Abstract
During chronic HIV-1 infection, regulatory CD4 T cells (Tregs) frequently represent the largest subpopulation of CD4 T cell subsets, implying relative resistant to HIV-1. When HIV-1 infection of CD4 T cells was explored in vitro and ex vivo from patient samples, Tregs possessed lower levels of HIV-1 DNA and RNA in comparison with conventional effector and memory CD4 T cells. Moreover, Tregs suppressed HIV-1 expression in other CD4 T cells in an in vitro co-culture system. This suppression was mediated in part via multiple inhibitory surface proteins expressed on Tregs. Antibody blockade of CTLA-4, PD-1, and GARP on Tregs resulted in increased HIV-1 DNA integration and mRNA expression in neighboring CD4 T cells. Moreover, antibody blockade of Tregs inhibitory proteins resulted in increased HIV-1 LTR transcription in co-cultured CD4 T cells. Thus, Tregs inhibit HIV-1 infection of other CD4 T cell subsets via interactions with inhibitory cell surface proteins.
Keywords: Regulatory CD4 T cells, HIV-1, HIV-1 integration, HIV-1 mRNA, CTLA-4, PD-1, GARP
1. Introduction
Regulatory CD4 T cells (Tregs) are essential for maintaining immune homeostasis, preventing autoimmunity, and regulating chronic inflammatory diseases (Campbell and Koch, 2011), and they are characterized by expression of the FoxP3 transcription factor. In the context of HIV-1 infection and AIDS, a significant volume of data has accumulated over the last decade about the role of Tregs, but there remains debate as to their help or hindrance during infection with HIV-1 (Whiteside, 2015). Some studies have concluded that Tregs facilitate control of AIDS development via inhibition of pathology associated with persistent immune activation which occurs during HIV-1 infection (Card et al., 2009; Chase et al., 2008; Lopez-Abente et al., 2016). By contrast, other data support the notion that Tregs restrict HIV-1-specific immune responses making viral eradication more difficult and resulting in persistently chronic infection (Legrand et al., 2006; Li et al., 2008). Therefore, there is not a simple answer to whether or not Tregs are beneficial or disadvantageous during individual cases of HIV-1 infection (Chevalier and Weiss, 2013; Phetsouphanh et al., 2014). In addition, CD4 Tregs function can be altered following direct HIV-1 infection (Tran et al., 2008). Understanding the role that Tregs play during HIV-1 infection, thus, remains a challenge (Imamichi and Lane, 2012).
Others have reported that Tregs were more susceptible to CXCR4 HIV-1 infections following polyclonal activation in vitro, but their definitions of Tregs by flow cytometry are not identical to our work and others (Moreno-Fernandez et al., 2009). In contrast, we previously reported that FoxP3-transduced primary human CD4 T cells function as Tregs in vitro and are partially resistant to HIV-1 infection by down-regulating HIV-1 LTR transcription via an NFAT-dependent pathway (Selliah et al., 2008). Moreover, FoxP3-expressing regulatory CD4 T cells also limited HIV-1 expression in neighboring non-Tregs CD4 T cells in co-culture. In the current study, we now demonstrate that Tregs themselves are relatively resistant to HIV-1 infection, and also suppress viral expression in adjacent CD4 T cells in a cell contact-dependent manner. Specifically, GARP (Glycoprotein A Repetitions Predominant), an important Tregs cell surface inhibitory protein, participates in suppression of HIV-1 expression in neighboring CD4 T cells. These results provide novel insights regarding alterable mechanisms involving the role of Tregs during HIV-1 infection.
2. Results/Discussion
2.1. Tregs express lower levels of HIV-1 than other effector and memory CD4 T cell subsets
It is well known that HIV-1 readily infects and is expressed in activated CD4 T cells, but resting CD4 T cells are largely resistant to HIV-1 infection. However, within a heterogeneous T cell population it is not entirely clear which CD4 T cell subsets are infected and how efficiently these subsets express HIV-1. To address this question and mimic natural infection, peripheral blood CD4 T cells were isolated from healthy blood donors and infected promptly in bulk with HIV-1 without any prior cell activation. The cells were maintained in culture for 5–7 days in the presence of moderate amounts of recombinant human IL-2 (30–50 U/ml) to maintain cell survival. The infected CD4 T cells were then phenotypically sorted by flow cytometry into Tregs – CD25highCD127low, conventional effector CD4 T cells (Tconvs) – CD25lowCD127high, memory CD4 T cells (Tmems) – CD25(−)CD45RO (+), and naïve CD4 T cells (Tnaives) – CD25(−)CD45RA(+) (Fig. 1A). Real-time RT-PCR confirmed that Tregs expressed the highest mRNA levels of the Tregs master transcription factor, FoxP3, with lower levels in CD4 Tconvs, Tmems, and Tnaives, in that decreasing order (Fig. 1B). Having sorted the CD4 T cell subsets, HIV-1 gag mRNA and total HIV-1 DNA levels were measured. As shown in Fig. 1B and C, both CD4 Tconvs and Tmems expressed the highest levels of HIV-1 mRNA, and contained the highest levels of HIV-1 DNA, but there were no statistically significant differences between these subsets. In contrast, Tregs had markedly lower HIV-1 mRNA and DNA levels. As expected, CD4 Tnaives contained viral mRNA and DNA, near the lower limits of detection (Fig. 1B and C). Furthermore, to address the kinetics of infection in the different CD4 T cell subsets, cells were immediately sorted from freshly isolated peripheral blood mononuclear cells by flow cytometry. The four CD4 T cells subsets were then separately infected with HIV-1 to observe potential changes of HIV-1 DNA levels in each subset. As shown in Fig. 1D, both Tconvs and Tmems showed the highest levels of HIV-1 DNA at 48 h and 96 h post infection, with similar levels detected at both time points. By comparison, Tregs and Tnaives had notably lower viral DNA levels but with similar levels detected at each time point for each respective subset. The relative levels of HIV-1 DNA in the different CD4 T cells infected in isolation (Fig. 1D) mirrored the results seen in the bulk infections (Fig. 1C). Moreover, there were no significant changes in HIV-1 DNA levels in the individual subsets noted at the 2 time points analyzed.
Fig. 1.
