Abstract
Patients with heart failure (HF) and left ventricular assist devices (LVAD) have dysregulated thrombo-inflammatory responses, mediated in part by platelets. While studies of platelet activation have been undertaken in HF, changes in the platelet transcriptome in HF patients following mechanical unloading with an LVAD have not been investigated. We prospectively enrolled and longitudinally followed advanced HF patients (n=32) for a mean of 57 months post-LVAD implantation. For comparison, healthy donors were also enrolled (n=20). Platelets were hyperactive in HF, as evidenced by significantly increased formation of circulating platelet-monocyte aggregate formation. Platelet transcriptome interrogation by next-generation RNA-sequencing identified that the expression of numerous genes (n=588) was significantly (FDR<0.05) altered in HF patients prior to LVAD implantation. Differentially expressed genes were predicted to have roles in angiogenesis, immune and inflammatory responses, apoptosis, and cardiac muscle contraction. 90 days following LVAD implantation, the majority (80%) of differentially expressed genes in HF patients normalized, as compared to the platelet transcriptomes of healthy donors. In conclusion, advanced HF is associated with marked alterations in the platelet transcriptome. While LVAD implantation to off load the failing heart results in resolution in the majority of differentially expressed genes, a subset of the platelet transcriptome remains persistently altered.
Keywords: Heart failure, left ventricular assist device, platelets, transcriptome, RNA
Introduction
Heart failure (HF) is a growing epidemic worldwide. HF is associated with a poor quality of life, incapacitating symptoms, hospitalizations, high socioeconomic costs, and premature death. Cardiovascular complications associated with HF include acute myocardial infarction, stroke, pulmonary embolism, and deep vein thrombosis.1 Although pharmacologic therapy has significantly improved the prognosis of many patients with HF, the historic static and low rate of heart transplantation leaves patients with advanced HF relatively few options.
Left ventricular assist devices (LVADs) are mechanical pumps that serve to unload the failing heart. LVADs are used most commonly either to bridge an ill patient to heart transplantation or, in some cases, as destination therapy. In all cases, blood is actively removed from the left ventricle, spun either in rotary or centrifugal fashion - thereby unloading the ventricle - and delivered to the aorta. Despite increasing both life expectancy and quality of life for HF patients, surgery to implant the LVAD and the LVAD itself present significant risks, including infection, bleeding, pump thrombosis, and stroke.2
Platelets are anucleate cells that circulate in the bloodstream. Platelets are canonically known to mediate key aspects of hemostasis and thrombosis, wound healing, and inflammation.3, 4 Dysregulated platelet functions are common during thrombo-inflammatory diseases and can result in either injurious bleeding or thrombotic events. As such, their importance in the pathophysiology of HF is widely recognized, albeit if still incompletely understood.5, 6 Much of the research in hemostasis and thrombosis during HF has focused on investigations into circulating, thrombo-inflammatory plasma factors and classic markers of platelet activation7, 8. To date, studies examining changes in the platelet molecular signature during HF and longitudinally following mechanical unloading with an LVAD are absent in the field.
RNA sequencing (RNA-seq) is an efficient method of analyzing the entirety of the platelet transcriptome, and has been increasingly applied in the study of both healthy donors and patients with thrombo-inflammatory diseases, including patients with myocardial infarction. Although platelets are anucleate, their transcriptome is not fixed. Rather, established and emerging evidence indicates that the platelet transcriptome is dynamically altered during settings of human aging, myocardial infarction, sepsis, and cancer.9–13
In the present study, we comprehensively examined the platelet transcriptome longitudinally in advanced HF patients prior to and at serial time points following LVAD implantation. We identified that the platelet transcriptome is altered in HF with hundreds of genes significantly up- or down-regulated and, further, LVAD implantation was associated with significant normalization of the platelet transcriptome.
Methods
Participant Recruitment
Heart failure (HF) patients and control donors without HF were prospectively recruited from the University of Utah Health Sciences Center. All patients and donors provided written informed consent and the study was IRB approved (# 60878). Heart failure patients were eligible for inclusion in the study if they had advanced HF and were scheduled for mechanical unloading via implantation of a durable continuous flow LVAD, as part of their recommended medical care. In HF patients, routine laboratory tests (including platelet counts and lactate dehydrogenase [LDH]), bleeding and thrombotic complications, and cardiovascular parameters were recorded through medical record review. Control donors without HF were eligible for inclusion in the study if they were free from cardiovascular disease, HF, cancer, liver/renal disease, or thromboembolic events and were not using anti-platelet agents. Pregnancy was an exclusion criterion in both cohorts.
Platelet Isolation
Whole blood (~40mLs) from patients was carefully collected by venipuncture into acid-citrate-dextrose (ACD, 3.2% ACD per volume) sterile vacutainer tubes. For HF patients, blood was obtained immediately prior to LVAD implantation (pre-LVAD), at ~30 days following LVAD implantation, and at ~90 following LVAD implantation. For healthy donors, whole blood was just collected at one time point. The whole blood was centrifuged at 150 x g for 20 minutes at room temperature (RT) to isolate platelet rich plasma. Platelets were isolated and leuko-depleted using our previously published methods that results in a highly-purified population of fewer than 3 leukocytes/107 platelets (>99.9% purity) as counted by hemocytometer.9, 14, 15 These highly purified, isolated platelets (< 3 leukocytes per 1 × 107 platelets) were resuspended in round-bottom polypropylene tubes (Becton Dickinson, Franklin Lakes, NJ) in serum-free, warmed (37°C) M199 (Lonza, Walkersville, MD).
