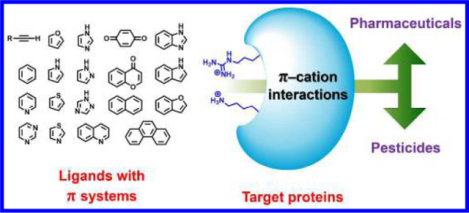
Abstract
The π-cation interaction that differs from the cation-π interaction is a valuable concept in molecular design of pharmaceuticals and pesticides. In this Perspective we present an up-to-date review (from 1995 to 2017) on bioactive molecules involving π-cation interactions with the recognition site, and categorize into systems of inhibitor-enzyme, ligand-receptor, ligand-transporter, and hapten-antibody. The concept of π-cation interactions offers use of π systems in a small molecule to enhance the binding affinity, specificity, selectivity, lipophilicity, bioavailability, and metabolic stability, which are physiochemical features desired for drugs and pesticides.
Keywords: π-cation interaction, π-cation bond, molecular recognition, molecular design, pharmaceutical, pesticide, drug design
Graphical Abstract
Introduction
Molecular recognition in biological systems relies on specific noncovalent interactions between partner molecules, including electrostatic interactions (e.g., salt bridge, hydrogen bond, and halogen bond), van der Waals interactions (e.g., dipole-dipole, dipole-induced dipole, and London dispersion forces), π-effects (cation-π, anion-π, polar-π, and π stacking) and hydrophobic effects. In particular, a positive charged molecule interacts with an electron-rich π system to form a strong π-effect termed the cation-π interaction or cation-π bond.1, 2 Such an interaction is influenced by the nature of the cation and π systems, binding geometry, and solvation effects. Geometric criteria typically require a distance of ≤ 6 Å and an angle 60° ≤ θ ≤ 90° between the π system and the cation center (Figure 1).1, 2 Remarkably, the cation-π interaction comprises a substantial electrostatic characteristics, and the bonding strength is comparable to salt bridge and hydrogen bond in aqueous solutions and at physiological conditions.3 Recent evidence indicates that the cation-π interaction is ubiquitous in nature and considerably counts on the ligand binding, which has brought attention to chemists, biologists, and material scientists.4, 5
Figure 1.
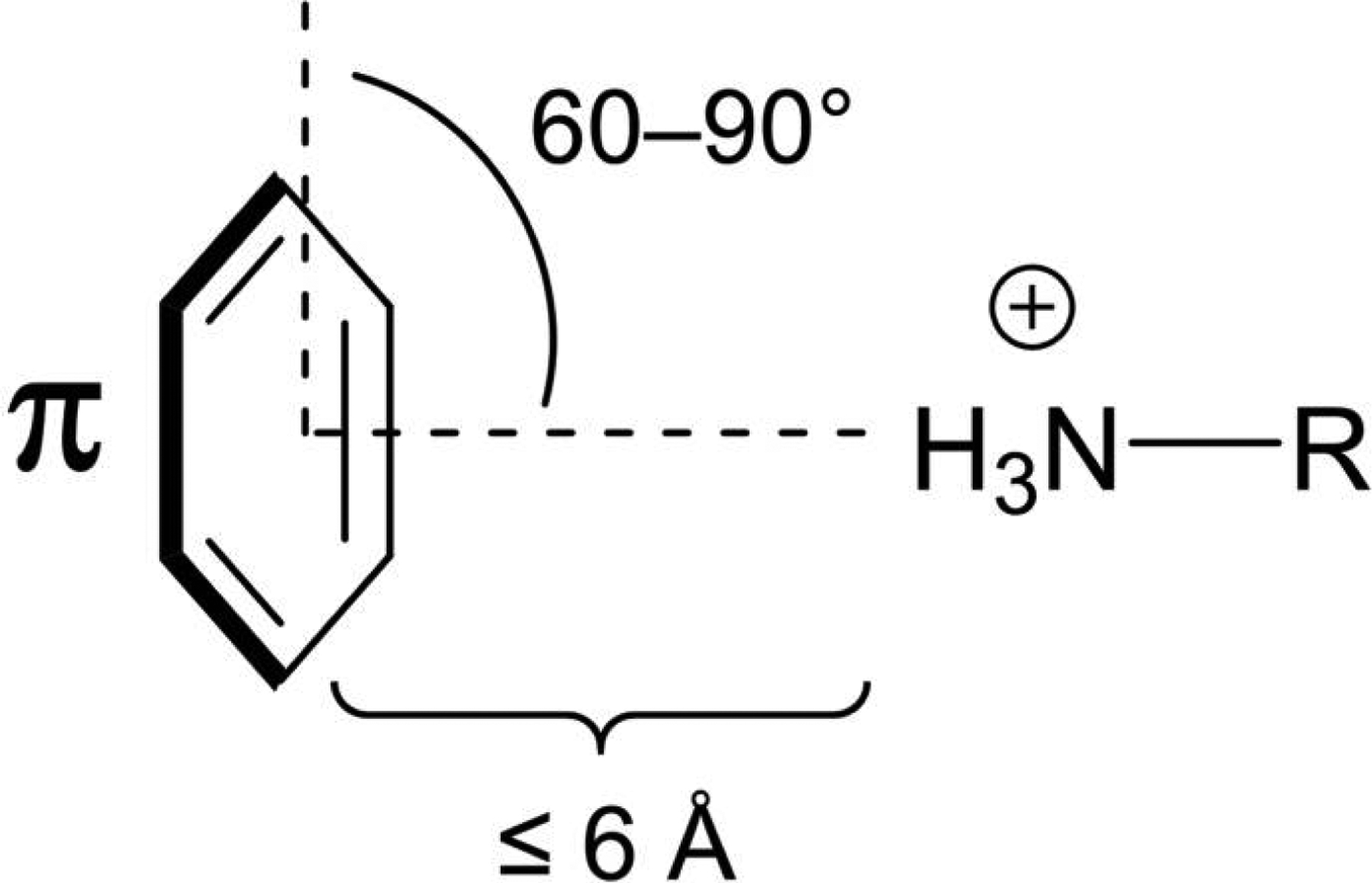
Basic geometric criteria for the π-cation interaction as exemplified with benzene and an ammonium cation.
Conceptually, there are two types of interactions between the π system and cationic partners depending on which one is the ligand or the ligand recognition site on a protein. In this context, cation-π interactions refer to a positive charged ligand interacting with aromatic amino acid residues (e.g., Phe, Tyr, or Trp) on target proteins. Such phenomenon has been extensively studied in the literature, as exemplified in classic cases that acetylcholine (ACh), nicotine, and muscarine with the quaternary ammonium specifically interact with Tyr and Trp residues of the mammalian acetylcholine receptors (AChRs)6–8 (Figure 2A), and neonicotinoids selectively bind to the insect nAChR.9, 10 Readers with interest are directed to the review articles by Dougherty et al. on the cation-π interaction11, 12 and by Casida et al. on applications in the insecticide discovery.13, 14 On the other hand, π-cation interactions, which were first recognized in a hapten-antibody system,15, 16 refer to a small-molecule ligand with π systems interacting with cationic amino acid residues (e.g., protonated Arg or Lys) in target proteins (Figure 2B). Applications of the π-cation interaction concept in design of bioactive molecules, however, have not been fully exploited.
Figure 2.
