The gut microbiome is pertinent to many aspects of animal health, and there is a great need for natural but tractable experimental systems to examine the processes shaping gut microbiome assembly. Here, the holothurian (sea cucumber) Sclerodactyla briareus was explored as an experimental system to study microbial colonization in the gut, as S. briareus individuals have the ability to completely eviscerate and rapidly regenerate their digestive organs. After induced evisceration, microbial community assembly was characterized over 20 days in regenerating animals. This study demonstrated that colonization of the sea cucumber gut was deterministic; despite immersion in a diverse consortium of environmental microbes, a specific subset of microbes proliferated in the gut, including taxa that likely conferred energetic and immune advantages to the host. Sea cucumbers have the potential to revolutionize our understanding of gut microbiome assembly, as rapid and repeatable gut tissue regeneration provides a promising and tractable experimental system.
KEYWORDS: holothurian, community assembly, gut microbiome, host-microbe, intestinal colonization, tissue regeneration
ABSTRACT
The gut microbiome has far-reaching effects on host organism health, so understanding the processes that underlie microbial community assembly in the developing gut is a current research priority. Here, a holothurian (also known as sea cucumber; phylum Echinodermata) host is explored as a promising model system for studying the assembly of the gut microbiome. Holothurians have a unique capacity for evisceration (expulsion of the internal organs), followed by rapid regeneration of the gut, decoupling host ontogeny from gut tissue development and permitting experimental manipulation of the gut microbiome in mature host individuals. Here, evisceration was induced in the sea cucumber Sclerodactyla briareus, and regenerating stomach and intestine microbiomes were characterized before and on days 0, 13, 17, and 20 after evisceration using Illumina sequencing of 16S rRNA genes. Regenerating stomach and intestine tissues had microbial communities significantly different from those of mature tissues, with much higher alpha diversity and evenness of taxa in regenerating tissues. Despite immersion in a diverse pool of sediment and seawater microbes in flowthrough seawater aquaria, regenerating gut microbiomes differed at each stage of regeneration and displayed a highly similar community structure among replicates, providing evidence for deterministic host selection of a specific microbial consortium. Moreover, regenerating gut tissues acquired a microbiome that likely conferred energetic and immune advantages to the sea cucumber host, including microbes that can fix carbon and degrade invading pathogens.
IMPORTANCE The gut microbiome is pertinent to many aspects of animal health, and there is a great need for natural but tractable experimental systems to examine the processes shaping gut microbiome assembly. Here, the holothurian (sea cucumber) Sclerodactyla briareus was explored as an experimental system to study microbial colonization in the gut, as S. briareus individuals have the ability to completely eviscerate and rapidly regenerate their digestive organs. After induced evisceration, microbial community assembly was characterized over 20 days in regenerating animals. This study demonstrated that colonization of the sea cucumber gut was deterministic; despite immersion in a diverse consortium of environmental microbes, a specific subset of microbes proliferated in the gut, including taxa that likely conferred energetic and immune advantages to the host. Sea cucumbers have the potential to revolutionize our understanding of gut microbiome assembly, as rapid and repeatable gut tissue regeneration provides a promising and tractable experimental system.
INTRODUCTION
In most animals, the gut microbiome is pertinent to almost all aspects of host health (1, 2), including metabolism and digestion, the immune system, and even behavior (3). Perturbations or abnormal development of the gut microbiome are associated with various gastrointestinal and metabolic diseases (1, 2); thus, a better understanding of how the gut microbiome develops in animals may be critical to the treatment of some chronic diseases. The assembly of the gut microbiome can be shaped by maternal transmission, host genetic factors, or a combination of host-microbe and microbe-microbe interactions (1, 4). For example, the gut microbiota can be altered by host nutrient secretions (5), host diet (6), or through microbe-microbe interactions such as metabolite secretion (7) or the reduction in oxygen availability by primary colonizing microbes (8). Many studies have used germfree animals to artificially induce gut microbial community assembly (9), and other studies track microbial community development from birth through infancy in humans (10–12). Here, a holothurian (sea cucumber) system was explored to study colonization and community assembly in the gut microbiome of mature hosts under natural conditions, using regeneration to separate the development of the gut from the ontogeny of the host organism. Sea cucumbers are echinoderms, closely related to chordates within the deuterostome phylogeny (13), making them a particularly attractive invertebrate model for gut microbiome research. Furthermore, invertebrate microbiomes may be simpler than vertebrate microbiomes, making them tractable systems, yet there is a dearth of information about the invertebrate gut microbiome (14).
Holothurians are well known for their remarkable ability to eviscerate most of their internal organs, including the digestive tract, using specialized mutable connective tissue (15). The adaptive significance of sea cucumber evisceration is still uncertain; while evisceration occurs in the presence of predators for some holothurians (16), others eviscerate seasonally (17), or evisceration may be a mechanism for the host to expel internal parasites (18). After expulsion of their internal organs, holothurians regenerate the lost viscera at a remarkable rate, in as little as 7 days, beginning with the digestive tract (15). Many holothurians are deposit feeders, consuming bacteria and organic matter in the sediment (19); they have a significant impact on organic matter decomposition and nutrient cycling in soft substrate marine ecosystems (20). Some of the microbes that enter the holothurian gut evade consumption and populate the sea cucumber foregut, where they flourish (19). Sea cucumbers can begin feeding as early as 16 days post-evisceration (21), making them an ideal system to study microbial colonization of the gut.
Immense quantities of sediment pass through the sea cucumber gut; thus, the gut microbiome may play essential roles in preventing pathogenic microbial colonization, metabolizing organic matter in the ingested sediment, or producing essential vitamins (22, 23). However, few studies have characterized the gut microbiome of sea cucumbers (24), and most presently available studies examined the microbiome of the commercially valuable sea cucumber Apostichopus japonicus (22, 25–30). Two studies examined the regenerating gut microbial communities of A. japonicus (23, 29), demonstrating that the composition of the gut microbiome changes rapidly during visceral regeneration, yet further study with increased replication is clearly warranted to understand the processes driving microbial community assembly in this system. The deposit-feeding sea cucumber Sclerodactyla briareus lives in muddy sediments along the continental shelf of the western Atlantic Ocean (31). S. briareus burrows into the sediment and feeds by extending its oral tentacles, which collect particles of sediment, microbes, and organic debris from the seawater or sediment surface (32). Evisceration of S. briareus can be rapidly induced with KCl, leading to anterior expulsion of the viscera (33) and regeneration of intestinal tissues within 20 days (21). Here, S. briareus is explored as a system for studying community assembly in the regenerating gut microbiome.
S. briareus individuals were collected from two different estuarine ponds with distinct sediment microbial communities, stimulated to eviscerate, and allowed to regenerate their lost viscera in a flowthrough seawater tank environment. The aims of this study were to (i) describe the pattern of microbial colonization of the holothurian stomach and intestine during regeneration on days 13, 17, and 20 post-evisceration, (ii) test whether newly formed stomach and intestine microbial communities resemble the initial condition of the mature microbiomes, (iii) examine the influence of the historical environment on microbial community assembly by regenerating S. briareus individuals collected from two different sediment environments in a common sediment environment, and (iv) determine whether the stomach and intestine microbiomes are randomly or deterministically assembled from the pool of potential colonizing microbes.
RESULTS
Sea cucumber visceral regeneration in 20 days.
The first S. briareus individual was dissected after 7 days, and it contained only white mesentery tissue and no visible stomach or intestine. By day 13, dissection revealed that all S. briareus individuals had yellow developing intestine tissue, surrounded by white mesentery tissue. On day 13, only one individual out of 13 had begun feeding, indicated by the presence of sediment in the intestine. By day 17, all dissected S. briareus individuals had regenerated a calcareous ring, stomach, and intestine, and 2 out of 6 individuals had begun feeding. By day 20, all S. briareus individuals had regenerated a stomach and intestine, and all 13 individuals had been feeding. While the stomach and intestine were regenerated by day 20, the regenerated intestine lacked the red color present in the mature intestine (Fig. 1A) that is indicative of blood vessels surrounding the intestine. The regenerated intestines were smaller in length and thinner in diameter than the mature intestine (Fig. 1B).
FIG 1.
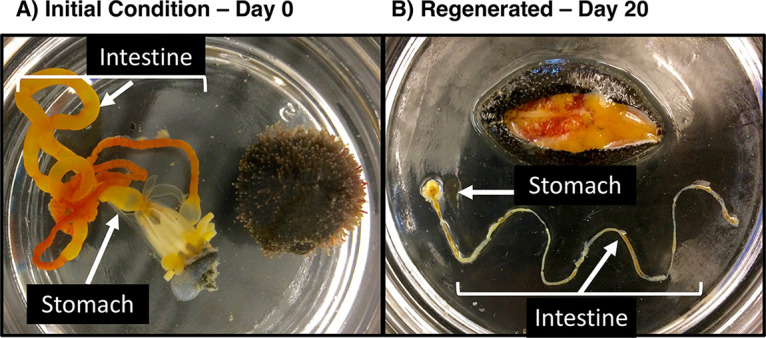
Photographs of eviscerated stomach and intestine tissues from a mature S. briareus individual (A) and a longitudinally dissected S. briareus individual revealing regenerated stomach and intestine tissues after 20 days in the experimental tank (B). There is evidence of feeding by the specimen in panel B, i.e., brown ingested material is visible in the stomach and intestine.
Microbial communities in stomach, intestine, and environmental samples.
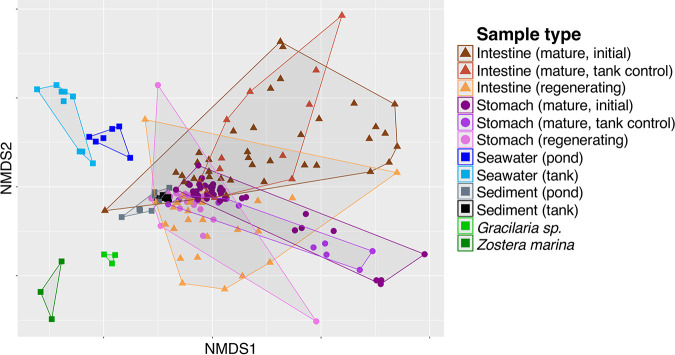
After sequence quality control and processing, the final data set contained 185 samples and 1,258,000 amplicon sequences, collected from 38 environmental samples and 51 S. briareus individuals. In this study, 16S rRNA gene sequences were grouped into amplicon sequence variants (ASVs), which are analogous to operational taxonomic units (OTUs) but do not impose arbitrary sequence similarity thresholds. As reflected by the divergent clusters among sample types in the nonmetric multidimensional scaling (NMDS) plot (Fig. 2), microbial communities from stomach, intestine, sediment, algae and seagrass, and seawater sources were significantly different (permutational multivariate analysis of variance [PERMANOVA], df = 6, total df = 184, pseudo-F = 11.86, P = 0.001) (Table 1). Overall, S. briareus stomach and intestine microbial communities were distinct from all environmental sources (Table 1), including both seawater and sediment microbial communities (Fig. 2). Sea cucumber microbiomes were also distinct from algae- and seagrass-associated microbial communities (Fig. 2), despite intimate association with these primary producers in the environment at each site. Overall, stomach and intestine microbial communities were more similar to sediment than to seawater or algae and seagrass microbial communities (Fig. 2); stomach samples shared a higher percentage of ASVs with sediment samples (12.9 to 42.7%) than with seawater (3.4 to 17.7%), and intestine microbial communities displayed the same trend (see Table S1 in the supplemental material). Stomach and intestine samples shared the lowest proportion of ASVs with the alga Gracilaria sp. and the seagrass Zostera marina (Table S1).