RNA Isolation, Sequencing, and Analysis
We performed RNA-sequencing on a subset of enrolled advanced HF patients (n=14) and healthy donors (n= 7). For LVAD patients and donors, equal numbers of isolated platelets were carefully lysed in TRIzol (5×108 platelets/mL), and DNAse-treated total RNA was isolated9, 16, 17. RIN scores were similar between HF patients and healthy control donors (4.8±0.2 vs 4.7±0.4, p=0.83). Sequencing was performed using HiSeq 50 Cycle Single-Read Sequencing (version 4, Illumina, San Diego, CA) after library preparation with Illumina TruSeq RNA Library Preparation Kit with poly(A) Selection. A minimum of 20 million reads were counted for each sample (sufficient for differential expression analysis). The length of reads was 200bp and reads were aligned (STAR) to the reference genome GRCh37/hg19 and a pseudo transcriptome containing splice junctions. In addition to RIN scores, we also used additional metrics to evaluate the quality of the sequencing data, including per sequence GC Content, assessment of alignment by both STAR, gene coverage by PICARD, feature counts for mapped reads, and FASTQC quality scores and sequencing histograms (Supplemental Table 1 and Supplemental Figures 2A-2B). Normalized counts (an index of RNA abundance) were used in all analyses, as is currently standard in our institutional Sequencing Core. We excluded transcripts with 5 or fewer normalized counts, and analyzed more than 11,000 transcripts meeting this threshold. Specifically, for the hypothesis that the platelet transcriptome was altered in HF patients versus age, sex, and race matched control donors, we tested 11,301 transcripts in analyses comparing the platelet transcriptome between HF patients and controls. For the hypothesis that the platelet transcriptome would normalize in HF patients following LVAD implantation, we tested 11,039 transcripts in longitudinal analyses within the same HF patients (e.g. pre-LVAD and post-LVAD). The Deseq2 analysis package, controlling for levels of PTPRC, was used to quantitate fragments per kilobase of transcript per million mapped reads (FPKMs). Pathway analyses were performed using the Database for Annotation, Visualization and Integrated Discovery (DAVID, https://david.ncifcrf.gov/). RNA sequencing data has been deposited in the NIH Sequence Read Archive PRJNA531691 (for control subjects) and PRJNA595551 (for heart failure subjects).
Protein Assays
Platelet poor plasma was harvested from whole blood via centrifugation. Plasma was stored at −80°C until assayed for circulating protein levels. The Human PDGF-BB ELISA kit (Sigma Aldrich, RAB0397), the Human CXCL4/PF4 Quantikine ELISA Kit (R&D Systems, DPF40), and the Human CCL5/RANTES Quantikine ELISA Kit (R&D Systems, DRN00B) were used according to the manufacturer’s protocols to measure the level of platelet-derived growth factor (PDGF), platelet factor 4 (PF4), and RANTES (regulated on activation, normal T cell expressed and secreted) in plasma.
Statistical Analyses
For RNA-seq based analyses, differential expression was determined using the Wald test (which allows for time-series analyses) in Deseq2. Within Deseq2, we also controlled for multiple comparisons using a false discovery rate (FDR) according to the method by Benjamini-Hochberg. Significant variance in expressed transcripts were pre-specified as those transcripts with an FDR <0.05.
For all other analyses, data were examined for normality using skewness and kurtosis tests. Groups were compared using the Student’s t-test or Wilcoxon Rank Sum (for continuous variables) and using the Chi-squared or Fisher’s exact test (for categorical variables), as appropriate (GraphPad Prism v7.0). A two-tailed p < 0.05 was considered statistically significant.
Results
Characteristics of Heart Failure Patients and Outcomes
We recruited and longitudinally followed 32 patients with advanced HF scheduled for LVAD implantation (LVAD) from May 2013 to April 2014. Advanced HF patients had a mean age of 57 (range of 18 to 76 years) and the majority of the patients were male and Caucasian, reflecting local demographics (Table 1). About one-quarter of HF patients had diabetes mellitus and more than 30% were diagnosed with hypertension. Control donors were well-matched to HF patients with regards to age and race, and similar to HF patients about 20% had diabetes (Table 1). In the entire cohort, a significantly greater proportion of HF patients were male, although for RNA-sequenced based comparisons shown below, HF patients and controls were similar with regards to biological sex (Table 3). The most common type of heart failure was ischemic cardiomyopathy (ICM, 40.6%), followed by idiopathic dilated cardiomyopathy (IDCM, 31.3%) (Table 2). Prior to LVAD implantation, HF patients had a mean (±SEM) ejection fraction of 18.2% (±1.1) and a mean (±SEM) cardiac index of 1.9 (±0.1).
Table 1. Clinical Characteristics of Heart Failure (HF) Patients.
Data are mean (±SEM) unless otherwise indicated. For heart failure patients, cardiac index and ejection fraction are prior to LVAD implantation.
| HF Patients (n=32) | Control Subjects (n=20) | P-value | |
|---|---|---|---|
| Age | 57.1 (±2.4) | 52.7 (±2.3) | 0.22 |
| BMI | 27.6 (±1.2) | 26.7 (±0.8) | 0.73 |
| Cardiac Index | 1.9 (±0.1) | N/A | -- |
| Ejection Fraction | 18.2 (±1.1) | N/A | -- |
| Male, n (%) | 28 (87.5) | 12 (60) | 0.04 |
| Caucasian, n (%) | 27 (84.4) | 16 (80) | 0.72 |
| Diabetes, n (%) | 8 (25) | 4 (20) | 0.74 |
| Hypertension, n (%) | 11 (34.4) | 5 (25) | 0.55 |
Table 3. Comparative clinical characteristics of heart failure patients and control subjects where platelet RNA-sequencing was performed.
Data are mean (±SEM) unless otherwise specified.
| Control Subjects (n=7) | Heart Failure Patients (n=14) | P-value | |
|---|---|---|---|
| Age (yrs) | 48.9 (±3.9) | 47.9 (±4.0) | 0.88 |
| Male, n (%) | 5 (71.4) | 11 (78.6) | 1.0 |
| White, n (%) | 7 (100) | 14 (100) | 1.0 |
Table 2. Characteristics and outcomes of HF patients.
INTERMACS score, the type of HF, LVAD device type, and outcomes for HF patients are shown below. Data are N (%) unless otherwise specified.
| HF Patients (n=32) | |
|---|---|
| INTERMACS score, mean ±SEM | |
| 2 | 12 (±37.5) |
| 3 | 14 (±43.8) |
| 4 | 6 (±18.8) |
| Type of Heart Failure | |
| ICM | 13 (40.6) |
| DCM | 4 (12.5) |
| INICM | 1 (3.1) |
| NCIM | 2 (6.3) |
| IDCM | 10 (31.3) |
| NIDCM | 1 (3.1) |
| NI+ISCCM | 1 (3.1) |
| Device Type | |
| HeartMateII | 20 (62.5) |
| HeartWare HVAD | 8 (25) |
| Jarvik 2000 | 3 (9.4) |
| BiVADs | 1 (3.1) |
| Purpose | |
| Bridge-to-Transplant | 20 (68.7) |
| Destination Therapy | 10 (31.3) |
| Other | 2 (6.3) |
| Anticoagulation Therapy | 29 (90.6) |
| Post-operative Complications | |
| Bleeding | 15 |
| Stroke | 5 |
| Pump Thrombosis | 3 |
| Hemorrhage | 1 |
| Multiorgan Dysfunction | 1 |
| Mortality | 5 (15.6) |
In accordance with accepted treatment strategies, more than 90% of HF patients (pre-LVAD) were prescribed anti-platelet/anti-thrombotic therapeutics either individually or in combination. The most common therapies were aspirin (58.6%), Coumadin (41.4%), and Plavix (17.2%). Approximately two thirds of patients were implanted with a HeartMate II LVAD (HMII). The majority of HF patients received an LVAD as a bridge to transplantation, and about 30% of patients received an LVAD as destination therapy (Table 2).