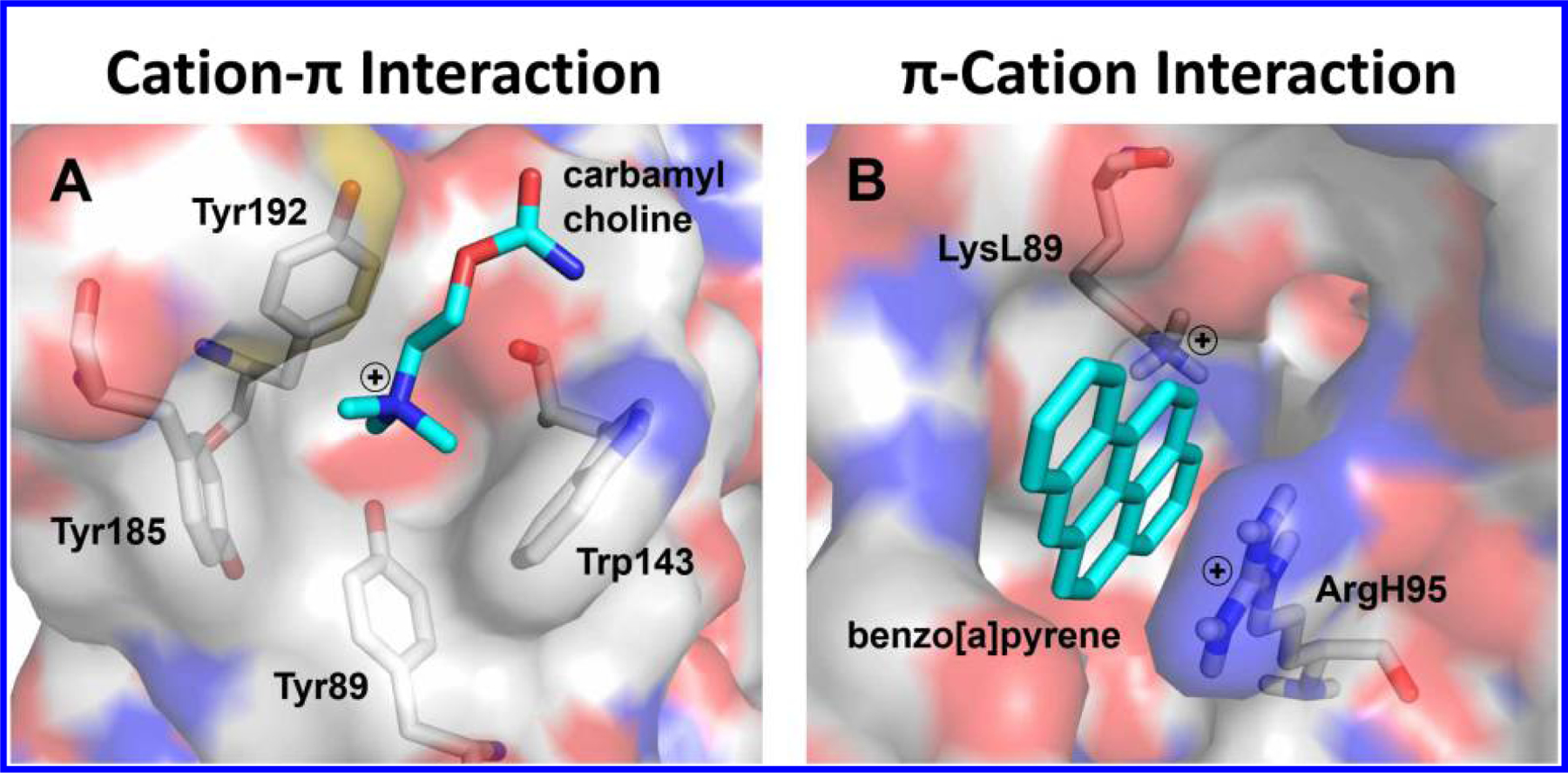
Comparison of the cation-π and π-cation interactions in ligand-protein recognition. (A) Carbamylcholine binding to acetylcholine binding protein. Model from PDB code 1UV6 (ref. 7). (B) Benzo[a]pyrene binding to monoclonal antibody 4D5. Model from reported data (ref. 16).
This Perspective therefore confines the scope on π-cation interactions and highlights the past development of bioactive small molecules that involve such interactions for ligand-protein recognition, primarily focusing on applications in analytical chemistry, biochemistry, medicinal chemistry, and pesticide chemistry. Some cases from our research group centering on inhibitor-enzyme and hapten-antibody interactions are elaborated. The π-cation interaction regarding protein folding, protein-protein interactions, and ligand-metal cofactor interactions is out of scope of this paper. A literature search was performed with the SciFinder database to cover a period from January 1995 to December 2017. We present our outlooks on potential applications of the π-cation interaction and bring attentions to use it as a tool in molecular design of pharmaceuticals and pesticides. The paper represents the first comprehensive review of the relevant literature on the topic of “π-cation interactions” in the past 22 years.
Inhibitor-Enzyme Interactions
Cytoplasmic enzymes without complex protein-protein or protein-membrane interactions are widely studied on the ligand-protein recognition in the literature. Glycogen synthase kinase-3β (GSK-3β), a serine/threonine protein kinase, is a therapeutic target for Alzheimer’s disease, dipolar disorder, type-2 diabetes, inflammation, and many metabolic diseases. Co-crystallization of GSK-3β with an inhibitor PF-04802367 revealed that the inhibitor binds at the ATP site of GSK-3β, where the triazole ring of PF-04802367 forms a strong π-cation interaction with Arg141 essential to its potency and selectivity.17 In silico screening and synthesis led to the pyrimidin-4-one based GSK-3β inhibitors, which make π-cation interactions with Arg141 at the ATP site of GSK-3β.18–21 Virtual screening and molecular docking also led to identification of GSK-3β inhibitors containing isoquinoline and quinazolinone scaffolds that favors π-cation interactions with Arg141 at the ATP site of GSK-3β.22 Benzothiazinones are allosteric modulators of GSK-3β showing π-cation interactions with Lys205.23 Aryl anilinomaleimide based inhibitors bind at the ATP site of GSK-3β and show π-cation interactions with Lys183.24 Most recently, we applied the concept of π-cation interactions in drug design, and discovered a series of highly selective and potent GSK-3β inhibitors based on the 6-C-glycosylflavone scaffold.25, 26 Comparative molecular modeling of GSK-3α/β isoforms indicated that the catechol B-ring of the 6-C-glycosylflavone inhibitors forms critical π-cation interactions with Lys183 at the substrate site of GSK-3β (Figure 3), but neither GSK-3α nor other 40 kinases tested, thereby achieving high GSK-3 isoform-selectivity and high kinase selectivity.26
Figure 3.
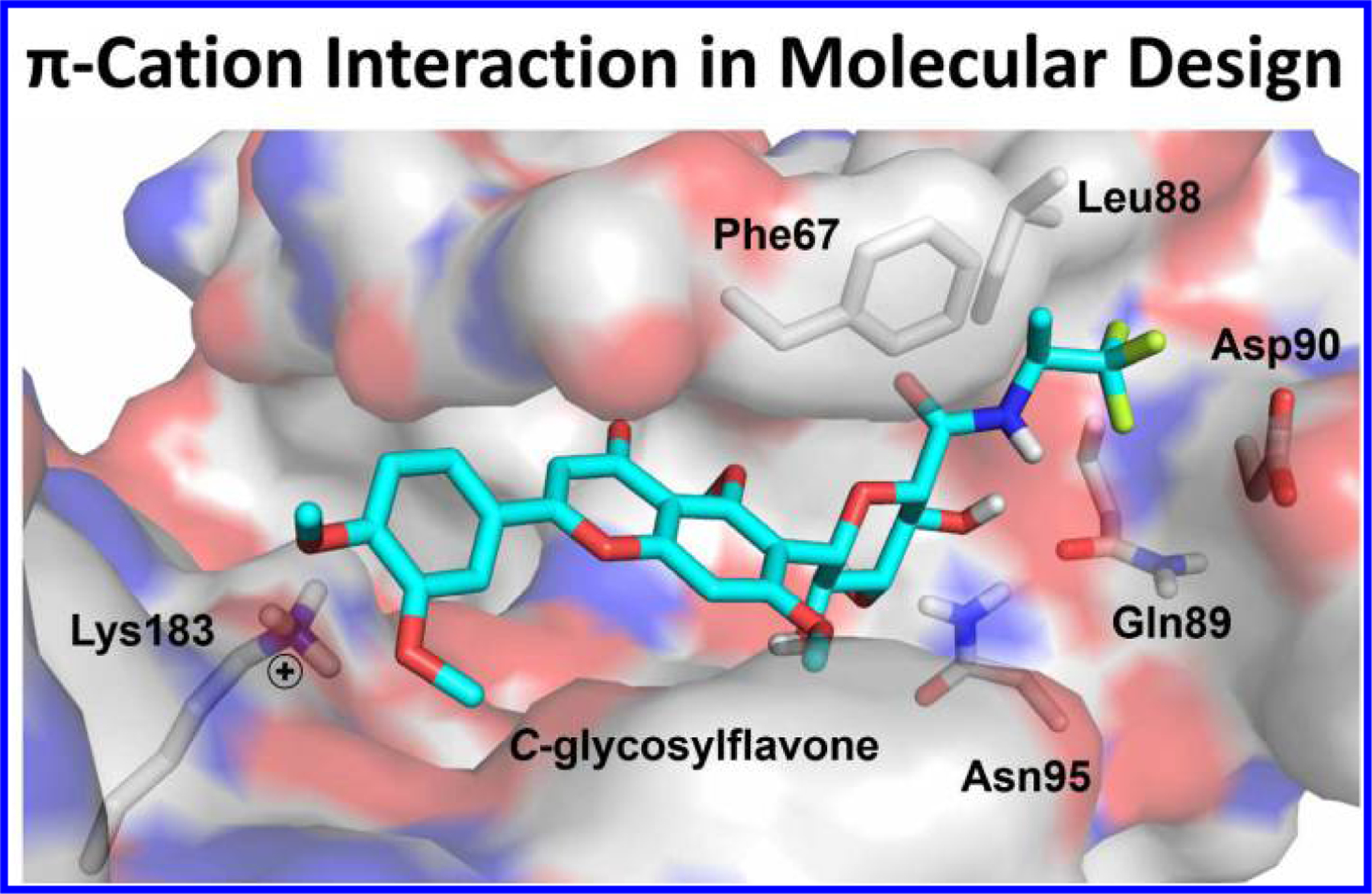
Application of π-cation interactions in computer-aided drug design of GSK-3β inhibitors. The new inhibitor containing a C-glycosylflavone scaffold binding to GSK-3β, and the catechol ring forms a π-cation interaction with Lys183. Model from reported data (ref. 26).
