Abstract
The presence of SARS-CoV-2 in raw wastewaters has been demonstrated in many countries affected by this pandemic. Nevertheless, virus presence and infectivity in treated wastewaters, but also in the receiving water bodies are still poorly investigated. In this study, raw and treated samples from three wastewater treatment plants, and three river samples within the Milano Metropolitan Area, Italy, were surveyed for SARS-CoV-2 RNA detection by means of real time RT-PCR and infectivity test on culture cells. SARS-CoV-2 RNA was detected in raw, but not in treated wastewaters (four and two samples, respectively, sampled in two dates). The isolated virus genome was sequenced, and belonged to the strain most spread in Europe and similar to another found in the same region. RNA presence in raw wastewater samples decreased after eight days, probably following the epidemiological trend estimated for the area. Virus infectivity was always null, indicating the natural decay of viral pathogenicity in time from emission. Samples from receiving rivers (three sites, sampled in the same dates as wastewaters) showed in some cases a positivity to real time RT-PCR, probably due to non-treated, or inefficiently treated discharges, or to the combined sewage overflows. Nevertheless, also for rivers infectivity was null. Risks for public health should be limited, although a precautionary approach to risk assessment is here advocated, giving the preliminary nature of the presented data.
Keywords: SARS-CoV-2, Wastewater, Infectivity, Genome, Milano
Graphical abstract
Highlights
-
•
SARS-CoV-2 RNA presence and infectivity in wastewaters and receptors was assessed.
-
•
Viral RNA was detectable in the inflow but not in the outflow wastewaters.
-
•
Viral RNA was present in receptors due to sewage overflows or inefficient treatment.
-
•
SARS-CoV-2 infectivity was null both in wastewaters and receptors.
-
•
A precautionary approach in the assessment of contagious risk is advocated.
1. Introduction
On March 11th, 2020, the World Health Organization (WHO) has officially declared the novel coronavirus (COVID-19) outbreak a global pandemic. This emergency saw an unprecedented investment of energies and resources to rapidly monitor SARS-CoV-2 circulation and trend. The high rate of asymptomatic infected individuals (Al-Tawfiq, 2020) has challenged the estimation of infection spread basing on clinical survey, and alternative approaches, such as wastewater-based epidemiology (WBE), were proposed (Medema et al., 2020; Randazzo et al., 2020). Since SARS-CoV-2 shedding in stools is supposed to be high (Gupta et al., 2020; Zhang et al., 2020), quantification of viral RNA in raw wastewater (WW) could be a reliable marker of infection prevalence in the population (Hovi et al., 2012; Wigginton et al., 2015). However, protocols for large scale wastewater-based epidemiology still need to be refined, mostly because of uncertainties in viral recovery rates during concentration and RNA extraction, and differences in the RNA detectability with available molecular markers (Hart and Halden, 2020).
Coronaviruses are enveloped viruses, and their persistence in the environment should be short. Nevertheless, little is known concerning SARS-CoV-2 survival and diffusion in WWs and in the receiving surface waters (Naddeo and Liu, 2020), which could depend mostly on environmental conditions. Similarly, infectivity test of SARS-CoV-2 in treated WW has not been extensively evaluated, and in some cases virus could be potentially still infectious (Singer and Wray, 2020; J. Wang et al., 2020; Wurtzer et al., 2020).
In Europe, northern Italy was one of the earliest and most infected area, with about 240,000 cases on July, 6th, 2020 (https://www.ecdc.europa.eu/en/geographical-distribution-2019-ncov-cases). In particular the Milano Metropolitan Area, including the Province of Milano and Monza e Brianza with 24.499 and 5.795 cases, respectively, experienced an infection rate (case divided by resident population) almost double (0.76% and 0.67% respectively) respect to the rest of Italy (0.40%) (https://github.com/pcm-dpc/COVID-19/blob/master/dati-province/dpc-covid19-ita-province.csv). In the present study, we evaluated the presence and infectivity of SARS-CoV-2 RNA in raw and treated WWs collected in three wastewater treatment plants (WWTPs), covering the entire Milano Metropolitan Area, and in three receptor rivers. The aim of the study was to provide a survey of viral dispersion and potential related health risk in the aquatic environment during the peak phase of the epidemic. Given this main aim, we adopted analytical protocols which targeted mostly the possibility to analyse rapidly fresh, viable water samples (i.e., qualitative detection of RNA and cell cultures), sacrificing detection sensitivity. Moreover, isolated genomic strains of SARS-CoV-2 were sequenced for confirmation and compared to those found in ill patients in the area and worldwide.
2. Materials and methods
2.1. Study area and WWTP characteristics
A total of 18 grab samples have been collected in three WWTPs, located in the two Provinces of Milano and Monza e Brianza (4 raw and 2 treated WWs) and in their receptor water bodies (Lambro River, Vettabbia Canal and Lambro Meridionale River, Fig. 1 ). All WWTPs and rivers were sampled on April, 14th and April, 22nd, 2020 almost at the same hour (1.00 p.m., instantaneous samples).
Fig. 1.
Map of Milano Metropolitan Area, with the two main river basins highlighted: 1) the Lambro Meridionale (LM) River, closed and sampled on Mirasole bridge; 2) the Lambro (L) River, closed and sampled on Melegnano bridge. The Vettabbia Canal is also indicated. Approximate locations of all WWTPs are indicated with squares whose size is proportional to their capacity (population equivalent). WWTP-A, the main plant of Monza and Brianza, WWTP-B and WWTP-C, the main plants of Milano, have been investigated in this study.
