide valuable information regarding the bacterial responses to subinhibitory concentrations of these compounds. Few studies have explored the genome-wide molecular responses of Escherichia coli and other bacteria to biocides. The E. coli expression profile following exposure to hydrogen peroxide has been studied by various groups, including ours (22, 23). A few researchers have explored the transcriptomics of different bacteria exposed to sodium hypochlorite (SOD) (24–26), ethanol (ETOH) (27, 28), povidone-iodine (25), benzalkonium chloride (BENZ) (24, 25), peracetic acid (PERA) (24, 29), and chlorhexidine (XID) (30, 31). Research on biocides has focused on a few compounds at a time under a range of experimental conditions with different protocols, strains, and media across research groups, making it difficult to compare data across these dimensions.
To bridge this gap, we first created a cohesive transcriptomics data set of E. coli MG1655 responses to subinhibitory concentrations of 10 commonly used biocides under otherwise identical conditions (Fig. 1A). We then identified both common and biocide-specific stress responses to each biocidal compound and performed differential expression and ontological analysis to elucidate the key players and their role in the bacterial response to specific biocides. We further validated the effects of those candidate genes by assessing the fitness effects of their knockouts through the use of survival assays and growth curves (Fig. 1B).
FIG 1.
Systems analysis of E. coli response to biocide stress. (A) Ten biocides frequently used in residential and commercial applications were selected. (B) Overview of the experimental and computational setting. E. coli MG1655 cells were grown for 12 h in minimal medium (M9 medium) with 0.4% glucose and then exposed for either a short term (30 min) or a long term (8 to 12 h) to the biocides benzalkonium chloride, chlorophene, chlorhexidine, hydrogen peroxide, glutaraldehyde, ethanol, isopropanol, peracetic acid, sodium hypochlorite, or povidone-iodine. Samples were taken for genome-wide transcriptomics profiling, determination of differentially expressed genes (DEGs), and Gene Ontology (GO) analysis. Biocide susceptibility was assayed on selected knockout mutants through the use of growth curves and survival assays.
RESULTS
The general, biocide-agnostic, stress response dominated gene expression.
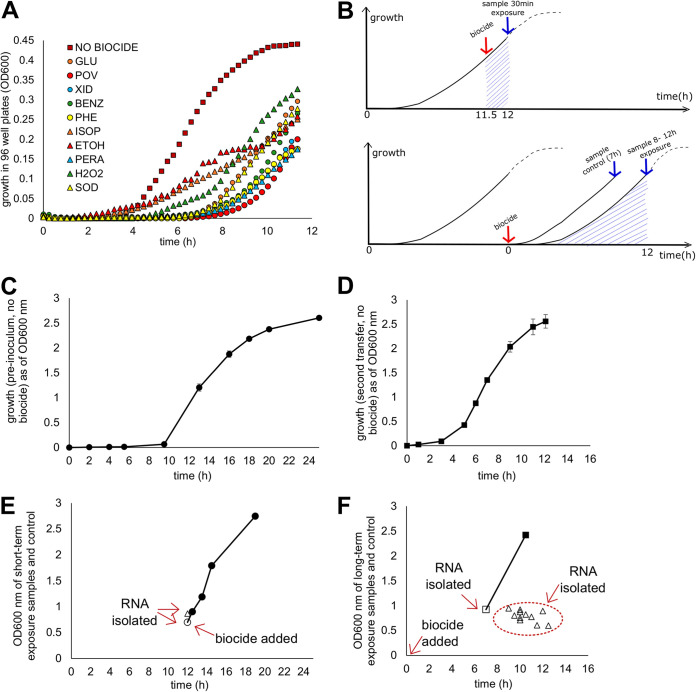
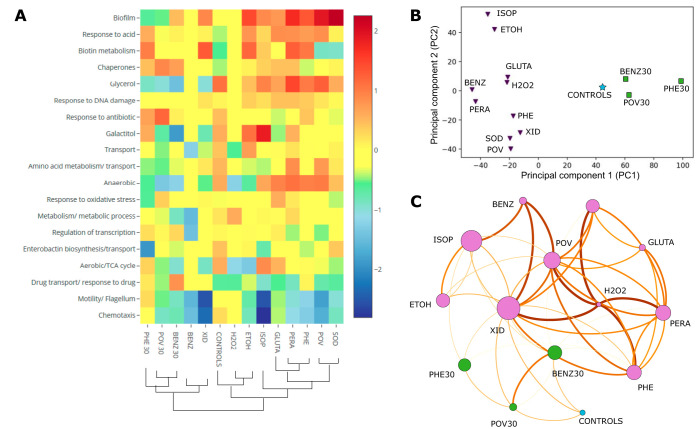
We evaluated the gene expression for Escherichia coli after continued short-term (30-min) and long-term (8-h to 12-h) exposure to 10 biocides (Fig. 1 and Table 1; see also Table S1 in the supplemental material and Materials and Methods for details). Transcriptomic samples were collected at mid-exponential growth (Fig. 2; see Materials and Methods for more information). We observed variation in the number of differentially expressed genes (DEGs) across conditions (205 to 396 genes in the short-term exposure versus 14 to 672 genes in the long-term exposure; Table S2). The short-term response to biocides was characterized by the upregulation of chaperones (14 DEGs; average log2 fold change [FC] in expression, 0.70) and response to drugs or antibiotics (22 DEGs; average log2 FC, 0.67). In comparison, the long-term response included the upregulation of biofilm formation (7 DEGs; average log2 FC, 1.13), the downregulation of chemotaxis (23 DEGs; average log2 FC, –1.27), and the downregulation of motility and flagellum assembly (45 DEGs; average log2 FC, –1.27) (Fig. 3A; Table 2; Fig. S1A).
TABLE 1.
Biocides utilized in this worka
| Biocide | Abbreviation | Concn (transcriptomics) | Neutralizer(s) (survival assay) | Group | Mode of action |
|---|---|---|---|---|---|
| Benzalkonium chloride | BENZ | 3.63 mg/liter | Lecithin (0.5%) and Tween 80 (1%) | Cationic agent (QAC) | Membrane damage |
| Chlorhexidine | XID | 1.48 μM | Lecithin (0.5%) and Tween 80 1%) | Cationic biguanide | Membrane damage |
| Chlorophene | PHE | 0.25 μM | Lecithin (0.5%) and Tween 80 (1%) | Halogenated phenolic | Inhibits membrane-bound proteins |
| Glutaraldehyde | GLUTA | 29 μM | Sodium bisulfite (1%) | Aldehyde | Protein denaturation and cross-linkage |
| Hydrogen peroxide | H2O2 | 272 μM | NA | Peroxygen | Oxidative |
| Ethanol | ETOH | 2.8% (vol/vol) | Dilution only | Alcohol | Membrane damage and protein denaturation |
| Isopropanol | ISOP | 2.7% (vol/vol) | Dilution only | Alcohol | Membrane damage and protein denaturation |
| Peracetic acid | PERA | 9 μM | Sodium thiosulfate (1%) and Tween 80 (1%) | Peroxygen | Oxidative |
| Povidone-iodine | POV | 12.5 μg/ml | Sodium thiosulfate (1%) | Halogen | Interacts with thiol groups on proteins |
| Sodium hypochlorite | SOD | 3.64 μM | Sodium thiosulfate (1%) | Chlorine | Oxidative |
The concentrations to which E. coli cells were exposed before samples were taken for transcriptomic assays are shown. The concentrations of biocides used for transcriptomics were determined to give, on average, 50% growth inhibition (according to the OD600) at 12 h compared to the growth of a control not exposed to a biocide. NA, not applicable (the assay was not performed); QAC, quaternary ammonium compound.
FIG 2.
Growth and sample collection protocol. (A) The growth curves for E. coli MG1655 in 96-well plates with and without the presence of each biocide at the concentration picked for transcriptomics assays. (B) Growth and sample collection protocol for samples exposed to biocide for the short (top) and long (bottom) term. (C) Typical growth of E. coli MG1655 in assay tubes without biocide, starting from a frozen culture. (D) Typical growth of E. coli MG1655 in assay tubes without biocide, starting from dilution of the preinoculum depicted in panel C. (E) Actual OD600 values for samples with a short-term exposure and the control at the time of collection (open circle, control; open triangle, samples with biocide exposure). Closed circles represent the trajectory of growth of the control after sample collection. (F) Actual OD600 values for samples with long-term exposure and the control at the time of collection (open square, control; open triangles, samples with biocide exposure). The closed square represents the trajectory of growth of the control after sample collection.
