ABSTRACT
Nuclear envelope (NE) budding is a recently described phenomenon wherein large macromolecular complexes are packaged inside the nucleus and extruded through the nuclear membranes. Although a general outline of the cellular events occurring during NE budding is now in place, little is yet known about the molecular machinery and mechanisms underlying the physical aspects of NE bud formation. Using a multidisciplinary approach, we identify Wash, its regulatory complex (SHRC), capping protein and Arp2/3 as new molecular components involved in the physical aspects of NE bud formation in a Drosophila model system. Interestingly, Wash affects NE budding in two ways: indirectly through general nuclear lamina disruption via an SHRC-independent interaction with Lamin B leading to inefficient NE bud formation, and directly by blocking NE bud formation along with its SHRC, capping protein and Arp2/3. In addition to NE budding emerging as an important cellular process, it shares many similarities with herpesvirus nuclear egress mechanisms, suggesting new avenues for exploration in both normal and disease biology.
KEY WORDS: WASH, Wash regulatory complex, Nuclear envelope budding, Nuclear exit, Arp2/3, Actin nucleation, Vesicle-mediated nucleocytoplasmic transport
Highlighted Article: Wash, the Wash regulatory complex, capping protein and Arp2/3 are identified as new components of nuclear envelope buds used for nuclear pore complex-independent export of large macromolecule complexes from the nucleus.
INTRODUCTION
Transport of macromolecules from the nucleus to the cytoplasm is essential for all developmental processes, including the regulation of differentiation and aging, and, when mis-regulated, is associated with diseases and cancer (Grünwald et al., 2011; Siddiqui and Borden, 2012; Tran et al., 2014). This indispensable process has been thought to occur exclusively through nuclear pore complexes (NPCs), channels that regulate what exits (and enters) the nucleus (Daneholt, 2001; Grünwald et al., 2011). Recently, nuclear envelope (NE) budding was identified as an alternative pathway for nuclear exit, particularly for large developmentally required ribonucleoprotein (megaRNP) complexes that would otherwise need to unfold/remodel to fit through the NPCs (Fradkin and Budnik, 2016; Hatch and Hetzer, 2014, 2012; Jokhi et al., 2013; Li et al., 2016; Parchure et al., 2017; Speese et al., 2012). In this pathway, large macromolecule complexes, such as megaRNPs, are encircled by the nuclear lamina (type-A and -B lamins) and the inner nuclear membrane (INM), pinched off from the INM, fuse with the outer nuclear membrane and release the megaRNPs into the cytoplasm (Fig. 1A–C). Strikingly, NE budding shares many features with the nuclear egress mechanism used by herpesviruses (Bigalke and Heldwein, 2016; Hagen et al., 2015; Lye et al., 2017; Parchure et al., 2017; Roller and Baines, 2017). As viruses often utilize pre-existing host pathways, the parallel between nuclear exit of herpesvirus nucleocapsids and that of megaRNPs and/or other large cargoes suggests that NE budding may be a general cellular mechanism (Fradkin and Budnik, 2016; Mettenleiter et al., 2013; Parchure et al., 2017; Roller and Baines, 2017). Indeed, this pathway has also been implicated in the removal of obsolete macromolecular complexes or other material (i.e. large protein aggregates or poly-ubiquitylated proteins) from the nucleus (Jokhi et al., 2013; Ramaswami et al., 2013; Rose and Schlieker, 2012).
Fig. 1.
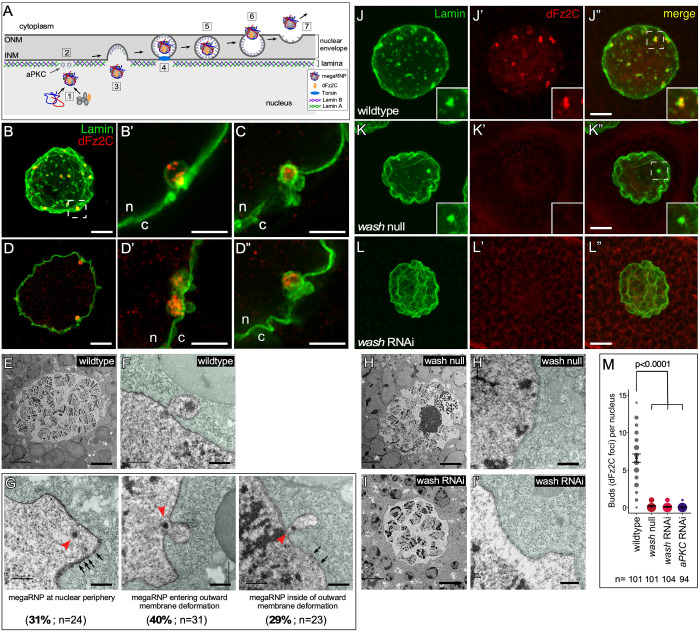
Wash mutant nuclei lack NE buds. (A) Schematic of NE budding steps: megaRNPs are assembled and Fz2C is incorporated (1), the nuclear lamina is modified by aPKC (2), megaRNPs enter the membrane deformation (3) and are encapsulated by inner nuclear membrane (INM) (4), scission of the INM (5), NE bud fusion with the outer nuclear membrane (ONM) (6) and megaRNP exit into cytoplasm (7). (B–D″) Super-resolution micrograph projection (B) or single slice (D) of wild-type larval salivary gland nucleus stained with antibodies to Lamin B and Fz2C. Large Lamin B- and Fz2C-positive puncta indicate NE buds (n, nucleus; c, cytoplasm). (B′) High magnification view of highlighted region of B. (C) High magnification view of NE buds. (D′,D″) High magnification view of NE buds in D. (E,F) TEM micrographs taken at 800× (E) and 8000× (F) of wild-type larval salivary gland nucleus (cytoplasm false colored in green). (G) TEM micrographs (8000×) of NE buds (red arrowheads) in wild-type larval salivary gland nuclei showing the distribution of phenotypes observed (black arrows denote NPCs). (H–I′) TEM micrographs (800×; H,I or 8000×; H′,I′) of wash null (H,H′) and wash RNAi (I,I′) larval salivary gland nuclei. (J–L″) Salivary gland nuclei from wild-type (J–J″), wash null (K–K″), and wash RNAi (L–L″) larvae stained for Lamin B and Fz2C. (M) Quantification of the number of NE buds per nucleus in larval salivary glands (the size of the spots is proportional to number of nuclei with the indicated number of buds). The mean±95% c.i. is shown. P-values are indicated (Kruskal–Wallis test). Scale bars: 5 µm (B,D,E,H,I,J–L″); 0.5 µm in (B′,C,D′,D″,F,G,H′,I′).
NE budding was first demonstrated in Drosophila synapse development, proving to be essential for neuromuscular junction (NMJ) integrity. In this context, a C-terminal fragment of the Wingless receptor Fz2, Fz2C, was shown to associate with megaRNPs that formed foci at the nuclear periphery and exited the nucleus by budding through the nuclear envelope (Speese et al., 2012). Failure of this process resulted in aberrant synapse differentiation and impaired NMJ integrity (Speese et al., 2012). In a subsequent study, the NE budding pathway was shown to be necessary for the nuclear export of megaRNPs containing mitochondrial RNAs: disruption of NE budding led to deterioration of mitochondrial integrity and premature aging phenotypes that were similar to those associated with lamin mutations (i.e. laminopathies) (Jokhi et al., 2013; Li et al., 2016). Similar endogenous perinuclear foci/buds have been observed in plants and vertebrates, as well as other Drosophila tissues (i.e. larval salivary gland nuclei; Fig. 1B,C), suggesting that cellular NE budding is a widely conserved process (Hadek and Swift, 1962; Hochstrasser and Sedat, 1987; LaMassa et al., 2018; Panagaki et al., 2018 preprint; Parchure et al., 2017; Speese et al., 2012; Szollosi and Szollosi, 1988).
The spectrum of processes requiring this non-canonical nuclear exit pathway and the molecular machineries needed for this process, which encompasses membrane deformations, traversal across a membrane bilayer and nuclear envelope remodeling for a return to homeostasis, are largely unknown. One class of proteins that are involved in membrane–cytoskeletal interactions and organization is the Wiskott–Aldrich Syndrome (WAS) protein family (Takenawa and Suetsugu, 2007). WAS protein subfamilies are involved in a wide variety of essential cellular and developmental processes, as well as in pathogen infection and disease (Burianek and Soderling, 2013; Campellone and Welch, 2010; Rottner et al., 2010; Rotty et al., 2013; Takenawa and Suetsugu, 2007). WAS family proteins polymerize branched actin through the Arp2/3 complex, and often function as downstream effectors of Rho family GTPases (Campellone and Welch, 2010; Takenawa and Suetsugu, 2007). We identified Wash as a new WAS subfamily that is regulated in a context-dependent manner: Wash can bind directly to Rho1 GTPase (in Drosophila) or it can function along with the multi-protein WASH regulatory complex [SHRC; comprised of SWIP, Strumpellin, FAM21 and CCDC53 (also known as WASHC4, WASHC5, WASHC2 and WASHC3, respectively, in mammals)] (Derivery et al., 2009; Duleh and Welch, 2010; Gomez and Billadeau, 2009; Jia et al., 2010; Linardopoulou et al., 2007; Liu et al., 2009; Park et al., 2013; Veltman and Insall, 2010; Verboon et al., 2018, 2015a,b). Wash regulation by Rho family GTPases outside of Drosophila has not yet been described (Jia et al., 2010); instead its regulation has been characterized in the context of its SHRC. WASH and its SHRC are evolutionarily conserved and their mis-regulation is linked to cancers and neurodegenerative disorders (Leirdal et al., 2004; Linardopoulou et al., 2007; McGough et al., 2014; Nordgard et al., 2008; Ropers et al., 2011; Ryder et al., 2013; Türk et al., 2017; Valdmanis et al., 2007; Zavodszky et al., 2014). Importantly, we have shown that Wash is present in the nucleus where it interacts directly with B-type lamins and, when mutant,affects global nuclear organization/functions, as well as causing an abnormal wrinkled nucleus morphology reminiscent of that observed in diverse laminopathies (Verboon et al., 2015b,c). Mammalian WASH proteins have also been shown to localize to the nucleus in developmental and cell-type specific manners (Verboon et al., 2015c; Xia et al., 2014).
Here, we show that Wash, its SHRC, capping protein and Arp2/3 are also involved in the NE budding pathway, as mutants for any of these components lack Fz2C foci/lamin buds and display the NMJ integrity and premature aging phenotypes previously associated with the loss of NE budding. In addition, we find that CCDC53 and SWIP (SHRC subunits) colocalize with Fz2C foci/lamin buds. We show that Wash is present in several independent nuclear complexes. The nuclear interactions of Wash with its SHRC are separate from those with B-type Lamin, leading to effects on different subsets of nuclear Wash functions. We also find that Wash-dependent Arp2/3 actin nucleation activity is required for proper NE budding. We propose that Wash and its SHRC play a physical and/or regulatory role in the process of NE budding.
RESULTS
Drosophila Wash mutants lack NE buds
Drosophila larval salivary glands undergo NE budding; Fz2C foci can be observed surrounded by Lamin B at the nuclear periphery using confocal microscopy (Fig. 1B–D″) (see also Fig. S1A) or transmission electron microscopy (TEM; Fig. 1E,F) (Hochstrasser and Sedat, 1987). Through TEM, we observe that these megaRNPs are adjacent to a curved evagination of the nuclear membrane, suggesting that there is likely a membrane deformation event that precedes megaRNP encapsulation by the INM (40%, n=78 NE-budding events) (Fig. 1G). Finally, Fz2C antibody is a biomarker for nuclear buds (Speese et al., 2012), and colocalizes with these Lamin foci (Fig. 1B–D″,J–J″).
While staining wash mutants for Lamin B, we observed notably fewer Lamin ‘buds’ (for either A- or B-type Lamins) than in wild-type nuclei (Verboon et al., 2015b). To determine whether this reduction of Lamin buds was a result of disrupted NE budding, we co-labeled wash mutant larval salivary gland nuclei for Lamin B and Fz2C. We generated wash mutant salivary glands in two different ways: (1) using an outcrossed homozygous null allele, washΔ185hz(outX) (hereafter referred to as ‘wash null’; Verboon et al., 2018), and (2) expressing an RNAi construct for wash (HMC05339) specifically in the salivary gland using the GAL4-UAS system (hereafter referred to as ‘wash RNAi’) (Fig. 1J–M). We find that wash null and wash RNAi larval salivary gland nuclei exhibit an average of 0.17 (95% c.i. 0.1–0.25; n=101) and 0.1 (95% c.i. 0.04–0.16; n=104) Fz2C foci/NE buds, respectively, compared to an average of 6.58 (95% c.i. 6.02–7.16) Fz2C foci/NE buds in wild type (n=101, P<0.0001) (Fig. 1J–M). The phenotypes that we observe are similar to those reported previously for aPKC knockdowns (0.01, 95% c.i. 0–0.03; n=94; Fig. 1M) (Fradkin and Budnik, 2016; Jokhi et al., 2013; Li et al., 2016; Parchure et al., 2017). TEM sections from wash null and wash RNAi knockdown nuclei showed a similar lack of NE buds (n=50, n=50, respectively; Fig. 1H–I′).
Wash mutants display phenotypes associated with loss of NE budding
As our results indicate that Wash is involved in NE budding, we would expect wash mutants to exhibit the phenotypes associated with the two processes shown previously to be reliant on NE budding: NMJ development and mitochondrial integrity. Motor neurons in larval body wall muscle have synaptic boutons that are necessary for muscle contractility (Menon et al., 2013). In wild-type larvae, these boutons are largely mature and, on average, contain 1.6 per 100 boutons (n=664) that exhibit an absence of the neuronal differentiation marker Discs large 1 (Dlg1, referred to here as DLG) known as the ‘ghost bouton’ phenotype (Fig. 2A,D). Motor neurons from wash null and wash RNAi larval body wall muscle have an increased number of ghost boutons: 8.5 ghost boutons per 100 boutons (n=570, P≤0.0001) and 8.4 ghost boutons per 100 boutons (n=649, P≤0.0001), respectively (Fig. 2B–D), consistent with phenotypes observed under conditions known to disrupt nuclear budding (Speese et al., 2012).