Enzymes relevant to oncology are promising targets for cancer chemotherapy. Rho-associated protein kinases (ROCK1 and ROCK2) play a key role in cancer metastasis. The aminoindane triazine analogues inhibit ROCK1 in various cancer cell lines where the aminoindane moiety forms strong π-cation interactions with Lys105 of ROCK1.27 The ROCK2 inhibitor CAY10576 showed π-cation interactions with Lys121.28 Mitogen-activated protein kinases (MAPKs) including p38 kinases and extracellular signal-regulated kinases (ERKs) are aberrant in many tumors. The imidazolinone inhibitors form strong π-cation interactions with Lys53 of p38 kinase and Arg65 of ERK2 kinase in molecular docking studies.29 The cyclin-dependent protein kinase-8 (CDK8) is involved in oncogenic control. The X-ray co-crystallographic analysis indicated that the aryl substituted thieno[2,3-c]pyridines are specific CDK8 inhibitors showing π-cation interactions with Arg356 at the ATP catalytic site.30 The phytochemical thymoquinone forms π-cation interactions with Lys540 of the polo-box domain of polo-like kinase 1 (Plk1) responsible for anticancer activity.31 Virtual screening of anticancer compounds also identified that many inhibitors of B-cell lymphoma extra-large proteins (Bcl-xL) show π-cation interactions between the aromatic rings of the inhibitors and the arginine residues (Arg100, Arg103, Arg132, Arg139 and Arg165) of Bcl-xL.32 Inhibition of Poly [ADP-ribose] polymerase-1 (PARP-1) shows efficacy improvement for many cytotoxic agents in cancer therapy. Hexahydrobenzonaphthyridinones are potential anticancer agents showing favorable π-cation interactions with Arg217 and Lys242 at the catalytic site of PARP-1.33 Ku86 enzyme and X-ray repair cross-complementing protein 4 (XRCC4) of the DNA-repair protein complex in tumors are burdens for cancer therapy. Lithospermic acid and salvianolic acid B, the traditional Chinese medicine (TCM) compounds, showed enzyme inhibitions, where the catechol ring of lithospermic acid interacts with Lys338 of Ku86 via π-cation forces,34 while the two catechol rings of salvianolic acid B form π-cation interactions with both Lys187 and Lys190 of XRCC4.35 Uroporphyrinogen decarboxylase (UROD) is implicated in resistance of cancer radiotherapy. The TCM compound scopolin containing the coumarin moiety forms π-cation interactions with Arg37 of UROD in molecular modeling.36 Molecular and quantum mechanics analyses on the 5-isoxazolylbenzimidazole analogues indicated that the binding affinity and selectivity are attributed to π-cation interactions between the aromatic moieties and Arg1173 of CREB-binding protein (CREBBP) bromodomains.37 CREBBP associated with leukemia. The CDC25 phosphatase inhibitor IRC-083864 as a potential anticancer agent shows π-cation interactions with Arg548 of CDC25.38
Many antiviral and antibacterial compounds have also shown π-cation interactions with their targets. The diketo-acid compound S-1360 is a potent inhibitor of the human immunodeficiency virus type 1 (HIV-1) integrase, where the triazole ring forms π-cation interactions with Lys159 at the active site.39 The tetrahydropyrimidine-2-one derivatives form π-cation interactions with Lys574 of glycoprotein 41 and inhibit HIV-1.40 Molecular docking of the anti-influenza drug oseltamivir (known as Tamiflu) derivatives suggested that the aniline π system of analogues interacts with Arg152, Arg225 and Arg293 on neuraminidase of the influenza A virus (H1N1), thereby increasing the binding affinity and antiviral activities in vitro.41, 42 Another class of antiviral compounds, naphthoquinones, inhibits H5N1 neuraminidase in part due to strong π-cation interactions with Arg224.43 The phytochemical hesperetin inhibits chikungunya virus (CHIKV) activity. In silico study on hesperetin suggested apparent π-cation interactions with Arg70 and Arg374 in CHIKV nsP1 and nsP4 enzymes, respectively.44 Virtual screening of the thiadiazole compounds also led to discovery of new CHIKV envelope glycoprotein inhibitors with specific π-cation interactions between Arg100 of E2 protein and Lys52 of E1 protein.45 Baicalein is a plant-derived flavone known to show anti-dengue virus (DENV) activity. In silico analysis showed that baicalein forms multiple π-cation interactions with Lys42 and Lys74 in DENV NS3/NS2B protein, with Lys401 in DENV NS5 protein, and with Arg2 in DENV E protein.46 Mechanistic investigations on the methicillin-resistant Staphylococcus aureus (MRSA) found that an antibiotic ceftaroline (known as Teflaro) binds at the allosteric site of penicillin binding protein 2A (PBP2A) and shows strong π-cation interactions with Lys273 and Lys316.47 X-ray co-crystallographic analysis indicated that new β-lactamase inhibitors containing the benzoate group form π-cation interactions with Arg340 of the Acinetobacter-derived cephalosporinase (ADC-7) and inhibit the multidrug resistant bacterium Acinetobacter baumannii.48
Utilization of π-cation interactions has demonstrated successes in molecular design. For example, structure-based drug design led to discovery of 2,8-diazaspiro[4.5]decan-1-one analogues as new prolyl hydroxylase domain-containing protein 2 (PHD2) inhibitors for anemia treatment.49 These analogues favor the π-cation interaction with Arg322 of PHD2 observed in the X-ray co-crystallographic complex.49 Virtual screening of TCM for anti-aging natural products led to identification that (S)-tryptophan-betaxanthin and rosmarinic acid are potent agonists of NAD-dependent deacetylase sirtuin-1 (Sirt1), where their aromatic moieties form π-cation interactions with Arg274 of Sirt1 in molecular simulation studies.50 γ-Aminobutyric acid aminotransferase (GABA-AT) regulates neurotransmission and is a therapeutic target in epileptic disorders. Aryl substituted 5,6-dihydropyrimidine-2(1H)-thiones are GABA-AT inhibitors and form π-cation interactions with Lys203A and Arg192A of GABA-AT.51 Quantitative structure-activity relationship (QSAR) studies on the quinoline-4-carboxamide analogues with cytochrome P450 2C9 (CYP2C9) enzyme demonstrated that the π-cation interaction of the naphthalene moiety with Arg108 increases the type II substrate binding affinity and prevents undesired drug-drug interactions and drug metabolism.52 The isoflavone puerarin is a tyrosinase inhibitor forming a π-cation interaction with Arg268.53 A synthetic aryl-copper complex [Cu(L1)(Phen)] shows anticancer activity and proteasome inhibition, where the phenanthroline ring of the complex forms π-cation interactions with Arg125 of 20S proteasome.54 Molecular dynamics study indicated that the phospholipase A2 (PLA2) inhibitors, PMS1062 analogues, form π-cation interactions with Arg7.55 Studies on aryl lactosamine derivatives binding to human galectin-3 indicated that the aromatic rings show favorable π-cation interactions with Arg144 and Arg186.56 Human macrophage migration inhibitory factor (MIF) is implicated in inflammatory and autoimmune diseases. The MIF inhibitor NVS-2 containing an anisyl group shows a π-cation interaction with Lys32 that leads to a striking increase of binding affinity.57 Studies on 5-aminosalicylic derivatives indicated their binding to myeloperoxidase (MPO), which involves π-cation interactions with Arg239.58 Succinate-coenzyme Q reductase (SQR) is a central enzyme in the respiratory chain and Krebs cycle, which is a target protein for pharmaceuticals and agricultural fungicides. Virtual screening and structure-based design led to discovery of many pyrazole-4-carboxamides as potent SQR inhibitors, where the pyrazole ring forms π-cation interactions with Arg46 of the C chain.59–62
Ligand-Receptor Interactions
Development of specific ligands to target protein receptors is continuously of great interest in drug discovery. Inspired by the classic cation-π interactions of ACh, nicotine, or muscarine with the AChRs, searching for novel aromatic ligands that specifically form π-cation interactions with cationic amino acid residues of protein receptors has been employed in the past years.