The three WWTPs globally collect 11 m3/s of sewage from of a population of about 2 million persons (Fig. 1), and are all equipped with secondary treatments and a tertiary disinfection step by peracetic acid or high intensity UV lamps. More in detail, WWTP-A (Monza), which treats about 2 m3/s of sewages from approximately 480.000 population equivalents (p.e.) and 4.500 industrial facilities, discharges into the river Lambro after a disinfection step with peracetic acid. WWTP-B (Milano) treats about 4 m3/s of sewage from approximately 1.050.000 p.e. from the west part of Milano. After a disinfection with high intensity UV lamps, it discharges into the stream Lambro Meridionale, which is a tributary of river Lambro. Finally, WWTP-C (Milano) treats about 5 m3/s of sewage from approximately 1.250.000 p.e. from the east part of Milano, carried to the plant by two main sewage collectors (Lines 1 and 2). After a disinfection with peracetic acid, it discharges into the Vettabbia channel, which is a tributary of river Lambro.
2.2. Sample collection and preparation
Sampling was done using separate stainless steel buckets to avoid cross contamination and transported in dark glass bottles to laboratory under refrigeration. Rivers were sampled from bridges. Water samples were filtered to exclude particulate and bacteria. One liter of water samples were pre-filtered on glass fiber filters (Whatman GF/F, 0.7 μm nominal pore size, 47 mm diameter) until filter clogging. Then 50 mL of pre-filtered water was taken and further filtered on nitrocellulose Millipore MCE filters (0.22 μm nominal pore size, 47 mm diameter) by using a vacuum pump. All glassware was disinfected by a diluted ipochlorite solution and rinsed with ultrapure water between every sample process. One initial, one intermediate and a final negative control for each one of the two filtering sessions (i.e., on April, 14th and 22nd, 2020), obtained by filtering 50 mL of molecular biology grade water, were included in the analysis. All controls resulted negative to real time RT-PCR detection, indicating that there was no cross contamination from SARS-CoV-2 during filtration. A previous study indicated that partitioning of enveloped viruses on the solid fraction of wastewaters could account up to 26% (Ye et al., 2016), so in the present case detection and isolation of SARS-CoV-2 was only focused on the filtered water fraction. Anyway, we performed a comparative test of viral RNA extraction and SARS-CoV-2 detection for the solid and liquid fractions of the same sample (3 samples collected on April, 14th, 2020), with the same protocols described below, and patterns of real time RT-PCR positivity were equivalent. Being the assay of infectivity one of the main aim of this work, we have preferred to avoid any preliminary concentration treatment of samples indeed, which could increase the chance to detect viral RNA, but at the risk of reducing infectivity of coronaviruses, since the addition of chemical compounds or mechanical stress could hamper their viability (Gundy et al., 2009; Ye et al., 2016).
2.3. SARS-CoV-2 RNA extraction and real time RT-PCR
Detection protocols already available for diagnostic routine were employed in the present case, to answer the demand for the rapid survey of WWs samples during the epidemic peak. A total of 200 μL of filtered sample was used for viral RNA extraction using the QIAMP Viral RNA mini kit (Qiagen, Hilden, Germany), according to manufacturer protocol. No additional treatment (e.g., DNAse addition) was done. The extraction solutions and protocols were especially formulated to remove any PCR inhibitor from the eluted RNA. We avoided to concentrate viral samples before RNA extraction, so that the risk of inhibitors carryover should be minimized. However, we added 1 μL of the internal plasmidic control included in the 2019-nCoV real-time RT-PCR kit panel (Product Ref. RR-0479-02, Liferiver, Shanghai, China), used for real time RT-PCR, to samples during RNA extraction, to check for PCR inhibition (Gibson et al., 2012) and extraction recovery efficiency (see Table S1 for Ct thresholds indicated by manufacturers below which inhibition should be excluded). Moreover, one negative control, consisting on 200 μL of molecular biology grade water, was included in each RNA extraction session.
To detect and assess the presence of the SARS-CoV-2, the above cited real time RT-PCR panel (CE-IVD, TGA and NMPA(CFDA) approved for diagnostic identification of SARS-CoV-2) was employed, containing primers and probes that target the nucleocapsid (N) gene, the ORF1ab gene and the E gene (Jung et al., 2020). Primers set was designed by manufacturer for both universal detection of SARS-like coronaviruses as well as specific detection of six 2019-nCoV strains. Primers sequences and PCR mix composition are proprietary information retained by manufacturer and not publicly available. Among the three screened genes, the ORF1ab region resulted one of the most sensitive region for RNA amplification and detection (Jung et al., 2020). The N gene coding for nucleocapsid is also largely employed for SARS-CoV-2 screening, while also the Envelope (E) region was indicated as a reliable candidate marker at this scope (Corman et al., 2020). Specificity of the selected genes for SARS-CoV-2 target was certified by the manufacturer. Real time RT-PCRs were performed on an Applied Biosystems™ 7500 Real-Time PCR Systems. Reaction conditions included two initial cycles at 45 °C for 10 min, and 95 °C for 3 min, followed by 45 cycles of amplification (95 °C for 15 s, 58 °C for 30 s). A 20 μL PCR mix was set for each sample by mixing 5 μL of RNA (or control) to 19 μL of mix and 1 μL of RT-PCR Enzyme Mix, and reactions run on sterile 96-well qRT-PCR plates.
The limit of detection (LOD 95%) of this kit was estimated to be 484 viral copies/mL on viral RNA extracted from cultured SARS-CoV-2 (X. Wang et al., 2020). Although protocols (i.e., RNA extraction) followed the same workflow of the above cited study, the presence of influent PCR inhibitors in WW and river samples, which are not a standard sample typology for diagnostic kits, could however not be excluded. Hence, discussion of results will acknowledge the possibility of false negative, although amplification of the added internal control resulted always within the quality control thresholds (Ct values) indicated by the manufacturer (Table S1 in Supplementary Material), suggesting that inhibition could be not relevant. Positive and negative controls provided in the real time RT-PCR kit were also included in the amplification plates, and resulted always within the quality control thresholds (Ct values) indicated by the manufacturer as well (Table S1 in Supplementary Material), proving efficient amplification and absence of contamination. The analyses were run in duplicate for each sample, and rerun in case of incongruencies between the two amplification profiles (i.e., cases in Table S2).
