Abstract
Z-disc-associated, alternatively spliced, PDZ motif-containing protein (ZASP) is a principal component of the sarcomere. The three prevalent isoforms of ZASP in skeletal muscle are generated by alternative splicing of exons 9 and 10. The long isoforms, either having (ZASP-L) or lacking exon 10 (ZASP-LΔex10), include an N-terminal PDZ domain, an actin-binding region (ABR) with a conserved motif (ZM), and three C-terminal LIM domains. The short isoform (ZASP-S) lacks the LIM domains. Mutations, A147T and A165V, within the ZM of ZASP-LΔex10 cause myofibrillar myopathy, but the mechanism is unknown. We have prepared these proteins, their ABR, and the respective mutant variants in recombinant form, characterized them biophysically, and analyzed their actin-binding properties by surface plasmon resonance and electron microscopy. All the proteins were physically homogeneous and monomeric and had circular dichroic spectra consistent with partially folded conformations. Comparison of the NMR HSQC spectra of ZASP-S and the PDZ domain showed that the ABR is unstructured. ZASP-S and its mutant variants and ZASP-LΔex10 all bound to immobilized G-actin with high affinity (Kd ≈ 10−8 to 10−9 M). Constructs of the isolated actin-binding region missing exon 10 (ABRΔ10) bound with lower affinity (Kd ≈ 10−7 M), but those retaining exon 10 (ABR+10) did so only weakly (Kd ≈ 10−5 M). ZASP-S, and the ABRΔ10, also induced F-actin and array formation, even in conditions of low ionic strength and in the absence of KCl and Mg2+ ions. Interestingly, the ZM mutations A147T and A165V did not affect any of the results described above.
Graphical Abstract

The sarcomere is the basic functional unit in striated muscle. The Z-disc constitutes the lateral boundaries of the sarcomere where antiparallel actin filaments from adjoining sarcomeres are cross-linked by α-actinin. The Z-disc plays a critical role in the structural integrity of the sarcomere and in the transmission of force during muscle contraction.1–4 For all its seeming structural simplicity when imaged in cross-section by transmission electron microscopy, the Z-disc is actually composed of a large number of proteins. Current maps show the approximate locations and interactions of over 20 protein species; at least 50 species have been generally assigned to the Z-disc, and some 227 have been reported at some time to be associated with this structure.1 By now, with the recognition of numerous signaling activities in the Z-disc,2,3 that number has almost certainly increased.
ZASP is a highly expressed component of the Z-disc. It plays an important role in Z-disc assembly5 and has an essential stabilizing role as a so-called linker-strut in the final functioning unit; mice deficient in ZASP die shortly after birth and show evidence of sarcomere disruption and muscle weakness.6 ZASP also serves in a signaling capacity through an interaction with protein kinase C (PKC).7,8 ZASP (Cypher in mouse) belongs to the ALP/Enigma family of proteins. All of these proteins have an N-terminal PDZ domain, (usually) three C-terminal LIM domains, and an intermediate region of variable length. Since the first description of a PDZ-LIM domain protein,9 the number of such proteins has grown greatly. There are by now extensive reviews for both the PDZ and the LIM domains describing their various structures, arrangements, binding partners, and means of interaction.7,8,10–14 Compared to the PDZ and LIM domains, very little is known about the intervening region. In the NCBI database this region is still identified as a domain of unknown function. It has been proposed that the interior region of ZASP interacts with α-actinin15 and is involved in the interaction with Ankrd2 and p53.16 More recently, it was proposed that the region between the PDZ and LIM domains interacts with skeletal muscle actin.17 Similarly, while an evolutionarily conserved 26-residue sequence, the so-called ZASP-like motif (ZM, or sZM in the case of striated muscle), is recognized in this region, and while a number of autosomal dominant mutations occurring directly in the ZM are clearly implicated in the eventual development of myofibrillar myopathies, nothing is known about what their effects are at the molecular level.8 The ZM mutants A147T and A165V are reported in individuals of European descent, and recently the mutation N155H was the first one to be identified in a Chinese pedigree.18
The human LDB3/ZASP locus contains 16 exons (mouse Cypher contains 17). Exons 1–3 code for the PDZ domain, exons 4–11 code for the intermediate region, and exons 12–16 for the three LIM domains. A recent compilation16 of the eight currently known isoforms of human ZASP (ZASP1–ZASP8) shows a remarkable variability in their exon compositions. All forms possess a PDZ domain, but not all have the LIM domains, though all that do, have three. The greater variability occurs in the region between the PDZ and LIM domains, where the number of exons ranges from 1 to 6, and these are expressed in various combinations. Three isoforms of ZASP are commonly recognized in skeletal muscle; a shorter form (ZASP-S, corresponding to the ZASP6 isoform) lacking exons 10–16 but having exon 9, a longer form lacking exon 10 (ZASP-LΔex10, corresponding to the ZASP2 isoform) and a yet longer form that includes exon 10 (ZASP-L; this form is not included in that recent compilation16). The mutation R268C, which also causes myofibrillar myopathy, is located in exon 9 that is expressed in ZASP-S but not in the longer isoforms.19
Little is known about the structure of ZASP apart from the mentioned sequence analysis and domain matching. Further, there are no molecular details for any of the known protein interactions, the perturbations of which can be disease related. To advance this understanding we have studied ZASP-LΔex10 and various shorter forms including ZASP-S, their actin-binding regions (ABR), and the respective A147T and A165V mutant variants. All the proteins studied were physically homogeneous and monomeric and had secondary structures consistent with partially folded conformations. Further structural analysis by NMR indicated that the N-terminal PDZ domain is folded, has the same structure in ZASP as in the isolated domain, and that the rest of the molecule, is highly mobile and only partially structured. The binding to immobilized monomeric G-actin was studied using surface plasmon resonance. We found that ZASP-LΔex10 and ZASP-S bind to skeletal muscle G-actin with high affinity, that ABRΔ10 binds with lower affinity, and that ABR+10 binds only weakly. The interaction of ZASP-S with actin was also observed by electron microscopy. We observed that ZASP-S and skeletal muscle actin form paracrystalline arrays. The mutations A147T, A165V, and R268C do not appear to influence binding to actin.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
ZASP-LΔex10
Baculovirus protein expression of ZASP-LΔex10 (NM_001080114; NP_001073583) was performed by Kinnakeet Biotechnology. Insect cell paste was resuspended in break buffer (20 mM HEPES buffer (pH 7.2) containing 2 M urea, 0.1 mM TCEP) and lysed by two passes through a French press and then sonicated briefly to reduce viscosity. The lysate was centrifuged at 25000g for 1 h, and the supernatant was loaded on a Ni-Sepharose column (GE Healthcare). The column was washed with 20 mM HEPES (pH 7.2) containing 0.1% Triton-X100 and 0.25 M NaCl, and the N-terminally His-tagged protein was eluted using a 0–0.4 M imidazole gradient in column buffer. Further purification was achieved with Q Sepharose in 25 mM Tris-HCl (pH 7.5), 0.1 mM TCEP, 0.05% β-octyl-glucoside, 4% sucrose and 2 M urea. A linear gradient of 0–0.5 M NaCl was used to elute the protein. The protein was then buffer exchanged by dialysis as described below for ZASP-S.