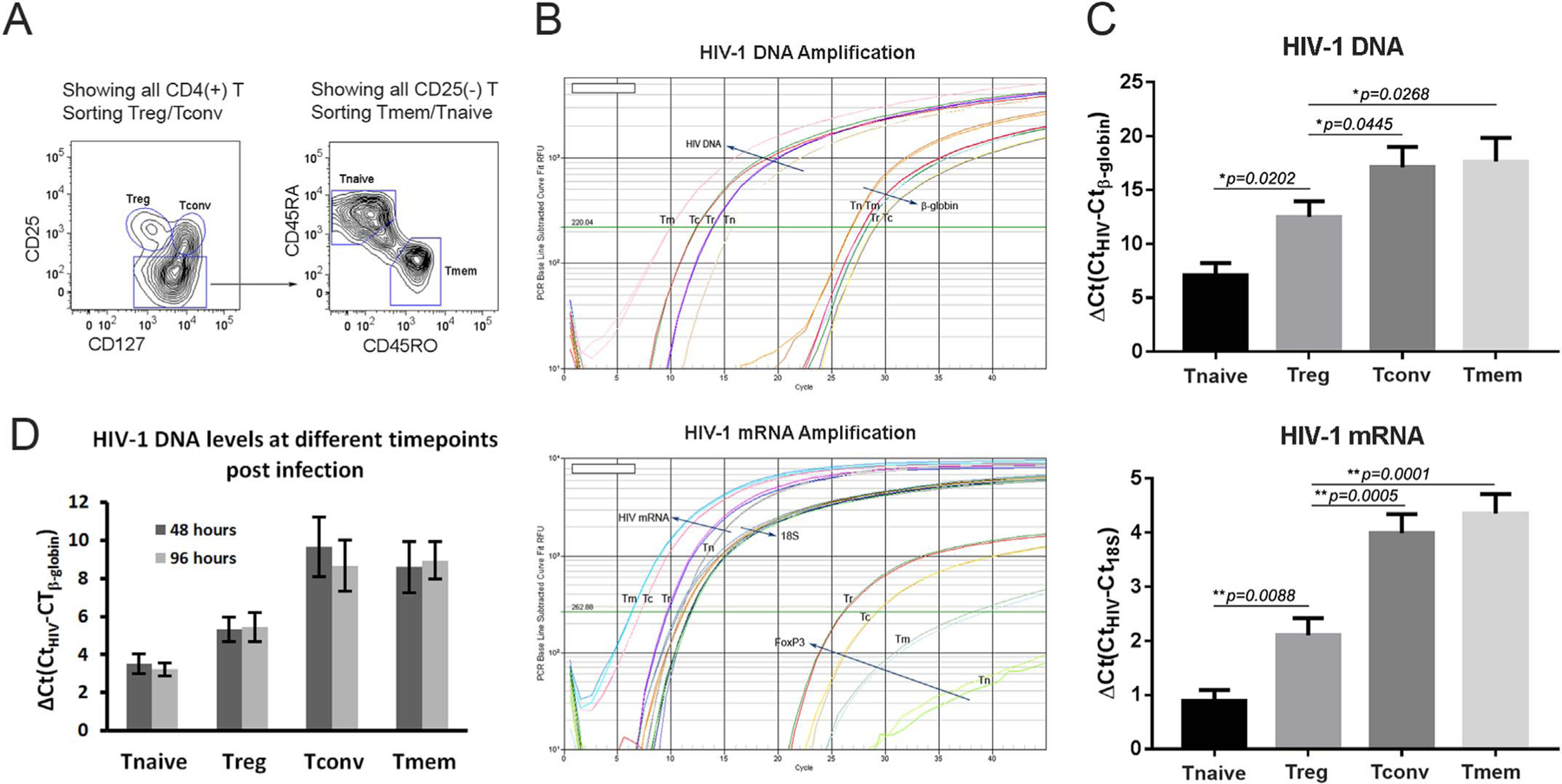
HIV-1 infection of CD4 T cells in vitro demonstrates lower levels of HIV-1 DNA and gag mRNA in Tregs compared to Tconvs and Tmems. (A) FLOW contour plot shows the cell sorting strategy. The in vitro bulk infected ((B) and (C)) or freshly isolated ((D)) CD4 T cells were stained and sorted into Tregs (CD25high CD127low), Tconvs (CD25low CD127high), Tmems (CD25(−) CD45RO(+)), and Tnaives (CD25(−)CD45RA(+)) populations. (B) Semi-quantitative real-time PCR plots of HIV-1 viral DNA (top) and mRNA (bottom) levels, relative to control β-globin DNA and 18 S RNA, for Tmems (Tm), Tconvs (Tc), Tregs (Tr), and Tnaives (Tn) populations. (C) Bar graphs depict the mean results of viral DNA (top) and mRNA (bottom) levels from three independent experiments. Standard error bars are shown along with statistically significant comparisons between CD4 T cell populations. (D) Bar graph shows the viral DNA levels over two different time points as indicated post infection (n=3).
In order to confirm the results obtained in vitro, HIV-1 levels in CD4 T cell subsets from HIV-1 infected patient samples were examined immediately ex vivo. Patient A was acutely infected [not receiving antiretroviral therapy (ART)] with an HIV-1 viral load of 360,000 copies/ml and a CD4 T cell count of 254 cells/μl. Patient B was chronically HIV-1 infected and receiving ART with a viral load of 69,000 copies/ml and a CD4 T cell count of 652 cells/μl. The patients’ peripheral blood samples were independently sorted to obtain the various CD4 T cell subsets, and each subset was assessed for HIV-1 mRNA and DNA by Real-time RT-PCR and PCR, respectively. FoxP3 mRNA levels were used to help confirm the CD4 T cell subsets based on sorting by cell surface phenotypes. Both HIV-1 infected patient samples demonstrated that CD4 Tmems and Tconvs possessed the highest levels of HIV-1 DNA and mRNA (Fig. 2). Consistent with the results from the in vitro infection experiments, CD4 Tregs had notably lower levels of HIV-1 DNA and mRNA (Fig. 2). As expected, very low levels of HIV-1 DNA, and undetectable levels of HIV-1 mRNA, were present in the CD4 Tnaives subset (Fig. 2). Therefore, both the in vitro infection data and the ex vivo HIV-1 patient results support the notion that Tregs, despite expressing the requisite receptors for HIV-1, are less likely to be infected by HIV-1.
Fig. 2.
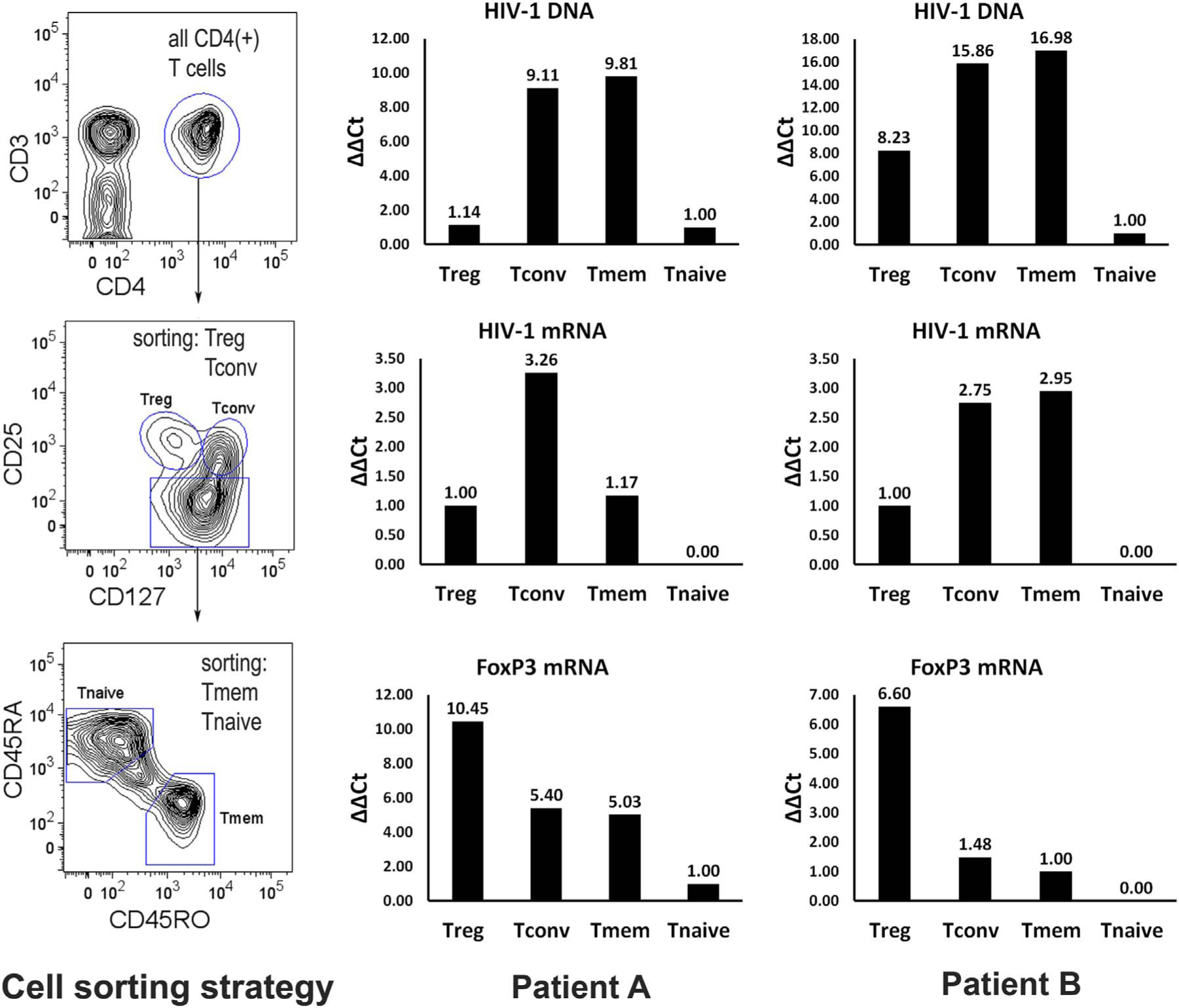
CD4 Tregs from HIV-1/AIDS patients express lower levels of HIV-1 DNA and viral mRNA than Tconvs and Tmems ex vivo. (Left panels) CD4 T cell populations were sorted from PBMC of newly infected (patient A) and chronically HIV-1 infected (patient B) individuals. Bar graphs depict HIV-1 total DNA (top), gag mRNA (middle), and host FoxP3 mRNA (bottom) for sorted Tregs (Tr), Tconvs (Tc), Tmems (Tm), and Tnaives (Tn) populations. Levels were measured in triplicate and the mean values are shown.
