Abstract
Development of the CRISPR-Cas9 gene-editing system has given rise to a new era of gene editing with wide applications in biology, medicine, agriculture, and other fields. However, the overexpression of Cas9 nuclease causes off-target effects and may trigger an immune response in vivo. Therefore, we constructed a self-restricting CRISPR-Cas9 system, where the target gene sequence corresponding to the guide RNA (gRNA) is inserted on either end of the Cas9 promoter. When double-strand breaks (DSBs) are induced in the target gene sequence, the Cas9 promoter is cut off and transcription ceases. With this system, expression of Cas9 protein at 60 h after transfection is only 10% that of the wild-type system, with about 70% promoter deletion efficiency. The target site editing efficiency and homologous recombination efficiency of the self-restricting system remain at about 50% and 30%, respectively, while the frequency of off-target indel formation decreased by 76.7%. Further, the number of indel types was also reduced from 13 to 2. Because this system does not include additional gRNA sequences, the possibility of introducing new off-target mutations is decreased. Importantly, this system is composed of a single plasmid, which could potentially be easily introduced in vivo using a viral vector or nanoparticles.
Graphical Abstract

Introduction
The CRISPR-Cas9 system is easy to utilize, convenient, and quick. Since it was first reported it has been widely used, quickly superseding previous generation zinc-finger nucleases (ZFNs) and transcription activator-like effector nucleases (TALENs).1, 2, 3, 4 Only the design of a single guide RNA (sgRNA) directed against the target sequence is required for use of CRISPR-Cas9. This guide RNA (gRNA) guides the Cas9 nuclease to bind to a specific gene sequence and induce a DNA double-strand break (DSB).5, 6, 7 Cells then perform self-repair in one of two ways, through homology-directed double-stranded DNA (dsDNA) repair (HDR) or non-homologous end joining (NHEJ).8,9 The CRISPR-Cas9 gene-editing technology is superior to other approaches because it can permanently alter expression of target genes. Thus, this powerful tool can be used to efficiently generate a gene knockout, knockin, or chromosome transposition.10
Several translational studies utilizing CRISPR-Cas9 in preclinical and clinical settings have been reported. In 2017, researchers used CRISPR-Cas9 to remove a portion of the mHTT gene in the brain cells of a mouse model of Huntington’s disease.11 The CRISPR-Cas9 gene knockin function was used to specifically insert the chimeric antigen receptor (CAR) gene targeting CD19 into the T cell receptor alpha chain (TRAC) locus of patients, enhancing the efficacy of CAR-T cells.12 Recently, the US Food and Drug Administration (FDA) has granted CTX001, which targets fetal hemoglobin, fast-track qualification for the treatment of sickle cell disease (SCD) (ClinicalTrials.gov: NCT03655678).
Although CRISPR-Cas9 has many potential gene-editing applications, off-target effects have hampered its clinical use.13,14 One of the major causes of the off-target effects of the CRISPR system is the continued high-intensity expression of Cas9 nuclease within cells.15,16 Thus, several strategies have been developed to reduce the off-target effects resulting from Cas9 overexpression.
-
1.
The transfection of CRISPR-Cas9 ribonucleoproteins (RNPs). In this strategy, the Cas9 protein and in vitro-transcribed sgRNA are pre-complexed and directly delivered into target cells via electroporation or lipid-mediated-transfection. Because the half-life of the Cas9-sgRNA RNP complex is shorter than the time required to transcribe plasmid or viral nucleic acid, the off-target rate is lower.17
Two major limitations need to be acknowledged with respect to the RNP approach.18 First, there is an inability to precisely control the amounts of RNPs transfected into cells. Second, efficient delivery of these molecules in vivo is difficult because gRNA is easily degraded and the RNP molecular complex is quite large.
-
2.
Suppression of Cas9 protein activity. The need for precision control of Cas9 over the dimensions of dose and time has created a demand for inhibitory anti-CRISPR molecules that can terminate Cas9 activity following on-target editing.19 Several protein-based anti-CRISPR molecules have been reported.20,21 Protein-based anti-CRISPRs can be highly potent, because they generally possess a greater number of SpCas9 interaction sites. However, the anti-CRISPR protein targeting SpCas9 is too large and easily degraded by proteases, limiting its use.22,23 Compared with anti-CRISPR proteins, small-molecule inhibitors can be cell permeable, reversible, proteolytically stable, and non-immunogenic. Recently, small-molecule inhibitors of CRISPR-Cas9 have been reported.24 It remains to be determined whether these small-molecule inhibitors interact with other targets in mammalian cells, and thus they need to be tested in vivo.24
-
3.
Tunable or inducible systems regulate Cas9 working time, which is helpful to decrease the amount of undesirable DNA cleavage in the genome. Placing Cas9 under the control of a tetracycline-responsive element (TRE) confers the possibility to achieve conditional expression of this gene in the presence of tetracycline/doxycycline (Tet/Dox).25 However, even in the off state, when Tet/Dox are absent, Cas9 expression still exhibits leakiness.26
Other models have used the properties of light-inducible heterodimerization proteins for the modulation of Cas9 activity. This system uses a split Cas9 variant that is fused to light-responsive proteins.27 Upon stimulation with blue light, Cas9 becomes active by reconstitution of the whole active Cas9 nuclease. Although such systems are to some extent reversible and adjustable for regulating Cas9 activity, they are limited to in vitro applications. For in vivo use, the need to penetrate deep tissues to induce Cas9 expression would require more invasive equipment.
-
4.
Knockout of the Cas9 gene. A self-limiting CRISPR-Cas9 system, simultaneous expression of two gRNAs, one targeting the Cas9 gene and the other for the target gene, has recently been reported.28,29 However, it is well-known that introducing an additional gRNA targeting the Cas9 gene will inevitably produce additional off-target effects. Thus, this method cannot effectively reduce off-target effects and requires further optimization.
In order to reduce the overexpression of Cas9 protein and reach an expression mode of less is good, as well as to avoid new off-target problems caused by the introduction of additional gRNAs, we constructed a self-restricting CRISPR-Cas9 (SR-CRISPR) system. In this study, we selected programmed cell death protein 1 (PD-1) as the target. Further, we verified that with this system not only is the editing efficiency of the target gene unchanged but off-target effects are also significantly reduced.
Results
Design of Self-Restricting CRISPR-Cas9 (SR-CRISPR) System
In this study, we selected PD-1 as the target and designed three corresponding gRNAs. Among them, gRNA3 was found to induce efficient cleavage of Cas9 and was selected for use in the construction of the SR-CRISPR gene-editing plasmid (Figure S1). The top and bottom strand of gRNA3 were annealed and cloned into a genome-editing plasmid (pX458), which was then named pX458 s3. Unique recognition sites for restriction enzymes AgeI and KpnI were adjacent to each end of the ca promoter. The pX458 s3 plasmid was digested with AgeI and KpnI, the annealed oligo (the target gene consensus sequence corresponding to the gRNA) (Table S1) was then inserted into the KpnI and AgeI sites directly preceding and following the Cas9 promoter, respectively (Figure 1A), to construct the pX458 s3-AgeI-KpnI (SR-CRISPR) plasmid (Figure 1B). Because the gRNA also targets the Cas9 gene promoter, transcription is terminated and overexpression is prevented. When transfected into HEK293T cells, this plasmid is designed to induce controlled expression of the Cas9 protein at a level that still allows the formation of a protein complex with the gRNA targeted to PD1. Thus, this should achieve the goal of reducing off-target effects, while at the same time allowing the Cas9/gRNA complex to cut the PD1 gene at the target site to form the indel mutation (Figure 1C).