Fig. 2.
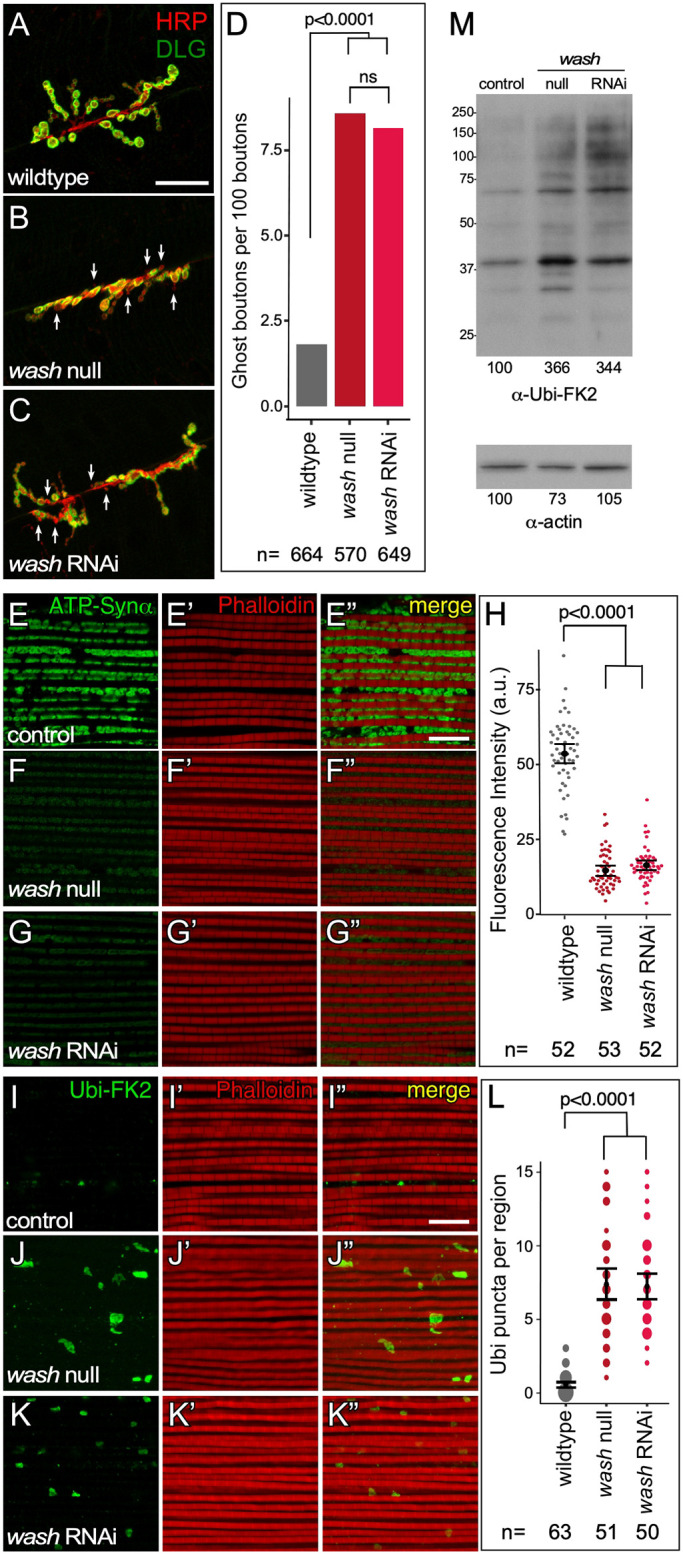
Wash mutant nuclei display NE bud associated phenotypes. (A–C) Confocal micrograph projection of synaptic boutons from wild-type (A), wash null (B), and wash RNAi (C) larval body wall muscle showing HRP and DLG expression. The ghost bouton phenotype designated by loss of DLG staining (arrows). (D) Quantification of ghost bouton frequency in larval body wall muscle neurons. (E–G″) Confocal micrograph projection of adult IFM from wild-type (E–E″), wash null (F–F″) and wash RNAi (G–G″) flies aged 21 days stained for the activity-dependent mitochondrial marker ATP-Synthetase α (ATP-Synα) and with phalloidin. (H) Quantification of mitochondrial fluorescence intensity in IFM. (I–K″) Confocal micrograph projection of adult IFM from wild-type (I–I″), wash null (J–J″), and wash RNAi (K–K″) flies aged 21 days stained with the poly-ubiquitin aggregate marker Ubi-FK2 and phalloidin. (L) Quantification of number of large Ubi-FK2 marks per muscle region in IFM (the size of the spots is proportional to number of instances of the indicated of punta per region seen for each fly line). (M) Western blot of adult IFM lysates from control, wash null, and wash RNAi flies aged for 21 days showing poly-ubiquitin aggregate protein levels, and actin loading control. P-values are indicated; ns, not significant [two-tailed Fisher's exact test (D); two-tailed Student's t-test (H); Kruskal–Wallis test (L)]. The mean±95% c.i. is shown for H and L. Scale bars: 20 µm (A–C); 10 µm (E–G″,I–K″).
We next looked at whether wash affects mitochondrial integrity in the indirect flight muscle (IFM) associated with the NE-budding process by using the activity-dependent mitochondrial marker ATP-Synthetase α. wash null and wash RNAi IFMs from adults aged 21 days show a 3.7- (n=53) and 3.2- (n=52) fold decrease, respectively (P<0.0001), in mitochondrial activity compared to a wild-type control (Fig. 2E–H). IFM from wash null and wash RNAi flies aged for 21 days also show an increase in poly-ubiquitin aggregates (a marker of mitochondrial damage), as muscle regions from these mutants show an average of 7.7 (n=51) and 7.4 (n=50) large poly-ubiquitin positive puncta, while IFM from control flies show just an average of 0.5 (n=63) (Fig. 2I–L). Tissue lysates from wash null and wash RNAi IFMs from adults aged for 21 days show a similar increase of poly-ubiquitin aggregates by western blotting, indicating mitochondrial damage (Fig. 2M). Taken together these data indicate that Wash plays a fundamental role in NE budding.
SHRC regulates Wash activity in NE budding and accumulates at NE budding sites
Drosophila Wash undergoes context-dependent regulation: it can function either as a direct effector of the Rho1 small GTPase or as part of the SHRC (Liu et al., 2009; Verboon et al., 2018, 2015a). To determine whether Wash interacts with Rho1 or SHRC in the NE budding context, we stained larval salivary glands with antibodies specific to Wash, Rho1 and each of the four SHRC components (CCDC53, Strumpellin, SWIP and FAM21) (Magie et al., 2002; Rodriguez-Mesa et al., 2012; Verboon et al., 2015a). Interestingly, while Wash and all four SHRC proteins are highly expressed in the nucleus, Rho1 is not (Fig. 3A–F″; Fig. S1B), indicating that the SHRC, but not Rho1, could regulate Wash activity in the NE budding context. Importantly, the SHRC components CCDC53 and SWIP are enriched at NE buds (Fig. 3G–M″), supporting a role for these proteins in NE budding. CCDC53 is particularly enriched at the base (neck) of NE buds (Fig. 3H–H″,K–K″).
Fig. 3.
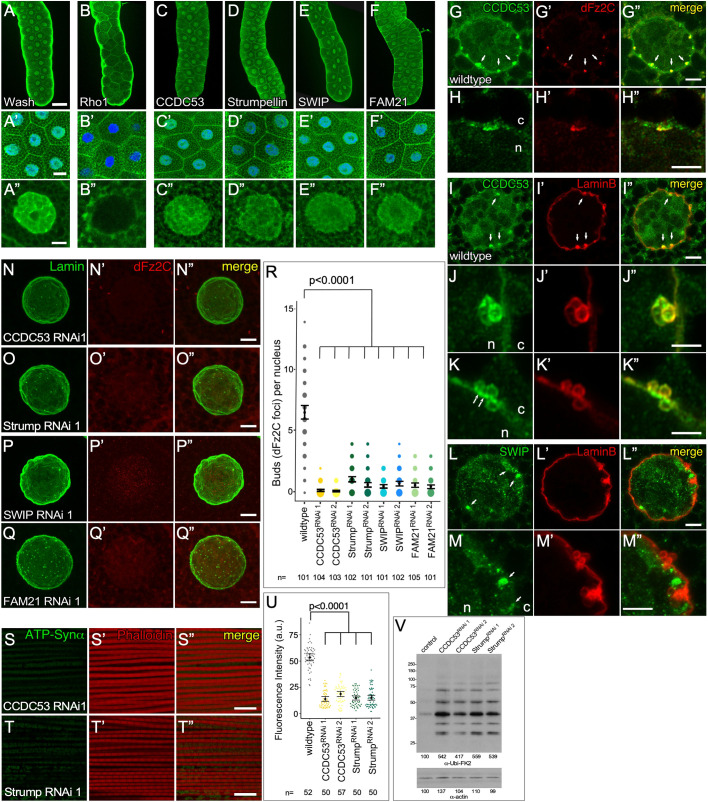
SHRC is expressed in the nucleus and accumulates at NE buds, and SHRC mutants lack NE buds and display downstream NE-budding-associated phenotypes. (A–F″) Confocal micrograph projection of wild-type larval salivary glands showing expression of Wash (A–A″), Rho1 (B–B″) and SHRC subunits [CCDC53 (C–C″), Strumpellin (D–D″), SWIP (E–E″), and FAM21 (F–F″)]. DAPI staining (blue) marks nuclei in B′–F′. (G–G″) Single slice super-resolution (Airyscan) micrograph of wild-type larval salivary gland nucleus showing CCDC53 enrichment at NE bud sites and colocalization with Fz2C (arrows). (H–H″) High magnification view of single budding site from G. (I–M″) Single slice super-resolution micrograph of wild-type larval salivary gland nucleus showing CCDC53 (I–I″) and SWIP (L–L″) enrichment at NE bud sites and colocalization with Lamin B puncta at the nuclear periphery (arrows). High magnification views of CCDC53 (J–K″) or SWIP (M–M″) colocalization with Lamin B at NE buds. (N–Q″) Confocal micrograph projections of CCDC53 RNAi (N–N″), Strumpellin RNAi (O–O″), SWIP RNAi (P–P″) and FAM21 RNAi (Q-Q″) larval salivary gland nuclei stained with Lamin B and Fz2C. (R) Quantification of NE buds per nucleus from two independent RNAi lines per SHRC subunit (the size of the spots is proportional to number of nuclei with the indicated number of buds). (S–T″) Confocal micrograph projections of adult IFM from CCDC53 RNAi (S–S″) and Strumpellin (T–T″) RNAi flies aged 21 days stained for the activity-dependent mitochondrial marker ATP-Synthetase α (ATP-Synα) and with phalloidin. (U) Quantification of ATP-Synα fluorescence intensity in adult IFM. (V) Western blot of adult IFM lysates from control, CCDC53 RNAi, and Strumpellin RNAi flies aged 21 days showing poly-ubiquitin aggregate protein levels, and actin loading control. P-values are indicated [two-tailed Student's t-test (U); Kruskal–Wallis test (R)]. The mean±95% c.i. is shown for R and U. Scale bars: 100 µm (A–F); 20 µm (A′–F′); 5 µm (A″–G″,I–I″,L–L″,N–Q″); 0.5 µm (H–H″,J–K″,M-M″); 10 µm (S–T″). n, nucleus; c, cytoplasm.
To determine whether SHRC activity is required for NE budding, we knocked down individual SHRC components by expressing RNAi constructs (two independent RNAi constructs for each subunit) specifically in the larval salivary gland using the GAL4-UAS system (Fig. 3N–Q″; Fig. S1C–F″). We co-labeled these SHRC knockdown larval salivary gland nuclei for Lamin B and Fz2C and found that all four SHRC knockdowns show a significant decrease in NE buds similar to that of wash null and wash RNAi larval salivary gland nuclei (Fig. 3N–Q″; Fig. S1C–F″). The number of Fz2C foci/NE buds per nucleus was quantified for each line (Fig. 3R) and we find that all lines show 0.1–1.1 (95% c.i. 0.07–1.3) foci/buds per nucleus on average (n>100 and P<0.0001 in each case) compared to 6.58 (95% c.i. 6.02–7.16) foci/buds in control nuclei (n=100, P<0.0001). Of note, the nuclei from SHRC-knockdown larval salivary glands are spherical and morphologically indistinct from wild-type nuclei, suggesting that the wrinkled nuclear phenotype observed in wash mutants (Verboon et al., 2015b) is SHRC independent.
Consistent with their loss of Fz2C foci/NE buds, SHRC component knockdowns also exhibit phenotypes associated with disrupted NE budding (Fig. 3S–V). Adult IFM from 21-day-old Strump or CCDC53 knockdown flies show a decrease in mitochondrial activity, as measured by determining ATP-Synthetase α levels (Fig. 3S–U; Fig. S1G–H″). Strump RNAi1 and RNAi2 show a 3.6- and 3.5-fold decrease in activity compared to wild type (n=50 and n=50, P<0.0001) and CCDC53 RNAi1 and RNAi2 show a 3.8- and 2.9-fold decrease in activity compared to wild type (n=50 and n=57, P<0.0001) (Fig. 3U). Western blots of IFM lysates from these knockdowns also show an increase in poly-ubiquitin aggregates, indicating mitochondrial damage (Fig. 3V). Taken together, our results are consistent with nuclear Wash functioning, together with its SHRC, in NE budding.
