Whole-cell biosensors based on reporter genes can be designed for heavy metal detection but often require the optimization of their sensitivity and specific adaptations for practical use in the field. Magnetotactic bacteria as cellular hosts for biosensors are interesting models, as their intrinsic magnetism permits them to be easily concentrated and entrapped to increase the arsenic-response signal. This paves the way for the development of sensitive and immobilized whole-cell biosensors tailored for use in the field.
KEYWORDS: arsenic, freeze-drying, magnetotactic bacteria, whole-cell biosensor
ABSTRACT
According to the World Health Organization, arsenic is the water contaminant that affects the largest number of people worldwide. To limit its impact on the population, inexpensive, quick, and easy-to-use systems of detection are required. One promising solution could be the use of whole-cell biosensors, which have been extensively studied and could meet all these criteria even though they often lack sensitivity. Here, we investigated the benefit of using magnetotactic bacteria as cellular chassis to design and build sensitive magnetic bacterial biosensors. Promoters potentially inducible by arsenic were first identified in silico within the genomes of two magnetotactic bacteria strains, Magnetospirillum magneticum AMB-1 and Magnetospirillum gryphiswaldense MSR-1. The ArsR-dependent regulation was confirmed by reverse transcription-PCR experiments. Biosensors built by transcriptional fusion between the arsenic-inducible promoters and the bacterial luciferase luxCDABE operon gave an element-specific response in 30 min with an arsenite detection limit of 0.5 μM. After magnetic concentration, we improved the sensitivity of the biosensor by a factor of 50 to reach 10 nM, more than 1 order of magnitude below the recommended guidelines for arsenic in drinking water (0.13 μM). Finally, we demonstrated the successful preservation of the magnetic bacterium biosensors by freeze-drying.
IMPORTANCE Whole-cell biosensors based on reporter genes can be designed for heavy metal detection but often require the optimization of their sensitivity and specific adaptations for practical use in the field. Magnetotactic bacteria as cellular hosts for biosensors are interesting models, as their intrinsic magnetism permits them to be easily concentrated and entrapped to increase the arsenic-response signal. This paves the way for the development of sensitive and immobilized whole-cell biosensors tailored for use in the field.
INTRODUCTION
Numerous environmental pollutants, including endocrine disruptors (1) and plastics (2) but also heavy metals (3), pose a significant human health risk worldwide (4). Among them is arsenic, one of the most important chemicals to monitor in drinking water for health and safety issues according to the World Health Organization (WHO). Despite its high toxicity and occurrence in many parts of the world, arsenic concentrations in risky areas are often not monitored because of the high cost and lack of portable analytical techniques (5).
The use of microbial whole-cell biosensors could be a complementary, easy-to-use, and inexpensive approach for the specific and semiquantitative detection of arsenic. Many bacterial whole-cell biosensors described in the literature are derived from natural resistance mechanisms, containing a genetic regulation system. This consists of the coupling between a transcriptional regulator and a promoter rerouted for biosensor technology to control the expression of a reporter gene. The four following induced bacterial metabolic pathways have been described in the presence of arsenic (6): the arsRDABC operon encoding a reductase and an efflux pump responsible for the export of inorganic arsenite out of the cell, methylation systems with an arsenite S-adenosylmethionine methyltransferase (ArsM) that can methylate arsenite, and two respiratory pathways based on either the oxidation of arsenite [As(III)] to arsenate [As(V)] (aio/arx genes) or the reduction of As(V) to As(III) (arr genes). The regulation of the ars operon is the most described (6). Its expression is genetically controlled by the ArsR regulator protein; in the absence of arsenic, this repressor binds DNA as a dimer and prevents transcription, while in the presence of arsenic, the affinity of the ArsR-metalloid complex for the genomic operator decreases, allowing transcription and therefore signal translocation.
Numerous arsenic biosensors based on the ArsR regulatory system were described using reporter genes encoding either light emission protein (e.g., luciferase) or fluorescent protein (e.g., green fluorescent protein) (7). Most of them are hosted in Escherichia coli cells, allowing arsenic detection in less than 1 h (8), with high specificity and a detection limit as low as 13 nM (0.1 μg/liter) (9). Much progress has been made to improve the performance of biosensors compared to their conventional chemical counterparts, but their commercialization and field use have not yet fully matured. Various semiautonomous in-line water analyzers using luminescent whole-cell biosensors have been described (10, 11), but cell preservation, immobilization, and concentration with highly sensitive measurement maintained in the field remain a major challenge.
In this study, we investigated the capacity of magnetotactic bacteria (MTB) to be tailored as luminescent biosensors. MTB comprise a group of taxonomically, physiologically, and morphologically diverse prokaryotes with the unique ability to synthesize ferromagnetic nanoparticles that allow them to orient and move along the lines of the Earth’s magnetic field (12–14). This magnetotactic property is due to the biomineralization of iron-rich magnetic nanocrystals embedded in lipidic vesicles forming an organelle called the magnetosome (13). The alignment of 15 to 20 magnetosomes in the cytoplasm acts like a compass needle to orient bacteria in geomagnetic fields, simplifying their search for preferred microaerobic environments (15). Many biotechnological applications exploiting this unique magnetic property are being developed in both the medical and environmental fields (16). Here, we extend the scope of applications to environmental monitoring and report that Magnetospirillum magneticum AMB-1 (17) and Magnetospirillum gryphiswaldense MSR-1 (18) can be genetically modified to allow arsenic-dependent expression of a transcriptional lux-based reporter gene. This study demonstrates the strong potential of MTB for the development of sensitive and magnetically guided whole-cell biosensors.
RESULTS AND DISCUSSION
Resistance of magnetotactic strains to arsenic.
To determine the optimal microorganism to host luciferase reporters, we focused on two magnetotactic model strains, Magnetospirillum magneticum AMB-1 and Magnetospirillum gryphiswaldense MSR-1. Both environmental strains are cultivable under laboratory conditions, are genetically modifiable (19, 20), and have been shown to be able to grow and absorb metal ions such as cadmium (21), cobalt, manganese, and copper (22).
To facilitate simultaneous screening of many conditions, we adapted and developed the cultivation of MTB in microplates. This allowed us to grow AMB-1 and MSR-1 strains in the presence of different concentrations of arsenite and follow cell growth (optical density at 600 nm [OD600]) for 20 h. Under these conditions, the resulting half maximal inhibitory concentrations (IC50) for arsenite were determined to be 259 ± 25 μM and 406 ± 23 μM for AMB-1 and MSR-1, respectively. In comparison, the same experiment performed on cultures of an E. coli strain leads to an IC50 of 11.2 mM. These values are much higher than the WHO recommended guideline for drinking water (0.13 μM) or the French legislation for industrial effluent (0.33 μM) and are thus compatible with the use of both magnetic strains as cellular chassis for arsenic biosensors.
In some areas, arsenic concentrations can reach levels so high that they could be lethal for the biosensors. Under such conditions, their use would give a false-negative result. As described for whole-cell E. coli biosensors (8), a control experiment is required using magnetotactic strains hosting transcriptional reporter fusions that are constitutively expressed (pBBr_Lux; Table 1 and 2). In the absence of a luminescence signal for these control strains, the sample would simply be diluted to reach nonlethal conditions for the bacteria.
TABLE 1.
List of plasmids used in this study
| Plasmid | Promoter | Reporter gene | Descriptiona | Reference or source |
|---|---|---|---|---|
| pBBR1MCS-2 | T3 | None | Empty plasmid, Kr | 56 |
| pBBr_Venus | T3 | mVenus | Constitutive expression of mVenus fluorescence, Kr | 57 |
| pBBr_Lux | T3 | luxCDABE | Constitutive expression of luminescence with the luxCDABE operon, Kr | This study |
| pArsEcoli_Venus | pArs + arsR | mVenus | Expression of an mVenus gene optimized for MTB driven by an arsenic-inducible promoter from E. coli, in Kr | 8 |
| pArsRAMB1_Venus | pArs + arsR1 | mVenus | Expression of an mVenus gene optimized for MTB driven by an arsenic-inducible promoter from AMB-1, Kr | This study |
| pArsRAMB1_LuxCDABE | pArs + arsR1 | luxCDABE | Expression of the luxCDABE operon driven by an arsenic-inducible promoter from AMB-1, Kr | This study |
| pArsMMSR1_Venus | pArs + arsR’ | mVenus | Expression of an mVenus gene optimized for MTB driven by an arsenic-inducible promoter from MSR-1, Kr | This study |
| pArsMMSR1_LuxCDABE | pArs + arsR′ | luxCDABE | Expression of the luxCDABE operon driven by an arsenic-inducible promoter from MSR-1, Kr | This study |
Kr, kanamycin resistance.
