Highlights
-
•
In this study, unlike CNN architectures, COVID-19 was determined from chest X-ray images with a smaller number of layers.
-
•
More COVID-19, pneumonia, and no-findings images were used than in previous studies. This increases the reliability of the system more.
-
•
As is known, reducing the size of the image may cause some information in the image to be lost. Given these facts, good classification accuracy has been achieved with capsule networks, even the image size has been reduced to 128 × 128 pixels.
Keywords: Coronavirus, Capsule networks, Deep learning, Chest x-ray images, Artificial neural network
Abstract
Coronavirus is an epidemic that spreads very quickly. For this reason, it has very devastating effects in many areas worldwide. It is vital to detect COVID-19 diseases as quickly as possible to restrain the spread of the disease. The similarity of COVID-19 disease with other lung infections makes the diagnosis difficult. In addition, the high spreading rate of COVID-19 increased the need for a fast system for the diagnosis of cases. For this purpose, interest in various computer-aided (such as CNN, DNN, etc.) deep learning models has been increased. In these models, mostly radiology images are applied to determine the positive cases. Recent studies show that, radiological images contain important information in the detection of coronavirus. In this study, a novel artificial neural network, Convolutional CapsNet for the detection of COVID-19 disease is proposed by using chest X-ray images with capsule networks. The proposed approach is designed to provide fast and accurate diagnostics for COVID-19 diseases with binary classification (COVID-19, and No-Findings), and multi-class classification (COVID-19, and No-Findings, and Pneumonia). The proposed method achieved an accuracy of 97.24%, and 84.22% for binary class, and multi-class, respectively. It is thought that the proposed method may help physicians to diagnose COVID-19 disease and increase the diagnostic performance. In addition, we believe that the proposed method may be an alternative method to diagnose COVID-19 by providing fast screening.
1. Introduction
The COVID-19 disease caused by the SARS-CoV-2 virus first appeared in Wuhan, China. COVID-19 affects the respiratory system, causing fever and cough, and in some serious cases, causes pneumonia [1]. Pneumonia is a type of infection that causes inflammation in the lungs, and besides the SARS-CoV-2 virus, bacteria, fungi, and other viruses often play a role in the emergence of this disease [2]. Conditions such as weak immune system, asthma, chronic diseases and elderliness increase the severity of pneumonia. Treatment of pneumonia varies depending on the organism causing the infection, but usually antibiotics, cough medicines, antipyretics, and pain killers are effective for treatment [3]. Depending on the symptoms, patients can be hospitalized and, in more severe cases, they can be taken to the intensive care unit. The COVID-19 outbreak is considered a serious disease due to its high permeability, and contagiousness [4]. In addition, this epidemic has a great impact on the healthcare system, by virtue of the high number of patients hospitalized in intensive care units, the length of treatment, and the lack of hospital resources [5], [6]. Thus, it is vital to diagnose the diseased at an early stage in order to prevent such scenarios.
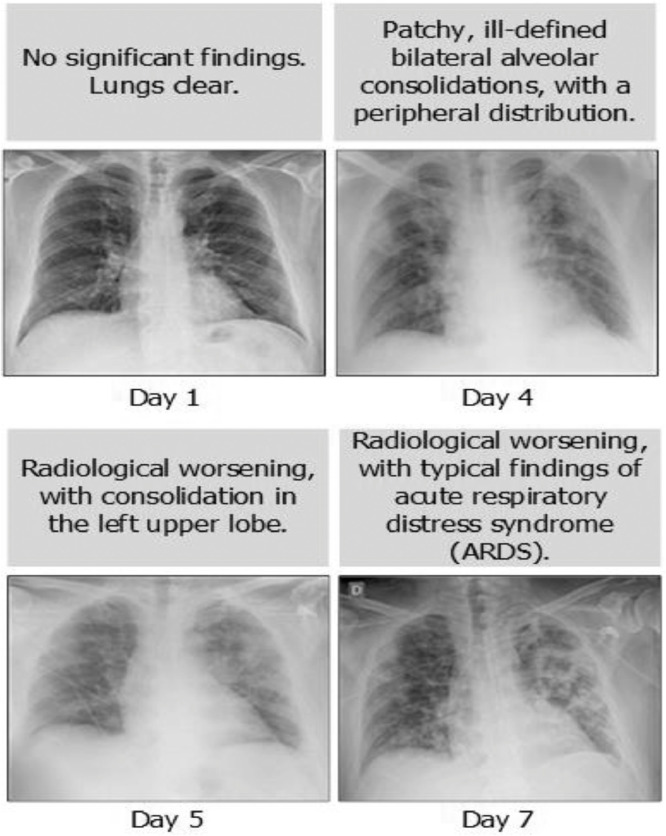
Computed tomography, and X-ray images play an important role in the early diagnosis and treatment of COVID-19 disease [7]. The fact that X-ray images are cheaper and faster, and patients are exposed to less radiation cause these images to preferred more than CT images [8], [9]. However, it is not easy to diagnose pneumonia manually. White spots on X-ray images need to be examined and interpreted in detail by a specialist. Yet, these with spots can be confused with tuberculosis or bronchitis, which can lead to misdiagnosis. In Fig. 1 , we provide a chest X-ray images of a COVID-19 patient taken on days 1, 4, 5, and 7.
Fig. 1.
Chest X-ray image of COVID-19 patient [10].
Manual examination of X-ray images provides accurate diagnosis of the disease in 60–70% [11], [12]. Additionally, 50–75% performance is achieved by manual analysis of CT images [11]. Therefore, artificial intelligence based solutions may provide a less costly and accurate diagnosis for both COVID-19 and other types of pneumonia. Deep learning models, one of the artificial intelligence techniques, are used successfully in the biomedical field. Diagnosis of cardiac arrhythmia [13], [14], brain injuries [15], [16], lung segmentation [17], [18], breast cancer [19], [20], skin cancer [21], [22], epilepsy [23], [40], [72], and pneumonia [24], [25], [26], [27], [28], [29] with deep learning models has increased the popularity of these algorithms in biomedical field.
Radiology images have been widely used recently for the diagnosis of COVID-19. Apostolopoulos et al. [30] developed a deep learning-based method for the diagnosis of COVID-19. In the study, both binary class and multi-class analysis process took place. A total of 224 COVID-19, 700 bacterial pneumonia, and 500 no-findings X-ray images were used. The performance of the developed model was measured with accuracy, sensitivity, and specificity values. The proposed model achieved an accuracy rate of 98.78% for the binary class (COVID-19 vs. No-findings), and 93.48% for the multi class (COVID-19 vs. No-findings vs. pneumonia). Similarly, Hemdan et al. [31] proposed a deep learning model for the diagnosis of COVID-19 disease, and compared the model with 7 different deep learning algorithms. The success of the proposed method has been determined with accuracy, precision, recall, and f1-score values. An average of 74.29% accuracy was obtained for binary class problem. Narin et al. [32] performed COVID-19 diagnosis using chest X-ray images by developing the ResNet50, InceptionV3, and InceptionResNetv2 deep learning models. In the study, the binary classification process was carried out, and the data were validated with 5 fold cross-validation. The performance of the models was evaluated with five different criteria, and an average of 98% accuracy was achieved with the ResNet50 model. Wang et al. [33] provided a deep learning model to diagnose COVID-19 disease. The performance of the proposed method was evaluated with sensitivity and accuracy metrics, and the results were compared with VGG19 and ResNet50 deep learning models. At the end of the study, average accuracy was achieved as 93.3%. Sethy et al. [34] extracted features from COVID-19′s X-ray images using the deep learning model and classified these features with SVM. The performance of this hybrid model, which was created by combining ResNet50 and SVM models, was measured with f1-scores and Kappa values. It was emphasized in the study that this method, which is compared with other methods, is more effective. Afshar et al. [35] used capsule networks to diagnose COVID-19 cases. The success of the proposed model was evaluated with specificity, sensitivity, and accuracy values and 98.3% accuracy was achieved. Mobiny et al. [36] developed capsule networks to diagnose COVID-19 cases with X-ray images. The developed model was compared with Inceptionv3, ResNet50, and DenseNet121 deep learning models and the proposed method has been more successful.