FIG 2.
Nonmetric multidimensional scaling (NMDS) plot of microbial communities from mature (initial and tank control) and regenerating stomach (circles) and intestine (triangles) tissues, seawater and sediment from the environment and the experimental tank (squares), and algae (Gracilaria sp.) and seagrass (Zostera marina) from the pond collection sites (squares). Note that regenerating stomach and intestine tissues include days 13, 17, and 20 of regeneration.
TABLE 1.
Pairwise comparisons of beta diversity among stomach, intestine, and environmental microbial communitiesa
| Multivariate pairwise test comparison | df | PERMANOVA |
PERMDISP |
||
|---|---|---|---|---|---|
| t | P | t | P | ||
| Mature stomach vs mature intestine | 95 | 5.79 | 0.001* | 5.58 | 0.001* |
| Regenerating stomach vs regenerating intestine | 44 | 1.24 | 0.068 | 2.57 | 0.04* |
| Mature stomach vs regenerating stomach | 65 | 4.02 | 0.001* | 1.64 | 0.18 |
| Mature intestine vs regenerating intestine | 74 | 3.36 | 0.001* | 0.34 | 0.79 |
| Mature stomach vs regenerating intestine | 75 | 4.10 | 0.001* | 5.16 | 0.001* |
| Mature intestine vs regenerating stomach | 64 | 3.36 | 0.001* | 2.41 | 0.03* |
| Seawater vs mature stomach | 62 | 4.57 | 0.001* | 3.79 | 0.004* |
| Seawater vs regenerating stomach | 31 | 2.83 | 0.002* | 1.98 | 0.08 |
| Seawater vs mature intestine | 61 | 3.28 | 0.001* | 0.15 | 0.92 |
| Seawater vs regenerating intestine | 41 | 2.62 | 0.001* | 0.49 | 0.68 |
| Sediment vs mature stomach | 68 | 4.16 | 0.001* | 3.47 | 0.003* |
| Sediment vs regenerating stomach | 37 | 1.76 | 0.004* | 1.40 | 0.32 |
| Sediment vs mature intestine | 67 | 3.35 | 0.001* | 0.57 | 0.62 |
| Sediment vs regenerating intestine | 47 | 1.72 | 0.007* | 0.80 | 0.53 |
| Sediment vs seawater | 34 | 2.23 | 0.001* | 0.36 | 0.78 |
| Algae, seagrass vs mature stomach | 53 | 3.82 | 0.001* | 1.69 | 0.16 |
| Algae, seagrass vs regenerating stomach | 22 | 2.77 | 0.001* | 0.57 | 0.66 |
| Algae, seagrass vs mature intestine | 52 | 2.58 | 0.001* | 1.02 | 0.36 |
| Algae, seagrass vs regenerating intestine | 32 | 2.34 | 0.001* | 1.42 | 0.31 |
| Algae, seagrass vs seawater | 19 | 2.34 | 0.001* | 1.91 | 0.29 |
| Algae, seagrass vs sediment | 25 | 2.43 | 0.001* | 0.51 | 0.68 |
Denominator degrees of freedom are listed for all pairwise comparisons, and significant P values (<0.05) are indicated with asterisks. PERMDISP tests were conducted to ensure that significant PERMANOVA results were not due to unequal dispersion of variability among groups. Algae and seagrass include the red algae Gracilaria sp. and the seagrass Zostera marina. t, t statistic of the pairwise PERMANOVA test.
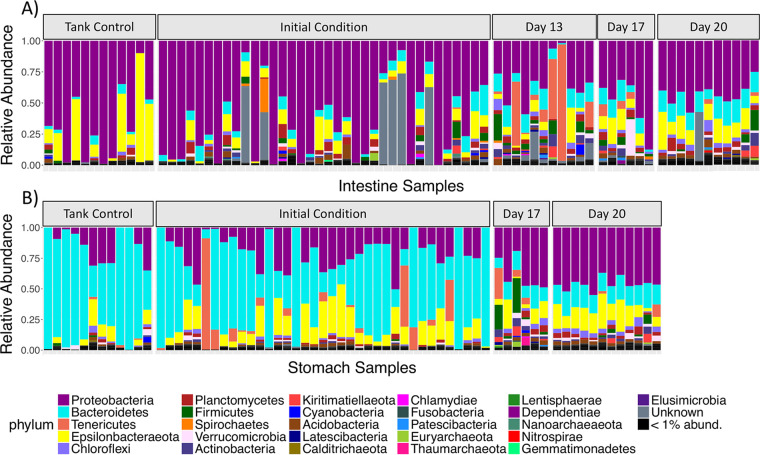
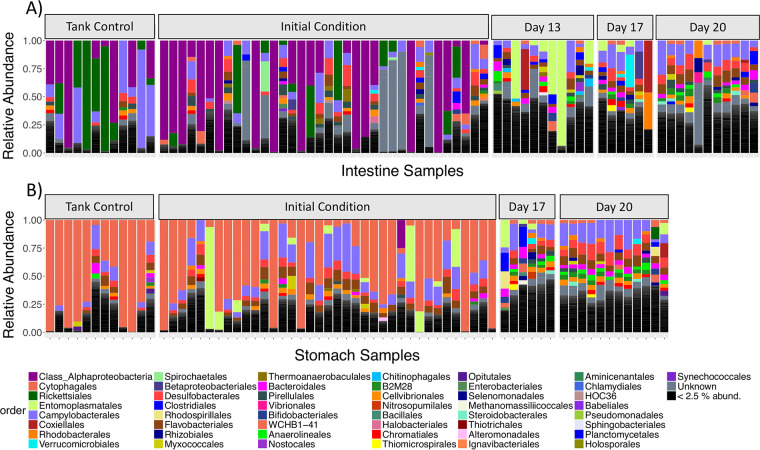
Mature stomach and intestine microbial communities displayed significantly different community structures, although the dispersion of variance was unequal between the two groups (Table 1), cautioning against the validity of the comparison. Nevertheless, the compositions of the mature stomach and intestine microbiomes were quite distinct at both the phylum and order levels (Fig. 3 and 4). The stomach microbiome of mature S. briareus individuals (initial condition and tank control individuals) was comprised mostly of the phylum Bacteroidetes (mean relative abundance, 47% in the stomach versus 8% in the intestine), while the mature intestinal microbiome contained mostly Proteobacteria (56% in the intestine versus 25% in the stomach) (Fig. 3). Both stomach and intestine microbial communities contained similar abundances of the phylum Epsilonbacteraeota (11 to 12%) (Fig. 3; Table 2). Mature stomach microbial communities were dominated by the bacterial orders Cytophagales (38%) and Campylobacterales (12%) from the phyla Bacteroidetes and Epsilonbacteraeota, respectively (Fig. 4B; Table 2). In contrast, mature intestinal microbial communities were dominated by the order Campylobacterales (11%) and by an unknown Alphaproteobacteria (36%) (Fig. 4A; Table 2). Interestingly, members of the order Rickettsiales reached a high abundance in the mature intestinal microbiome of select individuals, mostly tank controls, yet they were noticeably absent from the stomach and the regenerating intestinal microbiomes (Fig. 4; Table 2). The three most abundant ASVs from the mature stomach and intestine microbiomes included one uncultured Bacteroidetes from the family Cyclobacteriaceae, an unknown Alphaproteobacteria, and a Sulfurovum sp. from the phylum Epsilonbacteraeota.
FIG 3.
Relative abundance barplots of sea cucumber microbial communities from replicate intestine (A) and stomach (B) samples over regeneration time. Colors represent microbial phylum-level taxonomy.
FIG 4.
Relative abundance barplots of sea cucumber microbial communities from replicate intestine (A) and stomach (B) samples over regeneration time. Colors represent microbial order-level taxonomy. Note that the first taxon is a single ASV that was unclassified at the order level but is classified as a member of the Alphaproteobacteria.
TABLE 2.
Relative abundances of the top 10 phyla, classes, and orders of microbes found in all stomach and intestine samplesa
| Taxonomic grouping | Relative abundance (±SD) in: |
|||
|---|---|---|---|---|
| Stomach | Intestine | Sediment | Seawater | |
| Phylum | ||||
| Bacteroidetes | 0.47 (±0.32) | 0.08 (±0.06) | 0.16 (±0.04) | 0.39 (±0.03) |
| Proteobacteria | 0.25 (±0.17) | 0.56 (±0.28) | 0.45 (±0.02) | 0.24 (±0.04) |
| Epsilonbacteraeota | 0.12 (±0.10) | 0.11 (±0.14) | 0.10 (±0.04) | 0.02 (±0.01) |
| Tenericutes | 0.05 (±0.13) | 0.04 (±0.13) | <0.01 (±0.01) | <0.01 (±0.01) |
| Chloroflexi | 0.02 (±0.02) | 0.01 (±0.02) | 0.07 (±0.03) | <0.01 (±0.01) |
| Planctomycetes | 0.02 (±0.02) | 0.03 (±0.03) | 0.02 (±0.01) | 0.01 (±0.01) |
| Acidobacteria | 0.01 (±0.01) | 0.02 (±0.02) | 0.04 (±0.01) | <0.01 (±0.01) |
| Firmicutes | 0.01 (±0.04) | 0.02 (±0.04) | <0.01 (±0.01) | <0.01 (±0.01) |
| Verrucomicrobia | 0.01 (±0.01) | 0.01 (±0.03) | <0.01 (±0.01) | 0.16 (±0.05) |
| Actinobacteria | 0.01 (±0.01) | 0.02 (±0.02) | <0.01 (±0.01) | 0.05 (±0.02) |
| Class | ||||
| Bacteroidia | 0.47 (±0.32) | 0.08 (±0.06) | 0.14 (±0.04) | 0.39 (±0.03) |
| Gammaproteobacteria | 0.14 (±0.10) | 0.15 (±0.13) | 0.26 (±0.03) | 0.05 (±0.01) |
| Campylobacteria | 0.12 (±0.10) | 0.11 (±0.14) | 0.10 (±0.04) | 0.02 (±0.01) |
| Deltaproteobacteria | 0.07 (±0.05) | 0.05 (±0.05) | 0.18 (±0.01) | 0.02 (±0.01) |
| Mollicutes | 0.05 (±0.13) | 0.04 (±0.13) | <0.01 (±0.01) | <0.01 (±0.01) |
| Alphaproteobacteria | 0.04 (±0.05) | 0.36 (±0.37) | <0.01 (±0.01) | 0.17 (±0.04) |
| Anaerolineae | 0.02 (±0.02) | 0.01 (±0.02) | 0.06 (±0.03) | <0.01 (±0.01) |
| Planctomycetacia | 0.01 (±0.01) | 0.02 (±0.03) | <0.01 (±0.01) | 0.01 (±0.01) |
| Verrucomicrobiae | 0.01 (±0.01) | 0.01 (±0.03) | <0.01 (±0.01) | 0.16 (±0.05) |
| Clostridia | 0.01 (±0.03) | 0.02 (±0.03) | <0.01 (±0.01) | <0.01 (±0.01) |
| Order | ||||
| Cytophagales | 0.38 (±0.37) | 0.02 (±0.02) | 0.02 (±0.01) | <0.01 (±0.01) |
| Campylobacterales | 0.12 (±0.10) | 0.11 (±0.14) | 0.10 (±0.04) | 0.02 (±0.01) |
| Entomoplasmatales | 0.05 (±0.13) | 0.04 (±0.13) | <0.01 (±0.01) | <0.01 (±0.01) |
| Flavobacteriales | 0.05 (±0.04) | 0.03 (±0.03) | 0.04 (±0.02) | 0.28 (±0.09) |
| Desulfobacterales | 0.05 (±0.04) | 0.04 (±0.04) | 0.11 (±0.01) | <0.01 (±0.01) |
| Cellvibrionales | 0.02 (±0.02) | 0.02 (±0.02) | 0.02 (±0.01) | 0.02 (±0.01) |
| Bacteroidales | 0.02 (±0.02) | 0.02 (±0.02) | 0.06 (±0.01) | <0.01 (±0.01) |
| Pirellulales | 0.01 (±0.01) | 0.02 (±0.02) | <0.01 (±0.01) | <0.01 (±0.01) |
| Clostridiales | 0.01 (±0.03) | 0.02 (±0.03) | <0.01 (±0.01) | <0.01 (±0.01) |
| Rickettsiales | <0.01 (±0.01) | 0.08 (±0.20) | <0.01 (±0.01) | <0.01 (±0.01) |
The sediment and seawater columns of data display the abundances of these host-associated microbes in environmental samples.
