Ethanol production from carbon monoxide (CO) as a carbon and energy source by Clostridium ljungdahlii and “Clostridium autoethanogenum” is currently being commercialized. During gas fermentation, ethanol synthesis is NADH-dependent. However, ethanol oxidation and its regulatory mechanism remain incompletely understood. Energy metabolism analysis demonstrated that reduced ferredoxin is the sole source of NADH formation by the Rnf-ATPase system, which provides ATP for cell growth during CO fermentation. Therefore, ethanol production is tightly linked to biomass production (ATP production). Clarification of the mechanism of ethanol oxidation and biosynthesis can provide an important reference for generating high-ethanol-yield strains of C. ljungdahlii in the future.
KEYWORDS: CO fermentation, Clostridium ljungdahlii, acetate, acetogenesis, ethanol oxidation
ABSTRACT
Bioethanol production from syngas using acetogenic bacteria has attracted considerable attention in recent years. However, low ethanol yield is the biggest challenge that prevents the commercialization of syngas fermentation into biofuels using microbial catalysts. The present study demonstrated that ethanol metabolism plays an important role in recycling NADH/NAD+ during autotrophic growth. Deletion of bifunctional aldehyde/alcohol dehydrogenase (adhE) genes leads to significant growth deficiencies in gas fermentation. Using specific fermentation technology in which the gas pressure and pH were constantly controlled at 0.1 MPa and 6.0, respectively, we revealed that ethanol was formed during the exponential phase, closely accompanied by biomass production. Then, ethanol was oxidized to acetate via the aldehyde ferredoxin oxidoreductase pathway in Clostridium ljungdahlii. A metabolic experiment using 13C-labeled ethanol and acetate, redox balance analysis, and comparative transcriptomic analysis demonstrated that ethanol production and reuse shared the metabolic pathway but occurred at different growth phases.
IMPORTANCE Ethanol production from carbon monoxide (CO) as a carbon and energy source by Clostridium ljungdahlii and “Clostridium autoethanogenum” is currently being commercialized. During gas fermentation, ethanol synthesis is NADH-dependent. However, ethanol oxidation and its regulatory mechanism remain incompletely understood. Energy metabolism analysis demonstrated that reduced ferredoxin is the sole source of NADH formation by the Rnf-ATPase system, which provides ATP for cell growth during CO fermentation. Therefore, ethanol production is tightly linked to biomass production (ATP production). Clarification of the mechanism of ethanol oxidation and biosynthesis can provide an important reference for generating high-ethanol-yield strains of C. ljungdahlii in the future.
INTRODUCTION
Ethanol, an important chemical and cost-effective product, has widespread applications in the industrial and medical fields (1). Traditionally, bioethanol is produced from starch via microbial fermentation. Considering the rapid growth of the global population and the demand for food, lignocellulosic biomass can instead be utilized for producing bioethanol, known as a second-generation ethanol fuel. However, a pretreatment step and usage of an expensive exogenous hydrolytic enzyme are required to obtain soluble sugars before microbial fermentation, making the entire process less cost-effective (2). In this regard, syngas from biomass gasification is another satisfactory feedstock for ethanol production by Clostridium ljungdahlii, “Clostridium autoethanogenum,” and Clostridium carboxidivorans (3–13).
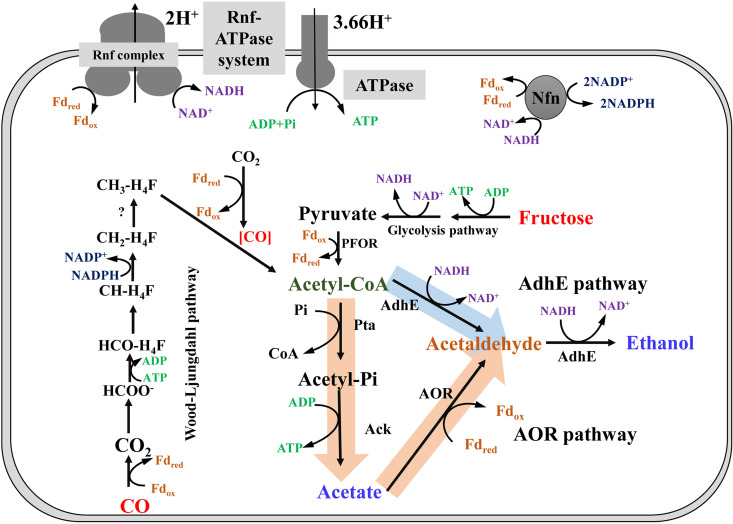
C. ljungdahlii and C. autoethanogenum were originally described as separate species; however, comparison of their genomic sequences has indicated that they belong to the same species (4, 14). These microorganisms can use carbon monoxide (CO) as a carbon and energy source for ethanol production (7, 11, 13). Two ethanol biosynthesis pathways have been reported (Fig. 1) (15). One is the classic ethanol formation pathway, starting with acetyl-CoA as the precursor, which is converted to ethanol by two redox reactions involving two bifunctional aldehyde/alcohol dehydrogenases encoded by two adhE genes. This pathway is named the AdhE pathway. In the other pathway, named the AOR pathway, acetate acts as a precursor, and acetaldehyde:ferredoxin oxidoreductase encoded by aor catalyzes the redox reaction of acetaldehyde formation (15). Ethanol synthesis from acetaldehyde overlaps in the two ethanol formation pathways. Deleting adhE1/2 significantly inhibits ethanol production during heterotrophic fermentation by C. ljungdahlii (16). Therefore, AdhE is important for ethanol formation (13, 15–17). Acetate supplementation and deletion of aor1 and aor2 in experiments with C. autoethanogenum using CO as the carbon source has revealed that the AOR pathway plays a critical role in the conversion of acids to alcohols. Interestingly, inactivation of aor2 alone elevates ethanol production under the same cultivation conditions (15). Taken together, although important aspects of ethanol production have been elucidated, little is known regarding the regulatory mechanisms of ethanol production in these organisms (18–21).
FIG 1.
Metabolic pathway of ethanol biosynthesis in C. ljungdahlii. Abbreviations used: Pta, pta, phosphotransacetylase; Ack, ack, acetate kinase; AdhE, adhE, aldehyde/alcohol dehydrogenase; AOR, aor, acetaldehyde:ferredoxin oxidoreductase; PFOR, pfor, pyruvate:ferredoxin oxidoreductase; LdhA, ldhA, lactate dehydrogenase; ALDC, aldc, acetolactate decarboxylase; 23BDH, bdh, 2,3-butanediol dehydrogenase; Fdred, reduced ferredoxin; Fdox, oxidized ferredoxin; Nfn, electron-bifurcating and ferredoxin-dependent transhydrognenase; Rnf complex, membrane-associated and energy-conserving reduced ferredoxin:NAD+ oxidoreductase. The Rnf-ATPase system is a system of two enzyme complexes in which the Rnf complex generates a proton gradient across the membrane by the oxidation of Fdred with NAD+. The second enzyme complex, the ATPase complex, consumes the proton gradient and phosphorylates ADP to ATP in the cytoplasm. AdhE pathway, ethanol formation by AdhE catalysis; AOR pathway, AOR participates in acetate and ethanol formation.
During acetaldehyde synthesis, the AOR pathway requires reduced ferredoxin (Fdred) as a cofactor, whereas the AdhE pathway requires NADH (Fig. 1). Thus, ethanol biosynthesis is dependent on energy metabolism in C. ljungdahlii and C. autoethanogenum. In autotrophic microbes growing on CO, Fdred produced via CO oxidization is the primary energy source. ATP, NADH, and NADPH are generated via oxidation of Fdred (17, 18, 22). Among these energy equivalents, ATP can also be generated by substrate-level phosphorylation through acetate formation. Considering ATP consumption by formyl-THF formation in the Wood-Ljungdahl pathway (WLP), there is no net ATP production by substrate-level phosphorylation during gas fermentation. Therefore, ATP is mainly generated by membrane-bound ATP synthase and a transmembrane proton (H+) gradient, which is established via an oxidation reaction of Fdred with Rnf complex (21, 23–27). On the other hand, NADH is generated as an electron acceptor. It must be coupled with another redox reaction to maintain the redox balance in cells. One of the main outlets for this is ethanol biosynthesis, which requires NADH as an electron donor during autotrophic growth (18, 28, 29). Although the detailed energy metabolic net is unclear for gas fermentation in C. ljungdahlii and C. autoethanogenum, it is obvious that ethanol formation is tightly associated with energy conservation, especially ATP generation (20).
