Abstract
Phylogenomic analysis of whole genome sequences of five benzylisoquinoline alkaloid (BIA)-producing species from the Ranunculales and Proteales orders of flowering plants revealed the sequence and timing of evolutionary events leading to the diversification of these compounds. (S)-Reticuline is a pivotal intermediate in the synthesis of many BIAs and our analyses revealed parallel evolution between the two orders, which diverged ∼122 million years ago (MYA). Berberine is present in species across the entire Ranunculales, and we found co-evolution of genes essential for production of the protoberberine class. The benzophenanthridine class, which includes the antimicrobial compound sanguinarine, is specific to the Papaveraceae family of Ranunculales, and biosynthetic genes emerged after the split with the Ranunculaceae family ∼110 MYA but before the split of the three Papaveraceae species used in this study at ∼77 MYA. The phthalideisoquinoline noscapine and morphinan class of BIAs are exclusive to the opium poppy lineage. Ks estimation of paralogous pairs indicates that morphine biosynthesis evolved more recently than 18 MYA in the Papaver genus. In the preceding 100 million years gene duplication, neofunctionalization and recruitment of additional enzyme classes, combined with gene clustering, gene fusion, and gene amplification, resulted in emergence of medicinally valuable BIAs including morphine and noscapine.
Key words: benzylisoquinoline alkaloids, plant genomes, gene tree analysis, metabolic pathway evolution, gene clusters, gene fusion
Annotated genomes of five benzylisoquinoline alkaloid (BIA)-producing species are used to conduct gene tree analysis and uncover the events leading to emergence of major BIA biosynthetic pathways in Ranunculales. Ks estimation of key paralogous pairs revealed that morphine biosynthesis evolved more recently than 18 million years ago in the Papaver genus. Contrary to previous reports, this study indicates that BIA biosynthesis evolved independently in the Ranunculales and Proteales orders.
Introduction
Benzylisoquinoline alkaloids (BIAs; all abbreviations are listed in Table 1) are a structurally diverse class of tyrosine-derived specialized metabolites occurring primarily in the order Ranunculales (Figure 1). Many of them possess pharmaceutical activities, among which the potent analgesics codeine and morphine, belonging to the morphinan subclass of BIAs, and the antitussive and anticancer noscapine, belonging to the phthalideisoquinoline subclass, are produced exclusively in opium poppy for the pharmaceutical industry.
Table 1.
List of Abbreviations.
| Abbreviation | Full name |
|---|---|
| 4′OMT | 3-HYDROXY-N-METHYLCOCLAURINE 4′-O-METHYLTRANSFERASE |
| 6OMT | NORCOCLAURINE 6-O-METHYLTRANSFERASE |
| 9OMT | (S)-SCOULERINE 9-O-METHYLTRANSFERASE |
| BBE | BERBERINE BRIDGE ENZYME |
| BIA | Benzylisoquinoline alkaloid |
| CFS | (S)-CHEILANTHIFOLINE SYNTHASE |
| CNMT | COCLAURINE-N-METHYLTRANSFERASE |
| CODM | CODEINE 3-O-DEMETHYLASE |
| COR | CODEINONE REDUCTASE |
| CS | (S)-CANADINE SYNTHASE |
| CYP80B | (S)-N-METHYLCOCLAURINE 3′-HYDROXYLASE |
| CYP82X1 | CYTOCHROME P450 CYP82X1 |
| CYP82X2 | CYTOCHROME P450 CYP82X2 |
| CYP82Y1 | CYTOCHROME P450 CYP82Y1 |
| DBOX | DIHYDROBENZOPHENANTHRIDINE OXIDASE |
| Ks | Number of synonymous substitutions per synonymous site |
| MSH | (S)-N-METHYLSTYLOPINE HYDROXYLASE |
| MYA | Million years ago |
| NCBI | National Center for Biotechnology Information |
| NCS | (S)-NORCOCLAURINE SYNTHASE |
| NISO | NEOPINONE ISOMERASE |
| P6H | PROTOPINE-6-HYDROXYLASE |
| PS | (−)-PLUVIATOLIDE SYNTHASE |
| PSAT1 | ACETYLTRANSFERASE 1 |
| PSMT2 | O-METHYLTRANSFERASE 2 |
| PSMT3 | O-METHYLTRANSFERASE 3 |
| PSCXE1 | CARBOXYLESTERASE 1 |
| PSSDR1 | SHORT-CHAIN DEHYDROGENASE/REDUCTASE |
| SALAT | SALUTARIDINOL-7-O-ACETYLTRANSFERASE |
| SALR | SALUTARIDINE REDUCTASE |
| SALSYN | SALUTARIDINE SYNTHASE |
| SanR | SANGUINARINE REDUCTASE |
| SPS | (S)-STYLOPINE SYNTHASE |
| STORR | (S)-TO-(R)-RETICULINE P450-OXIDOREDUCTASE |
| STOX | (S)-TETRAHYDROPROTOBERBERINE OXIDASE |
| T6ODM | THEBAINE 6-O-DEMETHYLASE |
| THS2 | THEBAINE SYNTHASE |
| TNMT | (S)-TETRAHYDROPROTOBERBERINE-N-METHYLTRANSFERASE |
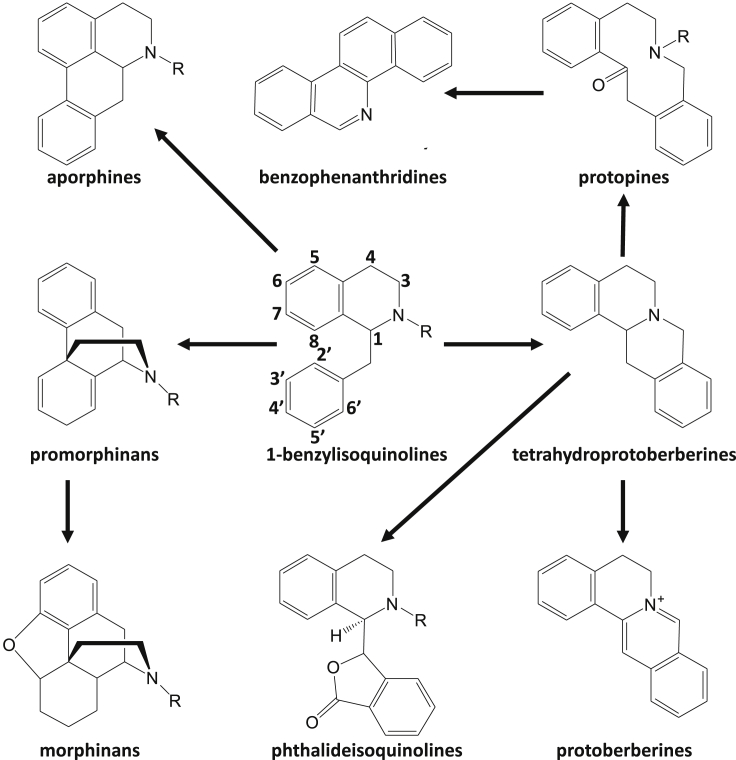
Figure 1.
Structural Scaffolds of Major Subclasses of Benzylisoquinoline Alkaloids Derived from 1-Benzylisoquinolines.
Numbering of different carbon positions is shown for the scaffold of 1-benzylisoquinolines.
The pathways of some common intermediates/end products and the corresponding genes have been elucidated from a few representative species from the order Ranunculales, providing a blueprint to investigate their evolutionary origins. In addition to these functionally characterized individual genes, draft genome assemblies of five BIA-producing species have been recently reported (Ming et al., 2013, Liu et al., 2017, Filiault et al., 2018, Gui et al., 2018, Guo et al., 2018, Hori et al., 2018).
Whole-genome sequence assemblies provide the full set of paralogous sequences of gene families for accurate comparison of orthologous relationships between species, and also allow estimation of species divergence times. We performed phylogenetic analyses of the gene families containing functionally characterized genes in combination with the complete paralog sets from the draft genomes to better understand and date key events in BIA pathway evolution.
The first BIA in the biosynthetic pathway, (S)-norcoclaurine, is understood to be formed through the condensation of tyrosine-derived dopamine and 4-hydroxyphenylacetaldehyde catalyzed by NORCOCLAURINE SYNTHASE (NCS) (Stadler et al., 1989, Samanani and Facchini, 2001). Intricate combinations of reactions including N- and O-methylations, hydroxylations, carbon–carbon couplings, reductions, and acetylations generate the diversity of BIAs found across the Ranunculales. Many structurally distinct subclasses of BIA are derived from (S)-reticuline, a 1-benzylisoquinoline alkaloid that serves as a central branch-point intermediate to other BIA subclasses such as protoberberines, benzophenanthridines, phthalideisoquinolines, and morphinans (Figures 1 and 2C; Hagel and Facchini, 2013). In this work, we focus our analyses on the genes that are involved in the biosynthetic pathways leading to the important BIAs berberine, sanguinarine, noscapine, and morphine, which represent these different structural subclasses.
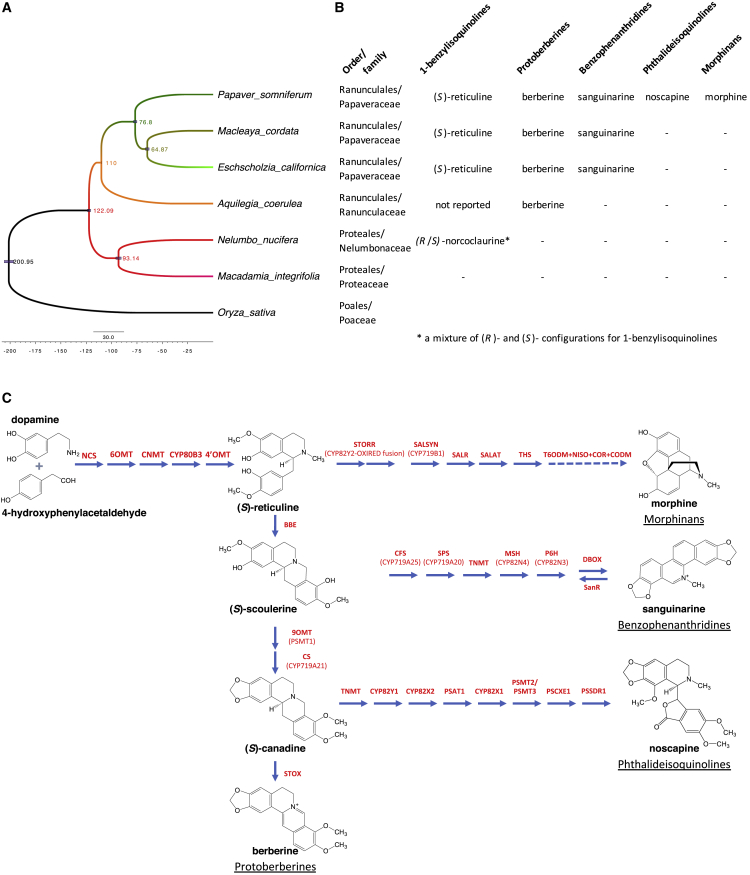
Figure 2.
The Occurrence of Benzylisoquinoline Alkaloids in Ranunculales and Proteales Species with Published Genome Assemblies.