2.4. Cell culture and virus isolation
In order to evaluate the infectivity of SARS-CoV-2, a viral isolation protocol was conducted through the utilization of VERO E6 cells (ATCC® CRL-1586™), an African green monkey kidney cell line. VERO cells were cultured in Dulbecco's Modified Eagle Medium with l-glutamine (DMEM, Gibco™ ThermoFisher Scientific), which were supplemented with 10% of heat-inactivated fetal bovine serum (FBS, Gibco™ ThermoFisher Scientific) and 1% Penicillin-Streptomycin [5,000 U/mL] (Pen-Strep, Gibco™ ThermoFisher Scientific). Each viral sample (2 mL) was incubated in duplicate in a 25 cm2 cell culture flask, together with 5 mL of DMEM medium, at 37 °C and at 5% CO2 atmospheric pressure for 72 h. At the end of the 72 h waiting period, 5 mL of new DMEM medium was added to the culture flask, which was again incubated at 37 °C with a CO2 level of 5% for 48 h. Finally, infectivity was assessed daily by screening cells for cytopathic effects (CPE) under reverse-phase light microscope. Limit of Detection (LOD) of infectivity for SARS-CoV-2 on VERO E6 cells is not available, although replication kinetics, adaptation and CPE on these cells has been verified and described (Ogando et al., 2020). Estimates of LOD based on CPE observation on VERO cells were found to be in the range of 1 × 101–1 × 10−2 TCID50 for a wide group of viral families (Gombold et al., 2014).
2.5. Whole genome sequencing and phylogenetic analysis
A single genome coming from the WWTP A (Table 1 ) was sequenced, to confirm real time RT-PCR specificity. Twenty microliters of the obtained genomic RNAs were retro-transcribed using 1 Random Primers pool (Product Ref. C1181, Promega, USA). Ten microliters of cDNA have been used to prepare a SARS-CoV-2 library trough the “Ion AmpliSeq SARS-CoV-2 Research Panel” able to cover more than 99% of virus genome (Thermo Fisher Scientific, Waltham USA). All libraries have been prepared by using Ion AmpliSeq™ technology (Thermo Fisher Scientific), and sequenced on Ion Torrent PGM sequencer performing 550 flows to get reads up to around 250 bp (Thermo Fisher Scientific).
Table 1.
Results of real time RT-PCR amplification (positivity +, negativity -), infectivity test (presence or absence of CPE), genome sequencing and caffeine analysis, as obtained for WWTPs and rivers for the two sampling dates. Genes code refers to the nucleocapsid (N) gene, the Orf1ab gene and the E gene. n.a.: analysis not performed.
| Date | Sample origin | Station | Treatment | Gene positivity |
Infectivity test | Genome code | Caffeine μg/L | ||
|---|---|---|---|---|---|---|---|---|---|
| Orf1ab | N | E | |||||||
| 14/04/2020 | WWTPs | A | Raw | + | − | + | No CPE | hCoV-19/Italy/HSacco-1/2020 | 35.31 |
| A | Treated | − | − | − | No CPE | n.a. | <0.02 | ||
| B line 1 | Raw | + | − | + | n.a. | n.a. | 28.71 | ||
| B line 2 | Raw | + | + | + | No CPE | n.a. | 32.81 | ||
| C | Raw | − | − | − | n.a. | n.a. | 26.31 | ||
| C | Treated | − | − | − | n.a. | n.a. | <0.02 | ||
| Rivers | Vettabbia | − | + | + | − | No CPE | n.a. | <0.02 | |
| Lambro Meridionale | − | + | + | − | No CPE | n.a. | 0.36 | ||
| Lambro | − | + | + | − | No CPE | n.a. | 0.28 | ||
| 22/04/2020 | WWTPs | A | Raw | − | − | − | No CPE | n.a. | 24.89 |
| A | Treated | − | − | − | No CPE | n.a. | 0.03 | ||
| B line 1 | Raw | + | + | − | No CPE | n.a. | 22.72 | ||
| B line 2 | Raw | − | − | − | No CPE | n.a. | 16.24 | ||
| C | Raw | − | − | − | No CPE | n.a. | 21.61 | ||
| C | Treated | − | − | − | No CPE | n.a. | 0.06 | ||
| Rivers | Vettabbia | − | − | − | − | No CPE | n.a. | <0.02 | |
| Lambro Meridionale | − | − | − | − | No CPE | n.a. | 0.41 | ||
| Lambro | − | + | + | − | No CPE | n.a. | 0.44 | ||
Genome assembly was obtained using a mapping-based approach. Low quality read bases were trimmed out using Trimmomatic software with the MAXINFO:50:0.3 parameter set. Then, SNP calling was performed using the Wuhan-Hu-1 strain genome (accession MN908947.3) as reference. The genome consensus sequence was obtained on the basis of the identified SNPs and the reference sequence. Reference bases were called in conserved positions with coverage above five, otherwise N were introduced.
A dataset of 3.995 SARS-CoV-2 genomic sequences was retrieved from GISAID database (Elbe and Buckland-Merrett, 2017). The multi-genome alignment including our genome, GISAID genomes and reference genome was obtained using the Purple pipeline (Gona et al., 2020), (available at https://skynet.unimi.it/index.php/tools/purple-tool/). The nucleotide distances among the strains were computed using the R library Ape and the 300 most similar sequences to our strain were selected for Maximum Likelihood phylogenetic analysis. The phylogenetic analysis was performed using RaxML8 software with 100 pseudo bootstraps, after model selection using ModelTest-NG. The obtained phylogenetic tree was visualized using iTol web tool.