In addition to X-ray images, there are COVID-19 diagnostic studies performed with computed tomography. Zheng et al. [37] developed a novel deep learning model and proposed model was tested with 499 CT images. At the end of the study, an average of 88.55% accuracy was achieved. In the study by Ying et al. [38], CT images were used to diagnose COVID-19 disease and distinguish the pneumonia. The proposed deep learning model was compared with some models in the literature and the performance of the model was determined with accuracy, precision, recall, AUC, and f1-scores.
Nowadays, with the prominence of deep learning models, huge data sets can be evaluated much more comfortably. As in many fields, the most preferred deep learning model in medicine is CNN-based models. Yet, CNN architectures have some limitations. One of these constrains is the max pooling. Max pooling is designed to transfer the most valuable information from the previous layer to the next layer. This causes small details in the data to be lost, and the data may not be transferred to other layers. Also, existing CNN models cannot maintain the part-whole relationship of the objects. In order to overcome these shortcomings of CNNs, Sabour et al. [39] proposed a new neural network called Capsule Networks in 2017. They suggest that with this proposed model, they overcome the shortcomings of existing CNN models. There are several studies on capsule networks and X-ray images. While CT images were used in the study of Mobiny et al. [36], in the study of Afshar et al. [35], X-ray images and 5 different thorax data sets were examined. The number of COVID-19 cases used in the study was not specified.
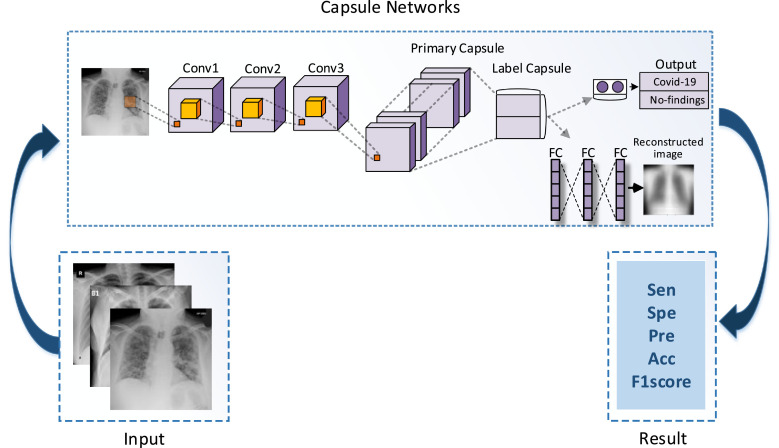
In our study, 231 COVID-19 images were examined. In addition, unlike other articles, a new network architecture was proposed in our study. In the study, binary class (COVID-19, and No-findings) and multi-class (COVID-19, and No-findings and Pneumonia) classification was carried out with capsule networks. COVID-19 and other images of different sizes have been resized to be input data to capsule networks. In the study, a novel model is proposed, which is different from the existing capsule networks. The flowchart of the study is shown in Fig. 2 .
Fig. 2.
Flowchart of the proposed method.
The rest of the study is organized as follows. In the Materials and Method section, data set and capsule network architecture are mentioned. In the Experimental Results section, the findings are discussed. In the Conclusion, general contributions of the study are presented.
2. Materials and method
2.1. The dataset
In this study, X-ray images of COVID-19 [41], and no-findings and pneumonia [42] were used. The database generated by Cohen [41] is constantly updated with images of COVID-19 patients from different parts of the world. During this study, 231 COVID-19 images were used. There is no detailed record of patient information in the database. In the database provided by Wang et al. [42], 1050 no-findings, and 1050 pneumonia images were used.
2.2. Capsule networks
Capsule networks have been developed to maintain the positions of objects and their properties in the image, and to model their hierarchical relationships [39]. In the convolution neural networks, valuable information in the data comes to the fore with the pooling layer. Since the data is transmitted to the next layer by pooling, it may not possible for the network to learn small details [43]. In addition, CNN produces a scalar value in neural output. Capsule networks create vectorial output of the same size but with different routings, thanks to the capsules, which contain many neurons. The routings of a vector represent the parameters of the images [44]. CNNs use scalar input activation functions such as ReLU, Sigmoid, and Tangent. On the other hand, capsule networks use a vectorial activation function called squashing given in Eq. (1);
| (1) |
In the Eq. (1), vj indicates the output of the capsule j, and sj indicates the total input of the capsule. vj shrinks long vectors towards 1 if there is an object in the image, and chokes short vectors towards 0 if there is no object in the image [45], [46].
Except for the first layer of capsule networks, the total input value of capsule sj is found by weighted sum of the prediction vectors (Uj|i) in the capsules located in the lower layers. The prediction vector (Uj|i) is calculated by multiplying a capsule in the lower layer by its output (Oi), and a weight matrix (Wij).
| (2) |
| (3) |
where bij is the coefficient determined by the dynamic routing process and is calculated as in Eq. (4);
| (4) |
In the Eq. (4), aij donates the log probability. The sum of the correlation coefficients between capsule i, and capsules in the top layer is 1 and the log prior probability is determined by Softmax [47]. In capsule networks, a margin loss has been proposed to determine whether objects of a particular class are present and can be calculated with the Eq. (5);
| (5) |
The value of Tk is 1 if and only if the class k is present. m + = 0.9 ve m− = 0.1 are the hyper parameters and denotes down-weighting of the loss [47]. The length of the vectors calculated in the capsule networks indicates the probability of being in that part of the image, while the direction of the vector contains the parameter information such as texture, color, position, size, etc. [46], [48]
2.2.1. Capsule network architecture
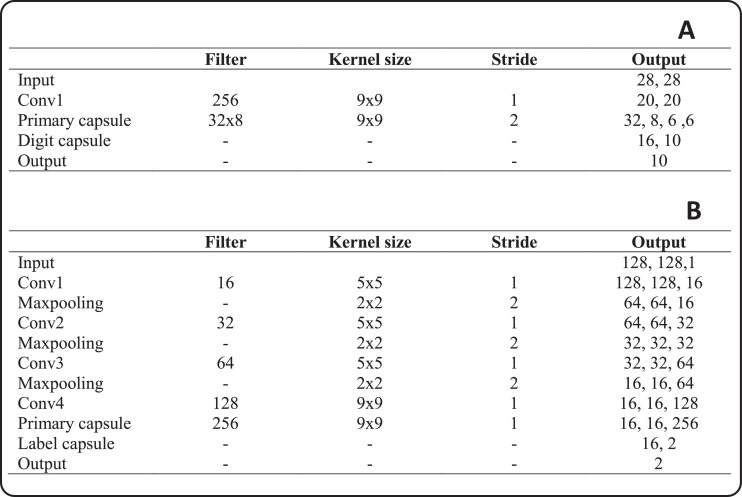
Original capsule networks were used to classify 28 × 28 size MNIST images. The network has one convolution layer, one primary layer, one-digit layer, and three fully connected layers. Convolution layers contains 256 kernels of size 9 × 9. This layer converts pixel densities to local features with the size of 20 × 20 to be used as inputs to primary capsules [49]. Second layer (Primary Caps) contains 32 different capsules and each capsule applied eighth 9 × 9 × 256 convolutional kernels. Both layers used the ReLU activation function. The last layer (Digit Caps) outputs 16D vectors that contain all the instantiation parameters required for reconstruction the object [39], [49].
