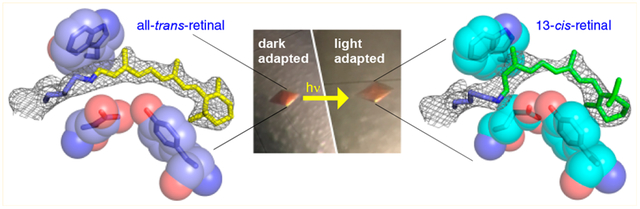
Abstract
Bacteriorhodopsin represents the simplest, and possibly most abundant, phototropic system requiring only a retinal-bound transmembrane protein to convert photons of light to an energy-generating proton gradient. The creation and interrogation of a microbial rhodopsin mimic, based on an orthogonal protein system, would illuminate the design elements required to generate new photoactive proteins with novel function. We describe a microbial rhodopsin mimic, created using a small soluble protein as a template, that specifically photoisomerizes all-trans to 13-cis retinal followed by thermal relaxation to the all-trans isomer, mimicking the bacteriorhodopsin photocycle, in a single crystal. The key element for selective isomerization is a tuned steric interaction between the chromophore and protein, similar to that seen in the microbial rhodopsins. It is further demonstrated that a single mutation converts the system to a protein photoswitch without chromophore photoisomerization or conformational change.
Graphical Abstract
INTRODUCTION
The conversion of light into chemical energy is an essential natural process. The key phototropic reaction is performed via exquisitely evolved chromophore-bound protein complexes.1 The retinal-bound opsins, known as rhodopsins, are the earliest such proteins known,2 exhibiting functional diversity that includes proton and ion pumps, ion channels,3 signaling,4 vision,5 and sensing.6 The apo-proteins, referred to as opsins, are seven transmembrane helix (“7TM”) integral membrane proteins and form an imine (Schiff base, SB) between retinal, or a closely related analog, and a buried lysine residue to generate the pigment.1c In bacteriorhodopsin, the specific photoisomerization of the resulting all-trans retinylidene chromophore induces a photocycle consisting of relatively short-lived intermediates. The specific retinal isomerization from all-trans-15-anti to 13-cis-15-anti results in a substantial change in the pKa of the chromophore’s imine (Figure 1), leading to the translocation of protons across the membrane.1b,7n This is in contrast to the chromophore’s solution behavior, where photon absorption leads to a wide distribution of retinal isomers.8 A vast literature that includes time-resolved spectroscopy, structural biology, most notably X-ray crystallography,9 and biochemical experiments10 describes the structure, spectroscopy, and lifetimes of the various bacteriorhodopsin intermediates and identifies the key protein residues essential for function.1b,9a,11 Nonetheless, fundamentally, the function of all rhodopsins depends on the specific bond isomerization of the bound retinylidene chromophore. Therefore, the creation of an orthogonal, simplified protein system, amenable to routine biophysical measurements, especially atomic-resolution structural interrogation, that recapitulates a specific bond isomerization similar to that seen in bacteriorhodopsin provides a unique opportunity for future investigations.
Figure 1.
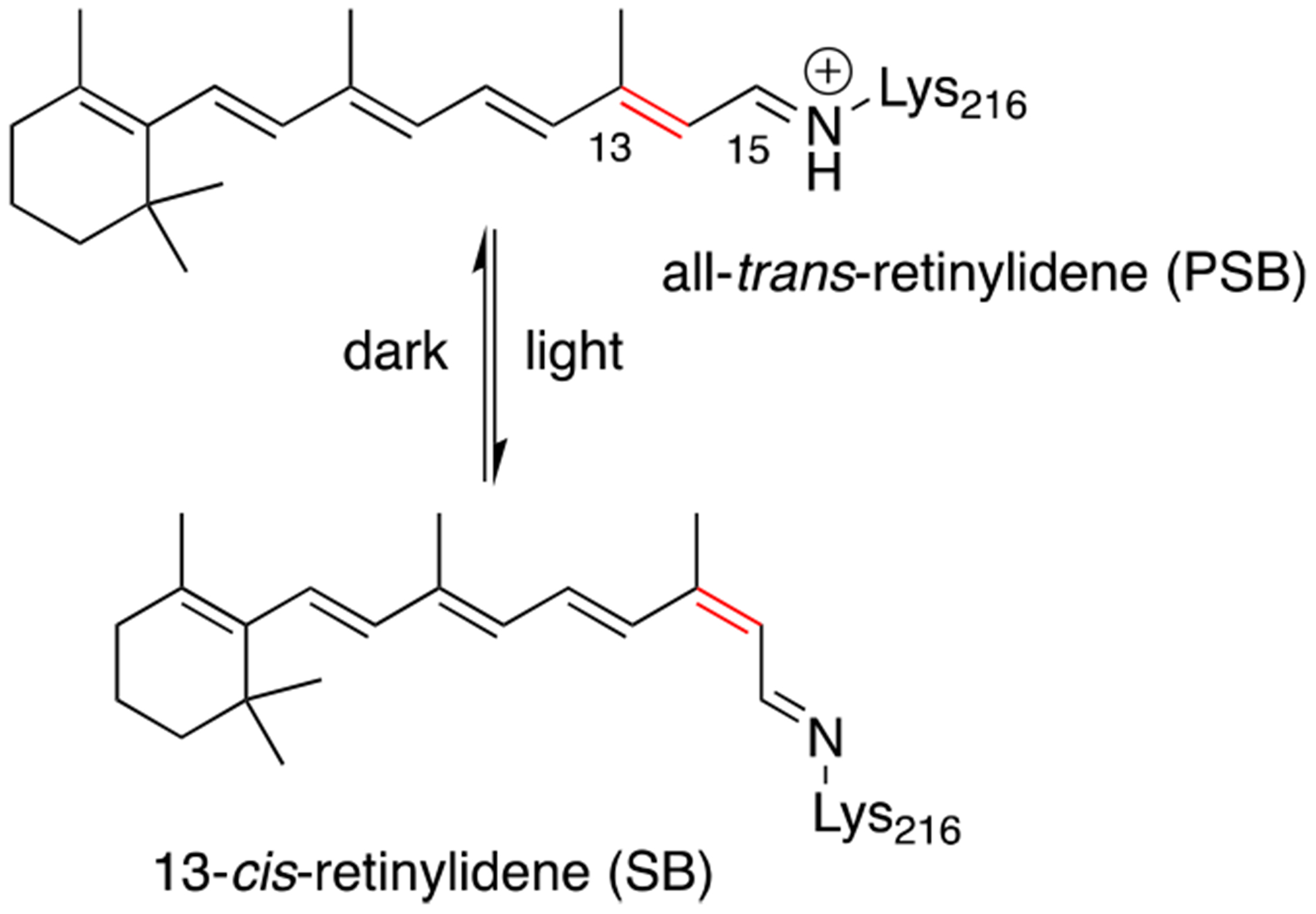
All-trans retinylidene chromophore of bacteriorhodopsin photoisomerized to the 13-cis isomer followed by thermal regeneration of the all-trans isomer.