In this work, adhE genes of C. ljungdahlii were deleted using CRISPR-Cas9 technology to eliminate the ethanol synthesis pathway. The gene deletion mutants and wild type (WT) were cultured in the presence of CO to elucidate the ethanol formation pathway in C. ljungdahlii grown autotrophically. The ethanol production dynamics were investigated and the regulatory mechanisms were analyzed.
RESULTS AND DISCUSSION
Deletion of adhE genes.
In C. ljungdahlii, ethanol synthesis is NADH-dependent, and AdhE is critical for both the AdhE and AOR pathways (Fig. 1). Two adhE genes (adhE1 [CLJU_c16510] and adhE2 [CLJU_c16520]) encoding AdhE enzymes were found in the genome of C. ljungdahlii (13). A CRISPR-Cas9-based genome editing system (30) was used to delete adhE genes, generating a ΔadhE1 mutant and a double deletion mutant, the ΔadhE1 ΔadhE2 mutant (i.e., the ΔadhE1+2 mutant). The ΔadhE1 mutant was described previously (30) and was used as the parental strain to generate the double mutant in this work. Genomic DNA of putative double mutants in selective plates was extracted, and then DNA fragments were PCR-amplified using the primers adhE1-f/adhE2-r. Positive bands were obtained and sequenced to confirm the deletion of adhE1 and adhE2 (see Fig. S1 in the supplemental material).
Batch fermentation of adhE deletion mutants of C. ljungdahlii on fructose.
During growth on fructose in yeast extract-tryptone-fructose (YTF) medium, two mutants (ΔadhE1 and the ΔadhE1 ΔadhE2 mutant) eventually achieved a similar cell density that was 76% lower than the WT, suggesting that adhE knockout affects cell energy metabolism (Fig. S2). In terms of the primary end products, the ΔadhE1 ΔadhE2 mutant produced 54% less ethanol (22 ± 2 mM) than the WT (48 ± 2 mM) (Table 1), which is in agreement with previous results obtained by Leang et al. (16). The ΔadhE1 mutant produced 30% less ethanol than the WT (30); however, after three sequential subcultures, this mutant restored its ability to produce ethanol and even generated 27% higher titers of ethanol (61 ± 4 mM) than the WT in this study (Table 1). The details of the mechanism of this phenomenon need further investigation. To further test the impact of adhE deletion, specific activities of AdhE and AOR were determined in the cell extracts of the WT and adhE deletion mutant strains collected at 48 h (Table 1). Trace-specific activities of AdhE were observed in the ΔadhE1 and ΔadhE1 ΔadhE2 mutants, indicating that adhE1 is the dominant functional gene during the catalysis reaction from acetaldehyde to ethanol. The specific activity of AdhE in the WT was 0.07 U/mg, exceeding the values for adhE mutants (Table 1). Concerning the specific activity of AOR, the value for the WT was 0.1 U/mg, which was much higher than that of the ΔadhE1 (0.01 U/mg) and ΔadhE1 ΔadhE2 mutants (0.007 U/mg) (Table 1). Analyses of these enzyme activities indicated that adhE deletions significantly decreased both AdhE and AOR activities in the mutants.
TABLE 1.
Enzyme activity assay at 48 h and the final product titers of C. ljungdahlii WT, ΔadhE1 mutant, and ΔadhE1 ΔadhE2 mutant strains in YTF and modified DSMZ 879 medium
| Strain | YTF medium |
Modified DSMZ 879 medium |
||||
|---|---|---|---|---|---|---|
| AdhE (U/mg) | AOR (U/mg) | Ethanol (mM) | Acetate (mM) | Ethanol (mM) | Acetate (mM) | |
| Wild type | 0.07 | 0.1 | 48 ± 2 | 83 ± 13 | 17 ± 0.7 | 37 ± 2 |
| ΔadhE1 mutant | <0.01 | 0.01 | 61 ± 4 | 62 ± 1 | 27 ± 0.8 | 29 ± 0.1 |
| ΔadhE1 ΔadhE2 mutant | <0.01 | <0.01 | 22 ± 2 | 84 ± 1 | NDa | 39 ± 4 |
ND, not detectable.
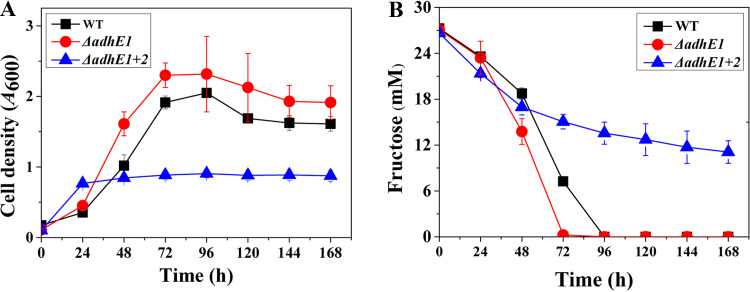
YTF medium, containing tryptone and yeast extract, is a nutrient-rich medium which possibly affects growth and ethanol formation during fermentation. Thus, a chemically defined medium (modified DSMZ 879 medium) was used to culture the WT and the adhE mutants. The growth and fermentation profiles of the WT and the ΔadhE1 strain were similar (Fig. 2, Fig. S3). The ΔadhE1 strain generated 59% higher ethanol titers (27 ± 0.8 mM) than the WT (17 ± 0.7 mM) (Table 1). Compared with the finding for the WT, the ΔadhE1 ΔadhE2 mutant displayed diminished growth and consumed less fructose during fermentation. In total, 11 ± 1.5 mM fructose was detected in the broth of the ΔadhE1 ΔadhE2 mutant, whereas the WT and ΔadhE1 strains completely exhausted fructose before 96 h of culture. Furthermore, ethanol was not detected in the broth for the ΔadhE1 ΔadhE2 mutant, indicating that deletion of two adhE genes eliminates ethanol biosynthesis completely.
FIG 2.
Growth and fructose uptake profiles of C. ljungdahlii WT (black squares), ΔadhE1 mutant (red circles) and ΔadhE1 ΔadhE2 mutant (blue triangles) strains grown on defined DSMZ 879 medium. (A) Growth profiles. (B) Fructose residue.
C. ljungdahlii possesses two adjacent adhE genes in its genome. RNA-seq results revealed that adhE1 was transcribed at a significantly higher level than adhE2 (see below), indicating that AdhE1 is the dominant functional enzyme involved in the conversion of acetaldehyde to ethanol. This is in agreement with the result for the AdhE activity assay, in which adhE1 deletion resulted in significantly decreased AdhE enzyme activity in cell extract (Table 1). Regarding ethanol synthesis, adhE1 deletion did not eliminate ethanol production, which is consistent with previous results (30). AOR activity (specific activity, 0.01 U/mg) was detected in the cell extract of the ΔadhE1 strain, suggesting that some acetaldehyde was generated. We speculated that acetaldehyde can be reduced by AdhE2 or other alcohol dehydrogenases, resulting in ethanol production. Interestingly, the ΔadhE1 strain produced more ethanol than the WT under the same fermentation conditions in this study. This phenotype was also observed during fermentation by the ΔadhE1 mutant of C. autoethanogenum on CO, which produced 171% to 183% higher titers of ethanol than the WT (15). Why the ΔadhE1 strain produced markedly higher levels of ethanol remains an enigma.
Because of the inactivation of both adhE genes, the ΔadhE1 ΔadhE2 mutant strain must use other metabolic pathways to recycle reducing equivalents. YTF medium contains 10 g/liter yeast extract and 16 g/liter tryptone, which can supply the required metabolites for fermentation by C. ljungdahlii. Then, reducing equivalents can be recycled through biochemical reactions, in which these metabolites participate. Therefore, the ΔadhE1 ΔadhE2 mutant strain grew well in YTF medium. In contrast, C. ljungdahlii, to a large extent, depends on ADHs to achieve NADH reoxidation in modified DSMZ 879 medium. Thus, the ΔadhE1 ΔadhE2 mutant strain displayed significant growth deficiency and lost its capacity for ethanol production in modified DSMZ 879 medium.