TABLE 2.
List of bacterial strains used in this study
| Strain | Plasmid | Description | Reference or source |
|---|---|---|---|
| AMB-1 | None | Wild type | 17 |
| MSR-1 | None | Wild type | 18 |
| AMB_pBBr | pBBR1-MCS2 | AMB-1 negative-control strain | |
| MSR_pBBr | pBBR1-MCS2 | MSR-1 negative-control strain | |
| AMB_Venus | pBBr_Venus | AMB-1 positive-control strain | This study |
| MSR_Venus | pBBr_Venus | MSR-1 positive-control strain | This study |
| AMB_Lux | pBBr_LuxCDABE | AMB-1 positive-control strain | This study |
| MSR_Lux | pBBr_LuxCDABE | MSR-1 positive-control strain | This study |
| AMB_pAEcoliV | pArsEcoli_Venus | AMB-1 fluorescent arsenic biosensor | This study |
| AMB_pAA1V | pArsR1AMB1_Venus | AMB-1 fluorescent arsenic biosensor | This study |
| MSR_pAA1V | pArsR1AMB1_Venus | MSR-1 fluorescent arsenic biosensor | This study |
| AMB_Pred | pArsR1AMB1_LuxCDABE | AMB-1 luminescent arsenic biosensor | This study |
| MSR_Pred | pArsR1AMB1_LuxCDABE | MSR-1 luminescent arsenic biosensor | This study |
| AMB_pAM′V | pArsM′MSR1_Venus | AMB-1 fluorescent arsenic biosensor | This study |
| MSR_pAM′V | pArsM′MSR1_Venus | MSR-1 fluorescent arsenic biosensor | This study |
| AMB_Pmet | pArsM′MSR1_LuxCDABE | AMB-1 luminescent arsenic biosensor | This study |
| MSR_Pmet | pArsM′MSR1_LuxCDABE | MSR-1 luminescent arsenic biosensor | This study |
| E_pAEcoliV | pArsEcoli_Venus | E. coli fluorescent arsenic biosensor | This study |
| E. coli WM3064 | None | Helper strain for conjugation of MTB | 40 |
As arsenic is not the only toxic compound in areas polluted by anthropogenic activities, we also estimated the resistance of both strains to a wide range of potential toxic metal(loid) ions [Cd(II), Co(II), Cu(II), Hg(II), Mo(VI), Ni(II), and Sb(III)] (see Table S2 in the supplemental material). These results correspond to the first systematic quantitative measurement of MTB sensitivity to a broad range of metal(loid) ions. A comparison with E. coli is difficult because different methods and strains give different values in the literature (23–25), but our results clearly indicate that both MTB strains are somewhat resistant to a variety of metal(loid)s. However, since AMB-1 appears to be more sensitive than MSR-1, the latter is likely a better candidate for use in highly polluted environments.
Search for arsenic resistance operons in AMB-1 and MSR-1.
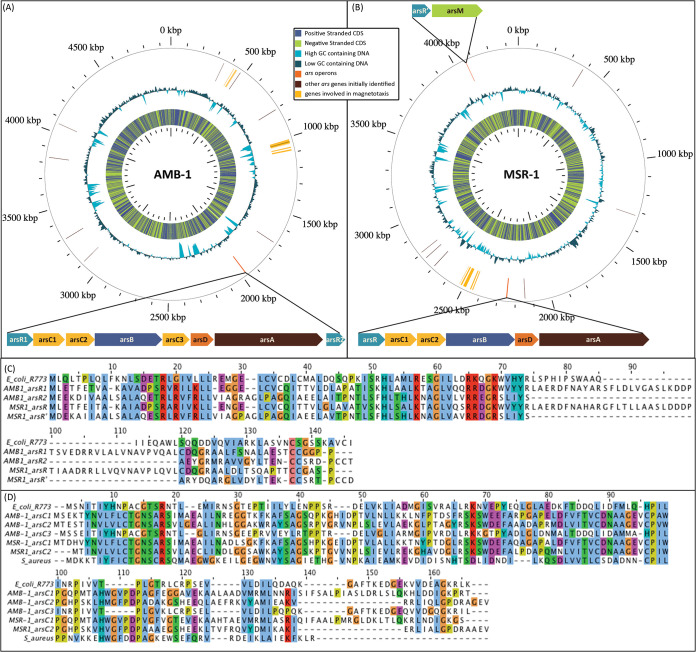
Heterologous expression of transcriptional reporter fusions in AMB-1 and MSR-1 was first investigated using the arsenic-inducible promoter Pars from E. coli, which has been fully characterized (8, 26). No arsenic-dependent signal was obtained from the resulting biosensors hosted in both magnetic strains (results obtained with AMB-1 are presented in Fig. S1), likely because of the phylogenic distance between Enterobacteria and magnetotactic Proteobacteria. As a consequence, an in-depth analysis of AMB-1 and MSR-1 genomes was performed, aiming at finding putative arsenic-inducible operons for the design of the magnet biosensor. We first focused on the identification of genes encoding putative ArsR regulators using a BLAST search against all the genome sequences, using the ArsR sequence from E. coli as a query. As a result, ten genes in AMB-1 and eight in MSR-1 were predicted to belong to the family of ArsR-StmB regulatory proteins, a large family of metal binding transcriptional regulators. To refine the identified targets, we analyzed whether the neighboring genes were related to known arsenic resistance operons and, finally, found three putative arsenic operons, one in the AMB-1 genome and two in the MSR-1 genome (Fig. 1).
FIG 1.
(A and B) Representation of (A) AMB-1 and (B) MSR-1 genome maps obtained with GView (46). The location of genes involved in magnetotaxis is shown in yellow (27). The locations of genes encoding proteins displaying sequence similarity with E. coli ArsR repressor are shown in brown. The locations of the putative operons involved in arsenic resistance (one in AMB-1 and two in MSR-1) are shown in orange with details on the putative arsenite-related genes and their organizations. (C) Sequence alignments obtained with Jalview (44) for the different ArsR repressors identified in the potential MTB operons and the E. coli ArsR repressor. (D) Sequence alignments obtained with Jalview for the arsenate reductase (ArsC) from MTB with ArsC from E. coli and S. aureus.
In MSR-1, one operon (nucleotide positions 1967558 to 1972950 in the genome) is likely involved in the methylation of arsenic with the ArsR′ repressor controlling the expression of arsM, a gene encoding an As(III) S-adenosylmethionine methyltransferase that could methylate arsenite up to trimethylarsenite, a volatile form of arsenic (6). Two similar operons likely involved in the export of arsenite after arsenate reduction (28, 29) were also identified in the MSR-1 (from nucleotide position 2246773 to 2251292) and AMB-1 (from nucleotide position 4062734 to 4063844) genomes. In both genomes, the operons contain, in different orders, arsR, arsD, arsA, arsB, and arsC with three copies of arsC in MSR-1 (arsC1, arsC2, and arsC3), two copies of arsC (arsC1 and arsC2) in AMB-1, and two copies of arsR (arsR1 and arsR2) in AMB-1. ArsR1 from AMB-1 and ArsR from MSR-1 share 75.4% sequence identity and are both homologous to E. coli ArsR (27.1% and 25.7% sequence identity, respectively). The ArsR2 sequence differs more significantly (19% sequence identity with E. coli ArsR) but is homologous to ArsR′ regulating the methylation operon in MSR-1 (80.8% identity), suggesting that there are two classes of repressors in magnetospirilla that differently regulate the Ars operons. arsC1 and arsC2 in AMB-1 and MSR-1 both encode an arsenate reductase close to ArsC from Staphylococcus aureus (with sequence identities between 21 and 29%), which is predicted to use thioredoxin as an electron shuttle (30), whereas ArsC3 found in the AMB-1 operon is highly similar to E. coli ArsC (sequence identity of 71.1%), which uses glutaredoxin as an electron source (58).