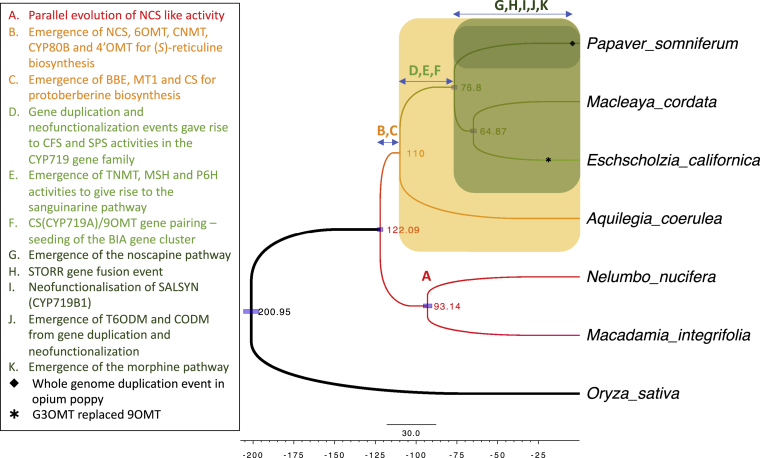
(A) Inferred phylogenetic tree with single-copy orthologous sequences across seven genomes (Supplemental Table 1). The scale bar shows 30.0 million years. Divergence timings are estimated using BEAST2 (Bouckaert et al., 2019) and shown at the nodes. Branch colors indicate species. Light-blue bars at the nodes indicate the range with 95% highest posterior density.
(B) Occurrence of selected alkaloids representing different benzylisoquinoline classes with respect to the species shown in Figure 2A.
(C) BIA biosynthetic pathways and enzymes characterized from species belonging to the order Ranunculales. Enzyme names are provided in the list of abbreviations in Table 1.
Berberine belongs to the protoberberine subclass of BIAs with a tetracyclic ring system. It is widely distributed among many different plant species, especially in Ranunculaceae, Papaveraceae, Berberidaceae, and Menispermaceae families in the order Ranunculales (Neag et al., 2018). (S)-Reticuline is first converted to (S)-scoulerine through a methylene-bridge-forming reaction catalyzed by BERBERINE BRIDGE ENZYME (BBE). After O-methylation, a methylenedioxy bridge is added to the isoquinoline moiety by (S)-canadine synthase (CS), a cytochrome P450 CYP719 protein. The product, (S)-canadine, is then converted to berberine in a final oxidation step.
Sanguinarine belongs to the benzophenanthridine subclass of BIAs that has so far only been reported from members of the Papaveraceae family within the order Ranunculales (Krane et al., 1984, Sariyar, 2002). Its biosynthetic pathway also involves the conversion of (S)-reticuline to the tetrahydroprotoberberine alkaloid (S)-scoulerine, which is followed by CYP719-catalyzed methylenedioxy bridge formation, N-methylation, cytochrome P450 CYP82N protein-catalyzed oxidative ring opening, and further hydroxylation to form its distinctive tetracyclic structural skeleton.
The phthalideisoquinoline alkaloid noscapine has a chemical structure that has a two-ring phthalide moiety connected to its isoquinoline moiety. Noscapine is derived through further chemical modifications from tetrahydroprotoberberines. Unlike other phthalideisoquinoline alkaloids, noscapine and some of its intermediates bear an additional hydroxyl/O-methyl group at the C-8 position of the isoquinoline benzyl ring (Figure 2C; Blaskó et al., 1982). In opium poppy (Papaver somniferum), all but two of the genes required for noscapine biosynthesis from (S)-reticuline occur as a cluster in the genome (Winzer et al., 2012). Noscapine occurs in some but not all species within the genus Papaver of the Papaveraceae, and the same is true for the morphinan subclass of BIAs (Sariyar, 2002). The biosynthesis of morphinans requires a two-step epimerization of (S)-reticuline to (R)-reticuline. The reactions are catalyzed by STORR, a unique bifunctional cytochrome P450-oxidoreductase fusion protein (Farrow et al., 2015, Winzer et al., 2015). The opium poppy genome has revealed that the noscapine biosynthetic genes are part of a larger BIA gene cluster that also includes STORR and all four other genes required for the production of the first morphinan alkaloid thebaine from (S)-reticuline (Guo et al., 2018).
Aside from opium poppy, the draft genomes of BIA-producing species have become available for Eschscholzia californica and Macleaya cordata of the Papaveraceae family (Liu et al., 2017, Hori et al., 2018) and Aquilegia coerulea of the Ranunculaceae family (Filiault et al., 2018). Other than these plant species in the order Ranunculales, sacred lotus (Nelumbo nucifera) from the order Proteales is also rich in BIA alkaloids, and genome assemblies of this species have also been reported (Ming et al., 2013, Gui et al., 2018). Even though BIA biosynthetic genes have yet to be functionally characterized in sacred lotus, the complete coverage of the gene space provided by the whole-genome assembly is a valuable and useful source for inferring potential orthologous history at the sequence level for BIA genes characterized in other plants. Although BIA structures are also present sporadically in some species of other plant orders such as Cornales, Laurales, Magnoliales, Piperales, and Sapindales, there are no draft genome assemblies yet available from such species. Furthermore, there are no gene sequences functionally characterized to be involved in BIA biosynthesis from these orders. Despite the lack of a comprehensive genome coverage of BIA-producing plant species in all of the above orders, the four draft genome assemblies of Ranunculales as well as that of sacred lotus have allowed us to perform thorough phylogenetic analyses on the gene families related to BIA biosynthesis and investigate the origin and orthologous history of key gene functions in these pathways.
We also estimate the time frame for the emergence of a metabolic pathway by dating the STORR gene fusion in the morphinan pathway and gene duplications leading to paralogs such as CYP82X1/CYP82X2/CYP82Y1 in the noscapine pathway and THEBAINE 6-O-DEMETHYLASE (T6ODM)/CODEINE 3-O-DEMETHYLASE (CODM) in the morphinan pathway in opium poppy (Winzer et al., 2012, Winzer et al., 2015, Guo et al., 2018). This analysis allows us to determine the order and timing of key events that give rise to the biosynthetic pathways to noscapine and morphine.
Results and Discussion
Phylogenomic Analysis of BIA-Producing Plants
Phylogenomic analysis was used to determine the evolutionary relationship and date divergence times of all five BIA-producing plants for which draft assembly genomes have been reported so far. Four genomes are from species in the order Ranunculales. Of these, P. somniferum, E. californica, and M. cordata belong to the Papaveraceae family, whereas A. coerulea is from the Ranunculaceae family. N. nucifera is a member of the Nelumbonaceae family in the order Proteales. Because of its taxonomic relationship with N. nucifera, Macadamia integrifolia was also included in our analysis, since it is the only other member of Proteales with a published genome assembly (Nock et al., 2016). However, no BIAs have been reported from this species. The genome of the monocot rice was used as an outgroup (Kawahara et al., 2013). We identified 117 single-copy orthologous groups in these seven genomes using OrthoFinder software (Supplemental Table 1) and then used these to estimate species divergence times with BEAST2 software (Figure 2A; Emms and Kelly, 2015, Bouckaert et al., 2019). The resulting species tree shows that the Ranunculales clade diverged from the Proteales ∼122 million years ago (MYA) and the three Papaveraceae species from A. coerulea of the Ranunculaceae family ∼110 MYA. In addition, P. somniferum is estimated to have split from the other two Papaveraceae species, E. californica and M. cordata, about 77 MYA. This provides the time frame for studying the BIA evolution in these plants.
Figure 2B summarizes the distribution of the key BIA metabolites, including the central intermediate (S)-reticuline and other important end products such as berberine, sanguinarine, noscapine, and morphine, in these five BIA-producing species as reported in the literature: the presence of the 1-benzylisoquinolines (S)-reticuline is well documented in all three Papaveraceae species (Hagel et al., 2015, Winzer et al., 2015, Liu et al., 2017). Berberine is one of the main BIAs found in A. coerulea (Winek et al., 1964), but very little else is known about its biosynthesis or intermediates such as (S)-reticuline in this species. Although not the dominant BIA in the three Papaveraceae species, berberine has been reported to occur in trace amounts in these plants (Schmidt et al., 2007, Hagel et al., 2015, Och et al., 2017). As such, they should all have the biosynthetic capability of producing berberine. Both E. californica and M. cordata are rich in benzophenanthridine alkaloids, including sanguinarine (Liu et al., 2017, Hori et al., 2018). Opium poppy has also been reported to accumulate sanguinarine in cell cultures (Desgagné-Penix et al., 2010), whereas noscapine and morphinans such as morphine and codeine are the most dominant BIAs found in aerial tissues such as stem and capsules (Winzer et al., 2012, Winzer et al., 2015). However, none of the above BIAs have been detected in sacred lotus, even though other BIAs belonging to aporphine, bisbenzylisoquinoline, and 1-benzylisoquinoline subclasses have been reported (Menéndez-Perdomo and Facchini, 2018). This suggests that alternative BIA biosynthetic pathways may be present in N. nucifera.
Biosynthesis of (S)-Reticuline Evolved in Ranunculales after Divergence from Proteales 122 MYA
The distribution of common BIA metabolites such as (S)-reticuline, berberine, and sanguinarine across Ranunculales plants suggests shared evolutionary history of their biosynthetic pathways. We used gene tree analysis to shed light on key evolutionary events shaping the emergence of new BIA pathways in these plants. Figure 2C shows our current biochemical and molecular understanding of BIA biosynthesis. This has overwhelmingly come from Ranunculales species, P. somniferum, E. californica, and Coptis japonica in particular. The biosynthetic pathways and all genes involved in the biosynthesis of berberine, sanguinarine, noscapine, and morphine have been functionally characterized (Figure 2C). We used the genomes of the five BIA-producing species in conjunction with other available data in the GenBank NR database to carry out gene tree analyses of these BIA pathway genes. As such, the gene search space has covered over 100 annotated plant genomes in addition to other single entries of both functionally characterized and uncharacterized sequences in the NR database. This has provided insight into the timing of gene duplications, gene fusion, and neofunctionalization events as well as the evolutionary relationship of these pathways in the context of BIA biosynthesis.
(S)-NORCOCLAURINE SYNTHASE (NCS) is the key enzyme catalyzing the first step in BIA biosynthesis, the condensation of dopamine and 4-hydroxyphenylacetaldehyde to form (S)-norcoclaurine (Figure 3). The NCS gene has been functionally characterized from a number of species in the order of Ranunculales (Figure 2C; Samanani et al., 2004, Minami et al., 2007, Lee and Facchini, 2010, Lichman et al., 2017).