Heart failure patients were followed for a mean of 57 months (range 51–63) following LVAD implantation. A total of 24 thromboembolic and/or bleeding complications in 18 patients occurred during this follow-up period (Table 2). All stroke and pump thrombosis complications were in patients with the HMII device. Overall, 5 patients (15.6%) died during the follow-up period, with 4 of the 5 deaths being due to bleeding or thrombosis and one patient dying from right ventricular failure. Of the 5 patients that died, two patients had the HMII, two patients had the HeartWare HVAD, and one patient had the Jarvik device.
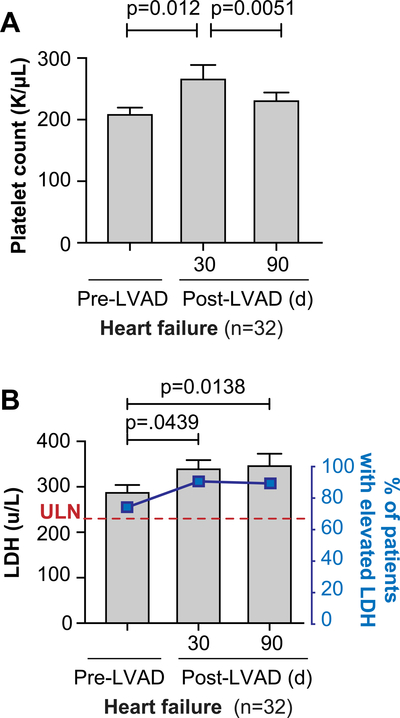
Platelet counts were within normal levels in HF patients pre-LVAD implantation. There was a significant increase in circulating platelet counts in HF patients 30 days following surgery for LVAD implantation (Figure 1A). Platelet counts in HF patients 90 days following surgery had returned to baseline. As previously observed 18, LDH levels in HF patients were elevated prior to LVAD implantation (74%), significantly increased 30 days following LVAD implantation, and remained elevated at 90 days following LVAD implantation (90%) (Figure 1B).
Figure 1. Platelet counts and lactate dehydrogenase (LDH) levels in patients with heart failure prior to and longitudinally following LVAD implantation.
(A) Circulating platelet counts were measured in patients with heart failure (n=32) prior to LVAD implantation (pre-LVAD) and again 30 days and 90 days post-LVAD implantation (post-LVAD). (B) Circulating levels of LDH were measured prior to LVAD implantation (pre-LVAD) and again 30 days and 90 days post-LVAD implantation (post-LVAD). The left axis, and the corresponding bars and p-values, reflect the measured units per liter (u/L). The dotted red line reflects the upper limit of the normal (ULN, which was 230 u/L based on our clinical laboratory’s reference range). The right axis shows the percentage of HF patients who had an LDH above the upper limit of normal (ULN). Numbers on the top refer to the p-value.
LVAD Implantation is Associated with Increased Platelet Activation
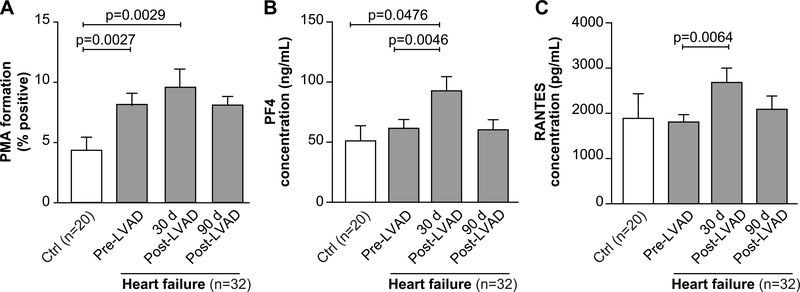
We assessed indices of platelet activation in HF patients prior to and longitudinally following LVAD implantation. Endogenous platelet activation measured by platelet-monocyte aggregate (PMA) formation, a sensitive marker of in vivo platelet activation 19–22, was significantly increased in HF patients, relative to control subjects, prior to LVAD implantation (Figure 2A). Thirty days following LVAD implantation, PMA formation in HF patients rose significantly but returned to near baseline levels 90 days following LVAD implantation (Figure 2A). This is consistent with increased platelet activation in HF patients.
Figure 2. Platelets are activated in patients with heart failure prior to and longitudinally following LVAD implantation.
(A) To assess endogenous platelet activation, whole blood was drawn from patients with heart failure (n=32) or matched control donors without HF (Ctrl, n=20). Baseline (e.g. unstimulated) whole blood flow cytometry was performed to measure levels of circulating platelet-monocyte aggregate (PMA) formation, a sensitive index of endogenous platelet activation. (B-C) From the same whole blood samples, plasma was harvested as described in the methods. Levels of (B) platelet factor 4 (PF4) and (C) RANTES (regulated upon activation, normal T cell expressed and secreted) were measured by commercially available ELISA. Numbers on the top refer to the p-value.
Circulating plasma levels of PF4 and RANTES, α-granule chemokines secreted by activated platelets, were similar between HF patients and controls before LVAD implantation (Figure 2B–C). Levels of PF4 and RANTES significantly increased in HF patients 30 days following LVAD implantation, but then returned to baseline levels 90 days following LVAD implantation (Figure 2B–C). Consistent with dynamic changes in circulating platelet counts (Figure 1A), these indices of platelet activation may be due to surgical stress. Interestingly, activation of platelet integrin αIIbβ3 was suppressed in HF patients, perhaps due to the use of anti-platelet and anti-thrombotic therapies (Supplemental Figure 1A).