Transcriptional analysis of the Ars operons.
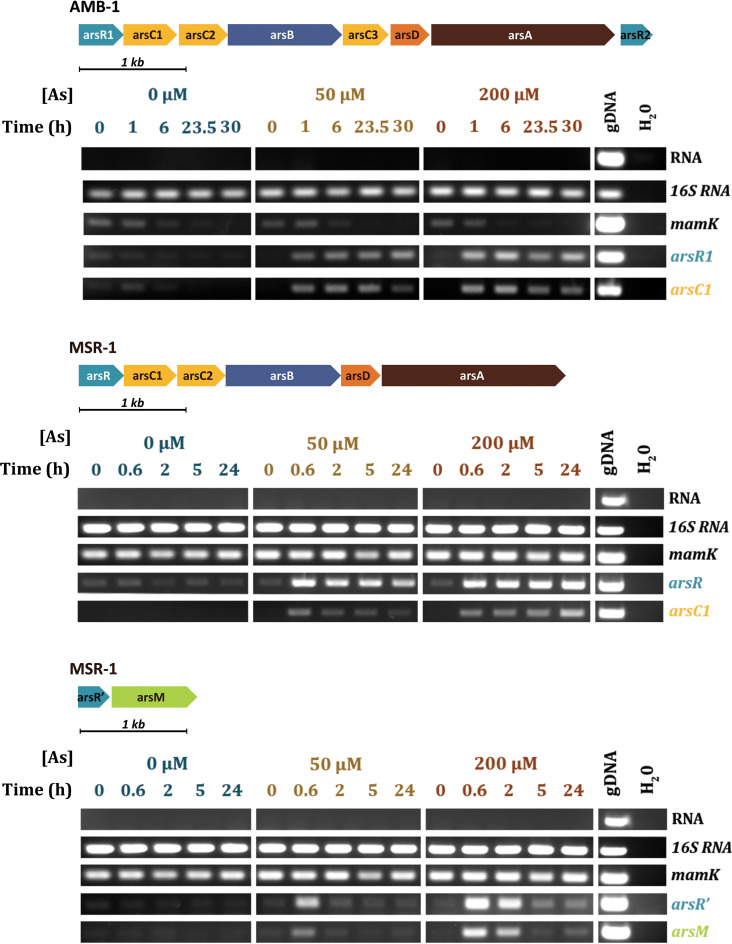
The arsenic responses of the three putative operons were investigated using transcriptional approaches. AMB-1 and MSR-1 cells were first grown in the presence of 0, 50, or 200 μM arsenite, and growth curves and magnetism of the cells were monitored for 41 to 54 h depending on the strain. No significant impact of the metalloid ion on the cell fitness and magnetic behaviors was observed between the three conditions (Fig. S2). Reverse transcription-PCR (RT-PCR) was used to measure transcripts from the three operons between 0 and 30 h after arsenite addition. arsR1 (or arsR) and arsC1 genes were followed for the reductase operon in both AMB-1 and MSR-1 and arsR′ and arsM for the methyltransferase operon of MSR-1 (Fig. 2). mamK transcripts were also used as an additional control of effective magnetosome synthesis since this gene encodes an actin-like protein required for magnetosome alignment that is not known to be related to any arsenic dependency (31). Although measurements of the coefficient of magnetism (Cmag) indicate that cell magnetism is maintained in both strains, the amount of mamK transcript is not constant in AMB-1, even in the absence of arsenite. These variations could be related to the unmeasured expression of MamK-like, a homologous actin-like protein encoded in a distinct genomic islet previously identified in the genome of AMB-1 (32). As the same phenomenon is observed under all conditions and the Cmag is constant, we conclude that arsenic has no influence over AMB-1 magnetism.
FIG 2.
RT-PCR experiments performed in the presence of various concentrations of arsenite for (top) arsR1 and arsC1 putatively involved in the reduction and export pathway of arsenic in AMB-1, (middle) arsR and arsC1 putatively involved in the reduction and export pathway of arsenic in MSR-1, and (bottom) arsR′ and arsM putatively involved in the methylation pathway in MSR-1.
For the AMB-1 reductase operon, induction of the transcription of arsR1 and arsC1 is clearly observed after 60 min of exposure to 50 or 200 μM arsenite (Fig. 2). As MSR-1 grows faster in our culture conditions, the first sampling was made after 40 min of exposure, revealing a similar result with the rapid induction of arsR and arsC1. For the two reductase operons, arsenite induction is maintained beyond 24 h with an increase in the quantity of transcripts over time.
The response of the methyltransferase operon from MSR-1 is quite different. Although the induction of arsM and arsR′ is rapidly detected 40 min following the addition of metalloid ions to the culture medium, no transcription is observed 2 h or 5 h after the addition of arsenite at 50 μM or 200 μM, respectively. Both the arsRDABC and arsR′M operons are thus transcribed very rapidly in MSR-1, but the methylation operon is repressed in the following hours, all the more rapidly, as the initial metalloid concentration is low. This behavior is likely related to an intracellular decrease in arsenic concentration and would suggest that ArsR′ has a lower affinity for arsenic than ArsR. Despite these differences, transcriptional experiments demonstrate that the promoters of the three operons identified by in silico analysis are all regulated as a function of arsenic concentration, and as a consequence, may be good candidates for biosensors with a reporter gene system.
Genetic construction of the biosensors.
We focused only on the reductase operon from AMB-1 and the methyltransferase operon from MSR-1 in the following experiments carried out to build the different biosensors. Four plasmid constructions were built by insertion of either arsR1 or arsR′ and their respective promoters followed by two different reporter genes. The gene encoding the fluorescent protein Venus was first chosen because of its small size and because it was known to be functional in MTB (33–35). However, preliminary experiments performed to measure the fluorescent signal in rich growth media, such as the one used for MSR-1, revealed a high background noise and determined that it would require additional washing to remove growth medium in order to obtain an optimal measurement. Since these additional procedures are not compatible with the design of an easy-to-use biosensor, we turned to the luxCDABE operon as a luminescent reporter (8) allowing an autonomous bioluminescence emission of the luciferase enzyme without needing to add any substrate in the medium. This autonomous luminescent reporter has never been used in MTB, and our results demonstrate that it is fully functional in magnetospirilla (see below). The corresponding plasmids harboring luxCDABE under the control of ParsR1 or ParsR′ (promoter and regulator gene) were mobilized into AMB-1 and MSR-1 by conjugation, resulting in four whole-cell biosensors named AMB-Pred (AMB-1 with promoter from reductase operon ParsR1 regulating luxCDABE), AMB-Pmet (AMB-1 with promoter from methylation operon ParsR′ regulating luxCDABE), MSR-Pred (MSR-1 with promoter from reductase operon ParsR1 regulating luxCDABE), and MSR-Pmet (MSR-1 with promoter from methylation operon ParsR′ regulating luxCDABE).
Sensitivity and specificity of the magnetic biosensors.
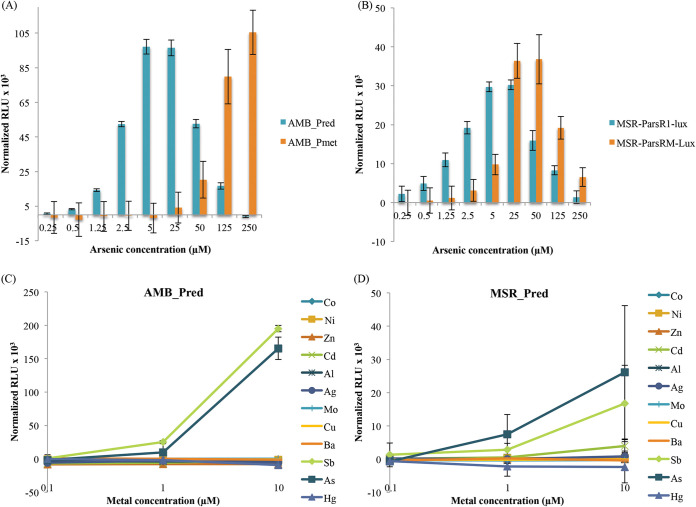
Dose responses of the biosensors were first characterized by monitoring the bioluminescence emission (normalized to the OD) over a range of arsenite concentrations in both MTB strains harboring the two different lux-based reporter plasmids (Fig. 3). The response time in both strains is short regardless of the promoter, resulting in a significant signal measurable only 30 min after addition of arsenite, which corresponds to one of the shortest detection times for an arsenic biosensor (7).