2.2.2. The proposed network architecture
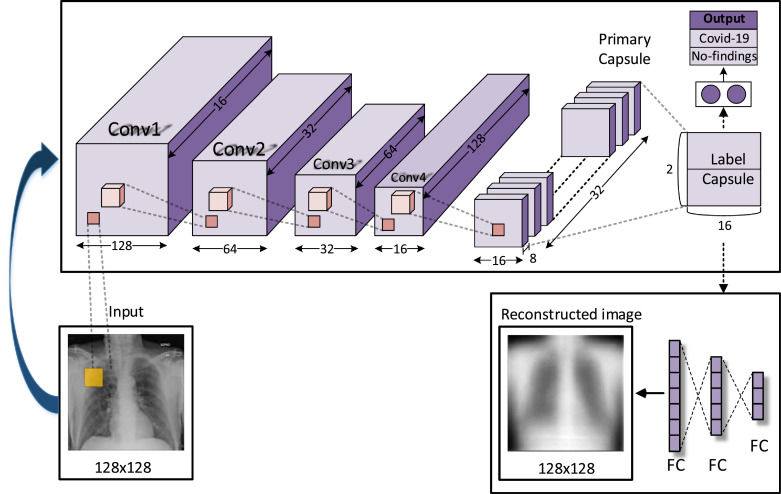
For the classification of 128 × 128 images, we propose a new network model with five convolution layers. The reason for adding more convolution layers is to provide more effective feature map to the primary layer as an input. Fig. 3 shows the proposed network architecture. In Fig. 4 , the details of the original capsule network and the proposed network architecture are given. First layer contains 16 kernels of size 5 × 5 with a stride of 1. Max-pooling with size and stride of 2 was applied to the first layer exit. The same structure was used in the next two layers. The kernel numbers of the second and third layers are 32 and 64, respectively. Fourth layer includes 128 kernels of size 9 × 9 with a stride of 1. The fifth layer is the primary layer and it contains 32 different capsules and each capsule applied 9 × 9 convolutional kernels with a stride of 1. The LabelCaps layer has 16-dimensional (16D) capsules for two classes and three classes, and ReLU activation function is used for all layers.
Fig. 3.
The Convolutional CapsNet architecture for classification of COVID-19, and No-findings images.
Fig. 4.
A- Original capsule network architecture, B- the proposed Convolutional CapsNet architecture.
2.2.2.1. Preprocessing and data augmentation
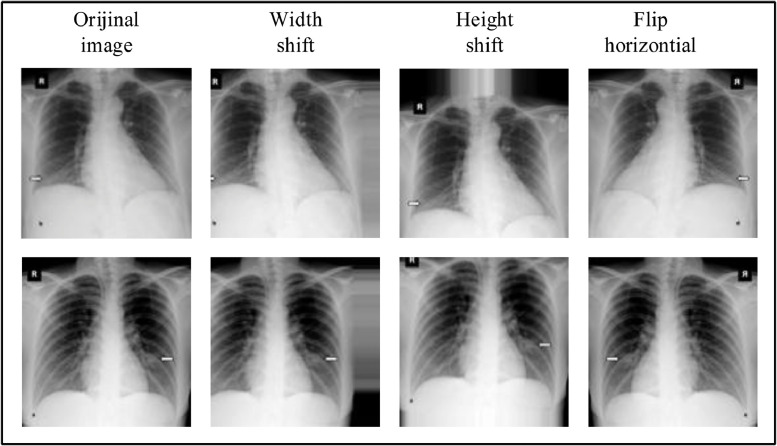
The input size for the proposed model is 128 × 128. Yet, since the lengths and widths of the images in the data set are not the same size, all images were resized to 128 × 128 pixels. Since the number of COVID-19 X-ray images is limited, data augmentation strategies have been applied to avoid overfitting problems. For data augmentation, width shift range (0.2), height shift range (0.2), and flip horizontal parameters were considered. As a result, COVID-19 X-ray images have neem increased from 231 to 1050. Fig. 5 shows an example of data augmentations applied to COVID-19 images.
Fig. 5.
Data augmentation process applied to COVID-19 images (left to right: original, width shift range:0.2, height shift range:0.2, and flip horizontal).
2.3. Performance evaluation
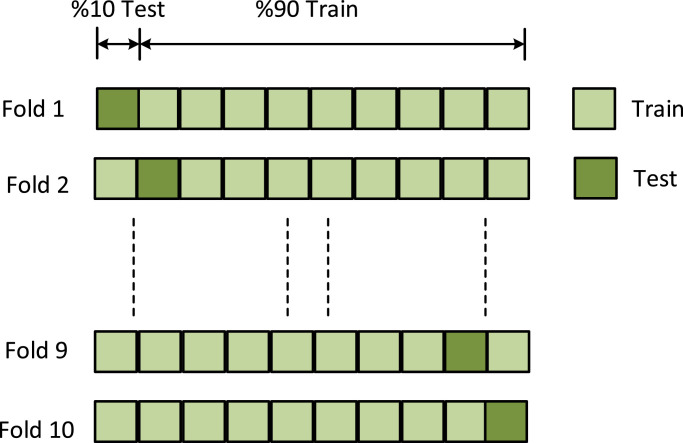
The performance of the proposed method was evaluated with 10 fold cross-validation. The data set was divided into 10 parts and 9 parts were used for training, while the remaining one was used for testing (Fig. 6 ). This process was carried out for all parts and the performance of the method was calculated by taking the average of all parts.
Fig. 6.
The graphical representation of training and test data.
The used parameters to compare performances are defined as follows;
-
•
True Positive (TP) shows the number of correctly identified COVID-19,
-
•
False negative (FN) indicates the number of incorrectly identified COVID-19,
-
•
True negative (TN) is the number of correctly identified No-findings,
-
•False positive (FP) is the number of incorrectly identified No-findings.
(6) (7) (8) (9) (10)
3. Experimental results and discussion
Since the data sets consist of images from different sources, all images were first resized to 128 × 128 pixels. The resolution of images is high and it is required a powerful system to analyze the images with the original size by using capsule networks. It is time consuming and costly to process the high quality images like in all classical deep learning architectures. For this reason, images have been resized to 128 × 128 pixels.
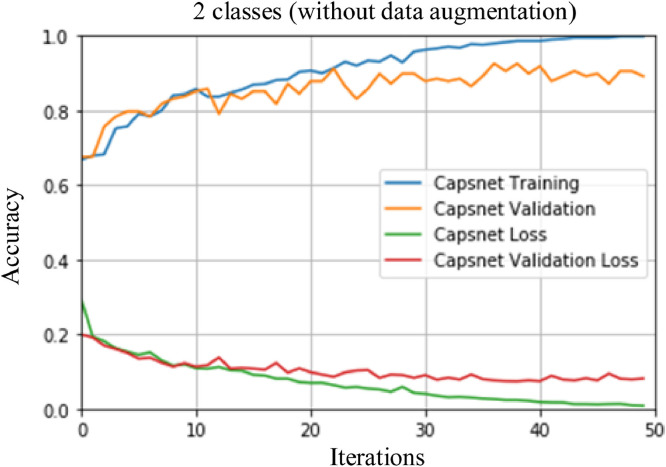
Two different scenarios were used for COVID-19 detection from X-ray images. In the first scenario, a binary class model (COVID-19 vs. No-findings) was performed. In the second scenario, a multi class model (COVID-19 vs. No-findings vs. Pneumonia) was proposed. During the training phase, the performance of method was evaluated with 10 fold cross-validation with the number of 50 epochs. The first scenario includes three different applications. In the first application, a training was conducted using 231 COVID-19 and 500 No-findings images. In the second application, data augmentation was applied. In the last application, the original capsule network architecture was used. The training and loss graph for a fold of the first application is shown in Fig. 7 .