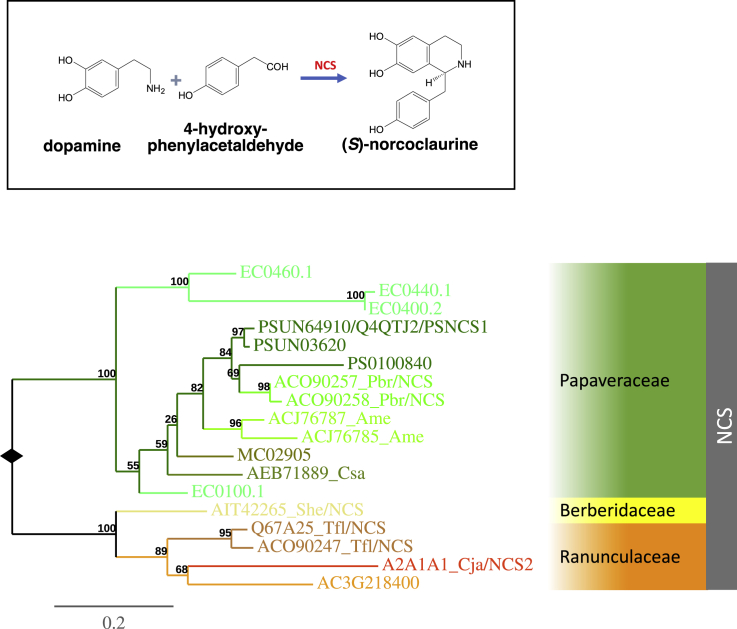
Figure 3.
Phylogenetic Analyses Show an NCS Clade across Species in the Ranunculales.
All functionally characterized NCSs are members of this NCS clade. Sequences from species of the same family form strongly supported subclades. The best-scoring maximum-likelihood tree was inferred using RAxML (Stamatakis, 2006) All branches are drawn to scale as indicated by the scale bar (substitutions/site), and the bootstrap values from an analysis of 100 replicates are shown at the nodes. The solid diamonds indicates the root of the tree. The NCS clade is indicated by the dark-gray bar. Branch colors indicate species. Each subclade that contains sequences from species of the same family is highlighted in color. ACxG, EC, MC, and PS prefixes in sequence identifiers denote sequences from the annotation datasets of A. coerulea, E. californica, M. cordata, and P. somniferum, respectively. For the remaining sequences, a three-letter code was used at the end of the sequence identifiers to indicate the respective species (see “General information for gene trees” in Supplemental Information). Additional sequence information is provided in Supplemental Table 5. The inset shows the reaction catalyzed by NCS.
The NCS gene tree shows a clade of sequences exclusively from families in the order Ranunculales (Figure 3). A single Papaveraceae subclade contains the functionally characterized PSNCS1 sequence from opium poppy and orthologous sequences from the other two genomes as well as other Papaveraceae sequences. Similarly, a single sequence from the A. coerulea genome (AC3G218400) forms a single group exclusively with sequences of other Ranunculaceae species, which in turn group with a sequence from the Berberidaceae species Sinopodophyllum hexandrum (Figure 3), suggesting monophyletic origin of this NCS in the common ancestor of Papaveraceae, Ranunculaceae, and Berberidaceae.
The four other enzymes required for the subsequent O-methylation, N-methylation, and hydroxylation steps in the biosynthesis of (S)-reticuline are NORCOCLAURINE 6-O-METHYLTRANSFERASE (6OMT), COCLAURINE-N-METHYLTRANSFERASE (CNMT), cytochrome P450 HYDROXYLASE (CYP80B), and 3-HYDROXY-N-METHYLCOCLAURINE 4′-O-METHYLTRANSFERASE (4′OMT) (Frenzel and Zenk, 1990a, Frenzel and Zenk, 1990b, Loeffler and Zenk, 1990). The genes encoding these enzymes have been characterized from the Ranunculaceae species C. japonica and the Papaveraceae species P. somniferum and E. californica (Pauli and Kutchan, 1998, Huang and Kutchan, 2000, Morishige et al., 2000, Choi et al., 2002, Ikezawa et al., 2003). Similar to the NCS gene tree, the respective trees for the genes encoding these enzymes show clades containing sequences exclusively from Ranunculales. Orthologous sequences from the genomes of all four species from the Ranunculales order are represented in these clades (Supplemental Figures 1–3). Furthermore, the clades of 6OMT/4′OMT and CNMT resemble that of the NCS gene, in that in each gene clade Papaveraceae sequences form a subclade that is sister to the Ranunculaceae subclade with a single gene sequence from S. hexandrum representing the Berberidaceae. In the CYP80 tree (Supplemental Figure 1), the CYP80B clade contains a single Papaveraceae subclade that groups with a Ranunculaceae subclade with no data being available from other Ranunculales families. In summary, this analysis suggests that all five genes encoding enzymes required for the biosynthesis of (S)-reticuline are likely to be present in the Ranunculales common ancestor before the divergence of the Papaveraceae and Ranunculaceae 110 MYA.
Five NCS-like sequences have been reported in the sacred lotus genome based on sequence homology and considered as norcoclaurine synthases (NnNCS), but only by association of the gene expression profiles and alkaloid content (Vimolmangkang et al., 2016). To understand the relationship between the Ranunculales NCSs and these NnNCSs, we extended our analyses to include sequences that fell into other clades in an expanded NCS gene tree (Supplemental Figure 4). The five paralogous sequences identified in sacred lotus appear in the extended tree but not in the Ranunculales NCS clade (Supplemental Figure 4), suggesting that NCS activity may have evolved independently in these two orders. However, it should be noted that gene function of these candidates remains to be fully characterized at the biochemical level. Furthermore, no orthologs of the other genes involved in (S)-reticuline biosynthesis are present in the sacred lotus genome except for the 6OMT gene (Supplemental Figures 1–3), which is consistent with the observation that (S)-reticuline is absent from this species. This suggests that the biosynthetic pathway for (S)-reticuline evolved in the order Ranunculales after the divergence of Proteales and Ranunculales about 122 MYA.
In sacred lotus, many 1-benzylisoquinoline alkaloids have both (R) and (S) stereochemical configurations (Menéndez-Perdomo and Facchini, 2018). In contrast, in Ranunculales only (S) enantiomers have been detected in the 1-benzylisoquinoline intermediates in early biosynthetic steps up to (S)-reticuline. This is consistent with the stereospecificity of NCS activity in Ranunculales (Lee and Facchini, 2010). Moreover, the major sacred lotus aporphines such as nuciferine are (R) enantiomers and often lack the hydroxyl group corresponding to the 4′ position of the 1-benzylisoquinoline scaffold, as shown in Figure 1. Therefore, it is possible the benzylisoquinolines in N. nucifera are not derived from the same NCS-catalyzed condensation of dopamine and 4-hydroxyphenylacetaldehyde. Instead, the biosynthesis of BIAs in this species may start with an enzyme with catalytic activity similar to that of NCS but with less stringent stereochemical requirement and/or different substrate specificity. This analysis therefore suggests that while BIA biosynthesis has a monophyletic origin within the order Ranunculales, it may have paraphyletic origins in angiosperms outside the order, such as sacred lotus. This is in contrast to the previous suggestion that BIA metabolism has a monophyletic origin across all angiosperms (Liscombe et al., 2005). Functional characterization of genes that are responsible for the NCS-like activities in sacred lotus as well as BIA-producing plants of other families will provide more definitive answers to the aforementioned.
The Common Pathway for Protoberberine Biosynthesis Evolved in the Common Ancestor of the Ranunculales about 110–122 MYA
Four enzymes are required for berberine biosynthesis from (S)-reticuline (Figure 2C). Of these, CS catalyzes the formation of a methylenedioxy bridge to produce (S)-canadine and is a member of the CYP719 subfamily of the cytochrome P450s (Rueffer and Zenk, 1994, Ikezawa et al., 2003).
Phylogenetic analysis shows the robustly supported CYP719A/B clade (Figure 4), containing sequences exclusively from Ranunculales. This clade contains all functionally characterized CYP719s, including CYP719A1 and CYP719A21, the CS enzymes that have been characterized from C. japonica and P. somniferum, respectively.
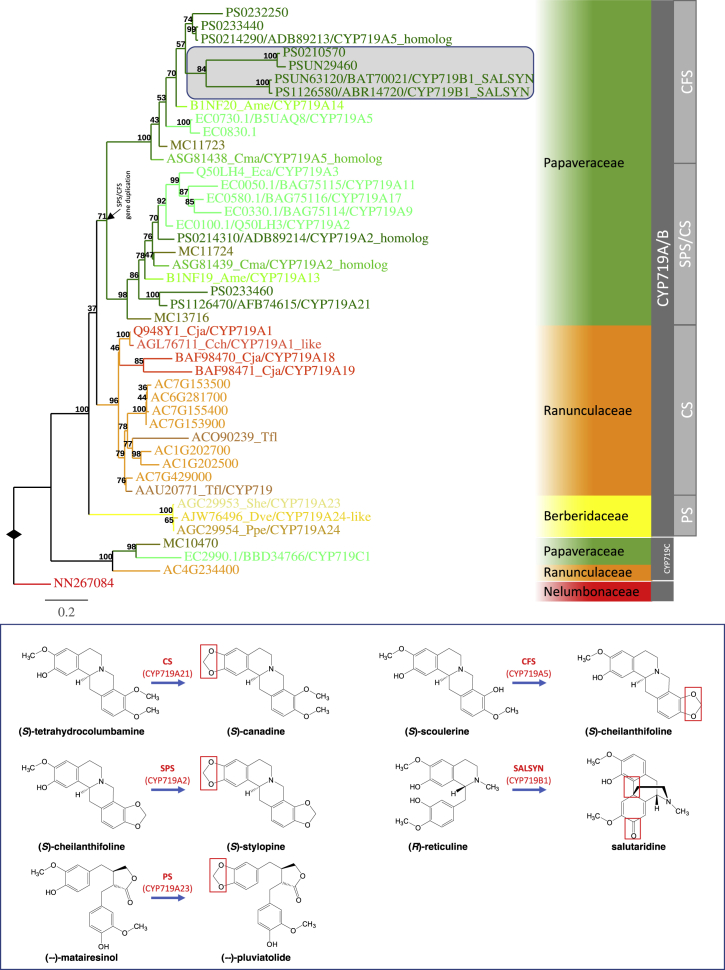
Figure 4.
CYP719 Tree Analysis Suggests Key Duplication Events for the Evolution of Multiple BIA Biosynthetic Pathways.
All functionally characterized CYP719 proteins fall into the CYP719A/B clade, which contains sequences exclusively from Ranunculales species. The CYP719A/B clade contains several strongly supported subclades. The CS subclade contains sequences exclusively from Ranunculaceae species, and all of its functionally characterized members exhibit CS activity. Both the SPS/CS and CFS subclades consist of sequences exclusively from Papaveraceae species. All functionally characterized SPS proteins fall into the SPS/CS subclade, whereas all functionally characterized CFS proteins are members of the CFS subclade. The SPS/CS subclade also contains the opium poppy CS sequence CYP719A21, and the trifunctional (S)-canadine synthase/(S)-stylopine synthase/(S)-nandinine synthase (CYP719A13) from Argemone mexicana. SALSYN (CYP719B1) falls into an opium poppy group inside the CFS subclade highlighted by the shaded box in the tree. The figure descriptors and legends are in the same format as described in Figure 3. Extended information of the gene tree is included in Supplemental Information. The black arrow indicates the occurrence of the gene-duplication event giving rise to SPP/CFS. The inset shows the reactions catalyzed by functionally characterized members of the CYP719 subfamily with the reactive groups highlighted in red boxes.