Batch fermentation of adhE deletion mutants on syngas.
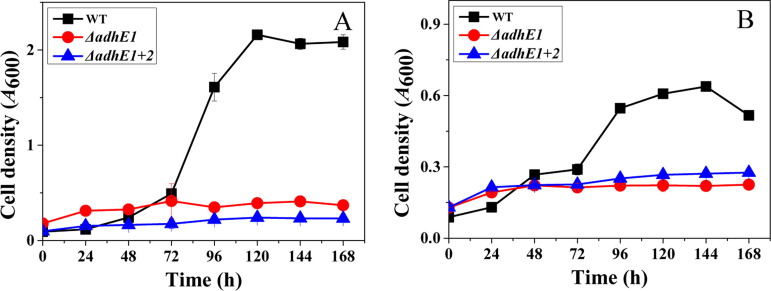
In the case of gas fermentation with CO or H2:CO2 as the carbon and energy sources, the ΔadhE1 and ΔadhE1 ΔadhE2 mutants exhibited significantly diminished growth and an inability to generate ethanol (Fig. 3). The WT grew well on CO and achieved a much higher cell density (A600, 2.1 ± 0.1) than the mutants (0.4 ± 0.02 for the ΔadhE1 mutant and 0.24 ± 0.02 for the ΔadhE1 ΔadhE2 mutant). In addition, 28 ± 5 mM ethanol was detected in the end products in the WT. The fermentation profiles of the WT and mutants grown on H2:CO2 were similar to those grown on CO (Table 2; Fig. S4 and S5). Analysis of syngas uptake indicated that the WT consumed more syngas than the adhE deletion mutants. Taken together, these results demonstrated that AdhE plays a critical role in growth and metabolism during gas fermentation by C. ljungdahlii.
FIG 3.
Growth profiles of C. ljungdahlii WT (black squares), ΔadhE1 mutant (red circles), and ΔadhE1 ΔadhE2 mutant (blue triangles) strains on syngas. (A) CO:CO2 (80:20, vol/vol). (B) H2:CO2 (60:40, vol/vol).
TABLE 2.
Headspace pressure change and the final product titers of C. ljungdahlii WT, ΔadhE1 mutant, and ΔadhE1 ΔadhE2 mutant on syngas
| Strain | CO:CO2 (vol/vol, 80/20) |
CO2:H2 (vol/vol, 40/60) |
CO:CO2 (vol/vol, 80/20) |
CO2:H2 (vol/vol, 40/60) |
||||
|---|---|---|---|---|---|---|---|---|
| Ethanol (mM) | Acetate (mM) | Ethanol (mM) | Acetate (mM) | SPb (MPa) | CO:CO2 (vol/vol) | SP (MPa) | CO2:H2 (vol/vol) | |
| Wild type | 28 ± 5 | 53 ± 5 | 10 ± 2 | 61 ± 2 | 0.06 ± 0.02 | 1:9 | 0.08 ± 0.02 | 3:4 |
| ΔadhE1 mutant | NDa | 19 ± 0.3 | ND | 13 ± 2 | 0.13 ± 0.04 | 1:1 | 0.11 ± 0.04 | 4:5 |
| ΔadhE1 ΔadhE2 mutant | ND | 15 ± 0.5 | ND | 20 ± 1 | 0.14 ± 0.03 | 1:1 | 0.11 ± 0.01 | 1:1 |
ND, not detectable.
SP, space pressure. The beginning space pressure is 0.2 MPa.
Fed-batch fermentation of WT under controlled pH and gas pressure.
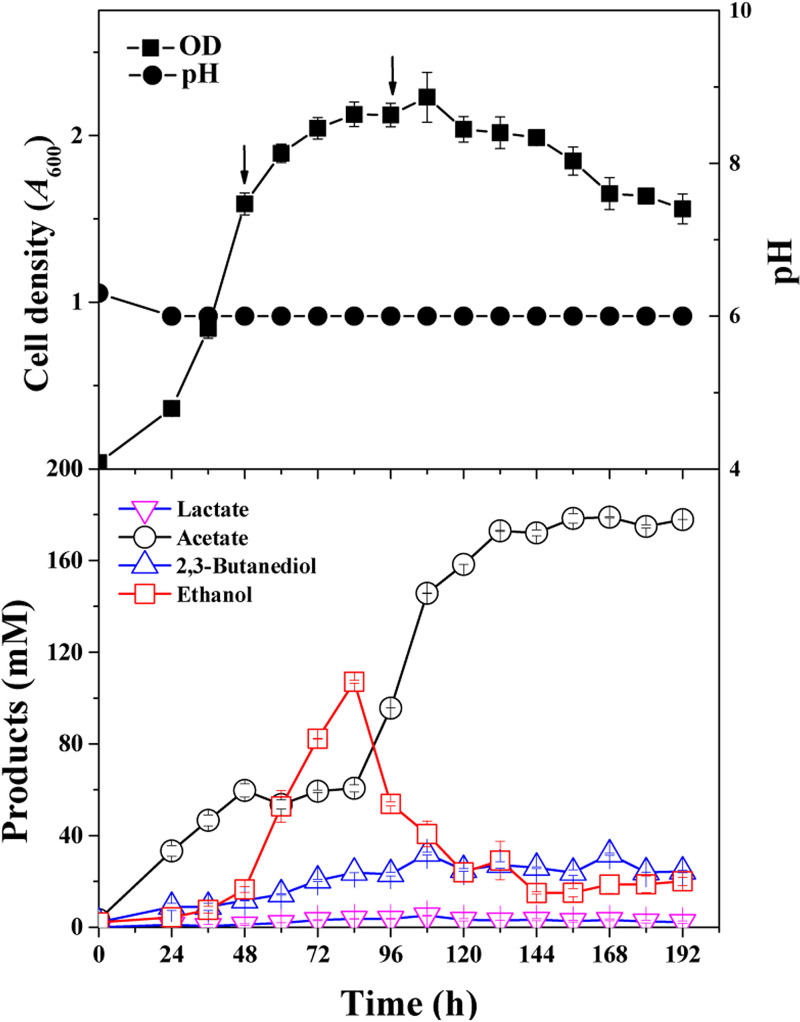
To provide a preferable growth condition for the WT, the fermentation process was controlled at pH 6.0 and at a pressure of 0.1 MPa with a constant CO:CO2 supply (vol/vol, 80/20). C. ljungdahlii exhibited excellent growth, and the density of cell growth reached an A600 of 2.2 ± 0.2. The end products included 177 ± 0.8 mM acetate, 24 ± 0.5 mM 2,3-butanediol, and trace amounts of lactate. Interestingly, the ethanol concentration (107 ± 0.6 mM) peaked at 84 h in the course of batch fermentation (Fig. 4). Analysis of ethanol synthesis and cell growth indicated that ethanol was largely produced during the exponential phase and oxidized during the stationary phase, which was also observed in previous studies (13, 15).
FIG 4.
Growth and product concentrations of C. ljungdahlii WT grown on CO:CO2 (vol/vol, 80/20) with pH 6.0 and gas pressure of 0.1 MPa. Arrows: these two time points represent the exponential and stationary growth phases, respectively. Samples were taken out of the bioreactor at these time points for ethanol oxidation and RNA-Seq analysis.
ATP is the universal energy equivalent in microbial growth, and demand for ATP is high during the exponential phase, in which biomass is quickly accumulated. During gas fermentation by C. ljungdahlii, NADH is produced as an electron carrier in large quantities, accompanied by ATP synthesis (13, 21). We hypothesized that ethanol was a preferred product as an additional NADH sink to maintain the redox balance during gas fermentation. During the stationary phase, biomass no longer accumulates, and less ATP and NADH are produced. However, NADH is still needed. Thus, ethanol oxidation occurs at this stage to provide energy and reducing equivalents to fulfill the survival of cells. The fed-batch fermentation result in this study supports our hypothesis.
