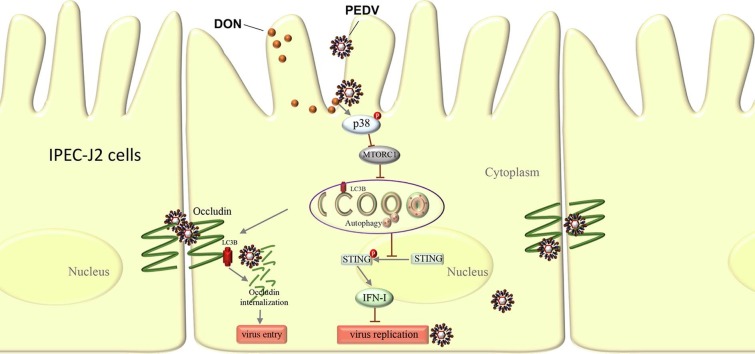
Graphical abstract
Keywords: Environmental Toxins, Deoxynivalenol, PEDV, Autophagy, MAPK p38 signaling
Abbreviations: ADFI, Average daily feed intake; ADG, Average daily gain; ATP, Adenosine triphosphate; CQ, Chloroquine; DON, Deoxynivalenol; IFN, Interferon; IPEC-J2, Porcine small intestinal epithelial cell line; LDH, Lactatedehydrogenase; LC3B, Light chain 3 β; LSCM, Laser scanning confocal microscope; MAPK, Mitogen-activated protein kinase; MTORC1, Mammalian target of rapamycin complex 1; PEDV, Porcine epidemic diarrhea virus; PI3K, Phosphoinositide 3 kinase; STING, Stimulator of interferon genes; siRNA, small interfering RNA; SQSTM1, Sequestosome 1; TCID50, Tissue culture infective dose; TGEV, Transmissible gastroenteritis virus; TEM, Transmission electron microscopy
Highlights
-
•
DON promotes PEDV entry and replication, and aggravates gut barrier injury.
-
•
CRISPR‐Cas9‐mediated LC3B knockout indicated a role of autophagy in the promotion.
-
•
p38 activation was required for the DON-enhanced autophagy in PEDV-infected cells.
-
•
Environmental toxins (eg, mycotoxins) might affect the prevalence of coronavirus.
Abstract
Mycotoxins are toxic metabolites produced by fungal species that commonly present in the global environment, especially in cereals and animal forages. The changing global environment may further increase the exposure to these toxins, posing a serious threat to humans and animals. Recently, coronavirus has become one of the most important pathogens threatening human and animal health. It is not clear whether environmental toxins, such as mycotoxins, will affect coronavirus infection. Given that pigs are among the animals most affected by coronavirus and highly homologous to humans, weaned piglets and IPEC-J2 cells were respectively chosen as in vivo and in vitro model to explore the impacts of deoxynivalenol (DON), the most abundant trichothecene mycotoxin in feed, on porcine epidemic diarrhea virus (PEDV) infection and the mechanisms involved. In vivo, twenty-seven piglets infected naturally with PEDV were randomly divided into three groups, receiving the basal diet containing 0, 750 and 1500 μg/kg DON, respectively. Significant increases in the diarrhea rates, gut barrier injury and PEDV proliferation of piglets’ small intestine were observed in experimental groups compared with the control. Additionally, the autophagosome-like vesicles and the autophagy-related proteins expression were also increased in experimental groups. In vitro, we observed that 0.1, 0.5 and 1.0 μM DON significantly promoted the entry and replication of PEDV in IPEC-J2 cells, along with the induction of a complete autophagy. CRISPR‐Cas9‐mediated knockout of LC3B indicated a vital role of autophagy in the promotion. Pretreatment with p38 signaling inhibitor could significantly block the induction of autophagy, indicating that DON could promote the PEDV infection by triggering p38-mediated autophagy. Our findings suggest that mycotoxin could influence the prevalence of coronavirus and provide new ideas for the prevention and control of coronavirus.
1. Introduction
Mycotoxins, the secondary metabolites produced by fungi, are capable of causing mycotoxicosis (diseases and death) in human and animals (Chen et al. 2018). Several environmental factors contribute to the presence of mycotoxins in globally feedstuffs and food commodities cereal, resulting in regular animals and human exposure (Alshannaq and Yu, 2017, Pascari et al., 2018). And this occurrence could further stimulate by the on-going global warming. Most of the reported effects of mycotoxins are negative in terms of intestinal health, where the barrier is damaged and accompanied by an increase of the gut pathogen (Heyndrickx et al., 2015, Luo et al., 2019). However, the interactions between gut pathogen and environmental mycotoxins are not clear.
Coronaviruses (CoVs) are RNA viruses that have become a major public health concern, severely damaging the respiratory, digestive and nervous systems of humans and animals (Khot, 2019, Yin and Wunderink, 2018, Zong et al., 2019). Human CoVs were considered as serious viruses since the emergence of the Severe Acute Respiratory Syndrome (SARS) in 2002–2003 (Cheng et al., 2007, Lai et al., 2005). The threat of coronaviruses was further highlighted with the emergence of the Middle East Respiratory Syndrome‐CoV (MERS‐CoV) outbreak in 2012 and the 2019 novel CoV (COVID-19) outbreak in 2019 (Ashour et al., 2019, Yang et al., 2020). Animal CoVs are known to cause important diseases in domestic animals or birds, leading to catastrophic economic loss (Lin et al., 2016, Zhou et al., 2018). Although rare, animal CoVs have the potential to infect humans and could further spread through many transmissions (Forni et al. 2017). Pig is one of the animals most susceptible to coronavirus infection. Swine coronaviruses mainly include transmissible gastroenteritis virus (TGEV), porcine deltacoronavirus (PDCoV), porcine epidemic diarrhea virus (PEDV) and the emerging HKU2-like porcine enteric alphacoronavirus (PEAV) (Niederwerder and Hesse, 2018, Wang et al., 2019b). Similar to other enteric coronavirus diseases, the porcine epidemic diarrhea (PED) is age-dependent but more destructive (Lee, 2015, Li et al., 2011, Song and Park, 2012), causing the intestinal epithelial cells necrosis, villous atrophy, vomit, diarrhea and dehydration in pigs of all ages, even the death of piglets (Wang et al. 2019a). The differences in the morbidity and mortality of weaned piglets among different farms suggested the presence of other non-infectious factors affecting the progress of PED. Therefore, we speculated that deoxynivalenol (DON), the most abundant trichothecene mycotoxin (Broekaert et al., 2017, Pierron et al., 2016), might aggravate PED in weaned piglets.
Autophagy is the major intracellular degradation system that is essential for survival, differentiation and homeostasis of cells (Matsuda et al., 2018, Pott and Maloy, 2018, Tang et al., 2015). It principally serves a regulatory mechanism to control the innate immune response against intracellular pathogens (McEwan, 2017, Riffelmacher et al., 2018, Yang et al., 2017). Dysregulation of autophagy contributes to the pathogenesis of an increasing number of diseases (Shen et al., 2018, Yang et al., 2018). Importantly, in certain viral infection settings, the self-cannibalistic or, paradoxically, even the pro-survival functions of autophagy may be deleterious (Chen et al. 2019). Evidences suggest that some viruses, including PEDV, may induce autophagy in order to utilize it for their replication when they infect a target cell (Chen et al., 2017, Guo et al., 2017, Harris et al., 2015). However, the impacts of DON on PEDV infection are equivocal, and questions about whether DON induces autophagy in target cells remain unanswered.
In this study, we evaluated the effects of DON on PEDV infection in vitro and in vivo, and found that DON promotes autophagosomes formation, thereby facilitating the entry and replication of PEDV. Our findings will provide novel perspective to advance the understanding in the pathogenesis of PEDV and new ideas for the prevention and control of coronavirus.
2. Materials and methods
2.1. Animal experiments
All experiments were conducted according to the standards of the European Guidelines for Animal Welfare and were approved by the Committee for the Care and Use of Experimental Animals of the Nanjing Agricultural University (Animal Ethics Number: SYXK (Su) 2011–0036). Animal experiments were carried out at a 1400-weaned piglets farm. Eighty weaned piglets (aged 3 weeks) were selected. These piglets were positive for PEDV naturally and negative for TGEV and porcine rotavirus as determined by fecal and blood diagnostics. Twenty-seven out of eighty piglets (BW = 5.5 ± 0.5 kg) were selected and randomly divided into three groups, with 3 replicates per group and 3 piglets per replicate. Piglets were assigned to 3 groups (PEDV Group: infected piglets received a basal diet; PEDV + 750 μg/kg DON Group: infected piglets received the basal diet containing 750 μg/kg feed DON; and PEDV + 1500 μg/kg DON Group: infected piglets received the basal diet containing 1500 μg/kg feed DON), for 14 days. All piglets were housed in the same facility but different rooms under biosafety conditions and allowed free access to water and feed during the experiment. Average daily gain (ADG) and average daily feed intake (ADFI) were evaluated to determine the growth performance of piglets. On dpi 14, the piglets were euthanized and tissue samples of duodenum, jejunum, ileum and mesenterium were collected.
2.2. Diarrhea rate and diarrhea index evaluation
Diarrhea in each piglet was recorded and scored daily according to the state of feces. Piglets with dry and cylindrical feces are scored 0 point. Piglets with soft and tangible feces are scored 1 point. Piglets with sticky and semi-solid feces are scored 2 points. Piglets with liquid and unformed feces are scored 3 points. Diarrhea rate = [number of diarrhea piglets per replicate / (number of piglets per replicate days)] 100%. Diarrhea index = diarrhea scores sum of piglets of per replicate / (number of piglets per replicate days).
2.3. Reagents and antibodies
Deoxynivalenol (DON, purity ≥ 98%, for experiments in vitro), chloroquine (CQ), rapamycin (Rapa) and rabbit anti-LC3B antibody were purchased from Sigma-Aldrich (St. Louis, USA). Deoxynivalenol (DON, purity ≥ 98%, for experiments in vivo) was purchased from Pribolab (Immunos, Singapore). SB203580 and AUD-S100 were purchased from MedChemExpress (New Jersey, USA). Rabbit anti-SQSTM1, anti-MAPKs, anti-JAK1, anti-pSTING/STING, anti-PI3K, anti-β-actin antibodies and horseradish peroxidase (HRP)-conjugated goat anti-rabbit secondary antibody were purchased from Cell Signaling Technology (Boston, USA). Rabbit anti-claudin1, anti-occludin, anti-ZO-1 and anti-p-MTORC1/MTORC1 antibodies were purchased from Abcam (Cambridge, UK). Porcine epidemic diarrhea virus (PEDV) strain CV777 was obtained from Jiangsu Academy of Agricultural Sciences (Nanjing, China). Rabbit anti-PEDV-N antibody was prepared by our lab. Poly (I:C) (LMW) / LyoVecTM was purchased from InvivoGen (San Diego, USA).