In the CYP719A/B clade, CYP719A1 forms a CS subclade exclusively with CYP719 sequences identified either from the genome of A. coerulea or from other Ranunculaceae species in the GenBank NR database, suggesting a monophyletic origin in Ranunculaceae. The subclade that includes functionally characterized (−)-PLUVIATOLIDE SYNTHASE (PS) contains sequences exclusively from the Berberidaceae family and is strongly supported. However, there is no bootstrap support for a sister relationship of the Berberidaceae PS clade to the Ranunculaceae CS subclade. Taking the species phylogeny of these taxonomic groups and the gene tree topology into account, we postulate that the sister relationship of the Ranunculaceae CS and the Berberidaceae PS subclades is obscured in the gene tree by long branches in members of the PS subclade. PS, such as CYP719A23 and CYP719A24, is capable of converting (−)-matairesinol into (−)-pluviatolide by catalyzing methylenedioxy bridge formation in the biosynthesis of the phenolic aryltetralin lignan podophyllotoxin in Berberidaceae species Podophyllum hexandrum and Podophyllum peltatum (Marques et al., 2013). The difference in substrate structures to BIAs may have required substantial changes at the sequence levels, which may have resulted in masking of the sister grouping with the Ranunculaceae CS subclade in our analysis.
All the remaining sequences group strongly together and are exclusively from Papaveraceae species, forming a sister relationship with the Ranunculaceae CS and Berberidaceae PS subclades. This Papaveraceae subgroup is further divided into two strongly supported subclades both containing sequences from the three Papaveraceae genomes. This indicates a gene duplication in the Papaveraceae lineage following divergence of the Papaveraceae and Ranunculaceae/Berberidaceae.
It has been reported that CYP719A13 from Argemone mexicana converts (S)-tetrahydrocolumbamine to (S)-canadine, (S)-cheilanthifoline to (S)-stylopine, and (S)-scoulerine to (S)-nandinine; thus, CYP719A13 can be involved in both sanguinarine and berberine formation in A. mexicana (Díaz Chávez et al., 2011). The opium poppy CS (CYP719A21) and the trifunctional (S)-canadine synthase/(S)-stylopine synthase/(S)-nandinine synthase CYP719A13 from A. mexicana fall in only one of the Papaveraceae subclades, suggesting that the CS activity was present in the common ancestor of Ranunculaceae and Papaveraceae, and this activity has been maintained in some Papaveraceae gene members after gene-duplication events.
BBE and (S)-SCOULERINE 9-O-METHYLTRANSFERASE (9OMT) catalyze the first two steps in the biosynthesis of berberine from (S)-reticuline. The former catalyzes a methylene-bridge-forming step to convert (S)-reticuline to (S)-scoulerine, resulting in a first tetrahydroprotoberberine structure, while the latter adds a methyl group to the 9-hydroxyl group of (S)-scoulerine (Dittrich and Kutchan, 1991, Takeshita et al., 1995). Gene tree analyses of these two genes show a tree topology similar to that of the (S)-reticuline biosynthetic genes, supporting a single-copy origin of BBE and 9OMT genes in the Ranunculales lineage (Supplemental Figure 5). Orthologs of both genes were found in all four Ranunculales genome assemblies, apart from E. californica lacking an ortholog of 9OMT. In this case, an activity similar to that of 9OMT was reported being carried out by a methyltransferase, G3OMT, belonging to a different subfamily (Purwanto et al., 2017). Taken together, we speculate that a functional replacement and gene loss of the original 9OMT may have occurred in the E. californica lineage.
In summary, since orthologs of BBE, 9OMT, and CS exist in the assembled genomes of all four species from the order Ranunculales, we propose that the protoberberine biosynthetic pathway in the Ranunculales evolved after the split of Ranunculales/Proteales around 122 MYA and prior to the divergence of Ranunculaceae and Papaveraceae at 110 MYA. The occurrence of berberine in species among most families in the order Ranunculaes is consistent with this proposal.
The Sanguinarine Pathway Emerged in the Papaveraceae Lineage 77 MYA
The first two steps involved in the conversion of tetrahydroprotoberberine (S)-scoulerine to the benzophenanthridine alkaloid sanguinarine are catalyzed by two members of the cytochrome P450 CYP719 subfamily, (S)-CHEILANTHIFOLINE SYNTHASE (CFS) and (S)-STYLOPINE SYNTHASE (SPS) (Figure 2C; Bauer and Zenk, 1989, Ikezawa et al., 2003, Ikezawa et al., 2009). All functionally characterized CFS enzymes fall into the aforementioned CFS subclade in the CYP719 tree that included sequences exclusively from Papaveraceae (Figure 4; Ikezawa et al., 2007, Ikezawa et al., 2009, Díaz Cháve et al., 2011, Yahyazadeh et al., 2017). On the other hand, all reported SPS are members of the sister SPS/CS subclade that is also composed exclusively of Papaveraceae sequences with all three genomes represented. This suggests that the neofunctionalization leading to CFS and SPS activities occurred soon after the gene-duplication event discussed above before the divergence of P. somniferum, E. californica, and M. cordata approximately 77 MYA. Interestingly, we found that the CFS and SPS genes are closely associated in the genomes of both P. somniferum and M. cordata, suggesting that they arose by tandem duplication.
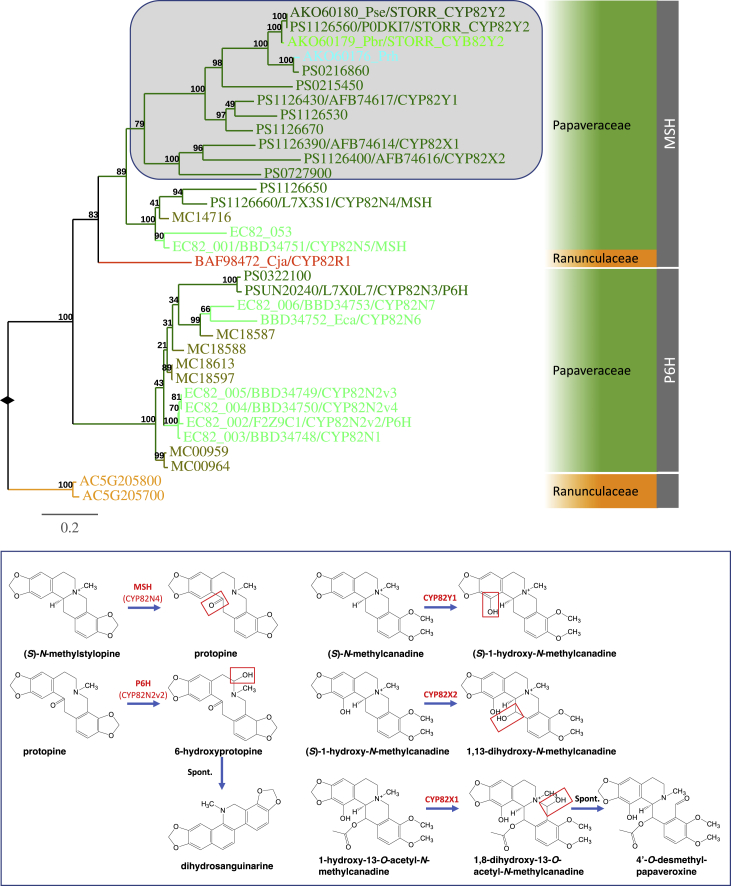
The major structural conversions from N-methylated tetrahydroprotoberberine to protopine and subsequently benzophenanthridine subclass of BIAs including sanguinarine are catalyzed by (S)-N-METHYLSTYLOPINE HYDROXYLASE (MSH) and PROTOPINE-6-HYDROXYLASE (P6H) (Figure 2C). Both belong to the CYP82N/R/X/Y subfamily of the cytochrome P450 enzymes (Rueffer and Zenk, 1987, Tanahashi and Zenk, 1990; Takemura et al., 2013, Beaudoin and Facchini, 2013). Gene tree analyses show that the members of the CYP82N/R/X/Y subfamily are to date exclusively from species of Ranunculaceae and Papaveraceae (Figure 5). Two sequences from the genome of A. coerulea form an outgroup to two sister clades. The MSH clade contains the functionally characterized MSH, CYP82N4, from opium poppy and sequences from the three Papaveraceae genomes as well as two other sequences from Papaver bracteatum and Papaver rhoeas. The functionally uncharacterized CYP82R1 is the only Ranunculaceae sequence in this clade and is sister to all Papaveraceae sequences. On the other hand, the P6H clade is composed of sequences exclusively from the three Papaveraceae genomes including the functionally characterized P6Hs, CYP82N2v2, and CYP82N3, from E. californica and P. somniferum, respectively. Thus, both MSH and P6H activities are also likely to have emerged from single ancestral orthologs before the divergence of P. somniferum, E. californica, and M. cordata 77 MYA. The presence of the CYP82R1 sequence from the Ranunculaceae species C. japonica in the MSH clade indicates that both MSH and P6H orthologs must have existed in the common ancestor of the Ranunculales. Both orthologs seem to have been lost in the lineage of A. coerulea.
Figure 5.
The CYP82N/R/X/Y Tree Shows Recruitment of CYP82 Enzymes into the Noscapine and Morphinan Pathways.
Members of CYP82N/R/X/Y subfamily from the Papaveraceae family fall into either the MSH or P6H clade. Both clades are strongly supported. The P6H clade contains all functionally characterized P6H proteins and sequences from each of the three Papaveraceae species E. californica, M. cordata, and P. somniferum. The MSH clade contains two sister subclades. One subclade contains all functionally characterized MSH enzymes, and all three Papaveraceae lineages are represented. The other comprises sequences from only Papaver species and is highlighted by the shaded box in the tree. Three CYP82 members, CYP82X1, CYP82X2, and CYP82Y1, involved in noscapine biosynthesis, fall into this subclade. CYP82Y2, the cytochrome P450 module of the STORR fusion protein, is also a member of this subclade. The figure descriptors and legends are in the same format as described in Figure 3. Extended information of the gene tree is included in Supplemental Information. The inset shows the reactions catalyzed by functionally characterized members of the CYP82N/R/X/Y subfamily with the reactive groups highlighted in red boxes.
In the sanguinarine pathway, (S)-stylopine is N-methylated by (S)-TETRAHYDROPROTOBERBERINE-N-METHYLTRANSFERASE (TNMT) after the conversion from (S)-scoulerine by CFS and then SPS, and before being hydroxylated by MSH to form protopine (Figure 2C; Rueffer et al., 1990, O'Keefe and Beecher, 1994, Liscombe and Facchini, 2007). In the TNMT clade of the N-methyltransferase tree, the four A. coerulea sequences form a robust orthologous group, sister to other sequences exclusively from Papaveraceae species including all TNMTs characterized to date (Supplemental Figure 3). Since sequences from the genomes of all three Papaveraceae species are represented, this TNMT activity must have evolved before the divergence of these three lineages.
In summary, our analyses suggest the pathways leading to the biosynthesis of the benzophenanthridine alkaloids such as sanguinarine evolved after gene duplication/neofunctionalization in the CYP719, TNMT, and CYP82N/R/X/Y families before the divergence of P. somniferum, E. californica, and M. cordata 77 MYA and after the split of Papaveraceae from Ranunculaceae around 110 MYA. Consistent with this, the benzophenanthridine subclass of BIAs and enzyme activities of CFS, SPS, TNMT, MSH, and P6H have so far only been detected in the Papaveraceae family (Krane et al., 1984, Rueffer and Zenk, 1987, Bauer and Zenk, 1989, Bauer and Zenk, 1991, Tanahashi and Zenk, 1990, Sariyar, 2002, Liscombe and Facchini, 2007).