We have had a long-standing interest in using protein mimics to understand protein–chromophore interactions that lead to wavelength regulation in rhodopsins.12 We now turn our attention to the factors that govern the isomerization events by investigating the minimal requirements of specific isomerization in a model system that mimics the hallmarks of the natural system. Herein, we report the re-engineering of human cellular retinoic acid binding protein II (CRABPII), a small, soluble β-sandwich cytosolic protein from the intracellular lipid binding protein family, to undergo specific photoisomerization of all-trans to 13-cis retinylidene and thermal isomerization back to the all-trans isomer in a single crystal, representing a complete photocycle of the microbial rhodopsins. Serendipitously, with a specific mutant, we observe a case of protein conformational change in response to photoexcitation at a wavelength in which protein amino acids do not absorb. This is in lieu of the expected chromophore isomerization, signifying the response of the protein to the chromophore excited state. This study establishes a new model system for studying the isomerization pathway of retinoids in real time, with well-characterized starting and ending points of isomerization.
RESULTS AND DISCUSSION
Choosing the Initial Protein Template
To initiate our investigation, we required a platform that would support photoinduced isomerization of the bound chromophore. As described in our previous studies, a number of CRABPII variants exhibited photoinduced changes in the protonation state of a retinylidene-bound protein, which ultimately dictated the pigmentation of the protein–chromophore complex.12e X-ray crystallography and UV–vis spectroscopy showed that imine bond isomerization from 15-syn to 15-anti is responsible for this change. As depicted in Figure 2a, CRABPII hexamutant M1 (R111K:Y134F:T54V:R132Q:P39Y:R59Y) binds retinal to initially form the 15-syn isomer. In this arrangement, the imine nitrogen atom is placed in a polar environment that supports a high pKa regime and thus yields a protonated Schiff base (PSB), leading to the observed colored pigment. Time-dependent 15-syn to 15-anti imine isomerization yields the thermodynamic product that places the nitrogen atom in a low pKa environment, leading to the colorless neutral Schiff base (SB). Additionally, green and UV light irradiation interconverts between the PSB and SB, respectively, in solution and in crystalline states.12e
Figure 2.
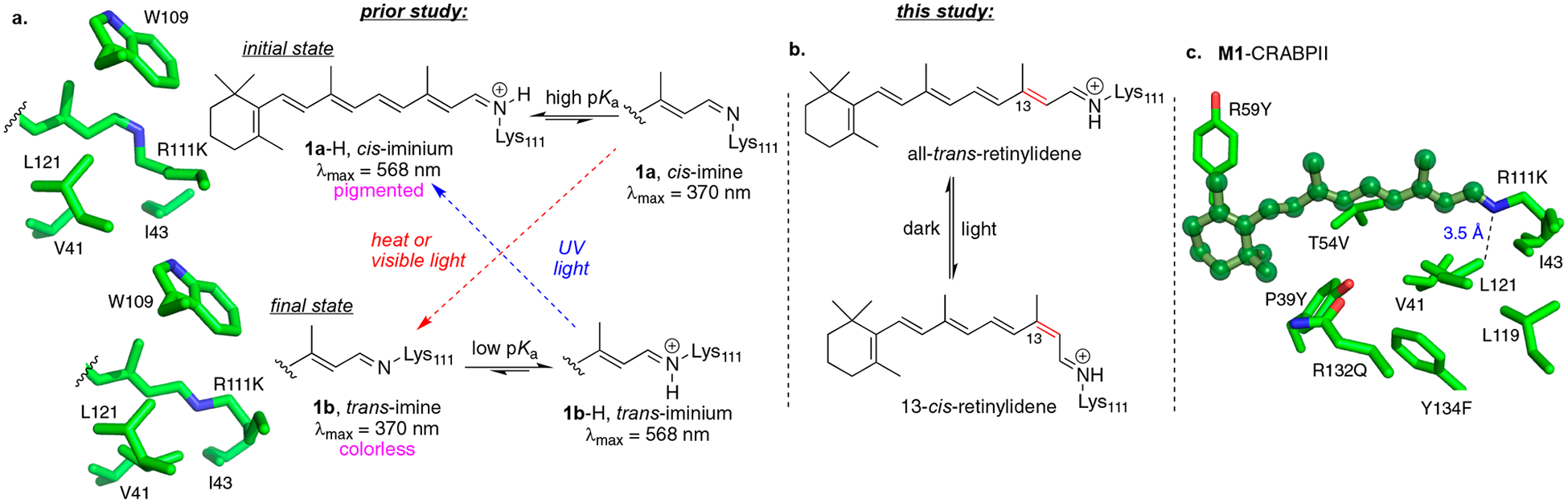
(a) Photocycle of CRABPII hexamutant M1 (R111K:Y134F:T54V:R132Q:P39Y:R59Y) bound to retinal. The photoinduced isomerization of the imine functionality leads to changes in its pKa and consequently its protonation state. Hydrophilic and hydrophobic environments of the imine nitrogen atom for the cis and trans geometry, respectively, are highlighted in the figure obtained from the crystal structure of each form (PDB IDs 4YFP and 4YFQ). (b) The present study aims to induce photoisomerization to the C13 isomer while maintaining the protonated iminium. (c) All-trans-retinal-bound M1-CRABPII, with key residues highlighted.
With the ability to photochemically and specifically isomerize the imine bond, the goal was set to design a protein capable of isomerizing the C13 double bond, reminiscent of the action of microbial rhodopsin (Figure 2b). Although caution is warranted with regard to mechanistic interpretations of a mimic that might not operate in the same manner as bacteriorhodopsin, a well-characterized and easy-to-manipulate mimic, derived from a structurally orthogonal template, has the potential to illustrate fundamental principles of retinal photoisomerization.
Using CRABPII mutant M1 as the initial template, it was first necessary to prevent the deprotonation of the iminium due to exclusive imine bond isomerization, as observed previously. Otherwise, imine isomerization to a low pKa state would result in a nonabsorbing, unproductive thermodynamic sink, obviating any other isomerization pathway because the absorption of the chromophore would shift away (hypsochromic) from the wavelength of irradiation.12a,e We envisioned that the placement of an acidic residue could stabilize the protonated species in any imine isomeric state. As illustrated in Figure 2c, the proximity of Leu121 to the imine nitrogen atom (3.5 Å) led to the introduction of the L121E mutation in M1, which successfully abolished the imine isomerization pathway. This was substantiated via irradiation of retinal-bound M1-L121E with either visible or UV light, which no longer exhibited the photochemical change seen in M1 (Figure 3a). Instead, a decrease in the absorbance of both the imine (387 nm) and the iminium (461 nm) bands, independent of the wavelength of irradiated light, was observed, presumably due to changes in the structure of the chromophore (vida infra).