2.4. Histological analysis
Jejunum tissues samples were fixed in 4% paraformaldehyde, embedded in paraffin and sectioned at a thickness of 4 μm. For histopathological examination, tissue slices were stained with hematoxylin and eosin, and observed under the microscope. For immunohistochemistry examination, tissue slices were incubated with antibodies against PEDV N protein, followed by incubation with the second antibody and streptavidin-peroxidase complex. The peroxidase conjugates were visualized using DAB solution.
2.5. Quantitative Real-Time PCR (qRT-PCR) analysis
qRT-PCR was performed using the StepOnePlus Real-Time PCR System (Applied Biosciences) as previously described (Liu et al. 2018b). Briefly, total RNA was isolated from the cells using an RNA Extraction Kit (Takara, Japan) and then reverse-transcribed to cDNA using the PrimeScript RT Master Mix Kit (Takara, Japan). All primers are listed in Table 1 . The relative expression was determined using the Δcycle threshold (ΔCt) method with GAPDH serving as a reference gene.
Table 1.
Primers used in this study.
| Primers | Sequence (5′ − 3′) | Purpose |
|---|---|---|
| GAPDH | F: TGGGCGTGAACCATGAGAA R: CCTCCACGATGCCGAAGT |
qRT-PCR for detection of GAPDH |
| PEDV-N | F: GTCTGA-CAACAGCGGCAAAA R: TTTCGCCCTTGGGAATTCTC |
qRT-PCR for detection of PEDV-N |
| PEDV-S | F: TGTTTATTCTGTCACGCCATGTT R: CCAGGCAACTCCCTAGTATTGCT |
qRT-PCR for detection of PEDV-S |
| IFN-α | F: ACTTCCACAGACTCACCCTCTATC R: ATGACTTCTGCCCTGATGATCT |
qRT-PCR for detection of IFN-α |
| IFN-β | F: TGCATCCTCCAAATCGCTCT R: ATTGAGGAGTCCCAGGCAAC |
qRT-PCR for detection of IFN-β |
| IFN-γ | F: TTTTGTCACTCTCCTCTTTCCA R: GACTTTGTGTTTTTCTGGCTCTTAC |
qRT-PCR for detection of IFN-γ |
| IFN-λ | F: GGTGCTGGCGACTGTGATG R: GATTGGAACTGGCCCATGTG |
qRT-PCR for detection of IFN-λ |
| shLC3B | GCTTGCAGCTCAATGCTAACCCTCGAGGGTTAGCATTGAGCTGCAAGC | Knockdown of LC3B |
| scrambled | GCGCGCTTTGTAGGATTCTCGCTCGAGCGAATCCTACAAAGCGCGC | Knockdown of scrambled |
| OccludinsiRNA | UAAGCUCUUGUACUCCUGCtt | Knockdown of Occludin |
| ControlsiRNA | UUCUCCGAACGUGUCACGUtt | Knockdown of scrambled |
2.6. Cell cultures
The porcine intestinal cell line (IPEC-J2) was stored in our laboratory and grown in DMEM/F12 (1:1) medium supplemented with 10% fetal bovine Serum (Invitrogen, Carlsbad, USA), 1% insulin-transferrin-selenium (ITS), 5 ng/mL epidermal growth factor (EGF; Sigma, USA) and 1% antibiotics at 37 °C in a humidified atmosphere containing 5% CO2.
2.7. Cell viability assay
Cell viability was monitored by 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2-H-tetrazolium bromide (MTT; Sigma, USA) assay as previously described (Liu et al. 2018a). Briefly, IPEC-J2 cells were cultured in 96-well plates at a density of 5 × 103 cells/well with corresponding treatments. Then, each well was added with 15 μL of MTT (5 mg/mL) for another 4 h at 37 °C. The supernatants were discarded and incubated with 150 μL DMSO to dissolve the precipitate. Absorbance was measured at 490 nm with a reference wavelength of 595 nm. All tests were performed three times.
2.8. Detection of virus attachment, entry, replication and release
For attachment assay, cells were cooled for 2 h at 4℃ and then infected with PEDV in the presence of different concentrations of DON for 2 h at 4℃. After washing with cold PBS, cells were cultured with fresh medium for 24 h at 37℃. For entry assay, cells were incubated with PEDV for 2 h at 4℃ and switched to 37℃ for 4 h in the presence of different concentrations of DON after washing three times. Then cells were washed with PBS and incubated for another 24 h at 37℃. For replication assay, cells were incubated with PEDV for 2 h at 37℃. Cells were washed with PBS and cultured with different concentrations of DON for another 24 h at 37℃. For release assay, cells were incubated with PEDV for 24 h at 37℃. After washing with PBS, cells were cultured with different concentrations of DON for another 4 h at 37℃. After different treatments, the supernatant was collected and titrated using TCID50 and the cells were harvested for immunoblotting, respectively.
2.9. LC3B−/− IPEC-J2 cell production by CRISPR/ Cas9 system
The small guide RNAs (sgRNAs) were designed using Breaking‐Cas (http:// bioinfogp.cnb.csic.es/tools/breakingcas/) online tool and synthesized (Invitrogen). The sgRNA was cloned pCas-Puro-U6 plasmid and the pCas-Puro-U6 plasmid Linear was obtained using the BbsI restriction enzyme (Thermo Fisher Scientific). The plasmids containing sgRNA were transfected into IPEC-J2 cells with GeneTran III (Biomiga) for 48 h, and then the transfected cells were selected using 5 μg/mL of puromycin. The selected cells were subjected to serial dilutions in 96‐well plate to obtain a single cell colony. After 14 days of colony formation, each single colony was picked and expanded. Genomic DNA was extracted from individual clones and sequenced to confirm the specificity of targeting.
2.10. Immunoblotting and coimmunoprecipitation analysis
The relative protein expression levels were measured by immunoblotting as previously described with minor modifications (Qian et al. 2018). Briefly, equal amounts of protein obtained from the lysed cells were loaded onto 12% SDS-PAGE gels and transferred onto PVDF membranes (Millipore, USA). After blocking with 5% BSA for 2 h, the PVDF membranes were incubated at 4 °C overnight with primary antibodies, followed by a 1-h incubation with secondary antibodies at room temperature. The expected protein bands were detected using Image Quant LAS 4000 (GE Healthcare Life Sciences, USA). The relative abundance of the target protein (normalized to β-actin) was quantified by densitometric analysis using the Image Pro-Plus 6.0 software. For coimmunoprecipitation (Co-IP) experiments, equal amounts of protein obtained from the lysed cells were incubated with polyclonal rabbit anti-LC3B, anti-Occludin or anti-IgG antibody (as a negative control) overnight after incubating with Dynabeads protein G (Therom, USA) for 1 h. Subsequent experimental procedures were the same as immunoblotting.
2.11. Fluorescence microscopy
Cells grown on coverslips were fixed with 4% paraformaldehyde for 20 min at 4 °C. After washing three times, cells were blocked with 1% BSA at room temperature and incubated with primary rabbit anti-occludin antibody and secondary FITC-conjugated goat anti-rabbit antibody (Invitrogen, USA), respectively. For the analysis of LC3B expression, cells grown on coverslips to 60–70% confluence were transfected with the pLVX-mRFP-EGFP-LC3B plasmid (provided by Prof. Qian Yang, Nanjing Agriculture University, Nanjing, China) using jetPRIME transfection reagent (Polyplus-transfection, Illkirch, France) according to the manufacturer’s protocols. Nuclei were stained with DAPI (Blue, Beyotime Biotechnology, China). Fluorescence microscopy was performed using a Zeiss LSM710 confocal microscope (Zeiss, Oberkochen, Germany).
2.12. RNA interference
Occludin-specific siRNA and control siRNA were designed and synthesized by Invitrogen (Thermo Fisher, USA). All primers are listed in Table 1. Cells were transfected with 100 nM occludin-specific or control siRNA duplexes by use of jetPRIME transfection reagent according to the manufacturer’s guidelines. Twenty-four hours after transfection, cells were washed with DMEM/F12 and cultured in DMEM/F12 with 4% FBS until further treatments.
2.13. Quantification of virus titer
Viral titers were determined by 50% endpoint dilution (50% tissue culture infective dose [TCID50]) assays on IPEC-J2 cells as previously described (Liu et al. 2018a). Briefly, IPEC-J2 cells cultured in 96-well plates were inoculated with 10-fold dilutions of the harvested culture supernatants for indicated time. Microscope was used to detect the viral antigen according to the cell damage. Viral titers were expressed as TCID50/mL by using the Reed-Muench method.
2.14. Statistical analysis
Statistical analyses were performed using Graph Pad Prism 7.0 by one-way analysis of variance (ANOVA), and the data were expressed as the means ± SD. P < 0.05 was regarded as significant.
3. Results
3.1. Low doses exposure of DON could aggravate intestinal injury and facilitate PEDV infection in weaned piglets
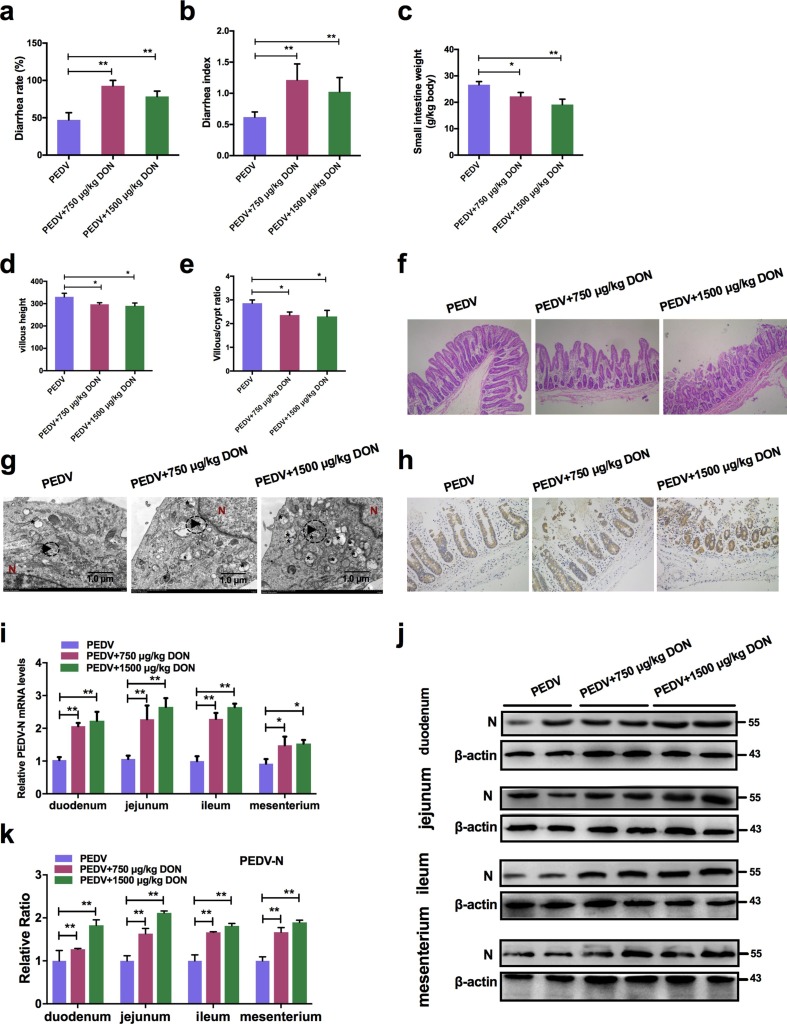
To evaluate the effects of DON exposure on PEDV-infected piglets, we performed animal experiments. Twenty-seven piglets infected naturally with PEDV were randomly divided into three groups: PEDV group received a basal diet, PEDV + 750 μg/kg DON group received the basal diet containing 750 μg/kg feed DON; PEDV + 1500 μg/kg DON group received the basal diet containing 1500 μg/kg feed DON. After 14 days, we observed that the diarrhea rates (Fig. 1 a) and diarrhea index (Fig. 1b) of piglets in experiment groups were increased compared with that in PEDV group (P < 0.01). The ADG (average daily gain, Table 2 ), ADFI (average daily feed intake, Table 2) and small intestine weight (Fig. 1c) of piglets in experiment groups were also lower than that in PEDV group (P < 0.05). The pathological results showed that DON exposure aggravated the villous atrophy (Fig. 1d, e) and barrier injury of PEDV-infected piglet jejunums (Fig. 1f). It's worth noting that these damages were not caused by DON alone, as 1500 μg/kg DON exposure alone did not significantly damage the intestinal tract (Pasternak et al. 2018), indicating that 750 and 1500 μg/kg DON were low doses exposure.