The Platelet Transcriptome is Altered in Patients with Heart Failure Prior to Mechanical Unloading
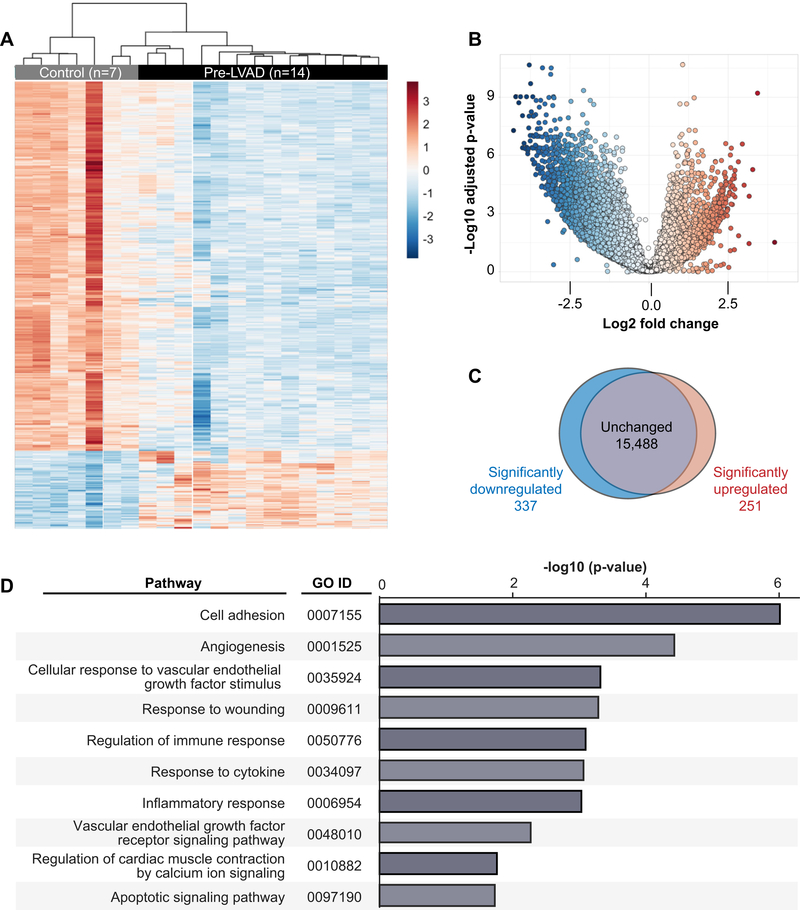
We next used RNA-seq to globally profile the platelet transcriptome in a subset of HF patients prior to mechanical unloading with LVAD and for comparison, age, sex, and race matched healthy controls. Table 3 shows clinical characteristics of the HF patients and controls where the platelet transcriptome was profiled by RNA sequencing. Globally, the platelet transcriptome was altered in HF (Figure 3A–C), with 588 transcripts that were significantly (FDR<0.05) differentially expressed in platelets isolated from HF patients (n=14) compared to healthy donors (n=7). Of these 588 transcripts, 337 were significantly downregulated and 251 were significantly upregulated (Figure 3C, Supplemental Table 2). Table 4 provides a list of the top 20 significantly (FDR<0.05) differentially expressed transcripts between HF patients and healthy donors. To gain more insight into the biological roles of differentially expressed transcripts in HF, we next performed pathway analyses. Significantly enriched pathways (FDR<0.05) included cell adhesion, angiogenesis and vascular endothelial growth factor (VEGF), regulation of cardiac muscle contraction by calcium ion signaling, immune/inflammatory responses, and apoptosis. Figure 3D shows the full list of all significantly enriched pathways identified in this analysis. Together, these results demonstrate that the platelet transcriptome is markedly altered in patients with HF, with many genes and pathways implicated in the pathobiology of HF.
Figure 3. The platelet transcriptome is altered in patients with heart failure prior to LVAD implantation.
Platelets were isolated from heart failure patients prior to LVAD implantation (pre-LVAD, n=14) or matched control donors (Control, n=7). The platelet transcriptome was analyzed by next generation RNA-sequencing and compared between heart failure patients and control donors. (A) Heat map, (B) volcano plot, and (C) Venn diagram of significantly (FDR<0.05) differentially expressed transcripts between heart failure patients and control donors. In the heat map, each row represents a differentially expressed gene and each column represents a unique patient or control donor. Red indicates transcripts that were significantly upregulated and blue indicates transcripts that were significantly down-regulated. (D) Gene ontology (GO) pathway analysis for significantly differentially expressed transcripts (n=588, FDR<0.05) in heart failure patients compared to control donors.
Table 4. List of the top 20 significantly differentially expressed transcripts in platelets isolated from heart failure patients.
The comparison is made between heart failure patients prior to LVAD implantation and healthy donors. Transcripts are ranked by the log2 fold-change in descending order.
| Gene ID | Gene Symbol | Gene Name | Log2 Fold-Change | p(adj) |
|---|---|---|---|---|
| ENSG00000170323 | FABP4 | Fatty Acid Binding Protein 4 | 2.74 | 7.75774E-06 |
| ENSG00000099260 | PALMD | Palmdelphin | 2.47 | 5.33646E-05 |
| ENSG00000132170 | PPARG | Peroxisome Proliferator Activated Receptor Gamma | 2.43 | 0.000616819 |
| ENSG00000137960 | GIPC2 | Semaphorin Cytoplasmic Domain Associated Protein 2 | 2.35 | 0.000483865 |
| ENSG00000132622 | HSPA12B | Heat Shock Protein Family A (Hsp70) Member 12B | 2.24 | 0.000541917 |
| ENSG00000066735 | KIF26A | Kinesin Family Member 26A | 2.23 | 0.000616819 |
| ENSG00000130595 | TNNT3 | Troponin T3, Fast Skeletal Type | 2.23 | 0.000483865 |
| ENSG00000074219 | TEAD2 | TEA Domain Transcription Factor 2 | 2.21 | 0.001184061 |
| ENSG00000188643 | S100A16 | S100 Calcium Binding Protein A16 | 2.20 | 0.000483865 |
| ENSG00000131831 | RAI2 | Retinoic Acid-Induced Protein 2 | 2.19 | 0.000829674 |
| ENSG00000152583 | SPARCL1 | SPARC-Like Protein 1 | 2.17 | 0.001184061 |