Figure 3.
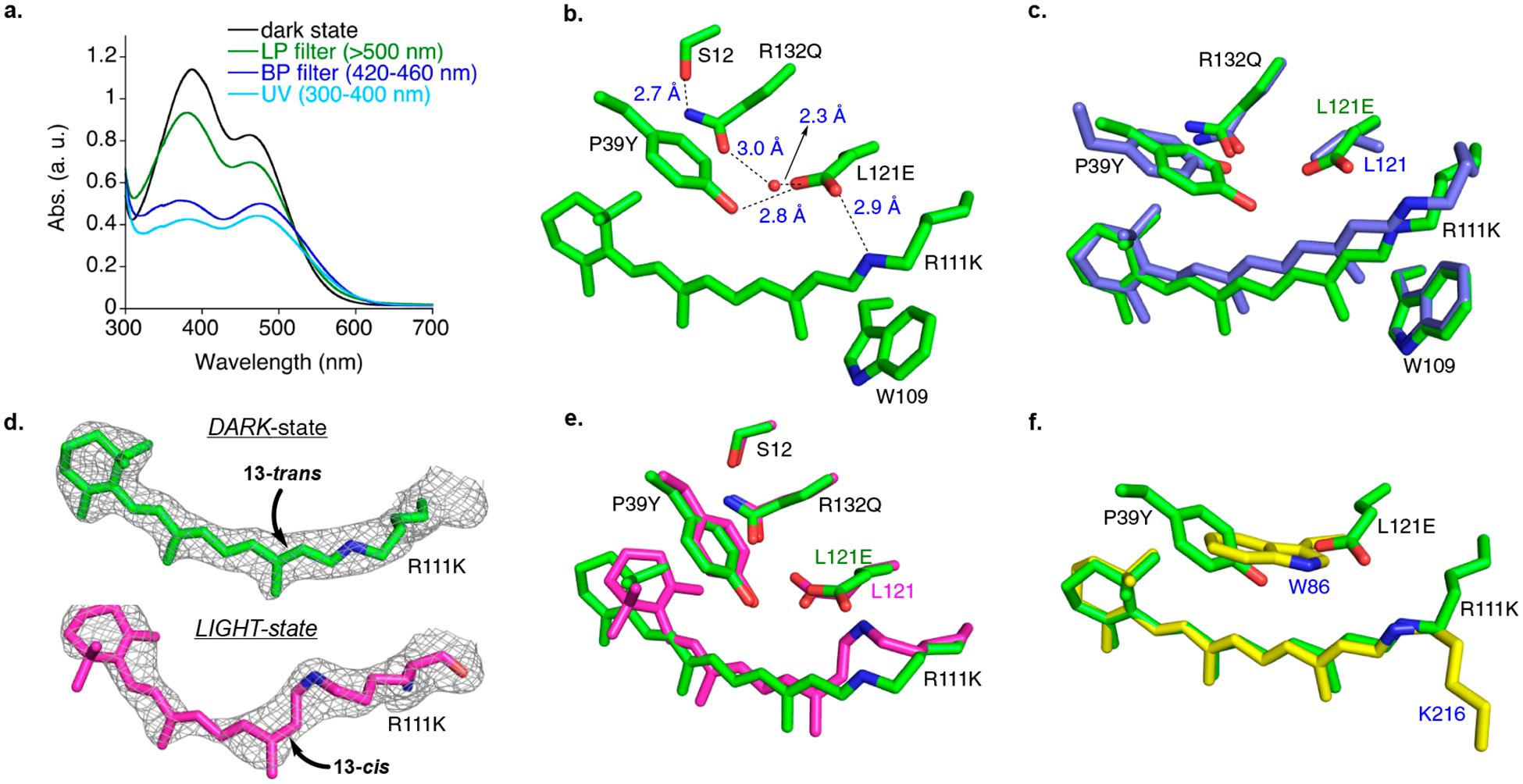
(a) UV–vis absorption spectrum of all-trans-retinal bound M1-L121E in the dark state (black spectrum) and after green light irradiation (long-pass filter, >500 nm, green spectrum); blue light irradiation (440 ± 20 nm, blue spectrum), and UV irradiation (UV band-pass filter, 300–400 nm, cyan spectrum). (b) Hydrogen bonding network between the imine hydrogen of R111K and L121E, P39Y, H2O-303, R132Q, and Ser12. (c) Overlay of M1 (blue carbons) with all-trans-retinal bound M1-L121E (green carbons). (d) Electron density (contoured at 1σ) of all-trans-retinylidene in M1-L121E in the dark state (green carbons) vs 13-cis-15-syn-retinal imine (magenta carbons) generated after 5 min of laser irradiation at 399 nm. (e) Overlay of all-trans-retinylidine-bound M1-L121E in the dark (green) with the 13-cis-15-syn imine (magenta) generated after 5 min of laser irradiation at 399 nm, showing the movement of Lys111 and Glu121 and the rotation of the β-ionone ring in 13-cis upon isomerization. (f) Overlay of all-trans-retinal bound M1-L121E imine (green) and all-trans-retinal bound bacteriorhodopsin (yellow carbons, PDB code 1C3W) illustrating how the P39YL121E interaction mimics Trp86 in bacteriorhodopsin relative to retinal. All heteroatoms are colored by type, with N being blue and O being red.
The structure of all-trans retinal-bound M1-L121E (Figure 3b) revealed a trans-iminium engaged as a salt bridge with the newly installed Glu121. The overlay of M1 and M1-L121E also reveals a serendipitous structural rearrangement (Figure 3c). Tyr39, which hydrogen bonds with Gln132 in M1, adopts a new conformation, presumably as a result of a stronger hydrogen bond with Glu121. In this conformation, Tyr39 sterically impinges on the bound retinal, causing a translation of the polyene in the vertical axis, sandwiching the chromophore between Trp109, located on the opposite side of the chromophore, and Tyr39 while tilting the chromophore relative to the trajectory in M1. Figure 3c shows that the structural changes affect the polyene portion of the chromophore, leaving the β-ionone ring in approximately the same position as seen in M1. Nonetheless, the interaction between Glu121 and the iminium maintains the protonation state. As expected, the salt bridge between Glu121 and the nitrogen atom of the trans-iminium gives a substantially blue-shifted spectrum (461 nm, compared to 568 nm for M1)12e by localizing the positive charge on the polyene (Figure S1).12g The L121E mutation plays a dual role in keeping the imine protonated during photoisomerization (pKa determination in Figure S2) and provides a steric interaction that orders the chromophore in the binding pocket by sandwiching the chromophore between Tyr39 and Trp109. (The mutation of L121 to aromatic residues and its consequences are described in Figures S3–S5.)