Biosynthesis of the Phthalideisoquinoline Alkaloid Noscapine Evolved in the Opium Poppy Lineage following Specific Gene Family Expansion/Neofunctionalization
With the exception of BBE and TNMT, all genes required for the synthesis of noscapine and the first morphinan alkaloid thebaine from the branch-point intermediate (S)-reticuline occur as a major BIA gene cluster in the opium poppy genome (Winzer et al., 2012, Guo et al., 2018). Of these, CYP82X1, CYP82X2, and CYP82Y1 are involved in converting the N-methyl tetrahydroprotoberberine alkaloid (S)-N-methylcanadine to the phthalideisoquinoline alkaloid noscapine (Figure 2C, Winzer et al., 2012, Dang and Facchini, 2014). All three are members of the cytochrome P450 CYP82N/R/X/Y subfamily. Gene tree analyses show they are all in a well-supported subgroup, which is sister to that containing orthologous MSH sequences in the MSH clade (Figure 5). No sequences from the E. californica or M. cordata genomes are present in this subgroup. This suggests the gene duplications and neofunctionalization resulting in CYP82X1, CYP82X2, and CYP82Y1 occurred in the opium poppy lineage after its split from the other two Papaveroideae species.
Likewise, gene tree analyses of PSAT1 and PSCXE1, two other genes encoding proteins involved in noscapine biosynthesis (Winzer et al., 2012), support the emergence of these activities after gene duplication and neofunctionalization specifically in the opium poppy lineage (Supplemental Figures 6 and 7) with no orthologous sequence being found in the genomes of E. californica and M. cordata.
Figure 7.
Key Evolutionary Events in Benzylisoquinoline Alkaloid Metabolism as Inferred from Gene Tree and Ks Analyses.
Timing of evolutionary events are indicated by letters on the same species tree as shown in Figure 2A. Blue arrows highlight the estimated divergence time span from the analyses. The Ranunculales lineage is highlighted by an orange box, the Papaveraceae lineage in light green, and the opium poppy lineage in dark green. The inset box shows a description of the evolutionary events with the font color corresponding to specific lineages.
Noscapine biosynthesis shares the first three enzymatic steps with the protoberberine biosynthetic pathway converting (S)-reticuline to (S)-canadine (Figure 2C). In the opium poppy genome the genes encoding two of these activities from (S)-scoulerine to (S)-canadine, the opium poppy 9OMT ortholog PSMT1 and CYP719A21, are adjacent to each other head-to-head in opposite directions and separated by 1.6 kb in the BIA gene cluster. We observed that the orthologs of both genes are present in the same orientation 80 kb apart in the M. cordata genome assembly with only one annotated hypothetical gene in between. We hypothesize that these two protoberberine pathway genes represent an ancient “seeding” gene pair for the BIA gene cluster to which other genes have been recruited during the evolution of noscapine and morphinan biosynthesis.
In addition to PSMT1, PSMT2 and PSMT3 are also involved in noscapine biosynthesis. They are all members of the same O-methyltransferase family, with PSMT2 and PSMT3 being more closely related to each other than to PSMT1 (Cabry et al., 2019). PSMT2 and PSMT3 share the same exon/intron structure (Winzer et al., 2012). However, our gene tree analyses show that they fall into separate clades with orthologs from Ranunculaceae/Berberidaceae species (Supplemental Figures 2, 5, and 8), suggesting they are derived from different copies in the common ancestor of the Ranunculales. As discussed above, PSMT1 activity, also required for berberine biosynthesis, evolved in the Ranunculales common ancestor. PSMT2 is the only functionally characterized member in its clade (Supplemental Figure 6). PSMT3 is a member of the 6OMT clade and is sister to the 6OMTs from other Papaver species that are required for the biosynthesis of the central intermediate (S)-reticuline. We therefore propose that PSMT3 arose in the opium poppy lineage through neofunctionalization following gene duplication from an ancestral 6OMT copy. It has been found that the formation of a heterodimer of PSMT2 and PSMT3 is required for the substrate-specific O-methylation reaction of the hydroxyl group introduced at the C-8 position by CYP82Y1 (Li and Smolke, 2016, Li et al., 2018, Park et al., 2018). In vitro assays showed that 6OMT can replace PSMT3 but not PSMT2 in the heterodimer with it maintaining the same substrate specificity, consistent with our analyses that PSMT3 and 6OMT are closely related.
All three O-methyltransferases are present within a 150-kb segment in the genome as part of the BIA gene cluster, with PSMT2 and PSMT3 being 77 kb apart, only separated by CYP82Y1 (Winzer et al., 2012, Guo et al., 2018). Despite the physical proximity and sharing the same exon/intron structure, our gene tree analyses show that PSMT2 and PSMT3 must have been recruited to the BIA gene cluster independently.
Taken together, our analysis suggests that the components of the noscapine biosynthetic pathway following conversion of (S)-canadine to (S)-N-methylcanadine by TNMT have evolved in the opium poppy lineage after divergence from E. californica and M. cordata 77 MYA. Analysis of the genomes from more species within the opium poppy lineage would be necessary to improve the resolution of the evolutionary time frame leading to the emergence of the noscapine biosynthetic pathway and the assembly of the BIA gene cluster.
Biosynthesis of the Morphinan Alkaloids Emerged in the Papaver Genus
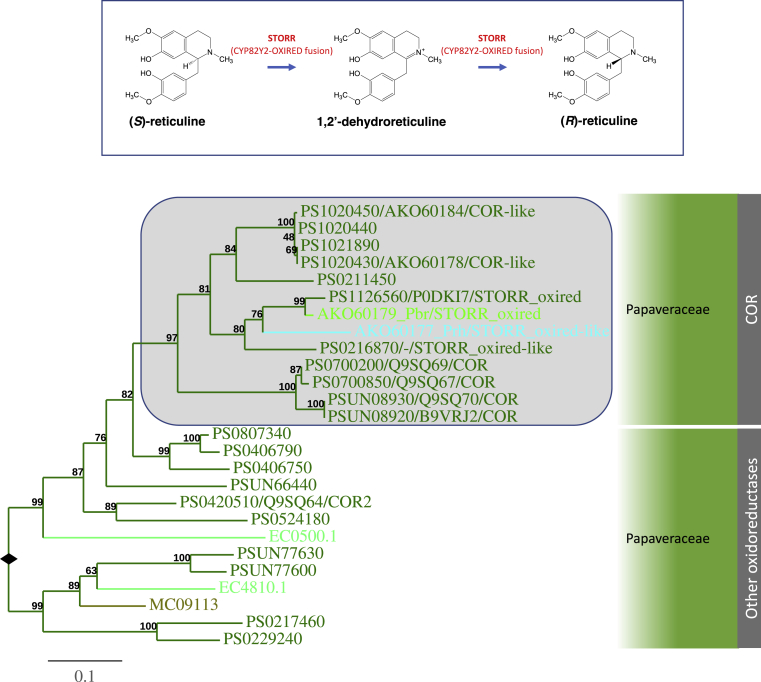
All five genes, including STORR, SALUTARIDINE SYNTHASE (SALSYN), SALUTARIDINOL-7-O-ACETYLTRANSFERASE (SALAT), SALUTARIDINE REDUCTASE (SALR), and THEBAINE SYNTHASE (THS), required for the first morphinan alkaloid thebaine from (S)-reticuline are part of the BIA gene cluster in opium poppy. The emergence of the STORR fusion is considered a key event for the evolution of morphinan biosynthesis in opium poppy, enabling the conversion of (S)-reticuline to (R)-reticuline (Figure 2C; Farrow et al., 2015, Winzer et al., 2015). The CYP82N/R/X/Y gene tree shows that CYP82Y2, the cytochrome P450 module of the opium poppy STORR fusion protein, groups strongly with those of STORR orthologs from Papaver setigerum and P. bracteatum. Both CYP82Y2 and its closest paralog, PS0216860, belong to the same subgroup as CYP82X1, CYP82X2, and CYP82Y1 which is sister to the MSH orthologs in the MSH clade (Figure 5). No sequences from the genomes of E. californica and M. cordata are present in this subgroup. Thus, we conclude that the gene duplication giving rise to the CYP82Y2 module of STORR occurred in the lineage leading to opium poppy after its split from E. californica and M. cordata.
The oxidoreductase module of the STORR fusion protein and its closest paralog PS0216870 are members of an oxidoreductase family closely related to the CODEINONE REDUCTASE (COR) proteins, the enzymes involved in the last steps of morphine biosynthesis (Figures 2C and 6). The COR clade contains sequences exclusively from the genus Papaver, mostly from the opium poppy genome. The analyses of both modules show specific gene family expansion in the opium poppy lineage for both families, and a fragmental genome duplication that occurred prior to the STORR fusion event.
Figure 6.
Opium Poppy Lineage-Specific Expansion of the Oxidoreductase Subfamily Gave Rise to the STORR Oxidoreductase Module.
The Papaveraceae oxidoreductase subfamily contains sequences mostly from opium poppy. A COR clade is nested within a much larger group containing only Papaver sequences and highlighted by the shaded box in the tree. The COR clade contains 11 sequences from the opium poppy genome including both the STORR oxidoreductase module and four COR isoforms. The figure descriptors and legends are the in same format as described in Figure 3. Extended information of the gene tree is included in Supplemental Information. The inset shows the reaction catalyzed by STORR.
SALSYN and SALAT are two enzymes involved in the structural conversions of the 1-benzylisoquinoline alkaloid (R)-reticuline to promorphinans (Figure 2C). Both are in groups containing exclusively Papaver sequences in their respective gene trees, further supporting our hypothesis that the morphinan pathway evolved within the opium poppy lineage (Figure 4 and Supplemental Figure 6). SALSYN (CYP719B1) exclusively converts the R-epimer of reticuline to salutaridine (Gesell et al., 2009), a reaction requiring intramolecular C–C phenol coupling. This reaction would have required extensive neofunctionalization, as other functionally characterized CYP719 proteins all catalyze the formation of methylenedioxy bridges (Figure 4). SALAT carries out the O-acetylation of salutaridinol and is one of the closest paralogs to PSAT1, an enzyme involved in the noscapine pathway (Supplemental Figure 6; Lenz and Zenk, 1995, Grothe et al., 2001). Dating the divergence time between SALAT and PSAT1 would require additional genomes in the opium poppy lineage, which would provide useful insight into the evolutionary timing and relationship of the morphinan and noscapine biosynthetic pathways.
While the emergence of STORR opened the gateway for promorphinan biosynthesis and ultimately the first morphinan alkaloid thebaine, the evolution of T6ODM and CODM was crucial for the production of the morphinan alkaloid morphine (Farrow and Facchini, 2013). T6ODM, CODM, and COR are not part of the BIA gene cluster in opium poppy, but each occurs in multiple locally linked copies (Guo et al., 2018). Both CODM and T6ODM are members of the 2-OXOGLUTARATE/Fe(II)-DEPENDENT DIOXYGENASE family that also went through recent rapid family expansion in the opium poppy lineage, similar to the COR clade (Supplemental Figure 9).