2.2. Tregs inhibit HIV-1 expression in other CD4 T cell subsets
Having confirmed our previous in vitro data that FoxP3-expressing CD4 T cells are relatively resistant to HIV-1 infection (Selliah et al., 2008), we wanted to clarify whether FoxP3-expressing CD4 T cells diminish HIV-1 expression within co-cultured neighboring CD4 T cell subsets (Selliah et al., 2008). For this purpose, freshly isolated peripheral blood CD4 T cells were first stimulated with PHA plus IL-2 for 16–24 h, then labeled with CFSE (5(6)-Carboxyfluorescein diacetate succinimidyl ester) and infected with the HIV-1 NL4–3 strain. The stimulation with PHA plus IL-2 was used to facilitate HIV-1 infection of freshly isolated CD4 T cells, and the CFSE labeling allowed for the identification of the responding cells from the co-cultured Tregs or Tconvs. The CFSE-labeled cells were cultured for 7 days after mixing with autologous CD4 Tregs at a 1:1 ratio. Similar studies were performed by mixing 1:1 with CD4 Tconvs as a control. As expected, the co-cultured CFSE-negative Tregs expressed notably lower p24 than the corresponding Tconvs (Fig. 3A and B). Moreover, responding CD4 T cells co-cultured with control Tconvs had the highest percentage of HIV-1 p24 expression. In comparison, responding CD4 T cells co-cultured with Tregs showed a significantly lower percentage of p24+ cells. These in vitro testing results are consistent with previously published data by others (Moreno-Fernandez et al., 2011) and confirm that Tregs repress HIV-1 expression in neighboring (co-cultured) CD4 T cells.
Fig. 3.

Tregs, but not Tconvs, decrease HIV-1 expression in co-cultured responder CD4 T cells. See Section 4 for details of co-culture and HIV-1 infection. PHA and IL-2 activated CD4 T cells (see Section 4) were infected with HIV-1 and co-cultured with Tregs or Tconvs. (A) Flow cytometry dot plots reveal the p24/gag protein levels of responder CD4 T cells (CFSE-labeled), as well as co-cultured (CFSE-negative) Tregs (above) or control Tconvs (below). (B) A bar graph depicts the mean values ± SEM (n=4), and statistically significant differences for the cell populations.
Because CFSE equally enters the daughter cells during cell division and can be used to monitor distinct generations of proliferating cells by dye dilution, we further analyzed HIV-1 p24 expression to explore if decreased infection levels correlated with decreased proliferation. As shown in Fig. S1 A, responding cells of each descendent generation co-cultured with Tregs had lower p24 expression than their counterparts co-cultured with Tconvs. Specifically, over 20% of 6th generation responding cells in the presence of Tconvs still expressed p24, whereas the corresponding cells co-cultured with Tregs had not yet reached the 6th generation. Thus, decreased HIV-1 expression is likely attributed to the ability of Tregs to suppress proliferation of CD4 T cells. Moreover, all offspring generations of cells of both groups demonstrated a tendency for decreased p24 expression over the consecutive generations. However, there was a sharper decline in p24 expression in responder cells co-cultured with Tregs (Fig. S1 A). This was quantified (MFI multiplied by percentage positive cells) with lower level HIV-1 infection, such that HIV-1 expression in G1 responding cells to Tregs decreased more rapidly than G1 responding cells to Tconvs (G1/G0 = 0.92×2061/2.15×2471 = 35.7% in responding cells to Tregs, versus, G1/G0 = 4.15×2277/5.85×2637 = 61.5% in responding cells to Tconvs (Fig. S1B)). Taken together, these data suggest that in addition to decreased proliferation leading to decreased HIV-1 infection, an additional inhibitory effect of Tregs contributes to lower levels of HIV-1 expression of co-cultured CD4 T cells.
2.3. Ex vivo expanded Tregs maintain their phenotype and function in vitro
To mechanistically explore how Tregs decrease HIV-1 expression in neighboring CD4 T cell subsets, a greater absolute number of Tregs were needed for experimentation. Since Tregs account for a very small percentage of human peripheral CD4 T cells, cell sorted Tregs were expanded with Dynabeads as in the Section 4 (along with Tconvs in parallel). Cell numbers increased by approximately 20–40-fold after 8–10 days of in vitro culture. To confirm the expanded cell phenotypes, T cell surface markers were examined by flow cytometry. As shown in Fig. 4A, the high expression of FoxP3 was maintained in expanded Tregs, along with Tregs-associated markers, including CD25, CTLA-4, PD-1, GARP, and CCR4. Thus, the expanded Tregs maintained their ex vivo phenotype and were easily distinguished from expanded Tconvs (Fig. 4B). Moreover, the expanded Tregs notably suppressed the proliferation of autologous PBMC in stark contrast to PBMC co-cultured with expanded Tconvs (Fig. 4C). Therefore, the expanded Tregs were deemed valuable for assessing mechanism of action of how Tregs suppress HIV-1 infection of neighboring (co-cultured) CD4 T cell populations.
Fig. 4.
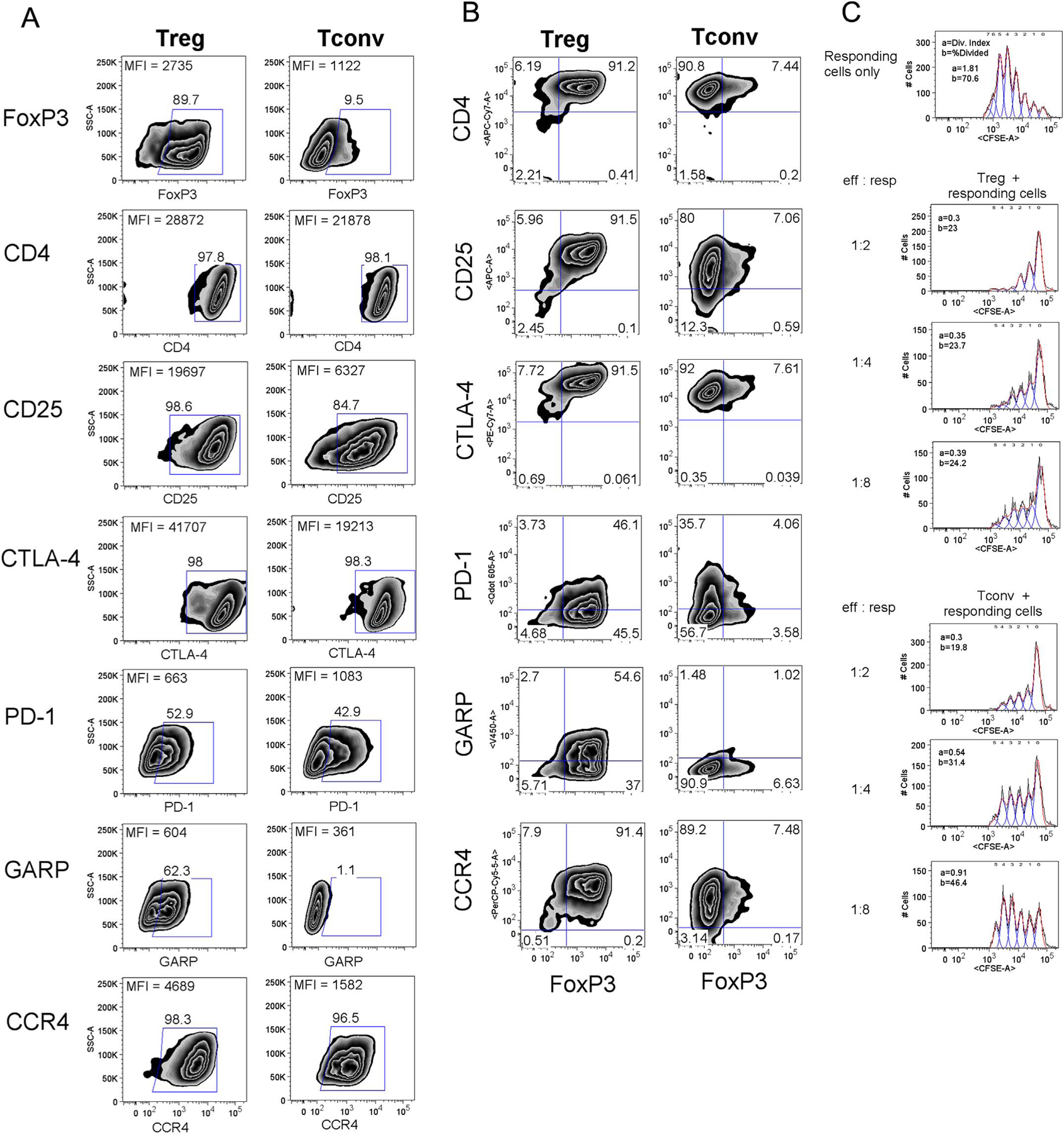
Ex vivo expanded Tregs retain cell surface phenotype and suppressive function. See Section 4 for expansion of Tregs. (A) and (B) Flow cytometry profiles show the relative levels of expression for cell surface markers of the ex vivo expanded Tregs and Tconvs. (C) Tregs (top panels) or control Tconvs (bottom panels) were co-cultured with responding PBMC at varying effector to responder ratios as detailed. Proliferation in response to anti-CD3/CD28 coated bead is denoted by CFSE dilution along the X-axis.