Solution Photoirradiation
Intrigued by the significant drop in the absorption of both SB and PSB bands upon irradiation of the M1-L121E/all-trans-retinal complex with either UV or green light (Figure 3a), the chromophore was extracted from photoirradiated samples and analyzed by HPLC (Figure S6). Three peaks were apparent in the HPLC traces for all reactions: photoirradiation with a UV band-pass filter (300–400 nm) and a blue band-pass filter (440 nm ±20 nm); green light irradiation with a band-pass filter (>500 nm) and a 399 nm laser. In the extracts of all four reactions, the peak that coelutes with authentic all-trans retinal was major, while the second peak coeluted with authentic 13-cis retinal and the third peak was assigned as 9,13-di-cis-retinal, based on the 1H NMR aldehyde chemical shift comparison with the reported isomer. (See the SI and Figures S6–S8 for details of each extract and the ratio of identified isomers.) The extinction coefficient of cis isomers is approximately 80% of that of the all-trans isomer, thus the drop in absorption noted above is partially attributed to the production of the 13-cis and 9,13-di-cis-isomers, along with some potential photobleaching of the chromophore. Nonetheless, the production of the 13-cis isomer was exciting, although HPLC analysis of the extracted sample revealed the all-trans-retinal as the major constituent. The presence of the all-trans-retinal could be due to either a low quantum yield of photoisomerization or thermal relaxation back to the thermodynamically more stable isomer during sample manipulation and preparation, resembling the thermal relaxation of the 13-cis-isomer back to the all-trans isomer in bacteriorhodopsin. To further clarify the nature of the reaction, we next investigated the isomerization in the crystalline state.
Crystal Photoirradiation.
To explore the isomerization pathway in the crystalline form, the irradiation of all-trans retinal bound M1-L121E crystals with a 399 nm laser was performed. (See Figures S9 and S10 in the SI for details.) The crystals were frozen in liquid nitrogen after 30 s and 5 min of irradiation, and their structures were determined. (See Figure S11 in the SI and Figure 3d for details.) In stark contrast to the results from HPLC analysis of the extracted chromophores, the electron density of the bound chromophore, after 5 min of laser irradiation, shows no evidence of all-trans retinal and instead is consistent with its quantitative photoconversion to the 13-cis-15-syn isomer (Figure 3d; see Figure S11 for the crystal structure of the 30 s irradiated sample). The observed photoisomerization of the bound retinylidene from 13-trans-15-anti to 13-cis-15-syn is similar to the “bicycle pedal mechanism” as first proposed by Warshel13 and also suggested for the dark adaptation of bacteriorhodopsin.11c,14 In his proposal, two adjacent double bonds undergo isomerization, as seen with the photoisomerization of M1-L121E. Although it is not possible to rule out different mechanisms with end-state observations from crystal structures, the final geometry of the photoisomerized M1-L121E is in contrast to the “hula-twist” proposal that predicts the isomerization of an olefin and its adjacent single bond.15 An alternate explanation for the formation of the 15-syn isomer could be the fast thermal adaptation of 15-anti to 15-syn, which is not detectable in the crystal within the time resolution of our method. As shown in Figure 3e, the overlay of prelaser and postlaser irradiation indicates substantial β-ionone ring rotation and polyene movement. Furthermore, the conformational changes in Glu121, which follows the movement of the imine nitrogen atom, and in Lys111 are apparent. Interestingly, the position of Tyr39 remains fixed before and after irradiation. Tyr39 may act as a “steric pillar” that dictates the path of isomerization in a specific direction (Figure 3e). The role of Tyr39 mimics that of Trp86 in bacteriorhodopsin, which sterically packs against the retinal and assists in directing the photoisomerization of the C13–C14 double bond.16 As shown in the overlay of M1-L121E and bacteriorhodopsin17 (Figure 3f), Glu121 and Tyr39 occupy a similar space relative to the retinal, similar to Trp86 in bacteriorhodopsin. Once again, the fortuitous yet critical role of the L121E mutation is apparent because it aids in orienting Tyr39 in a conformation that resembles Trp86 in bacteriorhodopsin, leading to specific 13-cis photoisomerization. To verify the importance of the steric buttressing effect of the P39Y mutation, it was mutated to Gln, resulting in M1-L121E:P39Q. (The equivalent P39F mutation gave uninterpretable results.) As anticipated, there is no evidence of isomerization to the 13-cis isomer either in solution or in the crystal (Figures S12 and S13), thus confirming the critical function of the tyrosine residue. These observations, however, cannot disregard electronic effects as a result of mutations such as P39Y, where the polarity of its immediate environment has changed. It is noteworthy that the isomerization in the crystalline state of M1-L121E/all-trans-retinal complex indicates complete conversion to the 13-cis isomer, suggesting the mixture obtained in solution to be the result of postirradiation thermal relaxation. Nonetheless, we cannot rule out the possibility that the difference in behavior of crystal vs solution-state isomerization could be due to the solid-state environment where the lattice interaction restricts the conformational flexibility of molecules.
Thermal Reversion to Original State
With photoisomerization to 13-cis confirmed, its thermal stability was next investigated. To this end, photochemically generated 13-cis-bound M1-L121E crystals were left in the dark at room temperature for 10 and 25 min. We anticipated the thermal reversion of the 13-cis isomer to the apparently more thermodynamically stable all-trans isomer in M1-L121E. This would complete the photocycle that is essentially identical to that of the microbial rhodopsins, though on a significantly slower time scale. Structures of all of the thermally equilibrated crystals (after 10 and 25 min of equilibration) revealed electron density consistent with all-trans-retinal bound M1-L121E, confirming the completion of a microbial rhodopsin photocycle (Figures 4a and S14–S16). The main characteristics of the structure, most notably the rotation and trajectory of the β-ionone ring, are clearly observed (Figure S16). This is consistent with the fact that the extraction of retinal-bound M1-L121E, after irradiation with visible light in solution, shows both all-trans (Figures S6 and S8) and 13-cis isomers, with the latter the result of irradiation and the former the result of thermal relaxation to the more stable all-trans form. Thus, a complete photocycle, analogous to a microbial rhodopsin, has been generated in an orthogonal protein and confirmed at atomic resolution.
Figure 4.