Figure 1.
Construction of a Self-Restricting CRISPR-Cas9 System
(A) Schematic diagram of the construction of SR-CRISPR plasmid (pX458 s3-AgeI-KpnI). The promoter of the Cas9 gene in the px458 s3 plasmid is flanked by the AgeI and KpnI restriction sites. The gRNA target sequence from the gene of interest (PD1) was inserted at both of these sites. In addition, the Cas9 gene and EGFP gene are linked in tandem by the T2A sequence and driven by the same CAG promoter in this plasmid. Cas9 is also fused with SV40 and nucleoplasmin nuclear localization signals (NLSs). The target gene gRNA, Cas9, and EGFP are all co-expressed from this plasmid. (B) Sequencing results of the AgeI and KpnI restriction site insertion sequences. The shaded parts are the insertion sequences. Red circles are PAM sites. (C) Schematic diagram of the self-restricting CRISPR-Cas9 system regulating the expression of Cas9 protein. The SR-CRISPR plasmid is transfected into HEK293T cells, and Cas9 is translated and forms a protein complex with gRNA to induce indel mutations of the PD1 gene at the target site. The protein complex simultaneously cuts the gRNA recognition sequence at both ends of the Cas9 promoter to prevent excessive Cas9 protein transcription.
Detection of Restricted Cas9 Expression
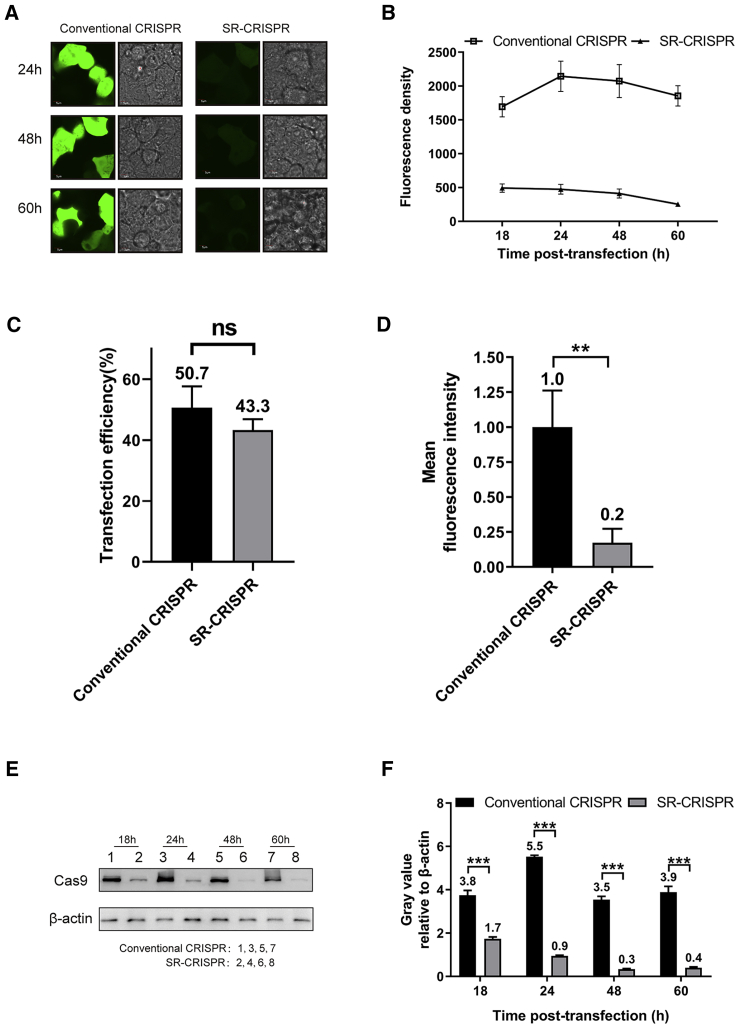
To verify that the SR-CRISPR system inhibits overexpression of Cas9 protein, we transfected the unmodified px458 s3 plasmid (conventional CRISPR) and the modified px458 s3-Agel-KpnI plasmid (SR-CRISPR) into HEK293T cells. The pX458 vector also expresses EGFP, and the Cas9 and EGFP genes are linked in tandem by a T2A sequence and driven by the same CAG promoter. As a result, the expression of EGFP can be utilized as a readout of Cas9 expression. At different time points after transfection (24, 48, and 60 h), the difference in intracellular fluorescence intensity between cells transfected with the SR-CRISPR plasmid and the control parental plasmid was significant (Figure 2A). We quantitated the fluorescence density at each time point, and the results showed that EGFP increased gradually in cells transfected with the parental vector, reaching the highest level at 24 h before leveling off. In contrast, the SR-CRISPR-transfected cells showed the highest level at 18 h, and the fluorescence density decreased gradually with the passage of time (Figure 2B). In addition, flow cytometric analysis of EGFP 60 h after transfection showed that the percentage of EGFP-expressing cells (transfection efficiency) between cells transfected with the SR-CRISPR plasmid and the control parental plasmid was similar (Figure 2C; Figure S2). However, the average fluorescence intensity of the SR-CRISPR-transfected cells was only 20% of that of the parental vector-expressing cells (defined as 1) (Figure 2D). Western blot analysis was conducted to confirm these findings (Figure 2E). At 18 h after transfection, the expression of Cas9 protein in the SR-CRISPR and parental vector-transfected cells was already significantly different, and by 60 h, the Cas9 expression level in the SR-CRISPR cells was only 10% that of the control. Consistent with the confocal results, Cas9 protein reached its peak at 24 h before staying consistent in control cells, whereas the protein expression level in the SR-CRISPR cells peaked earlier and gradually decreased with time (Figure 2F). Together, the results in Figure 2 show that, compared with the conventional CRISPR system, the self-regulating CRISPR-Cas9 system can greatly reduce the expression of Cas9 protein, thereby inhibiting Cas9 overexpression.
Figure 2.
The Expression of Cas9 Protein Is Significantly Reduced in the Self-Regulating CRISPR-Cas9 System
(A) Confocal microscopy at different time points of HEK293T cells transfected with parental pX458 s3 plasmid or SR-CRISPR plasmid. (B) Fluorescence density of EGFP at different time points in HEK293T cells transfected with parental pX458 s3 plasmid or SR-CRISPR plasmid. Bar graphs are shown as average of three independent experiments with error bars representing SEM. (C) Flow cytometric analysis of the transfection efficiency at 60 h in HEK293T cells transfected with parental pX458 s3 plasmid or SR-CRISPR plasmid. Bar graphs are shown as average of three independent experiments with error bars representing SEM. (D) Flow cytometric analysis of the average fluorescence intensity of EGFP at 60 h in HEK293T cells transfected with parental pX458 s3 plasmid or SR-CRISPR plasmid. Bar graphs are shown as average of three independent experiments with error bars representing SEM. (E) Western blot of Cas9 protein at different time points in HEK293T cells transfected with parental pX458 s3 plasmid or SR-CRISPR plasmid. (F) Quantitation of Cas9 relative to the internal control, β-actin, at different time points in HEK293T cells transfected with parental pX458 s3 plasmid or SR-CRISPR plasmid. The expression of Cas9 protein was analyzed by western blot, and the gray values of all bands were scanned and analyzed using ImageJ software. Results were normalized to the gray value of the internal control (β-actin) and transfection efficiency. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001. Bar graphs are shown as the average of three independent experiments with error bars representing SEM.