Similar to the mature stomach and intestine microbiomes, the regenerating stomach and intestine microbiomes were comprised mainly of the phyla Proteobacteria, Bacteroidetes, and Epsilonbacteraeota (Fig. 3). However, at lower taxonomic levels such as order, the regenerating stomach and intestine microbial communities were much more diverse than the mature microbiomes (Fig. 4). Rather than being dominated by a few taxonomic orders, the regenerating stomach and intestine communities were comprised of >10 orders, with relatively even representation across replicate samples (Fig. 4). These trends were also quite evident at the ASV level. Within mature stomach and intestine microbiomes, only five and three ASVs, respectively, made up 50% of the total microbial community, while in the regenerating stomach and intestine microbiomes, 54 and 67 ASVs, respectively, made up 50% of the community. The regenerating stomach microbiome was dominated by the orders Campylobacterales (13.7%), Desulfobacterales (7.3%), Flavobacteriales (6.2%), and Bacteroidales (4.3%), while regenerating intestine tissues were dominated by the orders Campylobacterales (10%), Flavobacteriales (5.3%), Desulfobacterales (4.8%), Coxiellales (3.5%), Rhodobacterales (3.4%), and Bacteroidales (3.0%) (Fig. 4).
Seawater and sediment microbial communities were also dominated by Proteobacteria and Bacteroidetes at the phylum level, but they were quite distinct from S. briareus microbial communities at lower taxonomic levels (Table 2; Fig. S1). Sediment microbial communities were dominated by the orders Campylobacterales (13%), Desulfobacterales (10%), Flavobacteriales (6%), and Bacteroidales (6%) (Fig. S1). Seawater samples were composed primarily of the orders Flavobacteriales (21%), Rhodobacterales (10%), the SAR11 clade (9%), Verrucomicrobiales (9%), and Synechococcales (7%) (Fig. S1). The dominant bacterial order found in the stomach, Cytophagales, was present at only 2% and <1% abundance in sediment and seawater microbial communities, respectively (Table 2). Likewise, the most abundant microbe in the intestine, an unknown alphaproteobacterium, comprised only 5% or less of sediment and seawater microbial communities (Table 2).
Tank residence, but not field collection site, affected gut microbial communities.
Despite collection from distinct ponds, the initial stomach and intestine microbial communities were not affected by collection site (PERMANOVA pairwise comparisons, P = 0.10 for stomach, P = 0.45 for intestine) (Table 3). Seawater samples collected from the two ponds also did not differ (Table 3), likely reflecting the connectivity of the two estuarine ponds. In contrast, sediment samples collected from Jehu and Hamblin Ponds were significantly different (Table 3). The tank environment contained a different pool of sediment and seawater microbes than the pond collection sites. The seawater collected from each of the two ponds differed significantly from the flowthrough tank seawater (Table 3). Surprisingly, although the tank sediment was sourced from Hamblin Pond, tank sediment sampled on days 13, 17, and 20 differed significantly from field-collected Hamblin and Jehu Pond sediment (Table 3).
TABLE 3.
Pairwise comparisons of beta diversity of samples by sitea
| Multivariate pairwise test comparison | df | PERMANOVA |
PERMDISP |
||
|---|---|---|---|---|---|
| t | P | t | P | ||
| Stomach | |||||
| Jehu vs Hamblin | 35 | 1.52 | 0.10 | 4.98 | 0.97 |
| Tank control vs Jehu | 28 | 2.29 | 0.01* | 1.12 | 0.35 |
| Tank control vs Hamblin | 29 | 1.11 | 0.25 | 0.98 | 0.46 |
| Intestine | |||||
| Jehu vs Hamblin | 34 | 0.95 | 0.45 | 1.04 | 0.44 |
| Tank control vs Jehu | 28 | 1.37 | 0.09 | 1.25 | 0.29 |
| Tank control vs Hamblin | 28 | 1.52 | 0.04* | 5.98 | 0.96 |
| Sediment | |||||
| Jehu vs Hamblin | 7 | 2.45 | 0.01* | 2.34 | 0.04* |
| Tank vs Jehu | 10 | 1.54 | 0.01* | 4.13 | 0.01* |
| Tank vs Hamblin | 9 | 3.17 | 0.01* | 2.02 | 0.23 |
| Seawater | |||||
| Jehu vs Hamblin | 4 | 5.64 | 0.12 | 1.04 | 0.60 |
| Tank vs Jehu | 10 | 2.30 | 0.02* | 9.32 | 0.008* |
| Tank vs Hamblin | 10 | 2.24 | 0.02* | 9.72 | 0.004* |
Pairwise comparisons were of Jehu and Hamblin Ponds with the experimental tank for mature (initial and tank control) stomach and intestine microbial communities and for sediment and seawater microbial communities. Denominator degrees of freedom are listed for all pairwise comparisons, and significant P values (<0.05) are indicated with asterisks. PERMDISP tests were conducted to ensure that significant PERMANOVA results were not due to unequal dispersion of variability among groups. t, t statistic of the pairwise PERMANOVA test.
Living in the flowthrough seawater tank for 20 days significantly altered the stomach and intestine composition of actively feeding S. briareus individuals (tank controls) compared to the initial condition of the field-collected mature microbiomes (Table 4, see tank control versus initial). However, the stomach microbiome of tank controls differed from the initial stomach microbiome only at Jehu Pond, not at Hamblin Pond (Table 3). The intestinal microbiome displayed the opposite trend; tank controls differed from initial intestinal microbiomes at Hamblin Pond but not at Jehu Pond (Table 3). Despite differences, the taxonomic composition of the tank control stomach and intestine microbial communities resembled that of the initial, field-collected microbiomes at the phylum and order levels (Fig. 3 and 4). Mature microbiomes from tank control and field-collected individuals also occupy overlapping regions in the NMDS plot (Fig. 2), indicating similarities in community structure. Despite the finding that tank residence altered mature stomach and intestine microbiomes, tank control microbiomes were significantly different from regenerating microbiomes on each day of regeneration (Table 4); thus, changes in the regenerating microbiomes were not simply the result of a tank effect.
TABLE 4.
Pairwise comparisons of beta diversity among regenerating stomach and intestinal microbial communities over timea
| Multivariate pairwise test comparison | df | PERMANOVA |
|
|---|---|---|---|
| t | P | ||
| Stomach | |||
| Initial (day 0) vs day 17 | 40 | 2.42 | 0.001* |
| Initial (day 0) vs day 20 | 45 | 3.59 | 0.001* |
| Day 17 vs day 20 | 15 | 1.49 | 0.018* |
| Tank control vs initial (day 0) | 45 | 1.85 | 0.023* |
| Tank control vs day 17 | 15 | 3.06 | 0.001* |
| Tank control vs day 20 | 20 | 4.39 | 0.001* |
| Intestine | |||
| Initial (day 0) vs day 13 | 43 | 2.50 | 0.001* |
| Initial (day 0) vs day 17 | 39 | 1.87 | 0.004* |
| Initial (day 0) vs day 20 | 43 | 2.78 | 0.001* |
| Day 13 vs day 17 | 14 | 1.14 | 0.15 |
| Day 13 vs day 20 | 18 | 1.85 | 0.002* |
| Day 17 vs day 20 | 14 | 1.07 | 0.215 |
| Tank control vs initial (day 0) | 44 | 1.55 | 0.035* |
| Tank control vs day 13 | 19 | 2.30 | 0.001* |
| Tank control vs day 17 | 15 | 1.69 | 0.005* |
| Tank control vs day 20 | 19 | 2.78 | 0.001* |
Denominator degrees of freedom are listed for all pairwise comparisons, and significant P values (<0.05) are indicated with asterisks. The overall PERMDISP tests for stomach and intestine were not significant; thus, pairwise tests for unequal dispersion of variability among groups were not conducted. t, t statistic of the pairwise PERMANOVA test.
Patterns in microbial colonization of the regenerating stomach and intestine.
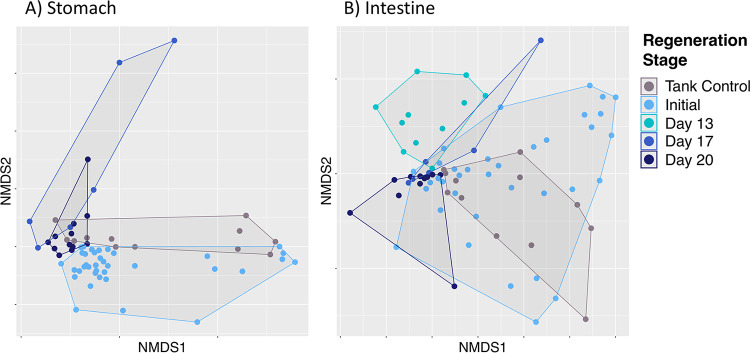
Regenerating stomach and intestine microbial communities were significantly different from those of the mature stomach and intestine at each stage of regeneration (Table 4; Fig. 5). Regenerating S. briareus individuals garnered a unique microbial composition compared to those with a fully developed digestive tract (initial condition) (Table 4), including individuals with a fully developed digestive tract placed in the same environment as the regenerating animals (tank controls) (Table 4), suggesting that the regeneration process fosters a novel microbial community. Further, the regenerating stomach and gut microbiomes were significantly different from those of both sediment and seawater sources (Table 1), suggesting host selection during tissue regeneration. While the mature stomach and intestine microbiomes of S. briareus individuals differed significantly from one another, tissue regeneration resulted in a convergence of microbial communities from the regenerating stomach and intestine tissues (Table 1).