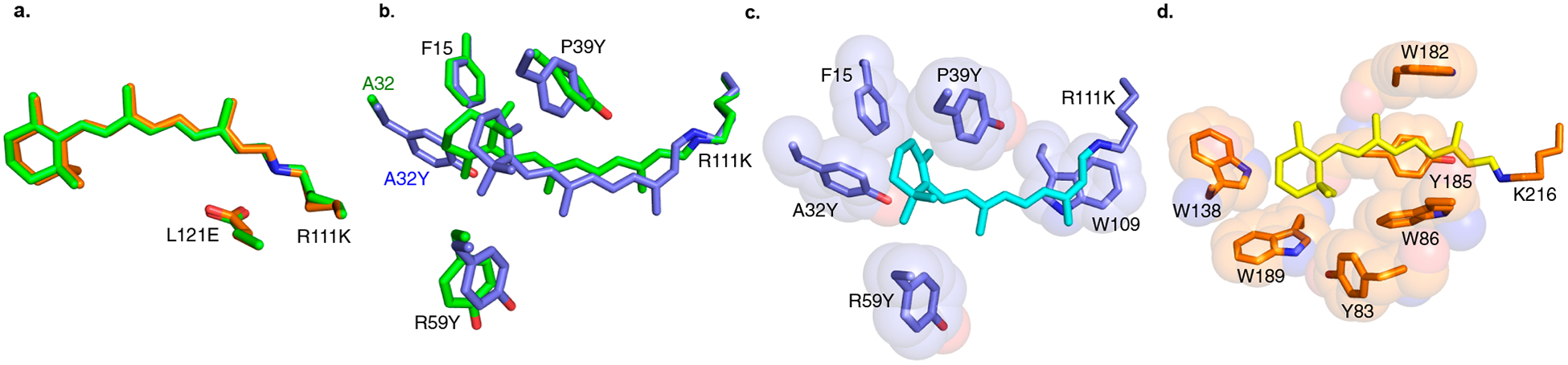
(a) Overlay of the all-trans-retinal/M1-L121E complex in the dark (green) and after laser irradiation followed by incubation in the dark (25 min, orange) highlighting the structural similarity of the chromophore before and after a complete photocycle. (b) Overlay of the all-trans-retinal/M1-L121E complex in the dark (green) with the 13-cis-retinal/M1-L121E:A32Y complex. (c) Aromatic residues located in the binding pocket of 13-cis-bound M1-L121E:A32Y. (d) Bacteriorhodopsin (PDB code 1C3W) binding pocket where the chromophore is sandwiched with aromatic residues.
Mechanistic Insights and Predictions
Our interpretation of the results described above naturally leads to two predictions regarding the mechanism. First, stabilizing or restricting the ionone ring conformation can stabilize distinct conformational states (either all-trans or 13-cis) of retinal. Second, both Glu121 and Tyr39 should be essential in giving a 13-cis isomerizing system. On the basis of the first prediction, we hypothesized that the stabilization of the rotated ionone ring conformation would also stabilize the binding of the 13-cis isomer in the binding pocket (Figure 3e). To this end, Ala32, located at the mouth of the binding pocket, was mutated to aromatic amino acids to persuade the rotation of the ring as a result of steric compression afforded by the larger amino acids. The incubation of 13-cis-retinal with M1-L121E:A32Y leads to an absorption indicative of an iminium. (See the UV–vis trace in Figure S17b.) Gratifyingly, the crystal structure of the latter complex (with 1.58 Å resolution) clearly shows the 13-cis isomer bound exclusively in the binding cavity (Figure 4b,c). The overlay of this structure with M1-L121E postirradiation (13-cis-isomer) shows both the β-ionone and polyene moieties to be quite similar (Figure S17), while the overlay of this structure with all-trans retinal-bound structures shows clear and obvious structural variations (Figure 4b). An analysis of the high-resolution structure indicates that the 13-cis isomer is stabilized by the packing of several aromatic residues, including Phe15, Tyr39, Tyr32, Tyr59, and Trp109 inside the binding pocket, mostly through “aromatic sandwiches” (Figure 4c). Interestingly, this is reminiscent of aromatic/chromophore interactions observed in a number of microbial rhodopsins,1b,9d,17 where the chromophore finds itself well-packed in a sphere of aromatic residues (Figure 4d). These results demonstrate that by increasing the pKa and packing of P39Y closer to those of the polyene (using the L121E mutation) and altering the β-ionone ring packing (via the A32Y mutation), a 13-cis “unfriendly” binding pocket (M1) is converted to a 13-cis-specific protein, nestling a well-ordered 13-cis retinylidene inside the binding pocket.
To test the second prediction, mutations were made to both Tyr39 and Glu121 residues. As described above, the P39Q mutation to give M1-L121E:P39Q resulted in a protein complex incapable of isomerizing the C13 double bond. This demonstrates the importance of the steric interaction between Tyr39 and the chromophore in directing isomerization to the 13 bond. To verify the role of the L121E mutation in the iminium stabilizing species, we examined the photochemical characteristics of the structurally conservative M1-L121Q mutant. As shown in Figure 5a, the irradiation of M1-L121Q in solution leads to behavior similar to that of mutants that undergo imine isomerization with two apparent pKa’s for each isomer. UV light irradiation (<400 nm) results in a loss of intensity in the 374 nm SB absorption, concomitant with a large increase in the PSB absorption at 522 nm. Conversely, irradiation with visible light (>500 nm) reduces the PSB absorption at 522 nm along with an increase in the 374 nm SB absorption (Figures S18 and S19).12e At first, this result was not unexpected because the loss of the salt bridge afforded through the interaction of the iminium with Glu121 could reestablish the imine cis/trans isomerization observed before, leading to two states with distinct pKa’s (Figure S20). Nonetheless, the crystal structure analysis reveals a surprising and unanticipated mechanism for the observed pKa change.
Figure 5.