Lamin meshes are required for nuclear budding
We were next interested in understanding the function of Wash and its regulation by the SHRC during NE budding. Since mutations in Lamin A/C (A-type) disrupt NE budding (Li et al., 2016; Parchure et al., 2017), and we had shown previously that Wash interacts directly with Lamin B (B-type) (Verboon et al., 2015b), we investigated whether NE-budding phenotypes were due to a structural defect in Lamin B organization. Lamin B and Lamin A/C form homotypic meshes underlying the INM (Shimi et al., 2015) and, due to this, Lamin B might also affect NE budding. Indeed, salivary gland nuclei from Lamin B RNAi knockdowns exhibit characteristic wrinkled nuclei with a decrease in Fz2C foci/NE buds, with 2.19 (95% c.i. 1.96–2.42) buds per nuclei (n=171) (Fig. 4A,B). Consistent with this, adult IFMs from Lamin B RNAi flies aged 21 days show a 3.1-fold decrease in mitochondrial activity compared with wild type (n=50, P<0.0001), as assayed by determining ATP-Synthetase α levels (Fig. 4C,D). Additionally, western blots of adult IFM lysates from Lamin B RNAi flies aged for 21 days show an increase in poly-ubiquitin aggregates, indicating mitochondrial damage (Fig. 4E). These data suggest that alteration of nuclear lamina structure can reduce, but not eliminate, NE budding.
Fig. 4.
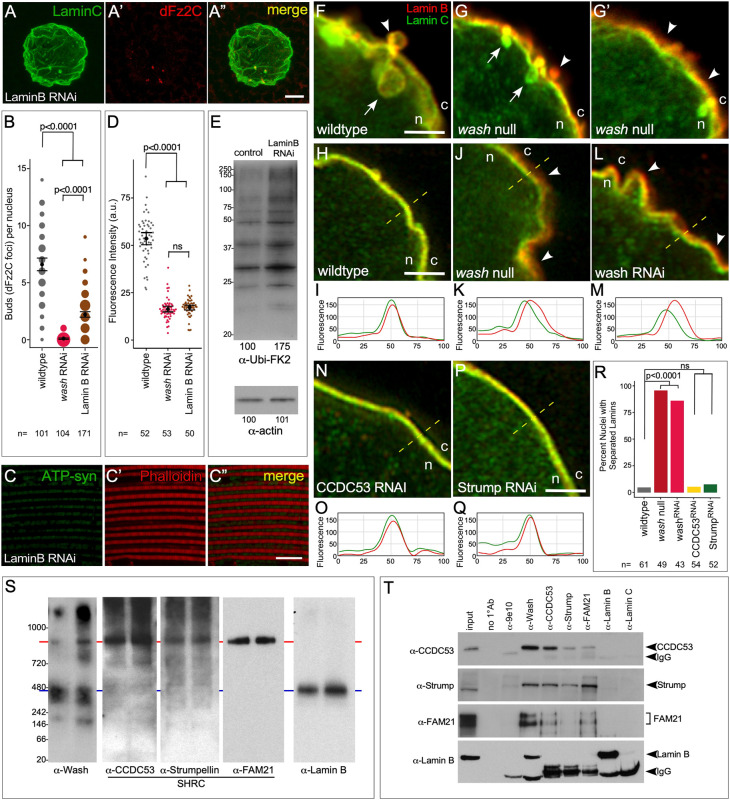
Alteration of the nuclear lamina structure can reduce, but does not eliminate, NE budding. (A) Confocal micrograph projection of Lamin B RNAi larval salivary gland nucleus co-labeled with antibodies to Lamin C and Fz2C. (B) Quantification of number of NE buds in control, wash RNAi, and Lamin B RNAi nuclei subunit (the size of the spots is proportional to number of nuclei with the indicated number of buds). (C) Confocal micrograph projection of adult IFM from Lamin B RNAi flies aged 21 days stained with ATP-Synthetase α (ATP-Syn) and with phalloidin. (D) Quantification of ATP-Syn α fluorescence intensity in adult IFM. (E) Western blot of adult IFM lysates from control and Lamin B RNAi flies aged 21 days showing poly-ubiquitin aggregate protein levels, and actin loading control. (F–G′) Single slice confocal micrographs of nuclear periphery from wild-type (F) and wash null (G,G′). In F and G, arrows indicate inward projecting NE buds and arrowheads indicate outward projecting NE buds. Arrowheads in G′ indicate areas of Lamin B/Lamin C separation. (H–Q) Single slice super-resolution micrographs of larval salivary gland nuclei (H,J,L,N,P) and line plots of regions indicated by dashed lines (I,K,M,O,Q) from wild-type (H,I), wash null (J,K), wash RNAi (L,M), CCDC53 RNAi (N,O), and Strumpellin RNAi (P,Q) showing Lamin B and Lamin C organization at the nuclear periphery. Arrowheads in J and L indicate areas of Lamin B/Lamin C separation. Lamin C also shows lower level uniform distribution within the nucleus. (R) Quantification of the percentage of salivary gland nuclei showing Lamin B and Lamin C separation. (S) Western blots from Blue Native PAGE of wild-type nuclear extracts probed with antibodies to Wash, SHRC subunits (CCDC53, Strumpellin and FAM21), and Lamin B. A putative ∼900 kDa complex with Wash and SHRC (red line) and ∼450 kDa complex with Wash and Lamin B (blue lines) are indicated. (T) Western blots of immunoprecipitations from wild-type nuclear extracts with no primary antibody included (no 1° AB), a non-specific antibody (9e10), Wash, CCDC53, Strumpellin, FAM21, Lamin B and Lamin C. Blots were probed with antibodies to CCDC53, Strumpellin, FAM21 and Lamin B as indicated. P-values are indicated [two-tailed Fisher's exact test (R); two-tailed Student's t-test (D); Kruskal–Wallis test (B)]. The mean± 95% c.i. is shown for B and D. Scale bars: 5 µm (A–A″); 1 µm (F–H,J,L,N,P); 10 µm (C–C″). n, nucleus; c, cytoplasm.
Wash, but not the SHRC, interacts with and disrupts homotypic Lamin meshes
Interestingly, our previous data suggested that the involvement of Wash with Lamin B might be separate from its involvement with the SHRC, as Wash mutants exhibit a wrinkled nuclear morphology (Verboon et al., 2015b), whereas SHRC mutants do not (Fig. 1K–L″ versus Fig. 3N–Q″). How the Lamin B and Lamin A/C networks are organized with respect to each other or the INM is not well understood (Shimi et al., 2015). However, factors that disrupt this homotypic meshwork may disrupt the structural platform at/upon which NE budding occurs. We next analyzed the structure of the nuclear lamina in Wash and SHRC mutants. Wild-type larval salivary gland nuclei co-labeled with antibodies to Lamin B and Lamin C show tight Lamin B and Lamin C colocalization around the nuclear periphery (Fig. 4F,H,I,R). Strikingly, in nuclei from wash null and wash RNAi larval salivary glands, the signals from the two Lamins are separated, with Lamin B lying closest to the INM (consistent with it encoding a CAAX domain) and Lamin C positioned closest to the chromatin (consistent with Lamin C association with chromatin) (Fig. 4G,G′,J–M,R). Importantly, nuclei from SHRC subunit knockdowns (CCDC53 RNAi and Strump RNAi) have Lamin B and Lamin C organization that is indistinguishable from wild type (Fig. 4N–R).
As an orthogonal means of determining whether Wash functions separately to the SHRC to influence the nuclear lamina, we separated protein complexes from fly Kc cell nuclear lysates using Blue Native PAGE. We observed that Wash is present in multiple nuclear complexes, suggesting it is involved in multiple nuclear processes (Fig. 4S). We find that one major Wash-containing complex (∼900 kDa) overlaps with a complex containing CCDC53, Strumpellin and FAM21 (Fig. 4S), whereas a separate Wash-containing complex (∼450 kDa) overlaps with a Lamin B-containing complex (Fig. 4S). To confirm that Wash forms distinct complexes with the SHRC components and with Lamin B in the nucleus, we immunoprecipitated Wash, three SHRC subunits (CCDC53, Strumpellin and FAM21), Lamin B and Lamin C from fly cell nuclear lysates and probed the resulting western blots for SHRC and Lamin B (Fig. 4T). In each case, Wash pulls down the corresponding SHRC subunit, the SHRC subunit pulls itself down, and SHRC complex members co-immunoprecipitated with each other (Fig. 4T). However, while Wash pulls down Lamin B, none of the SHRC subunits co-immunoprecipitated with Lamin B or Lamin C. These results are consistent with Wash forming distinct nuclear complexes with Lamin B and with the SHRC, and suggest that Wash may have two roles in NE budding: an indirect role on Lamin B to regulate the Lamin meshwork, and a direct role with its SHRC in the physical formation of NE buds.
Wash–SHRC interactions are required for NE budding, whereas Wash–Lamin B interaction is required for nuclear morphology
To further delineate Wash–Lamin B versus Wash–SHRC function in NE budding, we mapped the sites on the Wash protein that facilitate binding to Lamin B and CCDC53 (the interaction of Wash with this SHRC subunit has been reported to facilitate stable Wash–SHRC formation; Derivery et al., 2009; Jia et al., 2010; Rottner et al., 2010; Verboon et al., 2018), then made specific point mutations that functionally block these interactions (see Materials and Methods) (Fig. S2A). The final constructs, harboring point mutations in the context of the full-length Wash protein, designated washΔSHRC and washΔΔLamB, respectively, were examined for interaction specificity using GST pulldown assays (Fig. S2B). Transgenics generated with these Wash point mutations (see Materials and Methods), as well as a wild-type Wash rescue construct (washWT), were individually crossed into the wash null homozygous background so that the only Wash activity comes from the transgene under control of the endogenous wash promoter. The wild-type version of these transgenics (washWT) is expressed in both the nucleus and cytoplasm (Verboon et al., 2015b) and rescues previously described wash mutant phenotypes, including its premature ooplasmic streaming phenotype in oocytes (Fig. S2C,C′,G) (Liu et al., 2009; Verboon et al., 2018). The other two wash point mutation transgenic lines are similarly functional; as expected, washΔΔLamB rescues the premature ooplasmic streaming phenotype, whereas washΔSHRC did not (Fig. S2D–G). Interestingly, while the knockdown of Wash downregulates expression of the SHRC and vice versa (Derivery et al., 2009; Jia et al., 2010; Verboon et al., 2018), Wash and CCDC53 are still present in washΔSHRC mutant nuclei despite their inability to bind to each other (Fig. S2H–K″).
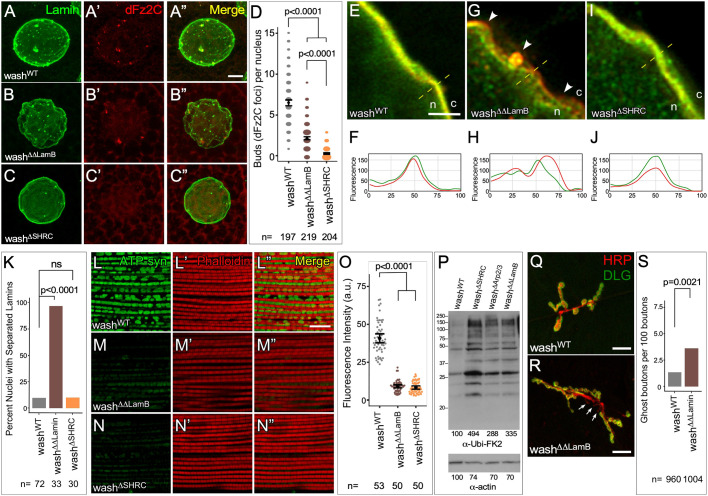
Salivary gland nuclei from washWT larvae show a rescue of NE budding: nuclei from these mutants show 6.58 (95% c.i. 6.22–6.95) buds per nuclei (n=102; P=0.895 compared to wild-type) and nuclear morphology is indistinguishable from that in wild type (Fig. 5A–A″,D). However, nuclei from washΔSHRC point mutants show 0.5±0.1 buds per nucleus (n=101, P<0.0001), but had nuclear morphology indistinguishable from washWT (Fig. 5C,D). These data strongly suggest that Wash, under the regulatory control of the SHRC, is required for NE budding. Remarkably, larval salivary gland nuclei from the washΔΔLamB point mutants (Fig. 5B–B″,D) have a less-severe intermediary phenotype with 1.5±0.1 NE buds per nucleus, a significantly greater number of NE buds than that exhibited by the washΔSHRC point mutant (n=104, P<0.0001). These results suggest that Wash has two independent effects on NE buds: an effect on Lamin activity (washΔΔLamB) and an effect on actin-related activity (washΔArp2/3 or washΔSHRC). The wash null mutation exhibits both effects. Using multivariate modeling (Poisson regression) of the count of buds versus disrupted lamin and actin activity, we find a much smaller effect on the count of nuclear buds in washΔΔLamB versus washΔArp2/3 or washΔSHRC, where the log of expected buds decreases by 0.99 (P<0.0001) for disrupted lamin activity versus 3.14 (P<0.0001) for disrupted actin activity. The significance of both disruptions reinforces our hypothesis that these are indeed independent effects. washΔΔLamB also exhibit a wrinkled nuclear morphology similar to wash null mutants, wash RNAi knockdowns and Lamin B RNAi knockdowns (Fig. 5B) (Verboon et al., 2015b). Super-resolution microscopy analysis of Lamin B and Lamin C organization in salivary gland nuclei from the wash point mutants shows that washΔΔLamB exhibits separated Lamin B and C meshes, while washWT and washΔSHRC has Lamin organization that was indistinguishable from wild type, consistent with the specificity of these mutations (Fig. 5E–K).
Fig. 5.