FIG 3.
Transcriptomic response of E. coli to 10 commonly used biocides. (A) DEGs (q < 0.05) from RNA-seq were organized into clusters based on GO biological process terms. The average between the log2 fold change in expression of DEGs belonging to the same category was calculated, and the biological processes were organized in descending order of differential regulation. (B) Principal-component analysis (PCA) for the DEGs identified (triplicates). Squares and triangles correspond to the samples with short- and long-term exposures, respectively. The star represents the result for the comparison between the two sets of controls. (C) Network connections between the biocide conditions. The node size depends on the number of DEGs for the specified biocide condition, and the connections between nodes (edges) are based on shared DEGs. A higher edge thickness indicates a higher number of shared DEGs compared to the total number of DEGs for the condition. Only the top three connections per condition are shown. BENZ, benzalkonium chloride; BENZ30, benzalkonium chloride exposure for 30 min; ETOH, ethanol; GLUTA, glutaraldehyde; H2O2, hydrogen peroxide; ISOP, isopropanol; PERA, peracetic acid; PHE, chlorophene; PHE30, chlorophene exposure for 30 min; POV, povidone-iodine; POV30, povidone-iodine exposure for 30 min; SOD, sodium hypochlorite; XID, chlorhexidine.
TABLE 2.
Biological processes that were the most affected for each conditiona
| Condition | Most affected biological process (score) |
|
|---|---|---|
| Upregulated | Downregulated | |
| BENZ | NA | Motility/flagellum (−1.56) |
| XID | Biotin metabolism (1.31) | Motility/flagellum (−2.52) |
| PHE | Biofilm (1.33) | Chemotaxis (−1.28) |
| GLUTA | Anaerobic growth (0.95) | Drug transport/response (−0.57) |
| H2O2 | Metabolism (0.62) | Aerobic growth (−1.15) |
| ETOH | Biofilm (1.46) | Aerobic growth (−1.33) |
| ISOP | Galactitol (1.86) | Chemotaxis (−2.80) |
| PERA | Biofilm (1.72) | Chemotaxis (−1.09) |
| POV | Biofilm (1.92) | Motility/flagellum (−1.81) |
| SOD | Biofilm (2.26) | Motility/flagellum (−0.98) |
| Long term | Biofilm (1.13) | Motility/flagellum (−1.27) |
| Long term | Glycerol (0.55) | Chemotaxis (−1.27) |
| BENZ30 | Drug transport/response (0.82) | Glycerol (−1.44) |
| PHE30 | Biotin metabolism (1.00) | Enterobactin (−1.92) |
| POV30 | Response to antibiotic (1.19) | Anaerobic growth (−1.34) |
| Short term | Chaperones (0.70) | Glycerol (−1.11) |
| Short term | Response to antibiotic (0.67) | Anaerobic growth (−1.05) |
The score corresponds to the average of the log2 fold change in expression for all the DEGs belonging to the corresponding pathway/biological process. The top up- and downregulated processes were also calculated for the average of all the conditions belonging to either short- or long-term exposure. NA, not applicable.
Principal-component analysis (PCA) of the DEG signatures revealed grouping of the samples into distinct groups that correlated with the time of exposure (short or long). While for the two alcohols, ethanol and isopropanol (ISOP), the various samples clustered together, as expected, the samples corresponding to short-term exposure formed a cluster separate from those exposed to the same biocide but for a longer duration (Fig. 3B; Fig. S1B). Among the most informative genes determined by sparse PCA, there were genes related to motility, respiration, the acid stress response, biofilm formation, and transport (Fig. S1C and D), all of which are processes controlled by genes that were differentially expressed distinctly between samples with short-term exposure and samples with long-term exposure. The network analysis recapitulated the clustering results together with common DEG associations (Fig. 3C), and an overview of the temporal dynamics is provided in Fig. 4.
FIG 4.
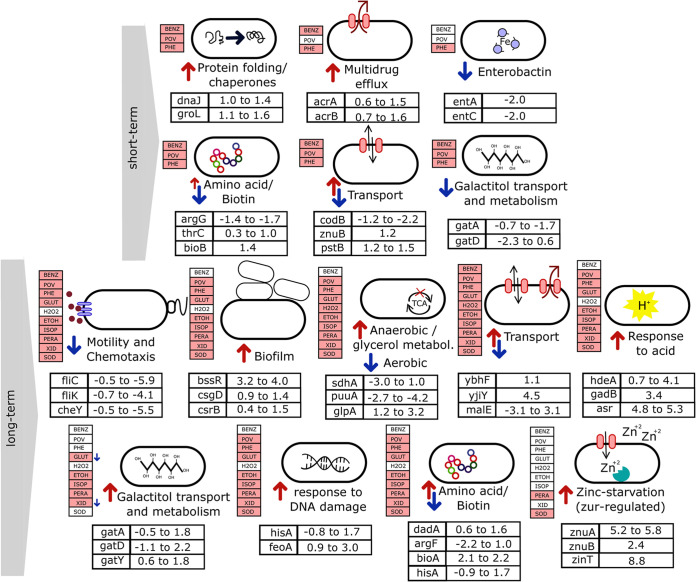
E. coli responses to short- and long-term biocide exposure. Upregulated processes (red arrows) and downregulated processes (blue arrows) after subinhibitory short-term (3 biocides) or long-term (10 biocides) biocide exposure. Biocides were colored when the biological process was affected (at least one DEG belonged to that category) after exposure to the biocide. Representative DEGs for each biological process are shown along with their log2 fold change in expression over all biocides. Detailed information with the complete list of DEGs can be found in Table S1 in the supplemental material. metabol., metabolism.
Protein folding was an early response to biocide stress.
The short-term exposure (30 min) of E. coli to biocides (BENZ for 30 min [BENZ30], chlorophene [PHE] for 30 min [PHE30], and POV for 30 min [POV30]) resulted in high levels of overexpression of several chaperones and cochaperonins, such as dnaK, dnaJ, groL, groS, htpG, hscA, cpxP, and clpB. Interestingly, this was not observed for cells exposed to the same biocides for more extended periods (Fig. 3A; Table S1). Exposure to povidone-iodine for 30 min resulted in the overexpression of ibpB (a molecular chaperone), which was among the genes for which the highest log2 FC (>6.1) was observed for the entire transcriptome data set (Table S3). Hence, the stress response to misfolded proteins appears to be an early response of the cell to stress, replaced later by other types of protective adaptations, such as biofilm formation. The exception was the chaperone gene spy, which was highly upregulated by long-term exposure to isopropanol. This chaperone has previously been shown to be highly upregulated in E. coli exposed to other alcohols (68).
Long-term exposure to biocides induced biofilm formation and shut down motility.
Long-term exposure (8 to 12 h) of E. coli to biocide stress resulted in the downregulation of genes related to motility and chemotaxis (Fig. 3A). Overall, this was a strong response for all biocides, except for glutaraldehyde (GLUTA) and H2O2 (Fig. 4). Downregulation was more expressive for cells exposed to chlorhexidine (XID) and isopropanol (ISOP) than for the cells exposed to the remaining conditions. For both biocides, the most downregulated gene was related to motility (Table S3). The promotion of biofilm formation occurs through the inhibition of motility (32), which is consistent with our results. Transcriptomic data revealed a strong indication of the induction of biofilm formation by long-term biocide exposure, even though different regulators were expressed in each group of biocides, such as bssR, csgD, and csrB. Our results indicate that although different biocide treatments may trigger the same outcome response (biofilm formation) by E. coli, diverse pathways and regulators are preferentially activated, depending on the agent utilized.
Long-term exposure to biocides rewired respiration pathways, induced anaerobiosis, and shut down the TCA cycle.
A significant number of genes related to the tricarboxylic acid (TCA) cycle were downregulated after the long-term exposure of E. coli to biocides (Fig. 3A; Table 3; Table S1), such as sdhBC and gltA. Accordingly, several genes appeared to be differentially regulated in the direction of an anaerobic state of the cells: cydAB, ompW, glpABC, and nrdD were upregulated, while katE, puuA, and acnA were downregulated (Table 3). Taken together, the evidence indicates that an anaerobic state of E. coli cells was induced by long-term biocide exposure.