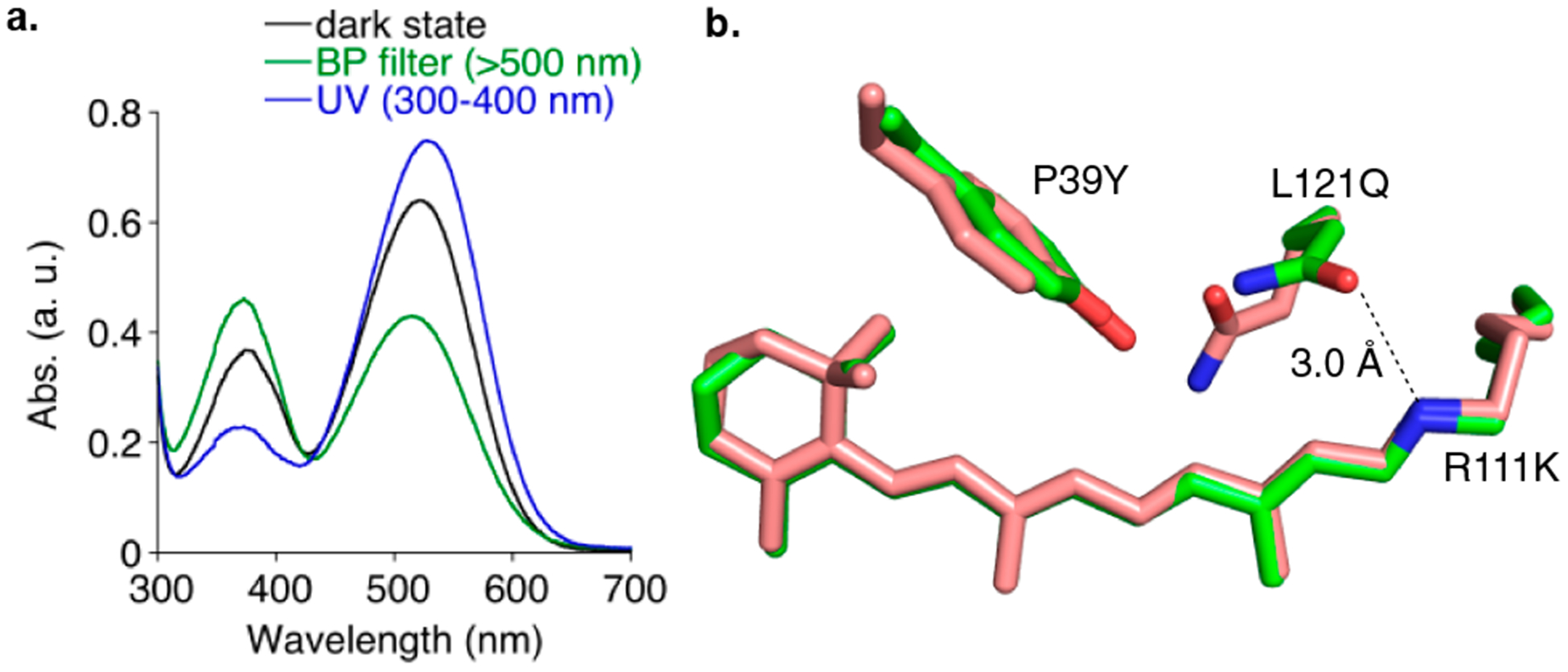
(a) UV–vis spectra of M1-L121Q before and after irradiation with green light, showing a high/low pKa system where green light irradiation leads to decreased PSB and increased SB absorption. (b) Overlay of the all-trans-retinal-bound M1-L121Q imine before (green carbons) and after (salmon carbons) laser irradiation at 532 nm, clearly showing the Gln121 movement. The all-trans retinal bound M1-L121Q structure features a direct hydrogen bond between the nitrogen atom of the imine and Gln121 (3.0 Å). Laser irradiation (532 nm) of crystals does not result in the isomerization of any bonds; however, L121Q swings away from the imine (now ~4.7 Å), leading to the low pKa form of the imine and SB formation.
Photoirradiation Leads to Side-Chain Motion
Not unexpectedly, all-trans retinal-bound M1-L121Q is structurally almost identical to all-trans retinal-bound M1-L121E, with Gln121 making a direct hydrogen bond with the iminium nitrogen atom (Figure 5b). Surprisingly, the irradiation of the all-trans bound crystal with the 399 nm laser showed no evidence of chromophore isomerization or conformational change. Instead, Gln121 adopts two conformations: the original conformation seen in the dark (55% occupancy) and a new position that is rotated away from the iminium (45% occupancy), resulting in a naked imine nitrogen atom with no other interacting residues (Figure S21b). Interestingly, irradiation with green light (532 nm laser irradiation) leads to the complete movement of Gln121 away from the imine nitrogen atom (Figures 5b and S21c). An imine under these circumstances would be expected to have a substantially suppressed pKa relative to the dark state, consistent with the solution result that shows an increase in the SB form upon light irradiation (Figure 5a).
In contrast to the expected isomerization of the chromophore’s double bond, leading to a change in the environment of the imine and its pKa, irradiation of the M1-L121Q retinal complex leads to a change in the conformation of an interacting amino acid side chain, thus effectively altering the environment. The mechanism of this process is unclear, but one possibility is that the photoinduced motion of the chromophore induces the conformational change in the side chain. It is important to point out that the motion is only for the side chain, not the main chain. This is reminiscent, albeit on a much longer time scale, of the subpicosecond X-ray laser time-resolved study of bacteriorhodopsin, which showed that the critical water molecule moves away from the chromophore in response to the change in the polarity of the excited state, before the chromophore itself displays substantial motion.11c In both cases, it is the surrounding atoms that move in response to the photophysics of the chromophore rather than the motion of the chromophore driving the conformational change in the rest of the protein system. Such subtle changes in the structure of the surroundings upon irradiation would be difficult to identify in the absence of high-resolution structural interrogation of the irradiated systems. This may explain the complex hydrogen bonding network holding the two key aspartates in place in bacteriorhodopsin. Perhaps this prevents the interacting residues from moving during photoexcitation, resulting in a low pKa form, which would sabotage the bacteriorhodopsin photocycle.11b,18 Another example of a similar phenomenon, where the exclusive motion of the protein is the result of chromophore absorption, is seen in the AppA BLUF domain using time-resolved FTIR.19
CONCLUSIONS
An artificially designed protein that recapitulates the photocycle of microbial rhodopsins, specifically and quantitatively photoisomerizing all-trans-retinal to 13-cis-retinal in a well-diffracting crystal, has been created. Because of its slow rate of isomerization, our engineered rhodopsin mimic can be exploited using relatively simple time-resolved spectroscopic and crystallographic methods. It also introduces an ideal system for time-resolved crystallography. Using this system, a mechanism for protein photoswitching that involves no net conformational change of the chromophore was identified at atomic resolution. This observation, which would be difficult to observe using other techniques, opens the door to a novel strategy for the design and study of photoactive and photoswitchable protein systems.
Supplementary Material
ACKNOWLEDGMENTS
All crystallographic data were collected at the Advanced Photon Source, an Office of Science User Facility operated for the U.S. Department of Energy (DOE) Office of Science by Argonne National Laboratory, supported by the U.S. DOE under contract no. DE-AC02-06CH11357. Use of the LS-CAT Sector 21 was supported by the Michigan Economic Development Corporation of the Michigan Technology Tri-Corridor (grant 085P1000817) and the MSU office of the Vice President for Research.
Funding
Generous support was provided by the NIH (GM101353).