Ethanol oxidation in CO fermentation.
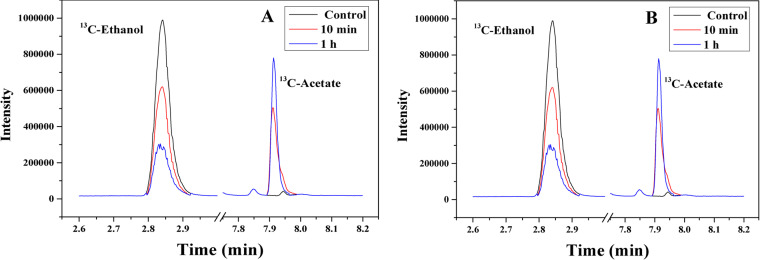
Gas chromatography-mass spectrometry (GC-MS) was used to analyze the products of ethanol oxidation using 13C-labeled ethanol and acetate, which have higher mass weights than unlabeled molecules. In a mixture containing stationary cells collected at 96 h and 13C-labeled acetic acid, only a trace amount of 13C-labeled ethanol was detected up to 12 h (Fig. 5 and Fig. S6). On the other hand, with the addition of 13C-labeled ethanol, a large amount of 13C-labeled acetic acid was detected. These results suggest that ethanol could be used by C. ljungdahlii cells to produce acetate in the absence of CO (no carbon and energy source).
FIG 5.
Detection of 13C labeled products after incubation of C. ljungdahlii WT cells in fresh medium with 13C-acetate (A) or 13C-ethanol (B) under anaerobic conditions.
AOR-harboring acetogens, such as C. ljungdahlii and C. autoethanogenum, are capable of reducing acetate to ethanol during gas fermentation. According to our work and some published results, alcohols, including ethanol and butanol, could also be converted into their corresponding primary carboxylic acids (13, 15). These results suggest the existence of a flexible metabolic mechanism that regulates the conversion between ethanol and acetate. The redox reaction of acetate to acetaldehyde with Fdred is thermodynamically unfavorable under standard conditions (ΔG°′ = 35 kJ/mol). However, this reaction can occur under physiological conditions with an intracellular pH of 6.0 and 1,000-fold higher intracellular acetate levels versus acetaldehyde (31). During gas fermentation by C. ljungdahlii, CO, as a carbon and energy source, is metabolized quickly for growth during the exponential phase (6, 32). Under this condition, Fdred, ATP, NADPH, and NADH can be quickly produced (Fig. 1). Acetate can be converted to ethanol through the AOR pathway for recycling of reducing equivalents to maintain the redox balance during metabolism. During the stationary phase, biomass production ceases, resulting in a reduction of ATP demand. Therefore, the oxidation of ethanol to acetate is driven by a thermodynamic balance, during which reducing equivalents are released. It is noted that no ATP was produced during ethanol oxidation. This speculation can explain why ethanol production increased during the exponential phase and decreased during the stationary phase (17, 18, 33). Chemostat technology can maintain a constant growth of strains at a specific dilution rate with pH control in the bioreactor. Under optimal conditions, biomass and ethanol are produced in large quantities, and ethanol oxidation does not occur (33).
The pathway of ethanol oxidation.
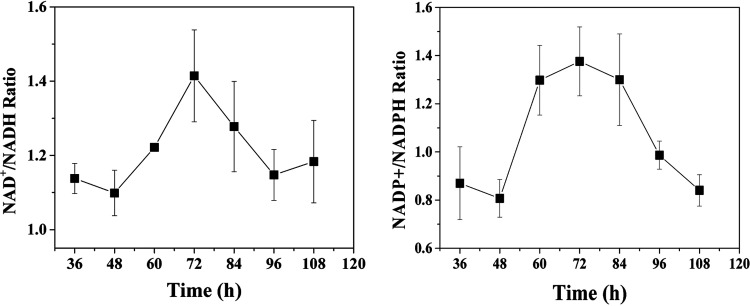
Ethanol can be biosynthesized through both the AdhE and AOR pathways (Fig. 1). Which pathway is used by C. ljungdahlii during ethanol oxidation? Though acetate and ethanol have identical stoichiometry regarding carbon balance as C2-compounds in end products, their energy metabolism is significantly different between the AdhE and AOR pathways during gas fermentation. Through the AdhE pathway, 2 mol of NADH and 1 mol of ATP are produced during ethanol oxidation to acetate, whereas through the AOR pathway, 1 mol of Fdred and 1 mol of NADH are produced. During CO fermentation, NADPH is formed by electron bifurcation of Fdred and NADH via Fd-dependent transhydrogenase (Nfn) (18). Therefore, the NAD+/NADH and NADP+/NADPH ratios were determined to figure out the pathway for ethanol oxidation to acetate (Fig. 6). It was found that the NAD+/NADH ratio increased during the exponential phase and decreased during the stationary phase, which was consistent with the ethanol titer profile in the broth, indicating that ethanol synthesis requires electron transfer from NADH and ethanol oxidation supplies electrons to NAD+. Furthermore, the NADP+/NADPH ratio curve was identical to the NAD+/NADH ratio curve throughout fermentation (Fig. 6B), revealing that the Fdox/Fdred ratio varied during ethanol oxidation. Therefore, our results indicated that ethanol is oxidized by the AOR pathway. This means ethanol generation and reuse share the same metabolic pathway during gas fermentation in C. ljungdahlii. Our observation provides evidence explaining why deletion of aor2 or adhE1 enhances the production of ethanol in C. autoethanogenum fermentation (15). Inactivation of aor2 or adhE1 blocks ethanol oxidation, thereby increasing the titers of ethanol in the broth.
FIG 6.
Time courses of NAD+/NADH and NADP+/NADPH ratios during fermentation in C. ljungdahlii WT grown on CO:CO2 (vol/vol, 80/20).
Transcriptional analysis of ethanol biosynthesis and oxidation.
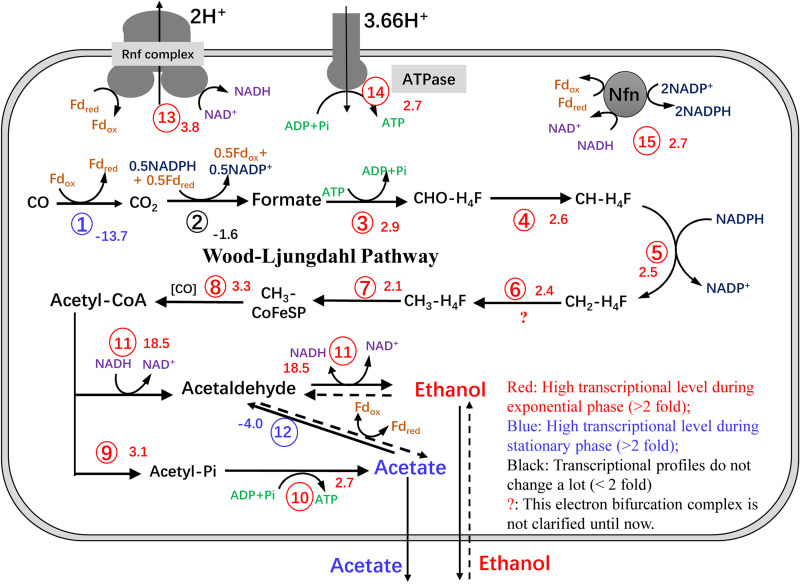
Because of ethanol production and oxidation, carbon and redox flows were distinctly different at the exponential (48 h) and stationary (96 h) phases. Comparative transcriptomics was performed using RNA-Seq technology to investigate intracellular flux patterns at the transcriptional level. Table S2 lists 49 genes involved in the central carbon and energy metabolic pathways. As speculated, C. ljungdahlii required ATP for biomass production; thus, the genes related to ATP formation, which include Rnf-ATPase system genes, AdhE pathway genes, acetate formation genes, and the Nfn gene, had higher transcriptional levels in the exponential phase than in the stationary phase. Although the WLP is an ATP-consuming pathway during formyl-THF formation, it is the sole CO fixation pathway and principal metabolic pathway involved in gas fermentation. Therefore, the expression levels of genes involved in the WLP are higher during the exponential phase. During the stationary phase, biomass production ceases, and the efficiency of carbon source uptake is decreased (Fig. 7).