FIG 5.
Nonmetric multidimensional scaling (NMDS) plot of microbial communities from stomach (A) and intestine (B) tissues over regeneration time.
The regenerating stomach microbiome of S. briareus did not differ by site (two-way PERMANOVA, df = 1, total df = 66, pseudo-F = 2.20, P = 0.073), while regeneration stage significantly influenced stomach microbial community composition (df = 3, total df = 66, pseudo-F = 8.33, P = 0.001) (Fig. 5A). The regenerating intestinal microbiome displayed the same trend: site did not affect microbial community composition (two-way PERMANOVA, df = 1, total df = 75, pseudo-F = 1.18, P = 0.284), but intestine communities differed significantly over regeneration time (df = 4, total df = 75, pseudo-F = 4.61, P = 0.001) (Fig. 5B). Further, the differences in stomach and intestine microbial communities over regeneration time were not due to unequal dispersion of variance (permutation tests for homogeneity of multivariate dispersions [PERMDISP], P = 0.064 for stomach, P = 0.145 for intestine).
Pairwise tests revealed that the stomach microbiome had a significantly different microbial community composition on each day that it was sampled during regeneration (initial versus day 17 versus day 20) (Table 4; Fig. 5A). Intestinal microbial communities on days 13, 17, and 20 were significantly different from those of the initial condition (Fig. 5B); however, intestinal microbial community composition did not differ significantly between days 13 and 17 and between days 17 and 20 (Table 4). In contrast, tank sediment and seawater microbial communities did not change over time (Fig. S1) (PERMANOVA, df = 2, total df = 8; pseudo-F = 1.25, P = 0.017 for sediment; pseudo-F = 6.35, P = 0.004 for seawater; but P > 0.10 for all pairwise comparisons between days), demonstrating that the regenerating sea cucumber gut displayed temporal changes despite static environmental source microbial pools.
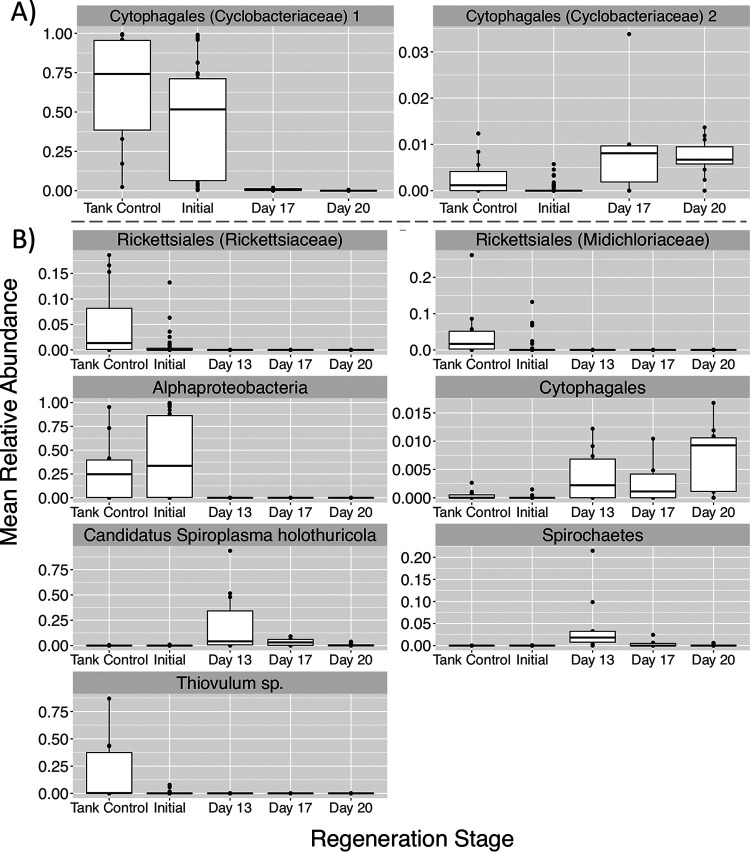
Differentially abundant ASVs during stomach and intestine regeneration.
Analysis of composition of microbiomes (ANCOM) tests identified differentially abundant ASVs over time in regenerating stomach and intestine microbial communities. Interestingly, Cytophagales was the dominant bacterial order in initial and tank control stomach microbial communities but was present only at very low abundances (<3%) in regenerating stomachs (Fig. 4B). In the stomach microbiome, two sequence variants from the family Cyclobacteriaceae (order Cytophagales) were differentially abundant over time (Fig. 6A). One Cyclobacteriaceae ASV was extremely abundant (>50% of the community) in tank control and initial condition stomachs but was nearly absent in regenerating stomachs (Fig. 6A), despite its presence in the tank sediment. The other Cyclobacteriaceae ASV, although far less abundant, increased over time in regenerating stomach microbial communities (Fig. 6A).
FIG 6.
Mean relative abundances of ASVs from stomach (n = 2 ASVs) (A) and intestine (n = 7 ASVs) (B) microbial communities that displayed significantly different abundances over regeneration time with ANCOM tests. The lowest taxonomic classification for each ASV is indicated above the plot.
In the intestine of regenerating S. briareus individuals, 7 ASVs displayed significantly different abundances over time. Notably, two ASVs from the order Rickettsiales (families Rickettsiaceae and Midichloriaceae) displayed high abundances in mature intestinal microbial communities but were nearly absent (<1% abundant) from regenerating intestinal communities (Fig. 6B). Likewise, one unknown Alphaproteobacteria ASV was very abundant in initial and tank control intestines (>25% of the community) but was absent from regenerating S. briareus intestines (Fig. 6B). In contrast, one ASV from the order Cytophagales was nearly absent in initial intestine microbial communities but increased in abundance during regeneration (Fig. 6B). Two ASVs, “Candidatus Spiroplasma holothuricola” (phylum Tenericutes) and a member of the phylum Spirochaetes, displayed high abundances on day 13 of intestine regeneration. Finally, one ASV in the genus Thiovulum was significantly more abundant in the intestines of tank control S. briareus individuals (Fig. 6B).
Balls of sediment were observed within the coelomic cavity of some regenerating S. briareus individuals on days 17 and 20 (Fig. S2). While the ASV classified as “Candidatus Spiroplasma holothuricola” was not detected at all in the tank sediment and was detected only at extremely low abundances (<0.00002%) in the sediment collected at the ponds, it was the predominant ASV detected in the samples of sediment found within the coelomic cavity of S. briareus individuals (Fig. S2). Other taxa found within the sediment inside S. briareus included Sulfurovum sp., Actinomarina sp., and a cyanobacterium in the family Phormidesmiaceae.
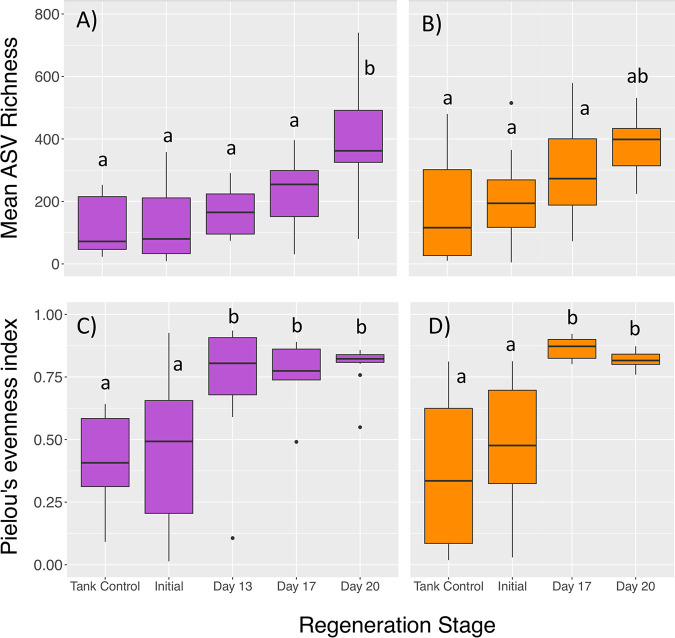
Regenerating stomach and intestine microbiomes display increased diversity and evenness.
During gut regeneration, microbial alpha diversity increased over time in both stomach and intestine tissues relative to the mature microbiomes (Fig. 7). Mean ASV richness was significantly affected by both sample type (stomach versus intestine) and regeneration day (two-way ANOVA, error df = 137; df = 1, F = 6.48, P = 0.012 for type; df = 4, F = 17.4, P < 0.001 for day). On average, stomach tissues from S. briareus individuals hosted a greater number of ASVs per sample (232 ± 19) than intestine tissues (179 ± 18). In the regenerating stomach and intestine, ASV richness increased over time relative to the initial condition, reaching a peak on day 20 (Fig. 7A and B). Microbial community evenness also increased during regeneration. Pielou’s evenness index did not differ in stomach versus intestine samples but was significantly affected by regeneration day (two-way ANOVA, error df = 137; df = 1, F = 0.21, P = 0.65 for type; df = 4, F = 18.9, P < 0.001 for day). Pielou’s evenness index was significantly higher on days 13, 17, and 20 than it was in initial condition and tank control microbial communities (Fig. 7C and D).
FIG 7.
Alpha diversity indices, including mean ASV richness for intestine (purple) (A) and stomach (orange) (B) microbial communities and mean Pielou’s evenness for intestine (purple) (C) and stomach (orange) (D) microbial communities over regeneration time. Different letters indicate significant differences in diversity indices with regeneration time (ANOVA pairwise comparisons, P < 0.05).
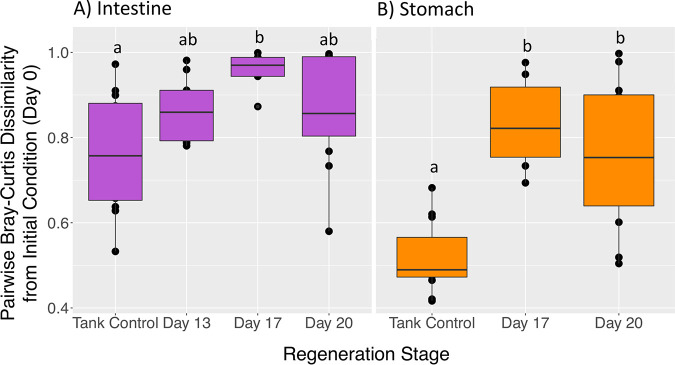
Similarity to the mature microbiome increased over time during gut regeneration.