2.4. Tregs inhibition of HIV-1 expression by other CD4 T cell subsets depends on the interaction via Tregs cell surface inhibitory proteins
Tregs are constitutively immunosuppressive cells critical for the control of autoimmunity and inflammatory pathology, and multiple mechanisms have been proposed for their ability to suppress a large number of distinct immune cell types (Shevach, 2009). Specifically, CTLA-4-dependent expression has been described as a core mechanism by which Tregs inhibit other lymphocyte function (Wing and Sakaguchi, 2012). Indeed, CTLA-4 had been reported to be a critical factor in reducing HIV-1 dissemination through dendritic cell-CD4 T cell synapses (Moreno-Fernandez et al., 2014). To assess this possibility, blockade of CTLA-4, as well as inhibition of PD-1 and GARP, were analyzed for disrupting the ability of Tregs to suppress HIV-1 expression of co-cultured bulk populations of CD4 T cells. After synchronous expansion, the responding CD4 T cells (originating from the CD25(−) population) were labeled with CFSE and infected with HIV-1 NL4–3. This was quickly followed by co-culture with Tregs or Tconvs at a 1:1 ratio in the absence or presence of antibodies directed to various Tregs cell surface proteins. Antibodies to Tregs cells surface proteins were tested in isolation or in combination (CTLA-4 + PD-1, or CTLA-4 + PD-1 + GARP). Infection with HIV was assessed by detection of total and integrated HIV-1 DNA levels, as well as HIV-1 mRNA levels.
Clinically, the application of highly active antiretroviral therapy (HAART) is able to control viral loads of HIV-1-infected patients to undetectable levels (≤50 copies per ml plasma). However, the existence of an HIV-1 reservoir severely hampers virus eradication in patients receiving HAART (Chavez et al., 2015). Thus, integrated HIV-1 DNA has been used to characterize the size of the HIV-1 reservoir, since integrated HIV-1 DNA levels correlate with viral outgrowth assays and other important parameters that characterize the viral reservoir (Eriksson et al., 2013; Graf and O’Doherty, 2013; Kiselinova et al., 2016). Importantly, HIV-1 viral DNA levels, particularly integrated DNA levels, have been previously reported to correlate proportionally with viral replication/plasma viremia in HIV-1/AIDS patients (Mexas et al., 2012). To precisely monitor HIV-1 DNA and RNA levels in the above described antibody blockade experiments, approaches were modified from “repetitive-sampling alu-gag PCR” for detecting the viral integrated DNA copy number (De Spiegelaere et al., 2014; Liszewski et al., 2009). A similar method was established by us using HIV-1 LTR-(NFκB)-gag PCR for detecting total DNA copy number, and measurement for relevant viral mRNA levels was performed as characterized by others (Pasternak et al., 2008).
Not previously studied in the setting of HIV-1 infection, antibody blockade of either PD-1 or GARP notably increased HIV-1 DNA (total and integrated) and mRNA levels in the responder cells (Fig. 5 and Supplemental Fig. S2). Interestingly, antibody blockade of CTLA-4 did not lead to increased HIV-1 integrated DNA or mRNA in co-cultured CD4 T cells. However, blockade of CTLA-4 and PD-1 together augmented HIV-1 integrated DNA and mRNA beyond that of PD-1 blockade alone (Fig. 5C and D). This synergism was not noted with antibody blockade of both CTLA-4 and PD-1 when the responding cells were co-cultured with Tconvs (Fig. 5C and D). Taken together, GARP, PD-1, and CTLA-4 all appear to be important cell surface molecules utilized by Tregs to suppress HIV-1 expression in neighboring CD4 T cell populations.
Fig. 5.