Fig. 1.
Low doses exposure of DON could aggravate intestinal injury and facilitate PEDV infection in weaned piglets. Piglets infected naturally with PEDV were fed with a basal diet containing 0, 750 or 1500 μg/kg DON. a Diarrhea rate of piglets. b Diarrhea index of piglets. c Small intestine weight of piglets. d Villus length of jejunums. e Villus/crypt ratio of jejunums. f Histopathological examination of jejunums (magnification, × 200). g TEM observation of jejunums. The virus particles (black arrowheads) were observed. The scale bar indicates 1.0 μm. h Immunohistochemistry examination. The PEDV nucleocapsid (N) (brown signals) expression in jejunums of piglets was measured (magnification, × 200). i Effects of DON on the mRNA levels of PEDV-N gene in duodenum, jejunum, ileum and mesenterium of piglets. j, k Effects of DON on the protein levels of PEDV-N in duodenum, jejunum, ileum and mesenterium of piglets. The data are expressed as mean ± SD (n ≥ 3). *P < 0.05, **P < 0.01.
Table 2.
Growth performance of piglets.
| Groups | n | Initial body weight | Final body weight | ADG | ADFI | F/G |
|---|---|---|---|---|---|---|
| kg | kg | g/day | g/day | |||
| I | 6 | 5.48 | 8.62a | 223.83a | 219.83a | 0.99b |
| II | 6 | 5.52 | 7.52b | 142.83b | 202.50b | 1.47ab |
| III | 6 | 5.45 | 7.20b | 125.0b | 188.5b | 1.60a |
| SEM | 0.15 | 0.16 | 15.10 | 5.04 | 0.19 |
The data with different little letters show significant difference in the same column (P < 0.05).
Next, we evaluated whether low doses exposure of DON could affect PEDV proliferation in piglets using transmission electron microscopy (TEM), immunohistochemistry, qRT-PCR and immunoblotting. Compared with PEDV group, the virus particles (black arrowheads) observed under TEM were increased in jejunum of piglets in DON groups (Fig. 1g). The immunohistochemistry results showed that the PEDV antigens represented by brown signals in enterocytes of piglets fed with the DON contamination diet were enhanced (Fig. 1h). In addition, we found that both the PEDV-N mRNA levels (Fig. 1i) and the PEDV-N protein levels (Fig. 1j, k) in jejunum of piglets in DON groups were increased significantly compared with that in PEDV group. These data suggested that low doses exposure of DON could aggravate intestinal injury and virus infection of PEDV-infected piglets.
3.2. Low concentrations exposure of DON could facilitate PEDV replication and entry in IPEC-J2 cells.
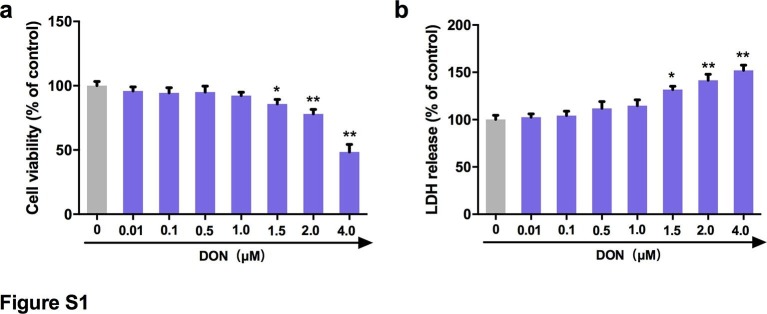
To eliminate the cytotoxic effects of DON, the viability of IPEC-J2 cells treated with different concentrations of DON was analyzed by enzymatic reduction of MTT. As shown in Fig. S1a, the viability of IPEC-J2 cells was decreased at concentrations of 1.5 to 4.0 μM (P < 0.05). The release of LDH in the supernatant was quantified by detection of LDH enzymatic activity to evaluate the effect of increasing concentrations of DON on the permeabilization of IPEC-J2 cell membrane. Significant increases were observed in the release of LDH after treatment with 1.5 to 4.0 μM DON (Fig. S1b). Therefore, 0.01, 0.1, 0.5 and 1.0 μM DON were regarded as low concentrations and used in subsequent experiments.
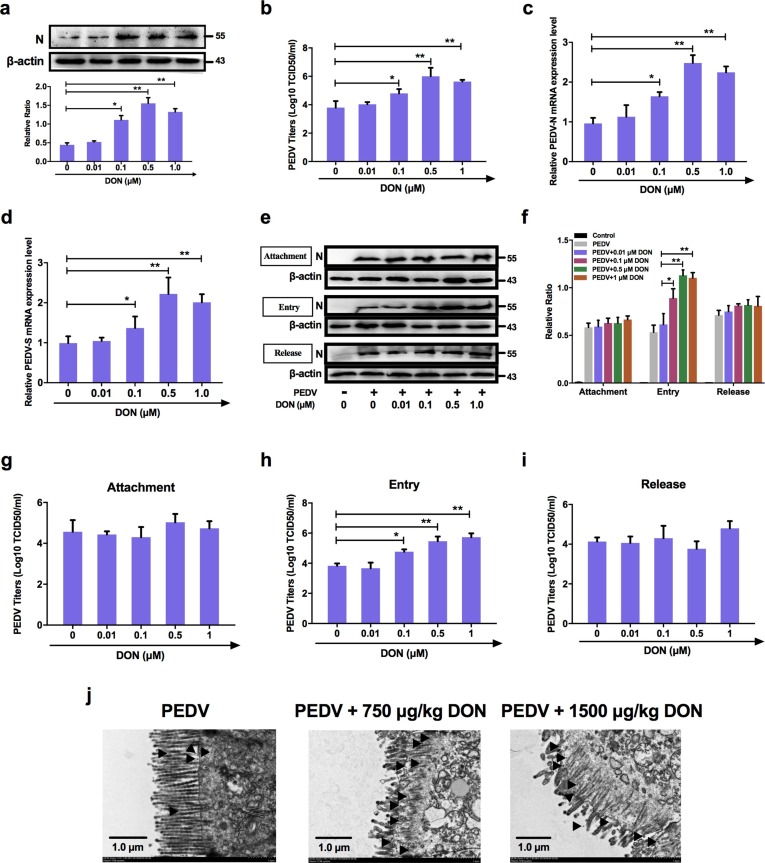
To explore whether DON can also affect PEDV infection in vitro, the effects of DON on PEDV replication were firstly assayed. IPEC-J2 cell monolayers were infected with PEDV for 2 h, and cultured with DON at concentrations between 0.01 and 1 μM for an additional 24 h. The data showed that, compared with the PEDV group, the protein level of PEDV-N (Fig. 2 a), the viral titer (Fig. 2b) and the mRNA levels of PEDV-N and -S genes (Fig. 2c, d) were significantly increased in PEDV-infected cells treated with 0.1, 0.5 or 1 μM DON, which were matching with the serum and tissue DON concentrations that promoted PEDV infection (Devreese et al., 2014, Goyarts and Danicke, 2006).
Fig. 2.
Low concentrations exposure of DON could promote the replication and entry of PEDV. For virus replication: a Cell lysates were subjected to immunoblotting with antibodies to PEDV-N protein or β-actin (loading control). b Cells were assayed for PEDV viral titers. qRT-PCR were performed to analyze the mRNA levels of PEDV-N (c) and -S (d) genes. For virus attachment, entry and release: e, f Cell lysates were subjected to immunoblotting with antibodies to PEDV-N protein or β-actin (loading control). Viral titers assays for PEDV attachment (g), entry (h) and release (i). j TEM observation of jejunums for PEDV entry. The data are expressed as mean ± SD (n = 3). *P < 0.05, **P < 0.01.
In addition to replication, the process by which a virus infects a host involves adhesion, invasion, and release. We then surveyed the relationship between DON exposure and virus attachment, entry and release in IPEC-J2 cells as described in Materials and Methods. As determined by immunoblotting, PEDV entry were increased in IPEC-J2 cells exposed to 0.1–1.0 μM DON, but there was little change in PEDV attachment and release (Fig. 2e, f). The results of virus titers were consistent with immunoblotting (Fig. 2g-i). At the same time, more virus particles (black arrowheads, Fig. 2j) invading the jejunum of piglets could be observed under TEM in experiment groups compared with the PEDV group. Taken together, these results suggest that low concentrations exposure of DON contributed to PEDV replication and entry in vitro and vivo