Wash point mutants show separation of phenotypes for specific Wash activities. (A–C″) Confocal micrograph projections of larval salivary gland nuclei from washWT (A–A″), washΔΔLamB (B–B″), and washΔSHRC (C–C″) stained for Lamin B and Fz2C. (D) Quantification of NE buds per nucleus in larval salivary gland nuclei (the size of the spots is proportional to number of nuclei with the indicated number of buds). (E–J) Single slice super-resolution micrographs of larval salivary gland nuclei (E,G,I) and line plots of regions indicated by dashed lines (F,H,J) from washWT (E,F), washΔΔLamB (G,H), and washΔSHRC (I,J) showing Lamin B and Lamin C organization at the nuclear periphery. Arrowheads in G indicate areas of Lamin B/Lamin C separation; n, nucleus; c, cytoplasm. (K) Quantification of the percentage of salivary gland nuclei showing Lamin B and Lamin C separation. (L–N″) Confocal micrograph projections of adult IFM from washWT (L–L″), washΔΔLamB (M–M″) and washΔSHRC (N–N″) flies aged 21 days stained for the activity-dependent mitochondrial marker ATP-Synthetase α (ATP-Syn) and with phalloidin. (O) Quantification of ATP-Syn α fluorescence intensity from adult IFMs. (P) Western blot of adult IFM lysates from washWT, washΔΔLamB, washΔSHRC and washΔArp2/3 flies aged 21 days showing poly-ubiquitin aggregate protein levels, and actin loading control. (Q,R) Confocal micrograph projection of synaptic boutons from washWT (Q) and washΔΔLamB (R) larval body wall muscle co-stained for HRP and DLG. Ghost boutons are indicated (arrows). (S) Quantification of ghost bouton frequency in larval body wall muscle neurons. P-values are indicated [two-tailed Fisher's exact test (K,S); two-tailed Student's t-test (O); Kruskal–Wallis test (D)]. The mean±95% c.i. is shown for D and O. Scale bars: 5 µm (A–C″); 1 µm (E,G,I); 10 µm (L–N″); 20 µm (Q,R).
As expected due to an overall reduction in NE buds, IFM from 21-day-old washΔSHRC and washΔΔLamB point mutants both show a decrease in mitochondrial activity, as assayed through assessing the level of ATP-Synthetase α (Fig. 5L–O). IFMs from washΔSHRC and washΔΔLamB flies show a 5.0- (n=50) and 4.5- (n=50) fold decrease in mitochondrial activity, respectively, when compared to IFMs from the washWT construct (P<0.0001 in each case). Additionally, western blots of IFM lysates from washΔSHRC and washΔΔLamB show an increase in poly-ubiquitin aggregates compared to the washWT construct, indicating mitochondrial damage (Fig. 5P). Western blots of IFM lysates from washΔSHRC show an increase in poly-ubiquitin aggregates compared to in the washWT construct, indicating mitochondrial damage (Fig. 5P). IFM lysates from washΔΔLamB show a less-pronounced intermediate increase in poly-ubiquitin aggregates (Fig. 5P). Larval body wall muscle from washΔΔLamB mutants exhibit an increased number of ghost boutons: 3.6 ghost boutons per 100 boutons (n=1004) compared to control (washWT; 1.4 ghost boutons per 100 boutons; n=960; P=0.0021), consistent with the intermediate Fz2C foci/NE bud phenotype displayed by these mutants (Fig. 5Q–S). Thus, these Wash point mutations clearly demonstrate that Wash has dual roles in NE budding.
Wash is required for the initial steps of NE bud formation
To understand how Wash and its SHRC fits into the physical NE budding pathway, we examined its relationship to previously proposed cellular events comprising NE budding. Based on the lack of NE buds observed in aPKC mutants and the requirement for lamin phosphorylation in herpesvirus nuclear egress, aPKC has been proposed to phosphorylate Lamin thereby seeding sites that can undergo NE budding (Speese et al., 2012). As both wash and aPKC mutations almost completely abolish NE budding, epistasis experiments are not feasible. In addition, knockdown of Drosophila Wash downregulates expression of the SHRC members and vice versa (Fig. S1I–L″; Verboon et al., 2018). However, because the SHRC member CCDC53 accumulates at budding sites, we looked at localization of CCDC53 in aPKC knockdown salivary gland nuclei. Consistent with the proposed role for aPKC, CCDC53 does not accumulate in foci, nor are NE buds present in this background, which suggests that Wash and the SHRC function after aPKC in this process (Fig. 6A–A″).
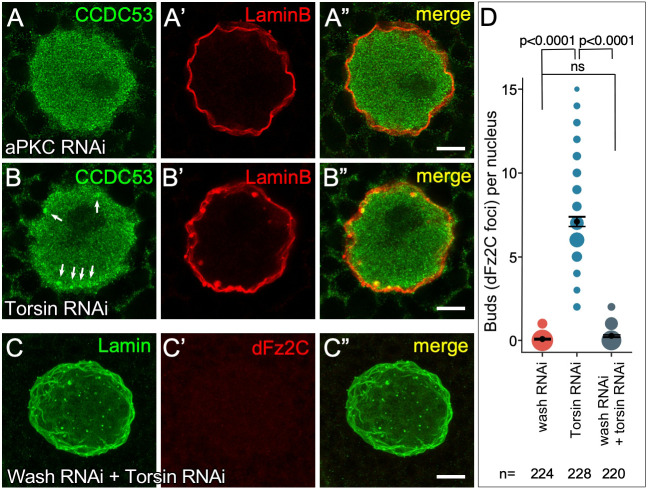
Fig. 6.
Wash acts prior to NE buds being pinched off from the inner nuclear membrane. (A–A″) Single slice super-resolution micrograph from an aPKC RNAi larval salivary gland nucleus showing loss of CCDC53 enrichment at NE bud sites and lack of colocalization with Lamin B at the nuclear periphery. (B–B″) Single slice super-resolution micrograph from a Torsin RNAi larval salivary gland nucleus showing CCDC53 enrichment at NE bud sites and colocalization with Lamin B at the nuclear periphery (arrows). (C–C″) Confocal micrograph projection of larval salivary gland nuclei from a Wash RNAi +Torsin RNAi double knockdown stained for Lamin B and Fz2C. (D) Quantification of number of Fz2C foci/NE buds per nucleus in wash RNAi, torsin RNAi, and wash RNAi+torsin RNAi nuclei (the size of the spots is proportional to number of nuclei with the indicated number of buds). P-values indicated (Kruskal–Wallis test). The mean±95% c.i. is shown. Scale bars: 5 µm.
Torsin, an AAA ATPase, has been proposed to function in NE bud scission from the inner nuclear membrane (INM) as Torsin accumulates at sites of contact between the megaRNP and INM, and torsin mutants exhibit accumulation of INM-tethered megaRNPs within the perinuclear space (Jokhi et al., 2013). To determine whether Wash and its SHRC are required for the initial steps of NE bud formation, and in particular before these buds pinch off from the INM, we also looked at the localization of CCDC53 in torsin RNAi knockdown salivary gland nuclei. Consistent with established torsin phenotypes, we find increased numbers of NE buds, and importantly, these buds colocalize with CCDC53 foci (Fig. 6B–B″), suggesting that torsin acts after Wash/SHRC role in this pathway. To verify this, we looked at epistasis by generating wash and torsin double RNAi knockdown larval salivary gland nuclei and co-staining them for Lamin B and Fz2C. We found that wash and torsin double RNAi larval salivary gland nuclei exhibited an average of 0.27 (95% c.i. 0.21−0.34) Fz2C foci/NE buds (n=220), compared to an average of 6.58 (95% c.i. 6.02−7.16) Fz2C foci/NE buds in wild type (n=100, P<0.0001) (Fig. 6C–D) and 0.1 (95% c.i. 0.04−0.16) and 7.09 (95% c.i. 6.79−7.37) Fz2C foci/NE buds in wash RNAi and torsin RNAi nuclei, respectively. Taken together, these results demonstrate that Wash and the SHRC act in the early steps of NE budding, between the modification of lamins proposed to establish NE budding sites and those buds being pinched off from the INM.
The Arp2/3 complex is required for NE budding
As WAS family proteins have been implicated in membrane–cortical cytoskeleton interactions leading to membrane deformations (i.e. protrusions and endocytosis) in the cytoplasm, we explored whether or not these might have a similar role in NE budding. Wash encodes several biochemical activities, including actin nucleation and actin and microtubule (MT) binding, bundling and crosslinking (Liu et al., 2009; Verboon et al., 2018, 2015a); however, when working in concert with the SHRC, Wash often functions upstream of the Arp2/3 complex to promote branched actin filament formation (Alekhina et al., 2017; Rottner et al., 2010). Mounting evidence has also demonstrated roles for actin and the Arp2/3 complex in diverse nuclear functions (Alekhina et al., 2017; Kyheröinen and Vartiainen, 2019; Percipalle and Vartiainen, 2019). If Wash requires its actin nucleation activity for NE budding, we would expect Arp2/3 complex subunit mutations to exhibit reduced NE bud formation and phenotypes associated with disrupted NE budding. We examined the expression and knockdown effects of two different Arp2/3 complex subunits. The Arp3 and Arpc1 subunits are expressed in fly Kc cell nuclear lysates, as assayed by western blotting (Fig. 7A,B). Staining larval salivary gland nuclei with antibodies to the Arpc1 subunit revealed that Arp2/3 is enriched around NE buds (Fig. 7C–D″). We generated RNAi knockdowns of Arp3 and Arpc1 in larval salivary glands as previously described and find that these nuclei had on average 0.79 (95% c.i. 0.62−0.97; n=102) and 1.03 (95% c.i. 0.84−1.23; n=102) buds, respectively, compared to 6.58 (95% c.i. 6.02−7.16) buds per nucleus in wild type (n=102; P<0.0001 in each case) (Fig. 7E–G). Additionally, nuclei from Arp3 and Arpc1 knockdown animals are spherical (similar to those in wild-type and SHRC knockdowns), consistent with Wash-dependent Arp2/3 activity being separate from nuclear morphology.
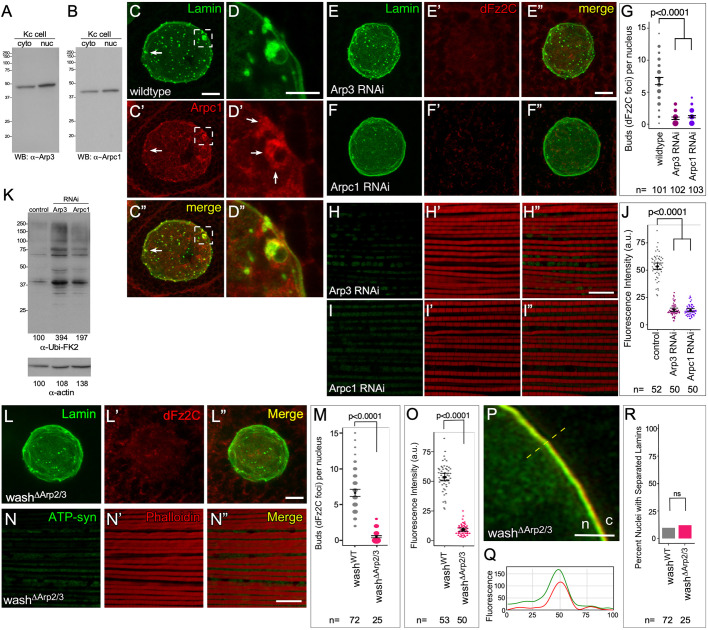
Fig. 7.
Arp2/3 activity is required for NE budding. (A,B) Western blot of Kc cell cytoplasmic and nuclear extracts probed with antibodies to Arp2/3 subunit Arp3 (A) or Arpc1 (B). (C–C″) Confocal micrograph projections of wild-type larval salivary gland nucleus showing Arpc1 colocalization with Lamin B puncta at the nuclear periphery (arrows, box). (D–D″) High magnification views of Arpc1 enrichment around NE bud boxed in C′ (arrows). (E–F″) Confocal micrograph projections of Arp3 RNAi (E–E″) and Arpc1 RNAi (F–F″) salivary gland nuclei stained for Lamin B and Fz2C. (G) Quantification of NE buds per nucleus in larval salivary glands (the size of the spots is proportional to number of nuclei with the indicated number of buds). (H–I″) Confocal micrograph projections of adult IFM from Arp3 RNAi (H–H″) and Arpc1 RNAi (I–I″) flies aged 21 days stained for ATP-Synthetase α (ATP-Syn) and with phalloidin. (J) Quantification of ATP-Syn α fluorescence intensity in adult IFM. (K) Western blot of adult IFM lysates from control, Arp3 RNAi and Arpc1 RNAi flies aged 21 days showing poly-ubiquitin aggregate protein levels, and actin loading control. (L–L″) Confocal micrograph projections of washΔArp2/3 larval salivary gland nucleus stained for Lamin B and Fz2C. (M) Quantification of NE buds per nucleus in larval salivary glands glands (the size of the spots is proportional to number of nuclei with the indicated number of buds). (N–N″) Confocal micrograph projections of adult IFM from washΔArp2/3 flies aged 21 days stained for ATP-Syn α and with phalloidin. (O) Quantification of ATP-Syn α fluorescence intensity in adult IFM. (P,Q) Single slice super-resolution micrographs of larval salivary gland nucleus (P) and line plot of region indicated (Q) from washΔArp2/3 showing Lamin B and Lamin C organization at the nuclear periphery. n, nucleus; c, cytoplasm. (R) Quantification of the percentage of salivary gland nuclei showing Lamin B and Lamin C separation. P-values indicated [two-tailed Fisher's exact test (P); two-tailed Student's t-test (H,M); Kruskal–Wallis test (E,K)]. The mean±95% c.i. is shown for G, J, M and O. Scale bars: 5 µm (C–C″,E–F″,L–L″); 1 µm (P); 10 µm (H–I″,N–N″); 0.5 µm (D–D″).
As expected given their reduction of Fz2C foci/NE buds, Arp3 and Arpc1 knockdowns also exhibit the phenotypes linked to disruption of NE budding. IFM from 21-day-old Arp3 and Arpc1 knockdown flies show a decrease in mitochondrial activity, as assayed by determining the level of ATP-Synthetase α (Fig. 7H–J). Arp3 and Arpc1 RNAi knockdown IFMs show a 3.1-(n=50) and 3.9-(n=50) fold decrease in mitochondrial activity, respectively (P<0.0001 in each case). Western blots from extract of these tissues also show an increase in poly-ubiquitin aggregates, indicating mitochondrial damage (Fig. 7K).