Fig. 7.
Training and loss graphs for a fold (without data augmentation) of the first application.
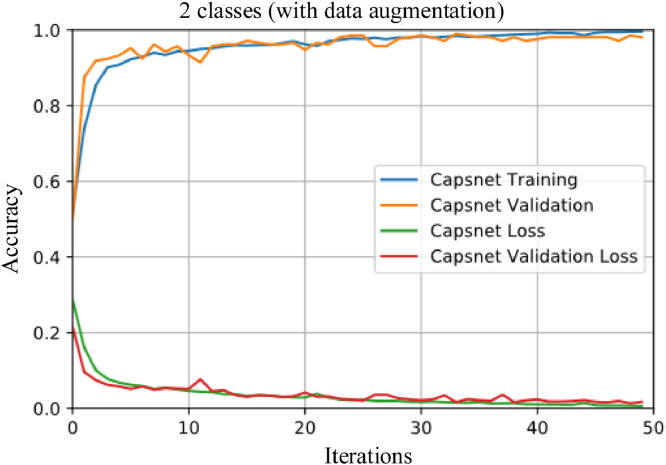
In the second application, 231 COVID-19 images were increased to 1050 with data augmentation process. 1050 No-findings images were also obtained from [41] for the second application. In the third application, 1050 COVID-19, and 1050 No-findings were used. Fig. 8 shows the training and loss graphs for a fold of the second application. In addition, the classification results obtained before and after data augmentation are given in Table 1 .
Fig. 8.
Training and loss graphs for a fold (with data augmentation) of the second application.
Table 1.
The classification results before and after data augmentation (A: the proposed Convolutional CapsNet method, B: original capsule network).
| Model | Data Aug. | Sen (%) | Spe (%) | Pre (%) | F1 (%) | Acc (%) | Processing time (per epoch) | Number of data (Covid19/No-findings) |
|---|---|---|---|---|---|---|---|---|
| A | No | 96.00 | 80.95 | 91.60 | 93.75 | 91.24 ± 5.35 | 16 | 231/500 |
| A | Yes | 97.42 | 97.04 | 97.06 | 97.24 | 97.24 ± 0.97 | 72 | 1050/1050 |
| B | Yes | 28.00 | 98.00 | 12.50 | 55.00 | 49.14 ± 0.99 | 500 | 1050/1050 |
As can be seen in Table 1, performance parameters have increased significantly with data augmentation. Although it is claimed that capsule networks have high performance with low data set, increasing the training data set is a parameter that significantly affects classification performance [50], [51]. According to Table 1, the original capsule network architecture is not sufficient for classifying X-ray images. In addition, the results showed that the original architecture was very poor and a lot of processing time was required for each epoch. Therefore, as shown in Fig. 3, a new network model is proposed in which the number of convolution layers is increased. With the proposed method, an accuracy of 91.24% was achieved without applying data augmentation. Then, data augmentation was applied, and the data set was restructured to be 1050. With the restructured data set, the accuracy rate of the training result increased by 6%, and the performance reached 97.24%. Table 2 shows the detailed results of all folds obtained by applying data augmentation.
Table 2.
10 fold results with data augmentation.
| Folds | Sen (%) | Spe (%) | Pre (%) | F1 (%) | Acc (%) |
|---|---|---|---|---|---|
| Fold 1 | 96.19 | 99.04 | 99.01 | 97.58 | 97.61 |
| Fold 2 | 99.04 | 97.14 | 97.19 | 98.11 | 98.09 |
| Fold 3 | 99.04 | 94.28 | 94.54 | 96.74 | 96.66 |
| Fold 4 | 98.09 | 95.23 | 95.37 | 96.71 | 96.66 |
| Fold 5 | 96.19 | 97.14 | 97.11 | 96.65 | 96.66 |
| Fold 6 | 98.09 | 98.09 | 98.09 | 98.09 | 98.09 |
| Fold 7 | 100.0 | 98.09 | 98.13 | 99.05 | 99.04 |
| Fold 8 | 97.14 | 98.09 | 98.07 | 97.60 | 97.61 |
| Fold 9 | 93.33 | 98.09 | 98.00 | 95.61 | 95.71 |
| Fold 10 | 97.14 | 95.23 | 95.32 | 96.22 | 96.19 |
| Mean±Std | 97.42 ± 1.81 | 97.04 ± 1.50 | 97.08 ± 01.42 | 97.24 ± 0.98 | 97.23 ± 0.97 |
In the second scenario, multi class (COVID-19 vs. No-findings vs. Pneumonia) X-ray images were classified using 10 fold cross-validation. The results are given in Table 3 . In addition, the training and loss graphs of the multi class training process are shown in Fig. 9 .
Table 3.
Multi class classification results.
| Folds | Sen (%) | Spe (%) | Pre (%) | F1 (%) | Acc (%) | Overall Acc (%) |
|---|---|---|---|---|---|---|
| Fold 1 | 81.59 | 90.41 | 82.16 | 81.66 | 87.33 | 81.59 |
| Fold 2 | 83.49 | 91.30 | 83.84 | 83.59 | 88.55 | 83.49 |
| Fold 3 | 89.52 | 94.66 | 89.88 | 89.53 | 92.91 | 89.52 |
| Fold 4 | 81.90 | 90.50 | 82.30 | 82.06 | 87.46 | 81.90 |
| Fold 5 | 83.49 | 91.53 | 83.79 | 83.50 | 88.79 | 83.49 |
| Fold 6 | 86.03 | 92.73 | 87.28 | 85.95 | 90.41 | 86.03 |
| Fold 7 | 85.71 | 92.65 | 86.07 | 85.84 | 90.23 | 85.71 |
| Fold 8 | 82.86 | 90.97 | 83.02 | 82.60 | 88.26 | 82.86 |
| Fold 9 | 83.17 | 91.30 | 83.36 | 82.97 | 88.60 | 83.17 |
| Fold 10 | 84.44 | 91.83 | 84.41 | 84.41 | 89.31 | 84.44 |
| Mean±Std | 84.22±2.24 | 91.79±1.21 | 84.61±2.32 | 84.21±2.24 | 89.19±1.57 | 84.22±2.24 |
Fig. 9.
Training and loss graphs for a fold (with data augmentation) for multi class problem.
Other hyper parameters of capsule networks are given in Table 4 . Three different r parameters (1,3,5) were considered to classify X-ray images. The best result was obtained with r = 5 in binary classification, and r = 3 in multiclass classification.
Table 4.
Hyper parameters of capsule networks.
| Routing | Optimizer | lr | Loss weight | Batch size | Epoch |
|---|---|---|---|---|---|
| 1, 3, 5 | Adam | 0.00005 | 0.392 8.192 |
16 | 50 |
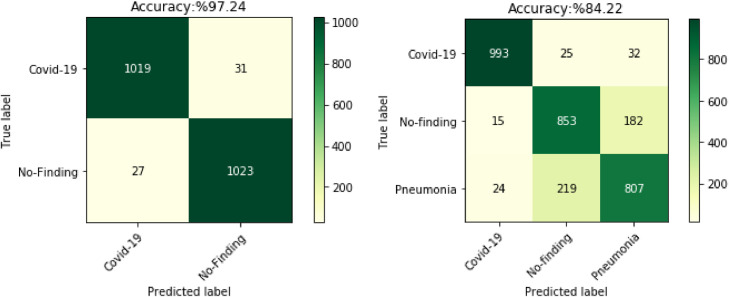
Confusion matrix is used to measure the accuracy of the model's predictions. Confusion matrixes of binary class and multi class classification results are given in Fig. 10 . As seen in Fig. 10a, the proposed method correctly classified 1019 of 1050 COVID-19 images in a binary class. In Fig. 10b, the proposed model accurately identified 993 COVID-19 images in a multi class. While COVID-19 was determined with 97.04% accuracy in binary classification, this ratio was 94.57% in multi class classification.