3.3. Alteration of occludin protein distribution induced by DON contributed to PEDV entry.
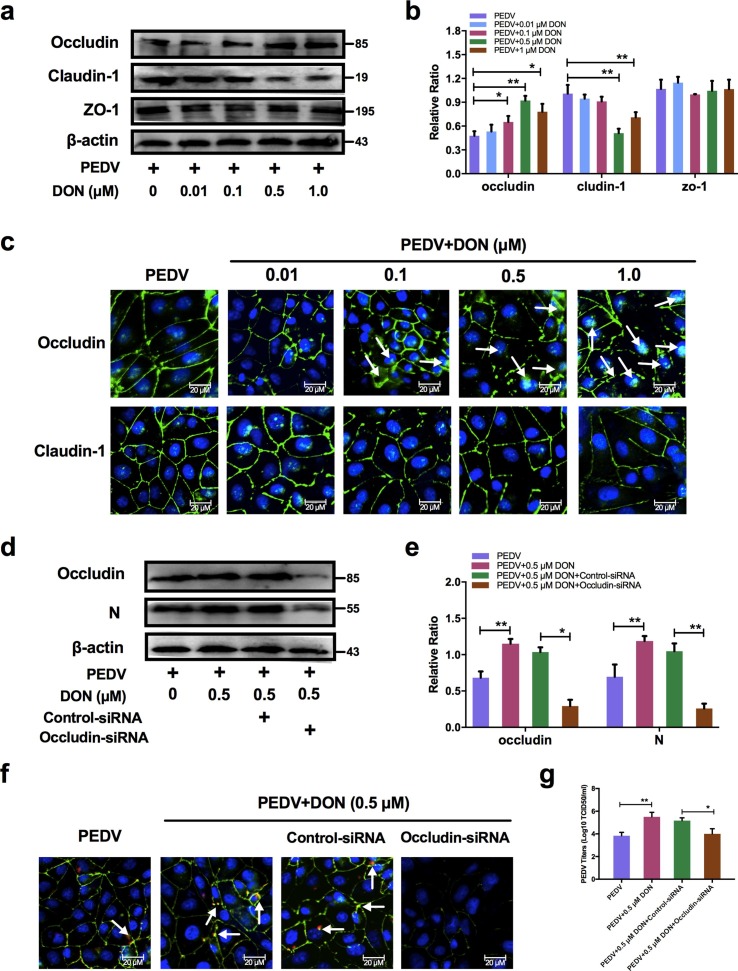
To explore the mechanism that low concentrations exposure of DON facilitated PEDV entry, we analyzed the levels of the tight junction proteins (ZO-1, occludin and claudin-1) in PEDV-infected IPEC-J2 cells exposed to DON. As determined by immunoblotting, the protein levels of ZO-1 had changed little and that of claudin-1 were significantly decreased by 0.5 and 1.0 μM DON, however, the protein levels of occludin were significantly increased by 0.1, 0.5 and 1.0 μM DON in PEDV-infected IPEC-J2 cells (Fig. 3 a, b). The cellular expression and distribution of occludin and claudin-1 were measured to further explore the relationship between tight junction proteins and DON-promoted PEDV infection in IPEC-J2 cells. Immunofluorescence analysis (IFA) results showed that tight junction formation in mock cells; PEDV infection induced the slight internalization of occludin, not claudin-1, indicating that occludin staining in the junctional area was decreased, and that in cytoplasm was increased; meaning that DON aggravated the internalization of occludin (Fig. 3c).
Fig. 3.
Occludin internalization was required for DON-promoted PEDV entry in IPEC-J2 cells. a, Cell lysates were subjected to immunoblotting (a, b, d. e) with antibodies to ZO-1, occludin, claudin-1, PEDV-N protein or β-actin (loading control). Cells were subjected to IFA (c, f) with antibodies to occludin (green), claudin-1 (green) and PEDV-N protein (red). Cell nuclei were stained with DAPI (blue). The scale bar indicates 20 μm. g Cells were assayed for PEDV viral titers. The data are expressed as mean ± SD (n = 3). *P < 0.05, **P < 0.01. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Subsequently, small interfering RNA (siRNA) duplexes targeting the occludin gene was used to further determine whether occludin internalization is required for PEDV infection promoted by DON in IPEC-J2 cells. As expected, immunoblotting showed that occludin siRNA-transfected cells exposed to DON at 0.5 μM were exhibited very low levels of PEDV entry compared with DON + PEDV group (Fig. 3d, e). IFA results validated that occludin knockdown induced a reduction of PEDV-N protein expression (Fig. 3f). The result of virus titers was consistent with immunoblotting (Fig. 3g). These data indicated that DON could facilitate PEDV entry via altering the cell junctional localization of the occludin.
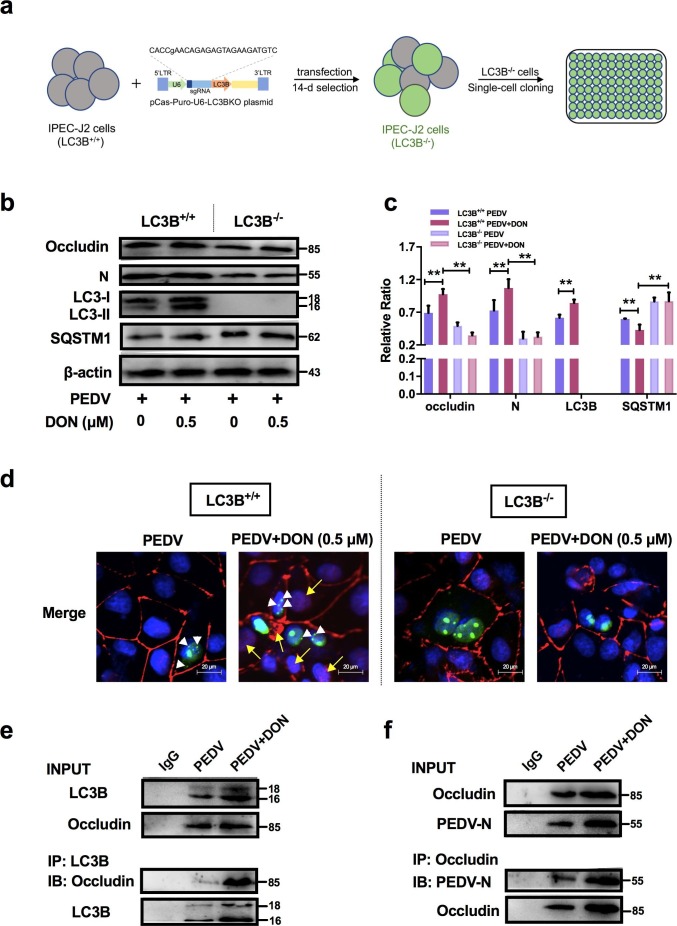
3.4. CRISPR‐Cas9‐mediated knockout of the LC3B in IPEC-J2 cells abolished the contribution of DON to occludin-mediated PEDV entry.
LC3B (light chain 3 β), a marker of autophagic activity, is present during the entirety of this autophagic process and is regulated by lots of signaling (Liu et al., 2018c, Ramkumar et al., 2017). To explore how DON altered the localization of occludin, we constructed the LC3B-/- IPEC-J2 cells by CRISPR‐Cas9 system to verify whether the LC3B contributes to the occludin localization (Fig. 4 a). The expression of autophagy-related proteins, occludin and PEDV-N were then measured. The results showed that the expression of LC3-II and the degradation of SQSTM1 were significantly increased in LC3B+/+ IPEC-J2 cells treated with DON, which were consistent with the changes of occludin expression. However, the increase in occludin expression and virus proliferation by DON was abolished in LC3B-/- IPEC-J2 cells (Fig. 4b, c). Next, we performed pEGFP transfection assays and observed that the colocalization of LC3B, occludin and virus induced by DON were arrested in LC3B-/- IPEC-J2 cells (Fig. 4d). The results of coimmunoprecipitation further confirmed that occludin could directly bind to LC3B and PEDV (Fig. 4e, f). Of note, the occludin bands in the PEDV group could also be observed in the IP test as occludin is involved in the PEDV invasion (Luo et al. 2017). These data suggested that DON could induce occludin internalization upon a canonical autophagy.
Fig. 4.
LC3B was required for occludin internalization-induced PEDV entry in IPEC-J2 cells exposed to DON. a Generation of LC3B‐knockout IPEC-J2 cells. b, c LC3B+/+ or LC3B-/- cell lysates were subjected to immunoblotting with antibodies to occludin, PEDV-N, LC3B, SQSTM1 or β-actin (loading control). d LC3B+/+ or LC3B-/- cells were subjected to LSCM with antibody to occludin (red, yellow arrowhead indicates occludin internalization) and plasmid to LC3B (green, white arrowheads). Cell nuclei were stained with DAPI (blue). The scale bar indicates 20 μm. e, f LC3B+/+ cell lysates were subjected to co-immunoprecipitation with antibodies to occludin, PEDV-N or LC3B. The data are expressed as mean ± SD (n = 3). *P < 0.05, **P < 0.01. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
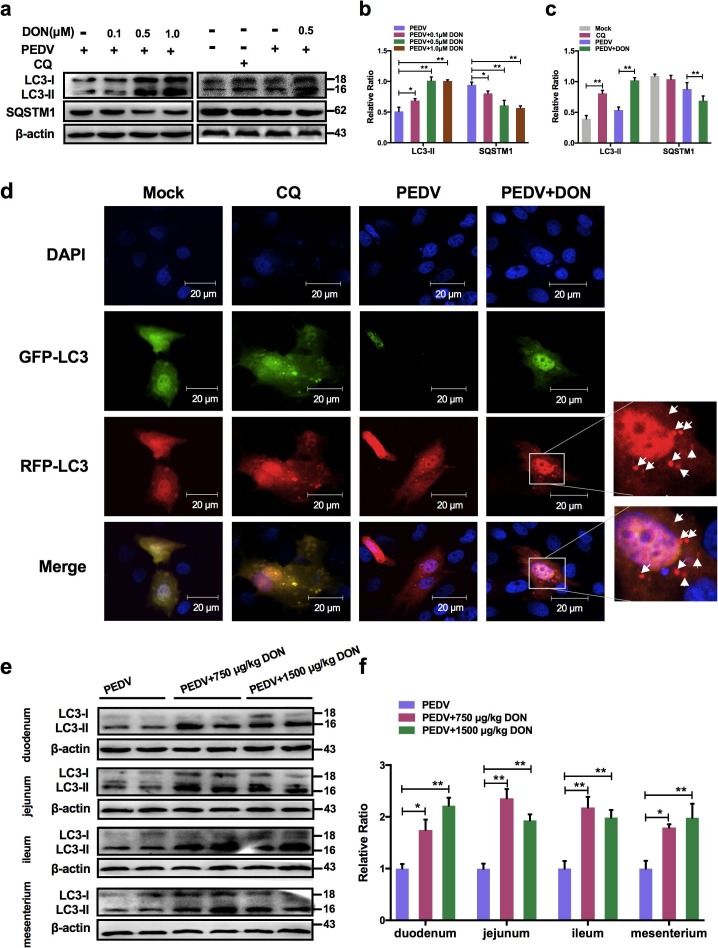
3.5. Low doses exposure of DON could trigger autophagy in the PEDV-infected IPEC-J2 cells and intestinal tissues of weaned piglets.
To determine the mechanism that DON promoted PEDV replication, the levels of autophagy-related protein LC3B was examined and the results showed that DON treatment led to a significant upregulation of LC3-II expression (Fig. 5 a, b). The expression of SQSTM1 was examined to further determine whether a complete autophagic flux occurred after DON exposure. We found that the protein level of SQSTM1 in PEDV-infected IPEC-J2 cells decreased after DON treatment (Fig. 5a, b, P < 0.05). The maximal effects of DON on the expression of autophagic markers were observed at an DON concentration of 0.5 μM, which is consistent with that in the virus replication result (Fig. 2). Moreover, the monomeric red fluorescent protein (mRFP)-Green fluorescent protein (GFP-LC3) tandem reporter construct was used to further measure DON-induced autophagic flux. In the acidic pH of the lysosome, lysosomal hydrolysis can attenuate the green fluorescence of this tandem autophagosome reporter, whereas it has no effect on red fluorescence. Therefore, autophagosomes have both GFP and mRFP signals, whereas autolysosomes have only mRFP signals (Xing et al. 2017). As shown in Fig. 5d, treatment with CQ, which inhibits the fusion of autophagosomes and lysosomes, resulted in yellow color-labeled autophagosomes, and RFP-LC3-labeled puncta structures were detected in PEDV-infected IPEC-J2 cells expressing the mRFP-GFP-LC3 reporter after incubation with 0.5 μM DON. The similar results could be observed in the immunoblotting experiment (Fig. 5a, c). These observations indicated that DON induced a complete autophagic flux in PEDV-infected IPEC-J2 cells.