ZASP-S
Escherichia coli cells expressing N-terminally His-tagged wild-type (WT) or mutant (A147T, A165V, R268C) ZASP-S (NM_001080116; NP_001073585) were broken with a French press in PBS buffer plus 0.2% Tween 20. The insoluble protein was washed with the break buffer defined above and extracted with 8 M guanidine-HCl and then diluted with an equal volume of 4 M guanidine-HCl. The extract was clarified by centrifugation, and protein in the supernatant, solubilized and unfolded, was fractionated by Ni-chelate affinity chromatography using Streamline Chelating resin (GE Healthcare) followed by gel filtration using Superdex 75. All chromatography buffers included 4 M guanidine-HCl. Proteins (25–50 mL) were diluted with 4 M urea to ∼1 mg/mL and folded by stepwise dialysis against 1 L each of 50 mM NaOAc (pH 5.0) containing 4 M, 2 M, 1 and 0 M urea. Any aggregated protein was removed by centrifugation at 150000g for 1 h followed by filtration using 0.2-μm Millex-GV (Millipore).
The untagged WT or mutant (A147T, A165V, R268C) ZASP-S and untagged WT ZASP-S missing exon 9 (ZASP-S 1–251) were extracted with 8 M guanidine-HCl and then gel filtrated on Superdex 200 in 4 M guanidine-HCl. The pooled protein was folded as described above. The yield of soluble protein, both tagged and nontagged, was about 50%. The proteins, if necessary, were further purified by cation-exchange chromatography in 2 M urea using S Sepharose (GE Healthcare). For purification of ZASP-S R268C, dithiothreitol was included in all buffers but was removed prior to gold labeling with monomaleimidyl-undecagold (vide infra).
ABRΔ10 and ABR+10 (N21)
Actin-binding region (ABR) constructs, both WT and mutant (A147T, A165V), either missing or not missing exon 10, and bearing a 21-residue His6-tag, were expressed partially soluble (no E. coli expression was observed for nontagged proteins). Cells were lysed in 20 mM HEPES (pH 7.5) containing 2 M urea using a French press. Following clarification by centrifugation, supernatants were fractioned by chromatography on Ni-Sepharose followed by gel filtration using Superdex 75. Further purification, if necessary, was achieved by anion exchange chromatography using Q Sepharose. All chromatography buffers included 2 M urea to prevent nonspecific binding to column matrices. Purified protein was then buffer exchanged into 50 mM NaOAc (pH 5).
ABRΔ10 (N34)
ABR constructs, both WT and mutant (A147T, A165V), missing exon 10 and bearing a 34-residue His6-tag were expressed in E. coli as insoluble aggregates. Following cell breakage, the proteins were extracted with 8 M guanidine-HCl and then bound to and eluted batch-wise from Streamline Chelating resin (GE Healthcare). Gel filtration was then done in 4 M guanidine-HCl using Superdex 75. Proteins were then folded from urea in 50 mM NaOAc (pH 5.0) as described for ZASP-S.
Protein Concentrations
The concentration of the purified ZASP-related proteins was determined by UV-absorbance at 280 nm using the absorbance values indicated in Table 1.
Table 1.
ZASP and Subdomains: Some Biophysical Parameters
| Mw (kDa)c |
|||||||
|---|---|---|---|---|---|---|---|
| proteina | His-tagb | sequence | AUC | speedd (rpm, × 103) | ν-bare (g/cm3) | A280f | affinityg Kd (M) |
| ZASP-LΔex10 | N24 | 69.37 | 60.0 (±1.3) | 14.5 | 0.724 | 0.95 | 3.9 × 10−8 |
| ZASP-S WT | N21 | 16.5 | 2.0 × 10−9 | ||||
| ZASP-S WT | none | 31.21 | 29.5 (±0.2) | 18.5 | 0.730 | 0.77 | 6.2 × 10−8 2.5 × 10−8h |
| ZASP-S 147 | none | 31.5 (±0.8) | 18.5 | 7.6 × 10−8 | |||
| ZASP-S 165 | none | 27.3 (±0.3) | 18.5 | 4.4 × 10−8 | |||
| ZASP-S 268 | none | 29.18 | 2.4 × 10−8 | ||||
| ZASP-S 1−251 | none | 27.00 | 0.68 | 2.5 × 10−8 | |||
| ABRΔ10 WT | N34 | 24.17 | 26.0 (±1.3) | 18.5 | 0.718 | 0.58 | 4.0 × 10−7 |
| ABRΔ10 147 | N34 | 33.6 (±0.8) | 18.5 | 2.6 × 10−7 | |||
| ABRΔ10 165 | N34 | 25.0 (±2.0) | 18.5 | 1.0 × 10−6 | |||
| ABRΔ10 WT | N21 | 22.80 | 24.4 (±1.0) | 20.0 | 0.718 | 0.58 | 4.6 × 10−7 3.6 × 10−8i |
| ABR+10 WT | N21 | 28.62 | 28.9 (±0.2) | 18.5 | 0.720 | 0.50 | 1.0 × 10−5 |
The proteins indicated are either wild-type sequence (WT) or the mutants A147T, A165V and R268C.
N-terminal His-tags: N21 = MGSSHHHHHHSSGLVPRGSHM, N24 = HHHHHHDYDIPTTENLYFQGAMGS, N34 = MGSSHHHHHHSSGLVPRGSHMASMTGGQQMGRIP.
The molecular weights of the constructs as derived from the gene sequence and determined (with error indicated) under native conditions by sedimentation equilibrium (AUC).
The speeds used for AUC analysis.
The partial specific volumes (ν-bar) were calculated from the amino acid compositions.
Absorbance was calculated from amino acid composition: values indicated are A280 nm of 1 mg/mL protein using a 1 cm path-length cell.
The binding of the proteins to immobilized G-actin was measured using SPR (Biacore). The binding affinities are given as dissociation constants (Kd) and are in molar (M) units.
Affinity determined with ZASP-S immobilized and G-actin as the analyte.
Affinity determined by titration calorimetry.
Actin
Lyophilized rabbit skeletal muscle actin was obtained from Cytoskeleton (Cat. no. AKL99). Skeletal muscle actin monomers (G-actin) and actin filaments (F-actin) were prepared as recommended by the supplier. Dilutions were done in general actin buffer (GAB) 5 mM Tris-HCl (pH 8.0), and 0.2 mM CaCl2. For G-actin (0.4 mg/mL in GAB), a volume of 180 μL was centrifuged in an A-110 airfuge rotor at 20 psi (∼100000g) at 21 °C for 1 h. A 50-μL aliquot was taken directly at the meniscus. The protein concentration was determined by absorbance at 280 nm using an extinction coefficient of 43 960 cm−1 M−1 and a blank obtained by ultrafiltration of the protein solution prior to centrifugation. Protein concentrations were typically ∼6 μM. Dilutions were performed with GAB unless specified otherwise.
Analytical Ultracentrifugation
A Beckman Optima XL-I analytical ultracentrifuge, absorption optics, an An-60 Ti rotor and standard double-sector centerpiece cells were used. Equilibrium measurements for the various constructs were made at 20 °C using the centrifuge speeds shown in Table 1. Concentration profiles were recorded every 4 h for 16 h, and then baselines were established by overspeeding at 45 000 rpm for 3 h. Data (the average of 8–10 scans collected using a radial step size of 0.001 cm) were analyzed using the standard Optima XL-I data analysis software. Sedimentation velocity experiments were performed at 40 000 rpm with scans recorded every 6 min for 3 h. Protein partial specific volumes (ν-bar), calculated from the amino acid compositions, and solvent densities were estimated using the program SEDNTERP (http://www.rasmb.bbri.org/). The ν-bar values are shown in Table 1.