Comparing the mean pairwise dissimilarity between S. briareus individuals from each day of regeneration to their initial conditions demonstrated that the regenerating intestinal microbiome became more similar to the initial condition over time. Tank control microbiomes were more similar to the initial condition than to regenerating microbiomes (Fig. 8). The intestinal microbiome reached maximum dissimilarity from the initial condition on day 17 and became more similar to the initial condition by day 20 (Fig. 8A). However, the regenerating stomach microbiome remained dissimilar from the initial condition (Fig. 8B).
FIG 8.
Mean pairwise Bray-Curtis dissimilarities between each day of regeneration and the initial condition (day 0) for intestine (purple) (A) and stomach (orange) (B) microbial communities. A lower dissimilarity index indicates greater similarity to the initial condition. Different letters indicate significant differences in dissimilarity indices with regeneration time (ANOVA pairwise comparisons, P < 0.05).
DISCUSSION
Complete intestinal regeneration and colonization by common gut symbionts.
The sea cucumber S. briareus followed a similar time course of regeneration in this study to that reported by Kille in 1935 (21), who found that ingested stomach and intestine contents were seen as early as 16 days after evisceration and “the first specimen was seen feeding by means of its small, unpigmented, regenerated tentacles 20 days after autotomy” (21). In this study, S. briareus regenerated intestine tissues by day 13 and stomach tissues by day 17, and all individuals began feeding on tank sediment by day 20. The regeneration process in A. japonicus is also rapid, with intestine tissues generated in as little as 10 days (29). With such rapid regeneration capabilities, sea cucumbers provide ample opportunity to study microbial colonization of stomach or intestine tissues during regeneration.
In this and other studies, the gut microbiome of sea cucumbers is comprised primarily of Alphaproteobacteria, Gammaproteobacteria, and Deltaproteobacteria, as well as the phylum Bacteroidetes (23, 25, 26, 28, 29, 34). Here, the stomach microbiome of S. briareus was comprised of nearly 50% Bacteroidetes, while the intestinal microbiome consisted of 50% Proteobacteria. While this study only compared the stomach and small intestine microbiota, different parts of the intestinal tract likely harbor distinct microbiota. The small intestine of the sea cucumber Holothuria glaberrima was also dominated by Proteobacteria and Bacteroidetes, while the large intestine was dominated by Firmicutes (24). In this study, the stomach microbiome was dominated by Bacteroidetes from the family Cyclobacteriaceae (order Cytophagales). These microbes exploit diverse ecological niches and are not known to be gut symbionts, but they may contribute to carbohydrate metabolism (35). The intestinal microbiome was dominated by a single Alphaproteobacteria ASV of unknown classification, with only 87% sequence similarity to an uncultured bacterium isolated from intertidal rocks (36), indicating that the diversity of invertebrate gut symbionts is far from being well characterized. In addition to Bacteroidetes and Alphaproteobacteria, members of the phylum Epsilonbacteraeota, formerly the class Epsilonproteobacteria (37), comprised 11% to 12% of the gut microbiome in S. briareus and were also detected in the intestinal microbiome of A. japonicus (30). The Epsilonbacteraeota associated with S. briareus belonged primarily to the order Campylobacterales, which associate with the healthy human gut (38) and dominate the gut microbiome of the sea urchin Lytechinus variegatus (39, 40).
Gut symbionts with potential energetic and immune system benefits were detected in the regenerating microbiome of S. briareus. In the gut microbiome of A. japonicus, functional genes responsible for carbohydrate metabolism, cell proliferation, and the immune system were upregulated during regeneration, suggesting selection of a beneficial microbiome by the host sea cucumber (23, 29). In this study, the most prevalent Campylobacterales ASV associated with S. briareus was a Sulfurovum sp., a strictly chemoautotrophic microbe that fixes carbon through sulfur oxidation (41, 42). Sulfurovum has been isolated from deep-sea hydrothermal vents (41, 43, 44), where it may provide supplemental nutrition to invertebrate hosts, including deep-sea crabs (45), hydrothermal vent shrimp (46), and the enigmatic gutless tube worm Riftia pachyptila (44). It is possible that chemoautotrophy may serve as a source of energy to S. briareus during the energetically expensive process of visceral regeneration, but further research is necessary to determine whether Sulfurovum provides the sea cucumber host with supplemental nutrition. Further, “Candidatus Spiroplasma holothuricola” was significantly enriched on day 13 of intestinal regeneration in S. briareus. The genome of “Ca. Spiroplasma holothuricola,” isolated from the hindgut of a deep-sea sea cucumber, contained CRISPR and restriction-methylation systems for destroying virus particles and had a streamlined genome, suggesting that it may be endosymbiotic to host cells (34). While it is possible that “Ca. Spiroplasma holothuricola” confers a protective immune system within the regenerating intestine, further research is required to determine the functional interactions between this microbe and its sea cucumber host.
The sea cucumber gut microbiome may also contain deleterious members. Rickettsiales are known as intracellular endosymbionts that parasitize a diversity of invertebrates, most notably arthropods; however, they have also been detected in the coelomic fluid of sea cucumbers (47). Alphaproteobacteria from the order Rickettsiales were abundant in the intestinal microbiomes of certain S. briareus individuals, and they were notably absent from the mature stomach and the regenerating stomach and intestine microbiomes. Given the high abundance of Rickettsiales in the intestinal microbiome of certain individuals, and absence in other individuals, it is possible that Rickettsiales are opportunistic parasites in the sea cucumber gut.
Deterministic host filtering of microbial taxa in the sea cucumber intestinal microbiome.
Sea cucumber stomach and intestine microbiomes were distinct from environmental sources, including sediment, seawater, and algal microbial communities. Selective consumption of some microbial taxa, and retention of others as symbionts, occurs in diverse marine invertebrates and likely in sea cucumbers as well (25). Stomach and intestine microbial communities of S. briareus were more similar to those of sediment than to seawater or algal and seagrass microbial communities, reflecting their role as a deposit feeder. Surprisingly, initial stomach and intestine microbial communities of S. briareus did not differ by collection site, even though Jehu and Hamblin Ponds contained distinct sediment microbial communities. S. briareus selected the same consortium of stomach and intestine microbes, despite distinct sediment microbiomes, providing evidence for host filtering of microbial symbionts. Similarly, one experiment that reciprocally transplanted the gut microbiomes of zebrafish and mice demonstrated that microbial community composition is shaped by host selection, regardless of the input community (9).
In addition to the deterministic process of selective host filtering from a diverse pool of environmental microbes (48), the evisceration experiment revealed evidence for deterministic assembly during the process of visceral regeneration. In contrast to stochastic colonization of host-associated microbial communities, which leads to high variability in community structure and taxonomic composition among replicate samples (49), regenerating sea cucumber stomach and intestine microbiomes displayed strikingly similar relative abundances of many bacterial orders across replicate samples on each day of regeneration (Fig. 4). While tank residence did alter the microbiomes of tank control individuals in this experiment relative to those of field-collected individuals, regenerating stomach and intestine microbiomes were always distinct from those of tank controls, indicating that changes in the regenerating microbiomes were not simply the result of tank effects. In future experiments, allowing sea cucumbers to regenerate gut microbiomes in their natural environment, or in a variety of distinct but replicated tank sediment environments, would provide additional evidence that the conversion of developing microbiomes is the result of deterministic processes. The microbiome of the sea cucumber Holothuria glaberrima also displayed significant differences between field-collected and aquarium-acclimated individuals, which were posited as differences due to food availability or composition (24). In this study, the sediment and seawater microbiomes of the tank environment differed significantly from those of field-collected sediment and seawater, which likely contributed to the differences in the microbiomes of actively feeding tank controls. Yet, the strikingly diverse and reproducible community composition among replicate regenerating sea cucumbers indicates that it is likely that the gut microbiome of S. briareus is shaped at least in part by deterministic factors. Further, the intestinal microbiome became more similar to the initial condition by day 20, portending a possible return to the initial consortium of microbial taxa associated with mature intestine tissues. During A. japonicus intestine regeneration, microbial community composition also became more similar to the mature microbiome after 21 days (29). In the human gut, microbial community assembly over the first 1 to 2 years is also nonrandom (10, 50), with a successional sequence of microbes displaying enrichment in different functional traits, such as oxygen tolerance, motility, and sporulation over time (8). It is possible that a combination of host-microbe and microbe-microbe interactions shapes the developing gut microbiome over time, especially as increasing microbial diversity may promote syntrophy or metabolic cross-feeding, which could generate new opportunities for microbial coexistence (51).
Enhanced microbial diversity and evenness of taxa in regenerating stomach and intestine.
Regenerating stomach and intestine microbial communities were distinct from mature microbial communities, with significantly different community structure, higher alpha diversity, and taxonomic divergence at the order level. Remarkably, only 3 to 5 ASVs made up 50% of all sequence reads in the mature stomach and intestine communities, while the combined abundances of the top 54 and 67 ASVs made up 50% of the regenerating stomach and intestine microbiomes, respectively, demonstrating the magnitude of this increased diversity and evenness in the regenerating gut microbiome. Similarly, the diversity of the intestinal microbiome in A. japonicus was elevated in regenerating intestines compared to the initial condition (23, 29). Regenerating stomach and intestine microbial communities in S. briareus individuals were distinct on each day, with the exception of intestinal microbial communities between days 13 and 17 and days 17 and 20. In contrast, tank seawater and sediment microbial communities did not display temporal differences. The intestinal microbiome of A. japonicus also displayed significant temporal differences over 21 days of regeneration (29). In S. briareus gut microbiomes, the number of ASVs increased gradually throughout regeneration time, while the evenness of taxa was much higher in all regenerating stomach and intestine microbial communities than in mature microbiomes. In a report by Wang et al. (23), alpha diversity also increased over time, reaching a maximum between days 15 and 25, and then declined. It is likely that given more time, the diversity of the stomach and intestine microbiomes would have decreased, as dominant, late-successional intestine symbionts become more numerous. Interestingly, one Bacteroidetes ASV from the order Cytophagales dominated initial stomach microbial communities but was nearly absent from regenerating stomachs, despite its presence in the tank sediment. Likewise, one unknown Alphaproteobacteria ASV was very abundant in initial intestines but absent from regenerating S. briareus intestines, indicating that regenerating microbiomes were not immediately colonized by the dominant symbionts of mature communities. This study likely captured the initial stages of intestinal microbiome assembly, consisting of a diverse community with high evenness among taxonomic groups, yet it was not carried out long enough for stabilization and proliferation of the dominant taxa that comprise the mature intestinal microbiome.