In the absence of other genome assemblies from species within the opium poppy lineage, we calculated the number of synonymous substitutions per synonymous site (Ks) between paralogous gene pairs to gain insight about the timing of these gene-duplication events. Previously, we found that the closest paralogs (PS0216860 and PS0216870) of the STORR P450 and oxidoreductase modules are located just 865 bp apart on a different chromosome of the opium poppy genome (Guo et al., 2018). From this we hypothesized that the STORR gene fusion event must have followed a segmental duplication. We have now calculated the Ks values of CYP82Y2/PS0216860 and STORR-oxired/PS0216870 paralog pairs with all 17 different substitution models with the KaKs_Calculator software (Wang et al., 2010). These are all in the range of 0.279456–0.387986 and 0.290438–0.356423, respectively (Supplemental Table 2). Based on a synonymous substitution rate (r) of 6.98 per billion years (109) (Guo et al., 2018), the time (T) for this duplication would be in the range of 20.0–27.8 and 20.8–25.5 MYA, respectively (T = Ks/2r). Thus, based on our reasoning that the segmental duplication event must have occurred prior to the STORR gene fusion, we propose that the emergence of the pathway to morphinan biosynthesis occurred more recently than 20 MYA.
The gene duplication of the common ancestor that eventually gave rise to both CODM and T6ODM through neofunctionalization can also provide an important time point for the evolution of morphine biosynthesis. We calculated the pairwise Ks between the three CODM and five T6ODM gene copies. All are between 0.245096 and 0.327976, pointing to a possible time of most recent ancestral gene duplication giving rise to these two enzyme activities at 17.6–23.5 MYA.
Similar to noscapine, our gene tree analysis of CYP82N/R/X/Y, CYP719, ACETYLTRANSFERASE, COR, and CODM/T6ODM indicate that the morphine biosynthetic pathway arose through gene duplications, gene fusion, and neofunctionalization events that built on activities derived from the existing protoberberine and sanguinarine biosynthetic pathways in the opium poppy lineage after the divergence from the E. californica and M. cordata lineages 77 MYA. Ks estimation of the paralogous pairs of the STORR modules and CODM/T6ODM has enabled us to speculate that the morphine pathway may have arisen more recently than 18 MYA, which is consistent with the proposed emergence of the Papaver genus around 19–29 MYA (Valtueña et al., 2012, Xie et al., 2014).
Concluding from our tree analyses, we propose a monophyletic origin for all five genes required for the biosynthesis of (S)-reticuline, the central intermediate of BIA metabolism in the order Ranunculales. The biosynthetic pathway leading to production of the protoberberine alkaloid, berberine, emerged before the divergence of the Papaveraceae and Ranunculaceae families between 110 and 122 MYA. BIA synthesis in N. nucifera, which belongs to the order Proteales, is very likely to have evolved through a parallel process (Figure 7). Emergence of the sanguinarine pathway occurred after the Papaveraceae–Ranunculaceae divergence 110 MYA but before the split of the three Papaveraceae species at 77 MYA. We found that the gene duplications necessary for subsequent gene fusion and neofunctionalization of essential genes involved in the production of both the phthalideisoquinoline alkaloid noscapine and morphinans including codeine and morphine occurred exclusively in the opium poppy lineage. We therefore propose that the biosynthetic pathways involved in both noscapine and morphinans emerged more recently in the opium poppy lineage after its divergence from the E. californica and M. cordata lineages at 77 MYA. Based on Ks estimation of the paralogous pairs of the two STORR modules and CODM/T6ODM, we further propose that morphine biosynthesis evolved more recently than 18 MYA in the Papaver genus.
Further validation of the conclusions drawn from our analysis of the available draft genome assemblies of five BIA-producing species will be possible once genome assemblies of other BIA-accumulating species from within the Papaveraceae and Ranunculaceae as well as those from other families in the Ranunculales such as Berberidaceae, Menispermaceae, Circaeasteraceae, and Lardizabaloideae become available. In this respect we note that in our analysis, we infer the monophyletic origin of BIA biosynthetic pathways in Ranunculales based on the understanding that the Papaveraceae diverged from the Berberidaceae, Menispermaceae, and Ranunculaceae families earlier than the latter three diverged from each other according to the Angiosperm Phylogeny Group. Ultimately, functional characterization of genes involved in BIA biosynthesis across the range of species that produce BIAs including N. nucifera, A. coerulea, and M. cordata will be necessary in order to be absolutely definitive about our understanding of evolutionary events that resulted in the emergence of BIA metabolism in the Ranunculales. Despite these limitations we consider that our analysis, representing as it does a complete coverage of the gene space provided by the whole-genome assembly of five relevant species, has allowed us to propose an orthologous history at the sequence level for the BIA genes and reveal key events in the evolution of BIA biosynthetic pathways.
This study demonstrates the effectiveness of using gene tree analysis of whole-genome assemblies to identify key evolutionary events leading to the emergence of the structural genes necessary for new biosynthetic pathways to evolve. The recruitment model of metabolic evolution proposes that the emergence of new biochemical pathways requires the co-evolution of structural genes and associated transcription factors resulting in a regulon of coordinately expressed genes (Shoji, 2019). The metabolic evolution of noscapine and morphinan biosynthesis is particularly interesting in this regard, as the regulon includes 15 genes in a localized genomic region of 600 kb in the opium poppy genome plus additional genes such as BBE and TNMT that are not specific to noscapine or morphinan biosynthesis and others that are specific only to morphine production including T6ODM, CODM, and COR. Now that we have established key evolutionary events leading to the emergence of the structural genes, the challenge is to integrate these events with those involved in gene regulation at both the transcription-factor and genome-organization levels giving rise to the BIA 15 gene cluster.
Methods
Estimation of Species Divergence Times of Five BIA-Producing Plants
Divergence of seven plant species was dated with BEAST2 v2.5.1 (Bouckaert et al., 2019) based on a concatenated dataset of orthologous groups from their draft genomes. The five BIA-producing plants include four species in the order Ranunculales and N. nucifera, a member of the Nelumbonaceae family in the order Proteales. The four Ranunculales species are P. somniferum, E. californica, and M. cordata from the Papaveraceae family and A. coerulea from the Ranunculaceae family. M. integrifolia, the only other member of Proteales with its genome assembly available, was included (Nock et al., 2016), whereas monocot rice was used as an outgroup (Kawahara et al., 2013). Annotation datasets of all seven genomes can be downloaded from publicly available sources (Supplemental Table 3).
OrthoFinder v2.0 (Emms and Kelly, 2015) was used to identify orthologous groups across the seven genomes with their annotated datasets. Protein sequence alignments were then constructed for each orthologous group with ClustalX (Thompson et al., 2002), and the conserved blocks in each alignment were evaluated and selected with Gblocks v0.91b by allowing gap positions within final blocks (Castresana, 2000). A final alignment was constructed by concatenating all conserved blocks, and used in the subsequent analyses. This was then used to estimate species divergence times based on a Bayesian approach with BEAST2.
For the divergence time estimation, we used the birth–death model for tree priors and a Jones–Taylor–Thornton substitution model with a strict clock rate. To calibrate divergence times, an exponential model was chosen for monocot–dicot split time (mean: 150 MYA; SD: 4 MYA) and Papaveraceae–Ranunculaceae split time (mean: 110 MYA; SD: 4 MYA) (Guo et al., 2018). The Markov chain Monte Carlo was repeated 500 000 000 times with 1000 steps to allow chain convergence. Tracer v1.7.1 (Rambaut et al., 2018) was employed to check chain convergence and ensure satisfactory effective sampling sizes for all parameters. The maximum clade credibility tree was calculated with TreeAnnotator v2.5.1 within the BEAST2 software package, summarizing the estimated mean age and 95% confidence intervals from post-burn-in (20%) trees.
Gene Tree Analyses
We used a phylogenetic approach to examine the orthologous relationships of members of all gene families that contain functionally characterized genes involved in the biosynthesis of berberine, sanguinarine, noscapine, and morphinan alkaloids in BIA-producing plants. All query sequences are listed in Supplemental Table 4. For each query sequence, local BLASTP searches were performed against the annotated protein datasets of all seven genomes described above (Altschul et al., 1997). The sequences with an expected value lower than e−20 or within top 100 hits of the lowest expected values were then retrieved from each annotated protein datasets of all seven species. Parallel BLASTP searches for each query were also carried out against the non-redundant protein sequences (NR) database at the NCBI. A maximum number of 400 sequences were retrieved with an expected value lower than e−20. These two sets of resulting sequences were then combined for each of the starting query sequences. Duplicated and partial sequences were removed together with other synthetic sequence entries in the NR database. Species identification associated with the NR database entries were verified and corrected where possible by checking the associated publications.
Protein sequence alignments were made firstly with ClustalX. Conserved blocks in each alignment were evaluated and selected with Gblocks analysis. The best-scoring maximum-likelihood tree was inferred in conjunction with bootstrap analyses of 100 replicates using RAxML v8.2.12 (Stamatakis, 2006). The substitution model was the same as used in the above BEAST2 analyses. The trees from this first round of analyses were then evaluated. A subset of sequences was selected for a further round of analyses if (1) it contained the query sequence and (2) it formed a group with above 50% bootstrap value support. Not all gene tree analyses yielded informative results in terms of strong bootstrap support. This may be due to either too little starting data (short sequences) or too few informative sites caused by extreme mutation rates along the lineages. As such, the gene families containing STOX, DBOX, SDR1, SALR, THS, and NISO did not give informative phylogenetic relationships, and were therefore not included in further analyses. For the gene families with a clear subset identified, a further round of Gblocks analyses was performed to allow more residues to be included in the conserved blocks in order to achieve higher bootstrap support value in the subsequent phylogenetic analyses with RAxML. This process was reiterated whenever further improvements could be made. The final sets of sequences selected for the gene analyses are summarized in Supplemental Table 5. In the final trees, groups with above 70% bootstrap value were considered as strongly supported.
Estimation of Divergence Times between Paralogs Using Ks Analyses
KaKs_Calculator v2.0 (Wang et al., 2010) was used to estimate the timing of the gene-duplication events between paralog gene pairs in opium poppy genome. These include the pairings of the STORR P450 and oxidoreductase modules and their corresponding closest paralogs (PS0216860 and PS0216870) as well as the pairings between the T6ODM and CODM copies. The coding sequences of the paralog pairs were aligned using the MUSCLE alignment tool (Edgar, 2004). The alignments were then converted into the input format required for the KaKs_Calculator software to estimate Ks values between paralog pairs.
The divergence times of the gene duplication leading to the paralog pair were then estimated with the formula T = Ks/2r, where T denotes divergence time, Ks the number of synonymous substitutions, and r the substitution rate. With the absence of the absolute substitution rate for the specific gene families, we used a synonymous substitution rate (r) of 6.98 per billion (109) years that was estimated in the opium poppy lineage after its divergence from the Ranunculaceae species A. coerulea (Guo et al., 2018).
Funding
I.A.G. received support from the Biotechnology and Biological Sciences Research Council, United Kingdom (grant BB/K018809/1) and the Garfield Weston Foundation, United Kingdom.
Author Contributions
Y.L. performed all species and gene tree analyses and Z.H. carried out the OrthoFinder analysis. Y.L., T.W., and I.A.G. analyzed and interpreted results and were major contributors in writing the manuscript. All authors read and approved the final manuscript.