Circular Dichroism
Spectra were recorded at 20 °C using a Jasco-715 spectropolarimeter equipped with a PTC-343W1 Peltier-type thermostatic cell holder. Measurements in the far-UV region (180–260 nm) were made using a 0.02 cm path-length cell and in the near-UV region (320–260 nm) a 1.0 cm cell. Protein solutions for both near- and far-UV were ∼1.0 OD at 280 nm, and a bandwidth of 1 nm was used. Solutions were centrifuged at 150000g, filtered with a Millex-GV 0.22-μm unit (Millipore), and degassed prior to use. Secondary structures were estimated from far-UV spectra by deconvolution using the online software DichroWeb (http://dichroweb.cryst.bbk.ac.uk/html/home.shtml). For thermal denaturation studies, spectra were monitored at 222 nm using a 1 cm path-length cell with a Teflon stopper (Hellma). Cooling circulating water was supplied using a Neslab RTE-100 thermostatic circulator. Proteins (∼0.1 OD 280 nm) were heated at 1–2 °C per min with a temperature slope of 20–90 °C. The step resolution was 1 °C, the response time 1 s, the bandwidth 2 nm, and the sensitivity 100 mdeg. Temperatures at the transition midpoints, i.e., the melting temperature (Tm), were estimated from first derivative plots of the melting curves.
Surface Plasmon Resonance
All experiments were performed on a BIAcore X100 (GE Healthcare) instrument at 25 °C. HBS-EP buffer (10 mM HEPES (pH7.4), 150 mM NaCl, 3 mM EDTA, 0.05% polysorbate 20) was used as the running buffer, and data were analyzed using Biacore X100 evaluation software (GE Healthcare). Cell 1 was left untreated to serve as a reference surface, and cell 2 was used as the experimental surface. G-actin was reconstituted with water to give a 10 mg/mL stock solution. The actin stock solution was diluted to 1 mg/mL in GAB and then further diluted with 10 mM NaOAc buffer (pH 3.8) and immobilized on CM5 sensor chips by the standard amine coupling method (Amine Coupling kit, GE Healthcare) at a flow rate of 5 mL/min. The immobilization levels of the proteins on the sensor chip surfaces were about 2000 RU. For kinetic analysis, analytes were prepared by serial dilution with HBS-EP buffer over a range of greater than 100-fold and injected over both the reference and experimental surfaces at a flow rate of 30 μL/min. Sensor chips were regenerated by a 60 s injection of 50 mM NaOH. Signals from the reference surface and an ensemble of buffer blank injections were subtracted to correct for nonspecific binding and injection artifacts. The corrected results were fitted globally using a 1:1 binding model and the determined association rate constants (ka) and dissociation rate constants (kd) used to calculate the equilibrium dissociation constants (Kd).
Limited Tryptic Digestion
His-tagged ZASP-S WT (∼1 mg/mL) in 25 mM Tris-HCl (pH 7) was digested with 0.1% (w/w) trypsin at 20 °C for up to 40 min. The sample was quenched with AEBSF, solid guanidine-HCl added to 4 M, and then gel filtrated on Superdex 75. Further purification of peak fractions was performed by reverse phase. Lyophilized samples resuspended in 50% acetonitrile, 0.1% TFA, and sinapinic acid (matrix) were analyzed on a Shimadzu Biotech Axima Assurance MALI-TOF mass spectrometer.
Titration Calorimetry
A MicroCal VP-ITC (GE Heathcare) was used. The ligand G-actin at 50 μg/mL was titrated with 4 μL injections of His-tagged ABRΔ10 (N21 His-tag) at 300 μg/mL with stirring at 459 rpm.
Nuclear Magnetic Resonance
Untagged 15N-labeled ZASP-S WT was refolded as described above. The protein was more than 90% pure. The folded protein was dialyzed into three different buffers: 50 mM NaOAc (pH 5.0), 20 mM sodium phosphate (pH 6.5), and 20 mM sodium phosphate (pH 6.5), 100 mM NaCl. The NMR spectra were typically recorded at 298 K using 950 MHz 1H frequency BRUKER spectrometers, equipped with triple resonance gradient CryoProbes. Spectra were processed using NMRPipe/NMRDraw software20 and analyzed using CCPN software.21
Gold Labeling
ZASP-S was labeled with gold particles in two ways, either at the N-terminal end through a His6-tag with 5 nm Ni-NTA-Nanogold, or at the C-terminal end through the unique cysteine in ZASP-S R268C with 1.4 nm monomaleimido Nanogold (Nanoprobes).
ZASP-Actin Complex Formation
For paracrystal formation, untagged WT ZASP-S was mixed with either G-actin or F-actin. ZASP-S was buffer changed into either 5 mM NaOAc (pH 5.0), or 5 mM Tris-HCl (pH 7.0), or HBS-EP prior to mixing the proteins, usually in a 1:1 molar ratio.
For decoration of F-actin, His-tagged WT ZASP-S labeled with 5 nm Ni-NTA-Nanogold, in 10 mM Tris-HCl (pH 7.0), 50 mM KCl, 2 mM MgCl2, 1 mM ATP was mixed in a 1:1–2:1 molar ratio with 4 μM F-actin in the same buffer (untagged WT ZASP-S served as a control). In initial survey experiments, the pH was varied from 5.0–9.0, as were the concentrations of NaCl (5–500 mM), KCl (50–125 mM), and MgCl2 (2–8 mM). In some early experiments the samples were fixed with 0.1% glutaraldehyde (EMS). Decoration of F-actin was also attempted with ZASP-S R268C labeled with 1.4 nm monomaleimido Nanogold.
For initiation of G-actin polymerization in the presence of gold-labeled ZASP-S, 5 nm Ni-NTA-Nanogold (Nanoprobes) and His-tagged ZASP-S were mixed in a 1:6 molar ratio and incubated for 1 h at 21 °C. F-actin formation was then initiated as described above with G-actin and the gold-ZASP probe in a 10:1 molar ratio, respectively.
For on-grid initiation of F-actin, untagged WT ZASP-S was buffer changed into 2.5 mM NaOAc (pH 5.0). A 4.5-μL drop of G-actin in GAB (6 μM) was applied to a glow-discharged grid. Into this was injected 0.5 μL of either ZASP-S (30 μM) or 2.5 mM NaOAc (pH 5.0) buffer and mixed. The grids were wicked, rinsed, and stained after 30, 60, 120, and 300 s.