FIG 3.
(A and B) Bioluminescence signal normalized by OD and background noise (signal without arsenic) obtained after 30 min of exposure to different arsenite concentrations for the (A) AMB-1 and (B) MSR-1 biosensors. (C and D) Bioluminescence signal normalized by OD and background noise (signal without metal[loid]s) obtained after 30 min of exposure to different metal(loid)s for the (C) AMB-1 and (D) MSR-1 biosensors. The error bars are standard deviations.
The sensitivities of the four biosensors toward arsenite differed significantly (Fig. 3A and B). Indeed, for biosensors built with the methyltransferase-linked ArsR′ promoter, a significant luminescent signal is only measurable for metalloid concentrations above 5 μM in MSR-1 and 125 μM in AMB-1. This low sensitivity is in line with the low affinity of ArsR′ for arsenic as hypothesized with reverse transcription-PCR experiments. On the other hand, as little as 0.5 μM arsenite can be detected in both AMB-Pred and MSR-Pred, the two biosensors designed using the AMB-1 identified reductase operon. These results demonstrate that the promoter selected from one strain can be functional in the other strain. In addition, since luminescence emission is linear for both strains within a concentration range from 0.5 to 5 μM arsenite, they can therefore be used for semiquantitative measurements of arsenite in environmental samples, either by fast detection in the laboratory or in the field using a portable luminometer.
Metalloid specificity of the biosensors was also characterized. As arsenite [As(III)] is not the only oxidation state of arsenic in water, we also tested the sensibility of the biosensors for arsenate [As(V)]. No signal was obtained for this form, either because of a low arsenate uptake in MTB cells or because of a low affinity of the ArsR repressor for arsenate as shown, for example, in Cupriavidus metallidurans CH34 (36). For other metal(loid)s, the bioluminescence emission was followed from AMB-Pred and MSR-Pred 30 min after the addition of different ion species in a broad range of concentrations (0.1 to 10 μM range) (Fig. 3C and D). A very-low-level response or no response was measured for all the common toxic metal(loid) ions tested here except for arsenite and antimony. Such a dual response has already been well documented in the literature for the arsenite resistance system in E. coli (37). As a result, the sensitivity and specificity measured for AMB-Pred and MSR-Pred strains are very similar to the characteristics obtained for biosensors hosted in E. coli (8).
Biosensor performances after magnetic concentration.
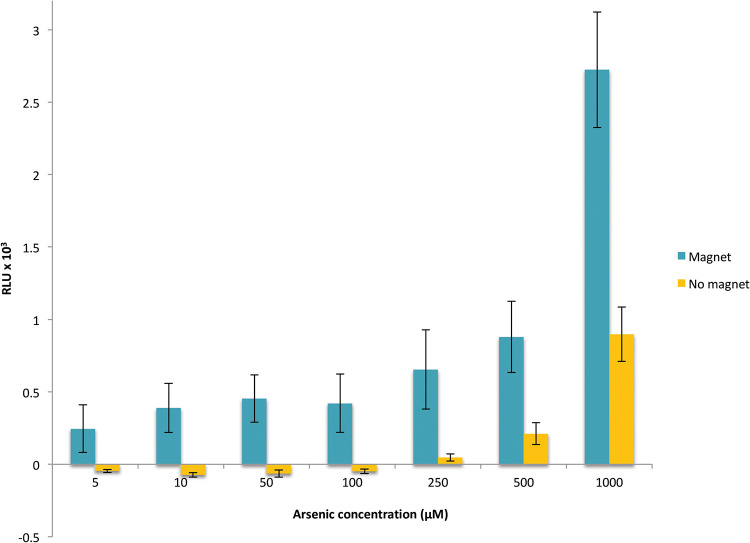
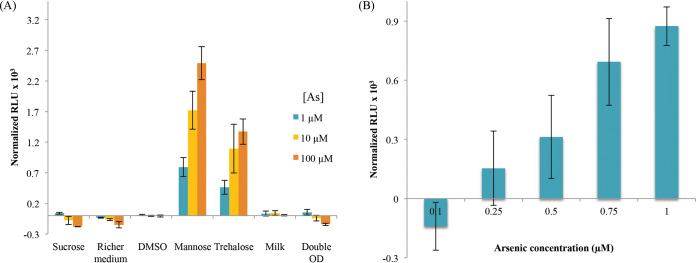
Our experiments clearly indicate that we developed functional unique magnetic, sensitive, and specific biosensors. However, their sensitivity (0.5 μM) is not low enough to detect arsenic in water intended for human consumption (0.13 μM arsenic) or even industrial effluents (0.32 μM arsenic). To improve the detection limit, we therefore exploited the magnetic character of our biosensors. The luminescence emission was monitored for MSR-Pred cells incubated for 30 min with 5 nM to 1.0 μM arsenite in the presence of a magnet. This easy-to-do magnetic concentration step increases the signal (Fig. 4), a definite advantage compared to biosensors developed in E. coli, for which a concentration step by filtration is sometimes necessary to improve the signal-to-noise ratio (10). In addition, the magnetic concentration allows a significant improvement in the sensitivity of the biosensor, allowing the detection limit to be extended from 0.5 μM to 10 nM arsenite, which is 10 times lower than the drinking water standard (Fig. 4). In addition, these promising results could be further improved by the construction of an automatic system, which would significantly reduce the experimental bias associated with manual sample handling after the magnetic concentration step.
FIG 4.
Bioluminescence signal normalized by OD and background noise (signal without arsenic) for the MSR-Pred biosensor obtained after 30 min of incubation with arsenite in the presence or absence of a magnet. The results are the averages of three independent experiments performed on each of biological triplicates. The error bars are standard errors of the mean.
Biosensor performances after freeze-drying.
In order to use whole-cell biosensors for on-site measurements, cells have to be conditioned in latent and portable form. Freeze-drying has been successfully used to preserve functional E. coli whole-cell luminescent biosensors (8) but has never been described for MTB. The challenge therefore is to find the right conditions in which most bacteria remain alive. Two concentrations of MSR-Pred cells (1011 or 2 × 1011) were mixed with different cryoprotectants as follows: sucrose, dehydrated milk, DMSO, mannose, or trehalose. Freeze-drying was performed for 24 h, and cells were revived by simply adding water to the aliquots. For conditions where mannose and trehalose were used as cryoprotectants, the biosensors were capable of giving a luminescence signal after incubation with arsenite at concentrations above 1 μM (Fig. 5A), whereas no signal was measured using other cryoprotectants. In both cases, cells remain magnetic after freeze-drying. For trehalose-treated cells, luminescence continued to increase over time, whereas with mannose, the signal was maximized after 5 h but subsequently decreased (Fig. S3). We therefore focused on trehalose and succeeded in improving the efficiency of freeze-drying by increasing the trehalose concentration from 10 g/liter to 50 g/liter and incubating the samples after revival at room temperature instead of 28°C. A weak signal at 0.5 μM arsenite is observed but with high variability that may be improved in the future using automatic procedures for the conservation experiments. At this stage, a detection limit of 0.75 μM arsenite in 1.5 h can be obtained with lyophilisates stored for 1 week at 4°C (Fig. 5B).
FIG 5.
(A) Bioluminescence signal normalized by OD and background noise (signal without arsenic) obtained after 3 h of exposure to different arsenite concentrations for the freeze-dried MSR-Pred biosensor under different cryoprotecting conditions. (B) Bioluminescence signal normalized by OD and background noise (signal without arsenic) obtained after 1.5 h of incubation with different arsenite concentrations for MSR-Pred freeze-dried with 50 g/liter trehalose and rehydrated after 1 week of storage at 4°C. The error bars are standard deviations.