TABLE 3.
Common genes involved in the biological processes affected by long-term biocide exposurea
| Process | Gene(s) | Description (reference) | Regulation | Condition(s) |
|---|---|---|---|---|
| Motility | fli series | Related to flagellum | Down | BENZ, ETOH, GLUTA, ISOP, PERA, PHE, POV, SOD, XID |
| flg series | Related to flagellum | Down | BENZ, ETOH, ISOP, PERA, PHE, POV, SOD, XID | |
| cheY, cheZ | Chemotaxis/motility | Down | BENZ, ETOH, ISOP, PERA, PHE, POV, SOD, XID | |
| motA, motB | Motility protein | Down | BENZ, ETOH, ISOP, PERA, PHE, POV, SOD, XID | |
| Biofilm | bssR | Regulator of biofilm formation | Up | PERA, PHE, POV, SOD |
| csgD | Transcriptional regulator | Up | GLUTA, ISOP, PHE, XID | |
| csrB | Regulatory RNA | Up | ETOH, ISOP, PERA, PHE, XID | |
| Aerobic/anaerobic growth | sdhB, sdhC, sdhA | Succinate dehydrogenase; part of the TCA cycle | Down | ETOH, H2O2, PERA, PHE, POV, SOD, XID |
| gltA | Citrate synthase; part of the TCA cycle | Down | ETOH, PERA, PHE, POV, SOD, XID | |
| cyA, cyB | Cytochrome components; induced under oxygen limitation conditions (58) | Up | POV, PERA, XID, SOD, PHE, ISOP, ETOH, GLUTA | |
| ompW | Outer membrane protein; upregulated during the transition from the aerobic to the anaerobic state (59) | Up | PERA, PHE, POV, SOD | |
| glpABC | Glycerol-3-phosphate dehydrogenase; functional under anaerobic conditions | Up | PERA, PHE, POV, SOD | |
| nrdD | DNA synthesis and repair; functional under anaerobic conditions (60) | Up | GLUTA, PHE, SOD, POV, PERA | |
| katE | Response to oxidative stress; upregulated in the aerobic state compared to its regulation during anaerobiosis (61) | Down | ETOH | |
| puuA | Putrescine utilization pathway; upregulated during the transition from the anaerobic to the aerobic state and downregulated in the absence of oxygen (62) | Down | ETOH, ISOP, SOD, XID PERA, PHE, POV | |
| acnA | Citric acid and glyoxylate cycles; repressed during anaerobiosis (63) | Down | ETOH, SOD | |
| Acid stress | hdeA, hdeB | Chaperone | Up | POV, XID, PHE, PERA, SOD, GLUTA |
| gadB, gadC | Glutamate decarboxylase | Up | XID, PHE, GLUTA | |
| asr | Acid shock protein | Up | ETOH, ISOP | |
| DNA damage | hisA | Isomerase | Up | PERA, PHE, POV, SOD, XID |
| feoA, feoB | Iron transport proteins | Up | ETOH, PERA, PHE, POV, SOD, XID | |
| deoABC | Thymidine phosphorylase | Up | ETOH, PERA, PHE, POV, SOD, XID | |
| Stress response | osmC | Has peroxidase activity and accumulates under some other stress conditions, such as osmotic stress (64) | Up | XID |
| uspGF | General stress factors with increased expression under several stress conditions (65) | Up | PERA, PHE | |
| gpmM | Role in oxidative stress response (66) | Up | PERA, POV | |
| wrbA | Unclear physiological functions in the cell | Up | PERA, PHE, POV, XID | |
| zinT | Protein that binds cadmium and zinc; was proposed to be a general stress factor with an unknown mechanism and a possible interaction role with ABC transporters (67) | Up | SOD | |
| Response to antibiotics | acrA | Multidrug efflux protein | Up | PHE, PHE30, BENZ30 |
| ybjG | Phosphatase | Up | BENZ30 | |
| phoU | Phosphate signaling protein | Up | BENZ30, POV30, ISOP, ETOH | |
| ybhG | Role in drug resistance (34) | Up | PHE, PHE30 | |
| sseA | 3-Mercaptopyruvate sulfurtransferase | Up | XID, ISOP |
A summary of the genes up- or downregulated after long-term exposure to biocides is shown. The complete list with the respective log2 fold change in expression and P values can be found in Table S1 in the supplemental material. When the reference is not explicit, the gene function was obtained from the EcoCyc database (55).
The acid stress response was ubiquitously present after long-term exposure to biocides and varied in its regulation.
The strong upregulation of genes associated with the response to acid functional category was observed across biocides after long-term exposure (Fig. 3A; Table S1). The genes hdeA and hdeB were upregulated upon exposure to some of the biocides, while gadB and gadC were upregulated upon exposure to others (Table 3). Additionally, asr was upregulated in both alcohols (Table 3). For cells exposed to povidone-iodine, hdeA was the most upregulated gene (Table S3). Similar to what was observed for biofilm regulators, different genes associated with the response to acid were preferentially overexpressed after exposure to each biocide. Still, the common outcome (in which genes involved in the functional category response to acid were upregulated) was consistently observed across treatments. None of the genes mentioned above was upregulated after short-term exposure to biocides (BENZ30, PHE30, and POV30), suggesting a role of the response to acid preferentially during the late stress response.
Biocide exposure upregulated the response to stress, DNA damage, and antibiotic response.
Several genes associated with the cellular response to DNA damage (33) were differentially expressed after biocide exposure. Among those, some were highly upregulated (log2 FC, >1), such as hisA, the feoAB operon, and the deoABC operon (Table 3).
Genes associated with the response to stress were also highly upregulated (log2 FC, >1) after the long-term exposure of E. coli to biocides, such as osmC, uspG, uspF, gpmM, wrbA, and zinT (Table 3). In particular, the zinT gene was highly upregulated after treatment with sodium hypochlorite, exhibiting the highest log2 FC (8.7) of the entire data set.
Cross-resistance between antimicrobials is a matter of increasing concern, and biocide exposure can potentially select for bacteria with increased tolerance to biocides and antibiotics (2–5). Among the genes that were differentially expressed after biocide exposure and that belong to the functional category response to antibiotic were acrA, encoding a multidrug efflux protein; ybjG, encoding a membrane protein; phoU, encoding a negative regulator; ybhG, encoding a putative membrane protein; and sseA, encoding the sulfurtransferase enzyme (Table 3).
Multidrug efflux pumps were not a general biocide response mechanism but specific to chlorophene, benzalkonium chloride, and chlorhexidine.
Multidrug efflux has been one of the most discussed processes related to biocide tolerance, mainly due to its importance for cross-resistance to antibiotics. Contrary to expectations, we found that this mechanism was upregulated only in the case of benzalkonium chloride, chlorhexidine, and chlorophene. Both the ybhF gene, encoding the multidrug ABC transporter, and the ybhG gene, encoding a membrane protein, which are suggested to play a role in the efflux of antibiotics, such as cefoperazone and chloramphenicol (34), were upregulated after short- and long-term exposure to PHE. The multidrug efflux protein gene mdtK was upregulated after exposure to both PHE and XID (Table S1).