Electron Microscopy
Typically, samples were applied to carbon-coated 400-mesh copper grids made hydrophilic by 10 s of glow-discharge in an atmosphere of 25% oxygen and 75% argon, allowed to adsorb for 60 s, rinsed on deionized water, stained for 30 s with 1% uranyl acetate, and then air-dried. For specimens involving soluble protein or small complexes, the stain solution was supplemented with 0.5 mM β-octyl-glucoside to improve wetting of the grid. For specimens labeled with 1.4 nm Nanogold, staining was done with methylamine vanadate (Nanoprobes) to improve visualization of the gold particles. Micrographs were recorded on a CM120 electron microscope (FEI) equipped with a CCD, typically at a nominal instrument magnification of 35000×. Calibration was done with the 2.3 nm spacing from tobacco mosaic virus particles. Computational diffraction of paracrystals and single F-actin was done using B-Soft.22 For cryoelectron microscopy (cryo-EM), F-actin was draped over holes on holey carbon films and then incubated with untagged WT ZASP-S in 25 mM NaOAc (pH 5.0) at various concentrations for various times prior to blotting and plunging. Specimens were examined with a CM200 electron microscope (FEI).
RESULTS
Expression and Purification of ZASP Proteins and Subdomains
We tested several ZASP constructs comprising different subdomains (Figure 1). All proteins were expressed in E. coli except ZASP-LΔex10, which was expressed in the baculovirus system. ZASP-L did express well in E. coli, but the protein was poorly soluble and attempts to fold it were unsuccessful. The ZASP-S constructs (Table 1) were insoluble but could be folded with high yield as described in Experimental Procedures. The ABR constructs with the longer N-terminal His-tag (N34) were insoluble when expressed in E. coli, whereas the constructs with the shorter (N21) His-tag (from pET-28) were partially soluble. The actin-binding and other biophysical properties of the ABR constructs described below were not altered by the length of the linker between the ABR peptide and the N-terminal His6-tag. Whether ABR constructs were directly purified as soluble protein, or folded and then purified, they exhibited limited stability when stored at 4 °C, tending to slowly aggregate. Therefore, all ABR-related proteins were only used when freshly prepared and centrifuged as mentioned below.
Figure 1.
Domain organization of ZASP and ZASP subdomains. Exons 1–3 and 5–16 are indicated, with the widths of the colored bars proportional to exon (amino acid) length (exon 4 is absent in all constructs studied here). Exons and their corresponding structural/functional domains: Exons 1–3 (PDZ domain); exons 5, 6, 7, 8, 10, 11 (ABR) with the ZASP-like motif (ZM) in exon 6 highlighted in yellow; exons 12–16 (three LIM domains). The domain structures of ZASP-LΔ10 and ZASP-S used in this study are shown below. The locations of the mutations A147T, N155H, A165V, and R268C are shown in the context of ZASP-S. Black bar = 50 amino acids.
Purification of the proteins, with or without a His-tag, usually preceded folding and was achieved using Ni-chelate chromatography (for constructs with a His-tag) and gel filtration in the presence of guanidine-HCl. Proteins were folded at ∼1 mg/mL by dialysis wherein guanidine was exchanged for urea and then buffer alone. Small amounts of aggregate were removed by low-speed centrifugation. Protein folding recoveries for all constructs were 50–80% with no differences between WT and mutant proteins. Folded proteins could be concentrated to greater than 5 mg/mL at pH 5.0. SDS-PAGE of His-tagged ZASP-S and variants is shown in Figure S1, and that of ZASP-S (1–251) is shown in Figure S4.
All purified ZASP proteins appeared most soluble in the low ionic strength pH 5.0 acetate buffer described. It is normal practice to gel filtrate samples following folding to resolve any physical heterogeneity especially from aggregated protein. All the ZASP proteins showed strong adsorption to the Superdex gel filtration media used. Sedimentation velocity analysis indicated that most constructs contained between 10–30% of fast moving (high s-values) material corresponding to aggregated protein, and the remainder had a single sedimenting boundary with low s-values corresponding to monomeric protein. On the basis of this finding, samples were routinely centrifuged at 150000g for 1 h prior to use to remove aggregates, this step serving essentially the same role as gel filtration.
Molecular Weight Determinations of ZASP-S and ABRs: Sedimentation Equilibrium Centrifugation
The native molecular weights of the ZASP proteins were determined by sedimentation equilibrium ultracentrifugation (Figure 2 and Table 1). All the purified proteins were ideal monomeric proteins at pH 5.0 with no tendency to self-associate over the concentration range of the analyses. This was also true whether or not the proteins were His-tagged. The mutant variants A147T and A165V in the context of either the ZASP-S or the smaller ABR constructs were also monomeric.
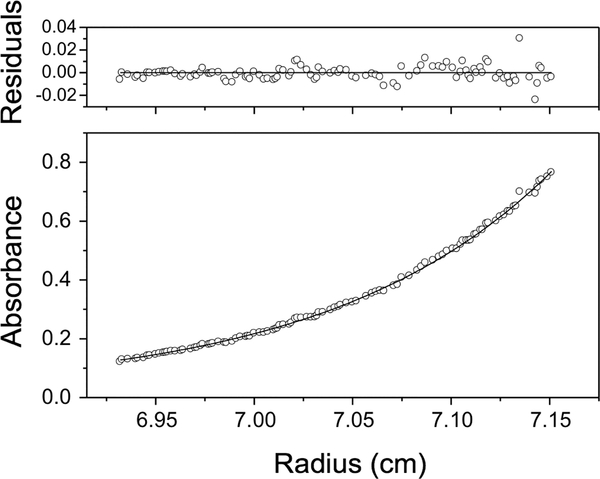
Figure 2.
Sedimentation equilibrium analysis of untagged WT ZASP-S. The analysis was at pH 7.0 and the panels show absorbance (bottom panel) and residuals (upper panel). Open circles show UV absorbance gradients in the centrifuge cell. The solid line indicates the calculated fit for an ideal single species. Residuals show the difference between the fitted and experimental values as a function of radial position.
To check the solubility and molecular weight of ZASP-S at various pH values, the protein (∼1.5 mg/mL) was analyzed between pH 5 and pH 7 (not shown). The protein was monomeric over this pH range; however, at pH 7 the sedimentation equilibrium scans (over 16 h at 20 °C) indicated the presence of small amounts (∼5%) of high molecular weight aggregates. There was no evidence of reversible association such as monomer–dimer or trimer systems. On the basis of sedimentation equilibrium and velocity studies of ZASP-S and the shorter ABR constructs, we conclude that the proteins are all monomeric but that small amounts of aggregate accumulate in preparations stored at 4 °C, especially at pH 7. This behavior was taken into account when performing biochemical and structural studies, and as a precaution the proteins were always centrifuged at 150000g for 1 h prior to use.
Conformation of ZASP-S: Circular Dichroism
As most constructs were folded from denaturated protein it was important to establish that any differences in biochemical properties, especially protein binding, were not due to conformational perturbations. Circular dichroism was used to study protein conformation and folding. The secondary structure was estimated by CD measurements in the far-UV (180–260 nm). The untagged WT ZASP-S spectrum (Figure 3A) appeared to be of mixed α-helix and β-sheet content which on deconvolution indicated ∼17% α-helix and 24% β-sheet. The N-terminal residues of ZASP-S (1–84) are derived from the PDZ domain (Figure 1), and its known structure (PDB: 4YDP) contributes ∼5% α-helix and 10% β-sheet to the overall structure. The secondary structures of the two ZASP-S mutants, A147T and A165V, were both very similar to WT protein (Figure 3A). The near-UV CD region (320–260 nm), derived from asymmetric orientations of tryptophan and tyrosine residues, is the conformational fingerprint region, and the spectral similarity of the WT and mutant proteins suggests very similar tertiary structures (Figure 3B). These spectra are characterized by a negative trough of ellipticity centered on 270 nm, presumably derived from tyrosine residues, and the flat baseline like spectrum of the WT protein denatured with 4 M guanidine-HCl (Figure 3B) emphasizes that the signal is derived from folded protein.