Fig. 10.
Confusion matrix of binary and multi class classification results.
3.1. Decoder part
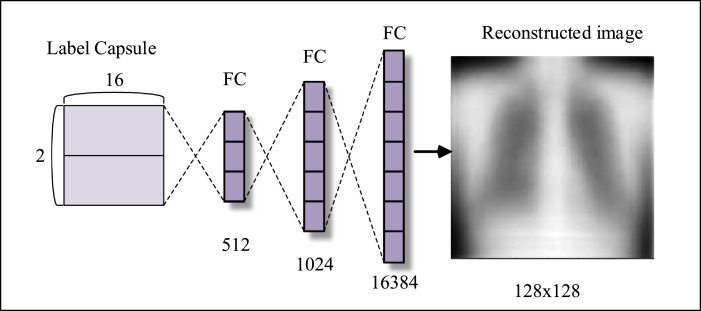
Capsule networks use the autoencoders structure to reconstruct data. Autoencoders consist of encoder and decoder [43]. In the proposed capsule network, the encoder consists of convolution layer, primary layer, and label layer, while the decoder part includes of three fully connected layers. The decoder tries to reconstruct the X-ray image using the properties generated in the encoder. In doing so, the encoder uses the difference of the mean square error between the reconstructed and input image. The low error indicates that the rebuilt image is similar to the input image [45], [52]. Fig. 11 shows the decoder structure of the proposed model.
Fig. 11.
Decoder structure of the proposed model.
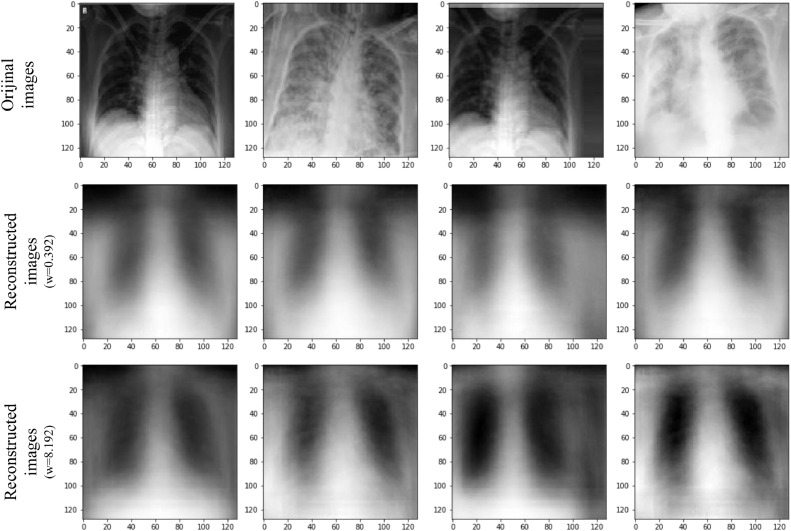
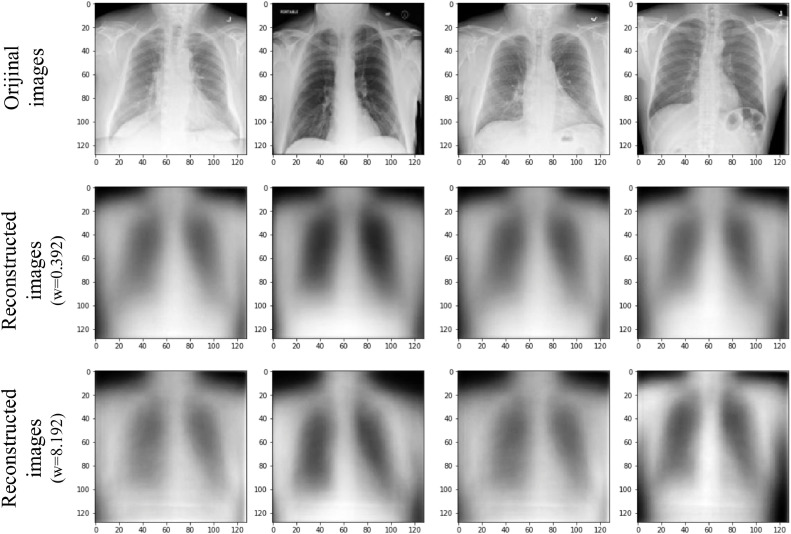
X-ray images encoded with the proposed capsule network, and reconstructed with the decoder are provide in Fig. 12, Fig. 13 , respectively. Two different weights are used for the reconstruction loss function in the decoder [53]. The first one is 0.392 (in MNIST dataset (0.0005 × 28 × 28) in the original CapsNet architecture, and the second is 8.192 (0.0005 × 128 × 128) calculated for 128 × 128 X-ray images.
Fig. 12.
Reconstructed COVID-19 X-ray images. First line: original images, second line: reconstructed images (weight of reconstruction loss (w) = 0.392), third line: reconstructed images (weight of reconstruction loss (w) = 8.192).
Fig. 13.
Reconstructed No-findings X-ray images. First line: original images, second line: reconstructed images (weight of reconstruction loss (w) = 0.392), third line: reconstructed images (weight of reconstruction loss (w) = 8.192).
In images reconstructed via decoder, blur is observed. This may be due to changes in the data in the training set or CapsNet's inability to distinguish the noise in the image [45], [53]. Also, when the weight of reconstruction loss is increased from 0.392 to 8.192, it is seen that the blurriness of the reconstructed images decreases, and images gain a little more clarity. An important advantage of capsule networks is that they can reconstruct images. In this way, it is not necessary to use methods such as heatmap [52], [54], [55], [56] to comprehend which part of image is effective in extracting features in CNN models. Thanks to the reconstruction ability of capsule networks, it is clearer which parts of the image are used for the classification process. Classification was carried out using the proposed method with the reconstructed chest X-ray images shown in Figs. 12, and 13. These parts can be considered as the parts where effective features are selected for the classification of the image. It is known that COVID-19 is difficult to diagnose because it overlaps with other lung infections [35]. Therefore, it is important to distinguish COVID-19 from other lung infections while extracting features from chest X-ray images. Although 128 pixels size images were used in this study, the proposed model was able to discriminate COVID-19, and No-findings with 97.24% accuracy, whereas COVID-19, no-findings, and pneumonia images with 84.22% accuracy. We believe that, when much larges images are used, the proposed model may reveal effective features that can distinguish different types of infections in the lung area.