FIG 7.
Schematic of the central metabolic pathways of C. ljungdahlii grown on CO:CO2 (vol/vol, 80/20). The sequence numbers represent the correlation enzyme or enzyme complex participating in the metabolic reactions listed in detail in Table S2. The positive values represent the increased change fold during the exponential phase. The negative values represent the decreased change fold during the exponential phase.
The comparative transcriptomics results suggest that the expression levels of aor2 and CO dehydrogenase complex genes participating in central metabolic pathways are higher during the stationary phase. This result further proves that the AOR pathway plays a critical role in ethanol oxidation. Based on gene function analysis, both pathways are responsible for important ferredoxin-dependent redox reactions. Fdred is the original energy source for CO fermentation. Genes related to Fdred formation are highly expressed, indicating that C. ljungdahlii requires energy for metabolism during the stationary phase. However, detailed functions of this CO dehydrogenase remain to be further studied beyond the transcriptional level. Notably, during H2:CO2 fermentation, primary energy is derived from the electron bifurcation reaction of the hydrogenase complex rather than from CO reduction during gas fermentation by C. ljungdahlii, and ethanol is also oxidized during H2:CO2 fermentation in C. autoethanogenum (15). Stoichiometric analysis of ATP gains based on gas fermentation by C. ljungdahlii revealed that the ATP yield is 0.75 mol ATP with 1 mol of acetate formed during autotrophic growth on H2:CO2, in contrast to 10 mol of ATP with 1 mol of acetate, 1 mol of 2,3-butanediol, and 4 mol of ethanol formed for cells grown on CO (34).
Formate dehydrogenases (FDHs) catalyze the first step in the carbonyl branch of the WLP. Interestingly, all genes related to the WLP are organized in a huge gene cluster (Clju_c37550 to -37670). fdh genes are not located in this cluster, and some are located distant from this cluster in the chromosome (13). The genome of C. ljungdahlii contains three gene clusters that encode potential FDHs. At the transcriptional level, the fdh gene expression level indicated by the reads per kilobase per million (RPKM) value is high. However, the fold change values are not greater than 2. We consider that their transcription profiles during the exponential and stationary phases do not significantly change. Furthermore, FDH forms a complex with an electron-bifurcating hydrogenase, and this complex accounts for 6% (m/m) of all cytoplasmic proteins involved in C. autoethanogenum growth on CO (18). These results suggest that FDHs play an important role in the metabolic processes of gas fermentation during both the exponential and stationary phases.
The expression profiles of nine genes related to central metabolic pathways were investigated during the exponential and stationary phases via quantitative real-time PCR to verify the RNA-Seq result (Fig. S7). The results indicated that the fold change trends of these key genes were consistent with the findings of RNA-Seq analysis.
Conclusions. The Rnf-ATPase system plays a critical role in NADH production, accompanied by ATP generation during gas fermentation by C. ljungdahlii (4, 11, 35, 36). Ethanol synthesis is a preferred strategy for recycling NADH/NAD+ to maintain the redox balance during autotrophic growth. Therefore, deletion of both adhE genes leads to significant growth deficiencies. In this study, an ethanol oxidation phenotype was clearly observed during fed-batch fermentation with pH and gas pressure control. Combined with the results of key gene knockout, we propose that ethanol synthesis and oxidation are closely related to the growth phase during gas fermentation. Ethanol is synthesized as an NADH sink to achieve redox balance during the exponential phase and is oxidized during the stationary phase in the classic batch fermentation of C. ljungdahlii. These findings provide experimental proof to explain some important previously divulged results. (i) Why does the aor2 deletion mutant of C. autoethanogenum produce up to 2.5-fold more ethanol than the WT strain under the same growth condition (15)? Our results indicate that AOR2, as well as AdhE1, is responsible for ethanol oxidation during the stationary phase. Inactivation of aor2 blocks the oxidation pathway, resulting in a high ethanol concentration in end products. (ii) Why is continuous fermentation (chemostat fermentation) suitable for ethanol production using syngas as a feedstock in C. autoethanogenum and C. ljungdahlii (17, 18, 33)? Our study results illustrated that biomass production is a prerequisite for ethanol production. Continuous fermentation technology maintains strain growth at a constant rate, and ATP is continuously produced. This prompts continuous ethanol biosynthesis rather than oxidation.
MATERIALS AND METHODS
Bacterial strains and media.
Escherichia coli strains were cultivated at 37°C in LB medium in the presence of appropriate antibiotics for general plasmid propagation and cloning. C. ljungdahlii DSM 13528 was purchased from the Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH, Braunschweig, Germany, and conserved by freezing mid-exponential phase cultures at −80°C with 20% glycerol. The cultures were cultivated at 37°C under anaerobic conditions. YTF medium was used in mutant construction, and a modified DSMZ 879 medium with a headspace of a gas mixture (CO:CO2, 80:20 or H2:CO2, 60:40) as the carbon source was used in gas fermentation (35, 37). The modified DSMZ 879 medium has the following composition (per liter): 1.0 g NH4Cl, 0.1 g KCl, 0.2 g MgSO4·7H2O, 0.8 g NaCl, 0.02 g CaCl2·2H2O, 0.1 g KH2PO4, 2.5 mg Na2WO4·2H2O, 1.0 g NaHCO3, 1.0 g cysteine-HCl·H2O, 1 g yeast extract, 0.5 g cysteine, 0.5 mg resazurin, 10 ml trace element solution, and 10 ml vitamin solution. Trace element solution contains 2.0 g nitrilotriacetic acid, 1.3 g MnCl2·H2O, 0.4 g FeSO4·7H2O, 0.2 g CoCl2·7H2O, 0.2 g ZnSO4·7H2O, 0.2 g Na2MoO4·2H2O, 0.02 g NiCl2·6H2O, and 0.1 g Na2SeO3·5H2O in 1 liter distilled water. The vitamin solution contains 2 mg biotin, 2 mg folic acid, 10 mg pyridoxine-HCl, 25 mg thiamine-HCl·2H2O, 5 mg riboflavin, 5 mg nicotinic acid, 5 mg d-Ca-pantothenate, 0.1 mg vitamin B12, 5 mg ρ-aminobenzoic acid, and 5 mg lipoic acid in 1 liter distilled water. Analytical-grade chemicals used in the medium were purchased from Sinopharm Chemical Reagent Co., Ltd. (Shanghai, China). All antibiotics were purchased from Sangon Co., Ltd. (Shanghai, China).
Plasmid construction and ΔadhE1 ΔadhE2 mutant deletion.
Using the adhE1-deleted C. ljungdahlii strain as the parent (30), we deleted the adhE2 gene, yielding a ΔadhE1 ΔadhE2 mutant. The strains and primers used in this study are listed in Tables 3 and 4. The CRISPR/Cas9 editing plasmid, pMTLcas-adhE2, was constructed for adhE2 deletion in C. ljungdahlii. The plasmid pMTLcas-adhE1 was digested with SalI/XhoI, yielding a linear vector without the original single guide RNA (sgRNA) and two homologous arms (HAs). The sgRNA that targets the 20-nucleotide (nt) target spacer of adhE2 was obtained by PCR-amplification using the primers adhE2gRNAs/gRNAaster with pMTLcas-adhE1 as the template. The two HAs that flank the coding region of adhE2 were PCR-amplified from C. ljungdahlii genomic DNA using the primers adhE2uas/adhE2us and adhE2ds/adhE2das. Then, the fragments of sgRNA and HAs were assembled through an overlap extension PCR, yielding the sgRNA-HA fragment. Finally, the above-mentioned linear vector (derived from pMTLcas-adhE1) and sgRNA-HA fragment were assembled using the ClonExpress MultiS one-step cloning kit (Vazyme Biotech Co. Ltd., Nanjing, China), which resulted in the target plasmid pMTLcas-adhE2.
TABLE 3.