ABBREVIATIONS
- 7TM
seven transmembrane helix
- SB
Schiff base
- CRABPII
human cellular retinoic acid binding protein II
- PSB
protonated Schiff base
- BLUF
blue light using FAD
- HPLC
high-pressure liquid chromatography
- M1
R111K:Y134F:T54V:R132Q:-P39Y:R59Y
Footnotes
The authors declare no competing financial interest.
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/jacs.8b12493.
Experimental data including protein expression and purification, UV–vis spectra, HPLC extraction, crystallization conditions, crystal irradiation, and X-ray data collection and refinement statics; crystallographic files such as atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes: 6MOP, 6MQZ, 6MQY, 6MQW, 6MQX, 6MPK, 6MOQ, 6MOR, 6MOV, 6MQI, 6MQJ, 6MOX, and 6MR0) (PDF)
REFERENCES
- (1).(a) Schotte F; Cho HS; Kaila VRI; Kamikubo H; Dashdorj N; Henry ER; Graber TJ; Henning R; Wulff M; Hummer G; Kataoka M; Anfinrud PA Watching a signaling protein function in real time via 100-ps time-resolved Laue crystallography. Proc. Natl. Acad. Sci. U. S. A 2012, 109, 19256. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Ernst OP; Lodowski DT; Elstner M; Hegemann P; Brown LS; Kandori H Microbial and Animal Rhodopsins: Structures, Functions, and Molecular Mechanisms. Chem. Rev 2014, 114, 126. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Sudo Y; Ihara K; Kobayashi S; Suzuki D; Irieda H; Kikukawa T; Kandori H; Homma M A microbial rhodopsin with a unique retinal composition shows both sensory rhodopsin II and bacteri<EDorhodopsin-like properties. J. Biol. Chem 2011, 286, 5967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Béjà O; Aravind L; Koonin EV; Suzuki MT; Hadd A; Nguyen LP; Jovanovich SB; Gates CM; Feldman RA; Spudich JL; Spudich EN; DeLong EF Bacterial Rhodopsin: Evidence for a New Type of Phototrophy in the Sea. Science 2000, 289, 1902. [DOI] [PubMed] [Google Scholar]
- (3).Kandori H. Ion-pumping microbial rhodopsins. Front. Mol. Biosci 2015, 2 DOI: 10.3389/fmolb.2015.00052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Gordeliy VI; Labahn J; Moukhametzianov R; Efremov R; Granzin J; Schlesinger R; Buldt G; Savopol T; Scheidig AJ; Klare JP; Engelhard M Molecular basis of transmembrane signalling by sensory rhodopsin II-transducer complex. Nature 2002, 419, 484. [DOI] [PubMed] [Google Scholar]
- (5).Govorunova EG; Koppel LA The Road to Optogenetics: Microbial Rhodopsins. Biochemistry 2016, 81, 928. [DOI] [PubMed] [Google Scholar]
- (6).Royant A; Nollert P; Edman K; Neutze R; Landau EM; Pebay-Peyroula E; Navarro J X-ray structure of sensory rhodopsin II at 2.1-Å resolution. Proc. Natl. Acad. Sci. U. S. A 2001, 98, 10131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Kandori H In Chemical Science of π-Electron Systems; Akasaka T, Osuka A, Fukuzumi S, Kandori H, Aso Y, Eds.; Springer: Tokyo, 2015; p 695. [Google Scholar]
- (8).Liu RSH; Asato AE Methods in Enzymology; Academic Press: 1982; Vol. 88, p 506. [Google Scholar]
- (9).(a) Hirai T; Subramaniam S Protein conformational changes in the bacteriorhodopsin photocycle: comparison of findings from electron and X-ray crystallographic analyses. PLoS One 2009, 4, No. e5769. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Nango E; Royant A; Kubo M; Nakane T; Wickstrand C; Kimura T; Tanaka T; Tono K; Song C; Tanaka R; Arima T; Yamashita A; Kobayashi J; Hosaka T; Mizohata E; Nogly P; Sugahara M; Nam D; Nomura T; Shimamura T; Im D; Fujiwara T; Yamanaka Y; Jeon B; Nishizawa T; Oda K; Fukuda M; Andersson R; Båth P; Dods R; Davidsson J; Matsuoka S; Kawatake S; Murata M; Nureki O; Owada S; Kameshima T; Hatsui T; Joti Y; Schertler G; Yabashi M; Bondar A-N; Standfuss J; Neutze R; Iwata S A three-dimensional movie of structural changes in bacteriorhodopsin. Science 2016, 354, 1552. [DOI] [PubMed] [Google Scholar]; (c) Kato HE; Zhang F; Yizhar O; Ramakrishnan C; Nishizawa T; Hirata K; Ito J; Aita Y; Tsukazaki T; Hayashi S; Hegemann P; Maturana AD; Ishitani R; Deisseroth K; Nureki O Crystal structure of the channelrhodopsin light-gated cation channel. Nature 2012, 482, 369. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Vogeley L; Sineshchekov OA; Trivedi VD; Sasaki J; Spudich JL; Luecke H Anabaena Sensory Rhodopsin: A Photochromic Color Sensor at 2. Science 2004, 306, 1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).(a) Maresca JA; Keffer JL; Miller KJ Biochemical Analysis of Microbial Rhodopsins Current Protocols in Microbiology; John Wiley & Sons: Hoboken, NJ, 2016; Vol. 41, 1F.4.1.. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Shibata M; Kandori H FTIR studies of internal water molecules in the Schiff base region of bacteriorhodopsin. Biochemistry 2005, 44, 7406. [DOI] [PubMed] [Google Scholar]; (c) Govorunova EG; Sineshchekov OA; Janz R; Liu X; Spudich JL NEURO-SCIENCE. Natural light-gated anion channels: A family of microbial rhodopsins for advanced optogenetics. Science 2015, 349, 647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).(a) Hatanaka M; Kashima R; Kandori H; Friedman N; Sheves M; Needleman R; Lanyi JK; Maeda A Trp86 → Phe Replacement in Bacteriorhodopsin Affects a Water Molecule near Asp85 and Light Adaptation. Biochemistry 1997, 36, 5493. [DOI] [PubMed] [Google Scholar]; (b) Otto H; Marti T; Holz M; Mogi T; Stern LJ; Engel F; Khorana HG; Heyn MP Substitution of amino acids Asp-85, Asp-212, and Arg-82 in bacteriorhodopsin affects the proton release phase of the pump and the pK of the Schiff base. Proc. Natl. Acad. Sci. U. S. A 1990, 87, 1018. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Nogly P; Weinert T; James D; Carbajo S; Ozerov D; Furrer A; Gashi D; Borin V; Skopintsev P; Jaeger K; Nass K; Båth P; Bosman R; Koglin J; Seaberg M; Lane T; Kekilli D; Brünle S; Tanaka T; Wu W; Milne C; White T; Barty A; Weierstall U; Panneels V; Nango E; Iwata S; Hunter M; Schapiro I; Schertler G; Neutze R; Standfuss J Retinal isomerization in bacteriorhodopsin captured by a femtosecond x-ray laser. Science 2018, 361, No. eaat0094. [DOI] [PubMed] [Google Scholar]; (d) Kobayashi T; Saito T; Ohtani H Real-time spectroscopy of transition states in bacteriorhodopsin during retinal isomerization. Nature 2001, 414, 531. [DOI] [PubMed] [Google Scholar]; (e) Wickstrand C; Dods R; Royant A; Neutze R Bacteriorhodopsin: Would the real structural intermediates please stand up? Biochim. Biophys. Acta, Gen. Subj 2015, 1850, 536. [DOI] [PubMed] [Google Scholar]; (f) Lanyi JK Annu. Rev. Physiol 2004, 66, 665. [DOI] [PubMed] [Google Scholar]; (g) Gozem S; Luk HL; Schapiro I; Olivucci M Theory and Simulation of the Ultrafast Double-Bond Isomerization of Biological Chromophores. Chem. Rev 2017, 117, 13502. [DOI] [PubMed] [Google Scholar]
- (12).(a) Berbasova T; Nosrati M; Vasileiou C; Wang W; Lee KSS; Yapici I; Geiger JH; Borhan B Rational Design of a Colorimetric pH Sensor from a Soluble Retinoic Acid Chaperone. J. Am. Chem. Soc 2013, 135, 16111. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Berbasova T; Santos EM; Nosrati M; Vasileiou C; Geiger JH; Borhan B Light-Activated Reversible Imine Isomerization: Towards a Photochromic Protein Switch. ChemBioChem 2016, 17, 407. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Crist RM; Vasileiou C; Rabago-Smith M; Geiger JH; Borhan B Engineering a rhodopsin protein mimic. J. Am. Chem. Soc 2006, 128, 4522. [DOI] [PubMed] [Google Scholar]; (d) Lee KSS; Berbasova T; Vasileiou C; Jia X; Wang W; Choi Y; Nossoni F; Geiger JH; Borhan B Probing Wavelength Regulation with an Engineered Rhodopsin Mimic and a C15-Retinal Analogue. Chem-PlusChem 2012, 77, 273. [Google Scholar]; (e) Nosrati M; Berbasova T; Vasileiou C; Borhan B; Geiger JH A Photoisomerizing Rhodopsin Mimic Observed at Atomic Resolution. J. Am. Chem. Soc 2016, 138, 8802. [DOI] [PMC free article] [PubMed] [Google Scholar]; (f) Wang W; Geiger JH; Borhan B The photochemical determinants of color vision: revealing how opsins tune their chromophore’s absorption wavelength. BioEssays 2014, 36, 65. [DOI] [PMC free article] [PubMed] [Google Scholar]; (g) Wang W; Nossoni Z; Berbasova T; Watson CT; Yapici I; Lee KS; Vasileiou C; Geiger JH; Borhan B Tuning the electronic absorption of protein-embedded all-trans-retinal. Science 2012, 338, 1340. [DOI] [PMC free article] [PubMed] [Google Scholar]; (h) Yapici I; Lee KSS; Berbasova T; Nosrati M; Jia X; Vasileiou C; Wang W; Santos EM; Geiger JH; Borhan B Turn-On” Protein Fluorescence: In Situ Formation of Cyanine Dyes. J. Am.Chem. Soc 2015, 137, 1073. [DOI] [PMC free article] [PubMed] [Google Scholar]; (i) Huntress MM; Gozem S; Malley KR; Jailaubekov AE; Vasileiou C; Vengris M; Geiger JH; Borhan B; Schapiro I; Larsen DS; Olivucci M Toward an Understanding of the Retinal Chromophore in Rhodopsin Mimics. J. Phys. Chem. B 2013, 117, 10053. [DOI] [PubMed] [Google Scholar]
- (13).Warshel A Bicycle-pedal model for the first step in the vision process. Nature 1976, 260, 679. [DOI] [PubMed] [Google Scholar]
- (14).Seltzer S MNDO barrier heights for catalyzed bicycle-pedal, hula-twist, and ordinary cis-trans isomerizations of protonated retinal Schiff base. J. Am. Chem. Soc 1987, 109, 1627. [Google Scholar]
- (15).(a) Liu RS; Asato AE The primary process of vision and the structure of bathorhodopsin: a mechanism for photoisomerization of polyenes. Proc. Natl. Acad. Sci. U. S. A 1985, 82, 259. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Liu RSH Photoisomerization by Hula-Twist: A Fundamental Supramolecular Photochemical Reaction. Acc. Chem. Res 2001, 34, 555. [DOI] [PubMed] [Google Scholar]
- (16).Rothschild KJ; Gray D; Mogi T; Marti T; Braiman MS; Stern LJ; Khorana HG Vibrational spectroscopy of bacteriorhodopsin mutants: chromophore isomerization perturbs trytophan-86. Biochemistry 1989, 28, 7052. [DOI] [PubMed] [Google Scholar]
- (17).Luecke H; Schobert B; Richter HT; Cartailler JP; Lanyi JK Structure of bacteriorhodopsin at 1.55 A resolution. J. Mol. Biol 1999, 291, 899. [DOI] [PubMed] [Google Scholar]
- (18).(a) Dunach M; Marti T; Khorana HG; Rothschild KJ UV-visible spectroscopy of bacteriorhodopsin mutants: substitution of Arg-82, Asp-85, Tyr-185, and Asp-212 results in abnormal light-dark adaptation. Proc. Natl. Acad. Sci. U. S. A 1990, 87, 9873. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Needleman R; Chang M; Ni B; Varo G; Fornes J; White SH; Lanyi JK Properties of Asp212— —Asn bacteriorhodopsin suggest that Asp212 and Asp85 both participate in a counterion and proton acceptor complex near the Schiff base. J. Biol. Chem 1991, 266, 11478. [PubMed] [Google Scholar]; (c) Rothschild KJ; Braiman MS; He YW; Marti T; Khorana HG Vibrational spectroscopy of bacteriorhodopsin mutants. Evidence for the interaction of aspartic acid 212 with tyrosine 185 and possible role in the proton pump mechanism. J. Biol. Chem 1990, 265, 16985. [PubMed] [Google Scholar]
- (19).Iwata T; Nagai T; Ito S; Osoegawa S; Iseki M; Watanabe M; Unno M; Kitagawa S; Kandori H Hydrogen Bonding Environments in the Photocycle Process around the Flavin Chromophore of the AppA-BLUF domain. J. Am. Chem. Soc 2018, 140, 11982. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