| ENSG00000198959 | TGM2 | Transglutaminase 2 | 2.17 | 0.000248434 |
| ENSG00000203883 | SOX18 | SRY-Box Transcription Factor 18 | 2.16 | 0.000483865 |
| ENSG00000130158 | DOCK6 | Dedicator Of Cytokinesis 6 | 2.14 | 0.000963129 |
| ENSG00000167680 | SEMA6B | Semaphorin 6B | 2.13 | 0.003055168 |
| ENSG00000177469 | CAVIN1 | Caveolae Associated Protein 1 | 2.12 | 0.000829674 |
| ENSG00000072163 | LIMS2 | LIM Zinc Finger Domain Containing 2 | 2.12 | 0.000829674 |
| ENSG00000241644 | INMT | Indolethylamine N-Methyltransferase | 2.12 | 0.001697573 |
| ENSG00000277494 | GPIHBP1 | Glycosylphosphatidylinositol Anchored High Density Lipoprotein Binding Protein 1 | 2.11 | 0.006910868 |
| ENSG00000065054 | SLC9A3R2 | SLC9A3 Regulator 2 | 2.10 | 0.00093032 |
The Expression of Numerous Platelet Transcripts Normalizes Following Mechanical Unloading of the Heart
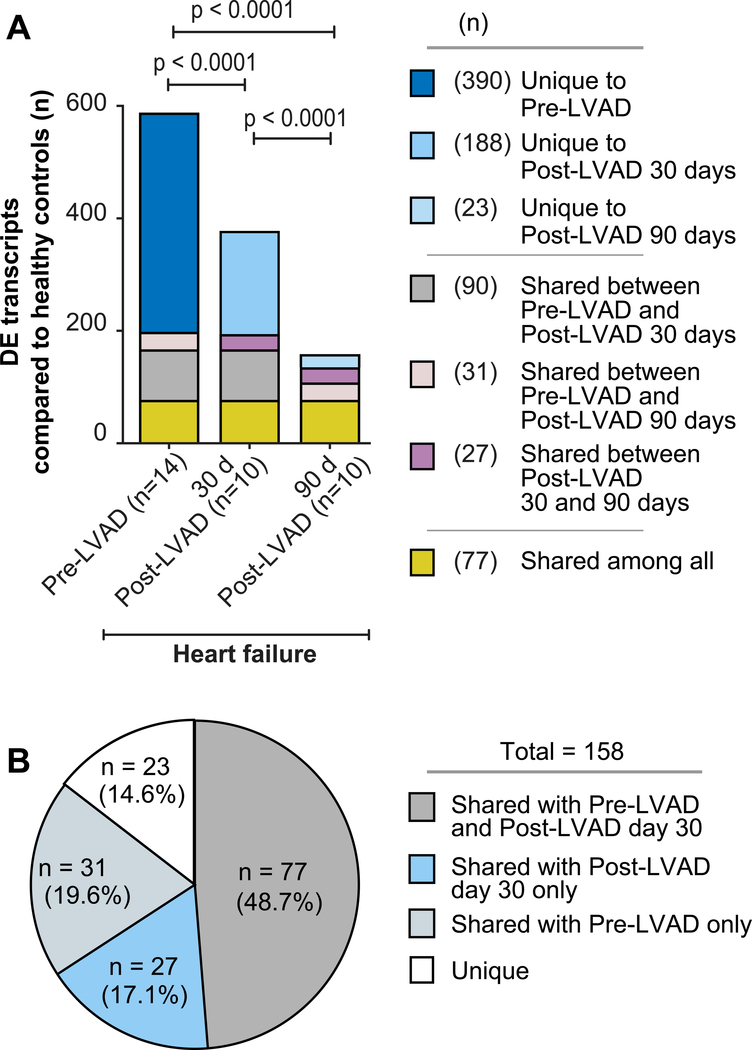
To understand how mechanical unloading of the heart by LVAD implantation affects the platelet transcriptome, we next performed longitudinal profiling of the platelet transcriptome in HF patients following LVAD placement. The platelet transcriptome was assessed by RNA-seq and all platelet samples from LVAD patients were run simultaneously, thereby adding substantial rigor to cross-comparative analyses. In HF patients, mechanical unloading of the heart by LVAD placement was associated with substantial normalization of the platelet transcriptome, as compared to control subjects. These changes were evident by 30 days following LVAD implantation, where the expression of a majority (390/588, 66%) of the differentially expressed platelet transcripts in HF patients pre-LVAD normalized when compared to control subjects (Figure 4A, Supplemental Table 3). By pathway analyses, platelet transcripts whose expression normalized were implicated in integrin signaling, shear stress, vasculogenesis, and inflammation (Supplemental Figure 3).
Figure 4. The expression of many transcripts in platelets normalizes in heart failure patients following LVAD implantation.
(A) Comparison of the number of significantly differentially expressed (DE) transcripts (FDR<0.05) in platelets from heart failure patients followed longitudinally over time (e.g. prior to LVAD implantation (n=14), 30 days (n=10), and again 90 days (n=10) following LVAD implantation), as compared to matched healthy controls (n=7). As denoted in the legend to the right, “Unique” refers to transcripts in platelets that were significantly differentially expressed in heart failure patients, compared to controls, at just one time point. “Shared” refers to transcripts in platelets that were durably and significantly differentially expressed in heart failure patients, compared to controls, at two or more time points. (B) Pie chart showing the distribution of significantly differentially expressed transcripts in platelets from heart failure patients 90 days following LVAD implantation, as compared to platelet transcriptomic assessments 30 days following LVAD implantation (Post-LVAD day 30) or prior to LVAD implantation (pre-LVAD). “Unique” and “Shared” denote the same as in panel A.
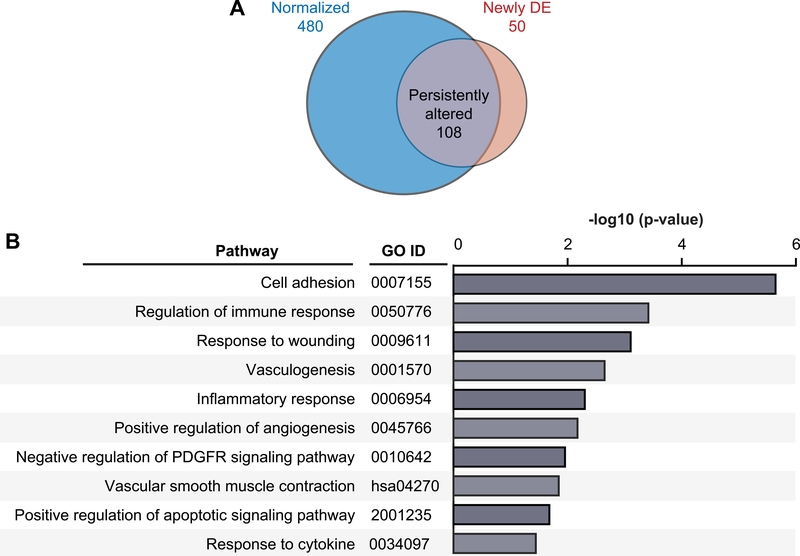
By 90 days after LVAD implantation, there was even further normalization (n=480/588, ~82%) of significantly differentially expressed transcripts in HF patients compared to baseline (Figure 4A–B). These transcripts were involved in processes relevant to HF, including adhesion, inflammation, immunity, and angiogenesis. There was a set of 108 transcripts in platelets that remained persistently altered 90 days after LVAD implantation, compared to transcriptomic assessment at 30 days following LVAD implantation (Figure 5A and Supplemental Table 4), and a small set of transcripts (n=50) that were newly differentially expressed at 90 days following LVAD implantation, compared to pre-LVAD conditions in HF patients (Figure 5 and Supplemental Table 5).