Figure 3.
Circular dichroic spectra of untagged WT and mutant (A147T, A165V) ZASP-S. The proteins (∼1.0 OD 280 nm) were at 20 °C. (A) Far-UV (0.02 cm path-length cell) and (B) near-UV (1 cm path-length cell). The secondary structure was estimated from A and B, which is the conformational fingerprint region. Units of the ordinate [θ]mwr have the dimensions: deg × dmol−1 × cm−2.
The thermal denaturation of untagged WT and mutant (A147T and A165V) ZASP-S was measured by helix–coil transitions at 222 nm. The midpoints of the transitions, i.e., the melting temperatures (Tm), were similar (Figure 4). The spectral changes upon denaturation were small due to relatively low helical content and the signal being noisy; therefore, the small observed Tm differences between the WT and variants are probably not significant.
Figure 4.
Thermal denaturation of untagged WT and mutant (A147T, A165V) ZASP-S. The proteins (∼0.1 OD 280 nm) were scanned using a 1 cm path-length cell over the temperature range indicated at 222 nm. The midpoints of the transitions are the melting temperatures (Tm) and were estimated from first derivative of the curves indicated. Units of the ordinate are mean residue ellipticity [θ]mwr and have the dimensions: deg × dmol−1 × cm−2.
On the basis of the concordance between the near- and far-UV CD spectra and the denaturation Tm values, there is unlikely to be any major conformational or structural differences between the WT and mutant variants. Detailed structural determinations may, of course, reveal small local conformational differences not detectable by the global measurements described.
Further Probing of ZASP-S Conformation
pH Titration
The near-UV CD spectra of His-tagged WT ZASP-S were obtained as a function of pH (Figure 5). There was a loss of structure at the lower pH values; however, after the His-tag was removed the protein was stable over the pH range studied. Whether this is due to a charge difference or the linker sequence at the N-terminus (21 residues) has not been studied. Regardless of the constructs used, all binding studies were performed at pH 7.0 and higher.
Figure 5.
Circular dichroic spectra of WT ZASP-S as a function of pH. Near-UV of ZASP-S (∼1 OD 280 nm with 1 cm path-length cell) spectra at the pH values indicated. The proteins were untagged except where indicated.
Limited Proteolytic Processing
As the ZASP-S secondary structure estimate suggests that some regions of the protein are unstructured, we performed limited proteolytic digestion with trypsin. The principle of this analysis is that flexible and exposed regions of a protein will be processed faster than folded ones.23 Using trypsin, both untagged and His-tagged WT ZASP-S at pH 7 were rapidly processed (Figure S4). The digested protein was bound and eluted from Ni-chelate resin, indicating that the initial cleavage occurred at the C-terminal end. Analysis of the main digestion product (marked with an arrow in Figure S4) by mass spectrometry indicated a main product with mass of 26 648 Da corresponding to residues 1–228 (the N-terminal Met, as predicted, had been processed). Minor processing was observed at residues 217, 218, and 224, indicating a hot spot region consistent with high arginine content. The processed C-terminal sequence includes exon 9 (residues 252–282). This exon is unique to ZASP-S and, based on the digestion results, appears to be solvent accessible and unstructured. The far- and near-UV CD spectra of the ZASP-S 1–251 were similar to ZASP-S despite the loss of apparently unstructured C-terminal residues (data not shown).
Conformation of ZASP-S: Nuclear Magnetic Resonance
The HSQC spectrum of ZASP-S (Figure 6) is indicative of a largely unstructured species with well-folded regions, as shown by the low dispersion of resonances in the center of the spectrum together with the highly dispersed peaks surrounding them. Comparison of the spectrum with that of the isolated PDZ domain24 shows that the dispersed peaks mostly belong to the PDZ domain. The minor differences in the chemical shifts of these resonances imply that the structure of the PDZ domain is not appreciably altered by the presence of the unstructured part of the protein. The uneven intensities of some of the resonances suggest the presence of different exchange regimes in different regions of the protein, minor degradation, or the presence of minor contaminants.
Figure 6.
Overlay of the 1H–15N HSQC spectrum of untagged WT ZASP-S recorded at pH 5.0 and 298 K (in black) and the reconstructed 1H–15N HSQC spectrum published24 for the ZASP PDZ domain at pH 6.5 and 298 K (in red). The assignment of the most dispersed peaks of the PDZ domain is shown. Compared to the PDZ domain, ZASP-S shows numerous additional unresolved peaks, demonstrating the unstructured nature of the actin-binding region.
Conformation of ABRΔ10 and ABR+10: Circular Dichroism
Several ABR constructs were studied (Figure 1), in these proteins the N-terminal PDZ and C-terminal LIM domains were deleted, and they all contained exons 5–8 and 11 (exon 6 includes the ZM region). The designations ABRΔ10 and ABR+10 refer to the absence or presence of exon 10 (Note: ZASP-S does not contain exon 10). ABRΔ10 was first expressed with a 34-residue N-terminal His-tag as an insoluble protein, and this was solubilized and folded as described in Experimental Procedures. The far-UV CD spectrum (Figure S3A), analogous to ZASP-S, appears to arise from mixed α-helix-β-sheet structure, and deconvolution of the spectra indicates ∼12% α-helix and 24% β-sheet. There may be significant unstructured protein present as indicated by the NMR analysis of ZASP-S. When either ABRΔ10 or ABR+10 is expressed with a shorter 21-residue His-tag, they are mostly soluble, and both proteins have similar far-UV spectra (Figure S3A). Also, for ABRΔ10, the spectral similarity (identity) of protein derived from soluble or insoluble expression suggests similar structures. The intensities of the near-UV spectra of the ABR proteins, although weak and noisy, are similar (Figure S3B) and consistent with the far-UV data. These data indicate there is little conformational change in the ABR upon insertion or deletion of exon 10.
Skeletal Muscle G-actin Binding Properties of ZASP and Its Subdomains
The actin-binding activity of the various ZASP constructs was studied by surface plasmon resonance. Rabbit skeletal muscle actin was reconstituted under buffer conditions favoring monomeric (G) protein. Any polymeric protein was then removed by centrifugation and the soluble protein was checked by dynamic light scattering prior to immobilization. The G-actin (ligand) was immobilized on the chips as described (Experimental Procedures). Dilution series of the various ZASP constructs (analytes) were used to construct senograms where the rates of binding (ka) and dissociation (kd) are displayed in real time (Figure S5). Table 1 summarizes the binding data as dissociation constants (Kd) determined as the ratio of ka/kd. The His-tagged ZASP-S constructs bound to G-actin with nanomolar affinity. There was no significant difference in binding affinities between the WT and mutant ZASP proteins. The same set of proteins minus the N-terminal His-tag behaved the same as their tagged counterparts except that the binding affinities were all about 10-fold lower. We have not fully explored the reason for this difference, although it does not appear to be conformation related, as the His-tagged and untagged proteins have essentially identical far-and near-UV CD spectra at pH 7 and similar melting temperatures, though it may be charge related. The His-tag adds positive charge at pH 7, and this may assist protein binding to the negatively charged actin (the net charge at pH 7 is −7).