Fig. 5.
Low concentrations exposure of DON could promote autophagosomes formation in PEDV-infected IPEC-J2 cells and piglets. Cell (a, b, c) or tissue (e, f) lysates were subjected to immunoblotting with antibodies to autophagy-related proteins (LC3B and SQSTM1) or β-actin (loading control). d Cells were subjected to LSCM with plasmid to autophagosomes (green spots) and autophagolysosomes (red spots, white arrowheads). Cell nuclei were stained with DAPI (blue). The scale bar indicates 20 μm. The data are expressed as mean ± SD (n = 3). *P < 0.05, **P < 0.01. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
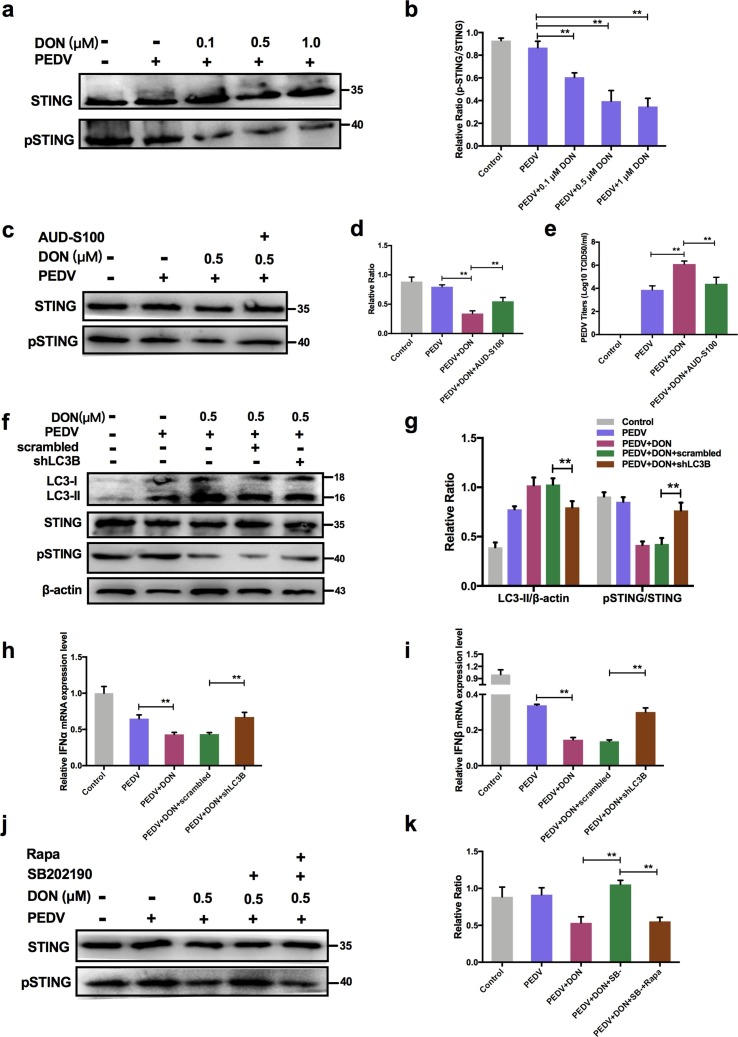
Similarly, the intestinal autophagy levels of piglets were measured by testing the autophagosome-like vesicles formation using TEM and the LC3-II/LC3-I ratio using immunoblotting. As shown in Fig. 1g, a larger number of double- or single-membrane vesicles (black asterisk) was observed in jejunum of piglets with the basal diet containing 750 and 1500 μg/kg DON compared with the PEDV group. The LC3-II/LC3-I ratios were also significantly increased in the duodenum, jejunum, ileum and mesenterium of piglets in experimental groups compared with that of piglets in PEDV group (Fig. 5e, f). These data indicated that DON could enhance autophagy in the PEDV-infected IPEC-J2 cells and piglets, which might be responsible for DON-promoted PEDV infection as autophagy can facilitate to PEDV proliferation (Guo et al. 2017).
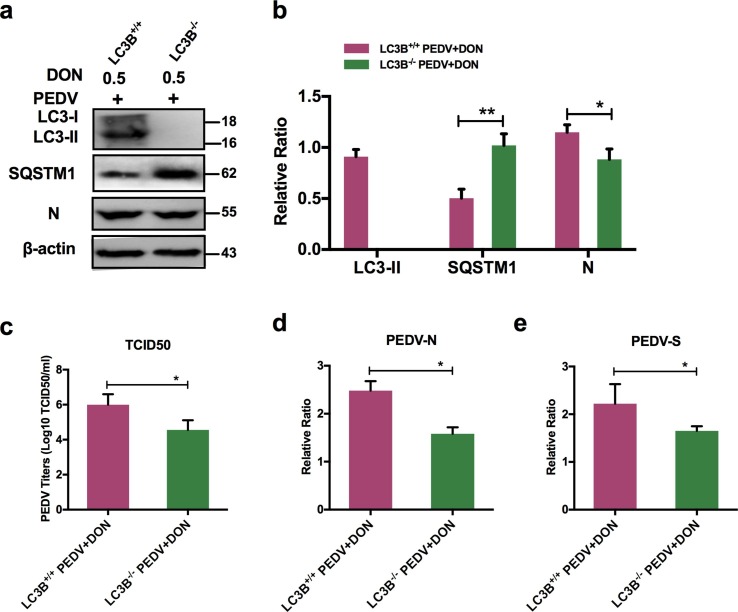
3.6. CRISPR‐Cas9‐mediated knockout of LC3B in IPEC-J2 cells suppressed the promotion of DON to PEDV replication.
To confirm the role of autophagy in DON-promoted PEDV replication, we compared the viral yield in LC3B+/+ and LC3B-/- IPEC-J2 cells exposed to DON. A significant decrease was observed in the protein expression of LC3B in LC3B-/- IPEC-J2 cells compared with that in LC3B+/+ cells (Fig. 6 a, b). Moreover, PEDV viral yield exhibited the same decrease, as demonstrated by the down-regulation of PEDV-N protein level (Fig. 6a, b), PEDV viral titers (Fig. 6c), and PEDV-N / -S mRNA levels (Fig. 6d, e), indicating that the decreased viral yield was due to the inhibition of autophagy. Collectively, these results suggested that the LC3B-medicated autophagy machinery was required for DON-promoted PEDV replication in IPEC-J2 cells.
Fig. 6.
Inhibition of autophagy could decrease DON-promoted viral yield in IPEC-J2 cells. a, b LC3B+/+ or LC3B-/- cell lysates were subjected to immunoblotting with antibodies to LC3B, SQSTM1, PEDV-N or β-actin (loading control). c LC3B+/+ or LC3B-/- cells were assayed for PEDV viral titers. RT-qPCR were performed to analyze the mRNA levels of PEDV-N (d) and -S (e) genes. The data are expressed as mean ± SD (n = 3). *P < 0.05, **P < 0.01.
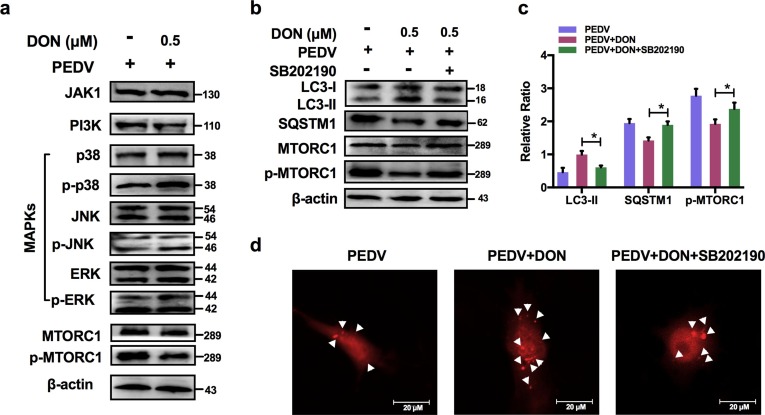
3.7. Activation of p38/MTORC1 signaling pathway was required for the upregulation of LC3B by DON in PEDV-infected IPEC-J2 cells.
To explore how DON induced autophagy, JAKs, PI3K and MAPKs signaling related-proteins were detected. The immunoblotting results revealed that there was no significance in JAK1, PI3K, p-JNK/JNK and p-ERK/ERK proteins expression after DON treatment. But, a significant increase in p-p38 was observed (Fig. 7 a). In addition, p-MTORC1 was significantly downregulated by DON. So, we supposed the activation of p-p38 might induce autophagy. To determine our hypothesis, the inhibitor of p-p38, SB202190, was supplied. The data showed that SB202190 inhibited the LC3II expression and SQSTM1 degradation increased by DON, upregulated the p-MTORC1 expression (Fig. 7b, c) and blocked the formation of autophagosomes (Fig. 7d). Therefore, DON induced autophagy via upregulating the p38/MTORC1 signaling pathway in PEDV-infected IPEC-J2 cells.
Fig. 7.