After the coronavirus outbreak, many different computer-aided studies are conducted to assist physicians for the diagnosis of COVID-19. Most of these studies applied X-ray and CT images to detect COVID-19 with artificial intelligence approaches. Apostolopoulos et al. [30] developed a deep learning model for the diagnosis of COVID-19. In the study, both binary class, and multi class classification analysis took place. The proposed model reached an accuracy of 98.75% for the binary class, and 93.48% for the multi class. In study of Ozturk et al. [52], authors proposed DarkCovidNet model to diagnose COVID-19 with X-ray images. They have achieved 98.08%, and 87.02% accuracies for binary and multiclass classification, respectively. Togacar et al. [57] developed MobileNet, a deep learning-based model, for COVID-19 detection. They have reached 99.27% accuracy rate using 295 COVID-19, 98 pneumonias, and 65 no-findings X-ray images. The researches also applied CT images to diagnose COVID-19. Ying et al. [38], provided VGG16, DenseNet, ResNet and DRE-Net to detect COVID-19 with 88 COVID-19 images, and 86 normal images. Wang et al. [33], obtained a classification accuracy of 89.50% using the InceptionNet deep learning model with CT images. ResNet18, and ResNet23 was considered to diagnose COVID-19 with CT images in the study of Butt et al. [59]. 189 COVID-19, 194 pneumonias, and 145 no findings CT images were used for 3 –class problem. At the end of the study, researches reached 86.70% accuracy rate. Panvar et al. [71] proposed a deep learning model, nCOVnet to determine the COVID-19 disease using X-ray images. Binary class problem was considered in the study, and the performance of the proposed method was determined with an accuracy of 97.60%
In this study, a deep learning model based on capsule networks was performed for the detection of COVID-19 disease. Totally 3150 (1050 COVID-19, 1050 Pneumonia, and 1050 No-Findings) X-ray images were used in the study. COVID-19 images were increased from 231 to 1050 by data augmentation method. We achieved an accuracy of 91.24%, and 97.24% for binary class problem without data augmented and with data augmented, respectively. In three-class problem, we obtained an accuracy ıf 84.22% with the data augmented. In addition, by comparing the performance of the model we proposed with some studies in the literature, we have provided the accuracy results for both binary class and multiple-class problem in Table 5 , and Table 6 , respectively.
Table 5.
Comparison of the proposed method with other methods in binary class classification.
| Study | Type of images | Number of cases | Methods | Data set | Acc (%) |
|---|---|---|---|---|---|
| Apostolopoulos et al. [30] | X-ray images | 224 – COVID-19, 504 – No-Findings |
VGG19 | [41], [60], [61] | 98.75 |
| Hemdan et al. [31] | X-ray images | 25 – COVID-19 25 – No-Findings |
VGG19, DenseNet121 | [41] | 90.00 |
| Narin et al. [32] | X-ray images | 50 – COVID-19, 50 – No-Findings |
ResNet50 | [41], [62] | 98.00 |
| Sethy et al. [34] | X-ray images | 25 – COVID-19, 25 – No-Findings |
ResNet50 | [41] | 95.38 |
| Ozturk et al. [52] | X-ray images | 127 –COVID-19, 500 – No-Findings |
DarkCovidNet | [41] | 98.08 |
| Panvar et al. [71] | X-ray images | 192 COVID-19 145 – No-Findings |
nCOVnet | [41] | 97.62% |
| Wang et al. [33] | CT images | 325 – COVID-19, 740 – Pneumonia |
InceptionNet | [66], [67], [68] | 89.50 |
| Zheng et al. [37] | CT images | 313 – COVID-19, 229 – No-Findings |
DeCovNet | [69] | 90.01 |
| Ying et al. [38] | CT images | 88 – COVID-19, 86 – No-Findings |
DRE-Net | [63], [64], [65] | 86.00 |
| The proposed method | X-ray images |
1050 – COVID-19, 1050 – No Findings |
CapsNet | [41], [42] | 97.24 |
Table 6.
Comparison of the proposed method with other methods in multi class classification.
| Study | Type of images | Number of cases | Methods | Data set | Acc (%) |
|---|---|---|---|---|---|
| Apostolopoulos et al. [30] | X-ray images | 224 – COVID-19, 714 – Pneumonia, 504 – No-Findings |
VGG19 | [41], [60], [61] | 93.48 |
| Ozturk et al. [52] | X-ray images | 127 – COVID-19, 500 - Pneumonia, 500 – No-Findings |
DarkCovidNet | [41] | 87.02 |
| Togacar et al. [57] | X-ray images | 295 – COVID-19, 98 – Pneumonia, 65 – No-Findings |
MobileNetV2 | [41], [70] | 99.27 |
| Butt et al. [59] | CT images | 189 – COVID-19, 194 – Pneumonia, 145 – No-Findings |
ResNet | – | 86.70 |
| The proposed method | X-ray images |
1050 – COVID-19, 1050 – Pneumonia, 1050 – No - Findings |
CapsNet | [41], [42] | 84.22 |
The advantages of the proposed model can be stated as follows;
-
•
In this study, unlike CNN architectures, COVID-19 was determined from chest X-ray images with a smaller number of layers (4 convolution layers + primary capsule layer). In addition, capsule networks can achieve successful results with several convolution layers while CNN architectures need to use more layers [30], [31], [52], [58]. The low number of layers causes the model to be less complex.
-
•
More COVID-19, pneumonia, and no-findings images were used than in previous studies. This increases the reliability of the system more.
-
•
As is known, reducing the size of the image may cause some information in the image to be lost. Given these facts, good classification accuracy has been achieved with capsule networks, even the image size has been reduced to 128 × 128 pixels. The image size used in many CNN-based studies is larger than 128 × 128 pixels [31], [32], [33].
The disadvantages of the proposed model can be stated as follows;
-
•
Capsule networks require a lot of hardware resources when processing large images, and accordingly processing time increases. For this reason, images with the small dimensions were used in the study.
-
•
Input image sizes must be the same to be classified with the capsule networks. The images in the dataset differ in both size and number of channels [41], [42]. This situation requires a serious preprocessing before the images are given to the model as input.
With the proposed model, it is aimed for physicians to make faster decisions on X-ray images in COVID-19 detection. The workload of physicians can be reduced with these computer-aided methods. In addition, rapid diagnosis is very important for physicians to deal with more patients efficiently. In the future studies, we want to use more data sets in order to validate the reliability and accuracy of the proposed model. Also, we plan to use different data types such as CT images to diagnose COVID-19.
4. Conclusion
In this study, an approach using capsule networks for classification of COVID-19, no-findings, and pneumonia X-ray images is proposed. To the best of our knowledge, there are several studies in which COVID-19 X-ray images are classified using capsule networks. In this study, the ability of classification of COVID-19 X-ray images of capsule networks was examined. The results showed that capsule networks can effectively classify even in a limited data set. Capsule networks are planned to be trained with larger data sets to achieve the level of success that can assist physicians in the diagnosis of coronavirus disease. A training process with large data sets is very important in determining the validity and reliability of the system.
Declaration of Competing Interest
The authors declare no conflicts of interest.
References
- 1.Guan W., Ni Z., Hu Y., Liang W., Ou C. et al. Clinical characteristics of coronavirus disease 2019 in China. New England Journal of Medicine. 2020;382(18):1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pereira R.M., Bertolini D., Teixeira L.O., Silla C.N., Jr., Costa Y.M.G. COVID-19 identification in chest X-ray images on flat and hierarchical classification scenarios. Comput Methods Programs Biomed. 2020;194 doi: 10.1016/j.cmpb.2020.105532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kwon E., Kim M., Choi E., Park Y., Kim C. Tamoxifen-induced acute eosinophilic pneumonia in a breast cancer patient. Int J Surg Case Rep. 2019;60:186–190. doi: 10.1016/j.ijscr.2019.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tolksdorf K., Buda S., Schuler E., Wieler L.H., Haas W. Influenza-associated pneumonia as reference to assess seriousness of coronabviurs disease (COVID-19) Euro Surveill. 2020;25(11) doi: 10.2807/1560-7917.ES.2020.25.11.2000258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grasselli G., Pesenti A., Cecconi M. Critical cate utilization for the COVID-19 outbreak in Lombardy, Italy. JAMA. 2020;323(16):1545–1546. doi: 10.1001/jama.2020.4031. [DOI] [PubMed] [Google Scholar]
- 6.Jiang X., Coffee M., Bari A., Wang J., Jiang X. Towards an artificial intelligence framework for data-driven prediction of coronavirus clinical severity. Computers, Materials & Continua. 2020;63(1):537–551. doi: 10.32604/cmc.2020.010691. [DOI] [Google Scholar]
- 7.Zu Z.Y., Jiang M.D., Xu P.P., Chen W., Ni Q.Q. Coronavirus disease 2019 (COVID-19): a perspective from China. Radiology. 2020 doi: 10.1148/radiol.2020200490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Self W.H., Courtney D.M., McNaughton C.D., Wunderink R.G., Kline J.A. High discordance of chest X-ray and computed tomography for detection of pulmonary opacities in ED patients: implications for diagnosing pneumonia. Am J Emerg Med. 2013;31(2):401–405. doi: 10.1016/j.ajem.2012.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rubin G.D., Ryerson C.J., Haramati L.B., Sverzellati N., Kanne J.P. The role of chest imaging in patient management during the covid-19 pandemic: a multinational consensus statement from the Fleischer society. Radiology. 2020;296(1):172–180. doi: 10.1148/radiol.2020201365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lorente, E. COVID-19 pneumonia - evolution over a week, https://radiopaedia.org/cases/covid-19-pneumonia-evolution-over-a-week-1. [Accessed 2 June 2020].