We wanted to further test the self-cutting efficiency of the self-regulating CRISPR-Cas9 system. To this end, we designed an upstream primer within the promoter and a downstream primer (Table S1) near the gRNA recognition sequence inserted at the KpnI digestion site of the pX458 s3-AgeI-KpnI plasmid (Figure S3A). As a result, in the absence of excision, the promoter can be amplified, and it cannot be amplified if excision has occurred. Genomes from the control and SR-CRISPR-transfected cells were subjected to real-time PCR amplification. As can be seen in Figure S3B, the cutting efficiency of the SR-CRISPPR promoter is about 70%. We performed PCR (Table S2) and next generation sequencing (NGS) (Figure S4A). It has been shown that many insertions and deletions occur near the predicted cutting site after promoter excision. Furthermore, the frequency of indel mutations upstream of the predicted cutting site was found to be markedly higher than that downstream (Figure S4B). The analysis of frequency distribution of indel mutations revealed that about 70% of indels were deletions. The frequency of deletions greater than 10 bp was an overwhelming majority (Figure S4C).
Analysis of Target Gene-Editing Efficiency
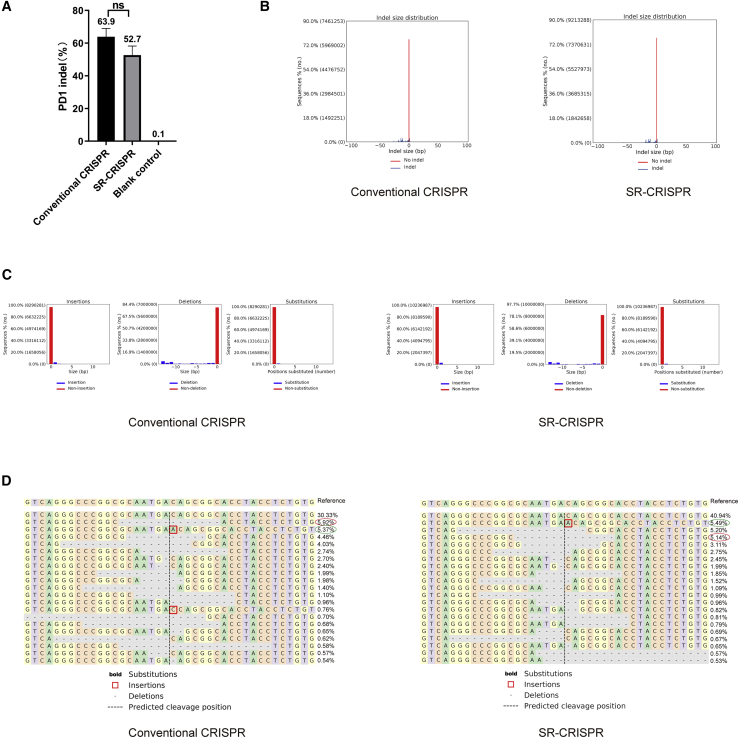
In order to test the editing efficiency of SR-CRISPR for the target gene, PD1, we transfected the parental pX458 s3 plasmid and the SR-CRISPR plasmid into HEK293T cells and extracted the cell genome 60 h later. Primers (Table S2) were designed on both sides of the target gene cutting site, and two rounds of PCR were carried out to prepare sequencing libraries. Sequencing was performed using the NGS method, and genome editing was analyzed. As shown in Figure 3A, the editing efficiency of the parental and SR-CRISPR vector-transfected groups is not significantly different, with both being about 50%. For both, the frequency of indel mutations on the left side of the predicted cutting site was found to be higher than that on the right side (Figure 3B). Interestingly, insertions and substitutions occurred mainly on the right side of the predicted cutting site with both vectors, and deletions occurred mainly on the left side of the predicted cutting site within the PD1 gene. Moreover, the frequency of deletions greater than 10 bp occurring on the left side of the predicted cutting site was found to be relatively high, and the frequency of deletions of less than 5 bp was relatively low (Figure 3C). Figure 3D shows that there were 23 indel types, 17 of which occurred in both vector-transfected groups with the high-frequency indel types of the two groups being roughly the same. For example, the highest frequency indel type in the control group (a 7-base deletion on both sides of the predicted cleavage site) is also ranked the third highest frequency indel type in the SR-CRISPR group. The highest frequency indel type in the SR-CRISPR group (an A base inserted at the first position to the right of the predicted cleavage site) was ranked the second highest frequency indel type in the control group. Together, these results indicate that there is no reduction in the PD1 editing efficiency of the self-regulating CRISPR-Cas9 system.
Figure 3.
Editing Efficiency of the PD1 Target Gene
(A) Comparison of the editing efficiency of the PD1 target gene by the parental and SR-CRISPR vectors. At 60 h after transfection, genomes were extracted for NGS to determine the editing efficiency of PD1. Results are the mean ± SEM of n = 3 independent biological replicates. (B) Frequency distribution of alleles with indels (blue) and without indels (red) of the parental (left) and SR-CRISPR (right)-transfected cells. (C) Specific classification of the parental (left) and SR-CRISPR (right)-induced modifications. Left panels: frequency distribution of sequence modifications that increase read length with respect to the reference amplicon, classified as insertions (positive indel size). Middle panels: frequency distribution of sequence modifications that reduce read length with respect to the reference amplicon, classified as deletions (negative indel size). Right panels: frequency distribution of sequence modifications that do not alter read length with respect to the reference amplicon, which are classified as substitutions (number of substituted positions shown). (D) Alignments of alleles with the reference allele. The predicted cut site is indicated by the vertical black line. The percentage and total number of each allele are shown to the right of each alignment. All alleles observed with frequency rate ≥0.20% are shown. ns, no statistical difference.
We wanted to further analyze the efficiency of homologous recombination of target sites in this self-regulating system. To this end, we co-transfected the homologous template (Table S3) with the parental and SR-CRISPR plasmids into HEK293T cells and extracted the genome 60 h later. The efficiency of target site homologous recombination was then analyzed using NGS. There was no significant difference in the efficiency of homologous recombination at the target site between the CRISPR vectors, with both being about 30%. As shown in Figure 4A, the percentage of the imperfect HDR was found to be extremely low (less than 1.0%). Figure 4B shows the imperfect HDR indel frequencies of the parental and SR-CRISPR vectors on both sides of the predicted cutting site. Both types of indel are the same, and the frequencies of each of the three types of indel mutations are similar (0.5%, 0.3%, and 0.2%) (Figure 4C). NHEJ repair occurs while HDR occurs, and the ratio of NHEJ in the control and SR-CRISPR vector groups was found to be higher than that of HDR, with the indels resulting from NHEJ also occurring on the left side of the PD1 gene cutting site (Figure 4D). Interestingly, although the efficiency of homologous recombination in the SR-CRSPR-transfected cells was not reduced compared with the control, the number of indel mutations due to NHEJ was decreased (Figure 4A). Moreover, fewer types of indels were generated by NHEJ in these cells (29 types) than in the control-transfected cells (34 types) (Figure 4E). Together, these results indicate that the self-regulating CRISPR-Cas9 system induces efficient homologous recombination editing of the PD1 target gene at a level similar to the classic CRISPR system.