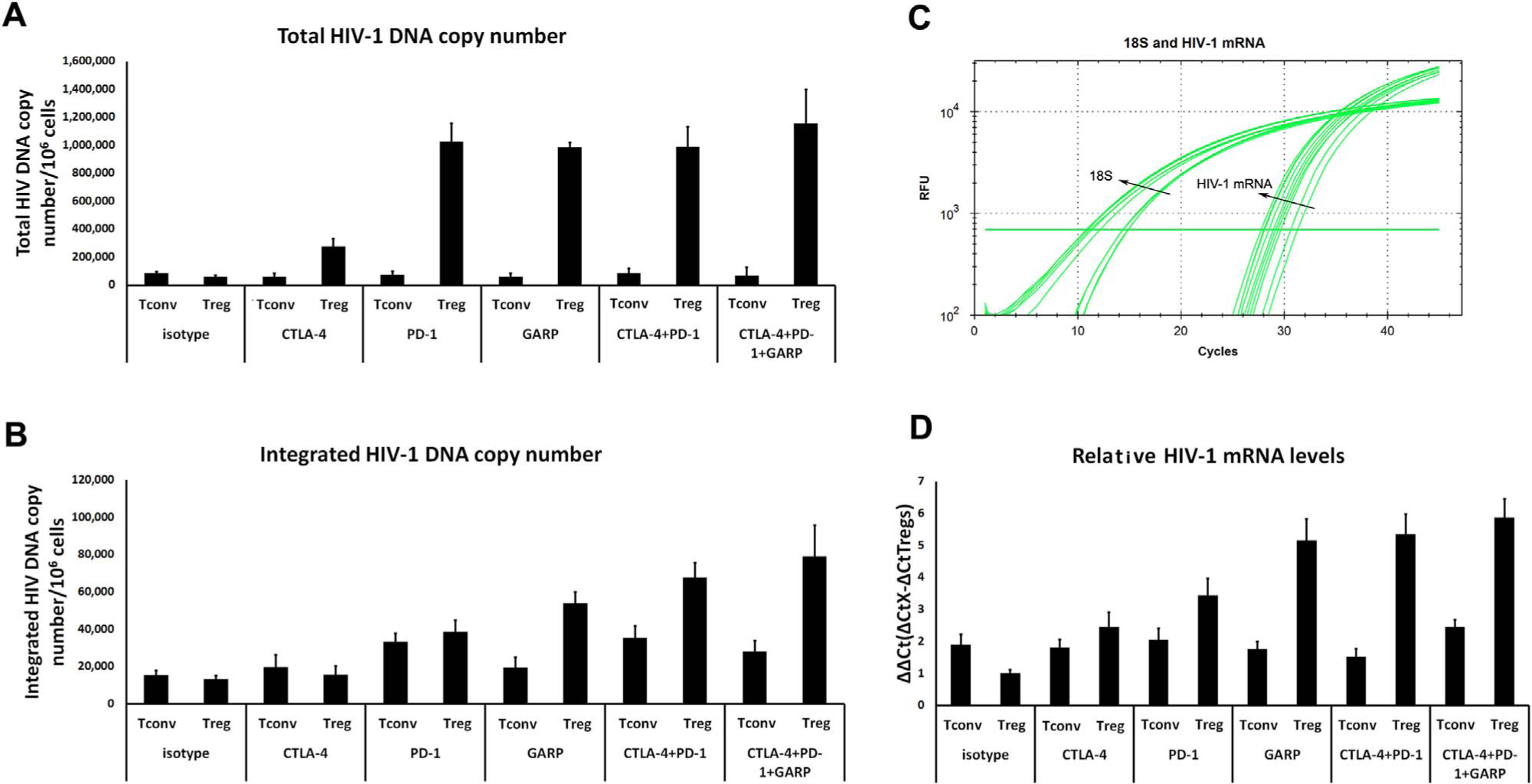
Single or combinational antibody blockade of Tregs cell surface proteins (CTLA-4, PD-1, GARP) increases HIV-1 DNA levels and mRNA levels in co-cultured responder CD4 T cells. Tregs or Tconvs (control) were co-cultured with responding CD4 T cells during HIV-1 infection in vitro in the presence of blocking antibodies to cell surface molecules (see Section 4). Bar graph depictions show HIV-1 total DNA copies (A) and integrated DNA copies (B) of responding CD4 T cells co-cultured with Tconvs (left bars in pairs) or Tregs (right bars in pairs) in the presence of anti-CTLA-4, anti-PD-1, anti-GARP, anti-CTLA-4 plus anti-PD-1, or anti-CTLA-4 plus anti-PD-1 plus anti-GARP antibodies (see Section 4 for copy number calculation). (C) Real-time PCR amplification curves in single and combinational antibody blockings for detection of HIV-1 gag mRNA (18 S RNA as an internal reference control) from cells shown in (A) and (B). (D) Bar graphs depict HIV-1 relative gag mRNA levels (ΔΔCt values) from the responding CD4 T cells in the presence of Tconvs (control) or Tregs, plus the different antibody conditions. All bar graphs represent means ± SEM from 3 independent experiments.
2.5. Tregs cell surface inhibitory proteins play roles in suppression of HIV-1 LTR transcription in other CD4 T cell subsets
We previously determined inhibition of cell proliferation by Tregs contributes to decreased HIV-1 infection in responding CD4 T cells (Fig. S1A). Moreover, the sharper decline in HIV-1 expression in responding CD4 T cells in the presence of Tregs compared to Tconvs (Fig. S1A, B) suggested there was likely another mechanism beyond the effect on cell proliferation that allows Tregs to inhibit HIV-1 expression in co-cultured CD4 T cells.
We previously reported that FoxP3(+) CD4 T cells gave rise to lower HIV-1 LTR transcription than FoxP3(−) CD4 T cells (Selliah et al., 2008). Along these lines, we similarly explored the following: a), if the in vitro expanded Tregs also have decreased HIV-1 LTR transcription, and b), if these Tregs are able to inhibit HIV-1 LTR transcription within the co-cultured CD4 T cells. Using transfected reporter gene constructs, HIV-1 LTR transcriptional activity was lower in Tregs and Tnaives compared to T convs and Tmems (Fig. 6A, C), as expected. Interestingly, the co-cultured responding CD4 T cells transfected with the HIV-1 LTR-reporter construct and co-cultured with Tregs demonstrated decreased HIV-1 transcription (Fig. 6B). This decrease in HIV-1 transcription was not seen when responding CD4 T cells were co-cultured with Tconvs (Fig. 6B). Moreover, the decrease in HIV-1 transcription in the presence of Tregs was partially corrected by the addition of antibodies directed against CTLA-4, PD-1, and GARP in combination (Fig. 6B, D). The antibodies did not alter HIV-1 transcription in responder CD4 T cells co-cultured Tconvs (Fig. 6B, D). As Tregs are also known to generate inhibitory cytokines, such as IL-10 and TGF-β, cytokines levels were analyzed in the supernatants from these experiments. Notably, the presence of the antibodies to CTLA-4, PD-1, and GARP did not notably alter the levels of IL-10 or TGF-β (< 6% difference, data not shown). Thus, the effect of the antibodies on the ability of Tregs to decrease HIV-1 transcription in co-cultured CD4 T cells was not likely mediated via IL-10 or TGF-β.
Fig. 6.
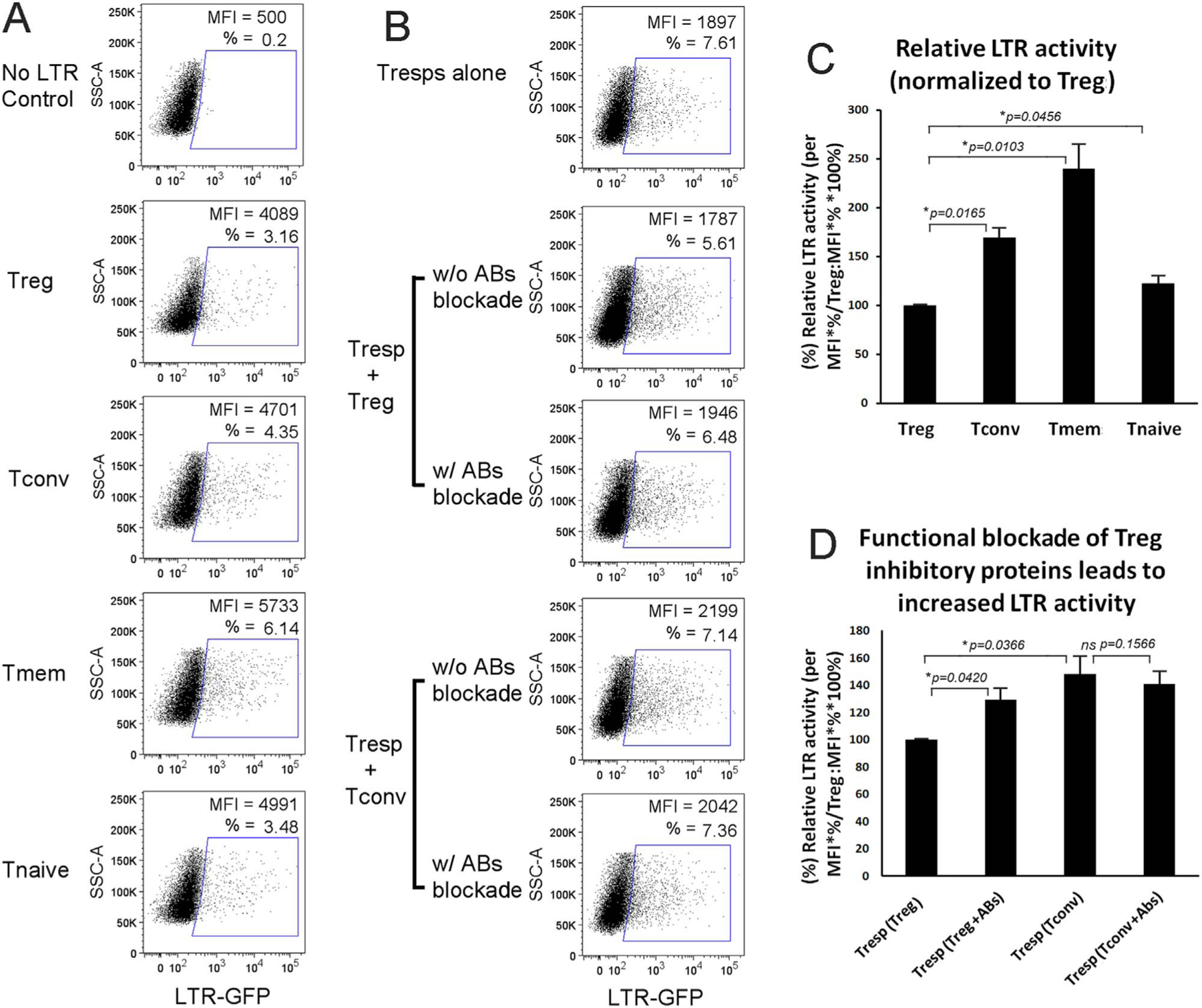
Tregs inhibit HIV LTR transcription within Tregs themselves or in neighboring responding CD4 T cells. (A) Flow cytometry plots show that LTR transcriptional activity in in vitro expanded Tregs is lower than in in vitro expanded T subsets Tconvs and Tmems, and (C) bar plots show the means ± SD values of relative percentages after normalization to Tregs from three independent experiments. (B) Flow cytometry dot plots show that Tregs, but not Tconvs, inhibit LTR transcription in their respective co-cultured responding CD4 T cells; and the presences of antibodies against Tregs cell surface proteins (anti-CTLA-4, PD-1, and -GARP1) partially blocked the inhibition; and (D) the means ± SD values of relative percentages normalized to Tregs from three independent experiments are summarized in the bar plots.