The N-methyl-D-aspartate (NMDA) receptor is a ligand-gated Ca2+ channel that mediates excitatory synaptic transmission in brains. It has been implicated in synaptic plasticity for motor and memory functions. Interestingly, a study on anesthetic aromatic compounds inhibiting the NMDA receptor indicated that the potency shows a higher linear correlation with π-cation electrostatic energy (R2 = 0.85) than hydrophobicity (R2 = 0.30) and molecular volume (R2 = 0.14).63 Such evidence substantiates the significance of π-cation interactions independent from hydrophobicity in molecular recognition, and implies potential in drug design. The type-3 serotonin receptor (5-HT3R) is a pentameric ligand-gated ion channel mediating neuronal depolarization and excitation in nervous systems and is a therapeutic target for anti-emetics. The granisetron analogues are 5-HT3R antagonists, in which the granisetron-bound crystal structure shows π-cation interactions with Arg55 of 5-HT binding protein.64 Potassium channel subfamily K member 2 (K2P2.1) is a dimeric voltage-gated ion channel essential in electrogenesis, ischemia and anesthesia. Co-crystallization of K2P2.1 with two selective activators, an N-aryl-sulfonamide and a thiophene-carboxamide, defined a π-cation interaction with Lys271 of K2P2.1 that controls binding selectivity.65
G protein-coupled receptors (GPCRs) are a class of transmembrane protein receptors involving tremendous cell signaling cascades, which are well-studied therapeutic targets for human diseases. Vasopressin V1A receptor (V1AR) is a GPCR that regulates platelet aggregation, glycogenolysis, and vascular contraction. Molecular modeling and QSAR study of 134 antagonists of V1AR containing benzoazepine, benzodiazepine, or 1-benzenesulfonyl-2,3-dihydro-1H-indole pharmacophores have demonstrated that π-cation interactions with Arg214 play an important role in the ligand binding to V1AR and thus contribute to the pharmacological effects.66 Cannabinoid receptors (CB1/2) belonging to the GPCR subfamily are therapeutic targets for the treatment of neuropathic pain, glaucoma, inflammation, and cardiovascular disease. New CB2 ligands with benzimidazole and benzothiophene moieties show π-cation interactions with Lys109 responsible for the CB2 selectivity.67
Receptor tyrosine kinases (RTKs) are cell-surface receptors regulating diverse cellular processes and their dysfunctions have implicated in oncogenesis and cancer progression. Tropomyosin receptor kinase A (TrkA) is a RTK aberrant in different types of cancer. Salicylhydrazone analogues show TrkA inhibition in cancer cells, where the benzylidenephenyl ring of these inhibitors forms π-cation interactions with Arg673 in docking studies.68 The biquinoline-pyridine hybrid compounds show potent inhibition against the epidermal growth factor receptor (EGFR, a RTK) and cytotoxicity against cancer cells, for which their aryl groups form π-cation interactions with Lys721 at the ATP catalytic site of EGFR.69
Protein receptors for protein tethering or protein-protein interactions are attractive targets for drug discovery. The urokinase receptor (uPAR), a cell membrane anchored protein, binds to protein partners and involves in plasminogen activation and cancer metastasis. Synthetic pyrrolinones inhibit uPAR against protein-protein interactions, where the π-cation interactions with Arg53 are critical as observed in the X-ray co-crystallographic structure.70 Rational design of pro-inflammatory cytokine interleukin-2 (IL-2) inhibitors to prevent the cytokine/receptor interaction resulted in the finding that 2-methyl-1H-indole derivatives show strong binding affinity to IL-2 via π-cation interactions with Arg38.71
Ligand-Transporter Interactions
The π-cation interaction has also been found between aromatic ligands and protein transporters. The cytotoxic flavonoid derivative 3d binds with human serum albumin (HSA), by which its π-cation interactions with Lys199, Arg218, and Arg222 in HSA are critical for drug deposition and delivery.72 The two antidiabetics, glipizide and gliclazide, bind HSA through π-cation interactions with Lys190 and Arg410, respectively.73 Some imidazolium and pyridinium based antimicrobials show π-cation interactions with Lys195 and Arg257 of HSA.74
Hapten-Antibody Interactions
In the past years, the π-cation interaction between aromatic haptens and antibodies has been applied in immunoassay development for environmental management, food safety and human health relevance. The ample utilities of such methods are applicable to biomedical and agricultural areas.
Polycyclic aromatic hydrocarbons (PAHs) are pollutants derived from incomplete combustion of organic substance such as biofuel, fossil fuel, wood, cigarette, and charcoal-broiled meat. Some PAHs such as benzo[a]pyrene (BaP) are carcinogenic, mutagenic, and immunosuppressive. In our early work to develop sensitive immunoassays for detection of PAHs, we reported that the monoclonal antibody 4D5 can recognize a wide range of PAHs including BaP.15 Mutagenesis and molecular modeling studies demonstrated that the aromatic π system of BaP forms strong interactions with LysL89 and ArgH95 at the binding site of the antibody 4D5 (Figure 2B).16 To the best of our knowledge, the study16 provided the first evidence with a mechanistic basis on π-cation interactions in hapten-antibody recognition, which stimulates active researches on PAHs immunoassay development and application in the following years. New PAH hapten derivatives, for example, were designed to produce monoclonal antibody B[a]P-13 with different immunospecificity.75 The fluorescence line-narrowing spectroscopy has been used for specific analyses of complex PAHs cross-reacted with monoclonal antibodies.76 Most recently, we engineered a ring-hydroxylating dioxygenase mutant for the catabolism of BaP by which the molecular mechanisms are involved in the π-cation and π-π stacking interactions.77
Polychlorinated biphenyls (PCBs) are ubiquitous, persistent and toxic organic pollutants in the environment. We found that PCBs specifically bind to the monoclonal antibody S2B1, where the PCB phenyl ring plays a pivotal role in π-cation interactions with ArgL46 at the active binding site.78–80
Future Perspectives
Taking advantage of π-cation interactions as a molecular design tool can improve many key features of bioactive chemicals: (1) binding affinity, (2) specificity and selectivity, (3) lipophilicity and bioavailability, and (4) metabolic stability. As elaborated above, the concept of π-cation interactions has been adopted by chemists in drug and pesticide discovery and environmental management (Table 1 and Figure 4). However, in the field of pesticides (including growth regulators), the community has not paid adequate attention as only four relevant papers on the development of SQR inhibitors59–62 were found in the past 22 years.
Table 1.
Reviewed π-Cation Interactions in Ligand-Protein Recognition from 1995 to 2017
| class | ligand | target protein | Functional relevance | ref. |
|---|---|---|---|---|
| inhibitor-enzyme | PF-04802367 | GSK-3β | Alzheimer | 17 |
| pyrimidin-4-ones | GSK-3β | Alzheimer | 18–21 | |
| imidazo-isoquinolines quinazolinones | GSK-3β | psychiatric disorder | 22 | |
| benzothiazinones | GSK-3β | diabetes | 23 | |
| aryl anilinomaleimides | GSK-3β | depression | 24 | |
| 6-C-glycosylflavones | GSK-3β | Alzheimer | 25, 26 | |
| aminoindane triazines | ROCK1 | cancer | 27 | |
| CAY10576 | ROCK2 | cancer | 28 | |
| imidazolinones | p38 kinase and ERK2 | cancer | 29 | |
| aryl thieno[2,3-c]pyridines | CDK8 | cancer | 30 | |
| thymoquinone | PBD-Plkl | cancer | 31 | |
| polyaromatic ligands | Bcl-xL | cancer | 32 | |
| hexahydrobenzonaphthyridinones | PARP-1 | cancer | 33 | |
| lithospermic acid | Ku86 | cancer | 34 | |
| salvianolic acid B | XRCC4 | cancer | 35 | |
| scopolin | UROD | cancer | 36 | |
| 5-isoxazolylbenzimidazoles | CREBBP | cancer | 37 | |
| IRC-083864 | CDC25 | cancer | 38 | |
| S-1360 | HIV-1 integrase | HIV/AIDS | 39 | |
| tetrahydropyrimidine-2(1H)-ones | glycoprotein 41 | HIV/AIDS | 40 | |
| oseltamivir analogues | neuraminidase | influenza virus | 41, 42 | |
| naphthoquinones | neuraminidase | influenza virus | 43 | |
| hesperetin | CHIKV proteins nsP1 and nsP4 | chikungunya virus | 44 | |
| thiadiazoles | CHIKV envelope glycoproteins E1 and E2 | chikungunya virus | 45 | |
| baicalein | DENV proteins NS3/NS2B, NS5 and E | dengue virus | 46 | |
| ceftaroline | PBP2A | Antimicrobial resistance | 47 | |
| S02030 analogues | ADC-7 | Antimicrobial resistance | 48 | |
| 2,8-diazaspiro[4.5]decan-1-ones | PHD2 | anemia | 49 | |
| (S)-tryptophan-betaxanthin rosmarinic acid | Sirtl | aging | 50 | |
| aryl 5,6-dihydropyrimidine-2(1H)-thiones | GABA-AT | epileptic disorder | 51 | |
| quinoline-4-carboxamides | CYP2C9 | drug-drug interaction, drug metabolism | 52 | |
| puerarin | tyrosinase | skin disorders | 53 | |
| aryl-copper complexes | 20S proteasome | cancer | 54 | |
| PMS1062 | PLA2 | inflammation | 55 | |
| aryl lactosamines | galectin-3 | fibrosis | 56 | |
| NVS-2 | MIF | inflammation | 57 | |
| 5-aminosalicylic derivatives | MPO | inflammation | 58 | |
| pyrazole-4-carboxamides | SQR | respiratory chain in mitochondria | 59–62 | |
| ligand-receptor | volatile benzene analogues | NMDA receptor | anesthetics | 63 |
| granisetrons | 5-HT3R | emetics | 64 | |
| N-aryl-sulfonamide thiophene-carboxamide | K2P2.1 | electrogenesis, ischemia and anesthesia | 65 | |
| benzoazepines | V1aR | platelet aggregation, | 66 | |
| benzodiazepines | glycogenolysis, | |||
| 1-benzenesulfonyl-2,3-dihydro-1H-indoles | vascular contraction | |||
| benzimidazoles benzothiophenes | CB1/2 receptor | neuropathic pain | 67 | |
| salicyl-hydrazones | TrkA | cancer | 68 | |
| biquinoline-pyridines | EGFR | cancer | 69 | |
| phenyl pyrrolinones | uPAR | cancer | 70 | |
| 2-methyl-1H-indoles | IL-2R | immune response | 71 | |
| ligand-transporter | flavonoid derivative 3d | HSA | drug delivery | 72 |
| glipizide and gliclazide | HSA | diabetes | 73 | |
| imidazolium/pyridinium analogues | HSA | antibiotics | 74 | |
| hapten-antibody | PAHs | mAb 4D5 | pollutant detection, cancer | 15, 16 |
| PAHs | mAb B[a]P-13 | pollutant detection, cancer | 75 | |
| PAHs | mAbs anti-PAH, 4D5, and 8E11 | pollutant detection, cancer | 76 | |
| PAHs | RHD | Pollutant bioremediation | 77 | |
| PCBs | mAb S2B1 | pollutant detection, cancer | 78–80 |
Figure 4.