Figure 4.
HDR Efficiency of the Target Gene PD1
(A) Comparison of the editing efficiency of the parental CRISPR system and the self-regulating system. We co-transfected the homologous template along with the parental or SR-CRISPR vectors into HEK293T and analyzed the editing efficiency at 60 h after transfection. Results are the mean ± SEM of n = 3 independent biological replicates. ∗∗∗p < 0.001. (B) Frequency distribution of alleles with indels (blue) and without HDR indels (red) in the parental (left) and SR-CRISPR (right)-transfected cells. (C) The homologous template (5′-GTCAGGGCCCGGCGCAATAAGCTTGCGGCACCTACCTCTG-3′) is shown as the reference allele with alignments below. (D) Frequency distribution of alleles with NHEJ indels (blue) and without NHEJ indels (red) in the parental (left) and SR-CRISPR (right) groups. (E) Visualization of the distribution of identified alleles around each cleavage site for the guide (5′-GTCAGGGCCCGGCGCAATGACAGCGGCACCTACCTCTGTG-3′). The vertical dashed line indicates the predicted cleavage site. The percentage and total number of each allele are shown to the right of each alignment. All alleles observed with frequency ≥0.20% are shown.
Analysis of Off-Target Effects
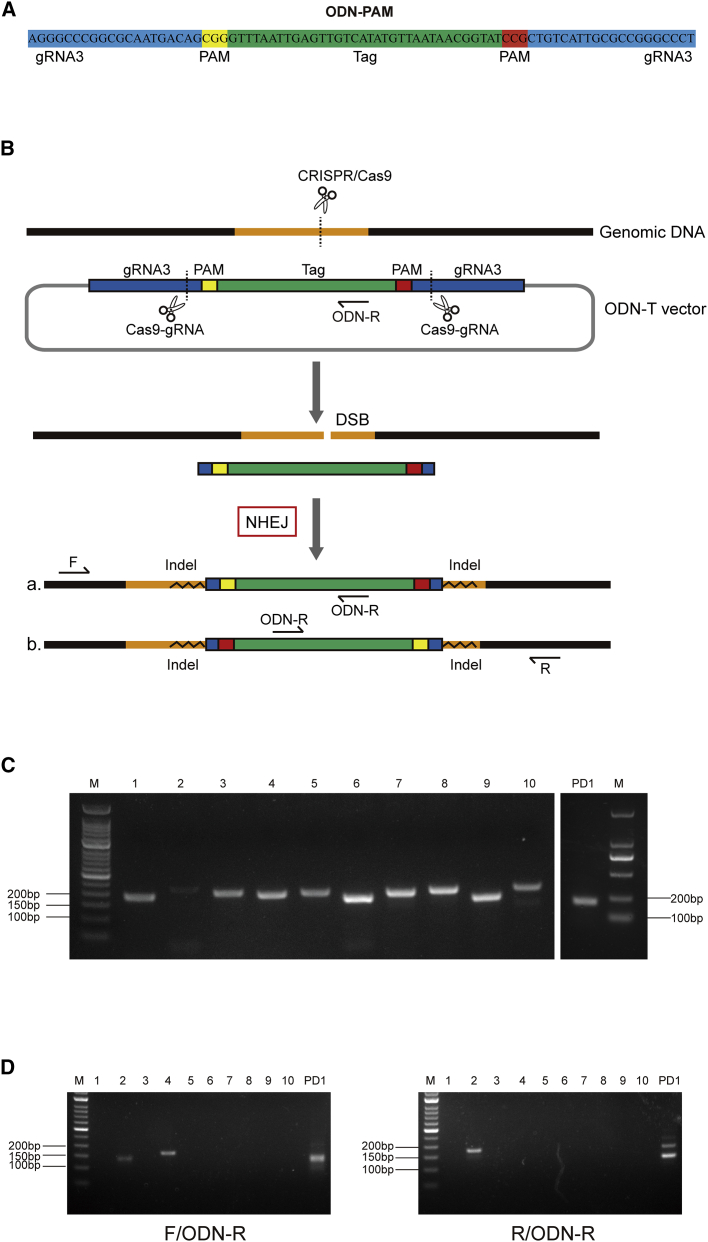
The Guide Design Resources website (http://zlab.bio/guide-design-resources) was used to predict off-target mutation sites based on the sequence of the gRNA. Using this tool, we selected the 10 predicted off-target sites with the highest scores as detection objects (Table S4). Homology-independent targeted insertion (HITI), broadly tested in vitro and in vivo, is a knockin NHEJ-based strategy.30, 31, 32 With this system the donor sequence is flanked by one gRNA target site. Based on CRISPR-Cas9-mediated HITI studies, the following strategies were adopted to verify off-target sites: an 80-bp fragment was synthesized (Table S1), ODN-PAM (Figure 5A), and ligated into the T vector (cloning vector). The two ends of this fragment are the same symmetrical sequence comprised of a target gene gRNA3 and the PAM site, which is close to a Tag sequence. We named the constructed vector ODN-T. The parental px458 s3 plasmid and ODN-T vector were co-transfected into HEK293T cells. The gRNA/Cas9 complex is expected to cleave DNA at the target site or at potential off-target sites in the genome, resulting in DSBs. The gRNA3 recognition sequence of the ODN-T vector will also be recognized and cleaved to generate free Tag fragments. The free fragment can be inserted into the genomic DNA by the NHEJ repair method, resulting in insertions a and b in opposite directions (Figure 5B). Primers (Table S5) were designed upstream and downstream of the target and potential cleavage sites. The expected PCR amplification product is 200–250 bp in size. The size of the electrophoresis band of the PCR amplification product at the target site and at 10 potential off-target sites was found to be consistent with expectations (Figure 5C). The upstream and downstream primers were then used to perform a second round of PCR with the ODN-R primer corresponding to the Tag. The electrophoresis results showed that the PCR products of the upstream and downstream primers at the target position produced bands similar to the expected target band size. Of 10 potential off-targets, only off-2 and off-4 had bands of the expected size, and with the ODN-R primer only off-4 amplified the expected bands with the upstream primers, but not with the downstream primers, indicating that the potential off-2 and off-4 are likely true off-target sites (Figure 5D).
Figure 5.
Determination of Off-Target Sites
(A) Sequence of ODN-PAM. (B) Schematic diagraming the principle of detection of the editing site by co-transfection of the pX458 s3 plasmid and ODN-T vector into HEK293T cells. (C) PCR electrophoretogram of potential off-target sites (left) and the target site (right). (D) PCR using the ODN-R primer with the upstream (left) or downstream (right) primers.