Activation of p38/MTORC1 signaling pathway was required for the activation of LC3B-mediated autophagy by DON in PEDV-infected IPEC-J2 cells. a Cell lysates were subjected to immunoblotting with antibodies to JAK1, PI3K, MAPKs, p-MTORC1/MTORC1 or β-actin (loading control). b, c Cell lysates were subjected to immunoblotting with antibodies to LC3B, SQSTM1, p-MTORC1/MTORC1 or β-actin (loading control) at the present of p-p38 inhibitor, SB202190. d Cells were subjected to LSCM with plasmid to autophagosomes (red spots, white arrowheads). The scale bar indicates 20 μm. The data are expressed as mean ± SD (n = 3). *P < 0.05, **P < 0.01. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
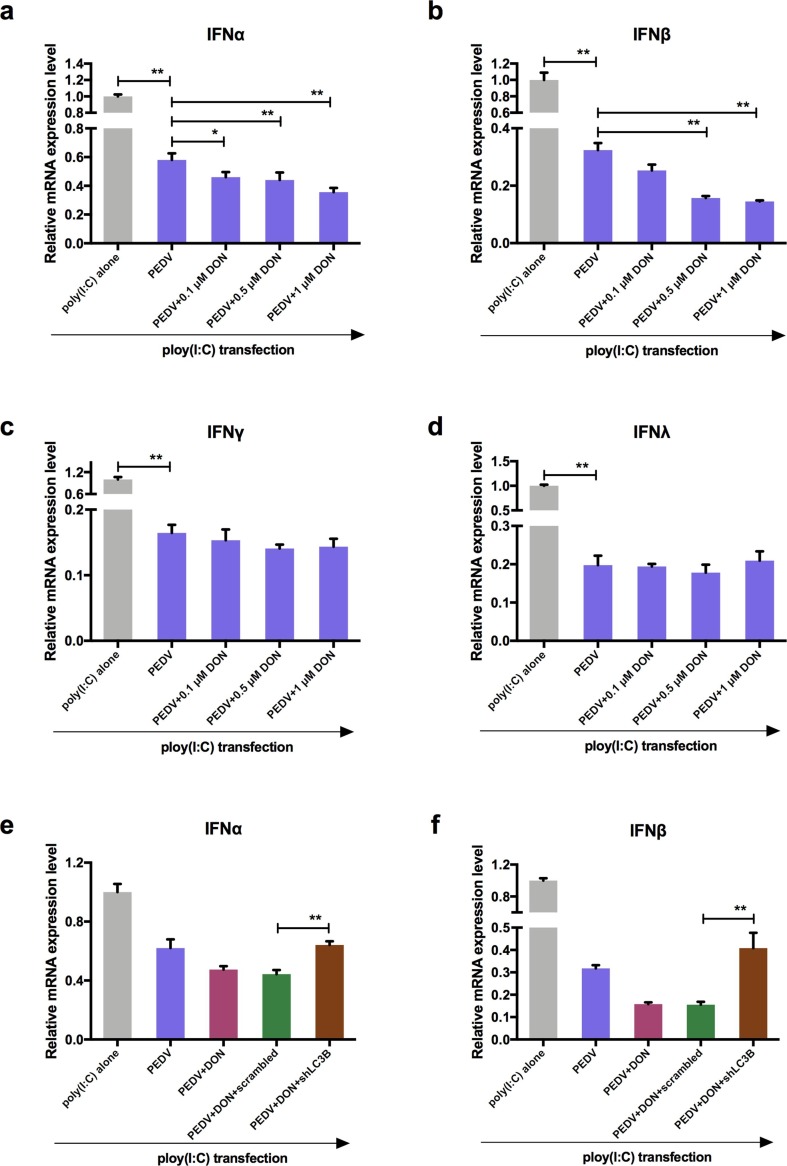
3.8. Low concentrations of DON facilitate PEDV to escape innate immune by activating autophagy in IPEC-J2 cells.
To explore how autophagy affected virus replication, the effects of DON on interferon (IFN-α, IFN-β, IFN-γ, and IFN-λ) expression in PEDV-infected IPEC-J2 cells were measured as autophagy is an important component of both innate and acquired immunity to pathogens (Sumpter and Levine 2010). Following poly (I:C) transfection, 0.1, 0.5 and 1 μM DON treatment specifically inhibited the expression of IFN-α and IFN-β compared with PEDV-infected cells (Fig. 8 a, b). However, there was no significance in the mRNA level of IFN-γ and IFN-λ (Fig. 8c, d) after DON treatment. The shLC3B was used to confirm whether autophagy played a role in downregulation of type I interfere. The results showed that DON-downregulated type I interferon could be blocked by shLC3B (Fig. 8e, f, P < 0.05), indicating that autophagy played a key role in the suppression of antiviral innate immune by DON.
Fig. 8.
Low concentrations exposure of DON could inhibit the host innate immunity by activating autophagy in PEDV-infected IPEC-J2 cells. RT-qPCR were performed to analyze the mRNA levels of IFN-α (a), IFN-β (b), IFN-γ (c) and IFN-λ (d) in cells transfected with poly (I: C). RT-qPCR were performed to analyze the mRNA levels of IFN-α (e) and IFN-β (f) in cells transfected with poly (I: C) at the absence or presence of scrambled or shLC3B. The data are expressed as mean ± SD (n = 3). *P < 0.05, **P < 0.01.
3.9. Autophagy-mediated STING pathway was required for PEDV escape innate immune in IPEC-J2 cells treated with DON.
To explore how autophagy participated in the regulation of innate immune, STING signaling was detected in PEDV-infected IPEC-J2 cells treated with DON. The immunoblotting results revealed that DON treatment inhibited the phosphorylation of STING (Fig. 9 a, b, P < 0.01), which was consistent with the changes in expression of IFN-α and IFN-β. AUD-S100 was used to confirm whether STING activation significantly regulate the viral titer in the infected cells. The results showed that the virus titer increased by DON significantly decreased when pSTING was inhibited by AUD-S100 (Fig. 9c, d and e), suggesting that STING phosphorylation participated in the regulation of innate immune by autophagy.(See Fig. 10 )
Fig. 9.
Activation of autophagy by DON suppressed the antiviral innate immune response via inhibiting STING signaling phosphorylation in PEDV-infected IPEC-J2 cells. a-d Cell treated with or without AUD-S100 lysates were subjected to immunoblotting with antibodies to p-STING and STING. e Cells treated with or without AUD-S100 were assayed for PEDV viral titers. f, g Cell lysates were subjected to immunoblotting with antibodies to LC3B, p-STING/STING or β-actin (loading control) at the absence or presence of scrambled or shLC3B. h, i RT-qPCR were performed to analyze the mRNA levels of IFN-α and IFN-β at the absence or presence of scrambled or shLC3B. j, k Cell lysates were subjected to immunoblotting with antibodies to p-STING, STING or β-actin (loading control) at the absence or presence of Rapa or SB202190.The data are expressed as mean ± SD (n = 3). *P < 0.05, **P < 0.01.
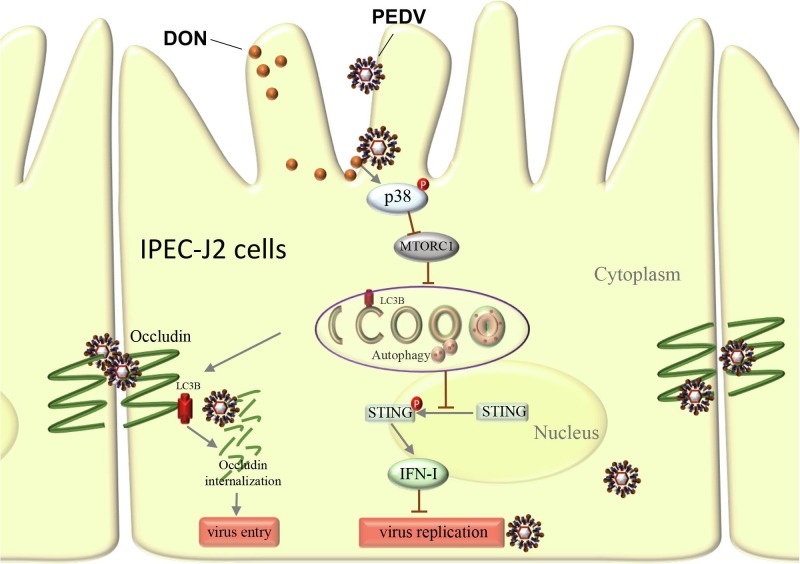
Fig. 10.
Schematic depicting role of LC3B during PEDV infection. DON exposure activates p38 signaling and triggers a complete autophagy in PEDV-infected IPEC-J2 cells. Approximately 2 hrs post-infection, LC3B induces occludin internalization to promote PEDV entry through its role as a positive regulator of autophagy. Later in infection (24 hpi.), the up-regulation of LC3B by DON contributes PEDV to escape innate immune via inhibiting the STING signaling phosphorylation, leading to production of large amounts of virus.
Given that the changes in p-STING were consistent with autophagy levels and IFN-I expressions, the scrambled and shLC3B plasmids were constructed and used to determine whether autophagy downregulated the expression of type I interfere via inhibiting STING signaling pathway. The data showed that shLC3B significantly blocked the inhibitory effects of DON on STING phosphorylation (Fig. 9f, g). In addition, the inhibition of type I interfere expression were also significantly blocked (Fig. 9h, i). In order to further verify whether the change of p-STING was affected by p38-madiated autophagy rather than directly by p38, SB202190 was used to detect the effects of p38 inhibition on p-STING levels. The results showed that p38 inhibitor, SB202190, indeed could inhibit the phosphorylation of STING, but the effects disappeared in the presence of autophagy activator, rapamycin (Rapa) (Fig. 9j, k). These data suggested that STING signaling was required to control the replication promotion of PEDV by DON and its antiviral activities were mediated by autophagy.
4. Discussion
Coronaviruses cause a variety of diseases in animals ranging from enteritis in cows and pigs and upper respiratory disease in birds to potentially lethal human respiratory infections (Fehr and Perlman 2015). Current coronavirus pathogenesis research target primarily virus life cycle and variation, little attention focus on non-infectious factors, such as environmental toxins. DON contamination in the environment is a serious problem all over the world (Escriva et al., 2015, Liao et al., 2018, Yu et al., 2017). Pigs are considered to be one of the most sensitive species and frequently exposed to DON owing to grains account for a large proportion in their feedstuffs. PED outbreak caused by PEDV has been distributed all over the world (Lee 2015). Thus, the co-existence of DON and PEDV occurs frequently in global pig farms. This study provided evidence for the first time that DON contamination in feed could promote PEDV infection in piglets and IPEC-J2 cells, documenting that autophagy activated by DON modulates the promotion. These results indicated that the future of human and animal CoV outbreaks will not only depend on how the viruses evolve, but will also depend on how we find and reduce the impact of non-infectious factors, and develop efficient prevention and treatment strategies based on their underlying mechanisms.
Intestinal mucosal, the first barrier to food contaminants, chemicals, and pathogens, plays an important role in regulating the immune response to stressors (Kagnoff, 2014, Perez-Lopez et al., 2016). Since the ability of DON to efficiently cross biological barriers, fast dividing cells such as intestinal epithelial cells will be more susceptible to the detrimental effects of DON (Danicke et al., 2010, Maresca, 2013). PEDV mainly infects pig small intestinal epithelial cells through the digestive tract, we therefore hypothesized that some substances in feed, such as DON contamination, that lead to pig intestinal epithelial cell stress might encourage the progress and spread of PED. Therefore, the intestinal porcine epithelial cell line IPEC-J2 was used as an in vitro model of swine small intestine epithelium. Experiments in vivo were performed on twenty-seven weaned piglets received the basal diet containing DON. In the present study, we found that low concentrations of DON could facilitate the entry and replication of PEDV in vivo and in vitro, suggesting that mycotoxin contamination might contribute to the infection and epidemic of coronavirus.