Primers used in this study
| Name | Sequence (5′–3′) |
|---|---|
| adhE2gRNAs | AATCTTAAGGAGGAGTTTTCGTCGACGACGAAGCACTCCTAAAGGCGTTTTAGAGCTAG |
| gRNAaster | ATAAAAATAAGAAGCCTGCAAATGCAGGCTTCTTATTTTTATAAAAAAAGCACCGACTC |
| adhE2uas | TTGCAGGCTTCTTATTTTTATATATTGGAAGTTGGTAAAGGATATATGGTAG |
| adhE2us | TTTATTTTTTCTTTCTTGTTTTCTTCTCCACAGTCATTAATTAACGACACCTCTTTCC |
| adhE2das | ACTGTGGAGAAGAAAACAAGAAAGAAAAAATAAATATATAATAAATTG |
| adhE2ds | AAGCTTGCATGTCTGCAGGCCTCGAGCACCATGCACTCTGGCTTAGATCCCCTAAAAG |
| adhE1-f | AAGGATATATGGTAGTATTTGCAGG |
| adhE2-r | GCTCAATTTTATTTACTGCAGTCAC |
TABLE 4.
Strains and plasmids used in this study
| Strain or plasmid name | Characteristics | Reference or source |
|---|---|---|
| Strains | ||
| C. autoethanogenum DSM 10061 (wild type) | DSMZ | |
| C. ljungdahlii DSM 13528 (wild type) | DSMZ | |
| ΔadhE1 mutant | DSM 13528 ΔadhE1 | 30 |
| ΔadhE1 ΔadhE2 mutant | DSM 13528 ΔadhE1 ΔadhE2 | This work |
| Plasmids | ||
| pMTLcas-adhE1 | pCB102 ori, catP, ColE1, tra, Pthl-Cas9, ParaE-sgRNA, adhE1 homologous arm | 30 |
| pMTLcas-adhE2 | pCB102 ori, catP, ColE1, tra, Pthl-Cas9, ParaE-sgRNA, adhE2 homologous arm | This work |
Then, the target plasmid was transferred into adhE1-deleted C. ljungdahlii by electroporation to delete adhE2 according to the protocol (30). The C. ljungdahlii mutant containing deletions in both adhE1 and adhE2 was named the ΔadhE1 ΔadhE2 mutant.
Batch fermentation with gas.
Batch fermentation was performed in a 250-ml screw-cap bottle with a 50-ml working volume in triplicate. The medium was assembled in a Coy anaerobic chamber (Grass Lake, MI, USA). After autoclaving, FeSO4, vitamins, cysteine-HCl, and NaHCO3 were added using syringes with a 0.2-μm filter. Then, gases in the headspace were substituted by CO:CO2 (80:20, vol/vol) or H2:CO2 (60:40, vol/vol) as required with a pressure of 0.2 MPa every 12 h. Fed-batch fermentation was carried out in a FUS-5L bioreactor (Gouqiang Biotech Co. Ltd., Shanghai, China) containing 2.5 liters of modified DSM 879 medium. The gases were controlled at a constant pressure of 0.1 MPa. The stirring rate was 250 rpm, and the pH was maintained at 6.0 automatically by adding 2 M KOH. A 300-ml preculture of C. ljungdahlii was inoculated into the bioreactor, and 5-ml samples were withdrawn every 12 h for cell density monitoring and product analysis.
Preparation of cell extracts and enzyme activity analysis.
Wild-type, ΔadhE1 mutant, and ΔadhE1 ΔadhE2 mutant strains were cultured in YTF medium with 5 g/liter fructose until the late-exponential phase. Cells were collected by centrifugation at 10,000 × g at 4°C under strictly anoxic conditions and were resuspended in 20 ml of anoxic 50-mM potassium phosphate (pH 7.4) containing 2 mM dithiothreitol (DTT). Lysozyme was added to the cell suspension, and the mixture was incubated at 37°C for 30 min. Then, it was moved to an anaerobic chamber for ultrasonication. Finally, cells debris was removed by centrifugation at 35,000 × g at 4°C for 1 h. The protein concentration was determined using the Bradford method (18).
Acetaldehyde:ferredoxin oxidoreductase activity and ethanol dehydrogenase activity were determined under strictly anoxic conditions at 37°C in 1.5-ml anaerobic cuvettes sealed with rubber stoppers (Hellma GmbH, Müllheim, Germany). The cuvettes were filled with pure N2 at 1.2 × 105 Pa as the gas phase before use to maintain anaerobic conditions during enzyme catalysis. The reactions were monitored photometrically at the specified wavelength. Ferredoxin reduction was monitored at 430 nm (Δεox-red ≈ 13.1 mM−1 cm−1), and NADH formation was monitored at 340 nm (Δεox-red ≈ 6.2 mM−1 cm−1). One unit (1 U) was defined as the transfer of 2 μmol electrons min−1. For the acetaldehyde:ferredoxin oxidoreductase activity assay, the mixture contained 50 mM Tris-HCl (pH 7.4), 2 mM DTT, 1.5 mM acetaldehyde, and 30 μM ferredoxin. For alcohol dehydrogenase activity determination, the assay mixture contained 50 mM Tris-HCl (pH 7.4), 2 mM DTT, 1.5 mM ethanol, and 1 mM NAD+ (17, 18).
Ferredoxin was obtained by heterologous expression of gene WP_013236834.1 from C. autoethanogenum in E. coli C41 (38–40). Gene amplification was performed by PCR with genomic DNA of C. autoethanogenum as the template. The following primers were used: 5′-CATGCCATGGCATATAAAATTACAGAGGAT-3′ (reverse primer, the NcoI restriction site is underlined); 5′-CCGCTCGAGGCTTTCTTCAACTGGTGCTC-3′ (forward primer, the XhoI restriction site is underlined). The PCR fragment was digested by restriction endonucleases and subsequently ligated into expression vector pET28b, which had been digested by the same restriction endonucleases. Finally, the constructed plasmid was transferred into E. coli C41 (DE3), which already harbored plasmids pRKISC and pCodonPlus for production of iron-sulfur proteins. Cell cultivation and ferredoxin purification steps were performed as previously described (38). Ferredoxin was stored at −20°C in an N2 atmosphere until use.
Gene expression analysis by RNA-Seq.
Comparative transcriptomics of cells between the exponential and stationary phases was performed to investigate gene expression profiles based on two biological replicates. Cell pellets cultured in a bioreactor were collected by centrifugation at 10,000 × g at −4°C for 48 h and 96 h and then were immediately frozen in liquid nitrogen and stored at −80°C. RNA isolation and high-throughput RNA sequencing (RNA-Seq) were conducted by Oebiotech Corp. (Shanghai, China). Total RNA was extracted using a mirVana microRNA (miRNA) isolation kit (Ambion, Santa Clara, CA, USA) following the manufacturer’s protocol. RNA integrity was evaluated using an Agilent 2100 bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). Samples with an RNA integrity number (RIN) of ≥7 were subjected to subsequent analysis. Libraries were constructed using a TruSeq stranded mRNA LTSample prep kit (Illumina, San Diego, CA, USA) according to the manufacturer’s instructions. Then, these libraries were sequenced on an Illumina sequencing platform (HiSeq 2500), and 150-bp/125-bp paired-end reads were generated. Based on reads per kilobase of transcript per million mapped reads (RPKM) normalization, the gene expression profiles were analyzed.
In order to confirm the authenticity of RNA-Seq data, the expressions of nine key genes located in the central metabolic pathway were evaluated using quantitative real-time PCR (qRT-PCR) on a LightCycler 480 system (Roche Applied Science, Indianapolis, USA) (35). cDNA synthesis was performed according to the instructions from TransGen Biotech Co., Beijing, China. The genes and primers used here are shown in Table S1. All quantitative PCRs were repeated in three biological and three technical replications.
Analytical method.
Cell growth was determined spectrophotometrically at 600 nm (A600) using a UV/visible Ultrospec 2100 Pro spectrophotometer (GE Healthcare, USA). The concentrations of fructose, ethanol, acetic acid, lactate, and 2,3-butanediol were determined using an Agilent 1100 high-performance liquid chromatography (HPLC) system with an Agilent Hi-Plex H column equipped with a refractive index detector and operated at 35°C. Column temperature was maintained at 55°C. Slightly acidified (5 mM H2SO4) water was used as the mobile phase at a flow rate of 0.7 ml/min. Molecular hydrogen (H2) and carbon dioxide (CO2) were detected using a gas chromatography device (GC-7820; Lunan Analysis Instruments Co. Ltd., Tengzhou, China) equipped with a TDX-01 column (2-m length, 3-mm inner diameter). The gas mixture was determined by a thermal conductivity detector with high-purity argon acting as the carrier gas. Changes in the headspace pressure of bottles during gas fermentation were measured using a Hakin pressure detector (Hakin Instruments Co. Ltd., Qingdao, China).