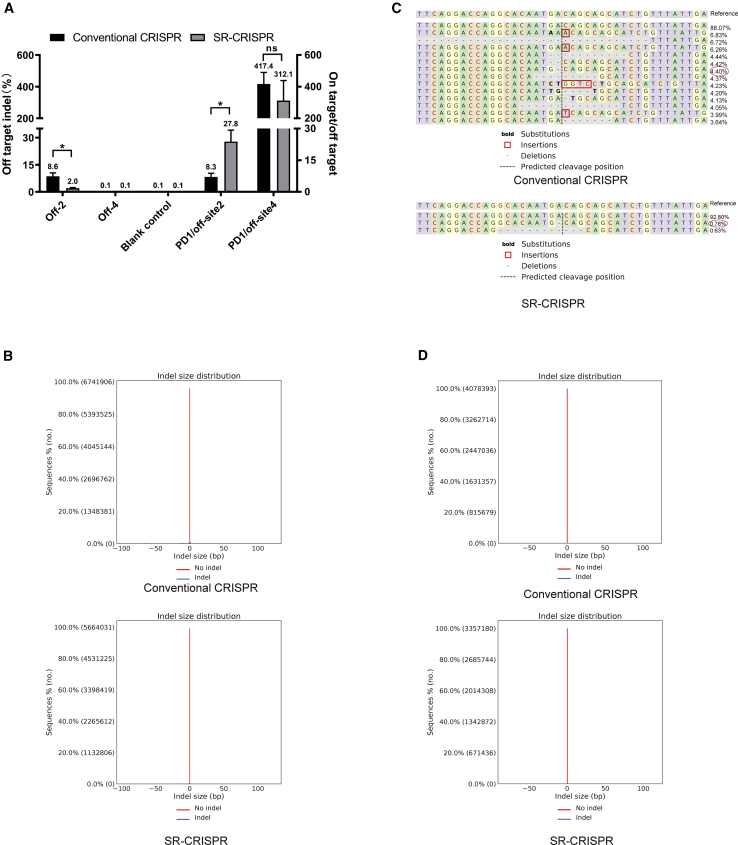
Therefore, in order to analyze the off-target effects produced by the self-regulating CRISPR-Cas9 system, we studied off-target sites 2 and 4 as representative sites, and the indel frequency generated by incorrect cutting was analyzed. Two rounds of PCR were performed to prepare sequencing libraries with primers designed on both sides of the predicted cutting sites of off-target 2 and 4 (Table S2). The off-target efficiency was analyzed using NGS. For the off-2 site, the indel frequency was reduced from 8.6% (control plasmid) to 2.0% (SR-CRISPR plasmid), a 76.7% reduction. We next calculated the ratio of target site PD1 to off-target sites 2 and 4. The PD1/off-2 site ratio in the modified group was 27.8, and the ratio in the unmodified group was 8.3 (Figure 6A). Thus, these results verify that off-2 is a real off-target site. Figure 6B shows that following transfection with the control vector, indel mutations occurred on both sides of the predicted cutting site of off-2, while transfection of the SR-CRISPR vector resulted in most of the indel mutations to the left of the predicted cutting site. The number of types of indel was also reduced from 13 (parental vector) to 2 (SR-CRISPR vector) (Figure 6C). Although one type of indel was novel (did not occur with the parental vector), its frequency was very low (0.63%). The other type of indel was reduced from 4.40% (parental vector) to 0.78% (SR-CRISPR vector). Although insertion, deletion, and replacement occurred following transfection with the parental vector, only deletion occurred with the SR-CRISPR plasmid (Figure 6C). In addition, for the off-4 site (Figure 6D), no significant difference in mutations was found between the SR-CRISPR and parental vector. This may be because off-target site 4 is a low-frequency off-target site, which also explains why only a single expected PCR band (upstream primer with primer ODN-R) was observed for this site. Together, the results in Figure 6 verify that the self-regulating CRISPR-Cas9 system can effectively reduce off-target effects.
Figure 6.
Off-Target Effects in HEK293T Cells
(A) Off-target effects of off-2 and off-4. We transfected the parental pX458 s3 plasmid (conventional CRISPR) and the pX458 s3-AgeI-KpnI plasmid (SR-CRISPR) into HEK293T cells, and extracted the genome 60 h after transfection to perform NGS. The ratio of PD1 target sites to off-target sites 2 and 4 was calculated. Results are the mean ± SEM of n = 3 independent biological replicates. ∗p < 0.05. (B) off-2: frequency distribution of alleles with indels (blue) and without indels (red) of the parental (top) and SR-CRISPR (bottom)-transfected cells. (C) off-2: alignments are shown in comparison with the reference allele. The predicted cut site is represented by the vertical black line. The percentage and total number of each allele are shown to the right of each alignment. All alleles observed with frequency ≥0.20% are shown. (D) off-4: frequency distribution of alleles with indels (blue) and without indels (red) in the parental (top) and SR-CRISPR (bottom)-transfected groups.
Discussion
The prospects for use of the CRISPR-Cas9 system in the clinical treatment of genetic diseases and tumors are immense; however, off-target effects are the prime obstacle limiting clinical application of this system.33 In recent years, researchers have optimized the specificity of the CRISPR-Cas9 system by improving gRNA specificity34, 35, 36, 37 and modifying the Cas9 protein in order to reduce these off-target effects.38,39
The continuous expression of Cas9 protein is an important cause of off-target effects;15,16 thus, several strategies have been developed to control Cas9 overexpression. Among these approaches, the knockout of the Cas9 gene is a simple and effective method for restricting Cas9 overexpression.
In a previous study, a gRNA sequence-targeting Cas9 gene was added to the gene-editing vector to destroy the Cas9 gene.28 However, it is well-known that introducing an additional gRNA will inevitably produce additional off-target effects. Therefore, reducing Cas9 expression with this method bears the risk of additional off-target effects in the genome. Our design uses the same gRNA sequence to destroy the target gene and prevent the continuous expression of Cas9. Because there is no increase in the number of gRNAs, there is no risk of additional off-target effects. Furthermore, the authors of the previous study did not conduct quantitative analysis of the reduction of off-target indels occurring as a result of the destruction of the Cas9 gene. In addition, the qualitative analysis results revealed that the target site editing efficiency was also reduced with this system compared with the conventional CRISPR. In contrast, our system did not result in reduced editing efficiency of the target site, and it actively reduced the formation of off-target indels by 76%.
Recently, Shen et al.29 built a synthetic L7Ae switch to regulate Cas9 expression. This system employs the K-turn system on one vector along with a gRNA sequence to recognize the target gene and a gRNA sequence to recognize the Cas9 gene on the other vector. The synthetic switch is intended to control the transcription and translation of Cas9 simultaneously.29 Nevertheless, these designs use two gRNA sequences, increasing the risk of off-target effects. In addition, two heterologous proteins from archaebacteria were also expressed in the cell, which made the operation cumbersome. The requirement for co-transfection of two plasmids greatly reduces the delivery efficiency and increases the difficulty of use in vivo. A comparative analysis of these data is shown in Figure S5. With respect to destroying the Cas9 gene, the formation of off-target indels was reduced by 37.29%. For the restriction of the translation system, the formation of off-target indels was reduced by 60.76%. Further, the combination of destroying the Cas9 gene and restricting translation can reduce the off-target indel by 88.53%. The SR-CRISPR system constructed for our study reduced the formation of off-target indels by 76%. Thus, although there is not a significant difference in the effectiveness of these systems, many risks are avoided with our approach.