What drives DON to promote virus entry? PEDV crosses the porcine intestinal mucosa to cause intestinal infection, and then results in an acute viral enteric disease, which means that PEDV must gain access to the tight junctions. As noted earlier, the alteration of tight junction proteins distribution might participate in virus entry, for example, occludin internalization contributes to PEDV entry (Luo et al. 2017). In the experiments reported here, we demonstrated that DON could aggravate occludin internalization in PEDV-infected cells. When the occludin gene was silenced by siRNA, the promotion of DON to PEDV entry in IPEC-J2 cells was disappeared simultaneously, indicating that occludin internalization contributes to the DON-induced PEDV entry. Autophagy is a selective degradation process of various subcellular structures and related to cell membrane integrity and membrane proteins distribution (Mansilla Pareja et al. 2017). CRISPR‐Cas9‐mediated knockout of the LC3B blocked the internalization of occludin, indicating the vital role of autophagy in alteration of occludin protein distribution.
In addition, autophagy can serve dual roles in virus infection with either pro- or anti- viral functions depending on the virus and the stage of the viral replication cycle (Paul and Münz 2016). It is not only required for an antiviral response against some virus infection (Moy et al. 2014), but also take an active part in the viral life cycle by, eg, facilitating its entry into and release from cells (Montespan et al. 2017). We found that DON increased significantly autophagy levels in PEDV-infected piglets and IPEC-J2 cells, which are consistent with the changes in viral infection levels. CRISPR‐Cas9‐mediated knockout of the LC3B blocked the promotion of DON to PEDV viral yield, indicated that autophagy, including DON-induced autophagy, was hijacked by viruses and manipulated to their own advantage (Guo et al. 2017). JAK1 (Fleming 2016), PI3K and MAPKs (Schmeisser et al., 2014, Schmeisser et al., 2013, Xu et al., 2015) signallings can regulate the induction of intracellular autophagy. MAPK p38 has a dual role in the regulation of autophagy, both as a positive and negative regulator (Sui et al. 2014). Like E Platinum BGC-823 cells (Hu et al. 2012), DON induced autophagy via suppression of mTORC1 by decreasing phosphorylation of MAPK p38 in PEDV-infected cells.
But, how did DON-activated autophagy promote PEDV replication? The primary role of autophagy in innate immune is regulating the expression of IFN-I (Martin et al., 2018, Tian et al., 2019). Upregulation of IFN-I can inhibit viral proliferation, whereas downregulation of it contributes to virus infection (Song et al. 2018). Stimulator of interferon genes (STING) is a critical component of the cellular innate immune response to pathogenic cytoplasmic DNA (Garcia-Belmonte et al. 2019). It can be activated by the enzyme cGAMP synthase (cGAS) and then activates interferon regulatory factors (IRFs) and NF-κB, which leads to the induction of type I interferon and other immune response genes. Inhibition of STING phosphorylation by DON decreased the IFN-I expression and facilitated PEDV to escape innate immune as the decrease of IFN-I can cause continuous infection (Deng et al. 2019). Consistent with the results of others (Prabakaran et al. 2018), attenuation of the STING signaling occurred through autophagy.
In conclusion, the present study showed that DON exposure could aggravate PED in weaned piglets and promote PEDV entry and replication, suggesting that mycotoxin contamination could influence the prevalence of coronavirus. Our findings provide the novel perspective to advance the understanding in the pathogenesis of PEDV and new ideas for the prevention and control of coronavirus. And the underlying molecular mechanism may reveal new pathways for developing potential novel antiviral strategies against PEDV infection.
Author contributions
KHH and CFW designed and provided guidance for the experiments. DDL performed the experiments and wrote the manuscript. LG, QW, JRS and XXC performed the experiments and acquired the data. DDL, KHH and CFW analyzed and interpreted the data. All authors contributed to the experiments.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This study was financially supported by the National Key Research and Development Program (2017YFD0501001, 2016YFD0501203), the National Natural Science Foundation of China (31772811), the Innovative Engineering Program of Jiangsu Postgraduate Training (KYCX18_0718, Jiangsu, China) and the Priority Academic Program Development of Jiangsu Higher Education Institutions (Jiangsu, China).
Handling Editor: Yong-Guan Zhu
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.envint.2020.105949.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Supplementary Fig. 1.
IPEC-J2 cell monolayers were cultured with various concentrations of DON for 48 h, cell viability (a) and LDH release (b) were assayed as described in Materials and Methods.
References
- Alshannaq, A., Yu, J.H. (2017) Occurrence, Toxicity, and Analysis of Major Mycotoxins in Food International journal of environmental research and public health 14 doi:10.3390/ijerph14060632. [DOI] [PMC free article] [PubMed]
- Ashour, H.M., Elkhatib, W.F., Rahman, M.M., Elshabrawy, H.A. (2020) Insights into the Recent 2019 Novel Coronavirus (SARS-CoV-2) in Light of Past Human Coronavirus Outbreaks Pathogens (Basel, Switzerland) 9 doi:10.3390/pathogens9030186. [DOI] [PMC free article] [PubMed]
- Broekaert N. In vivo contribution of deoxynivalenol-3-β-d-glucoside to deoxynivalenol exposure in broiler chickens and pigs: oral bioavailability, hydrolysis and toxicokinetics. Arch. Toxicol. 2017;91:699–712. doi: 10.1007/s00204-016-1710-2. [DOI] [PubMed] [Google Scholar]
- Chen C. Exposure to aflatoxin and fumonisin in children at risk for growth impairment in rural Tanzania. Environ. Int. 2018;115:29–37. doi: 10.1016/j.envint.2018.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, X., et al. (2019) Cisplatin induces autophagy to enhance hepatitis B virus replication via activation of ROS/JNK and inhibition of the Akt/mTOR pathway Free radical biology & medicine 131:225-236 doi:10.1016/j.freeradbiomed.2018.12.008. [DOI] [PubMed]
- Chen Y. Autophagy pathway induced by a plant virus facilitates viral spread and transmission by its insect vector. PLoS Pathog. 2017;13:e1006727. doi: 10.1371/journal.ppat.1006727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng V.C., Lau S.K., Woo P.C., Yuen K.Y. Severe acute respiratory syndrome coronavirus as an agent of emerging and reemerging infection. Clin. Microbiol. Rev. 2007;20:660–694. doi: 10.1128/cmr.00023-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danicke S., Hegewald A.K., Kahlert S., Kluess J., Rothkotter H.J., Breves G., Doll S. Studies on the toxicity of deoxynivalenol (DON), sodium metabisulfite, DON-sulfonate (DONS) and de-epoxy-DON for porcine peripheral blood mononuclear cells and the Intestinal Porcine Epithelial Cell lines IPEC-1 and IPEC-J2, and on effects of DON and DONS on piglets. Food Chem. Toxicol. 2010;48:2154–2162. doi: 10.1016/j.fct.2010.05.022. [DOI] [PubMed] [Google Scholar]
- Deng X. Coronavirus endoribonuclease activity in porcine epidemic diarrhea virus suppresses type I and type III interferon responses. J. Virol. 2019 doi: 10.1128/jvi.02000-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devreese M., Antonissen G., De Backer P., Croubels S. Efficacy of active carbon towards the absorption of deoxynivalenol in pigs Toxins (Basel) 2014;6:2998–3004. doi: 10.3390/toxins6102998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escriva L., Font G., Manyes L. vivo toxicity studies of fusarium mycotoxins in the last decade: a review. Food Chem. Toxicol. 2015;78:185–206. doi: 10.1016/j.fct.2015.02.005. [DOI] [PubMed] [Google Scholar]
- Fehr A.R., Perlman S. Coronaviruses: an overview of their replication and pathogenesis Methods in molecular biology. (Clifton, NJ) 2015;1282:1–23. doi: 10.1007/978-1-4939-2438-7_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming S.B. Viral Inhibition of the IFN-Induced JAK/STAT Signalling Pathway: Development of Live Attenuated Vaccines by Mutation of Viral-Encoded IFN-Antagonists. Vaccines. 2016;4 doi: 10.3390/vaccines4030023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forni D., Cagliani R., Clerici M., Sironi M. Molecular Evolution of Human Coronavirus Genomes. Trends Microbiol. 2017;25:35–48. doi: 10.1016/j.tim.2016.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Belmonte R., Perez-Nunez D., Pittau M., Richt J.A., Revilla Y. African Swine Fever Virus Armenia/07 Virulent Strain Controls Interferon Beta Production through the cGAS-STING Pathway. J. Virol. 2019;93 doi: 10.1128/jvi.02298-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyarts T., Danicke S. Bioavailability of the Fusarium toxin deoxynivalenol (DON) from naturally contaminated wheat for the pig. Toxicol. Lett. 2006;163:171–182. doi: 10.1016/j.toxlet.2005.10.007. [DOI] [PubMed] [Google Scholar]
- Guo X. Porcine Epidemic Diarrhea Virus Induces Autophagy to Benefit Its Replication Viruses. 2017;9:53. doi: 10.3390/v9030053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris K.G. RIP3 Regulates Autophagy and Promotes Coxsackievirus B3 Infection of Intestinal Epithelial Cells. Cell Host Microbe. 2015;18:221–232. doi: 10.1016/j.chom.2015.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyndrickx E., Sioen I., Huybrechts B., Callebaut A., De Henauw S., De Saeger S. Human biomonitoring of multiple mycotoxins in the Belgian population: Results of the BIOMYCO study. Environ. Int. 2015;84:82–89. doi: 10.1016/j.envint.2015.06.011. [DOI] [PubMed] [Google Scholar]
- Hu C. E Platinum, a newly synthesized platinum compound, induces autophagy via inhibiting phosphorylation of mTOR in gastric carcinoma BGC-823 cells. Toxicol. Lett. 2012;210:78–86. doi: 10.1016/j.toxlet.2012.01.019. [DOI] [PubMed] [Google Scholar]
- Kagnoff M.F. The intestinal epithelium is an integral component of a communications network. J. Clin. Investig. 2014;124:2841–2843. doi: 10.1172/jci75225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khot W.Y. Nadkar MY (2020) The. Novel Coronavirus Outbreak - A Global Threat The Journal of the Association of Physicians of India. 2019;68:67–71. [PubMed] [Google Scholar]
- Lai M.Y., Cheng P.K., Lim W.W. Survival of severe acute respiratory syndrome coronavirus Clinical infectious diseases : an official publication of the Infectious Diseases Society of. America. 2005;41:e67–e71. doi: 10.1086/433186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C. Porcine epidemic diarrhea virus: An emerging and re-emerging epizootic swine virus Virology journal. 2015;12:193. doi: 10.1186/s12985-015-0421-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W. (2012) New variants of porcine epidemic diarrhea virus, China. Emerg. Infect. Dis. 2011;18:1350–1353. doi: 10.3201/eid1808.120002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y., Peng Z., Chen L., Nussler A.K., Liu L., Yang W. Deoxynivalenol, gut microbiota and immunotoxicity: A potential approach? Food Chem. Toxicol. 2018;112:342–354. doi: 10.1016/j.fct.2018.01.013. [DOI] [PubMed] [Google Scholar]