3. Conclusion
In summary, Tregs were confirmed to be relatively resistant to HIV-1 since they possessed lower HIV-1 DNA and viral mRNA levels compared to Tconvs and Tmems, for both cells infected in vitro and those examined ex vivo from HIV-1+ individuals. Tregs were further noted to suppress HIV-1 infection of co-cultured responder CD4 T cells. Mechanistically, specific antibody blockade experiments demonstrated that Tregs cell surface inhibitory proteins, CTLA-4, PD-1, and GARP participated in suppressing HIV-1 infection of neighboring CD4 T cell populations, in part by decreasing HIV-1 LTR transcriptional activity. These results expand our knowledge of how Tregs aid in suppressing HIV-1 infection of neighboring CD4 T cells populations.
4. Materials and methods
4.1. Cell sorting
All procedures involving human subjects were approved by the Institutional Review Board of the University of Alabama at Birmingham (UAB). Informed consent was obtained from all healthy adult participants. Human CD4 T cells were isolated from the peripheral blood of volunteers by negative selection using a proprietary antibody mix (StemCell Technologies, Vancouver, BC) and red blood cell rosetting as previously described (Zhang et al., 2004). HIV-1+/AIDS patient PBMC were provided by the Center for AIDS Research (CFAR) at UAB. The CD4 T subpopulations of Tregs (CD4+, CD25high, CD127low), conventional effectors or Tconvs (CD4+, CD25+, CD127high), memory or Tmems (CD4+, CD25−, CD45RO+), and Tnaives (CD4+, CD25−, CD45RA+) were purified from patient PBMC directly by staining for CD4 T cell surface markers CD3 (PerCP), CD4 (FITC), CD25 (APC), CD127 (PE), CD45RA (V450), and CD45RO (Alexa Fluor 700), and sorting the stained cells on an ARIA cell sorter (BD Biosciences, San Jose, CA, USA) as shown in Fig. 2A. The post-HIV-1 infection of CD4 T cells in vitro were only staining by CD25, CD127, CD45RA, CD45RO; and individual subpopulations were sorted in Fig. 1A. Fluorescently conjugated antibodies were purchased from BD Biosciences and Biolegend (SanDiego, CA, USA).
4.2. CD4 T cell expansion and characterization
The sorted CD4 T cell subsets (Tregs, Tconvs) from healthy donors were expanded by use of the Dynabeads Human Tregs Expander kit (Life Technologies, NY, USA) and IL-2 (provided by the NIH AIDS Research and Reference Reagent Program, Germantown, MA, USA). The Dynabeads Tregs Expander kit and protocol were slightly altered with modified beads (anti-CD3/CD28 monoclonal antibodies) and a differing cell ratio of 2:1 (beads: cell). In brief, CD4 T cells were mixed with anti-CD3/CD28 antibody conjugated Dynabeads at a 2:1 ratio overnight prior to addition of human rIL-2 (500 U/ml). The cells were cultured for 8–10 days in vitro, at concentration of 1–2×106/ml with addition of IL-2 every other day, prior to removal of the Dynabeads for utilization. To identify the expanded CD4 T cell subsets, cells were analyzed by flow cytometry for the Tregs master transcription factor, FoxP3 (Alexa Fluor 488), and typically highly-expressed cell surface markers, such as CTLA-4 (PE-Cy7), PD-1 (Qdot 605), GARP (V450), and CCR4 (PerCP-Cy5.5). All fluorochrome-conjugated monoclonal antibodies were obtained from Biolegend (San Diego, CA, USA). Intracellular staining of FoxP3 was performed according to the product protocol guide as previously reported (Selliah et al., 2008). Functional suppression of expanded Tregs was monitored by modified proliferation assays (Quah et al., 2007). Briefly, Tregs were mixed with CFSE-labeled autologous PBMCs at varying ratios prior to co-culture in vitro for 4 days in the presence of anti-CD3/CD28-coated stimulating Dynabeads (at a 1:1 ratio to the PBMC). PBMC proliferation data were collected by flow cytometry (LSRII, BD Bioscience) and analyzed by FlowJo v9 software (FlowJo LLC, Ashland, Oregon, USA).
4.3. HIV-1 infection and blockade of the interaction between Tregs and responder CD4 T cells
The HIV-1 NL4–3 virus was produced by transfecting HEK 293T cells with plasmid pNL4–3, and resulting viral titers were determined through infection of the Jurkat-derived HIV-1 LTR J2574 reporter T cell line (Duverger et al., 2013). The spinoculation method was employed to infect the freshly isolated CD4 T cells. In short, cells were mixed with HIV-1 viral stock at a multiplicity of infection (MOI) of 0.01–0.1 and centrifuged at 1200×g for 2 h at room temperature. Alternatively, virus-cell mixtures were incubated for 2–4 h at 37 °C to infect PHA-IL-2-stimulated, or in vitro expanded, CD4 T cells (O’Doherty et al., 2000). The infected cells were cultured for 7 days in the presence of IL-2 at a concentration of 30–50 U/ml followed by cell sorting. In experiments with CFSE-, or Alexa Fluor 450-labeled responding cells plus Tregs mixed in co-culture, single or combinations of specific antibodies directed against CTLA-4, PD-1, and GARP were added to the media with final concentrations of 10 μg/ml each. All antibodies were purchased from R&D systems (Minneapolis, MN, USA). All infection and sorting procedures were strictly performed in a BL3 lab facility as part of the UAB CFAR.