As a next step, we plan to design a gRNA-insertion box within our vector (Figure S6), to greatly simplify the construction steps required to make it adaptable for other target genes. For primary cells or tissues, such as T cells, that are difficult to transfect, low transfection efficiency is indeed a challenge for genome editing with CRISPR-Cas9. However, one solution is to use viral vectors for transduction of Cas9 and gRNA, as shown to be effective in many related studies.40, 41, 42 Our SR-CRISPR system is composed of a single plasmid and could easily be introduced into a viral vector.
Single-base editing technology (base editing) is a new target gene modification technology developed based on the CRISPR-Cas system. This system makes use of either a cytosine base editor (CBE) or adenine base editor (ABE). In this system, a deactivated Cas protein lacking catalytic activity is fused with the deaminase, which can bind to single-stranded DNA (ssDNA) so as to cause base substitution within the target.43,44 Unfortunately, both CBE and ABE are known to cause severe off-target phenomena at the RNA and DNA levels.45,46 Recently, some experiments have found that the introduction of a specially designed RNA into cells to recruit endogenous adenosine deaminase to efficiently and accurately edit specific adenosines of targeted gene transcripts results in no obvious off-target phenomena.47,48 Thus, this indicates that similar to Cas9 off-target effects, the high-intensity expression of the deaminase and deactivated Cas9 fusion proteins is an important cause of off-target effects. Therefore, the idea of less is good with respect to Cas9 protein expression proposed in this paper could also be applied to optimization of the single-base editing system to reduce off-target effects.
In short, the self-restricting CRISPR-Cas9 system presented here can effectively control the expression of Cas9 protein, resulting in a moderate concentration of Cas9 protein in the cell and effective reduction of off-target effects without a significant loss of editing efficiency. A further study should be carried out to verify additional target genes and detect the genome-wide off-target effects with the SR-CRISPR system. In addition, it may be possible to further optimize this system in the future. For example, the wild-type Cas9 protein could be replaced by mutants (espcas9 and spcas9-HF1) that significantly reduce off-target effects. Also, the use of viral vectors or encapsulation within nanomaterials for delivery of this system may improve its efficiency even further. Thus, this study provides a foundation for promoting the clinical application of the CRISPR-Cas9 system in the treatment of genetic diseases and tumors.
Materials and Methods
Plasmid Construction
To construct the self-limiting SpCas9 plasmid, we modified in our laboratory the existing pX458 s3 vector targeting the PD1 gene. The Cas9 and EGFP genes of the pX458 vector are linked in tandem by a T2A sequence and driven by the same CAG promoter. Two complementary 27-bp oligonucleotides corresponding to the target gene were annealed to generate dsDNA with 4-bp overhangs on both ends. This dsDNA sequence was subcloned into AgeI and KpnI restriction sites preceding and following the CAG promoter, respectively. The resultant gRNA expression vector was designated pX458 s3-AgeI-KpnI (SR-CRISPR).
Transfection into HEK293T Cells
HEK293T cells (high-glucose DMEM, 10% fetal bovine serum [FBS]) were grown to confluency, then seeded in six-well plates at a density of 5 × 105 cells/well and cultured at 37°C until the cell density reached 80%–90%. The parental vector (pX458 s3) (2.5 μg/well) and SR-CRISPR vector (pX458 s3-AgeI-KpnI) (2.5 μg/well) were transfected into cells using Lipofectamine 2000 (Thermo Fisher Scientific, UK), and the indel mutation frequency was analyzed after 60 h.
Preparation of Libraries and Deep Sequencing
NGS primers of the target region were designed in the 200- to 300-bp size range. Primers were not permitted to reside within a 50-nt region to either side of the expected sgRNA cut site to ensure coverage of varied deletion sizes. A two-step PCR fusion method was performed to attach bar-coded sequencing adaptors for multiplex deep sequencing. Sequencing of the libraries was completed with an Illumina NovaSeq. The percentage of indels and substitutions was analyzed online (https://github.com/pinellolab/crispresso2).49 The genome-editing efficiency of the target region was calculated with the following formula: a/b, where a was the percentage of indels and substitutions, and b was the transfection efficiency.
Confocal Fluorescence Microscopy and Flow Cytometry
At 18, 24, 48, and 60 h after transfection, we assessed EGFP expression in the cells using a confocal fluorescence microscope (40×; FV1000; Olympus, Japan). We then calculated the fluorescence density at each time point and plotted the data to generate a graph of fluorescence density. At the 60-h time point, the fluorescence intensity of EGFP was detected by flow cytometry (Beckman, Brea, CA, USA), and the average value of three experimental results was calculated. The statistical difference between the parental vector and the SR-CRISPR vector was calculated using SPSS 20.0 (IBM, Chicago, IL, USA) and GraphPad Prism 8 (v.8.0.2.263; GraphPad Software, CA, USA) software.
Western Blot Analysis of Cas9 Protein Expression
HEK293T cells were lysed using radio immunoprecipitation assay (RIPA) lysate buffer (Beyotime, China), and samples were centrifuged at high speed to remove the insoluble fraction. Samples were then denatured with SDS protein loading buffer at 100°C for 5 min. Proteins were separated on a 10% SDS-PAGE gel followed by transfer to polyvinylidene fluoride (PVDF) membranes by electrophoresis. Western blot analysis was performed according to standard procedures. The primary antibody used to detect Cas9 was anti-CRISPR-Cas9 antibody (1:3,000 dilution; Abcam, Cambridge, UK), and the primary antibody for the internal control, β-actin, was anti-β-actin antibody (1:5,000 dilution; Fudebio-tech, China). The secondary antibody for both Cas9 and β-actin was HRP-conjugated Affinipure Goat Anti-Mouse IgG (h + 1) (1:10,000 dilution; Abcam, Cambridge, UK). In order to ensure consistency and accuracy of quantification, we incubated PVDF membranes of all samples in the same container with the same antibody for the same amount of time prior to development using Western Lightning Plus-ECL (Beyotime, China). An imaging system for western blots (Tanon, China) was used for imaging. The gray value of the band was analyzed using ImageJ software and normalized to the gray value of the internal control (β-actin) and transfection efficiency. Statistical differences in Cas9 protein expression were calculated with SPSS 20.0, and GraphPad Prism was used for graphing.
Real-Time PCR
Genomes were extracted using the Genomic DNA Extraction Kit (TAKARA) and subjected to real-time PCR amplification using Hieff qPCR SYBR Green Master Mix (Yeasen, China) with primers to specifically detect promoter cleavage efficiency. The Cas9 gene was selected as the internal control. The specificity of amplification products was determined from the melting curve analysis performed at the end of each run. The data for SR-CRISPR vector were normalized to that of the parental vector.
Statistical Analysis
The data reported here are means ± standard error of the mean (SEM) of three independent culture experiments. Statistical comparisons were performed using the two-tailed Student’s t test. p < 0.05 was considered statistically significant.
Author Contributions
W.-f.Z. designed the experiments; H.W. conducted the experiments and wrote the paper; H.L. and C.-y.G. conducted the experiments; Y.-s.L., Z.C., Y.-q.L., Q.L., Y.-z.J., H.-z.W., F.-l.W., C.-l.T., H.S., and H.-b.B. analyzed the data; H.-w.S. contributed to the drafting of the manuscript.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (grants 81703053 and 21771042), Guangdong Basic and Applied Basic Research Foundation (grants 2018A030313860, 2020A1515010889, and 2018A030313114), and the Innovative and Strong School Project of Guangdong Higher Education Institutions (grants 2017KZDXM049 and 2017KCXTD020).