- Lin C.M., Saif L.J., Marthaler D., Wang Q. Evolution, antigenicity and pathogenicity of global porcine epidemic diarrhea virus strains. Virus Res. 2016;226:20–39. doi: 10.1016/j.virusres.2016.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D., Lin J., Su J., Chen X., Jiang P., Huang K. Glutamine Deficiency Promotes PCV2 Infection through Induction of Autophagy via Activation of ROS-Mediated JAK2/STAT3 Signaling Pathway. J. Agric. Food Chem. 2018;66:11757–11766. doi: 10.1021/acs.jafc.8b04704. [DOI] [PubMed] [Google Scholar]
- Liu D., Su J., Lin J., Qian G., Chen X., Song S., Huang K. Activation of AMPK-dependent SIRT-1 by astragalus polysaccharide protects against ochratoxin A-induced immune stress in vitro and in vivo. International journal of biological macromolecules. 2018;120:683–692. doi: 10.1016/j.ijbiomac.2018.08.156. [DOI] [PubMed] [Google Scholar]
- Liu Z. p38beta MAPK mediates ULK1-dependent induction of autophagy in skeletal muscle of tumor-bearing mice Cell stress. 2018;2:311–324. doi: 10.15698/cst2018.11.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo S., Terciolo C., Bracarense A., Payros D., Pinton P., Oswald I.P. In vitro and in vivo effects of a mycotoxin, deoxynivalenol, and a trace metal, cadmium, alone or in a mixture on the intestinal barrier. Environ. Int. 2019;132:105082. doi: 10.1016/j.envint.2019.105082. [DOI] [PubMed] [Google Scholar]
- Luo X., Guo L., Zhang J., Xu Y., Gu W., Feng L., Wang Y. Tight Junction Protein Occludin Is a Porcine Epidemic Diarrhea Virus Entry Factor. J. Virol. 2017;91 doi: 10.1128/JVI.00202-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansilla Pareja M.E., Bongiovanni A., Lafont F., Colombo M.I. Alterations of the Coxiella burnetii Replicative Vacuole Membrane Integrity and Interplay with the Autophagy Pathway Frontiers in cellular and infection microbiology. 2017;7:112. doi: 10.3389/fcimb.2017.00112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maresca M. From the gut to the brain: journey and pathophysiological effects of the food-associated trichothecene mycotoxin deoxynivalenol Toxins (Basel) 2013;5:784–820. doi: 10.3390/toxins5040784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin P.K. Autophagy proteins suppress protective type I interferon signalling in response to the murine gut microbiota Nature microbiology. 2018;3:1131–1141. doi: 10.1038/s41564-018-0229-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda J. Antioxidant role of autophagy in maintaining the integrity of glomerular capillaries. Autophagy. 2018;14:53–65. doi: 10.1080/15548627.2017.1391428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwan D.G. Host-pathogen interactions and subversion of autophagy Essays in biochemistry. 2017;61:687–697. doi: 10.1042/ebc20170058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montespan C. Multi-layered control of Galectin-8 mediated autophagy during adenovirus cell entry through a conserved PPxY motif in the viral capsid. PLoS Pathog. 2017;13:e1006217. doi: 10.1371/journal.ppat.1006217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moy R.H. Antiviral autophagy restrictsRift Valley fever virus infection and is conserved from flies to mammals. Immunity. 2014;40:51–65. doi: 10.1016/j.immuni.2013.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niederwerder M.C., Hesse R.A. Swine enteric coronavirus disease: A review of 4 years with porcine epidemic diarrhoea virus and porcine deltacoronavirus in the United States and Canada. Transboundary and emerging diseases. 2018;65:660–675. doi: 10.1111/tbed.12823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascari X., Ramos A.J., Marin S., Sanchis V. Mycotoxins and beer. Impact of beer production process on mycotoxin contamination. A review Food research international (Ottawa, Ont) 2018;103:121–129. doi: 10.1016/j.foodres.2017.07.038. [DOI] [PubMed] [Google Scholar]
- Pasternak J.A., Aiyer V.I.A., Hamonic G., Beaulieu A.D., Columbus D.A., Wilson H.L. Molecular and Physiological Effects on the Small Intestine of Weaner Pigs Following Feeding with Deoxynivalenol-Contaminated Feed Toxins. (Basel) 2018;10 doi: 10.3390/toxins10010040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul P, Münz C (2016) Chapter Four - Autophagy and Mammalian Viruses: Roles in Immune Response, Viral Replication, and Beyond. In: Kielian M, Maramorosch K, Mettenleiter TC (eds) Advances in Virus Research, vol 95. Academic Press, pp 149-195. doi:https://doi.org/10.1016/bs.aivir.2016.02.002. [DOI] [PubMed]
- Perez-Lopez A., Behnsen J., Nuccio S.P., Raffatellu M. Mucosal immunity to pathogenic intestinal bacteria Nature reviews Immunology. 2016;16:135–148. doi: 10.1038/nri.2015.17. [DOI] [PubMed] [Google Scholar]
- Pierron A., Alassane-Kpembi I., Oswald I.P. Impact of two mycotoxins deoxynivalenol and fumonisin on pig intestinal health Porcine Health Management. 2016;2:21. doi: 10.1186/s40813-016-0041-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pott J., Maloy K.J. Epithelial autophagy controls chronic colitis by reducing TNF-induced apoptosis. Autophagy. 2018;14:1460–1461. doi: 10.1080/15548627.2018.1450021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prabakaran T. Attenuation of cGAS-STING signaling is mediated by a p62/SQSTM1-dependent autophagy pathway activated by TBK1. The EMBO journal. 2018;37 doi: 10.15252/embj.201797858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian G. Ochratoxin A induces cytoprotective autophagy via blocking AKT/mTOR signaling pathway in PK-15cells. Food and chemical toxicology : an international journal published for the British Industrial Biological Research Association. 2018;122:120–131. doi: 10.1016/j.fct.2018.09.070. [DOI] [PubMed] [Google Scholar]
- Ramkumar A. Classical autophagy proteins LC3B and ATG4B facilitate melanosome movement on cytoskeletal tracks. Autophagy. 2017;13:1331–1347. doi: 10.1080/15548627.2017.1327509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riffelmacher T., Richter F.C., Simon A.K. Autophagy dictates metabolism and differentiation of inflammatory immune cells. Autophagy. 2018;14:199–206. doi: 10.1080/15548627.2017.1362525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmeisser H., Bekisz J., Zoon K.C. New function of type I IFN: induction of autophagy Journal of interferon & cytokine research : the official journal of the International Society for Interferon and Cytokine Research. 2014;34:71–78. doi: 10.1089/jir.2013.0128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmeisser H. Type I interferons induce autophagy in certain human cancer cell lines. Autophagy. 2013;9:683–696. doi: 10.4161/auto.23921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen G. Autophagy as a target for glucocorticoid-induced osteoporosis therapy. Cellular and molecular life sciences : CMLS. 2018;75:2683–2693. doi: 10.1007/s00018-018-2776-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song D., Park B. Porcine epidemic diarrhoea virus: a comprehensive review of molecular epidemiology, diagnosis, and vaccines Virus genes. 2012;44:167–175. doi: 10.1007/s11262-012-0713-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J. Suppression of the toll-like receptor 7-dependent type I interferon production pathway by autophagy resulting from enterovirus 71 and coxsackievirus A16 infections facilitates their replication. Arch. Virol. 2018;163:135–144. doi: 10.1007/s00705-017-3592-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sui X. p38 and JNK MAPK pathways control the balance of apoptosis and autophagy in response to chemotherapeutic agents. Cancer Lett. 2014;344:174–179. doi: 10.1016/j.canlet.2013.11.019. [DOI] [PubMed] [Google Scholar]
- Sumpter R., Jr., Levine B. Autophagy and innate immunity: triggering, targeting and tuning Seminars in cell. & developmental biology. 2010;21:699–711. doi: 10.1016/j.semcdb.2010.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y. Autophagy protects intestinal epithelial cells against deoxynivalenol toxicity by alleviating oxidative stress via IKK signaling pathway Free radical biology & medicine. 2015;89:944–951. doi: 10.1016/j.freeradbiomed.2015.09.012. [DOI] [PubMed] [Google Scholar]
- Tian Y, Wang ML, Zhao J (2019) Crosstalk between Autophagy and Type I Interferon Responses in Innate Antiviral Immunity Viruses 11 doi:10.3390/v11020132. [DOI] [PMC free article] [PubMed]
- Wang Q., Vlasova A.N., Kenney S.P., Saif L.J. Emerging and re-emerging coronaviruses in pigs Current opinion in virology. 2019;34:39–49. doi: 10.1016/j.coviro.2018.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Fang L., Liu S., Ke W., Wang D., Peng G., Xiao S. Susceptibility of porcine IPI-2I intestinal epithelial cells to infection with swine enteric coronaviruses. Vet. Microbiol. 2019;233:21–27. doi: 10.1016/j.vetmic.2019.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing Y., Liqi Z., Jian L., Qinghua Y., Qian Y. Doxycycline Induces Mitophagy and Suppresses Production of Interferon-beta in IPEC-J2 Cells Frontiers in cellular and infection microbiology. 2017;7:21. doi: 10.3389/fcimb.2017.00021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D. Modification of BECN1 by ISG15 plays a crucial role in autophagy regulation by type I IFN/interferon. Autophagy. 2015;11:617–628. doi: 10.1080/15548627.2015.1023982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D., Livingston M.J., Liu Z., Dong G., Zhang M., Chen J.K., Dong Z. Autophagy in diabetic kidney disease: regulation, pathological role and therapeutic potential. Cellular and molecular life sciences : CMLS. 2018;75:669–688. doi: 10.1007/s00018-017-2639-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q. TRIM32-TAX1BP1-dependent selective autophagic degradation of TRIF negatively regulates TLR3/4-mediated innate immune responses. PLoS Pathog. 2017;13:e1006600. doi: 10.1371/journal.ppat.1006600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Y, et al. (2020) The deadly coronaviruses: The 2003 SARS pandemic and the 2020 novel coronavirus epidemic in China Journal of autoimmunity:102434 doi:10.1016/j.jaut.2020.102434. [DOI] [PMC free article] [PubMed]
- Yin Y., Wunderink R.G. MERS, SARS and other coronaviruses as causes of pneumonia Respirology. (Carlton, Vic) 2018;23:130–137. doi: 10.1111/resp.13196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M., Chen L., Peng Z., Nussler A.K., Wu Q., Liu L., Yang W. Mechanism of deoxynivalenol effects on the reproductive system and fetus malformation: Current status and future challenges. Toxicology in vitro : an international journal published in association with BIBRA. 2017;41:150–158. doi: 10.1016/j.tiv.2017.02.011. [DOI] [PubMed] [Google Scholar]
- Zhou P. Fatal swine acute diarrhoea syndrome caused by an HKU2-related coronavirus of bat origin. Nature. 2018;556:255–258. doi: 10.1038/s41586-018-0010-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zong Q.F., Huang Y.J., Wu L.S., Wu Z.C., Wu S.L., Bao W.B. Effects of porcine epidemic diarrhea virus infection on tight junction protein gene expression and morphology of the intestinal mucosa in pigs Polish journal of veterinary sciences. 2019;22:345–353. doi: 10.24425/pjvs.2019.129226. [DOI] [PubMed] [Google Scholar]