Figure 5. Pathway analyses of differentially expressed transcripts in platelets from heart failure patients prior to and 90 days following LVAD implantation.
(A) Venn diagram of platelet transcripts whose expression normalized (as compared to control subjects) in heart failure patients 90 days after LVAD implantation (blue, n=480), remained significantly persistently altered (grey, n=108; FDR<0.05) 90 days after LVAD implantation, or were newly significantly differentially expressed (orange, n=50; FD<0.05). (B) Gene ontology (GO) pathway analysis for all differentially expressed transcripts in heart failure patients that normalized 90 days after LVAD implantation (n=480). In all panels, “n” refers to transcript number. (DE = differentially expressed).
Interestingly, a number of new, significantly differentially expressed transcripts (n=188) present in platelets from HF patients 30 days following LVAD implantation normalized upon follow-up assessment at 90 days post-LVAD placement (Supplemental Table 6). These transcripts may have been altered in expression due to surgical stress, consistent with our observations of transient, increased platelet activation at this same time point (Figure 2A). Overall, 77 transcripts remained durably altered in HF patients at all three time points assessed (Figure 4A–B and Supplemental Table 7).
Discussion
Platelets mediate critical aspects of the pathophysiology that occurs in HF, including dysregulated thrombotic and inflammatory responses.2 Nevertheless, a comprehensive and longitudinal analysis of the human platelet transcriptome in HF patients, before and following mechanical unloading with LVAD, has not yet been undertaken. Emerging evidence demonstrates that the platelet transcriptome can be altered during disease and, in some settings, may provide important diagnostic, prognostic, and therapeutic clues. Therefore, an understanding of platelet transcriptomics in HF may yield new insights into the pathophysiology of the disease and how LVAD implantation may influence platelet gene expression and thus host responses.
In the present study, we identified that the platelet transcriptome is altered in patients with advanced HF with hundreds of genes being significantly up- and down-regulated. Moreover, the function of these genes were broadly implicated in adaptive and maladaptive physiological processes known to occur in HF, including inflammation, angiogenesis, and cardiac muscle contraction.
A unique aspect of our study is the longitudinal assessment of the platelet transcriptome in HF patients prior to and following mechanical unloading. To the best of our knowledge, this is the first study to assess not only the human platelet transcriptome in advanced HF, but also to examine how offloading the heart by LVAD implantation influences gene expression over time. Notably, we found that 90 days following LVAD implantation, the majority of significantly differentially expressed transcripts in platelets from HF patients had returned to similar levels observed in matched control subjects. There remained a set (n=77) of transcripts in platelets that were significantly differentially expressed prior to LVAD implantation and that were also differentially expressed post-LVAD. This clinical study obviously cannot define the mechanisms driving platelet transcriptomic changes following LVAD implantation. We postulate that offloading the failing human heart (the primary function of an LVAD) is contributing to these changes over time, but cannot exclude the possibility that other processes (e.g. platelet turnover following LVAD implantation, changes in flow dynamics, alterations in vWF structure and activity) may also be contributing. We hope that these findings serve to be hypothesis generating for future studies in the field, including comparative analyses between cell-specific and whole blood transcriptomics.
Concordant with changes in the platelet transcriptome, platelets from HF patients also exhibited evidence of increased endogenous PMA formation prior to LVAD surgery, with further significant, transient upregulation following LVAD implantation. The formation of PMAs is a very sensitive marker of platelet activation and in some settings of systemic inflammation, including cardiovascular disease, may be more sensitive than classic markers such as soluble p-selectin. This may explain, in part, why some – but not all - studies of platelet activation in heart failure did not observe evidence of increased platelet activation.2, 23 Consistent with this, we did not observe significantly increased indices of either soluble RANTES and PF4 in HF patients prior to LVAD. Soluble RANTES and PF4 did increase transiently following LVAD implantation (e.g. 30 days post-implantation) but then returned to pre-operative levels. This suggests acute upregulation due to surgical stress.
The treatment of HF may vary from medical therapy to transplantation. One strategy for patients with end-stage heart failure is mechanical unloading through implantation of an LVAD. This has been associated with an improved quality of life and extended life expectancy in patients who are waiting for, or unable to receive, a heart transplant.24 The limitation of wide-spread use of this therapy revolves around LVAD-mediated adverse events, mostly from thromboembolic and bleeding events.25–27 In addition to globally examining the platelet transcriptome in HF, we also endeavored to understand how transcriptional changes may provide insights into the pathobiology of HF. Many enriched pathways identified in our datasets, are implicated in the progression of heart failure. As one example, we also found that genes involved in the regulation of cardiac function and blood pressure were differentially expression in HF pathways. In addition, cardiac apoptosis is recognized as an important factor in the progression to heart failure.
This study should be viewed in the context of several limitations. We did not have sufficient material to examine the RNA expression of candidate genes by other platforms. Additionally, while HF patients and healthy donors were generally well-matched, we could not control for medications HF patients were taking, and were not powered to determine if medications impact the platelet transcriptome. Our event rate per patient enrolled was lower than would be allowed for statistically meaningful comparisons of platelet transcriptomics with major adverse outcomes such as pump thrombosis or major bleeding. In addition, at the time of data collection and analysis, the HeartMate II pump was still being actively implanted. Whether these findings would be observed with newer systems (e.g. the HeartMate 3) remains to be investigated.28
Conclusions
In conclusion, we report the first longitudinal, RNA-sequencing based transcriptomics study of platelets in advanced HF patients prior to and following mechanical unloading with an LVAD. We demonstrate that in HF, the platelet transcriptome is altered with hundreds of biologically relevant transcripts either up- or down-regulated. Moreover, LVAD implantation was associated with marked normalization of a majority of these transcripts.