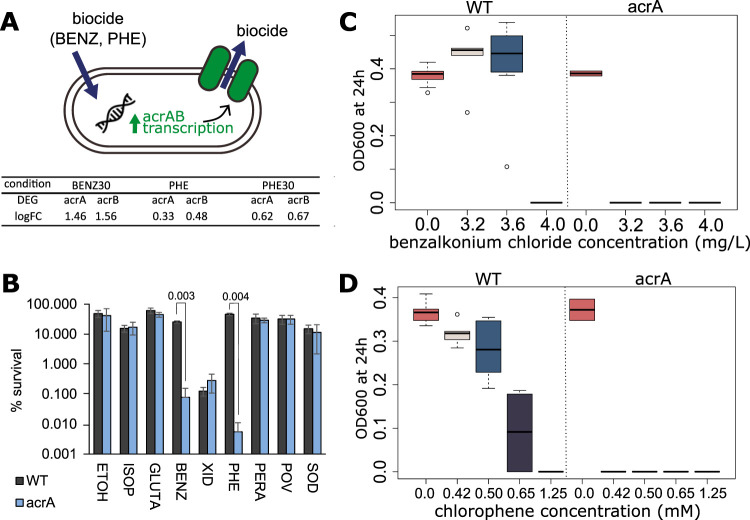
Besides those, AcrAB is one of the best-described and best-characterized pumps. The proteins contribute to antimicrobial resistance by decreasing the internal cell concentration of a wide range of compounds, such as antibiotics, detergents, and dyes (14). Also, acrB has been shown to play a role in enterobactin export (35). The multidrug efflux genes acrAB were upregulated after exposure to BENZ for 30 min (BENZ30) and after both short- and long-term exposure to chlorophene (PHE30 and PHE, respectively). The upregulation of acrAB may protect cells against harmful concentrations of such biocides by decreasing their level inside the cell (Fig. 5A). We evaluated the effect of exposure to BENZ, PHE, and seven additional biocides for an acrA knockout mutant. We observed that the absence of the efflux pump affected E. coli survival, which was specifically found for BENZ and PHE (Fig. 5B). We confirmed the increased susceptibility of the acrA mutant to the biocides BENZ and PHE with 96-well plate growth assays (Fig. 5C and D).
FIG 5.
Biocide susceptibility for the E. coli mutant with a knockout of the multidrug efflux protein gene acrA. (A) The genes for the multidrug efflux system AcrAB were upregulated after exposure to benzalkonium chloride and chlorophene. (B) Percent survival of the mutant acrA and parental strain (WT) exposed to the indicated biocides for 15 min. The concentrations were as follows: ETOH, 15% (vol/vol); ISOP, 11% (vol/vol); GLUTA, 125 μM; BENZ, 12.8 mg/liter; XID, 42 μM; PHE, 0.5 mM; PERA, 18 μM; POV, 33 μg/ml; SOD, 80 μM. Error bars represent the standard errors for biological duplicates, and the P values for significance are indicated at the top of the bars. (C and D) Growth at 24 h in the presence of the indicated concentration of the biocide in 96-well plates. (C) Benzalkonium chloride; (D) chlorophene. The definitions of the abbreviations are as follows: BENZ, benzalkonium chloride; XID, chlorhexidine gluconate; PHE, chlorophene; ETOH, ethanol; ISOP, isopropanol; GLUTA, glutaraldehyde; SOD, sodium hypochlorite; POV, povidone-iodine; SOD, sodium hypochlorite; WT, wild-type E. coli BW25113.
Zinc starvation genes played a role in E. coli survival after exposure to sodium hypochlorite and peracetic acid.
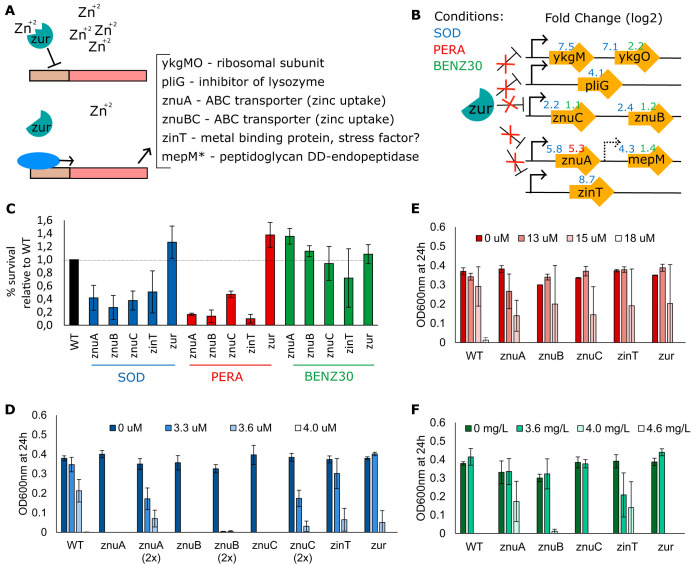
Zinc homeostasis in bacterial cells is maintained by uptake and export systems tightly regulated by their regulators, which include the Zur repressors and members of the Fur protein family of iron regulators. The metalloprotein Zur has a high affinity for zinc (36) and is proposed to regulate the zinc transporter genes znuABC (37), zinT (previously yodA) (38), pliG (39), and the putative ribosomal protein genes ykgMO (40) by binding to the DNA when zinc is abundant and repressing expression of the genes (Fig. 6A). We observed the overexpression of genes proposed to be regulated by zur after E. coli exposure to sodium hypochlorite (SOD), peracetic acid (PERA), and benzalkonium chloride (BENZ30) (Fig. 6B), even though no difference in expression of zur itself was detected. Expression levels were considerably higher for SOD and PERA, with log2 FC values being above 5 (Fig. 6B; Table S3).
FIG 6.
Exposure to biocides induces a zinc starvation response. (A) Zinc starvation disengages the transcriptional regulator zur from DNA binding (37), resulting in overexpression of zur-regulated genes (37–40). (B) Long-term exposure to sodium hypochlorite (SOD) and peracetic acid (PERA) and a short-term 30-min exposure to benzalkonium chloride (BENZ30) resulted in the overexpression of zur-regulated genes. (C) Percent survival relative to that of the wild type (E. coli BW25113) for the indicated mutants and E. coli MG1655 in SOD at 49 μM, PERA at 18 μM, and BENZ at 13 mg/liter. (D to F) Growth at 24 h in the presence of the indicated concentration of the biocide in 96-well plates. (D) Sodium hypochlorite; (E) peracetic acid; (F) benzalkonium chloride. Error bars represent standard errors for biological replicates.
We further investigated the survival of E. coli knockout mutants for znuABC, zinT, and zur after exposure to SOD, PERA, or BENZ for 15 min, followed by neutralization and plating into agar medium. The relative survival of the znuA, znuB, znuC, and zinT mutants was significantly lower than that of the wild-type strain after exposure to both SOD and PERA (Fig. 6C). On the other hand, a mutant for the repressor zur showed an increased ability to survive exposure to the biocides, which could have been a direct result of the derepression of the genes controlled by zur. Interestingly, we did not observe such an effect for mutants exposed to BENZ: no significant difference was observed between the mutants and the parent strain, except for the znuAB mutants, which were less sensitive to the biocide than the parent strain (Fig. 6C). We also assessed the growth of the mutants for 24 h in a plate reader in the presence of a range of concentrations of each biocide (Fig. 6D to F). Unlike the survival assay, this evaluation of growth did not include neutralization of the biocide and was an indication of growth inhibition (and not necessarily killing) by the biocides. All mutants showed increased sensitivity to SOD compared to the parent strain. However, the sensitivity of the znuABC mutant was considerably higher (Fig. 6D). Since the growth of the znuA, znuB, and znuC mutants was slower than that of the wild-type strain in minimal medium without biocide, we further assessed growth with an initial concentration of cells twice as high as that for the parent strain to minimize the effect of growth defects for evaluation of biocide susceptibility. The strains were still more susceptible to the biocide than the parent strain, even with a higher initial cell concentration (Fig. 6D). Differences between the mutants and the parent strain were not noticeable after exposure to PERA (Fig. 6E), and higher fluctuations between replicates indicated that the concentration of 15 μM was close to the limit for total growth inhibition for the cell concentration utilized in the assay. Growth in the presence of BENZ was similar to that of the wild type for most mutant strains except the znuA and zinT mutants (Fig. 6F).
DISCUSSION
Genome-wide transcriptional profiling of the biocide response usually entails experiments in single biocides, at single time points, and under different conditions, factors which complicate comparisons and the delineation of general versus specific responses to biocide exposure (22–31).
Even though it is widely known that stress conditions generate a temporally structured gene expression response in bacterial cells (41), most studies related to biocides are limited to observations at a single time point, limiting our understanding of the bacterial response to such chemicals. Here, our inclusion of samples at early and late exposure times allowed us to observe a temporal choreography of responses and differences between short- and long-term exposure to biocides. One of these was the upregulation of chaperones during the short-term exposure to biocides, indicating that the response to misfolded proteins is exclusively an early stress response. Accordingly, the expression of amino acid-related genes was markedly distinct between cells exposed short and long term to the same biocide, which emphasizes distinct protein production requirements with treatment for each length of time.