2.6. Caffeine quantification
Caffeine is a wide used tracer of urban-only pollution and can be correlated with the number of people connected in the sewage system (Viviano et al., 2017). Because it is a highly degradable molecule, especially in the wastewater treatment plants, detecting its presence in a surface water is a clear indication of the presence of untreated sources of wastewaters or the malfunctioning of the sewer network. Caffeine has been determined by a Liquid Chromatography-High Resolution Mass Spectrometry (LC-HRMS) method by using an Orbitrap Q Exactive mass analyser equipped with Ultimate 3000 HPLC system (Thermo Fisher Scientific, U.S.). The mass spectrometer operated in HESI positive ionization mode and a Full scan-data dependent scan acquisition mode (FS-ddms2) was applied. The liquid chromatography parameters and the mass spectrometer settings are detailed in Supplementary Material S2 (Tables S3–S5).
Identification has been carried out by comparison with the retention time, extracted accurate mass (m/z 195.0882 amu) of [M + H]+ ion and MS/MS spectrum of pure standard solutions (prepared in ultrapure water) which were used also for calibration. All acquired accurate masses were within 2 ppm from the theoretical one. No interferences have been detected. Quantitative determination has been carried out by external calibration. Accuracy has been determined by recovery of three spiked samples. The recoveries were all greater than 90%. Precision were estimated as the reproducibility of three replicates of real samples, one inlet of WWTP (WWTP A) and one river (River Lambro). The coefficients of variation were respectively 18% and 23%. LOD, calculated according to the ISO 6107-2: 2006 standard, was 0.02 μg/L.
3. Results and discussion
3.1. Presence and infectivity of SARS-CoV-2 in wastewaters
The amplification of SARS-CoV-2 RNA genes ORF1ab, N and E was successful in the raw WWs from all the WWTPs on April, 14th, 2020, and only in the raw WW of the WWTP-B plant on April, 22nd, 2020 (Table 1). Hence, the presence of detectable SARS-CoV-2 RNA in influent WWs is confirmed even in the case of the Milano metropolitan area (La Rosa et al., 2020), in congruence with growing evidences worldwide (Ahmed et al., 2020; Kokamemi et al., 2020; Lodder and de Roda Husman, 2020; Medema et al., 2020; Randazzo et al., 2020; Wu et al., 2020; Wurtzer et al., 2020). On the contrary, treated WWs did not result positive to amplification on any date (Table 1).
The null detection of viral RNA in the outflows of Milano and Monza WWTPs plants, equipped with tertiary treatments, is in agreement with the case of the Murcia region (Spain) where tertiary treatments abated SARS-CoV-2 concentration below the LOD (50 gene copies per reaction) in all the studied plants (Randazzo et al., 2020). We cannot exclude that viral copies were still present in the outflows of the studied WWTPs at low concentrations (Randazzo et al., 2020), under the 95% LOD of the multiplex reaction, or that any viral particles was not sampled in the 200 μL of filtered WWs used for RNA extraction. Moreover, the presence of PCR inhibitors in the matrixes could not be totally excluded.
It is also known that the targeted genes proved different sensitivity thresholds (Jung et al., 2020; Zhou et al., 2020), so that virus detection could be influenced by the sensitivity of the employed marker. Interestingly, ORF1ab gene resulted amplified in all positive samples, while both the other two genes (N and E) failed to be amplified in two out of five positive samples. Intriguingly, this is in contrast with growing evidences for a higher sensitivity of markers targeting the N region, respect to the ORF1ab (Jung et al., 2020; Zhou et al., 2020). Any speculation about the origin of this discrepancy cannot here be reliably advanced, and specific in-depth assessment of PCR efficiency, or mutations accumulation in genomes increasing the false negative rate (Phan, 2020; Su et al., 2020) should be done.
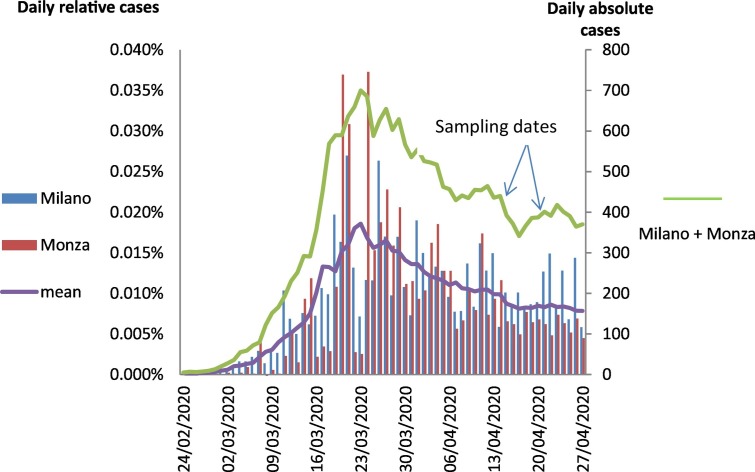
Interestingly, the positivity disappeared in most of the inlet samples on April, 22nd, indicating a possible decrease of the viral concentration. Sampling sessions were planned during the first stages of the epidemic decline. Fig. 2 shows that both Provinces recorded the same trend and magnitude of relative diagnosed cases in the study period. The infection started to increase since the end of February 2020, reached a maximum in the second half of March 2020, and then was slowly decreasing. However, it is worth of mention that these epidemiological data need standardized collection methods (Sims and Kasprzyk-Hordern, 2020) and are probably affected by unequal sampling efforts and asynchrony of records respect to the real infection dates. Moreover, the use of non-quantitative methods for RNA detection, and the reduced sampling frequency did not allow to assess any correlation between the viral RNA titers and the clinical epidemiological trend.
Fig. 2.
Trend of COVID-19 cases diagnosed in Province of Milano and Monza and Brianza (source: Protezione Civile, https://github.com/pcm-dpc/COVID-19/blob/master/dati-province/dpc-covid19-ita-province.csv). On the left y-axis, relative cases represent the new daily cases divided by the resident population in each Province. On the right y-axis, absolute cases represent the weekly window mean of the sum of the new daily absolute cases of both Provinces.