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtm.2020.06.012.
Contributor Information
Hong-wei Shao, Email: shaohw2000@163.com.
Wen-feng Zhang, Email: zwfsnowdream@126.com.
Supplemental Information
References
- 1.Chira S., Gulei D., Hajitou A., Zimta A.A., Cordelier P., Berindan-Neagoe I. CRISPR/Cas9: Transcending the Reality of Genome Editing. Mol. Ther. Nucleic Acids. 2017;7:211–222. doi: 10.1016/j.omtn.2017.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gupta R.M., Musunuru K. Expanding the genetic editing tool kit: ZFNs, TALENs, and CRISPR-Cas9. J. Clin. Invest. 2014;124:4154–4161. doi: 10.1172/JCI72992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sander J.D., Joung J.K. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat. Biotechnol. 2014;32:347–355. doi: 10.1038/nbt.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Khan F.A., Pandupuspitasari N.S., Chun-Jie H., Ao Z., Jamal M., Zohaib A., Khan F.A., Hakim M.R., ShuJun Z. CRISPR/Cas9 therapeutics: a cure for cancer and other genetic diseases. Oncotarget. 2016;7:52541–52552. doi: 10.18632/oncotarget.9646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A., Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mali P., Yang L., Esvelt K.M., Aach J., Guell M., DiCarlo J.E., Norville J.E., Church G.M. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gallagher D.N., Haber J.E. Repair of a Site-Specific DNA Cleavage: Old-School Lessons for Cas9-Mediated Gene Editing. ACS Chem. Biol. 2018;13:397–405. doi: 10.1021/acschembio.7b00760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jasin M., Rothstein R. Repair of strand breaks by homologous recombination. Cold Spring Harb. Perspect. Biol. 2013;5:a012740. doi: 10.1101/cshperspect.a012740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ding Y., Li H., Chen L.L., Xie K. Recent Advances in Genome Editing Using CRISPR/Cas9. Front. Plant Sci. 2016;7:703. doi: 10.3389/fpls.2016.00703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang S., Chang R., Yang H., Zhao T., Hong Y., Kong H.E., Sun X., Qin Z., Jin P., Li S., Li X.J. CRISPR/Cas9-mediated gene editing ameliorates neurotoxicity in mouse model of Huntington’s disease. J. Clin. Invest. 2017;127:2719–2724. doi: 10.1172/JCI92087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eyquem J., Mansilla-Soto J., Giavridis T., van der Stegen S.J., Hamieh M., Cunanan K.M., Odak A., Gönen M., Sadelain M. Targeting a CAR to the TRAC locus with CRISPR/Cas9 enhances tumour rejection. Nature. 2017;543:113–117. doi: 10.1038/nature21405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen S.J. Minimizing off-target effects in CRISPR-Cas9 genome editing. Cell Biol. Toxicol. 2019;35:399–401. doi: 10.1007/s10565-019-09486-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang D.C., Wang X. Off-target genome editing: A new discipline of gene science and a new class of medicine. Cell Biol. Toxicol. 2019;35:179–183. doi: 10.1007/s10565-019-09475-7. [DOI] [PubMed] [Google Scholar]
- 15.Fu Y., Foden J.A., Khayter C., Maeder M.L., Reyon D., Joung J.K., Sander J.D. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat. Biotechnol. 2013;31:822–826. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin Y., Cradick T.J., Brown M.T., Deshmukh H., Ranjan P., Sarode N., Wile B.M., Vertino P.M., Stewart F.J., Bao G. CRISPR/Cas9 systems have off-target activity with insertions or deletions between target DNA and guide RNA sequences. Nucleic Acids Res. 2014;42:7473–7485. doi: 10.1093/nar/gku402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brunetti L., Gundry M.C., Kitano A., Nakada D., Goodell M.A. Highly Efficient Gene Disruption of Murine and Human Hematopoietic Progenitor Cells by CRISPR/Cas9. J. Vis. Exp. 2018;134:57278. doi: 10.3791/57278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gundry M.C., Brunetti L., Lin A., Mayle A.E., Kitano A., Wagner D., Hsu J.I., Hoegenauer K.A., Rooney C.M., Goodell M.A., Nakada D. Highly Efficient Genome Editing of Murine and Human Hematopoietic Progenitor Cells by CRISPR/Cas9. Cell Rep. 2016;17:1453–1461. doi: 10.1016/j.celrep.2016.09.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nuñez J.K., Harrington L.B., Doudna J.A. Chemical and Biophysical Modulation of Cas9 for Tunable Genome Engineering. ACS Chem. Biol. 2016;11:681–688. doi: 10.1021/acschembio.5b01019. [DOI] [PubMed] [Google Scholar]
- 20.Hynes A.P., Rousseau G.M., Lemay M.L., Horvath P., Romero D.A., Fremaux C., Moineau S. An anti-CRISPR from a virulent streptococcal phage inhibits Streptococcus pyogenes Cas9. Nat. Microbiol. 2017;2:1374–1380. doi: 10.1038/s41564-017-0004-7. [DOI] [PubMed] [Google Scholar]
- 21.Pawluk A., Staals R.H., Taylor C., Watson B.N., Saha S., Fineran P.C., Maxwell K.L., Davidson A.R. Inactivation of CRISPR-Cas systems by anti-CRISPR proteins in diverse bacterial species. Nat. Microbiol. 2016;1:16085. doi: 10.1038/nmicrobiol.2016.85. [DOI] [PubMed] [Google Scholar]
- 22.Pawluk A., Amrani N., Zhang Y., Garcia B., Hidalgo-Reyes Y., Lee J., Edraki A., Shah M., Sontheimer E.J., Maxwell K.L., Davidson A.R. Naturally Occurring Off-Switches for CRISPR-Cas9. Cell. 2016;167:1829–1838.e9. doi: 10.1016/j.cell.2016.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rauch B.J., Silvis M.R., Hultquist J.F., Waters C.S., McGregor M.J., Krogan N.J., Bondy-Denomy J. Inhibition of CRISPR-Cas9 with Bacteriophage Proteins. Cell. 2017;168:150–158.e10. doi: 10.1016/j.cell.2016.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maji B., Gangopadhyay S.A., Lee M., Shi M., Wu P., Heler R., Mok B., Lim D., Siriwardena S.U., Paul B. A High-Throughput Platform to Identify Small-Molecule Inhibitors of CRISPR-Cas9. Cell. 2019;177:1067–1079.e19. doi: 10.1016/j.cell.2019.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cao J., Wu L., Zhang S.M., Lu M., Cheung W.K., Cai W., Gale M., Xu Q., Yan Q. An easy and efficient inducible CRISPR/Cas9 platform with improved specificity for multiple gene targeting. Nucleic Acids Res. 2016;44:e149. doi: 10.1093/nar/gkw660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de Solis C.A., Ho A., Holehonnur R., Ploski J.E. The Development of a Viral Mediated CRISPR/Cas9 System with Doxycycline Dependent gRNA Expression for Inducible In vitro and In vivo Genome Editing. Front. Mol. Neurosci. 2016;9:70. doi: 10.3389/fnmol.2016.00070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nihongaki Y., Kawano F., Nakajima T., Sato M. Photoactivatable CRISPR-Cas9 for optogenetic genome editing. Nat. Biotechnol. 2015;33:755–760. doi: 10.1038/nbt.3245. [DOI] [PubMed] [Google Scholar]