4.4. Detection of HIV-1 p24 gag protein, mRNA, and DNA
For flow cytometry assays, the infected cells were fixed with 0.1% paraformaldehyde, and then were incubated with an RD1-conjugated anti-p24 antibody (Coulter, Fullerton, CA, USA) as previously described (Zhang et al., 2004). PBS containing 0.1% saponin and 10% fetal calf serum (FCS) was used as the permeablizing and staining buffer. For real-time semi-quantitative PCR and RT-PCR assays, genomic DNA and total RNA were first extracted from sorted subsets of infected cells by using DNeasy blood and tissue kits and RNeasy mini kits, respectively (QIAGEN, Germantown, MD, USA). cDNA was generated with Super-Script III reverse transcriptase (Life Tech, Carlsbad, CA, USA) from total RNA. For detection of HIV-1 gag mRNA (converted to cDNA) abundance and HIV-1 DNA levels (ΔCt), two rounds of PCR amplification were carried out as described previously (Pasternak et al., 2008; Zhang et al., 2012). In brief, the first round of PCR was performed using a conventional PCR machine with the setting as follows: 94 °C for 3 min, followed by 18 cycles at 94 °C for 30 s, 55 °C for 30 s, and 72 °C for 1 min. These products were subsequently used as templates for the second, semi-nested, real-time (RT) PCR, performed with a iQ5/CFX96 thermal cycler (Bio-Rad, Hercules, CA, USA). β-globin DNA and 18 S RNA were used as internal controls to calculate HIV-1 DNA load and mRNA abundance, respectively. ΔCt was obtained by subtracting CtTx (treatment) from the appropriate respective internal reference Ct values (i.e., Ctβ-globin or Ct18S). ΔΔCt was calculated by subtracting ΔCtTx with the lowest experimental ΔCt (typically, ΔCtTnaives). To determine the absolute copy number of HIV-1 total and integrated proviral DNA, the approach described by Una O’Doherty and her colleagues (De Spiegelaere et al., 2014; Liszewski et al., 2009) was modified by setting repeated gag-alone, alu-gag, and LTR-(NFκB)-gag in the first round of PCR with the running parameter as 95 °C for 3 min, followed by 40 cycles of 95 °C for 15 s, 55 °C for 15 s, and 68 °C for 3.5 min. The repetitive reaction wells were increased up to 48. The first round PCR products were then added along with TaqMan detection chemistry (Life Technologies, NY, USA), specific primers, and probes, and second round real-time PCR was carried out using a Quntstudio6 PCR machine (Life Technologies, NY, USA), with RT-PCR settings as follows: 50 °C for 2 min, then 95 °C for 10 min, followed by 45 cycles of 95 °C for 15 s and 60 °C for 1 min. Copy number was calculated by the Poisson statistical method by placing the standards at limiting dilution as described (De Spiegelaere et al., 2014). In brief, the average Ct values and standard deviation (SD) values were calculated according to gag-alone Ct values. The Ct threshold values were then set up for alu-gag and NFκB-gag reactions as mean Ct minus 2×SD. This was followed by counting the positive reactions and calculating positive reaction percentages as probabilities (p). The mean amounts of integrated and total HIV DNA per replicate reaction were calculated as (λ)=-ln(1-p)/0.1. The primers and probes used in standard PCR and RT-PCR reactions are listed in Table 1.
Table 1.
List of oligonucleotides used in standard and real-time PCR in this study.
| Name | Sequences |
|---|---|
| HIV-1 DNA gag (first round PCR) | GTTCCTGCTATGTCACTTCC |
| HIV-1 DNA alu (first round PCR) | GCCTCCCAAAGTGCTGGGATTACAG |
| HIV-1 DNA nfkb (first round PCR) | GACATCGAGCTTGCTACAAGG |
| HIV-1 DNA R (qPCR primer) | TTAAGCCTCAATAAAGCTTGCC |
| HIV-1 DNA U5 (qPCR primer) | CTTCGGGCGCCACTGCTAGA |
| HIV-1 DNA RU5 (qPCR probe) | CCAGAGTCACACAACAGACGGGCACAC |
| HIV-1 mRNA ksl (first round PCR) | CTTAGGCATCTCCTATGGCAGGAA |
| HIV-1 mRNA mf83 (first round PCR) | GGATCTGTCTCTGTCTCTCTCTCCACC |
| HIV-1 mRNA mf83 (qPCR primer) | The same as above |
| HIV-1 mRNA mf84 (qPCR primer) | ACAGTCAGACTCATCAAGTTTCTCTATCAAAGCA |
| HIV-1 mRNA ks2 (qPCR probe) | TTCCTTCGGGCCTGTCGGGTCCC |
| β-Globin-F (qPCR primer) | CCCTTGGACCCAGAGGTTCT |
| β-Globin-R (qPCR primer) | CGAGCACTTTCTTGCCATGA |
| β-Globin-P (qPCR probe) | GCGAGCATCTGTCCACTCCTGATGCTGTTATGGGCGCTCGC |
| 18SRNA-F (qPCR primer) | TTTCGATGGTAGTCGCCG |
| 18SRNA-R (qPCR primer) | TGGATGTGGTAGCCGTTTC |
| 18SRNA-P (qPCR probe) | CCACGGGTGACGGGGAATCAGG |
| FoxP3-F (qPCR primer) | ACTGACCAAGGCTTCATCTG |
| FoxP3-R (qPCR primer) | CCTCCGGACAGCAAACAG |
| FoxP3-P (qPCR probe) | TCATCCGACAAGGGCTCCTGC |
4.5. HIV-1 LTR reporter assays
The in vitro expanded CD4 T cells were nucleofected with HIV-1 LTR GFP reporter plasmid (Zhang et al., 2012) by using a P3 primary cell 4D-Nucleofector kit and running the pre-set program of the 4D-nucleofector system from Lonza (Walkersville, MD, USA). In co-culture LTR reporter assays, the responding cells were labeled with Alexa Fluor 670 before nucleofection with a LTR GFP reporter plasmid; transfected cells were rested for 2 h and then mixed with Tregs/Tconvs cells at 1:1 ratio. GFP expression was detected 24–48 h after nucleofection by flow cytometry (LSRII, BD Bioscience).
4.6. Statistical analyses
Standard one-way ANOVA analyses were performed using multiple paired comparisons with GraphPad Prism 7 software (La Jolla, CA). A p-value < 0.05 was considered to be statistically significant.
Supplementary Material
Supplemental Material: Fig. S1. Responding CD4 T cells in co-culture with Tregs express lower levels of HIV-1 with each cell division in comparison to those cells in co-culture with Tconvs. (A) Responding CD4 T cell generations from cells shown in Fig. 3 were defined by CFSE-labeled proliferation analysis, and p24 expression is detailed in each subsequent cell generation. (B) Similar experiment was performed by using in vitro synchronously expanded CD4 T cells in which responding cells were labeled and traced by Alexa Fluor 450.
Supplemental Material: Fig. S2. Real-time PCR individual data curves for results summarized in bar graphs in Figures 5A and 5B. (A) Repetitive real-time PCR amplification curves in gag-alone (pink) vs. NFκB-gag (blue) in total viral DNA assays. (B) Repetitive real-time PCR amplification curves in gag-alone (pink) vs. alu-gag (blue) in integrated DNA assays. Tconvs (Tc) are shown in the first and third columns, and Tregs (Tr) are depicted in the second and fourth columns. Antibody blockings are represented in the following rows: isotype control (top row), anti-CTLA-4 (2nd from top), anti-PD-1 (3rd from top), anti-GARP (4th from top), anti-CTLA-4 plus anti-PD-1 (5th from top), and anti-CTLA-4 plus anti-PD1 plus anti-GARP (bottom row).
Acknowledgements
We acknowledge support for TOR by the NIH Basic Mechanisms in AIDS Pathogenesis Training Grant 5T32AI007493-17 and by an American Association of Immunologists (AAI) Careers in Immunology Fellowship. The work was supported by a grant from the Kaul Pediatric Research Institute to RQC. We also thank the Birmingham Center for AIDS Research (CFAR) Virology Core at the University of Alabama at Birmingham (UAB) for their contribution to the development of the project and provision of critical infrastructure and resources, as well as the UAB CFAR Flow Cytometry Core with support provided through NIH/NIAID grant P30 AI27767.
Footnotes
Appendix A. Supplementary material
Supplementary data associated with this article can be found in the online version at http://dx.doi.org/10.1016/j.virol.2017.12.036.