Conclusion. We have demonstrated here that magnetotactic bacteria (AMB-1 and MSR-1) are robust toward metal(loid) toxicity and can be genetically modified to produce metalloid biosensors. Arsenic was chosen as the first target analyte based on the genomic prediction of ars operons, for which transcription was subsequently shown to be induced by this toxic metalloid species. Beyond the scope of this study, this concept could be extended to other inducible metal systems known in E. coli, such as CadR, MerR, or NikR. Magnetic strains hosting lux-based reporter plasmids are able to give a specific bioluminescent response with a detection limit of 0.5 μM for arsenite in 30 min for fresh cells or 90 min for freeze-dried cells. In addition, the use of a cellular magnetic chassis represents a major breakthrough in the field, allowing an increase in the sensitivity threshold by a factor of 50 using a facile magnetic concentration. This improved sensitivity is lower than standards set by the WHO for drinking water. Thereby, these magnetic luminescent biosensors are more efficient than E. coli biosensors by their increased sensitivity and their robustness regarding revitalization after freeze-drying.
These magnetic biosensors can therefore be associated with a portable luminometer to be used for in situ measurements. They can be hosted in semiautonomous online water analyzers, their magnetism making it possible to avoid the problematic concentration steps on membrane, which are typically required for these analyzers to obtain a sufficient signal from nonmagnetic whole-cell biosensors. The sensitivity of these magnetic biosensors can also be improved by genetic manipulation by using more intricate regulation pathways such as has been demonstrated in E. coli (9, 38). In addition, as shown by Roda et al. (39), adapted microfluidic systems capable of directing, guiding, or fixing the cells in specific compartments can be envisaged with our system, which paves the way toward the development of ground-breaking technologies in the field of whole-cell biosensors.
MATERIALS AND METHODS
Strains and culture conditions.
Strains and plasmids are presented in Tables 1 and 2. Magnetospirillum magneticum AMB-1 strains were cultured at 28°C in 1.5 mM MagMin medium (40) with 10 mg/liter of kanamycin when required. Magnetospirillum gryphiswaldense MSR-1 strains were cultured at 28°C in flask standard medium (41) with 50 μM iron citrate and 10 mg/liter of kanamycin when required. The media for AMB-1 and MSR-1 were equilibrated at 2% oxygen (flushed in the medium for AMB-1 and headspace for MSR-1) before inoculation. Escherichia coli DH5α was cultured in Luria broth (LB) at 37°C supplemented with 50 mg/liter of kanamycin. E. coli WM3064 was cultured in LB with 50 mg/liter of kanamycin and 0.3 mM diaminopimelic acid (DAP). The magnetism of the different strains was spectrophotometrically estimated by calculating the coefficient of magnetism (Cmag) as previously described (31, 42). The supplemental material includes further description of plasmids and bacterial strains used in this study, as well as solid compounds used to make metal(loid) ion solutions, primers used for plasmid construction and transcriptional analysis, the response of the E. coli and AMB-1 biosensors built using arsenic-inducible promoters from E. coli, AMB-1 and MSR-1 growth curves measured in the presence of various concentrations of arsenite, and kinetics of the luminescent signal of MSR-Pred after freeze-drying with mannose or trehalose.
Bacterial metal and metalloid resistance.
Bacterial growth was monitored after the addition of different metal(loid)s [As(III), Cd(II), Co(II), Cu(II), Hg(II), Mo(VI), Ni(II), Sb(III), and Zn(II)] (Table S1) in a broad range of concentrations. One hundred microliters of bacterial culture (OD600 = 0.1 for AMB-1 and OD600 = 0.5 for MSR-1) and 100 μl of growth medium supplemented with different metal(loid) concentrations were mixed in 96-well plates and incubated at 30°C. The absorbance at 600 nm was measured with a microplate reader Infinite 200 PRO (Tecan) every 30 min for 20 h. Growth rates were calculated by linear regression, and the half maximal inhibitory concentrations (IC50) were estimated with a linear regression between the two metal(loid) concentrations flanking the half maximum growth rate. The experiment was repeated in triplicate on three different cultures. The given IC50 are calculated as the mean of the three independent experiments, and the error corresponds to the standard deviation of the three values.
Genomic analysis.
The AMB-1 (GenBank accession number NC_007626) and MSR-1 (chromosome MGMSR.2 https://mage.genoscope.cns.fr/microscope/genomic/overview.php?O_id=1225) genomes were analyzed with the MaGe platform (Microbial Genome Annotation & Analysis) (43). Identification of genes putatively involved in arsenic resistance was performed either by a search for annotated genes whose name contained “ars” or by a BLAST analysis using the E. coli ArsR protein sequence (UniProt ID P15905) as a template. Genes located around the identified targets were subsequently analyzed and searched via BLAST with ArsA, ArsB, ArsC, and ArsD protein sequences from the E. coli arsenic resistance operon and ArsC from the S. aureus arsenic resistance operon. Jalview (44) was used for the final alignments of ArsR and ArsC. Sequence homology percentages were calculated using Ident and the Sim software (45). The genomes were displayed with GView (46).
Transcriptional analysis.
AMB-1 and MSR-1 cells were grown in 1-liter and 200-ml flasks, respectively. When the cultures reached the mid-logarithmic growth phase, arsenite was added at 0, 50, or 200 μM. For each concentration, aliquots containing 1010 to 1011 cells were sampled at five different times ranging from 0 to 30 h. Total RNA was isolated using the ReliaPrep RNA cell miniprep system (Promega). Residual genomic DNA was removed with DNase I (Ambion). Extracted RNA was quantified with NanoDrop. For each sample, 200 ng of RNA was reverse-transcripted using a SuperScript VILO cDNA synthesis kit, and targeted genes were amplified by PCR with GoTaq enzyme (Promega) and designed primers (Table 3). For each pair of primers, a negative control was conducted with water, and a positive control was conducted with genomic DNA.
TABLE 3.
List of primers used for amplification of cDNA targets for RT-PCR and arsenic promoters for biosensor constructions
| Primer or promoter | Sequence | Target | Bacterium |
|---|---|---|---|
| Primers | |||
| ARN16S_AMB1_F1077 | GGGCACTCTGAAGAAACTGC | 16S RNA gene | AMB-1 |
| ARN16S_AMB1_R1185 | GTCACCACCATTGTAGCACG | 16S RNA gene | AMB-1 |
| mamK_AMB1_F390 | GGCTCAGGAGGTGGTTCATA | mamK | AMB-1 |
| mamK_AMB1_R588 | GAGCCGCTCATCCACATAGT | mamK | AMB-1 |
| arsR_AMB1_F2 | TGCTTGAGACATTCGAAACCG | arsR1 | AMB-1 |
| arsR_AMB1_R112 | GCACCGTGGTGATCTGGC | arsR1 | AMB-1 |
| arsC_AMB1_F23 | CCTGTTTTTGTGCACCGGC | arsC1 | AMB-1 |
| arsC_AMB1_R154 | GCACCGTCGGGTCGATATG | arsC1 | AMB-1 |
| ARNr16S_MSR1_F1048 | GTACCGTCATCATCGTCCCC | 16S RNA gene | MSR-1 |
| ARNr16S_MSR1_R1200 | CACACTGGGACTGAGACACG | 16S RNA gene | MSR-1 |
| mamK_MSR1_F773 | TCGAGATTTTGTTGCGGTCC | mamK | MSR-1 |
| mamK_MSR1_R899 | ACGGGCGCAGTTTATCTTTC | mamK | MSR-1 |
| arsR_MSR1_2119_F272 | CGACGACGATCCCACTATCG | arsR1 | MSR-1 |
| arsR_MSR1_2119_R384 | CTGAGACGTCAAGTCCAGGG | arsR1 | MSR-1 |
| arsC_MSR1_F43 | ATTCCGCCCGTTCGATCATG | arsC1 | MSR-1 |
| arsC_MSR1_R176 | TTGGTCTTCTTCAGCAGGGC | arsC1 | MSR-1 |
| arsR_MSR1_3975_F117 | GAGGAATTGGCGGTCACTCC | arsR′ | MSR-1 |
| arsR_MSR1_3975_R236 | TGATCGTAACGGGCCGAATAG | arsR′ | MSR-1 |
| arsM_MSR1_F595 | CGACCCGTACCGAATTGTTC | arsM | MSR-1 |
| arsM_MSR1_R719 | TCGCGACTTTCTGACTTGGG | arsM | MSR-1 |
| Promoters | |||
| pArs_AMB1_F | TATGGTCTCTGAGGTGCCATGCCGACCAAGCCGG | pArs-arsR1 | AMB-1 |
| pArs_AMB1_R | CATAGGTCTCCTCATGGTGGTCCCCCACAGCAG | pArs-arsR1 | AMB-1 |
| pArsM_MSR1_F | TATGGTCTCTGAGGGCCGGTGTCGATGATGTTG | pArs-arsR’ | MSR-1 |
| pArsM_MSR1_R | CATAGGTCTCCTCATGGCATCCTCCTGTTCAGTCG | pArs-arsR’ | MSR-1 |
Plasmid constructions.
pBBr_Lux, pBBr_Venus, and pArsEcoli_Venus plasmids were constructed by restriction enzyme cloning. The luxCDABE operon was inserted using a SpeI digestion, and the gene encoding the Venus protein was inserted as an EcoRI/SacI digestion. pArsEcoli was amplified by PCR and inserted by BamHI/HindIII digestion upstream of the Venus gene. The constructions for the other biosensors were made using a type IIS enzyme, BsaI (47). The backbone pBBR1-MCS2 was modified to harbor only two BsaI recognition sequences enclosing the lacZ gene. The arsenic promoters and arsR genes were amplified from the bacterial genomes (Table 3). The resulting constructions are included in Tables 1 and 2. The plasmids were transformed in MTB by conjugation using a helper strain, E. coli WM3064 (40).