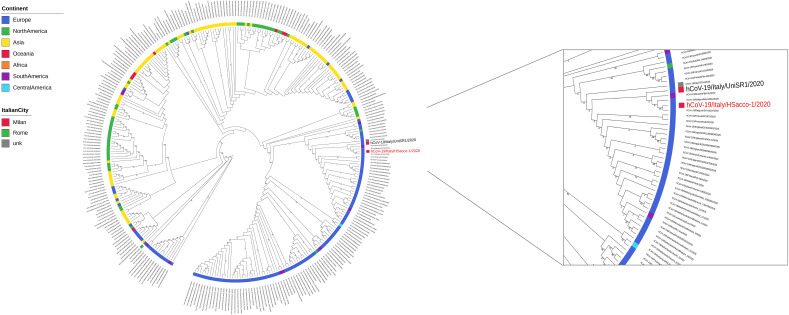
A single genome was sequenced, associated to a strain isolated in WWTP-A. The assembly procedure allowed to call 25.279 genome bases, corresponding to ~85% of the reference genome length. Phylogenetic analysis (Fig. 3 ) revealed that the sequenced strain is closely related to a SARS-CoV-2 strain isolated on March, 3rd, 2020 in Milan (GISAID code EPI_ISL_413489). We found two SNPs between the two strains, including a non-synonymous mutation on the ORF1 gene at position 2231 (L2231I). Moreover, this strain is within the main clade of European genomes, congruently with a common origin (Bai et al., 2020).
Fig. 3.
Maximum Likelihood phylogenetic tree including the the Hsacco-1 strain sequenced in this work and the 300 most similar SARS-CoV-2 strains retrieved from GISAID database. Geographic strains metadata, as retrieved from GISAID database, were mapped on the tree: the isolation continent is reported on the inner circle and, for the Italian strains only, the isolation city is reported on the external circle. The labels of the strains isolated in Milan are reported with larger size and the strain sequenced in this work is coloured in red. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Notably, the infectivity of SARS-CoV-2 in WWs, either raw or treated samples, resulted null, despite the presence of viral RNA in the samples (Table 1) and despite infectious virus are supposed to be present in stool and urine after emission (Singer and Wray, 2020). At this regard, it has been demonstrated that virus can be not infective also in human samples where declining, but still detectable, concentrations of SARS-CoV-2 RNA were determined (La Scola et al., 2020). Moreover, infectivity assays made on hospital WWs, before and after preprocessing disinfection, resulted negative as well, despite the detectability of SARS-CoV-2 RNA (J. Wang et al., 2020).
Enveloped virus are more susceptible to decay of their infectivity in wastewaters than non-enveloped viruses (Ye et al., 2016), especially when in presence of free active enzymes activity or predators like protozoan or metazoan (Kim and Unno, 1996). Survival of coronaviruses in wastewaters was estimated in a few studies. Temperature resulted one of the most influent parameters, and time to complete decay was estimated around 2 days at mild temperature (23–25 °C) and longer in colder conditions (over 22 days at 4 °C) (Gundy et al., 2009; Wang et al., 2005). However, most of the decay should occur immediately after the discharge in sewage, due to presence of solvents and detergents compromising the viral envelope, or to the absorption to the solid fraction (Gundy et al., 2009; Hart and Halden, 2020). In the present case-study, the time from stool emission to the arrival at the WWTP has been estimated to be about 6–8 h according to the mean corrivation times of WWs provided by the WWTP managers. In the sampling period, WWs temperature was in the range 18.5–19 °C and probably did not favour the survival of all, or most of SARS-CoV-2 viruses up to the inlet in the WWTPs. In this regard, the choice to avoid sample concentration to preserve virus infectivity could have also increased the chance of false negatives either for real time RT-PCR and infectivity test in the less concentrated samples, such as those of April, 22nd. Considering also the potential presence of free viral RNA in waters, a few studies have estimated a persistence from less than one hour in WWs (Limsawat and Ohgaki, 1997) to two days in sea water (Tsai et al., 1995), depending on environmental conditions and virus typology. Hence, any speculation about the role of wastewater treatments in reducing the viral concentration, respect to the natural decay of virus load in the sewage systems and WWTPs, cannot be advanced at this stage. At this regard, specific analyses targeting the persistence of SARS-CoV-2 RNA in WWs under different environmental conditions are ongoing. However, we strongly advocate for a precautionary approach in risk assessment in other contexts, being the presented data limited in time and related to highly efficient WWTPs (i.e., equipped with tertiary treatments).
3.2. Presence and infectivity of SARS-CoV-2 in surface waters
Positive detection of viral RNA was found in all receptors water bodies on April, 14th, 2020, but only in the Lambro River on April, 22nd, 2020 (Table 1), following probably the general epidemiological decreasing trend. The presence of SARS-CoV-2 RNA in surface waters, when compared to the negative detection found for treated WWs, could derive from different sources of viral RNA coexisting in the same basins. It is known that aliquots of non-treated sewage can be present in surface waters because of e.g., illicit discharges, malfunction of sewerage systems, and that their relative contribution to the river flow is enhanced during drought period (Mosley, 2015) such as in Spring, 2020. This hypothesis has been tested with our samples by determining the caffeine concentrations in rivers, which is a specific tracer of untreated domestic sewages (Viviano et al., 2017). Caffeine concentrations (0.3–0.4 μg/L) measured in Lambro and Lambro Meridionale rivers (Table 1) were higher than the background caffeine levels of Lambro river (0.08–0.09 μg/L) detected in a reference river section during a dry period (Viviano et al., 2017).