Concomitantly, the long-term bacterial response was directed toward an anaerobic state of the cell, the downregulation of motility and chemotaxis, and the overexpression of biofilm regulators. Recently, it was demonstrated that E. coli cells growing in an anaerobic state have a higher mutation rate (42), which, combined with the selective pressure of an environment with subinhibitory concentrations of biocides, may aid with the emergence and maintenance of antimicrobial-resistant strains.
The late response of the downregulation of motility could be a strategy to save energy, as the production of flagellum requires a considerable expenditure of energy that can be invested in other strategies for protection against stress, such as biofilm formation (32). It is important to highlight that cells grouped in biofilm structures have increased tolerance to biocides (43), and the consistent induction of a biofilm-forming state by bacteria exposed to subinhibitory concentrations of biocides represents a concern for the elimination of pathogens in hospitals and food production settings.
The induction of both biofilm regulators and acid stress-related genes occurred across most biocides after long-term exposure. However, in both cases, the pathway of choice was distinct between groups of biocides. Such an observation indicates that even though the long-term exposure to biocides may result in a similar output phenotype, E. coli utilizes various mechanisms of stress response at the gene expression level tailored to each compound encountered. Whether this mechanistic radiation is because of physical and chemical cross dependencies that internalize the correlation structure of the environmental setting that E. coli has evolved in remains to be determined.
The possibility of selection of bacterial strains cross-resistant to antibiotics once they have been exposed to subinhibitory concentrations of biocides and the occurrence of cross-resistance between biocides have been studied before (2–5, 44). Several genes implicated in increased tolerance to antibiotics were differentially expressed after exposure to biocides, including the genes for multidrug efflux pumps, such as the pump encoded by acrB, which we also correlated to decreased susceptibility to the biocides benzalkonium chloride and chlorophene.
The transcriptomics for E. coli exposed to specific biocides also allowed us to identify a group of DEGs regulated by zur, the negative regulator of the zinc starvation response, which was associated with biocide susceptibility. Bacterial mutants for the zinc transport systems encoded by znuABC and zupT were previously shown to be more sensitive to hydrogen peroxide than the wild type and attenuated during infection of mouse models, indicating a competitive advantage of zinc transport systems for infection (45). The expression of zur-regulated genes was also simultaneous with the overexpression of biofilm regulators and the downregulation of motility. Knockout mutants for znuABC have impaired motility (45), and mutants for zinT and ykgM have reduced biofilm formation (46). Overexpression of zinc transporters by prolonged exposure to a subinhibitory concentration of biocides may also contribute to biofilm formation and increased fitness in subsequent host infection.
Finally, aside from its intellectual merit on bacterial physiology, dissecting the bacterial response to biocide stress and understanding the potential mechanisms of tolerance and resistance are crucial when it comes to informing future policy and guidelines on biocide use.
MATERIALS AND METHODS
Biocides.
Benzalkonium chloride (MP Biomedicals), hydrogen peroxide (Macron), peracetic acid (Sigma-Aldrich), sodium hypochlorite (Sigma-Aldrich), glutaraldehyde (Amresco), chlorhexidine (Aldrich), chlorhexidine gluconate (Spectrum), and povidone-iodine (Sigma) stock solutions were prepared by dilution in sterile, demineralized water. Chlorophene (Aldrich) stock solution was prepared by dilution in ethanol (Sigma-Aldrich). To refer to each biocide, we use the abbreviations provided in Table 1. All solutions, as well as ethanol (Sigma-Aldrich) and isopropanol (Spectrum), were sterilized with 0.22-μm-pore-size filters and kept in the dark at 8°C. Working solutions were prepared daily by further dilutions in sterile, demineralized water.
Strain and culture conditions.
All bacterial samples used in this work were collected for RNA isolation at mid-exponential growth (Fig. 2). Escherichia coli MG1655 was obtained from frozen (−80°C) stock tubes and grown in minimal medium (M9 medium with 0.4% [wt/vol] glucose) for 12 h (mid-exponential growth) at 37°C (preinoculum). Next, the cells were diluted 1:100 into fresh M9 medium with 0.4% glucose containing 1 of the 10 biocides (Fig. 1 and 2). The biocides tested were benzalkonium chloride, isopropanol, ethanol, hydrogen peroxide, peracetic acid, sodium hypochlorite, glutaraldehyde, chlorophene, chlorhexidine, or povidone-iodine (Table 1). The subinhibitory concentrations utilized for each biocide were determined previously to provide, on average, 50% growth inhibition (according to the optical density at 600 nm [OD600]) after 12 h of exposure compared to the growth of a control without biocide when grown in a plate reader (Fig. 2A). Groth rates were not affected by most biocides except for ethanol and isopropanol. Instead, the cells showed an extended lag phase when growing in the presence of the biocides (Fig. 2A).The final biocide concentrations were as follows: benzalkonium chloride, 3.63 mg/liter; isopropanol, 2.7% (vol/vol); ethanol, 2.8% (vol/vol); hydrogen peroxide, 272 μM; peracetic acid, 9 μM; sodium hypochlorite, 3.64 μM; glutaraldehyde, 29 μM; chlorophene, 0.25 mM; chlorhexidine, 1.48 μM; and povidone-iodine, 12.5 μg/ml. The final concentration of ethanol for the experiments with chlorophene exposure was 0.5% (vol/vol). For sample collection, growth was stopped after 8 to 12 h, which corresponded to mid-exponential growth, when the OD600 reached values ranging from 0.6 to 0.95 (Fig. 2F). Tubes without biocide served as a control. For the control, growth was also stopped at the mid-exponential stage, at 7 h of growth. Cold 5% (vol/vol) phenol-ethanol was added (1.5 ml per 3 ml of sample) to each tube. The cells were pelleted by centrifugation at 4,000 rpm at 4°C for 10 min and stored at −80°C for up to 2 weeks. The preinoculum cells were also prepared as described above and stored for further analysis.
Additionally, preinoculum cells were also exposed to benzalkonium chloride, chlorophene, or povidone-iodine at the concentrations mentioned above, without further cell dilution, for 30 min (Fig. 1B and 2E) and stored as described above. Chlorophene was selected for the short-term-exposure evaluation due to the lack of information in the scientific literature regarding the microbial response to this biocide. Povidone-iodine and benzalkonium chloride were included for the short-term-exposure evaluation given the opposite claims regarding the development of bacterial resistance to these compounds. Adaptation to increasing concentrations of benzalkonium chloride as well as acquired cross-resistance to additional antimicrobials after prolonged exposure to the biocide has been reported (3, 44). Povidone-iodine, on the other hand, was claimed to have superior antimicrobial activity and has repeatedly been reported to be an antiseptic that does not select for resistance in bacterial strains (47, 48). All experiments were performed in triplicate.
RNA extraction and transcriptomics.
Total RNA was recovered from pelleted cells with an RNeasy minikit (Qiagen) and on-column DNase digestion (Qiagen). rRNA was removed with a capture oligonucleotide mix (MICROBExpress; Ambion). The total RNA concentration (3 μg) was previously optimized by our group to increase the ribosomal depletion yield for the kit (average amount of rRNA removed, above 99%). RNA cleanup was performed with a NucleoSpin RNA cleanup kit (Macherey-Nagel). For library preparation, a Kapa Stranded transcriptome sequencing (RNA-seq) library preparation kit for Illumina platforms (Kapa Biosystems) was used according to the manufacturer’s instructions. An extra step of size selection was performed using Agencourt AMPure XP magnetic beads (Beckman Coulter). The DNA concentration for each sample library was determined with a Qbit (v.2.0) fluorometer (Invitrogen). The DNA concentration of the final pooled library was determined with a Bioanalyzer DNA high-sensitivity assay (Agilent; DNA Technologies Core, University of California at Davis [UCDavis]). Sequencing was performed with a HiSeq 3000/4000 SR50 sequencer (DNA Technologies Core, UCDavis).
Transcriptomics data analysis.