To confirm that the interaction between Wash and Arp2/3 is required for its role in NE budding, we generated a point mutation in the conserved tryptophan residue (W498S) that, when mutated in mammalian WASP, abolished binding of WASP to Arp2/3 (Marchand et al., 2001) (Fig. S2A). This construct, harboring the point mutation in the context of the full-length Wash protein, designated washΔArp2/3, was examined for specificity using GST pulldown assays (Fig. S2B). washΔArp2/3 transgenics (see Materials and Methods) function as expected during oogenesis; it does not rescue the wash null ooplasmic streaming phenotype (Fig. S2F–G).
Larval salivary gland nuclei from washΔArp2/3 point mutants show significantly reduced NE buds, with only 0.51 (95% c.i. 0.35−0.53) buds per nucleus (n=102, P<0.0001), and exhibited nuclear morphology indistinguishable from washWT (Fig. 7L–M). Consistent with a requirement for its actin nucleation activity during NE budding, IFM from 21-day-old washΔArp2/3 point mutants show a 4.4-fold decrease in mitochondrial activity (n=50), as assayed by determining the level of ATP-Synthetase α (Fig. 7N–O). Additionally, western blots of IFM lysates from washΔArp2/3 show an increase in poly-ubiquitin aggregates, indicating mitochondrial damage (Fig. 5P). As might be expected, washΔArp2/3 does not exhibit separated Lamin B and Lamin C meshes (Fig. 7P–R). Taken together, our data are consistent with a model in which Wash and the SHRC work upstream of the Arp2/3 complex to promote NE budding, and that Wash functions with Lamin B and independently of the SHRC and Arp2/3 to affect nuclear morphology.
Capping protein is required for NE budding
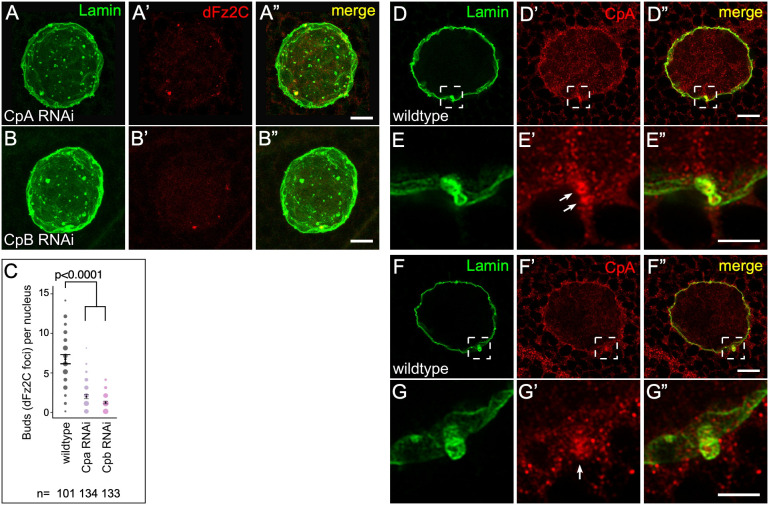
A subset of Wash–SHRC complexes are known to associate with the barbed end-binding heterodimeric capping protein (CapZα and CapZβ), which in turn have been shown to exhibit context-dependent functions ranging from promoting Arp2/3-dependent actin assemblies to inhibiting FAM21 activity (Amândio et al., 2014; Derivery et al., 2009; Edwards et al., 2014; Park et al., 2013; Rottner et al., 2010). To determine whether capping proteins (Cpa and Cpb in Drosophila) play a role in NE budding, we generated RNAi knockdowns of cpa and cpb in larval salivary glands. We find that these knockdown nuclei show a decrease in NE buds with on average 1.87 (95% c.i. 1.61−2.13; n=134) and 1.11 (95% c.i. 0.92−1.28; n=133) Fz2C foci/NE buds, respectively, compared to 6.58 (95% c.i. 6.02−7.16) Fz2C foci/NE buds per nucleus in wild type (n=102; P<0.0001 in each case) (Fig. 8A–C). Similar to SHRC and Arp2/3 knockdowns, nuclei from cpa and cpb knockdown animals are spherical rather than wrinkled. We also examined the expression of the Cpa subunit in larval salivary gland nuclei and found that, along with a general nuclear distribution, Cpa is enriched at lamin buds (Fig. 8D–G″), supporting a role for capping protein in NE budding.
Fig. 8.
Capping protein is required for NE budding. (A–B″) Confocal micrograph projections of CpA RNAi (A–A″) and CpB RNAi (B–B″) salivary gland nuclei stained for Lamin B and Fz2C. (C) Quantification of NE buds per nucleus in larval salivary glands (the size of the spots is proportional to number of nuclei with the indicated number of buds). (D–G″) Single slice super-resolution micrograph of wild-type larval salivary gland nucleus showing CpA enrichment around NE bud sites and colocalization with Lamin B puncta at the nuclear periphery (box; D,D′,F–F″). High magnification views of CpA enrichment around Lamin B at NE buds (arrows; E–E″,G–G″). P-values indicated [Kruskal–Wallis test (C)]. The mean±95% c.i. is shown for C. Scale bars: 5 μm (A–B″,D–D″,F–F″); 0.5 μm (E–E″,G–G″).
DISCUSSION
NE budding is an increasingly appreciated pathway for nuclear export of large macromolecular machineries, such as megaRNPs involved in the co-regulation of major developmental pathways or unwanted RNA or protein aggregates. We previously showed that Drosophila Wash is present in the nucleus where it is likely involved in a number of different nuclear processes (Verboon et al., 2015b). Here, we show that all members of the four-subunit Wash regulatory complex (SHRC), as well as the heterodimeric capping protein, are also present within the nucleus and, along with Wash, are necessary for NE budding. We show that Wash and SHRC act early in the NE budding pathway, and this process requires actin nucleation activity of Wash achieved through its interaction with Arp2/3. Loss of Wash, any of its SHRC subunits or Arp2/3 leads to the loss of Fz2C foci/NE buds, and mutants for these factors exhibit phenotypes associated with the two cellular processes shown to require NE budding: aberrant synaptic ‘ghost’ bouton formation leading to disrupted NMJ integrity, and mitochondrial degeneration associated with premature aging phenotypes (Li et al., 2016; Speese et al., 2012). We also show that the interaction of Wash with Lamin B results in a general disruption of the nuclear lamina and separation of the Lamin B/Lamin C homotypic meshes, leading to inefficient, rather than loss of, NE bud formation.
While the spectrum of processes that require this alternate nuclear egress mechanism is not yet known, SHRC components are linked to neurodegenerative disorders, including hereditary spastic paraplegias, Parkinson disease, amyotrophic lateral sclerosis (ALS) and Hermansky–Pudlak syndrome (McGough et al., 2014; Ropers et al., 2011; Ryder et al., 2013; Song et al., 2018; Türk et al., 2017; Valdmanis et al., 2007; Zavodszky et al., 2014). As an increasing number of neurodegenerative diseases and myopathies have been associated with the accumulation of RNA–protein aggregates in the nucleus, NE budding may be part of the endogenous cellular pathway for removing such aggregates/megaRNPs from the nucleus in normal cells (Laudermilch et al., 2016; Parchure et al., 2017; Ramaswami et al., 2013).
Nuclear buds or NPCs?
The parallels between the mechanism of NE budding and herpesvirus nuclear egress, as well as the presence of similar endogenous perinuclear foci/buds in other plant and animal nuclei, has suggested that NE budding is a conserved endogenous cellular pathway for nuclear export (Fradkin and Budnik, 2016; Panagaki et al., 2018 preprint; Parchure et al., 2017; Speese et al., 2012). Indeed, INM-encapsulated electron-dense granules have been identified in yeast and Torsin-deficient HeLa cells, and these show similarities to the Fz2C foci/NE buds observed in Drosophila muscle and salivary gland nuclei (Laudermilch et al., 2016; Parchure et al., 2017; Webster et al., 2014). While the full relationship between NPCs and NE buds is not yet known, one important difference is that the yeast and HeLa nuclear granules observed are much smaller (∼120 nm) than Fz2C foci/NE buds (∼500 nm) (see Fig. 1F,G). Our identification of Wash and SHRC, proteins with the capability of remodeling cortical cytoskeleton and/or membranes, in the physical aspects of NE budding lend support for NE budding being an alternative endogenous nuclear exit pathway.
The nuclear lamina and NE budding
NE budding has been proposed to occur at sites along the INM where the nuclear lamina is modified by aPKC phosphorylation (Fradkin and Budnik, 2016; Parchure et al., 2017; Speese et al., 2012). Both A- and B-type lamins play a role in NE budding and are thought to be the target of aPKC phosphorylation within the nuclear lamina, similar to the PKC-mediated phosphorylation of lamins that precedes lamina disassembly in mitotic NE breakdown (Güttinger et al., 2009), apoptosis (Cross et al., 2000) or during viral capsid nuclear egress (Park and Baines, 2006). Viral NE budding requires a virus-encoded nuclear egress complex (NEC), which has been implicated in the recruitment of kinases to the INM (Bigalke and Heldwein, 2016; Mettenleiter et al., 2013; Zeev-Ben-Mordehai et al., 2015). Cellular counterparts for these virally encoded NEC proteins have not yet been identified. It is also not yet known how this kinase activity is restricted to specific sites along the nuclear lamina or how those specific sites are selected.
We have previously shown that Wash interacts directly with Lamin B and that loss of nuclear Wash results in a wrinkled nuclear morphology reminiscent of that observed in laminopathies (Verboon et al., 2015b). We reasoned that Wash-mediated disruption of the nuclear lamina may account for its NE-budding phenotypes. Consistent with this idea, we find Lamin B knockdown nuclei and nuclei from a wash point mutant that disrupts the interaction of Wash with Lamin B (washΔΔLamB) exhibit a wrinkled nuclear morphology, reduced Fz2C foci/NE buds and NE-budding-associated phenotypes, albeit not as strong as those observed in Wash or SHRC mutants.
Lamin A/C and Lamin B isoforms form homotypic meshworks that interact among themselves (in as yet unknown ways), and that are somehow linked to integral membrane proteins of the INM and to the chromatin adjoining the INM (Shimi et al., 2015). Intriguingly, our data suggests that these lamin homotypic meshes are likely layered, rather than interwoven, and that Wash affects the anchoring of these lamin homotypic meshes to each other and/or the INM. Lamin knockdown or disruption of the Wash–Lamin B interaction leads to separated lamin isoform meshes and wrinkled nuclear morphology that are not observed in SHRC and Arp2/3 knockdown nuclei, suggesting that Wash can also affect NE budding by a means independent of disrupted global nuclear lamina integrity. Interestingly, the functions of Wash mediated with the SHRC and with Lamin B involve separate nuclear complexes. Consistent with this, we have shown previously that Drosophila Wash encodes several independent biochemical activities (actin nucleation, actin bundling, MT bundling and actin–MT crosslinking) and that the use of these activities is context dependent (Liu et al., 2009; Verboon et al., 2018, 2015a,b). In particular, when Wash interacts with Lamin, it does not require an association with SHRC or Arp2/3 (Verboon et al., 2015b). We suggest that the interaction of Wash with Lamin B is required for organizing the nuclear lamina and likely requires the actin bundling and/or cross-linking activities of Wash rather than its actin nucleation activity, such that wash mutants that cannot bind Lamin result in separation of the Lamin isoform meshes from each other. Taken together, our data suggest that loss of the interaction between Wash and Lamin B makes NE budding inefficient by generally disrupting the nuclear lamina, rather than directly disrupting NE bud formation. The role of aPKC in NE budding may also be somewhat indirect by generally disrupting the nuclear lamina thereby reducing the efficiency of NE bud formation. Alternatively, aPKC may target Wash: WASH phosphorylation by Src kinases has been shown to be necessary for regulating NK cell cytotoxicity (Huang et al., 2016).
A physical role for Wash in NE budding
For bud formation/envelopment of a megaRNP or macromolecular cargo to occur, the INM must interact with its underlying cortical nucleoskeleton to allow the INM deformation/curvature necessary to form the physical NE bud. Force must also be generated that allows the bud to extend into the perinuclear space, as well as for the scission of the nascent bud. In the cytoplasm, WAS family proteins are often involved in membrane–cortical cytoskeleton-coupled processes, including both ‘inward’ membrane deformations (i.e. endocytosis) and ‘outward’ membrane deformations (i.e. exocytosis and cell protrusions), that are required for signal/environment sensing and cell movement during normal development, as well as during pathological conditions (Burianek and Soderling, 2013; Campellone and Welch, 2010; Rottner et al., 2010; Stradal et al., 2004; Takenawa and Suetsugu, 2007). Mammalian WASH, in particular, has been implicated in endosome biogenesis and/or sorting in the cytoplasm, where it, along with its SHRC, drives Arp2/3-dependent actin assembly to influence endosome trafficking, remodels membrane, and facilitates membrane scission (Derivery et al., 2009; Duleh and Welch, 2010; Gomez and Billadeau, 2009; Rottner et al., 2010; Simonetti and Cullen, 2019). Thus, Wash encodes the biochemical properties needed to regulate the membrane deformation/curvature necessary to form the NE bud and/or play a role in generating the forces necessary to pinch off the NE bud from the INM. Consistent with Wash playing a role in the physical production of a NE bud, we find that Wash acts prior to Torsin, a protein that is implicated in NE bud scission from the INM, and it requires its actin nucleation activity.
We have identified Wash and its SHRC as new players in the cellular machinery required for the newly described endogenous NE budding pathway. Our data suggest that Wash is involved in two nuclear functions that can affect NE budding. (1) Wash is required to maintain the organization of Lamin isoforms relative to each other and the INM through its direct physical interaction with Lamin B. This Wash activity is SHRC and Arp2/3 independent, and is likely a non-specific mechanism because global disruption of the nuclear lamina/nuclear envelope would indirectly affect many nuclear processes, including NE budding. (2) Wash is specifically required for NE bud formation. This Wash activity is SHRC and Arp2/3 dependent. While the focus of NE budding research to date has centered on the composition of the megaRNPs and the spectrum of cellular/developmental processes requiring NE budding, Wash and the SHRC are likely involved in the physical aspects of NE budding. Thus, Wash and SHRC provide a molecular entry into the physical machinery that underlies NE budding. In the future, it will be exciting to further explore the roles of Wash in NE budding, and to determine how it functions to get macromolecular complexes through the INM, and how closely these nuclear roles parallel those in the cytoplasm.