As shown in Table 1, since the degradation in WWTPs is complete, with a removal greater than 99% in all the monitored plants, the WWTPs are not a source of caffeine in the receiving rivers. Consequently, the caffeine measured in rivers Lambro and Lambro Meridionale should have another origin. These findings suggest that a fraction of untreated sewage has been directly discharged in surface waters, due to non-collected domestic discharges or to the lack of separation of the urban runoff waters from the domestic effluents, which causes combined sewer overflows (CSOs). CSOs usually occur during high precipitation events in order to prevent damages on the WWTPs. However, even during prolonged drought, some CSO devices can be active because of possible failures of the sewerage systems (Salerno et al., 2018). In the case of these two rivers, the simultaneous measurement of caffeine and viral genetic material opens the possibility that the common origin is the untreated sewage.
The malfunctioning of CSOs can cause repeated contamination of receiving surface waters, and it is not uncommon even in sewages of many European countries (Rizzo et al., 2020) and United States (U.S. Environmental Protection Agency (EPA), 2004).
Nevertheless, the presence of SARS-CoV-2 RNA in the Vettabbia Canal on April, 14th, 2020 and the contextual lack of detection of caffeine in this water body suggest that also the sporadic release of virus traces by WWTP treated outlets cannot be completely excluded. At this regard, it has been shown that WWTPs, equipped with secondary treatment only, can release SARS-CoV-2 RNA in their effluents (Randazzo et al., 2020). Giving the wide presence of small and medium sized WWTPs in the river basins of this study (Fig. 1), we cannot exclude that this event occurred along the studied rivers and contributed to the spread of viral RNA in surface waters.
It is worth of mention that also in the positive cases concerning rivers, the infectivity of the SARS-CoV-2 was null (Table 1), suggesting a low risk of infection from river water. Nevertheless, as in the case of WWs, a precautionary approach in risk assessment is arguable, awaiting for accurate tests for viral persistence and infectivity in contaminated ecosystems. In particular, we cannot also exclude that viable viral particles were present also in Vettabbia and Lambro Meridionale even on April, 22nd, and that their concentrations were below the LOD of real time RT-PCR, or that they were not sampled in the unconcentrated 200 μL or 2 mL of filtered water used for RNA extraction and infectivity tests, respectively.
In conclusion, this study detected the presence of SARS-CoV-2 RNA in WWs and, firstly, in the receiving rivers in the Milano Metropolitan Area. The presence of SARS-CoV-2 genome in rivers indicated the partial efficiency of the current sewerage system of the Milano Metropolitan Area, which is a common occurrence in Europe and USA. Moreover, test for infectivity suggested that pathogenicity of virus in wastewaters and surface waters could be null. However, risks for public health should be evaluated under a precautionary approach, giving the preliminary nature of the presented results.
CRediT authorship contribution statement
Sara Giordana Rimoldi: Conceptualization, Investigation, Methodology, Formal analysis, Writing - original draft, Writing - review & editing. Fabrizio Stefani: Conceptualization, Investigation, Methodology, Formal analysis, Writing - original draft, Writing - review & editing. Anna Gigantiello: Investigation. Stefano Polesello: Conceptualization, Supervision, Writing - review & editing. Francesco Comandatore: Investigation. Davide Mileto: Investigation. Mafalda Maresca: Investigation. Concetta Longobardi: Investigation. Alessandro Mancon: Investigation. Francesca Romeri: Investigation. Cristina Pagani: Investigation. Francesca Cappelli: Investigation. Claudio Roscioli: Investigation. Lorenzo Moja: Writing - review & editing. Maria Rita Gismondo: Conceptualization, Supervision, Writing - review & editing. Franco Salerno: Conceptualization, Supervision, Writing - review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Sara Giordana Rimoldi and Fabrizio Stefani equally contributed to this paper. The authors want to thank the managers of the WWTPs for having contributed to this research and Sara Valsecchi (IRSA-CNR), Michele Rusconi, Jacopo Rigato, Alberto Sala, and Orietta Longoni (BrianzAcque) for collaboration in chemical analysis. The contribution of Igor Fochi and Salvatore Conti (Thermofisher Italia) in LC-HRMS analysis and genome sequencing, respectively, is also acknowledged. This research has been developed in the framework of the SWaRM-Net "Smart Water Resource Management - Networks" - "Smart Cities and Communities" Project (OR4) - Decreto Direttoriale MIUR 5 luglio 2012 n . 391/Ric. This paper is dedicated to the little Clara, which is born in the era of Covid-19.
Editor: Dr. Damia Barcelo
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.scitotenv.2020.140911.
Appendix A. Supplementary data
S1 Real time RT-PCR quality control criteria; S2 Caffeine quantification (chromatographic conditions).