Adapters and low-quality reads were removed from the raw reads by use of the Trimmomatic trimmer (49), followed by alignment to the reference files for E. coli MG1655 (50) by use of the Bowtie2 program (51). Then, the bam files generated by Bowtie2 were fed into the FeatureCounts program (52), yielding the counts for each gene in each replicate. We examined the proximity between replicates for different biocides by constructing a multidimensional plot over the counts for all the genes (53). Finally, differentially expressed genes (DEGs) were identified using the edgeR (53) and Deseq-2 (54) programs, with a false discovery rate of 0.05 being used as the threshold for calling differential expression. The common DEGs across these two approaches were reported as final DEGs. For gene expression analysis, the averages of the log2 fold change (log2 FC) in expression obtained by the two methods were used.
The expression data generated from RNA-seq were comprehensively analyzed for gene functionality, location in the cell, pathways, relationships between genes, and additional information with the EcoCyc (55) and STRING databases. The Cytoscape (v.3.4.0) program (56) with the STRING application (default settings of a confidence score cutoff of 0.4 and a maximum number of interactors of 0 were used) was used to build and analyze the gene networks for DEGs with absolute log2 FC values greater than 1. Each gene cluster generated by the program was manually screened for the processes or pathways present. Gene Ontology (GO) terms (biological process, molecular function, and cellular component) were captured from the EcoCyc database for all DEGs. The complete list of DEGs (q < 0.05) was used to build a heatmap with the biological processes affected by each biocide. All information was combined to express the processes most significantly affected by biocide exposure. Differentially expressed genes (DEGs) were classified and ranked according to the log2 FC. The DEGs that appeared under multiple conditions (equal to or more than three), as well as the DEGs with the highest absolute log2 FC values, were evaluated for the GO term biological process and included in the list of the most significant processes affected by biocide exposure.
Biocide exposure assays.
Mutants with knockouts of selected genes (acrA, znuABC, zinT, zur) were obtained from the Keio Collection (57). All strains were verified regarding the correct position of the kanamycin insert by PCR and sequencing. The primers used are listed in Table 4. The effects of biocide exposure on the knockout mutants were tested with growth curves and survival assays. For growth curve experiments, the mutants were grown overnight in minimal medium (M9 medium) with 0.4% (wt/vol) glucose. The OD600 was measured and adjusted to 0.1 ± 0.05. Two microliters of the cell cultures was transferred to each well of 96-well plates containing biocides at a range of concentrations diluted in M9 medium with glucose to a total of 200 μl/well. Both the Keio Collection parent strain E. coli BW25113 and strain MG1655 were used as controls and exposed to the same concentrations of the biocides for comparison. The plates were incubated for 24 h at 37°C with agitation in a plate reader (Synergy HT; BioTek). For survival assays, overnight cultures were diluted in M9 medium-glucose and grown to exponential phase (OD600, 0.5 ± 0.3). The OD600 was adjusted to 0.19 for a constant cell concentration across experiments. Cells were challenged with the biocides for 15 min at 37°C with agitation. After the addition of the appropriate neutralizer (Table 1), cells were diluted in 0.9% saline and plated on LB agar. Percent survival was calculated for each mutant by comparison to the survival of a control not exposed to the biocide. Results were normalized by considering the survival for the parent strain E. coli BW25113 (wild type [WT]) to be 100%.
TABLE 4.
Primers used in this worka
| Primer no. | Primer name | Primer sequence |
|---|---|---|
| 1 | kan_check_knockout | CCGTGATATTGCTGAAGAGC |
| 2 | kan_check_knockout_2 | GTTTCTGCGGACTGGCTTTC |
| 3 | check_acrA_knockout_fw | GTATGTACCATAGCACGACG |
| 4 | check_acrA_knockout_rv | CATGATGATAATGGCGATCAC |
| 5 | check_zur_knockout_fw | CATTACGGCAACAATAAGGG |
| 6 | check_zur_knockout_rv | AACCCGCAATGAATATCGC |
| 7 | check_zinT_knockout_fw | CTGAGAAAGCCATGCTCTCG |
| 8 | check_zinT_knockout_rv | TAGCTTGCGTTCAGTGGC |
| 9 | check_znuA_knockout_fw | CGGGCTATCTGTTGCACG |
| 10 | check_znuA_knockout_rv | CCAGCGACACATCAGAGA |
| 11 | check_znuB_knockout_fw | GGTGCTGAACAACTGGGT |
| 12 | check_znuB_knockout_rv | AGGTCGGATAAGGCGCTC |
| 13 | check_znuC_knockout_fw | TTGCACCTCCCCAGAGAG |
| 14 | check_znuC_knockout_rv | CAAACGAACCCAGCGGAC |
| 15 | ydcI fw | TTCTTGACGCCATCAACACTGCCG |
| 16 | ydcI rv | GCAAGGTCGTCTCTTTTTGTTGCTG |
| 17 | yccJ fw | GCTCATCACGTCGGTGAATGGG |
| 18 | yccJ rv | CCTTCTTCCCAAATCTTTTCCGCC |
| 19 | yjcZ fw | GGCACTGACGCAGATCGC |
| 20 | yjcZ rv | ACCTGCCTGCACCAGTAGG |
| 21 | znuA fw | GCGGACTTAGTCGTTTGGGTTGG |
| 22 | znuA rv | GCGTGGTCGTGATCATCATCATCG |
Primers 1 to 14 were used to check the correctness of the knockout strains from the Keio Collection (57) utilized in this work. Primers 3 to 14 were used to amplify the region expected to have the deletion according to the scheme shown in Fig. S2 in the supplemental material (primer PCR). Primers 1 and 2 (primer sequences) were used independently to verify the correct insertion of the kanamycin resistance gene and the deletion of the gene acrA, zur, zinT, znuA, znuB, or znuC in the strains. Primers 15 to 22 were used for quantitative PCR validation of the DNA sequencing data. See also Fig S3 in the supplemental material. fw, forward; rv, reverse.
Availability of data.
The data generated or analyzed during this study are included in this published article (and in the supplemental material. RNA-seq data are deposited in the NCBI Gene Expression Omnibus (GEO) database under accession number GSE124673.
Supplementary Material
ACKNOWLEDGMENTS
We thank the members of the I. Tagkopoulos lab for their comments and suggestions.
This work was supported by NSF awards 1254205 and 1743101 to I.T. and a fellowship from CAPES, Brazil (fellowship 99999.012999/2013-00), to B.M.P.P.
We declare that we have no competing interests.