Acknowledgments
No conflict of interest declared.
Published: February 5, 2020
Footnotes
Published by the Plant Communications Shanghai Editorial Office in association with Cell Press, an imprint of Elsevier Inc., on behalf of CSPB and IPPE, CAS.
Supplemental Information is available at Plant Communications Online.
Supplemental Information
References
- Altschul S.F., Madden T.L., Schäffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer W., Zenk M.H. Formation of both methylenedioxy groups in the alkaloid (S)-stylopine is catalyzed by cytochrome P-450 enzymes. Tetrahedron Lett. 1989;30:5257–5260. [Google Scholar]
- Bauer W., Zenk M.H. Two methylenedioxy bridge forming cytochrome P-450 dependent enzymes are involved in (S)-stylopine biosynthesis. Phytochemistry. 1991;30:2953–2961. [Google Scholar]
- Beaudoin G.A., Facchini P.J. Isolation and characterization of a cDNA encoding (S)-cis-N-methylstylopine 14-hydroxylase from opium poppy, a key enzyme in sanguinarine biosynthesis. Biochem. Biophys. Res. Commun. 2013;431:597–603. doi: 10.1016/j.bbrc.2012.12.129. [DOI] [PubMed] [Google Scholar]
- Blaskó G., Gula D.J., Shamma M. The phthalideisoquinoline alkaloids. J. Nat. Prod. 1982;45:105–122. [Google Scholar]
- Bouckaert R., Vaughan T.G., Barido-Sottani J., Duchêne S., Fourment M., Gavryushkina A., Heled J., Jones G., Kühnert D., De Maio N. Beast 2.5: an advanced software platform for Bayesian evolutionary analysis. PLoS Comput. Biol. 2019;15:e1006650. doi: 10.1371/journal.pcbi.1006650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabry M.P., Offen W.A., Saleh P., Li Y., Winzer T., Graham I.A., Davies G.J. Structure of papaver somniferum O-methyltransferase 1 reveals initiation of noscapine biosynthesis with implications for plant natural product methylation. ACS Catal. 2019;9:3840–3848. [Google Scholar]
- Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 2000;17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- Choi K.B., Morishige T., Shitan N., Yazaki K., Sato F. Molecular cloning and characterization of coclaurine N-methyltransferase from cultured cells of Coptis japonica. J. Biol. Chem. 2002;277:830–835. doi: 10.1074/jbc.M106405200. [DOI] [PubMed] [Google Scholar]
- Dang T.T., Facchini P.J. CYP82Y1 is N-methylcanadine 1-hydroxylase, a key noscapine biosynthetic enzyme in opium poppy. J. Biol. Chem. 2014;289:2013–2026. doi: 10.1074/jbc.M113.505099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desgagné-Penix I., Khan M.F., Schriemer D.C., Cram D., Nowak J., Facchini P.J. Integration of deep transcriptome and proteome analyses reveals the components of alkaloid metabolism in opium poppy cell cultures. BMC Plant Biol. 2010;10:252. doi: 10.1186/1471-2229-10-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díaz Cháve M.L., Rolf M., Gesell A., Kutchan T.M. Characterization of two methylenedioxy bridge-forming cytochrome P450-dependent enzymes of alkaloid formation in the Mexican prickly poppy Argemone mexicana. Arch. Biochem. Biophys. 2011;507:186–193. doi: 10.1016/j.abb.2010.11.016. [DOI] [PubMed] [Google Scholar]
- Dittrich H., Kutchan T.M. Molecular cloning, expression, and induction of berberine bridge enzyme, an enzyme essential to the formation of benzophenanthridine alkaloids in the response of plants to pathogenic attack. Proc. Natl. Acad. Sci. U S A. 1991;88:9969–9973. doi: 10.1073/pnas.88.22.9969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emms D.M., Kelly S. OrthoFinder: solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol. 2015;16:157. doi: 10.1186/s13059-015-0721-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrow S.C., Facchini P.J. Dioxygenases catalyze O-demethylation and O,O-demethylenation with widespread roles in benzylisoquinoline alkaloid metabolism in opium poppy. J. Biol. Chem. 2013;288:28997–29012. doi: 10.1074/jbc.M113.488585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrow S.C., Hagel J.M., Beaudoin G.A., Burns D.C., Facchini P.J. Stereochemical inversion of (S)-reticuline by a cytochrome P450 fusion in opium poppy. Nat. Chem. Biol. 2015;11:728–732. doi: 10.1038/nchembio.1879. [DOI] [PubMed] [Google Scholar]
- Filiault D.L., Ballerini E.S., Mandáková T., Aköz G., Derieg N.J., Schmutz J., Jenkins J., Grimwood J., Shu S., Hayes R.D. The Aquilegia genome provides insight into adaptive radiation and reveals an extraordinarily polymorphic chromosome with a unique history. eLife. 2018;7 doi: 10.7554/eLife.36426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frenzel T., Zenk M.H. S-adenosyl-L-methionine: 3′-hydroxy-N-methyl-(S)-coclaurine-4′-O-methyl transferase, a regio- and stereoselective enzyme of the (S)-reticuline pathway. Phytochemistry. 1990;29:3505–3511. [Google Scholar]
- Frenzel T., Zenk M.H. Purification and characterization of three isoforms of S-adenosyl-L-methionine: (R,S)-tetrahydrobenzylisoquinoline-N-methyltransferase from Berberis koetineana cell cultures. Phytochemistry. 1990;29:3491–3497. [Google Scholar]
- Gesell A., Rolf M., Ziegler J., Díaz Chávez M.L., Huang F.C., Kutchan T.M. CYP719B1 is salutaridine synthase, the C-C phenol-coupling enzyme of morphine biosynthesis in opium poppy. J. Biol. Chem. 2009;284:24432–24442. doi: 10.1074/jbc.M109.033373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grothe T., Lenz R., Kutchan T.M. Molecular characterization of the salutaridinol 7-O-acetyltransferase involved in morphine biosynthesis in opium poppy Papaver somniferum. J. Biol. Chem. 2001;276:30717–30723. doi: 10.1074/jbc.M102688200. [DOI] [PubMed] [Google Scholar]
- Gui S., Peng J., Wang X., Wu Z., Cao R., Salse J., Zhang H., Zhu Z., Xia Q., Quan Z. Improving Nelumbo nucifera genome assemblies using high-resolution genetic maps and BioNano genome mapping reveals ancient chromosome rearrangements. Plant J. 2018;94:721–734. doi: 10.1111/tpj.13894. [DOI] [PubMed] [Google Scholar]
- Guo L., Winzer T., Yang X., Li Y., Ning Z., He Z., Teodor R., Lu Y., Bowser T.A., Graham I.A. The opium poppy genome and morphinan production. Science. 2018;362:343–347. doi: 10.1126/science.aat4096. [DOI] [PubMed] [Google Scholar]
- Hagel J.M., Facchini P.J. Benzylisoquinoline alkaloid metabolism: a century of discovery and a brave new world. Plant Cell Physiol. 2013;54:647–672. doi: 10.1093/pcp/pct020. [DOI] [PubMed] [Google Scholar]
- Hagel J.M., Mandal R., Han B., Han J., Dinsmore D.R., Borchers C.H., Wishart D.S., Facchini P.J. Metabolome analysis of 20 taxonomically related benzylisoquinoline alkaloid-producing plants. BMC Plant Biol. 2015;15:220. doi: 10.1186/s12870-015-0594-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori K., Yamada Y., Purwanto R., Minakuchi Y., Toyoda A., Hirakawa H., Sato F. Mining of the uncharacterized cytochrome P450 genes involved in alkaloid biosynthesis in California poppy using a draft genome sequence. Plant Cell Physiol. 2018;59:222–233. doi: 10.1093/pcp/pcx210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang F.C., Kutchan T.M. Distribution of morphinan and benzo[c]phenanthridine alkaloid gene transcript accumulation in Papaver somninferum. Phytochemistry. 2000;53:555–564. doi: 10.1016/s0031-9422(99)00600-7. [DOI] [PubMed] [Google Scholar]
- Ikezawa N., Tanaka M., Nagayoshi M., Shinkyo R., Sakaki T., Inouye K., Sato F. Molecular cloning and characterization of CYP719, a methylenedioxy bridge-forming enzyme that belongs to a novel P450 family, from cultured Coptis japonica cells. J. Biol. Chem. 2003;278:38557–38565. doi: 10.1074/jbc.M302470200. [DOI] [PubMed] [Google Scholar]
- Ikezawa N., Iwasa K., Sato F. Molecular cloning and characterization of methylenedioxy bridge-forming enzymes involved in stylopine biosynthesis in Eschscholzia californica. FEBS J. 2007;274:1019–1035. doi: 10.1111/j.1742-4658.2007.05652.x. [DOI] [PubMed] [Google Scholar]
- Ikezawa N., Iwasa K., Sato F. CYP719A subfamily of cytochrome P450 oxygenases and isoquinoline alkaloid biosynthesis in Eschscholzia californica. Plant Cell Rep. 2009;28:123–133. doi: 10.1007/s00299-008-0624-8. [DOI] [PubMed] [Google Scholar]
- Kawahara Y., de la Bastide M., Hamilton J.P., Kanamori H., McCombie W.R., Ouyang S., Schwartz D.C., Tanaka T., Wu J., Zhou S. Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice (N. Y.) 2013;6:4. doi: 10.1186/1939-8433-6-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krane B.D., Fagbule M.O., Shamma M., Gozler B. The benzophenanthridine alkaloids. J. Nat. Prod. 1984;47:1–43. [Google Scholar]
- Lee E.J., Facchini P. Norcoclaurine synthase is a member of the pathogenesis-related 10/Bet v1 protein family. Plant Cell. 2010;22:3489–3503. doi: 10.1105/tpc.110.077958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz R., Zenk M.H. Acetyl coenzyme A:salutaridinol-7-O-acetyltransferase from Papaver somniferum plant cell cultures. The enzyme catalyzing the formation of thebaine in morphine biosynthesis. J. Biol. Chem. 1995;270:31091–31096. doi: 10.1074/jbc.270.52.31091. [DOI] [PubMed] [Google Scholar]
- Li Y., Smolke C.D. Engineering biosynthesis of the anticancer alkaloid noscapine in yeast. Nat. Commun. 2016;7:12137. doi: 10.1038/ncomms12137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Li S., Thodey K., Trenchard I., Cravens A., Smolke C.D. Complete biosynthesis of noscapine and halogenated alkaloids in yeast. Proc. Natl. Acad. Sci. U S A. 2018;115:E3922–E3931. doi: 10.1073/pnas.1721469115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichman B.R., Sula A., Pesnot T., Hailes H.C., Ward J.M., Keep N.H. Structural Evidence for the dopamine-first mechanism of norcoclaurine synthase. Biochemistry. 2017;56:5274–5277. doi: 10.1021/acs.biochem.7b00769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liscombe D.K., Facchini P.J. Molecular cloning and characterization of tetrahydroprotoberberine cis-N-methyltransferase, an enzyme involved in alkaloid biosynthesis in opium poppy. J. Biol. Chem. 2007;282:14741–14751. doi: 10.1074/jbc.M611908200. [DOI] [PubMed] [Google Scholar]