While the mature stomach and intestinal microbial communities of S. briareus displayed distinct community structure and composition, the regeneration processes resulted in a convergence of community structure and composition in regenerating stomach and intestinal microbiomes. Despite this convergence, regenerating gut microbial communities still displayed high diversity and evenness of taxa. In the human intestinal microbiome, alpha diversity is lowest at birth and increases as a function of time into adulthood (50, 52), opposite to the trend observed here. Sediment tends to have the highest microbial diversity of all free-living communities (53); thus, it is possible that the benthic lifestyle of S. briareus at the sediment-seawater interface permits colonization of the regenerating stomach and intestine by an exceptionally diverse microbial consortium, which is winnowed down over time until the mature stomach and intestinal microbial compositions are reached. Moreover, it is possible that during early regeneration and tissue colonization, the stomach and intestinal microbial communities resemble one another because the host environment has not yet differentiated enough to select for stomach- and intestine-specific microbiomes. Most microbiome studies have focused on the intestinal tract, ignoring the potential downstream influence of the stomach microbiome, and thus further research on the selection processes unique to each host environment is warranted.
Conclusion.
After induced evisceration, the sea cucumber S. briareus regenerated a functional stomach and intestine within 20 days. Mature stomach and intestine microbial communities were significantly different from regenerating microbiomes, which displayed a taxonomic and structural convergence yet differed significantly over regeneration time. The regenerating microbiome was colonized by a significantly more diverse microbial community with high evenness of taxa. Moreover, stomach and intestine microbiomes displayed highly repeatable community structure among replicates over time, suggesting deterministic host filtering of microbial symbionts from the regional species pool. Abundant and significantly enriched ASVs in the regenerating intestine may confer energetic and immune advantages to the sea cucumber host, and thus further investigation of the adaptive significance of these key microbes, including “Ca. Spiroplasma holothuricola” and Sulfurovum sp., is warranted. Rapidly regenerating holothurians are a promising and tractable model system to study microbial community assembly in the gut, with the capacity to advance our understanding of the fundamental processes shaping the gut microbiome.
MATERIALS AND METHODS
Experimental design.
Sea cucumbers (Sclerodactyla briareus) were collected on 1 September 2017 from two different sites adjacent to Waquoit Bay, a large estuary in Cape Cod, MA, USA. The two sites were Hamblin Pond (41.575°N, 70.505°W) and Jehu Pond (41.568°N, 70.498°W), which are connected across a distance of approximately 4,000 m by estuarine waterways. At Hamblin Pond, S. briareus individuals were entangled in abundant red algae (Gracilaria sp.), while at Jehu Pond, S. briareus individuals were collected from within a seagrass bed (Zostera marina). To characterize algal and seagrass microbial communities, three Gracilaria thallus fragments were collected from Hamblin Pond and three Zostera marina blades were collected at Jehu Pond. At each site, sediment samples (n = 4) were collected in 1.5-ml tubes, and 0.48 liters of seawater (n = 3) was filtered onto 0.7-μm filters (Whatman GF/F). Environmental samples were transported to the lab in a cooler with ice packs and frozen at –80°C until DNA extraction.
Sea cucumbers were transported live from the field to flowthrough seawater aquaria at the Marine Biological Laboratory in Woods Hole, MA. Sea cucumbers were acclimated to lab conditions for 2 days in two different aquaria containing sediment from the site where they were collected. The water temperature did not differ appreciably between the tank seawater and the collection location; the mean water temperature for incoming tank seawater, with data sourced from the Woods Hole, MA, NOAA buoy station (station BZBM3, 41°31’25″ N 70°40’16″ W) was 20.11°C (±0.40), while the mean water temperature nearby the collection sites in Waquoit Bay, sampled in September 2017, was 21.37°C (±0.76). After acclimation, S. briareus individuals were removed one at a time from the aquaria (Hamblin Pond, n = 20; Jehu Pond, n = 14) (see Fig. S3 in the supplemental material) and evisceration was induced by injecting approximately 2 ml 0.5 M KCl into the coelomic cavity through the oral (anterior) opening (33). After expulsion of the viscera, the stomach was aseptically dissected by cutting below the ring canal/pharynx and above the top of the intestine (Fig. 1A). The stomach was thicker walled and rounder than the intestinal tract below. For intestine samples, the thickest portion of the medial small intestine was removed; this region was usually yellow to orange in color, located approximately halfway down the gut (Fig. 1A). Although sampling multiple sections of the intestine would have been interesting, only one intestinal section was sampled to achieve high biological replication for the regeneration experiment. Five individuals from Jehu Pond did not eviscerate; they were dissected to examine initial stomach and intestine microbial communities. Twelve randomly chosen S. briareus individuals served as tank controls (Hamblin Pond, n = 7; Jehu Pond, n = 5). Tank control individuals entered the experimental tank without evisceration, continued feeding throughout the study, and were eviscerated on day 18 of the experiment using KCl, as described above, to acquire stomach and intestine microbial communities. Here, both initial condition (day 0) and tank control samples will together be referred to as “mature” stomach and intestine microbial communities to distinguish them from the regenerating tissues.
After sampling of the initial stomach and intestine microbial communities, eviscerated and tank control S. briareus individuals were assigned numbers using a random number generator, and each individual was placed into a numbered compartment within a flowthrough seawater table (Fig. S3). Sediment and pieces of Gracilaria sp., both sourced from Hamblin Pond, were placed into each compartment for food and habitat. This common garden experimental design was used so that sea cucumbers from both populations regenerated digestive organs in the presence of a common source pool of potential colonizing microbes. For stomach and intestine sampling on days 13, 17, and 20, individuals from each site were selected using a random number generator. Because newly formed guts could not be eviscerated, regenerated stomach and intestine tissues were sampled by aseptically dissecting S. briareus individuals along the body length (Fig. 1B). S. briareus individuals were anesthetized with magnesium chloride for at least 20 min prior to dissection. The first attempted sampling at 7 days post-evisceration yielded no regenerated tissues; samples were successfully collected on days 13, 17, and 20 postevisceration (Fig. S3). On day 13, only intestine tissues were visible (n = 13), but on days 17 (n = 6) and 20 (n = 13), both regenerated stomach and intestine tissues were visible and sampled. On days 13 and 20, S. briareus individuals sourced from both sites were sampled, but on day 17, only individuals from Hamblin Pond were sampled due to the greater number of eviscerated individuals from Hamblin Pond in the experiment (Fig. S3). Tank seawater (n = 3) and sediment (n = 3) samples were collected on each sampling day. On days 17 and 20, a small ball of sediment was observed within the coelomic cavity of several S. briareus individuals (Fig. S2). Out of curiosity, this sediment was sampled from two individuals on day 17 and two on day 20. All samples were transferred into sterile tubes and frozen at –80°C until DNA extraction. The final 16S amplicon data set contained the following: 76 intestine samples, 67 stomach samples, 17 sediment samples, 15 seawater samples, three Gracilaria sp. samples, three Zostera marina samples, and four samples of sediment found inside regenerating cucumbers.
Molecular methods.
For stomach samples that were visibly full of ingested contents, ingested sediment was removed prior to DNA extraction by aseptically separating the stomach tissue from the contents. Intestine samples, however, were left with ingested contents intact due to the fragile nature of the intestine, especially from early regeneration stages. To avoid contamination, sample acquisition was performed using standard aseptic technique, including flame sterilization and new scalpel blades between samples. DNA was extracted from S. briareus stomach and intestine tissues, sediment samples, seawater filters, Gracilaria sp. thallus fragments, and Zostera marina blades by using the MagAttract PowerSoil DNA kit (Qiagen) and the KingFisher flex system (Thermo Scientific) for automated DNA extraction. Amplification of the V4 region of the 16S rRNA gene was performed at Argonne National Laboratory using the Earth Microbiome Project universal primers 515f-806r (54), with the modified forward primer (55) to reduce bias against Crenarchaeota, Thaumarchaeota, and the SAR11 clade (56). DNA amplicons were sequenced on an Illumina MiSeq paired-end run (250 bp) at Argonne National Laboratory in accordance with the procedures described by Caporaso et al. (54).
16S rRNA sequence data analyses.
Amplicon sequences were processed with QIIME2 (57), where sequence reads were demultiplexed, paired-end reads were merged, and amplicon sequence variants (ASVs) were generated using the divisive amplicon denoising algorithm (DADA2) (58). Quality control within the DADA2 algorithm included chimera detection and removal, sequence error elimination, singleton exclusion, and sequence trimming (13 to 250 bp). Sequences were classified with the Silva 132 database and trimmed to the V4 region, and chloroplast and mitochondrial reads were subsequently removed. One highly abundant but unclassified ASV yielded BLAST results with 97.97% sequence similarity and 100% query coverage to “Candidatus Spiroplasma holothuricola” (34), so taxonomy was manually entered for that ASV. After examination of the alpha rarefaction curves (Fig. S4), samples were rarified to 6,800 sequences per sample, which removed only 4 samples from the data set.
Alpha diversity indices (ASV richness and Pielou’s evenness) were calculated in QIIME2 with q2-diversity, and two-way nonparametric Kruskal-Wallis analyses of variance (ANOVA) were used to statistically compare the diversity indices with the factors sample type (stomach versus intestine) and day of regeneration. Analysis of composition of microbiomes (ANCOM) tests (59) were implemented in QIIME2 to search for differentially abundant microbial taxa across regeneration time in the stomach and intestine. ASVs with significantly different abundances over time, indicated by ANCOM, were selected from the data set and graphed individually. Sequence data were imported into R (version 3.4.4) for subsequent analysis and visualization. Taxonomic barplots and nonmetric multidimensional scaling (NMDS) ordinations, based on Bray-Curtis dissimilarity matrices, were created with the R packages phyloseq (60) (version 1.22.3) and ggplot2 (61) (version 2.2.1).
Statistical differences in microbial community structure (beta diversity) were tested using permutational multivariate analysis of variance (PERMANOVA) based on Bray-Curtis dissimilarity matrices using the program PRIMER (version 6.1.11). One-way PERMANOVA tests compared (i) all samples with the fixed factor of sample type (stomach versus intestine versus environmental samples) and (ii) mature stomach and intestine microbial communities with the fixed factor of site (Jehu Pond versus Hamblin Pond versus tank controls). Two-way PERMANOVAs were used to compare regenerating stomach and intestine microbial communities over regeneration time (initial, day 13, day 17, and day 20), with site of origin and day as fixed factors. To verify that significant PERMANOVA outcomes were not a result of unequal dispersion of variability among groups, permutation tests for homogeneity of multivariate dispersions (PERMDISP) were also conducted in PRIMER (version 6.1.11) using the same factors. Finally, the difference in beta diversity between regenerating stomach and intestine microbial communities and the initial conditions was assessed by comparing mean pairwise Bray-Curtis distances between individuals from each group (days 13, 17, 20, and tank control) to the initial condition (day 0). This allowed a visual comparison of the magnitude of change in microbial communities relative to the initial condition throughout regeneration time.
Data availability.
Raw 16S rRNA sequences were deposited as FASTQ files to the Sequence Read Archive of the National Center for Biotechnology Information under accession no. SRP255421.
Supplementary Material
ACKNOWLEDGMENTS
This project would not have been possible without funding from the American Museum of Natural History’s Lerner-Gray Grants for Marine Research.