- 11.Kanne J.P., Little B.P., Chung J.H., Elicker B.M., Ketai L.H. Essentials for radiologists on COVID-19: an update—Radiology scientific expert panel. Radiology. 2020 doi: 10.1148/radiol.2020200527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xie X., Zhong Z., Zhao W., Zheng C., Wang F. Chest CT for typical 2019-nCoV pneumonia: relationship to negative RT-PCR testing. Radiology. 2020 doi: 10.1148/radiol.2020200343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rajput, K.S., Wibowo, S., Hao, C., and Majmudar, M.On arrhythmia detection by deep learning and multidimensional representation, arXiv, 190400138, 2019.
- 14.Parvaneh S., Rubin J., Babaeizadeh S., Xu-Wilson M. Cardiac arrhythmia detection using deep learning: a review. J Electrocardiol. 2019;57:70–74. doi: 10.1016/j.jelectrocard.2019.08.004. [DOI] [PubMed] [Google Scholar]
- 15.Menikdiwela M., Nguyen C., Shaw M. Deep learning on brain cortical thickness data for disease classification. Proceedings in 2018 Digital Image Computing: Techniques and Applications; Canberra, Australia; 2018. [DOI] [Google Scholar]
- 16.Basaia S., Agosta F., Wagner L., Canu E., Magnani G. Automated classification of Alzheimer's disease and mild cognitive impairment using a single MRI and deep neural networks. NeuroImage: Clinical. 2019;21 doi: 10.1016/j.nicl.2018.101645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Skourt B.A., Hassani A.E., Majda A. Lung CT image segmentation using deep neural networks. Procedia Comput Sci. 2018;127:109–113. doi: 10.1016/j.procs.2018.01.104. [DOI] [Google Scholar]
- 18.Gordienko Y., Gang P., Hui J., Zeng W., Kochura Y. Deep learning with lung segmentation and bone shadow exclusion techniques for chest x-ray analysis of lung cancer. Advances in Computer Science for Engineering and Education. 2018:638–647. doi: 10.1007/978-3-319-91008-6_63. [DOI] [Google Scholar]
- 19.Shen L., Margolies L.R., Rothstein J.H., Fluder E., McBride R., Sieh W. Deep learning to improve breast cancer detection on screening mammography. Sci Rep. 2019;9:12495. doi: 10.1038/s41598-019-48995-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hamed G., Marey M.A.ER., Amin S.ES., Tolba M.F. Proceedings of the International Conference on Artificial Intelligence and Computer Vision (AICV2020) 2020. Deep learning in breast cancer detection and classification. [DOI] [Google Scholar]
- 21.Kadampur M.A., Riyaee S.A. Skin cancer detection: applying a deep learning based model driven architecture in the cloud for classifying dermal cell images. Informatics in Medicine Unlocked. 2020;18 doi: 10.1016/j.imu.2019.100282. [DOI] [Google Scholar]
- 22.Nahata H., Singh S.P. Deep learning solutions for skin cancer detection and diagnosis. Machine Learning with Health Care Perspective. 2020:159–182. doi: 10.1007/978-3-030-40850-3_8. [DOI] [Google Scholar]
- 23.Toraman S. Kapsül ağları kullanılarak eeg sinyallerinin sınıflandırılması. Firat Universitesi Muhendislik Bilimleri Dergisi. 2020;32:203–209. doi: 10.35234/fumbd.661955. [DOI] [Google Scholar]
- 24.Jaiswal A.K., Tiwari P., Kumar S., Gupta D., Khanna A., Rodrigues J.J.P.C. Identifying pneumonia in chest X-rays: a deep learning approach. Measurement. 2019;145:511–518. doi: 10.1016/j.measurement.2019.05.076. [DOI] [Google Scholar]
- 25.Yadav S.S., Jadhav S.M. Deep convolutional neural network based medical image classification for disease diagnosis. J Big Data. 2019;6:113. doi: 10.1186/s40537-019-0276-2. [DOI] [Google Scholar]
- 26.Baltruschat I.M., Nickisch H., Grass M., Knopp T., Sallbach A. Comparison of deep learning approaches for multi-label chest X-ray classification. Sci Rep. 2019;9:6381. doi: 10.1038/s41598-019-42294-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Abiyev R.H., Ma’aitah M.K.S. Deep convolutional neural networks for chest diseases detection. J Healthc Eng. 2018 doi: 10.1155/2018/4168538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stephen O., Sain M., Maduh U.J., Jeong D. An efficient deep learning approach to pneumonia classification in healthcare. J Healthc Eng. 2019 doi: 10.1155/2019/4180949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chouhan V., Singh S.K., Khamparia A., Gupta D., Tiwari P. A novel transfer learning based approach for pneumonia detection in chest X-ray images. Applied Sciences. 2020;10(2):559. doi: 10.3390/app10020559. [DOI] [Google Scholar]
- 30.Apostolopoulos I.D., Mpesiana T.A. COVID-19: automatic detection from x-ray images utilizing transfer learning with convolutional neural networks. Physical and Engineering Sciences in Medicine. 2020;43:635–640. doi: 10.1007/s13246-020-00865-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hemdan, E.E., Shouman, M.A., and Karar, M.E.COVIDX-Net: a framework of deep learning classifiers to diagnose covid-19 in x-ray images, arXiv, 2003.11055, 2020.
- 32.Narin, A., Kaya, C., and Pamuk, Z.Automatic detection of coronavirus disease (COVID-19) using X-ray images and deep convolutional neural networks, arXiv, 2003.10849, 2020. [DOI] [PMC free article] [PubMed]
- 33.Wang, L., and Wong, A.COVID-Net: a tailored deep convolutional neural network design for detection of COVID-19 cases from chest radiography images, arXiv, 2003.09871, 2020. [DOI] [PMC free article] [PubMed]
- 34.Sethy, P.K., and Behera, S.K.Detection of coronavirus disease (COVID-19) based on deep features, Preprints, 2020. DOI: 10.20944/preprints202003.0300.v1 [DOI]
- 35.Afshar, P., Heidarian, S., Naderkhani, F., Oikonomou, A., Plaraniotis, K.N., and Mohammadi, A.COVID-CAPS: a capsule network-based framework for identification of COVID-19 cases from X-ray images, arXiv, 2004.02696, 2020. [DOI] [PMC free article] [PubMed]
- 36.Mobiny, A., Cicalese, P.A., Zare, S., Yuan, P., Abavisani, M., et al. [36], arXiv, 2004.07407, 2020.