The aforementioned actin binding studies were performed with G-actin immobilized on the chip to preclude any actin polymerization. However, the binding of G-actin to immobilized ZASP-S was also measured, and the binding affinity matched that of the reverse (standard) ligand-analyte configuration described above (Table 1).
ZASP-LΔex10, expressed in insect cells, was difficult to purify due to its low solubility in standard buffers and its adhesive tendencies toward both column matrices and protein contaminants. This necessitated protein purification in the presence of low (nondenaturing) concentrations of urea (∼1 M). Highly diluted ZASP-LΔex10 (essentially no urea present) had an affinity for G-actin very similar to that of untagged ZASP-S (Table 1).
The highly truncated ABRΔ10 and ABR+10 constructs were used to delineate the sequence boundaries required for interaction with actin. The ABRΔ10 construct, which shares exons 5–8 with ZASP-S, bound to actin with moderate affinity, about 10-fold lower than ZASP-S. The lower affinity was due to a faster dissociation rate (higher kd) compared to ZASP-S. However, the binding was still considered specific, based on its titratability and the shape of the sensograms. For confirmation, actin binding was also measured by titration calorimetry using G-actin as the ligand and ABRΔ10 as the titrant. The affinity was similar to that determined by SPR, supporting the specificity of binding. In contrast, the ABR+10 bound actin weakly (Kd ≈ 10−4 to 10−5 M). The reduced binding is unlikely to be due to the protein being unfolded as it was derived by soluble expression and has similar circular dichroic spectra as the active ABRΔ10.
Both the mutation (i.e., ZASP-S R268C) and deletion (i.e., ZASP-S 1–251) of exon 9 from ZASP-S had no effect on the actin binding affinity, indicating that this region may have some other binding or structural role, although based on limited tryptic digestion it may be a highly flexible region. ZASP-S 1–251 constructs with the addition of exon 11, or exons 10 and 11, are essentially equivalent to the ABRΔ10 and ABR+10 constructs, but with the N-terminal PDZ domain present. This agrees with the NMR data, which indicate that the N-terminal PDZ is not interacting substantially with the rest of the molecule.
ZASP-S Induces Actin Bundle Formation
The interaction of ZASP with actin was also observed by electron microscopy. Untagged ZASP-S and G-actin formed paracrystalline arrays (Figure 7A). These arrays formed in either the presence or absence of KCl and Mg2+ ions, and had the appearance of bundled F-actin. The arrays were usually about 50–100 nm in diameter and of indeterminate length, though typically very long. Power spectra calculated from the arrays show layer lines at 1/28 Å and 1/170 Å (Figure 7B), corresponding to the 27.5-Å actin-monomer and six-turn spacings, respectively (F-actin has 13/6 helical symmetry, with 13 subunits in 6 turns). Single filaments extending out from the arrays also showed the 28-Å spacing, as did pure F-actin controls. ZASP-S also formed arrays with (preformed) F-actin, and these were similar to those formed with G-actin, as were arrays formed by ZASP-S bearing either the A147T or A165V mutations (data not shown).
Figure 7.
Interaction of untagged WT ZASP with G-actin as observed by electron microscopy. (A) Arrays formed by interaction of ZASP-S with G-actin. (B) Averaged power spectrum calculated from straight regions of several arrays, such as indicated by the brace in (A). (C) G-actin preparation without addition of ZASP-S; a few residual F-actin not removed during high-speed centrifugation were sometimes observed. (D) Typical field of G-actin preparation 120 s after addition of one-half molar equivalent of ZASP-S. After 5 min arrays such as in (A) were observed. Bar = 100 nm in (A); 250 nm in (C and D).
Time-course observations of paracrystal formation, made by mixing untagged WT ZASP-S with G-actin directly on the microscope grid, showed that ZASP-S initiates F-actin (filament) formation. Newly formed F-actin appeared on the grid within seconds of mixing (Figure 7D) followed by the appearance of paracrystals. At longer times (minutes) essentially all filaments were incorporated into paracrystals. The observation that ZASP-S initiates F-actin formation prompted efforts to detect ZASP-S at the ends of F-actin; however, F-actin initiated by His-ZASP-S and subsequently labeled with the 5 nm Ni-NTA gold-atom cluster did not have the gold located preferentially at the filament termini. Preformed complexes of His-ZASP-S and the 5 nm Ni-NTA gold-atom cluster also did not end-label filaments.
An effort was also made to observe ZASP-S bound to single F-actin along their length. However, we found no way to either dissociate the arrays (and still retain decorated F-actin) or to decorate single F-actin with ZASP-S. From the relative masses of ZASP-S and monomeric actin (31 and 43 kDa, respectively), it was expected that ZASP-S could be detected on the filaments; however, as we did not, we assumed this was perhaps due to disorder in the position of ZASP. Attempts were therefore made to enhance detection by decorating F-actin with ZASP-S R268C labeled with a 1.4 nm monomaleimide gold-atom cluster but these were unsuccessful. The gold label did not associate with the filaments as judged by electron microscopy, and gold-labeled ZASP-S did not sediment into the sucrose cushion together with the F-actin during centrifugation (not shown). Attempts to detect N-terminally His-tagged ZASP-S on F-actin with the larger 5 nm Ni-NTA gold-atom cluster were similarly unsuccessful. Attempts to observe ZASP-S decoration of F-actin directly by cryo-EM have so far also been unsuccessful (data not shown).
DISCUSSION
ZASP-S and ABR Conformation and Structure
An initial goal of this study was to examine the ZASP mutations A147T and A165V, which drastically affect striated muscle structure and cause myofibrillar myopathy. Our aim was to relate these mutations to any structural perturbation of the protein and its molecular interactions. We focused on the subdomains of ZASP (Figure 1) as the full-length protein is difficult to produce in E. coli and only folds with low yield. Although we were able to produce soluble ZASP-LΔex10 in the baculovirus system the protein was still prone to aggregation, requiring low levels of urea to maintain solubility. The presence of the three C-terminal LIM domains, which are known to mediate multiple protein–protein interactions, probably limited the solubility of ZASP-LΔex10. Subsequently, we focused on ZASP-S, which is a naturally occurring form of the protein that includes the sites of the myofibrillar myopathy causing mutations (A147T, A165V, and R268C), and the ABRs of the longer ZASP isoforms in our studies (Figure 1).
With the methods described, we have produced the ZASP subdomains and their corresponding mutant forms. All the proteins were soluble and monomeric and had circular dichroic spectra consistent with partially folded conformations. Comparison of the NMR HSQC spectra of ZASP-S and the PDZ domain showed that the ABR is unstructured. This is reminiscent of the observation that the interior of the actinin-associated LIM protein (ALP) is unstructured,25 suggesting that this may be a general property of this family of proteins. Comparison of the WT and mutant versions of ZASP-S and ABRΔ10 by near- and far-UV CD (Figure 3) showed that the mutations result in no major structural perturbations. Thermal denaturation (Figure 4) also indicated that the stabilities of the WT and mutant proteins were very similar. Finally, the physical state of the proteins, as determined by analytical ultracentrifugation, indicated that all were stable monomers (Table 1). These results allow us to interpret with confidence any potential functional differences caused by the mutations.