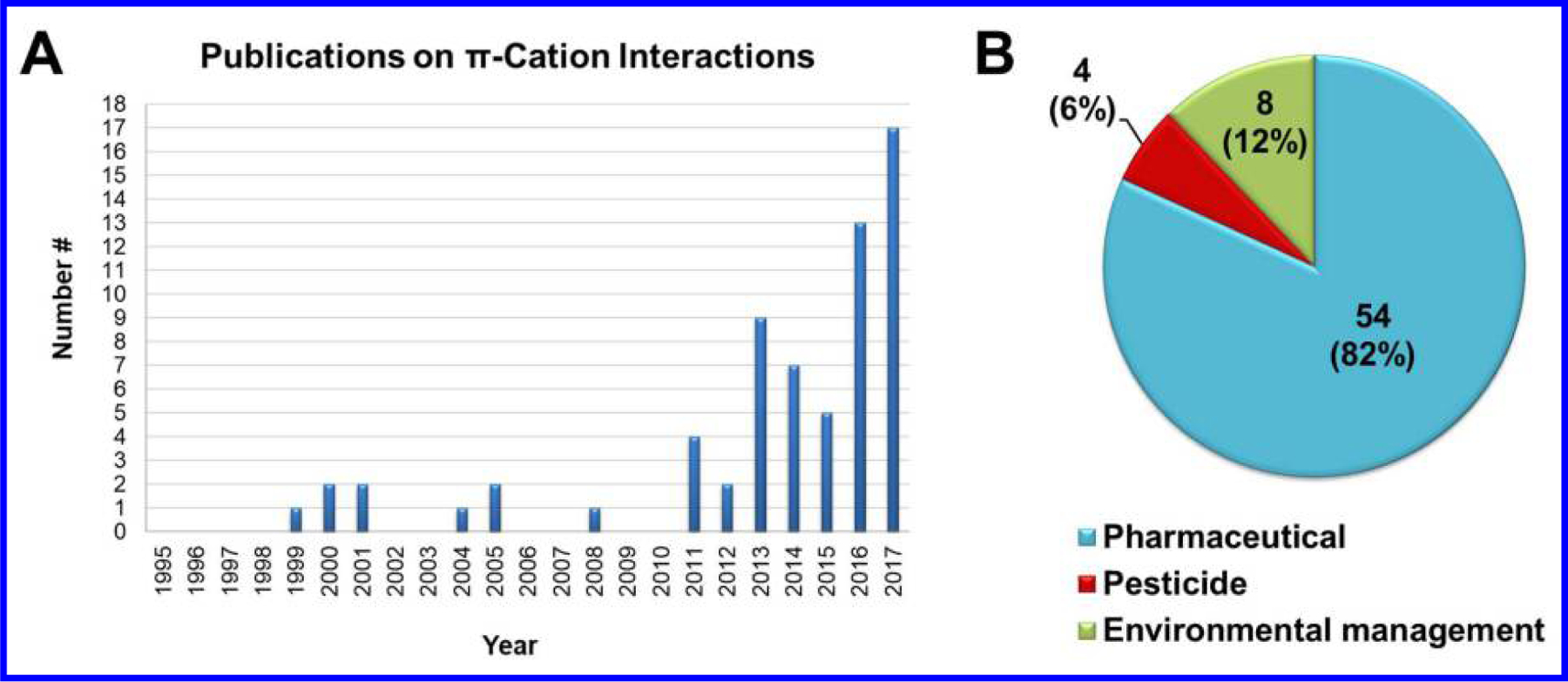
Publications on π-cation interactions in ligand-protein recognition from 1995 to 2017. (A) A chronology of publication counts. (B) Distribution of research focuses on pharmaceuticals, pesticides, and environmental management.
The π-cation interaction is universal in biology and would have an ample application in diverse areas. Pharmaceuticals and pesticides, although aiming at different markets, are developed in a similar process. In the early stages of R&D, both are involved in target protein identification, bioassay screening, hit-to-lead, and lead optimization. In terms of target proteins, humans, plants, insects, nematodes, fungi, and bacteria share a number of homologous proteins such as cytoplasmic enzymes, GPCRs, ion channels, signaling receptors, and carrier proteins.81, 82 Indeed, many drugs and pesticides show similar chemical structures and biological functions.83 The concept of designing bioactive molecules to target human proteins in neurological system, cellular respiratory chain, and fatty acid/amino acid biosynthesis is applicable to relevant target proteins in pest insects or weeds.
Modern drug or pesticide discovery begins with the screening of synthetic or natural chemical libraries by means of either in vitro bioassay or in silico approach against target proteins. Pre-selection of those chemicals with aromatic rings in consideration of π-cation interactions could plausibly increase screening hits and facilitate hit-to-lead process. Biopesticides are an emerging field of agricultural chemistry.84 Many natural products such as phytochemicals and TCM are known to their conjugated π systems in chemical structures and have shown successes in drug discovery.85 Prioritization of those aromatic natural products in screening might lead to promising biopesticides.
Regarding the process of molecular design and optimization, both pharmaceuticals and pesticides need to achieve high specificity and selectivity to minimize human and environmental toxicities and adverse effects. The illustrated cases here have underscored the advantage of π-cation interactions for the improvement of ligand-binding affinity and specificity. To design and optimize structures, the relative strength of π-cation interactions for the ligands can be estimated by simulation of the binding free energy. Moreover, lipophilicity is an important physicochemical parameter in chemical optimization.86 Application of π-cation interactions in drug/pesticide design not only offers the use of aromatic π systems in a small molecule for selective, specific and high affinity molecular recognition by the target protein, but also allows appropriate adjustment of ligand bioavailability (e.g., lipophilicity, pKa, and solubility), transport, distribution, and metabolism. Pesticides are often used for contact management such as seed treatment and aerial application, which requires effective chemical absorption into pests. A moderate lipophilicity is therefore desirable for pesticide absorption.
Overall, the π-cation interaction is common in molecular recognition. It is a rational and feasible concept for molecular design. In the past decade, there have been some applications of π-cation interactions, but the value is still underappreciated. We believe this Perspective would shed light upon continued studies in the field of pharmaceuticals and offer new insights to the agrochemical community.
Funding Sources
This work was supported in part by the NIH National Institute on Minority Health and Health Disparities grant 8G12MD007601 and by the USDA National Institute of Food and Agriculture Hatch project HAW5032-R.
Footnotes
The authors declare no competing financial interest.