References
- Ahmed W., Angel N., Edson J., Bibby K., Bivins A., O’Brien J.W., Choi P.M., Kitajima M., Simpson S.L., Li J., Tscharke B., Verhagen R., Smith W.J.M., Zaugg J., Dierens L., Hugenholtz P., Thomas K.V., Mueller J.F. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: a proof of concept for the wastewater surveillance of COVID-19 in the community. Sci. Total Environ. 2020 doi: 10.1016/j.scitotenv.2020.138764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Tawfiq J.A. Asymptomatic coronavirus infection: MERS-CoV and SARS-CoV-2 (COVID-19) Travel Med. Infect. Dis. 2020;35 doi: 10.1016/j.tmaid.2020.101608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai Y., Jiang D., Lon J.R., Chen X., Hu M., Lin S., Chen Z., Meng Y., Du H. 2020. Evolution and Molecular Characteristics of SARS-CoV-2 Genome. bioRxiv 2020.04.24.058933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K.W., Bleicker T., Brünink S., Schneider J., Schmidt M.L., Mulders D.G.J.C., Haagmans B.L., Van Der Veer B., Van Den Brink S., Wijsman L., Goderski G., Romette J.L., Ellis J., Zambon M., Peiris M., Goossens H., Reusken C., Koopmans M.P.G., Drosten C. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eurosurveillance. 2020;25:1–8. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbe S., Buckland-Merrett G. Data, disease and diplomacy: GISAID’s innovative contribution to global health. Glob. Challenges. 2017;1:33–46. doi: 10.1002/gch2.1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson K.E., Schwab K.J., Spencer S.K., Borchardt M.A. Measuring and mitigating inhibition during quantitative real time PCR analysis of viral nucleic acid extracts from large-volume environmental water samples. Water Res. 2012;46:4281–4291. doi: 10.1016/j.watres.2012.04.030. [DOI] [PubMed] [Google Scholar]
- Gombold J., Karakasidis S., Niksa P., Podczasy J., Neumann K., Richardson J., Sane N., Johnson-Leva R., Randolph V., Sadoff J., Minor P., Schmidt A., Duncan P., Sheets R.L. Systematic evaluation of in vitro and in vivo adventitious virus assays for the detection of viral contamination of cell banks and biological products. Vaccine. 2014;32:2916–2926. doi: 10.1016/j.vaccine.2014.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gona F., Comandatore F., Battaglia S., Piazza A., Trovato A., Lorenzin G., Cichero P., Biancardi A., Nizzero P., Moro M., Cirillo D.M. Comparison of core-genome MLST, coreSNP and PFGE methods for Klebsiella pneumoniae cluster analysis. Microb. Genomics. 2020;6(4) doi: 10.1099/mgen.0.000347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gundy P.M., Gerba C.P., Pepper I.L. Survival of coronaviruses in water and wastewater. Food Environ. Virol. 2009;1:10–14. doi: 10.1007/s12560-008-9001-6. [DOI] [Google Scholar]
- Gupta S., Parker J., Dolwani S., Smits S., Underwood J. 2020. Persistent Viral Shedding of SARS-CoV-2 in Faeces - A Rapid Review. medRxiv 2020.04.17.20069526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart O.E., Halden R.U. Computational analysis of SARS-CoV-2/COVID-19 surveillance by wastewater-based epidemiology locally and globally: feasibility, economy, opportunities and challenges. Sci. Total Environ. 2020;730 doi: 10.1016/j.scitotenv.2020.138875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hovi T., Shulman L.M., Van Der Avoort H., Deshpande J., Roivainen M., De Gourville E.M. Role of environmental poliovirus surveillance in global polio eradication and beyond. Epidemiol. Infect. 2012;140:1–13. doi: 10.1017/S095026881000316X. [DOI] [PubMed] [Google Scholar]
- Jung Y.J., Park G.-S., Moon J.H., Ku K., Beak S.-H., Kim Seil, Park E.C., Park D., Lee J.-H., Byeon C.W., Lee J.J., Maeng J., Kim S.J., Kim Seung Il, Kim B.-T., Lee M.J., Kim H.G. 2020. Comparative Analysis of Primer-probe Sets for the Laboratory Confirmation of SARS-CoV-2. BioRxiv 2020.02.25.964775. [DOI] [PubMed] [Google Scholar]
- Kim T.D., Unno H. The roles of microbes in the removal and inactivation of viruses in a biological wastewater treatment system. Water Sci. Technol. 1996;33:243–250. doi: 10.1016/0273-1223(96)00426-X. [DOI] [Google Scholar]
- Kokamemi B.A., Kurt H., Hacıoglu S., Yaralı C., Saatci A.M., Pakdemirli B., Yaykıran S. First Data-Set on SARS-CoV-2 Detection for Istanbul Wastewaters in Turkey. medRxiv. 2020 doi: 10.1101/2020.05.03.20089417. [DOI] [Google Scholar]
- La Rosa G., Iaconelli M., Mancini P., Bonanno Ferraro G., Veneri C., Bonadonna L., Lucentini L., Suffredini E. First detection of SARS-CoV-2 in untreated wastewaters in Italy. Sci. Total Environ. 2020;736:139652. doi: 10.1016/j.scitotenv.2020.139652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Scola B., Le Bideau M., Andreani J., Hoang V.T., Grimaldier C., Colson P., Gautret P., Raoult D. Viral RNA load as determined by cell culture as a management tool for discharge of SARS-CoV-2 patients from infectious disease wards. Eur. J. Clin. Microbiol. Infect. Dis. 2020;39:1059–1061. doi: 10.1007/s10096-020-03913-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limsawat S., Ohgaki S. Fate of liberated viral RNA in wastewater determined by PCR. Appl. Environ. Microbiol. 1997;63:2932–2933. doi: 10.1128/aem.63.7.2932-2933.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodder W., de Roda Husman A.M. SARS-CoV-2 in wastewater: potential health risk, but also data source. Lancet Gastroenterol. Hepatol. 2020;5:533–534. doi: 10.1016/S2468-1253(20)30087-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medema G., Heijnen L., Elsinga G., Italiaander R., Brouwer A. 2020. Presence of SARS-Coronavirus-2 in Sewage. medRxiv 2020.03.29.20045880. [DOI] [PubMed] [Google Scholar]
- Mosley L.M. Drought impacts on the water quality of freshwater systems; review and integration. Earth Sci. Rev. 2015;140:203–214. doi: 10.1016/j.earscirev.2014.11.010. [DOI] [Google Scholar]