A GC-mass spectrometry (GC-MS) system (Agilent 7890AGC) with a 5,975 C mass selective detector was used to analyze 13C-labeled ethanol and acetate. A 30-m HP-INNOWax column with a 0.25 mm inner diameter and 0.25 μm thickness was used, and helium was the carrier gas at 1 ml/min. The MS instrument was operated in the full scan mode to detect ethanol and acetate. One μl of sample was injected with the injection port temperature at 250°C. The temperature program was as follows: 50°C at 2 min, then increasing 10°C/min to 70°C and 25°C/min to 250°C, and then 250°C for 14 min. 13C-labeled ethanol or acetate was added into the modified DSM 879 medium to achieve a final concentration of 1 g/liter and was analyzed with a mass spectrometer (InertXL MSD; Agilent Technologies). The cells growing in CO under controlled pH and gas pressure were taken out of the bioreactor at the exponential and stationary phases. Cells were collected by centrifugation at 7,500 × g for 5 min at room temperature under anaerobic conditions. Then, the pellets were resuspended in fresh 13C-labeled ethanol medium and 13C-labeled acetate medium. The tubes were incubated at 37°C, and the reaction was stopped at different times by cooling on ice and centrifuging at 10,000 × g at 4°C, followed by GC/MS analysis. Carbon atoms in ethanol and acetate were marked by isotope. Thus, the characteristic peak of 13C-labeled acetate and ethanol could be tested by GC-MS to avoid ethanol and acetate in background cells.
The ratios of NADH/NAD+ and NADPH/NADP+ were measured using Amplite colorimetric NADH and NADPH assay kits (AAT Bioquest, Inc., Sunnyvale, CA, USA). The cell pellets were washed with 0.1 M ice-cold phosphate-buffered saline (PBS) and then suspended in lysate buffer at room temperature for 15 min prior to analysis. The reaction was carried out in 96-well plates at room temperature for 10 h. The concentrations of NADH, NADPH, NAD+, and NADP+ were monitored at 460 nm or 635 nm using a microplate reader (BioTek, Winooski, VT, USA).
Data availability.
RNA-Seq data were submitted to the ArrayExpress database (www.ebi.ac.uk/arrayexpress) under accession number E-MTAB-7753.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by grants from the National Natural Science Foundation of China (31800026), the Key Laboratory of Biofuel, Chinese Academy of Sciences (CASKLB2018X), QIBEBT Funding (QIBEBT ZZBS 201805), the State Key Laboratory of Microbial Technology Open Projects Fund (M2019-02), the Science and Technology Commission of Shanghai Municipality (17JC1404800), and the China Petrochemical Corporation (Sinopec).
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Clomburg JM, Crumbley AM, Gonzalez R. 2017. Industrial biomanufacturing: the future of chemical production. Science 355:aag0804. doi: 10.1126/science.aag0804. [DOI] [PubMed] [Google Scholar]
- 2.Liu CG, Xiao Y, Xia XX, Zhao XQ, Peng LC, Srinophakun P, Bai FW. 2019. Cellulosic ethanol production: progress, challenges and strategies for solutions. Biotechnol Adv 37:491–504. doi: 10.1016/j.biotechadv.2019.03.002. [DOI] [PubMed] [Google Scholar]
- 3.Molitor B, Richter H, Martin ME, Jensen RO, Juminaga A, Mihalcea C, Angenent LT. 2016. Carbon recovery by fermentation of CO-rich off gases: turning steel mills into biorefineries. Bioresour Technol 215:386–396. doi: 10.1016/j.biortech.2016.03.094. [DOI] [PubMed] [Google Scholar]
- 4.Marcellin E, Behrendorff JB, Nagaraju S, DeTissera S, Segovia S, Palfreyman RW, Daniell J, Licona-Cassani C, Quek LE, Speight R, Hodson MP, Simpson SD, Mitchell WP, Köpke M, Nielsen LK. 2016. Low carbon fuels and commodity chemicals from waste gases: systematic approach to understand energy metabolism in a model acetogen. Green Chem 18:3020–3028. doi: 10.1039/C5GC02708J. [DOI] [Google Scholar]
- 5.Liew F, Martin ME, Tappel RC, Heijstra BD, Mihalcea C, Köpke M. 2016. Gas fermentation: a flexible platform for commercial scale production of low-carbon-fuels and chemicals from waste and renewable feedstocks. Front Microbiol 7:694. doi: 10.3389/fmicb.2016.00694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liew F, Henstra AM, Winzer K, Köpke M, Simpson SD, Minton NP. 2016. Insights into CO2 fixation pathway of Clostridium autoethanogenum by targeted mutagenesis. mBio 7:e00427-16. doi: 10.1128/mBio.00427-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dürre P, Eikmanns BJ. 2015. C1-carbon sources for chemical and fuel production by microbial gas fermentation. Curr Opin Biotechnol 35:63–72. doi: 10.1016/j.copbio.2015.03.008. [DOI] [PubMed] [Google Scholar]
- 8.Adamberg K, Valgepea K, Vilu R. 2015. Advanced continuous cultivation methods for systems microbiology. Microbiology 161:1707–1719. doi: 10.1099/mic.0.000146. [DOI] [PubMed] [Google Scholar]
- 9.Munasinghe PC, Khanal SK. 2012. Syngas fermentation to biofuel: evaluation of carbon monoxide mass transfer and analytical modeling using a composite hollow fiber (CHF) membrane bioreactor. Bioresour Technol 122:130–136. doi: 10.1016/j.biortech.2012.03.053. [DOI] [PubMed] [Google Scholar]
- 10.Liu K, Atiyeh HK, Tanner RS, Wilkins MR, Huhnke RL. 2012. Fermentative production of ethanol from syngas using novel moderately alkaliphilic strains of Alkalibaculum bacchi. Bioresour Technol 104:336–341. doi: 10.1016/j.biortech.2011.10.054. [DOI] [PubMed] [Google Scholar]
- 11.Köpke M, Mihalcea C, Bromley JC, Simpson SD. 2011. Fermentative production of ethanol from carbon monoxide. Curr Opin Biotechnol 22:320–325. doi: 10.1016/j.copbio.2011.01.005. [DOI] [PubMed] [Google Scholar]
- 12.Munasinghe PC, Khanal SK. 2010. Syngas fermentation to biofuel: evaluation of carbon monoxide mass transfer coefficient (kLa) in different reactor configurations. Biotechnol Prog 26:1616–1621. doi: 10.1002/btpr.473. [DOI] [PubMed] [Google Scholar]
- 13.Köpke M, Held C, Hujer S, Liesegang H, Wiezer A, Wollherr A, Ehrenreich A, Liebl W, Gottschalk G, Durre P. 2010. Clostridium ljungdahlii represents a microbial production platform based on syngas. Proc Natl Acad Sci U S A 107:13087–13092. doi: 10.1073/pnas.1004716107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brown SD, Nagaraju S, Utturkar S, De Tissera S, Segovia S, Mitchell W, Land ML, Dassanayake A, Köpke M. 2014. Comparison of single-molecule sequencing and hybrid approaches for finishing the genome of Clostridium autoethanogenum and analysis of CRISPR systems in industrial relevant Clostridia. Biotechnol Biofuels 7:40. doi: 10.1186/1754-6834-7-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liew F, Henstra AM, Kӧpke M, Winzer K, Simpson SD, Minton NP. 2017. Metabolic engineering of Clostridium autoethanogenum for selective alcohol production. Metab Eng 40:104–114. doi: 10.1016/j.ymben.2017.