B.M.P.P. performed all experiments and analyses of the experimental data. X.W. performed all computational analyses. I.T. conceived the idea, analyzed the data, and supervised all aspects of the project. B.M.P.P., X.W., and I.T. wrote the manuscript.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Scientific Committee on Emerging and Newly Identified Health Risks and NIHR European Commission. 2009. Assessment of the antibiotic resistance effects of biocides. European Commission, Brussels, Belgium. [Google Scholar]
- 2.Braoudaki M, Hilton AC. 2004. Low level of cross-resistance between triclosan and antibiotics in Escherichia coli K-12 and E. coli O55 compared to E. coli O157. FEMS Microbiol Lett 235:305–309. doi: 10.1111/j.1574-6968.2004.tb09603.x. [DOI] [PubMed] [Google Scholar]
- 3.Langsrud S, Sundheim G, Holck AL. 2004. Cross-resistance to antibiotics of Escherichia coli adapted to benzalkonium chloride or exposed to stress-inducers. J Appl Microbiol 96:201–208. doi: 10.1046/j.1365-2672.2003.02140.x. [DOI] [PubMed] [Google Scholar]
- 4.Soumet C, Fourreau E, Legrandois P, Maris P. 2012. Resistance to phenicol compounds following adaptation to quaternary ammonium compounds in Escherichia coli. Vet Microbiol 158:147–152. doi: 10.1016/j.vetmic.2012.01.030. [DOI] [PubMed] [Google Scholar]
- 5.Tattawasart U, Hann A, Maillard J-Y, Furr J, Russell A. 2000. Cytological changes in chlorhexidine-resistant isolates of Pseudomonas stutzeri. J Antimicrob Chemother 45:145–152. doi: 10.1093/jac/45.2.145. [DOI] [PubMed] [Google Scholar]
- 6.Harbarth S, Soh ST, Horner C, Wilcox M. 2014. Is reduced susceptibility to disinfectants and antiseptics a risk in healthcare settings? A point/counterpoint review. J Hosp Infect 87:194–202. doi: 10.1016/j.jhin.2014.04.012. [DOI] [PubMed] [Google Scholar]
- 7.Tezel U, Pavlostathis SG. 2015. Quaternary ammonium disinfectants: microbial adaptation, degradation and ecology. Curr Opin Biotechnol 33:296–304. doi: 10.1016/j.copbio.2015.03.018. [DOI] [PubMed] [Google Scholar]
- 8.Food and Drug Administration. 2015. Safety and effectiveness of health care antiseptics; topical antimicrobial drug products for over-the-counter human. 21 CFR Part 310 Food and Drug Administration, Silver Spring, MD. [PubMed] [Google Scholar]
- 9.McBain A, Gilbert P. 2001. Biocide tolerance and the harbingers of doom. Int Biodeterior Biodegrad 47:55–61. doi: 10.1016/S0964-8305(01)00037-3. [DOI] [Google Scholar]
- 10.McMurry LM, Oethinger M, Levy SB. 1998. Triclosan targets lipid synthesis. Nature 394:531–532. doi: 10.1038/28970. [DOI] [PubMed] [Google Scholar]
- 11.McMurry LM, McDermott PF, Levy SB. 1999. Genetic evidence that InhA of Mycobacterium smegmatis is a target for triclosan. Antimicrob Agents Chemother 43:711–713. doi: 10.1128/AAC.43.3.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Food and Drug Administration. 2016. Safety and effectiveness of consumer antiseptics; topical antimicrobial drug products for over-the-counter human use. 21 CFR Part 310 Food and Drug Administration, Silver Spring, MD. [PubMed] [Google Scholar]
- 13.Russell A. 2003. Similarities and differences in the responses of microorganisms to biocides. J Antimicrob Chemother 52:750–763. doi: 10.1093/jac/dkg422. [DOI] [PubMed] [Google Scholar]
- 14.Blanco P, Hernando-Amado S, Reales-Calderon JA, Corona F, Lira F, Alcalde-Rico M, Bernardini A, Sanchez MB, Martinez JL. 2016. Bacterial multidrug efflux pumps: much more than antibiotic resistance determinants. Microorganisms 4:14. doi: 10.3390/microorganisms4010014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pagedar A, Singh J, Batish VK. 2012. Adaptation to benzalkonium chloride and ciprofloxacin affects biofilm formation potential, efflux pump and haemolysin activity of Escherichia coli of dairy origin. J Dairy Res 79:383–389. doi: 10.1017/S0022029912000295. [DOI] [PubMed] [Google Scholar]
- 16.Dukan S, Touati D. 1996. Hypochlorous acid stress in Escherichia coli: resistance, DNA damage, and comparison with hydrogen peroxide stress. J Bacteriol 178:6145–6150. doi: 10.1128/jb.178.21.6145-6150.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abdel Malek SMA, Badran YR. 2010. Pseudomonas aeruginosa PAO1 adapted to 2-phenoxyethanol shows cross-resistance to dissimilar biocides and increased susceptibility to antibiotics. Folia Microbiol (Praha) 55:588–592. doi: 10.1007/s12223-010-0094-6. [DOI] [PubMed] [Google Scholar]
- 18.Fox EM, Leonard N, Jordan K. 2011. Physiological and transcriptional characterization of persistent and nonpersistent Listeria monocytogenes isolates. Appl Environ Microbiol 77:6559–6569. doi: 10.1128/AEM.05529-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mavri A, Mozina SS. 2012. Involvement of efflux mechanisms in biocide resistance of Campylobacter jejuni and Campylobacter coli. J Med Microbiol 61:800–808. doi: 10.1099/jmm.0.041467-0. [DOI] [PubMed] [Google Scholar]
- 20.Pereira BMP, Tagkopoulos I. 2019. Benzalkonium chlorides: uses, regulatory status, and microbial resistance. Appl Environ Microbiol 85:e00377-19. doi: 10.1128/AEM.00377-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chapman JS. 2003. Disinfectant resistance mechanisms, cross-resistance, and co-resistance. Int Biodeterior Biodegrad 51:271–276. doi: 10.1016/S0964-8305(03)00044-1. [DOI] [Google Scholar]
- 22.Dragosits M, Mozhayskiy V, Quinones-Soto S, Park J, Tagkopoulos I. 2013. Evolutionary potential, cross-stress behavior and the genetic basis of acquired stress resistance in Escherichia coli. Mol Syst Biol 9:643. doi: 10.1038/msb.2012.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gunasekera TS, Csonka LN, Paliy O. 2008. Genome-wide transcriptional responses of Escherichia coli K-12 to continuous osmotic and heat stresses. J Bacteriol 190:3712–3720. doi: 10.1128/JB.01990-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ceragioli M, Mols M, Moezelaar R, Ghelardi E, Senesi S, Abee T. 2010. Comparative transcriptomic and phenotypic analysis of the responses of Bacillus cereus to various disinfectant treatments. Appl Environ Microbiol 76:3352–3360. doi: 10.1128/AEM.03003-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Murray JL, Kwon T, Marcotte EM, Whiteley M. 2015. Intrinsic antimicrobial resistance determinants in the superbug Pseudomonas aeruginosa. mBio 6:e01603-15. doi: 10.1128/mBio.01603-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang S, Deng K, Zaremba S, Deng X, Lin C, Wang Q, Tortorello ML, Zhang W. 2009. Transcriptomic response of Escherichia coli O157:H7 to oxidative stress. Appl Environ Microbiol 75:6110–6123. doi: 10.1128/AEM.00914-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goodarzi H, Bennett BD, Amini S, Reaves ML, Hottes AK, Rabinowitz JD, Tavazoie S. 2010. Regulatory and metabolic rewiring during laboratory evolution of ethanol tolerance in E. coli. Mol Syst Biol 6:378. doi: 10.1038/msb.2010.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Horinouchi T, Tamaoka K, Furusawa C, Ono N, Suzuki S, Hirasawa T, Yomo T, Shimizu H. 2010. Transcriptome analysis of parallel-evolved Escherichia coli strains under ethanol stress. BMC Genomics 11:579. doi: 10.1186/1471-2164-11-579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chang W, Toghrol F, Bentley WE. 2006. Toxicogenomic response of Staphylococcus aureus to peracetic acid. Environ Sci Technol 40:5124–5131. doi: 10.1021/es060354b. [DOI] [PubMed] [Google Scholar]
- 30.Hassan KA, Jackson SM, Penesyan A, Patching SG, Tetu SG, Eijkelkamp BA, Brown MH, Henderson PJ, Paulsen IT. 2013. Transcriptomic and biochemical analyses identify a family of chlorhexidine efflux proteins. Proc Natl Acad Sci U S A 110:20254–20259. doi: 10.1073/pnas.1317052110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nde CW, Jang H-J, Toghrol F, Bentley WE. 2009. Global transcriptomic response of Pseudomonas aeruginosa to chlorhexidine diacetate. Environ Sci Technol 43:8406–8415. doi: 10.1021/es9015475. [DOI] [PubMed] [Google Scholar]
- 32.Guttenplan SB, Kearns DB. 2013. Regulation of flagellar motility during biofilm formation. FEMS Microbiol Rev 37:849–871. doi: 10.1111/1574-6976.12018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Khil PP, Camerini-Otero RD. 2002. Over 1000 genes are involved in the DNA damage response of Escherichia coli. Mol Microbiol 44:89–105. doi: 10.1046/j.1365-2958.2002.02878.x. [DOI] [PubMed] [Google Scholar]