- 28.Chen Y., Liu X., Zhang Y., Wang H., Ying H., Liu M., Li D., Lui K.O., Ding Q. A self-restricted CRISPR system to reduce off-target effects. Mol. Ther. 2016;24:1508–1510. doi: 10.1038/mt.2016.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shen C.C., Hsu M.N., Chang C.W., Lin M.W., Hwu J.R., Tu Y., Hu Y.C. Synthetic switch to minimize CRISPR off-target effects by self-restricting Cas9 transcription and translation. Nucleic Acids Res. 2019;47:e13. doi: 10.1093/nar/gky1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lackner D.H., Carré A., Guzzardo P.M., Banning C., Mangena R., Henley T., Oberndorfer S., Gapp B.V., Nijman S.M.B., Brummelkamp T.R., Bürckstümmer T. A generic strategy for CRISPR-Cas9-mediated gene tagging. Nat. Commun. 2015;6:10237. doi: 10.1038/ncomms10237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Suzuki K., Izpisua Belmonte J.C. In vivo genome editing via the HITI method as a tool for gene therapy. J. Hum. Genet. 2018;63:157–164. doi: 10.1038/s10038-017-0352-4. [DOI] [PubMed] [Google Scholar]
- 32.He X., Tan C., Wang F., Wang Y., Zhou R., Cui D., You W., Zhao H., Ren J., Feng B. Knock-in of large reporter genes in human cells via CRISPR/Cas9-induced homology-dependent and independent DNA repair. Nucleic Acids Res. 2016;44:e85. doi: 10.1093/nar/gkw064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang J.H., Adikaram P., Pandey M., Genis A., Simonds W.F. Optimization of genome editing through CRISPR-Cas9 engineering. Bioengineered. 2016;7:166–174. doi: 10.1080/21655979.2016.1189039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ren X., Yang Z., Xu J., Sun J., Mao D., Hu Y., Yang S.J., Qiao H.H., Wang X., Hu Q. Enhanced specificity and efficiency of the CRISPR/Cas9 system with optimized sgRNA parameters in Drosophila. Cell Rep. 2014;9:1151–1162. doi: 10.1016/j.celrep.2014.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Labuhn M., Adams F.F., Ng M., Knoess S., Schambach A., Charpentier E.M., Schwarzer A., Mateo J.L., Klusmann J.H., Heckl D. Refined sgRNA efficacy prediction improves large- and small-scale CRISPR-Cas9 applications. Nucleic Acids Res. 2018;46:1375–1385. doi: 10.1093/nar/gkx1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cencic R., Miura H., Malina A., Robert F., Ethier S., Schmeing T.M., Dostie J., Pelletier J. Protospacer adjacent motif (PAM)-distal sequences engage CRISPR Cas9 DNA target cleavage. PLoS ONE. 2014;9:e109213. doi: 10.1371/journal.pone.0109213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kocak D.D., Josephs E.A., Bhandarkar V., Adkar S.S., Kwon J.B., Gersbach C.A. Increasing the specificity of CRISPR systems with engineered RNA secondary structures. Nat. Biotechnol. 2019;37:657–666. doi: 10.1038/s41587-019-0095-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shen B., Zhang W., Zhang J., Zhou J., Wang J., Chen L., Wang L., Hodgkins A., Iyer V., Huang X., Skarnes W.C. Efficient genome modification by CRISPR-Cas9 nickase with minimal off-target effects. Nat. Methods. 2014;11:399–402. doi: 10.1038/nmeth.2857. [DOI] [PubMed] [Google Scholar]
- 39.Ran F.A., Hsu P.D., Lin C.Y., Gootenberg J.S., Konermann S., Trevino A.E., Scott D.A., Inoue A., Matoba S., Zhang Y., Zhang F. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013;154:1380–1389. doi: 10.1016/j.cell.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Swiech L., Heidenreich M., Banerjee A., Habib N., Li Y., Trombetta J., Sur M., Zhang F. In vivo interrogation of gene function in the mammalian brain using CRISPR-Cas9. Nat. Biotechnol. 2015;33:102–106. doi: 10.1038/nbt.3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Holkers M., Maggio I., Henriques S.F., Janssen J.M., Cathomen T., Gonçalves M.A. Adenoviral vector DNA for accurate genome editing with engineered nucleases. Nat. Methods. 2014;11:1051–1057. doi: 10.1038/nmeth.3075. [DOI] [PubMed] [Google Scholar]
- 42.Xue W., Chen S., Yin H., Tammela T., Papagiannakopoulos T., Joshi N.S., Cai W., Yang G., Bronson R., Crowley D.G. CRISPR-mediated direct mutation of cancer genes in the mouse liver. Nature. 2014;514:380–384. doi: 10.1038/nature13589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Molla K.A., Yang Y. CRISPR/Cas-Mediated Base Editing: Technical Considerations and Practical Applications. Trends Biotechnol. 2019;37:1121–1142. doi: 10.1016/j.tibtech.2019.03.008. [DOI] [PubMed] [Google Scholar]
- 44.Yang B., Yang L., Chen J. Development and Application of Base Editors. CRISPR J. 2019;2:91–104. doi: 10.1089/crispr.2019.0001. [DOI] [PubMed] [Google Scholar]
- 45.Zuo E., Sun Y., Wei W., Yuan T., Ying W., Sun H., Yuan L., Steinmetz L.M., Li Y., Yang H. Cytosine base editor generates substantial off-target single-nucleotide variants in mouse embryos. Science. 2019;364:289–292. doi: 10.1126/science.aav9973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jin S., Zong Y., Gao Q., Zhu Z., Wang Y., Qin P., Liang C., Wang D., Qiu J.L., Zhang F., Gao C. Cytosine, but not adenine, base editors induce genome-wide off-target mutations in rice. Science. 2019;364:292–295. doi: 10.1126/science.aaw7166. [DOI] [PubMed] [Google Scholar]
- 47.Merkle T., Merz S., Reautschnig P., Blaha A., Li Q., Vogel P., Wettengel J., Li J.B., Stafforst T. Precise RNA editing by recruiting endogenous ADARs with antisense oligonucleotides. Nat. Biotechnol. 2019;37:133–138. doi: 10.1038/s41587-019-0013-6. [DOI] [PubMed] [Google Scholar]
- 48.Qu L., Yi Z., Zhu S., Wang C., Cao Z., Zhou Z., Yuan P., Yu Y., Tian F., Liu Z. Programmable RNA editing by recruiting endogenous ADAR using engineered RNAs. Nat. Biotechnol. 2019;37:1059–1069. doi: 10.1038/s41587-019-0178-z. [DOI] [PubMed] [Google Scholar]
- 49.Clement K., Rees H., Canver M.C., Gehrke J.M., Farouni R., Hsu J.Y., Cole M.A., Liu D.R., Joung J.K., Bauer D.E., Pinello L. CRISPResso2 provides accurate and rapid genome editing sequence analysis. Nat. Biotechnol. 2019;37:224–226. doi: 10.1038/s41587-019-0032-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.