I am extremely grateful to the Marine Biological Lab and to Dave Remsen for providing tank space and research facilities, to Bill Grossman for providing field collection assistance, and to Jack Gilbert and Neil Gottel for providing molecular lab space and support. Both Mike LaBarbera and Catherine Pfister provided helpful advice on experimental design as well as enthusiastic support for this project. Thanks to Katherine Ziska for field and lab research assistance and Stephanie Greenwald for performing 16S sequencing. I received helpful feedback on the manuscript from John Guittar, Heather Skeen, Mike LaBarbera, Tim Wootton, Joy Bergelson, Cathy Pfister, and the rest of the UChicago Microbial Ecology and Microbiome journal club.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Spor A, Koren O, Ley R. 2011. Unravelling the effects of the environment and host genotype on the gut microbiome. Nat Rev Microbiol 9:279–290. doi: 10.1038/nrmicro2540. [DOI] [PubMed] [Google Scholar]
- 2.Clemente JC, Ursell LK, Parfrey LW, Knight R. 2012. The impact of the gut microbiota on human health: an integrative view. Cell 148:1258–1270. doi: 10.1016/j.cell.2012.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Collins SM, Surette M, Bercik P. 2012. The interplay between the intestinal microbiota and the brain. Nat Rev Microbiol 10:735–742. doi: 10.1038/nrmicro2876. [DOI] [PubMed] [Google Scholar]
- 4.Foster KR, Schluter J, Coyte KZ, Rakoff-Nahoum S. 2017. The evolution of the host microbiome as an ecosystem on a leash. Nature 548:43–51. doi: 10.1038/nature23292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schluter J, Foster KR. 2012. The evolution of mutualism in gut microbiota via host epithelial selection. PLoS Biol 10:e1001424. doi: 10.1371/journal.pbio.1001424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rothschild D, Weissbrod O, Barkan E, Kurilshikov A, Korem T, Zeevi D, Costea PI, Godneva A, Kalka IN, Bar N, Shilo S, Lador D, Vila AV, Zmora N, Pevsner-Fischer M, Israeli D, Kosower N, Malka G, Wolf BC, Avnit-Sagi T, Lotan-Pompan M, Weinberger A, Halpern Z, Carmi S, Fu J, Wijmenga C, Zhernakova A, Elinav E, Segal E. 2018. Environment dominates over host genetics in shaping human gut microbiota. Nature 555:210–215. doi: 10.1038/nature25973. [DOI] [PubMed] [Google Scholar]
- 7.Venturelli OS, Carr AV, Fisher G, Hsu RH, Lau R, Bowen BP, Hromada S, Northen T, Arkin AP. 2018. Deciphering microbial interactions in synthetic human gut microbiome communities. Mol Syst Biol 14:e8157. doi: 10.15252/msb.20178157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guittar J, Shade A, Litchman E. 2019. Trait-based community assembly and succession of the infant gut microbiome. Nat Commun 10:512. doi: 10.1038/s41467-019-08377-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rawls JF, Mahowald MA, Ley RE, Gordon JI. 2006. Reciprocal gut microbiota transplants from zebrafish and mice to germ-free recipients reveal host habitat selection. Cell 127:423–433. doi: 10.1016/j.cell.2006.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koenig JE, Spor A, Scalfone N, Fricker AD, Stombaugh J, Knight R, Angenent LT, Ley RE. 2011. Succession of microbial consortia in the developing infant gut microbiome. Proc Natl Acad Sci U S A 108:4578–4585. doi: 10.1073/pnas.1000081107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ho TTB, Groer MW, Kane B, Yee AL, Torres BA, Gilbert JA, Maheshwari A. 2018. Dichotomous development of the gut microbiome in preterm infants. Microbiome 6:157. doi: 10.1186/s40168-018-0547-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stewart CJ, Ajami NJ, O'Brien JL, Hutchinson DS, Smith DP, Wong MC, Ross MC, Lloyd RE, Doddapaneni H, Metcalf GA, Muzny D, Gibbs RA, Vatanen T, Huttenhower C, Xavier RJ, Rewers M, Hagopian W, Toppari J, Ziegler A-G, She J-X, Akolkar B, Lernmark A, Hyoty H, Vehik K, Krischer JP, Petrosino JF. 2018. Temporal development of the gut microbiome in early childhood from the TEDDY study. Nature 562:583–588. doi: 10.1038/s41586-018-0617-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bourlat SJ, Juliusdottir T, Lowe CJ, Freeman R, Aronowicz J, Kirschner M, Lander ES, Thorndyke M, Nakano H, Kohn AB, Heyland A, Moroz LL, Copley RR, Telford MJ. 2006. Deuterostome phylogeny reveals monophyletic chordates and the new phylum Xenoturbellida. Nature 444:85–88. doi: 10.1038/nature05241. [DOI] [PubMed] [Google Scholar]
- 14.Petersen JM, Osvatic J. 2018. Microbiomes in natura: importance of invertebrates in understanding the natural variety of animal-microbe interactions. mSystems 3:e00179-17. doi: 10.1128/mSystems.00179-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.García-Arrarás JE, Greenberg MJ. 2001. Visceral regeneration in holothurians. Microsc Res Tech 55:438–451. doi: 10.1002/jemt.1189. [DOI] [PubMed] [Google Scholar]
- 16.Byrne M. 2001. The morphology of autotomy structures in the sea cucumber Eupentacta quinquesemita before and during evisceration. J Exp Biol 204:849–863. [DOI] [PubMed] [Google Scholar]
- 17.Byrne M. 1985. Evisceration behaviour and the seasonal incidence of evisceration in the holothurian Eupentacta quinquesemita (Selenka). Ophelia 24:75–90. doi: 10.1080/00785236.1985.10426621. [DOI] [Google Scholar]
- 18.Shinn GL. 1985. Reproduction of Anoplodium hymanae, a turbellarian flatworm (Neorhabdocoela, Umagillidae) inhabiting the coelom of sea cucumbers: production of egg capsules, and escape of infective stages without evisceration of the host. Biol Bull 169:182–198. doi: 10.2307/1541397. [DOI] [Google Scholar]
- 19.Amon RM, Herndl GJ. 1991. Deposit feeding and sediment. I. Interrelationship between Holothuria fubulosa (Holothurioida, Echinodermata) and the sediment microbial community. Marine Ecol 12:163–174. doi: 10.1111/j.1439-0485.1991.tb00250.x. [DOI] [Google Scholar]
- 20.MacTavish T, Stenton-Dozey J, Vopel K, Savage C. 2012. Deposit-feeding sea cucumbers enhance mineralization and nutrient cycling in organically-enriched coastal sediments. PLoS One 7:e50031. doi: 10.1371/journal.pone.0050031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kille FR. 1935. Regeneration in Thyone briareus Lesueur following induced autotomy. Biol Bull 69:82–108. doi: 10.2307/1537360. [DOI] [Google Scholar]
- 22.Bogatyrenko EA, Buzoleva LS. 2016. Characterization of the gut bacterial community of the Japanese sea cucumber Apostichopus japonicus. Microbiology 85:116–123. doi: 10.1134/S0026261716010033. [DOI] [PubMed] [Google Scholar]
- 23.Wang L, Zhao X, Xu H, Bao X, Liu X, Chang Y, Ding J. 2018. Characterization of the bacterial community in different parts of the gut of sea cucumber (Apostichopus Japonicus) and its variation during gut regeneration. Aquac Res 49:1987–1996. doi: 10.1111/are.13654. [DOI] [Google Scholar]
- 24.Pagán-Jiménez M, Ruiz-Calderón JF, Dominguez-Bello MG, García-Arrarás JE. 2019. Characterization of the intestinal microbiota of the sea cucumber Holothuria glaberrima. PLoS One 14:e0208011. doi: 10.1371/journal.pone.0208011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gao F, Li F, Tan J, Yan J, Sun H. 2014. Bacterial community composition in the gut content and ambient sediment of sea cucumber Apostichopus japonicus revealed by 16S rRNA gene pyrosequencing. PLoS One 9:e100092. doi: 10.1371/journal.pone.0100092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang G, Xu Z, Tian X, Dong S, Peng M. 2015. Intestinal microbiota and immune related genes in sea cucumber (Apostichopus japonicus) response to dietary β-glucan supplementation. Biochem Biophys Res Commun 458:98–103. doi: 10.1016/j.bbrc.2015.01.074. [DOI] [PubMed] [Google Scholar]
- 27.Sha Y, Liu M, Wang B, Jiang K, Sun G, Wang L. 2016. Gut bacterial diversity of farmed sea cucumbers Apostichopus japonicus with different growth rates. Microbiology 85:109–115. doi: 10.1134/S0026261716010112. [DOI] [Google Scholar]
- 28.Yamazaki Y, Meirelles PM, Mino S, Suda W, Oshima K, Hattori M, Thompson FL, Sakai Y, Sawabe T, Sawabe T. 2016. Individual Apostichopus japonicus fecal microbiome reveals a link with polyhydroxybutyrate producers in host growth gaps. Sci Rep 6:21631. doi: 10.1038/srep21631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang H, Wang Q, Liu S, Huo D, Zhao J, Zhang L, Zhao Y, Sun L, Yang H. 2019. Genomic and metagenomic insights into the microbial community in the regenerating intestine of the sea cucumber Apostichopus japonicus. Front Microbiol 10:1165. doi: 10.3389/fmicb.2019.01165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang G, Tian X, Dong S. 2019. Bacillus cereus and rhubarb regulate the intestinal microbiota of sea cucumber (Apostichopus japonicus Selenka): species-species interaction, network, and stability. Aquaculture 512:734284. doi: 10.1016/j.aquaculture.2019.734284. [DOI] [Google Scholar]
- 31.Brown WI, Shick JM. 1979. Bimodal gas exchange and the regulation of oxygen uptake in holothurians. Biol Bull 156:272–288. doi: 10.2307/1540917. [DOI] [Google Scholar]
- 32.Pearse AS. 1908. Observations on the behavior of the holothurian, Thyone briareus (Leseur). Biol Bull 15:259–288. doi: 10.2307/1535923. [DOI] [Google Scholar]
- 33.Smith GN Jr, Greenberg MJ. 1973. Chemical control of the evisceration process in Thyone briareus. Biol Bull 144:421–436. doi: 10.2307/1540018. [DOI] [Google Scholar]
- 34.He L-S, Zhang P-W, Huang J-M, Zhu F-C, Danchin A, Wang Y. 2017. The enigmatic genome of an obligate ancient Spiroplasma symbiont in a hadal holothurian. Appl Environ Microbiol 84:e01965-17. doi: 10.1128/AEM.01965-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pinnaka AK, Tanuku N. 2014. The family Cyclobacteriaceae, p 551–575. In Rosenberg E, DeLong EF, Lory S, Stackebrandt E, Thompson F (ed), The Prokaryotes: other major lineages of Bacteria and the Archaea. Springer, Berlin, Germany. [Google Scholar]