- 34.Yamanaka Y, Shimada T, Yamamoto K, Ishihama A. 2016. Transcription factor CecR (YbiH) regulates a set of genes affecting the sensitivity of Escherichia coli against cefoperazone and chloramphenicol. Microbiology 162:1253–1264. doi: 10.1099/mic.0.000292. [DOI] [PubMed] [Google Scholar]
- 35.Horiyama T, Nishino K. 2014. AcrB, AcrD, and MdtABC multidrug efflux systems are involved in enterobactin export in Escherichia coli. PLoS One 9:e108642. doi: 10.1371/journal.pone.0108642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Outten CE, O'Halloran TV. 2001. Femtomolar sensitivity of metalloregulatory proteins controlling zinc homeostasis. Science 292:2488–2492. doi: 10.1126/science.1060331. [DOI] [PubMed] [Google Scholar]
- 37.Patzer SI, Hantke K. 2000. The zinc-responsive regulator Zur and its control of the znu gene cluster encoding the ZnuABC zinc uptake system in Escherichia coli. J Biol Chem 275:24321–24332. doi: 10.1074/jbc.M001775200. [DOI] [PubMed] [Google Scholar]
- 38.Petrarca P, Ammendola S, Pasquali P, Battistoni A. 2010. The Zur-regulated ZinT protein is an auxiliary component of the high-affinity ZnuABC zinc transporter that facilitates metal recruitment during severe zinc shortage. J Bacteriol 192:1553–1564. doi: 10.1128/JB.01310-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gilston BA. 2012. Structural and mechanistic analysis of the zinc uptake regulator (Zur) protein from Escherichia Coli. PhD thesis. Northwestern University, Chicago, IL. [Google Scholar]
- 40.Hemm MR, Paul BJ, Miranda-Ríos J, Zhang A, Soltanzad N, Storz G. 2010. Small stress response proteins in Escherichia coli: proteins missed by classical proteomic studies. J Bacteriol 192:46–58. doi: 10.1128/JB.00872-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mitosch K, Rieckh G, Bollenbach T. 2019. Temporal order and precision of complex stress responses in individual bacteria. Mol Syst Biol 15:e8470. doi: 10.15252/msb.20188470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shewaramani S, Finn TJ, Leahy SC, Kassen R, Rainey PB, Moon CD. 2017. Anaerobically grown Escherichia coli has an enhanced mutation rate and distinct mutational spectra. PLoS Genet 13:e1006570. doi: 10.1371/journal.pgen.1006570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mah TF, O'Toole GA. 2001. Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol 9:34–39. doi: 10.1016/S0966-842X(00)01913-2. [DOI] [PubMed] [Google Scholar]
- 44.Bore E, Hébraud M, Chafsey I, Chambon C, Skjæret C, Moen B, Møretrø T, Langsrud Ø, Rudi K, Langsrud S. 2007. Adapted tolerance to benzalkonium chloride in Escherichia coli K-12 studied by transcriptome and proteome analyses. Microbiology 153:935–946. doi: 10.1099/mic.0.29288-0. [DOI] [PubMed] [Google Scholar]
- 45.Sabri M, Houle S, Dozois CM. 2009. Roles of the extraintestinal pathogenic Escherichia coli ZnuACB and ZupT zinc transporters during urinary tract infection. Infect Immun 77:1155–1164. doi: 10.1128/IAI.01082-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lim J, Lee K-M, Kim SH, Kim Y, Kim S-H, Park W, Park S. 2011. YkgM and ZinT proteins are required for maintaining intracellular zinc concentration and producing curli in enterohemorrhagic Escherichia coli (EHEC) O157:H7 under zinc deficient conditions. Int J Food Microbiol 149:159–170. doi: 10.1016/j.ijfoodmicro.2011.06.017. [DOI] [PubMed] [Google Scholar]
- 47.Durani P, Leaper D. 2008. Povidone-iodine: use in hand disinfection, skin preparation and antiseptic irrigation. Int Wound J 5:376–387. doi: 10.1111/j.1742-481X.2007.00405.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reimer K, Wichelhaus T, Schäfer V, Rudolph P, Kramer A, Wutzler P, Ganzer D, Fleischer W. 2002. Antimicrobial effectiveness of povidone-iodine and consequences for new application areas. Dermatology 204:114–120. doi: 10.1159/000057738. [DOI] [PubMed] [Google Scholar]
- 49.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kersey PJ, Allen JE, Armean I, Boddu S, Bolt BJ, Carvalho-Silva D, Christensen M, Davis P, Falin LJ, Grabmueller C, Humphrey J, Kerhornou A, Khobova J, Aranganathan NK, Langridge N, Lowy E, McDowall MD, Maheswari U, Nuhn M, Ong CK, Overduin B, Paulini M, Pedro H, Perry E, Spudich G, Tapanari E, Walts B, Williams G, Tello-Ruiz M, Stein J, Wei S, Ware D, Bolser DM, Howe KL, Kulesha E, Lawson D, Maslen G, Staines DM. 2016. Ensembl Genomes 2016: more genomes, more complexity. Nucleic Acids Res 44:D574–D580. doi: 10.1093/nar/gkv1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liao Y, Smyth GK, Shi W. 2014. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 53.Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Keseler IM, Mackie A, Santos-Zavaleta A, Billington R, Bonavides-Martínez C, Caspi R, Fulcher C, Gama-Castro S, Kothari A, Krummenacker M, Latendresse M, Muñiz-Rascado L, Ong Q, Paley S, Peralta-Gil M, Subhraveti P, Velázquez-Ramírez DA, Weaver D, Collado-Vides J, Paulsen I, Karp PD. 2017. The EcoCyc database: reflecting new knowledge about Escherichia coli K-12. Nucleic Acids Res 45:D543–D550. doi: 10.1093/nar/gkw1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. 2003. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio Collection. Mol Syst Biol 2:2006.0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cotter PA, Melville SB, Albrecht JA, Gunsalus RP. 1997. Aerobic regulation of cytochrome d oxidase (cydAB) operon expression in Escherichia coli: roles of Fnr and ArcA in repression and activation. Mol Microbiol 25:605–615. doi: 10.1046/j.1365-2958.1997.5031860.x. [DOI] [PubMed] [Google Scholar]
- 59.Xiao M, Lai Y, Sun J, Chen G, Yan A. 2016. Transcriptional regulation of the outer membrane porin gene ompW reveals its physiological role during the transition from the aerobic to the anaerobic lifestyle of Escherichia coli. Front Microbiol 7:799. doi: 10.3389/fmicb.2016.00799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sun X, Eliasson R, Pontis E, Andersson J, Buist G, Sjöberg B-M, Reichard P. 1995. Generation of the glycyl radical of the anaerobic Escherichia coli ribonucleotide reductase requires a specific activating enzyme. J Biol Chem 270:2443–2446. doi: 10.1074/jbc.270.6.2443. [DOI] [PubMed] [Google Scholar]
- 61.Schellhorn HE, Hassan HM. 1988. Transcriptional regulation of katE in Escherichia coli K-12. J Bacteriol 170:4286–4292. doi: 10.1128/jb.170.9.4286-4292.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Partridge JD, Scott C, Tang Y, Poole RK, Green J. 2006. Escherichia coli transcriptome dynamics during the transition from anaerobic to aerobic conditions. J Biol Chem 281:27806–27815. doi: 10.1074/jbc.M603450200. [DOI] [PubMed] [Google Scholar]
- 63.Gruer MJ, Guest JR. 1994. Two genetically-distinct and differentially-regulated aconitases (AcnA and AcnB) in Escherichia coli. Microbiology 140:2531–2541. doi: 10.1099/00221287-140-10-2531. [DOI] [PubMed] [Google Scholar]
- 64.Weber DJ, Rutala WA, Sickbert-Bennett EE. 2007. Outbreaks associated with contaminated antiseptics and disinfectants. Antimicrob Agents Chemother 51:4217–4224. doi: 10.1128/AAC.00138-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kvint K, Nachin L, Diez A, Nyström T. 2003. The bacterial universal stress protein: function and regulation. Curr Opin Microbiol 6:140–145. doi: 10.1016/s1369-5274(03)00025-0. [DOI] [PubMed] [Google Scholar]
- 66.Krisko A, Copic T, Gabaldón T, Lehner B, Supek F. 2014. Inferring gene function from evolutionary change in signatures of translation efficiency. Genome Biol 15:R44. doi: 10.1186/gb-2014-15-3-r44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.David G, Blondeau K, Schiltz M, Penel S, Lewit-Bentley A. 2003. YodA from Escherichia coli is a metal-binding, lipocalin-like protein. J Biol Chem 278:43728–43735. doi: 10.1074/jbc.M304484200. [DOI] [PubMed] [Google Scholar]
- 68.Quan S, Koldewey P, Tapley T, Kirsch N, Ruane KM, Pfizenmaier J, Shi R, Hofmann S, Foit L, Ren G, Jakob U, Xu Z, Cygler M, Bardwell JCA. 2011. Genetic selection designed to stabilize proteins uncovers a chaperone called Spy. Nat Struct Mol Biol 18:262–269. doi: 10.1038/nsmb.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.