REFERENCES
- (1).Dougherty DA, Cation-π interactions in chemistry and biology: A new view of benzene, Phe, Tyr, and Trp. Science 1996, 271, 163–168. [DOI] [PubMed] [Google Scholar]
- (2).Meyer EA; Castellano RK; Diederich F, Interactions with aromatic rings in chemical and biological recognition. Angew. Chem., Int. Ed 2003, 42, 1210–1250. [DOI] [PubMed] [Google Scholar]
- (3).Anslyn EV; Dougherty DA, Modern Physical Organic Chemistry. 1 ed.; University Science Books: Sausalito, CA, 2006. [Google Scholar]
- (4).Mahadevi AS; Sastry GN, Cation−π interaction: Its role and relevance in chemistry, biology, and material science. Chem. Rev 2013, 113, 2100–2138. [DOI] [PubMed] [Google Scholar]
- (5).Gebbie MA; Wei W; Schrader AM; Cristiani TR; Dobbs HA; Idso M; Chmelka BF; Waite JH; Israelachvili JN, Tuning underwater adhesion with cation–π interactions. Nature Chem. 2017, 9, 473–479. [DOI] [PubMed] [Google Scholar]
- (6).Zhong W; Gallivan JP; Zhang Y; Li L; Lester HA; Dougherty DA, From ab initio quantum mechanics to molecular neurobiology: A cation–π binding site in the nicotinic receptor. Proc. Natl. Acad. Sci. U.S.A 1998, 95, 12088–12093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Celie PHN; van Rossum-Fikkert SE; van Dijk WJ; Brejc K; Smit AB; Sixma TK, Nicotine and carbamylcholine binding to nicotinic acetylcholine receptors as studied in AChBP crystal structures. Neuron 2004, 41, 907–914. [DOI] [PubMed] [Google Scholar]
- (8).Xiu X; Puskar NL; Shanata JAP; Lester HA; Dougherty DA, Nicotine binding to brain receptors requires a strong cation–π interaction. Nature 2009, 458, 534–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Tomizawa M; Zhang N; Durkin KA; Olmstead MM; Casida JE, The neonicotinoid electronegative pharmacophore plays the crucial role in the high affinity and selectivity for the Drosophila nicotinic receptor: An anomaly for the nicotinoid cation−π interaction model. Biochemistry 2003, 42, 7819–7827. [DOI] [PubMed] [Google Scholar]
- (10).Tomizawa M; Talley TT; Maltby D; Durkin KA; Medzihradszky KF; Burlingame AL; Taylor P; Casida JE, Mapping the elusive neonicotinoid binding site. Proc. Natl. Acad. Sci. U.S.A 2007, 104, 9075–9080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Zacharias N; Dougherty DA, Cation–π interactions in ligand recognition and catalysis. Trends Pharmacol. Sci 2002, 23, 281–287. [DOI] [PubMed] [Google Scholar]
- (12).Dougherty DA, The Cation−π interaction. Acc. Chem. Res 2013, 46, 885–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Tomizawa M; Casida JE, Neonicotinoid insecticide toxicology: Mechanisms of selective action. Annu. Rev. Pharmacol. Toxicol 2005, 45, 247–268. [DOI] [PubMed] [Google Scholar]
- (14).Casida JE; Durkin KA, Neuroactive insecticides: Targets, selectivity, resistance, and secondary effects. Annu. Rev. Entomol 2013, 58, 99–117. [DOI] [PubMed] [Google Scholar]
- (15).Li K; Chen R; Zhao B; Liu M; Karu AE; Roberts VA; Li QX, Monoclonal antibody-based ELISAs for part-per-billion determination of polycyclic aromatic hydrocarbons: Effects of haptens and formats on sensitivity and specificity. Anal. Chem 1999, 71, 302–309. [DOI] [PubMed] [Google Scholar]
- (16).Pellequer J-L; Zhao B; Kao H-I; Bell CW; Li K; Li QX; Karu AE; Roberts VA, Stabilization of bound polycyclic aromatic hydrocarbons by a π-cation interaction. J. Mol. Biol 2000, 302, 691–699. [DOI] [PubMed] [Google Scholar]
- (17).Liang SH; Chen JM; Normandin MD; Chang JS; Chang GC; Taylor CK; Trapa P; Plummer MS; Para KS; Conn EL; Lopresti-Morrow L; Lanyon LF; Cook JM; Richter KEG; Nolan CE; Schachter JB; Janat F; Che Y; Shanmugasundaram V; Lefker BA; Enerson BE; Livni E; Wang L; Guehl NJ; Patnaik D; Wagner FF; Perlis R; Holson EB; Haggarty SJ; El Fakhri G; Kurumbail RG; Vasdev N, Discovery of a highly selective glycogen synthase kinase-3 inhibitor (PF-04802367) that modulates tau phosphorylation in the brain: Translation for PET neuroimaging. Angew. Chem., Int. Ed 2016, 55, 9601–9605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Uehara F; Shoda A; Aritomo K; Fukunaga K; Watanabe K; Ando R; Shinoda M; Ueno H; Kubodera H; Sunada S; Saito K-I; Kaji T; Asano S; Eguchi J.-i.; Yuki S; Tanaka S; Yoneyama Y; Niwa T, 6-(4-Pyridyl)pyrimidin-4(3H)-ones as CNS penetrant glycogen synthase kinase-3β inhibitors. Bioorg. Med. Chem. Lett 2013, 23, 6928–6932. [DOI] [PubMed] [Google Scholar]
- (19).Fukunaga K; Uehara F; Aritomo K; Shoda A; Hiki S; Okuyama M; Usui Y; Watanabe K; Yamakoshi K; Kohara T; Hanano T; Tanaka H; Tsuchiya S; Sunada S; Saito K-I; Eguchi J.-i.; Yuki S; Asano S; Tanaka S; Mori A; Yamagami K; Baba H; Horikawa T; Fujimura M, 2-(2-Phenylmorpholin-4-yl)pyrimidin-4(3H)-ones; A new class of potent, selective and orally active glycogen synthase kinase-3β inhibitors. Bioorg. Med. Chem. Lett 2013, 23, 6933–6937. [DOI] [PubMed] [Google Scholar]
- (20).Usui Y; Uehara F; Hiki S; Watanabe K; Tanaka H; Shouda A; Yokoshima S; Aritomo K; Adachi T; Fukunaga K; Sunada S; Nabeno M; Saito K-I; Eguchi J.-i.; Yamagami K; Asano S; Tanaka S; Yuki S; Yoshii N; Fujimura M; Horikawa T, Discovery of novel 2-(3-phenylpiperazin-1-yl)-pyrimidin-4-ones as glycogen synthase kinase-3β inhibitors. Bioorg. Med. Chem. Lett 2017, 27, 3726–3732. [DOI] [PubMed] [Google Scholar]
- (21).Kohara T; Nakayama K; Watanabe K; Kusaka S.-i.; Sakai D; Tanaka H; Fukunaga K; Sunada S; Nabeno M; Saito K-I; Eguchi J.-i.; Mori A; Tanaka S; Bessho T; Takiguchi-Hayashi K; Horikawa T, Discovery of novel 2-(4-aryl-2-methylpiperazin-1-yl)-pyrimidin-4-ones as glycogen synthase kinase-3β inhibitors. Bioorg. Med. Chem. Lett 2017, 27, 3733–3738. [DOI] [PubMed] [Google Scholar]
- (22).Darshit BS; Balaji B; Rani P; Ramanathan M, Identification and in vitro evaluation of new leads as selective and competitive glycogen synthase kinase-3β inhibitors through ligand and structure based drug design. J. Mol. Graph. Model 2014, 53, 31–47. [DOI] [PubMed] [Google Scholar]
- (23).Zhang P; Li S; Gao Y; Lu W; Huang K; Ye D; Li X; Chu Y, Novel benzothiazinones (BTOs) as allosteric modulator or substrate competitive inhibitor of glycogen synthase kinase 3β (GSK-3β) with cellular activity of promoting glucose uptake. Bioorg. Med. Chem. Lett 2014, 24, 5639–5643. [DOI] [PubMed] [Google Scholar]
- (24).Tantray MA; Khan I; Hamid H; Alam MS; Dhulap A; Kalam A, Synthesis of aryl anilinomaleimide based derivatives as glycogen synthase kinase-3β inhibitors with potential role as antidepressant agents. New J. Chem 2016, 40, 6109–6119. [Google Scholar]
- (25).Liang Z; Zhang B; Su WW; Williams PG; Li QX, C-Glycosylflavones alleviate tau phosphorylation and amyloid neurotoxicity through GSK3β inhibition. ACS Chem. Neurosci 2016, 7, 912–923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Liang Z; Li QX, Discovery of selective, substrate-competitive, and passive membrane permeable glycogen synthase kinase-3β inhibitors: Synthesis, biological evaluation, and molecular modeling of new C-glycosylflavones. ACS Chem. Neurosci 2018, DOI: 10.1021/acschemneuro.8b00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Shen M; Zhou S; Li Y; Pan P; Zhang L; Hou T, Discovery and optimization of triazine derivatives as ROCK1 inhibitors: Molecular docking, molecular dynamics simulations and free energy calculations. Mol. Biosyst 2013, 9, 361–374. [DOI] [PubMed] [Google Scholar]
- (28).Wang P; Yang Y; Shao Q; Zhou W, Selective inhibition of ROCK kinase isoforms to promote neuroregeneration after brain surgery. Med. Chem. Res 2016, 25, 40–50. [Google Scholar]
- (29).Rao SN, Tracking binding modes of 1,2,4-trisubstituted imidazolinone P38 MAP kinase and ERK-2 inhibitors. J. Mol. Graph. Model 2017, 76, 161–171. [DOI] [PubMed] [Google Scholar]