Biodetection experiments.
Bacterial cells were grown in their respective media with kanamycin until reaching the stationary phase. The bacterial cultures were dispatched in a 96-well plate. Each well was supplemented with 100 μl of culture and 100 μl of growth medium containing kanamycin and different metal(loid) concentrations [Ag(I), Al(III), As(III), Ba(II), Cd(II), Co(II), Cu(II), Hg(II), Mo(VI), Ni(II), Sb(III), or Zn(II)] (Table S1). The luminescence signal and OD600 were measured every 15 min over a 2-h period with a microplate reader Infinite 200 PRO (Tecan). The experiment was made on biological triplicates, with one replicate being an independent culture. For each replicate, the luminescence signal measured without arsenite, corresponding to the background signal associated with the culture, was subtracted from the signal obtained in the presence of arsenite. The displayed results are the means of relative luminescence units normalized by OD600 (normalized RLU) of the triplicates, and the standard deviation of the three values was calculated. The signal for one concentration is considered positive when the mean is at least twice the value of its standard deviation. Strains bearing pBBr_Lux were used as a positive control to evaluate the toxicity of the sample.
Response of the bacterial biosensors after magnetic concentration.
Cells from a culture of bacterial biosensor were equally distributed in 50-ml conical centrifuge tubes (10 ml by tube) before the addition of 0, 0.005, 0.01, 0.05, 0.1, 0.25, 0.5, or 1 μM arsenite. At the bottom of the tubes, a strong magnet (neodymium, N48) was placed to attract bacteria. After 30 min, the supernatant was removed, and the magnetic pellets were taken up in 200 μl of medium and deposited in a 96-well plate. The luminescence was read with a microplate reader Infinite 200 PRO (Tecan). The experiment was repeated three times with three independent cultures each time. For each condition (magnetically concentrated or not), the luminescence signal measured without arsenite was subtracted from the signal obtained in the presence of arsenite. The results are averaged between the three replicates of the three experiments, and the error is the standard error of the mean, , where σ is the standard deviation and n is the number of samples. A positive detection for a concentration is considered when the mean signal of the concentration is higher than twice the standard error.
Freeze-drying protocol and revival of the biosensors.
To test different cryoprotectants, a culture of the MSR-Pred strain (Tables 1 and 2), in stationary state, was concentrated to reach an OD of 20 or 40 followed by an equal dilution with medium containing the cryoprotectant at a final concentration of 12.5% (wt/vol) sucrose, 10% (vol/vol) DMSO, 10 g/liter mannose, 10 g/liter trehalose, and 10% (wt/vol) dehydrated milk. Solutions of cryoprotectant were previously sterilized using 0.2-μm-pore-size membrane filters. Cells mixed with cryoprotectant were frozen for 3 h at –80°C before freeze-drying for 24 h. The lyophilisates were revived by the addition of sterile water and incubated for 15 min and then diluted 5 times in growth medium with or without arsenite for luminescence measurements. The experiment was made on triplicates, with one replicate being a tube of lyophilized cells. For each replicate, the luminescence signal measured without arsenite, corresponding to the background signal associated with the culture, was subtracted from the signal obtained in the presence of arsenite. The displayed results are the mean of the relative luminescence units normalized by the OD600 (normalized RLU) of the triplicate, and the standard deviation of the three values was calculated. When improving freeze-drying conditions with trehalose, the same protocol was used to freeze-dry the bacteria concentrated at an OD of 20 and 5% trehalose and then revive them. The luminescence was measured on 4 lyophilized tubes in triplicate. The data were treated the same way to obtain the mean of the normalized RLU, and the standard deviation was calculated. The signal for one concentration is considered positive when the mean is at least twice the value of its standard deviation.
Data availability.
The sequence for the Magnetospirillum magneticum AMB-1 genome can be found under NCBI accession number NC_007626 (48). The Magnetospirillum gryphiswaldense MSR-1 v2 genome can be found under NCBI accession number HG794546 (49). The ArsR protein sequence was deposited under NCBI accession number P15905 (50). The nucleotide sequences of the structural genes for the arsenic pump of Escherichia coli, arsA and arsB, were deposited under NCBI accession numbers P08690 (51) and P08691 (52), respectively. The arsD gene encodes a second transacting regulatory protein of the plasmid-carried arsenical resistance operon has been deposited under NCBI accession number P46003 (54). The ArsC proteins of the arsenic resistance operon of Escherichia coli and Staphylococcus aureus plasmid pI258 have been deposited under NCBI accession numbers P08692 (53) and P0A006 (55), respectively.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by a Ph.D. grant from the CEA (Comissariat à l’Energie Atomique et aux Energies Alternative).
We thank Catherine Brutesco and Géraldine Adryanczyk for technical support, Hélène Javot, Daniel Garcia, Nicolas Ginet, Joris Tulumello, and Damien Faivre for fruitful scientific discussion, and Daniel Chevrier for proofreading the manuscript.
The manuscript was written through contributions of all authors. All authors approved the final version of the manuscript.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Vilela CLS, Bassin JP, Peixoto RS. 2018. Water contamination by endocrine disruptors: impacts, microbiological aspects and trends for environmental protection. Environ Pollut 235:546–559. doi: 10.1016/j.envpol.2017.12.098. [DOI] [PubMed] [Google Scholar]
- 2.Alimi OS, Farner Budarz J, Hernandez LM, Tufenkji N. 2018. Microplastics and nanoplastics in aquatic environments: aggregation, deposition, and enhanced contaminant transport. Environ Sci Technol 52:1704–1724. doi: 10.1021/acs.est.7b05559. [DOI] [PubMed] [Google Scholar]
- 3.Duruibe JO, Ogwuegbu MOC, Egwurugwu JN. 2007. Heavy metal pollution and human biotoxic effects. Int J Phys Sci 2:112–118. [Google Scholar]
- 4.Martins VV, Zanetti MOB, Pitondo-Silva A, Stehling EG. 2014. Aquatic environments polluted with antibiotics and heavy metals: a human health hazard. Environ Sci Pollut Res Int 21:5873–5878. doi: 10.1007/s11356-014-2509-4. [DOI] [PubMed] [Google Scholar]
- 5.Melamed D. 2004. Monitoring arsenic in the environment: a review of science and technologies for field measurements and sensors. https://www.epa.gov/remedytech/monitoring-arsenic-environment-review-science-and-technologies-field-measurements-and.