Supplementary Material
Figure S1. Integrin αIIbβ3 activation in platelets is durably suppressed in HF patients. Whole blood was drawn from patients with heart failure (n=32) or matched control donors without HF (Ctrl, n=20). Baseline (e.g. unstimulated) whole blood flow cytometry was performed to measure binding of the PAC-1 antibody to activated integrin αIIbβ3 on platelets.
Figure S2. Metric QC data on platelet RNA-seq data. Platelets were isolated as described in the methods and metric QC data assessed. (A) FastQC quality scores. (B) Picard normalized gene coverage.
Figure S3. Pathway analyses of transcripts in platelets that normalized following LVAD implantation. Pathway analysis was performed using the Generally Applicable Gene-set Enrichment (GAGE) package in R for analyses of transcripts in platelets whose expression normalized in heart failure patients 30 days following LVAD implantation. Normalization was defined as transcript expression in platelets from heart failure patients that did not significantly differ (FDR>0.05) compared to matched control donors.
Table S1. Metric QC data for platelet RNA-sequencing.
Table S2. List of significantly differentially expressed transcripts in platelets from HF patients prior to LVAD implantation, as compared to matched control subjects. The base mean is a measurement of relative RNA expression. Both p-nominal and p-adjusted values are shown. Transcripts are ordered based on p-adjusted values.
Table S3. List of transcripts in platelets from HF patients that were significantly differentially expressed prior to LVAD implantation and then normalized 30 days post LVAD implantation, as compared to matched control subjects. Transcripts are ordered alphabetically.
Table S4. List of transcripts in platelets from HF patients, assessed at 90 days following LVAD implantation, that remained significantly (FDR<0.05) differentially expressed compared to healthy controls.
Table S6. List of transcripts that were newly, but transiently, significantly differentially expressed in platelets from HF patients 30 days following LVAD implantation, as compared to pre-LVAD implantation and 90 days post-LVAD implantation. Transcripts are ordered alphabetically.
Acknowledgements
Funding Source: This work was supported by the NHLBI and NIA (HL142804, HL145237, HL130541, HL137606, AG059877, AG048022 to M.T.R.) and a VA Merit Award (I01 CX001696). This material is the result of work supported with resources and the use of facilities at the George E. Wahlen VA Medical Center, Salt Lake City, Utah. Research reported in this publication was supported by the National Center for Research Resources of the National Institutes of Health under Award Number U54 TR002355. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. This work was supported by the University of Utah Flow Cytometry Facility in addition to the National Cancer Institute through Award Number 5P30CA042014–24. The work was also supported by the Huntsman Cancer Institute High Throughput Genomics Shared Resource (HTG). We thank Ms. Diana Lim for her creativity and excellent figure preparation, Ms. Toni Blair for her assistance with clinical data collection, and Dr. Jesse Rowley for his expert assistance with aspects of RNA-sequencing.
Footnotes
Declaration of Interest Statement
The authors report no competing conflict of interest.
References
- 1.Kim JH, Shah P, Tantry US and Gurbel PA. Coagulation Abnormalities in Heart Failure: Pathophysiology and Therapeutic Implications. Curr Heart Fail Rep. 2016;13:319–328. [DOI] [PubMed] [Google Scholar]
- 2.Koliopoulou A, McKellar SH, Rondina M and Selzman CH. Bleeding and thrombosis in chronic ventricular assist device therapy: focus on platelets. Curr Opin Cardiol. 2016;31:299–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Koupenova M, Clancy L, Corkrey HA and Freedman JE. Circulating Platelets as Mediators of Immunity, Inflammation, and Thrombosis. Circ Res. 2018;122:337–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rondina MT, Weyrich AS and Zimmerman GA. Platelets as cellular effectors of inflammation in vascular diseases. Circ Res. 2013;112:1506–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chung I and Lip GY. Platelets and heart failure. Eur Heart J. 2006;27:2623–31. [DOI] [PubMed] [Google Scholar]
- 6.Schafer A, Eigenthaler M and Bauersachs J. Platelet activation in heart failure. Clin Lab. 2004;50:559–66. [PubMed] [Google Scholar]
- 7.Nascimbene A, Neelamegham S, Frazier OH, Moake JL and Dong JF. Acquired von Willebrand syndrome associated with left ventricular assist device. Blood. 2016;127:3133–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bartoli CR, Hennessy-Strahs S, Gohean J, Villeda M, Larson E, Longoria R, Kurusz M, Acker MA and Smalling R. A Novel Toroidal-Flow Left Ventricular Assist Device Minimizes Blood Trauma: Implications of Improved Ventricular Assist Device Hemocompatibility. Ann Thorac Surg. 2019;107:1761–1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Campbell RA, Franks Z, Bhatnagar A, Rowley JW, Manne BK, Supiano MA, Schwertz H, Weyrich AS and Rondina MT. Granzyme A in Human Platelets Regulates the Synthesis of Proinflammatory Cytokines by Monocytes in Aging. J Immunol. 2018;200:295–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Best MG, Vancura A and Wurdinger T. Platelet RNA as a circulating biomarker trove for cancer diagnostics. J Thromb Haemost. 2017;15:1295–1306. [DOI] [PubMed] [Google Scholar]
- 11.Eicher JD, Wakabayashi Y, Vitseva O, Esa N, Yang Y, Zhu J, Freedman JE, McManus DD and Johnson AD. Characterization of the platelet transcriptome by RNA sequencing in patients with acute myocardial infarction. Platelets. 2016;27:230–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rose JJ, Voora D, Cyr DD, Lucas JE, Zaas AK, Woods CW, Newby LK, Kraus WE and Ginsburg GS. Gene Expression Profiles Link Respiratory Viral Infection, Platelet Response to Aspirin, and Acute Myocardial Infarction. PLoS One. 2015;10:e0132259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Middleton EA, Rowley JW, Campbell RA, Grissom CK, Brown SM, Beesley SJ, Schwertz H, Kosaka Y, Manne BK, Krauel K, Tolley ND, Eustes AS, Guo L, Paine R 3rd, Harris ES, Zimmerman GA, Weyrich AS and Rondina MT. Sepsis alters the transcriptional and translational landscape of human and murine platelets. Blood. 2019;134:911–923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schwertz H, Rowley JW, Schumann GG, Thorack U, Campbell RA, Manne BK, Zimmerman GA, Weyrich AS and Rondina MT. Endogenous LINE-1 (Long Interspersed Nuclear Element-1) Reverse Transcriptase Activity in Platelets Controls Translational Events Through RNA-DNA Hybrids. Arterioscler Thromb Vasc Biol. 2018;38:801–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schwertz H, Rowley JW, Zimmerman GA, Weyrich AS and Rondina MT. Retinoic acid receptor-alpha regulates synthetic events in human platelets. J Thromb Haemost. 2017;15:2408–2418. [DOI] [PubMed] [Google Scholar]