- (30).Koehler MFT; Bergeron P; Blackwood EM; Bowman K; Clark KR; Firestein R; Kiefer JR; Maskos K; McCleland ML; Orren L; Salphati L; Schmidt S; Schneider EV; Wu J; Beresini MH, Development of a potent, specific CDK8 kinase inhibitor which phenocopies CDK8/19 knockout cells. ACS Med. Chem. Lett 2016, 7, 223–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Yin Z; Song Y; Rehse PH, Thymoquinone blocks pSer/pThr recognition by Plk1 polo-box domain as a phosphate mimic. ACS Chem. Biol 2013, 8, 303–308. [DOI] [PubMed] [Google Scholar]
- (32).Azam SS; Abro A; Tanvir F; Parvaiz N, Identification of unique binding site and molecular docking studies for structurally diverse Bcl-xL inhibitors. Med. Chem. Res 2014, 23, 3765–3783. [Google Scholar]
- (33).Halder AK; Saha A; Saha KD; Jha T, Stepwise development of structure–activity relationship of diverse PARP-1 inhibitors through comparative and validated in silico modeling techniques and molecular dynamics simulation. J. Biomol. Struct. Dyn 2015, 33, 1756–1779. [DOI] [PubMed] [Google Scholar]
- (34).Sun M-F; Chang T-T; Chang K-W; Huang H-J; Chen H-Y; Tsai F-J; Lin J-G; Chen CY-C, Blocking the DNA repair system by traditional Chinese medicine? J. Biomol. Struct. Dyn 2011, 28, 895–906. [DOI] [PubMed] [Google Scholar]
- (35).Sun M-F; Chen H-Y; Tsai F-J; Liu S-H; Chen C-Y; Chen CY-C, Search for novel remedies to augment radiation resistance of inhabitants of Fukushima and Chernobyl disasters: Identifying DNA repair protein XRCC4 inhibitors. J. Biomol. Struct. Dyn 2011, 29, 325–337. [DOI] [PubMed] [Google Scholar]
- (36).Tsou Y-A; Chen K-C; Lin H-C; Chang S-S; Chen CY-C, Uroporphyrinogen decarboxylase as a potential target for specific components of traditional Chinese medicine: A virtual screening and molecular dynamics study. PLoS ONE 2012, 7, e50087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Cortopassi WA; Kumar K; Paton RS, Cation–π interactions in CREBBP bromodomain inhibition: An electrostatic model for small-molecule binding affinity and selectivity. Org. Biomol. Chem 2016, 14, 10926–10938. [DOI] [PubMed] [Google Scholar]
- (38).Sarkis M; Miteva MA; Dasso Lang MC; Jaouen M; Sari M-A; Galcéra M-O; Ethève-Quelquejeu M; Garbay C; Bertho G; Braud E, Insights into the interaction of high potency inhibitor IRC-083864 with phosphatase CDC25. Proteins: Struct., Funct., Bioinf 2017, 85, 593–601. [DOI] [PubMed] [Google Scholar]
- (39).Huang M; Grant GH; Richards WG, Binding modes of diketo-acid inhibitors of HIV1 integrase: A comparative molecular dynamics simulation study. J. Mol. Graph. Model 2011, 29, 956–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Sepehri S; Gharagani S; Saghaie L; Aghasadeghi MR; Fassihi A, QSAR and docking studies of some 1,2,3,4-tetrahydropyrimidines: Evaluation of gp41 as possible target for anti-HIV-1 activity. Med. Chem. Res 2015, 24, 1707–1724. [Google Scholar]
- (41).Gema LR-S; Tolentino-Lopez LE; Martínez-Ramos F; Padilla-Martínez I; García-Machorro J; Correa-Basurto J, Targeting a cluster of arginine residues of neuraminidase to avoid oseltamivir resistance in influenza A (H1N1): A theoretical study. J. Mol. Model 2015, 21, 8. [DOI] [PubMed] [Google Scholar]
- (42).Neri-Bazán RM; García-Machorro J; Méndez-Luna D; Tolentino-López LE; Martínez-Ramos F; Padilla-Martínez II; Aguilar-Faisal L; Soriano-Ursúa MA; Trujillo-Ferrara JG; Fragoso-Vázquez MJ; Barrón BL; Correa-Basurto J, Design, in silico studies, synthesis and in vitro evaluation of oseltamivir derivatives as inhibitors of neuraminidase from influenza A virus H1N1. Eur. J. Med. Chem 2017, 128, 154–167. [DOI] [PubMed] [Google Scholar]
- (43).Sharma G; Vasanth Kumar S; Wahab HA, Molecular docking, synthesis, and biological evaluation of naphthoquinone as potential novel scaffold for H5N1 neuraminidase inhibition. J. Biomol. Struct. Dyn 2016, 1–10. [DOI] [PubMed] [Google Scholar]
- (44).Oo A; Hassandarvish P; Chin SP; Lee VS; Abu Bakar S; Zandi K, In silico study on anti-chikungunya virus activity of hesperetin. PeerJ 2016, 4, e2602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Rashad AA; Keller PA, Structure based design towards the identification of novel binding sites and inhibitors for the chikungunya virus envelope proteins. J. Mol. Graph. Model 2013, 44, 241–252. [DOI] [PubMed] [Google Scholar]
- (46).Hassandarvish P; Rothan HA; Rezaei S; Yusof R; Abubakar S; Zandi K, In silico study on baicalein and baicalin as inhibitors of dengue virus replication. RSC Advances 2016, 6, 31235–31247. [Google Scholar]
- (47).Rani N; Vijayakumar S; P.T.V L; Arunachalam A, Allosteric site-mediated active site inhibition of PBP2a using quercetin 3-O-rutinoside and its combination. J. Biomol. Struct. Dyn 2016, 34, 1778–1796. [DOI] [PubMed] [Google Scholar]
- (48).Caselli E; Romagnoli C; Powers RA; Taracila MA; Bouza AA; Swanson HC; Smolen KA; Fini F; Wallar BJ; Bonomo RA; Prati F, Inhibition of Acinetobacter-derived cephalosporinase: Exploring the carboxylate recognition site using novel β-lactamase inhibitors. ACS Infect. Dis 2017, DOI: 10.1021/acsinfecdis.7b00153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Deng G; Zhao B; Ma Y; Xu Q; Wang H; Yang L; Zhang Q; Guo TB; Zhang W; Jiao Y; Cai X; Zhang J; Liu H; Guan X; Lu H; Xiang J; Elliott JD; Lin X; Ren F, Novel complex crystal structure of prolyl hydroxylase domain-containing protein 2 (PHD2): 2,8-Diazaspiro[4.5]decan-1-ones as potent, orally bioavailable PHD2 inhibitors. Bioorg. Med. Chem 2013, 21, 6349–6358. [DOI] [PubMed] [Google Scholar]
- (50).Chen K-C; Jian Y-R; Sun M-F; Chang T-T; Lee C-C; Chen CY-C, Investigation of silent information regulator 1 (Sirt1) agonists from traditional Chinese medicine. J. Biomol. Struct. Dyn 2013, 31, 1207–1218. [DOI] [PubMed] [Google Scholar]
- (51).Sahu M; Siddiqui N; Iqbal R; Sharma V; Wakode S, Design, synthesis and evaluation of newer 5,6-dihydropyrimidine-2(1H)-thiones as GABA-AT inhibitors for anticonvulsant potential. Bioorg. Chem 2017, 74, 166–178. [DOI] [PubMed] [Google Scholar]
- (52).Peng C-C; Cape JL; Rushmore T; Crouch GJ; Jones JP, Cytochrome P450 2C9 type II binding studies on quinoline-4-carboxamide analogues. J. Med. Chem 2008, 51, 8000–8011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Zhang G; Guo X-H; Wang S-S; Li Y-Q; Li G-Z; Zhao W-J, Screening and identification of natural ligands of tyrosinase from Pueraria lobata Ohwi by a combination of ultrafiltration and LC-MS. Anal. Methods 2017, 9, 4858–4862. [Google Scholar]
- (54).Zuo J; Bi C; Fan Y; Buac D; Nardon C; Daniel KG; Dou QP, Cellular and computational studies of proteasome inhibition and apoptosis induction in human cancer cells by amino acid Schiff base–copper complexes. J. Inorg. Biochem 2013, 118, 83–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Plocki S; Aoun D; Ahamada-Himidi A; Tavarès-Camarinha F; Dong C-Z; Massicot F; Huet J; Adolphe-Pierre S; Chau F; Godfroid J-J; Gresh N; Ombetta JE; Heymans F, Molecular modeling, design, and synthesis of less lipophilic derivatives of 3-(4-tetradecyloxybenzyl)-4H-1,2,4-oxadiazol-5-one (PMS1062) specific for group II enzyme. Eur. J. Org. Chem 2005, 2005, 2747–2757. [Google Scholar]
- (56).Atmanene C; Ronin C; Téletchéa S; Gautier F-M; Djedaïni-Pilard F; Ciesielski F; Vivat V; Grandjean C, Biophysical and structural characterization of mono/di-arylated lactosamine derivatives interaction with human galectin-3. Biochem. Biophys. Res. Commun 2017, 489, 281–286. [DOI] [PubMed] [Google Scholar]