- 6.Yan G, Chen X, Du S, Deng Z, Wang L, Chen S. 2019. Genetic mechanisms of arsenic detoxification and metabolism in bacteria. Curr Genet 65:329–338. doi: 10.1007/s00294-018-0894-9. [DOI] [PubMed] [Google Scholar]
- 7.Kaur H, Kumar R, Babu JN, Mittal S. 2015. Advances in arsenic biosensor development: a comprehensive review. Biosens Bioelectron 63:533–545. doi: 10.1016/j.bios.2014.08.003. [DOI] [PubMed] [Google Scholar]
- 8.Prévéral S, Brutesco C, Descamps ECT, Escoffier C, Pignol D, Ginet N, Garcia D. 2017. A bioluminescent arsenite biosensor designed for inline water analyzer. Environ Sci Pollut Res Int 24:25–32. doi: 10.1007/s11356-015-6000-7. [DOI] [PubMed] [Google Scholar]
- 9.Wan X, Volpetti F, Petrova E, French C, Maerkl SJ, Wang B. 2019. Cascaded amplifying circuits enable ultrasensitive cellular sensors for toxic metals. Nat Chem Biol 15:540–548. doi: 10.1038/s41589-019-0244-3. [DOI] [PubMed] [Google Scholar]
- 10.Descamps ECT, Meunier D, Brutesco C, Prévéral S, Franche N, Bazin I, Miclot B, Larosa P, Escoffier C, Fantino J-R, Garcia D, Ansaldi M, Rodrigue A, Pignol D, Cholat P, Ginet N. 2017. Semi-autonomous inline water analyzer: design of a common light detector for bacterial, phage, and immunological biosensors. Environ Sci Pollut Res Int 24:66–72. doi: 10.1007/s11356-016-8010-5. [DOI] [PubMed] [Google Scholar]
- 11.Jouanneau S, Durand MJ, Thouand G. 2012. Online detection of metals in environmental samples: comparing two concepts of bioluminescent bacterial biosensors. Environ Sci Technol 46:11979–11987. doi: 10.1021/es3024918. [DOI] [PubMed] [Google Scholar]
- 12.Balkwill DL, Maratea D, Blakemore RP. 1980. Ultrastructure of a magnetotactic spirillum. J Bacteriol 141:1399–1408. doi: 10.1128/JB.141.3.1399-1408.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blakemore R. 1975. Magnetotactic bacteria. Science 190:377–379. doi: 10.1126/science.170679. [DOI] [PubMed] [Google Scholar]
- 14.Frankel RB, Blakemore RP, Wolfe RS. 1979. Magnetite in freshwater magnetotactic bacteria. Science 203:1355–1356. doi: 10.1126/science.203.4387.1355. [DOI] [PubMed] [Google Scholar]
- 15.Smith MJ, Sheehan PE, Perry LL, O’Connor K, Csonka LN, Applegate BM, Whitman LJ. 2006. Quantifying the magnetic advantage in magnetotaxis. Biophys J 91:1098–1107. doi: 10.1529/biophysj.106.085167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dieudonné A, Pignol D, Prévéral S. 2019. Magnetosomes: biogenic iron nanoparticles produced by environmental bacteria. Appl Microbiol Biotechnol 103:3637–3649. doi: 10.1007/s00253-019-09728-9. [DOI] [PubMed] [Google Scholar]
- 17.Matsunaga T, Sakaguchi T, Tadakoro F. 1991. Magnetite formation by a magnetic bacterium capable of growing aerobically. Appl Microbiol Biotechnol 35:651–655. doi: 10.1007/BF00169632. [DOI] [Google Scholar]
- 18.Schleifer KH, Schüler D, Spring S, Weizenegger M, Amann R, Ludwig W, Köhler M. 1991. The genus Magnetospirillum gen. nov. description of Magnetospirillum gryphiswaldense sp. nov. and transfer of Aquaspirillum magnetotacticum to Magnetospirillum magnetotacticum comb. nov. Syst Appl Microbiol 14:379–385. doi: 10.1016/S0723-2020(11)80313-9. [DOI] [Google Scholar]
- 19.Okamura Y, Takeyama H, Sekine T, Sakaguchi T, Wahyudi AT, Sato R, Kamiya S, Matsunaga T. 2003. Design and application of a new cryptic-plasmid-based shuttle vector for Magnetospirillum magneticum. Appl Environ Microbiol 69:4274–4277. doi: 10.1128/AEM.69.7.4274-4277.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schultheiss D, Schüler D. 2003. Development of a genetic system for Magnetospirillum gryphiswaldense. Arch Microbiol 179:89–94. doi: 10.1007/s00203-002-0498-z. [DOI] [PubMed] [Google Scholar]
- 21.Arakaki A, Takeyama H, Tanaka T, Matsunaga T. 2002. Cadmium recovery by a sulfate-reducing magnetotactic bacterium, Desulfovibrio magneticus RS-1, using magnetic separation. Appl Biochem Biotechnol 98–100:833–840. doi: 10.1385/ABAB:98-100:1-9:833. [DOI] [PubMed] [Google Scholar]
- 22.Tanaka M, Brown R, Hondow N, Arakaki A, Matsunaga T, Staniland S. 2012. Highest levels of Cu, Mn and Co doped into nanomagnetic magnetosomes through optimized biomineralisation. J Mater Chem 22:11919. doi: 10.1039/c2jm31520c. [DOI] [Google Scholar]
- 23.Adam V, Chudobova D, Tmejova K, Cihalova K, Krizkova S, Guran R, Kominkova M, Zurek M, Kremplova M, Jimenez AMJ, Konecna M, Hynek D, Pekarik V, Kizek R. 2014. An effect of cadmium and lead ions on Escherichia coli with the cloned gene for metallothionein (MT-3) revealed by electrochemistry. Electrochim Acta 140:11–19. doi: 10.1016/j.electacta.2014.06.091. [DOI] [Google Scholar]
- 24.Abskharon RNN, Hassan SHA, Gad El-Rab SMF, Shoreit A. 2008. Heavy metal resistant of E. coli isolated from wastewater sites in Assiut City, Egypt. Bull Environ Contam Toxicol 81:309–315. doi: 10.1007/s00128-008-9494-6. [DOI] [PubMed] [Google Scholar]
- 25.Scherer J, Nies DH. 2009. CzcP is a novel efflux system contributing to transition metal resistance in Cupriavidus metallidurans CH34. Mol Microbiol 73:601–621. doi: 10.1111/j.1365-2958.2009.06792.x. [DOI] [PubMed] [Google Scholar]
- 26.Brutesco C, Prévéral S, Escoffier C, Descamps ECT, Prudent E, Cayron J, Dumas L, Ricquebourg M, Adryanczyk-Perrier G, de Groot A, Garcia D, Rodrigue A, Pignol D, Ginet N. 2017. Bacterial host and reporter gene optimization for genetically encoded whole cell biosensors. Environ Sci Pollut Res Int 24:52–65. doi: 10.1007/s11356-016-6952-2. [DOI] [PubMed] [Google Scholar]
- 27.Uebe R, Schüler D. 2016. Magnetosome biogenesis in magnetotactic bacteria. Nat Rev Microbiol 14:621–637. doi: 10.1038/nrmicro.2016.99. [DOI] [PubMed] [Google Scholar]
- 28.Mobley HLT, Rosen BP. 1982. Energetics of plasmid-mediated arsenate resistance in Escherichia coli. Proc Natl Acad Sci U S A 79:6119–6122. doi: 10.1073/pnas.79.20.6119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Silver S, Keach D. 1982. Energy-dependent arsenate efflux: the mechanism of plasmid-mediated resistance. Proc Natl Acad Sci U S A 79:6114–6118. doi: 10.1073/pnas.79.20.6114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ji G, Garber EAE, Armes LG, Chen C-M, Fuchs JA, Silver S. 1994. Arsenate reductase of Staphylococcus aureus plasmid pI258. Biochemistry 33:7294–7299. doi: 10.1021/bi00189a034. [DOI] [PubMed] [Google Scholar]
- 31.Komeili A, Li Z, Newman DK, Jensen GJ. 2006. Magnetosomes are cell membrane invaginations organized by the actin-like protein MamK. Science 311:242–245. doi: 10.1126/science.1123231. [DOI] [PubMed] [Google Scholar]
- 32.Rioux J-B, Philippe N, Pereira S, Pignol D, Wu L-F, Ginet N. 2010. A second actin-like MamK protein in Magnetospirillum magneticum AMB-1 encoded outside the genomic magnetosome island. PLoS One 5:e9151. doi: 10.1371/journal.pone.0009151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boucher M, Geffroy F, Prévéral S, Bellanger L, Selingue E, Adryanczyk-Perrier G, Péan M, Lefèvre CT, Pignol D, Ginet N, Mériaux S. 2017. Genetically tailored magnetosomes used as MRI probe for molecular imaging of brain tumor. Biomaterials 121:167–178. doi: 10.1016/j.biomaterials.2016.12.013. [DOI] [PubMed] [Google Scholar]