Binding Studies with ZASP-S and ABR
Given that ZASP potentially interacts with numerous proteins in the Z-disc,1 it is a major undertaking to determine which interactions are directly affected by the mutations. This study examined the skeletal muscle actin-binding properties of ZASP based on our earlier investigations.17 As described, we have used SPR to measure the affinity of ZASP for G-actin. This approach, due to its sensitivity, has the advantage of allowing working with proteins at high dilution, thus, minimizing nonspecific (usually low-affinity) interactions. Also, as the G-actin (used as ligand) and ZASP-S (analyte) were both monomeric, the analysis involved a simple bimolecular interaction. Whereas the SPR measurements using the ZASP-S constructs were characterized by high affinity binding (Table 1) the ABR constructs exhibited lower binding. The ABRΔ10 construct binds to G-actin with moderate affinity (lower than ZASP-S), and the introduction of the A147T and A165V mutations into this construct had little or no effect on binding, analogous to the results with ZASP-S. On the other hand, the ABR+10 construct exhibited low affinity (Kd ≈ 10−5 M). The addition of the N-terminal PDZ domain to the ABRΔ10 did result in some enhancement in the affinity for actin, whereas the ABR+10 still bound only weakly (data not shown).
The A147T and A165V mutations do not appear to have any effect on actin binding when present in ZASP-L.17 Our results with the smaller constructs, using a more sensitive method, also show that these mutations do not affect the kinetics and affinity of actin binding. This suggests that the difference between the two long forms of ZASP is related to the presence or absence of exon 10. As the A147T and A165V mutations occur in the ZM motif, which is present in all constructs studied and which may form part of the actin-binding region,17 it is unlikely that modulation of actin binding has any direct role in myofibrillar myopathy. If the mutations disrupt critical associations with another Z disc associated protein,2 then this effect is more potent when ZASP is located in the Z band by virtue of its affinity for actin. If the protein cannot bind actin due to exon 10, then the mutations are effectively silent. The presence of exon 10 may block actin binding by simple steric interference or by reducing electrostatic potential of binding with the very basic actin due to reduction of the protein surface charge (exon 10 is acidic). It can be noted that taxonomic studies of protein complexes show that protein–protein interfaces are often polar and exploit electrostatic interactions.26 This hypothesis is supported by the observation that, although not part of the LIM domain, the inclusion of exon 10 impairs the affinity of ZASP for PKC relative to forms lacking exon 10.27 Aberrant splicing of ZASP resulting in enhanced expression of the exon 10-bearing isoforms in skeletal muscle correlated with the severity of muscle weakness in patients with myotonic dystrophy type 1, the most common adult-onset muscular dystrophy.28 Direct interaction of ZASP with PKC and skeletal muscle actin indicates that ZASP may mediate the effects of PKC in the remodeling of the actin cystoskeleton in striated muscle.7,8,29 Future studies will examine the role of reduced actin and PKC binding affinities of the exon 10-bearing ZASP isoforms in the disease mechanisms underlying muscle weakness of DM1 patients.
ZASP-Actin Complexes
As indicated above, the addition of ZASP-S to skeletal muscle G-actin resulted in the formation of F-actin (filaments). The F-actin formed in this way subsequently associated with one another to form arrays. Computational diffraction of these arrays showed that they had the same axial spacings as isolated F-actin. The PDZ domain was not required for array formation as ABRΔ10, both WT and mutant, formed similar arrays when mixed with F-actin (Figure S6). To our knowledge, the F-actin initiating activity of ZASP has not been reported previously. Conceivably, ZASP binds to G-actin monomers, thereby stabilizing them in a polymerization-competent conformation. ZASP was not detected at the ends of such F-actin. In this, it is similar to F-actin nucleators which dissociate after nucleation.30 ZASP may also be similar to nucleators such as palladin and nebulin that can induce actin polymerization under conditions otherwise unfavorable for polymerization.31,32 The formation of the F-actin arrays by ZASP has also not been reported. The arrays probably form as the result of many weak (secondary) interactions, either ZASP–ZASP or ZASP-actin. Whether the F-actin initiating activity of ZASP actually plays a role in vivo is not known. However, it is worth noting that ZASP and actin have been implicated in the formation of sarcoplasmic F-actin bundles and intranuclear rods, with an appearance similar to those reported here, in patients with myofibrillar myopathy.33
CONCLUSIONS
We have developed methods for the production of the ZASP subdomains involved in interactions with skeletal muscle G-actin. These were characterized by both biochemical and physical methods and shown to be monomeric and partially folded proteins with high affinity for G-actin. The myofibrillar myopathy causing ZASP mutations A147T and A165V affected neither protein folding and structure nor actin-binding affinity. Our results indicate that these mutations are most likely surface located residues which form part of the binding interface to an, as yet unidentified, key protein. The ZASP subdomains are all well suited for the structural studies required for establishing these molecular details.
Supplementary Material
ACKNOWLEDGMENTS
We thank Drs. Xiaoyan Lin and Sharolyn Kawakami-Schultz and Ms. Rachel Ohman for preparing some of the plasmids used in this study, the MRC NMR Centre for technical support, Dr. Naiqian Cheng for collecting preliminary cryo-EM data and Dr. Alasdair Steven for careful reading of the manuscript.
Funding
This research was supported by the Intramural Research Program of the National Institute of Arthritis and Musculoskeletal and Skin Diseases, and the Intramural Research Program of the National Institute of Neurological Disorders and Stroke, of the National Institutes of Health (USA), and the Biotechnology and Biological Sciences Research Council (UK).
ABBREVIATIONS
- HSQC
heteronuclear single quantum coherence spectroscopy
- LIM
is an acronym combining the first letters of the three transcription factors (lin-11, isl-1, and mec-3) in which the motif was first identified
- PDZ
is an acronym combining the first letters of the three proteins (post synaptic density protein, Drosophila disc large tumor suppressor, and zonula occludens-1 protein) in which the motif was first identified
- ZM
ZASP-like motif
Footnotes
The authors declare no competing financial interest.
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.biochem.7b00067.