References
- Campbell DJ, Koch MA, 2011. Phenotypical and functional specialization of FOXP3+ regulatory T cells. Nat. Rev. Immunol 11, 119–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Card CM, McLaren PJ, Wachihi C, Kimani J, Plummer FA, Fowke KR, 2009. Decreased immune activation in resistance to HIV-1 infection is associated with an elevated frequency of CD4(+)CD25(+)FOXP3(+) regulatory T cells. J. Infect. Dis 199, 1318–1322. [DOI] [PubMed] [Google Scholar]
- Chase AJ, Yang HC, Zhang H, Blankson JN, Siliciano RF, 2008. Preservation of FoxP3+ regulatory T cells in the peripheral blood of human immunodeficiency virus type 1-infected elite suppressors correlates with low CD4+ T-cell activation. J. Virol 82, 8307–8315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavez L, Calvanese V, Verdin E, 2015. HIV latency is established directly and early in both resting and activated primary CD4 T cells. PLoS Pathog. 11, e1004955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevalier MF, Weiss L, 2013. The split personality of regulatory T cells in HIV infection. Blood 121, 29–37. [DOI] [PubMed] [Google Scholar]
- De Spiegelaere W, Malatinkova E, Lynch L, Van Nieuwerburgh F, Messiaen P, O’Doherty U, Vandekerckhove L, 2014. Quantification of integrated HIV DNA by repetitive-sampling Alu-HIV PCR on the basis of poisson statistics. Clin. Chem 60, 886–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duverger A, Wolschendorf F, Zhang M, Wagner F, Hatcher B, Jones J, Cron RQ, van der Sluis RM, Jeeninga RE, Berkhout B, Kutsch O, 2013. An AP-1 binding site in the enhancer/core element of the HIV-1 promoter controls the ability of HIV-1 to establish latent infection. J. Virol 87, 2264–2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson S, Graf EH, Dahl V, Strain MC, Yukl SA, Lysenko ES, Bosch RJ, Lai J, Chioma S, Emad F, Abdel-Mohsen M, Hoh R, Hecht F, Hunt P, Somsouk M, Wong J, Johnston R, Siliciano RF, Richman DD, O’Doherty U, Palmer S, Deeks SG, Siliciano JD, 2013. Comparative analysis of measures of viral reservoirs in HIV-1 eradication studies. PLoS Pathog. 9, e1003174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graf EH, O’Doherty U, 2013. Quantitation of integrated proviral DNA in viral reservoirs. Curr. Opin. HIV AIDS 8, 100–105. [DOI] [PubMed] [Google Scholar]
- Imamichi H, Lane HC, 2012. Regulatory T cells in HIV-1 infection: the good, the bad, and the ugly. J. Infect. Dis 205, 1479–1482. [DOI] [PubMed] [Google Scholar]
- Kiselinova M, De Spiegelaere W, Buzon MJ, Malatinkova E, Lichterfeld M, Vandekerckhove L, 2016. Integrated and total HIV-1 DNA predict ex vivo viral outgrowth. PLoS Pathog. 12, e1005472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legrand FA, Nixon DF, Loo CP, Ono E, Chapman JM, Miyamoto M, Diaz RS, Santos AM, Succi RC, Abadi J, Rosenberg MG, de Moraes-Pinto MI, Kallas EG, 2006. Strong HIV-1-specific T cell responses in HIV-1-exposed uninfected infants and neonates revealed after regulatory T cell removal. PLoS One 1, e102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Gowans EJ, Chougnet C, Plebanski M, Dittmer U, 2008. Natural regulatory T cells and persistent viral infection. J. Virol 82, 21–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liszewski MK, Yu JJ, O’Doherty U, 2009. Detecting HIV-1 integration by repetitive-sampling Alu-gag PCR. Methods 47, 254–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Abente J, Correa-Rocha R, Pion M, 2016. Functional mechanisms of treg in the context of HIV infection and the janus face of immune suppression. Front Immunol. 7, 192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mexas AM, Graf EH, Pace MJ, Yu JJ, Papasavvas E, Azzoni L, Busch MP, Di Mascio M, Foulkes AS, Migueles SA, Montaner LJ, O’Doherty U, 2012. Concurrent measures of total and integrated HIV DNA monitor reservoirs and ongoing replication in eradication trials. AIDS 26, 2295–2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Fernandez ME, Joedicke JJ, Chougnet CA, 2014. Regulatory T Cells Diminish HIV Infection in Dendritic Cells - Conventional CD4(+) T Cell Clusters. Front. Immunol 5, 199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Fernandez ME, Rueda CM, Rusie LK, Chougnet CA, 2011. Regulatory T cells control HIV replication in activated T cells through a cAMP-dependent mechanism. Blood 117, 5372–5380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Fernandez ME, Zapata W, Blackard JT, Franchini G, Chougnet CA, 2009. Human regulatory T cells are targets for human immunodeficiency Virus (HIV) infection, and their susceptibility differs depending on the HIV type 1 strain. J. Virol 83, 12925–12933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Doherty U, Swiggard WJ, Malim MH, 2000. Human immunodeficiency virus type 1 spinoculation enhances infection through virus binding. J. Virol 74, 10074–10080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasternak AO, Adema KW, Bakker M, Jurriaans S, Berkhout B, Cornelissen M, Lukashov VV, 2008. Highly sensitive methods based on seminested real-time reverse transcription-PCR for quantitation of human immunodeficiency virus type 1 unspliced and multiply spliced RNA and proviral DNA. J. Clin. Microbiol 46, 2206–2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phetsouphanh C, Xu Y, Zaunders J, 2014. CD4 T cells mediate both positive and negative regulation of the immune response to HIV infection: complex role of T follicular helper cells and regulatory T cells in pathogenesis. Front. Immunol 5, 681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quah BJ, Warren HS, Parish CR, 2007. Monitoring lymphocyte proliferation in vitro and in vivo with the intracellular fluorescent dye carboxyfluorescein diacetate succinimidyl ester. Nat. Protoc 2, 2049–2056. [DOI] [PubMed] [Google Scholar]
- Selliah N, Zhang M, White S, Zoltick P, Sawaya BE, Finkel TH, Cron RQ, 2008. FOXP3 inhibits HIV-1 infection of CD4 T-cells via inhibition of LTR transcriptional activity. Virology 381, 161–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevach EM, 2009. Mechanisms of foxp3+ T regulatory cell-mediated suppression. Immunity 30, 636–645. [DOI] [PubMed] [Google Scholar]
- Tran TA, de Goer de Herve MG, Hendel-Chavez H, Dembele B, Le Nevot E, Abbed K, Pallier C, Goujard C, Gasnault J, Delfraissy JF, Balazuc AM, Taoufik Y, 2008. Resting regulatory CD4 T cells: a site of HIV persistence in patients on long-term effective antiretroviral therapy. PLoS One 3, e3305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteside TL, 2015. Clinical Impact of Regulatory T cells (Treg) in Cancer and HIV. Cancer Microenviron. 8, 201–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wing JB, Sakaguchi S, 2012. Multiple treg suppressive modules and their adaptability. Front. Immunol 3, 178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Clausell A, Robinson T, Yin J, Chen E, Johnson L, Weiss G, Sabbaj S, Lowe RM, Wagner FH, Goepfert PA, Kutsch O, Cron RQ, 2012. Host factor transcriptional regulation contributes to preferential expression of HIV type 1 in IL-4-producing CD4 T cells. J. Immunol 189, 2746–2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Genin A, Cron RQ, 2004. Overexpression of octamer transcription factors 1 or 2 alone has no effect on HIV-1 transcription in primary human CD4 T cells. Virology 321, 323–331. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Material: Fig. S1. Responding CD4 T cells in co-culture with Tregs express lower levels of HIV-1 with each cell division in comparison to those cells in co-culture with Tconvs. (A) Responding CD4 T cell generations from cells shown in Fig. 3 were defined by CFSE-labeled proliferation analysis, and p24 expression is detailed in each subsequent cell generation. (B) Similar experiment was performed by using in vitro synchronously expanded CD4 T cells in which responding cells were labeled and traced by Alexa Fluor 450.
Supplemental Material: Fig. S2. Real-time PCR individual data curves for results summarized in bar graphs in Figures 5A and 5B. (A) Repetitive real-time PCR amplification curves in gag-alone (pink) vs. NFκB-gag (blue) in total viral DNA assays. (B) Repetitive real-time PCR amplification curves in gag-alone (pink) vs. alu-gag (blue) in integrated DNA assays. Tconvs (Tc) are shown in the first and third columns, and Tregs (Tr) are depicted in the second and fourth columns. Antibody blockings are represented in the following rows: isotype control (top row), anti-CTLA-4 (2nd from top), anti-PD-1 (3rd from top), anti-GARP (4th from top), anti-CTLA-4 plus anti-PD-1 (5th from top), and anti-CTLA-4 plus anti-PD1 plus anti-GARP (bottom row).