- (57).Cisneros JA; Robertson MJ; Valhondo M; Jorgensen WL, A fluorescence polarization assay for binding to macrophage migration inhibitory factor and crystal structures for complexes of two potent inhibitors. J. Am. Chem. Soc 2016, 138, 8630–8638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Cabrera Pérez LC; Gutiérrez Sánchez M; Mendieta Wejebe JE; Hernández Rgodríguez M; Fragoso Vázquez MJ; Salazar JR; Correa Basurto J; Padilla Martínez II; Rosales Hernández MC, Novel 5-aminosalicylic derivatives as anti-inflammatories and myeloperoxidase inhibitors evaluated in silico, in vitro and ex vivo. Arab. J. Chem 2017, DOI: 10.1016/j.arabjc.2016.12.026. [DOI] [Google Scholar]
- (59).Zhu X-L; Xiong L; Li H; Song X-Y; Liu J-J; Yang G-F, Computational and experimental insight into the molecular mechanism of carboxamide inhibitors of succinate-ubquinone oxidoreductase. ChemMedChem 2014, 9, 1512–1521. [DOI] [PubMed] [Google Scholar]
- (60).Xiong L; Zhu X-L; Shen Y-Q; Kandergama Wasala Mudiyanselage Wishwajith W; Li K; Yang G-F, Discovery of N-benzoxazol-5-yl-pyrazole-4-carboxamides as nanomolar SQR inhibitors. Eur. J. Med. Chem 2015, 95, 424–434. [DOI] [PubMed] [Google Scholar]
- (61).Xiong L; Zhu X-L; Gao H-W; Fu Y; Hu S-Q; Jiang L-N; Yang W-C; Yang G-F, Discovery of potent succinate-ubiquinone oxidoreductase inhibitors via pharmacophore-linked fragment virtual screening approach. J. Agric. Food Chem 2016, 64, 4830–4837. [DOI] [PubMed] [Google Scholar]
- (62).Xiong L; Li H; Jiang L-N; Ge J-M; Yang W-C; Zhu XL; Yang G-F, Structure-based discovery of potential fungicides as succinate ubiquinone oxidoreductase inhibitors. J. Agric. Food Chem 2017, 65, 1021–1029. [DOI] [PubMed] [Google Scholar]
- (63).Raines DE; Gioia F; Claycomb RJ; Stevens RJ, The N-methyl-D-aspartate receptor inhibitory potencies of aromatic inhaled drugs of abuse: Evidence for modulation by cation-π interactions. J. Pharmacol. Exp. Ther 2004, 311, 14–21. [DOI] [PubMed] [Google Scholar]
- (64).Kesters D; Thompson AJ; Brams M; van Elk R; Spurny R; Geitmann M; Villalgordo JM; Guskov A; Helena Danielson U; Lummis SCR; Smit AB; Ulens C, Structural basis of ligand recognition in 5-HT3 receptors. EMBO Rep. 2013, 14, 49–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).Lolicato M; Arrigoni C; Mori T; Sekioka Y; Bryant C; Clark KA; Minor DL Jr, K2P2.1 (TREK-1)–activator complexes reveal a cryptic selectivity filter binding site. Nature 2017, 547, 364–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (66).Contreras-Romo MC; Martínez-Archundia M; Deeb O; Ślusarz MJ; Ramírez-Salinas G; Garduño-Juárez R; Quintanar-Stephano A; Ramírez-Galicia G; Correa-Basurto J, Exploring the ligand recognition properties of the human vasopressin V1a receptor using QSAR and molecular modeling studies. Chem. Biol. Drug Des 2014, 83, 207–223. [DOI] [PubMed] [Google Scholar]
- (67).Romero-Parra J; Mella-Raipán J; Palmieri V; Allarà M; Torres MJ; Pessoa-Mahana H; Iturriaga-Vásquez P; Escobar R; Faúndez M; Di Marzo V; Pessoa-Mahana CD, Synthesis, binding assays, cytotoxic activity and docking studies of benzimidazole and benzothiophene derivatives with selective affinity for the CB2 cannabinoid receptor. Eur. J. Med. Chem 2016, 124, 17–35. [DOI] [PubMed] [Google Scholar]
- (68).Alam MS; Choi S-U; Lee D-U, Synthesis, anticancer, and docking studies of salicylhydrazone analogues: A novel series of small potent tropomyosin receptor kinase A inhibitors. Bioorg. Med. Chem 2017, 25, 389–396. [DOI] [PubMed] [Google Scholar]
- (69).Sangani CB; Makawana JA; Duan Y-T; Yin Y; Teraiya SB; Thumar NJ; Zhu H-L, Design, synthesis and molecular modeling of biquinoline–pyridine hybrids as a new class of potential EGFR and HER-2 kinase inhibitors. Bioorg. Med. Chem. Lett 2014, 24, 4472–4476. [DOI] [PubMed] [Google Scholar]
- (70).Liu D; Xu D; Liu M; Knabe WE; Yuan C; Zhou D; Huang M; Meroueh SO, Small molecules engage hot spots through cooperative binding to inhibit a tight protein–protein interaction. Biochemistry 2017, 56, 1768–1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (71).Leimbacher M; Zhang Y; Mannocci L; Stravs M; Geppert T; Scheuermann J; Schneider G; Neri D, Discovery of small-molecule interleukin-2 inhibitors from a DNA-encoded chemical library. Chem. Eur. J 2012, 18, 7729–7737. [DOI] [PubMed] [Google Scholar]
- (72).Wei J; Jin F; Wu Q; Jiang Y; Gao D; Liu H, Molecular interaction study of flavonoid derivative 3d with human serum albumin using multispectroscopic and molecular modeling approach. Talanta 2014, 126, 116–121. [DOI] [PubMed] [Google Scholar]
- (73).Challier C; Beassoni P; Boetsch C; García NA; Biasutti MA; Criado S, Interaction between human serum albumin and antidiabetic compounds and its influence on the O2(1Δg)-mediated degradation of the protein. J. Photochem. Photobiol. B Biol 2015, 142, 20–28. [DOI] [PubMed] [Google Scholar]
- (74).Trush MM; Semenyuta IV; Vdovenko SI; Rogalsky SP; Lobko EO; Metelytsia LO, Synthesis, spectroscopic and molecular docking studies of imidazolium and pyridinium based ionic liquids with HSA as potential antimicrobial agents. J. Mol. Struct 2017, 1137, 692–699. [Google Scholar]
- (75).Scharnweber T; Fisher M; Suchànek M; Knopp D; Niessner R, Monoclonal antibody to polycyclic aromatic hydrocarbons based on a new benzo[a]pyrene immunogen. Fresenius. J. Anal. Chem 2001, 371, 578–585. [DOI] [PubMed] [Google Scholar]
- (76).Lin C; Chinnappan R; Acharya K; Pellequer J-L; Jankowiak R, On stabilization of a neutral aromatic ligand by π–cation interactions in monoclonal antibodies. Biophys. Chem 2011, 154, 35–40. [DOI] [PubMed] [Google Scholar]
- (77).Fu B; Xu T; Cui Z; Ng HL; Wang K; Li J; Li QX, Mutation of phenylalanine-223 to leucine enhances transformation of benzo[a]pyrene by ring-hydroxylating dioxygenase of Sphingobium sp. FB3 by increasing accessibility of the catalytic site. J. Agric. Food Chem 2018, 66, 1206–1213. [DOI] [PubMed] [Google Scholar]
- (78).Chiu Y-W; Chen R; Li QX; Karu AE, Derivation and properties of recombinant Fab antibodies to coplanar polychlorinated biphenyls. J. Agric. Food Chem 2000, 48, 2614–2624. [DOI] [PubMed] [Google Scholar]
- (79).Chiu Y-W; Li QX; Karu AE, Selective binding of polychlorinated biphenyl congeners by a monoclonal antibody: Analysis by kinetic exclusion fluorescence immunoassay. Anal. Chem 2001, 73, 5477–5484. [DOI] [PubMed] [Google Scholar]
- (80).Pellequer J-L; Chen S.-w. W.; Keum Y.-s.; Karu AE; Li QX; Roberts VA, Structural basis for preferential binding of non-ortho-substituted polychlorinated biphenyls by the monoclonal antibody S2B1. J. Mol. Recognit 2005, 18, 282–294. [DOI] [PubMed] [Google Scholar]
- (81).Santucci A; Bernardini G; Braconi D; Petricci E; Manetti F, 4-Hydroxyphenylpyruvate dioxygenase and its inhibition in plants and animals: Small molecules as herbicides and agents for the treatment of human inherited diseases. J. Med. Chem 2017, 60, 4101–4125. [DOI] [PubMed] [Google Scholar]
- (82).Ndikuryayo F; Moosavi B; Yang W-C; Yang G-F, 4-Hydroxyphenylpyruvate dioxygenase inhibitors: From chemical biology to agrochemicals. J. Agric. Food Chem 2017, 65, 8523–8537. [DOI] [PubMed] [Google Scholar]
- (83).Swanton CJ; Mashhadi HR; Solomon KR; Afifi MM; Duke SO, Similarities between the discovery and regulation of pharmaceuticals and pesticides: In support of a better understanding of the risks and benefits of each. Pest Manag. Sci 2011, 67, 790–797. [DOI] [PubMed] [Google Scholar]
- (84).Seiber JN; Coats J; Duke SO; Gross AD, Biopesticides: State of the art and future opportunities. J. Agric. Food Chem 2014, 62, 11613–11619. [DOI] [PubMed] [Google Scholar]
- (85).Newman DJ; Cragg GM, Natural products as sources of new drugs from 1981 to 2014. J. Nat. Prod 2016, 79, 629–661. [DOI] [PubMed] [Google Scholar]
- (86).Freeman-Cook KD; Hoffman RL; Johnson TW, Lipophilic efficiency: The most important efficiency metric in medicinal chemistry. Future Med. Chem 2013, 5, 113–115. [DOI] [PubMed] [Google Scholar]