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leang C, Ueki T, Nevin KP, Lovley DR. 2013. A genetic system for Clostridium ljungdahlii: a chassis for autotrophic production of biocommodities and a model homoacetogen. Appl Environ Microbiol 79:1102–1109. doi: 10.1128/AEM.02891-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mock J, Zheng YN, Mueller AP, Ly S, Tran L, Segovia S, Nagaraju S, Köpke M, Dürre P, Thauer RK. 2015. Energy conservation associated with ethanol formation from H2 and CO2 in Clostridium autoethanogenum involving electron bifurcation. J Bacteriol 197:2965–2980. doi: 10.1128/JB.00399-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang S, Huang H, Kahnt J, Mueller AP, Köpke M, Thauer RK. 2013. NADP-specific electron-bifurcating [FeFe]-hydrogenase in a functional complex with formate dehydrogenase in Clostridium autoethanogenum grown on CO. J Bacteriol 195:4373–4386. doi: 10.1128/JB.00678-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Valgepea K, Loi KQ, Behrendorff JB, Lemgruber RDP, Plan M, Hodson MP, Köpke M, Nielsen LK, Marcellin E. 2017. Arginine deiminase pathway provides ATP and boosts growth of the gas-fermenting acetogen Clostridium autoethanogenum. Metab Eng 41:202–211. doi: 10.1016/j.ymben.2017.04.007. [DOI] [PubMed] [Google Scholar]
- 20.Valgepea K, de Souza Pinto Lemgruber R, Meaghan K, Palfreyman RW, Abdalla T, Heijstra BD, Behrendorff JB, Tappel R, Köpke M, Simpson SD, Nielsen LK, Marcellin E. 2017. Maintenance of ATP homeostasis triggers metabolic shifts in gas-fermenting acetogens. Cell Syst 4:505–515. doi: 10.1016/j.cels.2017.04.008. [DOI] [PubMed] [Google Scholar]
- 21.Schuchmann K, Müller V. 2014. Autotrophy at the thermodynamic limit of life: a model for energy conservation in acetogenic bacteria. Nat Rev Microbiol 12:809–821. doi: 10.1038/nrmicro3365. [DOI] [PubMed] [Google Scholar]
- 22.Peters JW, Miller AF, Jones AK, King PW, Adams MWW. 2016. Electron bifurcation. Curr Opin Chem Biol 31:146–152. doi: 10.1016/j.cbpa.2016.03.007. [DOI] [PubMed] [Google Scholar]
- 23.Tremblay PL, Zhang T, Dar SA, Leang C, Lovley DR. 2012. The Rnf complex of Clostridium ljungdahlii is a proton-translocating ferredoxin:NAD+ oxidoreductase essential for autotrophic growth. mBio 4:e0040612. doi: 10.1128/mBio.00406-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fast AG, Papoutsakis ET. 2012. Stoichiometric and energetic analyses of non-photosynthetic CO2-fixation pathways to support synthetic biology strategies for production of fuels and chemicals. Curr Opin Chem Eng 1:380–395. doi: 10.1016/j.coche.2012.07.005. [DOI] [Google Scholar]
- 25.Fuchs G. 2011. Alternative pathways of carbon dioxide fixation: insights into the early evolution of life? Annu Rev Microbiol 65:631–658. doi: 10.1146/annurev-micro-090110-102801. [DOI] [PubMed] [Google Scholar]
- 26.Ragsdale SW, Pierce E. 2008. Acetogenesis and the Wood-Ljungdahl pathway of CO2 fixation. Biochim Biophys Acta 1784:1873–1898. doi: 10.1016/j.bbapap.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wood HG. 1991. Life with CO or CO2 and H2 as a source of carbon and energy. FASEB J 5:156–163. doi: 10.1096/fasebj.5.2.1900793. [DOI] [PubMed] [Google Scholar]
- 28.Daniell J, Köpke M, Simpson SD. 2012. Commercial biomass syngas fermentation. Energies 5:5372–5417. doi: 10.3390/en5125372. [DOI] [Google Scholar]
- 29.Membrillo-Hernández J, Lin EC. 1999. Regulation of expression of the adhE gene, encoding ethanol oxidoreductase in Escherichia coli: transcription from a downstream promoter and regulation by fnr and RpoS. J Bacteriol 181:7571–7579. doi: 10.1128/JB.181.24.7571-7579.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang H, Chai C, Li N, Rowe P, Minton NP, Yang S, Jiang W, Gu Y. 2016. CRISPR/Cas9-based efficient genome editing in Clostridium ljungdahlii, an autotrophic gas-fermenting bacterium. ACS Synth Biol 5:1355–1361. doi: 10.1021/acssynbio.6b00044. [DOI] [PubMed] [Google Scholar]
- 31.Thauer RK, Jungermann K, Decker K. 1977. Energy conservation in chemotrophic anaerobic bacteria. Bacteriol Rev 41:100–180. doi: 10.1128/MMBR.41.1.100-180.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martin ME, Richter H, Saha S, Angenent LT. 2016. Traits of selected Clostridium strains for syngas fermentation to ethanol. Biotechnol Bioeng 113:531–539. doi: 10.1002/bit.25827. [DOI] [PubMed] [Google Scholar]
- 33.Richter H, Molitor B, Wei H, Chen W, Aristilde L, Angenent LT. 2016. Ethanol production in syngas-fermenting Clostridium ljungdahlii is controlled by thermodynamics rather than by enzyme expression. Energy Environ Sci 9:2392–2399. doi: 10.1039/C6EE01108J. [DOI] [Google Scholar]
- 34.Zhu HF, Liu ZY, Zhou X, Yi JH, Lun ZM, Wang SN, Tang WZ, Li FL. 2020. Energy conservation and carbon flux distribution during fermentation of CO or H2/CO2 by Clostridium ljungdahlii. Front Microbiol 11:416. doi: 10.3389/fmicb.2020.00416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xie BT, Liu ZY, Tian L, Li FL, Chen XH. 2015. Physiological response of Clostridium ljungdahlii DSM 13528 of ethanol production under different fermentation conditions. Bioresour Technol 177:302–307. doi: 10.1016/j.biortech.2014.11.101. [DOI] [PubMed] [Google Scholar]
- 36.Valgepea K, de Souza Pinto Lemgruber R, Abdalla T, Binos S, Takemori N, Takemori A, Tanaka Y, Tappel R, Köpke M, Simpson SD, Nielsen LK, Marcellin E. 2018. H2 drives metabolic rearrangements in gas-fermenting Clostridium autoethanogenum. Biotechnol Biofuels 11:55. doi: 10.1186/s13068-018-1052-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Humphreys CM, McLean S, Schatschneider S, Millat T, Henstra AM, Annan FJ, Breitkopf R, Pander B, Piatek P, Rowe P, Wichlacz AT, Woods C, Norman R, Blom J, Goesman A, Hodgman C, Barrett D, Thomas NR, Winzer K, Minton NP. 2015. Whole genome sequence and manual annotation of Clostridium autoethanogenum, an industrially relevant bacterium. BMC Genomics 16:1085. doi: 10.1186/s12864-015-2287-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Demmer JK, Huang H, Wang S, Demmer U, Thauer RK, Ermler U. 2015. Insights into flavin-based electron bifurcation via the NADH-dependent reduced ferredoxin:NADP oxidoreductase structure. J Biol Chem 290:21985–21995. doi: 10.1074/jbc.M115.656520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nakamura M, Saeki K, Takahashi Y. 1999. Hyperproduction of recombinant ferredoxins in Escherichia coli by coexpression of the ORF1-ORF2-iscU-iscA-hscB-hscA-fdx-ORF3 gene cluster. J Biochem 126:10–18. doi: 10.1093/oxfordjournals.jbchem.a022409. [DOI] [PubMed] [Google Scholar]
- 40.Huang H, Hu L, Yu W, Li H, Tao F, Xie H, Wang S. 2016. Heterologous overproduction of 2[4Fe4S]- and [2Fe2S]-type clostridial ferredoxins and [2Fe2S]-type agrobacterial ferredoxin. Protein Expr Purif 121:1–8. doi: 10.1016/j.pep.2015.12.019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-Seq data were submitted to the ArrayExpress database (www.ebi.ac.uk/arrayexpress) under accession number E-MTAB-7753.