- 36.Couradeau E, Roush D, Guida BS, Garcia-Pichel F. 2017. Diversity and mineral substrate preference in endolithic microbial communities from marine intertidal outcrops (Isla de Mona, Puerto Rico. ). Biogeosciences 14:311–324. doi: 10.5194/bg-14-311-2017. [DOI] [Google Scholar]
- 37.Waite DW, Vanwonterghem I, Rinke C, Parks DH, Zhang Y, Takai K, Sievert SM, Simon J, Campbell BJ, Hanson TE, Woyke T, Klotz MG, Hugenholtz P. 2017. Comparative genomic analysis of the class Epsilonproteobacteria and proposed reclassification to Epsilonbacteraeota (phyl. nov.). Front Microbiol 8:682. doi: 10.3389/fmicb.2017.00682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rinninella E, Raoul P, Cintoni M, Franceschi F, Miggiano G, Gasbarrini A, Mele M. 2019. What is the healthy gut microbiota composition? A changing ecosystem across age, environment, diet, and diseases. Microorganisms 7:14. doi: 10.3390/microorganisms7010014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hakim JA, Koo H, Dennis LN, Kumar R, Ptacek T, Morrow CD, Lefkowitz EJ, Powell ML, Bej AK, Watts SA. 2015. An abundance of Epsilonproteobacteria revealed in the gut microbiome of the laboratory cultured sea urchin, Lytechinus variegatus. Front Microbiol 6:1047. doi: 10.3389/fmicb.2015.01047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hakim JA, Koo H, Kumar R, Lefkowitz EJ, Morrow CD, Powell ML, Watts SA, Bej AK. 2016. The gut microbiome of the sea urchin, Lytechinus variegatus, from its natural habitat demonstrates selective attributes of microbial taxa and predictive metabolic profiles. FEMS Microbiol Ecol 92:fiw146. doi: 10.1093/femsec/fiw146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Inagaki F, Takai K, Nealson KH, Horikoshi K. 2004. Sulfurovum lithotrophicum gen. nov., sp. nov., a novel sulfur-oxidizing chemolithoautotroph within the ϵ-Proteobacteria isolated from Okinawa Trough hydrothermal sediments. Int J Syst Evol Microbiol 54:1477–1482. doi: 10.1099/ijs.0.03042-0. [DOI] [PubMed] [Google Scholar]
- 42.Yamamoto M, Nakagawa S, Shimamura S, Takai K, Horikoshi K. 2010. Molecular characterization of inorganic sulfur-compound metabolism in the deep-sea epsilonproteobacterium Sulfurovum sp. NBC37-1. Environ Microbiol 12:1144–1153. doi: 10.1111/j.1462-2920.2010.02155.x. [DOI] [PubMed] [Google Scholar]
- 43.Mino S, Kudo H, Arai T, Sawabe T, Takai K, Nakagawa S. 2014. Sulfurovum aggregans sp. nov., a hydrogen-oxidizing, thiosulfate-reducing chemolithoautotroph within the Epsilonproteobacteria isolated from a deep-sea hydrothermal vent chimney, and an emended description of the genus Sulfurovum. Int J Syst Evol Microbiol 64:3195–3201. doi: 10.1099/ijs.0.065094-0. [DOI] [PubMed] [Google Scholar]
- 44.Giovannelli D, Chung M, Staley J, Starovoytov V, Le Bris N, Vetriani C. 2016. Sulfurovum riftiae sp. nov., a mesophilic, thiosulfate-oxidizing, nitrate-reducing chemolithoautotrophic epsilonproteobacterium isolated from the tube of the deep-sea hydrothermal vent polychaete Riftia pachyptila. Int J Syst Evol Microbiol 66:2697–2701. doi: 10.1099/ijsem.0.001106. [DOI] [PubMed] [Google Scholar]
- 45.Niemann H, Linke P, Knittel K, MacPherson E, Boetius A, Brückmann W, Larvik G, Wallmann K, Schacht U, Omoregie E, Hilton D, Brown K, Rehder G. 2013. Methane-carbon flow into the benthic food web at cold seeps—a case study from the Costa Rica subduction zone. PLoS One 8:e74894. doi: 10.1371/journal.pone.0074894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tokuda G, Yamada A, Nakano K, Arita NO, Yamasaki H. 2008. Colonization of Sulfurovum sp. on the gill surfaces of Alvinocaris longirostris, a deep-sea hydrothermal vent shrimp. Marine Ecol 29:106–114. doi: 10.1111/j.1439-0485.2007.00211.x. [DOI] [Google Scholar]
- 47.Enomoto M, Nakagawa S, Sawabe T. 2012. Microbial communities associated with holothurians: presence of unique bacteria in the coelomic fluid. Microbes Environ 27:300–305. doi: 10.1264/jsme2.me12020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhou J, Ning D. 2017. Stochastic community assembly: does it matter in microbial ecology? Microbiol Mol Biol Rev 81:e00002-17. doi: 10.1128/MMBR.00002-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Burke C, Steinberg P, Rusch D, Kjelleberg S, Thomas T. 2011. Bacterial community assembly based on functional genes rather than species. Proc Natl Acad Sci U S A 108:14288–14293. doi: 10.1073/pnas.1101591108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bäckhed F, Roswall J, Peng Y, Feng Q, Jia H, Kovatcheva-Datchary P, Li Y, Xia Y, Xie H, Zhong H, Khan MT, Zhang J, Li J, Xiao L, Al-Aama J, Zhang D, Lee YS, Kotowska D, Colding C, Tremaroli V, Yin Y, Bergman S, Xu X, Madsen L, Kristiansen K, Dahlgren J, Wang J, Jun W. 2015. Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe 17:690–703. doi: 10.1016/j.chom.2015.04.004. [DOI] [PubMed] [Google Scholar]
- 51.Goldford JE, Lu N, Bajić D, Estrela S, Tikhonov M, Sanchez-Gorostiaga A, Segrè D, Mehta P, Sanchez A. 2018. Emergent simplicity in microbial community assembly. Science 361:469–474. doi: 10.1126/science.aat1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Voreades N, Kozil A, Weir TL. 2014. Diet and the development of the human intestinal microbiome. Front Microbiol 5:494. doi: 10.3389/fmicb.2014.00494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Thompson LR, Earth Microbiome Project Consortium, Sanders JG, McDonald D, Amir A, Ladau J, Locey KJ, Prill RJ, Tripathi A, Gibbons SM, Ackermann G, Navas-Molina JA, Janssen S, Kopylova E, Vázquez-Baeza Y, González A, Morton JT, Mirarab S, Zech Xu Z, Jiang L, Haroon MF, Kanbar J, Zhu Q, Jin Song S, Kosciolek T, Bokulich NA, Lefler J, Brislawn CJ, Humphrey G, Owens SM, Hampton-Marcell J, Berg-Lyons D, McKenzie V, Fierer N, Fuhrman JA, Clauset A, Stevens RL, Shade A, Pollard KS, Goodwin KD, Jansson JK, Gilbert JA, Knight R, Rivera JLA, Al-Moosawi L, Alverdy J, Amato KR, Andras J, Angenent LT, Antonopoulos DA, Apprill A, Armitage D, Ballantine K, Bárta J, Baum JK, Berry A, Bhatnagar A, Bhatnagar M, Biddle JF, Bittner L, Boldgiv B, Bottos E, Boyer DM, Braun J, Brazelton W, Brearley FQ, Campbell AH, Caporaso JG, Cardona C, Carroll J, Cary SC, Casper BB, Charles TC, Chu H, Claar DC, Clark RG, Clayton JB, Clemente JC, Cochran A, Coleman ML, Collins G, Colwell RR, Contreras M, Crary BB, Creer S, Cristol DA, Crump BC, Cui D, Daly SE, Davalos L, Dawson RD, Defazio J, Delsuc F, Dionisi HM. 2017. A communal catalogue reveals Earth’s multiscale microbial diversity. Nature 551:457–463. doi: 10.1038/nature24621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, Fierer N, Knight R. 2011. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci U S A 108:4516–4522. doi: 10.1073/pnas.1000080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Parada AE, Needham DM, Fuhrman JA. 2016. Every base matters: assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ Microbiol 18:1403–1414. doi: 10.1111/1462-2920.13023. [DOI] [PubMed] [Google Scholar]
- 56.Walters W, Hyde ER, Berg-Lyons D, Ackermann G, Humphrey G, Parada A, Gilbert JA, Jansson JK, Caporaso JG, Fuhrman JA, Apprill A, Knight R. 2016. Improved bacterial 16S rRNA gene (V4 and V4-5) and fungal internal transcribed spacer marker gene primers for microbial community surveys. mSystems 1:e00009-15. doi: 10.1128/mSystems.00009-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, Al-Ghalith GA, Alexander H, Alm EJ, Arumugam M, Asnicar F, Bai Y, Bisanz JE, Bittinger K, Brejnrod A, Brislawn CJ, Brown CT, Callahan BJ, Caraballo-Rodríguez AM, Chase J, Cope EK, Da Silva R, Diener C, Dorrestein PC, Douglas GM, Durall DM, Duvallet C, Edwardson CF, Ernst M, Estaki M, Fouquier J, Gauglitz JM, Gibbons SM, Gibson DL, Gonzalez A, Gorlick K, Guo J, Hillmann B, Holmes S, Holste H, Huttenhower C, Huttley GA, Janssen S, Jarmusch AK, Jiang L, Kaehler BD, Kang KB, Keefe CR, Keim P, Kelley ST, Knights D, Koester I, Kosciolek T, Kreps J, Langille MGI, Lee J, Ley R, Liu Y-X, Loftfield E, Lozupone C, Maher M, Marotz C, Martin BD, McDonald D, McIver LJ, Melnik AV, Metcalf JL, Morgan SC, Morton JT, Naimey AT, Navas-Molina JA, Nothias LF, Orchanian SB, Pearson T, Peoples SL, Petras D, Preuss ML, Pruesse E, Rasmussen LB, Rivers A, Robeson MS, Rosenthal P, Segata N, Shaffer M, Shiffer A, Sinha R, Song SJ, Spear JR, Swafford AD, Thompson LR, Torres PJ, Trinh P, Tripathi A, Turnbaugh PJ, Ul-Hasan S, van der Hooft JJJ, Vargas F, Vázquez-Baeza Y, Vogtmann E, von Hippel M, Walters W, Wan Y, Wang M, Warren J, Weber KC, Williamson CHD, Willis AD, Xu ZZ, Zaneveld JR, Zhang Y, Zhu Q, Knight R, Caporaso JG. 2019. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol 37:852–857. doi: 10.1038/s41587-019-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP. 2016. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods 13:581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mandal S, Van Treuren W, White RA, Eggesbø M, Knight R, Peddada SD. 2015. Analysis of composition of microbiomes: a novel method for studying microbial composition. Microb Ecol Health Dis 26:27663. doi: 10.3402/mehd.v26.27663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.McMurdie PJ, Holmes S. 2013. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8:e61217. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wickham H. 2016. ggplot2: elegant graphics for data analysis. Springer-Verlag, New York, NY. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw 16S rRNA sequences were deposited as FASTQ files to the Sequence Read Archive of the National Center for Biotechnology Information under accession no. SRP255421.