MATERIALS AND METHODS
Reagents and resources
Specific information for all of the reagents and resources used in this study are given in Table S1.
Fly stocks and genetics
Flies were cultured and crossed at 25°C on yeast-cornmeal-molasses-malt extract medium. Flies used in this study are listed in Table S1. All fly stocks were treated with tetracycline and then tested by PCR to ensure that they did not harbor Wolbachia. RNAi knockdowns were driven in the salivary glands by the GAL4-UAS system using the P{Sgs3-GAL4.PD} driver (Bloomington Drosophila Stock Center, stock #6870). RNAi knockdowns were driven in the indirect flight muscle by the GAL4-UAS system using the P{w[+mC]=Mhc-GAL4.K}2 driver (Bloomington Drosophila Stock Center, stock #55133). RNAi knockdowns were driven in the larval body wall muscle by the GAL4-UAS system using the P{w[+mW.hs]=GawB}BG487 driver (Bloomington Drosophila Stock Center, stock #51634). The washΔ185 deletion allele was kept as a continuously outcrossed stock (Verboon et al., 2018).
Construction of Wash point mutant transgenic lines
The residues required for the interaction of Wash with Lamin and CCDC53 were mapped by successive GST pulldown assays using fragments of Wash protein, followed by specific point or substitution mutations in the context of the full-length Wash protein (as detailed in Figs 6A and 7J). Point and/or substitution mutations were confirmed by sequencing.
A 2.9-kb genomic fragment encompassing the entire wash gene was amplified by PCR, then subcloned into the Casper 4 transformation vector by adding KpnI (5′) and BamHI (3′) restriction sites. GFP was inserted N-terminal to the Wash ATG by PCR (GFP–WashWT). The Wash portion of this construct was swapped with the point and/or substitution mutations described above to generate GFP–WashΔSHRC, GFP–WashΔΔLamB and GFP–WashΔArp2/3.
These constructs were used to make germline transformants as previously described (Spradling, 1986). Transgenic lines that mapped to chromosome 2 and that had non-lethal insertions were kept. The resulting transgenic lines (P{w+; GFP-WashWT}, P{w+; GFP-WashΔSHRC}, P{w+; GFP-WashΔΔLamB}, and P{w+; GFP-WashΔArp2/3}) were recombined onto the washΔ185 null chromosome to assess the contribution of the particular Wash transgene. The resulting recombinants (washΔ185 P{w+; GFP-WashWT}) are essentially gene replacements, as wash activity is only provided by the transgene. These transgenes do not rely on overexpression, but rather on the spatial and temporal expression driven by the endogenous wash promoter itself. We analyzed a minimum of three lines per construct and checked all lines to confirm that the levels and spatial distribution of their expression is indistinguishable from that in wild type. The wild-type version of this transgene (washΔ185 P{w+; GFP-WashWT}) rescues the phenotypes associated with the outcrossed washΔ185 mutation (Liu et al., 2009; Verboon et al., 2018, 2015a,b).
Antibody production and characterization
Guinea pig antibodies were raised against bacterially double-tagged Fz2C protein at Pocono Rabbit Farm & Laboratory (Canadensis, PA) using their standard protocols. For expression of Fz2C, a DNA fragment covering amino acids 612–694 of Fz2 was generated by PCR and cloned into a modified ‘double-tag’ pGEX vector (Liu et al., 2009). Protein was purified as described previously (Rosales-Nieves et al., 2006). Western blotting was used to confirm antibody specificity using Fz2 purified protein, Kc cell, ovary and S2 whole-cell extracts. Antibody generation for Wash and SHRC subunits was described previously (Rodriguez-Mesa et al., 2012; Verboon et al., 2015a).
Lysate preparation
Drosophila cytoplasmic and nuclear extracts were made from Kc167 cells. Briefly, cells were grown to confluence in 500 ml spinflasks, pelleted for 5 min at 500 g, resuspended in 100 ml cold 1× PBS and re-pelleted for 5 min at 500 g. Cell pellets were flash frozen in liquid nitrogen. Cells were resuspended in sucrose buffer (0.32 M sucrose, 3 mM CaCl2, 2 mM MgAc, 0.1 mM EDTA, 1.5% NP40) with 2× protease inhibitors [Complete protease inhibitor (EDTA free; Sigma, St Louis, MO), 2 mM PMSF and 1 mM Na3VO4] and 2× phosphatase inhibitors [PhosSTOP; Sigma, St Louis, MO] at 100 µl per 105 cells and incubated on ice for 30 min. Lysate was dounce homogenized 10× on ice. Lysate was then centrifuged 10 min at 2900 g at 4°C, nuclei formed a pellet and supernatant was cytoplasmic extract. Lipids were removed from the top of cytoplasmic extract using a sterile swab, then the cytoplasmic fraction was removed and centrifuged for 10 min at 3300 g at 4°C. Cytoplasmic supernatant was removed and one-tenth of the supernatant volume of 11× RIPA was added. Cytoplasmic extract was aliquoted and flash frozen. Nuclear pellet was resuspended with sucrose buffer with protease/phosphatase inhibitors and NP40 and re-dounced. Nuclear lysate was then centrifuged for 10 min at 3300 g at 4°C, and supernatant was discarded. The nuclear pellet was resuspended in sucrose buffer without NP40 and centrifuged for 20 min at 3300 g at 4°C. The supernatant was discarded and nuclear pellet was resuspended in 2.5 ml of buffer (20 mM HEPES pH 7.9, 0.5 mM EDTA, 100 mM KCl and 10% glycerol) per liter of cells used. DNA was degraded using MNase in a 37°C water bath for 10 min. 20 μl of 500 mM EDTA per 500 μl lysate was added and incubated on ice for 5 min. Lysate was then nutated for 2 h at 4°C. Lysate was sonicated using a Sonic Dismembrator (Model 60; Fisher Scientific) at setting 3.5 with 10 s per pulse for 15 min. Lysate was clarified with a 15 min centrifugation at 25,000 g, aliquoted and flash frozen.
Adult indirect flight muscle (IFM) lysate was made from 21-day-old flies in RIPA buffer (HEPES 50 mM pH 7.5, NaCl 150 nM, 1% NP40, 0.5% deoxycholate sodium salt, EDTA 5 mM). IFMs were dissected from flies in cold PBS. PBS was removed and 200 μl of 1× RIPA buffer with 2× protease inhibitors (Complete EDTA free protease inhibitor; Sigma) per 20 IFMs was added. Lysates were homogenized with an Eppendorf microcentrifuge homogenizing pestle on ice. Lysates were then sonicated with a probe sonicator on setting 3 for three 10-s pulses and centrifuged at 25,000 g for 30 min at 4°C. Supernatant was removed and MgCl2 was added to 2 mM and 8 μl of Benzonase (Millipore) was added per 200 μl of lysate. Lysates were nutated at room temperature for 15 min. Lysates were centrifuged at 16,000 g for 30 min at 4°C. Supernatant was aliquoted and flash frozen.
Western blotting
Cytoplasmic and nuclear purity of lysates was assayed using β-Tubulin (E7, 1:2000, Developmental Studies Hybridoma Bank) and Lamin B (monoclonal 67.10, 1:1000, Developmental Studies Hybridoma Bank) antibodies. Lysate samples were normalized to a loading control (Actin Clone C4, 1:2500; MP Biochemicals) and then blotted according to standard procedures. The following antibodies were used: anti-Rho1 monoclonal (P1D9, 1:50, Developmental Studies Hybridoma Bank), anti-Arp3 (1:500; Stevenson et al., 2002), and anti-Arpc1 (1:500; Stevenson et al., 2002), and anti-mono and poly-ubiquitylated conjugates (FK2, 1:1000, Enzo Life Sciences, East Farmingdale, NY). Quantification of actin loading controls and Ubiquitin-FK2 expression was performed using ImageJ (NIH).
Immunoprecipitation
Nuclear lysate was incubated with primary antibody overnight at 4°C. Protein G–Sepharose (20 μl) was then added in 0.5 ml Carol buffer (50 mM HEPES pH 7.9, 250 mM NaCl, 10 mM EDTA, 1 mM DTT, 10% glycerol, 0.1% Triton X-100) plus 0.5 mg/ml bovine serum albumin (BSA) and protease inhibitors (Complete EDTA-free Protease Inhibitor cocktail; Sigma] and the reaction allowed to proceed for 2 h at 4°C. The beads were washed 1× with Carol buffer plus BSA and 2× with Carol buffer alone. Analysis was conducted using SDS-PAGE followed by western blotting. Antibodies used for immunoprecipitations are as follows: anti-9e10 (1:9; Developmental Studies Hybridoma Bank), anti-Wash monoclonal (1:6; P3H3; Rodriguez-Mesa et al., 2012), anti-CCDC53 (1:1000; Verboon et al., 2015a), anti-Strumpellin (1:1000; Verboon et al., 2015a), anti-FAM21 (1:1000; Verboon et al., 2015a), anti-Lamin B monoclonal (1:8; AD67.10, Developmental Studies Hybridoma Bank) and anti-Lamin C monoclonal (1:10; LC28.26, Developmental Studies Hybridoma Bank). Antibodies used for the IP western blots are as follows: mouse anti-CCDC53 polyclonal (1:1000), mouse anti-Strumpellin polyclonal (1:400), mouse anti-FAM21 polyclonal (1:400), and anti-Lamin B monoclonal 67.10 (1:200).
GST pulldown assays and Blue Native PAGE
GST pulldown assays were performed as previously described (Magie and Parkhurst, 2005; Magie et al., 2002; Rosales-Nieves et al., 2006). Blue Native Page was performed using a Novex Native PAGE Bis-Tris Gel System (Invitrogen) following manufacturer protocols. Briefly, Drosophila Kc cell nuclear extract was resuspended in 1× NativePAGE Sample Buffer (Invitrogen) with 1% digitonin and protease inhibitors, and incubated for 15 min on ice. Samples were centrifuged at 16,200 g for 30 min at 4°C, and supernatant was resuspended with G250 sample additive and Native PAGE sample buffer. These prepared samples were loaded on 3–12% Bis-Tris Native PAGE gels and electrophoresed using a 1× native PAGE running buffer system (Invitrogen). The cathode buffer included 1× cathode buffer additive (Invitrogen). Native mark protein standard (Invitrogen) was used as the molecular mass marker. Protein concentrations of adult fly mitochondrial preps were determined with a BCA protein assay kit (Thermo Scientific, USA) following the manufacturer's instructions. The following antibodies were used: mouse anti-Wash monoclonal (P3H3, 1:2; Rodriguez-Mesa et al., 2012), mouse anti-CCDC53 polyclonal (1:400; Verboon et al., 2015a), mouse anti-Strumpellin polyclonal (1:400; Verboon et al., 2015a), mouse anti-FAM21 polyclonal (1:400; Verboon et al., 2015a) and rabbit anti-Lamin B polyclonal (L6, 1:2500; Stuurman et al., 1996).
Electron microscopy
Drosophila third-instar larva salivary glands were processed for electron microscopy essentially as previously described (Pitt et al., 2000). Glands were dissected in 1× PBS then placed directly in fixative solutions [2.2% glutaraldehyde, 0.9% paraformaldehyde, 0.05 M cacodylate (pH 7.4), 0.09 M sucrose, 0.9 mM MgCl2]. Glands were fixed for 2.5 h at room temperature, followed by several rinses with 0.09 M sucrose and 0.05 M cacodylate (pH 7.4). Glands were post-fixed in 1% osmium, 0.8% potassium ferricyanide, 0.1 M cacodylate (pH 7.2) for 45 min at 4°C, followed by several rinses in 0.05 M cacodylate (pH 7.0). Glands were then treated with 0.2% tannic acid in 0.05 M cacodylate (pH 7.0) for 15 min at room temperature, followed by several rinses in distilled H2O. Glands were placed in 1% uranyl acetate in 0.1 M sodium acetate (pH 5.2) for 1 h at room temperature, and rinsed three times with 0.1 M sodium acetate (pH 5.2), followed by three rinses with distilled H2O. Specimens were dehydrated in a graded acetone series, embedded in Epon, and sectioned following standard procedures. Grids were viewed with a JEOL JEM-1230 transmission electron microscope and photographed with a Gatan UltraScan 1000 CCD camera.
Immunostaining of larval salivary glands
Salivary glands were dissected, fixed, stained and mounted as previously described (Verboon et al., 2015a). Primary antibodies were added at the following concentrations: mouse anti-Wash monoclonal (P3H3, 1:200; Rodriguez-Mesa et al., 2012), mouse anti-CCDC53 polyclonal (1:300; Verboon et al., 2015a), mouse anti-Strumpellin polyclonal (1:300; Verboon et al., 2015a), mouse anti-SWIP polyclonal (1:300; Verboon et al., 2015a), mouse anti-FAM21 polyclonal (1:300; Verboon et al., 2015a), mouse anti-Lamin B monoclonal (AD67.10 1:200, Developmental Studies Hybridoma Bank), mouse anti-Lamin C monoclonal (LC 28.26, 1:200, Developmental Studies Hybridoma Bank), guinea pig anti-Fz2C (1:2500, this study), rat anti-Arpc1 (1:500; Stevenson et al., 2002), and rat anti-Cpa (1:200; Amândio et al., 2014).