- 16.Campbell RA, Vieira-de-Abreu A, Rowley JW, Franks ZG, Manne BK, Rondina MT, Kraiss LW, Majersik JJ, Zimmerman GA and Weyrich AS. Clots Are Potent Triggers of Inflammatory Cell Gene Expression: Indications for Timely Fibrinolysis. Arterioscler Thromb Vasc Biol. 2017;37:1819–1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rowley JW, Oler AJ, Tolley ND, Hunter BN, Low EN, Nix DA, Yost CC, Zimmerman GA and Weyrich AS. Genome-wide RNA-seq analysis of human and mouse platelet transcriptomes. Blood. 2011;118:e101–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Madden JL, Drakos SG, Stehlik J, McKellar SH, Rondina MT, Weyrich AS and Selzman CH. Baseline red blood cell osmotic fragility does not predict the degree of post-LVAD hemolysis. ASAIO J. 2014;60:524–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shih L, Kaplan D, Kraiss LW, Casper TC, Pendleton RC, Peters CL, Supiano MA, Zimmerman GA, Weyrich AS and Rondina MT. Platelet-Monocyte Aggregates and C-Reactive Protein are Associated with VTE in Older Surgical Patients. Sci Rep. 2016;6:27478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rondina MT, Carlisle M, Fraughton T, Brown SM, Miller RR 3rd, Harris ES, Weyrich AS, Zimmerman GA, Supiano MA and Grissom CK. Platelet-monocyte aggregate formation and mortality risk in older patients with severe sepsis and septic shock. J Gerontol A Biol Sci Med Sci. 2015;70:225–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rondina MT, Brewster B, Grissom CK, Zimmerman GA, Kastendieck DH, Harris ES and Weyrich AS. In vivo platelet activation in critically ill patients with primary 2009 influenza A(H1N1). Chest. 2012;141:1490–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Burdess A, Michelsen AE, Brosstad F, Fox KA, Newby DE and Nimmo AF. Platelet activation in patients with peripheral vascular disease: reproducibility and comparability of platelet markers. Thromb Res. 2012;129:50–5. [DOI] [PubMed] [Google Scholar]
- 23.Michelson AD, Barnard MR, Krueger LA, Valeri CR and Furman MI. Circulating monocyte-platelet aggregates are a more sensitive marker of in vivo platelet activation than platelet surface P-selectin: studies in baboons, human coronary intervention, and human acute myocardial infarction. Circulation. 2001;104:1533–7. [DOI] [PubMed] [Google Scholar]
- 24.Pinney SP, Anyanwu AC, Lala A, Teuteberg JJ, Uriel N and Mehra MR. Left Ventricular Assist Devices for Lifelong Support. J Am Coll Cardiol. 2017;69:2845–2861. [DOI] [PubMed] [Google Scholar]
- 25.Koliopoulou A and Selzman CH. Stop the LVAD bleeding. J Thorac Dis. 2017;9:E437–E439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shah P, Tantry US, Bliden KP and Gurbel PA. Bleeding and thrombosis associated with ventricular assist device therapy. J Heart Lung Transplant. 2017;36:1164–1173. [DOI] [PubMed] [Google Scholar]
- 27.Stehlik J, Johnson SA and Selzman CH. Gold standard in anticoagulation assessment of left ventricular assist device patients?: how about bronze. JACC Heart Fail. 2015;3:323–6. [DOI] [PubMed] [Google Scholar]
- 28.Mehra MR, Goldstein DJ, Uriel N, Cleveland JC Jr., Yuzefpolskaya M, Salerno C, Walsh MN, Milano CA, Patel CB, Ewald GA, Itoh A, Dean D, Krishnamoorthy A, Cotts WG, Tatooles AJ, Jorde UP, Bruckner BA, Estep JD, Jeevanandam V, Sayer G, Horstmanshof D, Long JW, Gulati S, Skipper ER, O’Connell JB, Heatley G, Sood P, Naka Y and Investigators M. Two-Year Outcomes with a Magnetically Levitated Cardiac Pump in Heart Failure. N Engl J Med. 2018;378:1386–1395. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Integrin αIIbβ3 activation in platelets is durably suppressed in HF patients. Whole blood was drawn from patients with heart failure (n=32) or matched control donors without HF (Ctrl, n=20). Baseline (e.g. unstimulated) whole blood flow cytometry was performed to measure binding of the PAC-1 antibody to activated integrin αIIbβ3 on platelets.
Figure S2. Metric QC data on platelet RNA-seq data. Platelets were isolated as described in the methods and metric QC data assessed. (A) FastQC quality scores. (B) Picard normalized gene coverage.
Figure S3. Pathway analyses of transcripts in platelets that normalized following LVAD implantation. Pathway analysis was performed using the Generally Applicable Gene-set Enrichment (GAGE) package in R for analyses of transcripts in platelets whose expression normalized in heart failure patients 30 days following LVAD implantation. Normalization was defined as transcript expression in platelets from heart failure patients that did not significantly differ (FDR>0.05) compared to matched control donors.
Table S1. Metric QC data for platelet RNA-sequencing.
Table S2. List of significantly differentially expressed transcripts in platelets from HF patients prior to LVAD implantation, as compared to matched control subjects. The base mean is a measurement of relative RNA expression. Both p-nominal and p-adjusted values are shown. Transcripts are ordered based on p-adjusted values.
Table S3. List of transcripts in platelets from HF patients that were significantly differentially expressed prior to LVAD implantation and then normalized 30 days post LVAD implantation, as compared to matched control subjects. Transcripts are ordered alphabetically.
Table S4. List of transcripts in platelets from HF patients, assessed at 90 days following LVAD implantation, that remained significantly (FDR<0.05) differentially expressed compared to healthy controls.
Table S6. List of transcripts that were newly, but transiently, significantly differentially expressed in platelets from HF patients 30 days following LVAD implantation, as compared to pre-LVAD implantation and 90 days post-LVAD implantation. Transcripts are ordered alphabetically.