SDS-PAGE of WT and mutant (A147T, A165V) ZASP-S; sedimentation equilibrium analysis of ABR; circular dichroic spectra of ABR+10 and ABRΔ10; SDS-PAGE of ZASP-S following limited digestion with trypsin: comparison with ZASP-S 1–251; surface plasmon resonance analysis of untagged WT ZASP-S binding to immobilized G-actin; and electron microscopy of arrays formed by interaction of the ABRΔ10 with F-actin (PDF)
REFERENCES
- (1).Knoll R, Buyandelger B, and Lab M (2011) The sarcomeric Z-disc and Z-discopathies. J. Biomed. Biotechnol. 2011, 569628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Frank D, and Frey N (2011) Cardiac Z-disc signaling network. J. Biol. Chem. 286, 9897–9904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Luther PK (2009) The vertebrate muscle Z-disc: sarcomere anchor for structure and signalling. J. Muscle Res. Cell Motil. 30, 171–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Lange S, Ehler E, and Gautel M (2006) From A to Z and back? Multicompartment proteins in the sarcomere. Trends Cell Biol. 16, 11–18. [DOI] [PubMed] [Google Scholar]
- (5).Jani K, and Schock F (2007) Zasp is required for the assembly of functional integrin adhesion sites. J. Cell Biol. 179, 1583–1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Zhou Q, Chu PH, Huang C, Cheng CF, Martone ME, Knoll G, Shelton GD, Evans S, and Chen J (2001) Ablation of Cypher, a PDZ-LIM domain Z-line protein, causes a severe form of congenital myopathy. J. Cell Biol. 155, 605–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Zheng M, Cheng H, Banerjee I, and Chen J (2010) ALP/Enigma PDZ-LIM domain proteins in the heart. J. Mol. Cell Biol. 2, 96–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Sheikh F, Bang ML, Lange S, and Chen J (2007) ″Z″eroing in on the role of Cypher in striated muscle function, signaling, and human disease. Trends Cardiovasc. Med. 17, 258–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Wu RY, and Gill GN (1994) LIM domain recognition of a tyrosine-containing tight turn. J. Biol. Chem. 269, 25085–25090. [PubMed] [Google Scholar]
- (10).Li A, Ponten F, and dos Remedios CG (2012) The interactome of LIM domain proteins: the contributions of LIM domain proteins to heart failure and heart development. Proteomics 12, 203–225. [DOI] [PubMed] [Google Scholar]
- (11).Lee HJ, and Zheng JJ (2010) PDZ domains and their binding partners: structure, specificity, and modification. Cell Commun. Signaling 8, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Kadrmas JL, and Beckerle MC (2004) The LIM domain: from the cytoskeleton to the nucleus. Nat. Rev. Mol. Cell Biol. 5, 920–931. [DOI] [PubMed] [Google Scholar]
- (13).Nourry C, Grant SG, and Borg JP (2003) PDZ domain proteins: plug and play! Sci. Signaling 2003, RE7. [DOI] [PubMed] [Google Scholar]
- (14).van Ham M, and Hendriks W (2003) PDZ domains-glue and guide. Mol. Biol. Rep. 30, 69–82. [DOI] [PubMed] [Google Scholar]
- (15).Klaavuniemi T, and Ylanne J (2006) Zasp/Cypher internal ZM-motif containing fragments are sufficient to co-localize with alphaactinin–analysis of patient mutations. Exp. Cell Res. 312, 1299–1311. [DOI] [PubMed] [Google Scholar]
- (16).Martinelli VC, Kyle WB, Kojic S, Vitulo N, Li Z, Belgrano A, Maiuri P, Banks L, Vatta M, Valle G, and Faulkner G (2014) ZASP interacts with the mechanosensing protein Ankrd2 and p53 in the signalling network of striated muscle. PLoS One 9, e92259. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- (17).Lin X, Ruiz J, Bajraktari I, Ohman R, Banerjee S, Gribble K, Kaufman JD, Wingfield PT, Griggs RC, Fischbeck KH, and Mankodi A (2014) Z-disc-associated, alternatively spliced, PDZ motif-containing protein (ZASP) mutations in the actin-binding domain cause disruption of skeletal muscle actin filaments in myofibrillar myopathy. J. Biol. Chem. 289, 13615–13626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Zheng J, Chen S, Chen Y, Zhu M, and Hong D (2017) A novel mutation in the PDZ-like motif of ZASP causes distal ZASP-related myofibrillar myopathy. Neuropathology 37, 45–51. [DOI] [PubMed] [Google Scholar]
- (19).Selcen D, and Engel AG (2005) Mutations in ZASP define a novel form of muscular dystrophy in humans. Ann. Neurol. 57, 269–276. [DOI] [PubMed] [Google Scholar]
- (20).Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, and Bax A (1995) NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293. [DOI] [PubMed] [Google Scholar]
- (21).Fogh R, Ionides J, Ulrich E, Boucher W, Vranken W, Linge JP, Habeck M, Rieping W, Bhat TN, Westbrook J, Henrick K, Gilliland G, Berman H, Thornton J, Nilges M, Markley J, and Laue E (2002) The CCPN project: an interim report on a data model for the NMR community. Nat. Struct. Biol. 9, 416–418. [DOI] [PubMed] [Google Scholar]
- (22).Heymann JB, and Belnap DM (2007) Bsoft: image processing and molecular modeling for electron microscopy. J. Struct. Biol. 157, 3–18. [DOI] [PubMed] [Google Scholar]
- (23).Fontana A, Polverino de Laureto P, De Filippis V, Scaramella E, and Zambonin M (1997) Probing the partly folded states of proteins by limited proteolysis. Folding Des. 2, R17–26. [DOI] [PubMed] [Google Scholar]
- (24).Au Y, Atkinson RA, Guerrini R, Kelly G, Joseph C, Martin SR, Muskett FW, Pallavicini A, Faulkner G, and Pastore A (2004) Solution structure of ZASP PDZ domain; implications for sarcomere ultrastructure and enigma family redundancy. Structure 12, 611–622. [DOI] [PubMed] [Google Scholar]
- (25).Klaavuniemi T, Alho N, Hotulainen P, Kelloniemi A, Havukainen H, Permi P, Mattila S, and Ylanne J (2009) Characterization of the interaction between Actinin-Associated LIM Protein (ALP) and the rod domain of alpha-actinin. BMC Cell Biol. 10, 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Sheinerman FB, Norel R, and Honig B (2000) Electrostatic aspects of protein-protein interactions. Curr. Opin. Struct. Biol. 10, 153–159. [DOI] [PubMed] [Google Scholar]
- (27).Yamashita Y, Matsuura T, Kurosaki T, Amakusa Y, Kinoshita M, Ibi T, Sahashi K, and Ohno K (2014) LDB3 splicing abnormalities are specific to skeletal muscles of patients with myotonic dystrophy type 1 and alter its PKC binding affinity. Neurobiol. Dis. 69, 200–205. [DOI] [PubMed] [Google Scholar]
- (28).Nakamori M, Sobczak K, Puwanant A, Welle S, Eichinger K, Pandya S, Dekdebrun J, Heatwole CR, McDermott MP, Chen T, Cline M, Tawil R, Osborne RJ, Wheeler TM, Swanson MS, Moxley RT 3rd, and Thornton CA (2013) Splicing biomarkers of disease severity in myotonic dystrophy. Ann. Neurol. 74, 862–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Larsson C (2006) Protein kinase C and the regulation of the actin cytoskeleton. Cell. Signalling 18, 276–284. [DOI] [PubMed] [Google Scholar]
- (30).Dominguez R (2016) The WH2 Domain and Actin Nucleation: Necessary but Insufficient. Trends Biochem. Sci. 41, 478–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Gonsior SM, Gautel M, and Hinssen H (1998) A six-module human nebulin fragment bundles actin filaments and induces actin polymerization. J. Muscle Res. Cell Motil. 19, 225–235. [DOI] [PubMed] [Google Scholar]
- (32).Gurung R, Yadav R, Brungardt JG, Orlova A, Egelman EH, and Beck MR (2016) Actin polymerization is stimulated by actin cross-linking protein palladin. Biochem. J. 473, 383–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Claeys KG, Fardeau M, Schroder R, Suominen T, Tolksdorf K, Behin A, Dubourg O, Eymard B, Maisonobe T, Stojkovic T, Faulkner G, Richard P, Vicart P, Udd B, Voit T, and Stoltenburg G (2008) Electron microscopy in myofibrillar myopathies reveals clues to the mutated gene. Neuromuscul Disord 18, 656–666. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.