Immunostaining of larval body wall muscle
Flies were transferred daily and wandering third-instar larvae were collected and subsequently fileted in cold PBS. Body wall muscle filets were fixed for 10 min. The fixative used was: 16.7 mM KPO4 pH 6.8, 75 mM KCl, 25 mM NaCl, 3.3 mM MgCl2 and 6% formaldehyde. After three washes with 1× PBS with 0.1% Tween-20 (PTW), larval filets were permeabilized in 1× PBS plus 1% Triton X-100 for 2 h at room temperature, then blocked using 1× PBS, 0.1% Tween-20, 1% BSA and 0.05% azide (PAT) for 2 h at 4°C. Antibodies were added at the following concentrations: mouse anti-DLG (1:50) and rabbit anti-HRP (1:1400). The larval filets were incubated 48 h at 4°C. Primary antibody was then removed and filets were washed three times with 1× PBS, 0.1% Tween-20, 0.1% BSA, 2% normal goat serum (XNS) for 30 min each. Alexa Fluor-conjugated secondary antibodies (Invitrogen) diluted in 1× PBS, 0.1% Tween-20, 0.1% BSA (PbT) (1:1000) were then added and the filets were incubated overnight at 4°C. Larval filets were washed ten times with PTW at room temperature for 10 min each and were mounted on slides in SlowFade Gold medium (Invitrogen, Carlsbad, CA) and visualized using a Zeiss confocal microscope as described below. The total number of boutons was quantified in third-instar larval preparations double labeled with antibodies to HRP and DLG, at segments A2–A3 muscles 6–7. The number of ghost boutons was assessed by counting HRP-positive and DLG-negative boutons.
Actin visualization and immunostaining of indirect flight muscle
UAS controlled RNAi-expressing flies were crossed to MHC-GAL4 driver flies and female RNAi/MHC-GAL4 trans-heterozygous flies were collected and aged 21 days at 25°C. Fly thoraxes were dissected and cut along the ventral side in cold PBS. Thoraxes were fixed using 1:6 fixative and heptane for 15 min. The fixative used was: 16.7 mM KPO4 pH 6.8, 75 mM KCl, 25 mM NaCl, 3.3 mM MgCl2 and 6% formaldehyde. After three washes with PTW thoraxes were then cut along the dorsal side resulting in two halves and fixed again for 10 min using 1:6 fix/heptane for 15 min. After three washes with PTW, thoraxes were permeabilized in 1× PBS plus 1% Triton X-100 for 2 h at room temperature, then blocked using PAT for 2 h at 4°C. Antibodies were added at the following concentrations: mouse anti-ATP-Synthase α (15H4C4 1:100, Abcam, Cambridge, UK), and mouse anti-mono and poly-ubiquitylated conjugates (FK2 1:200, Enzo Life Sciences, East Farmingdale, NY). The thoraxes were incubated 48 h at 4°C. Primary antibody was then removed and thoraxes were washed three times with XNS for 30 min each. Alexa Fluor-conjugated secondary antibodies (Invitrogen, Carlsbad, CA) diluted in PbT (1:1000) and Alexa Fluor-conjugated phalloidin (1:50) were then added and the thoraxes were incubated overnight at 4°C. Thoraxes were washed ten times with PTW at room temperature for 10 min each and were mounted on slides in SlowFade Gold medium (Invitrogen, Carlsbad, CA) and visualized using a Zeiss confocal microscope as described below. To quantify ATP-Synthase α expression, we measured 512×512 pixel regions of the IFM and measured total fluorescence using ImageJ (NIH). Fluorescence measurements from separate experiments were normalized to the control genotype.
Confocal and super-resolution microscopy
Images of fixed tissues were acquired using a Zeiss LSM 780 spectral confocal microscope (Carl Zeiss Microscopy GmbH, Jena, Germany) fitted with a Zeiss 40×/1.0 oil Plan-Apochromat objective and a Zeiss 63×/1.4 oil Plan-Apochromat objective. FITC (Alexa Fluor 488) fluorescence was excited with the 488 nm line of an argon laser, and detection was between 498 and 560 nm. Red (Alexa Fluor 568) fluorescence was excited with the 561 nm line of a DPSS laser and detection was between 570 and 670 nm. The pinhole was set to 1.0 Airy Units. Confocal sections were acquired at 0.2–1.0 µm spacing. Super-resolution images were acquired using an Airyscan detector in Super Resolution mode and captured confocal images were then processed using the Airyscan Processing feature on the Zen software provided by the manufacturer (Carl Zeiss Microscopy GmbH, Jena, Germany).
Live image acquisition
To obtain live time-lapse images of oocytes, female flies were first fattened on yeast for 2 days. Females were then injected in the abdomen with 0.4% Trypan Blue (Thermo Fisher Scientific) diluted 1:5 in PBS, and allowed to sit for 1–2 h. Ovaries were dissected into individual egg chambers in halocarbon 700 oil (Halocarbon Products, River Edge, NJ) on a cover slip. Images were acquired on a Revolution WD system (Andor Technology Ltd., Concord, MA) mounted on a Leica DMi8 (Leica Microsystems Inc., Buffalo Grove, IL) with a 63×/1.4 NA objective lens with a 2× coupler and controlled by MetaMorph software. Images were acquired with 561 nm excitation using an Andor iXon Ultra 888 EMCCD camera (Andor Technology Ltd., Concord, MA). Time-lapse images were obtained by taking one single frame acquisition every 10 s for either 5 or 30 min.
Statistical analysis
All statistical analyses were performed using R-3.6.1. For count data (number of nuclear buds and number of ubiquitin puncta), gene knockdowns and point mutations were compared to the appropriate control, and statistical significance was calculated using a Kruskal–Wallis test for independence. For frequency of observation (ghost versus mature boutons, swirling versus not swirling, and lamin separated versus overlapping) a two-tailed Fisher's exact test was used. For all other analyses, a two-tailed Student's t-test was used to test for significance between conditions. For plots, the mean+95% c.i. was calculated using the Hmisc-4.2-0 package using 1000 bootstrap resamples. P<0.01 was considered significant. Modeling bud counts was performed using the glm function in R. Bud count was fitted using a Poisson regression to a binary score of whether the lamin (washΔΔLamB) or actin (washΔSHRC or washΔArp2/3) activity was disrupted. The wash null was considered to have both activities disrupted. The washΔSHRC and washΔArp2/3 were combined into a single actin disruption category because of what is biologically known about Wash, the SHRC and Arp2/3, as well as because there was no difference between these mutations on bud count (P=0.9).
Supplementary Material
Acknowledgements
We thank Jim Priess, Bobbie Schneider, and Steve MacFarlane for help with EM, and Helen McNeill, Yonit Tsatskis, Bina Sugumar, and Clara Prentiss for help with Blue Native PAGE. We thank Vivian Budnik, Lynn Cooley, Paul Fisher, Florence Janody, Bill Theurkauf, the Bloomington Stock Center, Kyoto Stock Center, Harvard Transgenic RNAi Project, Drosophila Genomics Resource Center, and Developmental Studies Hybridoma Bank for reagents used in this study.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: J.M.V., M.N., S.M.P.; Methodology: J.M.V., M.N., S.M.P.; Software: J.M.V., V.N.; Validation: J.M.V., M.N., K.A.D., J.R.D., S.M.P.; Formal analysis: J.M.V., M.N., K.A.D., J.R.D., V.N., S.M.P.; Investigation: J.M.V., M.N., K.A.D., J.R.D., S.M.P.; Writing - original draft: J.M.V., S.M.P.; Writing - review & editing: J.M.V., M.N., K.A.D., J.R.D., V.N., S.M.P.; Visualization: J.M.V., M.N., K.A.D., J.R.D., S.M.P.; Supervision: S.M.P.; Project administration: S.M.P.; Funding acquisition: S.M.P.
Funding
This work was supported, in part, by National Institutes of Health grant R01 AG023779 (to S.M.P.) and by the National Cancer Institute (NCI) Cancer Center Support grant P30 CA015704 (Pilot project and Shared Resources). Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at https://jcs.biologists.org/lookup/doi/10.1242/jcs.243576.supplemental
Peer review history
The peer review history is available online at https://jcs.biologists.org/lookup/doi/10.1242/jcs.243576.reviewer-comments.pdf
References
- Alekhina O., Burstein E. and Billadeau D. D. (2017). Cellular functions of WASP family proteins at a glance. J. Cell Sci. 130, 2235-2241. 10.1242/jcs.199570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amândio A. R., Gaspar P., Whited J. L. and Janody F. (2014). Subunits of the Drosophila actin-capping protein heterodimer regulate each other at multiple levels. PLoS ONE 9, e96326 10.1371/journal.pone.0096326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigalke J. M. and Heldwein E. E. (2016). Nuclear exodus: herpesviruses lead the way. Annu. Rev. Virol. 3, 387-409. 10.1146/annurev-virology-110615-042215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burianek L. E. and Soderling S. H. (2013). Under lock and key: spatiotemporal regulation of WASP family proteins coordinates separate dynamic cellular processes. Semin. Cell Dev. Biol. 24, 258-266. 10.1016/j.semcdb.2012.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campellone K. G. and Welch M. D. (2010). A nucleator arms race: cellular control of actin assembly. Nat. Rev. Mol. Cell Biol. 11, 237-251. 10.1038/nrm2867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross T., Griffiths G., Deacon E., Sallis R., Gough M., Watters D. and Lord J. M. (2000). PKC-delta is an apoptotic lamin kinase. Oncogene 19, 2331-2337. 10.1038/sj.onc.1203555 [DOI] [PubMed] [Google Scholar]
- Daneholt B. (2001). Packing and delivery of a genetic message. Chromosoma 110, 173-185. 10.1007/s004120000127 [DOI] [PubMed] [Google Scholar]
- Derivery E., Sousa C., Gautier J. J., Lombard B., Loew D. and Gautreau A. (2009). The Arp2/3 activator WASH controls the fission of endosomes through a large multiprotein complex. Dev. Cell 17, 712-723. 10.1016/j.devcel.2009.09.010 [DOI] [PubMed] [Google Scholar]
- Duleh S. N. and Welch M. D. (2010). WASH and the Arp2/3 complex regulate endosome shape and trafficking. Cytoskeleton (Hoboken) 67, 193-206. 10.1002/cm.20437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards M., Zwolak A., Schafer D. A., Sept D., Dominguez R. and Cooper J. A. (2014). Capping protein regulators fine-tune actin assembly dynamics. Nat. Rev. Mol. Cell Biol. 15, 677-689. 10.1038/nrm3869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fradkin L. G. and Budnik V. (2016). This bud's for you: mechanisms of cellular nucleocytoplasmic trafficking via nuclear envelope budding. Curr. Opin. Cell Biol. 41, 125-131. 10.1016/j.ceb.2016.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez T. S. and Billadeau D. D. (2009). A FAM21-containing WASH complex regulates retromer-dependent sorting. Dev. Cell 17, 699-711. 10.1016/j.devcel.2009.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grünwald D., Singer R. H. and Rout M. (2011). Nuclear export dynamics of RNA-protein complexes. Nature 475, 333-341. 10.1038/nature10318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Güttinger S., Laurell E. and Kutay U. (2009). Orchestrating nuclear envelope disassembly and reassembly during mitosis. Nat. Rev. Mol. Cell Biol. 10, 178-191. 10.1038/nrm2641 [DOI] [PubMed] [Google Scholar]
- Hadek R. and Swift H. (1962). Nuclear extrusion and intracisternal inclusions in the rabbit blastocyst. J. Cell Biol. 13, 445-451. 10.1083/jcb.13.3.445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagen C., Dent K. C., Zeev-Ben-Mordehai T., Grange M., Bosse J. B., Whittle C., Klupp B. G., Siebert C. A., Vasishtan D., Bäuerlein F. J. et al. (2015). Structural basis of vesicle formation at the inner nuclear membrane. Cell 163, 1692-1701. 10.1016/j.cell.2015.11.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatch E. M. and Hetzer M. W. (2012). RNP export by nuclear envelope budding. Cell 149, 733-735. 10.1016/j.cell.2012.04.018 [DOI] [PubMed] [Google Scholar]
- Hatch E. and Hetzer M. (2014). Breaching the nuclear envelope in development and disease. J. Cell Biol. 205, 133-141. 10.1083/jcb.201402003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochstrasser M. and Sedat J. W. (1987). Three-dimensional organization of Drosophila melanogaster interphase nuclei. II. Chromosome spatial organization and gene regulation. J. Cell Biol. 104, 1471-1483. 10.1083/jcb.104.6.1471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L., Zhu P., Xia P. and Fan Z. (2016). WASH has a critical role in NK cell cytotoxicity through Lck-mediated phosphorylation. Cell Death Dis. 7, e2301 10.1038/cddis.2016.212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia D., Gomez T. S., Metlagel Z., Umetani J., Otwinowski Z., Rosen M. K. and Billadeau D. D. (2010). WASH and WAVE actin regulators of the Wiskott-Aldrich syndrome protein (WASP) family are controlled by analogous structurally related complexes. Proc. Natl. Acad. Sci. USA 107, 10442-10447. 10.1073/pnas.0913293107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jokhi V., Ashley J., Nunnari J., Noma A., Ito N., Wakabayashi-Ito N., Moore M. J. and Budnik V. (2013). Torsin mediates primary envelopment of large ribonucleoprotein granules at the nuclear envelope. Cell Rep. 3, 988-995. 10.1016/j.celrep.2013.03.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyheröinen S. and Vartiainen M. K. (2019). Nuclear actin dynamics in gene expression and genome organization. Semin. Cell Dev. Biol. 102, 105-112. 10.1016/j.semcdb.2019.10.012 [DOI] [PubMed] [Google Scholar]
- LaMassa N., Arenas-Mena C. and Phillips G. R. (2018). Electron microscopic characterization of nuclear egress in the sea urchin gastrula. J. Morphol. 279, 609-615. 10.1002/jmor.20796 [DOI] [PubMed] [Google Scholar]