- Naddeo V., Liu H. Editorial Perspectives: 2019 novel coronavirus (SARS-CoV-2): what is its fate in urban water cycle and how can the water research community respond? Environ. Sci. Water Res. Technol. 2020;6:1213–1216. doi: 10.1039/d0ew90015j. [DOI] [Google Scholar]
- Ogando N.S., Dalebout T.J., Zevenhoven-Dobbe J.C., Limpens R.W., Meer Y. van der, Caly L., Druce J., Vries J.J.C. de, Kikkert M., Bárcena M., Sidorov I., Snijder E.J. 2020. SARS-Coronavirus-2 Replication in Vero E6 Cells: Replication Kinetics, Rapid Adaptation and Cytopathology. bioRxiv 2020.04.20.049924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan T. Genetic diversity and evolution of SARS-CoV-2. Infect. Genet. Evol. 2020;81 doi: 10.1016/j.meegid.2020.104260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randazzo W., Truchado P., Cuevas-Ferrando E., Simón P., Allende A., Sánchez G. SARS-CoV-2 RNA in wastewater anticipated COVID-19 occurrence in a low prevalence area. Water Res. 2020;181 doi: 10.1016/j.watres.2020.115942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzo A., Tondera K., Pálfy T.G., Dittmer U., Meyer D., Schreiber C., Zacharias N., Ruppelt J.P., Esser D., Molle P., Troesch S., Masi F. Constructed wetlands for combined sewer overflow treatment: a state-of-the-art review. Sci. Total Environ. 2020;727:138618. doi: 10.1016/j.scitotenv.2020.138618. [DOI] [PubMed] [Google Scholar]
- Salerno F., Viviano G., Tartari G. Urbanization and climate change impacts on surface water quality: enhancing the resilience by reducing impervious surfaces. Water Res. 2018;144:491–502. doi: 10.1016/j.watres.2018.07.058. [DOI] [PubMed] [Google Scholar]
- Sims N., Kasprzyk-Hordern B. Future perspectives of wastewater-based epidemiology: monitoring infectious disease spread and resistance to the community level. Environ. Int. 2020;139 doi: 10.1016/j.envint.2020.105689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer A.C., Wray R. 2020. Detection and Survival of SARS-coronavirus in Human Stool, Urine, Wastewater and Sludge. Preprints 2020060216. [DOI] [Google Scholar]
- Su Y.C.F., Anderson D.E., Young B.E., Zhu F., Linster M., Kalimuddin S., Low J.G.H., Yan Z., Jayakumar J., Sun L., Yan G.Z., Mendenhall I.H., Leo Y.-S., Lye D.C., Wang L.-F., Smith G.J.D. 2020. Discovery of a 382-nt Deletion During the Early Evolution of SARS-CoV-2. bioRxiv 2020.03.11.987222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai Y.L., Tran B., Palmer C.J. Analysis of viral RNA persistence in seawater by reverse transcriptase- PCR. Appl. Environ. Microbiol. 1995;61:363–366. doi: 10.1128/aem.61.1.363-366.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- U.S. Environmental Protection Agency (EPA) Water. Office of Water; Washington, DC, USA: 2004. Report to Congress on Impacts and Control of Combined Sewer Overflows and Sanitary Sewer Overflows. [DOI] [Google Scholar]
- Viviano G., Valsecchi S., Polesello S., Capodaglio A., Tartari G., Salerno F. Combined use of caffeine and turbidity to evaluate the impact of CSOs on river water quality. Water Air Soil Pollut. 2017;228:330. doi: 10.1007/s11270-017-3505-3. [DOI] [Google Scholar]
- Wang X.W., Li J.S., Jin M., Zhen B., Kong Q.X., Song N., Xiao W.J., Yin J., Wei W., Wang G.J., Si B.Y., Guo B.Z., Liu C., Ou G.R., Wang M.N., Fang T.Y., Chao F.H., Li J.W. Study on the resistance of severe acute respiratory syndrome-associated coronavirus. J. Virol. Methods. 2005;126:171–177. doi: 10.1016/j.jviromet.2005.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Feng H., Zhang S., Ni Z., Ni L., Chen Y., Zhuo L., Zhong Z., Qu T. SARS-CoV-2 RNA detection of hospital isolation wards hygiene monitoring during the Coronavirus Disease 2019 outbreak in a Chinese hospital. Int. J. Infect. Dis. 2020;94:103–106. doi: 10.1016/j.ijid.2020.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Yao H., Xu X., Zhang P., Zhang M., Shao J., Xiao Y., Wang H. Limits of detection of 6 approved RT–PCR kits for the novel SARS-Coronavirus-2 (SARS-CoV-2) Clin. Chem. 2020;2:6–7. doi: 10.1093/clinchem/hvaa099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigginton K.R., Ye Y., Ellenberg R.M. Emerging investigators series: the source and fate of pandemic viruses in the urban water cycle. Environ. Sci. Water Res. Technol. 2015;1:735–746. doi: 10.1039/c5ew00125k. [DOI] [Google Scholar]
- Wu F., Xiao A., Zhang J., Gu X., Lee W., Kauffman K., Hanage W., Matus M., Ghaeli N., Endo N., Duvallet C., Moniz K., Erickson T., Pr C., Thompson J., Alm E. 2020. SARS-CoV-2 Titers in Wastewater Are Higher Than Expected From Clinically Confirmed Cases. medRixiv 2020.04.05.20051540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurtzer S., Marechal V., Mouchel J.-M., Maday Y., Teyssou R., Richard E., Almayrac J.L., Moulin L. 2020. Evaluation of Lockdown Impact on SARS-CoV-2 Dynamics Through Viral Genome Quantification in Paris Wastewaters. medRxiv 2020.04.12.20062679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y., Ellenberg R.M., Graham K.E., Wigginton K.R. Survivability, partitioning, and recovery of enveloped viruses in untreated municipal wastewater. Environ. Sci. Technol. 2016;50:5077–5085. doi: 10.1021/acs.est.6b00876. [DOI] [PubMed] [Google Scholar]
- Zhang N., Gong Y., Meng F., Bi Y., Yang P., Wang F. 2020. Virus Shedding Patterns in Nasopharyngeal and Fecal Specimens of COVID-19 Patients. medRxiv 2020.03.28.20043059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Yunying, Pei F., Wang L., Zhao H., Li H., Ji M., Yang W., Wang Q., Zhao Q., Wang Y. 2020. Sensitivity Evaluation of 2019 Novel Coronavirus (SARS-CoV-2) RT-PCR Detection Kits and Strategy to Reduce False Negative. medRxiv 2020.04.28.20083956. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
S1 Real time RT-PCR quality control criteria; S2 Caffeine quantification (chromatographic conditions).