- Liscombe D.K., MacLeod B.P., Loukanina N., Nandi O.I., Facchini P.J. Evidence for the monophyletic evolution of benzylisoquinoline alkaloid biosynthesis in angiosperms. Phytochemistry. 2005;66:2501–2520. doi: 10.1016/j.phytochem.2005.04.044. [DOI] [PubMed] [Google Scholar]
- Liu X., Liu Y., Huang P., Ma Y., Qing Z., Tang Q., Cao H., Cheng P., Zheng Y., Yuan Z. The Genome of medicinal plant Macleaya cordata provides new insights into benzylisoquinoline alkaloids metabolism. Mol. Plant. 2017;10:975–989. doi: 10.1016/j.molp.2017.05.007. [DOI] [PubMed] [Google Scholar]
- Loeffler S., Zenk M.H. The hydroxylation step in the biosynthetic pathway leading from norcoclaurine to reticuline. Phytochemistry. 1990;29:3499–3503. [Google Scholar]
- Marques J.V., Kim K.W., Lee C., Costa M.A., May G.D., Crow J.A., Davin L.B., Lewis N.G. Next generation sequencing in predicting gene function in podophyllotoxin biosynthesis. J. Biol. Chem. 2013;288:466–479. doi: 10.1074/jbc.M112.400689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menéndez-Perdomo I.M., Facchini P.J. Benzylisoquinoline alkaloids biosynthesis in sacred lotus. Molecules. 2018;23 doi: 10.3390/molecules23112899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minami H., Dubouzet E., Iwasa K., Sato F. Functional analysis of norcoclaurine synthase in Coptis japonica. J. Biol. Chem. 2007;282:6274–6282. doi: 10.1074/jbc.M608933200. [DOI] [PubMed] [Google Scholar]
- Ming R., VanBuren R., Liu Y., Yang M., Han Y., Li L.T., Zhang Q., Kim M.J., Schatz M.C., Campbell M. Genome of the long-living sacred lotus (Nelumbo nucifera Gaertn.) Genome Biol. 2013;14:R41. doi: 10.1186/gb-2013-14-5-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morishige T., Tsujita T., Yamada Y., Sato F. Molecular characterization of the S-adenosyl-L-methionine:3'-hydroxy-N-methylcoclaurine 4'-O-methyltransferase involved in isoquinoline alkaloid biosynthesis in Coptis japonica. J. Biol. Chem. 2000;275:23398–23405. doi: 10.1074/jbc.M002439200. [DOI] [PubMed] [Google Scholar]
- Neag M.A., Mocan A., Echeverría J., Pop R.M., Bocsan C.I., Crişan G., Buzoianu A.D. Berberine: botanical occurrence, traditional uses, extraction methods, and relevance in cardiovascular, metabolic, hepatic, and renal disorders. Front. Pharmacol. 2018;9:557. doi: 10.3389/fphar.2018.00557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nock C.J., Baten A., Barkla B.J., Furtado A., Henry R.J., King G.J. Genome and transcriptome sequencing characterises the gene space of Macadamia integrifolia (Proteaceae) BMC Genomics. 2016;17:937. doi: 10.1186/s12864-016-3272-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Och A., Szewczyk K., Pecio Ł., Stochmal A., Załuski D., Bogucka-Kocka A. UPLC-MS/MS profile of alkaloids with cytotoxic properties of selected medicinal plants of the Berberidaceae and Papaveraceae families. Oxid Med. Cell Longev. 2017;2017:9369872. doi: 10.1155/2017/9369872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Keefe B.R., Beecher C. Isolation and characterization of S-adenosyl-L-methionine:tetrahydroberberine-cis-N-methyltransferase from suspension cultures of Sanguinaria canadensis L. Plant Physiol. 1994;105:395–403. doi: 10.1104/pp.105.1.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park M.R., Chen X., Lang D.E., Ng K.K.S., Facchini P.J. Heterodimeric O-methyltransferases involved in the biosynthesis of noscapine in opium poppy. Plant J. 2018;95:252–267. doi: 10.1111/tpj.13947. [DOI] [PubMed] [Google Scholar]
- Pauli H.H., Kutchan T.M. Molecular cloning and functional heterologous expression of two alleles encoding (S)-N-methylcoclaurine 3'-hydroxylase (CYP80B1), a new methyl jasmonate-inducible cytochrome P450-dependent mono-oxygenase of benzylisoquinoline alkaloid biosynthesis. Plant J. 1998;13:793–801. doi: 10.1046/j.1365-313x.1998.00085.x. [DOI] [PubMed] [Google Scholar]
- Purwanto R., Hori K., Yamada Y., Sato F. Unraveling additional O-methylation steps in benzylisoquinoline alkaloid biosynthesis in California poppy (Eschscholzia californica) Plant Cell Physiol. 2017;58:1528–1540. doi: 10.1093/pcp/pcx093. [DOI] [PubMed] [Google Scholar]
- Rambaut A., Drummond A.J., Xie D., Baele G., Suchard M.A. Posterior summarization in bayesian phylogenetics using tracer 1.7. Syst. Biol. 2018;67:901–904. doi: 10.1093/sysbio/syy032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rueffer M., Zenk M.H. Enzymatic formation of protopines by a microsomal cytochrome P-450 system of Corydalis vaginans. Tetrahedron Lett. 1987;28:5307–5310. [Google Scholar]
- Rueffer M., Zenk M.H. Canadine synthase from Thalictrum tuberosum cell cultures catalyses the formation of the methylenedioxy bridge in berberine synthesis. Phytochemistry. 1994;36:1219–1223. [Google Scholar]
- Rueffer M., Zumstein G., Zenk M.H. Partial purification and properties of S-adenosyl-l-methionine: (S)-tetrahydroprotoberberinecis-N-methyltransferase from suspension-cultured cells of Eschscholtzia and Corydalis. Phytochemistry. 1990;29:3727–3733. [Google Scholar]
- Samanani N., Facchini P.J. Isolation and partial characterization of norcoclaurine synthase, the first committed step in benzylisoquinoline alkaloid biosynthesis, from opium poppy. Planta. 2001;213:898–906. doi: 10.1007/s004250100581. [DOI] [PubMed] [Google Scholar]
- Samanani N., Liscombe D.K., Facchini P.J. Molecular cloning and characterization of norcoclaurine synthase, an enzyme catalyzing the first committed step in benzylisoquinoline alkaloid biosynthesis. Plant J. 2004;40:302–313. doi: 10.1111/j.1365-313X.2004.02210.x. [DOI] [PubMed] [Google Scholar]
- Sariyar G. Biodiversity in the alkaloids of Turkish Papaver species. Pure Appl. Chem. 2002;74:557–574. [Google Scholar]
- Schmidt J., Boettcher C., Kuhnt C., Kutchan T.M., Zenk M.H. Poppy alkaloid profiling by electrospray tandem mass spectrometry and electrospray FT-ICR mass spectrometry after [ring-13C6]-tyramine feeding. Phytochemistry. 2007;68:189–202. doi: 10.1016/j.phytochem.2006.10.003. [DOI] [PubMed] [Google Scholar]
- Shoji T. The recruitment model of metabolic evolution: jasmonate-responsive transcription factors and a conceptual model for the evolution of metabolic pathways. Front. Plant Sci. 2019;10:560. doi: 10.3389/fpls.2019.00560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadler R., Kutchan T.M., Zenk M.H. (S)-Norcoclaurine is the central intermediate in benzylisoquinoline alkaloid biosynthesis. Phytochemistry. 1989;28:1083–1086. [Google Scholar]
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Takemura T., Ikezawa N., Iwasa K., Sato F. Molecular cloning and characterization of a cytochrome P450 in sanguinarine biosynthesis from Eschscholzia californica cells. Phytochemistry. 2013;91:100–108. doi: 10.1016/j.phytochem.2012.02.013. [DOI] [PubMed] [Google Scholar]
- Takeshita N., Fujiwara H., Mimura H., Fitchen J.H., Yamada Y., Sato F. Molecular cloning and characterization of S-adenosyl-L-methionine:scoulerine-9-O-methyltransferase from cultured cells of Coptis japonica. Plant Cell Physiol. 1995;36:29–36. [PubMed] [Google Scholar]
- Tanahashi T., Zenk M.H. Elicitor induction and characterization of microsomal protopine-6-hydroxylase, the central enzyme in benzophenanthridine alkaloid biosynthesis. Phytochemistry. 1990;29:1113–1122. [Google Scholar]
- Thompson J.D., Gibson T.J., Higgins D.G. Multiple sequence alignment using ClustalW and ClustalX. Curr. Protoc. Bioinformatics. 2002 doi: 10.1002/0471250953.bi0203s00. Chapter 2, Unit 2.3. [DOI] [PubMed] [Google Scholar]
- Valtueña F.J., Preston C.D., Kadereit J.W. Phylogeography of a Tertiary relict plant, Meconopsis cambrica (Papaveraceae), implies the existence of northern refugia for a temperate herb. Mol. Ecol. 2012;21:1423–1437. doi: 10.1111/j.1365-294X.2012.05473.x. [DOI] [PubMed] [Google Scholar]
- Vimolmangkang S., Deng X., Owiti A., Meelaph T., Ogutu C., Han Y. Evolutionary origin of the NCSI gene subfamily encoding norcoclaurine synthase is associated with the biosynthesis of benzylisoquinoline alkaloids in plants. Sci. Rep. 2016;6:26323. doi: 10.1038/srep26323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Zhang Y., Zhang Z., Zhu J., Yu J. KaKs_Calculator 2.0: a toolkit incorporating gamma-series methods and sliding window strategies. Genomics Proteomics Bioinformatics. 2010;8:77–80. doi: 10.1016/S1672-0229(10)60008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winek C.L., Beal J.L., Cava M.P. Comparative chromatograghic identification of the alkaloids of ten Aquilegia species. J. Pharm. Sci. 1964;53:734–737. doi: 10.1002/jps.2600530707. [DOI] [PubMed] [Google Scholar]
- Winzer T., Gazda V., He Z., Kaminski F., Kern M., Larson T.R., Li Y., Meade F., Teodor R., Vaistij F.E. A Papaver somniferum 10-gene cluster for synthesis of the anticancer alkaloid noscapine. Science. 2012;336:1704–1708. doi: 10.1126/science.1220757. [DOI] [PubMed] [Google Scholar]
- Winzer T., Kern M., King A.J., Larson T.R., Teodor R.I., Donninger S.L., Li Y., Dowle A.A., Cartwright J., Bates R. Plant science. Morphinan biosynthesis in opium poppy requires a P450-oxidoreductase fusion protein. Science. 2015;349:309–312. doi: 10.1126/science.aab1852. [DOI] [PubMed] [Google Scholar]
- Xie H., Ash J.E., Linde C.C., Cunningham S., Nicotra A. Himalayan-Tibetan plateau uplift drives divergence of polyploid poppies: Meconopsis viguier (Papaveraceae) PLoS One. 2014;9:e99177. doi: 10.1371/journal.pone.0099177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yahyazadeh M., Ratmoyo P., Bittner F., Sato F., Selmar D. Cloning and Characterization of cheilanthifoline and stylopine synthase genes from Chelidonium majus. Plant Cell Physiol. 2017;58:1421–1430. doi: 10.1093/pcp/pcx077. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.