- Laudermilch E., Tsai P.-L., Graham M., Turner E., Zhao C. and Schlieker C. (2016). Dissecting Torsin/cofactor function at the nuclear envelope: a genetic study. Mol. Biol. Cell 27, 3964-3971. 10.1091/mbc.E16-07-0511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leirdal M., Shadidy M., Røsok Ø. and Sioud M. (2004). Identification of genes differentially expressed in breast cancer cell line SKBR3: potential identification of new prognostic biomarkers. Int. J. Mol. Med. 14, 217-222. 10.3892/ijmm.14.2.217 [DOI] [PubMed] [Google Scholar]
- Li Y., Hassinger L., Thomson T., Ding B., Ashley J., Hassinger W. and Budnik V. (2016). Lamin mutations accelerate aging via defective export of mitochondrial mRNAs through nuclear envelope budding. Curr. Biol. 26, 2052-2059. 10.1016/j.cub.2016.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linardopoulou E. V., Parghi S. S., Friedman C., Osborn G. E., Parkhurst S. M. and Trask B. J. (2007). Human subtelomeric WASH genes encode a new subclass of the WASP family. PLoS Genet. 3, e237 10.1371/journal.pgen.0030237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R., Abreu-Blanco M. T., Barry K. C., Linardopoulou E. V., Osborn G. E. and Parkhurst S. M. (2009). Wash functions downstream of Rho and links linear and branched actin nucleation factors. Development 136, 2849-2860. 10.1242/dev.035246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lye M. F., Wilkie A. R., Filman D. J., Hogle J. M. and Coen D. M. (2017). Getting to and through the inner nuclear membrane during herpesvirus nuclear egress. Curr. Opin. Cell Biol. 46, 9-16. 10.1016/j.ceb.2016.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magie C. R. and Parkhurst S. M. (2005). Rho1 regulates signaling events required for proper Drosophila embryonic development. Dev. Biol. 278, 144-154. 10.1016/j.ydbio.2004.10.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magie C. R., Pinto-Santini D. and Parkhurst S. M. (2002). Rho1 interacts with p120ctn and alpha-catenin, and regulates cadherin-based adherens junction components in Drosophila. Development 129, 3771-3782. [DOI] [PubMed] [Google Scholar]
- Marchand J.-B., Kaiser D. A., Pollard T. D. and Higgs H. N. (2001). Interaction of WASP/Scar proteins with actin and vertebrate Arp2/3 complex. Nat. Cell Biol. 3, 76-82. 10.1038/35050590 [DOI] [PubMed] [Google Scholar]
- McGough I. J., Steinberg F., Jia D., Barbuti P. A., McMillan K. J., Heesom K. J., Whone A. L., Caldwell M. A., Billadeau D. D., Rosen M. K. et al. (2014). Retromer binding to FAM21 and the WASH complex is perturbed by the Parkinson disease-linked VPS35(D620N) mutation. Curr. Biol. 24, 1670-1676. 10.1016/j.cub.2014.06.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon K. P., Carrillo R. A. and Zinn K. (2013). Development and plasticity of the Drosophila larval neuromuscular junction. Wiley Interdiscip. Rev. Dev. Biol. 2, 647-670. 10.1002/wdev.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mettenleiter T. C., Müller F., Granzow H. and Klupp B. G. (2013). The way out: what we know and do not know about herpesvirus nuclear egress. Cell. Microbiol. 15, 170-178. 10.1111/cmi.12044 [DOI] [PubMed] [Google Scholar]
- Nordgard S. H., Johansen F. E., Alnaes G. I. G., Bucher E., Syvänen A.-C., Naume B., Børresen-Dale A.-L. and Kristensen V. N. (2008). Genome-wide analysis identifies 16q deletion associated with survival, molecular subtypes, mRNA expression, and germline haplotypes in breast cancer patients. Genes Chromosomes Cancer 47, 680-696. 10.1002/gcc.20569 [DOI] [PubMed] [Google Scholar]
- Panagaki D., Neutze R. and Höög J. L. (2018). Nuclear envelope budding is an evolutionary conserved phenomenon. bioRxiv, 413153 10.1101/413153 [DOI] [Google Scholar]
- Parchure A., Munson M. and Budnik V. (2017). Getting mRNA-containing ribonucleoprotein granules out of a nuclear back door. Neuron 96, 604-615. 10.1016/j.neuron.2017.10.020 [DOI] [PubMed] [Google Scholar]
- Park R. and Baines J. D. (2006). Herpes simplex virus type 1 infection induces activation and recruitment of protein kinase C to the nuclear membrane and increased phosphorylation of lamin B. J. Virol. 80, 494-504. 10.1128/JVI.80.1.494-504.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park L., Thomason P. A., Zech T., King J. S., Veltman D. M., Carnell M., Ura S., Machesky L. M. and Insall R. H. (2013). Cyclical action of the WASH complex: FAM21 and capping protein drive WASH recycling, not initial recruitment. Dev. Cell 24, 169-181. 10.1016/j.devcel.2012.12.014 [DOI] [PubMed] [Google Scholar]
- Percipalle P. and Vartiainen M. (2019). Cytoskeletal proteins in the cell nucleus: a special nuclear actin perspective. Mol. Biol. Cell 30, 1781-1785. 10.1091/mbc.E18-10-0645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitt J. N., Schisa J. A. and Priess J. R. (2000). P granules in the germ cells of Caenorhabditis elegans adults are associated with clusters of nuclear pores and contain RNA. Dev. Biol. 219, 315-333. 10.1006/dbio.2000.9607 [DOI] [PubMed] [Google Scholar]
- Ramaswami M., Taylor J. P. and Parker R. (2013). Altered ribostasis: RNA-protein granules in degenerative disorders. Cell 154, 727-736. 10.1016/j.cell.2013.07.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Mesa E., Abreu-Blanco M. T., Rosales-Nieves A. E. and Parkhurst S. M. (2012). Developmental expression of Drosophila Wiskott-Aldrich Syndrome family proteins. Dev. Dyn. 241, 608-626. 10.1002/dvdy.23742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roller R. J. and Baines J. D. (2017). Herpesvirus nuclear egress. Adv. Anat. Embryol. Cell Biol. 223, 143-169. 10.1007/978-3-319-53168-7_7 [DOI] [PubMed] [Google Scholar]
- Ropers F., Derivery E., Hu H., Garshasbi M., Karbasiyan M., Herold M., Nurnberg G., Ullmann R., Gautreau A., Sperling K. et al. (2011). Identification of a novel candidate gene for non-syndromic autosomal recessive intellectual disability: the WASH complex member SWIP. Hum. Mol. Genet. 20, 2585-2590. 10.1093/hmg/ddr158 [DOI] [PubMed] [Google Scholar]
- Rosales-Nieves A. E., Johndrow J. E., Keller L. C., Magie C. R., Pinto-Santini D. M. and Parkhurst S. M. (2006). Coordination of microtubule and microfilament dynamics by Drosophila Rho1, Spire and Cappuccino. Nat. Cell Biol. 8, 367-376. 10.1038/ncb1385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose A. and Schlieker C. (2012). Alternative nuclear transport for cellular protein quality control. Trends Cell Biol. 22, 509-514. 10.1016/j.tcb.2012.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rottner K., Hänisch J. and Campellone K. G. (2010). WASH, WHAMM and JMY: regulation of Arp2/3 complex and beyond. Trends Cell Biol. 20, 650-661. 10.1016/j.tcb.2010.08.014 [DOI] [PubMed] [Google Scholar]
- Rotty J. D., Wu C. and Bear J. E. (2013). New insights into the regulation and cellular functions of the ARP2/3 complex. Nat. Rev. Mol. Cell Biol. 14, 7-12. 10.1038/nrm3492 [DOI] [PubMed] [Google Scholar]
- Ryder P. V., Vistein R., Gokhale A., Seaman M. N., Puthenveedu M. A. and Faundez V. (2013). The WASH complex, an endosomal Arp2/3 activator, interacts with the Hermansky-Pudlak syndrome complex BLOC-1 and its cargo phosphatidylinositol-4-kinase type IIα. Mol. Biol. Cell 24, 2269-2284. 10.1091/mbc.e13-02-0088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimi T., Kittisopikul M., Tran J., Goldman A. E., Adam S. A., Zheng Y., Jaqaman K. and Goldman R. D. (2015). Structural organization of nuclear lamins A, C, B1, and B2 revealed by superresolution microscopy. Mol. Biol. Cell 26, 4075-4086. 10.1091/mbc.E15-07-0461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqui N. and Borden K. L. B. (2012). mRNA export and cancer. Wiley Interdiscip. Rev. RNA 3, 13-25. 10.1002/wrna.101 [DOI] [PubMed] [Google Scholar]
- Simonetti B. and Cullen P. J. (2019). Actin-dependent endosomal receptor recycling. Curr. Opin. Cell Biol. 56, 22-33. 10.1016/j.ceb.2018.08.006 [DOI] [PubMed] [Google Scholar]
- Song L., Rijal R., Karow M., Stumpf M., Hahn O., Park L., Insall R., Schröder R., Hofmann A., Clemen C. S. et al. (2018). Expression of N471D strumpellin leads to defects in the endolysosomal system. Dis. Model. Mech. 11, dmm033449 10.1242/dmm.033449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speese S. D., Ashley J., Jokhi V., Nunnari J., Barria R., Li Y., Ataman B., Koon A., Chang Y.-T., Li Q. et al. (2012). Nuclear envelope budding enables large ribonucleoprotein particle export during synaptic Wnt signaling. Cell 149, 832-846. 10.1016/j.cell.2012.03.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spradling A. C. (1986). P Element-Mediated Transformation, Oxford: IRL Press. [Google Scholar]
- Stevenson V., Hudson A., Cooley L. and Theurkauf W. E. (2002). Arp2/3-dependent pseudocleavage [correction of psuedocleavage] furrow assembly in syncytial Drosophila embryos. Curr. Biol. 12, 705-711. 10.1016/S0960-9822(02)00807-2 [DOI] [PubMed] [Google Scholar]
- Stradal T. E. B., Rottner K., Disanza A., Confalonieri S., Innocenti M. and Scita G. (2004). Regulation of actin dynamics by WASP and WAVE family proteins. Trends Cell Biol. 14, 303-311. 10.1016/j.tcb.2004.04.007 [DOI] [PubMed] [Google Scholar]
- Stuurman N., Sasse B. and Fisher P. A. (1996). Intermediate filament protein polymerization: molecular analysis of Drosophila nuclear lamin head-to-tail binding. J. Struct. Biol. 117, 1-15. 10.1006/jsbi.1996.0064 [DOI] [PubMed] [Google Scholar]
- Szollosi M. S. and Szollosi D. (1988). ‘Blebbing’ of the nuclear envelope of mouse zygotes, early embryos and hybrid cells. J. Cell Sci. 91, 257-267. [DOI] [PubMed] [Google Scholar]
- Takenawa T. and Suetsugu S. (2007). The WASP-WAVE protein network: connecting the membrane to the cytoskeleton. Nat. Rev. Mol. Cell Biol. 8, 37-48. 10.1038/nrm2069 [DOI] [PubMed] [Google Scholar]
- Tran E. J., King M. C. and Corbett A. H. (2014). Macromolecular transport between the nucleus and the cytoplasm: advances in mechanism and emerging links to disease. Biochim. Biophys. Acta 1843, 2784-2795. 10.1016/j.bbamcr.2014.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Türk M., Schröder R., Khuller K., Hofmann A., Berwanger C., Ludolph A. C., Dekomien G., Müller K., Weishaupt J. H., Thiel C. T. et al. (2017). Genetic analysis of VCP and WASH complex genes in a German cohort of sporadic ALS-FTD patients. Neurobiol. Aging 56, 213.e211-213.e215. 10.1016/j.neurobiolaging.2017.04.023 [DOI] [PubMed] [Google Scholar]
- Valdmanis P. N., Meijer I. A., Reynolds A., Lei A., MacLeod P., Schlesinger D., Zatz M., Reid E., Dion P. A., Drapeau P. et al. (2007). Mutations in the KIAA0196 gene at the SPG8 locus cause hereditary spastic paraplegia. Am. J. Hum. Genet. 80, 152-161. 10.1086/510782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veltman D. M. and Insall R. H. (2010). WASP family proteins: their evolution and its physiological implications. Mol. Biol. Cell 21, 2880-2893. 10.1091/mbc.e10-04-0372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verboon J. M., Rahe T. K., Rodriguez-Mesa E. and Parkhurst S. M. (2015a). Wash functions downstream of Rho1 GTPase in a subset of Drosophila immune cell developmental migrations. Mol. Biol. Cell 26, 1665-1674. 10.1091/mbc.E14-08-1266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verboon J. M., Rincon-Arano H., Werwie T. R., Delrow J. J., Scalzo D., Nandakumar V., Groudine M. and Parkhurst S. M. (2015b). Wash interacts with lamin and affects global nuclear organization. Curr. Biol. 25, 804-810. 10.1016/j.cub.2015.01.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verboon J. M., Sugumar B. and Parkhurst S. M. (2015c). Wiskott-Aldrich syndrome proteins in the nucleus: aWASH with possibilities. Nucleus 6, 349-359. 10.1080/19491034.2015.1086051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verboon J. M., Decker J. R., Nakamura M. and Parkhurst S. M. (2018). Wash exhibits context-dependent phenotypes and, along with the WASH regulatory complex, regulates Drosophila oogenesis. J. Cell Sci. 131, jcs211573 10.1242/jcs.211573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster B. M., Colombi P., Jäger J. and Lusk C. P. (2014). Surveillance of nuclear pore complex assembly by ESCRT-III/Vps4. Cell 159, 388-401. 10.1016/j.cell.2014.09.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia P., Wang S., Huang G., Zhu P., Li M., Ye B., Du Y. and Fan Z. (2014). WASH is required for the differentiation commitment of hematopoietic stem cells in a c-Myc-dependent manner. J. Exp. Med. 211, 2119-2134. 10.1084/jem.20140169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavodszky E., Seaman M. N. J., Moreau K., Jimenez-Sanchez M., Breusegem S. Y., Harbour M. E. and Rubinsztein D. C. (2014). Mutation in VPS35 associated with Parkinson's disease impairs WASH complex association and inhibits autophagy. Nat. Commun. 5, 3828 10.1038/ncomms4828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeev-Ben-Mordehai T., Weberruss M., Lorenz M., Cheleski J., Hellberg T., Whittle C., El Omari K., Vasishtan D., Dent K. C., Harlos K. et al. (2015). Crystal structure of the herpesvirus nuclear egress complex provides insights into inner nuclear membrane remodeling. Cell Rep. 13, 2645-2652. 10.1016/j.celrep.2015.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.