- 37.Zheng, C., Deng, X., Fu, Q., Zhou, Q., Feng, J., et al. Deep learning-based detection for COVID-19 from chest CT using weak label, medRxiv, 2020. DOI: 10.1101/2020.03.12.20027185. [DOI]
- 38.Song, Y., Zheng, S., Li, L., Zhang, X., Zhang, X., et al. Deep learning enables accurate diagnosis of novel coronavirus (COVID-19) with CT images, medRxiv, 2020. DOI: 10.1101/2020.02.23.20026930. [DOI] [PMC free article] [PubMed]
- 39.Sabour, S., Frosst, N., and Hinton, G.E.Dynamic routing between capsules, arXiv, 1710.09829, 2017.
- 40.Cilasun M.H., Yalcin H. A deep learning approach to EEG based epilepsy seizure determination. Proceedings of the 24th Signal Processing and Communication Application Conference; Zonguldak, Turkey; 2016. [DOI] [Google Scholar]
- 41.Cohen, J.P.COVID-19 Image Data Collection, https://github.com/ieee8023/COVID-chestxray-dataset. [Accessed 2 June 2020].
- 42.Wang X., Peng Y., Lu L., Lu Z., Bagheri M., Summers R.M. Proceedings of Conference on Computer Vision and Pattern Recognition. 2017. ChestX-ray8: hospital-scale chest X-ray database and benchmarks on weakly-supervised classification and localization of common thorax diseases. [DOI] [Google Scholar]
- 43.Dombetzki L.A. An overview over capsule networks. Network Architectures and Services. 2018 [Google Scholar]
- 44.Sezer A., Sezer H.B. Capsule network-based classification of rotator cuff pathologies from MRI. Computers and Electrical Engineering. 2019;80 doi: 10.1016/j.compeleceng.2019.106480. [DOI] [Google Scholar]
- 45.Lukic V., Brüggen M., Mingo B., Croston J.H., Kasieczka G., Best P.N. Morphological classification of radio galaxies- capsule networks versus CNN. Mon Not R Astron Soc. 2019;487(2):1729–1744. doi: 10.1093/mnras/stz1289. [DOI] [Google Scholar]
- 46.Beser F., Kizrak M.A., Bolat B., Yildirim T. Recognition of sign language using capsule networks. Proceedings of the 26th Signal Processing and Communications Applications Conference; Izmir, Turkey; 2018. [DOI] [Google Scholar]
- 47.Xu Z., Lu W., Zhang Q., Yeung Y., Chen X. Gait recognition based on capsule network. J Vis Commun Image Represent. 2019;59:159–167. doi: 10.1016/j.jvcir.2019.01.023. [DOI] [Google Scholar]
- 48.Zhang X.Q., Zhao S.G. Cervical image classification based on image segmentation preprocessing and a CapsNet network model. Int J Imaging Syst Technol. 2018;29(1):19–28. doi: 10.1002/ima.22291. [DOI] [Google Scholar]
- 49.Qiao K., Zhang C., Wang L., Chen J., Zeng L. Accurate reconstruction of image stimuli from human functional magnetic resonance imaging based on the decoding model with capsule network architecture. Front Neuroinform. 2018;12 doi: 10.3389/fninf.2018.00062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mukhometzianov, R., and Carrillo, J.CapsNet comparative performance evaluation for image classification, arXiv, 1805.11195, 2018.
- 51.Sanchez A.J., Albarqouni S., Mateus D. Capsule networks against medical imaging data challenges. Lecture Notes in Computer Science. 2018:150–160. doi: 10.1007/978-3-030-01364-6_17. [DOI] [Google Scholar]
- 52.Ozturk T., Talo M., Yildirim E.A., Baloglu U.B., Yildirim O., Acharya R. Automated detection of COVID-19 cases using deep neural networks with X-ray images. Comput. Biol. Med. 2020:121. doi: 10.1016/j.compbiomed.2020.103792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mobiny A., Nguyen H.V. Fast CapsNet for lung cancer screening. Lecture Notes in Computer Science. 2018:741–749. doi: 10.1007/978-3-030-00934-2_82. [DOI] [Google Scholar]
- 54.Zhou, B., Khosla, A., Lapedriza, A., Oliva, A., and Torralba, A.Learning deep features for discriminative localization, arXiv, 1512.04150, 2015.
- 55.Yang W., Huang H., Zhang Z., Chen X., Huang K., Zhang S. Towards rich feature discovery with class activation maps augmentation for person re-identification. Proceedings of Conference on Computer Vision and Pattern Recognition; Long Beach, USA; 2019. [DOI] [Google Scholar]
- 56.Payer C., Stern D., Bischof H., Urschler M. Integrating spatial configuration into heatmap regression based CNNs for landmark localization. Med Image Anal. 2019;54:207–219. doi: 10.1016/j.media.2019.03.00. [DOI] [PubMed] [Google Scholar]
- 57.Togacar M., Ergen B., Comert Z. COVID-19 detection using deep learning models to exploit Social Mimic Optimization and structured chest X-ray images using fuzzy color and stacking approaches. Comput. Biol. Med. 2020:121. doi: 10.1016/j.compbiomed.2020.103805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang, S., Kang, B., Ma, J., Zeng, X., Xiao, M., et al. A deep learning algorithm using CT images to screen for Corona Virus 1 Disease(COVID-19), medRix, 2020. 10.1101/2020.02.14.20023028. [DOI] [PMC free article] [PubMed]
- 59.Xu, X., Jiang, X., Ma, C., Du, P., Li, X., et al.Deep learning system to screen coronavirus disease 2019 pneumonia, arXiv, 2002.09334, 2020. [DOI] [PMC free article] [PubMed]
- 60.COVID-19 X-rays. Available online: https://www.kaggle.com/andrewmvd/convid19-x-rays. [Accessed 6 June 2020].
- 61.Kermany D.S., Goldbaum M., Cai W., Valentim C.C.S., Liang H. Identifying medical diagnoses and treatable diseases by image-based deep learning. Cell. 2018;172(5):1122–1131. doi: 10.1016/j.cell.2018.02.010. [DOI] [PubMed] [Google Scholar]
- 62.Chest X-Ray Images. Available online: https://www.kaggle.com/paultimothymooney/chest-xray-pneumonia. [Accessed 6 June 2020].
- 63.Renmin Hospital of Wuhan University. [Accessed 15 June 2020].
- 64.The Third Affiliated Hospital and Sun Yat-Sen Memorial Hospital. [Accessed 15 June 2020].
- 65.The Sun Yat-Sen University in Guangzhou. [Accessed 15 June 2020].
- 66.Xi'an Jiaotong University First Affiliated Hospital. [Accessed 15 June 2020].
- 67.Nanchang University First Hospital. [Accessed 15 June 2020].
- 68.Xi'an No.8 Hospital of Xi'anMedical College. [Accessed 15 June 2020].
- 69.Union Hospital, Tongji Medical College, Huazhong University of Science and Technology. [Accessed 15 June 2020].
- 70.COVID-19 Radiography Database. Available online: https://www.kaggle.com/tawsifurrahman/covid19-radiography-database. [Accessed 16 June 2020].
- 71.Panwar H., Gupta P.K., Siddiqui M.K., Menendez R.M., Singh V. Application of deep learning for fast detection of COVID-19 in X-Rays using nCOVnet. Chaos, Solitons and Fractals. 2020;138 doi: 10.1016/j.chaos.2020.109944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Panahi S., Aram Z., Jafari S., Jun Ç.M., Sprott J.C. Modeling of epilepsy based on chaotic artificial neural network Chaos. Solitons and Fractals. 2017;105:150–156. doi: 10.1016/j.chaos.2017.10.028. [DOI] [Google Scholar]