- 34.Plan Sangnier A, Preveral S, Curcio A, Silva AKA, Lefèvre CT, Pignol D, Lalatonne Y, Wilhelm C. 2018. Targeted thermal therapy with genetically engineered magnetite magnetosomes@RGD: photothermia is far more efficient than magnetic hyperthermia. J Control Release 279:271–281. doi: 10.1016/j.jconrel.2018.04.036. [DOI] [PubMed] [Google Scholar]
- 35.Hafsi M, Preveral S, Hoog C, Hérault J, Perrier GA, Lefèvre CT, Michel H, Pignol D, Doyen J, Pourcher T, Humbert O, Thariat J, Cambien B. 2020. RGD-functionalized magnetosomes are efficient tumor radioenhancers for X-rays and protons. Nanomed Nanotechnol Biol Med 23:102084. doi: 10.1016/j.nano.2019.102084. [DOI] [PubMed] [Google Scholar]
- 36.Zhang Y-B, Monchy S, Greenberg B, Mergeay M, Gang O, Taghavi S, van der Lelie D. 2009. ArsR arsenic-resistance regulatory protein from Cupriavidus metallidurans CH34. Antonie Van Leeuwenhoek 96:161–170. doi: 10.1007/s10482-009-9313-z. [DOI] [PubMed] [Google Scholar]
- 37.Wang Q, Warelow TP, Kang Y-S, Romano C, Osborne TH, Lehr CR, Bothner B, McDermott TR, Santini JM, Wang G. 2015. Arsenite oxidase also functions as an antimonite oxidase. Appl Environ Microbiol 81:1959–1965. doi: 10.1128/AEM.02981-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jia X, Bu R, Zhao T, Wu K. 2019. Sensitive and specific whole-cell biosensor for arsenic detection. Appl Environ Microbiol 85:e00694-19. doi: 10.1128/AEM.00694-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roda A, Cevenini L, Borg S, Michelini E, Calabretta MM, Schüler D. 2013. Bioengineered bioluminescent magnetotactic bacteria as a powerful tool for chip-based whole-cell biosensors. Lab Chip 13:4881–4889. doi: 10.1039/c3lc50868d. [DOI] [PubMed] [Google Scholar]
- 40.Komeili A, Vali H, Beveridge TJ, Newman DK. 2004. Magnetosome vesicles are present before magnetite formation, and MamA is required for their activation. Proc Natl Acad Sci U S A 101:3839–3844. doi: 10.1073/pnas.0400391101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Heyen U, Schüler D. 2003. Growth and magnetosome formation by microaerophilic Magnetospirillum strains in an oxygen-controlled fermentor. Appl Microbiol Biotechnol 61:536–544. doi: 10.1007/s00253-002-1219-x. [DOI] [PubMed] [Google Scholar]
- 42.Lefèvre CT, Song T, Yonnet J-P, Wu L-F. 2009. Characterization of bacterial magnetotactic behaviors by using a magnetospectrophotometry assay. Appl Environ Microbiol 75:3835–3841. doi: 10.1128/AEM.00165-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vallenet D, Calteau A, Cruveiller S, Gachet M, Lajus A, Josso A, Mercier J, Renaux A, Rollin J, Rouy Z, Roche D, Scarpelli C, Médigue C. 2017. MicroScope in 2017: an expanding and evolving integrated resource for community expertise of microbial genomes. Nucleic Acids Res 45:D517–D528. doi: 10.1093/nar/gkw1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ. 2009. Jalview version 2: a multiple sequence alignment editor and analysis workbench. Bioinformatics 25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stothard P. 2000. The sequence manipulation suite: JavaScript programs for analyzing and formatting protein and DNA sequences. Biotechniques 28:1102–1104. doi: 10.2144/00286ir01. [DOI] [PubMed] [Google Scholar]
- 46.Petkau A, Stuart-Edwards M, Stothard P, Van Domselaar G. 2010. Interactive microbial genome visualization with GView. Bioinformatics 26:3125–3126. doi: 10.1093/bioinformatics/btq588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chaudhary VK, Shrivastava N, Verma V, Das S, Kaur C, Grover P, Gupta A. 2014. Rapid restriction enzyme-free cloning of PCR products: a high-throughput method applicable for library construction. PLoS One 9:e111538. doi: 10.1371/journal.pone.0111538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Matsunaga T, Okamura Y, Fukuda Y, Wahyudi AT, Murase Y, Takeyama H. 2005. Complete genome sequence of the facultative anaerobic magnetotactic bacterium Magnetospirillum sp. strain AMB-1. DNA Res 12:157–166. doi: 10.1093/dnares/dsi002. [DOI] [PubMed] [Google Scholar]
- 49.Wang X, Wang Q, Zhang W, Wang Y, Li L, Wen T, Zhang T, Zhang Y, Xu J, Hu J, Li S, Liu L, Liu J, Jiang W, Tian J, Li Y, Schüler D, Wang L, Li J. 2014. Complete genome sequence of Magnetospirillum gryphiswaldense MSR-1. Genome Announc 2(2):e00171-14. doi: 10.1128/genomeA.00171-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu J, Rosen BP. 1991. The ArsR protein is a trans‐acting regulatory protein. Mol Microbiol 5:1331–1336. doi: 10.1111/j.1365-2958.1991.tb00779.x. [DOI] [PubMed] [Google Scholar]
- 51.Chen C-M, Misrat TK, Silver S, Rosen BP. 1986. Nucleotide sequence of the structural genes for an anion pump (accession no. P08690). https://www.uniprot.org/uniprot/P08690. [PubMed]
- 52.Chen C-M, Misrat TK, Silver S, Rosen BP. 1986. Nucleotide sequence of the structural genes for an anion pump (accession no. P08691). https://www.uniprot.org/uniprot/P08691. [PubMed]
- 53.Chen C-M, Misrat TK, Silver S, Rosen BP. 1986. Nucleotide sequence of the structural genes for an anion pump (accession no. P08692). https://www.uniprot.org/uniprot/P08692. [PubMed]
- 54.Wu J, Rosen BP. 1993. The arsD gene encodes a second trans-acting regulatory protein of the plasmid-encoded arsenical resistance operon. Mol Microbiol 8:615–623. doi: 10.1111/j.1365-2958.1993.tb01605.x. [DOI] [PubMed] [Google Scholar]
- 55.Ji G, Silver S. 1992. Reduction of arsenate to arsenite by the ArsC protein of the arsenic resistance operon of Staphylococcus aureus plasmid pI258. Proc Natl Acad Sci U S A 89:9474–9478. doi: 10.1073/pnas.89.20.9474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kovach ME, Elzer PH, Steven Hill D, Robertson GT, Farris MA, Roop RM, Peterson KM. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175–176. doi: 10.1016/0378-1119(95)00584-1. [DOI] [PubMed] [Google Scholar]
- 57.Kremers G-J, Goedhart J, van Munster EB, Gadella TW. 2006. Cyan and yellow super fluorescent proteins with improved brightness, protein folding, and FRET Förster radius. Biochemistry 45:6570–6580. doi: 10.1021/bi0516273. [DOI] [PubMed] [Google Scholar]
- 58.Stevens SY, Hu W, Gladysheva T, Rosen BP, Zuiderweg ERP, Lee L. 1999. Secondary structure and fold homology of the ArsC protein from the Escherichia coli arsenic resistance plasmid R773. Biochemistry 38:10178–10186. doi: 10.1021/bi990333c. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequence for the Magnetospirillum magneticum AMB-1 genome can be found under NCBI accession number NC_007626 (48). The Magnetospirillum gryphiswaldense MSR-1 v2 genome can be found under NCBI accession number HG794546 (49). The ArsR protein sequence was deposited under NCBI accession number P15905 (50). The nucleotide sequences of the structural genes for the arsenic pump of Escherichia coli, arsA and arsB, were deposited under NCBI accession numbers P08690 (51) and P08691 (52), respectively. The arsD gene encodes a second transacting regulatory protein of the plasmid-carried arsenical resistance operon has been deposited under NCBI accession number P46003 (54). The ArsC proteins of the arsenic resistance operon of Escherichia coli and Staphylococcus aureus plasmid pI258 have been deposited under NCBI accession numbers P08692 (53) and P0A006 (55), respectively.